Abstract
Background
The placenta is a highly specialized temporary organ that is related to fetal development and pregnancy outcomes, and epidemiological data demonstrate an increased risk of placental abnormality after in vitro fertilization and embryo transfer (IVF-ET).
Methods
This study examines alterations in the transcriptome profile of first-trimester placentas from IVF-ET pregnancies and analyzes the potential mechanisms that play a role in the adverse perinatal outcomes associated with IVF-ET procedures. Four human placental villi from first-trimester samples were obtained through fetal bud aspiration from patients subjected to IVF-ET due to oviductal factors. An additional four control human placental villi were derived from a group of subjects who spontaneously conceived a twin pregnancy. We analyzed their transcriptomes by microarray. Then, RT-qPCR and immunohistochemistry were utilized to analyze several dysregulated genes to validate the microarray results. Biological functions and pathways were analyzed with bioinformatics tools.
Results
A total of 3405 differentially regulated genes were identified as significantly dysregulated (> 2-fold change; P < 0.05) in the IVF-ET placenta in the first trimester: 1910 upregulated and 1495 downregulated genes. Functional enrichment analysis of the differentially regulated genes demonstrated that the genes were involved in more than 50 biological processes and pathways that have been shown to play important roles in the first trimester in vivo. These pathways can be clustered into coagulation cascades, immune response, transmembrane signaling, metabolism, cell cycle, stress control, invasion and vascularization. Nearly the same number of up- and downregulated genes participate in the same biological processes related to placental development and maintenance. Procedures utilized in IVF-ET altered the expression of first-trimester placental genes that are critical to these biological processes and triggered a compensatory mechanism during early implantation in vivo.
Conclusion
These data provide a potential basis for further analysis of the higher frequency of adverse perinatal outcomes following IVF-ET, with the ultimate goal of developing safer IVF-ET protocols.
Electronic supplementary material
The online version of this article (10.1186/s12958-019-0494-7) contains supplementary material, which is available to authorized users.
Keywords: Placenta, In vitro fertilization and embryo transfer (IVF-ET), Gene expression, Human, Adverse perinatal outcomes
Background
The widespread application of therapeutic methods for subfertile patients, particularly in vitro fertilization and embryo transfer (IVF-ET), has remarkably increased the pregnancy rate [1]. The elective frozen-thawed single-blastocyst transfer approach, which has been widely adopted in multiple reproductive centers, has largely overcome the risks of multiple pregnancies [2, 3]. Recent census results show that the perinatal outcomes of the most recent groups of patients are better than those of previous groups of patients [4]. This positive trend is due to more appropriate embryo transfer strategies, milder ovarian stimulation, laboratory technological advances, and improved culture media [5]. However, even in singleton pregnancy, after adjustment for maternal confounding factors, the risk of multiple adverse outcomes during the perinatal period, including miscarriage, preterm birth, small for gestational age, low birth weight and gestational hypertension, are higher in IVF-ET pregnancies than in spontaneously conceived pregnancies [6–10].
The placenta, derived from the embryonic trophectoderm, is a highly specialized and adaptive temporary organ and is critical for embryonic development and perinatal outcomes [11]. Some animal experiments have shown that placental tissue is more sensitive to external disturbances than embryonic tissue [12, 13]. The possible explanation for this result is twofold. On the one hand, the trophectoderm, which subsequently develops into the placenta, is the first differentiated cell lineage of the embryo to address various changes in the external environment [14]. On the other hand, the trophectoderm, whose genomic imprints are unstable and vulnerable to the surrounding microenvironment during implantation, appears to be sensitive to in vitro culture conditions and prone to an imbalance in imprinted gene expression [15, 16]. Although the composition of the culture medium has continuously been improved, it is still not completely equivalent to the physiological conditions of the natural environment in vivo [17]. In addition, even with careful and gentle manipulation, the trophectoderm must withstand severe stress and interference from the environment, such as changes in temperature, pH, and oxygen tension; light exposure; pH variations during manipulation; and shear stress linked to repeated pipetting, which may interact and synergistically affect placental function and development [18–20].
Professionally, there is an urgent need to understand and determine the impact of IVF-ET on placental development as soon as possible. Although several well-designed studies, which are limited in terms of research design and material acquisition, have focused on the effects of IVF-ET on human placental function and development, these studies used term placentas. These data support the hypothesis that altered global gene expression leads to overrepresented biological pathways in the human term placenta after IVF-ET [21, 22]. Similarly, term and late-gestation placentas have been examined in animal models, and genome-wide mRNA expression revealed that IVF-ET significantly affects placental functions and triggers compensatory responses [23–25]. Due to alterations in the peri-implantation environment and to key functions in placental development, such as cell fusion, embryo implantation, immune tolerance, and tissue remodeling, investigating the influence of IVF-ET on first-trimester placental development and function is particularly important [26, 27]. In addition, according to maternal gestational physiology and the theory of fetal origins of adult disease [28, 29], related research is needed to determine IVF-ET effects on placental gene expression and biological function as soon as possible, which would contribute toward developing strategies to prevent these diseases.
In this study, we hypothesized that altered global gene expression occurs in placental tissue during the first trimester after IVF-ET compared with the first trimester after spontaneous conception. Specifically, we used a twin-to-singleton selective fetal reduction strategy to obtain first-trimester placentas in vivo. Our previous study using immunohistochemistry showed that IVF-ET affects first-trimester placental development and function [30]. Therefore, we carried out a microarray analysis to investigate the effects of IVF-ET treatment on gene expression and potential biological functions in human first-trimester placentas compared with human first-trimester placentas of spontaneous pregnancies. Furthermore, a better understanding of the placental mechanisms triggered by IVF-ET per se may be of future value in improving the safety of IVF-ET protocols.
Methods
Study subjects
From January to October 2018, twin-to-singleton selective fetal reduction was performed in 4 cases in the first trimester after IVF-ET. All patients from fresh embryo transfers underwent standard IVF-ET due to oviductal obstruction. The sperm quality test results of each male were normal. In this study, the reason for fetal reduction from twin to singleton was patient preference. The application of the IVF-ET clinical technology was licensed and authorized by the National Health Commission of the People’s Republic of China. The control group included 4 cases of unwanted spontaneously conceived monochorionic twins in the same period. Clinical data were collected at the obstetrics outpatient clinic at Beijing Jishuitan Hospital and organized in a database. The number of specimens needed for the analysis of variance in the microarray experiment, which was 4, was determined by the study’s statistical criteria. Information about the couple’s weights, smoking habits, alcohol consumption habits and hormone use was collected by questionnaire. Information on maternal age, gestational weeks, gravidity history and body mass index (BMI) was matched between the IVF-ET and spontaneously conceived groups.
Sample collection and ethics
For all cases in which a singleton was retained from a twin pregnancy, reductions were performed by the same senior physician by fetal bud aspiration under ultrasound guidance. We selected 4 cases for villi suction simultaneously. We used a special needle that was authorized by the State Intellectual Property Office of People’s Republic of China (No ZL 2015 20,035,543.7). The placental tissue was collected 30–35 days after embryo transfer, which is equivalent to 7–8 gestational weeks. The pregnancies in the control group of double chorionic twin pregnancies were diagnosed with early intrauterine pregnancy based on urine pregnancy tests and transvaginal ultrasound assessments. None of the patients had taken steroid hormone drugs for the previous 3 months and had a record of regular menstruation. The villi were collected during the conventional artificial abortion operation. The villi samples were isolated and purified immediately from the specimens according to the morphology within 30 min under an inverted microscope. All placental tissues were refloated with ice-cold PBS and then stored under RNase-free conditions at − 80 °C until future tissue homogenization and total RNA extraction. Another portion of the remaining specimens was transferred to 4% paraformaldehyde within 30 min after operation and incubated at 4 °C overnight. Then, the specimens were stored in 1% paraformaldehyde at 4 °C until routine dehydration and embedding in paraffin. All study participants were enrolled at the Beijing Jishuitan Hospital 2018, were of Asian ancestry and were living in Beijing.
RNA extraction
Tissue homogenization and total RNA extraction were accomplished at CapitalBio Corporation, Bejing, China, using the Macherey–Nagel NucleoSpin RNA II kit (Macherey–Nagel, Duren, Germany), where a significant reduction in genomic DNA and protein contamination was achieved. Total RNA was derived from approximately 200 mg of first-trimester placental tissue, including various placental types. A NanoDrop ND100 spectrophotometer (NanoDrop Technologies, USA) was used to evaluate the purity and concentration of isolated total RNAs. The intensity ratio of ribosomal 28S and 18S RNA was 1.5–1.8:1. The RNA was used in both the microarray assay and subsequent RT-qPCR analysis.
Microarray hybridization
Thirty nanograms of total RNA was amplified and transcribed into biotinylated cRNA according to the instructions provided in the MessageAmp II aRNA Amplification Kit (Ambion Inc., Carlsbad, Calif., USA). Samples of cDNA were hybridized to the Affymetrix Human Genome U133 Plus 2.0 Array. The cDNA of each sample was hybridized to one Array. This array contains approximately 54,000 oligonucleotide probes and covers more than 30,000 human genes mapped through UniGene or via RefSeq annotation (Affymetrix, Santa Clara, CA, USA). Then, the arrays were washed and stained on the Fluidics Station 450 (Affymetrix) and scanned on an Affymetrix GeneChip Scanner 3000 (Affymetrix) to analyze the hybridization data according to the manufacturer’s instructions. The data were collected from the scanned images and evaluated using the Affymetrix GeneChip Operating Software (GCOS 1.4). Differentially expressed genes between the IVF-ET and control groups were analyzed using Significance Analysis of Microarrays (SAM version 3.02, Stanford University, Stanford, CA).
Microarray data analysis
Our primary microarray data for the first-trimester placental samples have been submitted to the Gene Expression Omnibus of the National Center for Biotechnology Information [31]. The data can be obtained through GEO Series accession number GSE 122214 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122214). Data quality, linear modeling and significance analysis were performed by means of Bioconductor package linear models [32]. The cut-off value was determined to be a twofold difference in expression values. The proportion of false-positive corrections of < 0.05 was considered statistically significant. If a gene had multiple probe identification data in the results, the average fold change was calculated.
To assess the variability in placental tissues with nonparametric methods, hierarchical clustering and principal component analysis were performed to visualize global variation. Based on the difference in placental gene expression between IVF-ET pregnancy and spontaneously conceived pregnancy, a three-dimensional scatter plot was used to visualize the sample gene variation in the principal component analysis. The distance between different points was calculated using the number and covariance of differentially expressed genes. The complete-linkage hierarchical clustering algorithm was used to assess the variation among gene expression profiles.
Enrichment and integrated analysis of differentially regulated genes were performed by submitting data to Database for Annotation, Visualization and Integrated Discovery (DAVID), Ingenuity Pathway Analysis (IPA), Kyoko Encyclopedia of Genes and Genomes (KEGG), and KEGG Orthology Based Annotation System (KOBAS). According to Gene Ontology terms, DAVID divided blocks of functionally related differentially expressed genes based on biological process, molecular function and cellular component. The other three databases were used to visually integrate dysregulated genes and identify statistically significant enriched pathways. Up- and downregulated genes were investigated separately. Statistical controls were based on all human genes included in the Affymetrix Human Genome U133 Plus 2.0 Array. A false discovery rate (FDR) < 0.05 and p-value < 0.05 were used to determine statistical significance.
Quantitative real-time RT-PCR
Validation of the microarray results was performed by real-time RT-qPCR using the same samples for microarray. Synthesis of cDNA was achieved using the PrimeScript RT Reagent Kit (TaKaRa Biotechnology, Dalian, China) with 2 μg of RNA. RT-qPCR was performed with an ABI Prism 7300 Sequence Detection System (Applied Biosystems, Roche, USA) according to recommendations of the manufacturer. The following genes were used for microarray validation: alpha-fetoprotein (AFP); glial cells missing homolog 1 (Drosophila) (GCM1); leukocyte-associated immunoglobulin-like receptor 2 (LAIR2); phosphatase and tension homolog (PTEN); B-cell CLL/lymphoma 2 (BCL2); transferrin (TF); metallothionein 1 M (MT1M); epidermal growth factor receptor (EGFR); tubulin, beta 1 class VI (TUBB1); and vascular endothelial growth factor (VEGF). Considering their critical functions in the first-trimester placenta based on the biological analysis, these ten target genes, which showed differential expression in the microarray analysis, were selected and tested by RT-qPCR. Three technical and five biological replicates were run for all RT-qPCR analyses. Relative mRNA expression levels were calculated using the comparative threshold ΔΔCt method. Comparison of gene expression between the IVF-ET and control groups was performed using a two-tailed Student’s t-test to determine significant differences. P < 0.05 was considered significant. A list of all predesigned PCR primer pairs used in the RT-qPCR analysis is provided (Additional file 1: Table S1).
Immunohistochemistry (IHC)
The first-trimester placental tissues used for immunohistochemical staining originated from the same tissues used for the previous RNA extractions. The placental tissues that were stored in PFA/PBS were fixed and embedded in paraffin. Then, these samples were deparaffinized, rehydrated and sectioned. Immunohistochemical localization was performed to analyze the AFP, VEGF, TF, TUBB1, BCL2, GCM1, PTEN and LAIR2 gene products using 5-μm paraffin sections mounted on Superfrost Plus slides (Gerhard Menzel, Braunschweig, Germany). The primary antibodies utilized were as follows: AFP (1:25 dilution, ZM-0009; ZSGB-Bio, Beijing, China), VEGF (1:25 dilution, ZA0509, ZSGB-Bio, Beijing, China), TF (1:25 dilution, TA500848, ZSGB-Bio, Beijing, China), TUBB1 (1:25 dilution, TA506588, ZSGB-Bio, Beijing, China), BCL2 (1:25 dilution, ZA-0010; ZSGB-Bio, Beijing, China), GCM1 (1:100 dilution, ab187860; Abcam, Cambridge, UK), PTEN ((1:25 dilution, ZA-0635; ZSGB-Bio, Beijing, China), and LAIR2 (1:100 dilution, ab183145, Abcam, Cambridge, UK). Protein detection and negative control assessment were conducted simultaneously following the manufacturer’s instructions. A microscope magnification of × 400 was used. Two senior pathologists examined the stained sections in a double-blinded fashion and performed the quantifications manually. The positivity rates are described as stained cells vs the total number of cells.
Results
Characteristics of the patients
In terms of age, gestational age, parity and BMI, there were no significant differences between the women who were subjected to IVF-ET and those who spontaneously conceived (Table 1).
Table 1.
Characteristics of the patients in the two groups
| N | Mean age | Gestational age (day) | Parity | BMI (kg/m2) | |
|---|---|---|---|---|---|
| IVF-ET group | 4 | 30.66 ± 3.76 | 49.44 ± 3.14 | 1.10 ± 1.21 | 23.33 ± 3.17 |
| Control group | 4 | 28.86 ± 3.45 | 49.35 ± 3.23 | 2.21 ± 0.89 | 22.99 ± 2.35 |
| t | 0.833 | 0.637 | 0.858 | 1.311 | |
| p | 0.425 | 0.529 | 0.437 | 0.263 |
Microarray data
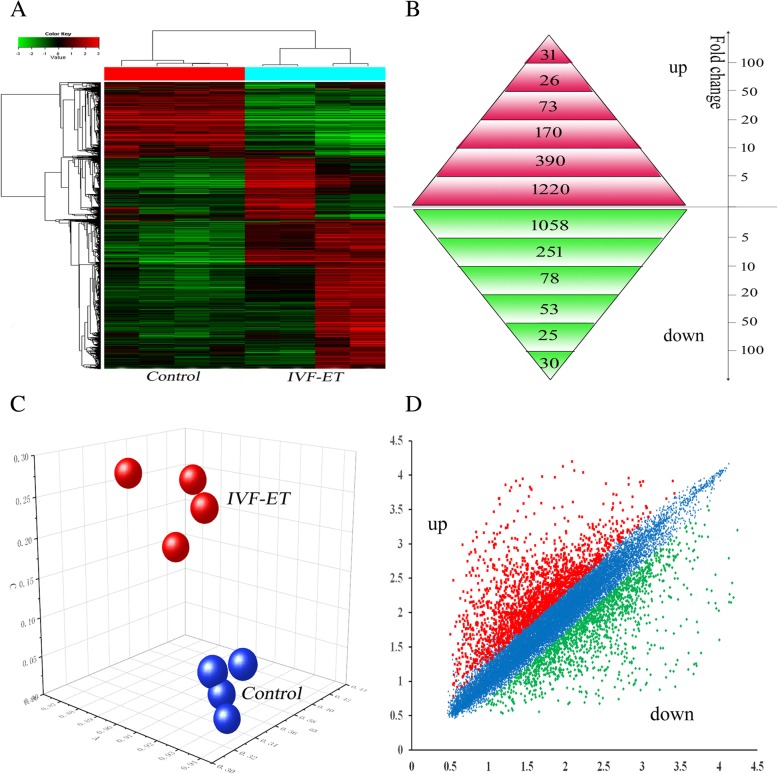
Transcriptomic profiling of 8 human first-trimester placental samples was performed on samples obtained from 4 IVF-ET and 4 spontaneously conceived pregnancies. A total of 3405 differentially expressed genes were validated according to a false discovery rate of 0.05. A total of 1910 genes were expressed at higher levels, and 1495 genes were expressed at lower levels in the first-trimester placentas produced by IVF-ET compared with those produced by natural conception. Interestingly, the numbers of upregulated (56.1%) and downregulated (43.9%) genes were well balanced (Fig. 1a, b, d).
Fig. 1.
Supervised hierarchical clustering and principal component analysis of the first-trimester placenta from the IVF-ET and control groups. a Supervised hierarchical clustering was performed with 3405 differentially expressed genes between the IVF-ET and naturally conceived pregnancies, allowing the distinction of the two sample groups. b Number of genes specifically modulated during the first trimester in the placenta after IVF-ET. c Principal component analysis performed on the placenta after IVF-ET and naturally conceived pregnancy in the first trimester based on the results of the microarray analysis. b: IVF-ET; N: Control). (d) Examples of the graphic display of expression profiling data obtained from the microarray analysis. The vertical axis is the log fold change in gene expression
Hierarchical clustering analysis of the microarray data demonstrated an expressional alteration and separation of placental dysregulated transcripts between the IVF-ET and control groups (Fig. 1a). Alternatively, the principal component analysis was applied to placental gene expression profiles and demonstrated the same separate pattern, where the two study groups clustered into two separate groups (Fig. 1c).
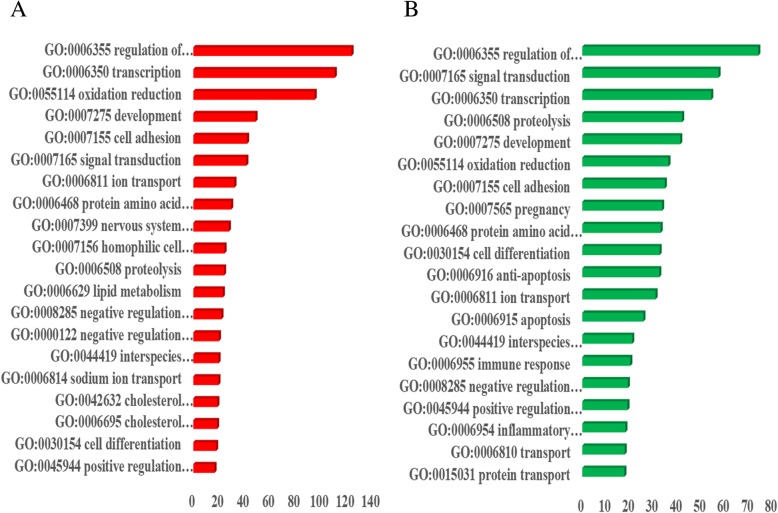
The functional enrichment annotation of differentially expressed genes demonstrated that more than 50 Gene Ontology (GO) categories and pathways were successfully clustered in the list of dysregulated transcripts. Many broad biological processes of upregulated genes in pregnancy progression were regulation of transcription DNA-dependent (n = 149, FDR P = 1.15 × 10− 124), transcription (n = 143, FDR P = 1.55 × 10− 111), oxidation reduction (n = 82, FDR P = 7.98 × 10− 96), development (n = 84, FDR P = 1.95 × 10− 49), and cell adhesion (n = 50, FDR P = 7.37 × 10− 43). The biological processes of the downregulated genes were regulation of transcription DNA-dependent (n = 102 FDR P = 8.38 × 10− 75), signal transduction (n = 103, FDR P = 4.54 × 10− 58), transcription (n = 87, FDR P = 5.65 × 10− 55), proteolysis (n = 53, FDR P = 8.86 × 10− 43), and development (n = 72, FDR P = 6.00 × 10− 42). Additional biological processes for the up- and downregulated genes are listed (Additional file 2: Table S2 Additional file 3: Table S3). Interestingly, almost the same numbers of up- and downregulated genes are allocated in the same biological process related to placental development and maintenance (Fig. 2). These differentially expressed genes were widely distributed to the nucleus, cytoplasm and cell membrane of the placenta. The GO analyses also highlight the disturbances in various gene and molecular functions and their link to pregnancy complications.
Fig. 2.
Functional enrichment analysis of differentially regulated genes in the placenta after IVF-ET compared with the control group during the first trimester according to biological processes. Red, upregulated genes; green, downregulated genes
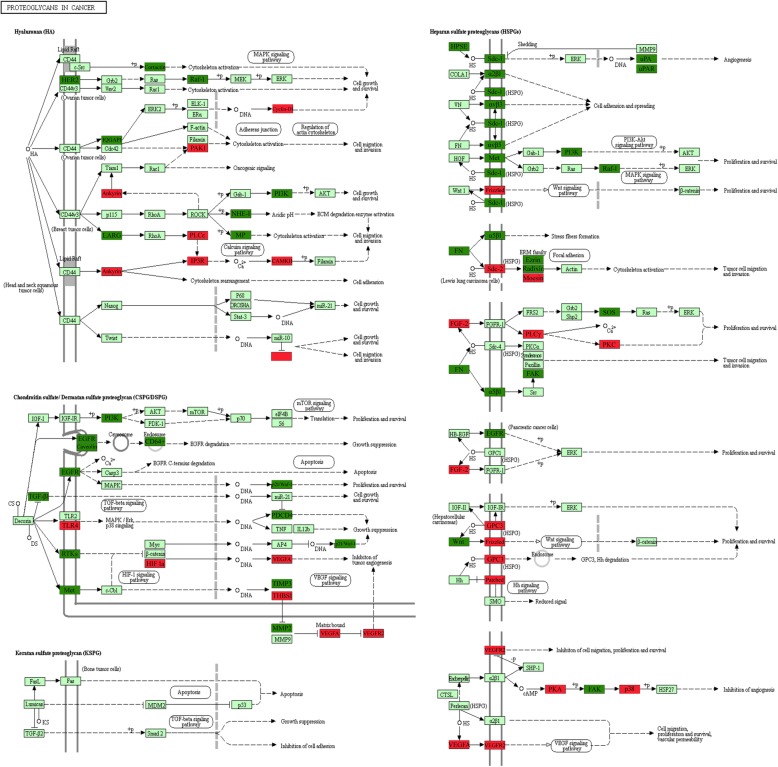
In the IPA, some networks were built based on the biological processes of differentially regulated placental gene expression in the IVF-ET group (Table 2). These abnormal pathways included metabolic pathways (n = 279, FDR P = 7.43 × 10− 22), complement and coagulation cascades (n = 44, FDR P = 5.74 × 10− 14), Proteoglycans in cancer (n = 58, FDR P = 4.17 × 10− 8), the PPAR signaling pathway (n = 23, FDR P = 1.08 × 10− 4), the Wnt signaling pathway (n = 35, FDR P = 1.53 × 10− 4), and the Jak-STAT signaling pathway (n = 36, FDR P = 5.50 × 10− 4), among others. For significant proteoglycans in cancer and the PPAR signaling pathway, we labeled the up- (red) and downregulated (green) genes in the signaling pathways (Fig. 3, Additional file 4: Figure S1).
Table 2.
Cluster of significant pathways related to IVF-ET
| No | Term | ID | Input number | P-Value |
|---|---|---|---|---|
| 1 | Metabolic pathways | hsa01100 | 279 | 7.43 × 10− 22 |
| 2 | Complement and coagulation cascades | hsa04610 | 44 | 5.74 × 10−14 |
| 3 | Proteoglycans in cancer | hsa05205 | 58 | 4.17 × 10−8 |
| 4 | Pathways in cancer | hsa05200 | 87 | 3.72 × 10−7 |
| 5 | Insulin resistance | hsa04931 | 33 | 1.08 × 10−5 |
| 6 | Carbon metabolism | hsa01200 | 33 | 1.46 × 10−5 |
| 7 | Axon guidance | hsa04360 | 44 | 2.23 × 10−5 |
| 8 | Ras signaling pathway | hsa04014 | 52 | 3.77 × 10−5 |
| 9 | Glycine, serine and threonine metabolism | hsa00260 | 17 | 4.59 × 10−5 |
| 10 | Mineral absorption | hsa04978 | 19 | 8.74 × 10−5 |
| 11 | PPAR signaling pathway | hsa03320 | 23 | 1.08 × 10−4 |
| 12 | Wnt signaling pathway | hsa04310 | 35 | 1.53 × 10− 4 |
| 13 | Transcriptional misregulation in cancer | hsa05202 | 41 | 1.72 × 10−4 |
| 14 | AGE-RAGE signaling pathway in diabetic complications | hsa04933 | 28 | 1.74 × 10−4 |
| 15 | HIF-1 signaling pathway | hsa04066 | 28 | 2.28 × 10−4 |
| 16 | mTOR signaling pathway | hsa04150 | 36 | 3.31 × 10−4 |
| 17 | Fatty acid metabolism | hsa01212 | 17 | 3.32 × 10−4 |
| 18 | Focal adhesion | hsa04510 | 44 | 3.81 × 10−4 |
| 19 | Regulation of actin cytoskeleton | hsa04810 | 46 | 3.91 × 10−4 |
| 20 | Fatty acid degradation | hsa00071 | 16 | 3.94 × 10−4 |
| 21 | Steroid hormone biosynthesis | hsa00140 | 19 | 3.94 × 10−4 |
| 22 | Bile secretion | hsa04976 | 21 | 4.29 × 10−4 |
| 23 | Aldosterone-regulated sodium reabsorption | hsa04960 | 15 | 4.65 × 10−4 |
| 24 | Jak-STAT signaling pathway | hsa04630 | 36 | 5.50 × 10−4 |
| 25 | ErbB signaling pathway | hsa04012 | 24 | 6.18 × 10−4 |
| 26 | Steroid biosynthesis | hsa00100 | 10 | 6.33 × 10−4 |
| 27 | Valine, leucine and isoleucine degradation | hsa00280 | 16 | 7.02 × 10−4 |
| 28 | Renal cell carcinoma | hsa05211 | 20 | 7.19 × 10−4 |
| 29 | Biosynthesis of amino acids | hsa01230 | 21 | 9.01 × 10−4 |
| 30 | Tyrosine metabolism | hsa00350 | 13 | 9.74 × 10−4 |
| 31 | Tryptophan metabolism | hsa00380 | 14 | 1.01 × 10−3 |
| 32 | EGFR tyrosine kinase inhibitor resistance | hsa01521 | 22 | 1.09 × 10−3 |
| 33 | Cytokine-cytokine receptor interaction | hsa04060 | 51 | 1.10 × 10−3 |
| 34 | Arginine biosynthesis | hsa00220 | 10 | 1.11 × 10− 3 |
| 35 | Proximal tubule bicarbonate reclamation | hsa04964 | 10 | 1.45 × 10−3 |
| 36 | Sphingolipid metabolism | hsa00600 | 15 | 1.46 × 10−3 |
| 37 | Rap1 signaling pathway | hsa04015 | 43 | 1.48 × 10−3 |
| 38 | Hippo signaling pathway-multiple species | hsa04392 | 11 | 1.64 × 10−3 |
| 39 | FoxO signaling pathway | hsa04068 | 30 | 1.79 × 10−3 |
| 40 | Glycerolipid metabolism | hsa00561 | 17 | 1.84 × 10−3 |
| 41 | Vitamin digestion and absorption | hsa04977 | 10 | 1.87 × 10−3 |
| 42 | Pentose phosphate pathway | hsa00030 | 11 | 2.05 × 10−3 |
| 43 | cAMP signaling pathway | hsa04024 | 40 | 2.13 × 10−3 |
| 44 | AMPK signaling pathway | hsa04152 | 28 | 2.26 × 10−3 |
| 45 | Arginine and proline metabolism | hsa00330 | 15 | 2.41 × 10−3 |
| 46 | Alanine, aspartate and glutamate metabolism | hsa00250 | 12 | 2.60 × 10−3 |
| 47 | Epithelial cell signaling in Helicobacter pylori infection | hsa05120 | 18 | 3.00 × 10−3 |
| 48 | beta-Alanine metabolism | hsa00410 | 11 | 3.13 × 10−3 |
| 49 | Insulin signaling pathway | hsa04910 | 30 | 3.13 × 10−3 |
| 50 | Propanoate metabolism | hsa00640 | 11 | 3.81 × 10−3 |
Fig. 3.
Differentially expressed genes were labeled in the proteoglycans in cancer pathway in the placenta after IVF-ET during the first trimester by KEGG. Red, upregulated genes; Dark green, downregulated genes; Light green, no change
Quantitative RT-PCR
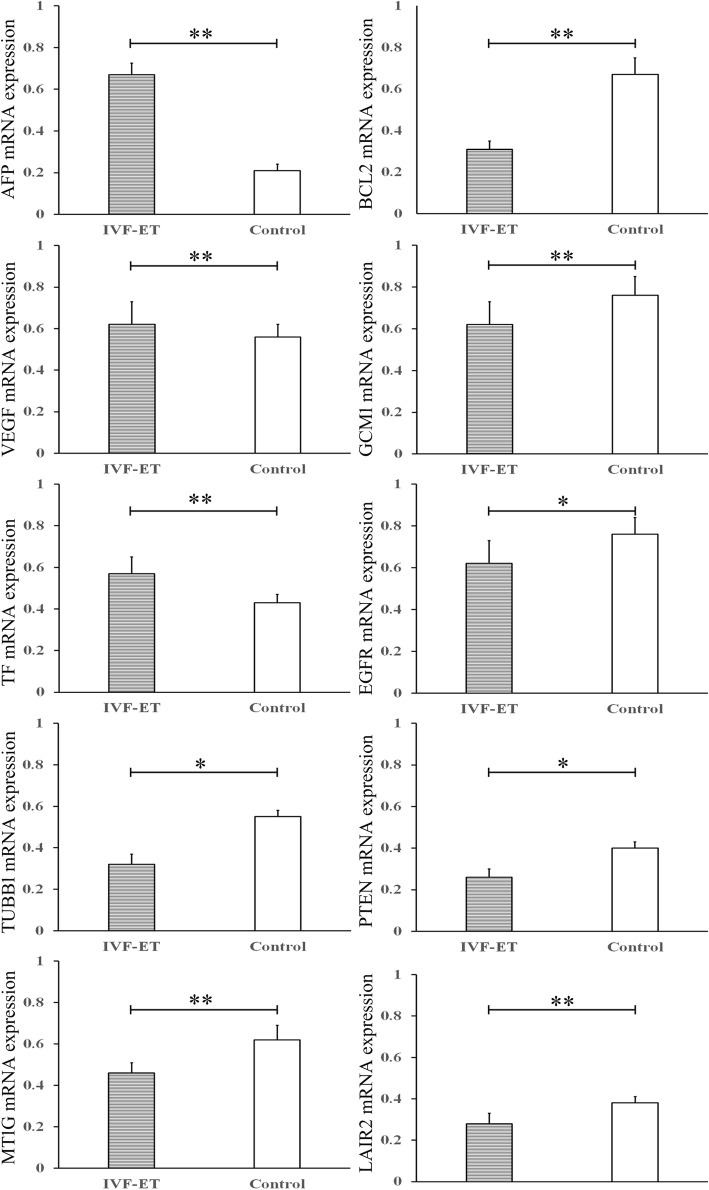
RT-qPCR analysis of ten selected genes was performed to validate the microarray data. These ten differentially regulated genes were selected according to their biological processes, which were mostly related to placental functions (Fig. 4). The RT-qPCR results confirmed the upregulation the AFP, VEGF, and TF genes and the downregulation of the BCL2, GCM1, EGFR, PTEN, LAIR2, TUBB1,and MT1G genes in the first-trimester placenta after IVF-ET compared with that after natural conception. We observed a significant concordance among the ten tested genes with the array hybridization obtained previously and confirmed more accurate gene expression differences.
Fig. 4.
Validation by RT-qPCR for selected genes from first-trimester placentas derived from IVF-ET compared with those derived from the control group. The mRNA levels of AFP, GCM1, LAIR2, PTEN, BCL2, TF, MT1M, EGFR, VEGF, and TUBB1 in the IVF-ET and control groups were examined by RT-qPCR. All data are expressed as the mean ± standard deviation, * indicates P < 0.05; ** indicates P < 0.01
Immunohistochemistry
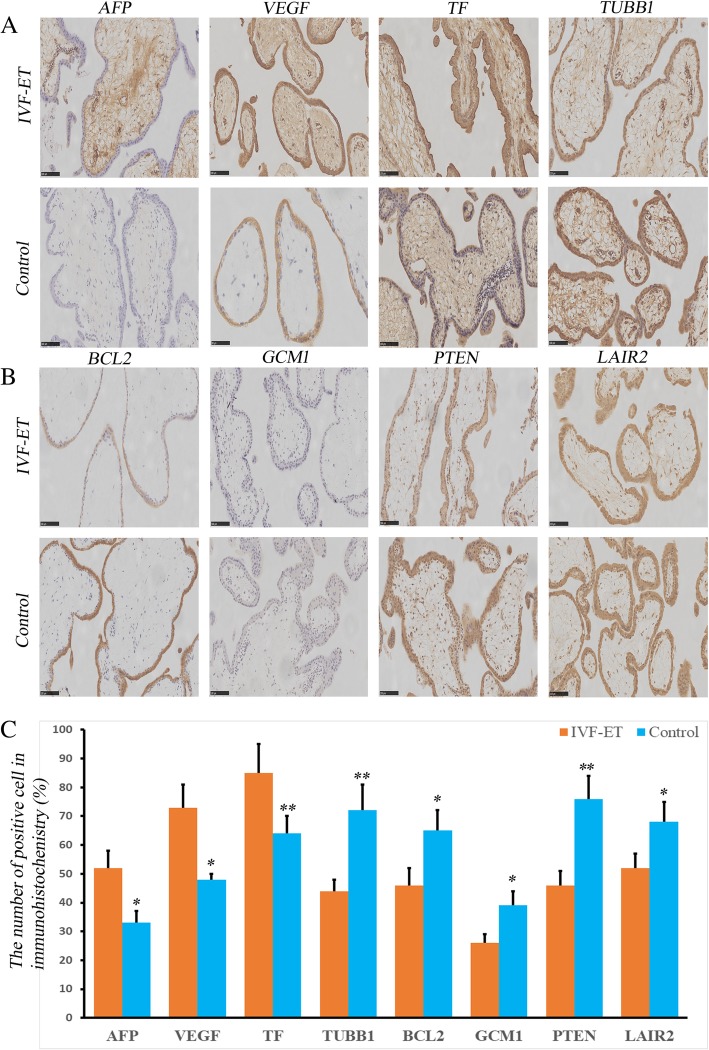
The localizations of the AFP, VEGF, TF, TUBB1, BCL2, GCM1, PTEN and LAIR2 proteins, which represent different biological processes in placentation, were investigated in the human first-trimester placenta (Fig. 5). These 8 genes were mainly localized to the cytoplasm, cell membrane and intercellular spaces of the human first-trimester placenta. In addition, both the IVF-ET group and control group showed expression of these 8 genes. A larger number of cells showed strong AFP, VEGF and TF staining, with cytoplasmic staining, in the first-trimester IVF-ET placental group than in the control group. The control group had higher expression of TUBB1, BCL2, GCM1, PTEN and LAIR2 than the IVF-ET group. The immunohistochemical expression of these genes is consistent with the results of the microarray and RT-qPCR.
Fig. 5.
Immunohistochemical analysis of the expression of 8 genes in the first-trimester placental tissues of the IVF-ET and control groups. a Immunohistochemical analysis of the expression levels of AFP, VEGF, TF and TUBB1 in the first-trimester placentas of the IVF-ET and control groups. b Immunohistochemical analysis of the expression levels of BCL2, GCM1, PTEN and LAIR2 in the first-trimester placentas of the IVF-ET and control groups. c Immunohistochemical analysis of the number of positively stained cells expressing the 8 genes in the IVF-ET and control groups. All data are expressed as the mean ± standard deviation, * indicates P < 0.05; ** indicates P < 0.01, Scale bar = 100 μm
Discussion
Epidemiological studies have demonstrated that even singleton pregnancies conceived after IVF-ET are more likely to develop prematurity, gestational hypertension, placental abruption, low birth weight and intrauterine growth retardation [6–10]. The risk of adverse outcomes originating from suboptimal placentation in pregnancies after IVF-ET is significantly greater than that in spontaneously conceived pregnancies [33, 34]. Several meta-analysis and animal-model studies have demonstrated that this phenomenon is due not only to the infertility diagnosis but also to IVF-ET itself [35–37]. Early placental implantation involves a series of important events, namely, early trophoblast migration, cell fusion, tissue remodeling, immune tolerance, metabolism and extracellular matrix degradation [38–40]. As these trophoblast properties disappear at the end of pregnancy [41, 42], it is particularly critical to study the effects of IVF-ET on first-trimester placental gene expression. Here, we demonstrated that IVF-ET pregnancies show altered expression of first-trimester placental genes that are critical to development and function compared with spontaneously conceived pregnancies. Genome-wide mRNA expression analysis of the first-trimester placenta after IVF-ET also revealed 50 significantly overrepresented biological pathways. Our findings provide evidence of functional placental pathology and a potential basis for understanding the increased risk associated with the first-trimester placenta following IVF-ET.
In the present study, we identified and confirmed that there was a significant difference in first-trimester placental gene expression between IVF-ET and spontaneous pregnancies. We report 3405 differentially expressed genes, including 1910 upregulated and 1495 downregulated genes. These genes were roughly clustered according to their biological functions: cellular process, physiological, biological regulation, metabolism, regulation of biological process, multicellular organismal process, catalytic activity, developmental process, binding and other functions. Our results are consistent with those of most recent studies and support the hypothesis that IVF-ET procedures affect placental gene expression, which subsequently may lead to a risk of adverse perinatal outcomes [43–45]. However, after comparing our data regarding the altered transcriptome in the first-trimester placenta with data from previous studies on the term placenta and endometrium during early implantation, we found that the IVF-ET procedure has a broader and more severe effect on early placentation [21, 22, 46]. Three term placentas from IVF-ET were examined using the same gene chip in our analysis, and only 18 differentially expressed genes and 6 biological functions were reported [21]. Another current study used 10 term placentas from IVF-ET for microarray analysis and identified 1866 differentially expressed genes, including 839 upregulated and 927 downregulated genes [22]. In addition, considering that modification of the maternal endometrium may affect trophoblast invasion and tissue remodeling, 641 differentially expressed genes were identified by microarray analysis from 23 samples from human endometrial biopsies [46]. Our results suggest that early placental tissue may undergo more extensive genetic and functional changes than term placental and other tissues from IVF-ET.
IPA of our data revealed that more than 50 pathways were disrupted in the first-trimester placenta derived from IVF-ET. Further analysis showed that these pathways are widely involved in key events of early placental development and function, whose disruption can lead to adverse outcomes [47–49]. We report the enrichment of multiple genes in the top 50 significantly overrepresented biological pathways. As expected, the first group of pathways comprise metabolic pathways. The IVF-ET process changes the physiological hypoxic environment of the trophoblast in vivo [50]. Metabolic disturbances may lead to metabolic reprogramming of the trophoblast in the first-trimester placenta [51]. Furthermore, the abnormal metabolic characteristics appear to be irreversible, even under normal conditions [52]. Abnormal metabolites, including prostaglandins, leukotrienes, lipid peroxidation and inflammatory mediation, may contribute to vasoconstriction and inflammatory syndrome, eventually resulting in adverse perinatal outcomes [53–55]. Complement and coagulation cascades contribute to homeostasis, thrombosis, defense infection, immune tolerance and apoptotic cell clearance [56, 57]. Abnormalities in these cascades may lead to recurrent pregnancy loss, vascular complications and preeclampsia [58, 59]. The PPAR signaling pathway plays an important role in trophoblast fusion, migration, angiogenesis, lipid metabolism and trophoblast cycle progression [60]. Abnormalities in the PPAR signaling pathway may contribute to increased apoptosis of trophoblasts, trophoblast fusion disorder and necrosis of syncytiotrophoblasts, resulting in severe preeclampsia and intrauterine growth restriction. [61, 62]. However, in most cases involving humans, no immediate abortion has been observed, and most women give birth successfully after IVF-ET, which supports the hypothesis that interference by IVF-ET can trigger a placental compensatory mechanism [63, 64].
Moreover, regarding the compensatory mechanism in the first-trimester placenta after IVF-ET, we analyzed altered gene expression as a whole, rather than at the individual gene level. Interestingly, we separately classified upregulated genes and downregulated genes from IVF-ET sources according to their biological functions and found that the biological functions affected by these genes were consistent between the up- and downregulated genes. In each of the affected biological function categories, the total numbers of upregulated and downregulated genes are similar. From the perspective of signaling pathways, we simultaneously demonstrated and provided the locations of the up- and downregulated genes. Our labeled pathway maps demonstrate that multiple local functional groups are simultaneously affected by the upregulation and downregulation of these genes, resulting in a nearly balanced biological function. The compensatory mechanism in placental development is also observed in animal models. In early pregnancy, the placenta of the IVF-ET mouse pregnancy is smaller than that of the naturally conceived pregnancy [65], and in the third trimester and delivery stage, the placenta is heavier than that of the control [66]. Likewise, even after adjusting for confounding factors in IVF-ET, the human placenta and neonatal weight ratio are often higher than those of the natural control [67]. However, a heavier placenta does not represent more efficient transport of oxygen and nutrients [68]. Another study demonstrated that the weight and size of the placenta is associated with coronary disease and life expectancy [69, 70]. Nevertheless, the human placental compensatory mechanism can only reach a certain point. More severe disruptions can lead to immediate and subsequent adverse outcomes.
To our knowledge, our research is the first to demonstrate alterations in human first-trimester placental gene expression in vivo during early implantation. However, we understand that there are a few limitations with the design of our study. The first limitation is the sample size, but the results were strengthened by the fact that the samples were similar in terms of age, BMI and gestational weeks. The second limitation is twin pregnancy. Although we obtained first-trimester placental tissues in vivo using a fetal reduction strategy, a twin pregnancy has a potential impact on placentation and does not fully reflect a singleton pregnancy. In this study, we focused only on the effect of IVF-ET on the first-trimester placental function and compensatory mechanism in vivo. Additionally, even though the samples were carefully examined under a microscope and stained by immunohistochemistry, we were still concerned about interference from maternal and embryonic cells. Nevertheless, we still believe that these altered gene expression patterns in the first-trimester placenta may partially explain the clinical characteristics and adverse perinatal outcomes of IVF-ET.
Conclusion
Our study describes the altered gene expression patterns in the early placenta of IVF-ET pregnancies compared to those in spontaneously conceived pregnancies. The abnormal biological functions and pathways associated with these differentially expressed genes provide a potential mechanism for understanding adverse perinatal outcomes following IVF-ET. Our contribution is the verification of a novel hypothesis that the increased risk of adverse perinatal outcomes in IVF-ET originates from early abnormal placentation. The initially disrupted placentation triggers a compensatory mechanism, and fortunately, most of these compensatory actions are successful. The innovation of our study is the examination of the first-trimester human placenta in vivo during the critical periods of implantation in IVF-ET, enabling relevant research to further investigate the target genes and pathways in vivo and not just in animal models. These data provide a potential basis for further analysis of the higher frequency of adverse perinatal outcomes following IVF-ET, with the ultimate goal of developing safer IVF-ET protocols.
Additional files
Table S1. Detail information on the selection of primers for real-time RT-PCR experiments (DOC 34 kb)
Table S2. The biological processes of up regulated genes in placental subjected to IVF-ET (DOC 68 kb)
Table S3. The biological processes of down regulated genes in placental subjected to IVF-ET (DOC 70 kb)
Figure S1. KEGG analysis of the differentially expressed genes, which are labeled, in the PPAR signaling pathway in the placenta after IVF-ET during the first trimester by KEGG. Red, upregulated; Blue, downregulated; Green, no change. (TIF 14811 kb)
Acknowledgments
The authors appreciate the help from all patients enrolled in the Department of Gynecology and Obstetrics, Beijing Jishuitan Hospital, Beijing, China. We gratefully acknowledge the CapitalBio Corporation for conducting the RNA extractions and microarrays. The authors are also grateful to the anonymous reviewers for their constructive comments on the manuscript.
Abbreviations
- AFP
Alpha-fetoprotein
- BCL2
B-cell CLL/lymphoma 2
- BMI
Body mass index
- DAVID
Database for annotation, visualization and integrated discovery
- EGFR
Epidermal growth factor receptor
- FDR
False discovery rate
- GCM1
Glial cells missing homolog 1 (Drosophila)
- IHC
Immunohistochemistry
- IPA
Ingenuity pathway analysis
- IVF-ET
In vitro fertilization and embryo transfer
- KEGG
Kyoto encyclopedia of genes and genomes
- KOBAS
KEGG orthology-based annotation system
- LAIR2
Leukocyte-associated immunoglobulin-like receptor 2
- MT1M
Metallothionein 1 M
- PTEN
Phosphatase and tension homolog
- RT-qPCR
Real-time quantitative PCR
- TF
Transferrin
- TUBB1
Tubulin beta 1 class VI
- VEGF
Vascular endothelial growth factor
Authors’ contributions
Dr. LFS designed the project and conducted the experimental study. LZ collected the samples, performed the experiments and wrote the manuscript. XLZ participated in designing the experiments and in writing and reviewing the manuscript. JFL was involved in performing some experiments. RZ were involved in the data collection and statistical analysis. RY and YW were involved in reviewing the manuscript. All authors read and approved the final manuscript.
Funding
The study was funded by the National Natural Science Foundation of China (No. 81070493). The funding bodies had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The primary microarray data of the placental samples have been submitted to the Gene Expression Omnibus of the National Center for Biotechnology Information. The data can be obtained through GEO Series accession number GSE 122214 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122214). Other data generated or analyzed during this study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Beijing Jishuitan Hospital (Permission no 201703–11), and written informed consent was obtained from each individual prior to recruitment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.De Geyter C, Calhaz-Jorge C, Kupka MS, Wyns C, Mocanu E, Motrenko T, et al. ART in Europe, 2014: results generated from European registries by ESHRE: the European IVF-monitoring consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE) Hum Reprod. 2018;33(9):1586–1601. doi: 10.1093/humrep/dey242. [DOI] [PubMed] [Google Scholar]
- 2.Ishihara O, Araki R, Kuwahara A, Itakura A, Saito H, Adamson GD. Impact of frozen-thawed single-blastocyst transfer on maternal and neonatal outcome: an analysis of 277,042 single-embryo transfer cycles from 2008 to 2010 in Japan. Fertil Steril. 2014;101(1):128–133. doi: 10.1016/j.fertnstert.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 3.Sunderam S, Boulet SL, Jamieson DJ, Kissin DM. Effects of patient education on desire for twins and use of elective single embryo transfer procedures during ART treatment: a systematic review. Reprod Biomed Soc Online. 2018;6:102–119. doi: 10.1016/j.rbms.2018.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Y, Chen W, Zhou L, Gao X, Xi X. Single embryo transfer improve the perinatal outcome in singleton pregnancy. J Matern Fetal Neonatal Med. 2019;27:1–6. doi: 10.1080/14767058.2019.1571029. [DOI] [PubMed] [Google Scholar]
- 5.Henningsen AK, Pinborg A. Birth and perinatal outcomes and complications for babies conceived following ART. Semin Fetal Neonatal Med. 2014;19(4):234–238. doi: 10.1016/j.siny.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Heo JS, Lee HJ, Lee MH, Choi CW. Comparison of neonatal outcomes of very low birth weight infants by mode of conception: in vitro fertilization versus natural pregnancy. Fertil Steril. 2019;111(5):962–970. doi: 10.1016/j.fertnstert.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Yu B, Vega M, Zaghi S, Fritz R, Jindal S, Buyuk E. Comparison of perinatal outcomes following frozen embryo transfer cycles using autologous versus donor oocytes in women 40 to 43 years old: analysis of SART CORS data. J Assist Reprod Genet. 2018;35(11):2025–2029. doi: 10.1007/s10815-018-1287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luke B, Gopal D, Cabral H, Diop H, Stern JE. Perinatal outcomes of singleton siblings: the effects of changing maternal fertility status. J Assist Reprod Genet. 2016;33(9):1203–1213. doi: 10.1007/s10815-016-0757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Declercq E, Luke B, Belanoff C, Cabral H, Diop H, Gopal D, et al. Perinatal outcomes associated with assisted reproductive technology: the Massachusetts outcomes study of assisted reproductive technologies (MOSART) Fertil Steril. 2015;103(4):888–895. doi: 10.1016/j.fertnstert.2014.12.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu L, Zhang Y, Liu Y, Zhang R, Wu Y, Huang Y, et al. Maternal and live-birth outcomes of pregnancies following assisted reproductive technology: a retrospective cohort study. Sci Rep. 2016;6:35141. doi: 10.1038/srep35141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toschi P, Czernik M, Zacchini F, Fidanza A, Loi P, Ptak GE. Evidence of placental autophagy during early pregnancy after transfer of in vitro produced (IVP) sheep embryos. PLoS One. 2016;11(6):e0157594. doi: 10.1371/journal.pone.0157594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet. 2008;17(1):1–14. doi: 10.1093/hmg/ddm280. [DOI] [PubMed] [Google Scholar]
- 13.Fauque P, Mondon F, Letourneur F, Ripoche MA, Journot L, Barbaux S, et al. In vitro fertilization and embryo culture strongly impact the placental transcriptome in the mouse model. PLoS One. 2010;5(2):e9218. doi: 10.1371/journal.pone.0009218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurowski A, Molotkov A, Soriano P. FGFR1 regulates trophectoderm development and facilitates blastocyst implantation. Dev Biol. 2019;446(1):94–101. doi: 10.1016/j.ydbio.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann M. R. W. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131(15):3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- 16.Yao J, Geng L, Huang R, Peng W, Chen X, Jiang X, et al. Effect of vitrification on in vitro development and imprinted gene Grb10 in mouse embryos. Reproduction. 2017;154(3):97–105. doi: 10.1530/REP-16-0480. [DOI] [PubMed] [Google Scholar]
- 17.Zandstra H, Brentjens LBPM, Spauwen B, Touwslager RNH, Bons JAP, Mulder AL, et al. Association of culture medium with growth, weight and cardiovascular development of IVF children at the age of 9 years. Hum Reprod. 2018;33(9):1645–1656. doi: 10.1093/humrep/dey246. [DOI] [PubMed] [Google Scholar]
- 18.Fauque P, Ripoche MA, Tost J, Journot L, Gabory A, Busato F, et al. Modulation of imprinted gene network in placenta results in normal development of in vitro manipulated mouse embryos. Hum Mol Genet. 2010;19(9):1779–1790. doi: 10.1093/hmg/ddq059. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh J, Mainigi M, Coutifaris C, Sapienza C. Outlier DNA methylation levels as an indicator of environmental exposure and risk of undesirable birth outcome. Hum Mol Genet. 2016;25(1):123–129. doi: 10.1093/hmg/ddv458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh J, Coutifaris C, Sapienza C, Mainigi M. Global DNA methylation levels are altered by modifiable clinical manipulations in assisted reproductive technologies. Clin Epigenetics. 2017;9:14. doi: 10.1186/s13148-017-0318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Cui Y, Zhou Z, Sha J, Li Y, Liu J. Altered global gene expressions of human placentae subjected to assisted reproductive technology treatments. Placenta. 2010;31(4):251–258. doi: 10.1016/j.placenta.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Nelissen EC, Dumoulin JC, Busato F, Ponger L, Eijssen LM, Evers JL, et al. Altered gene expression in human placentas after IVF/ICSI. Hum Reprod. 2014;29(12):2821–2831. doi: 10.1093/humrep/deu241. [DOI] [PubMed] [Google Scholar]
- 23.Duranthon V, Chavatte-Palmer P. Long term effects of ART: what do animals tell us? Mol Reprod Dev. 2018;85(4):348–368. doi: 10.1002/mrd.22970. [DOI] [PubMed] [Google Scholar]
- 24.Hossain MM, Tesfaye D, Salilew-Wondim D, Held E, Pröll MJ, Rings F, et al. Massive deregulation of miRNAs from nuclear reprogramming errors during trophoblast differentiation for placentogenesis in cloned pregnancy. BMC Genomics. 2014;15:43. doi: 10.1186/1471-2164-15-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Waal E, Mak W, Calhoun S, Stein P, Ord T, Krapp C, et al. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biol Reprod. 2014;90(2):22. doi: 10.1095/biolreprod.113.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher SJ. Why is placentation abnormal in preeclampsia? Am J Obstet Gynecol. 2015;213(4):S115–S122. doi: 10.1016/j.ajog.2015.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louwen F, Muschol-Steinmetz C, Reinhard J, Reitter A, Yuan J. A lesson for cancer research: placental microarray gene analysis in preeclampsia. Oncotarget. 2012;3(8):759–773. doi: 10.18632/oncotarget.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou K, Ding G, Huang H. Advances in research into gamete and embryo-fetal origins of adult diseases. Sci China Life Sci. 2019;62(3):360–368. doi: 10.1007/s11427-018-9427-4. [DOI] [PubMed] [Google Scholar]
- 29.Leijnse JEW, de Heus R, de Jager W, Rodenburg W, Peeters LLH, Franx A, et al. First trimester placental vascularization and angiogenetic factors are associated with adverse pregnancy outcome. Pregnancy Hypertens. 2018;13:87–94. doi: 10.1016/j.preghy.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Yang R, Liu YY, Zhao L, Wang Y, Li R, Liu P, et al. Human trophoblast cell during first trimester after IVF-ET differs from natural conceived pregnancy in development and function. Histol Histopathol. 2017;32(3):243–251. doi: 10.14670/HH-11-787. [DOI] [PubMed] [Google Scholar]
- 31.Yue XL, Gao ZQ. Identification of pathogenic genes of pterygium based on the gene expression omnibus database. Int J Ophthalmol. 2019;12(4):529–535. doi: 10.18240/ijo.2019.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapourani CA, Sanguinetti G. BPRMeth: a flexible Bioconductor package for modelling methylation profiles. Bioinformatics. 2018;34(14):2485–2486. doi: 10.1093/bioinformatics/bty129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maheshwari A, Pandey S, Shetty A, Hamilton M, Bhattacharya S. Obstetric and perinatal outcomes in singleton pregnancies resulting from the transfer of frozen thawed versus fresh embryos generated through in vitro fertilization treatment: a systematic review and meta-analysis. Fertil Steril. 2012;98(2):368–377. doi: 10.1016/j.fertnstert.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 34.Imudia AN, Awonuga AO, Kaimal AJ, Wright DL, Styer AK, Toth TL. Elective cryopreservation of all embryos with subsequent cryothaw embryo transfer in patients at risk for ovarian hyperstimulation syndrome reduces the risk of adverse obstetric outcomes: a preliminary study. Fertil Steril. 2013;99(1):168–173. doi: 10.1016/j.fertnstert.2012.08.060. [DOI] [PubMed] [Google Scholar]
- 35.Sharpe Richard M. Of mice and men: long-term safety of assisted reproduction treatments. Human Reproduction. 2018;33(5):793–796. doi: 10.1093/humrep/dey071. [DOI] [PubMed] [Google Scholar]
- 36.Donjacour A, Liu X, Lin W, Simbulan R, Rinaudo PF. In vitro fertilization affects growth and glucose metabolism in a sex-specific manner in an outbred mouse model. Biol Reprod. 2014;90(4):80. doi: 10.1095/biolreprod.113.113134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinborg A, Wennerholm UB, Romundstad LB, Loft A, Aittomaki K, Söderström-Anttila V, et al. Why do singletons conceived after assisted reproduction technology have adverse perinatal outcome? Systematic review and meta-analysis. Hum Reprod Update. 2013;19(4):87–104. doi: 10.1093/humupd/dms044. [DOI] [PubMed] [Google Scholar]
- 38.Turco MY, Gardner L, Kay RG, Hamilton RS, Prater M, Hollinshead MS, et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature. 2018;564(7735):263–267. doi: 10.1038/s41586-018-0753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng Y, Ma X, Deng L, Yao B, Xiong Y, Wu Y. Role of selectins and their ligands in human implantation stage. Glycobiology. 2017;27(5):385–391. doi: 10.1093/glycob/cwx009. [DOI] [PubMed] [Google Scholar]
- 40.Evans J. Hyperglycosylated hCG: a unique human implantation and invasion factor. Am J Reprod Immunol. 2016;75(3):333–340. doi: 10.1111/aji.12459. [DOI] [PubMed] [Google Scholar]
- 41.James JL, Carter AM, Chamley LW. Human placentation from nidation to 5 weeks of gestation. Part II: tools to model the crucial first days. Placenta. 2012;33(5):335–342. doi: 10.1016/j.placenta.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 42.Xie L, Sadovsky Y. The function of miR-519d in cell migration, invasion, and proliferation suggests a role in early placentation. Placenta. 2016;48:34–37. doi: 10.1016/j.placenta.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Litzky JF, Deyssenroth MA, Everson TM, Armstrong DA, Lambertini L, Chen J, et al. Placental imprinting variation associated with assisted reproductive technologies and subfertility. Epigenetics. 2017;12(8):653–661. doi: 10.1080/15592294.2017.1336589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu N, Barlow GM, Cui J, Wang ET, Lee B, Akhlaghpour M, et al. Comparison of genome-wide and gene-specific DNA methylation profiling in first-trimester chorionic villi from pregnancies conceived with infertility treatments. Reprod Sci. 2017;24(7):996–1004. doi: 10.1177/1933719116675056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomopoulos C, Tsioufis C, Michalopoulou H, Makris T, Papademetriou V, Stefanadis C. Assisted reproductive technology and pregnancy-related hypertensive complications: a systematic review. J Hum Hypertens. 2013;27(3):148–157. doi: 10.1038/jhh.2012.13. [DOI] [PubMed] [Google Scholar]
- 46.Senapati S, Wang F, Ord T, Coutifaris C, Feng R, Mainigi M. Superovulation alters the expression of endometrial genes critical to tissue remodeling and placentation. J Assist Reprod Genet. 2018;35(10):1799–1808. doi: 10.1007/s10815-018-1244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nevalainen J, Skarp S, Savolainen ER, Ryynänen M, Järvenpää J. Intrauterine growth restriction and placental gene expression in severe preeclampsia, comparing early-onset and late-onset forms. J Perinat Med. 2017;45(7):869–877. doi: 10.1515/jpm-2016-0406. [DOI] [PubMed] [Google Scholar]
- 48.Sibley CP. Treating the dysfunctional placenta. J Endocrinol. 2017;234(2):R81–R97. doi: 10.1530/JOE-17-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao F, Hu W, Li Y, Shen H, Hu J. Mono-2-ethylhexyl phthalate inhibits human extravillous trophoblast invasion via the PPARγ pathway. Toxicol Appl Pharmacol. 2017;327:23–29. doi: 10.1016/j.taap.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Puscheck Elizabeth E., Bolnick Alan, Awonuga Awoniyi, Yang Yu, Abdulhasan Mohammed, Li Quanwen, Secor Eric, Louden Erica, Hüttemann Maik, Rappolee Daniel A. Why AMPK agonists not known to be stressors may surprisingly contribute to miscarriage or hinder IVF/ART. Journal of Assisted Reproduction and Genetics. 2018;35(8):1359–1366. doi: 10.1007/s10815-018-1213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knyazev EN, Zakharova GS, Astakhova LA, Tsypina IM, Tonevitsky AG, Sukhikh GT. Metabolic reprogramming of trophoblast cells in response to hypoxia. Bull Exp Biol Med. 2019;166(3):321–325. doi: 10.1007/s10517-019-04342-1. [DOI] [PubMed] [Google Scholar]
- 52.Dunn WB, Brown M, Worton SA, Crocker IP, Broadhurst D, Horgan R, et al. Changes in the metabolic footprint of placental explant-conditioned culture medium identifies metabolic disturbances related to hypoxia and pre-eclampsia. Placenta. 2009;30(11):974–980. doi: 10.1016/j.placenta.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 53.Mitsuya K, Parker AN, Liu L, Ruan J, Vissers MCM, Myatt L. Alterations in the placental methylome with maternal obesity and evidence for metabolic regulation. PLoS One. 2017;12(10):e0186115. doi: 10.1371/journal.pone.0186115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murthi P, Wallace EM, Walker DW. Altered placental tryptophan metabolic pathway in human fetal growth restriction. Placenta. 2017;52:62–70. doi: 10.1016/j.placenta.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 55.Kiang JG, Krishnan S, Lu X, Li Y. Inhibition of inducible nitric-oxide synthase protects human T cells from hypoxia-induced apoptosis. Mol Pharmacol. 2008;73(3):738–747. doi: 10.1124/mol.107.041079. [DOI] [PubMed] [Google Scholar]
- 56.Jin X, Ma Q, Sun Z, Yang X, Zhou Q, Qu G, et al. Airborne fine particles induce hematological effects through regulating the crosstalk of the Kallikrein-Kinin, complement, and coagulation systems. Environ Sci Technol. 2019;53(3):2840–2851. doi: 10.1021/acs.est.8b05817. [DOI] [PubMed] [Google Scholar]
- 57.Levi M, Cohn DM, Zeerleder S. Hereditary angioedema: linking complement regulation to the coagulation system. Res Pract Thromb Haemost. 2019;3(1):38–43. doi: 10.1002/rth2.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Than NG, Romero R, Tarca AL, Kekesi KA, Xu Y, Xu Z, et al. Integrated systems biology approach identifies novel maternal and placental pathways of preeclampsia. Front Immunol. 2018;9:1661. doi: 10.3389/fimmu.2018.01661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scully M, Cataland S, Coppo P, de la Rubia J, Friedman KD, Kremer Hovinga J, et al. Consensus on the standardization of terminology in thrombotic thrombocytopenic purpura and related thrombotic microangiopathies. J Thromb Haemost. 2017;15(2):312–322. doi: 10.1111/jth.13571. [DOI] [PubMed] [Google Scholar]
- 60.Ruebner M, Langbein M, Strissel PL, Henke C, Schmidt D, Goecke TW, et al. Regulation of the human endogenous retroviral Syncytin-1 and cell-cell fusion by the nuclear hormone receptors PPARγ/RXRα in placentogenesis. J Cell Biochem. 2012;113(7):2383–2396. doi: 10.1002/jcb.24110. [DOI] [PubMed] [Google Scholar]
- 61.Ganss R. Maternal metabolism and vascular adaptation in pregnancy: the PPAR link. Trends Endocrinol Metab. 2017;28(1):73–84. doi: 10.1016/j.tem.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Allam HIG, Masri AAA. The potential therapeutic role of peroxisome ProliferatorActivated receptors agonist in Preeclamptic pregnant rats. J Coll Physicians Surg Pak. 2018;28(1):31–35. doi: 10.29271/jcpsp.2018.01.31. [DOI] [PubMed] [Google Scholar]
- 63.Higgins JS, Vaughan OR, Fernandez de Liger E, Fowden AL, Sferruzzi-Perri AN. Placental phenotype and resource allocation to fetal growth are modified by the timing and degree of hypoxia during mouse pregnancy. J Physiol. 2016;594(5):1341–1356. doi: 10.1113/JP271057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kouhkan A, Khamseh ME, Pirjani R, Moini A, Arabipoor A, Maroufizadeh S, et al. Obstetric and perinatal outcomes of singleton pregnancies conceived via assisted reproductive technology complicated by gestational diabetes mellitus: a prospective cohort study. BMC Pregnancy Childbirth. 2018;18(1):495. [DOI] [PMC free article] [PubMed]
- 65.Delle Piane L, Lin W, Liu X, Donjacour A, Minasi P, Revelli A, et al. Effect of the method of conception and embryo transfer procedure on mid-gestation placenta and fetal development in an IVF mouse model. Hum Reprod. 2010;25(8):2039–2046. doi: 10.1093/humrep/deq165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bloise E, Lin W, Liu X, Simbulan R, Kolahi KS, Petraglia F, et al. Impaired placental nutrient transport in mice generated by in vitro fertilization. Endocrinology. 2012;153(7):3457–3467. doi: 10.1210/en.2011-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haavaldsen C, Tanbo T, Eskild A. Placental weight in singleton pregnancies with and without assisted reproductive technology: a population study of 536,567 pregnancies. Hum Reprod. 2012;27(2):576–582. doi: 10.1093/humrep/der428. [DOI] [PubMed] [Google Scholar]
- 68.Mirbod P. Analytical model of the feto-placental vascular system: consideration of placental oxygen transport. R Soc Open Sci. 2018;5(4):180219. doi: 10.1098/rsos.180219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dahdouh S, Andescavage N, Yewale S, Yarish A, Lanham D, Bulas D, et al. In vivo placental MRI shape and textural features predict fetal growth restriction and postnatal outcome. J Magn Reson Imaging. 2018;47(2):449–458. doi: 10.1002/jmri.25806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barker DJ, Thornburg KL. Placental programming of chronic diseases, cancer and lifespan: a review. Placenta. 2013;34(10):841–845. doi: 10.1016/j.placenta.2013.07.063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Detail information on the selection of primers for real-time RT-PCR experiments (DOC 34 kb)
Table S2. The biological processes of up regulated genes in placental subjected to IVF-ET (DOC 68 kb)
Table S3. The biological processes of down regulated genes in placental subjected to IVF-ET (DOC 70 kb)
Figure S1. KEGG analysis of the differentially expressed genes, which are labeled, in the PPAR signaling pathway in the placenta after IVF-ET during the first trimester by KEGG. Red, upregulated; Blue, downregulated; Green, no change. (TIF 14811 kb)
Data Availability Statement
The primary microarray data of the placental samples have been submitted to the Gene Expression Omnibus of the National Center for Biotechnology Information. The data can be obtained through GEO Series accession number GSE 122214 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122214). Other data generated or analyzed during this study are available from the corresponding author upon reasonable request.