Abstract
Background
Emerging evidence suggests that metabolic alterations are a hallmark of cancer cells and contribute to tumor initiation and development. Cancer cells primarily utilize aerobic glycolysis (the Warburg effect) to produce energy and support anabolic growth. The type Iγ phosphatidylinositol phosphate kinase (PIPKIγ) is profoundly implicated in tumorigenesis, however, little is known about its role in reprogrammed energy metabolism.
Methods
Loss- and gain-of-function studies were applied to determine the oncogenic roles of PIPKIγ in colorectal cancer. Transcriptome analysis, real-time qPCR, immunohistochemical staining, Western blotting, and metabolic analysis were carried out to uncover the cellular mechanism of PIPKIγ.
Findings
In this study, we showed that PIPKIγ was frequently upregulated in colorectal cancer and predicted a poor prognosis. Genetic silencing of pan-PIPKIγ suppressed cell proliferation and aerobic glycolysis of colorectal cancer. In contrast, the opposite effects were observed by overexpression of PIPKIγ_i2. Importantly, PIPKIγ-induced prolific effect was largely glycolysis-dependent. Mechanistically, PIPKIγ facilitated activation of PI3K/Akt/mTOR signaling pathways to upregulate c-Myc and HIF1α levels, which regulate expression of glycolytic enzymes to enhance glycolysis. Moreover, pharmacological inhibition by PIPKIγ activity with the specific inhibitor UNC3230 significantly inhibited colorectal cancer glycolysis and tumor growth.
Interpretation
Our findings reveal a new regulatory role of PIPKIγ in Warburg effect and provide a key contributor in colorectal cancer metabolism with potential therapeutic potentials.
Fund
National Key Research and Development Program of China, Outstanding Clinical Discipline Project of Shanghai Pudong, Natural Science Foundation of China, and Science and Technology Commission of Shanghai Municipality.
Keywords: Colorectal cancer, Warburg effect, Phosphatidylinositol kinase, PIPKIγ, Tumor growth
Research in context.
Evidence before this study
Reprogramming metabolism is emerged as a hallmark of cancer. Warburg effect, also known as aerobic glycolysis, can support uncontrolled proliferation of cancer cells by providing abundant cellular buildings. Increased glycolysis contributes to nearly all aspects of the malignant characters of cancer cells.
Added value of this study
This study showed that PIPKIγ is profoundly implicated in colorectal cancer cell proliferation and aerobic glycolysis. PIPKIγ enhances Warburg effect by upregulation of c-Myc and HIF1α levels via activation of PI3K/Akt/mTOR signaling pathways. Pharmacological inhibition of PIPKIγ significantly suppressed tumor growth in vivo.
Implications of all the available evidence
This finding suggests that PIPKIγ is a critical glycolysis modulator and provide a potential target for anti-tumor therapy for colorectal cancer.
Alt-text: Unlabelled Box
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and ranks second in terms of mortality worldwide. Over 1.8 million new colorectal cancer cases and 881,000 deaths are estimated to occur in 2018, accounting for about 10% cancer cases and deaths [1]. Because increased early detection and application of colonoscopy with polypectomy, the clinical outcome of CRC patients has significantly improved during the past decades in many countries [2]. Surgical resection is the primary treatment option for CRC, but even with complete resection the tumor will be eventually recurred and developed to metastatic disease in many of these patients [3]. Therefore, the long-term survival outlook of CRC is still poor and highlights the development of more effective therapies for this life-threatening disease.
The type I phosphatidylinositol 4-phosphate 5-kinases (PIPKIs) are a family of enzymes that catalyze ATP-dependent phosphorylation of phosphatidylinositol 4-phosphate to generate phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. PI(4,5)P2 is subsequently converted into phosphatidylinositol (3,4,5)-trisphosphate [PI(3,4,5)P3] by PI3K. PI(4,5)P2 and PI(3,4,5)P3 are involved in a variety of cellular processes, such as vesicular trafficking, focal adhesion assembly, actin polymerization, endocytosis and agonist-induced calcium signaling [[4], [5], [6]]. PIPKIs comprise a family encoded by three genes that give rise to PIP kinase type Iα (PIPKIα), PIPKIβ and PIPKIγ [7]. In mammalian cells, the different isoforms of PIPKI share very conserved kinase domain but have a high level of sequence divergence at the C-terminus, which allows for their distinct localization and function via interactions with specific binding partners [8,9]. For example, talin recruits PIPKIγ to focal adhesions and the site-specific generation of PI(4,5)P2 enhances talin binding to β1-integrin [10,11]. Previously, many reports have revealed the important role of PIPKIγ in multiple oncogenic processes [12,13]. In breast cancer, PIPKIγ and talin couple phosphoinositide and adhesion signaling to control the epithelial to mesenchymal transition process [14]. In addition, PIPKIγ can regulate β-catenin nuclear importation and transcriptional activity to promote breast cancer malignant phenotypes [15]. In colorectal cancer, PIPKIγ positively regulates focal adhesion dynamics and cancer cell invasion [16]. In pancreatic cancer, we revealed that PIPKIγ, functioning downstream of EGFR signaling, is critical to the tumor growth and metastasis [17]. This indicates that the versatile roles of PIPKIγ during tumorigenesis.
Different form normal cells, cancer cells ferment glucose to lactate even in the presence of sufficient levels of oxygen [18]. This phenomenon is known as the Warburg effect or aerobic glycolysis. Warburg effect can provide cancer cell intermediary glucose metabolites to generate cellular buildings and reducing equivalents (such as NADPH) for rapid proliferation and avoiding apoptosis [19,20]. Notably, increased glycolysis is a common phenomenon in human cancers and correlates multiple tumorigenic phenotypes. Suppression of aerobic glycolysis can profoundly reduce tumourigenicity and improve chemosensitivity indicative of the importance of glycolysis to cancer cells [21]. Emerging evidences suggest that activation of oncogenes or inactivation of tumor suppressors contribute to elevated Warburg effect, such as MYC and TP53 [[22], [23], [24]]. Therefore, a comprehensive understanding of the links between glycolysis and CRC pathogenesis is of paramount significance for the development of new therapeutic agents in colorectal cancer.
In this study, we found that PIPKIγ is substantially upregulated in colorectal cancer cells and tumor tissues. Genetic silencing of pan-PIPKIγ inhibited the in vitro cell proliferation and in vivo tumor growth of CRC. Subsequently, transcriptomic data supported a novel regulatory role of PIPKIγ in glycolysis by activation of PI3K/AKT/mTOR/c-Myc-HIF1α axis. Finally, we showed that pharmacological inhibition of PIPKIγ with UNC3230 suppressed CRC glycolysis and xenograft tumor growth. Therefore, our findings define PIPKIγ as an important glycolysis regulator in CRC and suggest PIPKIγ as a promising therapeutic target for the clinical management of CRC.
2. Materials and methods
2.1. Cell culture and reagent
Human colorectal cancer cell lines LOVO, Caco-2, SW620, and SW480 were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Science (Shanghai, China); LS174T, HCT116, COLO205, and the normal colonic epithelial cell NCM460 were derived from the American Tissue Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in DMEM (Dulbecco's modified Eagle's medium, Gibco) or RPMI 1640 medium (Gibco, USA) medium supplemented with 10% fetal bovine serum (FBS, Gibco, USA), and the antibiotics penicillin (500 units/mL) and streptomycin (200 μg/mL). Cells were maintained at a humidified incubator with 5% CO2 atmosphere. 2-Deoxy-d-glucose (2-DG, D8375) and galactose (G0750) were purchased from Sigma (Shanghai, China). The specific inhibitor of PIPKIγ (UNC3230) was purchased from Tocris Bioscience (#5271/10, USA).
2.2. Generation of stable PIPKIγ knockdown cells
Lentiviral shRNA negative control and shRNA oligonucleotides targeting human PIPKIγ listed below were designed and synthesized by Genepharma (Shanghai, China). The sequences for the PIPKIγ shRNA were: sh-1, 5’-TGCGACGACGAGTTCATCATCATTCAAGAGATGATGATGAACTCGTCGTCGCTTTTTTC-3′; sh-2, 5’-TGCCTGGTCCTGGAAAGTTTCATTCAAGAGATGAAACTTTCCAGGACCAGGCTTTTTTC-3′; and sh-Ctrl, 5’-TGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTC-3′. The lenti-virius LV2 (pGLVU6/Puro) plasmids were transfected into human embryonic kidney 293 T cells using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer's manual. Colorectal cancer cells were then infected with lentivirus medium from the packaging cells 48 h after transfection in the presence of 8 μg/mL polybrene. After infection overnight, virus-containing medium was replaced by normal culture medium. The stable clones were selected with 2 μg/mL puromycin. Expression levels of PIPKIγ were confirmed by Western blotting analysis.
2.3. Transfection
For overexpression of PIPKIγ, the whole sequence of PIPKIγ_i2, mPIPKIγ_i2–1 and mPIPKIγ_i2–2 was synthesized by Genepharma (Shanghai, China) and then subcloned to the pcDNA3.1 plasmid. Six silent mutations were introduced in the sh-PIPKIγ target sequence in order to make it resistant to corresponding shRNA. The siRNA sequences for PIP4K2C were: si-1 sense, 5’-CCAGUCAUUUCAAGUUCAATT-3′; si-1 antisense, 5’-UUGAACUUGAAAUGACUGGTT-3′; si-2 sense, 5’-CCAACUAUCACCAGUACAUTT-3′; si-2 antisense, 5′- AUGUACUGGUGAUAGUUGGTT-3′; scrambled siRNA targeting no known gene sequence was used as the negative control. For transient expression of exogenous genes, 5 × 105 cells were plated in each well of 6-well plates and transfection was performed using with XtremeGENE 9 (Roche, USA) following the manufacturer's instructions.
2.4. Gene expression microarrays
For RNA preparation, total RNA from sh-Ctrl and sh-PIPKIγ cells were extracted according to standard protocol. RNA was processed and profiled on whole human Genome Microarray (4 × 44 K, Agilent) as recommended by the manufacturer. Background subtraction and normalization of probe set intensities were performed using Robust Multiarray Analysis (RMA). The microarray data is available at GEO database GSE130761. Gene set enrichment analysis (GSEA) was performed on the Broad Institute Platform and statistical significance (false discovery rate, FDR) was set at 0.25.
2.5. Real-time quantitative PCR
Total RNA was extracted from indicated cells using Qiagen RNeasy kits (Qiagen Valencia, CA). RNA quality and quantity were determined using Nanodrop™ spectrophotometer (NanoDrop products, Wilmington, CA). Next, 1 μg of total RNA was reverse transcribed using a First Strand cDNA Synthesis Kit (ThermoFisher Scientific) to synthesize complementary DNA (cDNA). Subsequently, the cDNA product was subjected to PCR amplification on 7500 real-time System (Applied Biosystems) to analyze the expression of mRNA; β-actin was used as an internal control. The PCR primers sequences used in this study are shown in supplementary Table 1. Relative quantification was performed using the comparative 2–ΔΔCt method.
2.6. Immunohistochemical analysis
The colorectal cancer tissue microarray used in this study was purchased from Zhuoli Biotech (#COC1504, http://www.zhuolibiotech.com/, Shanghai, China). For immunohistochemical analysis, paraffin-embedded tissue sections were deparaffinized in xylene and rehydrated through descending concentrations of ethanol. Then antigen retrieval was performed by boiling in 10 mM citrate buffer (pH 6.0) for 10 min, followed by treatment with 3% hydrogen peroxide to block endogenous peroxidase. After washing with 1 × PBS for three times, the sections were incubated with a primary antibody overnight at 4 °C. The antibodies used for immunohistochemistry were listed as follows: PIPKIγ (1: 200, Proteintech, 27,640-1-AP), PCNA (1:5000, Cell Signaling Technology, #13110), HIF1α (1: 300, Abcam, ab113642), c-Myc (1: 500, Abcam, ab32072), GLUT1 (1:200, Proteintech, 21,829-1-AP), LDHA (1:200, Proteintech, 19,987-1-AP), and PDK1 (1:200, Proteintech, 10,026-1-AP). The next day, the HRP-conjugated secondary antibody was added for 45 min at room temperature. The immunoreactivity was developed by 3,3′-diaminobenzidine (DAB). Finally, the sections were counterstained with hematoxylin and scoring was evaluated by two investigators blinded to the clinical information.
2.7. Western blot analysis
Cell lystates were separated using SDS-PAGE and then electrophoretically transferred onto PVDF membranes. After blocking with 5% defatted milk for 1 h at room temperature, the PVDF membranes were incubated with primary antibodies overnight at 4 °C, followed by incubation with HRP-conjugated secondary antibodies at room temperature for 45 min. β-actin antibody was used as loading control. Immunoblots were developed using the Pierce™ Western ECL Blotting substrate (ThermoFisher Scientific, Waltham, MA) and ChemiDoc Touch image system (Bio-Rad). The antibodies used were listed as follows: PIPKIγ (1: 2000, Abcam, ab109192), p-Akt (1:2000, Cell Signaling Technology, #4060), Akt (1:1000, Cell Signaling Technology, #4685), p-mTOR (1:1000, Cell Signaling Technology, #2971), mTOR (1:1000, Cell Signaling Technology, #2983), p-S6K (1:1000, Cell Signaling Technology, #9204), S6K (1:1000, Cell Signaling Technology, #9202), HIF1α (1: 1000, Abcam, ab113642), c-Myc (1: 1000, Abcam, ab32072), and β-actin (1:1000, Abcam, ab8227).
2.8. Measurement of glucose and lactate
Colorectal cancer cells with indicated genetic manipulations were cultured at normal condition for 2 days and culture medium was collected. Lactate production was measured using a commercial Lactate Assay Kit (Sigma) according to the manufacturer's protocol. For glucose uptake assay, the culture medium was replaced with phenol-red free DMEM with 10% FBS in continuous culture for 2 days. Glucose levels in the culture medium were measured using a Colorimetric Glucose Assay Kit (BioVision) as recommended by the manufacturer. All values were normalized on the basis of the total protein level (BCA protein assay, Thermo Fisher Scientific, USA).
2.9. Measurement of extracellular acidification rate (ECAR) and oxygen consumption ratio (OCR)
In vitro real-time ECAR and OCR was monitored with the Seahorse XF96 Flux Analyser (Seahorse Bioscience) in accordance to the manufacturer's instructions. In brief, colorectal cancer cells were seeded at a density of 2–3 × 104 per well. For ECAR assay, colorectal cancer cells were pre-incubated with unbuffered media for 1 h, followed by a sequential injection of 10 mM glucose, 1 mM oligomycin and 80 mM 2-DG to detect ECAR. For measurement of mitochondrial respiration, OCR was assessed by sequential injection of 1 mM oligomycin, carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, Sigma-Aldrich, C2920) and 2 mM antimycin A and rotenone (Sigma-Aldrich). The final output data were normalized by cell number or total protein level as demonstrated by BCA assay.
2.10. Cell proliferation and colony formation assay
Cell counting kit-8 (CCK-8, Dojindo Molecular Technologies, Japan) assay was performed to measure cell proliferation. Briefly, 2 × 103 cells were seeded and cultured in 96-well plates for 6 days. CCK-8 assay was carried out everyday according to the manufacturer's protocol. All experiments were performed independently in triplicate. Absorbance was recorded at 450 nm using a microplate reader. For colony formation assay, colorectal cancer cells were seeded in 6-well plate at a density of 1 × 103 cells per well. After continuous culture for 12–14 days, colonies formed were stained with 0.1% crystal violet. Each experiment was performed in triplicate and repeated twice.
2.11. Tumor xenograft experiment
Pathogen-free female athymic nude mice (5 weeks old, 18–20 g weigh) used in this study were managed at SPF Laboratory Animal Center in full accordance with the guidelines by the U.S. National Institutes of Health Guide for the Care and Use of Laboratory Animals. Tumor cells (SW480, 1 × 106) were injected subcutaneously into the right scapular region of nude. The tumor size was monitored every week, and the volume was calculated with the following formula: Volume = (width2 × length)/2. At the termination of the experiment, mice were sacrificed, and tumors were harvested, weighed, and fixed in formalin and embedded in paraffin or directly stored at −80 °C. For the pharmacological inhibition assay, mice were randomly divided into two groups when bore visible tumors (200 mm3); in the test group, mice were intraperitoneally injected with 5 mg/kg UNC3230 three times a week for three weeks; in the control group, mice were treated with saline containing 0.01% DMSO. All the animal studies were approved by the Animal Care and Use Committee of Shanghai East Hospital, Tongji University School of Medicine.
2.12. Statistical analysis
Results were presented in the form of means ± standard deviation (SD). Group difference was assessed using one-way ANOVA (SPSS 23.0) or the Student t-test (two-tailed). A two-sided p-value of <0.05 was considered statistically significant. *P < .05, **P < .01 and ***P < .001.
3. Results
3.1. PIPKIγ is highly expressed in colorectal cancer and predicts a poor prognosis
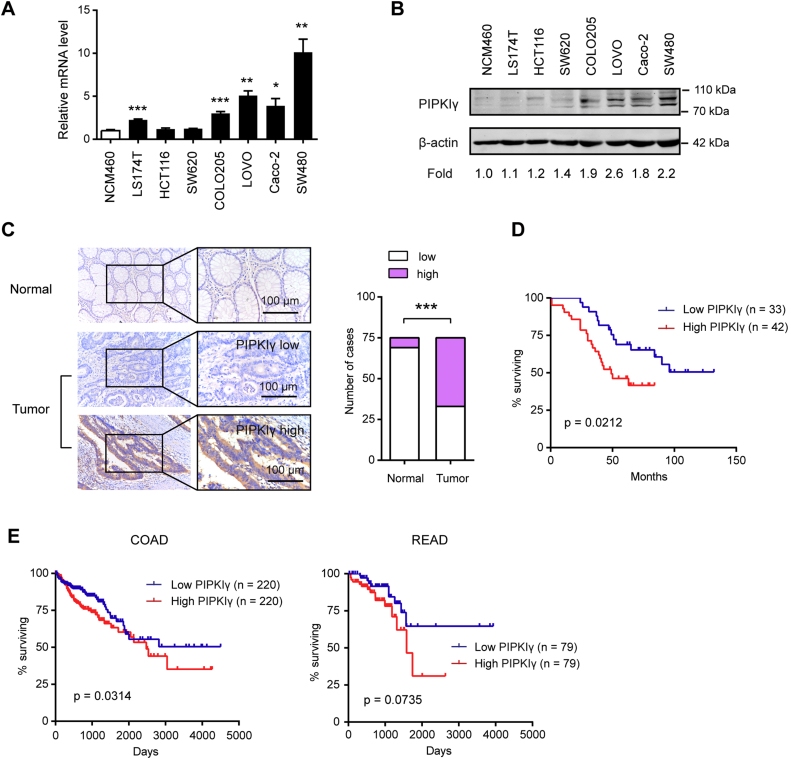
To determine the expression profile of PIPKIγ in colorectal cancer, real-time qPCR and Western blotting analysis were performed in colorectal cancer cell lines and the normal colonic epithelial cell NCM460. As shown in Fig. 1A and B, PIPKIγ mRNA and protein level were frequently overexpressed in colorectal cancer cell lines compared with the normal epithelial cell. Then, immunohistochemical analysis of a tissue microarray containing 75 matched tumor and non-tumor tissues were carried out to comprehensively characterize PIPKIγ expression in colorectal cancer tissues. The result showed that PIPKIγ was more highly expressed in colorectal cancer tissues compared to corresponding non-tumor tissues (Fig. 1C). Moreover, Kaplan-Meier curves showed that high PIPKIγ level was correlated with a reduced overall survival in colorectal cancer patients (Fig. 1D). Similar prognostic value of PIPKIγ was also conformed in colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) patients derived from the Cancer Genome Atlas (TCGA) cohort (Fig. 1E). Collectively, these data above suggested that PIPKIγ may act as oncogene in colorectal cancer tumorigenesis and progression.
Fig. 1.
High PIPKIγ expression level predicts a poor clinical outcome in colorectal cancer. (A) Real-time qPCR analysis of the mRNA level of PIPKIγ in colorectal cancer cell lines and the normal colonic epithelial cell NCM460. (B) Cell lysates of indicated cells were collected for immunoblotting analyses using PIPKIγ antibody; β-actin was loaded as a control. (C) IHC analysis was performed in a tissue microarray containing 76 matched tumor and non-tumor colorectal cancer tissues. Representative images of PIPKIγ and its expression intensity in non-tumor and tumor tissues were shown. Scale bar: 100 μm. (D) Kaplan-Meier analyses of overall survival of individuals with colorectal cancer based on PIPKIγ protein expression level. (E) Kaplan-Meier analyses of overall survival in colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) patients in the Cancer Genome Atlas (TCGA) cohort. The patients were dichotomously categorized on the basis of median PIPKIγ value into 2 groups. Subgroups were compared with the use of the log-rank test. *P < .05; **P < .01; ***P < .001.
3.2. PIPKIγ promotes tumor growth in colorectal cancer
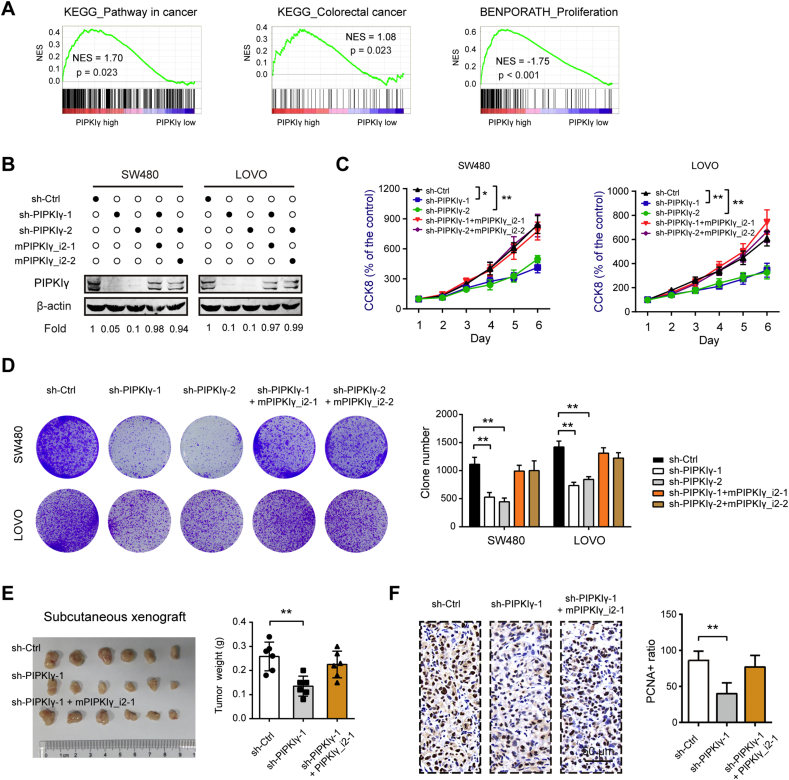
Next, COAD samples derived from TCGA cohort was stratified into 2 groups (high versus low) based on median PIPKIγ value. Gene set enrichment analysis (GSEA) revealed that PIPKIγ was profoundly implicated in several oncogenic pathways, such as KEGG_Pathway in cancer, KEGG_Colorectal cacer, and BENPORATH_Proliferation (Fig. 2A). To confirm the oncogenic roles of PIPKIγ in colorectal cancer, loss-of-function studies were performed in two cell lines, SW480 and LOVO, which had higher intrinsic PIPKIγ protein level. As shown in Fig. 2B, two specific shRNA targeting pan-PIPKIγ led to significant reduction in PIPKIγ protein. By CCK-8 assay (Fig. 2C) and colony formation assay (Fig. 2D), we found that silencing of PIPKIγ significantly inhibited the cell proliferation of colorectal cancer cells. To confirm the specific role of PIPKIγ in colorectal cancer, PIPKIγ knockdown was rescued by expressing a PIP5K1C mRNA made resistant to the shRNA by six silent mutations (mPIPKIγ) and coding for a wild-type PIPKIγ_i2 protein. Western blotting analysis demonstrated that re-expression of mPIPKIγ_i2 completely restored the protein level of PIPKIγ (Fig. 2B). As expected, mPIPKIγ_i2 largely blocked the growth-inhibiting effect induced by PIPKIγ knockdown (Fig. 2C, D). Furthermore, gain-of-function studies revealed that PIPKIγ_i2 overexpression dramatically enhanced colorectal cancer cell proliferation in vitro (Supplementary Fig. 1). Using the subcutaneous xenograft model, we demonstrated that silencing of pan-PIPKIγ in SW480 cells remarkably suppressed its tumor-forming capacity, which can be restored by re-expression of mPIPKIγ_i2 (Fig. 2E). Consistent with of the tumor-promoting role of PIPKIγ, PCNA staining revealed that PIPKIγ knockdown decreased tumor cell proliferation in vivo (Fig. 2F). Taken together, our results strongly suggested that in colorectal cells, PIPKIγ plausibly participates in the regulation of tumor growth.
Fig. 2.
PIPKIγ promotes colorectal cancer cell proliferation in vitro and tumor growth in vivo. (A) COAD samples derived from TCGA cohort was categorized into 2 groups (high versus low) based on median PIPKIγ value. Gene set enrichment analysis (GSEA) was performed to discover the difference between 2 groups. False discovery rate (FDR) was set at 0.25. NES represents normalized enrichment score. (B) Validation of pan-PIPKIγ knockdown and ectopic expression of mutant-PIPKIγ_i2 (resistant to PIPKIγ shRNA) in SW480 and LOVO cells using Western blotting. (C, D) The influence of PIPKIγ on colorectal cancer in vitro cell proliferation was determined by CCK-8 (C) and colony formation (D) assays, respectively. (E) sh-Ctrl, sh-PIPKIγ-1, and sh-PIPKIγ-1 + mPIPKIγ1_i2 SW480 cells were injected subcutaneously into the left forelimb of nude mice (n = 6 per group). Four weeks later, mice were sacrificed and tumor weights in each group were shown. (F) IHC analysis of PIPKIγ and PCNA expression from indicated subcutaneous xenograft. *P < .05 and **P < .01.
3.3. PIPKIγ regulates glycolysis in colorectal cancer
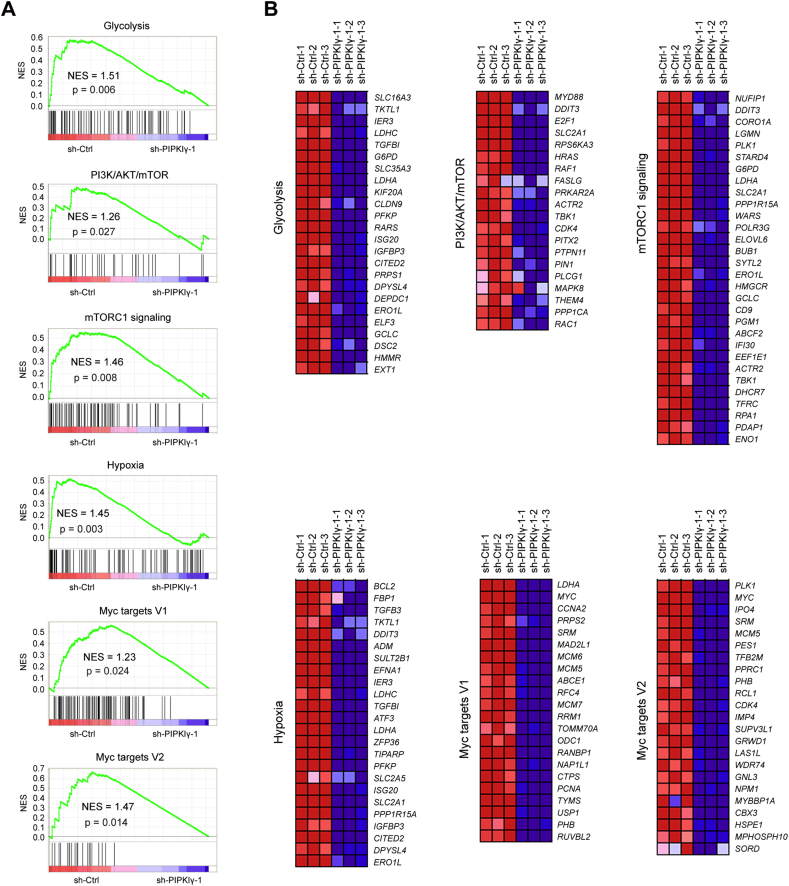
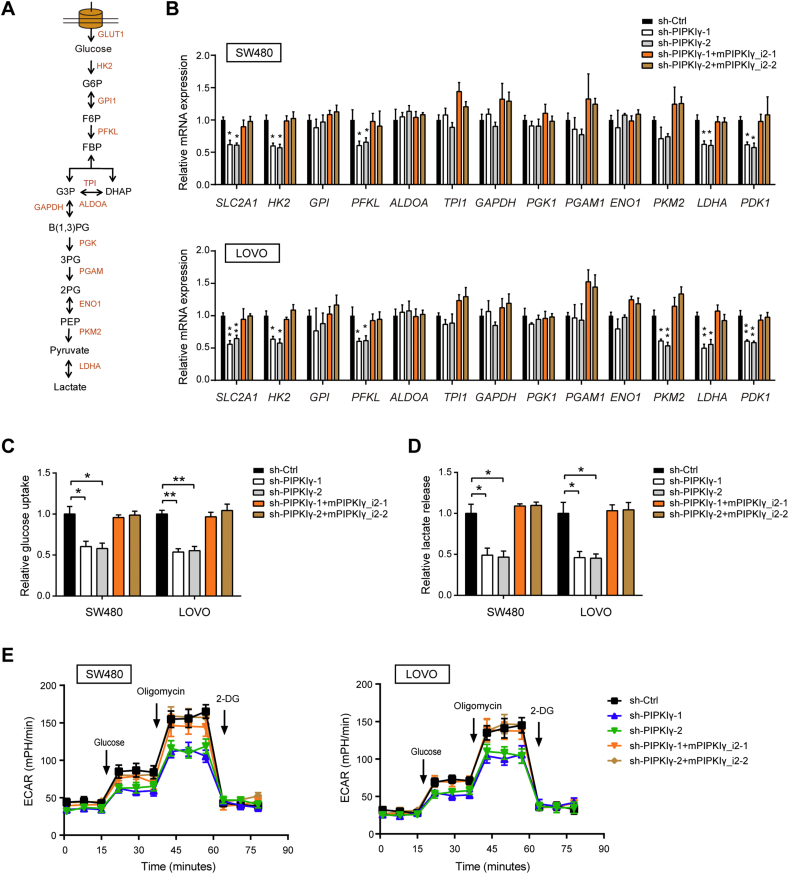
To elucidate the mechanism by which PIPKIγ promotes tumor growth, an Agilent gene expression microarray was used to determine the transcriptional changes after PIPKIγ knockdown. Gene set enrichment analysis showed that the gene sets related to glycolysis, PI3K/Akt/mTOR, mTORC1 signaling, hypoxia, Myc targets V1, and Myc targets V2 negatively correlated with PIPKIγ downregulation in SW480 cells (Fig. 3A). The top-scoring genes altered in the six gene sets included many glycolysis-related genes, such as SLC16A3, SLC2A1, PFKP, LDHA, and TKTL1, indicating that loss of PIPKIγ contributes to weakened glycolysis (Fig. 3B). To interrogate the potential regulatory role of PIPKIγ in tumor glucose metabolism, several experiments were performed to fully characterize metabolic alterations after PIPKIγ knockdown. Firstly, real-time qPCR analysis of glycolytic genes showed that SLC2A1, HK2, PFKL, PKM2, LDHA, and PDK1 were significantly downregualted by silencing of PIPKIγ (Fig. 4A and B); IHC analysis showed that PIPKIγ knockdown led to remarkable reduction in GLUT1, LDHA, and PDK1 protein expression in tumor tissues (Supplementary Fig. 2). Secondly, measurement of glucose and lactate in the cell culture medium demonstrated that PIPKIγ knockdown led to pronounced drop in glucose uptake and lactate production (Fig. 4C and D). Finally, the Seahorse XF96 Flux Analyser revealed that PIPKIγ knockdown decreased extracellular acidification rate (ECAR) with minimal implications to oxygen consumption ratio (OCR), suggesting that PIPKIγ mainly induces significant alterations to glycolysis but not TCA cycle (Fig. 4E and Supplementary Fig. 3). In line with the function of PIPKIγ in the regulation of cell proliferation, the decreased glycolytic metabolism induced by silencing of PIPKIγ can be completely restored by mPIPKIγ_i2. Moreover, overexpression of PIPKIγ significantly promoted the glycolytic activity of colorectal cancer cells as demonstrated by elevated glycolytic genes, increased glucose uptake and lactate production, and upregulated ECAR (Supplementary Fig. 4). Thus, these results strongly supported that PIPKIγ is involved in the Warburg effect of colorectal cancer cells.
Fig. 3.
Transcriptional changes induced by PIPKIγ knockdown. (A) GSEA plot of glycolysis, PI3K/Akt/mTOR, mTORC1 signaling, hypoxia, Myc targets V1, and Myc targets V2 pathways based on the sh-Ctrl versus sh-PIPKIγ-1 SW480 gene expression profiles. NES denotes normalized enrichment score. (B) Heat maps of the genes enriched in indicated pathways illustrate the changes in gene expression upon PIPKIγ knockdown. Red signal denotes higher expression and blue signal represents lower expression relative to the mean expression level within the group. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Glycolytic changes induced by PIPKIγ. (A) Summary of the glycolytic transporters, enzymes, and metabolites. (B-E) The impact of PIPKIγ on the expression of glycolytic enzymes (B), glucose uptake (C), lactate production (D), and extracellular acidification ratio (ECAR) (E) of SW480 and LOVO cells. *P < .05 and **P < .01.
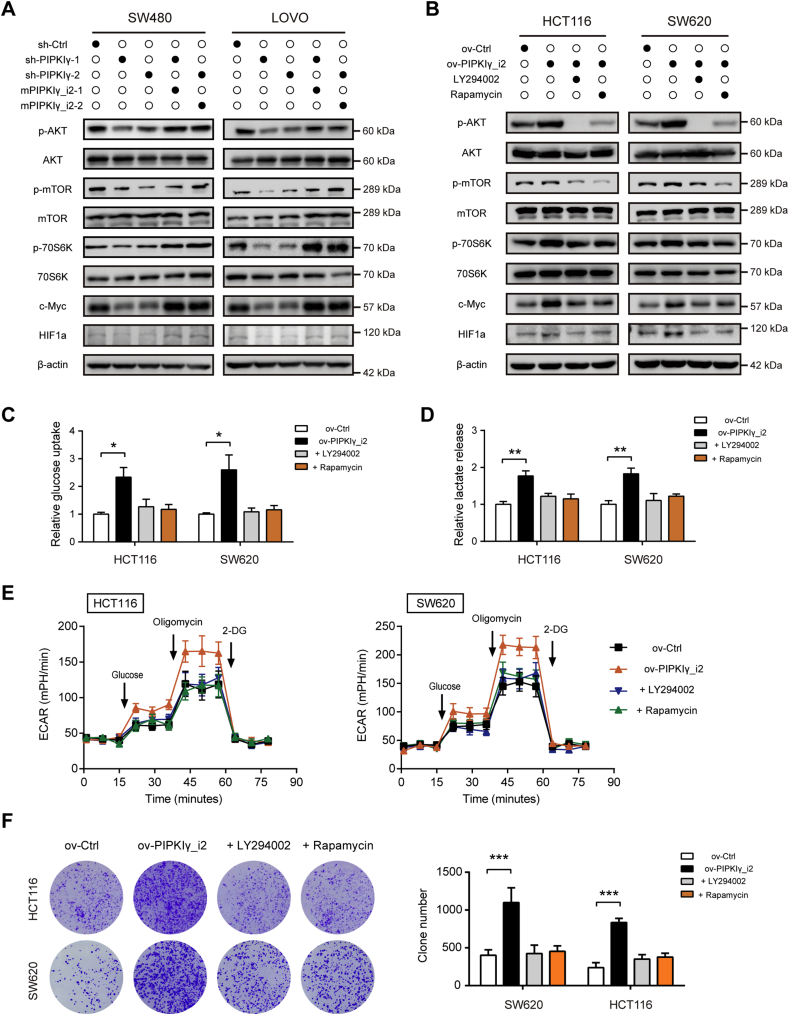
3.4. Loss of PIPKIγ causes inactivation of the PI3K/Akt/mTOR/c-Myc-HIF1α signaling pathway
As described above, our results suggested that PI3K/AKT/mTOR signaling, hypoxia, and Myc targets are regulated by PIPKIγ (Fig. 3A). HIF1α and c-Myc are two critical transcription factors implicated in the Warburg effect by regulating expression of glycolytic enzymes [23,25]. Interestingly, both of them can be regulated by the PI3K/Akt/mTOR pathway. Therefore, we hypothesized that PIPKIγ may regulate glycolysis by activation of the PI3K/Akt/mTOR/c-Myc-HIF1α signaling pathway. To test this hypothesis, we firstly examined PI3K/Akt/mTOR activity upon PIPKIγ knockdown. Western blotting analysis showed that silencing of PIPKIγ reduced the phosphorylation levels of Akt, mTOR and its major target S6K, which can be rescued by mPIPKIγ_i2 (Fig. 5A and Supplementary Fig. 5A). In addition, c-Myc and HIF1α protein level were also significantly downregulated by PIPKIγ knockdown (Fig. 5A and Supplementary Fig. 5A). In PIPKIγ_i2-overexpressing colorectal cancer cells, PI3K/Akt/mTOR activity was markedly increased compared to the control cells (Fig. 5B and Supplementary Fig. 5B). Of note, PIPKIγ-induced elevation of c-Myc and HIF1α was largely compromised by addition of PI3K inhibitor (LY294002) or mTOR inhibitor (Rapamycin) (Fig. 5B and Supplementary Fig. 5B). Consistently, inhibition of PI3K/Akt/mTOR signaling also blocked the glucose utilization (Fig. 5C), lactate secretion (Fig. 5D), ECAR (Fig. 5E), and survival advantage (Fig. 5F) induced by PIPKIγ. Collectively, these results suggested that PIPKIγ may activate PI3K/Akt/mTOR signaling pathway, which further increases c-Myc and HIF1α level to promote aerobic glycolysis in colorectal cancer cells.
Fig. 5.
PIPKIγ knockdown leads to inhibition of the PI3K/Akt/mTOR/c-Myc-HIF1α signaling pathway. (A) Western blot analysis for changes of PI3K/Akt/mTOR signaling pathway and c-Myc and HIF1α levels in sh-Ctrl, sh-PIPKIγ, and sh-PIPKIγ+mPIPKIγ_i2 cell lysates against indicated antibodies. (B) Cell lysates were harvested from ov-Ctrl and ov-PIPKIγ_i2 cells in the presence or absence of LY294002 or Rapamycin. Then proteins in PI3K/Akt/mTOR signaling pathway and c-Myc and HIF1α levels were analyzed by Western blotting. (C-F) The impact of PIPKIγ_i2 overexpression on the glucose uptake (C), lactate production (D), ECAR (E), and colony formation ability (F) of SW480 and LOVO cells in the presence of LY294002 or Rapamycin. *P < .05; **P < .01; ***P < .001.
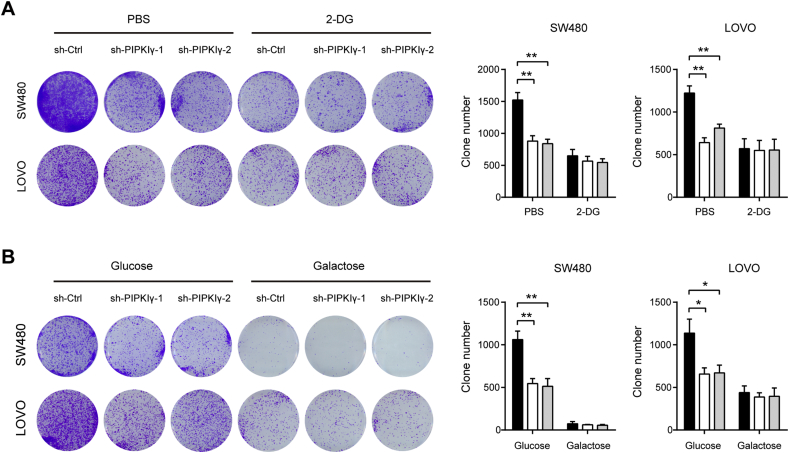
3.5. PIPKIγ-mediated growth advantage is glycolysis-dependent
The Warburg effect is emerged as a key contributor to tumor initiation and progression, and blocking the Warburg effect greatly inhibited tumorigenesis. Therefore, we investigated whether the Warburg effect is an important mechanism contributing to PIPKIγ-mediated oncogenic roles in colorectal cancer. To test this hypothesis, SW480 and LOVO cells were cultured in medium containing 5 mM 2-Deoxy-d-glucose (2-DG), which competitively inhibits the production of glucose-6-phosphate from glucose at the phosphoglucoisomerase level. In concordant with previous report, 2-DG clearly inhibited the anchorage-independent growth of colorectal cancer cells (Fig. 6A). Notably, 2-DG also abolished the suppressive effect of PIPKIγ knockdown on anchorage-independent growth of SW480 and LOVO cells (Fig. 6A). To further confirm our observation, we replaced glucose in the culture medium with galactose, which has a much lower rate than glucose entry into glycolysis. As a result, growth disadvantage induced by PIPKIγ knockdown was largely abolished by galactose (Fig. 6B). Taken together, these results strongly suggest that PIPKIγ-mediated Warburg effect promotes colorectal cancer tumorigenesis.
Fig. 6.
The prolific roles of PIPKIγ are glycolysis-dependent. (A) Colony formation ability of sh-Ctrl or sh-PIPKIγ SW480 and LOVO cells with or without 5 mM 2-DG. (B) Colony formation ability of sh-Ctrl or sh-PIPKIγ SW480 and LOVO cells with culture medium containing 25 mM glucose or galactose. *P < .05 and **P < .01.
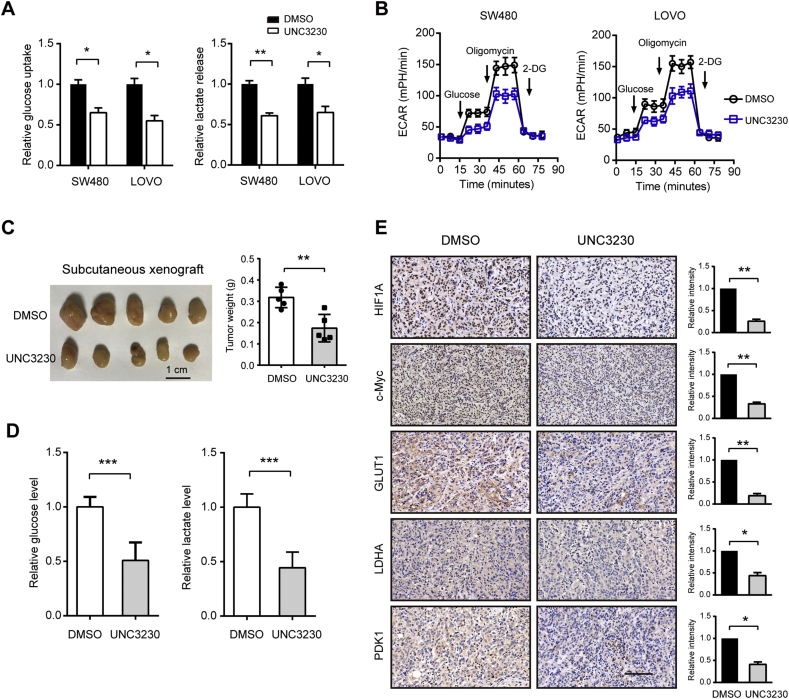
3.6. Pharmacological inhibition of PIPKIγ suppresses tumor growth
To test the therapeutic value of targeting PIPKIγ in colorectal cancer, we treated colorectal cancer cells with a selective PIPKIγ inhibitor, UNC3230. As a result, UNC3230 significantly inhibited the glycolytic phenotypes of SW480 and LOVO cells as revealed by reduced glucose uptake, lactate release, and ECAR (Fig. 7A and B). In xenograft tumors formed by SW480 cells, blocking PIPKIγ activity with UNC3230 significantly reduced tumor burden, glucose level, and lactate level in the subcutaneous model (Fig. 7C and D). In addition, immunohistochemical analysis of the xenograft tissues showed that c-Myc and HIF1α immunoreactivity were markedly downregulated by UNC3230 treatment. Similar observations were also found in the glucose transporter (GLUT1), LDHA, and PDK1 (Fig. 7E). Notably, UNC3230 can also efficiently inhibit PIP4K2C activity. We therefore performed a loss-of-function study of PIP4K2C in SW480 and LOVO cells. The results showed that knockdown of PIP4K2C showed no significant influence on CRC cell proliferation and glycolysis, indicating that the inhibitory role of UNC3230 was largely mediated by PIPKIγ (Supplementary Fig. 6). Collectively, these data above clearly showed that PIPKIγ activity is responsible for its oncogenic role in glycolysis and tumor growth.
Fig. 7.
Pharmacological inhibition of PIPKIγ suppresses tumor growth. (A) The impact of UNC3230 on the glucose uptake and lactate production in SW480 and LOVO cells. (B) The impact of UNC3230 on the ECAR of SW480 and LOVO cells. (C) SW480 cells (1 × 106) were injected subcutaneously into the left forelimb of nude mice (n = 5 per group). When bore visible tumors (200 mm3), mice were treated with saline with 0.01% DMSO or 5 mg/kg UNC3230 for 3 weeks. Three weeks later, mice were sacrificed and tumor weights in each group were shown. (D) The impact of UNC3230 on the glucose and lactate levels in xenograft tumor tissues. (E) IHC analysis of glycolytic regulators (c-Myc and HIF1α), transporter (GLUT1), and enzymes (LDHA and PDK1) in the xenograft tissues from control and UNC3230 group. Scale bar: 100 μm. *P < .05, **P < .01, and ***P < .001.
4. Discussion
Enhanced Warburg effect is a distinctive hallmark of cancer cells and often correlates oncogenic phenotypes and poor prognosis in cancer patients [26,27]. This metabolic character provides sufficient cellular buildings and energetic needs for cancer cells to promote proliferation and avoid apoptosis [20]. Interestingly, many human malignancies including CRC exhibit an increased glycolytic activity. Thus, revealing the critical contributor in the Warburg effect is of paramount importance to identify new therapeutic targets for colorectal cancer. In the present study, we identified PIPKIγ as a key regulator of Warburg effect in colorectal cancer and uncovered its underlying molecular mechanism. Through in vitro and in vivo studies, PIPKIγ was demonstrated to be a promising molecular target for CRC treatment.
PIPKIγ is a major phosphoinositide-generating enzyme that controls polyphosphoinositide metabolism. Increased expression of PIPKIγ is frequently noticed in human cancer cell lines and primary tumors. In pancreatic cancer, we previously showed that PIPKIγ is upregulated in all cancer cell lines detected and pY639-PIPKIγ exhibits remarkably strong staining in tumor tissues indicative of a pathogenic role for PIPKIγ during malignant transformation [17]. Moreover, pY639-PIPKIγ is also markedly elevated in invasive breast ductal carcinoma and correlates elevated histological grade, suggesting the important implications of PIPKIγ in tumor progression [28]. Using a tissue microarray containing 438 breast carcinomas tissues, Sun et al. showed a significant inverse correlation between strong PIPKIγ expression and overall patient survival [29]. Consistently, we found that PIPKIγ is commonly overexpressed in human colorectal cancer cell lines and tumor tissues. Analysis of a CRC tissue microarray and TCGA cohorts with clinical follow-ups showed that elevated PIPKIγ expression level positively correlated with reduced overall survival rate, indicating that PIPKIγ might act as a new prognostic factor or biomarker for colorectal cancer.
Through generation of PI(4,5)P2, PIPKIγ is critically important in a variety of biological processes, such as focal adhesion assembly [6,30], ciliogenesis [31], centriole duplication [32], and leukocyte recruitment [33]. Notably, PIPKIγ is also widely implicated in many oncogenic phenotypes, such as cell proliferation [13,34], migration [35], invasion [12,28], and the epithelial to mesenchymal transition process [14]. The dysregulated expression pattern of PIPKIγ prompted us to investigate its neoplastic activities in colorectal cancer. By both gain- and loss-of-function studies, we confirmed the growth-promoting effect of PIPKIγ in colorectal cancer cells. Through whole transcriptomic gene expression analysis, the altered glucose metabolism induced by PIPKIγ was revealed. During tumor growth, hypoxia and metabolic stress will be occurred in most solid tumors. To survive under this harsh microenvironment, cancer cells exhibit a metabolic shift from oxidative phosphorylation to glycolyis, which supports growth advantage by providing anabolic precursors and minimizing the reactive oxygen species in the mitochondria [19,36]. Our functional study showed that PIPKIγ can enhance the Warburg effect in colorectal cancer cells as demonstrated by glucose uptake, lactate production, extracellular acidification ratio, and expression of glycolytic enzymes. Importantly, blocking glycolysis by 2-DG or galactose largely compromised the growth-promoting effect of PIPKIγ. However, PIPKIγ had no significant impact on mitochondrial respiration as reveled by oxygen consumption ratio, suggesting its preferential roles in regulating aerobic glycolysis. In vitro, PIPKIγ knockdown led to 40–50% reduction in tumor glycolysis, suggesting that other oncogenic inputs involved in colorectal cancer cell glycolysis. In vivo, PIPKIγ knockdown resulted in approximately 50% reduction in tumor growth. Previously, we have demonstrated that PIPKIγ can regulate PD-L1 expression by activating NF-κB, suggesting that PIPKIγ might exhibit a role in the immune microenvironment [9] and the anti-tumor activity for targeting PIPKIγ might be enhanced in immune-competent models. Therefore, further works are warranted regarding the therapeutic value of targeting PIPKIγ in colorectal cancer.
The PI3K/Akt signaling pathways are often activated in human cancers [37,38]. These pathways are initiated by the generation of PI(3,4,5)P3 by PI3K-mediated phosphorylation of PI(4,5)P2. Previously, Thapa et al. clearly demonstrated the mechanism by which PIPKIγ couples with PI3K to activate PI3K/Akt signaling [34]. Of note, PI3K/Akt signaling and its downstream mTORC1 complex are central regulators of glycolysis [18]. Akt can enhance glucose transporter activity and promote glycolysis by activation of hexokinase and phosphofructokinase. Indeed, PIPKIγ knockdown markedly inhibited the activation of PI3K/Akt/mTORC1 signaling. Inhibition of PI3K using LY294002 or mTORC1 by rapamycin blocked enhanced glycolysis and growth advantage induced by PIPKIγ, suggesting that PI3K/Akt/mTORC1 signaling is responsible for PIPKIγ-mediated functions. It is well known that the Warburg effect can be regulated by several transcriptional factors [39], especially HIF1α and c-Myc [40,41]. HIF1α can increase expression of glycolytic enzymes such as LDHA, as well as PDK limit entry of pyruvate in TCA cycle by inhibiting the activity of pyruvate dehydrogenase [25]. Increased c-Myc can activate numerous genes involved in glycolysis and lactate production [23]. PIPKIγ knockdown suppressed, while overexpression increased HIF1α and c-Myc levels. Inhibition of PI3K/Akt/mTORC1 signaling downregulated PIPKIγ-induced HIF1α and c-Myc levels indicative of the role of PI3K/Akt/mTORC1/HIF1α-c-Myc aixs in the PIPKIγ-mediated glycolysis. Although the HIF1α and c-Myc levels are regulated by PI3K/Akt/mTORC1 signaling in colorectal cancer cells, we cannot fully exclude other inputs influenced by PIPKIγ in the contribution of increased HIF1α and c-Myc.
In conclusion, for the first time, we identified PIPKIγ as a novel regulator for aerobic glycoysis in human colorectal cancer cells. Our current results not only provide insight into the oncogenic roles of PIPKIγ in colorectal cancer, but also the molecular mechanisms by which PIPKIγ regulates aerobic glycolysis. However, further investigations are warranted concerning the roles of PIPKIγ in reprogrammed metabolism, including glutamine metabolism and fatty acid metabolism. Given pharmacological inhibition of PIPKIγ activity significantly suppressed tumor growth, our findings may provide alternative strategies for the treatment of colorectal cancer.
Funding sources
This work was supported by the National Key Research and Development Program of China (No. 2017YFC1308900); The Outstanding Clinical Discipline Project of Shanghai Pudong (PWYgy2018-02); the grant from the Natural Science Foundation of China (81502510); and the grant from Science and Technology Commission of Shanghai Municipality (17411968800).
Conflicts of interest statement
The authors declare no conflicts of interest.
Authors' contributions
Junli Xue and Wei Peng conceived the study plan. Wei Peng, Wei Huang, and Xiaoxiao Ge performed the experiments, analyzed the data and finished the manuscript writing. Liqiong Xue and Wei Zhao contributed to the in vivo experiments. Junli Xue supervised this study and edited the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.05.015.
Appendix A. Supplementary data
Supplementary material
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]; Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68(6):394-424. [DOI] [PubMed]
- 2.Atkin W., Wooldrage K., Brenner A., Martin J., Shah U., Perera S. Adenoma surveillance and colorectal cancer incidence: a retrospective, multicentre, cohort study. Lancet Oncol. 2017;18(6):823–834. doi: 10.1016/S1470-2045(17)30187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Atkin W, Wooldrage K, Brenner A, Martin J, Shah U, Perera S, et al. Adenoma surveillance and colorectal cancer incidence: a retrospective, multicentre, cohort study. The Lancet Oncology. 2017;18(6):823-34. [DOI] [PMC free article] [PubMed]
- 3.Angenete E. The importance of surgery in colorectal cancer treatment. Lancet Oncol. 2019;20(1):6–7. doi: 10.1016/S1470-2045(18)30679-X. [DOI] [PubMed] [Google Scholar]; Angenete E. The importance of surgery in colorectal cancer treatment. The Lancet Oncology. 2018. [DOI] [PubMed]
- 4.Schill N.J., Anderson R.A. Two novel phosphatidylinositol-4-phosphate 5-kinase type Igamma splice variants expressed in human cells display distinctive cellular targeting. Biochem J. 2009;422(3):473–482. doi: 10.1042/BJ20090638. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schill NJ, Anderson RA. Two novel phosphatidylinositol-4-phosphate 5-kinase type Igamma splice variants expressed in human cells display distinctive cellular targeting. The Biochemical journal. 2009;422(3):473-82. [DOI] [PMC free article] [PubMed]
- 5.Ling K., Doughman R.L., Iyer V.V., Firestone A.J., Bairstow S.F., Mosher D.F. Tyrosine phosphorylation of type Igamma phosphatidylinositol phosphate kinase by Src regulates an integrin-Talin switch. J Cell Biol. 2003;163(6):1339–1349. doi: 10.1083/jcb.200310067. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ling K, Doughman RL, Iyer VV, Firestone AJ, Bairstow SF, Mosher DF, et al. Tyrosine phosphorylation of type Igamma phosphatidylinositol phosphate kinase by Src regulates an integrin-talin switch. The Journal of cell biology. 2003;163(6):1339-49. [DOI] [PMC free article] [PubMed]
- 6.Ling K., Doughman R.L., Firestone A.J., Bunce M.W., Anderson R.A. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420(6911):89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]; Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420(6911):89-93. [DOI] [PubMed]
- 7.Heck J.N., Mellman D.L., Ling K., Sun Y., Wagoner M.P., Schill N.J. A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit Rev Biochem Mol Biol. 2007;42(1):15–39. doi: 10.1080/10409230601162752. [DOI] [PubMed] [Google Scholar]; Heck JN, Mellman DL, Ling K, Sun Y, Wagoner MP, Schill NJ, et al. A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Critical reviews in biochemistry and molecular biology. 2007;42(1):15-39. [DOI] [PubMed]
- 8.Barlow C.A., Laishram R.S., Anderson R.A. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 2010;20(1):25–35. doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends in cell biology. 2010;20(1):25-35. [DOI] [PMC free article] [PubMed]
- 9.Xue J., Chen C., Qi M., Huang Y., Wang L., Gao Y. Type Igamma phosphatidylinositol phosphate kinase regulates PD-L1 expression by activating NF-kappaB. Oncotarget. 2017;8(26):42414–42427. doi: 10.18632/oncotarget.17123. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xue J, Chen C, Qi M, Huang Y, Wang L, Gao Y, et al. Type Igamma phosphatidylinositol phosphate kinase regulates PD-L1 expression by activating NF-kappaB. Oncotarget. 2017;8(26):42414-27. [DOI] [PMC free article] [PubMed]
- 10.Lee S.Y., Voronov S., Letinic K., Nairn A.C., Di Paolo G., De Camilli P. Regulation of the interaction between PIPKI gamma and Talin by proline-directed protein kinases. J Cell Biol. 2005;168(5):789–799. doi: 10.1083/jcb.200409028. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee SY, Voronov S, Letinic K, Nairn AC, Di Paolo G, De Camilli P. Regulation of the interaction between PIPKI gamma and talin by proline-directed protein kinases. The Journal of cell biology. 2005;168(5):789-99. [DOI] [PMC free article] [PubMed]
- 11.Legate K.R., Takahashi S., Bonakdar N., Fabry B., Boettiger D., Zent R. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. EMBO J. 2011;30(22):4539–4553. doi: 10.1038/emboj.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]; Legate KR, Takahashi S, Bonakdar N, Fabry B, Boettiger D, Zent R, et al. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. The EMBO journal. 2011;30(22):4539-53. [DOI] [PMC free article] [PubMed]
- 12.Li L., Kolodziej T., Jafari N., Chen J., Zhu H., Rajfur Z. Cdk5-mediated phosphorylation regulates phosphatidylinositol 4-phosphate 5-kinase type I gamma 90 activity and cell invasion. FASEB J. 2019;33(1):631–642. doi: 10.1096/fj.201800296R. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li L, Kolodziej T, Jafari N, Chen J, Zhu H, Rajfur Z, et al. Cdk5-mediated phosphorylation regulates phosphatidylinositol 4-phosphate 5-kinase type I gamma 90 activity and cell invasion. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2019;33(1):631-42. [DOI] [PMC free article] [PubMed]
- 13.Li H., Xiao N., Wang Y., Wang R., Chen Y., Pan W. Smurf1 regulates lung cancer cell growth and migration through interaction with and ubiquitination of PIPKIgamma. Oncogene. 2017;36(41):5668–5680. doi: 10.1038/onc.2017.166. [DOI] [PubMed] [Google Scholar]; Li H, Xiao N, Wang Y, Wang R, Chen Y, Pan W, et al. Smurf1 regulates lung cancer cell growth and migration through interaction with and ubiquitination of PIPKIgamma. Oncogene. 2017;36(41):5668-80. [DOI] [PubMed]
- 14.Thapa N., Tan X., Choi S., Wise T., Anderson R.A. PIPKIgamma and Talin couple phosphoinositide and adhesion signaling to control the epithelial to mesenchymal transition. Oncogene. 2017;36(7):899–911. doi: 10.1038/onc.2016.267. [DOI] [PMC free article] [PubMed] [Google Scholar]; Thapa N, Tan X, Choi S, Wise T, Anderson RA. PIPKIgamma and talin couple phosphoinositide and adhesion signaling to control the epithelial to mesenchymal transition. Oncogene. 2017;36(7):899-911. [DOI] [PMC free article] [PubMed]
- 15.Schramp M., Thapa N., Heck J., Anderson R. PIPKIgamma regulates beta-catenin transcriptional activity downstream of growth factor receptor signaling. Cancer Res. 2011;71(4):1282–1291. doi: 10.1158/0008-5472.CAN-10-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schramp M, Thapa N, Heck J, Anderson R. PIPKIgamma regulates beta-catenin transcriptional activity downstream of growth factor receptor signaling. Cancer research. 2011;71(4):1282-91. [DOI] [PMC free article] [PubMed]
- 16.Wu Z., Li X., Sunkara M., Spearman H., Morris A.J., Huang C. PIPKIgamma regulates focal adhesion dynamics and colon cancer cell invasion. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024775. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu Z, Li X, Sunkara M, Spearman H, Morris AJ, Huang C. PIPKIgamma regulates focal adhesion dynamics and colon cancer cell invasion. PloS one. 2011;6(9):e24775. [DOI] [PMC free article] [PubMed]
- 17.Chen C., Wang X., Fang J., Xue J., Xiong X., Huang Y. EGFR-induced phosphorylation of type Igamma phosphatidylinositol phosphate kinase promotes pancreatic cancer progression. Oncotarget. 2017;8(26):42621–42637. doi: 10.18632/oncotarget.16730. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen C, Wang X, Fang J, Xue J, Xiong X, Huang Y, et al. EGFR-induced phosphorylation of type Igamma phosphatidylinositol phosphate kinase promotes pancreatic cancer progression. Oncotarget. 2017;8(26):42621-37. [DOI] [PMC free article] [PubMed]
- 18.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]; Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews Cancer. 2011;11(2):85-95. [DOI] [PubMed]
- 19.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029-33. [DOI] [PMC free article] [PubMed]
- 20.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]; Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annual review of cell and developmental biology. 2011;27:441-64. [DOI] [PubMed]
- 21.Shukla S.K., Purohit V., Mehla K., Gunda V., Chaika N.V., Vernucci E. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell. 2017;32(1) doi: 10.1016/j.ccell.2017.06.004. [71-87 e7] [DOI] [PMC free article] [PubMed] [Google Scholar]; Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV, Vernucci E, et al. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer cell. 2017;32(1):71-87 e7. [DOI] [PMC free article] [PubMed]
- 22.Levine A.J., Puzio-Kuter A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330(6009):1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]; Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330(6009):1340-4. [DOI] [PubMed]
- 23.Dang C.V., Le A., Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res Off J Am Assoc Cancer Res. 2009;15(21):6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15(21):6479-83. [DOI] [PMC free article] [PubMed]
- 24.Zhang C., Liu J., Liang Y., Wu R., Zhao Y., Hong X. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nature communications. 2013;4:2935. [DOI] [PMC free article] [PubMed]
- 25.Semenza G.L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123(9):3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]; Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. The Journal of clinical investigation. 2013;123(9):3664-71. [DOI] [PMC free article] [PubMed]
- 26.Jiang S.H., Li J., Dong F.Y., Yang J.Y., Liu D.J., Yang X.M. Increased serotonin signaling contributes to the warburg effect in pancreatic tumor cells under metabolic stress and promotes growth of pancreatic tumors in mice. Gastroenterology. 2017;153(1) doi: 10.1053/j.gastro.2017.03.008. [277-91 e19] [DOI] [PubMed] [Google Scholar]; Jiang SH, Li J, Dong FY, Yang JY, Liu DJ, Yang XM, et al. Increased Serotonin Signaling Contributes to the Warburg Effect in Pancreatic Tumor Cells Under Metabolic Stress and Promotes Growth of Pancreatic Tumors in Mice. Gastroenterology. 2017;153(1):277-91 e19. [DOI] [PubMed]
- 27.Iansante V., Choy P.M., Fung S.W., Liu Y., Chai J.G., Dyson J. PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat Commun. 2015;6:7882. doi: 10.1038/ncomms8882. [DOI] [PMC free article] [PubMed] [Google Scholar]; Iansante V, Choy PM, Fung SW, Liu Y, Chai JG, Dyson J, et al. PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nature communications. 2015;6:7882. [DOI] [PMC free article] [PubMed]
- 28.Chen C., Wang X., Xiong X., Liu Q., Huang Y., Xu Q. Targeting type Igamma phosphatidylinositol phosphate kinase inhibits breast cancer metastasis. Oncogene. 2015;34(35):4635–4646. doi: 10.1038/onc.2014.393. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen C, Wang X, Xiong X, Liu Q, Huang Y, Xu Q, et al. Targeting type Igamma phosphatidylinositol phosphate kinase inhibits breast cancer metastasis. Oncogene. 2015;34(35):4635-46. [DOI] [PMC free article] [PubMed]
- 29.Sun Y., Turbin D.A., Ling K., Thapa N., Leung S., Huntsman D.G. Type I gamma phosphatidylinositol phosphate kinase modulates invasion and proliferation and its expression correlates with poor prognosis in breast cancer. Breast Cancer Res BCR. 2010;12(1):R6. doi: 10.1186/bcr2471. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sun Y, Turbin DA, Ling K, Thapa N, Leung S, Huntsman DG, et al. Type I gamma phosphatidylinositol phosphate kinase modulates invasion and proliferation and its expression correlates with poor prognosis in breast cancer. Breast cancer research: BCR. 2010;12(1):R6. [DOI] [PMC free article] [PubMed]
- 30.Nader G.P., Ezratty E.J., Gundersen G.G. FAK, Talin and PIPKIgamma regulate endocytosed integrin activation to polarize focal adhesion assembly. Nat Cell Biol. 2016;18(5):491–503. doi: 10.1038/ncb3333. [DOI] [PubMed] [Google Scholar]; Nader GP, Ezratty EJ, Gundersen GG. FAK, talin and PIPKIgamma regulate endocytosed integrin activation to polarize focal adhesion assembly. Nature cell biology. 2016;18(5):491-503. [DOI] [PubMed]
- 31.Xu Q., Zhang Y., Wei Q., Huang Y., Hu J., Ling K. Phosphatidylinositol phosphate kinase PIPKIgamma and phosphatase INPP5E coordinate initiation of ciliogenesis. Nat Commun. 2016;7 doi: 10.1038/ncomms10777. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu Q, Zhang Y, Wei Q, Huang Y, Hu J, Ling K. Phosphatidylinositol phosphate kinase PIPKIgamma and phosphatase INPP5E coordinate initiation of ciliogenesis. Nature communications. 2016;7:10777. [DOI] [PMC free article] [PubMed]
- 32.Xu Q., Zhang Y., Xiong X., Huang Y., Salisbury J.L., Hu J. PIPKIgamma targets to the centrosome and restrains centriole duplication. J Cell Sci. 2014;127:1293–1305. doi: 10.1242/jcs.141465. Pt 6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu Q, Zhang Y, Xiong X, Huang Y, Salisbury JL, Hu J, et al. PIPKIgamma targets to the centrosome and restrains centriole duplication. Journal of cell science. 2014;127(Pt 6):1293-305. [DOI] [PMC free article] [PubMed]
- 33.Stadtmann A., Block H., Volmering S., Abram C., Sohlbach C., Boras M. Cross-talk between Shp1 and PIPKIgamma controls leukocyte recruitment. J Immunol. 2015;195(3):1152–1161. doi: 10.4049/jimmunol.1500606. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stadtmann A, Block H, Volmering S, Abram C, Sohlbach C, Boras M, et al. Cross-Talk between Shp1 and PIPKIgamma Controls Leukocyte Recruitment. Journal of immunology. 2015;195(3):1152-61. [DOI] [PMC free article] [PubMed]
- 34.Thapa N., Choi S., Tan X., Wise T., Anderson R.A. Phosphatidylinositol phosphate 5-kinase Igamma and Phosphoinositide 3-kinase/Akt Signaling couple to promote oncogenic growth. J Biol Chem. 2015;290(30):18843–18854. doi: 10.1074/jbc.M114.596742. [DOI] [PMC free article] [PubMed] [Google Scholar]; Thapa N, Choi S, Tan X, Wise T, Anderson RA. Phosphatidylinositol Phosphate 5-Kinase Igamma and Phosphoinositide 3-Kinase/Akt Signaling Couple to Promote Oncogenic Growth. The Journal of biological chemistry. 2015;290(30):18843-54. [DOI] [PMC free article] [PubMed]
- 35.Choi S., Thapa N., Hedman A.C., Li Z., Sacks D.B., Anderson R.A. IQGAP1 is a novel phosphatidylinositol 4,5 bisphosphate effector in regulation of directional cell migration. EMBO J. 2013;32(19):2617–2630. doi: 10.1038/emboj.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]; Choi S, Thapa N, Hedman AC, Li Z, Sacks DB, Anderson RA. IQGAP1 is a novel phosphatidylinositol 4,5 bisphosphate effector in regulation of directional cell migration. The EMBO journal. 2013;32(19):2617-30. [DOI] [PMC free article] [PubMed]
- 36.Martinez-Outschoorn U.E., Peiris-Pages M., Pestell R.G., Sotgia F., Lisanti M.P. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14(2):113. doi: 10.1038/nrclinonc.2017.1. [DOI] [PubMed] [Google Scholar]; Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nature reviews Clinical oncology. 2017;14(2):113. [DOI] [PubMed]
- 37.Liu P., Cheng H., Roberts T.M., Zhao J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8(8):627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nature reviews Drug discovery. 2009;8(8):627-44. [DOI] [PMC free article] [PubMed]
- 38.Bunney T.D., Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer. 2010;10(5):342–352. doi: 10.1038/nrc2842. [DOI] [PubMed] [Google Scholar]; Bunney TD, Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nature reviews Cancer. 2010;10(5):342-52. [DOI] [PubMed]
- 39.Li L., Liang Y., Kang L., Liu Y., Gao S., Chen S. Transcriptional regulation of the Warburg effect in cancer by SIX1. Cancer Cell. 2018;33(3) doi: 10.1016/j.ccell.2018.01.010. [368-85 e7] [DOI] [PubMed] [Google Scholar]; Li L, Liang Y, Kang L, Liu Y, Gao S, Chen S, et al. Transcriptional Regulation of the Warburg Effect in Cancer by SIX1. Cancer cell. 2018;33(3):368-85 e7. [DOI] [PubMed]
- 40.Dang C.V. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22-35. [DOI] [PMC free article] [PubMed]
- 41.Kroemer G., Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]; Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer cell. 2008;13(6):472-82. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material