Abstract
Background
Galectin 3 (LGALS3) gene expression is associated with poor survival in acute myeloid leukemia (AML) but the prognostic impact of LGALS3 protein expression in AML is unknown. LGALS3 supports diverse survival pathways including RAS mediated cascades, protein expression and stability of anti-apoptotic BCL2 family members, and activation of proliferative pathways including those mediated by beta Catenin. CD74 is a positive regulator of CD44 and CXCR4 signaling and this molecule may be critical for AML stem cell function. At present, the role of LGALS3 and CD74 in AML is unclear. In this study, we examine protein expression of LGALS3 and CD74 by reverse phase protein analysis (RPPA) and identify new protein networks associated with these molecules. In addition, we determine prognostic potential of LGALS3, CD74, and their protein networks for clinical correlates in AML patients.
Methods
RPPA was used to determine relative expression of LGALS3, CD74, and 229 other proteins in 231 fresh AML patient samples and 205 samples were from patients who were treated and evaluable for outcome. Pearson correlation analysis was performed to identify proteins associated with LGALS3 and CD74. Progeny clustering was performed to generate protein networks. String analysis was performed to determine protein:protein interactions in networks and to perform gene ontology analysis. Kaplan-Meir method was used to generate survival curves.
Findings
LGALS3 is highest in monocytic AML patients and those with elevated LGALS3 had significantly shorter remission duration compared to patients with lower LGALS3 levels (median 21.9 vs 51.3 weeks, p = 0.016). Pearson correlation of LGALS3 with 230 other proteins identifies a distinct set of 37 proteins positively correlated with LGALS3 expression levels with a high representation of proteins involved in AKT and ERK signaling pathways. Thirty-one proteins were negatively correlated with LGALS3 including an AKT phosphatase. Pearson correlation of proteins associated with CD74 identified 12 proteins negatively correlated with CD74 and 16 proteins that are positively correlated with CD74. CD74 network revealed strong association with CD44 signaling and a high representation of apoptosis regulators. Progeny clustering was used to build protein networks based on LGALS3 and CD74 associated proteins. A strong relationship of the LGALS3 network with the CD74 network was identified. For AML patients with both the LGALS3 and CD74 protein cluster active, median overall survival was only 24.3 weeks, median remission duration was 17.8 weeks, and no patient survived beyond one year.
Interpretation
The findings from this study identify for the first time protein networks associated with LGALS3 and CD74 in AML. Each network features unique pathway characteristics. The data also suggest that the LGALS3 network and the CD74 network each support AML cell survival and the two networks may cooperate in a novel high risk AML population.
Fund
Leukemia Lymphoma Society provided funds to SMK for RPPA study of AML patient population. Texas Leukemia provided funds to PPR and SMK to study CD74 and LGALS3 expression in AML patients using RPPA. No payment was involved in the production of this manuscript.
Keywords: LGALS3, CD74, RPPA, Proteomics, Acute myeloid leukemia
Research in context.
Evidence before this study
PubMed was used to search terms “galectin”, “Galectin 3”, “LGALS3”, and CD74 alone and in combination with terms AML and leukemia. We also used cBioPortal to investigate gene expression of LGALS3 and CD74 network proteins in TCGA AML databases.
Added value of this study
Prior to our study there was no proteomic study on LGALS3 or CD74 as prognostic factors alone or in the context of their active networks. This study determines prognostic potential for LGALS3, CD74, LGALS3 network, CD74 network, and combination of LGALS3/CD74 active networks in AML. The data identifies for the first time an at risk AML population based on the proteomic data.
Implications of all the available evidence
The data supports the use of new therapies to target LGALS3 and CD74 for the particular AML populations where these molecules impact survival.
Alt-text: Unlabelled Box
1. Introduction
Galectin 3 (LGALS3) is a beta-galactoside binding protein that participates in diverse cellular processes that support cell growth and cell survival [[1], [2], [3], [4], [5], [6], [7], [8], [9]]. There are at least fourteen known galectin family members of which ten are found in mammalian cells [1]. There are three families of galectins based on structure but LGALS3 is unique in that it is the only member of the chimeric group [1]. LGALS3 is the only galectin which can form pentamers and this enables the galectin to form lattices and thus participate in endocytotic processes [1]. LGALS3 is an excellent example of a molecule that acts as a tumor promoter in the context of the entire tumor microenvironment by promoting survival of malignant cells, supporting metastasis, suppressing immune surveillance, and modulating inflammatory expression of chemokines/cytokines [[1], [2], [3], [4], [5], [6], [7], [8], [9]]. LGALS3 supports cell survival by diverse mechanisms. The galectin has been shown to associate with BCL2 via a NWGR motif common to both proteins to help the anti-apoptotic molecule support mitochondrial integrity during stress challenge [[7], [8], [9]]. LGALS3 also supports cell proliferation via the WNT signaling pathway. LGALS3 can bind beta Catenin and Axin and also supports beta catenin protein stability by promoting Protein Kinase B (AKT) suppression of GSK3 beta [[10], [11], [12]]. LGALS3 is critical for RAS signaling and thus supports Mitogen Activated Protein Kinase (MAPK) and AKT cascades [1,2,[12], [13], [14], [15], [16], [17]]. LGALS3 positively regulates BCL2 and MCL-1 gene and protein expression in AML cells by supporting both ERK and AKT pathways [1,2,[12], [13], [14], [15], [16], [17]]. Suppression of LGALS3 by shRNA or with GCS-100 (an inhibitor of LGALS1 and LGALS3) blocks both AKT and ERK signaling pathways [15,17].
LGALS3 regulated pathways are involved in expression of genes and protein associated with cancer stem cells (CSC) and thus the galectin likely supports CSC [9]. Recent data suggests that LGALS3 supports malignant cell survival in AML [6,15,18]. In a cohort of Taiwanese AML patients, Cheng and colleagues reported that elevated LGALS3 mRNA was prognostic for poor survival outcome [18]. However, in that study the impact of LGALS3 protein expression or associations of the galectin with potential LGALS3 target proteins was not examined.
CD74 (also known as the invariant chain protein) is best known as a chaperone for major histocompatibility (MHC) Class II molecules involved in antigen presentation [19,20]. In addition to mediating MHC Class II molecule endocytosis, CD74 protects these molecules from proteolysis [[19], [20], [21]]. CD74 also has MHC Class II independent functions that involve the pro-inflammatory cytokine macrophage inhibitory factor (MIF) and cell surface signaling molecules CD44 and CXCR4 [[19], [20], [21]]. CD74 was found to bind MIF but CD74 alone is unable to initiate MIF signaling which requires either CD44 or CXCR4 [[19], [20], [21], [22], [23]]. CD74 dependent MIF signaling pathways include ERK, JNK, and AKT [[24], [25], [26], [27]]. CD74 dependent MIF signaling has been shown to suppress p53 function and to activate NF kappa B [26,28]. Regulation of NF kappa B by the MIF/CD74 axis may be critical for sustaining mitochondrial integrity [28]. CD74 is highly expressed in lymphocytes and macrophages and has therefore been implicated as a target for CLL therapy [29,30]. CD74 has been shown to play a role in AML microenvironment though via stromal cell derived CD74 [31]. Still, the role of CD74 in AML leukemic cells is unclear. A recent study by van Galen and colleagues using single cell analysis of gene expression in AML leukemic cell populations found that CD74, though not prominent in normal myeloid cells, was expressed at high levels in AML cells including primitive stem cells [32].
In the current study, we analyze the protein expression of LGALS3 and CD74 as well as 229 other proteins in AML blast cells derived from 231 patients using a powerful proteomic tool, Reverse Phase Protein Analysis (RPPA). The LGALS3 and CD74 interactomes, including novel associations revealed by the array was used to create networks for both these proteins. A particularly prognostic interaction with high activity within both the LGALS3 network and the CD74 network was revealed. Our findings suggest that LGALS3 network alone is prognostic for poor patient survival outcome and the network has a more potent negative impact on patient survival when the LGALS3 network interacts with the CD74 network. The data presented suggest strategies to target LGALS3 and/or CD74 may prove useful for the therapy of AML.
2. Materials and methods
2.1. Patient samples
Peripheral blood and bone marrow specimens were collected from 511 patients with newly diagnosed AML evaluated at The University of Texas M.D. Anderson Cancer Center (MDACC) between September 1999 and March 2007. Samples were acquired during routine diagnostic assessments in accordance with the regulations and protocols (Lab 01-473) approved by the Investigational Review Board of MDACC. Informed consent was obtained in accordance with the Declaration of Helsinki. Samples were analyzed under and Institutional Review Board–approved laboratory protocol (Lab 05–0654). Sample preparation was previously described [[33], [34], [35], [36], [37]]. Patient characteristics are listed in Table 1 and Table 2.
Table 1.
Patient demographics by category.
| Category Variables | Total | Normal | High | p Value |
|---|---|---|---|---|
| Number of cases | 205 | 154 | 51 | NA |
| Gender: Female | 46.3% | 48.1% | 41.2% | 0.394 |
| Gender: Male | 53.7% | 51.9% | 58.8% | |
| AHD ≥ 2 Mo: Yes | 31.2% | 31.2% | 31.4% | 0.95 |
| Prior Malignancy: Yes | 16.6% | 14.9% | 21.6% | 0.3 |
| Prior Chemo: Yes | 8.8% | 7.8% | 11.8% | 0.41 |
| Prior XRT: Yes | 8.3% | 5.8% | 15.7% | 0.027 |
| Infection: Yes | 27.8% | 26.0% | 33.3% | 0.35 |
| WHO Class: Not in other | 60.5% | 60.4% | 60.8% | 0.72 |
| WHO class: Multilineage Dysp | 15.6% | 14.3% | 19.6% | |
| WHO Class: therapy related | 8.3% | 9.1% | 5.9% | |
| WHO Class: AML w Char Gene Abnormality | 15.6% | 16.2% | 13.7% | |
| FAB: 0 | 5.9% | 7.1% | 2.0% | < 0.0001 |
| FAB: 1 | 12.7% | 16.2% | 2.0% | |
| FAB: 2 | 33.2% | 39.6% | 13.7% | |
| FAB: 4 | 28.3% | 24.0% | 41.2% | |
| FAB: 5 | 12.2% | 6.5% | 29.4% | |
| FAB: 6 | 2.0% | 1.3% | 3.9% | |
| FAB: 7 | 2.4% | 2.6% | 2.0% | |
| FAB: 8 | 2.9% | 2.6% | 3.9% | |
| FAB: 10 | 0.5% | 0.0% | 2.0% | |
| Cytogenetics: Favorable | 10.7% | 13.6% | 2.0% | 0.06 |
| Cytogenetics: Intermediate | 46.3% | 45.5% | 49.0% | |
| Cytogenetics: Unfavorable | 42.9% | 40.9% | 49.0% | |
| Response: CR | 57.6% | 55.8% | 62.7% | 0.31 for CR + PR vs resistant and excluding Fail |
| Response: PR | 2.4% | 3.2% | 0.0% | |
| Response: Resistant | 30.2% | 32.5% | 23.5% | |
| Response: Fail | 9.8% | 8.4% | 13.7% | |
| Alive | 19.0% | 21.4% | 11.8% | 0.11 |
| Relapse: No | 42.4% | 48.8% | 25.0% | 0.036 |
| Relapse: Yes | 61.9% | 57.0% | 75.0% |
Table 2.
Patient demographics by continuous variables.
| Continuous variable | Normal | High | p Value | |
|---|---|---|---|---|
| Age (years) | Mean | 58.5 | 60.6 | 0.84 |
| WBC | Mean | 38.144 | 50.524 | 0.145791 |
| Absolute Blast Count | Median | 5587 | 8164 | 0.96 |
| BM Blast | Mean | 60.232 | 55.471 | 0.229959 |
| BM Monocyte | Mean | 3.149 | 10.627 | 0.000026 |
| PB Blast | Mean | 45.050 | 32.93 | 0.008398 |
| PB MONO | Mean | 9.684 | 19.158 | 0.000212 |
| PB PROM | Mean | 0.724 | 2.804 | 0.077415 |
| HGB | Mean | 9.799 | 11.245 | 0.174850 |
| PLT | Mean | 72.208 | 89.059 | 0.209900 |
| LDH | Mean | 1808.071 | 2075.000 | 0.478537 |
| Albumin | Mean | 3.317 | 3.175 | 0.193330 |
| Bilirubin | Mean | 1.890 | 0.690 | 0.454687 |
| Creatinine | Mean | 1.040 | 1.151 | 0.227295 |
| Fibrinogen | Mean | 414.974 | 423.196 | 0.775193 |
| CD13 | Mean | 76.728 | 68.678 | 0.074873 |
| CD33 | Mean | 83.442 | 88.547 | 0.167026 |
| CD34 | Mean | 52.772 | 27.898 | 0.000071 |
| CD7 | Mean | 17.103 | 15.335 | 0.691007 |
| CD10 | Mean | 4.556 | 8.502 | 0.115709 |
| CD20 | Mean | 3.817 | 7.565 | 0.207067 |
| HLA.DR | Mean | 77.487 | 75.845 | 0.715248 |
| CD19 | Mean | 8.594 | 10.386 | 0.588748 |
2.2. Pathway analysis
String software (String 10.1; website: http://string-db.org) was used to determine protein associations [38].
2.3. RPPA method
Proteomic profiling was done on samples from patients with AML using RPPA. The method and validation of the technique are fully described in previous publications [[34], [35], [36], [37]]. Patient samples were printed in five serial dilutions onto slides along with normalization and expression controls. Slides were probed with strictly validated primary antibodies. Antibodies against 231 proteins were used for analysis (list provided in refs. [34, 37]). An IgG subtype specific secondary antibody was used to amplify the signal and finally a stable dye is precipitated. The stained slides were analyzed using the Microvigene software (Vigene Tech) to produce quantified data.
2.4. RPPA normalization and progeny cluster analysis
To determine relative protein expression patterns custom Reverse Phase Protein Arrays (RPPA) with peripheral blood or bone marrow samples from 511 Adult AML patients and 10 normal CD34+ bone marrow samples were created and probed with 231 validated antibodies. For RPPA, supercurve algorithms were used to generate a single value from the five serial dilutions [[34], [35], [36], [37]]. Loading control and topographical normalization procedures accounted for protein concentration and background staining variations. Analysis using unbiased clustering perturbation bootstrap clustering, and principle component analysis was then done as fully described in a previous publication [34]. For cluster analysis, methods were used as described in previous publications [[34], [35], [36], [37]]. As presented below, the range of expression was different depending on whether the samples were prepared from fresh vs. cryopreserved cells. Consequently, analysis was restricted to only the fresh samples. Proteins were divided into 31 Protein Functional Groups (ProFnGrp) based on known associations. A progeny clustering algorithm was used to determine the optimal number of protein clusters; recognizing groups of patients with correlated protein expression patterns. Principal component analysis (PCA) was done to map global differences and similarities between protein clusters and normal CD34+ samples. Protein networks were constructed using literature associations and correlation within the data set. Associations between clinical features, outcomes and signatures were determined. Hierarchical clustering was performed on a compilation of all protein clusters into one binary matrix to identify recurrent protein expression signatures that comprised similar combinations of protein constellations. From this we constructed a list of proteins that were over or under expressed in each signature [37]. A website containing “Leukemia Profile Atlases” is available at https://www.leukemiaatlas.org/.
Expression analysis of genes associated with LGALS3 and CD74 network proteins in de novo AML – Gene expression data from the TCGA AML data set derived from the 2013 New England Journal of Medicine publication is available using cBioPortal software [[39], [40], [41]]. The mRNA z-Scores (RNA Seq V2 RSEM) compare expression distribution of genes of interest in tumors that are diploid for the specific gene. Queries for LGALS3 were input using the TCGA AML 2013 New England Journal of Medicine dataset with a Z threshold of 2.0. Using Enrichment search for mRNA, comparison of LGALS3 was compared to genes from its RPPA network as well as to CD74 and genes from the Cd74 network. Significant differences in expression are identified by q-values derived from Benjamini-Hochberg procedure (see cBioPortal website; http://www.cbioportal.org/, refs. 40, 41).
2.5. Protein expression and gene expression validation in LGALS3 AML cell lines
OCI-AML3 cells were the kind gift from Mark Minden (Ontario Cancer Institute; Toronto, Canada). THP-1 was obtained from ATCC (Manassas, VA). LGALS3 knock down OCI-AML3 and THP-1 cell lines were previously described [15]. LGALS3 clone TRCN0000029308 targeting residues 606–626 on RefSeq NM_002306.3 was used. pLKO.1 control (plasmid 10879, Addgene, Cambridge, MA, USA) was used as negative control. Infected cells were selected with puromycin (Invivogen, San Diego, CA). Knockdown was verified by western blot analysis and real time PCR. For protein expression comparison in cell lines, immunoblot analysis was performed. Cells were boiled and sonicated in lysis buffer and protein (5 × 105 cell equivalents) was subjected to electrophoresis using SDS/PAGE. Immunoblot analysis was performed with antibodies against LGALS3 (Cell Signaling Technology, Beverly, MA), PPP2R2A/B/C/D (Santa Cruz Biotechnology, Dallas, TX), ATG7 (Cell Signaling Technology, Beverly, MA), and Tubulin (Sigma Aldrich, St. Louis, MO). Signals were detected by using the Odyssey Infrared Imaging System and quantitated by Odyssey software version 3.0 (both LI-COR Biosciences, Lincoln, NE, USA). Tubulin was used as a loading control. Real-time PCR (qRT-PCR) was used to assess gene expression in the cell lines. qRT-PCR was performed using an QuantStudio 3 PCR System (Life Technologies). Triplicate 20 ul reactions containing the equivalent of 7.5 ng total RNA were run using TaqMan Gene Expression Assays (Life Technologies) as directed by the manufacturer. Assays included ATG7 (Hs00197348_m1), LGALS3 (Hs00173587_m1), ITGAL (Hs00158218_m1), CCND3 (Hs00236949_m1), PRKCA (Hs00176973_m1), PARP1 (Hs00242302_m1), CD74 (Hs00269961_m1), MYC (Hs00153408_m1), CD44 (Hs01075862_m1), SSBP2 (Hs01044454_m1), PPP2R2A (Hs00953658_m1), CLPP (Hs00195655_m1) and B2M (Hs00187842_m1). ABL1 (Hs01104728_m1) was used as an endogenous control. QuantStudio Design and Analysis software (Life Technologies) was used to analyze the data.
2.6. Statistical analysis
For outcomes analysis patients were divided into two groups based on whether LGALS3 expression was within the range of the normal CD34+ cells, or was above normal. Comparison of the protein levels between paired samples was done by performing paired t-test. Association between protein expression levels and categorical clinical variables were assessed in R using standard t-tests, linear regression, or mixed effects linear models. Association between continuous variable and protein levels were assessed by using the Pearson and Spearman correlation and linear regression. Bonferroni corrections were done to account for multiple statistical parameters for calculating statistical significance. The Kaplan-Meir method was used to generate the survival curves. Univariate and multivariate Cox proportional hazard modeling was done to investigate association with survival with protein levels as categorized variables using the Statistica version 13.1 software (StatSoft, Tulsa OK). Although the mutational status of some molecular markers (NPM1, FLT3-ITD, DNMT3A and RAS mutations) are known for this dataset other more recently discovered prognostic markers (e.g., ASXL1, TET2, CEBPα, Wilms Tumor 1) are not; therefore, the multivariate analysis did not contain all known AML prognostic markers. Overall survival (OS) was determined based on the outcome of 205 newly diagnosed AML patients treated at UTMDACC and remission duration was based on the 118 patients that achieved remission.
3. Results
3.1. LGALS3 levels are elevated in some AML patient cell samples compared to normal CD34+ cells and are highest in monocytic leukemia patient
Levels of LGALS3 protein were significantly (p = 0.001) higher in the blast cells from the AML patients compared to normal CD34+ cells as shown in the histogram in Fig. 1A with 24.8% above the upper limit of the normal range (Fig. 1A). As mentioned above in “Materials and Methods”, expression was different between protein made from fresh cells compared to that made from cryopreserved cells, but did not differ between blood and bone marrow blasts (Fig. 1A). Consequently the remainder of the analysis was restricted to only the 231 fresh samples, of which 205 were treated and evaluable for outcome.
Fig. 1.
LGALS3 is elevated in a population of AML patients particularly those with monocytic AML. A) The range of LGALS3 protein expression in AML blast cells from 511 cases compared to normal counterpart CD34+ cells from 21 donors is shown stratified by time of protein preparation (Fresh on the day of collection or later from viably Cryopreserved cells). LGALS3 levels were significantly higher in cryopreserved cells compared to freshly prepared protein. Therefore, only data from the fresh samples was used for the rest of the analysis. Expression of LGALS3 in the fresh samples was compared in AML samples grouped by FAB class (B) or cytogenetic category (C).
The basic demographics of the normal range and high expression patients are shown in Tables 1 and 2. Patients with higher LGALS3 did not differ in gender, age, history of an antecedent hematological disorder, or World Health Organization classification (Table 1). Differences in LGALS3 expression varied substantially between French-American-British (FAB) AML sub-types determined by class with above normal expression most common among those with monocyte containing subtypes (M4 and M5, 70.6% vs 30.5%) and least common among those with early myeloid subtypes (M0, M1 and M2, 17.7% vs. 62.9%) (Table 1; Fig. 1B; p < 0.00001). Higher LGALS3 levels were also associated with significantly higher percentages of monocytes in the bone marrow and peripheral blood, and therefore with a lower percentage of blasts in the peripheral blood, however the absolute blast count did not differ based on LGALS3 expression level (Table 2). There was no statistical difference in LGALS3 expression between AML populations based on cytogenetic category although only 1 of 21 favorable cytogenetic cases had above normal LGALS3 expression (Fig. 1C; p = 0.42). There were no differences in expression of LGALS3 among patients with other mutations that were surveyed (i.e. NPM1, FLT3, RAS, DNMT3A, IDH1 or IDH2; data not shown).
As shown in Table 1, patients with above normal LGALS3 were slightly more likely to achieve remission (62.7% vs. 55.8%, p = 0.31). Elevated expression had no impact on OS (Fig. 2A). However, compared to those with normal range LGALS3, patients with higher than normal LGALS3 were significantly more likely to relapse (75% vs 57%, p = 0.036; Table 1) and had significantly shorter remission duration (21.9 vs. 51.3 weeks, p = 0.016; Fig. 2B). The higher relapse rate and shorter remission duration combined to make those with high LGALS3 have an inferior OS (median 44.4 vs 114.6 weeks, p = 0.015) among patients that achieved remission (Fig. 2C). LGALS3 level had no effect on OS among those that were resistant (p = 0.76; data not shown), or on survival after relapse (p = 0.66; Fig. 2D) suggesting that it did not affect the response to salvage therapy. Overall there was trend for a lower percentage of above normal LGALS3 patients to be alive at 8 years follow-up (11.8% vs 21.4%, p = 0.11; Table 1).
Fig. 2.
LGALS3 expression is prognostic for poor survival outcome in some AML populations. Kaplan Meir curves for overall survival (A) and remission duration (B) in the total AML patients studied are presented. Kaplan Meir curvrves for overall survival among the AML patient population that achieved complete remission (C) and for survival after relapse (D) are also included.
3.2. LGALS3 levels correlate with a variety of signaling molecules in blast cells from AML patients
RPPA was used to examine correlations of LGALS3 with 230 other proteins. As shown in Fig. 3A, 68 of 231 proteins showed statistically significant (p < 0.0001, R > 0.25) correlation with LGALS3, with positive correlation for 27 total and 10 phospho-proteins and negative correlation for 24 total and 7 phospho-proteins. The strongest positive correlation was with the autophagy protein ATG7. The phospho-proteins positively correlated with LGALS3 included survival kinases such as p-ERK (pY202/pY204), p-AKT (pT308), three phospho-protein variants of PKC delta (i.e. pT507, pS645, and pS664), and p-PKC alpha (pS657) (Fig. 3). LGALS3 expression also positively correlated with phosphorylation of the tyrosine kinase SRC (i.e. pY416 and pY527). The most negatively correlated protein was Single Stranded DNA Binding Protein 2 (SSBP2) (Fig. 3). Among the other proteins negatively correlated with LGALS3 was the members of the PP2A B55 family (PPP2R2A, PPP2R2B, PPP2R2C, and PPP2R2D).
Fig. 3.
LGALS3 expression is correlated with a distinct set of proteins in AML. Pearson correlation (R > 0.25, P < 0.0001) of LGALS3 with other proteins measured identifies a distinct set of positively and negatively associated proteins in the AML RPPA set (A).
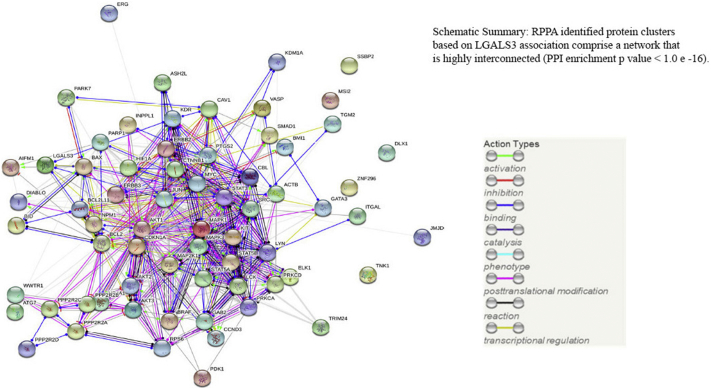
Protein network analysis was performed on the set of proteins associated with LGALS3 using String software (String 10.1; website: http://string-db.org; ref. 38). The network of LGALS3 proteins identified by RPPA are highly associated with a protein:protein enrichment p value <1.0e−16 (Fig. 4) by String. Numerous biological pathways (N = 588) and KEGG pathways (N = 86) associated with LGALS3 network were identified using the String software. Data are presented in Supplemental Table 1 and Supplemental Table 2, respectively. The top three biological pathways identified are protein phosphorylation (GO.0006468), cell surface receptor signaling pathway (GO.0007166), and regulation of cellular protein metabolic process (GO.0032268). The top KEGG pathway identified was PI3K-Akt signaling pathway (ID:4151) and proteins associated with AML (ID:5221) was 6th (Supplemental Table 2). ErbB signaling pathway (ID:4012) was 14th on the KEGG list. Though ErbB signaling is not generally considered in AML, ErbB is shown to be expressed in many of the AML samples in our cohort (data not shown) and thus this pathway may be worth study in AML.
Fig. 4.
String analysis reveals LGALS3 correlated proteins are highly interconnected in AML. String analysis of RPPA identified proteins in Fig. 3 reveals a high degree of protein:protein interaction (PPI) among the members of the LGALS3 network.
3.3. Suppression of LGALS3 in monocytic AML THP-1 cells induces expression of PP2A B subunit PPP2R2A
To assess whether LGALS3 acts directly on the various LGALS3 correlated proteins identified by RPPA, we utilized THP-1 transductant cells that express control shRNA (LKO) or LGALS3 shRNA that we have previously described [15]. At least in THP-1 cells, in most cases LGALS3 did not regulate protein expression of many of the LGALS3 associated proteins including ATG7, ITGAL, SSBP2, or ERG (Fig. 5; data not shown). At present, it is not clear whether these proteins act to regulate LGALS3 expression or if LGALS3 shares common regulators with these proteins. However, one exception was the PP2A B subunit family PPP2R2A/B/C/D. Suppression of LGALS3 resulted in near 2× fold increase in expression of the PP2A B subunits (Fig. 5) which is consistent with the negative correlation found between the proteins by RPPA (Fig. 3).
Fig. 5.

Suppression of LGALS3 by shRNA in THP-1 induces PPP2R2A/B/C/D expression but not ATG7. Protein lysates from control THP-1 (LKO) or THP-1 expressing LGALS3 shRNA were subject to electrophoresis and immunblot analysis performed. Antibodies against Tubulin, LGALS3, PPP2R2A/B/C/D, and ATG7 were used. Densitometry using LiCor software was performed and ratio of protein relative to Tubulin assessed relative to LKO THP-1 are listed.
3.4. Gene expression may drive association of LGALS3 with a number of proteins identified by RPPA as part of the LGALS3 network
To determine if correlations of LGALS3 network proteins with LGALS3 were similarly correlated with gene expression, we utilized cBioPortal software (http://www.cbioportal.org/) to query the TCGA AML database that derived from the 2013 New England Journal of Medicine publication [39]. Of the top nine unmodified proteins that were positively correlated with LGALS3 protein expression, expression of genes for eight proteins (ATG7, ITGAL, MAP2K1, MAPK1, JMJD6, CCND3, VASP, and PRKCA) were significantly higher (q value <0.05) in AML cells with elevated LGALS3 expression in the TCGA database (Fig. 6; Table 3).Expression of LCK was not correlated with LGALS3 (q value = 0.282; Table 3). Of the top nine unmodified proteins that were negatively correlated with LGALS3 protein expression, expression of genes for seven proteins (SSBP2, ERG, KIT, PPP2R2A, PARP1, MYC, and TRIM24) were significantly lower (q value <0.05) in AML cells with elevated LGALS3 expression (Fig. 6; Table 3). Expression of SMAD1 trended lower in cells with elevated LGALS3 (q value = 0.0726; Table 3). Expression of NR4A1 was actually higher in cells with elevated LGALS3 (q value = 0.0399; Table 3). At present, it is not clear if LGALS3 regulates gene expression of any of these genes, whether any of the network proteins may serve as a regulator of LGALS3 gene expression, or whether there is a yet unidentified common regulator to the genes in the LGALS3 RPPA network. To determine if LGALS3 may be involved in regulation of the gene expression of the proteins most positively correlated with LGALS3 expression, we utilized THP-1 cells that expressed control lentiviral plasmid (LKO) and THP-1 cells that expressed LGALS3 shRNA. qRT-PCR analysis of cDNA generated from RNA from these cells revealed that there was 90% reduction of LGALS3 expression by the shRNA (Fig. 7). However, suppression of LGALS3 did not result in a major alteration of expression of ATG7, ITGAL, CCND3, PRKCA, PARP1, MYC, SSBP2, or PPP2R2A (Fig. 7). At least in THP-1 cells, LGALS3 is not a direct regulator of any of these genes.
Fig. 6.
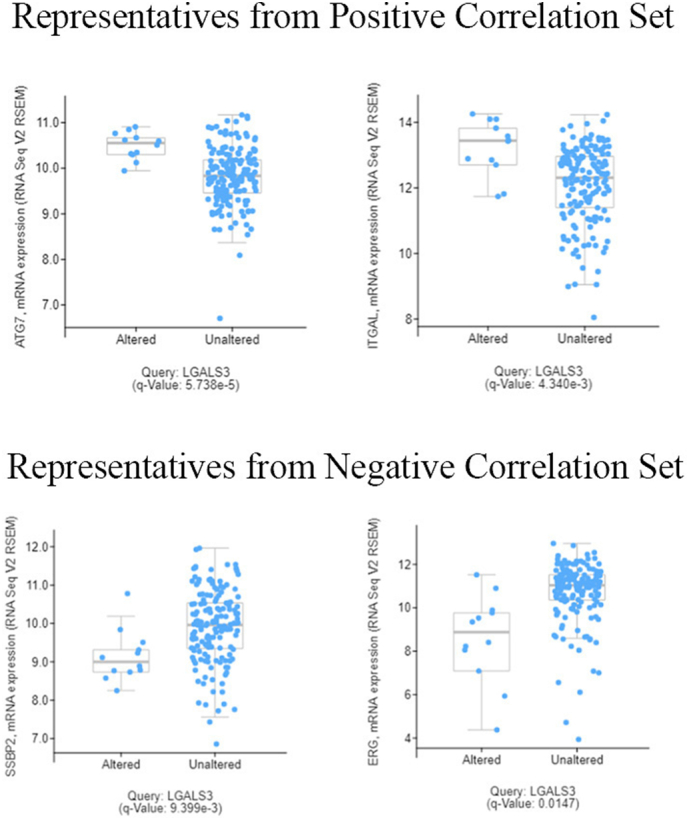
LGALS3 expression positively correlates with ATG7 and ITGAL and negatively correlates with SSBP2 and ERG in AML. CBioportal software was used to compare RNASeq measured gene expression of LGALS3 with ATG7, ITGAL, SSBP2, ERG, and other genes (listed in Table 3) in AML samples in the TCGA dataset from ref. [39].
Table 3.
Correlation of expression of LGALS3 network genes with LGALS3 in AML utilizing the RNASeq data in ref. [39]. The mRNA levels were measured by RNA Seq and the statistics used were q values derived from Benjamini-Hochberg procedure using CBioPortal software (described in refs. [40, 41]).
| Genes from Positive Correlation Group | q value relative to LGALS3 expression | Significant |
|---|---|---|
| ATG7 | 5.716 e−5 | Yes |
| ITGAL | 4.338 e−3 | Yes |
| JMJD6 | 0.0202 | Yes |
| MAPK1 | 0.0232 | Yes |
| CCND3 | 5.289 e−4 | Yes |
| VASP | 6.601 e−5 | Yes |
| PRKCA | 3.085 e−5 | Yes |
| LCK | 0.282 | No |
| Genes from Negative Correlation Group | q value relative to LGALS3 expression | Significant |
| SSBP2 | 9.399 e−3 | Yes |
| ERG | 0.0147 | Yes |
| PPP2R2A | 1.30 e−4 | Yes |
| KIT | 1.190 e−3 | Yes |
| MYC | 8.643 e−3 | Yes |
| TRIM24 | 0.0245 | Yes |
| PARP1 | 1.305 e−4 | Yes |
| NR4A1 | 0.0399 | Yes (but level is higher) |
| SMAD1 | 0.0726 | No |
| Genes from CD74 network | q value relative to LGALS3 expression | Significant |
| CD74 | 0.192 | No |
| CD44 | 0.0390 | Yes |
Fig. 7.

Suppression of LGALS3 does not alter gene expression of LGALS3 network protein genes or CD74 in THP-1 cells. RNA from THP-1 transductant cells with either LKO vector control shRNA or LGALS3 shRNA was isolated, cDNA produced, and mRNA levels of ATG7, LGALS3, ITGAL, CCND3, PRKCA, PARP1, CD74, MYC, CD44, SSBP2, PPP2R2A, CLPP, and B2M were determined by qRT-PCR and levels normalized to ABL-1 as described in “Materials and methods”.
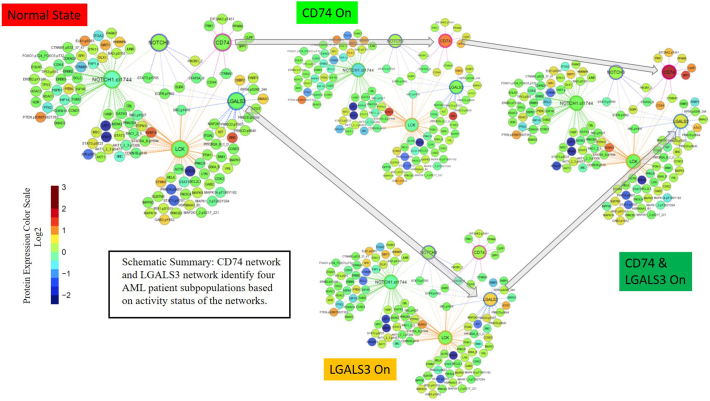
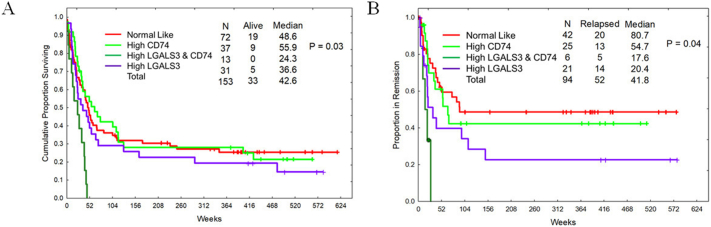
“Active” LGALS3 levels are associated with poor survival characteristics. Protein clusters in AML have been identified using RPPA profiles and bioinformatics data that groups these clusters by function, phenotype, and other parameters [[33], [34], [35], [36], [37]]. Since LGALS3 function varies depending on cellular location as well as the availability of potential targets, a more accurate assessment of LGALS3 contribution to survival will likely rely on “activity” of the galectin. The correlation studies identify a distinct set of proteins that are associated with LGALS3 (Fig. 3). Protein clustering analysis was performed as described in “Materials and Methods”. LGALS3 cluster was compared with a number of different protein clusters built using the RPPA data on this AML patient cohort [37]. Of the protein clusters analyzed, a relationship was found between LGALS3 cluster and a cluster involving the CD74 protein (Fig. 8). As shown in Fig. 8, clusters with LGALS3 active alone, or where LGALS3 and CD74 are both active, display elevated levels of LGALS3 with concomitant elevated expression of the LGALS3 positively correlated proteins and reduced expression of the LGALS3 negatively correlated proteins which were identified by RPPA and listed in Fig. 3. As shown in Fig. 8, in the cohort with active LGALS3 and CD74 active, expression of CD74 itself and some of the components in the CD74 network including CLPP, CD44, and Osteopontin (SPP1) are highly expressed compared to the normal state AML cohort. As shown in Fig. 9A, there is a difference in OS and remission duration between patients with normal state protein clustering and patients with “active” LGALS3. Compared to normal state patients, patients with “active” LGALS3 had shorter OS (36.6 versus 48.6 weeks, respectively; Fig. 9A) and shorter remission duration (20.4 versus 80.7 weeks, respectively; Fig. 9B). Patients with active CD74 network alone had a similar survival experience (Fig. 9A) but did exhibit a slightly shorter remission duration compared to the normal-like state patients (54.7 versus 80.7 weeks, respectively; Fig. 9B). For patients where both LGALS3 and CD74 are “active”, these patients display a very poor overall survival with no patients surviving beyond one year (24.3 versus 48.6 weeks for normal state patients; Fig. 9A). A similar adverse effect of “active” LGALS3 and “active” LGALS3 and CD74 was observed on remission duration (Fig. 9B). Patients with “active LGALS3 had shorter remission duration with a much lower percentage survival compared to patients with normal state protein clusters (17.6 versus 80.7 weeks, respectively; Figure 9B). AML patients with “active” LGALS3 and “active” CD74 displayed a very short remission duration with no patients remaining in remission beyond six months. These data suggest an active LGALS3 network influences AML patient survival especially if CD74 network is also active.
Fig. 8.
Progeny clustering identified an optimal number of 4 distinct protein clusters for this ProFnGrp. Protein networks were generated and showed interactions between “core-proteins” (large nodes) and other probed proteins (small nodes) from the data set. Clustering method has been described in our previous publication (ref. [37]) and further information on these protein networks can be found on our website “Leukemia Profile Atlases”, available at https://www.leukemiaatlas.org/. Progeny clustering identified one protein cluster with expression similar to that of the normal CD34+ samples which was designated as “normal-state” while three “leukemia-specific” protein patterns characterized by high expression individually of CD74, LGAL3, and a fourth state with both on.
Fig. 9.
Active LGALS3 and CD74 networks are associated with poor survival in AML patients. Kaplan Meir curves of AML patient populations defined by groups in Fig. 4 measuring overall survival (A) and remission duration (B).
3.5. CD74 protein expression in AML blast cells correlate with regulators of cell survival
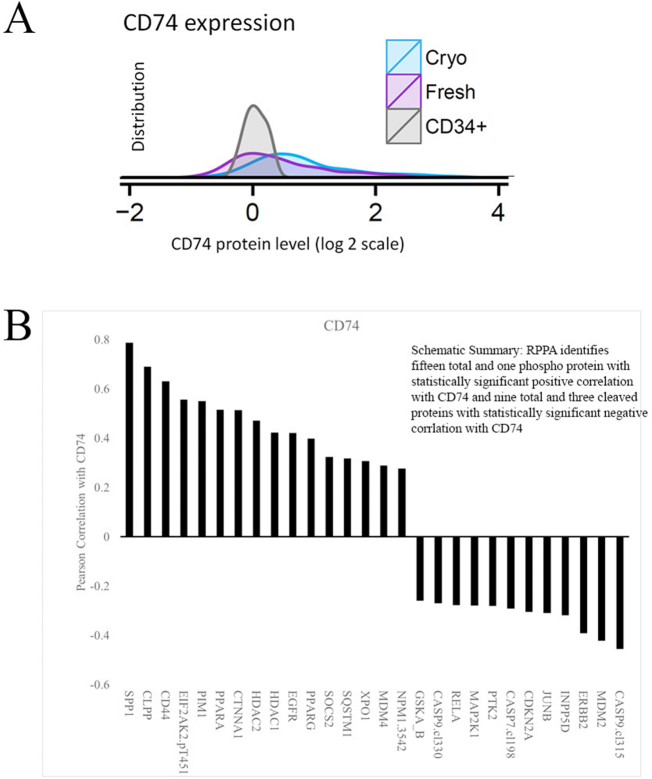
With the AML patient samples used for LGALS3 analysis, protein expression levels of CD74 in fresh and cryo frozen AML blast cells were compared to normal CD34+ cells by RPPA. Levels of CD74 protein were higher in 24.6% of the fresh blast cells from the AML patients compared to normal CD34+ cells as shown in the histogram in Fig. 10A. Next we determined correlation of CD74 with the other 230 proteins in our RPPA panel. Pearson correlation of proteins associated with CD74 identified 12 proteins negatively correlated with CD74 and 16 proteins that are positively correlated with CD74 (Fig. 10B). The strongest proteins correlated with CD74 are SPP1, CLPP, and CD44 (Fig. 10B). CD44 association is consistent with CD74 regulatory role of CD44 signaling [[19], [20], [21],42,43]. Association of CD74 with the mitochondrial protease CLPP is unknown. CLPP however is emerging as an important survival molecule in AML [44]. Also positively correlated with CD74 is Exportin 1(XPO1 also known as CRM1) which has been shown to have an adverse prognostic impact arising from its pro-survival functions in AML cells [45]. The proteins with the strongest negative correlation with CD74 are cleaved Caspase 9 and MDM2.
Fig. 10.
CD74 is elevated in AML patients associated with a distinct set of proteins in AML. A) The range of CD74 protein expression in AML blast cells from 511 cases compared to normal counterpart CD34+ cells from 21 donors is shown stratified by time of protein preparation (Fresh on the day of collection or later from viably Cryopreserved cells). Like LGALS3, CD74 levels were significantly higher in cryopreserved cells compared to freshly prepared protein. Like LGALS3, only data from the fresh samples was used for the rest of the analysis. (B)Pearson correlation of CD74 with other proteins measured identifies a distinct set of associated proteins in the AML RPPA set.
String analysis was performed on the network of proteins associated with CD74 by RPPA and this network was also highly associated with a protein:protein enrichment p value <1.0e-16 (Fig. 11). Many biological pathways (N = 535) and KEGG pathways (N = 83) were associated with CD74 network using String software (data are presented in Supplemental Table 3 and Supplemental Table 4, respectively). Many of the biological pathways identified are associated with immune regulation with four of the top ten including regulation of immune response (GO.0050776; 2nd on list), immune response-regulating signaling pathway (GO.0002764; 6th on list), innate immune response (GO.0045087; 7th on list), and immune response-reg. cell surface receptor sig. pathway (GO.0002768; listed 10th on list).
Fig. 11.
String analysis reveals CD74 correlated proteins are highly interconnected in AML. String analysis of RPPA identified proteins in Fig. 10B reveals a high degree of protein:protein interaction (PPI) among the members of the CD74 network.
The top KEGG pathway identified was Pathways in cancer (5200), with MicroRNAs in cancer (5206) listed 4th, and PI3K-Akt signaling pathway (4151) listed 5th.
4. Discussion
The current study represents the first proteomic analysis of LGALS3 and its potential network partners in AML. The previous study of LGALS3 mRNA expression from the Taiwan group suggested important prognostic potential of LGALS3 for poor survival outcome, but that study did not include other proteins that could potentially interact with LGALS3 in various biologic pathways [18]. LGALS3 is elevated in AML cells versus normal counterpart cells and is highest in monocytic AML cells (Fig. 1A and B). Measured alone, LGALS3 protein expression was important for OS in patients that achieved remission (Fig. 2A). These results are consistent with the RNA data reported by Cheng and colleagues [18]. The strong association between LGALS3 level and higher relapse rates and shorter remission duration, combined with the lack of an association between LGALS3 level and the rate of initial remission attainment, or the response to reinduction therapy, suggests a biologically unique effect for this protein. LGALS3 levels do not appear to affect chemosensitivity, but since high levels are associated with relapse, this suggests that the protein is functioning to promote leukemic cell recovery and regrowth after therapy. This suggests that anti-LGALS3 therapy might have more utility as a maintenance strategy in remission as opposed to being useful for reversing chemoresistance during induction and consolidation therapy. Maintenance therapy is generally not considered useful in AML, but this data suggests a maintenance therapy approach worth evaluating in the quarter of AML patients with high LGALS3 levels.
RPPA identified a distinct set of proteins associated with LGALS3 expression in the AML patients (Fig. 3). LGALS3 is associated with active AKT and MAPK signaling. The protein with the strongest positive correlation with LGALS3 is with ATG7, an autophagy protein that has recently been implicated in maintaining hematopoietic stem cells and serving as a survival factor in AML [46,47]. Recent studies implicate LGALS3 in regulation of autophagy via autophagasome formation, though whether the mechanism involves ATG7 is not clear [48]. Interestingly, phosphorylated PKC delta was positively correlated with LGALS3. PKC delta is viewed as a pro-stress kinase but recent studies suggest that the enzyme has pro-survival properties [[49], [50], [51]]. Kinehara and colleagues suggest that PKC delta may act in human pluripotent stem cells as part of a mechanism to regulate stem cell renewal [52]. The data also suggest a novel relationship between LGALS3 and PP2A. The negative correlation of LGALS3 expression with PPP2R2A/B/C/D could reflect LGALS3 suppression of PP2A. The PP2A isoform containing PPP2R2A dephosphorylates both AKT and PKC alpha [53]. Thus, potential suppression of the PP2A subunit by LGALS3 could account for elevated AKT and PKC alpha phosphorylation in samples where LGALS3 expression is elevated. Induction of PPP2R2A protein (Fig. 5) but not gene expression (Fig. 7) in THP-1 cells expressing LGALS3 shRNA suggests that LGALS3 acts directly on the PP2A subunits via a post-transcriptional mechanism in these cells. The TCGA data (Table 3) however suggests that there is a positive correlation between gene expression of LGALS3 and PPP2R2A suggesting that a common pathway may regulate the two genes.
PPP2RA/B/C/D was the only LGALS3 network protein demonstrated to be directly regulated by LGALS3 in the THP-1 cells (Fig. 5). In our previous study we saw potent suppression of AKT signaling by LGALS3 inhibition, so perhaps the mechanism involves LGALS3 suppression of the AKT phosphatase [15]. However, we did not see suppression of LGALS3 affect other network proteins in the THP-1 cells (data not shown). The role of other galectins such as LGALS1 in AML biology is not clear. LGALS1 may substitute for some LGALS3 functions particularly those involved in survival pathways as knock down of either LGALS1 or LGALS3 sensitized AML cells to BH3 mimetic drugs [15]. The failure of LGALS3 suppression to affect many of the RPPA identified proteins with the exception of PPP2R2A/B/C/D (Fig. 5) may reflect LGALS1 activity in these cells that may not be present in the primary AML cells. It is possible that many of the LGALS3 network proteins act to regulate LGALS3 rather than being regulated by the galectin. It is also possible that LGALS3 and some of the LGALS3 network proteins are subject to regulation by a yet unidentified common regulator(s). Further examination of the mechanism regulating the LGALS3 network is ongoing.
Network analysis from the data identifies a new extremely poor prognosis group based on the interaction between the LGALS3 and CD74 associated protein networks revealing potential biological pathways that may be critical in supporting AML cell survival. AML patients with both networks active are 8.5% of patients in the study (Fig. 9A) and thus this group may represent a sizeable population of AML patients. It is possible the two proteins regulate independent survival pathways that may have a synergistic effect on survival when both are active. The top ten biological processes associated with LGALS3 network include processes associated with cell metabolism (GO:0031325; GO:0032268; and GO:0032270), cell migration (GO:0030355), and response to growth factor stimulus (GO:0071363) and response to chemical stimulus (GO:0070887) (Supplemental Table 1). While it is unclear how LGALS3 might mechanistically influence leukemic cell recovery and growth after therapy, perhaps regulation of these cellular processes are important in addition to the well documented role of LGALS3 in regulation of cell cycle and cell proliferation [1,2,13,14]. Many of the CD74 network associated biological processes involved immune regulation (Supplemental Table 3) though it is unclear if CD74 network regulates potential immune response in AML. Many of the CD74 network associated biological processes did include those involved in regulation of cell death and apoptosis (Supplemental Table 3). Of the 31 proteins correlated with CD74 expression, 19 are associated with the biological pathway regulation of cell death (GO.0010941) and 16 are associated with the biological pathway negative regulation of apoptotic process (GO.0043066). The raises the question of what the cross-talk is between the LGALS3 and CD74 networks? Gene expression analysis of CD74, CD44, and CLPP in the THP-1 LKO cells versus THP-1 cells with LGALS3 shRNA showed no or only slight changes in these genes (Fig. 7). Protein expression of CD74, CD44, and CLPP were similar in THP-1 LKO and THP-1 LGALS3 shRNA cells (data not shown). While LGALS3 supports AKT activation via RAS, CD74 would be expected to support AKT via MIF mediated signaling involving CD44 and/or CXCR4 [[19], [20], [21], [22], [23], [24], [25], [26], [27]]. Though the functional roles of LGALS3 and CD74 in this process are very different, each network would contribute to activation and perhaps may explain why patients with both active networks do so poorly (Fig. 9A and B). Unfortunately, CXCR4 is not represented in the RPPA panel due to lack of validated antibody. However, CD44 is present and interestingly is most elevated in patients with active LGALS3 network and CD74 network (Fig. 8). LGALS3 has been shown to be critical for CD44 endocytosis so LGALS3 would be expected to promote CD44 surface expression [54]. In AML cells with LGALS3 supported CD44 surface expression, CD74 would be predicted to augment signaling mediated by CD44.
LGALS3 is well known as an immune regulatory molecule that suppresses host anti-tumor immune surveillance by diverse mechanisms [1,2,55]. LGALS3 blocks or at least dampens immune cell function by reducing surface expression of glycosylated T cell receptor in T cells and preventing NK cell receptor binding to antigen [1,2]. LGALS3 has emerged as a critical component in MSC in AML patients to impact response to therapy [56]. It is likely that LGALS3 secreted from MSC and other support cells in the AML microenvironment negatively impacts immune surveillance in AML patients. It is yet to be determined if LGALS3 derived from the leukemia cells plays a role as an immune response inhibitor in AML.
LGALS9 is emerging as an important immune checkpoint inhibitor molecule as a TIM-3 binding partner [2,57]. LGALS9 also regulates T cell function as a CD44 binding partner [58]. Whereas LGALS3 binding to CD44 promotes metastasis, LGALS9 binding to CD44 suppresses this process [59,60]. Future RPPA studies to determine the role of LGALS9 and galectins other than LGALS3 are warranted.
For the first time, an at risk AML population has been found that is associated with active LGALS3 and active CD74 networks (Fig. 9A and B). At present, it is unclear which if any proteins within the LGALS3 or CD74 networks is driving this phenomenon. CD44, SPP1, and CLPP are highly induced in the patient cohort with both networks active compared to patients with normal-like state (Fig. 8). CD44 would be assumed to be working via the CD74/CD44 axis [[19], [20], [21]]. SPP1 has recently been shown to be an important component in maintaining the tumor microenvironment in AML [61]. SPP1 is known to interact with various integrins and LGALS3 is a regulator of integrin function so perhaps integrin-mediated signaling is involved [1,2,62].
In summary, RPPA has revealed that LGALS3 is frequently elevated in AML patients especially those with monocytic leukemia. LGALS3 is prognostic as a single factor and in an active network is prognostic for shorter remission duration. A novel association of LGALS3 with CD74 has been found and AML patients with active LGALS3 and CD74 networks do extremely poorly. These results suggest that therapeutic strategies to target LGALS and/or CD74 should be modeled and studied in the laboratory. Therapeutic antibodies against CD74 such as Milatuzumab have been used in the clinic for various lymphoid leukemias, lymphoma, and multiple myeloma [29,30,63]. Targeting of LGALS3 by GCS-100 has been studied pre-clinically in many cancers including MM, AML, and K-RAS addicted solid tumor [[15], [16], [17]]. Specific LGALS3 inhibitory molecule GR-MD-02 is in the clinic for fibrosis [64]. An arsenal of agents to target CD74 and LGALS3 are available.
In summary, we have found that LGALS3 protein expression is prognostic for poor survival outcome which is consistent with RNA data from a previous study [18]. We have for the first time identified a LGALS3 protein network that is associated with poor outcome in AML patients, especially when the CD74 network is active. LGALS3 appears to be an upstream negative regulator of PP2A B subunit PPP2R2 family which suggests that the galectin has a role in inhibition of PP2A tumor suppressor function. In addition, we have found that CD74 may play an important role in AML cell survival, particularly in pathways involving CD44. Hopefully the data presented here will encourage the clinical development of agents to target LGALS3 and CD74 to benefit AML and other cancer patients where LGALS3 and CD74 networks are active.
Author contributions
PPR and SMK designed the study. YQ performed experiments for RPPA. SMK and KRC performed statistical analysis and analysis of RPPA data. MA analyzed clinical data. CWH and AAQ performed cluster analysis. VRR performed qRT-PCR experiments and analysis. RLG and SEH performed immunoblot experiments and analysis. PPR performed String and Gene Ontology analysis. PPR and SMK co-wrote the manuscript which was approved by all authors.
Acknowledgements
We would like to thank Jairo Matthews for assistance in obtaining the clinical samples for the study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.05.025.
Contributor Information
Peter P. Ruvolo, Email: pruvolo@mdanderson.org.
Steven M. Kornblau, Email: skornblau@mdanderson.org.
Appendix A. Supplementary data
Supplementary material
References
- 1.Ruvolo P.P. Galectin 3 as a guardian of the tumor microenvironment. Biochim Biophys Acta. 2016;1863:427–437. doi: 10.1016/j.bbamcr.2015.08.008. [DOI] [PubMed] [Google Scholar]; Ruvolo PP. Galectin 3 as a guardian of the tumor microenvironment. Biochim Biophys Acta. 2016; 1863: 427-437. [DOI] [PubMed]
- 2.Ruvolo P.P. Galectins as regulators of cell survival in the leukemia niche. Adv Biol Regul. 2019;71:41–54. doi: 10.1016/j.jbior.2018.09.003. [DOI] [PubMed] [Google Scholar]; Ruvolo PP. Galectins as regulators of cell survival in the leukemia niche. Adv Biol Regul. 2019; 71: 41-54. [DOI] [PubMed]
- 3.Yamamoto-Sugitani M., Kuroda J. Galectin-3 (Gal-3) induced by leukemia microenvironment promotes drug resistance and bone marrow lodgment in chronic myelogenous leukemia. Proc Natl Acad Sci U S A. 2011;108:17468–17473. doi: 10.1073/pnas.1111138108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yamamoto-Sugitani M, Kuroda J, et al. Galectin-3 (Gal-3) induced by leukemia microenvironment promotes drug resistance and bone marrow lodgment in chronic myelogenous leukemia. Proc Natl Acad Sci U S A. 2011; 108: 17468-17473. [DOI] [PMC free article] [PubMed]
- 4.Nakayama R., Kuroda J., Taniyama N. Suppression of SERPINA1-albumin complex formation by galectin-3 overexpression leads to paracrine growth promotion of chronic myelogenous leukemia cells. Leuk Res. 2014;38:103–108. doi: 10.1016/j.leukres.2013.07.026. [DOI] [PubMed] [Google Scholar]; Nakayama R, Kuroda J, Taniyama N, et al. Suppression of SERPINA1-albumin complex formation by galectin-3 overexpression leads to paracrine growth promotion of chronic myelogenous leukemia cells. Leuk Res. 2014; 38:103-108. [DOI] [PubMed]
- 5.Hu K., Gu Y., Lou L. Galectin-3 mediates bone marrow microenvironment-induced drug resistance in acute leukemia cells via Wnt/ß-catenin signaling pathway. J Hematol Oncol. 2015:81. doi: 10.1186/s13045-014-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu K, Gu Y, Lou L, et al. Galectin-3 mediates bone marrow microenvironment-induced drug resistance in acute leukemia cells via Wnt/ß-catenin signaling pathway. J Hematol Oncol. 2015;81. [DOI] [PMC free article] [PubMed]
- 6.Ruvolo P.P., Ruvolo V.R., Burks J.K. Role of MSC-derived galectin 3 in the AML microenvironment. Biochim Biophys Acta. 2018;1865:959–969. doi: 10.1016/j.bbamcr.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ruvolo PP, Ruvolo VR, Burks JK, et al. Role of MSC-derived galectin 3 in the AML microenvironment. Biochim Biophys Acta. 2018; 1865: 959-969. [DOI] [PMC free article] [PubMed]
- 7.Akahani S., Nangia-Makker P., Inohara H. Galectin-3: a novel antiapoptotic molecule with a functional BH1 (NWGR) domain of Bcl-2 family. Cancer Res. 1997;57:5272–5276. [PubMed] [Google Scholar]; Akahani S, Nangia-Makker P, Inohara H, et al Galectin-3: a novel antiapoptotic molecule with a functional BH1 (NWGR) domain of Bcl-2 family. Cancer Res. 1997; 57: 5272-5276. [PubMed]
- 8.Harazono Y., Nakajima K., Raz A. Why anti-Bcl-2 clinical trials fail: a solution. Cancer Metastasis Rev. 2014;33:285–294. doi: 10.1007/s10555-013-9450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harazono Y, Nakajima K, Raz A. Why anti-Bcl-2 clinical trials fail: a solution. Cancer Metastasis Rev. 2014; 33: 285-294. [DOI] [PMC free article] [PubMed]
- 9.Nangia-Makker P., Hogan V., Raz A. Galectin-3 and cancer stemness. Glycobiology. 2018;28:172–181. doi: 10.1093/glycob/cwy001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nangia-Makker P, Hogan V, Raz A. Galectin-3 and cancer stemness. Glycobiology. 2018; 28: 172-181. [DOI] [PMC free article] [PubMed]
- 10.Shimura T., Takenaka Y., Tsutsumi S. Galectin-3, a novel binding partner of beta-catenin. Cancer Res. 2004;64:6363–6367. doi: 10.1158/0008-5472.CAN-04-1816. [DOI] [PubMed] [Google Scholar]; Shimura T, Takenaka Y, Tsutsumi S, et al Galectin-3, a novel binding partner of beta-catenin. Cancer Res. 2004; 64: 6363-6367. [DOI] [PubMed]
- 11.Shimura T., Takenaka Y., Fukumori T. Implication of galectin-3 in Wnt signaling. Cancer Res. 2005;65:3535–3537. doi: 10.1158/0008-5472.CAN-05-0104. [DOI] [PubMed] [Google Scholar]; Shimura T, Takenaka Y, Fukumori T, et al Implication of galectin-3 in Wnt signaling. Cancer Res. 2005; 65: 3535-3537. [DOI] [PubMed]
- 12.Song S., Mazurek N., Liu C. Galectin-3 mediates nuclear beta-catenin accumulation and Wnt signaling in human colon cancer cells by regulation of glycogen synthase kinase-3beta activity. Cancer Res. 2009;69:1343–1349. doi: 10.1158/0008-5472.CAN-08-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]; Song S, Mazurek N, Liu C, et al Galectin-3 mediates nuclear beta-catenin accumulation and Wnt signaling in human colon cancer cells by regulation of glycogen synthase kinase-3beta activity. Cancer Res. 2009; 69: 1343-1349. [DOI] [PMC free article] [PubMed]
- 13.Elad-Sfadia G., Haklai R., Balan E. Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J Biol Chem. 2004;279:34922–34930. doi: 10.1074/jbc.M312697200. [DOI] [PubMed] [Google Scholar]; Elad-Sfadia G, Haklai R, Balan E, et al. Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J Biol Chem. 2004; 279: 34922-34930. [DOI] [PubMed]
- 14.Song S., Ji B., Ramachandran V. Overexpressed galectin-3 in pancreatic cancer induces cell proliferation and invasion by binding Ras and activating Ras signaling. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042699. [DOI] [PMC free article] [PubMed] [Google Scholar]; Song S, Ji B, Ramachandran V, et al. Overexpressed galectin-3 in pancreatic cancer induces cell proliferation and invasion by binding Ras and activating Ras signaling. PLoS One. 2012; 7: e42699. [DOI] [PMC free article] [PubMed]
- 15.Ruvolo P.P., Ruvolo V.R., Benton C.B. Combination of galectin inhibitor GCS-100 and BH3 mimetics eliminates both p53 wild type and p53 null AML cells. Biochim Biophys Acta. 2016;1863:562–571. doi: 10.1016/j.bbamcr.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ruvolo PP, Ruvolo VR, Benton CB, et al. Combination of galectin inhibitor GCS-100 and BH3 mimetics eliminates both p53 wild type and p53 null AML cells. Biochim Biophys Acta. 2016; 1863: 562-571. [DOI] [PMC free article] [PubMed]
- 16.Streetly M.J., Maharaj L., Joel S. GCS-100, a novel galectin-3 antagonist, modulates MCL-1, NOXA, and cell cycle to induce myeloma cell death. Blood. 2010;115:3939–3948. doi: 10.1182/blood-2009-10-251660. [DOI] [PMC free article] [PubMed] [Google Scholar]; Streetly MJ, Maharaj L, Joel S, et al. GCS-100, a novel galectin-3 antagonist, modulates MCL-1, NOXA, and cell cycle to induce myeloma cell death. Blood. 2010; 115: 3939-3948. [DOI] [PMC free article] [PubMed]
- 17.Seguin L., Camargo M.F., Wettersten H.I. Galectin-3, a Druggable vulnerability for KRAS-addicted cancers. Cancer Discov. 2017;7:1464–1479. doi: 10.1158/2159-8290.CD-17-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]; Seguin L, Camargo MF, Wettersten HI, et al. Galectin-3, a Druggable Vulnerability for KRAS-Addicted Cancers. Cancer Discov. 2017; 7: 1464-1479. [DOI] [PMC free article] [PubMed]
- 18.Cheng C.L., Hou H.A., Lee M.C. Higher bone marrow LGALS3 expression is an independent unfavorable prognostic factor for overall survival in patients with acute myeloid leukemia. Blood. 2013;121:3172–3180. doi: 10.1182/blood-2012-07-443762. [DOI] [PubMed] [Google Scholar]; Cheng CL, Hou HA, Lee MC, et al. Higher bone marrow LGALS3 expression is an independent unfavorable prognostic factor for overall survival in patients with acute myeloid leukemia. Blood. 2013; 121: 3172-3180. [DOI] [PubMed]
- 19.Schröder B. The multifaceted roles of the invariant chain CD74--more than just a chaperone. Biochim Biophys Acta. 2016;1863:1269–1281. doi: 10.1016/j.bbamcr.2016.03.026. [DOI] [PubMed] [Google Scholar]; Schröder B. The multifaceted roles of the invariant chain CD74--More than just a chaperone. Biochim Biophys Acta. 2016; 1863: 1269-1281. [DOI] [PubMed]
- 20.Penticuff J.C., Woolbright B.L., Sielecki T.M. MIF family proteins in genitourinary cancer: tumorigenic roles and therapeutic potential. Nat Rev Urol. 2019 doi: 10.1038/s41585-019-0171-9. Mar 26. [DOI] [PubMed] [Google Scholar]; Penticuff JC, Woolbright BL, Sielecki TM, et al MIF family proteins in genitourinary cancer: tumorigenic roles and therapeutic potential. Nat Rev Urol. 2019 Mar 26. doi: 10.1038/s41585-019-0171-9 [DOI] [PubMed]
- 21.Jankauskas S.S., Wong D.W.L., Bucala R. Evolving complexity of MIF signaling. Cell Signal. 2019;57:76–88. doi: 10.1016/j.cellsig.2019.01.006. [DOI] [PubMed] [Google Scholar]; Jankauskas SS, Wong DWL, Bucala R, et al Evolving complexity of MIF signaling. Cell Signal. 2019; 57: 76-88. [DOI] [PubMed]
- 22.Leng L., Metz C.N., Fang Y. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]; Leng L, Metz CN, Fang Y, et al MIF signal transduction initiated by binding to CD74. J Exp Med. 2003; 197: 1467-1476. [DOI] [PMC free article] [PubMed]
- 23.Shi X., Leng L., Wang T. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shi X, Leng L, Wang T, et al CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006; 25: 595-606. [DOI] [PMC free article] [PubMed]
- 24.Lue H., Kapurniotu A., Fingerle-Rowson G. Rapid and transient activation of the ERK MAPK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell Signal. 2006;18:688–703. doi: 10.1016/j.cellsig.2005.06.013. [DOI] [PubMed] [Google Scholar]; Lue H, Kapurniotu A, Fingerle-Rowson G, et al Rapid and transient activation of the ERK MAPK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell Signal. 2006; 18: 688-703. [DOI] [PubMed]
- 25.Lue H., Thiele M., Franz J. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 2007;26:5046–5059. doi: 10.1038/sj.onc.1210318. [DOI] [PubMed] [Google Scholar]; Lue H, Thiele M, Franz J, et al Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 2007; 26: 5046-5059. [DOI] [PubMed]
- 26.Gore Y., Starlets D., Maharshak N. Macrophage migration inhibitory factor induces B cell survival by activation of a CD74-CD44 receptor complex. J Biol Chem. 2008;283:2784–2792. doi: 10.1074/jbc.M703265200. [DOI] [PubMed] [Google Scholar]; Gore Y, Starlets D, Maharshak N, et al Macrophage migration inhibitory factor induces B cell survival by activation of a CD74-CD44 receptor complex. J Biol Chem. 2008; 283: 2784-2792. [DOI] [PubMed]
- 27.Lue H., Dewor M., Leng L. Activation of the JNK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on CXCR4 and CD74. Cell Signal. 2011;23:135–144. doi: 10.1016/j.cellsig.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lue H, Dewor M, Leng L, et al Activation of the JNK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on CXCR4 and CD74. Cell Signal. 2011; 23: 135-144. [DOI] [PMC free article] [PubMed]
- 28.De R., Sarkar S., Mazumder S. Macrophage migration inhibitory factor regulates mitochondrial dynamics and cell growth of human cancer cell lines through CD74-NF-κB signaling. J Biol Chem. 2018;293:19740–19760. doi: 10.1074/jbc.RA118.003935. [DOI] [PMC free article] [PubMed] [Google Scholar]; De R, Sarkar S, Mazumder S, et al Macrophage migration inhibitory factor regulates mitochondrial dynamics and cell growth of human cancer cell lines through CD74-NF-κB signaling. J Biol Chem. 2018; 293: 19740-19760. [DOI] [PMC free article] [PubMed]
- 29.Stein R., Qu Z., Cardillo T.M. Antiproliferative activity of a humanized anti-CD74 monoclonal antibody, hLL1, on B-cell malignancies. Blood. 2004;104:3705–3711. doi: 10.1182/blood-2004-03-0890. [DOI] [PubMed] [Google Scholar]; Stein R, Qu Z, Cardillo TM, et al. Antiproliferative activity of a humanized anti-CD74 monoclonal antibody, hLL1, on B-cell malignancies. Blood. 2004; 104: 3705-3711. [DOI] [PubMed]
- 30.Burton J.D., Ely S., Reddy P.K. CD74 is expressed by multiple myeloma and is a promising target for therapy. Clin Cancer Res. 2004;10:6606–6611. doi: 10.1158/1078-0432.CCR-04-0182. [DOI] [PubMed] [Google Scholar]; Burton JD, Ely S, Reddy PK, et al. CD74 is expressed by multiple myeloma and is a promising target for therapy. Clin Cancer Res. 2004; 10: 6606-6611. [DOI] [PubMed]
- 31.Abdul-Aziz A.M., Shafat M.S., Mehta T.K. MIF-induced stromal PKCβ/IL8 is essential in human acute myeloid leukemia. Cancer Res. 2017;77:303–311. doi: 10.1158/0008-5472.CAN-16-1095. [DOI] [PubMed] [Google Scholar]; Abdul-Aziz AM, Shafat MS, Mehta TK, et al MIF-Induced Stromal PKCβ/IL8 Is Essential in Human Acute Myeloid Leukemia. Cancer Res. 2017; 77: 303-311. [DOI] [PubMed]
- 32.van Galen P., Hovestadt V., Ii Wadsworth. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176:1265–1281. doi: 10.1016/j.cell.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]; van Galen P, Hovestadt V, Wadsworth Ii et al Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell. 2019; 176: 1265-1281 [DOI] [PMC free article] [PubMed]
- 33.Kornblau S.M., Qutub A., Yao H. Proteomic profiling identifies distinct protein patterns in acute myelogenous leukemia CD34+CD38- stem-like cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078453. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kornblau SM, Qutub A, Yao H, et al. Proteomic profiling identifies distinct protein patterns in acute myelogenous leukemia CD34+CD38- stem-like cells. PLoS One. 2013; 8: e78453. [DOI] [PMC free article] [PubMed]
- 34.Ruvolo P.P., Qiu Y., Coombes K.R. Phosphorylation of GSK3α/β correlates with activation of AKT and is prognostic for poor overall survival in acute myeloid leukemia patients. BBA Clin. 2015;4:59–68. doi: 10.1016/j.bbacli.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ruvolo PP, Qiu Y, Coombes KR, et al. Phosphorylation of GSK3α/β correlates with activation of AKT and is prognostic for poor overall survival in acute myeloid leukemia patients. BBA Clin. 2015; 4: 59-68. [DOI] [PMC free article] [PubMed]
- 35.Hu C.W., Kornblau S.M., Slater J.H. Progeny clustering: a method to identify biological phenotypes. Sci Rep. 2015;5 doi: 10.1038/srep12894. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu CW, Kornblau SM, Slater JH, et al. Progeny Clustering: A Method to Identify Biological Phenotypes. Sci Rep. 2015; 5: 12894. [DOI] [PMC free article] [PubMed]
- 36.Hoff F.W., Hu C.W., Qutub A.A. Shining a light on cell signaling in leukemia through proteomics: relevance for the clinic. Expert Rev Proteomics. 2018;6:1–10. doi: 10.1080/14789450.2018.1487781. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hoff FW, Hu CW, Qutub AA, et al. Shining a light on cell signaling in leukemia through proteomics: relevance for the clinic. Expert Rev Proteomics. 2018; 6: 1-10. [DOI] [PMC free article] [PubMed]
- 37.Hu C.W., Qiu Y., Ligeralde A. A quantitative analysis of heterogeneities and hallmarks in acute myelogenous leukaemia. Nat Biomed Eng. 2019 Apr 15 doi: 10.1038/s41551-019-0387-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; . Hu CW, Qiu Y, Ligeralde A, et al. A quantitative analysis of heterogeneities and hallmarks in acute myelogenous leukaemia. Nat Biomed Eng. 2019 Apr 15. doi: 10.1038/s41551-019-0387-2. [DOI] [PMC free article] [PubMed]
- 38.Szklarczyk D., Morris J.H., Cook H. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]; Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45: D362-368. [DOI] [PMC free article] [PubMed]
- 39.Cancer Genome Atlas Research Network, Ley T.J., Miller C. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cancer Genome Atlas Research Network, Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013; 368: 2059-2074. [DOI] [PMC free article] [PubMed]
- 40.Cerami E., Gao J., Dogrusoz U. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cerami E, Gao J, Dogrusoz U, et al The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2: 401-404. [DOI] [PMC free article] [PubMed]
- 41.Gao J., Aksoy B.A., Dogrusoz U. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013; 6: pl1. [DOI] [PMC free article] [PubMed]
- 42.Schwartz V., Lue H., Kraemer S. A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett. 2009;583:2749–2757. doi: 10.1016/j.febslet.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schwartz V, Lue H, Kraemer S, et al. A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett. 2009; 583: 2749-2757. [DOI] [PMC free article] [PubMed]
- 43.Shi X., Leng L., Wang T. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shi X, Leng L, Wang T, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006; 25: 595-606. [DOI] [PMC free article] [PubMed]
- 44.Cole A., Wang Z., Coyaud E. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2015;27:864–876. doi: 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cole A, Wang Z, Coyaud E, et al. Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell. 2015; 27: 864-876. [DOI] [PMC free article] [PubMed]
- 45.Kojima K., Kornblau S.M., Ruvolo V. Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood. 2013;121:4166–4174. doi: 10.1182/blood-2012-08-447581. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kojima K, Kornblau SM, Ruvolo V, et al. Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood. 2013; 121: 4166-4174. [DOI] [PMC free article] [PubMed]
- 46.Piya S., Kornblau S.M., Ruvolo V.R. Atg7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood. 2016;128:1260–1269. doi: 10.1182/blood-2016-01-692244. [DOI] [PMC free article] [PubMed] [Google Scholar]; Piya S, Kornblau SM, Ruvolo VR, et al. Atg7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood. 2016; 128: 1260-1269. [DOI] [PMC free article] [PubMed]
- 47.Riffelmacher T., Simon A.K. Mechanistic roles of autophagy in hematopoietic differentiation. FEBS J. 2017;284:1008–1020. doi: 10.1111/febs.13962. [DOI] [PubMed] [Google Scholar]; Riffelmacher T, Simon AK. Mechanistic roles of autophagy in hematopoietic differentiation. FEBS J 2017; 284: 1008-1020. [DOI] [PubMed]
- 48.Chen X., Khambu B., Zhang H. Autophagy induced by calcium phosphate precipitates targets damaged endosomes. J Biol Chem. 2014;289:11162–11174. doi: 10.1074/jbc.M113.531855. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen X, Khambu B, Zhang H, et al. Autophagy induced by calcium phosphate precipitates targets damaged endosomes. J Biol Chem. 2014; 289: 11162-11174. [DOI] [PMC free article] [PubMed]
- 49.Ruvolo V.R., Karanjeet K.B., Schuster T.F. Role for PKC δ in Fenretinide-mediated apoptosis in lymphoid leukemia cells. J Signal Transduct. 2010;2010 doi: 10.1155/2010/584657. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ruvolo VR, Karanjeet KB, Schuster TF, et al. Role for PKC δ in Fenretinide-Mediated Apoptosis in Lymphoid Leukemia Cells. J Signal Transduct. 2010; 2010: 584657. [DOI] [PMC free article] [PubMed]
- 50.Basu A., Pal D. Two faces of protein kinase Cδ: the contrasting roles of PKCδ in cell survival and cell death. Sci World J. 2010;10:2272–2284. doi: 10.1100/tsw.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]; Basu A, Pal D. Two faces of protein kinase Cδ: the contrasting roles of PKCδ in cell survival and cell death. Scientific World Journal. 2010; 10: 2272-2284. [DOI] [PMC free article] [PubMed]
- 51.Emoto Y., Manome Y., Meinhardt G. Proteolytic activation of protein kinase C delta by an ICE-like protease in apoptotic cells. EMBO J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Emoto Y, Manome Y, Meinhardt G, et al. Proteolytic activation of protein kinase C delta by an ICE-like protease in apoptotic cells. EMBO J. 1995; 14: 6148-6156. [DOI] [PMC free article] [PubMed]
- 52.Kinehara M., Kawamura S., Tateyama D. Protein kinase C regulates human pluripotent stem cell self-renewal. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054122. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kinehara M, Kawamura S, Tateyama D, et al. Protein kinase C regulates human pluripotent stem cell self-renewal. PLoS One. 2013; 8: e54122. [DOI] [PMC free article] [PubMed]
- 53.Ruvolo P.P., Qui Y.H., Coombes K.R. Low expression of PP2A regulatory subunit B55α is associated with T308 phosphorylation of AKT and shorter complete remission duration in acute myeloid leukemia patients. Leukemia. 2011;25:1711–1717. doi: 10.1038/leu.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ruvolo PP, Qui YH, Coombes KR, et al. Low expression of PP2A regulatory subunit B55α is associated with T308 phosphorylation of AKT and shorter complete remission duration in acute myeloid leukemia patients. Leukemia. 2011; 25:1711-1717. [DOI] [PMC free article] [PubMed]
- 54.Lakshminarayan R., Wunder C., Becken U. Galectin-3 drives glycosphingolipid-dependent biogenesis of clathrin-independent carriers. Nat Cell Biol. 2014;16:595–606. doi: 10.1038/ncb2970. [DOI] [PubMed] [Google Scholar]; Lakshminarayan R, Wunder C, Becken U, et al. Galectin-3 drives glycosphingolipid-dependent biogenesis of clathrin-independent carriers. Nat Cell Biol. 2014; 16: 595-606. [DOI] [PubMed]
- 55.Farhad M., Rolig A.S., Redmond W.L. The role of Galectin-3 in modulating tumor growth and immunosuppression within the tumor microenvironment. Oncoimmunology. 2018;7 doi: 10.1080/2162402X.2018.1434467. [DOI] [PMC free article] [PubMed] [Google Scholar]; Farhad M, Rolig AS, Redmond WL. The role of Galectin-3 in modulating tumor growth and immunosuppression within the tumor microenvironment. Oncoimmunology. 2018; 7: e1434467. [DOI] [PMC free article] [PubMed]
- 56.Kornblau S.M., Ruvolo P.P., Wang R.Y. Distinct protein signatures of acute myeloid leukemia bone marrow-derived stromal cells are prognostic for patient survival. Haematologica. 2018;103:810–821. doi: 10.3324/haematol.2017.172429. [DOI] [PMC free article] [PubMed] [Google Scholar]; . Kornblau SM, Ruvolo PP, Wang RY, et al Distinct protein signatures of acute myeloid leukemia bone marrow-derived stromal cells are prognostic for patient survival. Haematologica. 2018; 103: 810-821. [DOI] [PMC free article] [PubMed]
- 57.Zhu C., Anderson A.C., Schubart A. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]; . Zhu C, Anderson AC, Schubart A, et al The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005; 6: 1245-1252. [DOI] [PubMed]
- 58.Wu C., Thalhamer T., Franca R.F. Galectin-9-CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity. 2014;41:270–282. doi: 10.1016/j.immuni.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu C, Thalhamer T, Franca RF, et al Galectin-9-CD44 interaction enhances stability and function of adaptive regulatory T cells. Immunity. 2014; 41: 270-282. [DOI] [PMC free article] [PubMed]
- 59.Zhao Q., Guo X., Nash G.B. Circulating galectin-3 promotes metastasis by modifying MUC1 localization on cancer cell surface. Cancer Res. 2009;69:6799–6806. doi: 10.1158/0008-5472.CAN-09-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhao Q, Guo X, Nash GB, et al Circulating galectin-3 promotes metastasis by modifying MUC1 localization on cancer cell surface. Cancer Res. 2009; 69: 6799-6806. [DOI] [PMC free article] [PubMed]
- 60.Nobumoto A., Nagahara K., Oomizu S. Galectin-9 suppresses tumor metastasis by blocking adhesion to endothelium and extracellular matrices. Glycobiology. 2008;18:735–744. doi: 10.1093/glycob/cwn062. [DOI] [PubMed] [Google Scholar]; Nobumoto A, Nagahara K, Oomizu S, et al Galectin-9 suppresses tumor metastasis by blocking adhesion to endothelium and extracellular matrices. Glycobiology. 2008;18: 735-744. [DOI] [PubMed]
- 61.Xiao P., Sandhow L., Heshmati Y. Distinct roles of mesenchymal stem and progenitor cells during the development of acute myeloid leukemia in mice. Blood Adv. 2018;2:1480–1494. doi: 10.1182/bloodadvances.2017013870. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xiao P, Sandhow L, Heshmati Y, et al. Distinct roles of mesenchymal stem and progenitor cells during the development of acute myeloid leukemia in mice. Blood Adv. 2018; 2: 1480-1494. [DOI] [PMC free article] [PubMed]
- 62.Kazanecki C.C., Uzwiak D.J., Denhardt D.T. Control of osteopontin signaling and function by post-translational phosphorylation and protein folding. J Cell Biochem. 2007;102:912–924. doi: 10.1002/jcb.21558. [DOI] [PubMed] [Google Scholar]; Kazanecki CC, Uzwiak DJ, Denhardt DT. Control of osteopontin signaling and function by post-translational phosphorylation and protein folding. J Cell Biochem. 2007; 102: 912-924. [DOI] [PubMed]
- 63.Berkova Z., Tao R.H., Samaniego F. Milatuzumab - a promising new immunotherapeutic agent. Expert Opin Investig Drugs. 2010;19:141–149. doi: 10.1517/13543780903463854. [DOI] [PubMed] [Google Scholar]; Berkova Z, Tao RH, Samaniego F. Milatuzumab - a promising new immunotherapeutic agent. Expert Opin Investig Drugs. 2010; 19: 141-149. [DOI] [PubMed]
- 64.Harrison S.A., Dennis A., Fiore M.M. Utility and variability of three non-invasive liver fibrosis imaging modalities to evaluate efficacy of GR-MD-02 in subjects with NASH and bridging fibrosis during a phase-2 randomized clinical trial. PLoS One. 2018;13 doi: 10.1371/journal.pone.0203054. [DOI] [PMC free article] [PubMed] [Google Scholar]; Harrison SA, Dennis A, Fiore MM, et al Utility and variability of three non-invasive liver fibrosis imaging modalities to evaluate efficacy of GR-MD-02 in subjects with NASH and bridging fibrosis during a phase-2 randomized clinical trial. PLoS One. 2018; 13: e0203054 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material