Abstract
Genome editing technology is a technique for targeted genetic modifications, enabling the knockout and addition of specific DNA fragments. This technology has been widely used in various types of biomedical research, clinics and agriculture. In terms of disease research, constructing appropriate animal models is necessary. Combining reproductive technology with genome editing, many animal disease models have been generated for basic and clinical research. In addition, precisely targeted modifications allow genome editing to flourish in the field of gene therapy. Many mutations refractory to traditional gene therapy could be permanently corrected at the DNA level. Thus, genome editing is undoubtedly a promising technology for gene therapy. In this review, we mainly introduce the applications of genome editing in constructing animal disease models and gene therapies, as well as its future prospects and challenges.
Keywords: Genome editing, Disease models, Gene therapy, CRISPR-Cas9
1. Introduction
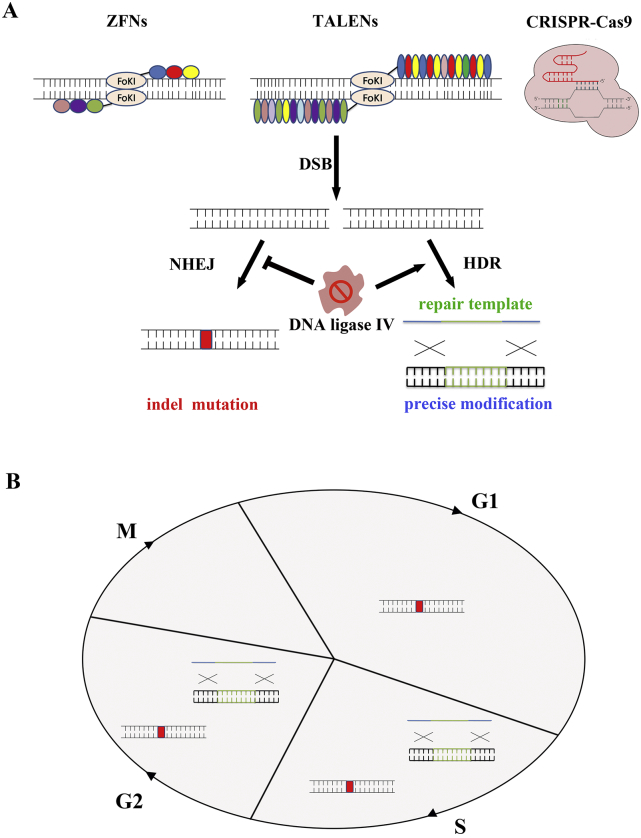
Genome editing technology refers to an operation technique that can make precise modifications to the genome by engineered nucleases. It is regarded as an ideal platform to knock out/in and replace the specific DNA fragment, and make accurate genome editing on the genome level. There are four major varieties of programmed nucleases: meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and the clustered regularly interspaced short palindromic repeat-associated nuclease Cas9 (CRISPR-Cas9). These nucleases mainly generate DNA double-strand breaks (DSBs) at specific sites in the genome by targeted recognition and cleavage. In the absence of a repair template, DSBs will activate endogenous DNA repair mechanisms, non-homologous end joining (NHEJ). NHEJ repairs the lesion by directly rejoining the two DSB ends which would lead to insertions and/or deletions (indels) at the DSB site. When these mutations occurred within the gene coding region, indels often cause frame shifts and result in the knockout of gene. As a result, loss-of-function mutations could permanently exist in targeted cells. Moreover, simultaneously introducing two targeted DSBs could realize specific deletion or inversion, duplication, and translocations/chromosomal rearrangements at nuclease cleavage sites [1]. In the case of repair templates, DSBs would be repaired by homologous directed repair (HDR), which happens at lower frequencies than NHEJ. The sequence differences present in the donor templates could be integrated into the editing locus to modify DNA permanently. It could be an appropriate way to achieve predictable insertion, substitution or deletion of target genes [2]. In addition, inhibition of NHEJ key molecules (such as DNA ligase IV) could promote the efficiency of HDR [3,4] (Fig. 1A). While HDR could only occur in S/G2, NHEJ is merely limited in mitosis phase [5] (Fig. 1B).
Fig. 1.
Genome editing exploit endogenous DNA repair mechanism. A) Genome editing nucleases (ZFNs, TALENs and CRISPR-Cas9) induce DSBs at targeted sites. DSBs are repaired by NHEJ or, in the presence of donor template, HDR. NHEJ will induce indels at editing site and HDR could insert predicted DNA fragments. The inhibition of NHEJ key enzyme DNA ligase IV could increase the efficiency of HDR. B) NHEJ and HDR would occur in different stages of cell cycle.
To date, meganucleases, ZFNs, TALENs and CRISPR-Cas9 have been developed for site-specific genome editing through taking advantages of their different characteristics (Table 1). Meganucleases, ZFNs and TALENs cleave specific DNA sites via protein-DNA interactions [6]. Different targets need diverse of engineered protein for each experiment. It is doubtless time-consuming and costly. Whereas CRISPR-Cas9 system bases on simple base pairing rules between a specific guide RNA (gRNA) and the target genome site, offers simple yet effective methods of genome editing [6]. CRISPR-Cas element is a defense system found in the prokaryotic immune system that prevents the replication of foreign viruses or plasmids in the host genome [6]. This system could be classified into three types of CRISPR mechanisms (Type I-III). For type I and type III CRISPR, a variety of Cas proteins are involved in the recognition and destruction of target genes. CRISPR-Cas9 belongs to the type II system which utilizes reduced number of Cas proteins, thereby much simpler for engineering [7].The CRISPR-Cas9 system consists of three major elements: Cas9 protein with endonuclease properties, CRISPR RNA (crRNA) for specific targeting, and trans-activating crRNA (tracrRNA) [8]. The crRNA and tracrRNA can form double-stranded RNA through the principle of base-complementary pairing, and combine with Cas9 protein to the formation of a complex for specific cleavage in targeted spot. The specific recognition function of crRNA is dependent on the protospacer adjacent motifs (PAM) in the downstream 3′ end of the target sequence, a necessary condition for the CRISPR-Cas9 system to express the genome editing function [6]. In 2012, Jinek et al. designed a single-stranded guide RNA (sgRNA) that replaced the crRNA-tracrRNA complex, which also directed the Cas9 protein to specifically cleave the target gene [9].
Table 1.
Comparison of different engineered nuclease platforms.
| ZFNs | TALENs | Cas9 | Meganuclease | |
|---|---|---|---|---|
| Recognition location | Typically 9–18 bp per monomer, 18–36 bp per pair | Typically 14–20 bp per monomer, 28–40 bp per pair | Typically 20 bp guide sequence + PAM sequence | Between 14 and 40 bp |
| Targeting restrictions | Difficult for non-G-rich sites | 5′ targeted base must be a T | Targeted site should precede a PAM sequence | Typically low efficiency for targeting novel sites |
| Specificity | Tolerating few positional mismatches | Tolerating few positional mismatches | Tolerating positional and multiple consecutive mismatches | Tolerating few positional mismatches |
| Difficulties of engineering | Requiring substantial protein engineering | Requiring complex molecular cloning methods | Using easy cloning methods and oligo synthesis | Requiring substantial protein engineering |
| Difficulties of in vivo delivery | Relatively easy as small size of expression elements suitable for varieties of viral vectors | Difficult due to the large size of functional components | Commonly used SpCas9 with large size may cause packaging problems for viral vectors such as AAV | Relatively easy as small size of expression elements suitable for varieties of viral vectors |
Genome editing has been utilized in different kinds of biomedical research. Constructing an appropriate animal model of disease is essential for studying the mechanism of human disease, and it also plays an important role in drug development and organ transplantation. With the development of germline genome editing, more and more animal disease models are being generated for clinical requirements. Recently, CRISPR system truly revolutionizes the field of genome editing. In 2013, Hwang et al. first provided the largest set of endogenous genes modified by CRISPR-Cas9 and demonstrated stability of this nuclease in vivo for toilless and efficient genetic editing of zebrafish [10]. In mammals, CRISPR-Cas system allowed the one-step generation of mice carrying mutations in multiple genes [11]. In the field of gene therapy, genome editing is also a very useful technology. The programmed therapeutic elements have the potential to directly correct genetic mutations in targeted tissues and cells for treating diseases which are refractory to traditional therapies. In this review, we will introduce recent progress in genome editing mediated generation of disease animal models and gene therapy, as well as the challenges and prospects of genome editing.
2. Genome Editing for Disease Modeling
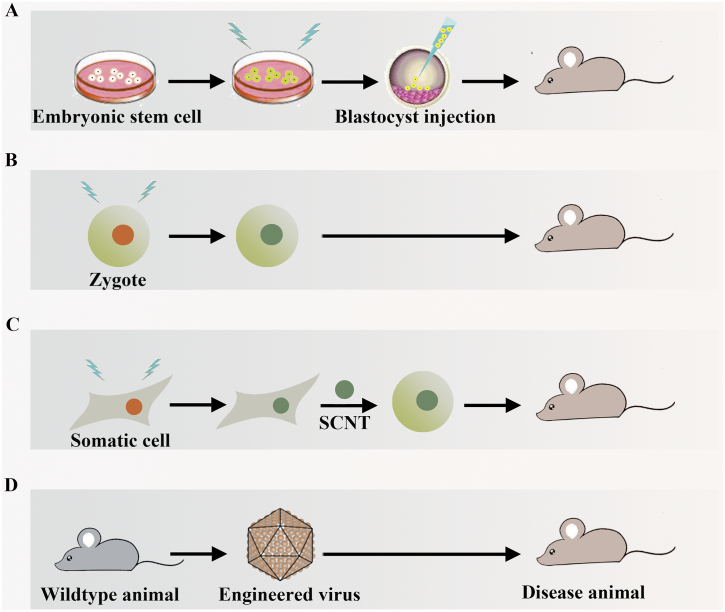
Disease animal models have been essential resources in advancing the biomedicine field. With the help of genome editing technologies, many applicable models with specific mutations which could mimic clinical phenotypes have been generated (Fig. 2). Disease models are generally constructed through bringing in site-specific modifications in embryonic stem (ES) cells. In addition, disease animals could be generated by editing induced mutated zygotes or editing somas combined somatic cell nuclear transfer (SCNT) technology. Moreover, diseases could be introduced by targeted destruction through applying engineered viruses loaded editing elements. Following viruses' injection in adult animals, tissues specific expression of editing elements provides a tool that allows rapid and accurate modification of genes, thereby circumventing embryonic lethality.
Fig. 2.
Schematic overview of constructing disease animal model in four major ways. A) Cultured embryonic stem cells (ESCs) can be used to introduce morbigenous mutations using genome editing tools. The edited ESCs can be injected into host blastocysts, whereafter are implanted into pseudo-pregnant to produce disease animal; B) Animal zygote is directly edited and the edited zygote is developed into diseased model; C) Disease animal could be generated by combining somatic cell genome editing and somatic cell nuclear transfer (SCNT) technology; D) Genome editing elements are packaged by viral vectors. Disease animal can be generated by administration of engineered virus.
2.1. Cancer Models
Cancer usually caused by complicated mechanisms containing complex genetic mutations. A variety of pathogenic transformations in tumor suppressor genes and oncogenes could results in tumorigenesis [12]. One of the most effective ways to study these mutational functions is to establish a cancer model that carries the mutated gene. Building a cancer model in a traditional way is a time-consuming and laborious process [13,14]. Either using patient-derived cells or cooperative studies with patients, the key problem is that these methods are researching late-stage samples, and could only finitely model primary oncogenic events [15]. With the help of genome editing tools, numerous studies have been carried out through modifying key genes for generating accurate and specific cancer models.
The WNT signaling pathway is frequently altered in hepatocellular carcinoma (HCC). 32.8% and 1.6% of HCCs contain β-catenin and Adenomatous Polyposis Coli (Apc) mutations [16]. Transmitting TALENs directed against these two driver genes could generate efficient and physiologic liver cancer mouse models [17]. Except for mammals, Van Nieuwenhuysen et al. focused on constructing familial adenomatous polyposis (FAP) model in X. tropicalis by TALENs-mediated Apc mutation [18]. After CRISPR-Cas9 showed up and was widely used, more and more cancer models were constructed. In a study by Sanchez-Rivera et al., the authors used CRISPR-Cas9 to edit tumor suppressor genes and resulted in the generation of lung adenocarcinomas in mice. They described an approach for functional exploration of candidate genes in mouse cancer models [19]. P53, Lkb1 and Kras are important genes related to lung cancer, and studies have shown that Lkb1 mutation can aggravate lung cancer [20]. Platt et al. injected viruses expressing the sgRNAs targeting p53, Lkb1, and Kras into the lung of the Cas9 knock-in mouse [21]. Significant tumor nodules appeared in the lung tissues of mice after injection and increased significantly with time. Compared with the traditional genetically altered mouse model, this novel mouse lung cancer model not only achieved multi-gene knockout of specific tissues, but also could probe the influence of multiple genes on the phenotype of lung cancer. Similarly aimed at disrupting tumor suppressor genes p53 and Pten in the liver through CRISPR-Cas9, mice could rapidly develop to liver cancers [22]. Part of lung cancers exist genomic rearrangements. Maddalo et al. described an efficient method to induce specific chromosomal rearrangements in vivo. By applying viral-mediated delivery of CRISPR system to somatic cells of adult mice, they generated a mouse model of Eml4-Alk-driven lung cancer [23]. In the same year, another group utilized CRISPR-Cas9 induced CD74-ROS1 translocation event and the EML4-ALK and KIF5B-RET inversion events thus provided a tractable approach for the study of genomic rearrangements in various types of lung cancers [24]. In 2015, Maresch et al. established a pancreatic cancer model by simultaneously editing multiple genes network sets applying multiplex delivery of CRISPR-Cas9 elements to the pancreas of adult mice. They also realized chromosomal deletions and complex chromosomal rearrangements and offered opportunities to study complex structural variation [25]. As CRISPR-Cas9 could edit multiple targets at the same time, it could be a versatile tool for generating other tumor models with complexity similar to cancer patient conditions, such as brain tumor model [26], acute myeloid leukemia [27], and so on.
2.2. Cardiovascular Disease Models
For better understanding the pathophysiology of human cardiovascular diseases, the key barrier is that in vitro models (such as cultured cardiomyocyte cell lines) are relatively lacking for cardiovascular systems. By contrast, in vivo models are more faithful to human cardiovascular conditions. Genome editing technologies have being used in creating a wide variety of cardiovascular conditions animal models [28]. Some zebrafish models have been created to mimic vascular development [29,30], cardiac development [31,32], cardiac regeneration [33], and inherited cardiomyopathy [34]. In a report on rodents, Carroll et al. injected Cas9 expression plasmids regulated by the Myh6 promoter into mouse zygotes to generate transgenic mice. This kind of transgenic mice robustly expressed Cas9 exclusively in heart cardiomyocytes. Authors delivered sgRNAs targeted Myh6 by adeno-associated virus (AAV) vector, and induced cardiac-specific genome editing. Finally, they caused hypertrophic cardiomyopathy in transgenic mice [35]. The limitation of rodents is less faithful to some human cardiovascular conditions (for example, myocardial infarction, dyslipidemia, electrophysiological disorders). Thanks to the development of germline genome editing, there have been many bigger mammalian models including rats, rabbits, pigs and even nonhuman primates (NHP). The variety of models will improve our understanding of different kinds of cardiovascular disorders. Pig models are physiologically, anatomically and genetically similar to humans. It is seemed as an ideal model for studying cardiovascular structure. In 2011, a research combined ZFNs with SCNT technology to generate pigs with disabled mutation in peroxisome proliferator-activated receptor gamma (Ppar-γ). The Ppar-γ knocked out pig model provided a useful tool to study the role of PPAR-γ in cardiovascular diseases [36]. Marfan syndrome (MFS) is an autosomal-dominant disorder with the symptoms of cardiovascular and skeletal abnormalities. It is confirmed caused by the mutation of heterozygous fbrillin-1 (FBN1). Umeyama et al. successfully established FBN1 mutant pigs with ZFNs mediated gene disruption. The phenotypes resembling human with MFS indicated the value of FBN1 mutant pigs as a model for better understanding the pathogenesis of MFS and developing treatments [37].
2.3. Ophthalmic Disease Models
There are >600 kinds of hereditary eye diseases and systemic hereditary diseases with ocular manifestations. The involved parts include the cornea, iris, lens and the vitreous, retina, optic nerve of the posterior segment of the eye, which can cause various retina diseases such as Meesmann epithelial corneal dystrophy (MECD) [38], congenital aniridia [39], congenital cataract [40,41], retinitis pigmentosa (RP) [42] and Leber hereditary opticneuropathy (LHON) [43]. Most of the ophthalmic diseases are not fully elucidated and there is no effective treatment, thus seriously affecting the patient's vision and quality of life. The application of genome editing technology to target animal genome and establish an animal model of ocular hereditary diseases can clarify the relationships between target genes and disease phenotypes, and may finally provide effective methods for studying the pathogenesis of hereditary eye diseases.
Congenital ocular coloboma (COC) is one of the main causes of visual impairment and blindness in children [44]. Studies have shown that mutations in the paired-box gene 6 (PAX6) are associated with COC [45], and PAX6 is a congenital adia-free pathogenic gene [39]. Nakayama et al. constructed the Xenopuslaevis PAX6 mutant strain using TALENs technology, and they used it to study the effects of PAX6 gene on early development stage of the eye and pathological changes of no iris malformation [46]. The results proved that the lesion characteristics of these mutant Xenopuslaevis were very similar to those of patients carrying the PAX6 mutation gene, so this animal model provided a powerful tool for studying the early defects of ocular development and the developmental basis of human iris-free malformation.
Cataract is the primary cause of loss of vision in humans, although with the improvement of surgical methods and implant intraocular lens implant (IOL) materials, surgical treatment can restore the vision of cataract patients. It's very necessary to construct suitable animal models for studying the safety of IOLs and the interaction between the lens and the drugs. Some research results suggested that the GJA8 gene encoding gap junction protein 50 was associated with autosomal dominant congenital cataract [40,41]. Researchers co-injected Cas9/sgRNA mRNA into rabbit zygote to construct a GJA8 knockout rabbit cataract model [47]. As a result, the gene mutation efficiency of GJA8 site reached 98.7% and 100% in embryos and young rabbit tissues, respectively. They achieved efficient gene editing of the rabbit genome through Cas9/sgRNA system, and provided a good disease model for cataract-related research. There are also many other examples of genome editing technology used in constructing hereditary eye disease models. For example, Leber congenital amaurosis (LCA) causes early-onset blindness due to mutations in the human Kcnj13 gene. Zhong et al. injected the CRISPR-Cas9 system to mouse embryos to produce Kcnj13 mutant mice, and the establishment of this model could well mimic human KCNJ13-associated LCA disease [48]. In addition, TALENs mediated paired-like homeodomain 2 (PITX2) gene deficiency in zebrafish demonstrated congenital defects like human phenotypes: abnormal development of the cornea, iris and iridocorneal angle [49]. Homma et al. used CRISPR-Cas9 to disrupt long-wavelength-sensitive (LWS) opsins of medaka and produced color-blind fish [50]. Retinoblastoma is a pediatric cancer of the eye which is caused by biallelic mutation of the Retinoblastoma 1 (RB1) gene. Naert et al. presented the first genuine genetic non-mammalian retinoblastoma model through triple multiplex CRISPR-Cas9 injections. The rb1/rb1double mosaic mutant tadpoles could rapidly develop retinoblastoma with detectable occasionally presence of pinealoblastoma (trilateral retinoblastoma). This model provided an ideal platform for novel drug screening [51].
2.4. Metabolism Disease Models
Life is metabolizing all the time so metabolic abnormalities can lead to a variety of diseases. Appropriate animal models will help us to understand the pathogenesis. By injecting TALENs elements into rat zygotes for specific knockout of Leptin receptor (Lepr), Chen and other colleagues obtained three lines of rats containing mutations in the Lepr locus. These strains showed phenotypes of obesity and metabolic disorders. They generated a Lepr mutant obese rat model that exhibited a high efficiency of germline transmission [52]. In another study, a viable Arnt2 mutation triggered by CRISPR-Cas9 caused hyperphagic obesity, diabetes and hepatic steatosis in mice. These findings confirmed the importance of ARNT2 in the homeostatic feeding response [53]. Fumarate hydratase (FH) plays a necessary role in the Krebs tricarboxylic acid (TCA) cycle, which catalyses the hydration of fumarate into malate. FH deficiency would result in metabolic disorder with poor neurological outcomes. Yu's work utilized TALENs for efficient genome editing in rat zygotes and obtained FH knockout rat offsprings. They established a novel rat model for further functional FH studies [54].
Genome editing technology can also be used to construct lipid abnormal disease models. Familial hypercholesterolemia is an autosomal monogenic dominant genetic disease that causes disorders of lipid metabolism in humans. Its pathogenesis stems from the low density lipoprotein receptor (LDLR) gene defect, which can cause clinical symptoms such as atherosclerosis. In 2012, Carlson et al. used TALENs targeting technology to knock out the LDLR of the pig fibroblast genome, and obtained LDLR−/− cloned pigs after SCNT, which had important biomedical value for mimicking lipid metabolic syndrome [55]. In addition, with the help of CRISPR-Cas9 system, Huang et al. simultaneously transferred sgRNA targeting apolipoprotein E (ApoE) and LDLR to porcine embryonic fibroblasts simultaneously through Cas9 vector. This was the first genetically modified porcine model to simulate lipid metabolism disorders [56]. Niemann-Pick disease type C1 (NPC1) is a lysosomal storage disease by abnormal accumulation of unesterified cholesterol and glycolipids in late endosomes and lysosomes. It is primarily caused by mutations in NPC1. For developing high-throughput drug screens, a high-capacity in vivo platform is indispensable. Tseng's achievements established a zebrafish NPC1 model created by CRISPR-Cas9 induced NPC1 mutations. This NPC1 model would be valuable for compound optimization and prioritizing subsequent in vivo testing [57].
Wilson disease (WD) is an autosomal recessive hereditary disorder of copper metabolism caused by sequence variations in the ATP7B gene. ATP7B is an important protein contributes to trans-membrane transport of copper. Recently, based on CRISPR-Cas9 mediated single amino acid substitute, Jiang et al. produced a rabbit WD model. At the onset of WD, the copper content in the livers of rabbits increased nine-fold, a level similar to patients with WD. So it would be a potential WD model for applications in pathological analysis, clinical treatment and gene therapy research [58]. In addition to the above, some studies focused on building other metabolism disease models relying on genome editing tools. NCKX3 knockout mice showed the significance of NCKX3 in regulating calcium hemostasis [59]. Another work generated ABCD1 mutant zebrafish to emphasize the necessity of ABCD1 for very long chain fatty acid metabolism [60].
2.5. Neuropathic and Muscle Disease Models
Neurodegenerative diseases (ND) have become one of the most terrible diseases because there are no precise diagnostic tools or established treatments, and their prevalence is rising as human life expectancy increases. These diseases including Huntington's disease (HD), Alzheimer's disease (AD), Parkinson's disease (PD), etc. [[61], [62], [63]]. A common feature of these neurological diseases is delayed neurological symptoms and degeneration, which preferentially affect nerve cells in the brain. For better understanding the mechanisms of HD, in 2018, Yan et al. applied CRISPR-Cas9 technology to accurately insert the human Huntington's mutation gene containing 150CAG repeat into the endogenous gene locus of pig HTT, and established a genome-edited pig expressing human mutant HTT via SCNT. This was the first large animal model established in the world to simulate genetic mutations in patients with neurodegenerative diseases [64]. As a major breakthrough in the field of neurodegenerative diseases, the establishment of Huntington's gene knock-in pigs could promote the development of new drugs for neurodegenerative diseases. By introducing missense mutation at Scn8a loci, Jones's team generated a mouse model of early onset epileptic encephalopathy with a pair of TALENs. Data demonstrated that this mouse model would be useful for development of pathogenesis and therapy of early onset seizure disorders [65].
Duchenne muscular dystrophy (DMD) is a severe X-linked muscular dystrophy caused by mutations of DMD gene. Present mdx (X-linked muscular dystrophy) mice could only partially model human disease conditions. Their small size imposes limitations on chronic muscular lesions and muscle weakness. Considering of this, Larcher et al. generated Dmdmdx rats through TALENs targeting exon 23 of DMD. These edited rats showed significantly reducing in muscle strength and decreasing in spontaneous motor activity. Rats are small animals but 10 times bigger than mice, Dmdmdx rats could be a new faithful small animal model of DMD [66]. Recently, Sui's team generated a rabbit model of DMD by co-injection of Cas9 mRNA and sgRNA into rabbit zygotes. This CRISPR system targeted exon 51 of DMD and the DMD knockout rabbits harbored the typical phenotypes of DMD. Moreover, specific pathology in the diaphragm and heart was similar to DMD patients. This novel model could be more valuable for preclinical studies [67].
NHP have an innate superiority compared to other animal models due to similarities with humans in genetics, physiology, developmental biology, social behavior and cognition. NHP could be ideal models, especially for nervous system diseases [68]. Though manipulating genes in monkeys is far more difficult than in other animals, the development of genome editing accelerates studies in establishing nonhuman primate models. Rett syndrome (RTT) is an X-linked neuro developmental disorder on the autism spectrum. Loss-of-function mutations of methyl-CpG-binding protein 2 (MECP2) will lead to RTT. In Chen's study, three pairs of TALENs were designed to target multiple sites on exon 3 of MECP2 and all three TALENs plasmids either individually or in combination were injected into one-cell monkey zygotes. The results showed that MECP2 mutant males were embryonic lethality, while mutant female appeared physiological and behavioral disorders. Importantly, these abnormalities were similar to human RTT patients. This RTT animal model provided more opportunities to explore disease mechanisms and find possible treatment options [69]. In the other study, the exon 4 and exon 46 of DMD were targeted by CRISPR-Cas9 to generate DMD monkey models [70].
2.6. Other Disease Models
Genome editing technologies enable us to deeply understand the mechanisms of many other diseases in greater detail. Primary immunodeficiencies comprise a multifarious group of rare and chronic diseases. Part of immune system missing or functions improperly threatens patients' lives. Serve combined immunodeficiency (SCID) is the most severe immunodeficiency [71]. In 2016, Japanese scientists optimized ZFNs and TALENs to create indels at interleukin-2 receptor subunit gamma (IL2RG) locus in pronuclear stage marmoset monkey embryos. Detectable double-stranded DNA mismatches resulted in inactivation of IL2RG concomitant with immunodeficiency. They demonstrated highly efficient generation of founder NHP with SCID phenotypes [72]. Another application took advantage of CRISPR-Cas9 mediated multiple genes editing, hence got one-step strategy for generating different kinds of immunodeficient mouse models [73]. In virtue of engineered nucleases mediated genome modifications, there have been other animal disease models for simulating Hermansky Pudlaksyndrome [74], human smallpox [75], Laron syndrome [76], colitis [77], Netherton syndrome [78] and so on. The advancements in genome editing technologies will further expand the use of animal models in biomedicine and beyond.
3. Applications of Genome Editing Technologies in Gene Therapy
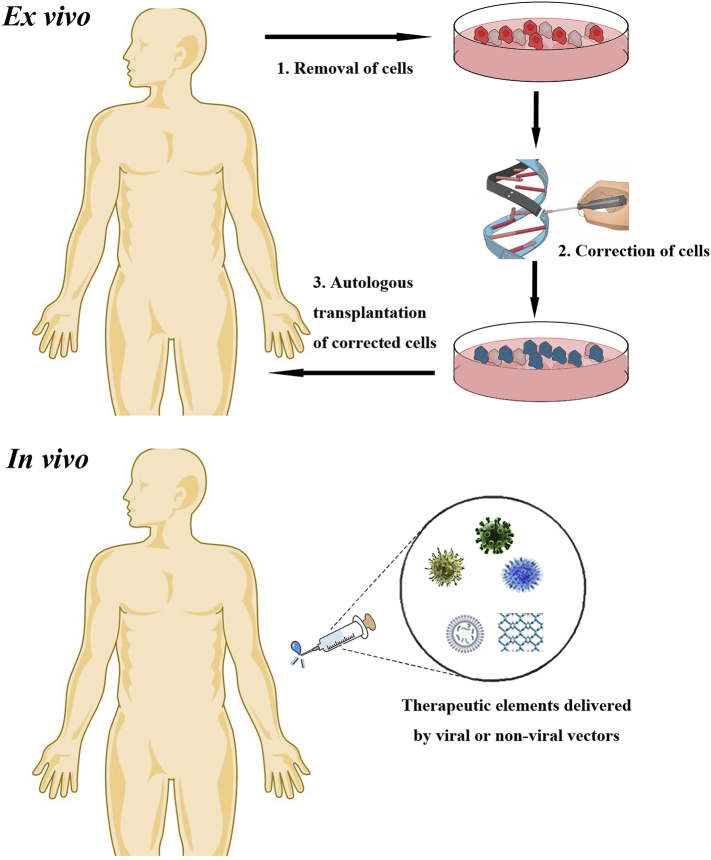
Genome editing technologies are not only used for generating disease animal models but also destined to enter the therapeutic area. There are plentiful means for genome editing based therapy: a) inactivation or correction of harmful mutations; b) introduction of protective mutations; c) insertion of therapeutic exogenous genes; d) destruction of viral DNA. Many proof-of-principle researches have displayed successful examples of gene therapy depending on genome editing. Acquiring therapeutic modifications requires delivery of engineered nucleases to target cells, which can be achieved either ex vivo or in vivo (Fig. 3).
Fig. 3.
Ex vivo and in vivo genome editing therapy. Top: in ex vivo editing therapy, cells are isolated from the patient to be treated, edited and then re-engrafted. Bottom: for in vivo editing therapy, engineered nucleases are delivered by viral or non-viral approaches and directly injected into the patient for systemic or targeted tissues effect.
3.1. Ex vivo Therapy
In ex vivo editing therapy, target cell population is removed from the body, modified by editing elements and then transferred back into host. Modifying on isolated specific cells could reduce the risk of off-target. In addition, many ex vivo strategies could control the specific dosage of programmed molecules delivered into cells [1]. However, ex vivo editing still faces some drawbacks. For example, cultured cells could engraft deficiently after reintroduction into patients as a result of decreased efficacy. Besides, isolated cells must be ensured survival outside the body whereas cells from some tissues fail to survive or lose characterizations prerequisite for their function. Thus, ex vivo therapies concentrate mainly on adult stem cell populations assured to survive and manipulation, such as hematopoietic system [1]. Furthermore, autologous induced pluripotent stem cell (iPSC) with no immune-barrier could be another ideal platform for ex vivo genome editing.
Specifically editing hematopoietic stem/progenitor cells (HSPCs) is a promising way for treating immunodeficiency diseases, such as SCID. In 2014, a strategy of correcting the defective IL2RG in HSPCs combined virus infection and mRNA electroporation to deliver corrected donor template and ZFNs, therefore provided a new avenue for treating SCID [79]. Similarly, three years later another group used CRISPR-Cas9 knock-in strategy to rescue this disease [80]. Disorders of β-globin gene could cause β-thalassemia and sickle cell disease (SCD). In 2016, Dever et al. reported the first CRISPR-Cas9 platform for achieving homologous recombination at the HBB gene in HSPCs. By combining Cas9 ribonucleoproteins and AAV donor delivery, they could purify a population of HSPCs with >90% targeted integration [81]. Recently, Li et al. put forward a treating strategy that re-activation of fetal γ-globin expression in blood cells. They employed CRISPR-Cas9 to disrupt a repressor binding region of γ-globin promoter and transduced HSPCs ex vivo. Finally, they achieved a pronounced switch from β- to γ-globin expression in adult red blood cells. Their HSPCs transduction approach could simplify thalassemia gene [82]. CCR5 ablation was conferred as a therapy strategy for HIV -1 cure. In human CD34+ HSPCs, efficiently edited by CRISPR-Cas9 could achieve long-term ablation of CCR5 and the resistance of HIV-1 infection. This strategy provided evidence for translating edited HSPCs transplantation for an HIV therapy to the clinic [83]. The lack of cancer-restricted surface markers would limit the use of chimeric antigen receptor (CAR)-T cells in antigen-specific immunotherapy. Kim et al. generated CD33 knocked out human HSPCs through CRISPR-Cas9 and enabled specific targeting of AML with CAR-T cells without myelotoxicity. In addition, CD33 knock out HSPCs retained normal myeloid function. These results provided a novel genetic engineering method for antigen-specific immunotherapy to refrain from destruction of normal myeloid cells [84]. Another study focused on CRISPR-Cas9 mediated multiplex gene editing in CAR-T cells. Although the safety and efficacy of these edited CAR-T cells needed be further tested, they were still promising reagents for cancer gene therapy [85].
T cells are the main target of HIV, knocking out CCR5 could endow T cells the resistance of HIV. Tebas's team enrolled patients with HIV and infused the patients with ZFNs modified CCR5 knock out CD4+ T cells. The results showed that the blood level of HIV DNA decreased in most patients. The infusion of autologous edited CD4+ T cells with CCR5 permanently dysfunctional was safe within the limits of this study [86]. In 2017, Yu et al. came up with a dual protection strategy which simultaneously ablated CCR5 and CXCR4 genes in human CD4+ T cells by co-delivery of two single-guide RNAs loaded with Cas9. This work might promote a functional cure for HIV infection [87]. In 2016, China preceded the first clinical CRISPR treatment at West China Hospital (Chengdu, China). T cells were extracted from a metastatic non-small-cell lung cancer patient and edited by CRISPR-Cas9 to disable an important immunosuppressor, programmed death-1 (PD-1). Chinese scientists hypothesized that modified immune cells could kill cancer cells after they were returned to the patient [88]. Henceforth, more institutions brought CRISPR based PD-1 knocked out of T cells into the clinic (for examples, see clinicaltrials.gov).
Combining iPSC with genome editing technologies provide more opportunities for ex vivo mediated gene therapy. A research published in 2017 showed that the correction of Hirschsprung-associated mutations in iPSCs via CRISPR-Cas9 could restore neural crest cell function thus promoted the study of mutations in enteric neural crest cells [89]. Three studies respectively applied ZFNs [90], TALENs [91] and CRISPR-Cas9 [92] modifying iPSCs to attain gene-correction of β-thalassemia. Park et al. realized functional correction of large factor VIII gene chromosomal inversions in hemophilia A patient derived iPSCs by means of CRISPR-Cas9. Endothelial cells differentiated from corrected iPSCs could express functional factor VIII without detectable off-target mutations. This work demonstrated the possibilities for cell-based hemophilia A therapy [93]. In the final cell product of iPSCs, tumorigenicity is a major hurdle to be introduced into the clinic. Execution of tumorigenicity must be considered for transplantation [94].
3.2. In vivo Therapy
In vivo editing involves direct delivery of engineered nucleases to targeted cells in the body. The primary advantage of in vivo editing over ex vivo approach is more extensive affected cell population could be modified or corrected. In addition, in vivo genome editing involves direct delivery of therapeutic tools to abnormal cells in their native tissues. These properties ensure in vivo editing treatment a wider range of applications [1]. Unquestionably, precisely targeting cells and tissues and efficiently delivering genome editing elements are prerequisite. The main delivery methods contain viral vectors and no-viral vectors. Non-viral vectors could transfer large capacity of genetic payloads, however endonucleases in physiological fluids and extracellular space might degrade engineered editing components result in decreased efficacy [95]. Relatively, viral vectors are more commonly used in therapeutic genome editing in vivo. There are three main types of viral delivery system: lentiviral vectors, adenoviral vectors and AAV vectors. The random integration property of lentiviral vector restricts the in vivo adhibition [96]. To date, there have been many successful preclinical therapeutic examples of in vivo editing therapy and some has entered the clinical trials stages (for examples, see clinicaltrials.gov).
Adenovirus is an unintegrated virus which can produce large amounts of recombinant virus in infected differentiated and nondividing cells. Engineered adenoviral vector contained therapeutic elements without viral structural proteins could target many kinds of cells. To introduce loss-of-function mutations into the endogenous PCSK9 gene, Ding et al. used adenoviral vector loaded CRISPR-Cas9 to target PCSK9 in mouse liver. They observed decreased plasma PCSK9 levels, increased hepatic LDL receptor levels and decreased plasma cholesterol levels (by 35%–40%) in mice blood. This approach could provide a potential prevention of cardiovascular disease [97]. The epidermal growth factor receptor (EGFR) was characterized as an important mediator involved in cancer progress [98]. Co-delivery Cas9 and EGFR mutation-specific sgRNA via adenoviral vector led to the specific disruption at oncogenic mutation site. The strategy of CRISPR-Cas9 based protective disruption significantly enhanced cancer cell killing and reduced tumor size in a xenograft mouse model of human lung cancer. It also could be a powerful remedial strategy to treat cancers with oncogenic mutations [99]. In another research which took advantage of adenoviral vector, CRISPR-Cas9 mediated genome editing could ameliorate the α1-antitrypsin deficiency phenotype in a humanized mouse model [100].
AAV is an unintegrated virus which exhibits more attractive features than adenoviral vector. The wild-type AAV is in naturally replication-defective without risk of any known disease. What's more, different serotypes of AAV with disparate tropisms target various tissues, such as liver, muscle, eye and so on. It is certified as an ideal viral vector for gene therapy [101]. In 2012, a gene therapy based on AAV vector for lipoprotein lipase deficiency contributed the first viral gene therapy product and acquired marketing approval in Europe. The first successful case of AAV mediated in vivo editing therapy was demonstrated in a mouse model of hemophilia B. ZFNs induced DSBs could efficiently be delivered directly to mouse liver, and when co-transfected with repair templates, F9 gene replacement would be realized at edited locus. This approach could restore F9 activity to 2–3% of normal which might regain hemostasis [102]. Building on this study, another group achieved F9 cDNA insertion by means of AAV harbored CRISPR-Cas9 elements [103]. Sharma et al. present another strategy of specific site integration for hemophilia B treatment. As albumin (Alb) is a protein with high expression in the liver, ZFNs packed by AAV could specifically integrate F9 within Alb gene and realize long-term expression of F9 at therapeutic level [104].
For treating genetic deficiency of the Ornithine transcarbamylase (OTC) enzyme, we applied a dual AAV system simultaneously expression Cas9 and donor DNA to repair gene mutation site in newborn OTC deficiency (OTCD) mouse livers. We found that mutations could be corrected in 10% of hepatocytes and prominently increased survival when challenged with a high-protein diet. These results showed the recovery of OTC function following the AAV mediated in vivo genome editing treatment [105]. In delaE50-MD dog model of DMD, providing disease dogs with therapeutic AAV mediated genome editing could restore the dystrophin level from 3% to 90%. These curative effects in the canine model proved potential of gene therapy for DMD [106]. Moreover, a paper published recently recommended an AAV-CRISPR approach for acquiring long-term restoration of dystrophin in the mdx mouse model. There had been other successful AAV mediated in vivo therapy examples for Mucopolysaccharidosis Type II (MPS II) [107], hereditary tyrosinemia type 1 (HT1) [108], Hutchinson–Gilford progeria syndrome (HGPS) [109,110] and the like. Through combing viral and non-viral delivery of CRISPR system, lipid nanoparticle–loaded delivery of Cas9 mRNA with AAV encoding sgRNA and repair template, Yin et al. generated an efficient strategy for treating human hereditary tyrosinemia [111]. Instead of genetic diseases, Li et al. designed and constructed an artificial virus delivery of CRISPR elements. They discovered a novel approach for ovarian cancer gene therapy [112]. By using a folate receptor-targeted liposome (F-LP) to deliver plasmid DNA co-expressing Cas9 and sgRNA targeting the ovarian cancer-related DNA methyltransferase 1 (DNMT1), we found a potential therapeutic regimen for ovarian cancer. The lipid-mediated CRISPR-Cas9 delivery system may be a useful technology for precise genome editing therapeutics [113]. With the development of gnome editing technologies, the applications scope of gene therapy must be further expanded.
4. Challenges and Prospects
As a more and more important auxiliary approach of biological research, genome editing expands applications range of gene therapy. Constructing an appropriate animal model of disease is essential for studying the mechanism of human diseases or treating diseases, besides also plays an important role in drug development and organ transplantation. The traditional methods of constructing an animal model of disease relies on the establishment of embryonic stem cells, thus animal models are limited to such as mice that easily acquire embryonic stem cells. In addition, the gene integration generated by transgenic technology is random along with low efficiency and applicability. Genome editing at specific sites would address these barriers. Along with the progress of germline editing technologies, more disease models which are close to clinical phenotypes have been established [28]. It would accelerate the investigation of the molecular mechanisms underlying pathogenesis. Great progress in developing engineered nucleases, such as ZFNs, TALENs and CRISPR-Cas9 has paved the avenue for genome editing to enter clinical practice. Recent years, an ever-increasing number of biological institutions focused on targeted genome editing technologies. Although ZFNs and TALENs reached the clinical stage before CRISPR-Cas9, simple CRISPR-Cas system certainly drove the rapid development of genome editing [114]. Taking advantage of CRISPR-Cas9 mediated correction of human genetic diseases, many preclinical studies are on the way [115]. In addition, cancer genomes commonly exist multifarious genetic mutations, CRISPR-Cas system could be utilized as a powerful editing tool for genetic screens of cancer and a promising approach to cancer therapy [116].
Although there has been great progress in the field of genome editing, some hurdles must be cleared for these editing platforms to finally reach the clinic. The efficiency of off-target of engineered nucleases need be concerned. Off-target effects may lead to unexpected and uncontrollable genome change thus increase the risk of malignant transformation. Appropriate sgRNAs design is very necessary, and Doench et al. created human and mouse genome-wide libraries by using devised sgRNAs design rules. They improved computational design rules and created optimized sgRNA libraries to maximize on-target activity and minimize off-target effects [117]. Through engineering a hairpin secondary structure onto the spacer region of sgRNAs could increase specificity of CRISPR effectors [118]. In addition, some other studies focused on developing Cas9 variants to enhance the specificity, such as introduction of point mutation [119], reconstructing nuclease domain [120], evolving broadened PAM compatibility [121] and so on. More importantly, it's indispensable to identify genome-wide off-target effects through a highly sensitive strategy [122]. Developed sequencing technology is a universally applicable method for unbiasedly identifying off-target situations in cells and organisms [123]. Accurately defining and quantifying off-target effects could better foster development of in vivo genome editing therapeutics. Preexisting immunity of host against engineered nucleases can decrease the efficacy of treatment and may pose significant safety issues. Study showed that S. pyogenes Cas9 could active T cells within the adult human population [124] and the immunogenicity of Cas9 had been demonstrated in mice [125]. Except for nucleases, host immune responses to delivery vehicles also influence therapeutic effects.
Viral vectors loaded with therapeutic elements play important roles in gene therapy. In recent decades, its combination with genome editing nucleases has greatly expanded the applications. Especially the emergence of AAV in gene therapy, many serotypes of AAV with specific tropisms for different tissues increase organs specificities of genome editing elements [101]. Despite the remarkable evolutions of viruses mediated genome editing, there are still some limitations that hinders clinical applications. The random integration of lentiviral vector would increase the risk of mutagenesis and oncogenesis [126]. For non-integrated adenoviral, the safety is a major problem. A patient's death occurred in a trial of adenoviral mediated gene therapy for OTCD [127]. Another non-integrated virus, AAV, exits the limitation of packing capacity. AAV could only encapsulate up to 5 kb genome size which is not applicable for big editing elements such as commonly used S. pyogenes Cas9 (4.2 kb). Addressing this obstacle, many studies focus on seeking solutions of augmenting the payload (for example, a) applying a smaller Cas9 for efficient genome editing [128]; b) dual AAV system for delivering editing elements and donor DNA [105]; c) shorter but functional regulatory elements [129]. Instead of loading capacity, existed immunity system against specific serotypes of AAV must be concerned in clinical applications. Non-viral vectors could deliver editing elements to targeted sites without packing capacity limitation and avoid immune response [130]. However, the potential degradation of functional element by endonucleases in the host would decrease the therapeutic efficiency. The combination of viral and non-viral vectors might be an ideal approach for genome editing mediated precise medicine. In summary, considering advances in the non-viral and viral delivery methods, as well as deeper understanding of pathophysiology mechanisms of human diseases, we are optimistic that genome editing certainly presents tantalizing opportunities for tackling a number of diseases that are beyond the reach of previous therapies in the near future.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (No. 2018ZX09733001-005-002), the National Natural Science Foundation of China (No. 81602699), the Science and Technology Major Project of Sichuan province (No.2017SZDZX0011), the Sichuan Science and Technology Program (Nos. 2018GZ0311, 2019YFG0266), the China Postdoctoral Science Foundation Funded Project (No. 2017M612968), the Salubris Academician Workstation for Innovative Biopharmaceuticals (No. 2017B090904017), and the 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (No. ZYJC18028).
Contributor Information
Yang Yang, Email: yang2012@scu.edu.cn.
Zhiyao He, Email: heyaode@163.com.
References
- 1.Cox D.B.T., Platt R.J., Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osakabe Y., Osakabe K. Genome editing with engineered nucleases in plants. Plant Cell Physiol. 2014;56:389–400. doi: 10.1093/pcp/pcu170. [DOI] [PubMed] [Google Scholar]
- 3.Chu V.T., Weber T., Wefers B., Wurst W., Sander S. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 4.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha K., Ma C., Lin H., Tang L., Lian Z. The anaphase promoting complex impacts repair choice by protecting ubiquitin signalling at DNA damage sites. Nat Commun. 2017;8 doi: 10.1038/ncomms15751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu Patrick D., Lander Eric S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S.W., Kim S., Kim J.M., Kim J.-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 8.Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saadatpour Z., Bjorklund G., Chirumbolo S., Alimohammadi M., Ehsani H. Molecular imaging and cancer gene therapy. Cancer Gene Ther. 2016 doi: 10.1038/cgt.2016.62. [DOI] [PubMed] [Google Scholar]
- 13.Stell A., Biserni A., Torre S.D., Rando G., Ramachandran B. Cancer modeling: modern imaging applications in the generation of novel animal model systems to study cancer progression and therapy. Int J Biochem Cell Biol. 2007;39:1288–1296. doi: 10.1016/j.biocel.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Van Dyke T., Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–144. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Perales S., Cano F., Lobato M.N., Rabbitts T.H. MLL gene fusions in human leukaemias: in vivo modelling to recapitulate these primary tumourigenic events. Int J Hematol. 2007;87:3–9. doi: 10.1007/s12185-007-0001-3. [DOI] [PubMed] [Google Scholar]
- 16.Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S., Li L., Kendrick S.L., Gerard R.D., Zhu H. TALEN-mediated somatic mutagenesis in murine models of cancer. Cancer Res. 2014;74:5311–5321. doi: 10.1158/0008-5472.CAN-14-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Nieuwenhuysen T., Naert T., Tran H.T., Van Imschoot G., Geurs S. TALEN-mediated apc mutation in Xenopus tropicalis phenocopies familial adenomatous polyposis. Oncoscience. 2015;2:555–566. doi: 10.18632/oncoscience.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez-Rivera F.J., Papagiannakopoulos T., Romero R., Tammela T., Bauer M.R. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji H., Ramsey M.R., Hayes D.N., Fan C., McNamara K. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 21.Platt Randall J., Chen S., Zhou Y., Yim Michael J., Swiech L. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddalo D., Manchado E., Concepcion C.P., Bonetti C., Vidigal J.A. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi P.S., Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maresch R., Mueller S., Veltkamp C., Öllinger R., Friedrich M. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat Commun. 2016;7 doi: 10.1038/ncomms10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuckermann M., Hovestadt V., Knobbe-Thomsen C.B., Zapatka M., Northcott P.A. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heckl D., Kowalczyk M.S., Yudovich D., Belizaire R., Puram R.V. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strong A., Musunuru K. Genome editing in cardiovascular diseases. Nat Rev Cardiol. 2016;14:11–20. doi: 10.1038/nrcardio.2016.139. [DOI] [PubMed] [Google Scholar]
- 29.Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 30.Ramchandran R., Novodvorsky P., Watson O., Gray C., Wilkinson R.N. Klf2ash317 mutant Zebrafish do not recapitulate morpholino-induced vascular and haematopoietic phenotypes. Plos One. 2015;10 doi: 10.1371/journal.pone.0141611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ota S., Hisano Y., Ikawa Y., Kawahara A. Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells. 2014;19:555–564. doi: 10.1111/gtc.12154. [DOI] [PubMed] [Google Scholar]
- 32.Fujii H., Kotani H., Taimatsu K., Ohga R., Ota S. Efficient multiple genome modifications induced by the crRNAs, tracrRNA and Cas9 protein complex in Zebrafish. Plos One. 2015;10 doi: 10.1371/journal.pone.0128319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao J., Navis A., Cox B.D., Dickson A.L., Gemberling M. Single epicardial cell transcriptome sequencing identifies Caveolin 1 as an essential factor in zebrafish heart regeneration. Development. 2016;143:232–243. doi: 10.1242/dev.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou J., Tran D., Baalbaki M., Tang L.F., Poon A. An internal promoter underlies the difference in disease severity between N- and C-terminal truncation mutations of Titin in zebrafish. Elife. 2015;4 doi: 10.7554/eLife.09406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carroll K.J., Makarewich C.A., McAnally J., Anderson D.M., Zentilin L. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc Natl Acad Sci U S A. 2016;113:338–343. doi: 10.1073/pnas.1523918113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang D., Yang H., Li W., Zhao B., Ouyang Z. Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011;21:979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umeyama K., Watanabe K., Watanabe M., Horiuchi K., Nakano K. Generation of heterozygous fibrillin-1 mutant cloned pigs from genome-edited foetal fibroblasts. Sci Rep. 2016;6 doi: 10.1038/srep24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corden L.D., Swensson O., Swensson B., Smith F.J., Rochels R. Molecular genetics of Meesmann's corneal dystrophy: ancestral and novel mutations in keratin 12 (K12) and complete sequence of the human KRT12 gene. Exp Eye Res. 2000;70:41–49. doi: 10.1006/exer.1999.0769. [DOI] [PubMed] [Google Scholar]
- 39.Ton C.C., Hirvonen H., Miwa H., Weil M.M., Monaghan P. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 40.Quinlan R.A., Zhu Y., Yu H., Wang W., Gong X. A novel GJA8 mutation (p.V44A) causing autosomal dominant congenital cataract. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.X. Jiang J. Gap junctions or hemichannel-dependent and independent roles of connexins in cataractogenesis and lens development. Curr Mol Med. 2010;10:851–863. doi: 10.2174/156652410793937750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossmiller B., Mao H., Lewin A.S. Gene therapy in animal models of autosomal dominant retinitis pigmentosa. Mol Vis. 2012;18:2479–2496. [PMC free article] [PubMed] [Google Scholar]
- 43.Newman N.J. Hereditary optic neuropathies: from the mitochondria to the optic nerve. Am J Ophthalmol. 2005;140(517) doi: 10.1016/j.ajo.2005.03.017. [e511–517.e519] [DOI] [PubMed] [Google Scholar]
- 44.Younes S., Tahri H. Severe congenital ocular coloboma. Pan Afr Med J. 2014;19:1. doi: 10.11604/pamj.2014.19.1.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo H., Dai L., Huang Y., Liao Q., Bai Y. A large novel deletion downstream of PAX6 gene in a Chinese family with ocular coloboma. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakayama T., Fisher M., Nakajima K., Odeleye A.O., Zimmerman K.B. Xenopus pax6 mutants affect eye development and other organ systems, and have phenotypic similarities to human aniridia patients. Dev Biol. 2015;408:328–344. doi: 10.1016/j.ydbio.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan L., Sui T., Chen M., Deng J., Huang Y. CRISPR/Cas9-mediated GJA8 knockout in rabbits recapitulates human congenital cataracts. Sci Rep. 2016;6 doi: 10.1038/srep22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong H., Chen Y., Li Y., Chen R., Mardon G. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci Rep. 2015;5:8366. doi: 10.1038/srep08366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendee K.E., Sorokina E.A., Muheisen S.S., Reis L.M., Tyler R.C. PITX2 deficiency and associated human disease: insights from the zebrafish model. Hum Mol Genet. 2018;27:1675–1695. doi: 10.1093/hmg/ddy074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Homma N., Harada Y., Uchikawa T., Kamei Y., Fukamachi S. Protanopia (red color-blindness) in medaka: a simple system for producing color-blind fish and testing their spectral sensitivity. BMC Genet. 2017;18:10. doi: 10.1186/s12863-017-0477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naert T., Colpaert R., Van Nieuwenhuysen T., Dimitrakopoulou D., Leoen J. CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci Rep. 2016;6 doi: 10.1038/srep35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y., Lu W., Gao N., Long Y., Shao Y. Generation of obese rat model by transcription activator-like effector nucleases targeting the leptin receptor gene. Sci China Life Sci. 2016;60:152–157. doi: 10.1007/s11427-016-5049-y. [DOI] [PubMed] [Google Scholar]
- 53.Turer E.E., San Miguel M., Wang K.-w., McAlpine W., Ou F. A viable hypomorphic Arnt2 mutation causes hyperphagic obesity, diabetes and hepatic steatosis. Dis Model Mech. 2018;11 doi: 10.1242/dmm.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu D., Zhong Y., Li X., Li Y., Li X. Generation of TALEN-mediated FH knockout rat model. Oncotarget. 2016;7:61656–61669. doi: 10.18632/oncotarget.11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carlson D.F., Tan W., Lillico S.G., Stverakova D., Proudfoot C. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang L., Hua Z., Xiao H., Cheng Y., Xu K. CRISPR/Cas9-mediated ApoE−/− and LDLR−/− double gene knockout in pigs elevates serum LDL-C and TC levels. Oncotarget. 2017;8:37751–37760. doi: 10.18632/oncotarget.17154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng W.-C., Loeb H.E., Pei W., Tsai-Morris C.-H., Xu L. Modeling Niemann-pick disease type C1 in zebrafish: a robust platform forin vivoscreening of candidate therapeutic compounds. Dis Model Mech. 2018;11 doi: 10.1242/dmm.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang W., Liu L., Chang Q., Xing F., Ma Z. Production of Wilson disease model rabbits with homology-directed precision point mutations in the ATP7B gene using the CRISPR/Cas9 system. Sci Rep. 2018;8:1332. doi: 10.1038/s41598-018-19774-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang H., Ahn C., Shin E.-K., Lee J.-S., An B.-S. NCKX3 was compensated by calcium transporting genes and bone resorption in a NCKX3 KO mouse model. Mol Cell Endocrinol. 2017;454:93–102. doi: 10.1016/j.mce.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 60.Strachan L.R., Stevenson T.J., Freshner B., Keefe M.D., Miranda Bowles D. A zebrafish model of X-linked adrenoleukodystrophy recapitulates key disease features and demonstrates a developmental requirement for abcd1 in oligodendrocyte patterning and myelination. Hum Mol Genet. 2017;26:3600–3614. doi: 10.1093/hmg/ddx249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan H.-C., Ho L.-I., Chi C.-S., Chen S.-J., Peng G.-S. Polyglutamine (PolyQ) diseases: genetics to treatments. Cell Transplant. 2014;23:441–458. doi: 10.3727/096368914X678454. [DOI] [PubMed] [Google Scholar]
- 62.Fan H.-C., Chi C.-S., Cheng S.-N., Lee H.-F., Tsai J.-D. Targeting new candidate genes by small molecules approaching neurodegenerative diseases. Int J Mol Sci. 2015;17:26. doi: 10.3390/ijms17010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan H.-C., Chen S.-J., Harn H.-J., Lin S.-Z. Parkinson's disease: from genetics to treatments. Cell Transplant. 2013;22:639–652. doi: 10.3727/096368912X655082. [DOI] [PubMed] [Google Scholar]
- 64.Yan S., Tu Z., Liu Z., Fan N., Yang H. A Huntingtin knockin pig model recapitulates features of selective neurodegeneration in Huntington's disease. Cell. 2018;173:989–1002. doi: 10.1016/j.cell.2018.03.005. [e1013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones J.M., Meisler M.H. Modeling human epilepsy by TALEN targeting of mouse sodium channelScn8a. Genesis. 2014;52:141–148. doi: 10.1002/dvg.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Larcher T., Lafoux A., Tesson L., Remy S., Thepenier V. Characterization of dystrophin deficient rats: a new model for Duchenne muscular dystrophy. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sui T., Lau Y.S., Liu D., Liu T., Xu L. A novel rabbit model of Duchenne muscular dystrophy generated by CRISPR/Cas9. Dis Model Mech. 2018;11 doi: 10.1242/dmm.032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y., Niu Y., Ji W. Genome editing in nonhuman primates: approach to generating human disease models. J Intern Med. 2016;280:246–251. doi: 10.1111/joim.12469. [DOI] [PubMed] [Google Scholar]
- 69.Chen Y., Yu J., Niu Y., Qin D., Liu H. Modeling Rett syndrome using TALEN-edited MECP2 mutant cynomolgus monkeys. Cell. 2017;169:945–955. doi: 10.1016/j.cell.2017.04.035. [e910] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Y., Zheng Y., Kang Y., Yang W., Niu Y. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum Mol Genet. 2015;24:3764–3774. doi: 10.1093/hmg/ddv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ott de Bruin L.M., Volpi S., Musunuru K. Novel genome-editing tools to model and correct primary immunodeficiencies. Front Immunol. 2015;6:250. doi: 10.3389/fimmu.2015.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sato K., Oiwa R., Kumita W., Henry R., Sakuma T. Generation of a nonhuman primate model of severe combined immunodeficiency using highly efficient genome editing. Cell Stem Cell. 2016;19:127–138. doi: 10.1016/j.stem.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 73.Zhou J., Shen B., Zhang W., Wang J., Yang J. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol. 2014;46:49–55. doi: 10.1016/j.biocel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 74.Wefers B., Meyer M., Ortiz O., Hrabe de Angelis M., Hansen J. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci U S A. 2013;110:3782–3787. doi: 10.1073/pnas.1218721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Q., Fan C., Zhou S., Guo Y., Zuo Q. Bioluminescent imaging of vaccinia virus infection in immunocompetent and immunodeficient rats as a model for human smallpox. Sci Rep. 2015;5 doi: 10.1038/srep11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cui D., Li F., Li Q., Li J., Zhao Y. Generation of a miniature pig disease model for human Laron syndrome. Sci Rep. 2015;5 doi: 10.1038/srep15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu C., Xiao L., Li F., Zhang H., Li Q. Generation of outbred Ace2 knockout mice by RNA transfection of TALENs displaying colitis reminiscent pathophysiology and inflammation. Transgenic Res. 2014;24:433–446. doi: 10.1007/s11248-014-9855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kasparek P., Ileninova Z., Haneckova R., Kanchev I., Jenickova I. A viable mouse model for Netherton syndrome based on mosaic inactivation of the Spink5 gene. Biol Chem. 2016;397:1287–1292. doi: 10.1515/hsz-2016-0194. [DOI] [PubMed] [Google Scholar]
- 79.Genovese P., Schiroli G., Escobar G., Di Tomaso T., Firrito C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schiroli G., Ferrari S., Conway A., Jacob A., Capo V. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aan0820. [DOI] [PubMed] [Google Scholar]
- 81.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li C., Psatha N., Sova P., Gil S., Wang H. Reactivation of γ-globin in adult β-YAC mice after ex vivo and in vivo hematopoietic stem cell genome editing. Blood. 2018;131:2915–2928. doi: 10.1182/blood-2018-03-838540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu L., Yang H., Gao Y., Chen Z., Xie L. CRISPR/Cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol Ther. 2017;25:1782–1789. doi: 10.1016/j.ymthe.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim M.Y., Yu K.-R., Kenderian S.S., Ruella M., Chen S. Genetic inactivation of CD33 in hematopoietic stem cells to enable CAR T cell immunotherapy for acute myeloid leukemia. Cell. 2018;173:1439–1453. doi: 10.1016/j.cell.2018.05.013. [e1419] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu X., Zhang Y., Cheng C., Cheng A.W., Zhang X. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 2017;27:154–157. doi: 10.1038/cr.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu S., Yao Y., Xiao H., Li J., Liu Q. Simultaneous knockout of CXCR4 and CCR5 genes in CD4+ T cells via CRISPR/Cas9 confers resistance to both X4- and R5-tropic human immunodeficiency virus type 1 infection. Hum Gene Ther. 2018;29:51–67. doi: 10.1089/hum.2017.032. [DOI] [PubMed] [Google Scholar]
- 88.Normile D. China sprints ahead in CRISPR therapy race. Science. 2017;358:20–21. doi: 10.1126/science.358.6359.20. [DOI] [PubMed] [Google Scholar]
- 89.Lai F.P.-L., Lau S.-T., Wong J.K.-L., Gui H., Wang R.X. Correction of Hirschsprung-associated mutations in human induced pluripotent stem cells via clustered regularly interspaced short palindromic repeats/Cas9, restores neural crest cell function. Gastroenterology. 2017;153:139–153. doi: 10.1053/j.gastro.2017.03.014. [e138] [DOI] [PubMed] [Google Scholar]
- 90.Ma N., Shan Y., Liao B., Kong G., Wang C. Factor-induced reprogramming and zinc finger nuclease-aided gene targeting cause different genome instability in beta-thalassemia induced pluripotent stem cells (iPSCs) J Biol Chem. 2015;290:12079–12089. doi: 10.1074/jbc.M114.624999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu P., Tong Y., X-z Liu, T-t Wang, Cheng L. Both TALENs and CRISPR/Cas9 directly target the HBB IVS2–654 (C > T) mutation in β-thalassemia-derived iPSCs. Sci Rep. 2015;5 doi: 10.1038/srep12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song B., Fan Y., He W., Zhu D., Niu X. Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev. 2015;24:1053–1065. doi: 10.1089/scd.2014.0347. [DOI] [PubMed] [Google Scholar]
- 93.Park C.-Y., Kim Duk H., Son Jeong S., Sung Jin J., Lee J. Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17:213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 94.Kawamata S., Kanemura H., Sakai N., Takahashi M., Go M. Design of a tumorigenicity test for induced pluripotent stem cell (iPSC)-derived cell products. J Clin Med. 2015;4:159–171. doi: 10.3390/jcm4010159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 96.Kotterman M.A., Chalberg T.W., Schaffer D.V. Viral vectors for gene therapy: translational and clinical outlook. Annu Rev Biomed Eng. 2015;17:63–89. doi: 10.1146/annurev-bioeng-071813-104938. [DOI] [PubMed] [Google Scholar]
- 97.Ding Q., Strong A., Patel K.M., Ng S.-L., Gosis B.S. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ali R., Wendt M.K. The paradoxical functions of EGFR during breast cancer progression. Signal Transduct Target Ther. 2017;2 doi: 10.1038/sigtrans.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koo T., Yoon A.R., Cho H.Y., Bae S., Yun C.O. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017;45:7897–7908. doi: 10.1093/nar/gkx490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bjursell M., Porritt M.J., Ericson E., Taheri-Ghahfarokhi A., Clausen M. Therapeutic genome editing with CRISPR/Cas9 in a humanized mouse model ameliorates alpha1-antitrypsin deficiency phenotype. EBioMedicine. 2018;29:104–111. doi: 10.1016/j.ebiom.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mingozzi F., High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 102.Li H., Haurigot V., Doyon Y., Li T., Wong S.Y. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ohmori T., Nagao Y., Mizukami H., Sakata A., Muramatsu S.-i. CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci Rep. 2017;7:4159. doi: 10.1038/s41598-017-04625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sharma R., Anguela X.M., Doyon Y., Wechsler T., DeKelver R.C. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126:1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang Y., Wang L., Bell P., McMenamin D., He Z. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Amoasii L., Hildyard J.C.W., Li H., Sanchez-Ortiz E., Mireault A. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Laoharawee K., DeKelver R.C., Podetz-Pedersen K.M., Rohde M., Sproul S. Dose-dependent prevention of metabolic and neurologic disease in murine MPS II by ZFN-mediated in vivo genome editing. Mol Ther. 2018;26:1127–1136. doi: 10.1016/j.ymthe.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang D., Li J., Song C.-Q., Tran K., Mou H. Cas9-mediated allelic exchange repairs compound heterozygous recessive mutations in mice. Nat Biotechnol. 2018;36:839–842. doi: 10.1038/nbt.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beyret E., Liao H.-K., Yamamoto M., Hernandez-Benitez R., Fu Y. Single-dose CRISPR–Cas9 therapy extends lifespan of mice with Hutchinson–Gilford progeria syndrome. Nat Med. 2019 doi: 10.1038/s41591-019-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Santiago-Fernandez O., Osorio F.G., Quesada V., Rodriguez F., Basso S. Development of a CRISPR/Cas9-based therapy for Hutchinson-Gilford progeria syndrome. Nat Med. 2019 doi: 10.1038/s41591-018-0338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li L., Song L., Liu X., Yang X., Li X. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano. 2017;11:95–111. doi: 10.1021/acsnano.6b04261. [DOI] [PubMed] [Google Scholar]
- 113.He Z.Y., Zhang Y.G., Yang Y.H., Ma C.C., Wang P. In vivo ovarian Cancer gene therapy using CRISPR-Cas9. Hum Gene Ther. 2018;29:223–233. doi: 10.1089/hum.2017.209. [DOI] [PubMed] [Google Scholar]
- 114.Cornu T.I., Mussolino C., Cathomen T. Refining strategies to translate genome editing to the clinic. Nat Med. 2017;23:415–423. doi: 10.1038/nm.4313. [DOI] [PubMed] [Google Scholar]
- 115.Men K., Duan X., He Z., Yang Y., Yao S. CRISPR/Cas9-mediated correction of human genetic disease. Sci China Life Sci. 2017;60:447–457. doi: 10.1007/s11427-017-9032-4. [DOI] [PubMed] [Google Scholar]
- 116.Fan P., He Z.-Y., Xu T., Phan K., Chen G.G. Exposing cancer with CRISPR-Cas9: from genetic identification to clinical therapy. Transl Cancer Res. 2018;7:817–827. [Google Scholar]
- 117.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kocak D.D., Josephs E.A., Bhandarkar V., Adkar S.S., Kwon J.B. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat Biotechnol. 2019 doi: 10.1038/s41587-019-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vakulskas C.A., Dever D.P., Rettig G.R., Turk R., Jacobi A.M. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24:1216–1224. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Akcakaya P., Bobbin M.L., Guo J.A., Malagon-Lopez J., Clement K. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 2018;561:416–419. doi: 10.1038/s41586-018-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wienert B., Wyman S.K., Richardson C.D., Yeh C.D., Akcakaya P. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364:286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wagner D.L., Amini L., Wendering D.J., Burkhardt L.M., Akyuz L. High prevalence of streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med. 2019;25:242–248. doi: 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- 125.Chew W.L., Tabebordbar M., Cheng J.K., Mali P., Wu E.Y. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baum C., Kustikova O., Modlich U., Li Z., Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum Gene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 127.Raper S.E., Chirmule N., Lee F.S., Wivel N.A., Bagg A. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 128.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Duan D. Systemic AAV micro-dystrophin gene therapy for Duchenne muscular dystrophy. Mol Ther. 2018;26:2337–2356. doi: 10.1016/j.ymthe.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.He Z.Y., Men K., Qin Z., Yang Y., Xu T. Non-viral and viral delivery systems for CRISPR-Cas9 technology in the biomedical field. Sci China Life Sci. 2017;60:458–467. doi: 10.1007/s11427-017-9033-0. [DOI] [PubMed] [Google Scholar]