Abstract
Purpose: Smoking is a strong relative risk factor for lung cancer. Extracellular vesicles (EVs), particularly exosomes, have been implicated in cancers. In this study, we characterized smoking induced extracellular vesicles in smokers with non-small cell lung cancer (NSCLC).
Methods: EVs were isolated from bronchoalveolar lavage (BAL) from smokers and NSCLC patients. EV microRNAs (miRNAs) were analyzed by using a TaqMan microRNA assays. Vesicle mRNAs and long non-coding RNAs (lncRNAs) were measured with quantitative RT-PCR. Tumor associated antigens were examined by Western Blot.
Results: Higher levels of local site EVs are found in the lung of smokers and NSCLC patients. Further, over 90% of lung EVs are round vesicles of approximately 50-200 nm, ie., exosomes. There are 21 EV miRNAs up regulated, while 10 miRNAs under regulated, in smokers when compared to controls (relative fold > 2, p < 0.05). These miRNAs were further observed to be dysregulated in NSCLC patients when compared to smokers. Bioinformatic analysis demonstrated that Proteoglycans, Fatty acid biosynthesis, ErbB, Hippo, TGF-beta, Wnt, Rap1, AMPK and Ras pathways were the most prominent pathways enriched in NSCLC EV miRNA signatures. In addition, messenger RNA transcripts including EGFR, KRAS, ALK, MET, LKB1, BRAF, PIK3CA, RET, and ROS1 were significantly higher expressed in lung EVs in smokers and NSCLC patients compared to controls. Long non-coding RNAs, including MALAT1, HOTAIR, HOTTIP, AGAP2-AS1, ATB, TCF7, FOXD2-AS1, HOXA11-AS, PCAF1, and BCAR4, were over expressed in EVs from smokers and NSCLC patients. Furthermore, protein levels of tumor associated antigens including BAGE, PD-L1, MAGE-3, and AKAP4 were significantly dysregulated in EVs of smokers and NSCLC patients compared to healthy controls.
Conclusions: In conclusion, these data demonstrated an intrinsic relationship of smoking dysregulated EVs and EVs contained RNA, proteins which may involve in the development of NSCLC.
Keywords: extracellular vesicles, smoker, NSCLC, tumor associated antigens
Introduction
Smoking is by far the leading risk factor for lung cancer 1-4. In the United States, cigarette smoking is linked to about 80% to 90% of lung cancers. Even people don't smoke; breathing in the smoke of others (called secondhand smoke or environmental tobacco smoke) can increase the risk of developing lung cancer. Secondhand smoke is further thought to cause more than 7,000 deaths from lung cancer each year 5, 6. Non-small-cell lung carcinoma (NSCLC) accounts for about 85% of all lung cancers, which are relatively insensitive to chemotherapy compared to small cell carcinoma 1-4.
Recent studies indicated that membrane-bound extracellular vesicles (EVs; 30-400nm) are important mediators of intercellular communication among different tissues and organs 7. EVs, found in most bodily fluids, contain proteins, RNA and lipids 7. Exosomes (~50-200nm) are released through the fusion of multivesicular bodies with the plasma membrane 8. Exosomes have a role in physiological and pathological intercellular signaling and in a wide spectrum of biological functions 8. It is currently unknown how smoking affects lung EVs pattern and their microRNA (miRNA) expression. In vitro studies suggest that tobacco smoke increased the release of pro-coagulant tissue factor positive extracellular vesicles by monocytes and macrophages 9, while studies on human subjects rather indicated a decrease in plasma EVs 10, 11. However, EVs are derived from many different cell types and vary extensively in size. Specifically, EVs encompassing the size range of exosomes, microvesicles and apoptotic bodies have not been investigated extensively in the context of smoking populations. Therefore, we determined the effects of habitual smoking on EVs profile in bronchoalveolar lavage (BAL) fluid and alterations in the NSCLC-related miRNAs, mRNAs, long non- coding RNAs (lncRNAs). We further examined differential expression of tumor associated antigens in EVs from smokers and NSCLC patients.
Materials and Methods
Ethics statement
This work received approval from the institution ethics committee of Tongji Medical College, Huazhong University of Science and Technology.
Patients
Clinicopathologic characteristics of smokers and lung cancer patients are summarized in Supplementary materials. Healthy subjects were patients undergoing endoscopy for unexplained upper abdominal complaints, had no reflux symptoms or endoscopic abnormalities, and did not use PPIs. Inclusion criteria for smoker patients required at least a 10 pack-year history of smoking, while the nonsmoker control subjects were self-reported never smokers. Smoking subjects were excluded if they had any significant comorbid conditions such as pregnancy or other acute or chronic disease such as pre-existing asthma, interstitial lung diseases or cardiovascular diseases. NSCLC patients have histologically or cytologically confirmed non-small cell lung cancer not amenable to curative intent therapy or stage IV NSCLC. Exclusion criteria includes patients who are concurrently receiving chemotherapy, biologics, immunotherapy for cancer treatment; as well as systemic anti-cancer treatment or investigational treatment. NSCLC subjects were also excluded if patients have a history of other malignancies within the last 5 years, except for non-melanoma skin cancer or carcinoma in situ of the cervix.
Bronchoalveolar lavage
BAL was performed by using standard protocol. The BAL consisted of 2 aliquots of 50 mL of normal saline instilled and aspirated manually using a 60 mL syringe. All subjects underwent conventional bronchoscopy. The BAL fluid was centrifuged at 1500 rpm for 10 minutes at 4°C. The supernatant was aliquoted was stored at -80°C for further EVs analysis.
Isolation of extracellular vesicles
EVs were isolated as described before 12. In brief, BAL fluid was subjected to successive centrifugations. EV pellets were resuspended in 0.32 M sucrose and centrifuged at 100,000×g for 1 hour.
Extracellular vesicles characterization by FACS analysis
BAL fluid was added directly to Dynabeads (2 mL/mL beads) coated with anti-MHC class II (MHCII) antibodies (clone HKB1; Invitrogen/Dynal).
RNA processing and MicroRNA expression profiling
Total RNA was extracted by using a mirVana miRNA isolation kit (Applied Biosystems, Foster City, CA). TaqMan microRNA assays (Applied Biosystems) was utilized for vesicle miRNA profiling. DIANA- mirPath 13 was introduced in biological pathways.
RT-qPCR
Total RNA quantity and quality was tested using the Agilent Bioanalyzer RNA 6000 Chip (Agilent, Santa Clara, CA). Reverse transcribed using random hexamers and reverse transcriptase and RT-qPCR was performed using gene-specific primers and SYBR green master mix on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Gene expression was normalized to the average of HPRT.
Immunoblotting
EVs (10µg) samples were lysed and subjected to SDS-PAGE and immunoblotted with antibodies against HLA-class I, AKAP4, BAGE, PD-L1, MAGE-3, Annexin-A2, and Annexin V (Cell Signaling Technology).
Statistical analysis
Mann-Whitney test was used to compare gene expression levels. Data were shown as mean ± SE. P < 0.05 were recognized statistically significant.
Results
Higher EV concentration with smoking
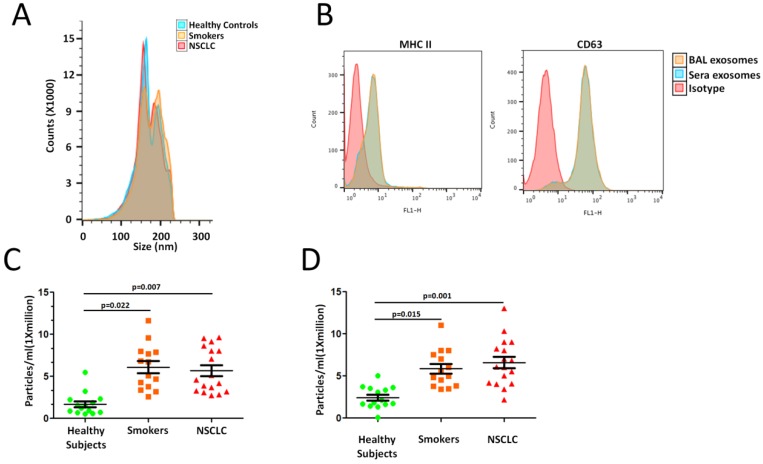
To assess differences in lung EVs with smoking, we designed cross sectional cohorts including smokers and NSCLC patients. Isolated BAL EVs were validated according to the International Society of Extracellular Vesicles guidelines 14. Known EV markers were present in each of the EV samples. The electron microscopy image shows intact, round vesicles of approximately 50-200 nm. A similar size distribution was also confirmed by nanoparticle tracking analysis (NTA) with a peak around 80 nm (Figure 1A). No significant differences were seen between the groups. These data confirm the size, morphology and protein markers that are characteristic of EVs.
Figure 1.
Higher EV concentration with smoking. A. Exosomal phenotyped EVs were examined by flow cytometric analysis. B. Cross-sectional cohort analysis of EV concentration in 14 smokers and 17 NSCLC patients compared to 14 healthy subjects. C. EVs concentrations when isolated by differential ultracentrifugation protocols.
We sought to determine whether there were differences in lung EV concentration between smokers and NSCLC patients. An analysis of a cross- sectional cohort consisting of both groups revealed a significantly higher EV concentration in 14 smokers and 17 NSCLC patients compared to 14 healthy subjects (Smokers vs. Controls, p=0.022; NSCLC vs. Controls, p=0.007; Figure 1C). Clinicopathologic characteristics of smokers and NSCLC patients are summarized in Supplementary Table 1.
To further confirm our results obtained using precipitation isolation methods, we used a protocol recently reported by Kowal et al. to compare EVs recovered from different steps of the differential ultracentrifugation protocols 15. Smokers and NSCLC patients had higher concentration of EVs isolated at both medium (10,000 x g) and ultracentrifugation (120,000 x g) speeds (Smokers vs. Controls, p=0.015; NSCLC vs. Controls, p=0.001; Figure 1D). Thus, smoking is associated with higher levels of local site EVs in the lung, which further indicated higher EV population, may play an important role in NSCLC development.
Dysregulated microRNAs expression in lung EVs from smokers and NSCLC patients
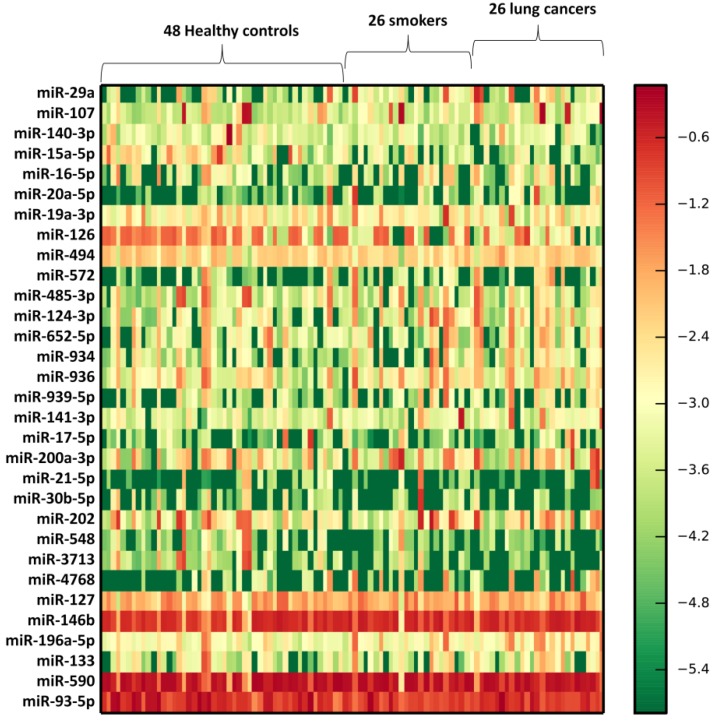
We profiled the expression of 366 miRNAs by using TaqMan miRNA arrays in an independent cohort of smokers and NSCLC patients (Supplementary Table 2). Vesicle miRNAs were normalized to the BAL volume. The results shown that 102 miRNAs (27.8%) could be detected. Further, 31 vesicle miRNAs were differentially expressed in lung EVs from smokers compared to healthy subjects, including 21 up-regulated vesicle miRNAs and 10 under-regulated miRNAs (p<0.05) (Table 1). Further, these 31 vesicle miRNAs shown similar expression change patterns in NSCLC exosomes when compared to those from healthy subjects (Table 1 and Figure 2). Further bioinformatics study demonstrated that 49 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were significantly enriched (p<0.05, FDR corrected) among differentially expressed vesicle miRNAs. Among them, Proteoglycans in cancer (p=7.197E-13), Fatty acid biosynthesis (p=5.120E-10), ErbB (p=7.282E-09), Hippo (p=2.989E-07), TGF-beta (p=6.440E-07), Pluripotency of stem cells (p=9.569 E-07), Pathways in cancer (p=8.143E-06), Wnt (p= 2.151E-05), Renal cell carcinoma (p=4.700E-05), Regulation of actin cytoskeleton (p=4.976E-05), Ubiquitin mediated proteolysis (p=1.023E-04), Rap1 (p=1.353 E-04), Glioma (p=1.353E-04), AMPK (p=5.012E-04), FoxO (p=5.472E-04), and Ras (p=5.472E-04) were significantly enriched (Table 2), suggesting that smoking induced exosomal miRNAs may dysregulate specific biologic pathways, which predisposed to NSCLC.
Table 1.
Exosomal microRNAs were differentially expressed in smokers and lung cancer patients.
| Smokers vs. Controls | Smokers vs. Controls | NSCLC vs. Smokers | NSCLC vs. Smokers | |
|---|---|---|---|---|
| miR-Name | Fold change | Adjusted p-value | Fold change | Adjusted p-value |
| miR-29a | 5.924 | 0.00117 | 1.954 | 0.00651 |
| miR-107 | 25.386 | 0.00753 | 2.502 | 0.00693 |
| miR-140-3p | 3.373 | 0.00776 | 4.415 | 0.00758 |
| miR-15a-5p | 3.117 | 0.00530 | 5.054 | 0.00832 |
| miR-16-5p | 2.752 | 0.00498 | 1.584 | 0.00856 |
| miR-20a-5p | 4.272 | 0.00984 | 2.971 | 0.00856 |
| miR-19a-3p | 7.593 | 0.00283 | 1.555 | 0.00896 |
| miR-126 | 19.105 | 0.00434 | 0.969 | 0.00901 |
| miR-494 | 2.527 | 0.00183 | 0.550 | 0.00920 |
| miR-572 | 3.013 | 0.00279 | 3.165 | 0.00977 |
| miR-485-3p | 4.099 | 0.00068 | 3.484 | 0.01044 |
| miR-124-3p | 6.447 | 0.04052 | 2.709 | 0.01051 |
| miR-652-5p | 8.507 | 0.00990 | 1.005 | 0.01184 |
| miR-934 | 5.409 | 0.00632 | 1.482 | 0.01249 |
| miR-936 | 2.534 | 0.00737 | 1.621 | 0.01374 |
| miR-939-5p | 3.331 | 0.00361 | 2.423 | 0.01441 |
| miR-141-3p | 2.967 | 0.00034 | 7.238 | 0.01452 |
| miR-17-5p | 3.319 | 0.00367 | 1.797 | 0.01453 |
| miR-200a-3p | 8.561 | 0.00136 | 1.086 | 0.01798 |
| miR-21-5p | 4.142 | 0.00709 | 0.873 | 0.01933 |
| miR-30b-5p | 5.924 | 0.00789 | 2.310 | 0.01988 |
| miR-202 | 0.093 | 0.00201 | 0.149 | 0.02065 |
| miR-548 | 0.25 | 0.00794 | 0.155 | 0.02155 |
| miR-3713 | 0.066 | 0.00503 | 0.059 | 0.02222 |
| miR-4768 | 0.358 | 0.00065 | 0.492 | 0.02229 |
| miR-127 | 0.127 | 0.00442 | 0.168 | 0.02281 |
| miR-146b | 0.254 | 0.00921 | 0.176 | 0.02477 |
| miR-196a-5p | 0.039 | 0.00665 | 0.130 | 0.02683 |
| miR-133 | 0.108 | 0.00482 | 0.149 | 0.03087 |
| miR-590 | 0.284 | 0.00677 | 0.085 | 0.03176 |
| miR-93-5p | 0.143 | 0.01742 | 0.163 | 0.00065 |
Figure 2.
Heatmap of BAL exosomal differential miRNA profiles in smokers and NSCLC patients. Heatmap representation of the mean fold change in smokers and NSCLC patients related differential miRNA signature. Two-dimensional grid matrix displaying 31 exosomal miRNAs in BAL was obtained by the functional heat-map in R. Columns refer to time course comparison: 48 healthy controls, 26 smokers and 26 NSCLC patients. Rows stand for the 31 differential miRNAs. Each entry of the grid refers to relative fold (log2) between the expression level of a given exosomal miRNA. The color of each entry is determined by the value of that fold difference, ranging from green (negative values) to red (positive values).
Table 2.
Biologic pathways enriched by differentially expressed smoking specific miRNAs.
| KEGG pathway | p-value | #genes | #miRNAs |
|---|---|---|---|
| Proteoglycans in cancer (hsa05205) | 7.197E-13 | 110 | 21 |
| Fatty acid biosynthesis (hsa00061) | 5.120E-10 | 5 | 5 |
| ErbB signaling pathway (hsa04012) | 7.282E-09 | 56 | 21 |
| Hippo signaling pathway (hsa04390) | 2.989E-07 | 76 | 21 |
| TGF-beta signaling pathway (hsa04350) | 6.440E-07 | 47 | 19 |
| Signaling pathways regulating pluripotency of stem cells (hsa04550) | 9.569E-07 | 74 | 21 |
| Pathways in cancer (hsa05200) | 8.143E-06 | 182 | 21 |
| Wnt signaling pathway (hsa04310) | 2.151E-05 | 74 | 21 |
| Renal cell carcinoma (hsa05211) | 4.700E-05 | 41 | 20 |
| Regulation of actin cytoskeleton (hsa04810) | 4.976E-05 | 106 | 21 |
| Ubiquitin mediated proteolysis (hsa04120) | 1.023E-04 | 71 | 21 |
| Rap1 signaling pathway (hsa04015) | 1.353E-04 | 103 | 21 |
| Glioma (hsa05214) | 1.353E-04 | 35 | 21 |
| AMPK signaling pathway (hsa04152) | 5.012E-04 | 64 | 20 |
| FoxO signaling pathway (hsa04068) | 5.472E-04 | 69 | 20 |
| Ras signaling pathway (hsa04014) | 5.472E-04 | 105 | 21 |
| Focal adhesion (hsa04510) | 6.650E-04 | 100 | 21 |
| mTOR signaling pathway (hsa04150) | 7.077E-04 | 37 | 18 |
| Gap junction (hsa04540) | 7.258E-04 | 45 | 19 |
| Thyroid hormone signaling pathway (hsa04919) | 8.249E-04 | 56 | 21 |
| Neurotrophin signaling pathway (hsa04722) | 8.352E-04 | 65 | 20 |
| Prolactin signaling pathway (hsa04917) | 9.840E-04 | 37 | 18 |
| Estrogen signaling pathway (hsa04915) | 9.963E-04 | 45 | 20 |
| Endocytosis (hsa04144) | 1.485E-03 | 96 | 22 |
| Circadian rhythm (hsa04710) | 1.690E-03 | 22 | 15 |
| Biotin metabolism (hsa00780) | 1.750E-03 | 2 | 3 |
| cGMP-PKG signaling pathway (hsa04022) | 1.750E-03 | 79 | 20 |
| Prostate cancer (hsa05215) | 1.829E-03 | 46 | 19 |
| Hedgehog signaling pathway (hsa04340) | 2.952E-03 | 30 | 15 |
| MAPK signaling pathway (hsa04010) | 3.214E-03 | 114 | 20 |
| Adrenergic signaling in cardiomyocytes (hsa04261) | 3.596E-03 | 65 | 20 |
| Sphingolipid signaling pathway (hsa04071) | 3.858E-03 | 57 | 19 |
| Non-small cell lung cancer (hsa05223) | 4.639E-03 | 30 | 20 |
| PI3K-Akt signaling pathway (hsa04151) | 4.970E-03 | 146 | 21 |
| Viral carcinogenesis (hsa05203) | 7.453E-03 | 81 | 20 |
| Oxytocin signaling pathway (hsa04921) | 7.453E-03 | 76 | 21 |
| Insulin signaling pathway (hsa04910) | 8.038E-03 | 67 | 20 |
| Glycosaminoglycan biosynthesis - heparan sulfate / heparin (hsa00534) | 8.620E-03 | 12 | 15 |
| Cell cycle (hsa04110) | 8.620E-03 | 57 | 20 |
| cAMP signaling pathway (hsa04024) | 8.620E-03 | 90 | 21 |
| Chronic myeloid leukemia (hsa05220) | 1.432E-02 | 38 | 20 |
| Adherens junction (hsa04520) | 1.547E-02 | 37 | 21 |
| Melanoma (hsa05218) | 1.685E-02 | 36 | 19 |
| Amphetamine addiction (hsa05031) | 1.685E-02 | 31 | 21 |
| Melanogenesis (hsa04916) | 1.794E-02 | 47 | 18 |
| Choline metabolism in cancer (hsa05231) | 1.953E-02 | 50 | 20 |
| T cell receptor signaling pathway (hsa04660) | 2.128E-02 | 50 | 20 |
| HIF-1 signaling pathway (hsa04066) | 2.981E-02 | 49 | 20 |
Messenger RNA transcripts were differentially expressed in lung exosomes in smokers and NSCLC
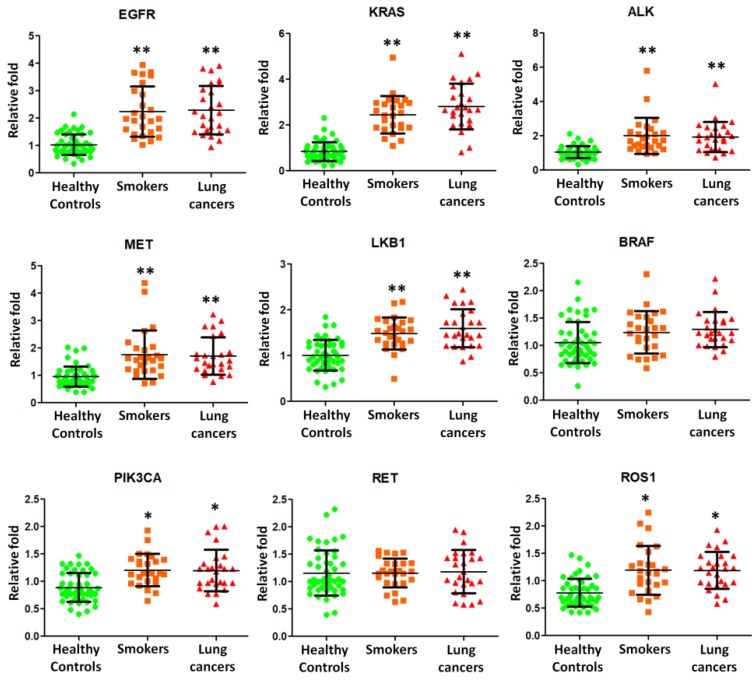
We then employed quantitative RT-PCR to detect the expression of mRNA transcripts isolated from exosomes. Nine potential BAL biomarkers in lung cancers, including epidermal growth factor receptor (EGFR), kirsten ras (KRAS), anaplastic lymphoma kinase (ALK), MET, liver kinase B1 (LKB1), B-type raf kinase (BRAF), catalytic subunit of phosphoinositide 3-kinase (PIK3CA), RET, and c-ros oncogene 1 (ROS1), were selected for quantitative RT-PCR experiments using BAL exosomal RNA from 26 smokers, 26 NSCLC patients, and 48 healthy controls. EGFR, KRAS, ALK, MET, LKB1, PIK3CA, and ROS1 were found to be present and significantly over expressed in smokers and NSCLC lung exosome (Figure 3).
Figure 3.
Quantitative RT-PCR experiments were performed on EGFR, KRAS, ALK, MET, LKB1, BRAF, PIK3CA, RET, and ROS1 mRNAs by using BAL exosomal RNA isolated from 26 smokers, 26 NSCLC patients, and 48 healthy controls.
Long non-coding RNAs were differentially expressed in lung exosomes isolated from smokers and NSCLC
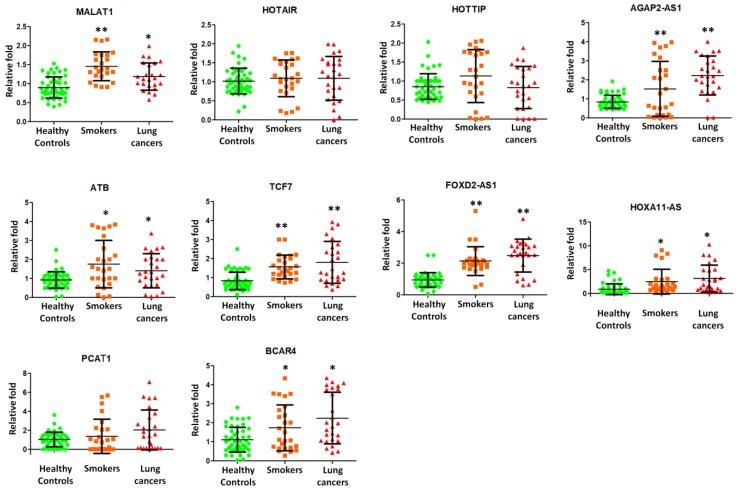
Further, we attempted to examine the presence of long non-coding RNAs (lncRNAs) in exosome and elucidate their roles in smokers and NSCLC patients. We then employed quantitative RT-PCR to detect the expression of lncRNAs isolated from lung exosomes. Nine potential lung cancers associated lncRNAs, including MALAT1, HOTAIR, HOTTIP, AGAP2-AS1, ATB, TCF7, FOXD2-AS1, HOXA11-AS, PCAF1, and BCAR4 were selected for quantitative RT-PCR experiments in 26 smokers, 26 NSCLC patients, and 48 healthy subjects. MALAT1, AGAP2-AS1, ATB, TCF7, FOXD2-AS1, HOXA11-AS, PCAF1, and BVAR4 were found to be present on the exosomes and significantly over expressed in smokers and NSCLC patients (Figure 4).
Figure 4.
Quantitative RT-PCR experiments were performed on nine lncRNAs including MALAT1, HOTAIR, HOTTIP, AGAP2-AS1, ATB, TCF7, FOXD2-AS1, HOXA11-AS, PCAF1, and BCAR4 by using BAL exosomal RNA isolated from 26 smokers, 26 NSCLC patients, and 48 healthy controls.
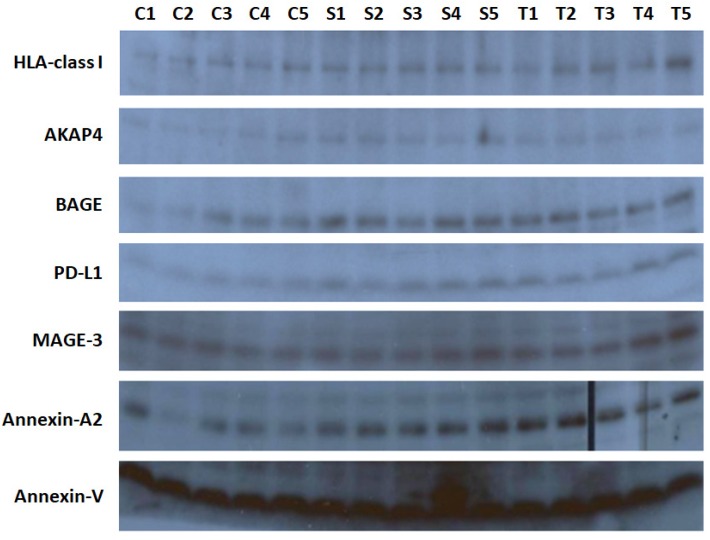
Smoking alters tumor associated antigens in EVs
Tumor associated antigens are associated with NSCLC 16. Therefore, we analyzed for dysregulation of HLA-class I, AKAP4, BAGE, PD-L1, MAGE-3, and Annexin-A2 in lung exosomes of 5 healthy controls, 5 smokers, and 5 NSCLC patients. Clinicopathologic characteristics of smokers and NSCLC patients are summarized in Supplementary Table 3. We used western-blot to test proteins from lysed EVs. Significant elevation of HLA-class I, BAGE, PD-L1, and Annexin-A2 protein level in smokers and NSCLC patients compared to those from healthy subjects (Figure 5). However, no difference was observed in expression of AKAP4 and MAGE-3 protein levels among the three groups (Figure 5). These data indicated that EV cargo is affected by smoking and may reflect the tumor associated antigens presentation in lung exosomes from smokers and NSCLC.
Figure 5.
Western blot to test HLA-class I, AKAP4, BAGE, PD-L1, MAGE-3, and Annexin-A2 protein levels in lung exosomes isolated from 5 healthy controls, 5 smokers, and 5 NSCLC patients. Annexin-V was loaded as a positive control.
Discussion
In this study, for the first time, we have verified the presence of EVs isolated from lung fluid from smokers and NSCLC patients. Smoking is associated with higher levels of local site EVs in the lung. Further, we revealed alterations in the vesicle miRNAs, mRNAs, lncRNAs profiles from smokers and NSCLC, suggesting the importance of EV contents in NSCLC development.
Somatic mutations in EGFR, KRAS, ALK, MET, LKB1, BRAF, PIK3CA, RET, and ROS1 genes are frequently detected in lung cancers. EGFR is known to be a good tumor target in lung cancers. KRAS plays the critical role in growth factor receptor signaling and KRAS gene mutations are usually detected in lung adenocarcinomas 17. Both EGFR and KRAS genes are responsible for encoding proteins that are embedded in cellular membranes. EGFR or KRAS mutations caused the protein production that is constantly turned on. If these mutations occur in lungs, lung cancer would develop. Rikova et al., 18 reported the MET receptor as the most important members of tyrosinated phosphopeptides in NSCLC tissue samples. V600E (50%), G469A (39%), and D594G (11%) on the BRAF kinase were identified as mutations in lung adenocarcinomas 19. EML4-ALK fusion was found in NSCLC by Soda et al.20 in 2007 as a novel potential oncogenic kinase. In our current study, we found that EGFR, KRAS, ALK, MET, LKB1, PIK3CA, and ROS1 were present on the exosome surface and further over expressed in lung exosome from smokers and NSCLC patients (Figure 2). Therefore, smoking induced exosomal mRNA molecules may play an important role in NSCLC.
LncRNA MALAT1 promoted brain metastasis by inducing EMT in lung cancer 21, 22, which indicated a poor prognosis in NSCLC. Up regulation of lncRNA HOTAIR was reported to enhance aggressive tumor phenotypes in human NSCLC 23. AGAP2-AS1 negatively controled lung cancer development by inhibiting LATS2 and KLF2 expression 24. lncRNA ATB over expression demonstrated a poor prognosis in NSCLC 25. TCF7 enhanced invasion by increasing EpCAM expression 26. FOXD2-AS1 promoted NSCLC cell growth by Wnt/β-catenin pathway 27. HOXA11-AS regulated DOCK8 and TGF-beta pathway, further promoted NSCLC development 28. PCAT-1 played as an oncogenic molecule in NSCLC progression 29. Increased expression of long non-coding RNA BCAR4 is predictive of poor prognosis in NSCLC patients 30. In this study, we, for the first time, found that MALAT1, AGAP2-AS1, ATB, TCF7, FOXD2-AS1, HOXA11-AS, PCAF1, and BVAR4 were present on the exosomes and significantly higher expressed in smokers and NSCLC patients (Figure 4). These results demonstrated that smoking dysregulated exosomal lncRNA molecules, which may predispose to NSCLC.
Tumor associated antigens (TAAs) are thought to be an ideal target for cancer immunotherapy 31, 32. Yasuda et al., identified a tumour associated antigen, annexin A2, in lung cancer patients with asbestos exposure 33. They further found that the immune response in lung cancer was modulated by annexin A2 and it was also associated with high circulating levels of an inflammatory cytokine IL-6. Mirandola et al 34 discovered that Sperm protein 17, AKAP4 and Pituitary tumor transforming gene 1 were potential immunotherapeutic targets in lung cancer patients. BAGE and MAGE-3 are expressed in lung cancer tissues 35. MAGE-3 is aberrantly expressed in a wide variety of tumors, including NSCLC. Several CD8+ T-cell epitopes of MAGE-3 have been identified in vitro 35. Cancer cells can up regulate the surface expression of programmed death-ligand 1 (PD-L1), further interacts with programmed death-1 (PD-1) receptor on T cells to elicit the immune checkpoint response 36. Exosomal PD-L1 carries active defense function to suppress T cell killing of breast cancer cells 37. In this study, we found that HLA-class I, BAGE, PD-L1, and Annexin-A2 protein levels are over expressed in smokers and NSCLC patients (Figure 5). These data indicated that EV harbors tumor associated antigens presentation in smokers and NSCLC.
There are still some limitations. Sample size is small because obtaining more blood samples with inclusion criteria in this study is a challenge in our center. Results have to be validated in a larger sample-size cohort including potential confounding clinical variables. Furthermore, there was no functional investigation of the identified miRNAs.
In conclusion, this pilot study demonstrated that smoking induced lung EVs in smokers and NSCLC patients. Furthermore, vesicle miRNAs, mRNAs, lncRNAs and protein presentations were dysregulated in smoking and NSCLCs. We propose that smoking induced exosomes carried tumor associated miRNAs, mRNAs, lncRNAs and tumor associated antigens, may participate in lung cancer development.
Supplementary Material
Supplementary tables.
Acknowledgments
This study is supported by the Natural Science Foundation of Hubei Province, China (Grant No. 2018CFB590 to G.Y.) and the Natural Science Foundation of China (NSFC grant 81700091 to F.W.).
Authors' contributions
FW, ZYY, LY, and JSF performed the experiments and data analysis. JJX, YJ, JZY, and DZ collected the subject's samples and clinical data. GHY conceived the study and supervised this study. All the authors contributed to the manuscript writing and revision.
Abbreviations
- EVs
Extracellular vesicles
- NSCLC
non-small cell lung cancer
- miRNAs
microRNAs
- lncRNAs
long non-coding RNAs
- BAL
bronchoalveolar lavage
- NTA
nanoparticle tracking analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- EGFR
epidermal growth factor receptor
- KRAS
kirsten ras
- ALK
anaplastic lymphoma kinase
- LKB1
liver kinase B1
- BRAF
B-type raf kinase
- PIK3CA
catalytic subunit of phosphoinositide 3-kinase
- ROS1
c-ros oncogene 1
References
- 1.Morabia A, Wynder EL. Cigarette smoking and lung cancer cell types. Cancer. 1991;68:2074–8. doi: 10.1002/1097-0142(19911101)68:9<2074::aid-cncr2820680939>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 2.Matsuura K, Kudoh S, Yoshimura N, Kimura T, Mitsuoka S, Hirata K. et al. Smoking history and prior surgical resection predict sensitivity to gefitinib in advanced non-small-cell lung cancer. Osaka city medical journal. 2006;52:1–8. [PubMed] [Google Scholar]
- 3.Igawa S, Sasaki J, Otani S, Shirasawa M, Niwa H, Kusuhara S. et al. Smoking History as a Predictor of Pemetrexed Monotherapy in Patients with Non-Squamous Non-Small Cell Lung Cancer. Oncology. 2016;91:41–7. doi: 10.1159/000446438. [DOI] [PubMed] [Google Scholar]
- 4.Gadgeel SM. Smoking history: tumor biology and clinical pharmacokinetic implications in patients with lung cancer. Clinical lung cancer. 2006;7:369–70. [PubMed] [Google Scholar]
- 5.Hori M, Tanaka H, Wakai K, Sasazuki S, Katanoda K. Secondhand smoke exposure and risk of lung cancer in Japan: a systematic review and meta-analysis of epidemiologic studies. Japanese journal of clinical oncology. 2016;46:942–51. doi: 10.1093/jjco/hyw091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan P, Buffler PA, Reynolds P, Wu AH, Wichmann HE, Agudo A. et al. Secondhand smoke exposure in adulthood and risk of lung cancer among never smokers: a pooled analysis of two large studies. International journal of cancer. 2004;109:125–31. doi: 10.1002/ijc.11682. [DOI] [PubMed] [Google Scholar]
- 7.Yoon YJ, Kim OY, Gho YS. Extracellular vesicles as emerging intercellular communicasomes. BMB reports. 2014;47:531–9. doi: 10.5483/BMBRep.2014.47.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willms E, Johansson HJ, Mager I, Lee Y, Blomberg KE, Sadik M. et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Scientific reports. 2016;6:22519. doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Yu D, Williams KJ, Liu ML. Tobacco smoke induces the generation of procoagulant microvesicles from human monocytes/macrophages. Arteriosclerosis, thrombosis, and vascular biology. 2010;30:1818–24. doi: 10.1161/ATVBAHA.110.209577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant R, Ansa-Addo E, Stratton D, Antwi-Baffour S, Jorfi S, Kholia S. et al. A filtration-based protocol to isolate human plasma membrane-derived vesicles and exosomes from blood plasma. Journal of immunological methods. 2011;371:143–51. doi: 10.1016/j.jim.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 11.Casey RG, Joyce M, Roche-Nagle G, Cox D, Bouchier-Hayes DJ. Young male smokers have altered platelets and endothelium that precedes atherosclerosis. The Journal of surgical research. 2004;116:227–33. doi: 10.1016/S0022-4804(03)00343-3. [DOI] [PubMed] [Google Scholar]
- 12.Freeman DW, Noren Hooten N, Eitan E, Green J, Mode NA, Bodogai M, Altered Extracellular Vesicle Concentration, Cargo and Function in Diabetes Mellitus. Diabetes; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T. et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic acids research. 2015;43:W460–6. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C. et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. Journal of extracellular vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B. et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E968–77. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasumoto K, Hanagiri T, Takenoyama M. Lung cancer-associated tumor antigens and the present status of immunotherapy against non-small-cell lung cancer. General thoracic and cardiovascular surgery. 2009;57:449–57. doi: 10.1007/s11748-008-0433-6. [DOI] [PubMed] [Google Scholar]
- 17.Aviel-Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: a review. Clinical lung cancer. 2006;8:30–8. doi: 10.3816/CLC.2006.n.030. [DOI] [PubMed] [Google Scholar]
- 18.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, Kris MG. et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:2046–51. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S. et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D. et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:1984–92. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 22.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. Journal of neuro-oncology. 2015;121:101–8. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N. et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochemical and biophysical research communications. 2013;436:319–24. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Sun M, Zang C, Ma P, He J, Zhang M. et al. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell death & disease. 2016;7:e2225. doi: 10.1038/cddis.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ke L, Xu SB, Wang J, Jiang XL, Xu MQ. High expression of long non-coding RNA ATB indicates a poor prognosis and regulates cell proliferation and metastasis in non-small cell lung cancer. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2017;19:599–605. doi: 10.1007/s12094-016-1572-3. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Wang D. Long noncoding RNA TCF7 promotes invasiveness and self-renewal of human non-small cell lung cancer cells. Human cell. 2017;30:23–9. doi: 10.1007/s13577-016-0147-5. [DOI] [PubMed] [Google Scholar]
- 27.Rong L, Zhao R, Lu J. Highly expressed long non-coding RNA FOXD2-AS1 promotes non-small cell lung cancer progression via Wnt/beta-catenin signaling. Biochemical and biophysical research communications. 2017;484:586–91. doi: 10.1016/j.bbrc.2017.01.141. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, He RQ, Dang YW, Zhang XL, Wang X, Huang SN. et al. Comprehensive analysis of the long noncoding RNA HOXA11-AS gene interaction regulatory network in NSCLC cells. Cancer cell international. 2016;16:89. doi: 10.1186/s12935-016-0366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B, Hou X, Zhan H. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in non-small cell lung cancer cells. International journal of clinical and experimental medicine. 2015;8:18482–7. [PMC free article] [PubMed] [Google Scholar]
- 30.Gong J, Zhang H, He L, Wang L, Wang J. Increased Expression of Long Non-Coding RNA BCAR4 Is Predictive of Poor Prognosis in Patients with Non-Small Cell Lung Cancer. The Tohoku journal of experimental medicine. 2017;241:29–34. doi: 10.1620/tjem.241.29. [DOI] [PubMed] [Google Scholar]
- 31.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nature reviews Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 32.Old LJ. Cancer/testis (CT) antigens - a new link between gametogenesis and cancer. Cancer immunity. 2001;1:1. [PubMed] [Google Scholar]
- 33.Yasuda M, Hanagiri T, Shigematsu Y, Onitsuka T, Kuroda K, Baba T. et al. Identification of a tumour associated antigen in lung cancer patients with asbestos exposure. Anticancer research. 2010;30:2631–9. [PubMed] [Google Scholar]
- 34.Mirandola L, Figueroa JA, Phan TT, Grizzi F, Kim M, Rahman RL. et al. Novel antigens in non-small cell lung cancer: SP17, AKAP4, and PTTG1 are potential immunotherapeutic targets. Oncotarget. 2015;6:2812–26. doi: 10.18632/oncotarget.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho MY, Tang SJ, Sun KH, Yang W. Immunotherapy for lung cancers. Journal of biomedicine & biotechnology. 2011;2011:250860. doi: 10.1155/2011/250860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. The Journal of clinical investigation. 2015;125:3384–91. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell research; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.