Abstract
Noble rot is a latent infection of grape berries caused by the necrotrophic fungus Botrytis cinerea, which develops under specific climatic conditions. The infected berries undergo biochemical and metabolic changes, associated with a rapid withering, which altogether offer interesting organoleptic features to sweet white wines. In this paper, we provide RNAseq datasets (raw and normalized counts as well as differentially expressed genes lists) of the transcriptome profiles of both grapevine berries (Vitis vinifera cv. Garganega) and B. cinerea during the establishment of noble rot, artificially induced in controlled conditions. The sequencing data are available in the NCBI GEO database under accession number GSE116741. These data were exploited in a comprehensive meta-analysis of gene expression during noble rot infection, gray mold and post-harvest withering. This highlighted an important common transcriptional reprogramming in different botrytized grape berry varieties and led to the identification of key genes specifically modulated during noble rot infection, which are described in the article entitled “Specific molecular interactions between Vitis vinifera and Botrytis cinerea are required for noble rot development in grape berries” Lovato et al., 2019.
Keywords: Grapevine berries, Vitis vinifera, Botrytis cinerea, Transcriptomic, RNA-Seq, Noble rot
Specifications table
| Subject area | Plant Biology |
| More specific subject area | Plant-pathogen interaction |
| Type of data | Raw and normalized counts; differentially expressed genes |
| How data was acquired | Sequencing data were acquired through Illumina HiSeq500 sequencing of TruSeq libraries prepared from RNA extracted from Vitis vinifera cv. Garganega berries artificially botrytized in vitro |
| Data format | Tables |
| Experimental factors | RNA used for library preparation and sequencing was isolated from mature Vitis vinfera cv. Garganega berries infiltrated in vitro with Botrytis cinerea to induce noble rot in controlled environmental conditions |
| Experimental features | RNA-seq data was obtained from 3′mRNA sequencing to estimate gene abundance in count per million (CPM) represent the expression level of each grapevine or B. cinerea transcript |
| Data source location | Not applicable |
| Data accessibility | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116741 |
| Related research article | Lovato A., Zenoni S., Tornielli GB., Colombo T., Vandelle E., Polverari A., Specific molecular interactions between Vitis vinifera and Botrytis cinerea are required for noble rot development in grape berries”, Postharvest Biology and Technology |
Value of the data
|
1. Data
Data reported here describe the sequencing results obtained from grapevine (V. vinifera cv. Garganega) berries artificially botrytized in vitro and harvested 12 days after infection [1]. Water-infiltrated berries and B. cinerea mycelium grown in vitro were used as controls for plant and fungus differential gene expression, respectively. Each set of samples includes three biological replicates. Data are available at the NCBI GEO database under accession number GSE116741.
From RNAseq analysis, around 50 million raw reads were generated for each sample (Table 1).
Table 1.
Statistics on Vitis vinifera and Botrytis cinerea TruSeq sequencing.
| Samples | # of sequences in fastq files | # of sequences in filtered fastq files [IlluQC.pl - NGS QC Toolkit] | % of High Quality reads [i.e.: passing filtering] | # of Aligned Pairs | % of Aligned Pairs |
|
|---|---|---|---|---|---|---|
| 1) | Botrytis cinerea biol. repl. 1 | 51.596.498 | 48.464.674 | 93,93% | 44.728.327 | 92,30% |
| 2) | Botrytis cinerea biol. repl. 2 | 48.796.078 | 45.945.913 | 94,16% | 42.621.946 | 92,80% |
| 3) | Botrytis cinerea biol. repl. 3 | 57.657.830 | 54.115.696 | 93,86% | 50.145.232 | 92,70% |
| 4) | B. cinerea-infected berries biol. repl. 1 | 47.398.134 | 44.179.114 | 93,21% | 30.345.824 | 68,50% |
| 5) | B. cinerea-infected berries biol. repl. 2 | 58.054.526 | 54.322.693 | 93,57% | 35.755.979 | 65,60% |
| 6) | B. cinerea-infected berries biol. repl. 3 | 68.936.519 | 64.588.098 | 93,69% | 45.693.676 | 70,50% |
| 7) | Water-injected berries biol. repl. 1 | 65.333.494 | 60.514.894 | 92,62% | 44.998.907 | 74,20% |
| 8) | Water-injected berries biol. repl. 2 | 65.244.803 | 61.257.338 | 93,89% | 46.173.353 | 75,20% |
| 9) | Water-injected berries biol. repl. 3 | 70.761.848 | 65.929.242 | 93,17% | 48.576.393 | 73,50% |
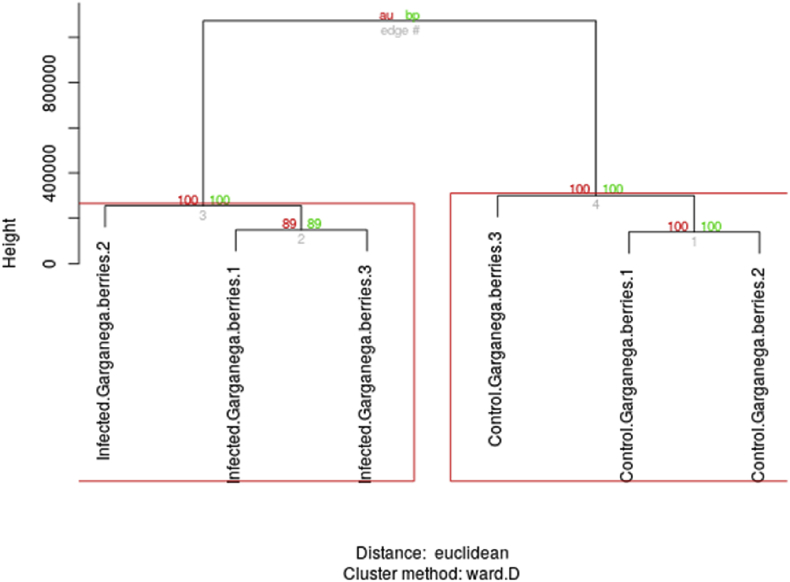
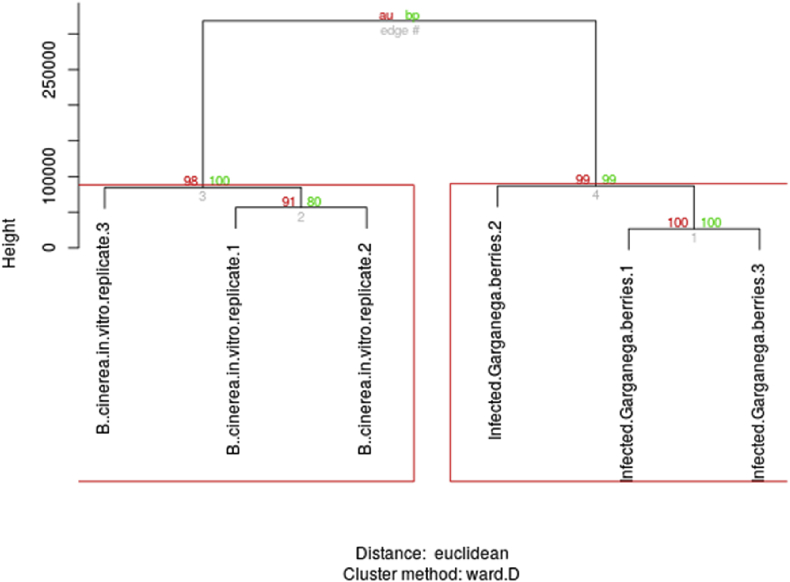
The reproducibility attested by the correlation across samples of the same condition and unsupervised partitioning of samples in the correct subgroups, when based on either V. vinifera normalized gene expression levels across samples (Fig. 1) or B. cinerea normalized counts (Fig. 2) argue that the relative changes in expression levels between conditions are due to the underlying biology, rather than a side effect related to differences in the biomass of the fungus respect to the plant, and support the relevance of the results from the differential expression analysis.
Fig. 1.
Unsupervised clustering of samples based on V. vinifera cv. Garganega normalized gene counts. Infected.Garganega.berries, grape berries artificially infected in vitro with B. cinerea; Control.Garganega.berries, grape berries water-infiltrated. Numbers 1 to 3 refer to biological replicates.
Fig. 2.
Unsupervised clustering of samples based on B. cinerea normalized gene counts. B.cinerea.in.vitro, B. cinerea mycelium grown in vitro on synthetic medium; infected.Garganega.berries, grape berries artificially infected in vitro with B. cinerea. Numbers 1 to 3 refer to biological replicates.
2. Experimental design, materials and methods
2.1. Garganega berry withering
Bunches of V. vinifera cv. Garganega berries were harvested in October 2014 in Monteforte d’Alpone (Verona, Italy) and were transferred to the Pasqua Vigneti e Cantine winery (Verona, Italy) for withering. Healthy grapes were collected at the commercial ripening stage (soluble solids content = 18.5 ± 0.25%) and were placed in perforated plastic boxes known as plateaux (∼5 kg in each) in a ventilated withering facility under natural conditions (17–20 °C, 78–82% relative humidity).
Randomly selected replicate berries were analyzed weekly to determine the soluble solids content using a DBR35 digital refractometer (Giorgio Bormac, Carpi, Italy). Three dedicated plateaux were weighed weekly using a CH50K50 electronic balance (Kern, Balingen, Germany) to determine the sampling time corresponding to a berry soluble solids content of 26.6% and a percentage weight loss of ∼30%.
2.2. B. cinerea inoculum preparation and in vitro growth conditions
The B05.10 strain of B. cinerea Pers.: Fr. (teleomorph Botryotinia fuckeliana (de Bary) Whetz.), isolated in 1999 from grape berries [2] and later sequenced [3], [4], was cultured on solid potato dextrose agar (Formedium, Hunstanton, UK) in the dark for 7 d at room temperature followed by 5 d light exposure to promote fungal sporulation. Conidia were collected from sporulating cultures in 0.1% (v/v) water-diluted Tween-20, counted using the Fast-Read 102 microscope counting chamber (Biosigma, Cona, Italy) and diluted to 1 × 105 conidia mL−1.
Liquid B. cinerea B05.10 cultures were incubated in flasks by inoculating 125 mL of potato dextrose broth (Formedium) with 7 × 106 conidia mL−1. After 7 d of incubation at 22 °C shaking at 120 rpm, mycelia were recovered by filtration and subsequently frozen.
2.3. Artificial noble rot induction
Samples of 1000 withered Garganega (with pedicels) were surface-sterilized for 5 min in 70% ethanol and dried in sterility. Of the 1000 sterilized Garganega berries, 500 berries were injected, until berry saturation (i.e. appearance of the first drop on berry surface), with ∼0.1 mL of a B. cinerea conidial suspension (1 × 105 conidia mL−1) using a 1-mL syringe with a needle and reaching berry mesocarp (almost the centre of the berry). In the same way, the remaining 500 berries were injected with sterile water as negative controls.
Garganega berries were incubated in sterile 24-well plates under controlled conditions (15 °C/15 h dark and 18 °C/9 h light) to induce noble rot. B. cinerea colonization was monitored daily in the infected berries.
Three biological replicates (∼100 berries each) of infected and uninfected Garganega berries were collected, deseeded and frozen at the same pourri plein/withering stage 12 d after injection.
2.4. RNA extraction
Infected or uninfected berry pericarps representing each grapevine variety as well as B. cinerea mycelia from in vitro cultures were ground under liquid nitrogen and total RNA was isolated from 200 mg of powdered sample using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, USA). The RNA quantity, integrity and purity were confirmed using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and a Bioanalyzer Chip RNA 7500 series II (Agilent Technologies, Santa Clara, CA, USA).
2.5. RNA-Seq analysis
For each sample, an unstranded library was prepared from 2.5 μg of total RNA using the TruSeq Library Prep Kit v2 (Illumina, San Diego, CA, USA). Sequencing on an Illumina HiSeq500 device (101 bp, paired-end) was carried out at the Functional Genomics Centre (FGC), Department of Biotechnology, University of Verona, Verona, Italy.
The preprocessing and analysis of RNA-Seq data were carried out using high-performance computing resources made available by CINECA (Class C ISCRA project: IsC33_NobleRot). Due to de-multiplexing, the raw reads for each individual sample were represented by multiple FASTQ files in the RNA-Seq dataset (GEO ID: GSE116741). As the first data preprocessing step, all raw reads related to the same sample were merged in a unique FASTQ file. Because the reads were obtained by paired-end sequencing, the same merging procedure was applied to both sets of raw reads (R1 and R2) on each sample. Raw reads were quality controlled using IlluQC software as part of the NGS QC Toolkit [5] with default parameters (quality cutoff = 20; required percentage of nucleotides in reads with quality score at least equal to quality cutoff = 70%). The detailed description of individual parameters can be found in the NGS QC Toolkit reference manual (http://59.163.192.90:8080/ngsqctoolkit/NGSQCToolkitv2.3.3_manual.pdf).
Filtered reads were mapped against a mixed reference genome (Botrytis cinerea + Vitis vinifera) using TopHat v2.0.11 [6]. Mapped reads were summarized at the gene level into a count matrix using the htseq-count tool from the HTSeq library [7]. Gene models provided to the htseq-count tool to score reads mapping unambiguously to a single gene were obtained from the Grape Genome Database release V1 (http://genomes.cribi.unipd.it/DATA/) for Vitis vinifera [8] and from the Ensembl database release 28 (genome assembly: ASM15095v2) for B. cinerea [9]. Genomic data were downloaded on August 5, 2015.
The gene-level count matrix was used for differential expression analysis with edgeR [10]. Only genes showing at least three mapped reads in at least three samples of the count matrix were tested for differential expression (GEO ID: GSE116741). Modulated transcripts among the different conditions were defined using the following criteria: (i) |log2 FC| > 1; and (ii) Benjamini and Hochberg adjusted p-value < 0.05.
Following the summarization of mapped reads into a count-matrix, where counts represent the total number of reads aligning to each gene, two subsets of read counts, relative to V. vinifera genes (Supplemental File 1) or B. cinerea genes (Supplemental File 2), were separately normalized across samples before proceeding to the analysis of differential gene expression among the different conditions. In particular, library sizes, used to measure the relative abundance of each gene in each RNA sample and thus to express it as normalized gene counts suitable for the comparison across samples, were estimated based only on V. vinifera gene counts on one hand or only on B. cinerea gene counts on the other hand, yielding two tables of normalized gene counts (Supplemental Files 3 and 4). Each of those two tables served as input to assay separately the differential expression across conditions for genes of V. vinifera (Supplemental File 5) or B. cinerea (Supplemental File 6).
Acknowledgements
This work was supported by Regione Veneto - POR - Fondo Sociale Europeo 2007–2013 (Ob. Competitività Regionale e Occupazione - Reg. 1081/2006. Asse IV “Capitale Umano”. Progetto 1695/101/9/1148/2013) and a Joint Project (Joint Project 2016; prot.: JPVR16L2AF; Title: “Noble Rot development in harvested grapes: transcriptomic and metabolic effects on different genotypes”) funded by the University of Verona.
We are grateful to the wineries “Pasqua Vigneti e Cantine SpA” and “Azienda Agricola Cavalchina” for providing grapevine berry samples and allowing the setting up of the experiments in their withering facilities.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.104150.
Contributor Information
Elodie Vandelle, Email: elodiegenevieve.vandelle@univr.it.
Annalisa Polverari, Email: annalisa.polverari@univr.it.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supplemental File 1. Raw gene-level count matrices of Vitis vinifera cv. Garganega samples analyzed by RNA-Seq.
Supplemental File 2. Normalized gene-level count matrices of Vitis vinifera cv. Garganega samples analyzed by RNA-Seq.
Supplemental File 3. Raw gene-level count matrices of B. cinerea cultures analyzed by RNA-Seq.
Supplemental File 4. Normalized gene-level count matrices of B. cinerea cultures analyzed by RNA-Seq.
Supplemental File 5. List of genes differentially expressed in Vitis vinifera cv. Garganega berries undergoing noble rot.
Supplemental File 6. List of differentially expressed genes in B. cinerea during noble rot development in Vitis vinifera cv. Garganega berries.
References
- 1.Lovato A., Zenoni S., Tornielli G.B., Colombo T., Vandelle E., Polverari T. Specific molecular interactions between vitis vinifera and botrytis cinerea are required for noble rot development in grape berries. Postharvest. Biol. Technol. 2019;156:110924. doi: 10.1016/j.postharvbio.2019.05.025. [DOI] [Google Scholar]
- 2.Quidde T., Büttner P., Tudzynski P. Evidence for three different specific saponin-detoxifying activities in Botrytis cinerea and cloning of a gene coding for a putative avenacinase. Eur. J. Plant Pathol. 1999;105:273–283. doi: 10.1023/A:1008796006051. [DOI] [Google Scholar]
- 3.Amselem J., Cuomo C.A., Van Kan J.A.L., Viaud M., Benito E.P., Couloux A., Coutinho P.M., de Vries R.P., Dyer P.S., Fillinger S., Fournier E., Gout L., Hahn M., Kohn L., Lapalu N., Plummer K.M., Pradier J.-M., Quévillon E., Sharon A., Simon A., ten Have A., Tudzynski B., Tudzynski P., Wincker P., Andrew M., Anthouard V., Beever R.E., Beffa R., Benoit I., Bouzid O., Brault B., Chen Z., Choquer M., Collémare J., Cotton P., Danchin E.G., Da Silva C., Gautier A., Giraud C., Giraud T., Gonzalez C., Grossetete S., Güldener U., Henrissat B., Howlett B.J., Kodira C., Kretschmer M., Lappartient A., Leroch M., Levis C., Mauceli E., Neuvéglise C., Oeser B., Pearson M., Poulain J., Poussereau N., Quesneville H., Rascle C., Schumacher J., Ségurens B., Sexton A., Silva E., Sirven C., Soanes D.M., Talbot N.J., Templeton M., Yandava C., Yarden O., Zeng Q., Rollins J.A., Lebrun M.-H., Dickman M. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002230. e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Kan J.A.L., Stasse J.H.M., Mosbach A., Van Der Lee T.A.J., Faino L., Farmer A.D., Papasotiriou D.G., Zhou S., Seidl M.F., Cottam E., Edel D., Hahn M., Schartz D.C., Dietrich R.A., Widdison S., Scalliet G. A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 2017;18:75–89. doi: 10.1111/mpp.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel R.K., Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030619. e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anders S., Pyl P.T., Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vitulo N., Forcato C., Carpinelli E.C., Telatin A., Campagna D., D'Angelo M., Zimbello R., Corso M., Vannozzi A., Bonghi C., Lucchin M., Valle G. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014;14:99. doi: 10.1186/1471-2229-14-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kersey P.J., Lawson D., Birney E., Derwent P.S., Haimel M., Herrero J., Keenan S., Kerhornou A., Koscielny G., Kähäri A., Kinsella R.J., Kulesha E., Maheswari U., Megy K., Nuhn M., Proctor G., Staines D., Valentin F., Vilella A.J., Yates A. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;8(Database issue):D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.