Abstract
PII signal transduction proteins are widely spread among all domains of life where they regulate a multitude of carbon and nitrogen metabolism related processes. Non-diazotrophic cyanobacteria can utilize a high variety of organic and inorganic nitrogen sources. In recent years, several physiological studies indicated an involvement of the cyanobacterial PII protein in regulation of ammonium, nitrate/nitrite, and cyanate uptake. However, direct interaction of PII has not been demonstrated so far. In this study, we used biochemical, molecular genetic and physiological approaches to demonstrate that PII regulates all relevant nitrogen uptake systems in Synechocystis sp. strain PCC 6803: PII controls ammonium uptake by interacting with the Amt1 ammonium permease, probably similar to the known regulation of E. coli ammonium permease AmtB by the PII homolog GlnK. We could further clarify that PII mediates the ammonium- and dark-induced inhibition of nitrate uptake by interacting with the NrtC and NrtD subunits of the nitrate/nitrite transporter NrtABCD. We further identified the ABC-type urea transporter UrtABCDE as novel PII target. PII interacts with the UrtE subunit without involving the standard interaction surface of PII interactions. The deregulation of urea uptake in a PII deletion mutant causes ammonium excretion when urea is provided as nitrogen source. Furthermore, the urea hydrolyzing urease enzyme complex appears to be coupled to urea uptake. Overall, this study underlines the great importance of the PII signal transduction protein in the regulation of nitrogen utilization in cyanobacteria.
Keywords: PII signaling protein, GlnB, cyanobacteria, nitrogen regulation, nitrate uptake, ammonium uptake, urea uptake, ABC transporters
Introduction
The emergence of the oxygenic photosynthesis by ancestors of present cyanobacteria (Soo et al., 2017) laid the ground for the evolution of present days life on planet earth. Until today, cyanobacteria occupy a high variety of illuminated habitats, where they represent one of the most abundant primary producers (Whitton, 2012). Accordingly, cyanobacteria are essential contributors to the global carbon cycle. Many cyanobacterial strains have acquired the ability to fix atmospheric nitrogen, making them key players in the global nitrogen turnover (Herrero and Flores, 2008). Nitrogen represents a necessary macronutrient for all living organisms and therefore constitutes an important growth-limiting factor in most ecosystems (Vitousek and Howarth, 1991). The regulation of nitrogen metabolism in cyanobacteria mainly depends on the fine-tuned network of the signal transduction protein PII, the global nitrogen transcription factor NtcA and the NtcA co-activator PipX (Vegapalas et al., 1992; Espinosa et al., 2006, 2007, 2014; Forchhammer, 2008; Luque and Forchhammer, 2008; Forcada-Nadal et al., 2018).
PII signal-transduction proteins are widespread in all three domains of life, where they represent one of the largest and most ancient families of signaling proteins (Chellamuthu et al., 2013; Forchhammer and Luddecke, 2016). PII proteins are involved in the regulation of various nitrogen- and carbon-anabolic processes (Forchhammer, 2004, 2008). Canonical PII proteins are homo-trimeric with three characteristic loop regions, designated as B-, C-, and T-loops, which compose the effector molecule binding sites (Cheah et al., 1994; Xu et al., 2003; Forchhammer, 2004, 2008; Llacer et al., 2007; Fokina et al., 2010a; Zhao et al., 2010; Zeth et al., 2014). The large surface exposed T-loop is the prevailing protein interaction module of PII. The PII proteins sense the energy status of the cell by the competitive binding of ADP or ATP (Zeth et al., 2014). Binding of ATP and synergistic binding of 2-oxoglutarate (2-OG) allows PII to sense the current carbon/nitrogen status of the cell (Fokina et al., 2010a). 2-OG is an intermediate of the TCA-cycle that provides the carbon skeleton for inorganic nitrogen incorporation by the glutamine synthetase/glutamate synthase (GS/GOGAT) cycle. Due to this, 2-OG links carbon and nitrogen metabolism and acts as an indicator for the intracellular carbon/nitrogen balance (Muro-Pastor et al., 2001; Fokina et al., 2010a). Besides effector molecule binding, post-translational modification of PII represents a second level of regulation (Forchhammer et al., 2004; Merrick, 2014). Depending on nitrogen availability, cyanobacterial PII can be phosphorylated at the apex of the T-loop at position Ser49 (Forchhammer and Tandeau de Marsac, 1995; Forchhammer and Hedler, 1997). In other prokaryotes, like E. coli, PII is modified by uridylylation in response to nitrogen availability instead of phosphorylation (Jiang et al., 1998). Binding of effector molecules as well as post-translational modifications lead to various PII conformations. Depending on the conformational state, PII can interact with a variety of interaction partners and thereby regulate the cellular C/N balance (Radchenko and Merrick, 2011; Forchhammer and Luddecke, 2016). In cyanobacteria, PII indirectly regulates the global nitrogen control transcriptional factor NtcA through binding of the NtcA co-activator PipX (Espinosa et al., 2007). In common with other bacteria, the cyanobacterial PII protein can control the acetyl-CoA levels by interacting with the biotin carboxyl carrier protein (BCCP) of acetyl-CoA carboxylase (ACC) (Hauf et al., 2016). Furthermore, PII regulates arginine biosynthesis by interacting with the enzyme N-acetylglutamate kinase (NAGK), which catalyzes the rate-limiting step of this pathway (Caldovic and Tuchman, 2003; Heinrich et al., 2004; Llacer et al., 2007; Watzer et al., 2015). If sufficient energy and nitrogen is available, indicated by a high intracellular ATP and low 2-OG level, non-phosphorylated PII interacts with NAGK, enhancing its catalytic efficiency and relieving it from feedback inhibition by arginine (Heinrich et al., 2004; Maheswaran et al., 2004; Llacer et al., 2007). At high intracellular arginine levels, the carbon/nitrogen storage polymer cyanophycin (multi-L-arginyl-poly-L-aspartate) accumulates in Synechocystis sp. strain PCC 6803 (hereafter Synechocystis) (Maheswaran et al., 2006; Watzer et al., 2015). A PII variant was identified with a single amino acid substitution, Ile86 to Asn86 [thereafter referred as PII(I86N)], which constitutively binds NAGK in vitro (Fokina et al., 2010b). Replacing the wild-type PII with a I86N variant in Synechocystis generated a mutant strain, which strongly overproduced arginine and cyanophycin (Watzer et al., 2015). On the other hand, the PII(I86N) strain showed a growth defect in ammonium-supplemented medium (Watzer et al., 2015).
Cyanobacteria use nitrogen sources in a hierarchical order, with ammonium being the preferred nitrogen source. As a consequence, when ammonium is provided together with other suitable nitrogen sources, ammonium will be utilized first (Muro-Pastor et al., 2005). In most natural habitats, the ammonium availability is low, so that high affinity ammonium permeases are required for efficient ammonium uptake (Rees et al., 2006). In Synechocystis, the Amt1 permease is mainly responsible for ammonium uptake (Montesinos et al., 1998). However, elevated intracellular ammonium concentrations are toxic to the cells (Drath et al., 2008; Dai et al., 2014), and therefore, ammonium transport must be tightly controlled. The ammonium transporter family (Amt) is widespread among all domains of life (Wirén and Merrick, 2004). In E. coli, the PII homolog GlnK regulates the ammonium permease AmtB by direct protein–protein interaction. Under ammonium excess conditions, ADP-complexed GlnK blocks an uncontrolled influx of ammonium by inserting the apex of the T-loop into the cytoplasmic exit pores of AmtB (Conroy et al., 2007).
For the assimilation of nitrate, an active nitrate transporter, a nitrate reductase (NR) and a nitrite reductase (NiR) are required (Ohashi et al., 2011). Two types of nitrate transporter systems have been found among cyanobacteria, a high-affinity nitrate/nitrite permease NrtP and the ABC-type transporter NrtABCD (NRT) (Omata et al., 1993; Luque et al., 1994; Sakamoto et al., 1999; Ohashi et al., 2011). NRT is a bispecific nitrate/nitrite transporter showing high affinity for both substrates (Maeda and Omata, 1997). Intracellular nitrate is first reduced to nitrite by NR and subsequently reduced to ammonium by NiR. Subsequently, ammonium is assimilated in the GS/GOGAT cycle (Flores and Herrero, 1994). Both, NR and NiR, use photosystem I reduced ferredoxin as an electron donor, indicating a coupling of photosynthesis and nitrate assimilation (Manzano et al., 1976; Flores and Herrero, 1994). Addition of ammonium to nitrate adapted cells results in an immediate inhibition of nitrate uptake and a repression of proteins involved in nitrate assimilation (NR and NiR). The ammonium-induced inhibition of NRT is regulated by the PII protein and the C-terminal domain of NrtC (Kobayashi et al., 1997; Lee et al., 1998). Phosphomimetic variants of PII and a PII phosphatase (PphA) deletion mutant, in which PII is constitutively phosphorylated, showed ammonium promoted inhibition of nitrate uptake like the wild-type (Lee et al., 2000; Kloft and Forchhammer, 2005). However, this response was abolished in a PII deficient mutant (Kloft and Forchhammer, 2005).
The ability to utilize urea as nitrogen source is widely distributed among bacteria, fungi and algae (Baker et al., 2009; Solomon et al., 2010; Esteves-Ferreira et al., 2018). In common with other bacteria, the cyanobacteria Synechocystis and Anabaena sp. PCC 7120 possess a high affinity urea ABC-type transporter, which is capable of urea import at concentrations lower than 1 μM (Valladares et al., 2002). The gene cluster urtABCDE, encoding all subunits of this ABC-type urea transporter, is transcriptionally controlled by the global transcription factor NtcA (Valladares et al., 2002).
The present study was inspired by the phenotype of the cyanophycin-accumulating strain variant Synechocystis PII(I86N), which was impaired in ammonium utilization. Starting with analyzing a possible regulation of the cyanobacterial Amt1 permease by PII, we found additional evidence for a direct regulation of the nitrate/nitrite transporter NrtABCD and the urea transporter UrtABCDE by the PII signaling protein during this study.
Materials and Methods
Cultivation Conditions
Standard cloning procedures were performed in Escherichia coli NEB 10-beta (NEB). Strains were grown in LB-medium at 37°C with constant shaking at 300 rpm.
Synechocystis strains were grown photoautotrophically in BG-11 medium supplemented with 5 mM NaHCO3 and nitrate, ammonium, or urea as nitrogen source (Rippka et al., 1979). BG-11 agar plates were produced by adding 1.5% (w/v) Bacto-agar (Difco), 0.3% (w/v) sodium thiosulfate pentahydrate and 10 mM TES-NaOH pH 8 (Roth) to liquid BG-11 medium. Antibiotics were added when required. Cultivation of liquid cultures for physiological experiments occurred in 50, 100, or 500 mL Erlenmeyer flasks, at 28°C and with constant shaking of 120 rpm. Cultures were continuously illuminated with a photon flux rate of 40–50 μE. Growth rates were determined by measuring the optical density at 750 nm (OD750).
For induction of nitrogen starvation conditions, exponentially growing cells (OD750 0.4–0.8) were harvested by centrifugation (3,000 × g for 10 min at room temperature), washed and resuspended in BG-11 medium lacking a suitable nitrogen source (BG-110).
For spot assays (drop plate method), Synechocystis cultures were adjusted to an OD750 of 1. A dilution series to the power of 10 was made using BG-110. Then, 5 μL of every dilution step (100–10-4) were dropped on BG-11 agar plates. Plates were cultivated at 28°C with constant illumination of 40 μmol of photons s-1 m-2.
Bacterial Two-Hybrid Assay
Plasmids were constructed by PCR amplification using high-fidelity Q5 polymerase (NEB) and oligonucleotides with overlapping regions. Genomic Synechocystis DNA or plasmids served as templates. PCR fragments were inserted in linearized bacterial two-hybrid vectors pUT18 and pKT25 containing the genes for either the T18 or T25 subunit of the adenylate cyclase CyaA (Karimova et al., 2001) by isothermal, single-reaction DNA assembly according to Gibson et al. (2009). Since the multiple cloning site of pKT25 is located downstream of the T25 subunit, allowing only N-terminal localization of the tag, we constructed plasmid pKT25n (Table 1; pKT25n_fw and pKT25n_rev) to achieve a C-terminal fusion of the tag to the gene of interest. Therefore, plasmid pKT25 was linearized using PCR and the gene of interest was fused upstream of the T25 subunit. Primers, plasmids, and strains used in this study are listed in Tables 1–3, respectively.
Table 1.
Oligonucleotides used in this study.
| Primer | Sequence (5′–3′ direction) |
|---|---|
| pKT25n_fw | ACCATGCAGCAATCGCATCAG |
| pKT25n_rev | CATAGCTGTTTCCTGTGTGAAATTG |
| glnB_fw | TGTGTGGAATTGTGAGCGGATAACAATTTCACACAGGAAACAGCTATGAAAAAAGTAGAAGCGATTATTC |
| glnB_rev | CTCGCTGGCGGCTGAATTCGAGCTCGGTACCCGGGGATCAATAGCTTCGGTATCCTTTTC |
| pipX_fw | TTCACACAGGAAACAGCTATGAGTAACGAAATTTACCTTAAC |
| pipX_rev | GATGCGATTGCTGCATGGTAAAAGTGTTTTTATGTAACTTTG |
| pipX_pKT25n_fw | AAGTTACATAAAAACACTTTTACCATGCAGCAATCGCATCAG |
| pipX_pKT25n_rev | TAAGGTAAATTTCGTTACTCATAGCTGTTTCCTGTGTGAAATTG |
| amt1_pKT25_fw | CTGGCGCGCACGCGGCGGGCTGCAGGGTCGACTCTAGAGATGTCTAATTCGATATTGTCTAAAC |
| amt1_pKT25_rev | AAAACGACGGCCGAATTCTTAGTTACTTAGGTACCCGGGGATCTTATTCAGGGACAGTGG |
| amt1_fw | AACAATTTCACACAGGAAACAGCTATGTCTAATTCGATATTGTCTAAAC |
| amt1_rev | TGATGCGATTGCTGCATGGTTTCAGGGACAGTGGCACCG |
| amt1_pKT25n_fw | TCTCCGGTGCCACTGTCCCTGAAACCATGCAGCAATCGCATC |
| amt1_pKT25n_rev | ACAATATCGAATTAGACATAGCTGTTTCCTGTGTGAAATTGTTATCCGC |
| nrtC_pKT25_fw | CGCGCACGCGGCGGGCTGCAGGGTCGACTCTAGAGGATCCCCCCTTCATTGAAATTGATCATGTTG |
| nrtC_pKT25_rev | AGTCACGACGTTGTAAAACGACGGCCGAATTCTTAGTTATTGATTAACTTGATCAATTTGGTCGATGAG |
| nrtC_fw | AATTTCACACAGGAAACAGCTATGCCCTTCATTGAAATTGATCATG |
| nrtC_rev | CTGATGCGATTGCTGCATGGTTTGATTAACTTGATCAATTTGG |
| nrtC_pKT25n_fw | AAATTGATCAAGTTAATCAAACCATGCAGCAATCGCATCAG |
| nrtC_pKT25n_rev | TCAATTTCAATGAAGGGCATAGCTGTTTCCTGTGTGAAATTG |
| nrtD_pKT25_fw | CGCACGCGGCGGGCTGCAGGGTCGACTCTAGAGGATCCCCAAACAATGAATGTCAATGACCCTATCC |
| ntrD_pKT25_rev | CCCAGTCACGACGTTGTAAAACGACGGCCGAATTCTTAGTTAAGACCCTTCCATGGATTCCACTGAGGGGGTAG |
| nrtD_fw | AATTTCACACAGGAAACAGCTATGCAAACAATGAATGTCAATGACCCTATC |
| nrtD_rev | CTGATGCGATTGCTGCATGGTAGACCCTTCCATGGATTCCACTGAG |
| nrtD_pKT25n_fw | TGGAATCCATGGAAGGGTCTACCATGCAGCAATCGCATCAG |
| nrtD_pKT25n_rev | ATTGACATTCATTGTTTGCATAGCTGTTTCCTGTGTGAAATTG |
| urtD_pKT25_fw | CGCACGCGGCGGGCTGCAGGGTCGACTCTAGAGGATCCCACCAGCAAAATCTTAGAAATTCAAG |
| urtD_pKT25_rev | CCCAGTCACGACGTTGTAAAACGACGGCCGAATTCTTAGCTAATCTCCATCCTCATCAAC |
| urtD_fw | ACACAGGAAACAGCTATGACCAGCAAAATCTTAGAAATTCAAGAC |
| urtD_rev | GATGCGATTGCTGCATGGTATCTCCATCCTCATCAACACTG |
| urtD_pKT25n_fw | AGTGTTGATGAGGATGGAGATACCATGCAGCAATCGCATCAG |
| urtD_pKT25n_rev | TTCTAAGATTTTGCTGGTCATAGCTGTTTCCTGTGTGAAATTG |
| urtE_fw | GCGCGCACGCGGCGGGCTGCAGGGTCGACTCTAGAGGATGCTATGTTATCCTTTCCCCCATTCTTG |
| urtE_rev | CCCAGTCACGACGTTGTAAAACGACGGCCGAATTCTTAGTTATACTGCCAAAAATTTTTGGATAAC |
| urtE_fw | ACAATTTCACACAGGAAACAGCTATGGCTATGTTATCCTTTCCC |
| urtE_rev | TGATGCGATTGCTGCATGGTTACTGCCAAAAATTTTTGGATAACC |
| urtE_pKT25n_fw | TATCCAAAAATTTTTGGCAGTAACCATGCAGCAATCGCATCAG |
| urtE_pKT25n_rev | GGGAAAGGATAACATAGCCATAGCTGTTTCCTGTGTGAAATTG |
Table 3.
Strains used in this study.
| Strains | Description | References |
|---|---|---|
| E. coli NEB 10-beta | Cloning strain | NEB |
| E. coli BTH101 | Bacterial two-hybrid host strain | Euromedex |
| Synechocystis sp. PCC 6803 | Wild type | Pasteur Culture Collection |
| Synechocystis sp. PII(I86N) | Genomic PII(I86N) mutant | Watzer et al., 2015 |
| Synechocystis sp. ΔPII | Chromosomal deletion of glnB | Hisbergues et al., 1999 |
| Synechocystis sp. ΔPII + PII-Venus | Synechocystis sp. ΔPII transformed with pVZ322 encoding a PII-Venus fusion | Hauf et al., 2016 |
Escherichia coli BTH101 cells were co-transformed with plasmid pUT18 and plasmid pKT25 or pKT25n (Table 2). Plasmids pKT25 or pKT25n contained the possible PII interaction partner fused N- or C-terminal to the T25 subunit. Plasmid pUT18 contains a gene-fusion of the PII-encoding glnB gene or a genetically modified glnB gene containing the I86N mutation [Ile (5′ATC) at codon position 86 to Asn (5′AAC)] (Watzer et al., 2015), the R9L mutation [Arg (5′CGC) at codon position 9 to Leu (5′CTG)] (Fokina et al., 2010a) or the S49D mutation [Ser (5′TCG) at codon position 49 to Asp (5′GAT)] (Lee et al., 2000) with the T18 subunit. The glnB gene and the modified glnB genes were always fused N-terminal with the T18 subunit. Co-transformants were plated on LB-plates (supplemented with 100 μg mL-1 ampicillin and 50 μg mL-1 kanamycin) and cultivated for 2 days at 30°C.
Table 2.
Plasmids used in this study.
| Plasmid | Tag localization | Description | References |
|---|---|---|---|
| pPD-NFLAG | N-terminal | Encoding the N-terminal 3xFLAG tag | Hollingshead et al., 2012 |
| pPD-CFLAG | C-terminal | Encoding the C-terminal 3xFLAG tag | Chidgey et al., 2014 |
| pKT25 | Encoding T25 fragment of adenylate cyclase CyaA (amino acids 1–224) | Karimova et al., 2001 | |
| pKT25n | Derived from pKT25. Upstream of the T25 fragment | This study | |
| pUT18 | Encoding T18 fragment of adenylate cyclase CyaA (amino acids 225–399) | Karimova et al., 2001 | |
| pUT18 glnB | N-terminal | Derived from pUT18. Encoding glnB | This study |
| pUT18 glnB (I86N) | N-terminal | Derived from pUT18. Encoding glnB containing the I86N mutation | This study |
| pKT25n pipX | C-terminal | Derived from pKT25. Sequence encoding pipX. Positive control | This study |
| pKT25 pipX | N-terminal | Derived from pKT25. Sequence encoding pipX. Positive control | This study |
| pKT25n amt1 | C-terminal | Derived from pKT25. Sequence encoding amt1 | This study |
| pKT25 amt1 | N-terminal | Derived from pKT25. Sequence encoding amt1 | This study |
| pKT25n nrtC | C-terminal | Derived from pKT25. Sequence encoding nrtC | This study |
| pKT25 nrtC | N-terminal | Derived from pKT25. Sequence encoding nrtC | This study |
| pKT25n nrtD | C-terminal | Derived from pKT25. Sequence encoding nrtD | This study |
| pKT25 nrtD | N-terminal | Derived from pKT25. Sequence encoding nrtD | This study |
| pKT25n urtD | C-terminal | Derived from pKT25. Sequence encoding urtD | This study |
| pKT25 urtD | N-terminal | Derived from pKT25. Sequence encoding urtD | This study |
| pKT25n urtE | C-terminal | Derived from pKT25. Sequence encoding urtE | This study |
| pKT25 urtE | N-terminal | Derived from pKT25. Sequence encoding urtE | This study |
To reduce the level of heterogeneity, five clones from each plate were picked to inoculate 5 mL LB-medium (containing 100 μg mL-1 ampicillin and 50 μg mL-1 kanamycin). Cultures were cultivated overnight at 37°C. Overnight cultures were diluted 1:100 in 3 mL of fresh LB-medium (containing 100 μg mL-1 ampicillin and 50 μg mL-1 kanamycin) and grown to an OD600 of 0.7. Three μL of each culture were plated on X-Gal (containing 100 μg mL-1 ampicillin, 50 μg mL-1 kanamycin, 1 mM IPTG, 40 μg mL-1 X-Gal) and MacConkey (containing 100 μg mL-1 ampicillin, 50 μg mL-1 kanamycin, 1 mM IPTG, 1% maltose) reporter plates. Reporter plates were incubated for 3–4 days at 25°C.
Construction and Cultivation of Synechocystis PII-3xFLAG Tag Strains
The previously described pPD-NFLAG and pPD-CFLAG plasmids were used to construct Synechocystis PII (GlnB) fusion proteins with an N- or C-terminal 3xFLAG tag, respectively (Hollingshead et al., 2012; Chidgey et al., 2014). Together with a kanamycin resistance cassette, this construct was inserted in the Synechocystis wild-type genome by homologous recombination, replacing the psbAII gene (Chidgey et al., 2014). Transformants were selected and segregated by kanamycin resistance. For pull-down experiments, 2 L batch cultures were inoculated at OD750 = 0.2 in BG-11 medium and propagated as described above, with magnetic stirring at 120 rpm and bubbling with 2% CO2 (v/v) supplemented ambient air. Cell harvesting was performed at OD750 = 0.6 by mixing the cultures with ice in a 2:1 ratio (w/w) for rapid metabolic inactivation, and centrifugation at 7,477 × g for 10 min. Cell pellets were subsequently washed with nitrogen-free BG-110 at 4°C and snap frozen in liquid nitrogen. For experimental controls, the wild-type strain was similarly cultivated and subjected to pull-down assays followed by mass spectrometry analyses as described below. Two independent replicates were prepared per condition.
Preparation of Cell Extracts and Anti-FLAG Pull-Down
Frozen cell pellets were washed with 5 mL IP buffer, containing 25 mM MES/NaOH; pH 6.5, 5 mM CaCl2, 10 mM MgCl2, and 20% (v/v) glycerol and subsequently resuspended in 3 mL IP buffer, including protease inhibitors (complete EDTA-free; Roche) for cell lysis. Therefore, an equal volume of glass beads (0.1–0.15 mm diameter) was added and cells were disrupted using a FastPrep-24 Ribolyser (MP Biomedicals) with five cycles of 20 s at 6.5 m s-1 and 4°C. After centrifugation for 5 min at 3,314 × g and 4°C, the supernatant was transferred and adjusted with IP buffer to a final volume of 6 mL. For membrane protein solubilization, 1.5% (w/v) dodecyl-β-D-maltoside (DDM; Carl Roth) was added and incubated under agitation for 1 h at 4°C, before insoluble cell debris was removed by centrifugation for 20 min at 20,000 × g and 4°C. The soluble cell extracts (from 2 L cultures) were subjected to the PII-FLAG pull-down procedure: for this, 400 μL of resuspended anti-FLAG-M2-agarose (Sigma) was added into empty SPE-columns and washed twice with each 1 mL IP buffer, supplemented with 0.04% (w/v) DDM, before the supernatants were incubated for 5 min and removed by gravity flow. Repeated washing steps with each 5 mL DDM supplemented IP buffer were performed similarly, until the flow through appeared colorless. Elution of coupled proteins from the anti-FLAG resin was performed by incubation in 600 μL DDM supplemented IP buffer containing 0.3 μg/mL 3xFLAGTM peptide (Sigma-Aldrich) for 40 min.
Proteomics Workflow, NanoLC-MS/MS Analysis, and Data Processing
Eluates from the pull-down workflow were subjected to acetone/methanol precipitation and resulting protein pellets were resuspended in denaturation buffer for subsequent tryptic digestion as described elsewhere (Spät et al., 2015). Resulting peptide mixtures were subjected to stage tip purification (Rappsilber et al., 2007). Protein detection by mass spectrometry (MS) was performed as described previously (Spät et al., 2015): in brief, peptide mixtures were loaded onto a 15 cm reversed-phase C18 nanoHPLC column on an EASY-LC system (Proxeon Biosystems) and separated in a 90 min segmented linear gradient. Eluted peptides were ionized by the on-line coupled ESI source and injected in a LTQ Orbitrap XL mass spectrometer (Thermo Scientific). MS spectra were acquired in the positive-ion mode. Per scan cycle, one initial full (MS) scan was followed by fragmentation of the 15 most intense multiply charged ions by collision induced dissociation (CID) for MS/MS scans. The scan range was 300–2,000 m/z for precursor ions at resolution 60,000 and sequenced precursors were dynamically excluded for fragmentation for 90 s. The lock mass option was enabled for real time mass recalibration (Olsen et al., 2005). All raw spectra were processed with the MaxQuant software (version 1.5.2.8) (Cox and Mann, 2008) at default settings. Peak lists were searched against a target-decoy database of Synechocystis sp. PCC 6803 with 3,671 protein sequences, retrieved from Cyanobase (Nakao et al., 2010) (July 2014), plus the sequence of the N- or C-terminal 3xFLAG tagged PII fusion protein (MDYKDDDDKDYKDDDDKDYKDDDDKAAAKKVEAIIRPF KLDEVKIALVNAGIVGMTVSEVRGFGRQKGQTERYRGSEYT VEFLQKLKIEIVVDEGQVDMVVDKLVSAARTGEIGDGKIFIS PVDSVVRIRTGEKDTEAI or MKKVEAIIRPFKLDEVKIALVN AGIVGMTVSEVRGFGRQKGQTERYRGSEYTVEFLQKLKIEIV VDEGQVDMVVDKLVSAARTGEIGDGKIFISPVDSVVRIRTG EKDTEAIASYKDDDDKDYKDDDDKDYKDDDDK), respectively, as well as 245 common contaminants with the following database search criteria: trypsin was defined as a cleaving enzyme and up to two missed cleavages were allowed; carbamido-methylation of cysteines was defined as a fixed modification and methionine oxidation and protein N-terminal acetylation as variable modifications. Peptide and protein false discovery rates retrieved from MaxQuant were limited to 1%, each. Raw data acquired by mass spectrometry was deposited at the ProteomeXchange Consortium via the Pride partner repository (Vizcaino et al., 2013) under the identifier PXD013411.
Determination of Nitrate, Nitrite, Ammonium, and Urea in Cell-Free Culture Medium
To determine the nitrate, ammonium, or urea uptake, exponentially growing cells (OD750 = 0.4–0.8) were harvested by centrifugation (3,000 × g for 10 min at room temperature) and washed twice with BG-110 medium lacking combined nitrogen sources. Subsequently, the cultures were adjusted to OD750 = 1 with BG-110 medium. The assays were started by adding either 200 μM NaNO3, 200 μM NH4+ or 150 μM urea, respectively. Cells were cultivated under constant shaking of 120 rpm and illumination of 40–50 μE. 1 mL aliquots of the cell suspensions were taken and subsequently centrifuged (13,000 × g for 5 min at room temperature) to remove the cells.
For nitrate quantification, the absorbance at 210 nm was measured in the cell-free medium. Since both nitrate and nitrite absorb at 210 nm, the apparent nitrate values were corrected for the presence of nitrite (Kloft and Forchhammer, 2005). Nitrite concentration of the cell-free medium was determined using the modified Griess reaction according to Fiddler (1977).
The ammonium concentration of cell-free medium was measured by using the Nessler reaction (Vogel et al., 1989). Urea was quantified by using the urea nitrogen (BUN) colorimetric detection kit (Invitrogen).
Microscopy Procedures and Image Evaluation
Fluorescence microscopy was performed using a Leica DM5500B microscope with a 100×/1.3 oil objective. For the detection of Venus fluorescence, an ET500/20× excitation filter and an ET535/30m emission filter were used referred as YFP-channel. To detect cyanobacterial autofluorescence, an excitation filter BP 535/50 and a suppression filter BP 610/75 were used. Image acquisition was done with a Leica DFC360FX black-and-white camera. Captured images were colored with the Leica Application Suite Software (LAS AF) provided by Leica Microsystems. Bright-field images were exposed for 6 ms, Venus fluorescence images for 150 ms and autofluorescence images for 100 ms. Microscope slides covered with dried 2% (w/v) agarose solution were used to immobilize the cells during all microscopical examinations.
Quantitative image evaluation by fluorescence intensity measurements was performed using the open-source software ImageJ (Fiji) (Schindelin et al., 2012). To estimate the fluorescence intensities in different cell compartments, a linear profile of the gray values across the cells through the plasma membrane and cytoplasm was determined. The average gray level of the cytoplasm was subtracted from the maximum gray level of the plasma membrane signal to yield the “Δ fluorescence” value, which indicates the cytoplasmic membrane-localized signal. An example of this profile data quantification is given in Supplementary Figure 1.
Results
PII Signaling Mutants Show Increased Sensitivity to Ammonium
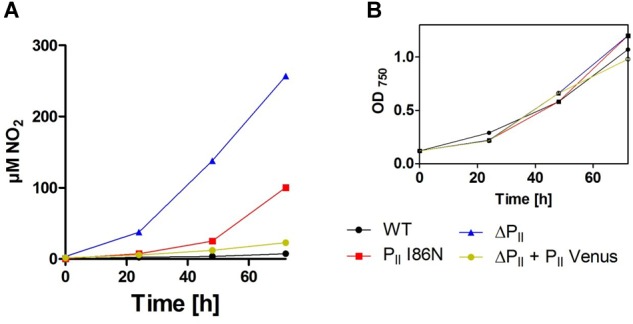
PII homologs of the GlnK subfamily from heterotrophic bacteria are known to regulate cellular ammonium influx by direct protein–protein interaction with the ammonium permease AmtB (Huergo et al., 2012). The observation of impaired ammonium utilization in the Synechocystis strain, which harbors the PII(I86N) variant, suggested that this PII variant either blocked ammonium uptake or abolished proper ammonium uptake regulation with consequent ammonium sensitivity. The Ile 86 to Asn point mutation in this PII variant causes a specific alteration of the T-loop conformation, which facilitates interaction with NAGK and thus, results in an over-activation of this PII target (Fokina et al., 2010b). Conversely, other PII targets might by negatively affected by the altered T-loop conformation. To gain further insights into the role of PII in ammonium uptake, we tested ammonium utilization of a PII deletion mutant (ΔPII) (Hisbergues et al., 1999) and of the ΔPII strain complemented with PII-Venus (PII-Venus-comp.) (Hauf et al., 2016). The complementation strain was previously generated by introducing a shuttle vector (pVZ322), encoding a PII-Venus fusion under control of the native glnB promoter into the Synechocystis ΔPII mutant (Hauf et al., 2016). The Venus fluorescent protein was fused to the C-terminus of PII to avoid sterical hindrance of the T-loop.
When grown in presence of 5 mM ammonium in liquid medium, the Synechocystis wild-type, the ΔPII and the PII-Venus-comp. strains showed similar growth rates and ammonium consumption (Figure 1A,B). However, with an elevated ammonium concentration of 10 mM, only the wild-type and PII-Venus-comp. strains could maintain normal growth, while the ΔPII mutant arrested growth (Figure 1C). Ammonium utilization of wild-type and PII-Venus-comp. were similar under this condition (Figure 1D) while the ΔPII strain ceased ammonium utilization (Figure 1D). The ammonium-sensitive phenotype of the ΔPII strain, in particular the response toward 10 mM ammonium, resembled the previously reported phenotype of the Synechocystis PII(I86N) strain (Watzer et al., 2015). One possibility to explain the inability of the PII mutant strains to grow in 10 mM ammonium-supplemented medium is an increased sensitivity toward toxic effects of ammonium. Although ammonium represents the preferred nitrogen source, it becomes toxic for many photosynthetic organisms at higher concentrations (Drath et al., 2008). To find out whether the reduced growth and impaired ammonium utilization results from ammonium intoxication, we assayed the growth of the various strains in the presence of increasing concentrations of ammonium using the spot assay method (Figure 2). Over a range from 5 to 40 mM ammonium, the wild-type and PII-Venus-comp. strains showed a similar growth and ammonium tolerance. However, the PII(I86N) mutant showed impaired growth at 25 mM while the ΔPII mutant was already strongly affected at a concentration of 10 mM ammonium (Figure 2). This demonstrated that PII is indeed required to cope with elevated ammonium concentrations. The PII(I86N) mutant can only partially replace the wild-type PII protein, whereas PII-Venus perfectly complemented the ΔPII strain.
FIGURE 1.
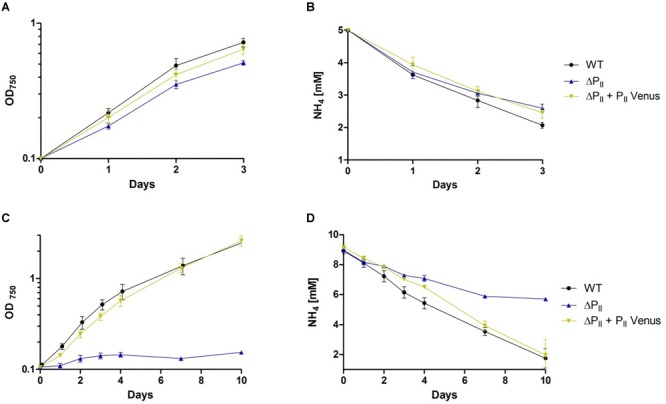
Ammonium supplemented growth and ammonium utilization of Synechocystis sp. wild-type, ΔPII and PII-Venus-comp. (ΔPII + PII-Venus) strains. Values are the means of three biological replicates. (A) Growth curve in presence of 5 mM ammonium. (B) Ammonium concentration in culture supernatants of the various Synechocystis strains grown in BG-11 medium with 5 mM ammonium. (C) Growth curve in presence of 10 mM ammonium. (D) Ammonium concentration in culture supernatants of the various Synechocystis strains grown in BG-11 medium with 10 mM ammonium.
FIGURE 2.
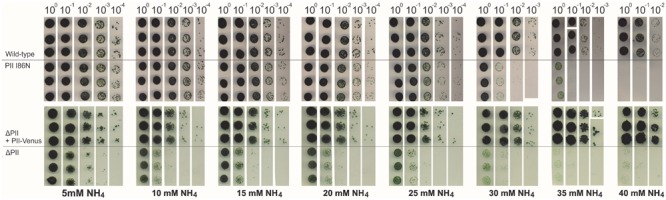
Ammonium sensitivity of the various Synechocystis strains (as in Figure 1) as determined by the drop plate method with three biological replicates per strain. Starved cultures were adjusted to an OD750 of 1.0 and diluted tenfold in series. The dilutions were dropped onto BG-11 agar plates containing increasing concentrations of ammonia (5–40 mM).
Identification of Novel PII Interaction Partners by FLAG-Tag PII Pull-Down Assays
Uncontrolled influx of ammonium and resulting intoxication of Synechocystis ΔPII and PII(I86N) mutants is likely a cause for the elevated ammonium sensitivity in these strains. This suggested the involvement of PII in the regulation of Synechocystis ammonium permeases. In order to verify such an interaction, we performed pull-down assays using a C-terminal 3xFLAG-tagged PII fusion protein that was expressed in Synechocystis under control of the strong psbA promoter. Protein extracts were prepared in presence of detergent, to solubilize membrane-bound proteins, and were subjected to anti-FLAG-tag immunoprecipitation. The proteome composition of the immunoprecipitate was subsequently analyzed by MS to reveal the proteins that co-purify with FLAG-tag PII. As an initial experiment, the immunoprecipitation was performed with cells grown in BG-11 medium, containing nitrate as unique nitrogen source. To discriminate between PII interaction partners and unspecific background proteins, the non-transformed wild-type strain was used as an experimental control. Each experiment was further validated by an independent replicate.
As a proof of concept, we were able to identify PII as the most abundant protein in the PII 3xFLAG pull-down (thereafter PII pull-down). Furthermore, some reported interaction partners, such as PipX, were either exclusively identified in the PII pull-down, or, as in the case of the PII phosphatase PphA, were strongly enriched compared to the background control (Supplementary Figure 2) (Kloft and Forchhammer, 2005; Espinosa et al., 2007; Forcada-Nadal et al., 2014). The identification of a large number of background proteins in the experiments depends on the wash-stringency, the abundance of individual proteins and the high sensitivity of MS-detection. To distinguish between specific PII-enriched interaction partners and background, we determined relative protein abundance ratios between the PII and control experiments by using label free quantification (LFQ) (Cox et al., 2014). Proteins with a significantly higher abundance in the PII pull-down were identified by significance analysis (p-value = 0.01). Since both independent replicates from nitrate-grown cultures yielded very similar results, we correlated the data (Figure 3). Using this approach, the major ammonium permease Amt1 (Montesinos et al., 1998) indeed appeared among the significantly enriched proteins, supporting our suggestion that Amt1 represents a PII binding target in cyanobacteria. Surprisingly, we could also identify subunits of other nitrogen import complexes besides Amt1 among the significantly enriched proteins, in particular subunits of the bispecific NrtABCD nitrate/nitrite and the UrtABCDE urea transporter complexes. For the NRT and URT transporters, all subunits except the periplasmic substrate binding proteins NrtA and UrtA were either exclusively present or strongly enriched in the PII pull-downs. This suggests that the cytoplasmic components of the transporters together with the pore forming subunit are still associated in a complex, detached from the loosely associated periplasmic binding proteins (NrtA or UrtA).
FIGURE 3.
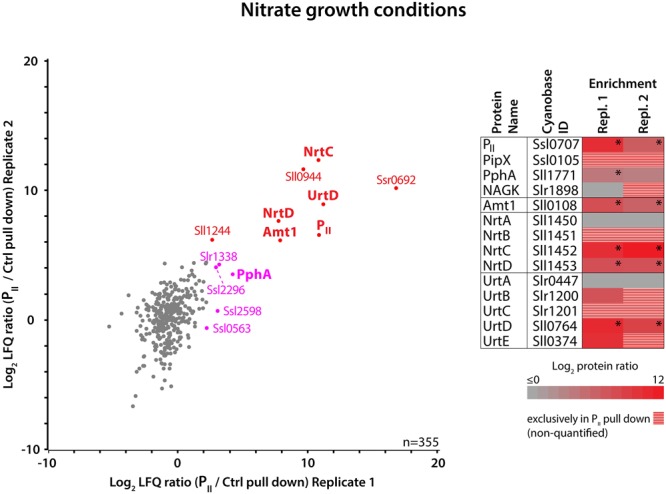
Proteins enriched in anti-FLAG-tag PII immunoprecipitates from nitrate growth conditions. (Left) Scatterplot of the log2 transformed LFQ protein ratios (PII/control pull-down experiment) of 355 quantified proteins detected in two independent replicates. PII pull-down specific outliers which are significantly enriched in both replicates are indicated in red, and significant outliers enriched only in Replicate 1 are indicated in magenta (p-value = 0.01). (Right) Heat map representation of the PII-specific enrichment of nitrogen metabolism related proteins. Log2 transformed LFQ protein ratios from both replicates are color coded and stars indicate significance. Proteins with missing LFQ ratios, detected exclusively in the PII but not in the control pull-down, are striped in red/gray.
To further validate a potential interaction between PII and the URT transporter complex, we repeated the experiment with cells grown in urea-supplemented medium. In agreement with the results from the PII pull-down at nitrate growth conditions, we detected an enrichment of the PII target PipX, the ammonium permease Amt1, subunits of the NRT nitrate/nitrite transporter, and most prominent, subunits of the URT urea transporter (Supplementary Figure 3). By correlating the data from both independent replicates (Figure 4), it became evident that UrtD and UrtE are the strongest enriched proteins under this condition besides PII. This suggests that the presence of urea in the medium influences the interaction of the URT transporter complex with PII.
FIGURE 4.
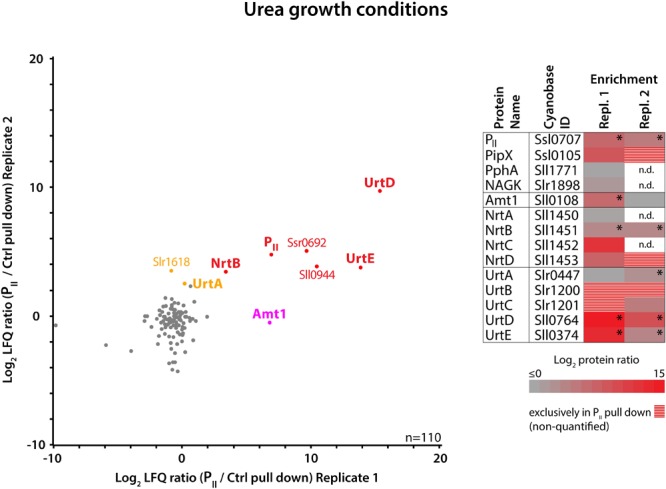
Proteins enriched in anti-FLAG-tag PII immunoprecipitates from urea growth conditions. (Left) Scatterplot of the log2 transformed LFQ protein ratios (PII/control pull-down experiment) of 110 quantified proteins detected in two independent replicates. PII pull-down specific outliers which are significantly enriched in both replicates are indicated in red, and significant outliers enriched in only one replicate are indicated in magenta (Replicate 1) or orange (Replicate 2) (p-value = 0.01). (Right) Heat map representation of the PII-specific enrichment of nitrogen metabolism related proteins. Log2 transformed LFQ protein ratios from both replicates are color coded and stars indicate significance. Proteins with missing LFQ ratios, detected exclusively in the PII but not in the control pull-down, are striped in red/gray, and n.d. indicates no detection in PII and control pull-down.
Besides proteins associated with nitrogen import, we also identified two small hypothetical proteins to be significantly enriched in all experiments, Sll0944 and Ssr0692. Interestingly, both proteins are under transcriptional control of the global nitrogen regulator NtcA (Giner-Lamia et al., 2017), and reveal opposed dynamics in response to nitrogen availability. Under nitrogen-limiting conditions, Sll0944 was found to be strongly up-regulated at transcriptome and proteome levels, whereas Ssr0692 was strongly down-regulated (Spät et al., 2015; Giner-Lamia et al., 2017). Intriguingly, Ssr0692 seems to be up-regulated under carbon limitation (Battchikova et al., 2010) and might be associated to carbon fixation, as it was identified as a potential interactor of NdhH (Sato et al., 2007). The protein Sll0944 is highly conserved in cyanobacteria and possesses a domain of unknown function (DUF1830). Overall, our results imply a direct linkage of Sll0944 and Ssr0692 to nitrogen metabolism with a regulatory involvement of PII. Both proteins are currently under investigation to validate a potential interaction with PII and to determine their function.
Surprisingly, the well-studied PII interactor NAGK was not clearly enriched in these experiments. A low percentage of PII-NAGK complexes was expected to be present under nitrate or urea growth conditions (Burillo et al., 2004), although the strongest interaction occurs under conditions of nitrogen oversupply. Possibly, the C-terminal 3xFLAG-tag fusion of this PII variant could negatively affect complex formation with NAGK. Therefore, pull-down experiments were also performed with an N-terminal 3xFLAG-tag fusion to PII. There, we could clearly identify NAGK in the pull-down of nitrate grown cells in two independent replicates (Table 4). The identification of NAGK in both N-terminal PII-fusion pull-down replicates implies that the localization of the 3xFLAG-tag has an influence on the interaction between PII and its targets. Comparing the results from N- or C-terminal FLAG-tag fused PII pull-down experiments can give additional information to identify potential PII interaction partners. In contrast to the experiments utilizing the C-terminal FLAG-tag, where subunits of the heterotrimeric urease complex UreABC were not enriched, to our surprise, the N-terminal FLAG-tagged PII protein co-purified the entire urease-complex including associated urease accessory proteins D, F, and G. This remarkable finding implies a possible direct interaction between the urease transport complex and the succeeding enzymatic machinery urea metabolism, indicating possible metabolic channeling of urea (Sweetlove and Fernie, 2018).
Table 4.
Identified proteins in pull-down experiments at nitrate growth conditions utilizing the N-terminal 3xFLAG-tagged PII fusion protein: Displayed are the identified proteins from two independent replicates (Repl. I and Repl. II).
| Protein name/ | Cyanobase | Mol. weight | ||
|---|---|---|---|---|
| complex | ID | (kDa) | iBAQ intensity |
|
| Repl. I | Repl. II | |||
| PII/N-3xFLAG PII | Ssl0707 | 15.43 | 4.2∗107 | 1.1∗109 |
| PipX | Ssl0105 | 10.45 | 3.1∗106 | 9.1∗107 |
| PphA | Sll1771 | 28.47 | 4.2∗104 | 9.1∗105 |
| NAGK | Slr1898 | 31.53 | 9.4∗104 | 6.5∗105 |
| Amt1 | Sll0108 | 53.58 | 1.2∗108 | 2.9∗108 |
| NrtA | Sll1450 | 48.97 | 1.1∗105 | 1.4∗106 |
| NrtB | Sll1451 | 29.72 | 8.8∗105 | 5.1∗106 |
| NrtC | Sll1452 | 75.10 | 1.4∗106 | 6.1∗106 |
| NrtD | Sll1453 | 36.56 | 1.9∗105 | 2.9∗106 |
| UrtA | Slr0447 | 48.36 | 1.0∗105 | 2.6∗105 |
| UrtB | Slr1200 | 41.68 | 2.4∗105 | 1.6∗106 |
| UrtC | Slr1201 | 45.08 | 3.5∗105 | 4.9∗106 |
| UrtD | Sll0764 | 41.19 | 8.1∗105 | 3.9∗106 |
| UrtE | Sll0374 | 27.42 | 6.5∗105 | 6.4∗106 |
| UreA | Slr1256 | 11.06 | – | 4.9∗105 |
| UreB | Sll0420 | 11.38 | 2.0∗105 | 5.2∗107 |
| UreC | Sll1750 | 61.04 | 9.9∗106 | 3.5∗107 |
| UreD | Sll1639 | 27.16 | 1.2∗105 | 6.4∗105 |
| UreF | Slr1899 | 20.23 | 3.1∗104 | 5.2∗105 |
| UreG | Sll0643 | 22.01 | 1.3∗105 | 1.2∗106 |
| Sll0944 | Sll0944 | 18.15 | 8.4∗105 | 9.8∗107 |
| Ssr0692 | Ssr0692 | 5.85 | 2.8∗105 | 9.3∗106 |
Protein intensities were calculated using the MaxQuant iBAQ algorithm (Schwanhausser et al., 2011).
Bacterial Two-Hybrid Assay
To further confirm the interaction of PII with the identified nitrogen transporters, we performed bacterial two-hybrid assays using the Bordetella pertussis adenylate cyclase two-hybrid system (BACTH) (Battesti and Bouveret, 2012). We tested the cytoplasmic localized ATP-binding proteins of the ABC-type transporters NrtC, NrtD, UrtD, UrtE (Omata et al., 1993; Valladares et al., 2002) as well as Amt1 for possible interactions with various PII proteins. For each interaction candidate, an N- and a C-terminal fusion to the T25 fragment was constructed. In case of a positive interaction with a PII-T18 fusion, cAMP is formed within a cAMP deficient E. coli host cell. The interactions were tested by plate assays on X-Gal and MacConkey reporter plates for highest detection sensitivity. The PII – PipX and leucine zipper interactions were used as positive controls. PII-T18 fusions with an empty pKT25 vector were used as negative controls. Next to wild-type PII-T18 fusion, we also included T18 fusions of the PII(I86N), PII(S49D), and PII(R9L) variants to find out how these different PII variants, including a phosphomimetic T-loop variant and variants with different effector binding properties would differ in potential PII interactions. Figure 5 shows the observed interactions on X-Gal plates in the bacterial two-hybrid assays.
FIGURE 5.
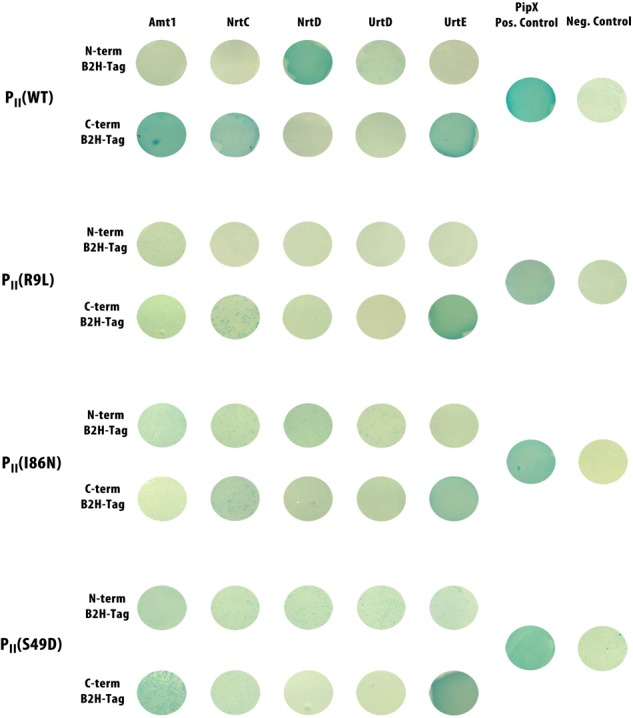
Bacterial-two-hybrid interactions on X-Gal plates of wild-type PII and the PII variants PII(R9L), PII(I86N), PII(S49D) with different transporter subunits. Interaction of PII with C-terminal tagged PipX was used as a positive control. PII with an empty pKT25 vector was used as negative control. Positive interactions are indicated by a blue coloration of the colonies.
All PII variants showed interaction with the known PII interaction partner PipX. Wild-type PII showed clear interaction with Amt1 C-terminally fused to the T25 fragment, while the variants PII(I86N) and PII(R9L) did not interact. The loss of interaction of PII(I86N) is in agreement with our previous conclusions regarding the impaired ammonium utilization of Synechocystis sp. strain PII(I86N). The phosphomimetic variant PII(S49D) showed weak interaction with Amt1. In case of the nitrate transport ATP-binding subunits NrtC and NrtD, the BACTH assay revealed clear interaction of wild-type PII with both subunits. Intriguingly, PII interacted only with NrtC when it was fused at its C-terminus with T25, whereas in the case of NrtD, PII interacted with the N-terminal fusion. In contrast to wild-type PII, the PII(R9L) variant was completely unable to interact with any of the Nrt-proteins, while weak interactions were observed with variants PII(S49D) and PII(I86N). With the ATP-binding subunits of the urea transporter, no interaction was observed with the UrtD protein, neither with wild-type PII nor with any of the tested variants. By contrast, the C-terminal T25 fusion of UrtE showed clear interaction with wild-type and all PII variants. The fact that positive interactions only occurred with selective combinations of PII variants and N-or C-terminal T25 fusions of target proteins supports the conclusion that the observed interactions are indeed specific.
PII Deletion Mutant and PII(I86N) Mutant Excrete Nitrite
The pull-down and BACTH assays shown above clearly indicated interaction of wild-type PII protein with components of the uptake systems for ammonium, nitrate, and urea. The interaction with Amt1 likely resembles the known interaction of the PII protein GlnK from heterotrophic bacteria with AmtB (Conroy et al., 2007; Gruswitz et al., 2007) and perfectly explains the physiological response of Synechococcus PCC 7942 (hereafter Synechococcus) and Synechocystis PII mutants toward ammonium. Interaction of PII with components of the NrtABCD nitrate/nitrite transporter had in fact been suggested previously by several physiological studies, which documented altered nitrate utilization properties in various Synechococcus and Synechocystis PII mutants (Kobayashi et al., 1997; Lee et al., 1998, 2000; Kloft and Forchhammer, 2005). A characteristic phenotype of a PII deletion mutant grown in the presence of nitrate is the uncontrolled uptake of nitrate and subsequent excretion of nitrite into the medium (Kloft and Forchhammer, 2005). Reduction of nitrate requires two electrons whereas six electrons are needed for the reduction of nitrite to ammonium. Because of the lower electron demand for nitrate reduction, nitrite reduction becomes limiting when insufficient reductant is available. Therefore, when nitrate uptake is uncontrolled, nitrite accumulates and is excreted. We wondered if the Synechocystis PII(I86N) strain shows a similar nitrite excretion phenotype than the PII deficient mutant when grown on nitrate and if PII-Venus complements the nitrite excretion phenotype of the PII-deficient mutant.
To answer this question, Synechocystis wild-type strain, PII(I86N), ΔPII and the PII-Venus comp. strains were grown with nitrate as sole nitrogen source under constant illumination (40 μmol photons m-2 s-1) and samples were removed for nitrite determination. Under these conditions, the various strains showed similar growth rates (Figure 6). Both mutant strains expected to be impaired in proper control of the NRT complex, the ΔPII mutant and the PII(I86N) strain, indeed excreted nitrite into the medium. Notably, the PII(I86N) mutant excreted much lower nitrite amounts than the PII deletion mutant, in agreement with the partial interaction of the PII(I86N) variant observed in the BACTH assays with NrtC and NrtD subunits. By contrast, PII-Venus perfectly complemented the nitrite excretion phenotype. Together, these results indicated that the BACTH interactions of PII with the NrtC and NtcD subunits are physiologically meaningful.
FIGURE 6.
(A) Nitrite excretion of Synechocystis sp. wild-type, PII(I86N), ΔPII and PII-Venus-comp. (ΔPII + PII-Venus) complementation grown in BG11 medium supplemented with nitrate. (B) Corresponding growth curve to the experiment shown in panel (A).
PII Is Responsible for Inhibition of Nitrate Uptake
To gain further insights in the PII-dependent regulation of nitrate uptake, we measured nitrate consumption rates of the various PII-mutants under different conditions. For this purpose, cells from exponential phase of growth in standard BG-11 medium were washed and subsequently incubated in BG-110 medium containing 200 μM NO3 as nitrogen source. Nitrate utilization was quantified by measuring the nitrate concentration in the culture supernatant over time. Under constant illumination of 40 μmol photons m-2 s-1, Synechocystis wild-type and the PII-Venus comp. strain showed lower nitrate consumption rates, while both, the PII(I86N) and ΔPII mutant, showed higher nitrate removal (Figure 7A). In agreement with our previous results, we observed excretion of nitrite by the PII(I86N) and ΔPII mutants (Figure 7B). Interestingly, while the nitrate utilization rate of the PII(I86N) and ΔPII mutant appeared almost identical, the two mutants differed in their rate of nitrite excretion. While nitrite excretion of the ΔPII mutant accelerated over time, the PII(I86N) initially excreted nitrite faster but then slowed it down over time, explaining the lower amounts of nitrite in the exponentially growing culture (Figure 6) as compared to the ΔPII mutant.
FIGURE 7.
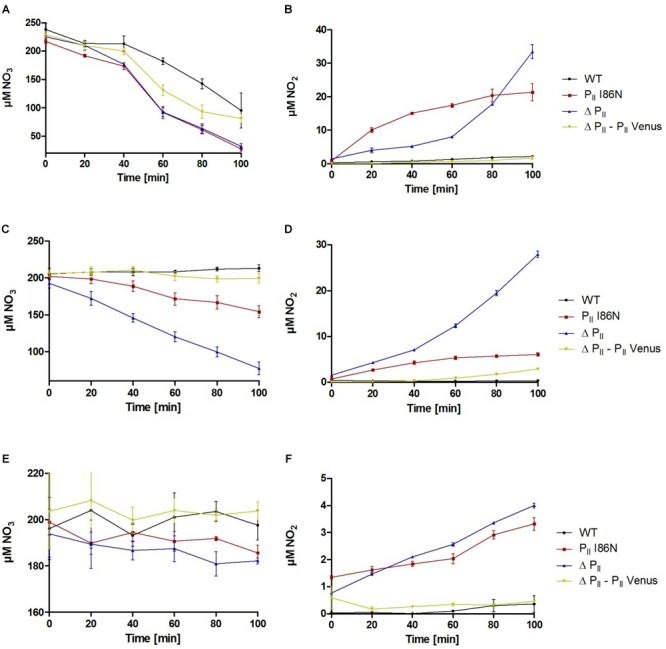
Nitrate utilization and nitrite excretion of various Synechocystis strains as indicated. Exponentially growing and nitrate supplemented Synechocystis cells were washed and resuspended in a medium containing 200 μM nitrate. Shown are the means of three biological replicates. (A) Nitrate utilization and (B) nitrite excretion under constant illumination of 40 μmol photons m-2 s-1. (C) Nitrate utilization and (D) nitrite excretion in presence of 2 mM ammonium and under constant illumination of 40 μmol photons m-2 s-1. (E) Nitrate utilization and (F) nitrite excretion in the dark.
Addition of ammonium to nitrate grown cells leads to an immediate inhibition of nitrate uptake. This ammonium-dependent inhibition of nitrate uptake requires the presence of PII protein or PII phosphomimetic variants (Lee et al., 1998). Here, we tested if the PII(I86N) mutant was able to perform the ammonium-dependent nitrate uptake inhibition. For this purpose, nitrate consumption rates were determined after the addition of 2 mM ammonium. Both, Synechocystis wild-type and the PII-Venus comp. strains showed a complete inhibition of nitrate uptake in response to ammonium (Figure 7C). By contrast, the PII(I86N) and ΔPII mutant strains maintained nitrate uptake after ammonium addition. However, nitrate uptake in the PII(I86N) mutant was clearly diminished as compared to the ΔPII mutant (Figure 7C). The uncontrolled nitrate uptake of the PII(I86N) and ΔPII mutant caused excretion of nitrite. Due to the diminished uptake of nitrate by the PII(I86N) mutant, its nitrite excretion was correspondingly lower as compared to the ΔPII mutant (Figure 7D).
In addition to the absence of ammonium, active nitrate transport also requires photosynthetic CO2 fixation (Romero et al., 1985). Nitrogen assimilation is tightly regulated by light/dark transitions. A transition from light to dark causes an immediate inhibition of nitrate uptake and an inhibition of the ammonium assimilating GS (Romero et al., 1985; Marques et al., 1992). To reveal a role of PII in this response, we tested the dark-switch-off of nitrate uptake in the various PII mutant strains. Although the overall nitrate consumption was very low in darkness (Figure 7E) clear differences could be resolved. Notably, the PII(I86N) and ΔPII mutant strains showed slow but constant nitrate uptake as compared to the wild-type and the PII-Venus comp. strain, which did not take up any nitrate. Corresponding to the uptake of nitrate, PII(I86N) and ΔPII mutant excreted small amounts of nitrite (Figure 7F), supporting the notion that the PII(I86N) and ΔPII mutants are unable to completely switch-off nitrate uptake in the dark.
PII Signaling Mutants Show Impaired Urea Utilization
To validate the physiological relevance of the interaction of PII with the UrtE subunit of the urea transporter UrtABCDE, we analyzed urea utilization in the different PII mutants. For this purpose, exponentially growing cells were washed and incubated in BG-110 medium, containing 150 μM urea as sole nitrogen source with constant illumination of 40 μmol photons m-2 s-1. Under these experimental conditions, Synechocystis wild-type and PII(I86N) mutant strains utilized similar amounts of urea (Figure 8A), whereas both, the ΔPII and PII-Venus comp. strains consumed considerably higher amounts of urea (Figure 8A). The wild-type property of the PII(I86N) variant is in agreement with the interaction of the PII(I86N) protein with the UrtE subunit, which appeared to be T-loop independent according to BACTH assays. Urea is hydrolyzed to CO2 and two molecules of ammonium by the enzyme urease (Mobley and Hausinger, 1989; Esteves-Ferreira et al., 2018). We wondered if an uncontrolled influx of urea could lead to an excretion of ammonium. Therefore, we measured ammonium concentrations in culture supernatant of those cells, which were incubated with 150 μM urea as the sole nitrogen source. Indeed, we detected ammonium excretion of the ΔPII mutant proportional to its urea utilization (Figure 8B). In contrast, only minor amounts of ammonium could be detected in the culture supernatant of the Synechocystis wild-type, PII(I86N) and PII-Venus comp. strain (Figure 8B).
FIGURE 8.
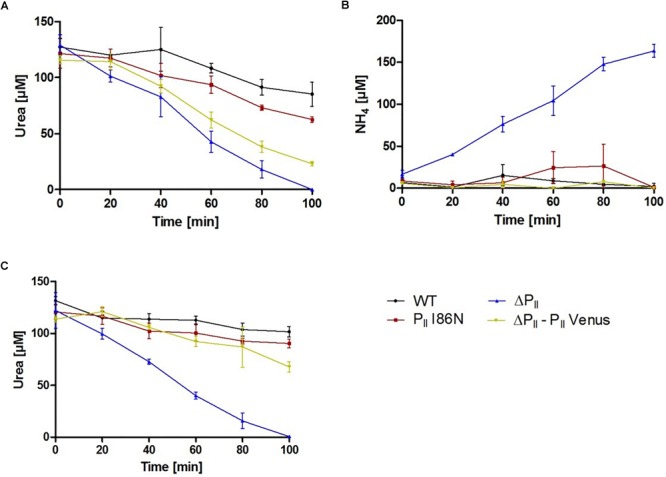
Urea utilization and ammonium excretion of various Synechocystis strains as indicated. Exponentially growing and nitrate supplemented Synechocystis cells were washed and resuspended in a medium containing 150 μM urea. Shown are the means of three biological replicates. (A) Urea utilization and (B) ammonium excretion under constant illumination of 40 μmol photons m-2 s-1. (C) Urea utilization in presence of 2 mM ammonium and under constant illumination of 40 μmol photons m-2 s-1.
Synechocystis utilizes available nitrogen sources in a hierarchical order. In presence of ammonium, the uptake of external nitrogen sources is blocked (Muro-Pastor et al., 2005). To test ammonium-dependent inhibition of urea utilization, we determined urea consumption in presence of 2 mM ammonium. The Synechocystis wild-type, PII(I86N) and PII-Venus comp. strains showed a clear inhibition in urea utilization in response to the addition of ammonium. In contrast, urea uptake in the ΔPII mutant was completely unaffected by ammonium addition (Figure 8A,C). This demonstrates that PII also controls urea uptake, as suggested by the interaction assays.
PII Localization Changes Upon Addition of Nitrate, Ammonium, and Urea
Our previous pull-down experiments and bacterial two-hybrid assays clearly showed interaction of Synechocystis PII with Amt1 as well as with nitrate and urea transport components. All interactions appeared to be physiologically relevant. Since all these transporters are localized to the plasma membrane, we suspected that this interaction might affect the cellular localization of PII. Therefore, we monitored PII localization using the Synechocystis PII-Venus comp. strain grown under different nitrogen regimes.
Nitrate-replete cells in the mid-exponential growth phase (OD750 ∼ 0.5) showed heterogeneous distribution of PII-Venus fluorescence in the cell. The majority of cells displayed a strong fluorescence signal in their center and peripheral cell boundary (Figure 9A,B,D). The intense signal in the center corresponds to the central cytoplasmic space, inside the multiple thylakoid layers. In this region, PII might interact with different soluble proteins, like NAGK or PipX. The area with low Venus fluorescence is occupied by the thylakoid membranes. The PII-Venus fluorescence at the cell boundary corresponds to the plasma membrane, where Amt1, NrtABCD and UrtABCDE are located (Hahn and Schleiff, 2014) (Figure 9C).
FIGURE 9.
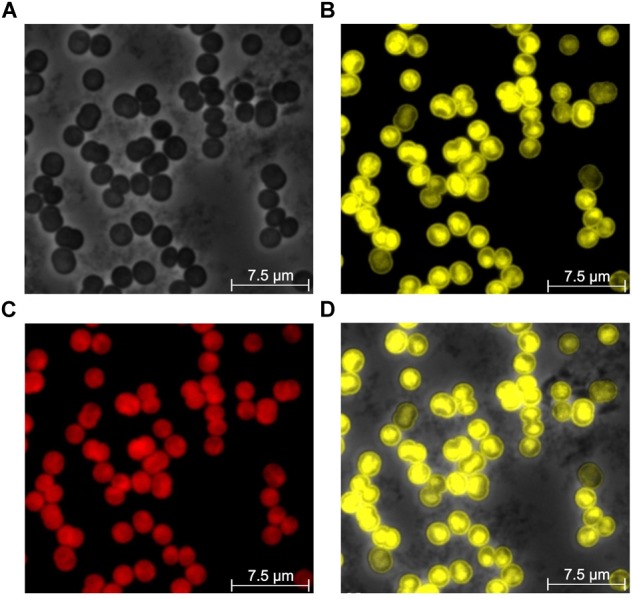
Localization of PII-Venus during exponential growth (OD750 of 0.5) of Synechocystis (A) Phase contrast image of cells. (B) Venus fluorescence. (C) Autofluorescence of thylakoid membranes. (D) Overlay of Venus fluorescence and phase contrast images.
When cells are shifted to nitrogen-depleted conditions, they activate the NtcA regulon, including the glnB gene and various uptake systems for nitrogen compounds. During prolonged starvation, they undergo chlorosis, which includes a reduction of thylakoid membranes (Forchhammer and Schwarz, 2018). The chlorotic cells rapidly respond to the addition of combined nitrogen sources. Therefore, investigation of PII-Venus fluorescence in chlorotic cells and following the addition of combined nitrogen sources was expected to reveal further insights in the in vivo dynamics of PII interactions. To monitor the PII localization in the chlorotic, nitrogen depleted state, cells were grown to an OD750 of 0.4–0.6, washed, and resuspended in BG-110. After 1 week of nitrogen starvation, the PII-Venus signal was evenly distributed throughout the whole cell and its localization on the plasma membrane was not as distinct as during nitrate-supplemented exponential growth (Figure 10A). To test the localization following the addition of different nitrogen sources, 1 week nitrogen starved Synechocystis cultures were supplemented with 5 mM NO3, 5 mM NH4+ or 5 mM urea, respectively. Immediately thereafter, a change in the PII localization became visible. The majority of cells showed a clearly visible, distinct plasma membrane localized PII-Venus fluorescence, while the remaining cytosol showed homogenously distributed fluorescence. The re-localization of PII to the plasma membrane appeared most clearly in response to ammonium addition as compared to nitrate or urea (Figure 10B–D).
FIGURE 10.
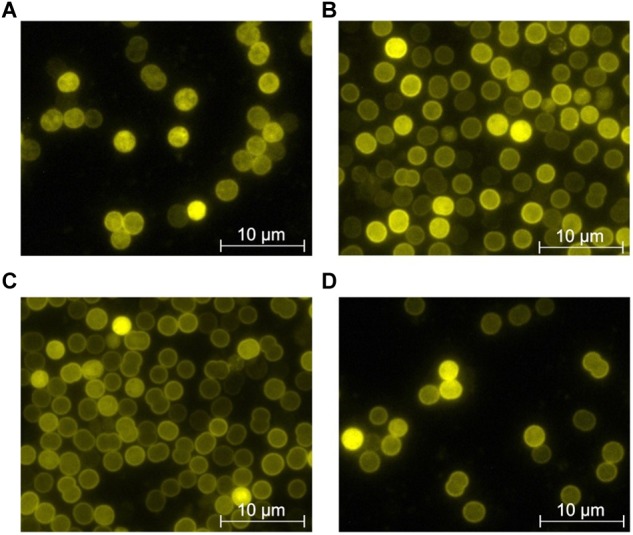
PII-Venus localization under different nitrogen supplemented conditions. (A) PII-Venus fluorescence of 1 week nitrogen starved Synechocystis cells. (B–D) PII-Venus fluorescence of Synechocystis during resuscitation from nitrogen starvation. One week nitrogen starved Synechocystis cultures were resuscitated by adding 5 mM nitrate (B), 5 mM ammonium (C), or 5 mM urea (D). Fluorescence images were taken 1 h after addition of the nitrogen source.
Since the fluorescence distribution is subject to a certain degree of heterogeneity with the bulk of single cells observed in the microscope, we attempted to quantitatively describe the average distribution of PII-Venus in a representative set of cells. Therefore, a profile of the fluorescence intensities across median cell sections was determined from 54 individual cells per investigated time point. From these profiles, the relative fluorescence intensity distribution between plasma membrane localized signal to the average cytoplasmic signal was determined and displayed as box-plot (Figure 11). Under nitrogen depletion, the majority of the cells showed a higher cytoplasmic PII-Venus localization than plasma membrane localization. Addition of a nitrogen source directly induced a re-localization of PII toward the plasma membrane. In the first 2 h after addition of nitrate or urea, PII-Venus showed a similar localization change (Figure 11A,C,D). Two hours after the addition of nitrate, the PII-derived signal from the plasma membrane decreased again and re-appeared in the cytoplasmic space (Figure 11A), whereas in urea treated cells, PII continued to accumulate at the plasma membrane (Figure 11C). Addition of ammonium induced the most prominent migration of PII-Venus to the plasma membrane in the first hour as compared to nitrate or urea, but thereafter, PII-Venus started to move back to the cytoplasm (Figure 11B).
FIGURE 11.
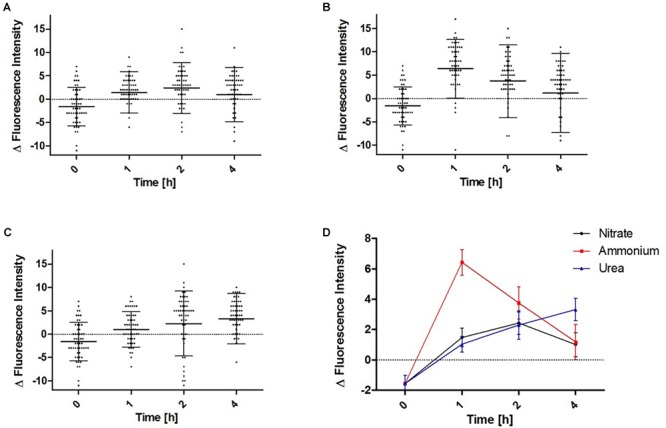
Quantification of the PII-Venus migration toward the plasma membrane in response to the addition of different nitrogen sources to nitrogen starved Synechocystis sp. cultures. A linear profile of the fluorescence intensities across midcell was recorded and evaluated as shown in Supplementary Figure 1. The maximum fluorescence intensity at the plasma membrane was normalized to the average intensity of the cytoplasm and then, the difference between plasma localized signal and average cytoplasmic signal was calculated (Δ fluorescence intensity). When Δ fluorescence intensity values are below 0, this indicates that the plasma membrane signal is weaker than the average cytoplasmic signal, while values above 0 indicate that the plasma membrane signal is higher than the average cytoplasmic signal. Resuscitation of 1 week nitrogen starved Synechocystis sp. cultures were induced by adding either 5 mM NO3 (A), 5 mM NH4 (B), or 5 mM urea (C). 50–60 cells were measured per time point. Dots indicate single cell measurements; whiskers showing the standard-deviations; thick black lines show the arithmetic mean. (D) Direct comparison of the mean Δ fluorescence intensities from NO3, NH4, and urea induced resuscitation.
Discussion
The present investigation unraveled a so far unrecognized global function of the cyanobacterial PII signaling protein in controlling the uptake of the major nitrogen sources ammonium, nitrate and urea. Previous studies already suggested occasionally involvement of PII in the regulation of ammonium and nitrate transport (Kobayashi et al., 1997; Lee et al., 1998, 2000; Kloft and Forchhammer, 2005; Conroy et al., 2007), however, this issue was never addressed systematically using a combined biochemical, genetic and physiological approach. Starting from a series of pull-down experiments using FLAG-tagged PII variants, we observed a putative interaction of PII with various nitrogen transport systems. The interactions were verified by bacterial-two hybrid assays and the biological significance of the interaction was validated by physiological experiments. Finally, the proposed dynamic interaction of PII with cytoplasmic membrane bound transporters was corroborated by fluorescence microscopy.
Significance Statement
PII signaling proteins play versatile roles in the coordination of carbon- and nitrogen anabolism in prokaryotes and plant chloroplast. In different phylogenetic lineages, PII controls a variety of different target proteins. In cyanobacteria, the PII paralog GlnB has been shown to control global nitrogen-responsive gene expression by interacting with the transcriptional co-activator PipX. Furthermore, PII controls nitrogen storage metabolism in cyanobacteria by regulating the key enzyme of arginine synthesis, N-acetylglutamate kinase. Finally, a key enzyme in fatty acid metabolism, the acetyl-CoA carboxylase, was shown to be a target of PII. Several lines of evidences suggested that PII might also be involved in the control of ammonium and nitrate uptake, however, direct involvement of PII has not yet been shown. In this study, we revealed the interactome of PII from the cyanobacterium Synechocystis by immunoprecipitating FLAG-tagged PII protein. We found prominent enrichment of components of ammonium, nitrate and urea uptake systems. Direct protein–protein interaction was confirmed by bacterial-two hybrid analysis and the physiological relevance was verified by analyzing ammonium-, nitrate-, and urea-uptake in various PII mutant strains of Synechocystis. This study, therefore, demonstrates that PII is the master regulator of the most prominent nitrogen transport systems in cyanobacteria.
Synechocystis PII Regulates Ammonium Uptake by Interacting With the Amt1 Ammonium Permease
In E. coli, the PII homolog GlnK regulates AmtB by direct protein–protein interaction to control the influx of ammonium (Conroy et al., 2007). In the GlnK-AmtB complex, the nucleotide binding pockets of GlnK are occupied with ADP and the T-loop of GlnK adopts a vertically extended conformation that closes the ammonium gas channel (Conroy et al., 2007; Maier et al., 2011; Forcada-Nadal et al., 2018). Since the conformation of the T-loop changes upon binding of ATP-Mg2+-2-oxoglutarate, the GlnK-AMT complex only forms under conditions of low 2-OG concentrations, which allows formation of the alternative GlnK-ADP complex (Radchenko et al., 2014).
The present results suggest that in Synechocystis, PII regulates the major ammonium permease Amt1 in a similar manner than GlnK the AmtB channel. A weak interaction of Amt1 could still be detected with the phosphomimetic variant S49D. The reduced affinity indicates that the negative charge at position 49 reduces the affinity to Amt1. Therefore, S49 phosphorylated wild-type PII is expected to have an even weaker affinity to Amt1 (two negative charges of phosphoserine as compared to one negative charge of aspartate). In this respect, phosphorylation of S49 would be analogous to uridylylation of Y51 in enterobacterial GlnK proteins, which prevents AmtB interaction (Conroy et al., 2007). Under conditions of strong PII phosphorylation (nitrogen-poor conditions or high CO2 to nitrate ratio), the Amt1 channel would remain open, allowing unrestricted uptake of ammonium ions. When cells are shifted to excess ammonium conditions, PII becomes dephosphorylated (Ruppert et al., 2002) and diminished 2-OG levels allow formation of the PII-ADP complex, which then can close the Amt1 pores.
In BACTH assays, the PII(R9L) variant was completely impaired in Amt1 interaction. This variant is also unable to interact with NAGK, presumably due to a stabilizing function of the R9 residue at the interface to the binding partner (Fokina et al., 2010a). The failure of this variant to interact with Amt1 indicates that also for Amt1 interaction, the lower face of PII with the protruding T-loops is involved in this protein–protein interaction, in agreement with the structure of the GlnK-AmtB complex.
The failure of the PII(I86N) variant to interact with Amt1 corresponds to the loss of its affinity toward ADP (Fokina et al., 2010b). PII(I86N) has one order of magnitude higher affinity toward ATP than toward ADP and thus, is expected to reside almost exclusively in the ATP-bound state, which would abrogate the interaction with Amt1.
Despite that ammonium uptake in the Synechocystis PII(I86N) variant is no more under PII control, this strain exhibits a higher ammonium tolerance than the ΔPII mutant. This difference could be explained by different ammonium assimilation properties. In the PII deletion mutant, the ammonium-scavenging arginine synthesis pathway is not active due to lacking activation of the key enzyme NAGK by PII (Maheswaran et al., 2006). By contrast, Synechocystis PII(I86N) highly enhances NAGK activity, resulting in high intracellular arginine concentrations that leads to accumulation of cyanophycin granules (Watzer et al., 2015). This pathway will foster the metabolic removal of excess ammonium and thus results in increased ammonium tolerance.
Analysis of the subcellular localization of PII in response to ammonium stimuli agrees with the above-depicted concept. Only 1 h after the addition of ammonium to chlorotic cells, PII shows a strong migration toward the plasma membrane. Providing ammonium to nitrogen-starved cells should cause a strong decrease of the cellular 2-OG levels. This leads to the observed accumulation of PII to the cytoplasmic membrane, preventing an excess uptake of too much ammonium. After a while, a new equilibrium will be established leading to a partial re-localization of PII to its cytoplasmic targets, such as NAGK. Indeed, Synechocystis cells recovering from nitrogen chlorosis start to produce cyanophycin already after a few hours (Watzer and Forchhammer, 2018).
PII Regulates Nitrate Uptake by Interacting With NrtD and NrtC Subunits
Several previous studies documented an involvement of PII in the regulation of nitrate/nitrite uptake. Our data provide novel insights regarding the mechanism of PII-mediated regulation of the NrtABCD transporter. It appears that the NrtABCD complex is directly regulated by interaction of PII with the cytoplasmic ATPase subunits NrtC and NrtD, as indicated by the co-isolation of wild-type PII with both of these proteins. The inability of the PII(I86N) complemented strain to regulate nitrate utilization as well as the reduced interaction of PII(I86N) with NrtC and NrtD subunits in BACTH assays could in principle be explained by the altered T-loop conformation or by the different ligand binding properties of this variant, discussed above. As shown by BACTH assays and by recent in vitro studies (Watzer and Forchhammer, unpublished), the PII(I86N) variant is indeed able to interact with PipX. This interaction requires a vertically extended conformation of the T-loop (Llacer et al., 2010; Forcada-Nadal et al., 2018). Although PII-PipX complexes are favored by the ADP-state of PII, efficient PipX-PII interaction also occurs with the PII-ATP complex. As explained above, the PII(I86N) variant is trapped in the ATP-bound state. The fact that this PII variant binds PipX indicates, that the T-loop of PII(I86N) is flexible enough to adopt various conformations. Therefore, it is likely that formation of the PII-NrtC or NrtD complex requires the ADP-bound state of PII, which is disabled in the PII(I86N) variant. As for NAGK and Amt1, the PII(R9L) variant is unable to interact with any of the NRT subunits, indicating that PII interaction involves the lower part of the PII body, where the T-loop emanates.
The phosphomimetic variant PII(S49D) shows, albeit weaker, interaction with the NRT subunits. This positive interaction explains previous reports, that Synechocystis and Synechococcus strains expressing these phosphomimetic variants, display a wild-type like regulation of nitrate/nitrite utilization (Lee et al., 2000; Kobayashi et al., 2005). Why the diminished interaction measured by BACTH assays does not properly reflect the physiological behavior of the PII(S49D) expressing strain (full complementation) could be due to different sensitivities of the assays. Kobayashi et al. (2005) used for the complementation of the Synechocystis deficient mutant plasmid-borne PII constructs, which cause higher expression levels as in the wild-type genotype. Therefore, a weaker interaction of PII with the NRT subunits could still be sufficient to inhibit NRT. The mechanism of NRT inhibition by PII still awaits structure-functional explanation. Our BACTH assays indicate that PII interacts with both ATPase subunits of NRT. Of these, the NrtC subunit seems to play a particular role: Truncation of the regulatory domain of NrtC results in an ammonium-insensitive NRT, despite the presence of a functional PII system. The regulatory domain of NrtC possesses a putative binding site for nitrate (Koropatkin et al., 2006). Probably PII, through binding to NrtC and NrtD, must act in concert with the regulatory NrtC domain, to stop nitrate uptake. If one of the two regulators is missing, ammonium inhibition would not work. It is tempting to speculate that the regulatory domain of NrtC directly senses the nitrate-state of the cells (Koropatkin et al., 2006) to prepare NRT for inhibition by the PII-ADP complex, whereas PII in the 2-OG-Mg-ATP complex (signaling high C/low N state) would not interact and, therefore, not inhibit NRT. This dual regulatory model combines information from the nitrate status with the global C/N and energy state sensing of PII to tune NRT activity to the actual need. This model also explains the light-response of NRT: Low energy conditions favor the ADP-complex of PII, explaining why in the dark, complete switch-off of NRT requires PII.
PII Regulates Urea Uptake by Interacting With the UrtE Subunit
In addition to Amt1 and NRT, our data also identified the ABC-type urea transporter (UrtABCDE) as a novel PII target, with the UrtE subunit being the direct interaction partner of PII. No BACTH interaction was observed for UrtD. This implies that the UrtD protein identified in the FLAG-tag pull-down resulted from pull-down of the entire URT complex. As deduced from the urea-utilization phenotype of the different PII variant strains, as well as form the properties of BACTH interactions, the interaction of PII with UrtE is distinct from the mode of Amt1 or NrtC and NrtD interaction. PII interaction with UrtE seems to be T-loop independent, since the phosphomimetic S49D variant as well as the PII(I86N) and PII(R9L) variants interacted in BACTH assays like wild-type PII. The ability of PII(R9L) to interact with UrtE indicates that the interaction may involve a region of PII distinct from the usual interaction face used by NAGK, PipX or Amt1.
In agreement with a different binding mode between PII and UrtE, the PII(I86N) variant could substitute wild-type PII regarding the control of urea utilization. This suggests that the PII-ATP complex is able to interact with UrtE in a non-conventional manner. The control of urea uptake prevents its futile hydrolysis and the consequent release of ammonium. Although the PII-Venus variant displayed higher urea uptake rates than the PII deficient mutant, it could prevent concomitant ammonium excretion, indicating that also the PII-Venus variant successfully regulates urea utilization. Consistently, all the tested PII variant strains (wild-type, I86N and PII-Venus) were able to carry out ammonium inhibition of urea uptake. Careful examination of the data shows that the PII-Venus strain displayed slightly higher urea uptake rates than the other two strains. It is possible that due to the bulky fluorescent proteins fused to PII the interaction with PII does not tune down urea uptake as efficient as in the case of the non-tagged PII variants. Nevertheless, the PII-Venus construct is functional, which also agrees with the re-localization of PII-Venus to the plasma membrane upon addition of urea to nitrogen-starved cells. Under certain conditions, the entire multisubunit urease complex appeared in PII-FLAG-tag pull-down experiments. This observation deserves further investigation. It suggests either a direct interaction of the urease complex with PII or a possible metabolic channeling between the urea uptake system and the multisubunit urease complex, leading to a co-immunoprecipitation of the entire super-complex. Further studies are required to reveal the molecular mechanism of urea metabolism and the involvement of PII in this process.
Conclusion
All together, the present study has expanded the insights in PII regulatory interactions in Synechocystis. According to this study, the PII regulatory network includes all relevant nitrogen uptake systems in Synechocystis. It is highly probable that the same holds true for other cyanobacteria. In Synechococcus elongatus, (Chang et al., 2013) genetic and physiological studies suggested that in addition to NRT, PII is likely involved in the control of a cyanate ABC transporter (CynABC), which is not present in Synechocystis. Localization of PII-YFP fusions to the cytoplasmic membrane could also be seen in fluorescent images from S. elongatus (Espinosa et al., 2018). Control of transport proteins appears to be a highly conserved property of the PII family members, as exemplified by the widely distributed GlnK-Amt interaction. Recently, a more distantly related member of the PII superfamily, SbtB, an accessory component of the sodium-dependent bicarbonate transporter SbtA, was shown to be involved in regulation of bicarbonate metabolism in Synechocystis (Selim et al., 2018). Moreover, many putative non-characterized PII family members are genetically linked to transport and channel proteins. However, except for GlnK-AmtB, no structures are known for PII-protein complexes with membrane channel proteins. Whether the interactions of the PII members with the channels follow a universal mode, or whether different types of interaction exist, remains to be demonstrated. However, the differences observed between the interactions of PII variants with UrtE, NrtC/D and Amt1 suggest that the interactions could be as versatile as observed for the various soluble PII interactors (Forcada-Nadal et al., 2018).
Data Availability
The datasets generated for this study can be found in the PRIDE ProteomeXchange consortium, PXD013411.
Author Contributions
BW designed, conducted, and evaluated the cyanobacterial growth experiments, quantifications of nitrate, nitrite, ammonium, and urea utilization and carried out the microscopic experiments. RS constructed Synechocystis strains expressing tagged PII. PS and OH carried out anti-FLAG pull-down experiments. PS and BM carried out and evaluated nanoLC-MS/MS analyses. MK designed and constructed bacterial two-hybrid vectors. NN carried out and evaluated bacterial two-hybrid assays. KF designed and supervised the study and wrote the manuscript with BW. All authors gave input and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by grants from the DFG (Fo195/9-2) and the research training group GRK 1708. RS was supported by the project LO1416 of the Czech Ministry of Education.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01428/full#supplementary-material
References
- Baker K. M., Gobler C. J., Collier J. L. (2009). Urease gene sequences from algae and heterotrophic bacteria in axenic and nonaxenic phytoplankton cultures. J. Phycol. 45 625–634. 10.1111/j.1529-8817.2009.00680.x [DOI] [PubMed] [Google Scholar]
- Battchikova N., Vainonen J. P., Vorontsova N., Keranen M., Carmel D., Aro E. M. (2010). Dynamic changes in the proteome of Synechocystis 6803 in response to CO2 limitation revealed by quantitative proteomics. J. Proteome Res. 9 5896–5912. 10.1021/pr100651w [DOI] [PubMed] [Google Scholar]
- Battesti A., Bouveret E. (2012). The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58 325–334. 10.1016/j.ymeth.2012.07.018 [DOI] [PubMed] [Google Scholar]
- Burillo S., Luque I., Fuentes I., Contreras A. (2004). Interactions between the nitrogen signal transduction protein PII and N-acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J. Bacteriol. 186 3346–3354. 10.1128/jb.186.11.3346-3354.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldovic L., Tuchman M. (2003). N-acetylglutamate and its changing role through evolution. Biochem. J. 372(Pt 2) 279–290. 10.1042/BJ20030002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Takatani N., Aichi M., Maeda S.-I., Omata T. (2013). Evaluation of the effects of PII deficiency and the toxicity of PipX on growth characteristics of the PII-less mutant of the cyanobacterium Synechococcus elongatus. Plant Cell Physiol. 54 1504–1514. 10.1093/pcp/pct092 [DOI] [PubMed] [Google Scholar]
- Cheah E., Carr P. D., Suffolk P. M., Vasudevan S. G., Dixon N. E., Ollis D. L. (1994). Structure of the Escherichia coli signal transducing protein PII. Structure 2 981–990. [DOI] [PubMed] [Google Scholar]
- Chellamuthu V. R., Alva V., Forchhammer K. (2013). From cyanobacteria to plants: conservation of PII functions during plastid evolution. Planta 237 451–462. 10.1007/s00425-012-1801-0 [DOI] [PubMed] [Google Scholar]
- Chidgey J. W., Linhartova M., Komenda J., Jackson P. J., Dickman M. J., Canniffe D. P., et al. (2014). A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26 1267–1279. 10.1105/tpc.114.124495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy M. J., Durand A., Lupo D., Li X. D., Bullough P. A., Winkler F. K., et al. (2007). The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. U.S.A. 104 1213–1218. 10.1073/pnas.0610348104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., Mann M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13 2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Dai G. Z., Qiu B. S., Forchhammer K. (2014). Ammonium tolerance in the cyanobacterium Synechocystis sp. strain PCC 6803 and the role of the psbA multigene family. Plant Cell Environ. 37 840–851. 10.1111/pce.12202 [DOI] [PubMed] [Google Scholar]
- Drath M., Kloft N., Batschauer A., Marin K., Novak J., Forchhammer K. (2008). Ammonia triggers photodamage of photosystem II in the cyanobacterium Synechocystis sp strain PCC 6803. Plant Physiol. 147 206–215. 10.1104/pp.108.117218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa J., Forchhammer K., Burillo S., Contreras A. (2006). Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 61 457–469. 10.1111/j.1365-2958.2006.05231.x [DOI] [PubMed] [Google Scholar]
- Espinosa J., Forchhammer K., Contreras A. (2007). Role of the Synechococcus PCC 7942 nitrogen regulator protein PipX in NtcA-controlled processes. Microbiology 153 711–718. [DOI] [PubMed] [Google Scholar]
- Espinosa J., Labella J. I., Cantos R., Contreras A. (2018). Energy drives the dynamic localization of cyanobacterial nitrogen regulators during diurnal cycles. Environ. Microbiol. 20 1240–1252. 10.1111/1462-2920.14071 [DOI] [PubMed] [Google Scholar]
- Espinosa J., Rodriguez-Mateos F., Salinas P., Lanza V. F., Dixon R., de la Cruz F., et al. (2014). PipX, the coactivator of NtcA, is a global regulator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 111 E2423–E2430. 10.1073/pnas.1404097111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves-Ferreira A. A., Inaba M., Fort A., Araujo W. L., Sulpice R. (2018). Nitrogen metabolism in cyanobacteria: metabolic and molecular control, growth consequences and biotechnological applications. Crit. Rev. Microbiol. 44 541–560. 10.1080/1040841X.2018.1446902 [DOI] [PubMed] [Google Scholar]
- Fiddler R. N. (1977). Collaborative study of modified AOAC method of analysis for nitrite in meat and meat products. J. Assoc. Off. Anal. Chem. 60 594–599. [PubMed] [Google Scholar]
- Flores E., Herrero A. (1994). “Assimilatory nitrogen metabolism and its regulation,” in The Molecular Biology of Cyanobacteria ed. Bryant D. A. (Dordrecht: Springer; ) 487–517. [Google Scholar]
- Fokina O., Chellamuthu V. R., Forchhammer K., Zeth K. (2010a). Mechanism of 2-oxoglutarate signaling by the Synechococcus elongatus PII signal transduction protein. Proc. Natl. Acad. Sci. U.S.A. 107 19760–19765. 10.1073/pnas.1007653107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokina O., Chellamuthu V. R., Zeth K., Forchhammer K. (2010b). A novel signal transduction protein PII variant from Synechococcus elongatus PCC 7942 indicates a two-step process for NAGK-PII complex formation. J. Mol. Biol. 399 410–421. 10.1016/j.jmb.2010.04.018 [DOI] [PubMed] [Google Scholar]
- Forcada-Nadal A., Forchhammer K., Rubio V. (2014). SPR analysis of promoter binding of Synechocystis PCC6803 transcription factors NtcA and CRP suggests cross-talk and sheds light on regulation by effector molecules. FEBS Lett. 588 2270–2276. 10.1016/j.febslet.2014.05.010 [DOI] [PubMed] [Google Scholar]
- Forcada-Nadal A., Llacer J. L., Contreras A., Marco-Marin C., Rubio V. (2018). The PII-NAGK-PipX-NtcA regulatory axis of cyanobacteria: a tale of changing partners, allosteric effectors and non-covalent interactions. Front. Mol. Biosci. 5:91. 10.3389/fmolb.2018.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K. (2004). Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol. Rev. 28 319–333. 10.1016/j.femsre.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Forchhammer K. (2008). PII signal transducers: novel functional and structural insights. Trends Microbiol. 16 65–72. 10.1016/j.tim.2007.11.004 [DOI] [PubMed] [Google Scholar]
- Forchhammer K., Hedler A. (1997). Phosphoprotein PII from cyanobacteria-analysis of functional conservation with the PII signal-transduction protein from Escherichia coli. Eur. J. Biochem. 244 869–875. [DOI] [PubMed] [Google Scholar]
- Forchhammer K., IrmLer A., Kloft N., Ruppert U. (2004). PII signalling in unicellular cyanobacteria: analysis of redox-signals and energy charge. Physiol. Plant. 120 51–56. 10.1111/j.0031-9317.2004.0218.x [DOI] [PubMed] [Google Scholar]
- Forchhammer K., Luddecke J. (2016). Sensory properties of the PII signalling protein family. FEBS J. 283 425–437. 10.1111/febs.13584 [DOI] [PubMed] [Google Scholar]
- Forchhammer K., Schwarz R. (2018). Nitrogen chlorosis in unicellular cyanobacteria - a developmental program for surviving nitrogen deprivation. Environ. Microbiol. 21 1173–1184. 10.1111/1462-2920.14447 [DOI] [PubMed] [Google Scholar]
- Forchhammer K., Tandeau de Marsac N. (1995). Phosphorylation of the PII protein (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942: analysis of in vitro kinase activity. J. Bacteriol. 177 5812–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., III, Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Giner-Lamia J., Robles-Rengel R., Hernández-Prieto M. A., Muro-Pastor M. I., Florencio F. J., Futschik M. E. (2017). Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 45 11800–11820. 10.1093/nar/gkx860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruswitz F., O’Connell J., Stroud R. M. (2007). Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. Proc. Natl. Acad. Sci. U.S.A. 104 42–47. 10.1073/pnas.0609796104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A., Schleiff E. (2014). “The cell envelope,” in The Cell Biology of Cyanobacteria eds Flores E., Herrero A. (Norfolk: Caister Academic Press; ) 29–87. [Google Scholar]
- Hauf W., Schmid K., Gerhardt E. C., Huergo L. F., Forchhammer K. (2016). Interaction of the nitrogen regulatory protein GlnB PII with biotin carboxyl carrier protein (BCCP) controls acetyl-CoA levels in the cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 7:1700. 10.3389/fmicb.2016.01700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich A., Maheswaran M., Ruppert U., Forchhammer K. (2004). The Synechococcus elongatus PII signal transduction protein controls arginine synthesis by complex formation with N-acetyl-L-glutamate kinase. Mol. Microbiol. 52 1303–1314. 10.1111/j.1365-2958.2004.04058.x [DOI] [PubMed] [Google Scholar]
- Herrero A., Flores E. (2008). The Cyanobacteria: Molecular Biology, Genomics, and Evolution. Norfolk: Caister Academic Press. [Google Scholar]
- Hisbergues M., Jeanjean R., Joset F., de Marsac N. T., Bedu S. (1999). Protein PII regulates both inorganic carbon and nitrate uptake and is modified by a redox signal in Synechocystis sp PCC 6803. FEBS Lett. 463 216–220. [DOI] [PubMed] [Google Scholar]
- Hollingshead S., Kopečná J., Jackson P. J., Canniffe D. P., Davison P. A., Dickman M. J., et al. (2012). Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J. Biol. Chem. 287 27823–27833. 10.1074/jbc.M112.352526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huergo L. F., Pedrosa F. O., Muller-Santos M., Chubatsu L. S., Monteiro R. A., Merrick M., et al. (2012). PII signal transduction proteins: pivotal players in post-translational control of nitrogenase activity. Microbiology 158(Pt 1) 176–190. 10.1099/mic.0.049783-0 [DOI] [PubMed] [Google Scholar]
- Jiang P., Peliska J. A., Ninfa A. J. (1998). Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 37 12782–12794. 10.1021/bi980667m [DOI] [PubMed] [Google Scholar]
- Karimova G., Ullmann A., Ladant D. (2001). Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3 73–82. [PubMed] [Google Scholar]
- Kloft N., Forchhammer K. (2005). Signal transduction protein PII phosphatase PphA is required for light-dependent control of nitrate utilization in Synechocystis sp. strain PCC 6803. J. Bacteriol. 187 6683–6690. 10.1128/JB.187.19.6683-6690.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Rodriguez R., Lara C., Omata T. (1997). Involvement of the C-terminal domain of an ATP-binding subunit in the regulation of the ABC-type nitrate/nitrite transporter of the cyanobacterium Synechococcus sp. PCC 7942. J. Biol. Chem. 272 27197–27201. 10.1074/jbc.272.43.27197 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Takatani N., Tanigawa M., Omata T. (2005). Posttranslational regulation of nitrate assimilation in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 187 498–506. 10.1128/JB.187.2.498-506.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin N. M., Pakrasi H. B., Smith T. J. (2006). Atomic structure of a nitrate-binding protein crucial for photosynthetic productivity. Proc. Natl. Acad. Sci. U.S.A. 103 9820–9825. 10.1073/pnas.0602517103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. M., Flores E., Forchhammer K., Herrero A., de Marsac N. T. (2000). Phosphorylation of the signal transducer PII protein and an additional effector are required for the PII mediated regulation of nitrate and nitrite uptake in the cyanobacterium Synechococcus sp PCC 7942. Eur. J. Biochem. 267 591–600. 10.1046/j.1432-1327.2000.01043.x [DOI] [PubMed] [Google Scholar]
- Lee H. M., Flores E., Herrero A., Houmard J., de Marsac N. T. (1998). A role for the signal transduction protein PII in the control of nitrate/nitrite uptake in a cyanobacterium. FEBS Lett. 427 291–295. [DOI] [PubMed] [Google Scholar]
- Llacer J. L., Contreras A., Forchhammer K., Marco-Marin C., Gil-Ortiz F., Maldonado R., et al. (2007). The crystal structure of the complex of PII and acetylglutamate kinase reveals how PII controls the storage of nitrogen as arginine. Proc. Natl. Acad. Sci. U.S.A. 104 17644–17649. 10.1073/pnas.0705987104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llacer J. L., Espinosa J., Castells M. A., Contreras A., Forchhammer K., Rubio V. (2010). Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc. Natl. Acad. Sci. U.S.A. 107 15397–15402. 10.1073/pnas.1007015107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque I., Flores E., Herrero A. (1994). Nitrate and nitrite transport in the cyanobacterium Synechococcus sp PCC 7942 are mediated by the same permease. Biochim. Biophys. Acta Bioenerg. 1184 296–298. [Google Scholar]
- Luque I., Forchhammer K. (2008). “Nitrogen assimilation and C/N balance sensing,” in The Cyanobacteria. Molecular Biology, Genomics and Evolution eds Herrero A., Flores E. (Norfolk: Caister Academic Press; ) 335–382. [Google Scholar]
- Maeda S. I., Omata T. (1997). Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp strain PCC 7942 involved in the transport of nitrate and nitrite. J. Biol. Chem. 272 3036–3041. 10.1074/jbc.272.5.3036 [DOI] [PubMed] [Google Scholar]
- Maheswaran M., Urbanke C., Forchhammer K. (2004). Complex formation and catalytic activation by the PII signaling protein of N-acetyl-L-glutamate kinase from Synechococcus elongatus strain PCC 7942. J. Biol. Chem. 279 55202–55210. 10.1074/jbc.M410971200 [DOI] [PubMed] [Google Scholar]
- Maheswaran M., Ziegler K., Lockau W., Hagemann M., Forchhammer K. (2006). PII-regulated arginine synthesis controls accumulation of cyanophycin in Synechocystis sp strain PCC 6803. J. Bacteriol. 188 2730–2734. 10.1128/Jb.188.7.2730-2734.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier S., Schleberger P., Lu W., Wacker T., Pfluger T., Litz C., et al. (2011). Mechanism of disruption of the Amt-GlnK complex by PII-mediated sensing of 2-oxoglutarate. PLoS One 6:e26327. 10.1371/journal.pone.0026327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano C., Candau P., Gomez-Moreno C., Relimpio A. M., Losada M. (1976). Ferredoxin-dependent photosynthetic reduction of nitrate and nitrite by particles of Anacystis nidulans. Mol. Cell. Biochem. 10 161–169. [DOI] [PubMed] [Google Scholar]
- Marques S., Merida A., Candau P., Florencio F. J. (1992). Light-mediated regulation of glutamine-synthetase activity in the unicellular cyanobacterium Synechococcus sp PCC6301. Planta 187 247–253. [DOI] [PubMed] [Google Scholar]
- Merrick M. (2014). Post-translational modification of PII signal transduction proteins. Front. Microbiol. 5:763 10.3389/fmicb.2014.00763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley H. L. T., Hausinger R. P. (1989). Microbial ureases - significance, regulation and molecular characterization. Microbiol. Rev. 53 85–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos M. L., Muro-Pastor A. M., Herrero A., Flores E. (1998). Ammonium/methylammonium permeases of a cyanobacterium. Identification and analysis of three nitrogen-regulated amt genes in Synechocystis sp. PCC 6803. J. Biol. Chem. 273 31463–31470. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor M. I., Reyes J. C., Florencio F. J. (2001). Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 276 38320–38328. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor M. I., Reyes J. C., Florencio F. J. (2005). Ammonium assimilation in cyanobacteria. Photosynth. Res. 83 135–150. [DOI] [PubMed] [Google Scholar]
- Nakao M., Okamoto S., Kohara M., Fujishiro T., Fujisawa T., Sato S., et al. (2010). CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res. 38 D379–D381. 10.1093/nar/gkp915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Shi W., Takatani N., Aichi M., Maeda S., Watanabe S., et al. (2011). Regulation of nitrate assimilation in cyanobacteria. J. Exp. Bot. 62 1411–1424. 10.1093/jxb/erq427 [DOI] [PubMed] [Google Scholar]
- Olsen J. V., de Godoy L. M. F., Li G. Q., Macek B., Mortensen P., Pesch R., et al. (2005). Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4 2010–2021. 10.1074/mcp.T500030-MCP200 [DOI] [PubMed] [Google Scholar]
- Omata T., Andriesse X., Hirano A. (1993). Identification and characterization of a gene-cluster involved in nitrate transport in the cyanobacterium Synechococcus sp PCC 7942. Mol. Gen. Genet. 236 193–202. 10.1007/Bf00277112 [DOI] [PubMed] [Google Scholar]
- Radchenko M., Merrick M. (2011). The role of effector molecules in signal transduction by PII proteins. Biochem. Soc. Trans. 39 189–194. 10.1042/BST0390189 [DOI] [PubMed] [Google Scholar]
- Radchenko M. V., Thornton J., Merrick M. (2014). Association and dissociation of the GlnK-AmtB complex in response to cellular nitrogen status can occur in the absence of GlnK post-translational modification. Front. Microbiol. 5:731. 10.3389/fmicb.2014.00731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2 1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Rees A. P., Woodward E. M. S., Joint I. (2006). Concentrations and uptake of nitrate and ammonium in the Atlantic ocean between 60 degrees N and 50 degrees S. Deep Sea Res. Part II Top. Stud. Oceanogr. 53 1649–1665. 10.1016/j.dsr2.2006.05.008 [DOI] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111 1–61. [Google Scholar]
- Romero J. M., Lara C., Guerrero M. G. (1985). Dependence of nitrate utilization upon active CO2 fixation in Anacystis nidulans - a regulatory aspect of the interaction between photosynthetic carbon and nitrogen-metabolism. Arch. Biochem. Biophys. 237 396–401. [DOI] [PubMed] [Google Scholar]
- Ruppert U., IrmLer A., Kloft N., Forchhammer K. (2002). The novel protein phosphatase PphA from Synechocystis PCC 6803 controls dephosphorylation of the signalling protein PII. Mol. Microbiol. 44 855–864. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Inoue-Sakamoto K., Bryant D. A. (1999). A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 181 7363–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Shimoda Y., Muraki A., Kohara M., Nakamura Y., Tabata S. (2007). A large-scale protein–protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 14 207–216. 10.1093/dnares/dsm021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 676–682. 10.1038/Nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., et al. (2011). Global quantification of mammalian gene expression control. Nature 473 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Selim K. A., Haase F., Hartmann M. D., Hagemann M., Forchhammer K. (2018). PII-like signaling protein SbtB links cAMP sensing with cyanobacterial inorganic carbon response. Proc. Natl. Acad. Sci. U.S.A. 115 E4861–E4869. 10.1073/pnas.1803790115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon C. M., Collier J. L., Berg G. M., Glibert P. M. (2010). Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquat. Microb. Ecol. 59 67–88. 10.3354/ame01390 [DOI] [Google Scholar]
- Soo R. M., Hemp J., Parks D. H., Fischer W. W., Hugenholtz P. (2017). On the origins of oxygenic photosynthesis and aerobic respiration in cyanobacteria. Science 355 1436–1439. 10.1126/science.aal3794 [DOI] [PubMed] [Google Scholar]
- Spät P., Macek B., Forchhammer K. (2015). Phosphoproteome of the cyanobacterium Synechocystis sp PCC 6803 and its dynamics during nitrogen starvation. Front. Microbiol. 6:248. 10.3389/fmicb.2015.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L. J., Fernie A. R. (2018). The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun. 9:2136. 10.1038/s41467-018-04543-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares A., Montesinos M. L., Herrero A., Flores E. (2002). An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 43 703–715. 10.1046/j.1365-2958.2002.02778.x [DOI] [PubMed] [Google Scholar]
- Vegapalas M. A., Flores E., Herrero A. (1992). NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol. Microbiol. 6 1853–1859. 10.1111/j.1365-2958.1992.tb01357.x [DOI] [PubMed] [Google Scholar]
- Vitousek P. M., Howarth R. W. (1991). Nitrogen limitation on land and in the sea - how can it occur. Biogeochemistry 13 87–115. [Google Scholar]
- Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., et al. (2013). The proteomics identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41 D1063–D1069. 10.1093/nar/gks1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A. I., Furniss B. S., Vogel A. I. (1989). Vogel’s Textbook of Practical Organic Chemistry. New York, NY: Longman Scientific & Technical. [Google Scholar]
- Watzer B., Engelbrecht A., Hauf W., Stahl M., Maldener I., Forchhammer K. (2015). Metabolic pathway engineering using the central signal processor PII. Microb. Cell Fact. 14:192. 10.1186/s12934-015-0384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzer B., Forchhammer K. (2018). Cyanophycin synthesis optimizes nitrogen utilization in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Appl. Environ. Microbiol. 84:e01298-18. 10.1128/AEM.01298-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton B. A. (2012). Ecology of Cyanobacteria II: Their Diversity in Space and Time. New York, NY: Springer. [Google Scholar]
- Wirén N. V., Merrick M. (2004). “Regulation and function of ammonium carriers in bacteria, fungi, and plants,” in Molecular Mechanisms Controlling Transmembrane Transport eds Boles E., Krämer R. (Berlin: Springer; ) 95–120. [Google Scholar]
- Xu Y. B., Carr P. D., Clancy P., Garcia-Dominguez M., Forchhammer K., Florencio F., et al. (2003). The structures of the PII proteins from the cyanobacteria Synechococcus sp PCC 7942 and Synechocystis sp. PCC 6803. Acta Crystallogr. D Biol. Crystallogr. 59(Pt 12) 2183–2190. 10.1107/S0907444903019589 [DOI] [PubMed] [Google Scholar]
- Zeth K., Fokina O., Forchhammer K. (2014). Structural basis and target-specific modulation of ADP sensing by the Synechococcus elongatus PII signaling protein. J. Biol. Chem. 289 8960–8972. 10.1074/jbc.M113.536557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M. X., Jiang Y. L., Xu B. Y., Chen Y., Zhang C. C., Zhou C. Z. (2010). Crystal structure of the cyanobacterial signal transduction protein PII in complex with PipX. J. Mol. Biol. 402 552–559. 10.1016/j.jmb.2010.08.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the PRIDE ProteomeXchange consortium, PXD013411.


