Abstract
Background
Milk quality in dairy cattle is routinely assessed via analysis of mid-infrared (MIR) spectra; this approach can also be used to predict the milk’s cheese-making properties (CMP) and composition. When this method of high-throughput phenotyping is combined with efficient imputations of whole-genome sequence data from cows’ genotyping data, it provides a unique and powerful framework with which to carry out genomic analyses. The goal of this study was to use this approach to identify genes and gene networks associated with milk CMP and composition in the Montbéliarde breed.
Results
Milk cheese yields, coagulation traits, milk pH and contents of proteins, fatty acids, minerals, citrate, and lactose were predicted from MIR spectra. Thirty-six phenotypes from primiparous Montbéliarde cows (1,442,371 test-day records from 189,817 cows) were adjusted for non-genetic effects and averaged per cow. 50 K genotypes, which were available for a subset of 19,586 cows, were imputed at the sequence level using Run6 of the 1000 Bull Genomes Project (comprising 2333 animals). The individual effects of 8.5 million variants were evaluated in a genome-wide association study (GWAS) which led to the detection of 59 QTL regions, most of which had highly significant effects on CMP and milk composition. The results of the GWAS were further subjected to an association weight matrix and the partial correlation and information theory approach and we identified a set of 736 co-associated genes. Among these, the well-known caseins, PAEP and DGAT1, together with dozens of other genes such as SLC37A1, ALPL, MGST1, SEL1L3, GPT, BRI3BP, SCD, GPAT4, FASN, and ANKH, explained from 12 to 30% of the phenotypic variance of CMP traits. We were further able to identify metabolic pathways (e.g., phosphate and phospholipid metabolism and inorganic anion transport) and key regulator genes, such as PPARA, ASXL3, and bta-mir-200c that are functionally linked to milk composition.
Conclusions
By using an approach that integrated GWAS with network and pathway analyses at the whole-genome sequence level, we propose candidate variants that explain a substantial proportion of the phenotypic variance of CMP traits and could thus be included in genomic evaluation models to improve milk CMP in Montbéliarde cows.
Electronic supplementary material
The online version of this article (10.1186/s12711-019-0473-7) contains supplementary material, which is available to authorized users.
Background
About 40% of the bovine milk produced worldwide is processed into cheese; because of this, the cheese-making properties (CMP) of bovine milk are economically important for the dairy industry. Direct measurement of CMP is costly and time-consuming, and cannot be obtained on a very large scale. However, mid-infrared (MIR) spectrometry, which is already widely employed to predict milk composition, has been shown to provide indirect measures of CMP that are sufficiently reliable to be used in genetic analyses [1]. Indeed, because of their strong dependence on milk composition traits [2], milk CMP, especially cheese yields and coagulation properties, can be routinely assessed at low cost from MIR spectra [3]. The information obtained from high-throughput MIR spectra can then be combined with genotypic data from cows that are generated for the purpose of genomic selection to provide a unique resource for large-scale genomic analyses of CMP aimed at identifying the genes involved in the genetic determinism of these traits.
Genomic regions containing quantitative trait loci (QTL) that affect traits of interest, such as CMP, can be identified by genome-wide association studies (GWAS). By combining the results of genotyping for genomic selection with reference data from the 1000 Bull Genomes Project, it becomes possible to carry out GWAS on imputed whole-genome sequences (WGS) that should contain the causative mutations for traits of interest [4]. However, even if these analyses are carried out at the sequence level, GWAS alone is generally not sufficient to identify causative genes, let alone causative variants for complex and polygenic traits. Indeed, due to the long-range linkage disequilibrium (LD) in dairy cattle, many variants with almost identical P-values that are potentially located in more than one gene or in intergenic regions are generally found in a QTL region, which complicates identification of the causative mutations. Moreover, complex traits are typically influenced by many genomic regions, most of which explain only a small proportion of the phenotypic variance and are thus difficult to detect by GWAS. Finally, GWAS performed on a single trait and single marker cannot take either the pleiotropic effects of variants or the interactions between them into account. Thus, a GWAS-based approach is a good starting point for identifying QTL regions but needs to be supplemented by additional analyses to capture a larger proportion of the genetic variance and to understand in depth the genetic architecture of complex traits, such as CMP. In the last decade, methods have been developed that build on GWAS results by using gene network analysis to highlight co-associated genes for a set of correlated traits [5, 6]. Once the gene network is built, it is then possible to carry out in silico functional analyses, based on databases from bovine or other organisms’ genomes, to identify key regulators that modulate gene expression or to highlight the enrichment of gene-sets linked to certain metabolic pathways. Gene network approaches have been applied to milk CMP [7], fatty acid composition [8, 9], and protein composition [10, 11] but, to date, there has been no joint analysis of CMP and milk composition in spite of the close relationship between the two groups of traits. Moreover, all previous studies examined only a limited number of cows (164 to 1100 cows) and genomic variants (50 K or HD SNP chips).
The goal of the FROM’MIR project is to analyze CMP and milk composition traits predicted from MIR spectra in the Montbéliarde dairy breed from the Franche-Comté region, which boasts the highest production of protected designation of origin (PDO) cheeses in France. Nine CMP traits (three measures of cheese yield, five coagulation traits, and one acidification trait) and 27 milk composition traits (protein, fatty acid, mineral, citrate, and lactose contents) were predicted with a relatively high degree of accuracy from more than 6.6 million MIR spectra of milk samples collected from 410,622 cows. Of these cows, 19,586 were genotyped with a SNP chip. A prior study revealed medium-to-high heritabilities for CMP traits as well as high genetic correlations among CMP traits and between CMP and some milk composition traits [3].
The objectives of the current study were first, to fine-map QTL for CMP and milk composition traits via GWAS of imputed WGS from 19,586 cows, and second, to explore the GWAS results using association weight matrices (AWM) [5] and partial correlation and information theory (PCIT) [6] analyses, in order to identify gene networks and metabolic and regulatory pathways that are associated with milk cheese-making and composition traits.
Methods
Animals, MIR spectra, and 50 K genotypes
For this study, we did not perform any experiments on animals; thus, no ethical approval was required. Details of the animals, milk analyses, and prediction equations were described in a prior study by Sanchez et al. [3]. Briefly, prediction equations were developed for nine CMP traits from 416 milk samples for which both reference measurements for those CMP traits and MIR spectra were taken. The CMP traits, described in Table 1, included three laboratory cheese yields (CYFRESH, CYDM, and CYFAT-PROT), five coagulation traits for pressed cooked cheese (PCC) and soft cheese (SC) (K10/RCTPCC, K10/RCTSC, aPCC, aSC, and a2SC), and milk pH after adding starter for PCC (pH0_PCC). The accuracies of MIR predictions, assessed by the coefficient of determination (R2), varied between 0.54 and 0.89 depending on the CMP trait (Table 1). Milk composition was also predicted using equations that were developed in previous projects (0.44 < R2 < 1; Table 1). Milk proteins and fatty acids were predicted with equations that were developed in the PhénoFinlait project [12–14], whereas for minerals and citrate content we used equations that were generated by the Optimir project [15]. Lactose was predicted by a Foss equation.
Table 1.
Means, standard deviations (SD) for cheese-making properties and milk composition traits in the genotyped population (N = 19,586), and accuracy of MIR predictions equations (R2 val)
| Trait | Description and unit | Mean | SD | R2 val |
|---|---|---|---|---|
| Cheese-making propertiesa | ||||
| CYFRESH | 100 × (g curd/g milk), in % | 37.7 | 4.95 | 0.82 |
| CYDM | 100 × (g DM curd/g DM milk), in % | 66.8 | 3.31 | 0.89 |
| CYFAT-PROT | (g milk fat + g milk protein)/kg curd, in g kg−1 | 189.7 | 14.3 | 0.54 |
| aPCC | Curd firmness at rennet coagulation time (RCT), in firm index (FI) | 18.8 | 1.72 | 0.76 |
| K10/RCTPCC | Curd organization index standardized for RCT | 0.37 | 0.06 | 0.68 |
| aSC | Curd firmness at RCT, in FI | 18.9 | 1.80 | 0.76 |
| a2SC | Curd firmness at 2 times RCT, in FI | 22.8 | 1.41 | 0.69 |
| K10/RCTSC | Curd organization index standardized for RCT | 0.37 | 0.07 | 0.72 |
| pH0_PCC | Initial value of pH | 6.52 | 0.04 | 0.62 |
| Protein composition | ||||
| PC | Protein content, in g/100 g milk | 3.36 | 0.20 | 1.00 |
| α-LA | α-lactalbumin, in g/100 g protein | 4.01 | 0.20 | 0.59 |
| β-LG | β-lactoglobulin, in g/100 g protein | 12.4 | 1.09 | 0.74 |
| αs1-CN | αs1-casein, in g/100 g protein | 32.2 | 0.18 | 0.88 |
| αs2-CN | αs2-casein, in g/100 g protein | 9.73 | 0.19 | 0.82 |
| β-CN | β-casein, in g/100 g protein | 29.7 | 0.68 | 0.92 |
| κ-CN | κ-casein, in g/100 g protein | 8.74 | 0.24 | 0.80 |
| ΣCN | Total caseins, in g/100 g protein | 80.8 | 0.74 | 0.98 |
| ΣWP | Total whey proteins, in g/100 g protein | 16.9 | 1.15 | 0.54 |
| Fatty acid composition | ||||
| FC | Fat content, in g/100 g milk | 3.73 | 0.32 | 1.00 |
| SFA | Saturated fatty acids, in g/100 g fat | 70.6 | 3.05 | 1.00 |
| MUFA | Mono-unsaturated fatty acids, in g/100 g fat | 26.5 | 2.68 | 0.97 |
| UFA | Unsaturated fatty acids, in g/100 g fat | 30.0 | 2.93 | 0.98 |
| PUFA | Poly-unsaturated fatty acids, in g/100 g fat | 3.33 | 0.39 | 0.76 |
| Σ C4-C10 | Sum of C4 to C10 fatty acids, in g/100 g fat | 11.6 | 0.71 | 0.95 |
| Σ C4-C12 | Sum of C4 to C12 fatty acids, in g/100 g fat | 14.2 | 0.93 | 0.95 |
| C14:0 | Myristic acid, in g/100 g fat | 11.1 | 1.05 | 0.94 |
| C16:0 | Palmitic acid, in g/100 g fat | 28.8 | 2.53 | 0.94 |
| C18:0 | Stearic acid, in g/100 g fat | 10.5 | 1.42 | 0.84 |
| C18:1 | Oleic acid, in g/100 g fat | 23.2 | 2.59 | 0.96 |
| Minerals | ||||
| Ca | Calcium, in mg/kg milk | 1165 | 69.6 | 0.82 |
| P | Phosphorous, in mg/kg milk | 1014 | 62.5 | 0.75 |
| Mg | Magnesium, in mg/kg milk | 100.9 | 5.5 | 0.77 |
| K | Potassium, in mg/kg milk | 1496 | 69.3 | 0.68 |
| Na | Sodium, in mg/kg milk | 338.3 | 29.1 | 0.44 |
| Other compounds | ||||
| Lactose | Lactose, in g/kg milk | 49.3 | 1.4 | 0.92 |
| Citrate | Citrate, in g/kg milk | 0.83 | 0.11 | 0.90 |
aFor pressed cooked cheese (PCC) and soft cheese (SC)
Prediction equations were applied to the original dataset, which comprised 6,670,769 milk samples originating from 410,622 Montbéliarde cows. Data from cows with at least three test-day records during the first lactation (1,442,371 test-day records from 189,817 cows) were adjusted for non-genetic effects in a mixed model with the Genekit software [16]. Herd × test-day × spectrometer, age at calving, and stage of lactation were included in this model as fixed effects, while animal genetic and permanent environmental effects were assumed to be random. Test-day data adjusted for fixed effects were then averaged over a lactation for each cow. A subset of 19,586 cows for which MIR spectra were available had also been genotyped for the purpose of genomic selection by using the BovineSNP50 (50 K, 6505 cows) or the EuroG10 K BeadChip (Illumina Inc., San Diego, 13,081 cows). Means and standard deviations of the traits for this subset are in Table 1. Using FImpute software [17], all genotypes were imputed to the 50 K-SNP level. A total of 43,801 autosomal SNPs were retained after quality control filters were applied. These filters were taken directly from the French national evaluation system [18]: individual call rate higher than 95%, SNP call rate higher than 90%, minor allele frequency (MAF) higher than 1% in at least one major French dairy cattle breed, and genotype frequencies in Hardy–Weinberg equilibrium with P > 10−4.
Imputation to whole-genome sequences
The 50 K SNP genotypes of the 19,586 cows were then imputed to whole-genome sequences (WGS). A two-step approach was applied in order to improve the accuracy of imputed genotypes of the WGS variants [19]: from 50 to 777 K high-density (HD) SNPs using FImpute software [17], and then, from imputed HD SNPs to WGS, using Minimac software [20]. In spite of a longer computing time, Minimac was preferred over FImpute to impute on WGS because it infers allele dosages in addition to the best-guess genotypes. Compared to the best-guess genotypes, allele dosages are expected to be more correlated to true genotypes [21] and to lead to a better targeting of causative mutations in GWAS analyses [22]. Imputations from 50 K to the HD SNP level were performed using a within-breed reference set of 522 Montbéliard bulls that were genotyped with the Illumina BovineHD BeadChip (Illumina Inc., San Diego, CA) [23]. WGS variants were imputed from HD SNP genotypes using WGS variants of 2333 Bos taurus animals, from the 6th run of the 1000 Bull Genomes Project [21, 24]. These animals represent 51 cattle breeds and include 54 Montbéliard individuals, most of them being major ancestor bulls with a high cumulated contribution to the breed (80.6%). We applied the protocol defined by the “1000 Bull Genomes” consortium [4, 25]: (1) short reads were filtered for quality and aligned to the UMD3.1 reference sequence [4, 26], and small genomic variations (SNPs and indels) were detected using SAMtools 0.0.18 [27]; (2) raw variants were filtered to produce 26,738,438 autosomal variants as described in Boussaha et al. [26]; and (3) filtered variants were annotated with the Ensembl variant effect predictor (VEP) pipeline v81 [28] and effects of amino-acid changes were predicted using the SIFT tool [29].
The precision of imputation from HD SNP to sequence was assessed using the coefficient of determination (R2) calculated with Minimac software [20]. In order to remove variants with low imputation accuracies, only variants with an R2 higher than 20% and a MAF higher than 1% were retained for further association analyses, i.e. 8,551,748 variants with a mean R2 of 76% (Fig. 1).
Fig. 1.
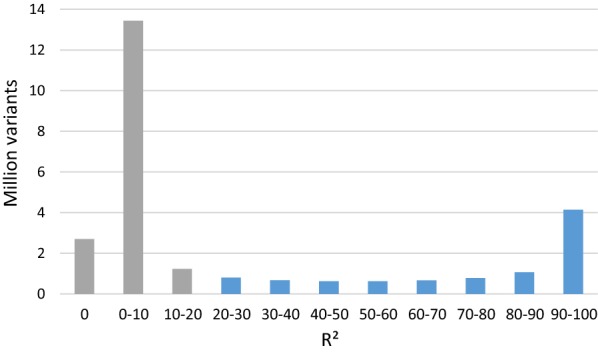
Distribution of imputation accuracies (coefficient of determination, R2) for 26.7 million sequence variants imputed with Minimac. Variants in blue, with R2 > 0.20, were retained for GWAS analyses
Whole-genome sequence association analyses
We performed single-trait association analyses between all 8,551,748 variants and the 36 CMP and milk composition traits described in Table 1. All association analyses were performed using the mlma option of the GCTA software (version 1.24), which applies a mixed linear model that includes the variant to be tested [30]:
| 1 |
where is the vector of pre-adjusted phenotypes, averaged per cow; is the overall mean; is the additive fixed effect of the variant to be tested for association; is the vector of predicted allele dosages, varying between 0 and 2; is the vector of random polygenic effects, with the genomic relationship matrix (GRM), calculated using the HD SNP genotypes [31], and is the polygenic variance, estimated based on the null model and then fixed while testing for the association between each variant and the trait of interest; and is the vector of random residual effects, with the identity matrix and the residual variance.
The Bonferroni correction was applied to the thresholds in order to account for multiple testing. We used a very stringent correction, which considered all 8.5 million tests as independent. Therefore, the 5% genome-wide threshold of significance corresponded to a nominal P value of 5.8 × 10−9 (− log10(P) = 8.2). When a given trait was significantly affected by multiple variants, the variants that were located less than 1 Mbp apart were grouped in the same QTL region. The bounds of QTL regions were then determined by considering the positions of variants that were included in the upper third of the peak. For each trait, the percentage of phenotypic variance explained by each QTL was calculated as follows: , with the phenotypic variance of the trait, and and are the frequency and the estimated allelic substitution effect, respectively, of the variant with the most significant effect in the QTL region.
Co-associated gene network analysis
Co-associated genes were detected from the GWAS results using the AWM approach [5, 6]. We first constructed two × matrices with variants row-wise ( = 8,551,748) and traits column-wise ( = 36). The first matrix contained variants’ z-score standardized additive effects, whereas the second one contained the P-values associated with those effects. Among the CMP traits, CYDM was selected as the key phenotype because it has the highest economic importance to the cheese-making process. The AWM was constructed following the procedure described in Ramayo-Caldas et al. [32]. SNPs were included in the analysis if their P-value for CYDM was less than or equal to 0.001. Due to the large number of traits analyzed, we calculated correlation coefficients between SNP additive effects for different traits and then selected the set of traits correlated with CYDM (|r| ≥ 0.25). Next, we explored the dependency among traits and we estimated that on average, six other phenotypes were associated with these SNPs at the same P-value (P ≤ 0.001). Other variants with significant effects on at least six traits were finally included in the analysis. Based on VEP annotation [33], we then selected only the SNPs that were located within or close to (within 10 kb of) genes. Among these, we retained only one variant per gene, i.e. the SNP that was associated with the largest number of traits or, in case of a tie, the variant for which the sum of P-values for the traits was the lowest.
Subsequently, to identify significant gene–gene interactions, partial correlations were computed using the PCIT algorithm developed by Reverter and Chan [34]; the algorithm was implemented in an R package designed for this purpose [35]. We visualized the gene network with Cytoscape 3.6.1 [36], with each node representing a gene and each edge representing a significant interaction. The centrality parameters of each node were assessed using the CentiScaPe 2.2 plug-in for Cytoscape [37]. For each node, we calculated the number of adjacent genes (degree parameter) and the relative node contribution (eigenvector parameter), with the latter value being higher (or lower) if the gene was connected to highly (or poorly) connected genes.
Identification of key regulators
Potential key regulators of the gene network were identified using two approaches. First, we used the iRegulon 1.3 plug-in for Cytoscape [38] to identify transcription factors (TF) in silico; this method was based on human datasets but included orthologous regions of ten other vertebrate genomes, including Bos taurus. Two types of data were used to identify regulatory regions that were shared by the genes identified in the network: (1) TF binding site motifs in the cis-regulatory regions, and (2) thousands of ChIP-Seq (chromatin immunoprecipitation followed by high-throughput sequencing) datasets from the ENCODE project [39] corresponding to targets of known TF. More details are in Janky et al. [38]. We then applied an information loss-less approach [6] that explored the connectivity of all regulators in the network, including TF, miRNA, and lnRNA. As recommended by Reverter and Fortes [6], we tested trios of TF genes to find the minimal set of TF genes with maximal coverage of the network.
Gene-set enrichment analysis
Next, we searched in the gene network for enrichment in gene ontology (GO) terms and pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG), using the ClueGO 2.5.1 plug-in for Cytoscape [40]. In order to avoid selecting GO terms that were too general (too many genes) or too specific (too few genes), we selected the 4th to 8th levels of the GO hierarchy. A gene set was considered to be enriched if the P-value associated with the hypergeometric test was lower than 0.05, after application of the Benjamini–Hochberg correction for multiple testing. GO terms and KEGG pathways were subsequently clustered in functional groups if the kappa statistic was higher than 0.4.
Results
GWAS analyses
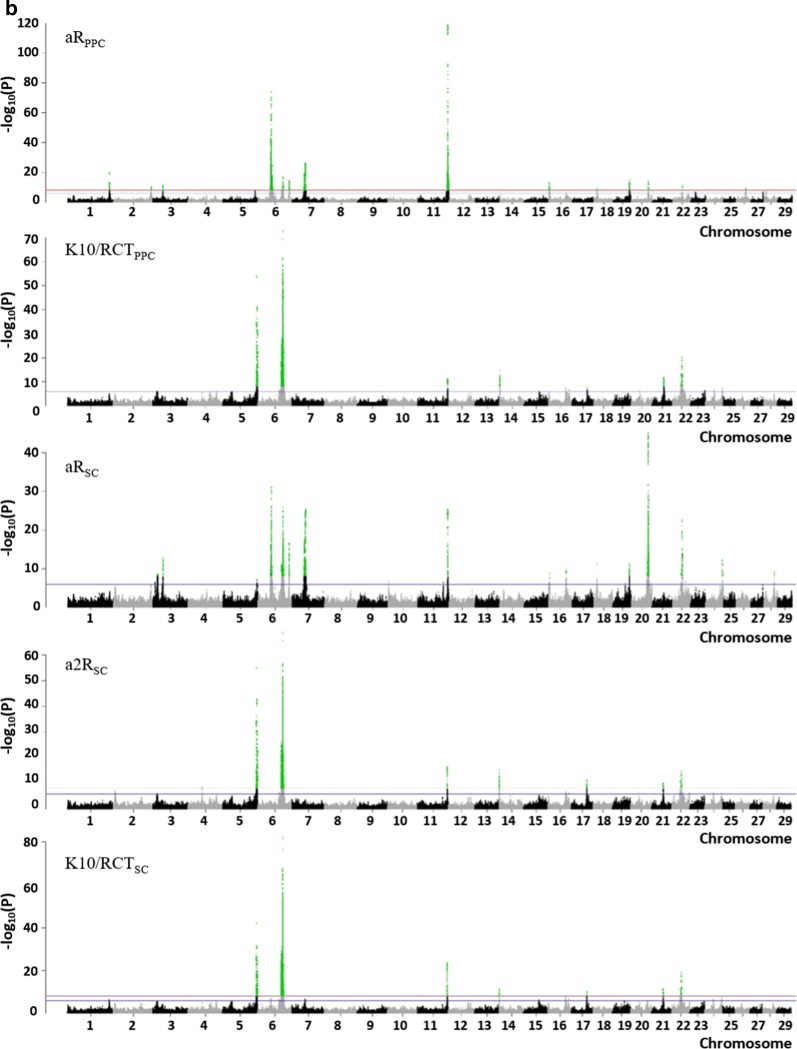
GWAS that was carried out on 8,551,748 imputed WGS variants for the 36 CMP and milk composition traits revealed 236,332 significant variant × trait combinations (− log10(P) > 8.2), corresponding to 79,803 different variants. Due to the high maximal − log10(P) value for a large number of genomic regions (up to 560 for one of the QTL detected on chromosome 11), the number of variants with significant effects (− log10(P) > 8.2) was sometimes very large in a given region. Thus, to best target candidate variants, we selected only the variants that were located in the upper third of the peaks, as described in the Methods section. In doing so, we defined 59 QTL regions, which contained 6757 distinct variants (Table 2). In each of the QTL regions, we designated “candidate variants” as the variants that were located within the confidence intervals of the QTL and the “best candidate variant” (described in Table 2) as the variant within a gene (or its upstream/downstream regions) with the most significant effects.
Table 2.
The 59 QTL regions identified by GWAS, the most likely candidate variant, and number of traits affected by the QTL
| QTL region | Best candidate varianta | Number of traitsd | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | BTA | From (bp) | To (bp) | # variants | #genes | bp | Variant ID | Rank | R2 | MAF | Effect | SE | − log10P | Gene | Functional annotation | Trait most strongly affected | CMP | Proteins | Fatty acids | Minerals |
| 1 | 1 | 144,389,419 | 144,398,814 | 6 | 1 | 144,395,375 | rs136069703 | 1 | 0.97 | 0.38 | 29.5 | 0.951 | 210.4 | SLC37A1 | Intron | P | 3 | 1 | 1 | 4 |
| 2 | 2 | 5,718,384 | 6,567,522 | 111 | 5 | 5,865,070 | rs136830094 | 1 | 0.91 | 0.22 | − 0.02 | 0.003 | 15.6 | INPP1 | Downstream | Lactose | 0 | 1 | 0 | 2 |
| 3 | 2 | 131,808,301 | 131,888,417 | 88 | 1 | 131,816,616 | rs133677653 | 1 | 0.91 | 0.49 | − 0.05 | 0.003 | 41.4 | ALPL | Intron | κ-CN | 1 | 3 | 0 | 1 |
| 4 | 3 | 7,442,755 | 8,149,715 | 56 | 5 | 7,935,102 | rs383033753 | 7 | 0.50 | 0.14 | − 0.65 | 0.096 | 10.7 | FCGR2B | Intron | CYDM | 2 | 0 | 1 | 0 |
| 5 | 3 | 15,514,034 | 15,928,379 | 401 | 7 | 15,525,599 | rs110073735 | 1 | 0.99 | 0.03 | − 2.78 | 0.410 | 10.9 | EFNA1 | Intron | Lactose | 1 | 0 | 0 | 3 |
| 6 | 3 | 34,235,208 | 34,355,357 | 35 | 4 | 34,327,146 | rs210558120 | 1 | 0.34 | 0.43 | 0.26 | 0.035 | 12.9 | KIAA1324 | Intron | aSC | 2 | 0 | 0 | 0 |
| 7 | 4 | 49,033,707 | 49,153,995 | 39 | 2 | 49,033,707 | rs380575157 | 1 | 0.47 | 0.20 | 0.02 | 0.001 | 30.7 | CBLL1 | Upstream | pH0_PCC | 2 | 3 | 0 | 2 |
| 8 | 4 | 75,743,094 | 79,803,738 | 59 | 5 | 77,825,429 | rs385069094 | 1 | 0.57 | 0.01 | − 2.34 | 0.360 | 10.1 | GCK | Intron | Mg | 0 | 1 | 0 | 3 |
| 9 | 4 | 92,588,016 | 92,966,245 | 227 | 7 | 92,624,543 | rs379514460 | 1 | 0.34 | 0.03 | − 29.0 | 4.226 | 11.2 | FSCN3 | Upstream | Ca | 0 | 0 | 0 | 1 |
| 10 | 5 | 29,947,476 | 31,423,430 | 315 | 25 | 29,947,476 | rs442522314 | 2 | 0.63 | 0.09 | − 2.26 | 0.383 | 8.4 | COX14 | Downstream | Lactose | 0 | 0 | 0 | 1 |
| 11 | 5 | 93,892,583 | 93,945,738 | 9 | 1 | 93,943,700 | rs210744452 | 1 | 0.77 | 0.05 | 0.11 | 0.010 | 27.7 | MGST1 | Intron | FC | 2 | 0 | 7 | 0 |
| 12 | 5 | 117,126,900 | 119,221,867 | 51 | 3 | 117,972,265 | rs525880746 | 1 | 0.79 | 0.04 | 42.4 | 2.186 | 83.0 | GRAMD4 | Upstream | Ca | 7 | 4 | 1 | 6 |
| 13 | 6 | 37,857,989 | 38,326,250 | 268 | 8 | 38,326,250 | rs382477515 | 1 | 0.37 | 0.42 | − 0.10 | 0.017 | 9.5 | IBSP | Upstream | β-CN | 0 | 1 | 0 | 0 |
| 14 | 6 | 46,555,489 | 47,082,793 | 89 | 3 | 46,876,802 | rs110408618 | 1 | 1.00 | 0.17 | 27.8 | 1.189 | 120.5 | SEL1L3 | Intron | K | 4 | 5 | 1 | 4 |
| 15 | 6 | 87,125,482 | 87,961,577 | 143 | 7 | 87,392,899 | rs382350292 | 3 | 0.53 | 0.23 | 0.03 | 0.002 | 76.4 | CSN3 | Downstream | K10/RSC | 8 | 8 | 0 | 1 |
| 16 | 6 | 108,967,622 | 109,145,154 | 91 | 5 | 109,026,822 | rs208213463 | 1 | 0.93 | 0.47 | − 8.42 | 0.904 | 19.9 | GAK | Intron | Ca | 2 | 2 | 0 | 2 |
| 17 | 7 | 958,741 | 1,146,962 | 359 | 4 | 975,515 | rs383246531 | 1 | 0.93 | 0.37 | − 1.56 | 0.180 | 17.4 | 47695b | Intron | Lactose | 0 | 0 | 0 | 2 |
| 18 | 7 | 41,576,519 | 46,646,915 | 399 | 23 | 46,452,656 | rs209828204 | 6 | 0.87 | 0.07 | 1.36 | 0.127 | 26.0 | FSTL4 | Downstream | aPCC | 2 | 2 | 0 | 1 |
| 19 | 9 | 102,731,669 | 103,130,410 | 207 | 3 | 102,885,120 | rs134445867 | 15 | 0.90 | 0.03 | − 0.14 | 0.023 | 9.3 | MPC1 | Intron | C4_C10 | 0 | 0 | 1 | 0 |
| 20 | 10 | 1,984,741 | 2,326,212 | 86 | 1 | 2,096,282 | rs385793060 | 67 | 0.99 | 0.35 | − 2.04 | 0.380 | 7.1 | 47622b | Upstream | Na | 0 | 0 | 0 | 1 |
| 21 | 10 | 48,359,318 | 50,266,445 | 213 | 1 | 49,459,919 | rs109896326 | 1 | 1.00 | 0.14 | 0.30 | 0.046 | 9.8 | RORA | Intron | UNSAT | 0 | 0 | 4 | 0 |
| 22 | 10 | 99,714,774 | 99,843,583 | 20 | 0 | 99,714,774 | rs440530756 | 1 | 0.22 | 0.02 | − 0.18 | 0.028 | 9.8 | – | Intergenic | PUNSAT | 0 | 0 | 2 | 0 |
| 23 | 11 | 9,023,594 | 9,684,851 | 11 | 3 | 9,684,851 | rs384459785 | 8 | 0.62 | 0.07 | − 2.82 | 0.385 | 12.6 | POLE4 | Intron | CYFAT-PROT | 3 | 0 | 3 | 0 |
| 24 | 11 | 14,152,677 | 15,493,112 | 132 | 6 | 14,284,886 | rs384594145 | 48 | 0.90 | 0.10 | − 0.01 | 0.001 | 18.7 | XDH | Upstream | pH0_PCC | 1 | 0 | 0 | 1 |
| 25 | 11 | 86,907,041 | 86,916,295 | 9 | 1 | 86,912,990 | rs481567394 | 1 | 0.21 | 0.05 | 0.02 | 0.003 | 16.7 | ATP6V1C2 | Intron | pH0_PCC | 1 | 0 | 0 | 0 |
| 26 | 11 | 103,273,963 | 103,322,890 | 214 | 4 | 103,301,982 | rs109907194 | 48 | 0.89 | 0.46 | − 1.09 | 0.017 | 559.7 | PAEP | Intron | ΣWP | 8 | 7 | 2 | 3 |
| 27 | 12 | 68,616,690 | 77,578,414 | 337 | 7 | 70,162,028 | rs721489054 | 22 | 0.83 | 0.17 | 0.22 | 0.019 | 29.0 | ABCC4 | Intron | C14:0 | 0 | 0 | 7 | 2 |
| 28 | 13 | 20,094,707 | 22,437,171 | 291 | 4 | 21,053,894 | rs378591536 | 7 | 0.34 | 0.07 | − 0.24 | 0.034 | 12.1 | 23216b | Intron | C14:0 | 0 | 0 | 1 | 0 |
| 29 | 13 | 45,394,264 | 48,611,254 | 75 | 4 | 46,734,011 | rs379821485 | 18 | 0.76 | 0.15 | − 0.01 | 0.001 | 10.6 | RF0026 | Downstream | pH0_PCC | 1 | 0 | 1 | 0 |
| 30 | 13 | 52,289,279 | 55,114,121 | 80 | 9 | 54,938,610 | rs110422533 | 2 | 1.00 | 0.39 | − 6.60 | 1.007 | 10.3 | GID8 | Synonymous | K | 0 | 0 | 2 | 1 |
| 31 | 13 | 64,648,620 | 64,870,118 | 19 | 3 | 64,812,464 | rs43717459 | 1 | 0.33 | 0.10 | − 0.24 | 0.027 | 18.9 | ACSS2 | Intron | C4–C10 | 0 | 0 | 1 | 0 |
| 32 | 13 | 79,225,285 | 79,326,265 | 6 | 1 | 79,326,265 | rs109862148 | 3 | 0.38 | 0.07 | − 0.04 | 0.007 | 8.9 | FAM65C | Downstream | αs2-CN | 0 | 1 | 0 | 0 |
| 33 | 14 | 1,622,956 | 1,881,116 | 27 | 9 | 1,629,753 | rs109035586 | 1 | 0.36 | 0.33 | 0.17 | 0.007 | 122.7 | GPT | Upstream | FC | 6 | 6 | 10 | 4 |
| 34 | 15 | 39,854,757 | 40,283,508 | 140 | 2 | 39,885,845 | rs134953698 | 2 | 0.79 | 0.18 | − 0.04 | 0.005 | 10.4 | ARNTL | Intron | PUNSAT | 0 | 0 | 1 | 0 |
| 35 | 15 | 52,993,384 | 54,612,060 | 341 | 10 | 53,943,342 | rs381948106 | 1 | 0.47 | 0.04 | 17.5 | 2.747 | 9.7 | RAB6A | Intron | P | 0 | 0 | 2 | 2 |
| 36 | 16 | 1,607,369 | 3,050,452 | 10 | 0 | 1,609,129 | rs42450079 | 1 | 0.54 | 0.28 | − 0.04 | 0.005 | 17.0 | – | Intergenic | κ-CN | 2 | 6 | 0 | 0 |
| 37 | 16 | 60,539,357 | 63,891,341 | 178 | 3 | 60,646,127 | rs137615589 | 72 | 0.75 | 0.14 | 0.02 | 0.004 | 9.8 | 38238b | Upstream | aSC | 2 | 1 | 0 | 0 |
| 38 | 16 | 67,700,538 | 67,811,269 | 167 | 2 | 67,758,163 | rs42465711 | 1 | 1.00 | 0.4 | 0.31 | 0.044 | 11.9 | SWT1 | Intron | CYDM | 2 | 1 | 2 | 1 |
| 39 | 16 | 70,170,086 | 71,493,899 | 59 | 1 | 71,432,479 | rs109766366 | 5 | 0.79 | 0.43 | − 0.23 | 0.032 | 12.3 | PROX1 | Intron | UNSAT | 0 | 0 | 4 | 2 |
| 40 | 17 | 29,348,215 | 30,211,790 | 203 | 9 | 29,938,428 | rs207509104 | 21 | 0.55 | 0.36 | 0.10 | 0.015 | 10.3 | LARP1B | Synonymous | C4–C10 | 0 | 0 | 1 | 0 |
| 41 | 17 | 52,753,338 | 53,240,467 | 283 | 6 | 53,072,959 | rs448501071 | 5 | 1.00 | 0.06 | − 0.32 | 0.019 | 62.4 | BRI3BP | Intron | C4–C10 | 2 | 3 | 2 | 1 |
| 42 | 18 | 10,566,605 | 11,091,131 | 68 | 5 | 11,002,789 | rs41867427 | 2 | 0.61 | 0.06 | 14.3 | 2.071 | 11.3 | CRISPLD2 | Intron | aSC | 2 | 0 | 0 | 1 |
| 43 | 19 | 51,304,834 | 51,538,272 | 67 | 2 | 51,383,847 | rs136067046 | 1 | 0.83 | 0.32 | 0.20 | 0.013 | 49.4 | FASN | Upstream | C14:0 | 0 | 0 | 9 | 0 |
| 44 | 19 | 55,229,384 | 57,240,571 | 267 | 10 | 57,151,350 | rs42848485 | 1 | 0.63 | 0.46 | 9.07 | 1.079 | 16.4 | FADS6 | Intron | Ca | 2 | 1 | 1 | 1 |
| 45 | 19 | 60,407,923 | 62,177,206 | 142 | 0 | 61,135,270 | rs41923848 | 1 | 0.91 | 0.13 | − 2.28 | 0.210 | 26.6 | – | Intergenic | Lactose | 0 | 3 | 0 | 3 |
| 46 | 20 | 58,245,970 | 58,457,768 | 87 | 1 | 58,446,058 | rs137085630 | 22 | 0.99 | 0.06 | − 1.01 | 0.036 | 176.5 | ANKH | Intron | Citrate | 5 | 4 | 2 | 5 |
| 47 | 21 | 40,120,343 | 44,138,058 | 63 | 2 | 41,638,428 | rs137153434 | 25 | 0.21 | 0.12 | 0.07 | 0.009 | 12.7 | G2E3 | Upstream | PC | 3 | 1 | 0 | 1 |
| 48 | 22 | 32,877,755 | 33,466,544 | 46 | 2 | 32,877,755 | rs208141216 | 10 | 0.30 | 0.08 | 0.08 | 0.007 | 25.7 | FAM19A4 | Intron | PC | 8 | 1 | 1 | 2 |
| 49 | 22 | 55,186,094 | 55,273,619 | 72 | 1 | 55,254,221 | rs43597796 | 1 | 0.93 | 0.49 | 0.00 | 0.001 | 12.6 | ATP2B2 | Intron | pH0_PCC | 1 | 1 | 0 | 0 |
| 50 | 22 | 61,257,725 | 61,312,492 | 18 | 1 | 61,284,069 | rs109001472 | 1 | 1.00 | 0.47 | − 0.09 | 0.008 | 28.3 | KLF15 | Intron | C4–C10 | 0 | 0 | 2 | 0 |
| 51 | 24 | 50,420,365 | 50,550,020 | 114 | 2 | 50,465,348 | rs383068825 | 4 | 0.53 | 0.34 | − 10.7 | 1.282 | 16.0 | SKA1 | Downstream | K | 1 | 1 | 0 | 2 |
| 52 | 24 | 58,744,952 | 58,825,217 | 23 | 3 | 58,817,202 | rs208779762 | 1 | 0.93 | 0.43 | − 6.57 | 0.807 | 15.4 | LMAN1 | Upstream | P | 1 | 1 | 0 | 1 |
| 53 | 25 | 2,994,081 | 3,261,509 | 28 | 3 | 3,241,838 | rs137696417 | 16 | 0.61 | 0.18 | − 0.03 | 0.007 | 6.5 | ADCY9 | Downstream | κ-CN | 0 | 1 | 0 | 0 |
| 54 | 25 | 25,642,563 | 29,605,418 | 82 | 11 | 26,498,356 | rs137150057 | 1 | 1.00 | 0.34 | − 0.11 | 0.020 | 11.3 | FAM57B | 5′UTR | C18:0 | 1 | 1 | 6 | 0 |
| 55 | 26 | 20,727,700 | 21,427,109 | 61 | 4 | 21149234 | rs136334180 | 11 | 1.00 | 0.31 | 0.39 | 0.035 | 27.6 | SCD | Upstream | UNSAT | 0 | 0 | 11 | 0 |
| 56 | 26 | 32,773,808 | 33,925,908 | 95 | 2 | 33,233,277 | rs385554497 | 36 | 0.27 | 0.02 | 1.55 | 0.270 | 8.1 | ACSL5 | intron | aPCC | 1 | 0 | 0 | 0 |
| 57 | 27 | 36,165,492 | 36,235,730 | 11 | 1 | 36,212,352 | rs208675276 | 1 | 0.54 | 0.41 | 0.65 | 0.040 | 60.4 | GPAT4c | 5′ UTR | C16:0 | 3 | 0 | 7 | 1 |
| 58 | 28 | 6,008,464 | 7,038,810 | 141 | 3 | 6,027,037 | rs382911338 | 1 | 0.37 | 0.18 | − 0.01 | 0.001 | 17.1 | PCNX2 | Intron | pH0_PCC | 1 | 2 | 0 | 1 |
| 59 | 29 | 9,253,006 | 9,622,389 | 145 | 3 | 9,343,362 | rs133715120 | 6 | 0.61 | 0.33 | − 2.76 | 0.264 | 24.9 | EED | Intron | Lactose | 0 | 3 | 1 | 4 |
aWhen a gene was present in the confidence interval of the QTL, the best candidate variant was the genic variant with the most significant effects (intergenic variants were discarded)
b47695, 47622, 23216, and 38238 for ENSBTAG00000047695, ENSBTAG00000047622, ENSBTAG00000023216, and ENSBTAG00000038238, respectively
cAlso named AGPAT6
dNumber of milk cheese-making (CMP), protein, fatty acid, and mineral composition traits with significant effects
These QTL regions varied in size (from 9.2 kbp to 8.9 Mbp) and contained from 6 to 401 variants; they were distributed on all Bos taurus autosomes (BTA) with the exception of BTA8 and 23 (Fig. 2 and [see Additional file 1: Figure S1]). In almost all the QTL regions (56), we identified variants that were located in one or more candidate genes. Around 60% (i.e. 4312 of 7393) of the variants detected in the QTL regions were located within or in the upstream/downstream region of 264 genes [see Additional file 2: Table S1]. Fifty-one of these variants were predicted to be responsible for an amino-acid change in the protein, whereas most of them (2972) were located in introns (Table 3).
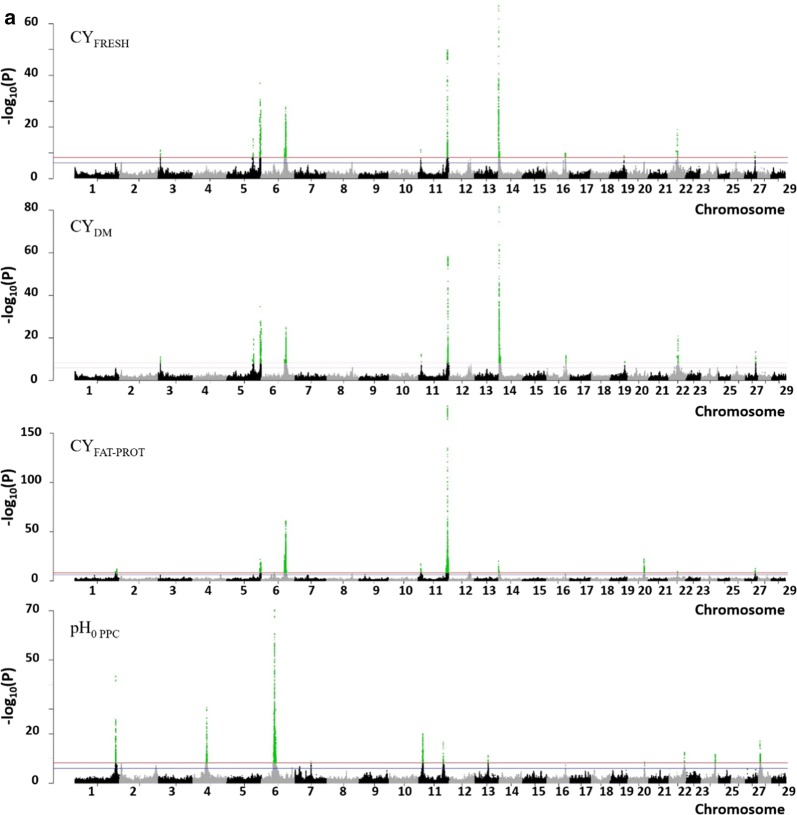
Fig. 2.
−log10(P) values plotted against the position of variants on Bos taurus autosomes for cheese-making traits. a Cheese yields (CY) and pH0 PPC, b coagulation traits
Table 3.
Functional annotations of variants included in the 59 QTL regions
| Functional annotation | Number of variants | % |
|---|---|---|
| Intergenic | 3081 | 41.7 |
| Intronic | 2972 | 40.2 |
| Upstream | 604 | 8.2 |
| Downstream | 584 | 7.9 |
| 3′ UTR | 26 | 0.35 |
| 5′ UTR | 10 | 0.14 |
| Synonymous | 65 | 0.88 |
| Missense | 51 | 0.69 |
| Total | 7393 | 100 |
We found the most significant effects around 103.3 Mbp on BTA11 (− log10(P) = 560), 144.4 Mbp on BTA1 (− log10(P) = 210), 58.4 Mbp on BTA20 (− log10(P) = 177), 1.6 Mbp on BTA14 (− log10(P) = 123), and 46.9 Mbp on BTA6 (− log10(P) = 120). In each of these five QTL, we identified variants that were located in candidate genes, which were, respectively, PAEP, SLC37A1, ANKH, GPT, and SEL1L3. All the variants were located in introns of the genes, with the exception of the best candidate variant of the GPT gene, which was found in the upstream region. Four other QTL had more moderate but nevertheless strong effects (− log10(P) between 60 and 83), on BTA5 (118 Mbp), BTA6 (87.4 Mbp), BTA17 (53.1 Mbp), and BTA27 (36.2 Mbp), with the best candidate variants located in GRAMD4 (upstream region), CSN3 (downstream region), BRI3BP (upstream region), and GPAT4 (3’UTR region), respectively. We also found candidate variants (− log10(P) between 25 and 50) in 11 other candidate genes, on BTA2 (ALPL), BTA4 (CBLL1), BTA5 (MGST1), BTA7 (FSTL4), BTA12 (ABCC4), BTA19 (FASN), BTA22 (FAM19A4 and KLF15), BTA25 (FAM57B), BTA26 (SCD), and BTA29 (EED). Finally, many other variants were identified in various genomic regions that had more moderate but significant effects after application of the Bonferroni correction (− log10(P) > 8.2); most of these were located in genes. All the QTL regions are described in detail in Table 2.
On average, each QTL had significant effects on about six traits. Only 13 QTL affected a single trait, while the other 46 QTL had pleiotropic effects on two to 26 traits. The QTL that affected the largest number of traits was located at about 1.6 Mbp on BTA14. For most traits, including FC, the variant with the strongest effect was not the well-known K232A polymorphism in the DGAT1 gene [see Additional file 3: Table S2]. More than half of the QTL (33), and in particular those with the most significant effects, had effects on CMP traits. Almost all of the QTL with significant effects on CMP traits presented significant pleiotropic effects on milk composition traits, as well. In contrast, the remaining 26 QTL affected milk composition (protein, fatty acid, mineral, citrate, or lactose content) but not CMP. Among traits, we observed large differences in both the number of QTL detected (ranging from 6 to 19) and in the total percentage of phenotypic variance (ranging from 4.7 to 62.4%) that was explained by the detected QTLs, and simply estimated by the sum of percentages per QTL. Overall, the larger the number of detected QTL for a given trait, the lower the percentage of phenotypic variance that was explained by each. For example, in our study, the most polygenic trait, aSC, was influenced by 19 QTL, each of which explained only 0.2 to 1.9% of the phenotypic variance. In contrast, we detected only six QTL for ΣWP but the QTL with the most important effect explained 56% of the phenotypic variance of this trait. As expected, the most heritable traits were those that presented the highest values of the total phenotypic variance explained by the QTL. The trait for which the largest amount of total phenotypic variance was explained by the QTL was β-LG (62%), which was also the most heritable trait analyzed in our study. For CMP traits, which are moderately heritable, from 12% (curd firmness) to 30% (curd firming time) of the phenotypic variability was explained by the QTL (i.e. from 27 to 65% of the genetic variance). Cheese yields presented intermediate results, as the detected QTL captured about 20% of their phenotypic variance, i.e. about 50% of their genetic variance. For CMP traits, the QTL that contributed the most were those detected in the regions of the PAEP, casein, and DGAT1 genes. However, other QTL regions on BTA5, 6, 16, 20, and 22 also generated noteworthy contributions. For protein composition traits, the highest-contributing QTL region was the PAEP gene region (up to 59% for β-LG). The region of the casein genes had a more moderate contribution (0.7–5.6%, depending on the trait), while the lesser-known QTL detected on BTA20 (at about 58 Mbp) explained 18, 9, and 7% of the phenotypic variance of α-LA, αs1-CN, and κ-CN, respectively. For fatty acid content, the QTL that we detected explained a much smaller part of the phenotypic variability. The top-contributing QTL were the DGAT1 gene region on BTA14 (12% for FC), FASN on BTA19 (1.5% for C14:0), GPAT4 on BTA27 (3.2% for C16:0), and SCD on BTA26 (2% for C18:1). In contrast to fatty acids but similarly to proteins, a relatively large part of the phenotypic variance in mineral content was explained by QTL that were located in the region of the SLC37A1 gene (3, 5, and 10% for Mg, K, and P, respectively) and the ANKH gene (20% for Mg). Two other regions influenced mineral content to a lesser extent: those at 117 Mbp on BTA5 (GRAMD4) and at 46 Mbp on BTA6 (SEL1L3).
Gene network
Using the AWM procedure, we reduced the set of 8.5 million variants tested in the GWAS to a set of 38,858 variants that had the most significant effects (P-value ≤ 0.001) on the key phenotype (CYDM). Seven CMP (CYFRESH, CYFAT-PROT, and the five coagulation traits) and eight milk composition traits (PC, FC, UNSAT, PUNSAT, C18:1, Ca, Mg, and P) were correlated with CYDM (r ≥ 0.25). On average, each of the 38,858 variants had significant effects (P-value ≤ 0.001) on six of the correlated traits. We also retained 2322 additional variants that had significant effects on at least six of the correlated phenotypes. Thus, the final dataset included 41,180 variants, which had significant effects on CYDM or on at least six correlated traits. Of these 41,180 variants, 15,330 were located in 736 genes (± 10 kb); the PCIT approach subsequently revealed 59,168 significant interactions among these genes. Thus, by merging the AWM and the PCIT approaches, the GWAS results on milk CMP and composition traits could be interpreted as a gene network of 736 nodes and 59,168 edges. The list of the 736 genes selected by AWM is in [see Additional file 4: Table S3].
For most of the traits, correlation coefficients from the z-score additive effects of the 736 variants retained by the AWM procedure were close to the correlation coefficients obtained from pedigree for the 16 phenotypes (Table 4). This suggested that the additive effects of the variants retained in the AWM analysis explained a large and representative part of the genetic relationships among the traits.
Table 4.
Genomic correlations calculated using additive effects of the 736 SNPs selected by the AWM (above the diagonal) and genetic correlations estimated from pedigree or taken from Sanchez et al. [3] (below the diagonal)
| CYFRESH | CYDM | CYFAT-PROT | K10/RCTPCC | aPCC | K10/RCTSC | aSC | a2SC | PC | FC | C18:1 | UFA | PUFA | Ca | Mg | P | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYFRESH | 1.00 | − 0.87 | − 0.84 | 0.25 | − 0.84 | 0.52 | 0.85 | 0.84 | 0.96 | − 0.82 | − 0.78 | − 0.80 | 0.70 | 0.52 | 0.83 | |
| CYDM | 0.97 | − 0.88 | − 0.85 | 0.27 | − 0.85 | 0.55 | 0.86 | 0.85 | 0.95 | − 0.80 | − 0.76 | − 0.78 | 0.71 | 0.55 | 0.84 | |
| CYFAT-PROT | − 0.82 | − 0.84 | 0.79 | − 0.61 | 0.84 | − 0.73 | − 0.79 | − 0.72 | − 0.72 | 0.57 | 0.51 | 0.58 | − 0.55 | − 0.54 | − 0.75 | |
| K10/RCTPCC | − 0.72 | − 0.73 | 0.65 | − 0.31 | 0.99 | − 0.69 | − 0.99 | − 0.98 | − 0.74 | 0.57 | 0.52 | 0.44 | − 0.80 | − 0.75 | − 0.86 | |
| aPCC | 0.77 | 0.78 | − 0.65 | − 0.76 | − 0.41 | 0.79 | 0.29 | 0.20 | 0.01 | 0.08 | 0.14 | 0.02 | − 0.12 | 0.29 | 0.35 | |
| K10/RCTSC | − 0.74 | − 0.76 | 0.71 | 0.80 | − 0.78 | − 0.75 | − 0.98 | − 0.95 | − 0.71 | 0.53 | 0.47 | 0.41 | − 0.75 | − 0.76 | − 0.87 | |
| aSC | 0.76 | 0.78 | − 0.67 | − 0.73 | 0.76 | − 0.77 | 0.63 | 0.64 | 0.32 | − 0.19 | − 0.14 | − 0.15 | 0.36 | 0.72 | 0.62 | |
| a2SC | 0.72 | 0.75 | − 0.64 | − 0.72 | 0.77 | − 0.77 | 0.74 | 0.97 | 0.75 | − 0.59 | − 0.53 | − 0.45 | 0.79 | 0.68 | 0.87 | |
| PC | 0.74 | 0.75 | − 0.52 | − 0.80 | 0.94 | − 0.81 | 0.91 | 0.89 | 0.77 | − 0.59 | − 0.55 | − 0.46 | 0.84 | 0.76 | 0.86 | |
| FC | 0.91 | 0.87 | − 0.57 | − 0.53 | 0.55 | − 0.47 | 0.51 | 0.48 | 0.60 | − 0.89 | − 0.87 | − 0.87 | 0.69 | 0.41 | 0.75 | |
| C18:1 | − 0.38 | − 0.45 | 0.22 | 0.20 | − 0.22 | 0.13 | − 0.18 | − 0.17 | − 0.23 | − 0.57 | 0.99 | 0.93 | − 0.52 | − 0.19 | − 0.56 | |
| UFA | − 0.34 | − 0.40 | 0.17 | 0.13 | − 0.14 | 0.05 | − 0.10 | − 0.09 | − 0.21 | − 0.55 | 0.47 | 0.93 | − 0.50 | − 0.16 | − 0.51 | |
| PUFA | − 0.47 | − 0.42 | 0.30 | 0.00 | − 0.02 | − 0.04 | 0.01 | 0.04 | − 0.29 | − 0.59 | 0.71 | 0.74 | − 0.42 | − 0.13 | − 0.50 | |
| Ca | 0.41 | 0.40 | − 0.25 | − 0.46 | 0.45 | − 0.37 | 0.39 | 0.42 | 0.41 | 0.26 | − 0.30 | − 0.30 | − 0.35 | 0.76 | 0.71 | |
| Mg | 0.54 | 0.58 | − 0.44 | − 0.58 | 0.59 | − 0.58 | 0.54 | 0.54 | 0.40 | 0.20 | − 0.25 | − 0.25 | − 0.05 | 0.60 | 0.70 | |
| P | 0.40 | 0.41 | − 0.31 | − 0.54 | 0.50 | − 0.53 | 0.42 | 0.40 | 0.40 | 0.29 | − 0.31 | − 0.30 | − 0.39 | 0.34 | 0.58 |
Among the 736 genes, 86 were located within QTL regions that had been highlighted by the GWAS analysis with a most-stringent threshold; these included the best candidate genes for 25 QTL. The remaining 650 genes were unique to the AWM analysis and had not been detected by GWAS. In contrast, 178 genes located within the confidence intervals of QTL detected with GWAS were not found in AWM analyses.
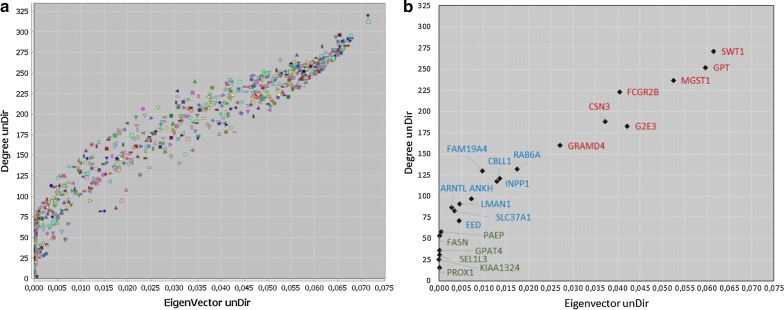
For each node of the gene network, we calculated the number of adjacent genes and the relative node contribution. Figure 3 lists the values of these parameters for the nodes of the gene network that were also best candidate genes in the GWAS analyses. This revealed genes that were highly connected with other genes in the network (SWT1, GPT, MGST1, FCGR2B, CSN3, G2E3, and GRAMD4), genes that were moderately connected (RAB6A, FAM19A4, INPP1, CBLL1, ANKH, LMAN1, ARNTL, SLC37A1, and EED), and genes that were poorly connected (PAEP, FASN, GPAT4, SEL1L3, KIAA1324, and PROX1).
Fig. 3.
Centiscape scatter plot view: number of adjacent genes (degree) and the relative node contribution (eigenvector) a for the 736 genes of the gene network and b for the best candidate genes
In silico functional analyses
Key regulators in the gene network were identified in silico using two approaches. From the analyses of binding site motifs and ChIP-Seq datasets, first we identified eight TF that presented a significant normalized enrichment score (NES). Each of these TF targeted from 136 to 261 genes in the gene network (Table 5), and all eight together targeted more than half of the network genes (416). Using an information loss-less approach, we then identified among the 736 genes the trios of regulators (TF, miRNA, and lnRNA) that had the best coverage of the whole gene network, i.e. trios that demonstrated the largest number of interactions with genes of the network with the least amount of overlap. With this second approach, we found 61 regulators, each with two to 276 significant interactions with genes of the network. The trios that covered the largest number of genes were ASXL3—HIC2—RNF2 and HIC2—ZPFM2—bta-mir-200c. These two trios interacted with the majority of the genes of the network, i.e. 529 and 528 genes, respectively.
Table 5.
Transcription factors (TFs) identified as key regulators of milk cheese-making and composition traits from both binding-site motifs and ChIP-Seq datasets, which presented significant normalized enrichment scores (NES)
| TF | NES | Number of binding site motifs | Number of ChIP-Seq datasets | Number of target genes | Chromosome | Gene start (bp) | Gene end (bp) | Gene description |
|---|---|---|---|---|---|---|---|---|
| HSPA1L | 4.90 | 5 | 1 | 261 | 23 | 27,334,344 | 27,338,328 | Heat shock 70 kDa protein 1-like |
| SMAD5 | 4.63 | 4 | 2 | 253 | 7 | 49,155,483 | 49,217,780 | SMAD family member 5 |
| HNF1B | 4.56 | 3 | 5 | 242 | 19 | 14,287,673 | 14,349,579 | HNF1 homeobox B |
| SMAP2 | 4.30 | 7 | 1 | 236 | 3 | 106,311,859 | 106,358,978 | Small ArfGAP2 |
| TFAP2A | 4.29 | 3 | 1 | 233 | 23 | 45,480,546 | 45,499,034 | Transcription factor AP-2 alpha |
| BCL11A | 4.25 | 5 | 1 | 195 | 11 | 43,071,977 | 43,174,031 | B Cell CLL/Lymphoma 11A |
| SMAD3 | 4.02 | 3 | 1 | 170 | 10 | 13,958,174 | 13,980,371 | SMAD family member 3 |
| RXRA | 3.49 | 2 | 1 | 136 | 11 | 105,990,344 | 106,015,000 | Retinoid X receptor alpha |
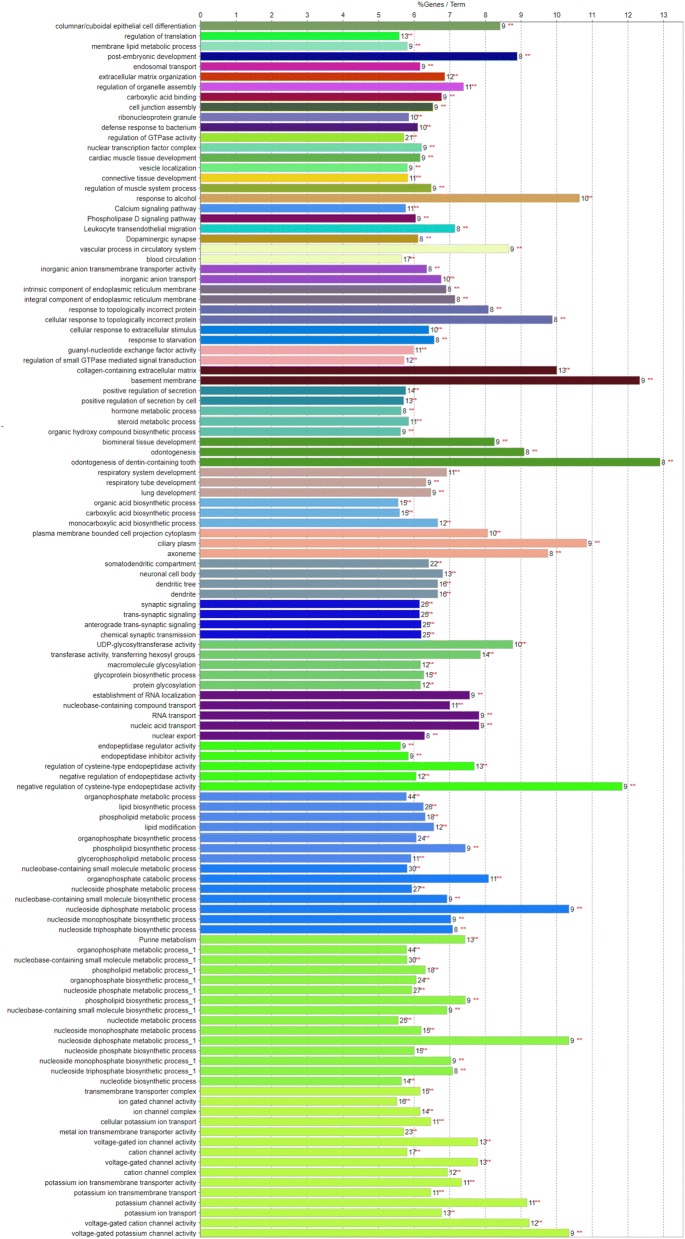
Genes of the network were found to be enriched in five KEGG pathways and 115 GO terms (corrected P-value between 2.10−17 and 2.10−4), which clustered into 44 functional groups (Fig. 4 and [see Additional file 5: Table S4]). The largest group comprised 15 GO terms; it contained 31 genes of the gene network and was related to the metabolic processes associated with potassium transport. The next three groups, with 28 GO terms and one KEGG pathway all related to phosphate and phospholipid metabolism, contained 66 genes of the network. Among these, there were many of the genes that had been highlighted by GWAS as having the most significant effects on milk CMP and composition traits: CSN1S1, DGAT1, FASN, GPAT4, INPP1, PPARA, PROX1, and SCD. Other groups, (for details [see Additional file 5: Table S4]), had a functional relationship with milk composition through endopeptidase activity (16 genes, including CSN2 and GRAMD4), protein glycosylation (19 genes), carboxylic acid biosynthesis (24 genes including FASN, PAEP, PPARA, PROX1, and SCD), inorganic anion transport (10 genes including ANKH and SLC37A1), and Ca- (11 genes) and phospholipase- (9 genes) signaling pathways.
Fig. 4.
Description of the five KEGG pathways and 105 GO terms that were significantly enriched among genes of the network and which clustered in 44 functional groups
Discussion
GWAS and gene network analyses are complementary
The GWAS approach used here—performed on whole-genome sequences from a large number of animals for complex cheese-making traits as well as fine-scale milk composition traits—led to the identification of 59 QTL regions. In order to limit the detection of false positives, we retained only the QTL that still demonstrated significant effects after applying the Bonferroni correction (P-value < 5.8 × 10−9) and therefore those that presented the strongest effects overall. The downside of this approach was that all the QTL in our analysis explained, on average, less than 50% of the total genetic variation of each trait (i.e. 20% of the phenotypic variance), and this value was probably overestimated. Indeed, when the true effect is small or when the P-value threshold is very low, the detection power is limited and a significant effect may be overestimated, leading to an overestimation of SNP variance. Some QTL were identified with very good resolution (narrow peaks), such as the 12 QTL for which only one candidate gene was identified within the confidence interval. Other QTL regions were larger and more gene-rich (up to 25 genes within the confidence interval), and identification of the best candidate gene was not straightforward. To address these two shortcomings—specifically, to capture the missing genetic variability and to better identify functional candidate genes within QTL regions—we carried out additional analyses, which complemented our GWAS results. The AWM-PCIT approach enabled us to identify a more comprehensive gene network of 736 genes from lower significant GWAS results (P-value < 0.001) by taking co-associations between traits into account. When we used the additive effects of variants that were located in these genes to calculate correlations between traits, the values obtained were similar to the genetic correlations we calculated from pedigree [3], suggesting that the gene network adequately explained the genetic relationships between traits. Finally, in silico functional analyses of genes of the network helped us to identify metabolic pathways and key regulators with functional links to milk cheese-making and composition traits. This last step, in addition to establishing functional links between the gene network and the analyzed traits, enabled us to identify candidate genes in some QTL regions. Therefore, by combining the results obtained through these different approaches, we are able to propose candidate genes for the main QTL regions, and for each, the best candidate for the causative variant, or at least, a variant in high LD with the causative variant.
Functional candidate genes
As expected, we confirmed the strong effects of the cluster of casein genes and the PAEP gene regions on protein composition as well as milk CMP. The QTL detected in the casein genes region explained up to 20% of the curd-firming time while the PAEP gene region explained up to 8.5% of cheese yields. The best candidate gene variants, i.e. variants with the most significant effects on traits, were located in the downstream region of the CSN3 gene, which encodes κ-CN (at 87,392,899 bp on BTA6), and in an intronic region of the PAEP gene, which encodes β-LG (at 103,301,982 pb on BTA11). The missense variants that are respectively responsible for the κ-CN [41] and β-LG [42] A/B polymorphisms had much weaker effects: they were ranked 100th and 56th, respectively, among the variants. The region of the DGAT1 gene on BTA14 had also large effects on milk composition (12% for FC) and on CMP (6.4% for CYDM). In spite of its low MAF in Montbéliarde cows (0.015), the K232A DGAT1 mutation [43] was the top-ranked variant for traits that were linked with some protein and phosphorous contents, and coagulation traits (1st for PC, α-LA, β-CN, and P; 2nd for a2SC, K10/RCTSC, and K10/RCTPCC) and it was one of the 736 variants retained by the AWM. However, in this gene-rich region, the GPT gene, which we found to be highly connected, i.e. presenting significant gene–gene interactions with many other genes of the AWM gene network, appeared to be also a good candidate for FC, CYDM, CYFRESH, and fatty acid composition. The best candidate variant, located in the upstream region of GPT (glutamic-pyruvic transaminase) at 1,629,753 bp (rs109035586), was ranked 1st for 12 traits, including FC, cheese yields, fatty acid composition, αS1-CN, and CITRATE. Interestingly, two polymorphisms in the GPT gene, including a missense variant that is located very close to the best candidate variant detected in our study (1,629,600 bp), were also recently found to be associated with fat percentage in a concordance analysis carried out on imputed whole-genome sequences of Holstein bulls [44]. This variant was also highly significant in our study but was ranked 44th among variants with significant effects on FC.
In addition to the three well-known QTL regions described above, we also found evidence that other genomic regions have highly significant effects on the traits analyzed; specifically, our analysis highlighted the SLC37A1, ALPL, MGST1, SEL1L3, FASN, ANKH, BRI3BP, SCD, and GPAT4 genes, which we had also previously detected in a sequence-based GWAS on milk protein and fatty acid composition [45, 46]. We confirm here their effects on milk composition and note their effects on CMP. As previously found, the MGST1, FASN, SCD, and GPAT4 genes mainly affected fatty acids whereas the SLC37A1, ALPL, SEL1L3, BRI3BP, and ANKH genes had effects mainly on proteins and minerals. As a consequence, and in accordance with genetic correlations that we had previously estimated from this dataset [3], the former set of genes exclusively influenced cheese yields whereas the latter set had greater effects on coagulation traits. Strong effects of ALPL, ANKH, and SEL1L3, which we had previously identified for protein composition [45], were confirmed for milk composition and CMP. In each of these regions, the current analysis reduced the size of the confidence intervals of the QTL and, in six of them, only one gene was found that encoded a known protein (SLC37A1, ALPL, MGST1, SEL1L3, ANKH, and GPAT4).
On BTA17, we found two QTL regions associated with de novo milk fatty acid synthesis, i.e. synthesis within the mammary epithelial cells of fatty acids C4:0 to C10:0. The first was within the LARP1B (La ribonucleoprotein domain family member 1B) gene, for which the best candidate was a synonymous variant located at 29,938,428 bp. This result corroborates the discovery of Duchemin et al. [47], who identified LARP1B as a causative gene for de novo synthesis of milk fatty acids through the imputation of sequence variants in this region. These authors noted a splice-region variant at 29,940,555 bp, which was close to the variant that we detected here. However, in spite of its high MAF (0.40), we excluded this variant because it was not significant in our study (P-value = 10−4 vs. 5.10−11 for the variant located at 29,938,428 bp). This region had limited effects in our study and affected only short FA traits. Instead, further along the same chromosome, we identified another region with much more significant effects on de novo fatty acid synthesis that also affected CMP and protein and mineral composition. The best candidate gene for this region was BRI3BP (BRI3 binding protein), with the most significant variant located at 53,072,959 bp in an intron of BRI3BP. This variant had been previously highlighted for its effects on FA composition in an independent population [48] and, in another study, we recently confirmed its effects on both CMP and milk composition traits [46]. Thus, it is a serious candidate for the causative variant behind the strong effects that we observed in the region. Although the BRI3BP gene was not an obvious functional candidate, it has been also described as affecting de novo fatty acid synthesis in a recent GWAS performed on imputed sequence variants in this region [49]. The most significant variant found by the authors of this study was also intronic (53,078,216 bp) but that particular variant was ranked 31st among variants with significant effects on C4–C10.
Finally, we identified other candidate genes that contained variants with non-negligible effects on milk composition and CMP traits. Among these, both GWAS and AWM analyses highlighted FCGR2B, KIAA1324, CBLL1, GRAMD4, ARNTL, RAB6A, ENSBTAG00000038238, SWT1, G2E3, FAM19A4, LMAN1, and EED. The FCGRB2, KIAA1324, G2E3, LMAN1, and EED genes have been previously identified as candidate genes for milk yield or milk composition [50–54], whereas the functional link between the other genes and bovine milk composition and cheese-making traits remains to be discovered.
Co-association gene network
The SLC37A1 (solute carrier family 37 member 1, a phosphorous antiporter) and ANKH (inorganic pyrophosphate transport regulator) genes, which encode transmembrane proteins involved in ion transport, both play a role in the inorganic anion transport that was revealed by the GO analysis. Thus, these genes are good candidates for having an effect on CMP and milk composition, with the strongest effects obtained for phosphorous (about 11% ) and citrate (about 32% ) contents, respectively. For each of these genes, we propose here an intronic candidate variant, located at 58,446,058 bp for ANKH and at 144,395,375 bp for SLC37A1. Very close to but distinct from those identified in previous studies [45, 53, 55], this variant is more significant in spite of a slightly lower imputation accuracy.
A set of genes, including those detected previously (DGAT1, FASN, GPAT4, CSN1S1, PAEP, and SCD) and those noted here for the first time (INPP1, PPARA, PROX1), appeared to play a role in phosphate and phospholipid metabolism as well as in the biosynthesis of carboxylic acids, which are fatty acid precursors. PROX1 (prospero homeobox 1) and PPARA (peroxisome proliferator activated receptor alpha) encode transcription factors; the former interacted with only 16 genes while the latter interacted with 128 genes within the network, including with FASN, SCD, GPAT4, and DGAT1. PPARA belongs to a superfamily of hormone receptors (PPAR) that regulate the transcription of genes involved in different lipid metabolism pathways [56]. FASN (fatty acid synthase) and SCD (stearoyl-coenzyme A desaturase 1) encode key enzymes in de novo fatty acid synthesis and fatty acid desaturation, respectively, and GPAT4 (glycerol-3-phosphate acyltransferase 4) is paralogous to DGAT1 (diacylglycerol O-acyltransferase 1), with the two genes occupying adjacent nodes of the mammary triglyceride synthesis chain [57]. In addition to their effects on protein composition, the PAEP and CSN1S1 genes, which encode milk β-LG and αs1-CN proteins, respectively, are also associated with genes involved in fatty acid metabolism. These results suggest a close link between milk fatty acid and protein metabolism. In goats, variants that are responsible for a decrease in CSN1S1 gene expression were also associated with a decrease in fat content, probably due to disruption of the structure and secretion of fat globules [58]. A similar relationship was pointed out in cattle by Knutsen et al. [49], who found a major effect of the PAEP gene region on the C4:0 content of bovine milk, and Pausch et al. [53], who identified strong pleiotropic effects of variants located in the CSN1S1 gene on fat and protein content. In addition, a strong association between PAEP and omega-3 fatty acids was observed by Boichard et al. [48]. All of these genes, which contain the top-ranked variants for, in particular, cheese yields and fatty acid composition, thus represent good candidates. Alone, they explained the largest part of the phenotypic variance captured in the present study for CYDM and FC, i.e. around 16% out of 20%.
In addition to the PPARA TF, we highlight here other genes for putative regulators as well, such as ASXL3 (additional sex combs like 3, transcriptional regulator) and bta-mir-200c, which interact with many genes of the network (276 and 240, respectively). Both are good candidates for key regulators in the network, as the protein encoded by ASXL3 has been shown to negatively regulate lipogenesis and bta-mir-200c miRNA has been found to be highly expressed in the mammary gland [59–61] and present in milk whey [62]. Interestingly, all of the regulators that we identified in our study were different from the TF found in previous studies that applied similar approaches to study milk proteins [10] or fatty acids [9]. Unlike these studies, we analyzed here milk protein, fatty acid, and mineral composition as well as cheese-making traits all together, which might explain the identification of different regulatory pathways. However, in spite of this, some of the significantly enriched GO terms or KEGG pathways that we highlight here were concordant with those previously reported for CMP traits (Ca signaling pathway) [7], milk protein content (potassium ion transport) [10], or fatty acid content (hormone and steroid metabolic processes) [9].
Causative variants
The approach that we used, which combines GWAS and post-GWAS analyses, was successful both in confirming previously reported candidate genes and in identifying new candidates that appear to be functionally linked to the analyzed traits. This was possible because our analyses were based on a large sample size, sequence-level genotypes, and detailed phenotypes for milk components in addition to complex CMP traits. However, for most of these genes, the top-ranked variant identified here was different both from what we had found before in an analysis of milk protein and fatty acid composition and from what had been detected in previous studies. Since the first GWAS on WGS imputed from the 1000 Bull Genomes reference population, in 2014 [4], to date published GWAS based on this approach have generally converged towards the same candidate genes but rarely towards the same best candidate variants in these genes. Using data from humans, Faye et al. [63] showed that when the causal variant is less accurately genotyped or imputed than one of its highly correlated neighboring variants, the neighboring variant can capture the association better than the causal variant. However, in our study, the HD SNP, imputed more accurately than sequence variants, were rarely the top variants of the peaks, with the noticeable exception in the SCD gene. For SLC37A1, the peak variant was more significant than variants already proposed in other studies and slightly better imputed. Nevertheless, we can anticipate that by accumulating bovine sequence data from different breeds and different populations, future runs of the 1000 Bull Genome Project will lead to better identification of causative variants by GWAS. More specifically, the expansion of the bovine sequence database should increase the accuracy of imputed genotypes and thus the probability of identifying the right variant. In addition, if GWAS analyses can be carried out in different breeds, meta-analyses should lead to a better resolution due to the linkage disequilibrium at shorter distances between breeds than within breed, and thus to a better discrimination of causal variants.
Conclusions
By combining GWAS and AWM approaches at the whole-genome sequence level on milk cheese-making and composition traits predicted from MIR spectra, this study highlights candidate genes with major effects that are functionally related to milk composition. For most of these, we are able to propose some candidate variants that are likely to be either causative or in linkage disequilibrium with causative variants. In addition to providing a better understanding of the metabolic pathways involved in the genetic determinism of cheese-making traits, this study should make it possible to select a set of variants that explain a large part of the genetic variability of cheese-making traits. The increase in the number of cows for which both genotypes and phenotypes are available allows better detection of variants which could be included in genomic prediction to more accurately select animals with high genetic merit for CMP and finally improve the efficiency of the cheese-making process, which is of vital economic importance in the dairy industry.
Additional files
Additional file 1: Figure S1. −log10(P) plotted against the position of variants on Bos Taurus autosomes for milk composition. Manhattan Plot obtained from GWAS for milk composition traits.
Additional file 2: Table S1. Description of the 264 genes located in confidence intervals of the QTL detected by GWAS for milk cheese-making and composition traits. Name and position of candidate genes identified by GWAS.
Additional file 3: Table S2. Percentage of the phenotypic variance of milk CMP and composition traits explained by each QTL. Individual effects of QTL on all CMP and composition traits, expressed as a percentage of the phenotypic variance of the trait.
Additional file 4: Table S3. Description of the 736 genes selected by the gene network analysis for milk cheese-making and composition traits. Name and position of candidate genes identified by AWM.
Additional file 5: Table S4. Gene ontology (GO) terms and KEGG pathways for genes selected by AWM. Name, description and list of genes of the gene ontology terms and KEGG pathways identified in the gene network analysis.
Acknowledgements
The authors gratefully acknowledge the breeders who participated in the FROM’MIR project; colleagues from the Conseil-Elevage 25-90 and INRA who coordinated farm sampling and data collection, and performed cheese-making reference measurements; the members of the scientific committee for advising on and managing this work; and the contribution of the 1000 Bull Genomes consortium.
Authors’ contributions
MPS performed GWAS, network, and pathway analyses and wrote the manuscript. YRC and AM provided support in developing computing programs, ST performed imputation analyses, and MBo managed sequence analyses of the 1000 Bull Genomes Project. MEJ developed the MIR prediction equations for milk cheese-making properties. VW, CL, SF, ADB, MBr, and DB designed and managed the FROM’MIR project. All authors read and approved the final manuscript.
Funding
This study was funded by the French Ministry of Agriculture, Agro-food, and Forest, the French Dairy Interbranch Organization (CNIEL), the Regional Union of Protected Designation Cheeses of Franche-Comté (URFAC), and the Regional Council of Bourgogne-Franche-Comté, under the project FROM’MIR.
Availability of data and material
The data (genotypes and phenotypes) that enabled the findings of this study were made available by UMOTEST, CEL25-90, and HSCEL. However, restrictions apply to the availability of these data: they were used under license for the current study, and are not publicly available.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marie-Pierre Sanchez, Email: marie-pierre.sanchez@inra.fr.
Yuliaxis Ramayo-Caldas, Email: yuliaxis.ramayo-caldas@inra.fr.
Valérie Wolf, Email: valerie.wolf@cel2590.fr.
Cécile Laithier, Email: cecile.laithier@idele.fr.
Mohammed El Jabri, Email: Mohammed.ElJabri@idele.fr.
Alexis Michenet, Email: alexis.michenet@allice.fr.
Mekki Boussaha, Email: mekki.boussaha@inra.fr.
Sébastien Taussat, Email: sebastien.taussat@allice.fr.
Sébastien Fritz, Email: sebastien.fritz@allice.fr.
Agnès Delacroix-Buchet, Email: agnes.delacroix-buchet@inra.fr.
Mickaël Brochard, Email: MBrochard@umotest.com.
Didier Boichard, Email: didier.boichard@inra.fr.
References
- 1.De Marchi M, Toffanin V, Cassandro M, Penasa M. Invited review: mid-infrared spectroscopy as phenotyping tool for milk traits. J Dairy Sci. 2014;97:1171–1186. doi: 10.3168/jds.2013-6799. [DOI] [PubMed] [Google Scholar]
- 2.Wedholm A, Larsen LB, Lindmark-Månsson H, Karlsson AH, Andrén A. Effect of protein composition on the cheese-making properties of milk from individual dairy cows. J Dairy Sci. 2006;89:3296–3305. doi: 10.3168/jds.S0022-0302(06)72366-9. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez MP, El Jabri M, Minéry S, Wolf V, Beuvier E, Laithier C, et al. Genetic parameters for cheese-making properties and milk composition predicted from mid-infrared spectra in a large dataset of Montbéliarde cows. J Dairy Sci. 2018;101:10048–10061. doi: 10.3168/jds.2018-14878. [DOI] [PubMed] [Google Scholar]
- 4.Daetwyler HD, Capitan A, Pausch H, Stothard P, Van Binsbergen R, Brøndum RF, et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet. 2014;46:858–867. doi: 10.1038/ng.3034. [DOI] [PubMed] [Google Scholar]
- 5.Fortes MRS, Reverter A, Zhang Y, Collis E, Nagaraj SH, Jonsson NN, et al. Association weight matrix for the genetic dissection of puberty in beef cattle. Proc Natl Acad Sci USA. 2010;107:13642–13647. doi: 10.1073/pnas.1002044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reverter A, Fortes MB. Breeding and genetics symposium: building single nucleotide polymorphism-derived gene regulatory networks: towards functional genomewide association studies. J Anim Sci. 2013;91:530–536. doi: 10.2527/jas.2012-5780. [DOI] [PubMed] [Google Scholar]
- 7.Dadousis C, Pegolo S, Rosa GJM, Gianola D, Bittante G, Cecchinato A. Pathway-based genome-wide association analysis of milk coagulation properties, curd firmness, cheese yield, and curd nutrient recovery in dairy cattle. J Dairy Sci. 2017;100:1223–1231. doi: 10.3168/jds.2016-11587. [DOI] [PubMed] [Google Scholar]
- 8.Buitenhuis B, Janss LL, Poulsen NA, Larsen LB, Larsen MK, Sorensen P. Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genom. 2014;15:1112. doi: 10.1186/1471-2164-15-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pegolo S, Dadousis C, Mach N, Ramayo-Caldas Y, Mele M, Conte G, et al. SNP co-association and network analyses identify E2F3, KDM5A and BACH2 as key regulators of the bovine milk fatty acid profile. Sci Rep. 2017;7:17317. doi: 10.1038/s41598-017-17434-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pegolo S, Mach N, Ramayo-Caldas Y, Schiavon S, Bittante G, Cecchinato A. Integration of GWAS, pathway and network analyses reveals novel mechanistic insights into the synthesis of milk proteins in dairy cows. Sci Rep. 2018;8:566. doi: 10.1038/s41598-017-18916-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambra R, Penagaricano F, Kropp J, Khateeb K, Weigel KA, Lucey J, et al. Genomic architecture of bovine kappa-casein and beta-lactoglobulin. J Dairy Sci. 2013;96:5333–5343. doi: 10.3168/jds.2012-6324. [DOI] [PubMed] [Google Scholar]
- 12.Ferrand M, Miranda G, Guisnel S, Larroque H, Leray O, Lahalle F, et al. Determination of protein composition in milk by mid-infrared spectrometry. In Proceedings of the VI ICAR reference laboratory network meeting: 28 May 2012; Cork; 2012
- 13.Ferrand-Calmels M, Palhiere I, Brochard M, Leray O, Astruc JM, Aurel MR, et al. Prediction of fatty acid profiles in cow, ewe, and goat milk by mid-infrared spectrometry. J Dairy Sci. 2014;97:17–35. doi: 10.3168/jds.2013-6648. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez MP, Ferrand M, Gele M, Pourchet D, Miranda G, Martin P, et al. Short communication: genetic parameters for milk protein composition predicted using mid-infrared spectroscopy in the French Montbeliarde, Normande, and Holstein dairy cattle breeds. J Dairy Sci. 2017;100:6371–6375. doi: 10.3168/jds.2017-12663. [DOI] [PubMed] [Google Scholar]
- 15.Gengler N, Soyeurt H, Dehareng F, Bastin C, Colinet F, Hammami H, et al. Capitalizing on fine milk composition for breeding and management of dairy cows. J Dairy Sci. 2016;99:4071–4079. doi: 10.3168/jds.2015-10140. [DOI] [PubMed] [Google Scholar]
- 16.Ducrocq V. Genekit, BLUP software. June 2011 version. Jouy-en-Josas: INRA GABI; 1998. [Google Scholar]
- 17.Sargolzaei M, Chesnais JP, Schenkel FS. A new approach for efficient genotype imputation using information from relatives. BMC Genom. 2014;15:478. doi: 10.1186/1471-2164-15-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boichard D, Guillaume F, Baur A, Croiseau P, Rossignol M, Boscher MY, et al. Genomic selection in French dairy cattle. Anim Prod Sci. 2012;52:115–120. doi: 10.1071/AN11119. [DOI] [Google Scholar]
- 19.van Binsbergen R, Bink MC, Calus MP, van Eeuwijk FA, Hayes BJ, Hulsegge I, et al. Accuracy of imputation to whole-genome sequence data in Holstein Friesian cattle. Genet Sel Evol. 2014;46:41. doi: 10.1186/1297-9686-46-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brondum RF, Guldbrandtsen B, Sahana G, Lund MS, Su G. Strategies for imputation to whole genome sequence using a single or multi-breed reference population in cattle. BMC Genom. 2014;15:728. doi: 10.1186/1471-2164-15-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pausch H, MacLeod I, Fries R, Emmerling R, Bowman PJ, Daetwyler HD, et al. Evaluation of the accuracy of imputed sequence variant genotypes and their utility for causal variant detection in cattle. Genet Sel Evol. 2017;49:24. doi: 10.1186/s12711-017-0301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoze C, Fouilloux MN, Venot E, Guillaume F, Dassonneville R, Fritz S, et al. High-density marker imputation accuracy in sixteen French cattle breeds. Genet Sel Evol. 2013;45:33. doi: 10.1186/1297-9686-45-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouwman AC, Veerkamp RF. Consequences of splitting whole-genome sequencing effort over multiple breeds on imputation accuracy. BMC Genet. 2014;15:105. doi: 10.1186/s12863-014-0105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouwman AC, Daetwyler HD, Chamberlain AJ, Ponce CH, Sargolzaei M, Schenkel FS, et al. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat Genet. 2018;50:362–367. doi: 10.1038/s41588-018-0056-5. [DOI] [PubMed] [Google Scholar]
- 26.Boussaha M, Michot P, Letaief R, Hoze C, Fritz S, Grohs C, et al. Construction of a large collection of small genome variations in French dairy and beef breeds using whole-genome sequences. Genet Sel Evol. 2016;48:87. doi: 10.1186/s12711-016-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1082. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu WX, Liu Y, Lu X, Niu XY, Ding XD, Liu JF, et al. A genome-wide association study identifies two novel promising candidate genes affecting Escherichia coli F4ab/F4ac susceptibility in swine. PLoS One. 2012;7:e32127. doi: 10.1371/journal.pone.0032127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramayo-Caldas Y, Renand G, Ballester M, Saintilan R, Rocha D. Multi-breed and multi-trait co-association analysis of meat tenderness and other meat quality traits in three French beef cattle breeds. Genet Sel Evol. 2016;48:37. doi: 10.1186/s12711-016-0216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reverter A, Chan EK. Combining partial correlation and an information theory approach to the reversed engineering of gene co-expression networks. Bioinformatics. 2008;24:2491–2497. doi: 10.1093/bioinformatics/btn482. [DOI] [PubMed] [Google Scholar]
- 35.Watson-Haigh NS, Kadarmideen HN, Reverter A. PCIT: an R package for weighted gene co-expression networks based on partial correlation and information theory approaches. Bioinformatics. 2010;26:411–413. doi: 10.1093/bioinformatics/btp674. [DOI] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol. 2014;10:e1003731. doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-into decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grosclaude F, Mahé MF, Mercier JC, Ribadeau-Dumas B. Localisation des substitutions d’acides aminés différenciant les varianst A et B de la caséine kappa bovine. Ann Genet Sel Anim. 1972;4:515–521. doi: 10.1186/1297-9686-4-4-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganai NA, Bovenhuis H, van Arendonk JA, Visker MH. Novel polymorphisms in the bovine beta-lactoglobulin gene and their effects on beta-lactoglobulin protein concentration in milk. Anim Genet. 2009;40:127–133. doi: 10.1111/j.1365-2052.2008.01806.x. [DOI] [PubMed] [Google Scholar]
- 43.Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, et al. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002;12:222–231. doi: 10.1101/gr.224202. [DOI] [PubMed] [Google Scholar]
- 44.Weller JI, Bickhart DM, Wiggans GR, Tooker ME, O’Connell JR, Jiang J, et al. Determination of quantitative trait nucleotides by concordance analysis between quantitative trait loci and marker genotypes of US Holsteins. J Dairy Sci. 2018;101:9089–9107. doi: 10.3168/jds.2018-14816. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez MP, Govignon-Gion A, Croiseau P, Fritz S, Hozé C, Miranda G, et al. Within-breed and multi-breed GWAS on imputed whole-genome sequence variants reveal candidate mutations affecting milk protein composition in dairy cattle. Genet Sel Evol. 2017;49:68. doi: 10.1186/s12711-017-0344-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez MP, Wolf V, El Jabri M, Beuvier E, Rolet-Répécaud O, Gaüzère Y, et al. Short communication: confirmation of candidate causative variants on milk composition and cheesemaking properties in Montbéliarde cows. J Dairy Sci. 2018;101:10076–10081. doi: 10.3168/jds.2018-14986. [DOI] [PubMed] [Google Scholar]
- 47.Duchemin SI, Bovenhuis H, Megens HJ, Van Arendonk JAM, Visker MHPW. Fine-mapping of BTA17 using imputed sequences for associations with de novo synthesized fatty acids in bovine milk. J Dairy Sci. 2017;100:9125–9135. doi: 10.3168/jds.2017-12965. [DOI] [PubMed] [Google Scholar]
- 48.Boichard D, Govignon-Gion A, Larroque H, Maroteau C, Palhiere I, Tosser-Klop G, et al. Genetic determinism of milk composition in fatty acids and proteins in ruminants, and selection potential. Prod Anim. 2014;27:283–298. [Google Scholar]
- 49.Knutsen TM, Olsen HG, Tafintseva V, Svendsen M, Kohler A, Kent MP, et al. Unravelling genetic variation underlying de novo-synthesis of bovine milk fatty acids. Sci Rep. 2018;8:2179. doi: 10.1038/s41598-018-20476-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kemper KE, Reich CM, Bowman PJ, vander Jagt CJ, Chamberlain AJ, Mason BA, et al. Improved precision of QTL mapping using a nonlinear Bayesian method in a multi-breed population leads to greater accuracy of across-breed genomic predictions. Genet Sel Evol. 2015;47:29. doi: 10.1186/s12711-014-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang J, Gao Y, Hou Y, Li W, Zhang S, Zhang Q, et al. Whole-genome resequencing of Holstein bulls for indel discovery and identification of genes associated with milk composition traits in dairy cattle. PLoS One. 2016;11:e0168946. doi: 10.1371/journal.pone.0168946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C, Sun D, Zhang S, Wang S, Wu X, Zhang Q, et al. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS One. 2014;9:e96186. doi: 10.1371/journal.pone.0096186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pausch H, Emmerling R, Gredler-Grandl B, Fries R, Daetwyler HD, Goddard ME. Meta-analysis of sequence-based association studies across three cattle breeds reveals 25 QTL for fat and protein percentages in milk at nucleotide resolution. BMC Genom. 2017;18:853. doi: 10.1186/s12864-017-4263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopdell TJ, Tiplady K, Struchalin M, Johnson TJJ, Keehan M, Sherlock R, et al. DNA and RNA-sequence based GWAS highlights membrane-transport genes as key modulators of milk lactose content. BMC Genom. 2017;18:968. doi: 10.1186/s12864-017-4320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kemper KE, Littlejohn MD, Lopdell T, Hayes BJ, Bennett LE, Williams RP, et al. Leveraging genetically simple traits to identify small-effect variants for complex phenotypes. BMC Genom. 2016;17:858. doi: 10.1186/s12864-016-3175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoonjans K, Staels B, Auwerx J. The peroxisome proliferator activated receptors (PPARs) and their effects on lipid metabolism and adipocyte differentiation. Biochim Biophys Acta. 1996;1302:93–109. doi: 10.1016/0005-2760(96)00066-5. [DOI] [PubMed] [Google Scholar]
- 57.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res. 2004;43:134–176. doi: 10.1016/S0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 58.Martin P, Leroux C. Caprine gene specifying alpha(s1)-casein: a highly suspicious factor with both multiple and unexpected effects. Prod Anim. 2000;13:125–132. [Google Scholar]
- 59.Li R, Dudemaine PL, Zhao X, Lei C, Ibeagha-Awemu EM. Comparative analysis of the miRNome of bovine milk fat, whey and cells. PLoS One. 2016;11:e0154129. doi: 10.1371/journal.pone.0154129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z, Liu H, Jin X, Lo L, Liu J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genom. 2012;13:731. doi: 10.1186/1471-2164-13-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le Guillou S, Marthey S, Laloe D, Laubier J, Mobuchon L, Leroux C, et al. Characterisation and comparison of lactating mouse and bovine mammary gland miRNomes. PLoS One. 2014;9:e91938. doi: 10.1371/journal.pone.0091938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X, Gao C, Li H, Huang L, Sun Q, Dong Y, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010;20:1128–1137. doi: 10.1038/cr.2010.80. [DOI] [PubMed] [Google Scholar]
- 63.Faye LL, Machiela MJ, Kraft P, Bull SB, Sun L. Re-ranking sequencing variants in the post-GWAS era for accurate causal variant identification. PLoS Genet. 2013;9:e1003609. doi: 10.1371/journal.pgen.1003609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. −log10(P) plotted against the position of variants on Bos Taurus autosomes for milk composition. Manhattan Plot obtained from GWAS for milk composition traits.
Additional file 2: Table S1. Description of the 264 genes located in confidence intervals of the QTL detected by GWAS for milk cheese-making and composition traits. Name and position of candidate genes identified by GWAS.
Additional file 3: Table S2. Percentage of the phenotypic variance of milk CMP and composition traits explained by each QTL. Individual effects of QTL on all CMP and composition traits, expressed as a percentage of the phenotypic variance of the trait.
Additional file 4: Table S3. Description of the 736 genes selected by the gene network analysis for milk cheese-making and composition traits. Name and position of candidate genes identified by AWM.
Additional file 5: Table S4. Gene ontology (GO) terms and KEGG pathways for genes selected by AWM. Name, description and list of genes of the gene ontology terms and KEGG pathways identified in the gene network analysis.
Data Availability Statement
The data (genotypes and phenotypes) that enabled the findings of this study were made available by UMOTEST, CEL25-90, and HSCEL. However, restrictions apply to the availability of these data: they were used under license for the current study, and are not publicly available.