Abstract
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world. Circular RNAs (circRNAs) are a new class of endogenous functional non-coding RNAs (ncRNAs), and have been demonstrated to play important roles in the development of HCC. This study aimed to explore the significance of circRNAs in HCC progression. HCC-associated circRNA expression profiles GSE94508 and GSE97332 were downloaded from the Gene Expression Omnibus database (GEO), and 87 differentially expressed circRNAs (DECs) between HCC tissues and paired non-cancer tissues were identified, including 76 up-regulated and 11 down-regulated circRNAs. Gene ontolog (GO) and pathway analyses of the host genes of these DECs suggested that these host genes were enriched in cell adhesion, cytosol, and protein binding, and were associated with tight junction and Wnt signaling pathways. CircRNA-miRNA interaction prediction identified 20 miRNAs that predispose to interact with DECs. Among these, four essential miRNAs, hsa-miR-7-5p, hsa-miR-145-5p, hsa-miR-203a-3p, and hsa-miR-192-5p, were reported to play pivotal roles in HCC progression by targeting multiple genes. Pathway analysis suggested that putative target genes of these essential miRNAs were involved in HCC-associated signaling pathways, such as Wnt, TGF-β, and Ras; whereas protein-protein network (PPI) analysis demonstrated that some validated target genes of these miRNAs, such as PIK3CA, AKT1, MYC, JUN, SMAD4, and SRC, were hub target genes as they have more counts of interacting protein. In the meantime, the deregulation of some DECs was validated in HCC cell line HepG2 compared with normal liver cell line L02 by quantitative real-time polymerase chain reaction (qRT-PCR) and the Sanger sequencing. This study identified a set of DECs in HCC, and provided a comprehensive understanding of the roles of these DECs in HCC progression.
Keywords: circular RNA, miRNA sponge, hepatocellular carcinoma, bioinformatic analysis
Introduction
Hepatocellular carcinoma (HCC), the fifth common cancer in the world, ranks the third in cancer-related death 1. Currently, the diagnosis of HCC is still poor, and therapeutic options are limited. Many HCC patients are not diagnosed until they are in advanced stages when curative therapies are too late to be applied; while for those patients who are eligible for surgical resection, liver transplantation or local ablation, although their life qualities could be improved by these therapies, their overall survival rates are still low. Therefore, novel biomarkers and therapeutic approaches are urgently needed to improve the overall survival of HCC patients.
Circular RNAs (circRNAs) are a new class of abundant endogenous functional non-coding RNAs (ncRNAs), and have been demonstrated to play important roles in different types of cancer, including gastric cancer, lung cancer, and HCC 2-4. They form covalently closed continuous loop structures with neither polarity nor polyadenylated tails, which make them more stable and abundant than their canonical linear transcripts from the same genes in cells 5, 6. Except for tissues, circRNAs can also be found in different body fluids, such as saliva, blood, and urine, suggesting that they may serve as non-invasive circulating biomarkers for cancer diagnosis 7. Studies showed that circRNAs may regulate gene expression at transcriptional, post-transcriptional, and translational levels. Specifically, circular exonic circRNAs, which predominantly exist in cytoplasm, harbor microRNA (miRNA) response elements (MREs), and may function as miRNA sponges by competitively binding to specific miRNAs to reduce their expression and function, resulting in enhanced expression of target genes of these miRNAs; while circular intronic RNAs or exon-intron circRNAs are mainly accumulate in the nucleus, and may regulate gene transcription and post-transcription 5, 8-10. These suggest that circRNAs may participate in various biological processes, such as cell proliferation, migration, invasion and metastasis, as well as apoptosis, and thus contribute to cancer progression.
Most recently, some circRNAs have been reported to be deregulated in HCC tissues, and the deregulation of these circRNAs may not only associate with clinicopathological features of HCC patients, but also impact on HCC progression by targeting specific miRNAs and proteins. Several circRNAs, such as circZKSCAN1, hsa_circ_0004018, and hsa_circ_0005075, have been identified as promising biomarkers for HCC diagnosis; whereas circMTO1 and circ-ITCH may serve as potential prognostic biomarkers for poor survival of HCC patients 11-15. Furthermore, some circRNAs have been reported to interfere with HCC progression by modulating the proliferation, apoptosis, migration and invasion via targeting different miRNAs and proteins. For example, circMTO1 inhibits the proliferation of HCC cells by sponging miR-9 and increasing the expression of miR-9 target gene p21, a cell cycle inhibitory protein; hsa_circ_100338 promotes the migration and invasion of HCC cells by targeting miR-141; while ciRS-7 facilitates the proliferation and invasion of HCC cells by competitively binding to hsa-miR-7-5p (miR-7) and promoting the expression of multiple miR-7 target genes, including CCNE1, PIK3CD, and EGFR 14, 16-19. These results indicate that circRNAs may play critical roles in HCC progression.
In the current study, we downloaded the HCC-associated circRNA expression profiles GSE94508 and GSE97332 from the Gene Expression Omnibus database (GEO), analyzed the differentially expressed circRNAs (DECs) in both profiles between paired HCC and non-tumor liver tissues, and investigated their possible target genes and pathways involved in HCC using bioinformatic approaches, to provided possible functions and underlying mechanisms of these DECs in HCC tumorigenesis and progression.
Materials and methods
Microarray analysis of gene expression
Two circRNA expression profiles, GSE94508 and GSE97332, were downloaded from the GEO (www.ncbi.nlm.nih.gov/geo/), both of which were completed on the Agilent-069978 Arraystar Human CircRNA microarray V1 GPL19978 platform 20. The GSE94508 dataset contained 5 pairs of HCC and paracancerous liver tissues, and the GSE97332 dataset contained 7 pairs of HCC and matched non-tumor liver tissues.
Identification of DECs
GEO2R 21 was used to analysis the raw data TXT files downloaded from GEO. The absolute value of log fold change > 1.0 and p value <0.05 were used as cut-off criteria. CircRNAs with statistical significance between HCC and non-tumor tissues in GSE94508 and GSE97332 were screened, separately. Then, circRNAs up-regulated or down-regulated in both profiles were selected and identified.
Functional enrichment analysis
Gene ontolog (GO) annotation, including biological process, cellular component, and molecular function, was analyzed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID; https://david. ncifcrf.gov) 22-24. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Panther (http.//www.pantherdb.org/) pathway analyses were used to classify genomic and gene functional information 25-26. The significant enrichment results were obtained with p values <0.05.
Prediction of circRNA-miRNA and miRNA-mRNA interactions
Online tools miRDB (http.//www.mirdb.org/) and circinteractome (https.//circinteractome.nia.nih.gov/) were used to predict the possible interactions between circRNAs and miRNAs 27-29; miRDB and TargetScan (http.//www.targetscan.org/mamm_31/) were applied for predicting target genes of the four essential miRNAs; while mirTarBase (http.//mirtarbase.mbc.nctu.edu.tw/php/index.php) was used to obtain experimentally strongly supported targets genes (by Reporter assay, Western blot, or qPCR) of these miRNAs 30-32.
Construction of protein-protein interaction (PPI) network
Experimentally supported target genes of the four essential miRNAs were put into the Search Tool for the Retrieval of Interacting Genes database (STRING; https.//string-db.org/), and an interaction network chart with a combined score > 0.4 was saved and exported 33. Subsequently, top 50 genes with more counts of interacting protein were selected, and the PPI network was visualized using Cytoscape software (version 3.6.1; http.//cytoscape.org/) 34. The values of gene interactions predicted by STRING were also imported into Cytoscape to identify hub genes among potential targets.
Cell culture
HCC cell line HepG2 was cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco, USA), and human normal liver cell line L02 was cultured in 1640 in a humidified 37℃ incubator with 5% CO2. Both media were supplemented with 10% FBS (Gibco.USA), 2 mmol/L L-glutamine and 100U/mL penicillin/100 μg/mL streptomycin (Life Technologies, CA, USA).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNAs from HepG2 and L02 cells were extracted using TRIzol reagent (Life Technologies, CA, USA) according to the manufacture's protocol. RNA quality and quantity was meatured using Synergy H4 Hybrid Multi-Mode Microplate Reader (BioTek Instruments Inc, Winooski, VT, USA). Total RNAs were reversely transcribe using HiScript Q RT SuperMix for qPCR with gDNA wiper (Vazyme Biotech, Nanjing, China), and qPCR assays were performed in triplicate using AceQ qPCR SYBR Green Master Mix kit (Vazyme Biotech, Nanjing, China) on the ABI QuantStudio 3 real-time PCR system (ThermoFisher Scientific, USA). The divergent primers used for detecting circRNAs were synthesized from Shanghai Generay Biotech (Shanghai, China), and β-actin was used as an internal control. The sequences of primers for qPCR were listed in Table 1, and the specificity of PCR products was evaluated by dissociation curve analysis and the Sanger sequencing (Sango Biotech, Shanghai, China).
Table 1.
Circular RNA (circRNA) primers used for qPCR
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| has_circ_0001806 | CCATCCCATCAGTTCATCCT | TTCACCTCCAAAGAGCATCC |
| has_circ_0003528 | GTAACCAGCAGCCTGGACTC | GCAACTTGCTGACCAGAACA |
| has_circ_0008583 | TTACGGGAGCAGATGATGAA | CCAAGAAGGAAGATGGGCTA |
| has_circ_0009910 | CAGGTTCTGGACGTCAAAGG | TCACCTCAGCCATGTGTCTC |
| has_circ_0032704 | TTGTTCCTCATCGCAGCAGT | ATAGAGGCGCACGTCAAACT |
| has_circ_0065214 | TCATGTCTGTGGGACTCTGC | GGGCGAGTAATCCTTCACAG |
| has_circ_0007762 | CATTCAGATGGCACCTTGAC | GTGCCCACATAGAGCCACTT |
| β-actin | AGAAAATCTGGCACCACACC | CAGAGGCGTACAGGGATAGC |
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, CA, USA). All Data were shown as mean ± SEM. Student's t-test was used to compare the differences of circRNA expression between L02 and HepG2 cells, and a p value <0.05 was considered statistically significant.
Results
Selection of DECs
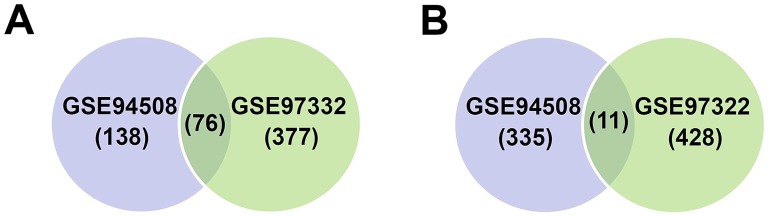
Two circRNA expression profiles, including GSE94508 and GSE97332, were downloaded from GEO, and GEO2R method was applied to analysis DECs between HCC liver tissues and paired non-tumor tissues. Results showed that many circRNAs were differentially expressed in HCC tissues. In GSE94508, 560 circRNAs were identified to be differentially expressed, including 214 up-regulated and 346 down-regulated circRNAs; while in GSE97332, 892 differentially expressed circRNAs were identified, including 453 up-regulated and 439 down-regulated circRNAs. Among them, 87 circRNAs, including 76 up-regulated and 11 down-regulated circRNAs, were observed in both circRNA expression profiles (Figure 1A and 1B, and Table 2).
Figure 1.
Differentially expressed circular RNAs (DECs) in hepatocellular carcinoma (HCC) compared with paired non-tumor liver tissues. (A-B) Venn diagram represents the overlap of the identified DECs in HCC tissues from the results of microarray GSE94508 (5 pairs) and GSE97332 (7 pairs). (A) Up-regulated circRNAs in HCC tissues. (B) Down-regulated circRNAs in HCC tissues.
Table 2.
Differentially expressed circRNAs (DECs) in hepatocellular carcinoma (HCC)
| No. | CircRNA ID | Chromosome location | Gene symbol | Accession number |
|---|---|---|---|---|
| Up-regulated circRNAs | ||||
| 1 | hsa_circ_0000291 | chr11:35163017-35163328+ | CD44 | NM_001202557 |
| 2 | hsa_circ_0000511 | chr14:20811282-20811431- | RPPH1 | NR_002312 |
| 3 | hsa_circ_0000554 | chr14:74551677-74551959+ | LIN52 | NM_001024674 |
| 4 | hsa_circ_0000644 | chr15:81195758-81195954- | KIAA1199 | NM_018689 |
| 5 | hsa_circ_0000673 | chr16:11940357-11940700- | RSL1D1 | NM_015659 |
| 6 | hsa_circ_0000747 | chr17:27209637-27210251+ | FLOT2 | NM_004475 |
| 7 | hsa_circ_0000981 | chr2:20240809-20240905- | LAPTM4A | NM_014713 |
| 8 | hsa_circ_0001228 | chr22:37868480-37870715- | MFNG | NM_002405 |
| 9 | hsa_circ_0001279 | chr3:33109721-33110462- | GLB1 | NM_001079811 |
| 10 | hsa_circ_0001338 | chr3:128824688-128825122- | RAB43 | NM_001204883 |
| 11 | hsa_circ_0001489 | chr5:59770958-59771235+ | PDE4D | NM_001165899 |
| 12 | hsa_circ_0001806 | chr8:68018139-68028357+ | CSPP1 | NM_024790 |
| 13 | hsa_circ_0001828 | chr8:142139086-142139265+ | DENND3 | NM_014957 |
| 14 | hsa_circ_0001834 | chr9:2017333-2017502+ | SMARCA2 | NM_003070 |
| 15 | hsa_circ_0001901 | chr9:138773785-138774005- | CAMSAP1 | NM_015447 |
| 16 | hsa_circ_0001917 | chrX:41519691-41530783- | CASK | NM_001126055 |
| 17 | hsa_circ_0001955 | chr15:64495280-64508912- | CSNK1G1 | NM_022048 |
| 18 | hsa_circ_0002191 | chr9:97535283-97563284+ | C9orf3 | NM_001193331 |
| 19 | hsa_circ_0002702 | chr9:35546426-35548532+ | RUSC2 | NM_001135999 |
| 20 | hsa_circ_0003528 | chr5:134032815-134044578+ | SEC24A | NM_021982 |
| 21 | hsa_circ_0003645 | chr16:19656207-19663412+ | C16orf62 | NM_020314 |
| 22 | hsa_circ_0003892 | chr19:11230767-11238761+ | LDLR | NM_000527 |
| 23 | hsa_circ_0003923 | chr2:238933982-238940895+ | UBE2F | NM_080678 |
| 24 | hsa_circ_0003945 | chr9:33953282-33956144- | UBAP2 | NM_018449 |
| 25 | hsa_circ_0003958 | chr7:27668989-27672064- | HIBADH | NM_152740 |
| 26 | hsa_circ_0004004 | chr5:172359438-172362313+ | ERGIC1 | NM_001031711 |
| 27 | hsa_circ_0004976 | chr2:25990451-25994409- | ASXL2 | NM_018263 |
| 28 | hsa_circ_0005397 | chr17:30500849-30503232+ | RHOT1 | NM_001033568 |
| 29 | hsa_circ_0005785 | chr12:110819556-110834257- | ANAPC7 | NM_016238 |
| 30 | hsa_circ_0006517 | chr3:188202379-188242575+ | LPP | NM_005578 |
| 31 | hsa_circ_0006789 | chrX:118544152-118544325+ | SLC25A43 | NM_145305 |
| 32 | hsa_circ_0007196 | chr3:50144199-50147121+ | RBM5 | NM_005778 |
| 33 | hsa_circ_0008274 | chr13:96485180-96489456- | UGGT2 | NM_020121 |
| 34 | hsa_circ_0008310 | chr17:7849045-7849304+ | CNTROB | NM_001037144 |
| 35 | hsa_circ_0008563 | chr1:21599191-21599404- | ECE1 | NM_001113347 |
| 36 | hsa_circ_0008583 | chr3:196817782-196846401- | DLG1 | NM_004087 |
| 37 | hsa_circ_0009910 | chr1:12049221-12052747+ | MFN2 | NM_014874 |
| 38 | hsa_circ_0012107 | chr1:44290402-44303983+ | ST3GAL3 | NM_174963 |
| 39 | hsa_circ_0013048 | chr1:82302569-82372915+ | LPHN2 | NM_012302 |
| 40 | hsa_circ_0014879 | chr1:160206924-160231148- | DCAF8 | NR_028103 |
| 41 | hsa_circ_0016404 | chr1:212977661-212977993+ | TATDN3 | NM_001146171 |
| 42 | hsa_circ_0018004 | chr10:27024168-27024508+ | PDSS1 | NM_014317 |
| 43 | hsa_circ_0026143 | chr12:49722709-49723237+ | TROAP | NM_005480 |
| 44 | hsa_circ_0027478 | chr12:69109406-69125499+ | NUP107 | NM_020401 |
| 45 | hsa_circ_0028196 | chr12:110826316-110834257- | ANAPC7 | NM_016238 |
| 46 | hsa_circ_0029325 | chr12:125270902-125284788- | SCARB1 | NM_005505 |
| 47 | hsa_circ_0031132 | chr14:21464367-21464870+ | METTL17 | NM_022734 |
| 48 | hsa_circ_0032704 | chr14:76173360-76187046+ | TTLL5 | NM_015072 |
| 49 | hsa_circ_0033408 | chr14:102842986-102931626+ | TECPR2 | NM_014844 |
| 50 | hsa_circ_0036005 | chr15:67004005-67008836+ | SMAD6 | NR_027654 |
| 51 | hsa_circ_0038718 | chr16:27351506-27353580+ | IL4R | NM_000418 |
| 52 | hsa_circ_0039053 | chr16:30510394-30510810+ | ITGAL | NM_002209 |
| 53 | hsa_circ_0043438 | chr17:37583953-37584043- | MED1 | NM_004774 |
| 54 | hsa_circ_0045006 | chr17:59152280-59161925+ | BCAS3 | NM_001099432 |
| 55 | hsa_circ_0045862 | chr17:76082583-76083174+ | TNRC6C | NM_001142640 |
| 56 | hsa_circ_0048937 | chr19:6934997-6937659+ | EMR1 | NM_001974 |
| 57 | hsa_circ_0049997 | chr19:17626981-17628198+ | PGLS | NM_012088 |
| 58 | hsa_circ_0051220 | chr19:41884185-41884424+ | TMEM91 | NM_001098821 |
| 59 | hsa_circ_0051732 | chr19:48660270-48660397- | LIG1 | NM_000234 |
| 60 | hsa_circ_0060055 | chr20:33866724-33872064- | EIF6 | NM_181468 |
| 61 | hsa_circ_0062682 | chr22:26936754-26937684- | TPST2 | NM_001008566 |
| 62 | hsa_circ_0064288 | chr3:11348416-11350535+ | ATG7 | NM_006395 |
| 63 | hsa_circ_0065214 | chr3:47466974-47476627- | SCAP | NM_012235 |
| 64 | hsa_circ_0067934 | chr3:170013698-170015181+ | PRKCI | NM_002740 |
| 65 | hsa_circ_0072088 | chr5:32379220-32388780- | ZFR | NM_016107 |
| 66 | hsa_circ_0073271 | chr5:88044886-88047860- | MEF2C | NM_002397 |
| 67 | hsa_circ_0074903 | chr5:168110970-168112932- | SLIT3 | NM_003062 |
| 68 | hsa_circ_0078738 | chr6:170033042-170058454- | WDR27 | NM_182552 |
| 69 | hsa_circ_0082182 | chr7:128317617-128323309+ | FAM71F2 | NM_001128926 |
| 70 | hsa_circ_0082564 | chr7:137569740-137570248- | CREB3L2 | NM_194071 |
| 71 | hsa_circ_0083766 | chr8:27382878-27394372+ | EPHX2 | NM_001979 |
| 72 | hsa_circ_0091331 | chrX:109310574-109352374+ | TMEM164 | NM_032227 |
| 73 | hsa_circ_0092283 | chr22:36681395-36681695- | MYH9 | NM_002473 |
| 74 | hsa_circ_0092298 | chr5:178287348-178287568+ | ZNF354B | NM_058230 |
| 75 | hsa_circ_0092310 | chr6:43467278-43467478+ | TJAP1 | NM_001146018 |
| 76 | hsa_circ_0092327 | chr19:17972481-17972901+ | RPL18A | NM_000980 |
| Down-regulated circRNAs | ||||
| 1 | hsa_circ_0003859 | chr19:41122794-41125398+ | LTBP4 | NM_001042544 |
| 2 | hsa_circ_0004913 | chr17:62248459-62265775- | TEX2 | NM_018469 |
| 3 | hsa_circ_0005428 | chr1:47761436-47770668- | STIL | NM_001048166 |
| 4 | hsa_circ_0007762 | chr6:147581750-147599340+ | STXBP5 | NM_001127715 |
| 5 | hsa_circ_0008160 | chr21:38439561-38441924- | PIGP | NR_028352 |
| 6 | hsa_circ_0036044 | chr15:68434283-68457142+ | PIAS1 | NM_016166 |
| 7 | hsa_circ_0038929 | chr16:29810948-29811369+ | KIF22 | NM_007317 |
| 8 | hsa_circ_0043302 | chr17:36353600-36353765- | LOC440434 | NR_036750 |
| 9 | hsa_circ_0051637 | chr19:47285639-47285806- | SLC1A5 | NM_005628 |
| 10 | hsa_circ_0056548 | chr2:135878388-135881816+ | RAB3GAP1 | NM_001172435 |
| 11 | hsa_circ_0078279 | chr6:151226785-151293194+ | MTHFD1L | NM_001242767 |
GO and pathway analysis of the host genes of DECs
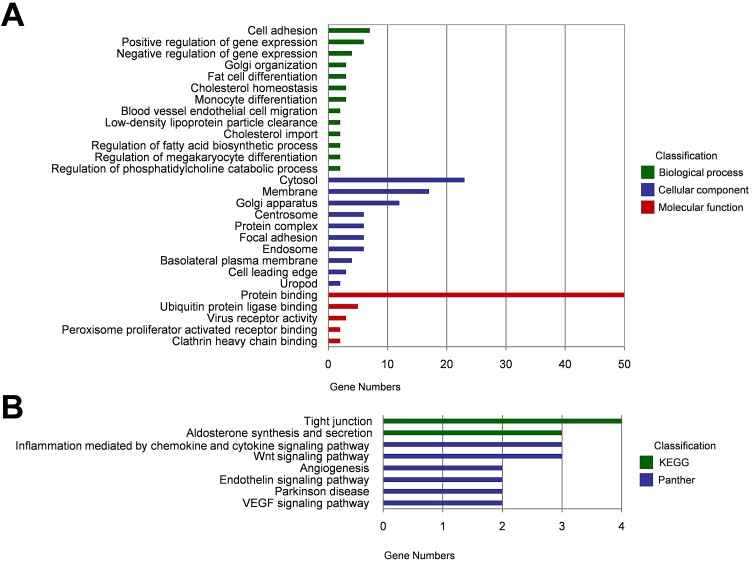
Since some circRNAs function by regulating their parental genes, GO analysis was carried out to annotate the host genes of the 87 DECs. Biological process analysis showed that these host genes were enriched in cell adhesion, positive/ negative regulation of gene expression, and Golgi organization. Cellular component analysis indicated that these host genes were remarkably enriched in cytosol, membrane and Golgi apparatus. For molecular function, the most significantly enriched GO term was protein binding (Figure 2A). KEGG and Panther pathway analysis demonstrated that tight junction, aldosterone synthesis and secretion, inflammation mediated by chemokine and cytokine signaling pathway, and Wnt signaling pathway may involved in circRNA-mediated regulatory network in the pathogenesis of HCC (Figure 2B).
Figure 2.
Functional annotation of the host genes of DECs. (A) Gene ontolog (GO) enrichment analyses of the host genes of DECs, including biological process, cellular component, and molecular function. (B) KEGG and Panther pathway enrichment analyses of the host genes of DECs.
Prediction of circRNA-miRNA interactions
Increasing evidences demonstrate that circRNAs may competitively bind to miRNAs and increase the expression of the target genes of these miRNAs. For the 87 DECs, prediction by online tools miRDB and circinteractome revealed 343 and 1085 circRNA-miRNA interactions, respectively. Furthermore, 20 consensus miRNAs from both prediction tools were identified, and DECs potentially bind to these miRNAs were presented in Table 3. Results showed that one specific circRNA may bind to more miRNAs, while different circRNAs could interact with one specific miRNA, indicating that circRNAs may impact on HCC progression by modulating various miRNAs.
Table 3.
Consensus microRNAs (miRNAs) and DECs potentially interact with them.
| MiRNAs | DECs predicted to interact with |
|---|---|
| hsa-miR-1248 | hsa_circ_0032704, hsa_circ_0000673, hsa_circ_0039053, hsa_circ_0016404, hsa_circ_0028196, hsa_circ_0003958, hsa_circ_0004976, hsa_circ_0008583, hsa_circ_0003528, hsa_circ_0001806, hsa_circ_0078279, hsa_circ_0005428 |
| hsa-miR-1236-3p | hsa_circ_0032704, hsa_circ_0016404, hsa_circ_0027478, hsa_circ_0065214, hsa_circ_0001806, hsa_circ_0065214, hsa_circ_0002191, hsa_circ_0014879, hsa_circ_0007762, hsa_circ_0003859 |
| hsa-miR-330-5p | hsa_circ_0001338, hsa_circ_0000291, hsa_circ_0005397, hsa_circ_0003892, hsa_circ_0007196, hsa_circ_0082182, hsa_circ_0002702, hsa_circ_0043302, hsa_circ_0026143, hsa_circ_0001955 |
| hsa-miR-615-5p | hsa_circ_0008563, hsa_circ_0045006, hsa_circ_0092283, hsa_circ_0009910, hsa_circ_0002702, hsa_circ_0001228, hsa_circ_0006789, hsa_circ_0000511, hsa_circ_0003859 |
| hsa-miR-1299 | hsa_circ_0039053, hsa_circ_0002702, hsa_circ_0003645, hsa_circ_0002191, hsa_circ_0056548, hsa_circ_0007762, hsa_circ_0001806, hsa_circ_0001955, hsa_circ_0083766 |
| hsa-miR-486-3p | hsa_circ_0002702, hsa_circ_0003859, hsa_circ_0001806, hsa_circ_0000554, hsa_circ_0000291, hsa_circ_0001806, hsa_circ_0033408, hsa_circ_0092310 |
| hsa-miR-370-3p | hsa_circ_0004976, hsa_circ_0003528, hsa_circ_0074903, hsa_circ_0003945, hsa_circ_0001338, hsa_circ_0026143, hsa_circ_0062682 |
| hsa-miR-589-5p | hsa_circ_0005428, hsa_circ_0000291, hsa_circ_0003892, hsa_circ_0008583, hsa_circ_0002191, hsa_circ_0078279 |
| hsa-miR-145-5p | hsa_circ_0001955, hsa_circ_0028196, hsa_circ_0083766, hsa_circ_0001489, hsa_circ_0027478, hsa_circ_0005428 |
| hsa-miR-1286 | hsa_circ_0000747, hsa_circ_0026143, hsa_circ_0038718, hsa_circ_0051732, hsa_circ_0003958, hsa_circ_0002702 |
| hsa-miR-7-5p | hsa_circ_0028196, hsa_circ_0065214, hsa_circ_0005785, hsa_circ_0003892, hsa_circ_0001489, hsa_circ_0092327 |
| hsa-miR-637 | hsa_circ_0016404, hsa_circ_0039053, hsa_circ_0026143, hsa_circ_0004913, hsa_circ_0003892, hsa_circ_0005397 |
| hsa-miR-377-3p | hsa_circ_0036044, hsa_circ_0003859, hsa_circ_0072088, hsa_circ_0008274, hsa_circ_0003645 |
| hsa-miR-203a-3p | hsa_circ_0039053, hsa_circ_0013048, hsa_circ_0032704, hsa_circ_0078279 |
| hsa-miR-1296-5p | hsa_circ_0001955, hsa_circ_0001489, hsa_circ_0001279, hsa_circ_0082564 |
| hsa-miR-192-5p | hsa_circ_0003528, hsa_circ_0078738, hsa_circ_0007196 |
| hsa-miR-1256 | hsa_circ_0033408, hsa_circ_0002191, hsa_circ_0001955 |
| hsa-miR-1289 | hsa_circ_0049997, hsa_circ_0004976, hsa_circ_0008160 |
| hsa-miR-215-5p | hsa_circ_0003528, hsa_circ_0078738 |
| hsa-miR-1261 | hsa_circ_0072088, hsa_circ_0009910 |
Target genes of some essential miRNAs and their function analysis
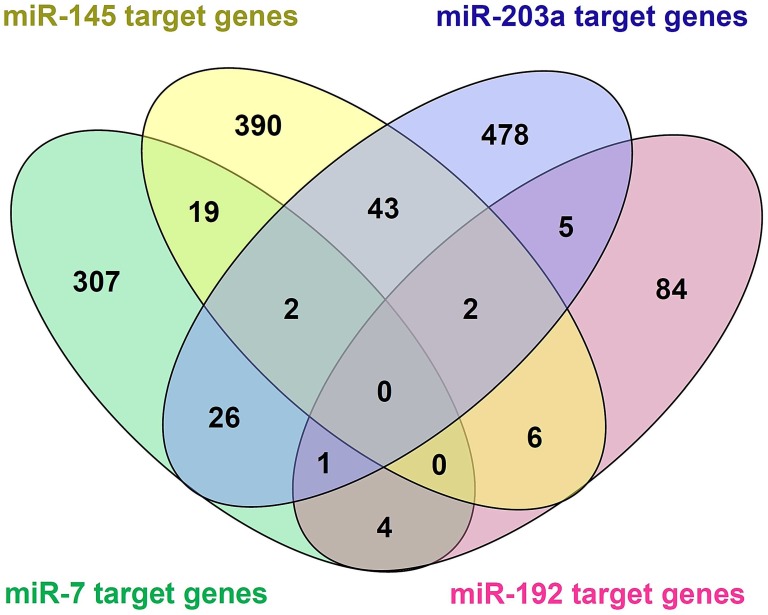
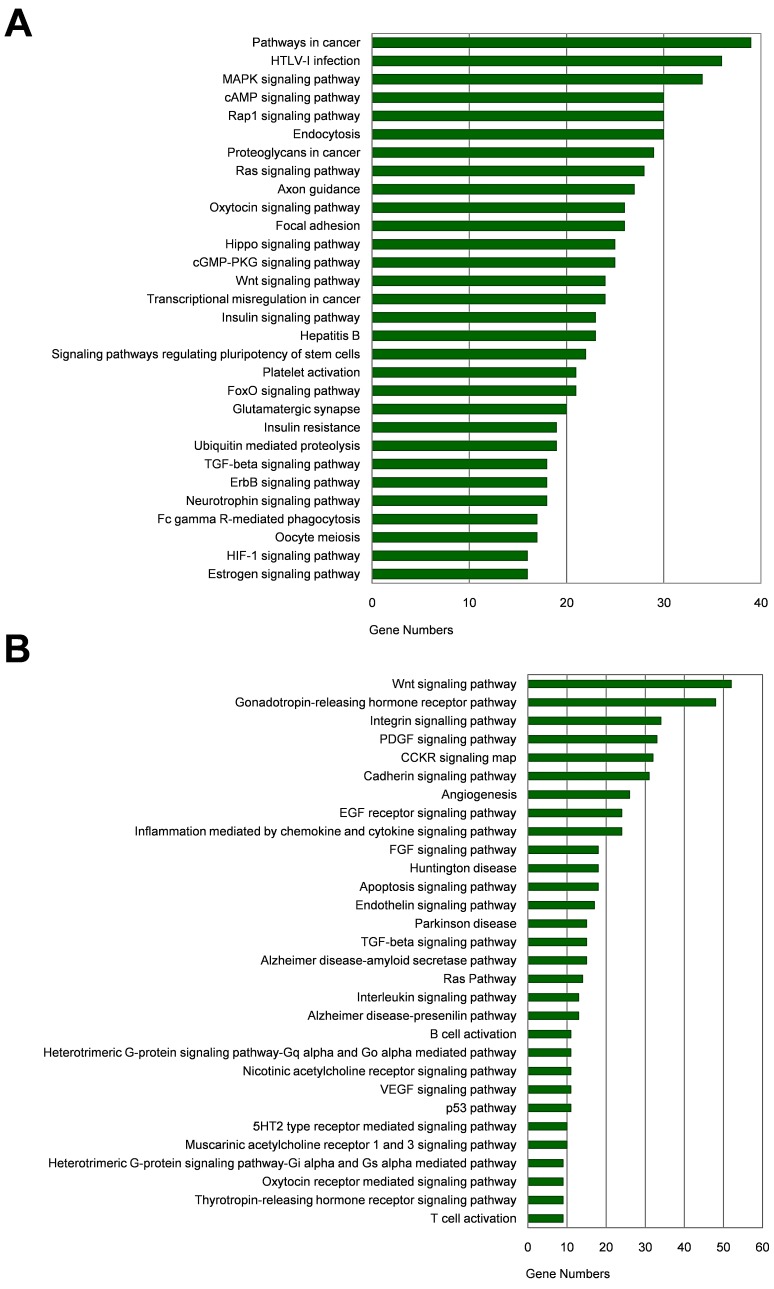
Among the 20 consensus miRNAs, four essential miRNAs, including miR-7, hsa-miR-145-5p (miR-145), hsa-miR-203a-3p (miR-203a), and hsa-miR-192-5p (miR-192), were reported to be down-regulated in HCC, and play pivotal roles in HCC progression by targeting multiple protein-coding genes. Using online tools miRDB and TargetScan, 1480 target genes of these four miRNAs were predicted, and then presented in Venn diagram (Figure 3). Two common target genes (SLC4A4 and ZDHHC9) were found for miR-7, miR-145 and miR-203a; two target genes (CPEB4, DYRK1A) were found for miR-145, miR-203a, and miR-192; and one target gene (MECP2) for miR-7, miR-203a and miR-192; while none of these genes was predicted to be targeted by all the four miRNAs. KEGG and Panther pathway analyses revealed that these target genes were involved in many HCC-associated signaling pathways, such as Wnt, TGF-β, and Ras, suggesting their possible roles in HCC pathogenesis (Figure 4).
Figure 3.
Venn diagram of target genes of the four essential microRNAs (miRNAs), including hsa-miR-7-5p (miR-7), hsa-miR -145-5p (miR-145), hsa-miR-203a-3p (miR-203), and hsa-miR-192-5p (miR-192). 1480 target genes of the four miRNAs were predicted using online tools miRDB and TargetScan.
Figure 4.
Pathway analysis of target genes of the four essential miRNAs. (A-B) KEGG pathway (A) and Panther pathway (B) functional analyses of these target genes.
Construction of essential miRNAs-centered regulatory network
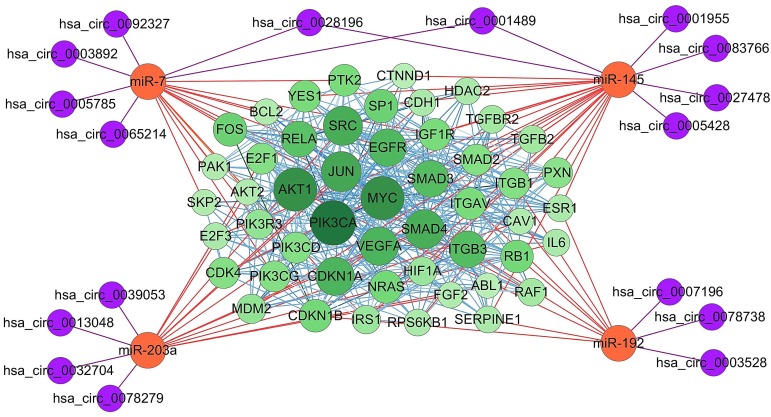
To analysis the significance of target genes of the four essential miRNAs, their experimentally strongly supported targets genes were obtained from mirTarBase. Among these, some genes, such as ABCC1 and CUL5, have been demonstrated to be targeted by more than one essential miRNAs, suggesting a complex regulatory network involving the four essential miRNAs and their target genes. Furthermore, protein-protein interaction (PPI) was analyzed using online tool STRING, and the PPI network of the top 50 genes which have more counts of interacting protein was visualized using Cytoscape software (Figure 5). Subsequently, four essential miRNAs and their potential interacting circRNAs were combined into the regulatory network. In this figure, proteins with bigger circles showed more interactions with other proteins than those with smaller circles.
Figure 5.
Essential miRNAs-centered circRNA-miRNA-mRNA network. The experimentally supported target genes of the four essential miRNAs were obtained using online tool mirTarBase. MiRNA-mRNA interaction was represented as orange edges, protein-protein interaction was represented as blue edges, and circRNA-miRNA interaction was represented as purple edges. PIK3CA, AKT1, MYC, JUN, SMAD4, and SRC were shown as hub target genes as they have more counts of interacting protein.
Validation of DECs
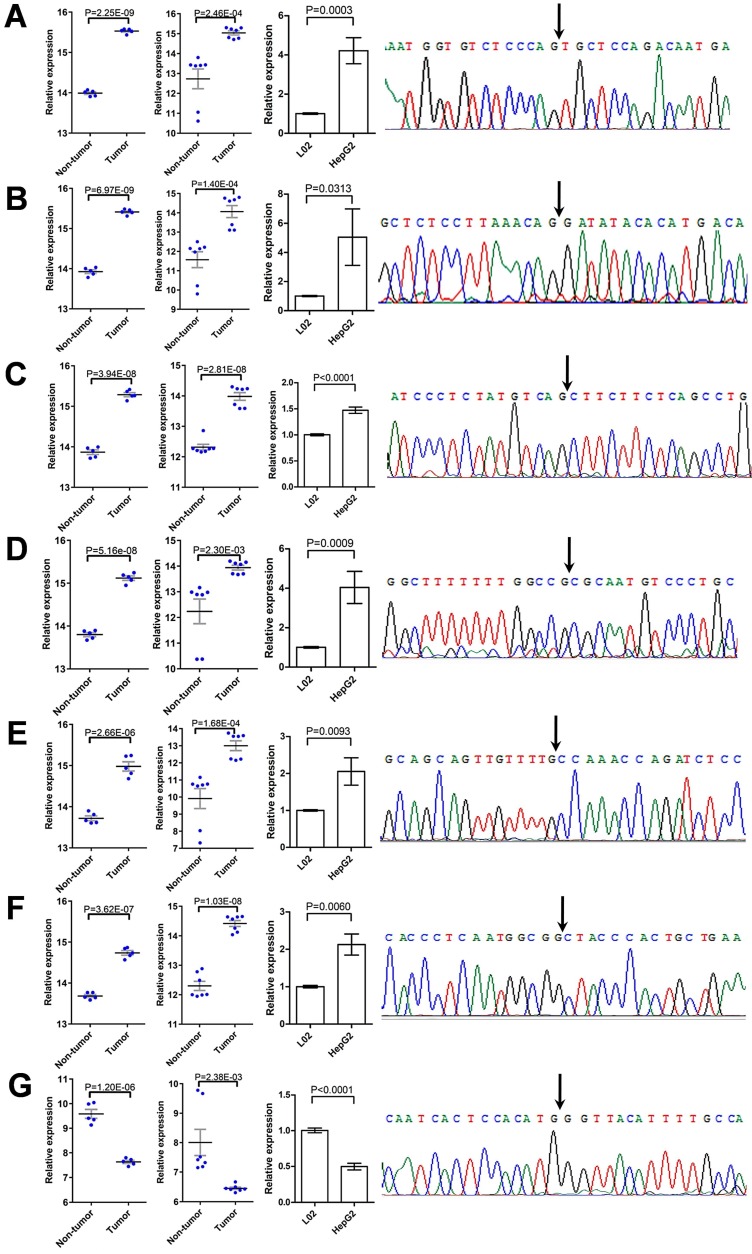
To verify the deregulation of DECs, nine of them were randomly chosen for further validation. The expression of these DECs in L02 and HepG2 cells were detected using qRT-PCR, and the specificity of qPCR products was evaluated by dissociation curve analysis and the Sanger sequencing. Among them, the expression of two circRNAs, including hsa_circ_0001489 and hsa_circ_0039053, was low in both cell lines, and was failed to be detected. Six circRNAs were validated to be greatly up-regulated in HepG2 cells compared with L02 cells, including hsa_circ_0001806, hsa_circ_0003528, hsa_circ_0008583, hsa_circ_0009910, hsa_circ_0032704, and hsa_circ_0065214 (Figure 6A-6F); while hsa_circ_0007762 was found to be greatly down-regulated in HepG2 cells compared with L02 cells (Figure 6G). The expression pattern of these circRNAs in HCC cell lines was similar to that in HCC specimens from the two GEO HCC datasets.
Figure 6.
Validation of DECs. (A-G) The deregulation of seven DECs, including hsa_circ_0001806 (A), hsa_circ_0003528 (B), hsa_circ_0008583 (C), hsa_circ_0009910 (D), hsa_circ_0032704 (E), hsa_circ_0065214 (F), and hsa_circ_0007762 (G), was validated by qRT-PCR and the Sanger sequencing. For each DEC, from left to right: comparison of relative expression of the DEC between HCC tissues and paired non-tumor tissues, as extracted from GSE94508 (five pairs) and GSE97332 (seven pairs), respectively; comparison of relative expression of the DEC between HCC cell line HepG2 and normal liver cell line L02, as determined by qRT-PCR; and the specificity of the divergent PCR products, as verified by the Sanger sequencing of the DEC in back-splice junction. Data were shown as mean ± SEM, and student's t-test was used to compare the differences of circRNA expression between two groups.
Discussion
Emerging evidences indicate that circRNAs are frequently deregulated in HCC tissues. Some deregulated circRNAs may serve as biomarkers for HCC diagnosis and prognosis, while others could regulate HCC progression through diverse mechanisms by functioning as miRNA sponges, RNA-binding protein sponges, or transcriptional regulators 4. In the current study, we identified 87 DECs from the HCC circRNA expression profiles of GSE94508 and GSE97332 downloaded from GEO database, and analyzed the functions and possible underlying mechanisms of these DECs in HCC tumorigenesis and progression using bioinformatic methods. In the meantime, the deregulation of some DECs was validated in HCC cells by qRT-PCR and the Sanger sequencing.
Among the 87 DECs identified in our study, four up-regulated circRNAs, including hsa_circ_0000673, hsa_circRNA_0072088, hsa_circ_0009910, and hsa_circ_0067934, have been reported to play important roles in the progression of different cancers, such as HCC, gastric cancer (GC), colorectal cancer (CRC), and esophageal squamous cell carcinoma (ESCC) 35-40. Two circRNAs, hsa_circ_0000673 and hsa_circRNA_0072088, were up-regulated in HCC and lung cancer, separately, and play tumor-promoting roles by modulating miR-767-3p/SET axis and miR-4302/znf121/MYC axis, respectively 35, 37. However, they were down-regulated, and may act as tumor suppressors in other cancers. Hsa_circ_0000673 was reported to inhibit tumor suppressor gene RUNX3 by targeting miR-532-5p, leading to inhibition of GC progression; while hsa_circRNA_0072088 was found to promote FOXO4 by targeting miR-532-3p, resulting in suppression of CRC development 36, 38. Other two circRNAs, hsa_circ_0009910 and hsa_circ_0067934, were demonstrated to be up-regulated in osteosarcoma and ESCC, separately, and play oncogenic roles in these cancers. Hsa_circ_0009910 was capable of sponging miR-499a and enhancing the expression of miR-499 target gene IL6R, and thus accelerating carcinogenesis of osteosarcoma; while hsa_circ_0067934 could promote the proliferation and migration of ESCC cells, and its expression level in ESCC was associated with tumor differentiation and TNM stages 39, 40. Interestingly, our study demonstrated the up-regulation of hsa_circ_009910 in HCC, indicating its possible roles in HCC progression. The precise function and mechanisms of this circRNA in HCC is currently under investigation.
Recent research works suggest that some circRNAs may regulate the transcription of their parental genes 41. In the current study, the possible GO functional terms and signaling pathways of the host genes of these DECs were characterized. Results showed that these host genes were enriched in cell adhesion, cytosol, and protein binding, all of which were associated with HCC progression. Pathway analysis from KEGG and Panther demonstrated that tight junction and Wnt signaling pathway are among the most relevant pathways for HCC. Tight junction signaling contributes to pathogenesis of GC and CRC, plays functional roles in epithelial-to-mesenchymal transition and viral entry, and could be used as potential targets for gastrointestinal and liver disease; while Wnt signaling pathway is well known to impact on hepatocarcinogenesis and HCC progression 42-45. These results suggested that these DECs may participate in HCC progression by regulating their parental genes.
Many miRNAs have been demonstrated to regulate HCC progression by modulating the expression of one or more HCC-associated genes 46, 47. However, the functions of miRNAs can be interfered by their possible interaction with circRNAs and lncRNAs. One of the most interesting examples is the interaction between ciRS-7 and miR-7. CiRS-7 is a special and unique natural circRNA which harbors more than 70 binding sites for miR-7. By binding to miR-7, ciRS-7 facilitates the expression of miR-7 target genes, such as CCNE1, PIK3CD, and EGFR, and thus blocks the inhibitory effect of miR-7 on HCC 17-19. Similarly, circMTO1 and circHIPK3 were found to modulate HCC progression by competitively binding to miR-9 and miR-124-3p, respectively, and thus increasing the expression of multiple oncogenic genes, such as p21, Norch1, AQP3 14, 48. In the current study, the interactions between DECs and miRNAs were predicted. Among all the DECs, hsa_circ_0005428 contains the most number of interactions and could potentially bind to 16 different miRNAs, whereas hsa_circ_0001834 and hsa_circ_000511 have only one potential miRNA binding site. Among the top 20 miRNAs which predispose to interact with DECs, each one contains at least two potential DECs binding sites. Specially, hsa-miR-1248 has as many as 12 DECs binding sites. Given the importance of miRNAs in HCC progression, it is possible that DECs could impact on HCC progression by targeting specific miRNAs to increasing the expression of their target genes.
Among the top 20 miRNAs, four essential miRNAs, including miR-7, miR-145, miR-203, and miR-192, were reported to be down-regulated in HCC, and could inhibit HCC progression by modulating the expression of multiple target genes; while their modulation on target genes may be blocked by their interaction with non-coding RNAs. For example, interaction between miR-7 and ciRS-7 or lncRNA KCNQ1OT1 led to reduced expression of miR-7 and increased expression of miR-7 target genes CCNE1, PIK3CD, EGFR, and ABCC1, resulting in promotion of HCC progression 18, 19, 49; whereas interaction between miR-145 and lincRNA-ROR or pseudogene OCT4-pg4 led to reduction of this miRNA and increase of miR-145 target genes RAD18, ZEB2, and OCT4, resulting in aggravation of HCC 50-52. MiR-203 could interact with three lncRNAs, including CRNDE, UCA1 and HULC, and the interaction of which greatly elevated the expression of miR-203 target genes BCAT1, SNAI2, and ADAM9, and thus facilitating HCC progression 53-55; while miR-192, however, could be blocked by lncRNA HOTTIP, and reduced expression of this miRNA led to exacerbated expression of its target gene GLS1, resulting in enhanced progression of HCC 56. Regarding to their pivotal roles in HCC progression, these four essential miRNAs were selected for further analysis. With their predicted and validated target genes, pathway and protein-protein interaction network analyses were carried out. KEGG and Panther pathway analyses suggested that the predicted target genes of these miRNAs were involved in various pathways, such as Wnt, TGF-β, oxytoxin, and Ras, all of which are closely related to hepatogenesis and HCC progression. A four essential miRNAs-centered circRNA-miRNA-mRNA network was established by combination of the potential interacting DECs of the four miRNAs with the PPI network based on the top 50 experimentally supported target genes of these miRNAs. Results suggested that some target genes of these miRNAs, such as PIK3CA, AKT1, MYC, JUN, SMAD4, and SRC, may impact on cell function by interacting with different molecules in cells and interfering with various signaling pathways, while specific circRNAs may control the expression of these genes by indirectly targeting essential miRNAs. As expected, we found that the expression of two circRNAs, including hsa_circ_0032704 and hsa_circ_0065214, was greatly increased in HCC tissues and cell lines, suggesting that these two circRNAs may function in HCC progression by targeting HCC suppressor genes miR-203a and miR-7, respectively, leading to enhanced expression of oncogenic proteins and promotion of HCC progression. Currently, their functions and molecular mechanisms, including possible interactions with miRNAs, are under investigation.
In conclusion, this study revealed aberrantly expressed circRNAs in HCC, and discussed their possible roles in HCC progression. Further investigations are needed to fully elucidate the functions and underlying mechanisms of these deregulated circRNAs and their regulatory networks in tumorigenesis and HCC progression.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81402840, 31402016, and 31572467); the National Key R&D Program of China (2018YFA0108400), the Natural Science Foundation of Jiangsu Province, China (BK20130495); the Training Project of Young Backbone Teachers of Jiangsu University, and Jiangsu University Senior Professional Science Foundation (11JDG120).
Abbreviations
- HCC
Hepatocellular carcinoma
- circRNAs
circular RNAs
- ncRNAs
non-coding RNAs
- GEO
the Gene Expression Omnibus database
- DECs
differentially expressed circRNAs
- miRNA
microRNA
- MREs
miRNA response elements
- GO
Gene ontolog
- PPI
protein-protein network
- qRT-PCR
quantitative real-time polymerase chain reaction
- miR-7
hsa-miR-7-5p
- DAVID
the Database for Annotation, Visualization, and Integrated Discovery
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- STRING
the Search Tool for the Retrieval of Interacting Genes database
- miR-145
hsa-miR-145-5p
- miR-203a
hsa-miR-203a-3p
- miR-192
hsa-miR-192-5p
- GC
gastric cancer
- CRC
colorectal cancer
- ESCC
esophageal squamous cell carcinoma
References
- 1.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis. from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Liu H, Hou L. et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu W, Bi ZY, Chen ZL. et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:18–27. doi: 10.1016/j.canlet.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Qiu LP, Wu YH, Yu XF. et al. The Emerging Role of Circular RNAs in Hepatocellular Carcinoma. J Cancer. 2018;9:1548–1559. doi: 10.7150/jca.24566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeck WR, Sorrentino JA, Wang K, Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA; 2013. p. 19. 141-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–9342. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu S, Yang X, Li X. et al. Circular RNA. A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Salzman J, Chen RE, Olsen MN. et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 10.Yu B, Shan G. Functions of long noncoding RNAs in the nucleus. Nucleus. 2016;7:155–166. doi: 10.1080/19491034.2016.1179408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao Z, Luo J, Hu K. et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu L, Yao T, Chen Q. et al. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8:58405–58416. doi: 10.18632/oncotarget.16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang X, Li G, Liu H. et al. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine (Baltimore) 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han D, Li J, Wang H. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 15.Guo W, Zhang J, Zhang D. et al. Polymorphisms and expression pattern of circular RNA circ-ITCH contributes to the carcinogenesis of hepatocellular carcinoma. Oncotarget. 2017;8:48169–48177. doi: 10.18632/oncotarget.18327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang XY, Huang ZL, Xu YH. et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Zhang M, Zheng X. et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu L, Gong X, Sun L. et al. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS One. 2016;11:e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Yang X, Xiong Q, Wu Y. et al. Quantitative Proteomics Reveals the Regulatory Networks of Circular RNA CDR1as in Hepatocellular Carcinoma Cells. J Proteome Res. 2017;16:3891–3902. doi: 10.1021/acs.jproteome.7b00519. [DOI] [PubMed] [Google Scholar]
- 20.Barrett T, Suzek TO, Troup DB. et al. NCBI GEO. mining millions of expression profiles-database and tools. Nucleic Acids Res. 2005;33:D562–566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T, Troup DB, Wilhite SE. et al. NCBI GEO. mining tens of millions of expression profiles-database and tools update. Nucleic Acids Res. 2007;35:D760–765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools. paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gene Ontology Consortium. GO: The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi H, Dong Q, Muruganujan A. et al. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics. 2016;32:1316–1322. doi: 10.1093/bioinformatics/btw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudekula DB, Panda AC, Grammatikakis I. et al. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biology. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis BP, Shih IH, Jones-Rhoades MW. et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 31.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Chou CH, Shrestha S, Yang CD. et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen L, Kuhn M, Stark M. et al. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W, Wen D, Gong L. et al. Circular RNA hsa_circ_0000673 promotes hepatocellular carcinoma malignance by decreasing miR-767-3p targeting SET. Biochem Biophys Res Commun. 2018;500:211–216. doi: 10.1016/j.bbrc.2018.04.041. [DOI] [PubMed] [Google Scholar]
- 36.Chang P, Wang F, Li Y. Hsa_circ_0000673 is down-regulated in gastric cancer and inhibits the proliferation and invasion of tumor cells by targetting miR-532-5p. Biosci Rep. 2018;38:BSR20180538. doi: 10.1042/BSR20180538. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Liu W, Ma W, Yuan Y. et al. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500:846–851. doi: 10.1016/j.bbrc.2018.04.172. [DOI] [PubMed] [Google Scholar]
- 38.Bian L, Zhi X, Ma L. et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p / FOXO4 axis. Biochem Biophys Res Commun. 2018;505:346–352. doi: 10.1016/j.bbrc.2018.09.073. [DOI] [PubMed] [Google Scholar]
- 39.Deng N, Li L, Gao J. et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. 2018;495:189–196. doi: 10.1016/j.bbrc.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 40.Xia W, Qiu M, Chen R. et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 42.Zeisel MB, Dhawan P, Baumert TF. Tight junction proteins in gastrointestinal and liver disease. Gut. 2018 doi: 10.1136/gutjnl-2018-316906. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekhar V, Pollicino T, Diaz G. et al. Infection with hepatitis C virus depends on TACSTD2, a regulator of claudin-1 and occludin highly downregulated in hepatocellular carcinoma. PLoS Pathog. 2018;14:e1006916. doi: 10.1371/journal.ppat.1006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu LJ, Xie SX, Chen YT. et al. Aberrant regulation of Wnt signaling in hepatocellular carcinoma. World J Gastroenterol. 2016;22:7486–7499. doi: 10.3748/wjg.v22.i33.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blagotinsek K, Rozman D. Targeting Signalling Pathways in Hepatocellular Carcinoma. Curr Pharm Des. 2017;23:170–175. doi: 10.2174/1381612822666161006160005. [DOI] [PubMed] [Google Scholar]
- 46.Xu X, Tao Y, Shan L. et al. The Role of MicroRNAs in Hepatocellular Carcinoma. J Cancer. 2018;9:3557–3569. doi: 10.7150/jca.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie KL, Zhang YG, Liu J. et al. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics. 2014;4:1176–1192. doi: 10.7150/thno.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen G, Shi Y, Liu M. et al. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu H, Yang L, Li L. et al. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ ABCC1 axis. Biochem Biophys Res Commun. 2018;503:2400–2406. doi: 10.1016/j.bbrc.2018.06.168. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Shen Z, Zhi Y. et al. Long non-coding RNA ROR promotes radioresistance in hepatocelluar carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch Biochem Biophys. 2018;645:117–125. doi: 10.1016/j.abb.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 51.Li C, Lu L, Feng B. et al. The lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2. Sci Rep. 2017;7:4637. doi: 10.1038/s41598-017-04113-w. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Wang L, Guo ZY, Zhang R. et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis. 2013;34:1773–1781. doi: 10.1093/carcin/bgt139. [DOI] [PubMed] [Google Scholar]
- 53.Ji D, Jiang C, Zhang L. et al. LncRNA CRNDE promotes hepatocellular carcinoma cell proliferation, invasion, and migration through regulating miR-203/ BCAT1 axis. J Cell Physiol. 2019;234:6548–6560. doi: 10.1002/jcp.27396. [DOI] [PubMed] [Google Scholar]
- 54.Xiao JN, Yan TH, Yu RM. et al. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143:981–990. doi: 10.1007/s00432-017-2370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan D, Shen S, Fu S. et al. miR-203 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting oncogene ADAM9 and oncogenic long non-coding RNA HULC. Anticancer Agents Med Chem. 2016;16:414–423. doi: 10.2174/1871520615666150716105955. [DOI] [PubMed] [Google Scholar]
- 56.Ge Y, Yan X, Jin Y. et al. fMiRNA-192 and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]