Abstract
Background
Gastric cancer (GC) ranks the fifth most common cancer, and chemotherapy is one of the most common treatments for GC. However, chemoresistance limits the effectiveness of chemotherapy and leads to treatment failure. This study aims to investigate the biological effect of miR-567 on gastric tumourigenesis and chemoresistance and reveal the possible mechanism.
Methods
We measured the expression of miR-567 in 37 paired normal and stomach tumour specimens, as well as GC cell lines by Real-time PCR. The functional effects of miR-567 were validated using in vitro and in vivo assays. Dual-luciferase report assays and Chromatin immunoprecipitation (ChIP) assay were conducted for target evaluation, western blot assay was used to confirm the relationships.
Findings
Our data showed that miR-567 was downregulated in gastric tissues and gastric cancer cells compared with normal tissues and gastric epithelial cells. In vitro, Gain- and lose-of-function assays showed miR-567 not only weakened cells proliferative ability, but also sensitized GC cells to 5-FU and oxaliplatin. In vivo, miR-567 overexpression significantly repressed the tumourigenesis of GC cells compared with the vector control. Mechanistic analysis showed that PIK3AP1 activated AKT phosphorylation in GC. Meanwhile, miR-567 directly targeted PIK3AP1 to inactivate PI3K/AKT/c-Myc pathway and c-Myc inversely regulated miR-567 expression, thus forming a miR-567-PIK3AP1- PI3K/AKT-c-Myc feedback loop explaining the function of miR-567.
Interpretation
Our studies revealed that miR-567 acts as a tumour suppressor gene and suppresses GC tumorigenesis and chemoresistance via a miR-567-PIK3AP1- PI3K/AKT-c-Myc feedback loop. These results suggest that miR-567 may serve as a target for chemoresistance and a potential prognostic biomarker for GC.
Keywords: microRNA-567, Gastric cancer, Chemoresistance, Tumour growth, Prognostic biomarker
Abbreviations: miR-567, microRNA-567; miRNA, microRNA; GC, gastric cancer; FBS, fetal bovine serum; ChIP, Chromatin immunoprecipitation; IHC, immunohistochemistry; NC, negative control; PCR, polymerase chain reaction; siRNA, small-interfering RNAs; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; UTR, untranslated region; AKT, protein kinase B
Highlights
-
•
miR-567 sensitized GC cells to 5-FU and oxaliplatin, suggesting that it can be a target for chemoresistance.
-
•
miR-567 functions as a suppressor in GC progression and may serve as a novel prognostic and therapeutic biomarker for GC.
-
•
miR-567 directly targeted PIK3AP1 to inactivate PI3K/AKT/c-Myc and regulated its own expression.
-
•
These findings uncover a plausible mechanism for AKT/PI3K signalling activation in cancer progression.
Research in context.
Evidence before this study
miR-567 was previously reported to target FGF5 and inhibit cell proliferation, migration and invasion in osteosarcoma. Meanwhile, researches have revealed that miR-567 acted as a tumour suppressor gene and inhibited the carcinogenesis of breast cancer.
Added value of this study
In our study, miR-567 was markedly downregulated in tumour tissues and GC cells compared with normal tissues and gastric epithelial cells. Also, miR-567 not only significantly inhibited cell proliferation in vitro and delayed xenograft tumour growth in vivo, but also sensitized GC cells to 5-FU and oxaliplatin. Furthermore, mechanistic analyses showed that miR-567 involved in a miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop explaining the biological function of miR-567 in GC.
Implications of all the available evidence
miR-567 is demonstrated to be a novel suppressor in GC tumorigenesis and drug resistance for the first time and might present as a molecular biomarker for GC progression.
Alt-text: Unlabelled Box
1. Introduction
Gastric cancer (GC) is the fifth most incident and the third most common cause of cancer-related death in the world [1]. The development of GC is a complex multistep process, including numerous genetic and epigenetic changes. GC is also affected by environmental factors, such as helicobacter pylori (Hp) infection [2]. In the past few year, advances in surgical techniques, radiotherapy, chemical therapy and targeted molecular therapy have improved the prognosis of GC [3,4]. However, early diagnosis of GC is a significant challenge, and advanced or recurrent GC patients have poor survival rates because of chemotherapy resistance [[5], [6], [7], [8]]. Therefore, gastric cancer is a threat to global health and there is an urgent need for early diagnosis marker as well as solving the chemoresistance problem. Recently, a growing number of miRNAs have been proved to be closely correlated with GC [6], but the specific molecular mechanisms underlying the their function in cancer cells are still under investigation.
microRNAs (miRNAs or miRs), a class of small non-coding RNAs of 19–25 nucleotides in length, are believed to regulate gene expression by binding miRNAs to the 3′untranslated region (3′UTR) of mRNAs, thereby leading to mRNA degradation or blocking of mRNA translation [[9], [10], [11], [12]]. It has been estimated that over 30% of human genes are regulated by miRNAs [13,14]. Acting as tumour suppressor genes or oncogenes, miRNAs are involved in many cellular processes, such as differentiation, proliferation, metastasis and chemoresistance [[15], [16], [17], [18]]. For example, miR-424-5p, miR-125b, miR-21 and miR-17-92 have been recognized as regulators of GC cell tumourigenesis and drug resistance [[19], [20], [21], [22], [23]].
miR-567 was previously reported to target FGF5 and inhibit cell proliferation, migration and invasion in osteosarcoma [24]. Meanwhile, researches have relvealed that miR-567 acted as a tumour suppressor gene and inhibited the carcinogenesis of breast cancer [[25], [26], [27], [28]]. However, the biological role of miR-567 and its molecular mechanism underlying GC progression still remains unknown. In our study, we found miR-567 was obviously down-regulated in GC tissue and GC cells compared with adjacent non-cancer tissues and gastric epithelial cells, suggesting it playing a suppressive role in GC tumour progression. Indeed, we discovered that miR-567 inhibits GC cell proliferation and enhanced chemotherapeutic sensitization to 5-FU and oxaliplatin. In addition, our findings showed an atypical miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop, which inhibited GC cell proliferation and sensitized GC cells to 5-FU and oxaliplatin. Altogether, these results provide a miR-567-mediated mechanism to modulate GC cell growth and chemotherapy resistance. This pathway suppresses proliferation and sensitizes CRC cells to 5-FU and oxaliplatin. All in all, these results provide a mechanism by which miR-567 modulates CRC cell growth.
2. Materials and methods
2.1. Cell culture and treatment
A series of GC cell lines (GES-1, MKN45, BGC823, AGS, MGC803, BGC803, MKN28) were obtained from Foleibao Biotechnology Development (Shanghai, China). The cells were cultured in RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (NBCS) (PAA Laboratories, Inc., Pasching, Austria). All of these cell lines were incubated in a humidified chamber with 5% CO2 at 37 °C. For inhibitor treatment, 10 mmol/L PI3K inhibitor LY294002 (Cell Signal Technology, Danvers, MA) was added in the cultured cells every two days.
PIK3AP1 plasmids, miR-567 mimic, anti-miR-567 oligos and all siRNA oligos including PIK3AP1 and c-Myc specific siRNAs were purchased from GenePharma (Shanghai, China). GC cells at exponential growth phase were plated into 6-well plates for 24 h at a density of 0.5 × 105 cells/mL, and transfected with 1 mg of siRNA or 4 μg cDNA using Lipofectamine 2000 reagent for 24 h (Invitrogen; Carlsbad, Calif, USA) in reduced serum medium (OPTI-MEM-I; Invitrogen) according to the manufacturer's protocol.
2.2. Clinical samples
Fresh primary GC specimens with paired normal gastric tissues were obtained from the Tumour Tissue Bank of Nanfang Hospital. There are at least 1 cm space between the cancer tissues and the paired normal tissues. In each case, pathological diagnosis was made after elective surgery for GC in Nanfang Hospital during 2009 and 2014. All experiments performed are endorsed by the Ethics Committee of Southern Medical University and complied with the Declaration of Helsinki. No informed consent was required because data were going to be analysed anonymously.
2.3. Animals
All animal experiments were carried out with the approval of the Southern Medical University Animal Care and Use Committee in accordance with the guidelines for the ethical treatment of animals. Nude nu/nu mice were maintained in a barrier facility in racks filtered with high-efficiency particulate air filter. The animals were fed with an autoclaved laboratory rodent diet. The mice in this study were purchased from the Experimental Animal Centre of Southern Medical University, which is certified by the Guangdong Provincial Bureau of Science. All animal experiments involved ethical and humane treatment under a license from the Guangdong Provincial Bureau of Science.
2.4. Western blot analysis
Protein expression was assessed by immunoblot analysis of cell lysates (20–40 μg) in RIPA buffer in the presence of rabbit antibodies to GAPDH (1:1000; Santa Cruz, California, USA); rabbit antibody to PIK3AP1 (1:1000; Proteintech); rabbit antibodies to PI3K, p-PI3K, p-AKT(Ser473)(#4060), AKT(#4691) (1:1000; CST, Danvers, MA); mouse antibodies to CCND1(1:1000; Proteintech); rabbit antibodies to c-Myc (1:1000; Abclone). Relative protein abundance of phosphorate proteins was determined by normalisation with levels of corresponding total protein. Relative protein abundance of total protein was determined by normalisation with levels of corresponding endogenous control protein.
2.5. Statistical analysis
Data were analysed using SPSS version 19.0 software (SPSS; Chicago, USA). Statistical significance of difference between groups was determined by a two-tailed paired Student's t-test. Kaplan-Meier plots were performed to investigate the prognostic relevance of PIK3AP1 in univariate analysis. Statistical significance was established at P < .05.
3. Results
3.1. miR-567 is down-regulated in GC tissues and cell lines
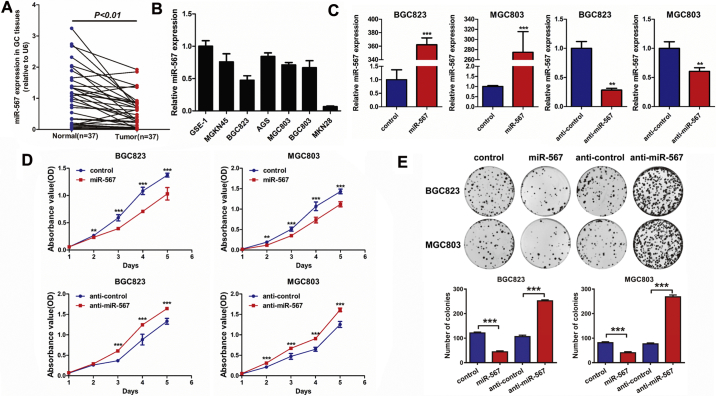
Real-time PCR was used to measure the expression of miR-567 in GC tissues and gastric cell lines. We detected a significant decrease of miR-567 mRNA expression in GC tissues compared with corresponding non-cancer tissues, which is at least 1 cm away from the cancer region (Fig. 1A). We further compared the miR-567 expression levels in GC cell lines with normal gastric epithelium cell line GES-1, which is non-malignant and none-tumourigenic cell lines [29,30]. A relatively lower mRNA expression was observed in GC cell lines (Fig. 1B).
Fig. 1.
miR-567 is down-regulated in GC tissues and cell lines, and inhibits GC cell proliferation and colony formation. (A) The expression level of miR-567 in GC tissues compared with corresponding non-cancer tissues. (B) Relative expression levels of miR-567 in gastric cancer cell lines and a normal gastric epithelial cell line, GES-1. (C) Real-time PCR was performed to detect the mRNA expression of miR-567 in MGC803 and BGC823 cells, both transfected with miR-567 mimic or anti-miR-567, Student's t-test, mean ± SD, **P < .01; ***P < .001. Cell Counting Kit-8 (CCK-8) assay (D) and colony formation assay (E) of GC cells were performed after transfected with miR-567 mimic or anti-miR-567, Student's t-test, mean ± SD, *P < .05; **P < .01; ***P < .001.
3.2. Exogenous miR-567 suppresses GC cell proliferation, colony formation, chemoresistance to 5-FU and oxaliplatin in vitro
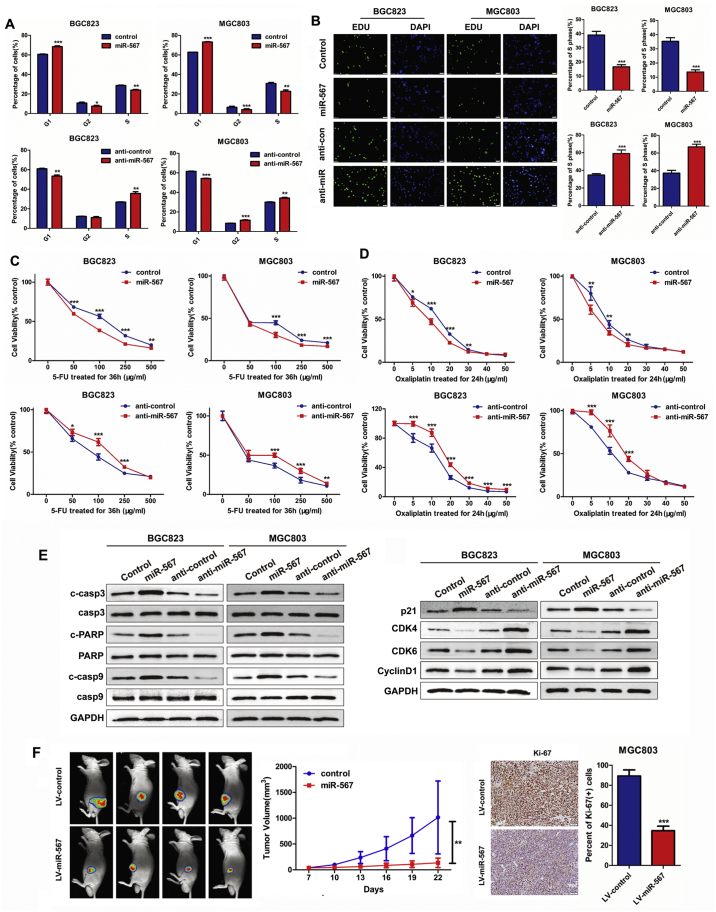
MGC803 and BGC823 cell lines were selected for further experiments as they have moderate endogenous expression levels of miR-567. We transfected miR-567 mimic oligonucleotides and anti-miR-567 into MGC803 and BGC823 cell lines to evaluate their effects on cellular behaviours. Real-time PCR was performed to detect the transfection efficiency (Fig. 1C). Subsequently, we examined the influence of miR-567 on GC cells using cell counting kit-8 (CCK-8) assay (Fig. 1D), colony formation assay (Fig. 1E), cell-cycle assay (Fig. 2A& S1) and EdU incorporation assay (Fig. 2B). The results showed that overexpressed miR-567 significantly suppressed cell growth and G1 to S cell cycle transition in MGC803 and BGC823 cells, which suggested that miR-567 inhibited cell proliferation by arresting the tumour cells at G1/G0 phase. By contrast, suppression of miR-567 markedly promoted cell proliferation and induced G1/S transition. GC cell lines overexpressing miR-567 exhibited significantly increased sensitivity to 5-FU and oxaliplatin, while inhibition of miR-567 expression caused the opposite effects (Fig. 2C & D). To further investigate the mechanism behind miR-567 suppression of chemoresistance and cell proliferation, we examined the protein expression of pro-apoptosis and cell cycle biomarkers in GC cells after miR-567 overexpression and knockdown, as shown in Fig. 2E, miR-567 increased the protein expression of cleave caspase-3, cleave caspase-9 and cleave PARP, suggesting miR-567 promoted chemosensitivity by inducing cell apoptosis. Furthermore, we found miR-567 expression was positively correlated with the protein expression of p21, while negatively correlated with the protein expression of CDK4, CDK6 and CyclinD1 in GC cells, indicating miR-567 may suppress GC cell proliferation by regulating the expression of cell cycle marker P21, CDK4, CDK6 and CyclinD1.
Fig. 2.
miR-567 suppresses GC cell proliferation and increases drug sensitivity in vitro and in vivo. FACS assays (A) and EdU incorporation assays (B) of GC cells were performed after transfected with miR-567 mimic or anti-miR-567, Student's t-test, mean ± SD, *P < .05; **P < .01; ***P < .001. (C&D) Dose-response curves of MGC803 and BGC823 treated with 5-FU for 36 h or oxaliplatin for 24 h, the cells were previously transfected with miR-NC, miR-567, anti-miR-NC or anti-miR-567. Parametric generalized linear model with random effects, Student's t-test, mean ± SD, *P < .05; **P < .01; ***P < .001. (E) Western blot experiments were used to analyse the expression of c-casp3, casp3, c-PARP, PARP, c-casp9, casp9, p21, CDK4, CDK6 and CyclinD1 after miR-567 knockdown and overexpression in MGC803 and BGC823 cells. (F) In vivo image detection of the xenograft tumour growth. Growth curve was measured and drawn, *P < .05. The tumour sections were under IHC staining using antibodies against Ki-67. Quantification of Ki67 staining of the xenograft tumours (right). ***P < .001.
3.3. Endogenous overexpression of miR-567 inhibits GC growth and progression in vivo
We used a lentivirus (LV)-based system to investigate the biological function of miR-567 in vivo. LV-miR-567 was used to infect MGC803 cells and establish GC cell lines with stable miR-567 overexpression. Real-time PCR assay confirmed a remarkable increase of miR-567 expression in LV-miR-567-transfected cells compared with LV-miR-NC-transfected cells (Fig. S2; P < .05). We used LV-miR-567-overexpressing MGC803 (MGC803/LV-miR-567) cells and control cells to perform a tumourigenesis assay in nude mice. Tumours in the MGC803/LV-miR-567 group grew more slowly than those in the MGC803/LV-NC group (Fig. 2F, left panel). Moreover, tumour in the MGC803/LV-miR-567 group grew more quickly than those in control group (P < .01). Immunohistochemistry (IHC) confirmed that the tumours of MGC803/LV-miR-567 group also showed significantly lower Ki-67 index compared with control group, suggesting that miR-567 overexpression markedly decreased cell proliferation (Fig. 2F, right panel).
3.4. PIK3AP1 is the direct target of miR-567
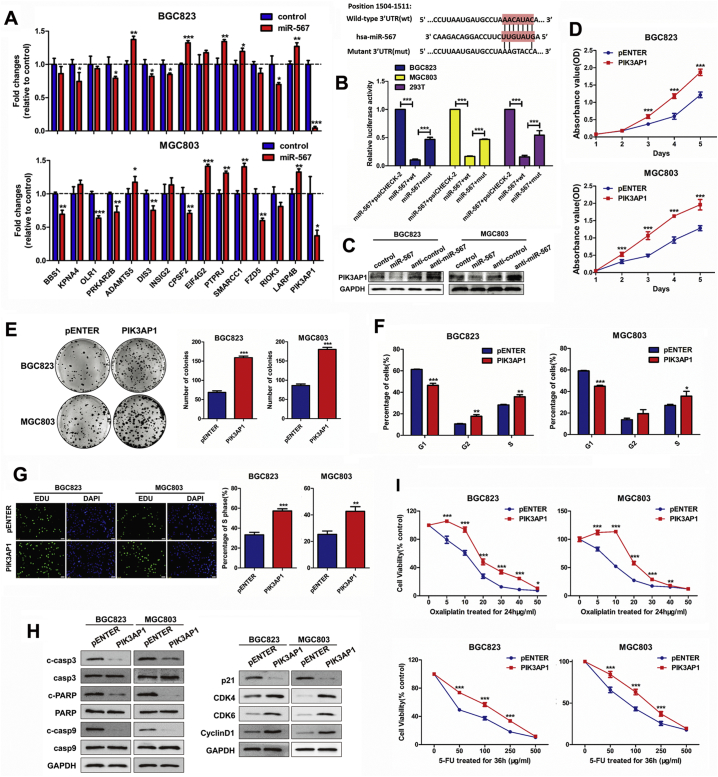
In order to identify the potential target gene of miR-567, we performed a bioinformatic analysis using eight databases (miRWalk, Mircot4, miRanda, miRDB, miRMap, PITA, RNAhybrid and Targetscan). After selecting the cancer-related genes that have not been reported in gastric cancer, we screened out 15 candidate genes from the common target genes predicted in these databases. Subsequently, real-time PCR assay showed that PIK3AP1 showed the most significant fold decrease among these genes after miR-567 overexpression (Fig. 3A). Therefore, PIK3AP1 were identified as a potential target of miR-567.
Fig. 3.
PIK3AP1 is the direct target of miR-567 and promotes GC cell proliferation and increases drug sensitivity. (A) Real-time PCR analysis were performed to detect the mRNA expression of candidate genes in MGC803 and BGC823 cells transfected with miR-567 mimic, Student's t-test, mean ± SD. (B) miR-567 and its putative binding sequences in the 3′UTR of PIK3AP1. A mutation was generated in the complementary site that bound to the seed region of miR-567. Luciferase reporter assay was used to determine miR-567 direct targeting the PIK3AP1 3′UTR, Student's t-test, mean ± SD, ***P < .001. (B) Western blot analysis were performed to detect the protein expression of PIK3AP1 in MGC803 and BGC823 cells, both transfected with miR-567 mimic or anti-miR-567. CCK-8 assay (C), colony formation assay (D), FACS assays (E) and EdU incorporation assays (F) of GC cells were performed after transfected with PIK3AP1 or pENTER vector, Student's t-test, mean ± SD, *P < .05; **P < .01. (H) Western blot experiments were used to analyse the expression of pro-apoptosis proteins, cell cycle maker and related proteins in PI3K/AKT pathway after miR-567 knockdown and overexpression in MGC803 and BGC823 cells. (I) Dose-response curves of MGC803 and BGC823 treated with 5-FU for 36 h or oxaliplatin for 24 h, the cells were previously transfected with PIK3AP1 and pENTER. Parametric generalized linear model with random effects, Student's t-test, mean ± SD, *P < .05; **P < .01; ***P < .001.
The analysis showed that miR-567 targeted PIK3AP1 by binding to the 3′-UTR region of PIK3AP1 (Fig. 3B above). Subsequently, dual-luciferase reporter assays were performed to determine whether miR-567 could directly target the 3′UTR region of PIK3AP1. 3′UTR fragment (wt) of PIK3AP1 containing miR-567 binding site and their mutant fragments (mut) were cloned into luciferase report vectors psiCHECK-2. The empty psiCHECK-2, wt or mut 3′UTR vector and miR-567 were co-transfected into MGC803, BGC823 and 293 T cell lines. The result showed that miR-567 markedly attenuated the luciferase activity of wide-type PIK3AP1 3′-UTR, whereas the effect was abrogated after the 3′-UTR binding site of PIK3AP1 was mutated (Fig. 3B below). Moreover, western blot assay showed that both PIK3AP1 protein expression decreased in miR-567-overexpressing GC cells but increased after miR-567 inhibition (Fig. 3C & S3). These findings reveal the potential significance of miR-567 as a determinant of PIK3AP1 expression in GC cells.
3.5. PIK3AP1 promotes GC cell proliferation and chemosensitivity
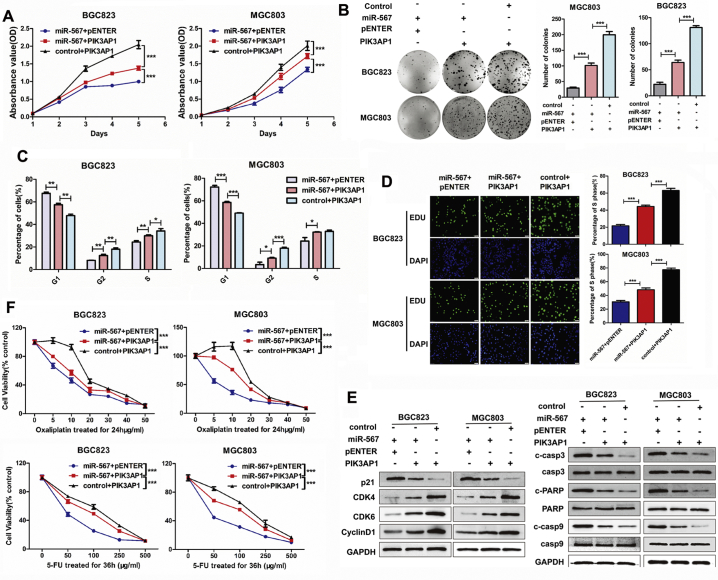
Given that miR-567 targeted PIK3AP1 and suppressed its expression, we investigated the biological function of PIK3AP1 in GC cells. Data from Kaplan-Meier plotter database were utilized to visualize the association between PIK3AP1 expression and overall survival in patients with GC. The results showed patients with high PIK3AP1 expression in tumours had a trend towards poorer survival when compared with patients showed low PIK3AP1 expression (Fig.S4; HR = 1.3, P = .018). CCK8 assay (Fig. 3D), colony formation assay (Fig. 3E), cell-cycle analysis (Fig. 3F & S5) and EdU incorporation assays (Fig. 3G) showed that overexpression of PIK3AP1 significantly enhanced the proliferative ability of MGC803 and BGC823 cells and increased the cells in S phase and G2 phase. Furthermore, PIK3AP1 overexpression induced chemoresistance to 5-FU and oxaliplatin in MGC803 and BGC823 cells (Fig. 3I). As shown in Fig. 4H, western blot assay showed PIK3AP1 decreased the protein expression of cleave caspase-3, cleave caspase-9 and cleave PARP, suggesting PIK3AP1 suppressed chemosensitivity by prevent cell apoptosis. Moreover, PIK3AP1 expression was negatively correlated with the protein expression of p21, while positively correlated with the protein expression of CDK4, CDK6 and CyclinD1 in GC cells, suggesting miR-567 promoted GC cell proliferation by mediated the expression of cell cycle markers.
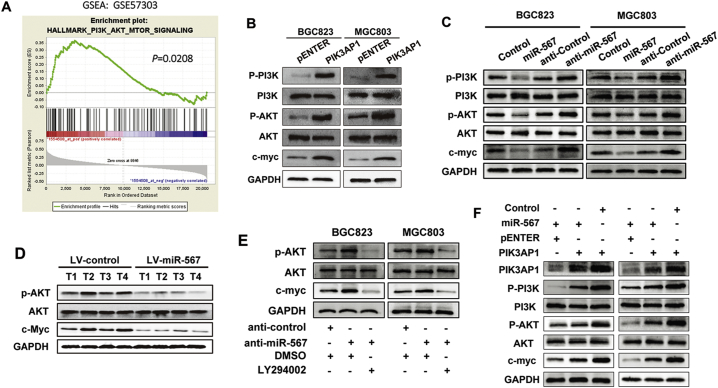
Fig. 4.
miR-567 negatively regulates with PI3K/AKT-c-Myc signalling in GC. (A) GSEA demonstrated enrichment of PI3K-AKT-mTOR signalling pathway in high PIK3AP1 expression GC group. (B) Western blot experiments were used to analyse the expression of relevant proteins in PI3K/AKT signal pathway after PIK3AP1 overexpression in MGC803 and BGC823 cells. (C) Western blot experiments were used to analyse the expression of relevant proteins in PI3K/AKT signal pathway after miR-567 knockdown and overexpression in MGC803 and BGC823 cells. (D) Western blot experiments were used to analyse the expression of relevant proteins in PI3K/AKT signal pathway in LV-miR-567-transfected cells and LV-miR-NC-transfected cells. (E) The upregulation of p-AKT and c-Myc induced by anti-miR-567 were abrogated after administration of LY294002 in MGC803 and BGC823 cells. (F) The downregulation of p-AKT and c-Myc induced by introduction of PIK3AP1 were abrogated after administration of miR-567 in MGC803 and BGC823 cells.
3.6. miR-567 negatively regulates PI3K/AKT-c-Myc pathway in GC cells
To identify the oncogenic signalling related to PIK3AP1, we applied GSEA to GC cases (GSE57303). The result showed that PIK3AP1 was positively associated with PI3K/AKT signalling in the GC cohort (Fig. 4A; P = .0208). As expected, western blot assay showed PIK3AP1 significantly increase phosphorylation levels of p-Akt, p-PI3K and PI3K/AKT downstream target c-Myc, indicating that PIK3AP1 regulated PI3K/AKT signalling pathway (Fig. 4B). Thus, we speculated that miR-567 is an upstream regulator of PI3K/AKT signalling. Indeed, Western blot assay showed low phosphorylation levels of p-Akt and p-PI3K, with no change in the total protein amount of Akt and PI3K, and down-regulation of c-Myc in GC cells treated with miR-567 mimic compared with control cells. Also, we observed opposite results of p-Akt, p-PI3K and c-Myc expression in GC cells treated with anti-miR-567 (Fig. 4C). Western blot results confirmed that miR-567 also regulated PI3K/AKT-c-Myc pathway in tumours from MGC803/LV-miR-567 cells (Fig. 4D). Furthermore, treatment with LY294002 was sufficient to neutralize the strengthened role of anti-miR-567 in PI3K/AKT-c-Myc pathway (Fig. 4E). In contrast, introduction of PIK3AP1 could rescue to activate the suppressed pathway (Fig. 4F). These results suggested an inhibitory role of miR-567 in GC proliferation, at least in part, by inhibiting PI3K/AKT-c-Myc pathway.
3.7. PIK3AP1 is essential to miR-567-mediated suppression of GC cell behaviour and oncogenic signalling
Rescue experiments showed that transiently transfecting PIK3AP1 into miR-567-overexpressing GC cells significantly restored miR-567-mediated suppression of proliferation and promoted G1/S and S/G2 cell cycle transition (Fig. 5A–D & S6). Furthermore, we observed that PIK3AP1 overexpression rescued miR-567-mediated suppression of CDK4, CDK6, and CyclinD1, but not p21 (Fig. 5E). Meanwhile, CCK-8 aasay showed that efficiently attenuated miR-567-induced sensitivity to 5-FU and oxaliplatin (Fig. 5F). Consistent with these result, we observed PIK3AP1 markedly decreased miR-567-induced apoptosis and reduced the protein expression of cleave caspase-3, cleave caspase-9 and cleave PARP (Fig. 5E). These findings indicate that PIK3AP1 overcomes cell proliferation suppression and drug sensitivity induced by miR-567.
Fig. 5.
miR-567 suppresses the PI3K/AKT-c-Myc pathway via targeting PIK3AP1. CCK-8 assay (A), colony formation assay (B), FACS assays (C) and EdU incorporation assays (D) showed miR-567-inhibited cell proliferation was counteracted after administration of PIK3AP1, Student's t-test, mean ± SD, *P < .05; **P < .01; ***P < .001. (E) The miR-567-induced downregulation of relevant proteins in cell cycle marker (CDK4, CDK6 and CyclinD1) as well as upregulation of pro-apoptosis proteins and cell cycle marker p21 were abrogated after administration of PIK3AP1 in MGC803 and BGC823 cells. (F) PIK3AP1 overexpression reversed miR-567-induced cell sensitivity to 5-FU compared to control cells. Student's t-test, mean ± SD, ***P < .001.
3.8. c-Myc inhibits miR-567 expression by binding to its promoter region
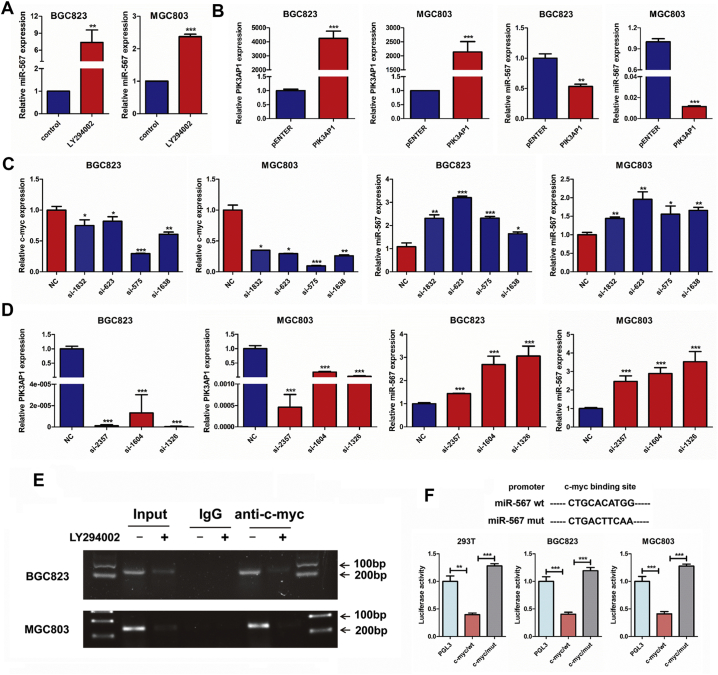
To examine whether miR-567 was inversely regulated by c-Myc, we used small-interfering RNAs (siRNA) to suppress c-Myc expression in MGC803 and BGC823 cells. Real-time PCR analysis showed increasing miR-567 expression after c-Myc knock-down, indicating that c-Myc is an upstream regulator of miR-567 (Fig. 6C). Moreover, Subsequent real-time PCR analysis showed that LY294002 and PIK3AP1 siRNAs significantly increased miR-567 expression (Fig. 6A & D), while PIK3AP1 overexpression reduced miR-567 expression (Fig. 6B), indicating PIK3AP1 and PI3K/AKT pathway can inversely regulated miR-567 expression. Taken together, we discover a mechanism that miR-567 expression is suppressed by c-Myc, PI3K/AKT pathway and PIK3AP1, thereby forming a complex miR-567-PIK3AP1-PI3K/AKT-c-Myc regulatory feedback loop.
Fig. 6.
c-Myc inhibits miR-567 by binding to its promoter region. (A–D) Real-time PCR was performed to detect the mRNA expression of miR-567 in MGC803 and BGC823 cells treated with LY294002, PIK3AP1, c-Myc siRNAs or PIK3AP1 siRNAs, Student's t-test, mean ± SD, *P < .05; **P < .01; ***P < .001. (E) PCR gel showing amplification of c-Myc-binding sites after ChIP using antibody against c-Myc. The gel figures were accompanied by the locations of molecular weight markers. (F) Schematic representation of the structure of c-Myc TFBS mutant and c-Myc wild-type (WT) that bound to the promoter regions of miR-567. Relative luciferase activity of the indicated promoter vectors in 293 T, MGC803 and BGC823 cells transfected with c-Myc plasmids, Student's t-test, mean ± SD, **P < .01; ***P < .001.
Chromatin immunoprecipitation (ChIP) assay was further used to determine whether c-Myc bound to the miR-567 promoter in MGC803 and BGC823 cells. Indeed, DNA from the immunoprecipitated chromatin showed an obvious enrichment of this specific region compared with negative control (IgG) pulldown, and the enrichment as abrogated when cell were treated with LY294002 (Fig. 6E). To investigate the transcriptional regulatory mechanism of miR-567 expression, we used JASPAR (http://jaspar.genereg.net) to analyse a 2-kb region upstream of the miR-567. One putative c-Myc-binding site (from 1504 to 1511) was predicted in human miR-567 promoter region (Fig. 6F). A reduction of the wild-type miR-567 promoter luciferase activity was observed on upregulation of c-Myc in 293 T, BGC823 and MGC803 cell lines (Fig. 6F; P < .001). Our data indicated c-Myc was involved in miR-567 transcription via binding to its promoter, and this process is negatively regulated by LY294002 which decrease c-Myc expression by suppressing PI3K/AKT pathway. Therefore, as the upstream regulators of c-Myc, AKT signalling and PIK3AP1 can inversely regulate miR-567 expression.
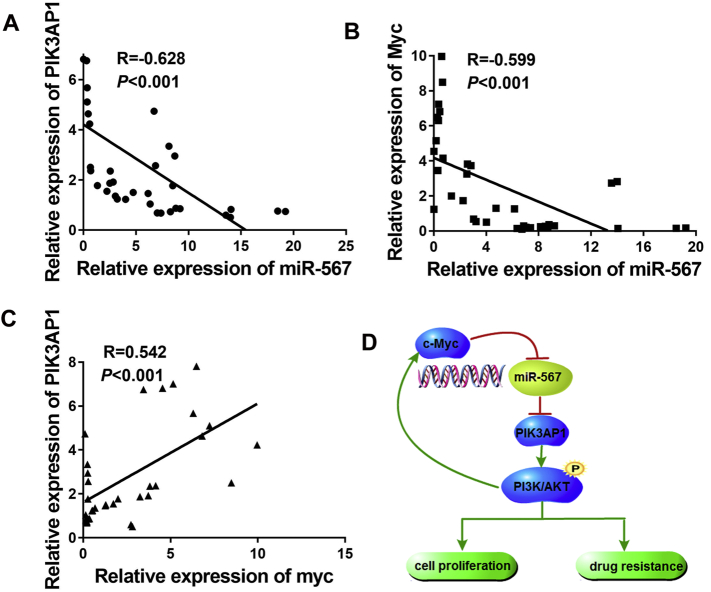
Finally, in order to prove our conclusion with more persuasive evidence, we conducted a real-time PCR assay, detecting the mRNA expression of miR-567, PIK3AP1 and c-Myc in 37 GC tissues and 37 pared adjacent normal tissue. Analysis of the results showed that miR-567 expression is negatively correlated with PIK3AP1 and c-Myc expression (Fig. 7A & B), but PIK3AP1 is positively correlated with c-Myc expression (Fig. 7C). Thus, the relationship among miR-567, PIK3AP1 and c-Myc is clearly identified.
Fig. 7.
Schematic representation of overall summary. (A) Real-time PCR assay were performed to detect the mRNA expression of miR-567 and PIK3AP1 in GC tissues. (B) Real-time PCR assay were performed to detect the mRNA expression of miR-567 and c-Myc in GC tissues. (C) Real-time PCR assay were performed to detect the mRNA expression of PIK3AP1 and c-Myc in GC tissues. (D) A schematic for an atypical miR-567-PIK3AP1–PI3K/AKT-c-Myc feedback loop.
4. Discussion
Although cancer cell proliferation and chemoresistance are the overwhelming causes of cancer mortality, a comprehensive picture of modular and cellular determinants governing these processes remains largely unknown. Multiple lines of evidence have proven that abnormal expression of miRNAs are linked to cancers tumourigenesis and drug resistance [32,33]. In our study, miR-567 was found be markedly downregulated in tumour tissues and GC cells compared with normal tissues and gastric epithelial cells. Subsequent experiments showed that miR-567 not only significantly inhibited cell proliferation in vitro and delayed xenograft tumour growth in vivo, but also sensitized GC cells to 5-FU and oxaliplatin via a miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop. In short, our study firstly demonstrates that miR-567 is a novel suppressor gene in GC tumourigenesis and drug resistance and might present as a molecular biomarker for GC progression.
As a downstream target of miR-567 indicated in our study, PIK3AP1 is essential for miR-567-mediated suppression of GC cell behaviour and oncogenic signalling. PIK3AP1 is an adapter protein originally isolated from B cells. After tyrosine-phosphorylated on its four YxxM, PIK3AP1 binds and recruits PI3K to the membrane upon B-cell receptor (BCR) oligormerization to facilitate generation of PIP3 from PIP2, this process is essential for BCR-induced AKT phosphorylation [31,32]. In natural killer (NK) cells, PIK3AP1 plays a similar role in immunoreceptor tyrosine-based activation motif (ITAM)-mediated AKT phosphorylation [33]. These studies suggest PIK3AP1 is the upstream regulator of PI3K/AKT pathway, which is consistent with the GSEA analysis and experimental result in our study. In Fig. 3A, although BBS1, OLR1, PRKAR2B, DIS3, CPSF2 and FZD5 also showed different fold decrease after miR-567 overexpression, PIK3AP1 displayed the most significant fold decrease compared with decrease of other gene. Moreover, previous study and GSEA analysis suggested PIK3AP1 was associated with PI3K/AKT pathway, which was crucial for cell proliferation, metabolism and survival [34,35]. Thus, we speculated that PIK3AP1 played an important role in miR-567-mediated GC tumourigenesis and chemoresistance, and chose PIK3AP1 as the potential target of miR-567. Indeed, subsequent experiments proved that PIK3AP1 was essential to miR-567-mediated suppression of GC tumourigenesis and drug resistance.
In our study, c-Myc inhibited miR-567 expression by binding to its promoter region, thus formed a miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop, by which miR-567 suppressed GC tumourigenesis and drug resistance. c-Myc is an oncogenic transcription factor playing a pivotal role in the control of cell proliferation, apoptosis and drug resistance [[36], [37], [38]]. Mutated c-Myc is observed in many cancers and resulted in persistent expression of c-Myc protein, which causes abnormal expression of many genes. A number of candidate c-Myc target genes regulate cell energy metabolism, cell cycle progression (particular in G1 phase) and chemoresistance [37,38]. Meanwhile, c-Myc has been reported to promote drug resistance to 5-Fu and oxaliplatin in colon cancer stem cells (CSCs) via regulating the expression of ATP-binding cassette transporters [38], suggesting its role in chemoresistance to 5-Fu and oxaliplatin in gastric cancer. Indeed, our study showed that the miR-567-PIK3AP1-PI3K/AKT-c-Myc pathway involving c-Myc is closely associated with resistance to 5-Fu and oxaliplatin. Moreover, AKT activation is associated with the activation of c-Myc expression. The glycogen synthetase kinase 3(GSK3) and forkhead transcription factor FKHRL1, both the downstream target of AKT signalling pathway, contribute to the upregulation of c-Myc expression [39]. Therefore, blocking PI3K/AKT pathway will lead the reduction of c-Myc expression. As expected, we observed a markedly decrease of c-Myc protein expression in MGC803 and BGC823 cells after treatment with LY294002 (an inhibitor of the PI3K/AKT signalling pathway). In all, these results provide a mechanism explaining the association between c-Myc and GC tumourigenesis and drug resistance, and emphasize the important role of c-Myc in GC progression.
Studies have shown that a single microRNA may contain multiple transcription factor binding sites, thus being regulating by different transcription factors [40,41]. In our study, miR-567 expression level between different pairs of c-Myc siRNAs or PIK3AP1 siRNAs was not consistent with their knock-down efficiency, for example, si-575 (c-Myc siRNAs) got the best knock-down efficiency but the level of miR-567 do not increase a lot especially in MGC803 cell. This phenomenon confused us and we speculated it was probably because of the off-target effects of siRNA. For example, si-575 might not target c-Myc specifically and reduce the expression of other genes at the same time, which negatively regulated miR-567 expression, thus resulting in the unexpected increase results of miR-567 expression. Nevertheless, in our study, the c-Myc siRNAs and PIK3AP1 siRNAs showed overall efficient knockdown effects on miR-567 expression, proving that c-Myc and PIK3AP1 can inversely regulate miR-567 expression.
5. Conclusions
As summarized in our working model in Fig. 7D, miR-567 inhibits GC cell proliferation and enhances chemotherapeutic sensitization to 5-FU and oxaliplatin via a positive feedback loop. As an upstream regulator of PIK3AP1, miR-567 blocks PI3K/AKT pathway by directly targeting PIK3AP1. c-Myc is regulated by AKT signalling and binds to the promoter region of miR-567, thus leading to transcription arrest of miR-567. Therefore, we find a novel miR-567-PIK3AP1-PI3K/AKT-c-Myc feedback loop, which explains the mechanism underlying miR-567-mediated tumourigensis and chemotherapy resistance. Taken together, our study suggests that miR-567 might act as a biomarker and molecular target for the prevention of GC progression.
Acknowledgments
Acknowledgements
Not applicable.
Ethical approval and consent to participate
All experiments performed are endorsed by the Ethics Committee of Southern Medical University and complied with the Declaration of Helsinki. No informed consent was required because data were going to be analysed anonymously. All animal experiments were carried out with the approval of the Southern Medical University Animal Care and Use Committee in accordance with the guidelines for the ethical treatment of animals. All animal experiments involved ethical and humane treatment under a license from the Guangdong Provincial Bureau of Science.
Consent for publication
All authors consent for publication.
Availability of supporting data
Availability in supplementary data.
Funding sources
This work was supported by the National Natural Science Foundation of China (Nos. 81572813, 81773082, 81702903, 81872423), Guangdong Natural Science Foundation (2017A030310038,2018B030311036), Fork Ying Tung Education Foundation (161035), Higher Education Fund Project of Guangzhou (2012C070) and Special Funds for the Cultivation of Guangdong College Students' Scientific and Technological Innovation. (“Climbing Program” Special Funds) (pdjhb0102). The funding institutions had no role in the study design, data collection, data analysis, interpretation or writing of the manuscript.
Declaration of interests
The authors have declared that no conflict of interest exists.
Author contributions
L. Z. led study design and prepared the manuscript; F. -F. Z, K. -T. L and X. -Q. Y carried out the experiments; H. W and J. W performed statistical analysis; W. -D. L assisted in tissue sample collection; R. Z performed data analysis and interpretation; L. -J. X provided data collection. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.05.003.
Appendix A. Supplementary data
Supplementary material
References
- 1.Figueiredo C., Camargo M.C., Leite M., Fuentes-Panana E.M., Rabkin C.S., Machado J.C. Pathogenesis of gastric cancer: genetics and molecular classification. Curr Top Microbiol Immunol. 2017;400:277–304. doi: 10.1007/978-3-319-50520-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leal M., Wisnieski F., de Oliveira Gigek C. What gastric cancer proteomic studies show about gastric carcinogenesis? Tumour Biol. 2016;37(8):9991–10010. doi: 10.1007/s13277-016-5043-9. [DOI] [PubMed] [Google Scholar]
- 3.Newton A., Datta J., Loaiza-Bonilla A., Karakousis G., Roses R. Neoadjuvant therapy for gastric cancer: current evidence and future directions. J Gastrointest Oncol. 2015;6(5):534–543. doi: 10.3978/j.issn.2078-6891.2015.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X., Wang Y., Qiu M. Postoperative chemoradiotherapy in gastric cancer: a phase I study of radiotherapy with dose escalation of oxaliplatin, 5-fluorouracil, and leucovorin (FOLFOX regimen) Med Oncol. 2011;28(Suppl. 1):S274–S279. doi: 10.1007/s12032-010-9741-7. [DOI] [PubMed] [Google Scholar]
- 5.Hu M., Li K., Maskey N. 15-PGDH expression as a predictive factor response to neoadjuvant chemotherapy in advanced gastric cancer. Int J Clin Exp Pathol. 2015;8(6):6910–6918. [PMC free article] [PubMed] [Google Scholar]
- 6.Shekari N., Baradaran B., Shanehbandi D., Kazemi T. Circulating microRNAs: valuable biomarkers for diagnosis and prognosis of gastric cancer. Curr Med Chem. 2018;25(6):698–714. doi: 10.2174/0929867324666171003123425. [DOI] [PubMed] [Google Scholar]
- 7.Park J., Kim S., Kim J. Risk factors for early metachronous tumor development after endoscopic resection for early gastric cancer. PLoS One. 2017;12(9) doi: 10.1371/journal.pone.0185501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi W., Gao J. Molecular mechanisms of chemoresistance in gastric cancer. World J Gastrointest Oncol. 2016;8(9):673–681. doi: 10.4251/wjgo.v8.i9.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y., Shen X., Tang T., Wu C. Weak regulation of many targets is cumulatively powerful - an evolutionary perspective on microRNA functionality. Mol Biol Evol. 2017;34(12):3041–3046. doi: 10.1093/molbev/msx260. [DOI] [PubMed] [Google Scholar]
- 10.Alamoudi A., Alnoury A., Gad H. miRNA in tumour metabolism and why could it be the preferred pathway for energy reprograming. Brief Funct Genomics. 2018;17(3):157–169. doi: 10.1093/bfgp/elx023. [DOI] [PubMed] [Google Scholar]
- 11.Murphy C., Singewald N. Potential of microRNAs as novel targets in the alleviation of pathological fear. Genes Brain Behav. 2018;17(3):e12427. doi: 10.1111/gbb.12427. [DOI] [PubMed] [Google Scholar]
- 12.Djami-Tchatchou A., Ntushelo K. Expression profile of stress-responsive Arabidopsis thaliana miRNAs and their target genes in response to inoculation with Pectobacterium carotovorum subsp. carotovorum. Pak J Biol Sci. 2017;20(3):147–153. doi: 10.3923/pjbs.2017.147.153. [DOI] [PubMed] [Google Scholar]
- 13.Filipowicz W., Bhattacharyya S., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 14.Friedman J., Jones P. MicroRNAs: critical mediators of differentiation, development and disease. Swiss Med Wkly. 2009;139(33–34):466–472. doi: 10.4414/smw.2009.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brás-Rosário L., Matsuda A., Pinheiro A. Expression profile of microRNAs regulating proliferation and differentiation in mouse adult cardiac stem cells. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0063041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukita S., Yamada T., Takahashi K. MicroRNAs 106b and 222 improve hyperglycemia in a mouse model of insulin-deficient diabetes via pancreatic β-cell proliferation. EBioMedicine. 2017;15:163–172. doi: 10.1016/j.ebiom.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palma Flores C., García-Vázquez R., Gallardo Rincón D. MicroRNAs driving invasion and metastasis in ovarian cancer: opportunities for translational medicine (review) Int J Oncol. 2017;50(5):1461–1476. doi: 10.3892/ijo.2017.3948. [DOI] [PubMed] [Google Scholar]
- 18.Cai J., Fang L., Huang Y. Simultaneous overactivation of Wnt/β-catenin and TGFβ signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat Commun. 2017;8 doi: 10.1038/ncomms15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J., Liu H., Hou L. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei C., Du F., Sun L. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017;8(10) doi: 10.1038/cddis.2017.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J., Wang J., Jiang X. MiR-125b promotes cell migration and invasion by targeting PPP1CA-Rb signal pathways in gastric cancer, resulting in a poor prognosis. Gastric Cancer. 2015;18(4):729–739. doi: 10.1007/s10120-014-0421-8. [DOI] [PubMed] [Google Scholar]
- 22.Zheng P., Chen L., Yuan X. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. 2017;36(1):53. doi: 10.1186/s13046-017-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cioffi M., Trabulo S.M., Sanchez-Ripoll Y. The miR-17-92 cluster counteracts quiescence and chemoresistance in a distinct subpopulation of pancreatic cancer stem cells. Gut. 2015;64(12):1936–1948. doi: 10.1136/gutjnl-2014-308470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D., Zhang C., Li X., Zhang H., Pang Q., Wan A. MicroRNA-567 inhibits cell proliferation, migration and invasion by targeting FGF5 in osteosarcoma. EXCLI J. 2018;17:102–112. doi: 10.17179/excli2017-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shao Y., Liang B., Long F., Jiang S. Diagnostic MicroRNA biomarker discovery for non-small-cell lung Cancer adenocarcinoma by integrative bioinformatics analysis. Biomed Res Int. 2017;2017 doi: 10.1155/2017/2563085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F., Wong S., Chan L., Cho W., Yip S., Yung B. Multiple regression analysis of mRNA-miRNA associations in colorectal cancer pathway. Biomed Res Int. 2014;2014:676724. doi: 10.1155/2014/676724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cava C., Bertoli G., Ripamonti M. Integration of mRNA expression profile, copy number alterations, and microRNA expression levels in breast cancer to improve grade definition. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0097681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertoli G., Cava C., Diceglie C. MicroRNA-567 dysregulation contributes to carcinogenesis of breast cancer, targeting tumor cell proliferation, and migration. Breast Cancer Res Treat. 2017;161(3):605–616. doi: 10.1007/s10549-016-4079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He X., Yang J., Ding Y., Liu W., Shen Q., Xia H. Increased epithelial and serum expression of macrophage migration inhibitory factor (MIF) in gastric cancer: potential role of MIF in gastric carcinogenesis. Gut. 2006;55(6):797–802. doi: 10.1136/gut.2005.078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z., Ji Z., Wang Y. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology. 2014;147(5):1043–1054. doi: 10.1053/j.gastro.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Okada T., Maeda A., Iwamatsu A., Gotoh K., Kurosaki T. BCAP: the tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation. Immunity. 2000;13(6):817–827. doi: 10.1016/s1074-7613(00)00079-0. [DOI] [PubMed] [Google Scholar]
- 32.Pongas G., Cheson B. PI3K signaling pathway in normal B cells and indolent B-cell malignancies. Semin Oncol. 2016;43(6):647–654. doi: 10.1053/j.seminoncol.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 33.MacFarlane A., Yamazaki T., Fang M., Sigal L., Kurosaki T., Campbell K. Enhanced NK-cell development and function in BCAP-deficient mice. Blood. 2008;112(1):131–140. doi: 10.1182/blood-2007-08-107847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C., Liang X., Wang J. Protein O-fucosyltransferase 1 promotes trophoblast cell proliferation through activation of MAPK and PI3K/Akt signaling pathways. Biomed Pharmacother. 2017;88:95–101. doi: 10.1016/j.biopha.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 35.Hauge M., Bruserud Ø., Hatfield K. Targeting of cell metabolism in human acute myeloid leukemia--more than targeting of isocitrate dehydrogenase mutations and PI3K/AKT/mTOR signaling? Eur J Haematol. 2016;96(3):211–221. doi: 10.1111/ejh.12690. [DOI] [PubMed] [Google Scholar]
- 36.Griffin N., Sharma G., Zhao X. ADA3 regulates normal and tumor mammary epithelial cell proliferation through c-MYC. Breast Cancer Res. 2016;18(1):113. doi: 10.1186/s13058-016-0770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciribilli Y., Singh P., Spanel R., Inga A., Borlak J. Decoding c-Myc networks of cell cycle and apoptosis regulated genes in a transgenic mouse model of papillary lung adenocarcinomas. Oncotarget. 2015;6(31):31569–31592. doi: 10.18632/oncotarget.5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., Wang P., Lu M., Zhang S. C-Myc regulation of ATP-binding cassette transporter reverses chemoresistance in CD133(+) colon cancer stem cells. Sheng Li Xue Bao. 2016;68(2):171–178. [PubMed] [Google Scholar]
- 39.Domínguez-Cáceres M., García-Martínez J., Calcabrini A. Prolactin induces c-Myc expression and cell survival through activation of Src/Akt pathway in lymphoid cells. Oncogene. 2004;23(44):7378–7390. doi: 10.1038/sj.onc.1208002. [DOI] [PubMed] [Google Scholar]
- 40.Su N., Wang Y., Qian M., Deng M. Combinatorial regulation of transcription factors and microRNAs. BMC Syst Biol. 2010;4:150. doi: 10.1186/1752-0509-4-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo J., Xiang G., Pan C. Discovery of microRNAs and transcription factors co-regulatory modules by integrating multiple types of genomic data. IEEE Trans Nanobioscience. 2017;16(1):51–59. doi: 10.1109/TNB.2017.2649560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Availability in supplementary data.