Abstract
Aim: Sunitinib remains the frontline treatment for metastatic clear-cell renal cell carcinoma (ccRCC). Drug resistance is inevitable and related mechanism warrant insightful elaboration.
Methods: In silico data mining of GEO and TCGA datasets was performed to identify potential target micro-RNA. In vitro and in vivo studies were performed to validate findings.
Results: Reproduction of GEO datasets revealed miR-15b significantly upregulated in sunitinib- resistant ccRCC. Five out of seven ccRCC cell lines demonstrated significantly overexpressed miR-15b after sunitinib treatment. Vector-mediated overexpression of miR-15b significantly induced resistance to sunitinib in ccRCC cells. Overexpression of miR-15b significantly induced less population in G1 phase of cell cycle and less apoptosis in cells treated sunitinib. Expression of genes negatively correlated with miR-15b in TCGA ccRCC (KIRC) dataset were cross-referenced with predicted targets of miR-15b and CCNC was selected as potential target for resistance mediation. Overexpression of miR-15b suppressed CCNC expression and protein (Cyclin C) levels. Cyclin C-associated proteins CDK19 and CDK8 were also suppressed following miR-15b overexpression. Silencing of CCNC mimicked overexpression of miR-25 inducing cell cycle progression passing G1 phase and less apoptosis in ccRCC cells treated by sunitinib. Overexpression of miR-15b also counteracted suppression of migration and colony formation by sunitinib in ccRCC cell lines. In vivo mouse xenograft models showed recovered tumor growth with miR-15b expression in mice treated with sunitinib.
Conclusion: We here show miR-15b as a possible culprit for sunitinib resistance in ccRCC. Targeting miR-15b could potentially overcome drug resistance and related mechanism warrants further investigation.
Keywords: clear-cell renal cell carcinoma, miR-15b, sunitinib, resistance
Introduction
Renal cell carcinoma is one of the most common urological malignancies worldwide 1. The clear cell renal cell carcinoma (ccRCC) is the most common subtype of it and therefore studied and known most clearly in biologically molecular mechanisms such as gene mutation, epigenetic modification and the influence of non-coding RNA 2. The VHL gene is regarded as one of the most important tumor suppressor genes in ccRCC 3. The loss of protein encoded by this gene can induce the overexpression of HIF1, promoting the proportion of VEGF, EGFR and TGF-а, all of which can stimulate the cell proliferation and angiogenesis 4. Anti-angiogenic tyrosine kinase inhibitors (TKIs) and antibodies against VEGF and its receptors together with associated molecules stand frontline treatment for metastatic ccRCC, which consists of ~30% of all cases and confers a poor prognosis 5. Among all TKIs, sunitinib is recommended as first line agents in all major guidelines 6. Unfortunately, >50% of patients are innately non- responders and almost all initial responders develop resistance over a period of approximate 2 years 7. Therefore, genetic culprits responsible for resistance may not only serve as biomarker but also provide potential therapeutic target to overcome the conundrum.
MicroRNAs (miRs) are non-coding RNAs that play critical roles in a variety of biological processes. In recent years, miRs have also been shown to play important roles in cancer development mainly by silencing tumor suppressor genes 8. In ccRCC various miRs associated with different stages of cancer development including oncogenic miRs mediating cell cycle progression, apoptosis, etc. whereas tumor-suppressive miRs combating biologic processes like angiogenesis 9. In the TKI era, several miRs have been shown to mediate sunitinib sensitivity and resistance 10, 11. Nonetheless, none of the miRs can now be applicable clinically and the complete regulatory network of miRs associated with sunitinib remains largely unclear.
In the current study, we aim to find potential miRs associated with sunitinib resistance in ccRCC. With reproduction of multiple GEO datasets, we attempted to pinpoint common miRs related to sunitinib resistance and performed in vitro and in vivo validation for our findings. We aim to provide pilot evidence for marker development monitoring sunitinib sensitivity in ccRCC patients.
Methods
In silico reproduction of GEO datasets
Three datasets of interest submitted to GEO platform were selected (GSE116251, GSE37766, and GSE66346). All datasets encompassed miRNA-array in either ccRCC patients or mouse models treated with sunitinib and were intended by the submitter to investigate sunitinib-resistance miRs. Significantly overexpressed miRs in all datasets were retrieved and common intersects were analyzed for target miR.
Cell lines, miR overexpression and siRNA
RCC cell lines 786-O, 769-P, A498, ACHN, Caki-1, Caki-2 and OS-RC-2 were obtained from the National Infrastructure of Cell Line Resorce. Sunitinib treatments were added at 5 µM in all single-dose in vitro assays. Overexpression of miR-15b was realized with transfection of miR-15-5p with Lipofectamine system when cells reached 70% confluence for 6 h before medium was refreshed (pmiR-15b). Sequence for siRNA construction against CCNC was reference from TRC (TRCN0000020190) and standard transfection protocol was followed.
Realtime RT-PCR
Total RNA was extracted with Trizol. After concentration was determined, RNAs were converted to cDNA using reverse-transcription PrimeScriptTM kit. Quantitative PCR was performed on ABI StepOne Plus System. Relative expression was calculated by expression level normalized to GAPDH level with the formula, 2-△△Ct. Realtime PCR for miR-15b was performed using CPK1019-200rxns hsa-mir-15b-3p Real-time RT-PCR Detection Kit. Primers for CCNC were as follows: forward 5'-CCT TGC ATG GAG GAT AGT GAA TG-3', reverse 5'- AAG GAG GAT ACA GTA GGC AAA GA-3'.
Western blotting
Cells were lysed using RIPA buffer and total protein was extracted. After concentration was determined, protein was loaded with buffer onto SDS-PAGE gel with subsequent electrophoresis. Protein was then transferred to PVDF membrane which was blocked with 5% non-fat milk. Membranes were then incubated with primary antibodies at 4 °C, including rabbit anti-GAPDH (5174, Cell Signaling Technology, Shanghai, China), CCNC (Anti-Cyclin C antibody, ab85927, Abcam, Shanghai), PARP (Cell Signaling 9541S), CDK8 (Cell Signaling) and CDK19 (Abcam). Corresponding secondary antibody was applied for 4 h. he protein bands were detected using an enhanced chemiluminescence (ECL) kit.
Proliferation
Cell proliferation was studied using crystal violet (CV) assay. Briefly, cells seeded in 96-well plates were stained using crystal violet at set time points. Cells were than treated with methanol and rinsed for excessive CV and plates were read on a plate reader.
Cell cycle and apoptosis
The flow cytometry was used to profile cell cycle and apoptosis. The HEPES binding buffer containing Annexin V and PI was applied to the cells for 15 min at room temperature. Samples were then analyzed with BD FACSCanto flow cytometer to determine percentages of apoptotic cells (Annexin V+/PI-). Cell cycle was also studied in trypsinized cells using flow cytometry.
Migration
Transwell assay was used to study cell migration. Briefly, cells were resuspended at the density of 1×106/ml in 300μl of serum free medium and seeded in the upper chamber of Transwell without any coating. The lower chambers were then filled with 500μl of complete medium and cells migrated through the membrane were stained with CV and counted.
Colony formation
Colony formation assay was used to profile anchorage-independent growth. Cells were seeded in in medium containing 10% FBS with 0.4% agarose, which was layered on the top of 0.6% agar in medium supplemented with 20% FBS on 60-mm plates. After 2 weeks of culture at 37 °C, plates were stained with 0.005% of crystal violet for 1 h. Colonies were counted microscopically and the relative colony numbers were measured.
Xenograft mouse model
RCC xenograft mouse models using 786-O cells have been well-stablished by our group. Three groups were set and sunitinib was administrated by oral gavage in a total volume of 150 μL, whilst pmiR-15b and control plasmid was overexpression was delivered to 786-O cells before implantation. All mice were sacrificed on Day 35 and tumors were extracted. Tumor size was calculated with the formula, Length * Width2 * 0.5236.
Statistical analysis
Comparisons between groups were analysed with the 2-tailed Student's t-test. The P value of 0.05 was accepted as statistically significant.
Results
Overexpression of miR-15b is associated with sunitinib resistance
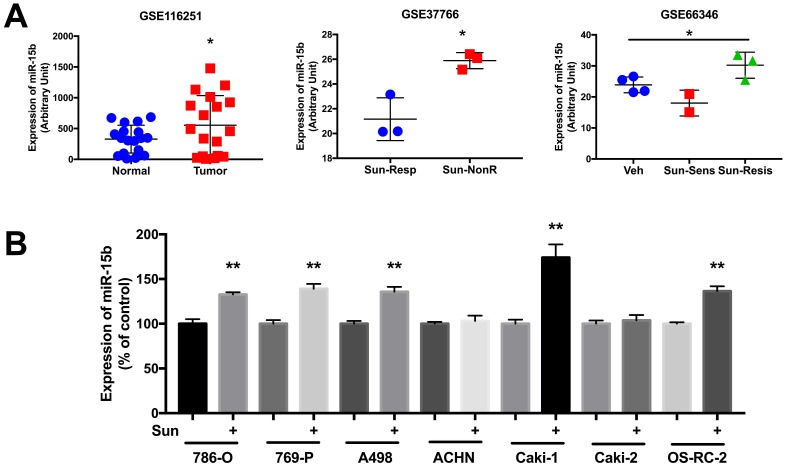
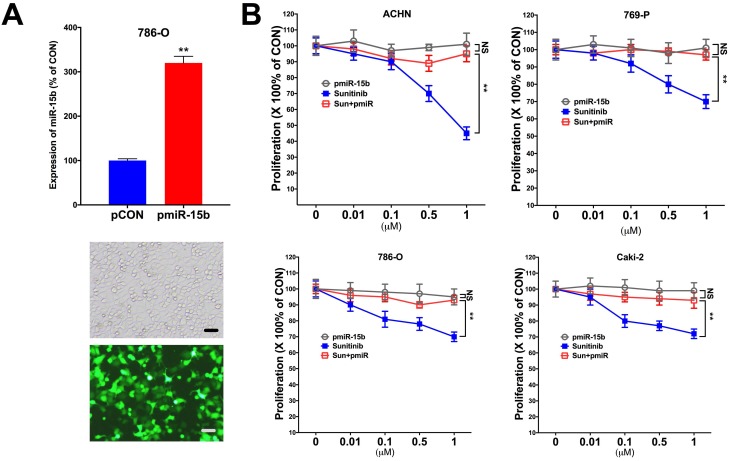
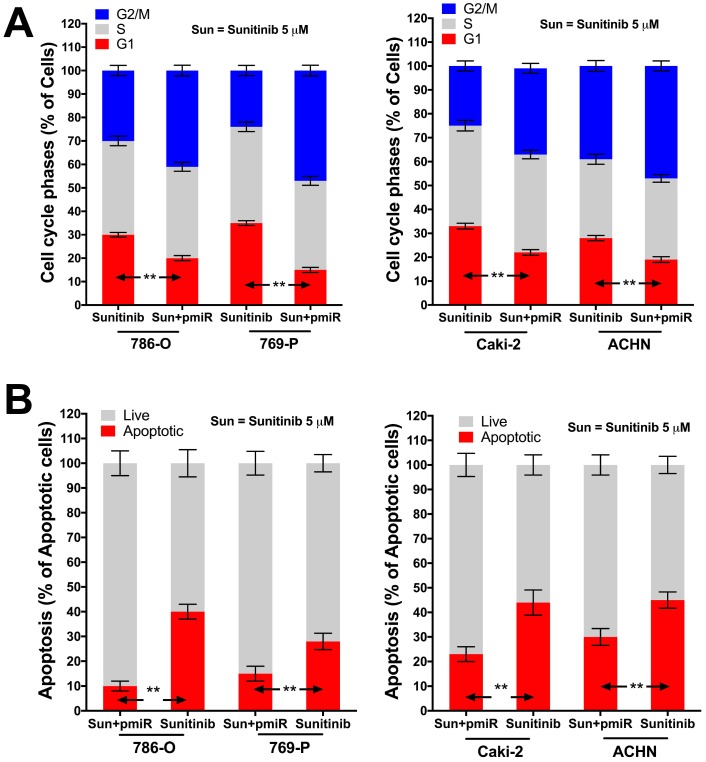
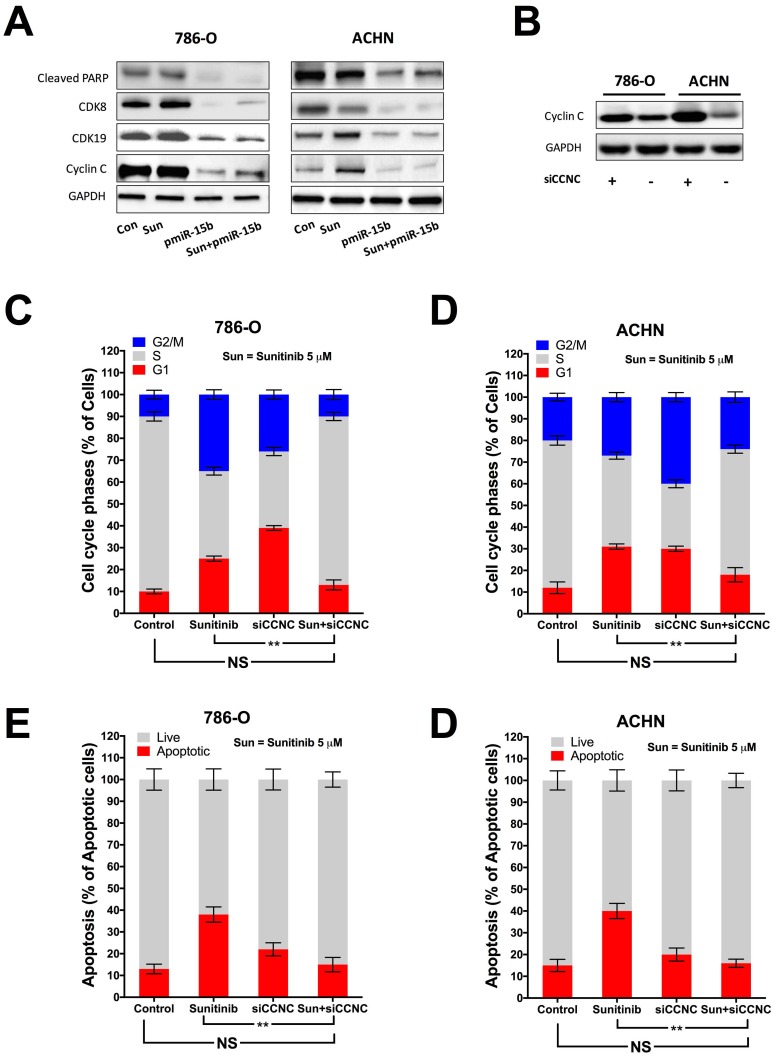
In silico reproduction of the GEO datasets (GSE116251, GSE37766, and GSE66346) revealed significantly upregulated miR-15b expression in renal cell carcinoma versus normal kidney tissue, in non-responder to sunitinib versus responder, and in sunitinib-resistant cell ccRCC cell lines versus vehicle control (Fig 1A). We then investigated expression of miR-15b in 7 RCC cell lines treated with sunitinib and found that 5 out of 7 demonstrated significant higher expression of miR-15b following treatment (Fig 1B). Here we showed that miR-15b could be a potential culprit for sunitinib resistance. We then applied plasmid- mediated overexpression of miR-15b (Fig 2A) and examined proliferation in 4 cell lines that showed overexpressed miR-15b shown in Fig 1B. We found that pmiR-15b significantly restored inhibitory effect of sunitinib in those cell lines (Fig 2B). We then applied flow cytometry and found that overexpression of miR-15b significantly reduced population in G1 phase induced by sunitinib in 4 RCC cell lines (Fig 3A). Similar effect was also noted in the 4 cell lines with regard to decreased cell apoptosis in cells with miR-15b overexpression compared with sunitinib alone (Fig 3B).
Figure 1.
miR-15b is overexpressed clear-cell renal cell carcinoma (ccRCC) and in sunitinib -resistant ccRCC. A) Reproduced from GEO dataset with accessions GSE116251, GDS5815 and GSE37766 showing miR-15b levels in normal kidney tissue, sunitinib-untreated and treated ccRCC; B) Expression of miR-15b expression in RCC cell lines treated and untreated with sunitinib.
Figure 2.
Overexpression of miR-15 mediates resistance to sunitinib. A) Overexpression of mir-15b tagged with GFP showed satisfactory efficiency; B) Detected at 72 h of treatment and transfection in 4 RCC cell lines showing proliferation at different doses of sunitinib (**P < 0.01).
Figure 3.
miR-15b restores anti-tumor effect of sunitinib (5 µM at 72 h) on cell cycle and apoptosis. A) Overexpression of miR-15b restores inhibitory effect of sunitinib on cell cycle progression at G1 phase; B) Overexpression of miR-15b restores effect of sunitinib on inducing apoptosis (**P < 0.01).
miR-15b targets CCNC to mediate sunitinib resistance
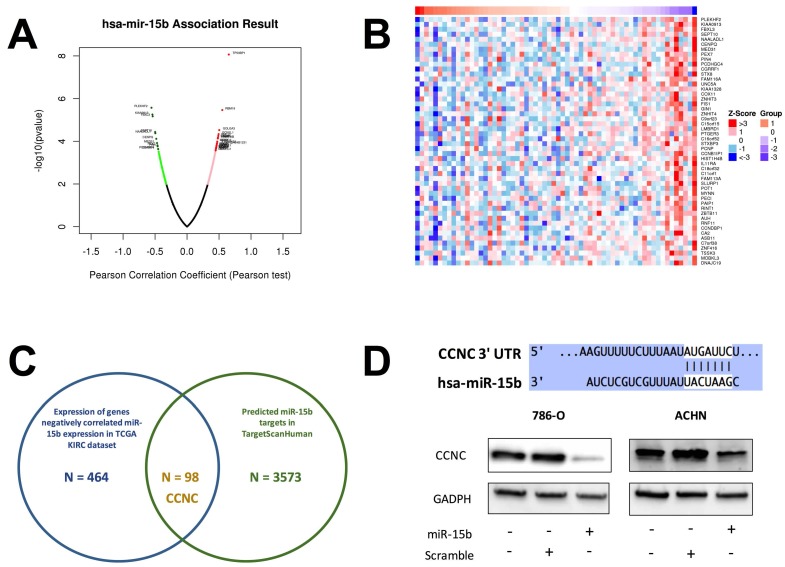
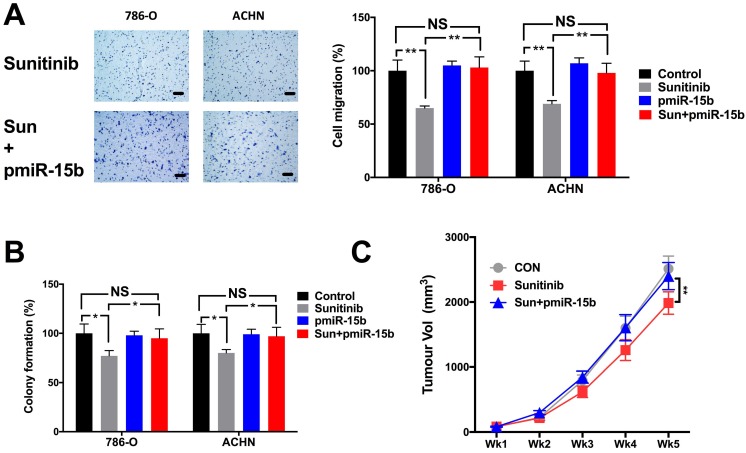
Expression of miR-15b in KIRC dataset was first correlated with expressions of genes detected by RNA seq (Fig 4A). Genes expressed with negative correlations were ranked by enrichment (Fig 4B) and such gene set was cross-referenced with genes listed in predicted targets of miR-15b (Fig 4C). Supplement of miR-15b resulted in lowered CCNC level in ccRCC cell lines (Fig 4D). We then investigated Cyclin C related proteins in 2 ccRCC cell lines treated with sunitinib and found that supplement of miR-15 notably decreased levels of CDK19, CDK8 and apoptotic indicator cleaved PARP (Fig 5A). By silencing CCNC, cells displayed similar alterations in cell cycle population to miR-15b overexpression, showing less population in G1 phase following sunitinib treatment (Fig 5C-D). Sunitinib treatment with CCNC silencing also induced less apoptotic cells compared with sunitinib alone in both ccRCC cell lines (Fig 5E-F). We next studied cell motility and found that miR-15b restored cell migration of 786-O and ACHN cell lines treated with sunitinib (Fig 6A). Anchorage-independent growth was profiled using colony formation assays. We found that inhibitory effect by sunitinib was effaced by supplement of miR-15b (Fig 6B). Xenograft mouse models using 786-O cells revealed similar results regarding tumor volume at wk5. We found tumors with both sunitinib and miR-15b showed restored growth as control (Fig 6C). Interestingly, those tumors appeared to grow at a similar rate to control starting from wk1.
Figure 4.
CCNC is a target of miR-15b in ccRCC. A) Reproduction of TCGA KIRC dataset of showed expression of genes significantly correlated with miR-15 expression with red color representing negative and green color representing positive correlation; B) Heat map showing expression of genes with negative correlation with miR-15b; C) Cross-referencing gene set in B) with miR-15b targets queried from TargetScanHuman revealing CCNC as potential target; D) Cartoon showing sequences match between miR-15b and CCNC with scramble and pmiR-15b lowering CCNC (Cyclin C) level in ccRCC cells.
Figure 5.
Knockdown (KD) of CCNC mimics miR-15b resisting sunitinib in ccRCC. A) miR-15 supplement decreasing levels of elements of CCNC signaling; B) Silencing CCNC using siRNA showing satisfactory effect; Cell cycle analysis showing restored G1 population following CCNC-KD in C) 786-O and D) ACHN cells; Apoptosis analysis showing restored apoptosis following CCNC-KD in C) 786-O and D) ACHN cells (**P < 0.01).
Figure 6.
miR-15b mediates sunitinib resistance in solid tumor growth of ccRCC. Supplement of pmiR-15b significantly restored inhibitory effect of sunitinib in A) cell migration; B) colony formation; and C) xenograft tumor model (**P < 0.01).
Discussion
In the current study, we show that miR-15b is associated with sunitinib resistance in ccRCC. Although miR-15b is associated with several tumors, its role in ccRCC has not been reported.
Our findings regarding miR15-b in ccRCC are intriguing as the miR is not only overexpressed in tumor tissue compared with normal kidney, but also upregulated following sunitinib treatment. Such finding is in line with some reports that miR-15 is initially associated with tumorigenesis in other types of cancer. Weng et al. report that miR-15b-5p/PDK4 axis regulates osteosarcoma proliferation through modulation of the Warburg effect 12. Wang et al. report miR-15b inhibits the progression of glioblastoma cells through targeting insulin-like growth factor receptor 1 13. Chen et al. report that miR-15b-5p facilitates the tumorigenicity by targeting RECK and predicts tumour recurrence in prostate cancer 14. Wang et al. report that miR-15b is amongst the 8 miRs involved in Hepatitis B virus (HBV)-unrelated hepatocellular carcinoma 15. Sun et al. however report that SIRT1 suppresses colorectal cancer metastasis by transcriptional repression of miR-15b-5p and SIRT1/miR-15b-5p/ACOX1 axis in colorectal cancer metastasis and suggested a potential target for metastatic colorectal cancer therapy 16. Kao et al. report that tumor suppressor microRNAs contribute to the regulation of PD-L1 expression in malignant pleural mesothelioma 17. Luo et al. report that MicroRNA-15b-5p targets ERK1 to regulate proliferation and apoptosis in rat PC12 cells 18. Zhao et al. report that Overexpression of miR-15b-5p promotes gastric cancer metastasis by regulating PAQR3 19. Wang et al. report that miR-15b promotes proliferation and invasion of non‑small cell lung carcinoma cells by directly targeting TIMP2 20. Last but not least, Li et al. report that OIP5, a target of miR-15b-5p, regulates hepatocellular carcinoma growth and metastasis through the AKT/mTORC1 and β-catenin signaling pathways 21. miR-15 has also been associated with sensitivity to other anti-cancer agents. Duan et al. report that multidrug resistance in osteosarcoma is associated with downregulation of miR-15b, and miR-15b reconstitution can reverse chemotherapy resistance in osteosarcoma 22. Zhao et al. report that miR-15b-5p resensitizes colon cancer cells to 5-fluorouracil by promoting apoptosis via the NF-κB/XIAP axis 23. Wang et al. report that miR-15b inhibits cancer-initiating cell phenotypes and chemoresistance of cisplatin by targeting TRIM14 in oral tongue squamous cell cancer 24. Those studies, taken together with our findings support a rather complex role of miR-15b in cancer development and treatment and indicate that role of miR-15b can hardly be extrapolated from one type of cancer to another.
In our study, overexpression of miR-15b displays potent antagonizing effect against sunitinib. As the miR was selected within the intersection of 3 experiments each representing a unique disease course in ccRCC treated by sunitinib. Expression of miR-15b following sunitinib treatment was detected after a short period and we suggest multiple resistance mechanism exist at different duration of treatment. Also, the role of miR-15b in mediating sunitinib sensitivity may also alter following prolonged exposure to drug and that is in part the reason several over sunitinib-resistance associated non-coding RNAs are reported in studies using different model and treatment durations. Qu et al. report exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA by showing sunitinib- resistant RCC with locked nucleic acids targeting lncARSR or an AXL/c-MET inhibitor restored sunitinib response 25. Xiao et al. report that mir-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A by demonstrating miR-144-3p overexpression enhanced cell proliferation, clonogenicity, migration, invasion, and chemoresistance in ccRCC cells. Yamaguchi et al. report sunitinib resistant RCC cells showed miR-575, miR-642b-3p, and miR-4430 was significantly increased, while that of miR-18a-5p, miR-29b-1-5p, miR-431-3p, and miR-4521 was significantly decreased 26. Lukamowicz-Rajska et al. report that miR-99b-5p expression and response to tyrosine kinase inhibitor treatment in clear cell renal cell carcinoma patients 27. Goto et al. reported that antitumor miR-101- mediated UHRF1 pathways may be suppressed by sunitinib treatment 28. Merhautova et al. report that miR-155 and miR-484 are associated with time to progression in metastatic renal cell carcinoma treated with sunitinib 29. Therefore, distinct mechanisms exist for sunitinib resistance and the sophisticated networks reflect evolution of cancer cells under selection stress.
In all, we here show for the first time that miR-15b is associated with sunitinib resistance in RCC. The current study serve as a pilot exploration and therapeutic potential of miR-15b to overcome drug resistance is now in progress by our group.
Acknowledgments
This study was sponsored in parted by National Natural Science Foundation of China (No. 81502189 and No. 81874123).
References
- 1.Capitanio U, Montorsi F. Renal cancer. The Lancet. 2016;387:894–906. doi: 10.1016/S0140-6736(15)00046-X. [DOI] [PubMed] [Google Scholar]
- 2.Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T. et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell. 2018;173:595–610.e11. doi: 10.1016/j.cell.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell TJ, Turajlic S, Rowan A, Nicol D, Farmery JHR, O'Brien T. et al. Timing the Landmark Events in the Evolution of Clear Cell Renal Cell Cancer: TRACERx Renal. Cell. 2018;173:611–23.e17. doi: 10.1016/j.cell.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choueiri TK, Longo DL, Motzer RJ. Systemic Therapy for Metastatic Renal-Cell Carcinoma. New England Journal of Medicine. 2017;376:354–66. doi: 10.1056/NEJMra1601333. [DOI] [PubMed] [Google Scholar]
- 6.Lalani A-KA, McGregor BA, Albiges L, Choueiri TK, Motzer R, Powles T, Systemic Treatment of Metastatic Clear Cell Renal Cell Carcinoma in 2018: Current Paradigms, Use of Immunotherapy, and Future Directions. European Urology; 2018. [DOI] [PubMed] [Google Scholar]
- 7.Méjean A, Ravaud A, Thezenas S, Colas S, Beauval J-B, Bensalah K. et al. Sunitinib Alone or after Nephrectomy in Metastatic Renal-Cell Carcinoma. New England Journal of Medicine. 2018;379:417–27. doi: 10.1056/NEJMoa1803675. [DOI] [PubMed] [Google Scholar]
- 8.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature Reviews Drug Discovery. 2017;16:203–22. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 9.Joosten SC, Smits KM, Aarts MJ, Melotte V, Koch A, Tjan-Heijnen VC. et al. Epigenetics in renal cell cancer: mechanisms and clinical applications. Nature Reviews Urology. 2018;15:430–51. doi: 10.1038/s41585-018-0023-z. [DOI] [PubMed] [Google Scholar]
- 10.Xiao W, Lou N, Ruan H, Bao L, Xiong Z, Yuan C. et al. Mir-144-3p Promotes Cell Proliferation, Metastasis, Sunitinib Resistance in Clear Cell Renal Cell Carcinoma by Downregulating ARID1A. Cellular Physiology and Biochemistry. 2017;43:2420–33. doi: 10.1159/000484395. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Wang T, Chen C, Wu Z, Bai P, Li S, Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. Journal of Cellular Biochemistry; 2018. [DOI] [PubMed] [Google Scholar]
- 12.Weng Y, Shen Y, He Y, Pan X, Xu J, Jiang Y. et al. The miR-15b-5p/PDK4 axis regulates osteosarcoma proliferation through modulation of the Warburg effect. Biochemical and Biophysical Research Communications. 2018;503:2749–57. doi: 10.1016/j.bbrc.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Liu H, Tian L, Wang F, Han L, Zhang W. et al. miR-15b Inhibits the Progression of Glioblastoma Cells Through Targeting Insulin-like Growth Factor Receptor 1. Hormones and Cancer. 2016;8:49–57. doi: 10.1007/s12672-016-0276-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R, Sheng L, Zhang H-J, Ji M, Qian W-Q. miR-15b-5p facilitates the tumorigenicity by targeting RECK and predicts tumour recurrence in prostate cancer. Journal of Cellular and Molecular Medicine. 2018;22:1855–63. doi: 10.1111/jcmm.13469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, Dong F, Xu Z, Sharma S, Hu X, Chen D, MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer; 2017. p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L-N, Zhi Z, Chen L-Y, Zhou Q, Li X-M, Gan W-J. et al. SIRT1 suppresses colorectal cancer metastasis by transcriptional repression of miR-15b-5p. Cancer Letters. 2017;409:104–15. doi: 10.1016/j.canlet.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Kao SC, Cheng YY, Williams M, Kirschner MB, Madore J, Lum T. et al. Tumor Suppressor microRNAs Contribute to the Regulation of PD-L1 Expression in Malignant Pleural Mesothelioma. Journal of Thoracic Oncology. 2017;12:1421–33. doi: 10.1016/j.jtho.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 18.Luo H, Li Y, Liu B, Yang Y, Xu Z-QD. MicroRNA-15b-5p targets ERK1 to regulate proliferation and apoptosis in rat PC12 cells. Biomedicine & Pharmacotherapy. 2017;92:1023–9. doi: 10.1016/j.biopha.2017.05.140. [DOI] [PubMed] [Google Scholar]
- 19.Zhao C, Li Y, Chen G, Wang F, Shen Z, Zhou R. Overexpression of miR-15b-5p promotes gastric cancer metastasis by regulating PAQR3. Oncology Reports. 2017;38:352–8. doi: 10.3892/or.2017.5673. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Zhan Y, Jin J, Zhang C, Li W. MicroRNA-15b promotes proliferation and invasion of non-small cell lung carcinoma cells by directly targeting TIMP2. Oncology Reports. 2017;37:3305–12. doi: 10.3892/or.2017.5604. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Zhang J, Lee M-J, Yu G-R, Han X, Kim D-G. OIP5, a target of miR-15b-5p, regulates hepatocellular carcinoma growth and metastasis through the AKT/mTORC1 and β-catenin signaling pathways. Oncotarget; 2017. p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan Z, Gao Y, Shen J, Choy E, Cote G, Harmon D. et al. miR-15b modulates multidrug resistance in human osteosarcomain vitroandin vivo. Molecular Oncology. 2017;11:151–66. doi: 10.1002/1878-0261.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao C, Zhao Q, Zhang C, Wang G, Yao Y, Huang X, miR-15b-5p resensitizes colon cancer cells to 5-fluorouracil by promoting apoptosis via the NF-κB/XIAP axis. Scientific Reports; 2017. p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Guo H, Yao B, Helms J. miR-15b inhibits cancer-initiating cell phenotypes and chemoresistance of cisplatin by targeting TRIM14 in oral tongue squamous cell cancer. Oncology Reports. 2017;37:2720–6. doi: 10.3892/or.2017.5532. [DOI] [PubMed] [Google Scholar]
- 25.Qu L, Ding J, Chen C, Wu Z-J, Liu B, Gao Y. et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653–68. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi N, Osaki M, Onuma K, Identification of MicroRNAs Involved in Resistance to Sunitinib in Renal Cell Carcinoma Cells. Anticancer Research; 2017. p. 37. [DOI] [PubMed] [Google Scholar]
- 27.Lukamowicz-Rajska M, Mittmann C, Prummer M, Zhong Q, Bedke J, Hennenlotter J, MiR-99b-5p expression and response to tyrosine kinase inhibitor treatment in clear cell renal cell carcinoma patients. Oncotarget; 2016. p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto Y, Kurozumi A, Nohata N, Kojima S, Matsushita R, Yoshino H, The microRNA signature of patients with sunitinib failure: regulation of <i>UHRF1</i> pathways by <i>microRNA-101</i> in renal cell carcinoma. Oncotarget; 2016. p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merhautova J, Hezova R, Poprach A, Kovarikova A, Radova L, Svoboda M. et al. miR-155 and miR-484 Are Associated with Time to Progression in Metastatic Renal Cell Carcinoma Treated with Sunitinib. BioMed Research International. 2015;2015:1–5. doi: 10.1155/2015/941980. [DOI] [PMC free article] [PubMed] [Google Scholar]