Key Points
Question
What genetic variants are associated with corneal hysteresis and corneal resistance factor, and do these variants confer susceptibility to keratoconus?
Findings
This genome-wide association study of 9029 unique participants identified 9 loci associated with corneal hysteresis and corneal resistance factor in the European Prospective Investigation into Cancer and Nutrition–Norfolk and TwinsUK studies. Four loci have been previously reported as associated with corneal disease, and the other 5 showed a suggestive association with keratoconus in our study; some loci were not previously reported for central corneal thickness.
Meaning
The findings appear to support the role of corneal hysteresis and corneal resistance factor as corneal biomarkers that may provide complementary information to central corneal thickness about corneal properties.
This case-control genome-wide association study identifies genetic variants associated with corneal biomechanical properties and examines whether these variants are associated with keratoconus in adults.
Abstract
Importance
Keratoconus is an important cause of visual loss in young adults, but little is known about its genetic causes. Understanding the genetic determinants of corneal biomechanical factors may in turn teach us about keratoconus etiology.
Objectives
To identify genetic associations with corneal biomechanical properties and to examine whether these genetic variants are associated with keratoconus.
Design, Setting, and Participants
A stage 1 discovery and replication genome-wide association study (GWAS) of corneal biomechanical properties was performed in 2 cross-sectional populations (6645 participants from the European Prospective Investigation into Cancer and Nutrition [EPIC]–Norfolk Eye Study and 2384 participants from the TwinsUK study). In stage 2, the association of genetic determinants identified in stage 1 with keratoconus was examined in a case-control study. A total of 752 patients with keratoconus were compared with 974 TwinsUK participants (undergoing direct sequencing) or 13 828 EPIC-Norfolk participants (undergoing genotyping and imputation) who were not part of the stage 1 analysis. Data were collected from March 1, 1993, through March 13, 2017, and analyzed from November 1, 2015, through February 1, 2018.
Exposures
In stage 1, allele dosage at genome-wide single-nucleotide polymorphisms (SNPs); in stage 2, allele dosage at SNPs with genome-wide significance (P < 5 × 10−8) in stage 1 and not previously reported as associated with corneal disease.
Main Outcomes and Measures
In stage 1, corneal hysteresis (CH) and corneal resistance factor (CRF), measured with the Ocular Response Analyzer (ORA); in stage 2, association with keratoconus compared with controls.
Results
Among 6645 participants in the discovery cohort (3635 women (54.7%); mean age, 69 years [range, 48-92 years]), 7 genome-wide significant loci associated with CH or CRF were identified that were independently replicated. Two further suggestive loci were identified after meta-analysis. To date, 5 of the identified loci, at ANAPC1, ADAMTS8, ADAMTS17, ABCA6, and COL6A1, have not previously been reported as associated with corneal disease. The ABCA6 locus (rs77542162) was associated with keratoconus using the TwinsUK (odds ratio [OR], 0.50; 95% CI, 0.27-0.92; P = .03) and EPIC-Norfolk controls (OR, 0.39; 95% CI, 0.22-0.70; P = .002). The other loci were associated with keratoconus using TwinsUK (OR per effect allele for ADAMTS8, 0.51 [95% CI, 0.37-0.71; P = 7.9 × 10−5]; for COL6A1, 1.65 [95% CI, 1.05-2.59; P = .03]) or EPIC-Norfolk (OR per effect allele for ANAPC1, 0.78 [95% CI, 0.68-0.89; P = 3.7 × 10−4]; for ADAMTS17, 0.82 [95% CI, 0.68-0.99; P = .04]) controls.
Conclusions and Relevance
Five loci that are associated with corneal biomechanical properties and that have suggestive associations with keratoconus were reported. These findings suggest the role of type VI collagen, extracellular matrix, and connective-tissue development for corneal biomechanics and keratoconus and the role of CH and CRF as biomarkers for keratoconus.
Introduction
Keratoconus is an important cause of visual loss in young adults and is one of the most common indications for corneal transplant.1,2 Linkage studies have identified several genetic regions associated with keratoconus,3 but genome-wide association studies (GWASs) for keratoconus have been underpowered and had limited success to date.4,5 A key clinical feature of keratoconus is progressive corneal thinning,2 and examination of central corneal thickness (CCT) as an endophenotype has proved a successful approach for identifying genetic associations with keratoconus. Variants identified in a GWAS meta-analysis of CCT were subsequently found to be associated with keratoconus, including at FOXO1 (OMIM 136533)6 and COL5A1 (OMIM 120215).7
Corneal hysteresis (CH) and corneal resistance factor (CRF) are measures derived by the Ocular Response Analyzer (ORA; Reichert Technologies). Corneal hysteresis and CRF are thought to provide a more complete characterization of corneal biomechanical properties than CCT.8 Several studies8,9,10,11,12 have reported that CH and CRF are lower in patients with keratoconus than in control individuals, and this difference is evident even in mild disease13 and suspected disease.14 We hypothesize that genes associated with CH and CRF in healthy participants are also associated with keratoconus. To date, no GWAS has been reported for CH or CRF. The aim of our study was to perform a GWAS of CH and CRF in 2 independent European populations and examine any identified loci for association with keratoconus in an independent cohort.
Methods
The discovery GWAS for CH and CRF was performed in participants from the European Prospective Investigation into Cancer and Nutrition (EPIC)–Norfolk Eye Study,15 and replication of results was sought in the TwinsUK study.16 We then performed a meta-analysis of the GWAS results from both studies. Single-nucleotide polymorphisms (SNPs) associated with CH and CRF and not previously reported in association with traits of interest were subsequently genotyped in an independent cohort of patients with keratoconus and tested for association by comparing them with controls from EPIC-Norfolk and TwinsUK who were not part of the CH or CRF quantitative trait GWAS. The EPIC-Norfolk Eye Study followed the principles of the Declaration of Helsinki and the Research Governance Framework for Health and Social Care. The study was approved by the Norfolk Local Research Ethics Committee and East Norfolk and Waveney NHS Research Governance Committee. All participants gave written, informed consent. The St Thomas’ Hospital local research ethics committee approved the study, and all the twin participants volunteered to join the TwinsUK Registry and gave informed consent to attend the hospital for phenotyping and for their data to be used for scientific research.
EPIC-Norfolk Eye Study
The EPIC-Norfolk Eye Study methods have been described in detail15,17,18 and are presented in the eMethods in the Supplement. Corneal hysteresis and CRF were measured with the ORA, which uses a short pulse of air to indent the cornea and measures inward and outward applanation pressures with an electro-optical system.8 Corneal hysteresis is the difference between the inward (P1) and outward (P2) applanation forces,8 and CRF is a linear combination of P1 and P2 that has been designed to maximize correlation with CCT and minimize correlation with intraocular pressure (IOP).19 Three ORA readings were taken per eye, and the best signal values were used, which were based on the pressure waveform assessed by the ORA software. Genotyping methods are described in the eMethods in the Supplement.
TwinsUK Study
The TwinsUK population is from an adult twin registry at St Thomas’ Hospital in London (eMethods in the Supplement). Participants received 2 ORA readings per eye, and the mean value was calculated. Genotyping and sequencing methods are described in the eMethods in the Supplement.
CH and CRF Quantitative Trait GWAS
The discovery GWAS was performed in EPIC-Norfolk Eye Study participants. Associations between genotype and CH or CRF (mean value of both eyes) were examined using linear regression adjusted for age and sex, with imputed allele dosage data and assuming an additive genetic model. Analyses were performed using SNPTEST, version 2.5.1 (Oxford University Innovation) in SNPs with a minor allele frequency (MAF) of at least 1%. The GWAS for CH and CRF were also performed in TwinsUK participants similarly to that in the EPIC-Norfolk participants but further adjusted for interrelatedness of participants, including family structure, using genome-wide efficient mixed-model analysis (GEMMA [http://www.xzlab.org/software.html]).20 The GWAS results from EPIC-Norfolk and TwinsUK were combined using inverse variance–weighted fixed-effect meta-analysis with METAL software.21
Keratoconus Cohort and Association Testing
For SNPs not previously reported as associated with corneal disease, we examined for association with keratoconus. In total, 752 patients with keratoconus were recruited from specialist clinics at Moorfields Eye Hospital, London, United Kingdom. The mean (SD) age of patients at the time of recruitment was 49 (14) years, and 535 (71.1%) were men. Self-reported ethnicity for all patients was European descent. Recruitment was approved by the Moorfields Eye Hospital Research Ethics Committee, and all participants provided written informed consent. Participants with keratoconus were examined as they attended specialist hospital clinics. Each participant was examined using tomography (Pentacam; Oculus), and the presence of keratoconus was confirmed using established criteria based on corneal thinning and corneal distortion.22 A previous bilateral keratoplasty for keratoconus was also accepted as confirmation of disease status. Patients with syndromic disease and keratoconus (eg, Down syndrome, Leber optic atrophy) were excluded. Patients with keratoconus underwent genotyping using custom-designed polymerase chain reaction assays (KASP)23 at LGC Genomics, Hoddesdon, United Kingdom. Genotypes were graphically assessed as Cartesian plots using SNPviewer (LGC Genomics, GmbH). Two separate cohorts were used as control groups. The primary control group was a subset of TwinsUK participants who underwent whole-genome sequencing but did not contribute to the CH or CRF GWAS (n = 974). For additional robustness, EPIC-Norfolk participants who were not included in the CH or CRF GWAS were considered as a second control group (n = 13 828), and for SNPs that were not directly targeted by the genotyping chip, imputed genotype was considered. Although keratoconus status was not specifically ascertained in the TwinsUK or the EPIC-Norfolk study, no participants had self-reported keratoconus in response to general questionnaires on their medical history. Case-control comparisons used logistic regression adjusted for age and sex, assuming an additive genetic model.
For corneal tissue gene expression analysis, we examined the relative expression in human cornea of the closest gene to each locus we identified as associated with CH, CRF, or keratoconus (eMethods in the Supplement). Data were analyzed from November 1, 2015, through February 1, 2018.
Results
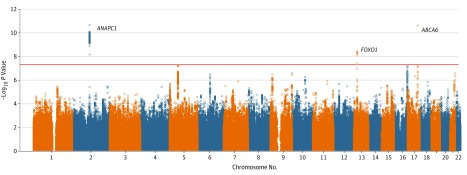
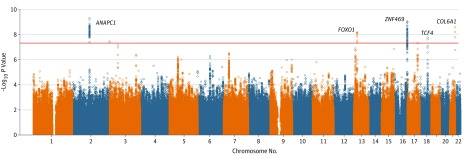
Among the 6645 EPIC-Norfolk participants with genotypic and ORA data, the mean age was 69 years (range 48-92); 3635 (54.7%) were women and 3010 (45.3%) were men. Mean (SD) CH was 10.1 (1.6) mm Hg, and mean (SD) CRF was 10.3 (1.7) mm Hg. Corneal hysteresis and CRF were strongly correlated (r2 = 0.76; P < .001). The EPIC-Norfolk GWAS results (Table 1 and eFigure 1 in the Supplement) demonstrated overlap of the SNPs associated with CH and CRF. Three genome-wide significant loci for CH (Figure 1) and 5 for CRF (Figure 2) were found. Loci in ANAPC1 (OMIM 608473) and near FOXO1 were associated with CH and CRF. In addition, a missense variant in ABCA6 (OMIM 612504) was associated with CH, and variants in or near ZNF469 (OMIM 612078), TCF4 (OMIM 602272), and COL6A1 (OMIM 120220) were associated with CRF. All genome-wide significant SNPs for the discovery CH and CRF GWAS are shown in eTables 1 and 2 in the Supplement, respectively.
Table 1. Top SNP at Each Significant Locus for Genome-Wide Association Study of Corneal Hysteresis and Corneal Resistance Factora.
| Variant Information | EPIC-Norfolk Results | TwinsUK Results | Meta-analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chromosome (Position) | Nearest Gene | Effect Allele/Other Allele | Effect Allele Frequency | Difference per Effect Allele, mm Hg | P Value | Difference per Effect Allele, mm Hg | P Value | Difference per Effect Allele, mm Hg | P Value |
| Corneal Hysteresis | ||||||||||
| rs202110867 | 2 (112550325) | ANAPC1 | G/C | 0.236 | 0.21 (0.15 to 0.27) | 2.3 × 10−11 | 0.20 (0.09 to 0.31) | 5.3 × 10−4 | 0.21 (0.15 to 0.26) | 4.4 × 10−14 |
| rs56009602b | 11 (130419717) | ADAMTS8 | T/C | 0.046 | 0.33 (0.19 to 0.46) | 1.3 × 10−6 | 0.35 (0.11 to 0.59) | .005 | 0.34 (0.23 to 0.46) | 4.3 × 10−9 |
| rs2721051 | 13 (41110884) | FOXO1 | T/C | 0.108 | −0.25 (−0.33 to 0.17) | 4.1 × 10−9 | −0.24 (−0.40 to −0.08) | .003 | −0.25 (−0.32 to −0.17) | 4.5 × 10−11 |
| rs12448211b | 16 (88296907) | ZNF469 | G/A | 0.363 | −0.15 (−0.20 to 0.09) | 7.2 × 10−8 | −0.11 (−0.21 to −0.01) | .03 | −0.14 (−0.19 to −0.09) | 1.7 × 10−8 |
| rs77542162 | 17 (67081278) | ABCA6 | G/A | 0.021 | 0.61 (0.43 to 0.79) | 2.6 × 10−11 | 0.23 (−0.14 to 0.59) | .23 | 0.54 (0.38 to 0.70) | 6.6 × 10−11 |
| Corneal Resistance Factor | ||||||||||
| rs77797441 | 2 (112539384) | ANAPC1 | T/C | 0.223 | 0.22 (0.15 to 0.29) | 5.1 × 10−10 | 0.18 (0.06 to 0.30) | .004 | 0.21 (0.15 to 0.27) | 8.0 × 10−12 |
| rs6445054b | 3 (172274219) | FNDC3B | T/C | 0.823 | 0.19 (0.12 to 0.27) | 4.3 × 10−7 | 0.20 (0.06 to 0.33) | .005 | 0.19 (0.13 to 0.26) | 1.4 × 10−8 |
| rs56009602b | 11 (130419717) | ADAMTS8 | T/C | 0.046 | 0.36 (0.22 to 0.51) | 1.2 × 10−6 | 0.40 (0.15 to 0.65) | .002 | 0.38 (0.25 to 0.50) | 3.3 × 10−9 |
| rs2721051 | 13 (41110884) | FOXO1 | T/C | 0.108 | −0.27 (−0.36 to 0.18) | 6.8 × 10−9 | −0.27 (−0.44 to −0.10) | .002 | −0.27 (−0.35 to −0.19) | 5.2 × 10−11 |
| rs72755233b | 15 (100152748) | ADAMTS17 | A/G | 0.111 | 0.24 (0.15 to 0.33) | 1.6 × 10−7 | 0.10 (−0.08 to 0.28) | .29 | 0.23 (0.15 to 0.32) | 4.5 × 10−8 |
| rs12719932 | 16 (88330349) | ZNF469 | A/G | 0.363 | −0.18 (0.12 to 0.24) | 9.4 × 10−10 | −0.13 (0.03 to 0.24) | .01 | −0.17 (0.12 to 0.22) | 5.0 × 10−11 |
| rs11659764 | 18 (53335512) | TCF4 | A/T | 0.056 | −0.35 (−0.47 to 0.23) | 2.6 × 10−8 | −0.23 (−0.45 to −0.02) | .04 | −0.32 (−0.43 to −0.21) | 3.9 × 10−9 |
| rs182804464 | 21 (47420667) | COL6A1 | G/C | 0.016 | −0.69 (−0.92 to 0.47) | 2.8 × 10−9 | −0.93 (−1.47 to −0.39) | 7.3 × 10−4 | −0.73 (−0.94 to −0.52) | 1.0 × 10−11 |
Abbreviations: EPIC, European Prospective Investigation into Cancer and Nutrition; SNP, single-nucleotide polymorphism.
Results are shown for the discovery cohort EPIC-Norfolk (n = 6645), replication cohort TwinsUK (n = 2384), and meta-analysis of both populations. All genome-wide significant SNPs for EPIC-Norfolk and the meta-analysis are presented in eTables 1-4 in the Supplement.
Indicates genome-wide significance only after meta-analysis.
Figure 1. Manhattan Plot for Corneal Hysteresis Genome-Wide Association Study in the European Prospective Investigation Into Cancer and Nutrition–Norfolk Study Population.
The horizontal red line represents the genome-wide significance threshold (P = 5 × 10−8).
Figure 2. Manhattan Plot for Corneal Resistance Factor Genome-Wide Association Study in the European Prospective Investigation Into Cancer and Nutrition–Norfolk Study Population.
The horizontal red line represents the genome-wide significance threshold (P = 5 × 10−8).
In the TwinsUK cohort, 2384 participants had genotypic and ORA data; mean (SD) age was 58 (12) years; and 2265 (95.0%) were women and 119 (5.0%) were men. With the exception of the ABCA6 locus (rs77542162), all other SNPs associated with CH or CRF in the EPIC-Norfolk study were replicated in the TwinsUK population (Table 1). The nonreplication of the ABCA6 SNP (rs77542162) may be attributed to its low MAF in both cohorts (2%) and to the fact this SNP was directly genotyped in the EPIC-Norfolk population but imputed in the TwinsUK population.
The meta-analysis of the CH and CRF GWAS from the EPIC-Norfolk and TwinsUK populations identified 3 additional genome-wide significant loci (Table 1 and eTables 3 and 4 in the Supplement). A variant in ADAMTS8 (OMIM 605175; rs56009602) was associated with CH (difference per effect allele, 0.34 [95% CI, 0.23-0.46] mm Hg; P = 4.3 × 10−9) and CRF (difference per effect allele, 0.38 [95% CI, 0.25-0.50] mm Hg; P = 3.3 × 10−9), and a missense variant in ADAMTS17 (OMIM 607511; rs72755233) was associated with CRF (difference per effect allele, 0.23 [95% CI, 0.15-0.32] mm Hg; P = 4.5 × 10−8). A variant in FNDC3B (OMIM 611909; rs6445054) was associated with CRF (difference per effect allele, 0.19 [95% CI, 0.13-0.26] mm Hg; P = 1.4 × 10−8). Locus-specific Manhattan plots for all identified loci from the meta-analysis are presented in eFigures 2 and 3 in the Supplement. For all loci, no significant expression quantitative trait locus effects were observed in any of the available Genotype-Tissue Expression project24 tissues beyond what we would have expected under multiple testing.
Four of the 9 loci associated with CH or CRF have previously been associated with keratoconus (at FOXO1, ZNF469, and FNDC3B)6 or Fuchs endothelial corneal dystrophy (FECD; at TCF4),25 supporting our hypothesis that CH and CRF are etiologically and genetically linked to corneal disease. We then examined the remaining 5 loci for association with keratoconus in 752 cases compared with 974 participants from TwinsUK undergoing independent direct sequencing or 13 828 participants from EPIC-Norfolk undergoing genotyping and imputation, none of whom were included in the CH and CRF quantitative analyses. We did not hypothesize that the TCF4 locus would be associated with keratoconus, given its reported association with a different corneal disorder (FECD); we therefore did not genotype the TCF4 locus in the patients with keratoconus. For all loci except ANAPC1, we used the most significant SNP from the CH and CRF quantitative analyses; the leading 2 ANAPC1 SNPs (rs77797441 and rs202110867, 1 each from the CH and CRF quantitative analyses) could not be successfully genotyped in the patients with keratoconus. Therefore, another SNP (rs114755846) in high linkage disequilibrium with these SNPs and almost as significantly associated with CH (difference per effect allele, 0.20 [95% CI, 0.14-0.26] mm Hg; P = 4.4 × 10−11) and CRF (difference per effect allele, 0.20 [95% CI, 0.13-0.27] mm Hg; P = 9.0 × 10−10) was adopted for the keratoconus validation.
Table 2 and eFigure 4 in the Supplement present the results for associations with keratoconus. The ABCA6 locus was nominally associated with keratoconus with TwinsUK and EPIC-Norfolk controls. The ADAMTS8 and COL6A1 loci were nominally associated with keratoconus using TwinsUK controls, and the ANAPC1 and ADAMTS17 loci were nominally associated with keratoconus using EPIC-Norfolk controls. The direction of effect at all 5 loci was the same using TwinsUK or EPIC-Norfolk controls (eFigure 4 in the Supplement). All directions of effect with keratoconus were as expected from the association with CH or CRF (ie, odds ratios [ORs] were >1.00 for alleles that were associated with lower CH and CRF). The power for detecting an association for each test is presented in eFigure 4 in the Supplement. After applying a stringent Bonferroni-corrected threshold for 10 tests of association with keratoconus (P = .005), associated 4 loci were at ADAMTS8 (using TwinsUK controls; OR per effect allele, 0.51 [95% CI, 0.37-0.71]; P = 7.9 × 10−5), ANPAC1 (using EPIC-Norfolk controls; OR per effect allele, 0.78 [95% CI, 0.68-0.89]; P = 3.7 × 10−4), and ABCA6 (using 6 EPIC-Norfolk controls; OR per effect allele, 0.39 [95% CI, 0.22-0.70]; P = .002).
Table 2. Associations of Loci With Keratoconusa.
| Chromosome No. | Gene | SNP | Effect Allele/Other Allele | Effect Allele Frequency in Cases | TwinsUK Controls | EPIC-Norfolk Controls | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect Allele Frequency | Odds Ratio (95% CI) | P Value | Effect Allele Frequency | Odds Ratio (95% CI) | P Value | Information Score | |||||
| 2 | ANAPC1 | rs114755846 | T/A | 0.195 | 0.215 | 0.89 (0.76-1.03) | .12 | 0.235 | 0.78 (0.68-0.89) | 3.7 × 10−4 | 0.996 |
| 11 | ADAMTS8 | rs56009602 | T/C | 0.029 | 0.055 | 0.51 (0.37-0.71) | 7.9 × 10−5 | 0.036 | 0.85 (0.61-1.17) | .31 | 0.862 |
| 15 | ADAMTS17 | rs72755233 | A/G | 0.093 | 0.108 | 0.85 (0.69-1.04) | .12 | 0.109 | 0.82 (0.68-0.99) | .04 | Directly genotyped |
| 17 | ABCA6 | rs77542162 | G/A | 0.008 | 0.016 | 0.50 (0.27-0.92) | .03 | 0.021 | 0.39 (0.22-0.70) | .002 | Directly genotyped |
| 21 | COL6A1 | rs182804464 | G/C | 0.021 | 0.013 | 1.65 (1.05-2.59) | .03 | 0.017 | 1.23 (0.83-1.81) | .30 | 0.978 |
Abbreviations: EPIC, European Prospective Investigation into Cancer and Nutrition; SNP, single-nucleotide polymorphism.
Results include 752 patients with keratoconus compared with TwinsUK controls undergoing sequencing (n = 974) and EPIC-Norfolk controls undergoing genotyping or imputation (n = 13 828). Effect allele frequencies are presented for the patients with keratoconus, TwinsUK controls, and EPIC-Norfolk controls separately.
Our gene expression analyses (eTable 5 in the Supplement) showed that ANAPC1 was expressed at a moderate level across all corneal cell types analyzed. ABCA6 was very highly expressed in the stroma compared with other corneal cell types. COL6A1 was the most highly expressed transcript in all cell types investigated (excluding stroma), with especially high levels in the endothelium. Relatively low levels of ADAMTS8 and ADAMTS17 expression raised the possibility that these genes are upregulated only in more distensible corneas.
Discussion
Our GWAS of CH and CRF and subsequent meta-analysis identified 9 loci, 7 of which were nominally associated in both the EPIC-Norfolk and TwinsUK populations. Of these loci, 3 were previously associated with CCT and keratoconus (FOXO1, ZNF469, and FNDC3B).6 Another locus (near TCF4) has previously been associated with FECD25 but was not reported to be associated with CCT or keratoconus. Five of the loci (ANAPC1, ADAMTS8, ADAMTS17, ABCA6, and COL6A1) have not been previously associated with CCT or keratoconus, to the best of our knowledge. Although we found evidence of association for these 5 loci with keratoconus, the significance levels varied considerably when using different control groups, raising the possibility of chance findings.
The finding that loci previously associated with CCT and keratoconus are also associated with CH and CRF supports the use of the ORA as a noninvasive instrument to measure corneal biomechanics. TCF4 has not been previously implicated for CCT, despite a much larger sample (>20 000) contributing to the CCT GWAS meta-analysis.6 This finding suggests that CH and CRF may measure different attributes of corneal biomechanics than CCT. The locus near TCF4 is known to confer major susceptibility to FECD,25 and, to date, no CCT GWAS hits have demonstrated association with FECD. Given that CH and CRF are derived by measuring the response of the cornea to mechanical distortion, it has been suggested that CH and CRF provide a dynamic measure of corneal biomechanics rather than the static geometric measurement of CCT.8 Our data support the concept that CH and CRF complement CCT when measuring corneal biomechanics and may provide insight into corneal diseases other than keratoconus, such as FECD.
The most statistically significant SNP that we identified as associated with CH and CRF was a common intronic variant in ANAPC1. ANAPC1 has been implicated in regulation of cell-cycle progression and has not been previously associated with ocular traits. However, a locus in ANAPC1 has been associated with bone mineral density,26 suggesting that a common process may underlie bone and corneal biomechanics.
The ABCA6 SNP (rs77542162) associated with CH was relatively uncommon, with a MAF of 2.1%, and results in a missense mutation. This variant was directly genotyped in the EPIC-Norfolk population as part of the exome chip content on the Affymetrix Axiom UK Biobank platform (Thermo Fisher Scientific, Inc). Expression of ABCA6 has been shown to be increased in dermal fibroblasts of patients with pseudoxanthoma elasticum.27 Pseudoxanthoma elasticum is a heritable disease of connective tissues that is typically associated with retinal abnormalities,28 although it has also been associated with keratoconus.29
The COL6A1 SNP (rs182804464) associated with CRF was also uncommon (MAF, 1.6%). COL6A1 encodes type VI collagen, which is present in human corneal stroma.30 Variation in COL6A1 has mainly been associated with myopathies, including the autosomal dominant Bethlem myopathy,30 but has not to date been associated with ocular disease.
The most significant SNP at the FOXO1 locus, rs2721051, was also the most significantly associated with CCT6 and, subsequently, with keratoconus (OR, 1.62; 95% CI, 1.40-1.88; P = 2.7 × 10−10).6,23 The directions of effect are consistent, with the T allele being associated with a lower CH and CRF in the present study and a thinner CCT in the previously reported GWAS,6 which is in agreement with an increased risk of keratoconus. The rs2721051 SNP is in a conserved region and DNase I hypersensitivity site approximately 20 000 base pairs upstream of FOXO1 and therefore may have a regulatory function.
The most significant SNP at the ZNF469 locus was the upstream rs12719932 variant. This SNP is in moderate linkage disequilibrium with the most significant SNP at the same locus in the previously reported CCT GWAS6 (rs6540223; r2 = 0.71; D′ = 0.86). The T allele at rs6540223 was associated with a thicker CCT and, unexpectedly, an increased risk of keratoconus (OR, 1.25; 95% CI, 1.11-1.40; P = 1.9 × 10−4).6 In our study, the T allele at rs6540223 was associated with a higher CH (difference per effect allele, 0.13 [95% CI, 0.07-0.18] mm Hg; P = 3.8 × 10−6) and CRF (difference per effect allele, 0.16 [95% CI, 0.10-0.22] mm Hg; P = 6.5 × 10−8), although not at genome-wide significant levels. This finding is consistent with the T allele being associated with a thicker CCT, because CCT is positively correlated with CH and CRF.10
After meta-analysis of the results from the EPIC-Norfolk and TwinsUK studies, we identified 2 further loci associated with CH or CRF, in ADAMTS17 (rs72755233; MAF = 11.1%) and in ADAMTS8 (rs56009602; MAF = 4.6%). Both genes are members of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family. The ADAMTS17 variant codes for an amino acid substitution (Thr446Asn), which, according to ClinVar estimations, is unlikely to have any serious clinical consequences. ADAMTS17 has been associated with ocular features of Weill-Marchesani syndrome, including ectopia lentis and spherophakia, suggesting a role in ocular connective-tissue properties.31 ADAMTS8 has previously been associated with IOP.32 However, measures of IOP are made via the cornea, and genetic associations with measured IOP may be related to corneal biomechanics rather than actual IOP inside the anterior chamber.
To determine the potential clinical relevance of our identified CH and CRF loci, we examined for association with keratoconus, a corneal disease associated with low CH and CRF. The ABCA6 locus was nominally associated with keratoconus using TwinsUK or EPIC controls. Less consistent results were seen for the other 4 loci, each of which were associated with keratoconus using one control group but not the other. For example, the relatively uncommon T allele at rs56009602 in ADAMTS8 was strongly associated with a reduced risk of keratoconus using TwinsUK controls (OR per effect allele, 0.51 [95% CI, 0.37-0.71]; P = 7.9 × 10−5) but not using EPIC-Norfolk controls (OR per effect allele 0.85 [95% CI, 0.61-1.17]; P = .31). The associations with keratoconus may be due to chance, or the inconsistency may be stochastic and related to the limited statistical power (eFigure 4 in the Supplement) and different study methods in the control groups (sequencing vs genotyping and imputation). The directions of effect were the same for each locus for both control groups (eFigure 4 in the Supplement), and all directions of effect with keratoconus were in keeping with the direction expected from the CH and CRF GWAS (the allele associated with lower CH and CRF was associated with an increased risk of keratoconus).
We supplemented the published evidence reporting lower CH in corneas with keratoconus8,9,10,11,12,13,14 and showed that the genetic determinants of CH and CRF may also contribute to pathological processes in keratoconus. How CH and CRF might be mechanically linked to the development of keratoconus remains unclear. Increased collagen distensibility has been hypothesized to be an important etiological factor for keratoconus.33 The distensibility of a membrane is determined by its elastic properties in addition to the membrane mass and the forces acting on the membrane. Corneal hysteresis is thought to represent the viscoelastic dampening properties of the cornea8 and may therefore provide an in vivo measurement of a component of corneal distensibility.
Strengths and Limitations
The strengths of our study include the large sample size to provide sufficient power to perform the first GWAS for CH and CRF to our knowledge. A limitation of our study was that the control populations were not genotyped using the same method as our keratoconus cases. However, to minimize bias, we included comparisons with 2 separate control groups, one of which (the TwinsUK group) underwent sequencing, further reducing the chance of results due to artifact or genotyping irregularities. Overall, the keratoconus study was inadequately powered to identify associations consistently with the TwinsUK and EPIC-Norfolk controls for 4 of 5 loci (eFigure 4 in the Supplement). In addition, some loci were imputed and not directly genotyped in EPIC-Norfolk and may therefore have been inaccurately called, especially for rarer variants.
Conclusions
This study’s findings appear to provide evidence for 9 loci associated with corneal biomechanical properties, including 4 previously reported loci. One of these loci, at ABCA6, was associated with keratoconus using 2 independent control populations. Suggestive associations with keratoconus were found for the other 4 loci at ANAPC1, ADAMTS8, ADAMTS17, and COL6A1. Collectively, these findings suggest the importance of type VI collagen, extracellular matrix, and connective-tissue development for corneal biomechanical properties and keratoconus. This study also supports the role of the ORA measuring corneal properties, which are complementary to the information provided by CCT.
eMethods. Participants and Analyses
eFigure 1. Q-Q Plots for the CH (Left) and CRF (Right) Discovery GWAS in EPIC-Norfolk
eFigure 2. LocusZoom Plots for Significant Loci From the GWAS Meta-analysis of Corneal Hysteresis in EPIC-Norfolk and TwinsUK
eFigure 3. LocusZoom Plots for Significant Loci From the GWAS Meta-analysis of Corneal Resistance Factor in EPIC-Norfolk and TwinsUK
eFigure 4. Associations of Loci With Keratoconus
eTable 1. Genome-wide Significant SNPs From Corneal Hysteresis GWAS in EPIC-Norfolk
eTable 2. Genome-wide Significant SNPs From Corneal Resistance Factor GWAS in EPIC-Norfolk
eTable 3. Genome-wide Significant SNPs From Meta-analysis of Corneal Hysteresis GWAS in EPIC-Norfolk and TwinsUK
eTable 4. Genome-wide Significant SNPs From Meta-analysis of Corneal Resistance Factor GWAS in EPIC-Norfolk and TwinsUK
eTable 5. Corneal Tissue Specific Expression Profiles
eReferences.
References
- 1.Gain P, Jullienne R, He Z, et al. Global survey of corneal transplantation and eye banking. JAMA Ophthalmol. 2016;134(2):167-173. doi: 10.1001/jamaophthalmol.2015.4776 [DOI] [PubMed] [Google Scholar]
- 2.Mas Tur V, MacGregor C, Jayaswal R, O’Brart D, Maycock N. A review of keratoconus: diagnosis, pathophysiology, and genetics. Surv Ophthalmol. 2017;62(6):770-783. doi: 10.1016/j.survophthal.2017.06.009 [DOI] [PubMed] [Google Scholar]
- 3.Bykhovskaya Y, Margines B, Rabinowitz YS. Genetics in keratoconus: where are we? Eye Vis (Lond). 2016;3(1):16. doi: 10.1186/s40662-016-0047-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdon KP, Macgregor S, Bykhovskaya Y, et al. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Invest Ophthalmol Vis Sci. 2011;52(11):8514-8519. doi: 10.1167/iovs.11-8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Bykhovskaya Y, Haritunians T, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. 2012;21(2):421-429. doi: 10.1093/hmg/ddr460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y, Vitart V, Burdon KP, et al. ; NEIGHBOR Consortium . Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013;45(2):155-163. doi: 10.1038/ng.2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Bykhovskaya Y, Canedo ALC, et al. Genetic association of COL5A1 variants in keratoconus patients suggests a complex connection between corneal thinning and keratoconus. Invest Ophthalmol Vis Sci. 2013;54(4):2696-2704. doi: 10.1167/iovs.13-11601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luce DA. Determining in vivo biomechanical properties of the cornea with an ocular response analyzer. J Cataract Refract Surg. 2005;31(1):156-162. doi: 10.1016/j.jcrs.2004.10.044 [DOI] [PubMed] [Google Scholar]
- 9.Shah S, Laiquzzaman M, Bhojwani R, Mantry S, Cunliffe I. Assessment of the biomechanical properties of the cornea with the ocular response analyzer in normal and keratoconic eyes. Invest Ophthalmol Vis Sci. 2007;48(7):3026-3031. doi: 10.1167/iovs.04-0694 [DOI] [PubMed] [Google Scholar]
- 10.Touboul D, Roberts C, Kérautret J, et al. Correlations between corneal hysteresis, intraocular pressure, and corneal central pachymetry. J Cataract Refract Surg. 2008;34(4):616-622. doi: 10.1016/j.jcrs.2007.11.051 [DOI] [PubMed] [Google Scholar]
- 11.Mollan SP, Wolffsohn JS, Nessim M, et al. Accuracy of Goldmann, ocular response analyser, Pascal and TonoPen XL tonometry in keratoconic and normal eyes. Br J Ophthalmol. 2008;92(12):1661-1665. doi: 10.1136/bjo.2007.136473 [DOI] [PubMed] [Google Scholar]
- 12.Ortiz D, Piñero D, Shabayek MH, Arnalich-Montiel F, Alió JL. Corneal biomechanical properties in normal, post-laser in situ keratomileusis, and keratoconic eyes. J Cataract Refract Surg. 2007;33(8):1371-1375. doi: 10.1016/j.jcrs.2007.04.021 [DOI] [PubMed] [Google Scholar]
- 13.Fontes BM, Ambrósio R Jr, Jardim D, Velarde GC, Nosé W. Corneal biomechanical metrics and anterior segment parameters in mild keratoconus. Ophthalmology. 2010;117(4):673-679. doi: 10.1016/j.ophtha.2009.09.023 [DOI] [PubMed] [Google Scholar]
- 14.Saad A, Lteif Y, Azan E, Gatinel D. Biomechanical properties of keratoconus suspect eyes. Invest Ophthalmol Vis Sci. 2010;51(6):2912-2916. doi: 10.1167/iovs.09-4304 [DOI] [PubMed] [Google Scholar]
- 15.Riboli E, Kaaks R. The EPIC Project: rationale and study design. European Prospective Investigation into Cancer and Nutrition. Int J Epidemiol. 1997;26(suppl 1):S6-S14. doi: 10.1093/ije/26.suppl_1.S6 [DOI] [PubMed] [Google Scholar]
- 16.Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort Profile: TwinsUK and healthy ageing twin study. Int J Epidemiol. 2013;42(1):76-85. doi: 10.1093/ije/dyr207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayat SA, Luben R, Keevil VL, et al. Cohort profile: A prospective cohort study of objective physical and cognitive capability and visual health in an ageing population of men and women in Norfolk (EPIC-Norfolk 3). Int J Epidemiol. 2014;43(4):1063-1072. doi: 10.1093/ije/dyt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khawaja AP, Chan MPY, Hayat S, et al. The EPIC-Norfolk Eye Study: rationale, methods and a cross-sectional analysis of visual impairment in a population-based cohort. BMJ Open. 2013;3(3):1-10. doi: 10.1136/bmjopen-2013-002684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luce D. Methodology for corneal compensated IOP and corneal resistance factor for an ocular response analyzer. Invest Ophthalmol Vis Sci. 2006;47(13):2266. [Google Scholar]
- 20.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190-2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes JAP, Tan D, Rapuano CJ, et al. ; Group of Panelists for the Global Delphi Panel of Keratoconus and Ectatic Diseases . Global consensus on keratoconus and ectatic diseases. Cornea. 2015;34(4):359-369. doi: 10.1097/ICO.0000000000000408 [DOI] [PubMed] [Google Scholar]
- 23.Liskova P, Dudakova L, Krepelova A, Klema J, Hysi PG. Replication of SNP associations with keratoconus in a Czech cohort. PLoS One. 2017;12(2):e0172365. doi: 10.1371/journal.pone.0172365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580-585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baratz KH, Tosakulwong N, Ryu E, et al. E2-2 protein and Fuchs’s corneal dystrophy. N Engl J Med. 2010;363(11):1016-1024. doi: 10.1056/NEJMoa1007064 [DOI] [PubMed] [Google Scholar]
- 26.Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491-501. doi: 10.1038/ng.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendig D, Langmann T, Kocken S, et al. Gene expression profiling of ABC transporters in dermal fibroblasts of pseudoxanthoma elasticum patients identifies new candidates involved in PXE pathogenesis. Lab Invest. 2008;88(12):1303-1315. doi: 10.1038/labinvest.2008.96 [DOI] [PubMed] [Google Scholar]
- 28.Finger RP, Charbel Issa P, Ladewig MS, et al. Pseudoxanthoma elasticum: genetics, clinical manifestations and therapeutic approaches. Surv Ophthalmol. 2009;54(2):272-285. doi: 10.1016/j.survophthal.2008.12.006 [DOI] [PubMed] [Google Scholar]
- 29.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42(4):297-319. doi: 10.1016/S0039-6257(97)00119-7 [DOI] [PubMed] [Google Scholar]
- 30.Zimmermann DR, Trüeb B, Winterhalter KH, Witmer R, Fischer RW. Type VI collagen is a major component of the human cornea. FEBS Lett. 1986;197(1-2):55-58. doi: 10.1016/0014-5793(86)80297-6 [DOI] [PubMed] [Google Scholar]
- 31.Morales J, Al-Sharif L, Khalil DS, et al. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am J Hum Genet. 2009;85(5):558-568. doi: 10.1016/j.ajhg.2009.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springelkamp H, Iglesias AI, Mishra A, et al. ; NEIGHBORHOOD Consortium . New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum Mol Genet. 2017;26(2):438-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edmund C. Assessment of an elastic model in the pathogenesis of keratoconus. Acta Ophthalmol (Copenh). 1987;65(5):545-550. doi: 10.1111/j.1755-3768.1987.tb07038.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Participants and Analyses
eFigure 1. Q-Q Plots for the CH (Left) and CRF (Right) Discovery GWAS in EPIC-Norfolk
eFigure 2. LocusZoom Plots for Significant Loci From the GWAS Meta-analysis of Corneal Hysteresis in EPIC-Norfolk and TwinsUK
eFigure 3. LocusZoom Plots for Significant Loci From the GWAS Meta-analysis of Corneal Resistance Factor in EPIC-Norfolk and TwinsUK
eFigure 4. Associations of Loci With Keratoconus
eTable 1. Genome-wide Significant SNPs From Corneal Hysteresis GWAS in EPIC-Norfolk
eTable 2. Genome-wide Significant SNPs From Corneal Resistance Factor GWAS in EPIC-Norfolk
eTable 3. Genome-wide Significant SNPs From Meta-analysis of Corneal Hysteresis GWAS in EPIC-Norfolk and TwinsUK
eTable 4. Genome-wide Significant SNPs From Meta-analysis of Corneal Resistance Factor GWAS in EPIC-Norfolk and TwinsUK
eTable 5. Corneal Tissue Specific Expression Profiles
eReferences.