Abstract
Background
Triple negative breast cancers (TNBCs) have a poor prognosis and are not amenable to endocrine- or HER2-targeted therapies. The malignant and invasive feature of TNBCs is correlated with its high cancer stem cell population. Recent results from us and others have unveiled an oncogenic role for the PRMT5-KLF4 axis in regulating tumor progression by orchestrating the stemness in mammary tumor cell as well as genome stability. Methylation of KLF4 by PRMT5 leads to KLF4 stabilization, resulting in promoting mitogenesis.
Methods
We have developed a small molecule inhibitor, WX2–43, that specifically intercepts the interaction between PRMT5 and KLF4, thereby enhancing KLF4 degradation.
Findings
Results from our characterization demonstrate that WX2–43 binds to the region between amino acids L400-M500 on PRMT5. Degradation of KLF4 down-regulates KLF4-mediated genes transcription. We have characterized the potent effect for WX2–43 in inhibiting PRMT5-KLF4 binding that, in turns, suppresses tumor progression and induces tumor cell death in both TNBC cultured-cell and animal models.
Interpretation
WX2–43-mediated inhibition of KLF4 methylation by PRMT5 could be a potential strategy for anti-TNBC treatment.
Fund
This work was supported, in whole or in part, by National Institutes of Health grants CA202963 and CA202948 (Wan), R21HL109654 (Xie), P30DA035778 (Xie and Bahar) and P41GM103712 (Bahar).
Keywords: WX2-43, PRMT5, KLF4, Methylation and breast cancer
Research in context.
Evidence before this study
Triple negative breast cancer (TNBCs) has poor prognosis and is not amenable to endocrine or targeted therapies. One of the common features for TNBC is its increased cancer stem cell population. KLF4 was originally characterized as a critical stem cell factor but its impact in orchestrating cancer stem cell expansion remains unclear. We previously identified PRMT5 as a critical upstream regulator for KLF4, although if PRMT5-KLF4 axis dictates KLF4-mediated genome stability and cancer stemness need to be determined. We observed that methylation of KLF4 by PRMT5 leads to KLF4 stabilization that, in turn, results in increased tumor mitogenesis. Although enzyme activity-based PRMT5 inhibitors have been developed, lacking in substrate specificity limits its advantage for application.
Added value of this study
Abnormal accumulations of PRMT5 and KLF4 are observed in triple negative breast cancer. Aberrant methylation of KLF4 could be a key factor causing the KLF4 protein accumulation in tumor specimens. Stabilization of KLF4 due to PRMT5 could disrupt DNA damage checkpoint and promotes cancer stem population, which enhance tumorigenesis. Development of a small-molecule inhibitor that specifically blocks the interaction between PRMT5 and KLF4 provides a new strategy to inhibit PRMT5-KLF4 promoted tumor progression.
Implications of all the available evidence
Our developed small-molecule inhibitor that specifically intercepts the interaction between PRMT5 and KLF4 provides a new avenue to suppress breast tumor progression, especially TNBCs.
Alt-text: Unlabelled Box
1. Introduction
Krüppel-like factor 4 (KLF4) acts as a pivotal transcription factor and regulates various biological processes such as cell growth, proliferation, apoptosis, metabolism and stem cell self-renewal, and maintenance of pluripotency [1]. KLF4 with other three transcriptional factors, OCT3/4, SOX2, and c-MYC could switch somatic cells into stem cells [2]. Surprisingly, results from recent genetic and pathological studies have uncovered an ambivalent nature for KLF4 in tumorigenesis as either a tissue-specific tumor suppressor or an oncogene with mystery underlying mechanism [3]. KLF4 has been reported to have tumor-suppressive properties in lung cancer, gastric cancer, colorectal cancers, urothelial cancer, cervical cancer, bladder, prostate and stomach, while it acts as an oncogenic factor in breast, oral, osteosarcoma, melanoma and skin squamous cell carcinomas [[4], [5], [6], [7]]. KLF4 has a pivotal function in cancer stem cells maintenance and differentiation in various types of cancer including breast, colon, and osteosarcoma [3,[8], [9], [10], [11]]. Interestingly in colon cancer, KLF4 has been recognized as a well-characterized tumor suppressor, it still functions as an oncogene for cancer stem cell [8,10,12]. Moreover, KLF4 plays a critical role in regulating genome integrity, which functions as a radioprotective factor in the gastrointestinal system and a determinant for life or death in response to DNA damage [4,[13], [14], [15], [16], [17], [18]].
The critical role of KLF4 in cancer stem cell and genome integrity maintenance leads it to be an attractive target for cancer therapy. Recent study reported that using simvastatin to downregulate KLF4 expression could abolish doxorubicin-induced cancer stem cells and reverses the doxorubicin-enhanced tumorigenesis in vitro and in vivo [9]. Kenpaullone, a time-dependent KLF4 expression suppressor, can reduce breast cancer self-renewal of breast cancer stem cells and cell motility [3]. Nevertheless, specific inhibitors for KLF4 actions remain unavailable.
Protein arginine methyltransferases (PRMTs) catalyze mono and dimethylation of the guanidino group of arginine residues using S-adenosyl methionine (SAM) as a methyl donor. PRMT5 is an arginine-based methyltransferase that adds two methyl groups to the guanidine nitrogen atoms of arginine. Dimethylation can be divided into asymmetric and symmetric type. Asymmetric demethylation (ADMAs) transfers two methyl groups onto one of the terminal nitrogen atoms of the guanidino group, while Symmetric demethylation (SDMA), only adds one methyl group onto each of the terminal nitrogen atoms. PRMT5 belongs to symmetric arginine dimethyltransferase, whose aberrant accumulation is associated with tumorigenesis, cell survival and cancer stem cell [19,20]. It is upregulated in various human malignancies, including lymphomas, lung cancer, breast cancer, and colorectal cancer [[21], [22], [23], [24], [25], [26]]. In breast cancer, PRMT5 has a critical function in controlling breast cancer stem cell function via histone methylation and substrate gene expression, such as FoxP1, KLF4, which induce breast cancer chemotherapy drug resistance [4,[25], [26], [27]]. As an emerging target for potential cancer treatment, a number of groups have discussed small-molecule inhibitors of PRMT5 [[21], [22], [23], [24],28,29]. Recently documented PRMT5 inhibitors such as EPZ015666 (GSK3235025) have wide targeting spectrum, which has elicited its single-agent activity in solid tumors and chronic myeloid cell as well as leukemia cells [23,24]. A phase I study of GSK3326595 regarding its safety and activity is continuing in solid tumors and non-Hodgkin lymphoma (NCT02783300). An ongoing phase I dose escalation study of GSK3326595 aims to identify the safety and activity of the PRMT5 inhibitor GSK3326595 in solid tumors and non-Hodgkin lymphoma (NCT02783300). This study was initiated 2 years' ago and is expected to be completed in 2019. However, the current PRMT5 inhibitors designed against its enzymatic activity lacks specificity for the individual downstream substrate.
We have recently demonstrated that KLF4 is a rapidly turned over protein with its half-life governed by VHL-VBC ubiquitin-protein ligase. We have further revealed that KLF4 could be methylated by PRMT5, whose methylation on amino acids R374, R376, and R377 counteracts VHL/VBC-mediated KLF4 ubiquitylation [4]. We further observed that adding up methyl group to KLF4 alters KLF4 protein conformation that, in turn, prevents KLF4 ubiquitylation VHL/VBC E3 ligase through affecting the recognition of KLF4 by VHL/VBC, thereby stabilizing KLF4. In response to DNA damage signal, PRMT5-mediated KLF4 methylation causes failure of DNA damage-induced KLF4 accumulation that, in turn, reduces p21-mediated DNA damage response. In tumorigenesis, downregulation of KLF4 methylation leads to increased Bax levels and reduced p21 expression levels that further suppress several critical oncogenes, including cyclin D2, cyclin B1, Myc, EGF, and IGF1, resulting in inhibition of tumor initiation and tumor progression. Results from TCGA and proteomic studies have indicated that PRMT5 upregulation closely associated with KLF4 accumulation in mammary cancer [4]. Thus, inhibition of KLF4 function through dissociating interaction between PRMT5 and KLF4 could be an ideal approach to suppress breast cancer. In the present study, we report a novel in silico identified and in vitro validated small-molecule that blocks PRMT5-KLF4 binding and inhibits breast cancer progression. Our development of a small-molecule inhibitor that specifically intercepts the interaction between PRMT5 and KLF4 provides a promising candidate drug that suppresses breast cancer growth. Our findings could potentially lead to the development of more specific PRMT5 inhibitor for anti-TNBC treatment.
2. Experimental procedures
2.1. Construction of the 3D structural model of PRMT5 and its substrates
The 3D structure of human KLF4 was constructed by using reported KLF4 crystal structures (PDB ID: 2WBS, 2WBU, and 4M9E). The 3D crystal structure of human p53 (PDB ID: 3Q01) was retrieved from Protein Data Bank (www.rcsb.org). The sequence (residues from 112 to 199) of small nuclear ribonucleoprotein-associated protein B (Drosophila melanogaster) was downloaded from UniprotKB (https://www.uniprot.org/) and constructed using I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) [30]. The 3D structures modeling of KLF4-PRMT5 binding was conducted on the basis of our previous report [4], and the 3D structural models of PRMT5-p53 and PRMT5-SMB were constructed by using Z-DOCK server (http://zdock.umassmed.edu/) [31].
2.2. Development of KLF4 methylation inhibitor
We adopted the MOLCAD module implemented in SYBYL-X 1.3 to explore the potential binding pockets based on the complex of KLF4-PRMT5. Sequentially, we performed virtual docking screenings against our 3D chemical database using our reported protocol in order to identify small molecules that can interfere with PRMT5/KLF4-binding [32]. Briefly, the docking parameters used were as follows: (a) the “number of starting conformations per ligand” was set to 3, and the “number of max conformations per fragment” was set to 20; (b) the “maximum number of rotatable bonds per molecule” was set to 100; (c) flags were turned on at “pre-dock minimization”, “post-dock minimization”, “molecule fragmentation”, and “soft grid treatment”; (d) “activate spin alignment method with density of search” was set to 9.0; and (e) the “number of spins per alignment” was set to 12.
2.3. Cell lines and cell culture
HEK293T, MCF10A, MCF12A, 184-B5, MDA-MB-468, MDA-MB-453, HCC1937, HCC38, MCF-7, and MDA-MB-231 were obtained from the American Type Culture Collection (Manassas, VA). The KLF4+/+ MEF and KLF4−/− MEF were gifts from Dr. Engda Hagos (Colgate University). HEK293T, MDA-MB-468, MDA-MB-453, HCC1937, HCC38, MCF-7, MDA-MB-231, KLF4+/+ MEF and KLF4−/− MEF were maintained in DMEM supplemented with 5 or 10% FBS, 1× antibiotic/antimycotic solution (100 units/ml streptomycin and 100 units/ml penicillin) (all from Invitrogen). MCF10A, MCF12A, 184-B5 were maintained in monolayer in DMEM/F12 supplemented with 5% horse serum (Gibeco), 10 μg/ml insulin (Sigma), 20 ng/ml EGF (Sigma), 0.5 μg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma) and 1× antibiotic/antimycotic solution.
2.4. Plasmids and transfection
The plasmids GST-PRMT1, GST-PRMT2, GST-PRMT6, GST-PRMT7, GST-PRMT9 and HA-PRMT4 were purchased from Addgene. HA-PRMT3 and HA-PRMT8 were purchased from Sino Biological. The PRMT3, PRMT8 and PRMT4 genes were subcloned into pGEX-4 T3 for GST fused protein expression. The plasmids pLenti6/FLAG-KLF4WT, pLenti6/FLAG-KLF43K, GST fused PRMT5 full and 1-400Aa, 1-500Aa, 500-637Aa fragments were generated as previously reported [4]. To construct the mutant of PRMT5 (EDE/AAA, Glu392A, Asp419A, and Glu435A), The primers were used as follows: 5′-caaatgagcccagaagcgcactgacaatgatgtct-3′; 5′-cggcatttgggtttttcgccacagcatacagcttt-3′; 5′-agacatcattgtcagtgcgcttctgggctcatttg-3′; 5′-aaagctgtatgctgtggcgaaaaacccaaatgccg-3′; 5′-accgtagtctcatcagccatgagggaatgggtg-3′; 5′-cacccattccctcatggctgatgagactacggt-3′. The point mutations were completed by using the QuikChange Multi Site-Directed Mutagenesis Kit (Agilent). For transfection, cells were plated to form a 50–70% confluent culture and then transfected using Lipofectamine 2000 (Invitrogen).
2.5. Lentiviral infection
pLenti6/FLAG-KLF4WT, pLenti6/FLAG-KLF43K were co-transfected with packaging plasmids pVSV-G, pRRE, and pRSV-REV into HEK293T cells by using Lipofectamine 2000. The packaged lentiviral particles were collected, mixed with polybrene, and then added to target cells. The stable cell lines were established by culturing the cells in the medium containing antibiotic blasticidin (10 μg/ml).
2.6. RNA inference
The synthesized siRNA sequences are as following: Luciferase, 5′-CGT ACGCGGAATACTTCGA-3′; SiPRMT5–1#, 5’-GCCCAGTTTGAGATGCCTTAT-3′. SiPRMT5–2#, 5′-CCGCTATTGCACCTTGGAA-3′. siRNAs (Sigma) specifically targeted to PRMT5 or luciferase were synthesized and transfected into cells using Lipofectamine 2000. Cells were collected at 48 h–72 h post-transfection for cell survival assay or western blot assay.
2.7. Western blotting and immunoprecipitation assay
Cells were harvested and lysed in radioimmune precipitation assay (RIPA) lysis buffer (Cell Signaling Technology) containing protease inhibitor mixture (Sigma) or 1× SDS loading buffer. The protein concentration was determined using Bio-Rad protein assay reagent. Western blotting was performed using anti-KLF4 (Santa Cruz Biotechnology), PRMT5 (Millipore, 07–405), p21 (BD Bioscience, 556,430), FLAG (Sigma), β-actin (Sigma), SmD3 (Abgent), SmB (clone 12F5, Sigma), p53 (Clone DO-1, Santa Cruz Biotechnology) and HRP-conjugated goat anti-mouse or antirabbit secondary antibody (Promega). The SYM10 antibody against Symmetric Di-Methyl Arginine motif (SDMA) was purchased from Millipore. Signals were detected with ECL reagents (Millipore). For immunoprecipitation assay, cell lysate was incubated with KLF4 (Santa Cruz Biotechnology), PRMT5 (Millipore, 07–405), SmD3 (Abgent), p53 (Clone DO-1, Santa Cruz Biotechnology) antibody overnight at 4 °C on a rotator, followed by the addition of protein A/G plus agarose (Santa Cruz Biotechnology) to the reaction for 2 h at 4 °C. After five washes with lysis buffer supplemented with protease inhibitor mixture, complexes were released by boiling for 5 min in 2× SDS-PAGE loading buffer. Western blotting was used to detect SDMA modification.
2.8. Recombinant GST-KLF4 and GST -PRMT5 pull-downs experiment
GST-fused KLF4 and PRMT5 were expressed in the E. coli strain BL21 and purified using glutathione-Sepharose 4B resin following manufacturer's protocol. 2 μg of purified recombinant protein was incubated at 4 °C for 1 h with 500 μg of cell lysates from MDA-MB-231 cells or 2 μg His tagged KLF4 purified from E. coli strain BL21. Glutathione resin was added and interacting protein was pulled down. After five washes with 1×TBS buffer, the resin was eluted with 1×SDS loading buffer.
2.9. Biotin-WX2-43 pull-downs assay
2 μg/ml biotin-WX2-43 was incubated at 4 °C for 1 h with 20 μl 1 μg/μl GST-PRMT5 fragments or GST-PRMTs (GST-PRMT1~9) purified from E. coli. Avidin-agarose resin (Sigma) was then added and incubated at 4 °C for 1 h. Biotin-WX2-43 interacting protein was pulled down by centrifuge. After five washes with 1× TBS buffer, the resin was eluted with 1xSDS loading buffer.
2.10. WX2-43 binding ELISA assay
Purified human GST-PRMT5 proteins were coated in a 96-well microplate at 2 μg/ml (100 μl/well) for overnight in 4 °C and washed three times with washing buffer (PBS with 0.2% Tween-20). Wells were blocked with 1% BSA in PBS, incubated for 1 h, and then rinsed with Wash Buffer. Biotinylated-WX2-43 (100 μl) with a linear range concentration of 0.1 nM-10 μM diluted in 1% BSA / PBS were added into the plates and further incubated for 2 h at room temperature. Plates were washed three times with washing buffer (PBS with 0.2% Tween-20). After rinsing, 100 μl of Streptavidin-HRP at a concentration of 50 ng/ml in 1% BSA in PBS was added and incubated for 20 min at room temperature. Then plates were washed three times with washing buffer (PBS with 0.2% Tween-20). 100 μl of Substrate TMB (3,3′,5,5’-Tetramethylbenzidine, Sigma) Solution was added and incubated for 20 min at room temperature. 50 μl of the Stop Solution was added to each well. The absorbance of light with the wavelength of 450 nm was measured with 800TS absorbance reader (Biotek). For freedom WX2-43 competitive ELISA, 5 μM biotin-WX2-43 in 1% BSA / PBS was mixed with freedom WX2-43 at 1:0 (5 μM biotin-WX2-43 : 0 μM WX2-43), 1:2 (5 μM biotin-WX2-43: 10 μM WX2-43), 1:5 (5 μM biotin-WX2-43: 25 μM WX2-43), 1:10 (5 μM biotin-WX2-43: 50 μM WX2-43), 1:20 (5 μM biotin-WX2-43 : 100 μM WX2-43) (mol: mol) and then incubated with GST-PRMT5 coated 96-well plates for 2 h.
2.11. Animal experiments assay
All animal experiments were performed according to the protocols approved by the Institutional Animal Ethics Committee of Northwestern University. The investigation conforms to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (2011, 8th Ed.). In the orthotopic model, 5 × 106 of MDA-MB-231 cells mixed with matrigel (1:1 volume) were injected into the mammary fat pads of 8-week-old female Crl: Nu-Foxn1(nu) nude mice. 1 week later when the tumor was palpable, WX2-43 and vehicle (10% N,N-Dimethylacetamide in saline) were intraperitoneally administrated twice per week at the dose of 40 mg/kg, 80 mg/kg. Tumor volume was measured twice a week. After 4 weeks continued administration, mice were sacrificed and tumor tissues were collected for western blot.
2.12. Flow cytometry analysis
The flow cytometry analysis was conducted as the previous report [4], the breast cancer cells at the logarithmic growth phase were digested with 0.25% trypsin and washed with PBS for three times, followed by being re-suspended in 100 μl PBS, and then stained with anti-CD44-PE and anti-CD24-FITC or stained with their isotype controls at room temperature for 40 min. The samples were then washed by PBS three times and finally re-suspended in 200 μl PBS. Flow cytometry analysis was performed on a BD Accuri™ C6 Flow Cytometer (BD Bioscience). The expression ratios of CD44 to CD24 (CD44/CD24) in different subtypes of breast cancer cell lines were calculated from the percentage of CD44 and CD24 positive subpopulations in the flow cytometry analysis.
2.13. Aldefluor assay
The ALDEFLUOR kit (StemCell Technologies, Durham, NC, USA) was used to analyze the population with high ALDH enzymatic activity according to the manufacturer's instructions. Briefly, the cells were incubated in the ALDEFLUOR assay buffer containing ALDH substrate (BAAA, 1 mmol/l per 1 × 106 cells), and incubated for 40 min at 37 °C. In each experiment, an aliquot of cells was incubated under identical conditions, using 50 mmol/l of diethylamino benzaldehyde (DEAB), a specific ALDH inhibitor, as a negative control.
2.14. Chemistry and compounds
PRMT5 inhibitor epz015666 was purchased from the Selleckchem Company. The compounds to be tested as possible blockers of PRMT5-KLF4 complexation, including WX2-43, were synthesized in-house. All compounds were prepared in DMSO stock solution at a concentration of 10 mM and stored at −20 °C before use. The compounds were then diluted in Iscove's modified Dulbecco's medium (GBICO) to make a working solution, as needed. The final DMSO concentration was <0.1% in the working solution.
2.15. Statistical analysis
The significance of the differences in the assays was analyzed by Student's t-test or one or two-way ANOVA, followed by Tukey's multiple comparisons test. A value of P < .05 was considered significant.
3. Results
3.1. Abnormal accumulations of PRMT5 and KLF4 are observed in triple negative breast cancer
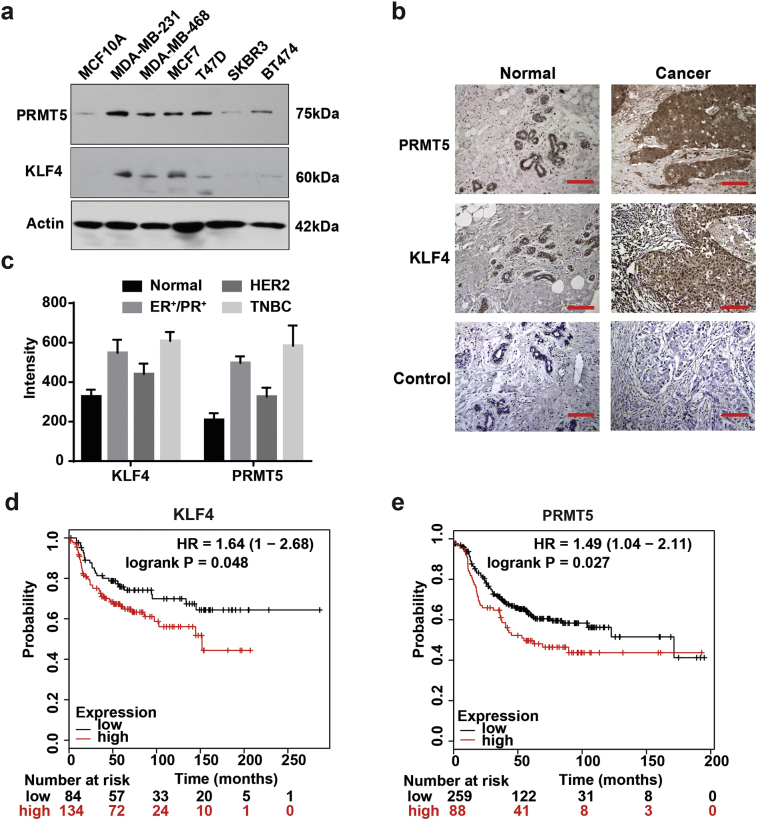
We have recently reported that deregulated KLF4 methylation by PRMT5 results in genome instability and mammary tumorigenesis. To decipher the abnormal accumulation of PRMT5 and KLF4 in breast cancer, we have measured the protein expression levels of KLF4 and PRMT5 in various molecular subtypes of breast cancer cells as well as human breast tumor tissue. As shown in Fig. 1, while KLF4 and PRMT5 expressions are relatively low in mammary epithelial cell lines MCF10A and moderate in HER2 positive breast cancer cell lines SKBR3 and BT474, significant accumulation of KLF4 is observed in triple negative and ER positive type of breast cancer cell lines MDA-MB-231, MDA-MB-468, MCF7 and T47D, especially in triple negative breast cancer (TNBC) cells MDA-MB-231 (Fig. 1a). We further analyzed the expression levels of both PRMT5 and KLF4 in various types of breast patient specimens using a tissue array with BC081120c, which contains 10 adjacent normal breast tissue (normal), 62 ER/PR positive breast cancer (ER+/PR+), 19 HER2 positive breast cancer (HER2) and 23 TNBC tissues. As shown in Fig. 1b–c, in general, both PRMT5 and KLF4 are overexpressed in various breast cancer tissues but severe PRMT5 and KLF4 protein accumulation were obviously detected in the TNBC. In addition, the Kaplan-Meier survival analyses for PRMT5 and KLF4 indicates that patients with overexpression of PRMT5 correlate well with poor prognosis in comparison with patients with low expression of PRMT5 or KLF4 in TNBC patients (Fig. 1d & e). Thus, the abnormal accumulation of PRMT5 and KLF4 protein could be oncogenic causation for mammary tumor initiation and progression. Targeting PRMT5-KLF4 axis could be a novel strategy anti-TNBC therapy.
Fig. 1.
Abnormal accumulation of PRMT5 and KLF4 are observed in triple negative breast cancer. (a) Expression of PRMT5 and KLF4 in mammary gland epithelial cell and various types of breast cancer cell lines. (b) Tissue arrays of 10 adjacent normal breast tissue (normal), 62 ER/PR positive breast cancer (ER+/PR+), 19 HER2 positive breast cancer (HER2) and 23 triple negative breast cancer (TNBC) were subjected to immunohistochemistry with anti-PRMT5 or anti-KLF4 and visualized by DAB staining. Representative normal and cancer tissue staining are shown. Scar bar, 100 μm. (c) Summary of B. (d-e) Kaplan-Meier survival assay of KLF4 (d) and PRMT5 (e) in TNBC. TNBC patients with PRMT5 or KLF4 overexpression show shorter survival time in comparison with patients with lower PRMT5 or KLF4 expression levels.
3.2. Methylation of KLF4 contributes to cell survival in response to DNA damage and cancer stem cell maintenance
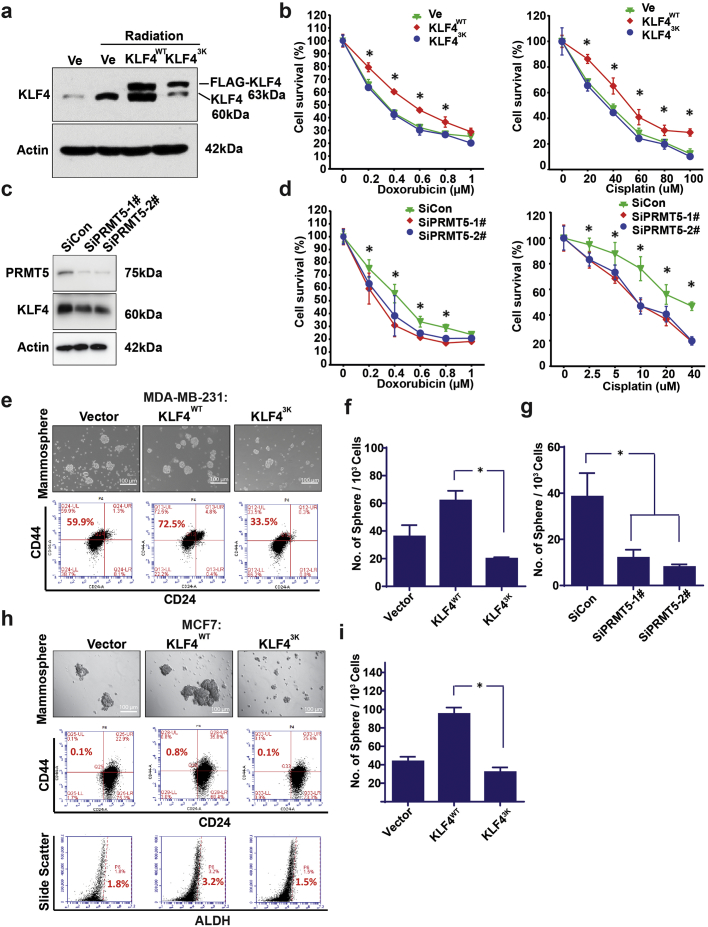
The current principal clinical therapy for TNBCs is relayed on chemotherapy following by radiation therapy [33]. High subpopulation of cancer stem cell existing in TNBC tissue has been a major issue affecting the therapeutic efficiency [27]. To ask if accumulation of KLF4 due to the PRMT5-mediated methylation regulates cancer cell survival and enhances cancer stem cell expansion, we have measured the impact of KLF4 methylation, by functional interference of KLF4 methylation through expression of a KLF4-methylation deficient mutant, KLF4-3 K (replacement of arginines 374, 376 and 377 by lysines), in cancer cell survival as well as maintenance of cancer stemness. As shown in Fig. 2a, we observed that the KLF4 protein levels are regulated in response to radiation in a methylation-dependent manner. While both endogenous and exogenous KLF4 are upregulated in response to g-radiation, interference of PRMT5-mediated KLF4 methylation by expression of KLF4-methylation deficient mutant significantly attenuates the radiation-induced upregulation of KLF4. We further observed that elevated expression of KLF4 leads to increased cancer cell survival against chemo-drug Doxorubicin and Cisplatin but suppression of KLF4 methylation results in significant attenuation of cancer cell survival (Fig. 2b). Meanwhile, knockdown PRMT5 expression by siRNA also decreases KLF4 expression and sensitizes MDA-MB-231 cells to chemo-drug Doxorubicin and Cisplatin (Fig. 2c & d). In addition, we have further examined the role of KLF4 methylation in the maintenance of cancer stem cell using 3D mammosphere formation assay. As shown in Fig. 2e–i, in both triple negative and ER positive breast cancer cells, elevated expression of KLF4 largely enhances cancer stem cell population (CD44+/CD24− or ALDH positive), but prevention of KLF4 methylation leads to significant suppression of breast cancer stem cells. Taken together, the above results clearly show that the methylation of KLF4 by PRMT5 is critical to regulating cell survival and cancer stem cell maintenance.
Fig. 2.
Methylation of KLF4 contributes to cell survival in response to DNA damage and cancer stem cell maintenance. (a) Expression of endogenous and exogenous wild-type as well as KLF4-methylation deficient mutant (KLF4-3K) eight hours after cellular treatment with 10Gy radiation. Functional interference in KLF4 methylation by expression of KLF4-3 K leads to dropped endogenous KLF4 expression levels. (b) Cell viability assay of MDA-MB-231 with overexpression of wild-type KLF4 or KLF4-3 K mutant before and after treatment with cisplatin or doxorubicin at the indicated concentration. Only the wild-type KLF4 showed its capability in promoting cell survival. The artrisks indicate significant difference (p < .05) between KLF4 and KLF4-3 K at the indicated concentration. Data are mean ± SEM; two-tailed t-test was used for the statistical analysis. (c) Expression of PRMT5 and KLF4 after PRMT5 knockdown by siRNA. (d) Cell viability assay of MDA-MB-231 with PRMT5 siRNA knockdown before and after treatment with cisplatin or doxorubicin at the indicated concentration. Loss of PRMT5 decrease cell survival. The artrisks indicate significant difference (p < .05) between SiCon and SiPRMT5–1#/−2# at the indicated concentration. Data are mean ± SEM; one-way ANOVA was used for the statistical analysis. (e-i) Elevated expression of wild-type KLF4 increases cancer stem cell expansion in both MCF7 and MDA-MB-231 cells, while expression of KLF4-3 K mutant results in suppression of cancer stem cell renewal. The cancer stem cell self-renewal of the mammosphere were estimated by measuring CD44/CD24 or ALDH cancer stem cell marker using flow cytometry. (g) knockdown PRMT5 by siRNA decrease mammosphere in MDA-MB-231 cells. (f, i) mammosphere count from e and h. The artrisks in f, g and i indicate significantly difference (p < .05). Data are mean ± SEM; one-way ANOVA was used for the statistical analysis.
3.3. Development of an inhibitor that specifically blocks the interaction between PRMT5 and KLF4
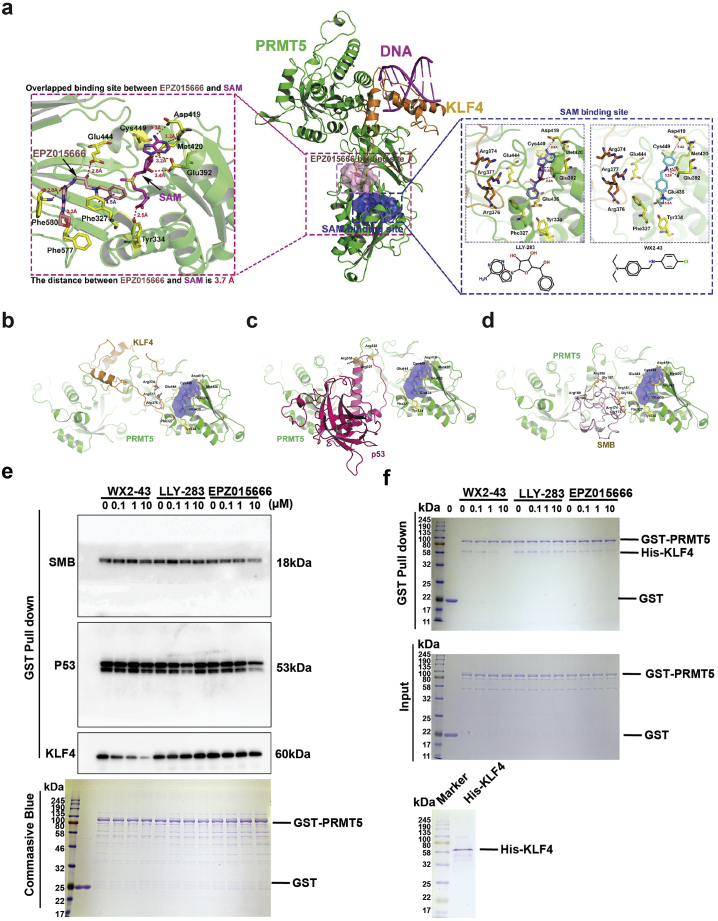
Based on the critical effect of KLF4 methylation in promoting breast tumor progression, cancer cell survival after chemotherapy and cancer stem cell expansion, we decided to develop small molecule inhibitor that could efficiently block PRMT5-KLF4 complexation as a new anti-breast cancer therapeutic approach. As shown in Fig. 3a (left), we first looked into the detailed information of the binding sites in PRMT5. We found that the key residues involved in the binding site of EPZ015666 included Phe327, Glu444, Phe577, Phe580. Moreover, we observed that EPZ015666 forms strong hydrogen bond with Glu444 (2.6 Å), Phe577 (3.3 Å), and Phe580 (2.8 Å), while interacting with Phe327 (3.5 Å) via a strong π- π interaction. The key PRMT5 residues involved in the binding site of SAM included Phe327, Tyr334, Glu392, Asp419, Met420, Glu435, Glu444, and Cys449. Furthermore, we observed that SAM formed a strong hydrogen bond with Asp419 (3.3 Å), Met420 (3.2 Å), Glu392 (3.2/3.4 Å), and Tyr334 (2.5 Å). The distance between EPZ015666 and SAM is only 3.7 Å, indicating that the binding sites between EPZ015666 and SAM overlap. To explore in-depth the mechanism of KLF4 methylation by PRMT5, we have recently conducted 3D modeling and molecular dynamics studies which revealed the molecular mechanism of interaction between PRMT5 and KLF4 as well as the properties of the binding cavity [4]. As shown in Fig. 3a (middle), we observed that the methylation regions include arginine 374, 376, and 377 of KLF4 interacted with PRMT5 through the binding cavity of EPZ015666 (overlapping), and the methylation region was near the SAM binding pocket, which the SAM binding cavity may act as an allosteric pocket for the protein-protein interface between PRMT5 and KLF4. Based on the established 3D database docking and modeling approaches, we have carried out a virtual screening of PRMT5-KLF4 modulator(s) using the SAM binding pocket. A 3D virtual compound library containing 540,000 compounds was used for the virtual screening. We have identified several small molecules that exhibited strong hydrogen-bonding and hydrophobic interactions with the PRMT5-KLF4 surface cavity, and then validated the compound effect on KLF4 and p21 expression (Fig. S1). Among these compounds, WX2–43 (MW = 288.82, ClogP = 7.9) was ranked as the top candidate (docking score: 7.90) (Fig. S2). The docking poses and structure of our chemical lead WX2–43 are shown in Fig. 3a (right). The key residues (Phe327, Tyr334, Glu392, Asp419, Met420, Glu435, Glu444, and Cys449) in WX2–43 binding are located near the PRMT5/KLF4 interface and are known to play important roles in the reorganization of SAM and LLY-283 [34]. We found that WX2–43 (Fig. 3a, right), LLY-283(Fig. 3a, right), and SAM (Fig. 3a, left) share a similar binding mode and interactions. Especially, WX2–43 formed hydrogen bonds with Glu392 (3.3 ± 0.1 Å), Asp419 (3.4 Å), and Glu435 (3.5 Å), while making strong hydrophobic contacts with Cys449 (4.7 Å), Phe327, and Met420. Similarly, LLY-283 formed interactions with Glu392 (2.6 ± 0.1 Å), Asp419 (2.8 Å), Met420, Cys449, etc. However, WX2–43 showed a novel and flexible structure, which may behave differently from SAM or LLY-283.
Fig. 3.
Development of an inhibitor that specifically blocks the interaction between PRMT5 and KLF4. (a) Strategy to design and screen specific inhibitors that block KLF4 methylation. The 3D structural model of PRMT5-KLF4 complex and the binding surface cavity has been simulated and a putative KLF4-binding cavity on the PRMT5 surface was defined by the Connolly solvent-accessible surface calculation with a 1.4-Å radius. The detailed interaction between the selected compound and PRMT5 is described. Among candidate compounds with potent IC50, one candidate compound named WX2–43 satisfies well the criteria of targeting specificity and selectivity. (b-d) The 3D structural models between PRMT5 and its substrates (KLF4, p53, and SMB) in the presence of WX2–43. (b) PRMT5-KLF4. (c) PRMT5-p53. (d) PRMT5-SMB. (e) GST proteins or GST fused PRMT5 proteins were incubated with MDA-MB-231 cell lysates in the presence of WX2–43, LLY-283 and EPZ015666 at the indicated dose for 4 h. The GST or GST fused PRMT5 proteins were pulldown and the binding proteins SMB, p53 and KLF4 were detected by western blot. The pulldown GST proteins or GST fused PRMT5 proteins were stained with coomassie blue (lower panel). WX2–43 inhibits PRMT5 pulldown KLF4 in does responded manner. (f) WX2–43 inhibits binding between GST-PRMT5 and His-KLF4. GST proteins or GST fused PRMT5 proteins were incubated with His tagged KLF4 in the presence of WX2–43, LLY-283 and EPZ015666 at the indicated dose for 4 h. The input GST-PRMT5 and His-tagged KLF4 were shown in the right panel. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further explore the role of WX2-43 for regulating the substrates of PRMT5, we constructed three protein-protein complexes including PRMT5-KLF4, PRMT5-p53, and PRMT5-SMB, as shown in Figure 3b–d. First, as we mentioned before, WX2-43 in our PRMT5-KLF4 model located close to (~9 Å), and can intercept, three key residues in KLF4, including Arg374, Arg376, and Arg377 (Supplemental Movie). Previously, we already demonstrated that PRMT5 interacts with KLF4 and catalyzes the methylation of these three arginine residues (Fig. 3b). Based on our model, we hypothesized that WX2-43 did not directly interrupt the protein-protein interface of PRMT5-KLF4 complex, it may regulate the PRMT5-KLF4 via an allosteric effect. Moreover, according to our 3D model of PRMT5-p53 (Fig. 3c), the distance between WX2-43 and two important arginines residues Arg333 and Arg337 involved in p53 methylation [35], was about 25 Å, indicating that the disruption of WX2-43 on PRMT5-p53 is not as strong as that of PRMT5-KLF4. Finally, based on our 3D structural model of PRMT5-SMB (Drosophila), as shown in Fig. 3d, We found that the distance between WX2-43 and the four key arginines of SMB involved in methylation [36] (R4-7: Arg174, Arg181, Arg189, and Arg196) were from 16 to 30 Å, indicating that the disruption of WX2-43 on PRMT5-SMB is not as strong as that of PRMT5-KLF4. The disruptions have been confirmed by a set of biological experiments presented below.
3.4. Characterization of blockade effect of WX2-43 on the PRMT5-KLF4 interaction and PRMT5-mediated KLF4 methylation
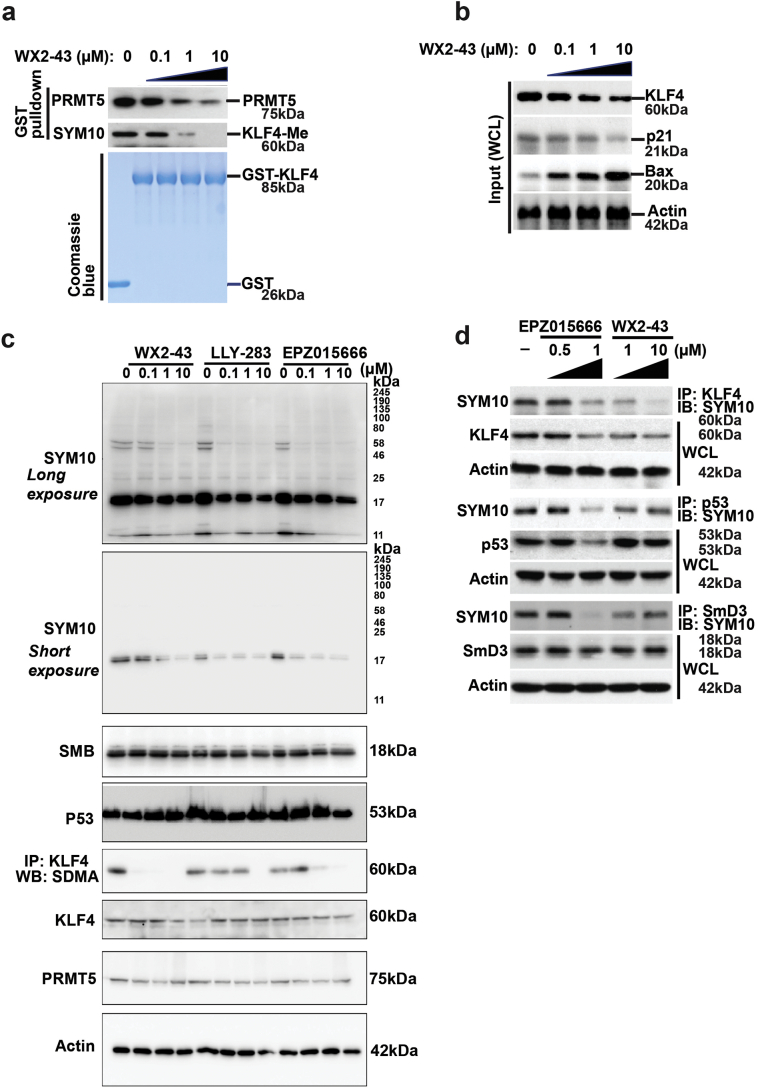
To further validate whether WX2-43 could block the binding between PRMT5 with its substrates including KLF4, P53 and SMB, we first detected the capacity of WX2-43 in dissociation of interaction between PRMT5 with KLF4, P53 and SMB by using functional cell lysate-based GST-fused PRMT5 pull-down assay. Interesting, all three compounds including WX2-43, LLY-283 and EPZ-0156666 have no capacity to dissociate PRMT5-p53 and PRMT5-SMB binding, while only the compounds WX2-43 has an obvious capacity to break PRMT5-KLF4 binding in MDA-MB-231 cell lysates in a dose-dependent manner (Fig. 3e). Meanwhile, we further using purified His-tagged KLF4 protein instead of MDA-MB-231 lysates identified only the WX2-43, other than LLY-283 and EPZ-0156666 could prevent PRMT5-KLF4 binding (Fig 3f). To further detect the capacity of WX2-43 in the dissociation of interaction between PRMT5 with KLF4, we further use functional cell lysate-based GST-KLF4 pull-down assay following by measurement of the KLF4 methylation status as well as binding of PRMT5 to KLF4 by western blotting. As shown in Fig. 4a, we observed that GST-KLF4 could efficiently pull down PRMT5 proteins from MDA-MB-231 cell lysates and the GST-KLF4 protein also could be methylated by endogenous PRMT5 presented in cell lysates. We further observed that pre-addition of the WX2-43 to the lysates significantly decreased the efficiency for KLF4 to pull-down PRMT5 and suppresses KLF4 methylation in a dose-dependent manner. We next measured the effect of WX2-43 on the KFL4 downstream target p21 and Bax. As shown in Fig. 4b, we observed that pretreatment of MDA-MB-231 cells with WX2-43 for 24 h, significantly decreased KLF4 protein levels and its transcriptional activation in upregulating p21 expression and downregulating Bax gene expression. These data are in support of the interference of WX2-43 with the methylation of KLF4 by PRMT5, thus promoting KLF4 ubiquitylation and degradation.
Fig. 4.
Characterization of blockade effect of WX2–43 on the PRMT5-KLF4 interaction and PRMT5-mediated KLF4 methylation. (a) WX2–43 efficiently intercepts the interaction between KLF4 and PRMT5, and blocks KLF4 methylation. (b) Blockade of KLF4 methylation further leads to downregulation of p21 and upregulation of Bax. (c) Specificity of WX2–43, LLY-283 and EPZ015666. MDA-MB-231 cells were treated with WX2–43, LLY-283 and EPZ015666 at indicated dose for 24 h and then expressions of SDMA, p53, KLF4, SMB were detected. All three compounds could highly efficiency abrogated the whole cell SDMA modification at 1 μM level. WX2–43 shows less efficiency at 0.1 μM level to inhibit total SDMA modification. However, WX2–43 shows more capability in inhibiting SDMA modification on KLF4. (d) Comparison of the substrate specificity for WX2–43 and that of the non-specific PRMT5 inhibitor EPZ015666.
To compare our newly developed PRMTY5-KLF4 specific inhibitor with recently reported non-specific PRMT5 inhibitor LLY-283 and EPZ015666, we have tested all three compounds side by side (Fig. 4c and d). By staining of SYM10 antibody against Symmetric Di-Methyl Arginine Motif (SDMA), the whole cell symmetric di-methylation modification could be measured. All three compounds could highly efficiency abrogated the whole cell SDMA modification at 1 μM level, but WX2-43 shows less potent at 0.1 μM dose (Fig. 4c). However, when immunoprecipitated KLF4 and detected the SDMA modification, WX2-43 shows more capacity to inhibit KLF4 modified by PRMT5, which indicated WX2-43 is more KLF4 specific PRMT5 inhibitor. WX2-43 is discovered here, on the other hand, based on specific blockade of interaction between PRMT5 and KLF4, and is therefore anticipated to suppress KLF4-mediated downstream effects exclusively. We further compared WX2-43 with EPZ015666 inhibits the catalytic activity of PRMT5 in general and thereby inhibits the methylation of all PRMT5 substrates indiscriminately. As shown in Fig. 4d, while WX2-43 specifically inhibits KLF4 methylation, EPZ015666 reduces the methylation of all three substrates. Based on these results, we have identified a specific PRMT5-KLF4 binding inhibitor. Thus, WX2-43 could be defined as a specific PRMT5-KLF4 inhibitor. Taken together, the above experiments validated that WX2-43, identified with guidance from in-silico studies, is an effective compound that specifically blocks PRMT5-KLF4 binding, and affects KLF4 stability and suppresses KLF4 cellular activity.
3.5. WX2-43 is a potent PRMT5 specific inhibitor
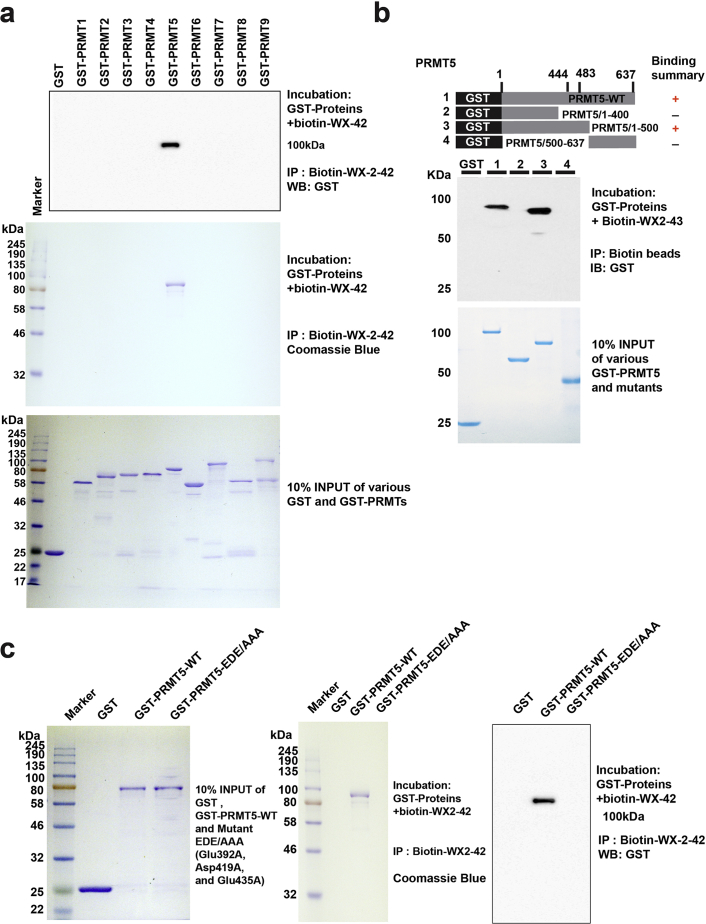
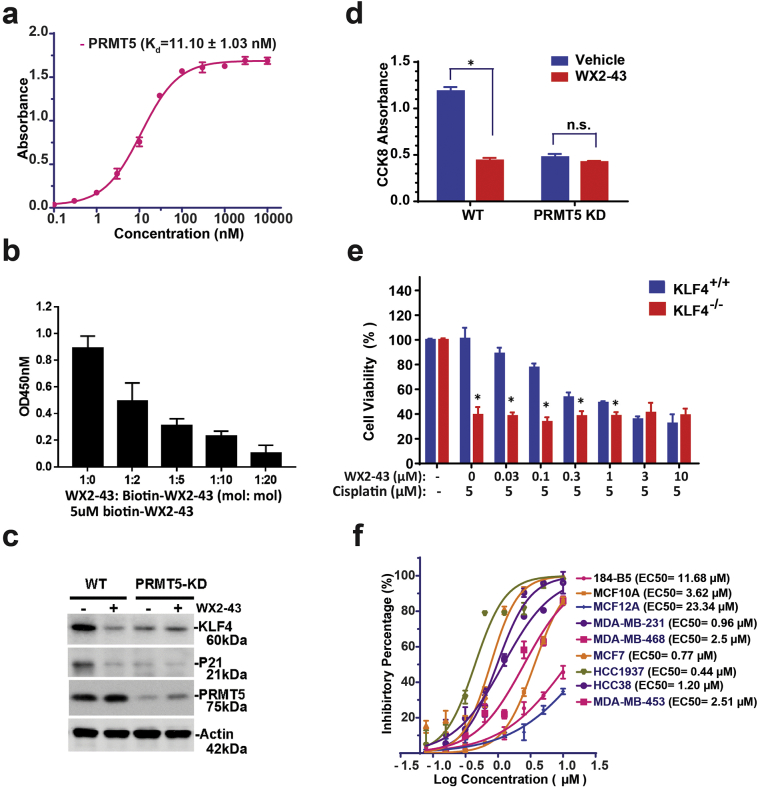
Given the presence of nine members in the protein arginine methyltransferases family, we next ask if our newly developed PRMT5 inhibitor also affect other members. Previous works have sketched the non-redundant feature for the nine members (PRMT1~9) (Fig. S3) [37]. To date, we expressed all nine GST fused PRMTs and then do the Biotin-labelled WX2-43 pull down. Interestingly, only the PRMT5 shows affinity to WX2-43 (Fig. 5a). To further study how WX2-43 binds to PRMT5 that in turn interrupts PRMT5-KLF4 binding, we have generated a series of GST-PRMT5 fragment deletions and then detected the exact molecular motif that binds to WX2-43 by using pulldown assay [38]. To date, we subjected biotinylated-WX2-43 conjugated with Streptavidin-agarose to a set of GST-PRMT5 fragments including 1-400Aa, 1-500Aa, 500-637Aa and the full-length, and then pull-down biotinylated-WX2-43 coupled Streptavidin-agarose following by measuring retaining of GST-PRMT5 fragment fusion protein abundance by Coomassie blue staining and western blotting (Fig. 5a). As shown in Fig. 5b, we only observed the GST-PRMT5 1–500 and the full length PRMT5 could be pulled down by biotin-WX2-43, indicating that the amino acids stretch between L400-M500 is involved in binding WX2-43. This result is consistent with the core amino acids predicted in our molecular docking model, including Glu392, Met420, Asp419, Glu435, Glu444 localized in this area. We further mutated the PRMT5 Glu392, Asp419 and Glu 435 to alanines (EDE/AAA) and then did the biotin-WX2-43 pulldown. Mutated these three amino acids abrogated WX2-43 binds to PRMT5, which indicated there three amino acids facilitate WX2-43 binding (Fig. 5c). We further measured the binding co-efficiency of biotin-WX2-43 with GST-PRMT5 using ELISA and found the binding Kd is 11.0 ± 1.03 nM (Fig. 6a). In addition, the binding of biotin-WX2–43 to PRMT5 was observed to be dose-dependent by free WX2-43 in the competition ELISA assay, suggesting the binding of biotin-WX2-43 to PRMT5 is specific (Fig. 6b).
Fig. 5.
WX2-43 is a potent PRMT5 specific inhibitor. (a) WX2–43 only binds to PRMT5 in the human PRMT family. The GST fused PRMTs family members were incubated with Biotin-labelled WX2–43 for 1 h and then pulldown with Avidin coupled agarose. The pulldown PRMTs family members were detected by western blot or coomassie staining. (b) WX2–43 specifically inhibits PRMT5-KLF4 binding and function. GST-fusion PRMT5 fragments were expressed in E.coli following by purification. GST-PRMT5 fragments were then incubated with biotin-WX2–43 and pulled down by streptavidin-agarose. The pulldown PRMT5 fragments were viewed by coomassie blue staining and detected by antibodies against GST tag. WX2–43 specifically binds to the amino acid stretch between L400-M500 on PRMT5. (c) Mutation of Glu392, Asp419 and Glu 435 to alanine (EDE/AAA) abrogate WX2–43 binds to PRMT5. GST fused PRMT5 wildtype or mutant (EDE/AAA) were incubated with Biotin-labelled WX2–43 for 1 h and then pulldown with Avidin coupled agarose. The pulldown proteins were detected by western blot or coomassie staining. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
WX2–43 is an inhibitor that specifically blocks KLF4-mediated transcription and cell survival. (a) Measurement of the dissociation constant Kd for WX2–43 that binds to PRMT5 by ELISA. GST-PRMT5 was coated and incubated with biotin-WX2–43. The binding biotin-WX2–43 was detected by HRP-streptavidin and TMB. (b) Free WX2–43 and biotin-labelled WX2–43 competitively binds to immobile GST-PRMT5. GST-PRMT5 was coated and incubated with the mixture of free WX2–43 and biotin-WX2–43 at indicated vol/vol. Binding of biotin-WX2–43 to GST-PRMT5 was detected by HRP-streptavidin and TMB. (c, d) Specificity of WX2–43 on inhibition of PRMT5/KLF4-mediated target and cell viability. MDA-MB-231 or MDA-MB-231-ShPRMT5 cells were treated with WX2–43 for 24 h, and the expression of PRMT5, KLF4, P21 as well as cell viability were measured. The artrisk indicates significant difference (p < .05) between the vehicle and WX2–43 treatment group. Data are mean ± SEM; two-tailed t-test was used for the statistical analysis. (e) WX2–43 only shows its effect on KLF4+/+ MEFs but not on KLF4 knockout MEFs. KLF4+/+ and KLF4−/− MEFs were treated with cisplatin at 5 μM and WX2–43 at the indicated concentration for 24 h, the cell viability was then measured by CCK-8. The artrisks indicate significant difference (p < .05) between KLF4+/+ and KLF4−/− MEFs. Data are mean ± SEM; two-tailed t-test was used for the statistical analysis. (f) Selectivity of WX2–43 between MCF7, TNBC cell line MDA-MB-231, MDA-MB-468, MDA-MB-453, HCC1937, HCC38 and mammary gland epithelial cell line MCF10A, MCF12A and 184-B5.
3.6. WX2-43 is an inhibitor that specifically blocks KLF4-mediated transcription and cell survival
We next examined whether WX2-43 inhibits KLF4 function via specifically affecting PRMT5-KLF4 axis. As shown in Fig. 6c–e, we observed that WX2-43 significantly decreases KLF4 abundance and its targeted gene p21 expression as well as cell proliferation, and this effect is very similar to that of PRMT5 knockdown, indicating the WX2-43 has no addictive effect on KLF4-p21 expression and cell proliferation other than via PRMT5. To further determine whether WX2-43 inhibits cell viability especially via KLF4, we have tested cell viability in response to cisplatin by using the KLF4 knockout MEFs (KLF4−/− MEFs) cells. We observed that, while wild-type KLF4 MEFs has obvious dose response to WX2-43 inhibition, lack of KLF4 in MEFs attenuates the dose responsive manner, suggesting the effect of WX2-43 is through KLF4 (Fig. 6e). We previously observed that the KLF4 expression levels in mammary gland epithelial cells are much lower in comparison with TNBC cells. We thus wondered if the response to WX2-43 is different between MCF10A and MDA-MB-231. We tested the IC50 of WX2-43 in mammary epithelial cell lines including 184-B5, MCF10A, MCF12A and breast cancer cell lines MDA-MB-231, MDA-MB-468, MCF7, HCC-1937, HCC38 and MDA-MB-453, and observed that triple negative breast cancer cell lines such as MDA-MB-231, HCC1937 cells are much more sensitive to WX2-43 than normal epithelial cancer cell lines MCF10A, MCF12A and 184-B5. The observed WX2-43 killing selectivity between mammary gland epithelial and TNBC cells suggest that WX2-43 could be a selective inhibitor for anti-TNBC therapy (Fig. 6f).
3.7. WX2-43 is a potent inhibitor that suppresses breast tumor progression
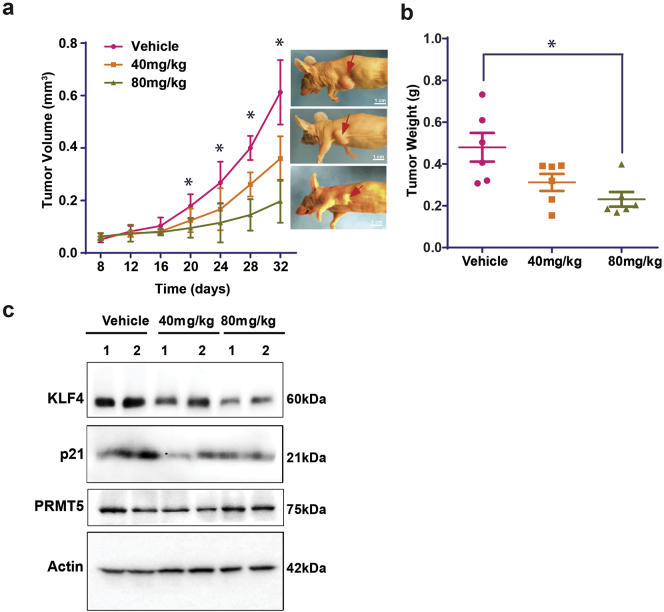
The above characterization has shown that WX2-43 is a potent inhibitor that specifically disrupts PRMT5-KLF4 interaction and selectively targets TNBC cells for growth inhibition. To determine the physiological and clinical relevance of WX2-43 inhibitor in breast cancer progression, we have examined the impact of WX2-43 in TNBC using human breast cancer xenograft model [39]. MDA-MB-231 cells were subcutaneously injected into athymic nude mice, WX2-43 where intraperitoneal (i.p.) administrated and tumor growth was monitored. The administration of 40, 80 mg/kg WX2-43 to mice had no obvious toxic effect and mice gained weight normally. 80 mg/kg WX2-43 reduced approximately by 60%, and 40 mg/kg WX2-43 reduced by about 40% both the size and weight of breast cancer xenograft tumor in comparison with vehicle (Fig. 7a-b). We further measured the expression levels of KLF4, p21 and PRMT5 in the isolated grafted tumors using Western blotting, and confirmed the effect of WX2-43 on inhibiting KLF4 function upon WX2-43 administration (Fig. 7c). The result from the in vivo analysis shows that WX2-43 is a potent inhibitor that suppresses TNBC progression. Altogether, our results indicate that PRMT5-KLF4 plays a critical role in promoting breast tumor initiation and progression through regulating cell genome stability and stem cell expansion. Blockade of the PRMT5-KLF4 oncogenic axis by our newly developed PRMT5-KLF4 inhibitor could be a novel strategy to treat TNBC.
Fig. 7.
WX2–43 is a potent inhibitor that suppresses breast tumor progression. (a, b) MDA-MB-231 cells were implanted into the mammary fat pad of nude mice. WX2–43 were treated twice a week at the indicated dose. Tumor volume was measured once a week and mice were sacrificed 32 days after injection. Tumor volume growth curve and tumor weight were measured. The artrisk indicates significant difference (p < .01) between vehicle, 40 mg/kg and 80 mg/kg treatment group. Data are mean ± SEM; one-way ANOVA was used for the statistical analysis. (c) Measurement of expression of PRMT5, KLF4 and P21 in xenograft tumor. After tumor harvest at 32 days, the tumors were lysed and detected by Western blotting using indicated antibodies.
4. Discussion
KLF4 is a pivotal cellular fate determinant, whose deregulation has been linked to a variety of human disease through orchestrating various cellular processes [1]. Previous studies from TCGA and pathological analyses unveiled the correlation of abnormal regulation of KLF4 with poor prognosis of breast cancer, although the underlying mechanism has been reminded until recently. We have demonstrated that the management of KLF4 protein levels is accurate and controlled by a sophisticated interplay between ubiquitylation and protein methylation [4,40]. We have shown the KLF4 protein turnover is regulated by VHL/VBC-mediated ubiquitylation following by degradation. Intriguingly, we have observed that the KLF4 protein degradation is counteracted by protein methylation, wherein the addition of methyl group to the KLF4 my arginine methyl-transferase PRMT5 stabilizes KLF4 stability through antagonizing its ubiquitylation due to interruption the binding between KLF4 with its ubiquitin E3 ligase VHL because of alteration of its intra-molecular confirmation [4,40]. Results from our pathological analyses have drawn attention to the unexpected accumulation of both PRMT5 and KLF4 in breast cancer tissue and their significant correlation [4]. Elevation of KLF4 or PRMT5 leads to malignant mammary epithelial cell transformation and tumor progression in preclinical models [4]. Evidence from multiple layers clearly implies that the suppression of KLF4 function by blockade of its methylation catalyzed by PRMT5 could be a valuable approach to inhibit breast tumor progression, especially for TNBC. In the present study, the development of a specific KLF4 methylation inhibitor provides a promising strategy to suppress breast tumor progression by targeting PRMT5-KLF4 interaction. Our characterization has further confirmed the specificity, selectivity as well as efficacy in killing TNBC cells in both cultured-cell and animal models.
PRMT5 is the major symmetric arginine methyltransferase in mammals [19]. Previous pathological studies have demonstrated the impact of PRMT5 in promoting various oncogenic processes [25,41]. Targeting PRMT5 recently became a hot spot for anti-cancer drug development. Various approaches such as the high-throughput screen, pharmacophore- and docking-based virtual screening have been utilized to search for PRMT5 small-molecule inhibitor. Such efforts have been further encouraged by the recently reported development of PRMT5 inhibitors by several companies [42]. Approximately 50 candidate PRMTs inhibitors have been documented, including EPZ015666 (IC50 = 22 nM), Compound 9–1 (IC50 = 35 μM), compound 17 (IC50 = 0.33 μM), Compounds CMP5 and HLCL-61 [[21], [22], [23],28,29]. Among these, EPZ015666 shows outstanding selectivity for PRMT5 with promising potency (IC50 = 0.022 μM) but limited to other PRMTs. PRMT5 is reported to modulate transcription through the methylation of histone H2 arginine 3 (H2R3), histone H3 arginine 8 (H3R8) and histone H4 arginine 3 (H4R3). The major effects of these inhibitors are more rely on histone methylation and transcriptional regulation. WX2-43, our newly developed PRMT5-KLF4 specific inhibitor is unique and precise because of its specificity on the blockade of PRMT5-facilitated KLF4 methylation, which determines KLF4 protein stability and KLF4-mediated oncogenesis. WX2-43, in preclinical models, has shown its attractive potency in killing TNBC cells with IC50 at 11 nM, and EC50 at 2 μM. Importantly, WX2-43 is not a PRMT5 pan-methyltransferase inhibitor but only specifically targets PRMT5-mediated KLF4 methylation. Therefore, WX2-43 is more specific than other inhibitors such as EPZ015666 to target the PRMT5-KLF4 axis for suppressing breast tumor progression. The sister inhibitor of EPZ015666, GSK3326595, is ongoing phase 1 trial in solid tumors and non-Hodgkin lymphoma, which implies the emerging clinic value of PRMT5 inhibitor. Likely, WX2-43 has its advantage and potential in targeting specificity in comparison with other PRMT5 pan-methyltransferase inhibitors in future’s anti-breast cancer treatment.
We demonstrated the role of PRMT5-KLF4 axis in regulating DNA damage response and breast tumorigenesis [4,43]. In response to genotoxic stress, precise control of KLF4 abundance determines the cellular fate either undergoing survival or committing to death through regulating the ratio between p21 and Bax [4]. Under the circumstance of oncogenesis, our chip assay revealed the elevated KLF4 levels leads to upregulation of a series of oncoproteins such as EGF, IGF1, Myc and CDKs [4]. These observations imply that manipulation of KLF4 by blockade of PRMT5-KLF4 axis could potentially synergize KLF4 with other critical targeted drugs such as CDK4/6 inhibitor and PARP inhibitors in potent killing breast cancer cells. We observed, in high KLF4 expression set breast cancer cell lines, the elevated KLF4 levels leads to reduced drug response [4]. Our work inspires a novel clinical intervention strategy in combination of PRMT5-KLF4 inhibitor with PARP inhibitor, talaraparib/olaparib, or CDK4 inhibitor for anti-TNBC therapy.
The following are the supplementary data related to this article.
The process of 3D structural modelling of PRMT5-KLF4 complex and the virtual screening characterized WX2-43 as a PRMT5-KLF4 novel inhibitor. We first constructed the complex of PRMT5-KLF4 with the missing region of our interest (residues 370-383; PKPKRGRRSWPRKR). Then we carried out virtual screening against a compound library using the binding pocket of SAM in PRMT5. WX2-43, one of our active compounds, was confirmed and found to specifically inhibit KLF4 methylation. Moreover, WX2-43 shows high potency and novel structure.
Supplementary material
Author contributions
Y.W. and X.X are the PIs who managed the whole project. Y.W., Z.Z., Z.F. and X.X. designed the experiments and wrote the manuscript. Z.Z. performed the experiments. D.H. provided some plasmids and participated in some experiments. Z.F. and P.Y. performed the development of PRMT5-KLF4 inhibitor. M.G. participated in structure simulation of PRMT5-KLF4. M.C. and W.J.G. provided consultation on some drug related experiments. I.B. supervised M.G. in molecular modeling of PRMT5-KLF4.
Declaration of competing interests
Dr. Xiangqun Xie issued a patent US8772541.
Acknowledgments
We thank members from Wan and Xie laboratories for their kind discussion of this manuscript. This work was supported, in whole or in part, by National Institutes of Health (NIH, United States) grants R01-CA202963 and R01-CA202948 (Wan), R21HL109654 (Xie), P30DA035778 (Xie and Bahar) and P41GM103712 (Bahar).
Contributor Information
Xiang-qun Xie, Email: xix15@pitt.edu.
Yong Wan, Email: yong.wan@northwestern.edu.
References
- 1.Tetreault M.P., Yang Y., Katz J.P. Kruppel-like factors in cancer. Nat. Rev. Cancer. 2013;13(10):701–713. doi: 10.1038/nrc3582. [DOI] [PubMed] [Google Scholar]; Tetreault MP, Yang Y, Katz JP. Kruppel-like factors in cancer. Nat Rev Cancer 2013; 13(10): 701-13. [DOI] [PubMed]
- 2.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]; Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126(4): 663-76. [DOI] [PubMed]
- 3.Yu F., Li J., Chen H. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30(18):2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yu F, Li J, Chen H, et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 2011; 30(18): 2161-72. [DOI] [PMC free article] [PubMed]
- 4.Hu D., Gur M., Zhou Z. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat. Commun. 2015;6:8419. doi: 10.1038/ncomms9419. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu D, Gur M, Zhou Z, et al. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun 2015; 6: 8419. [DOI] [PMC free article] [PubMed]
- 5.Rowland B.D., Bernards R., Peeper D.S. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 2005;7(11):1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]; Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol 2005; 7(11): 1074-82. [DOI] [PubMed]
- 6.Zhang L., Zhang L., Xia X., He S., He H., Zhao W. Kruppel-like factor 4 promotes human osteosarcoma growth and metastasis via regulating CRYAB expression. Oncotarget. 2016;7(21):30990–31000. doi: 10.18632/oncotarget.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang L, Zhang L, Xia X, He S, He H, Zhao W. Kruppel-like factor 4 promotes human osteosarcoma growth and metastasis via regulating CRYAB expression. Oncotarget 2016; 7(21): 30990-1000. [DOI] [PMC free article] [PubMed]
- 7.Riverso M., Montagnani V., Stecca B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene. 2017;36(23):3322–3333. doi: 10.1038/onc.2016.481. [DOI] [PMC free article] [PubMed] [Google Scholar]; Riverso M, Montagnani V, Stecca B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene 2017; 36(23): 3322-33. [DOI] [PMC free article] [PubMed]
- 8.Leng Z., Tao K., Xia Q. Kruppel-like factor 4 acts as an oncogene in colon cancer stem cell-enriched spheroid cells. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056082. [DOI] [PMC free article] [PubMed] [Google Scholar]; Leng Z, Tao K, Xia Q, et al. Kruppel-like factor 4 acts as an oncogene in colon cancer stem cell-enriched spheroid cells. PLoS One 2013; 8(2): e56082. [DOI] [PMC free article] [PubMed]
- 9.Li Y., Xian M., Yang B., Ying M., He Q. Inhibition of KLF4 by statins reverses adriamycin-induced metastasis and cancer stemness in osteosarcoma cells. Stem Cell Rep. 2017;8(6):1617–1629. doi: 10.1016/j.stemcr.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li Y, Xian M, Yang B, Ying M, He Q. Inhibition of KLF4 by Statins Reverses Adriamycin-Induced Metastasis and Cancer Stemness in Osteosarcoma Cells. Stem Cell Reports 2017; 8(6): 1617-29. [DOI] [PMC free article] [PubMed]
- 10.Oshima N., Yamada Y., Nagayama S. Induction of cancer stem cell properties in colon cancer cells by defined factors. PLoS One. 2014;9(7) doi: 10.1371/journal.pone.0101735. [DOI] [PMC free article] [PubMed] [Google Scholar]; Oshima N, Yamada Y, Nagayama S, et al. Induction of cancer stem cell properties in colon cancer cells by defined factors. PLoS One 2014; 9(7): e101735. [DOI] [PMC free article] [PubMed]
- 11.Nishi M., Sakai Y., Akutsu H. Induction of cells with cancer stem cell properties from nontumorigenic human mammary epithelial cells by defined reprogramming factors. Oncogene. 2014;33(5):643–652. doi: 10.1038/onc.2012.614. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nishi M, Sakai Y, Akutsu H, et al. Induction of cells with cancer stem cell properties from nontumorigenic human mammary epithelial cells by defined reprogramming factors. Oncogene 2014; 33(5): 643-52. [DOI] [PMC free article] [PubMed]
- 12.Muller M., Hermann P.C., Liebau S. The role of pluripotency factors to drive stemness in gastrointestinal cancer. Stem Cell Res. 2016;16(2):349–357. doi: 10.1016/j.scr.2016.02.005. [DOI] [PubMed] [Google Scholar]; Muller M, Hermann PC, Liebau S, et al. The role of pluripotency factors to drive stemness in gastrointestinal cancer. Stem Cell Res 2016; 16(2): 349-57. [DOI] [PubMed]
- 13.Talmasov D., Xinjun Z., Yu B. Kruppel-like factor 4 is a radioprotective factor for the intestine following gamma-radiation-induced gut injury in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;308(2):G121–G138. doi: 10.1152/ajpgi.00080.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Talmasov D, Xinjun Z, Yu B, et al. Kruppel-like factor 4 is a radioprotective factor for the intestine following gamma-radiation-induced gut injury in mice. Am J Physiol Gastrointest Liver Physiol 2015; 308(2): G121-38. [DOI] [PMC free article] [PubMed]
- 14.Yoon H.S., Yang V.W. Requirement of Kruppel-like factor 4 in preventing entry into mitosis following DNA damage. J. Biol. Chem. 2004;279(6):5035–5041. doi: 10.1074/jbc.M307631200. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yoon HS, Yang VW. Requirement of Kruppel-like factor 4 in preventing entry into mitosis following DNA damage. J Biol Chem 2004; 279(6): 5035-41. [DOI] [PMC free article] [PubMed]
- 15.Yoon H.S., Chen X., Yang V.W. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J. Biol. Chem. 2003;278(4):2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem 2003; 278(4): 2101-5. [DOI] [PMC free article] [PubMed]
- 16.Yoon H.S., Ghaleb A.M., Nandan M.O., Hisamuddin I.M., Dalton W.B., Yang V.W. Kruppel-like factor 4 prevents centrosome amplification following gamma-irradiation-induced DNA damage. Oncogene. 2005;24(25):4017–4025. doi: 10.1038/sj.onc.1208576. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yoon HS, Ghaleb AM, Nandan MO, Hisamuddin IM, Dalton WB, Yang VW. Kruppel-like factor 4 prevents centrosome amplification following gamma-irradiation-induced DNA damage. Oncogene 2005; 24(25): 4017-25. [DOI] [PMC free article] [PubMed]
- 17.Zhou Q., Hong Y., Zhan Q., Shen Y., Liu Z. Role for Kruppel-like factor 4 in determining the outcome of p53 response to DNA damage. Cancer Res. 2009;69(21):8284–8292. doi: 10.1158/0008-5472.CAN-09-1345. [DOI] [PubMed] [Google Scholar]; Zhou Q, Hong Y, Zhan Q, Shen Y, Liu Z. Role for Kruppel-like factor 4 in determining the outcome of p53 response to DNA damage. Cancer Res 2009; 69(21): 8284-92. [DOI] [PubMed]
- 18.Ghaleb A.M., Katz J.P., Kaestner K.H., Du J.X., Yang V.W. Kruppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene. 2007;26(16):2365–2373. doi: 10.1038/sj.onc.1210022. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ghaleb AM, Katz JP, Kaestner KH, Du JX, Yang VW. Kruppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene 2007; 26(16): 2365-73. [DOI] [PMC free article] [PubMed]
- 19.Bedford M.T., Clarke S.G. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell 2009; 33(1): 1-13. [DOI] [PMC free article] [PubMed]
- 20.Stopa N., Krebs J.E., Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell. Mol. Life Sci. 2015;72(11):2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci 2015; 72(11): 2041-59. [DOI] [PMC free article] [PubMed]
- 21.Zhu K., Jiang C., Tao H., Liu J., Zhang H., Luo C. Identification of a novel selective small-molecule inhibitor of protein arginine methyltransferase 5 (PRMT5) by virtual screening, resynthesis and biological evaluations. Bioorg. Med. Chem. Lett. 2018;28(9):1476–1483. doi: 10.1016/j.bmcl.2018.03.087. [DOI] [PubMed] [Google Scholar]; Zhu K, Jiang C, Tao H, Liu J, Zhang H, Luo C. Identification of a novel selective small-molecule inhibitor of protein arginine methyltransferase 5 (PRMT5) by virtual screening, resynthesis and biological evaluations. Bioorg Med Chem Lett 2018; 28(9): 1476-83. [DOI] [PubMed]
- 22.Mao R., Shao J., Zhu K. Potent, selective, and cell active protein arginine methyltransferase 5 (PRMT5) inhibitor developed by structure-based virtual screening and hit optimization. J. Med. Chem. 2017;60(14):6289–6304. doi: 10.1021/acs.jmedchem.7b00587. [DOI] [PubMed] [Google Scholar]; Mao R, Shao J, Zhu K, et al. Potent, Selective, and Cell Active Protein Arginine Methyltransferase 5 (PRMT5) Inhibitor Developed by Structure-Based Virtual Screening and Hit Optimization. J Med Chem 2017; 60(14): 6289-304. [DOI] [PubMed]
- 23.Chan-Penebre E., Kuplast K.G., Majer C.R. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015;11(6):432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]; Chan-Penebre E, Kuplast KG, Majer CR, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol 2015; 11(6): 432-7. [DOI] [PubMed]
- 24.Jin Y., Zhou J., Xu F. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J. Clin. Invest. 2016;126(10):3961–3980. doi: 10.1172/JCI85239. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jin Y, Zhou J, Xu F, et al. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J Clin Invest 2016; 126(10): 3961-80. [DOI] [PMC free article] [PubMed]
- 25.Chiang K., Zielinska A.E., Shaaban A.M. PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 2017;21(12):3498–3513. doi: 10.1016/j.celrep.2017.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chiang K, Zielinska AE, Shaaban AM, et al. PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell Rep 2017; 21(12): 3498-513. [DOI] [PMC free article] [PubMed]
- 26.Wang Z., Kong J., Wu Y. PRMT5 determines the sensitivity to chemotherapeutics by governing stemness in breast cancer. Breast Cancer Res. Treat. 2018;168(2):531–542. doi: 10.1007/s10549-017-4597-6. [DOI] [PubMed] [Google Scholar]; Wang Z, Kong J, Wu Y, et al. PRMT5 determines the sensitivity to chemotherapeutics by governing stemness in breast cancer. Breast Cancer Res Treat 2018; 168(2): 531-42. [DOI] [PubMed]
- 27.Ren D., Zhu X., Kong R. Targeting brain-adaptive cancer stem cells prohibits brain metastatic colonization of triple-negative breast cancer. Cancer Res. 2018;78(8):2052–2064. doi: 10.1158/0008-5472.CAN-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ren D, Zhu X, Kong R, et al. Targeting Brain-Adaptive Cancer Stem Cells Prohibits Brain Metastatic Colonization of Triple-Negative Breast Cancer. Cancer Res 2018; 78(8): 2052-64. [DOI] [PMC free article] [PubMed]
- 28.Alinari L., Mahasenan K.V., Yan F. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood. 2015;125(16):2530–2543. doi: 10.1182/blood-2014-12-619783. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alinari L, Mahasenan KV, Yan F, et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood 2015; 125(16): 2530-43. [DOI] [PMC free article] [PubMed]
- 29.Kong G.M., Yu M., Gu Z. Selective small-chemical inhibitors of protein arginine methyltransferase 5 with anti-lung cancer activity. PLoS One. 2017;12(8) doi: 10.1371/journal.pone.0181601. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kong GM, Yu M, Gu Z, et al. Selective small-chemical inhibitors of protein arginine methyltransferase 5 with anti-lung cancer activity. PLoS One 2017; 12(8): e0181601. [DOI] [PMC free article] [PubMed]
- 30.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat. Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods 2015; 12(1): 7-8. [DOI] [PMC free article] [PubMed]
- 31.Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30(12):1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014; 30(12): 1771-3. [DOI] [PMC free article] [PubMed]
- 32.Chen J.Z., Wang J., Xie X.Q. GPCR structure-based virtual screening approach for CB2 antagonist search. J. Chem. Inf. Model. 2007;47(4):1626–1637. doi: 10.1021/ci7000814. [DOI] [PubMed] [Google Scholar]; Chen JZ, Wang J, Xie XQ. GPCR structure-based virtual screening approach for CB2 antagonist search. J Chem Inf Model 2007; 47(4): 1626-37. [DOI] [PubMed]
- 33.Denkert C., Liedtke C., Tutt A., von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389(10087):2430–2442. doi: 10.1016/S0140-6736(16)32454-0. [DOI] [PubMed] [Google Scholar]; Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017; 389(10087): 2430-42. [DOI] [PubMed]
- 34.Bonday Z.Q., Cortez G.S., Grogan M.J. LLY-283, a potent and selective inhibitor of arginine methyltransferase 5, PRMT5, with antitumor activity. ACS Med. Chem. Lett. 2018;9(7):612–617. doi: 10.1021/acsmedchemlett.8b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bonday ZQ, Cortez GS, Grogan MJ, et al. LLY-283, a Potent and Selective Inhibitor of Arginine Methyltransferase 5, PRMT5, with Antitumor Activity. ACS Med Chem Lett 2018; 9(7): 612-7. [DOI] [PMC free article] [PubMed]
- 35.Jansson M., Durant S.T., Cho E.C. Arginine methylation regulates the p53 response. Nat. Cell Biol. 2008;10(12):1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]; Jansson M, Durant ST, Cho EC, et al. Arginine methylation regulates the p53 response. Nat Cell Biol 2008; 10(12): 1431-9. [DOI] [PubMed]
- 36.Anne J. Arginine methylation of SmB is required for Drosophila germ cell development. Development. 2010;137(17):2819–2828. doi: 10.1242/dev.052944. [DOI] [PubMed] [Google Scholar]; Anne J. Arginine methylation of SmB is required for Drosophila germ cell development. Development 2010; 137(17): 2819-28. [DOI] [PubMed]
- 37.Herrmann F., Pably P., Eckerich C., Bedford M.T., Fackelmayer F.O. Human protein arginine methyltransferases in vivo--distinct properties of eight canonical members of the PRMT family. J. Cell Sci. 2009;122:667–677. doi: 10.1242/jcs.039933. Pt 5. [DOI] [PubMed] [Google Scholar]; Herrmann F, Pably P, Eckerich C, Bedford MT, Fackelmayer FO. Human protein arginine methyltransferases in vivo--distinct properties of eight canonical members of the PRMT family. J Cell Sci 2009; 122(Pt 5): 667-77. [DOI] [PubMed]
- 38.Gao Y., Yang P., Shen H. Small-molecule inhibitors targeting INK4 protein p18(INK4C) enhance ex vivo expansion of haematopoietic stem cells. Nat. Commun. 2015;6:6328. doi: 10.1038/ncomms7328. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gao Y, Yang P, Shen H, et al. Small-molecule inhibitors targeting INK4 protein p18(INK4C) enhance ex vivo expansion of haematopoietic stem cells. Nat Commun 2015; 6: 6328. [DOI] [PMC free article] [PubMed]
- 39.Zhou Z., Luo A., Shrivastava I. Regulation of XIAP turnover reveals a role for USP11 in promotion of tumorigenesis. EBioMedicine. 2017;15:48–61. doi: 10.1016/j.ebiom.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhou Z, Luo A, Shrivastava I, et al. Regulation of XIAP Turnover Reveals a Role for USP11 in Promotion of Tumorigenesis. EBioMedicine 2017; 15: 48-61. [DOI] [PMC free article] [PubMed]
- 40.Gamper A.M., Qiao X., Kim J. Regulation of KLF4 turnover reveals an unexpected tissue-specific role of pVHL in tumorigenesis. Mol. Cell. 2012;45(2):233–243. doi: 10.1016/j.molcel.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gamper AM, Qiao X, Kim J, et al. Regulation of KLF4 turnover reveals an unexpected tissue-specific role of pVHL in tumorigenesis. Mol Cell 2012; 45(2): 233-43. [DOI] [PMC free article] [PubMed]
- 41.Huang S., Chi Y., Qin Y. CAPG enhances breast cancer metastasis by competing with PRMT5 to modulate STC-1 transcription. Theranostics. 2018;8(9):2549–2564. doi: 10.7150/thno.22523. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang S, Chi Y, Qin Y, et al. CAPG enhances breast cancer metastasis by competing with PRMT5 to modulate STC-1 transcription. Theranostics 2018; 8(9): 2549-64. [DOI] [PMC free article] [PubMed]
- 42.Gros L., Delaporte C., Frey S. Identification of new drug sensitivity genes using genetic suppressor elements: protein arginine N-methyltransferase mediates cell sensitivity to DNA-damaging agents. Cancer Res. 2003;63(1):164–171. [PubMed] [Google Scholar]; Gros L, Delaporte C, Frey S, et al. Identification of new drug sensitivity genes using genetic suppressor elements: protein arginine N-methyltransferase mediates cell sensitivity to DNA-damaging agents. Cancer Res 2003; 63(1): 164-71. [PubMed]
- 43.Hu D., Zhou Z., Davidson N.E., Huang Y., Wan Y. Novel insight into KLF4 proteolytic regulation in estrogen receptor signaling and breast carcinogenesis. J. Biol. Chem. 2012;287(17):13584–13597. doi: 10.1074/jbc.M112.343566. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu D, Zhou Z, Davidson NE, Huang Y, Wan Y. Novel insight into KLF4 proteolytic regulation in estrogen receptor signaling and breast carcinogenesis. J Biol Chem 2012; 287(17): 13584-97. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The process of 3D structural modelling of PRMT5-KLF4 complex and the virtual screening characterized WX2-43 as a PRMT5-KLF4 novel inhibitor. We first constructed the complex of PRMT5-KLF4 with the missing region of our interest (residues 370-383; PKPKRGRRSWPRKR). Then we carried out virtual screening against a compound library using the binding pocket of SAM in PRMT5. WX2-43, one of our active compounds, was confirmed and found to specifically inhibit KLF4 methylation. Moreover, WX2-43 shows high potency and novel structure.
Supplementary material