Abstract
Background
The role played by the humoral immune response in animals vaccinated against a mycobacterial disease such as paratuberculosis, is not well understood. Sheep vaccinated against Mycobacterium avium subsp. paratuberculosis (MAP) can still become infected and in some cases succumb to clinical disease. The strength and location of the humoral immune response following vaccination could contribute to the ability of sheep to clear MAP infection. We examined the peripheral antibody response along with the localised humoral response at the site of paratuberculosis infection, the ileum, to better understand how this contributes to MAP infection of sheep following vaccination and exposure.
Results
Through assessing MAP specific serum IgG1 and IgG levels we show that the timing and strength of the humoral immune response directly relates to prevention of infection following vaccination. Vaccinated sheep that subsequently became infected had significantly reduced levels of MAP specific serum IgG1 early after vaccination. In contrast, vaccinated sheep that did not subsequently become infected had significantly elevated MAP specific serum IgG1 following vaccination. Furthermore, at 12 months post MAP exposure, vaccinated and subsequently uninfected sheep had downregulated expression of genes related to the humoral response in contrast to vaccinated infected sheep where expression levels were upregulated.
Conclusions
The timing and strength of the humoral immune response following vaccination against paratuberculosis in sheep directly relates to subsequent infection status. An initial strong IgG1 response following vaccination was crucial to prevent infection. Additionally, vaccinated uninfected sheep were able to modulate that response following apparent MAP clearance, unlike vaccinated infected animals where there was apparent dysregulation of the humoral response, which is associated with progression to clinical disease.
Keywords: Antibody, Paratuberculosis, Vaccination, Sheep, Humoral immunity, Ileum, ELISA, Gene expression, Infection
Background
Humoral immunity is believed to play a role in the protective response against intracellular mycobacterial pathogens, such as Mycobacterium avium subsp. paratuberculosis (MAP), the causative agent of paratuberculosis in ruminants [1, 2]. The pathogenesis of paratuberculosis was traditionally characterised by an increase in the humoral response measured through antibody production at the end stages of disease [3, 4]. The switch from an initial dominant cell mediated immune (CMI) response to a humoral response is often thought to signify a breakdown of disease control by the host and progression to clinical disease [5]. While it is undisputed that Interferon gamma (IFNγ) production is essential for overcoming mycobacterial infection [6], the pattern of a protective immune response to MAP infection is actually not so clear [7], with some studies showing MAP-specific antibody responses in sheep as early as two weeks post exposure (wpe) [8] and occurring at the same time as an IFNγ response [9].
The protection provided by vaccines against mycobacterial pathogens, particularly MAP, is often incomplete [10–12]. Sheep and cattle vaccinated against MAP have reduced incidence of clinical disease and faecal shedding, however commercial vaccines fail to prevent infection in all animals [13–15]. Understanding the mechanisms behind how some vaccinated animals successfully clear infection when others do not, would allow development of new vaccines to specifically target a protective immune response in all animals. Traditional markers to assess vaccine efficacy such as IFNγ and total antibody response in isolation are not able to differentiate between animals protected by vaccination and those that are not [16]. Therefore, there is a need to explore alternate or additional markers of vaccine protection to truly understand a protective vaccine response. To this point, most work on correlates of vaccine-induced protection against paratuberculosis has focused on the CMI response, however recent studies have suggested a role for B cells as well [16, 17].
B cells are pivotal in the activation and modulation of both CMI and humoral immune responses [18, 19]. B cells function as antigen presenting cells but also produce antibodies enabling immune complexes that can regulate the function of effector cells such as macrophages [17, 20–23]. In ruminants, the proliferative capacity of peripheral B cells is reduced in animals where vaccination fails to provide protection against MAP [16]. This response was noted as early as 13 weeks’ post MAP exposure. Additionally, a study by Begg and Griffin [24], found significantly higher percentages of B cells in the gut of vaccinated animals that survived MAP challenge compared to diseased animals. Therefore, although peripheral B cells may be functionally impaired, the humoral response at the site of infection might be more important to vaccine-induced protection.
The activity and survival of B cells at the site of infection has been examined in relation to disease progression for mycobacterial infections, but not in response to vaccination. B cells in the tissue can be activated by several different mechanisms, including direct antigen contact, ligation of the CD40 receptor by T cells and binding of B cell activating factor (BAFF) [17]. CD40 ligand (CD40L) deficiency predisposes humans to opportunistic infections by intracellular bacteria [25] and can be correlated with severe tuberculosis in macaques [26]. In contrast, the expression of BAFF by circulating populations of CD4+ T cells is associated with active tuberculosis [27]. Other indicators of the humoral immune response, including B cell surface markers (CD81), cytokines that promote B cell survival (MIFF) and transcription factors (JUN), have all been implicated in the progression or prevention of mycobacterial infections [28–30]. These apparent contradictions suggest that detailed examination of B cell functionality and the humoral response at the site of infection is required. Furthermore, the ability to understand whether the humoral response does provide vaccine-elicited protection against MAP requires an understanding of the host’s ability to successfully mount this response at the site of infection.
We hypothesise that differences in B cell functionality may be correlated with infection status following exposure to MAP in vaccinated sheep. Therefore, we characterised the role of humoral immunity in sheep that were vaccinated and then became infected compared to vaccinated sheep that did not become infected. We assessed MAP-specific IgG1 and IgG levels and compared these with other aspects of B cell functionality, such as cell survival, differentiation, activation and receptor signalling, by gene expression in the gut tissues of these animals.
Results
Animal trial
Infection outcome in vaccinated and non-vaccinated sheep at the conclusion of the animal trial was determined by culture of live MAP from the intestine (Table 1.). In the vaccinated animals exposed to MAP, two were found to be infected with live bacteria at 52 wpe and 18 had no culturable MAP at this time. In contrast, in the non-vaccinated sheep, 10 were found to be infected with MAP and 10 were uninfected at 52 wpe.
Table 1.
Numbers of animals included in IgG1 and tissue gene expression analyses
| Treatment | Infection status | n for ELISA | n for gene expression |
|---|---|---|---|
| Vaccination | control | 5 | 3 |
| uninfected | 18 | 4 | |
| infected | 2 | 2 | |
| Non-vaccination | control | 5 | 3 |
| uninfected | 10 | 3 | |
| infected | 10 | 3 |
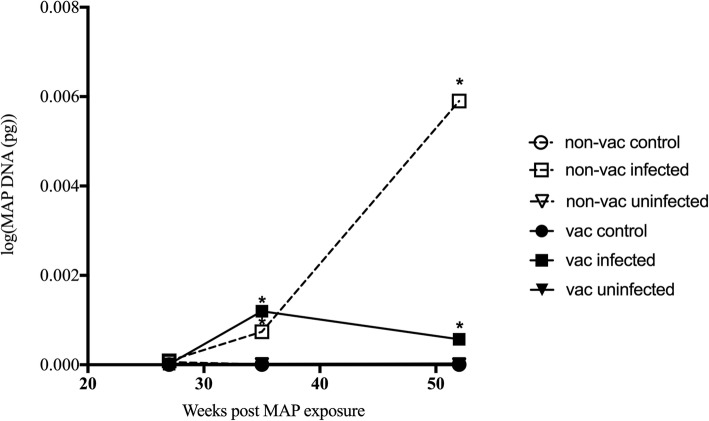
Vaccinated and non-vaccinated infected sheep shed significantly more MAP in the faeces from 35 weeks post MAP exposure till the end of the trial, compared to both uninfected and control animals (p < 0.05) (Fig. 1).
Fig. 1.
Quantity of MAP DNA shed in the faeces. Gudair™ vaccinated (vac) (6 weeks prior to MAP exposure) and non-vaccinated (non-vac) sheep were either exposed or left unexposed (control) to MAP. MAP exposed sheep were grouped based on infection status (infected and uninfected), determined by tissue culture at necropsy. The quantity (pg) of MAP DNA shed in the faeces was determined by direct faecal PCR at 3 timepoints throughout the trial. * denotes groups significantly different to all other groups not marked with an asterisk (p < 0.05)
Serum antibody
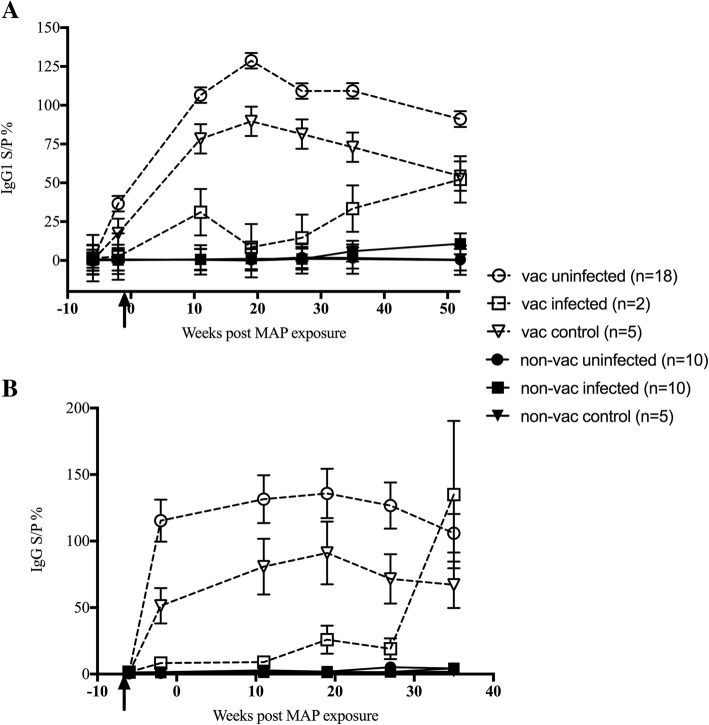
Vaccination significantly enhanced MAP-specific IgG1 levels in serum of sheep (p < 0.001) (Fig. 2a). In the vaccinated exposed animals, serum MAP-specific IgG1 was significantly higher in the cohort found to be uninfected at necropsy, compared to the infected animals. This polarised response was first evident prior to MAP exposure, as early as 1-month post vaccination (p < 0.05). Serum MAP-specific IgG1 peaked in the vaccinated uninfected animals at 19 wpe and then tended to wane until the final sampling at 52 wpe. In contrast, the vaccinated infected animals had significantly lower levels of MAP-specific IgG1 (p < 0.05). The IgG1 response over time was also dissimilar to the vaccinated uninfected sheep, with the vaccinated infected animal’s MAP-specific IgG1 peaking at 11 wpe, decreased at 19 weeks and then increasing until the final sampling at 52 wpe.
Fig. 2.
MAP-specific serum antibody response in sheep. Gudair™ vaccinated (vac) (6 weeks prior to MAP exposure) and non-vaccinated (non-vac) sheep were either exposed or left unexposed (control) to MAP. MAP exposed sheep were grouped based on infection status (infected and uninfected), determined by tissue culture at necropsy. MAP-specific IgG1 (a) and MAP-specific IgG (b) levels in serum were determined by ELISA. Data presented are the predicted mean and standard error, from the linear mixed model analysis. The arrow indicates the timepoint of vaccination. The animal number in each group is denoted in the legend
Non-vaccinated animals had very low levels of MAP-specific IgG1 in serum compared to vaccinates. There were no significant differences between the non-vaccinated controls and the two non-vaccinated exposed groups (infected or uninfected) at any time point. However, in the non-vaccinated infected group, similar to the vaccinated infected sheep, MAP-specific IgG1 levels in serum tended to increase at the final sampling time point.
A similar pattern to the serum MAP-specific IgG1 response was also seen in the MAP-specific IgG data (Fig. 2b). Vaccinated animals had significantly greater MAP-specific IgG levels than non-vaccinates (p < 0.001). As with the MAP-specific IgG1, vaccinated uninfected sheep had higher MAP-specific IgG levels, which remained high throughout the animal trial. Interestingly, the vaccinated infected sheep had very low MAP-specific IgG levels that were not significantly different from the non-vaccinated sheep, until 11 wpe. At 19 wpe, MAP-specific IgG levels began to increase in these vaccinated infected animals, with a rapid increase from 27 to 35 wpe.
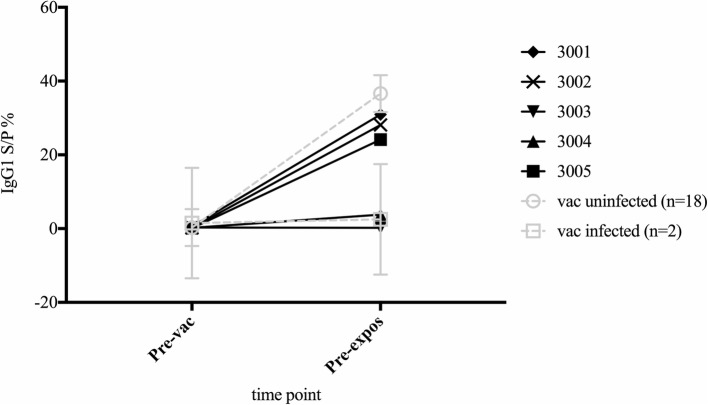
The polarised pre-exposure responses seen in the vaccinated infected and uninfected sheep was mimicked in the IgG1 response of vaccinated control animals. The vaccinated control sheep could be differentiated into high and low IgG1 responder groups at the pre-exposure time point (Fig. 3.).
Fig. 3.
MAP specific-IgG1 serum antibody levels in vaccinated sheep prior to MAP exposure. MAP-specific serum IgG1 levels in vaccinated unexposed sheep (black lines, numbers are individual sheep identifiers) were examined at pre-vaccination and pre-exposure (4 weeks post-vaccination, 2 weeks prior to MAP exposure) time points. The group average values for the vaccinated exposed sheep, categorised as vaccinated infected (tissue culture positive) (n = 2) and vaccinated uninfected (tissue culture negative) (n = 18) are also shown (dashed grey lines)
Gene expression in the ileum
Gene expression analysis of sheep ileal tissue was conducted to assess functionality of B cells at the site of predilection for MAP infection. Fold changes were determined in comparison to a baseline group established from specific research questions (Table 2). This method of analysis was adopted to ensure that the correct controls (baseline groups) were being used to understand the impacts of vaccination, exposure and the spectrum of disease.
Table 2.
Analysis matrix for sheep B cell-related gene expression in intestinal tissue for specific research questions
| Research question | Baseline group | Comparison group (s) |
|---|---|---|
| Q1. What gene expression changes are associated with disease outcome in non-vaccinated animals? Are these similar in vaccinated animals? | Non-vaccinated control |
Non-vaccinated infected Non-vaccinated uninfected Vaccinated infected Vaccinated uninfected |
| Q2. What gene expression changes are associated with disease outcome in vaccinated animals | Vaccinated control |
Vaccinated infected Vaccinated uninfected |
| Q3. What gene expression changes are associated with infection in vaccinated animals | Vaccinated uninfected | Vaccinated infected |
| Q4. What gene expression changes are associated with vaccination nonresponse in infected animals | Non-vaccinated infected | Vaccinated infected |
In relation to research question 1, gene expression responses were divergent in vaccinated and non-vaccinated animals that were uninfected at necropsy (Table 3). The majority of genes were down-regulated in the vaccinated uninfected animals, apart from Lyn and NFIL3. In contrast, in the non-vaccinated uninfected animals only 4 of the 11 genes examined were down-regulated. This dissimilarity of response was also seen in the vaccinated infected and the non-vaccinated infected animals, where only ERG1 and GRB2 were down regulated in the non-vaccinated infected, but 4 of the 11 genes were down regulated in the vaccinated infected. Interestingly, vaccinated uninfected animals had significantly decreased expression of CD84 (fold change 0.112, CI 0.021–0.605, p < 0.05) and BAFF (fold change 0.06, CI 0.004–0.89, p < 0.05) compared to non-vaccinated infected sheep. These same trends were seen in MIF expression, with an increase in expression in non-vaccinated infected animals and decreased expression in vaccinated controls and vaccinated uninfected sheep.
Table 3.
Gene expression changes (fold change) in the ileum of the different treatment groups compared to non-vaccinated control sheep
| Non-vaccinated uninfected (n = 10) | Non-vaccinated infected (n = 10) | Vaccinated control (n = 5) | Vaccinated uninfected (n = 18) | Vaccinated infected (n = 2) | ||
|---|---|---|---|---|---|---|
| aIgG1 (SP%) | 0.49 | 10.75 | 54.39 | 91.09 | 52.2 | |
| Up regulated | LYN | 2.1 | 1.2 | 1.4 | 1.6 | |
| GRB2 | 1.8 | 1.4 | ||||
| ERG1 | ||||||
| JUN | 1.4 | 1.1 | ||||
| NFIL3 | 2.2 | 1.4 | 2.8 | |||
| LCK | 1.5 | 3.2 | ||||
| CD81 | 1.5 | 1.9 | ||||
| CD84 | 3.8 a | |||||
| CD40LG | 2.5 | 5 | 5 | |||
| BAFF | 1.5 | 5 b | 2.2 | |||
| MIF | 1.1 | 2.4 | 1.3 | |||
| Down regulated | LYN | −1.8 | ||||
| GRB2 | −1.2 | −3 | −1.1 | |||
| ERG1 | −1.7 | −3.8 | −3.3 | −1.4 | −1.7 | |
| JUN | −1.4 | −1.9 | − 1.1 | |||
| NFIL3 | −1.1 | −1.6 | ||||
| LCK | −2.8 | −1 | − 1.1 | |||
| CD81 | −3.6 | −1.6 | − 1.2 | |||
| CD84 | −1.1 | −2.1 | −2.4 a | −1.1 | ||
| CD40LG | −1.1 | − 1 | ||||
| BAFF | −1.2 | -1 b | ||||
| MIF | −2.9 | −1.7 | ||||
aIgG1 SP% of each group at the same timepoint as gene expression analysis was performed. Significant differences between groups are indicated by bolding (p < 0.05) with a significantly different to a and b significantly different to b
In research question 2, only the vaccinated animals were examined due to their dissimilarity in expression responses from non-vaccinated sheep. MAP exposure in vaccinated sheep was associated with upregulation of the majority of the genes of interest (Table 4). CD84 was the only differentially regulated gene, where vaccinated uninfected sheep had decreased expression compared to vaccinated infected sheep. The differences between vaccinated infected and uninfected sheep were further explored in research question 3. The majority of the genes examined were up-regulated in the vaccinated infected sheep compared to the vaccinated uninfected sheep. This increased expression was strongest for CD40LG, BAFF, MIF, CD84 and NFIL3 (Table 5). In contrast, both EGR1 and Lck tended to have reduced expression in the vaccinated infected animals compared to the vaccinated uninfected sheep.
Table 4.
Gene expression changes (fold change) in the ileum of the vaccinated infected and uninfected sheep compared to vaccinated control sheep
| Vaccinated uninfected (n = 18) | Vaccinated infected (n = 2) | ||
|---|---|---|---|
| IagG1 (SP%) | 91.09 | 52.2 | |
| Up regulated | LYN | 2.56 | 2.86 |
| GRB2 | 2.65 | 4.33 | |
| ERG1 | 2.41 | 1.89 | |
| JUN | 1.75 | 1.99 | |
| NFIL3 | 2.21 | 4.47 | |
| LCK | 2.72 | 2.63 | |
| CD81 | 2.29 | 3.14 | |
| CD84 | 1.84 | ||
| CD40LG | 1.05 | 5 | |
| BAFF | 1.13 | 2.52 | |
| MIF | 1.67 | 3.82 | |
| Down regulated | LYN | ||
| GRB2 | |||
| ERG1 | |||
| JUN | |||
| NFIL3 | |||
| LCK | |||
| CD81 | |||
| CD84 | −1.12 | ||
| CD40LG | |||
| BAFF | |||
| MIF | |||
aIgG1 SP% of each group at the same timepoint as gene expression analysis was performed
Table 5.
Gene expression changes (fold change) in the ileum of sheep to answer specific research questions
| Vaccinated infected compared to vaccinated uninfected (Q3) |
Vaccinated infected compared to non-vaccinated infected (Q4) |
||
|---|---|---|---|
| Up regulated | LYN | 1.12 | 1.36 |
| GRB2 | 1.64 | 1.69 | |
| ERG1 | 2.17 | ||
| JUN | 1.14 | ||
| NFIL3 | 2.03 | 1.28 | |
| LCK | |||
| CD81 | 1.37 | ||
| CD84 | 2.07 | ||
| CD40LG | 5.37 | ||
| BAFF | 2.22 | ||
| MIF | 2.29 | ||
| Down regulated | LYN | ||
| GRB2 | |||
| ERG1 | −1.27 | ||
| JUN | −1.36 | ||
| NFIL3 | |||
| LCK | −1.03 | −3.37 | |
| CD81 | −2.17 | ||
| CD84 | −4.31 | ||
| CD40LG | −1.68 | ||
| BAFF | −7.52 | ||
| MIF | −1.82 | ||
In research question 4, the differences in gene expression between non-vaccinated and vaccinated sheep that were infected at necropsy was explored in more detail. The majority of the genes of interest were down regulated in the vaccinated infected animals compared to the non-vaccinated infected sheep (Q4 Table 2). This decreased expression was especially evident in BAFF, CD84 and Lck (Table 5).
Discussion
The exact role of B cells and antibodies in host immunity against intracellular pathogens has been a long-debated topic. We show here that B cell functionality is important in vaccine-induced clearance of infection in animals exposed to MAP. A rapid and potent IgG1 antibody response was seen in all uninfected vaccinated sheep. Furthermore, at 12 months post exposure, these uninfected animals had a dampening of the humoral response at the site of paratuberculosis predilection, in contrast to the late switch to a humoral dominated response often associated with the transition to clinical paratuberculosis. On the other hand, vaccinated infected sheep had a slower IgG1 response to vaccination and had up regulated expression of genes related to the humoral immune response in the ileum at 12 months post exposure.
The serum MAP-specific IgG1 response in vaccinated uninfected sheep was elevated compared to the vaccinated controls and vaccinated infected sheep. This significant difference was already evident prior to MAP exposure suggesting a failure of the host to effectively mount an early IgG1 response post vaccination. The route of entry of MAP into the host macrophage can impact the ability of the host cell to kill it [31, 32]. Opsonised bacteria are more likely to interact with the Fc receptor (FcR) on the surface of monocytes and macrophages [33]. Phagocytosis of opsonised MAP via the FcR increases trafficking of intracellular bacteria to the lysosome and increases killing [33–36]. IgG1 has the highest affinity for the Fc receptor out of all IgG subclasses [37], and it stands to reason that animals that have a high IgG1 response to vaccination would be better equipped to eliminate MAP from the gut [19, 38]. The polarised IgG1 response in the vaccinated control animals, suggests that MAP specific IgG1 levels after vaccination, even in animals without MAP exposure, could be a useful predictor of vaccine efficacy.
The initial low level of serum MAP-specific IgG1 in vaccinated infected sheep, followed by a sharp increase, could signify the change from a Th1 to Th2 mediated response. The switch from a Th1 to Th2 immune response in paratuberculosis has been associated with progression to clinical disease [5]. It is possible that the lack of an early antibody response in these animals aids early tissue invasion and leads to persistent infection, as animals with an inadequate IgG1 response appeared to be incapable of eliminating or controlling MAP. A similar late increase was also seen in the non-vaccinated infected animals, although to a much smaller magnitude. In stark contrast to these two groups, the vaccinated control and vaccinated uninfected animals appeared to have a gradual reduction in MAP-specific IgG1 in the serum towards the end of the trial. This reduction could signify a waning of the initial strong antibody response, either due to clearance of bacteria and decreased chronic antigenic stimulation, or the return to a more balanced immune state. The reduction of antibody levels, and presumably B cell response, could also lead to reductions in B cell related immunopathologies associated with an excessive response [39].
In the non-vaccinated sheep, there was no significant difference in levels of either IgG or IgG1 between the control, infected or uninfected animals at any sampling point. Low levels of MAP-specific IgG and IgG1 antibody are not unexpected in MAP exposed animals during the early stages of disease [4]. The differences seen between the vaccinated and non-vaccinated uninfected animals in terms of MAP-specific IgG1 could be the result of differences in the stimulus to induce protective immune responses. The immune response generated by a vaccine such as Gudair® is much stronger than that generated by natural exposure to MAP, due to continual antigen presentation via depot formation and the immune enhancing properties of the adjuvant [40].
The comparison of gene expression changes in the gut of sheep from multiple treatment groups created a dilemma as to what makes the best baseline group to determine fold change. As such, several different research questions were presented to explore the effects of vaccination, exposure to MAP and infection status on the expression of B cell related genes in the gut of ruminants.
Initially, to examine the impacts of both vaccination and infection outcome, the non-vaccinated control sheep were used as the baseline (Q1 Table 2). Vaccination alone decreased B cell-related gene expression and a similar pattern was found in vaccinated animals that resisted or recovered from infection (vaccinated uninfected). Previous work has suggested that early loss of B cell functionality could contribute to vaccine non-response [16], this lack of B cell activation in the current study could be due to samples being taken at a much later time point as well as differences in sample type (blood vs ileal tissue). There was an up-regulation of genes in ileal tissues in infected animals (both vaccinated and non-vaccinated) compared to healthy controls. Overall the results from this study suggests that a B cell response in the later stages of disease, at the site of infection, is not beneficial. Similarly, to the serum antibody results, a strong initial B cell mediated response could be beneficial, whilst a late response signifies progression to disease. The pattern of reduced expression in the vaccinated uninfected animals is likely to be due to clearance of MAP and subsequent return to an inactive or homeostatic immune state, which would be a response similar to the vaccinated unexposed animals where there is also no MAP to activate expression of B cell related genes.
In vaccinated animals, increased expression of CD40 ligand (CD40LG), JUN and B cell activating factor (BAFF) were related to infection status (Q2 Table 2). Engagement of CD40LG with the CD40 receptor on B cells is required for initiation and maintenance of the humoral immune response [41]. In the initial stages of humoral immunity after exposure, CD40/CD40LG signalling is required for the generation of high titres of class switched, high affinity antibodies [41]. During the progression of the immune response, signalling through these receptors encourages the development of memory B cells [39, 41]. The ligation of CD40 has been associated with production of AP-1 early response transcription factor, which is a heterodimer of cFOS and cJUN [42]. Overexpression of cJUN has been associated with prevention of apoptosis, highly increased proliferation and even immortalisation of B cells [43]. BAFF also plays a pivotal role in promoting the survival of plasmablasts, especially after CD40/CD40LG activation [39]. Proliferation and class switch triggered by CD40/CD40LG and then the prevention of apoptosis by BAFF and AP-1 would push the host’s immune response to one that is Th2 dominated [44]. The reduced expression of this gene in the vaccinated uninfected animals suggests that, at this timepoint, the humoral response in these animals is being down regulated or suppressed.
Interestingly, only Lck and EGR1 were down-regulated in the vaccinated infected sheep compared to the vaccinated uninfected animals, although this was not statistically significant. The expression of both of these genes influences B cell maturation, differentiation and antigen receptor signalling [45, 46]. Host tyrosine kinases, like Lck, are key host molecules utilised by intracellular pathogens to prevent killing [47]. In agreement with our work, the suppression of members of the Src family in chicken B cells renders them largely unresponsive to stimulation through the antigen receptor [45]. EGR1 expression is induced upon B cell antigen receptor signalling and accelerates B cell maturation [46]. The dysfunction of the B cell antigen receptor as a possible result of Lck gene down regulation could result in the reduced expression of EGR1. Therefore, it is possible that either upon vaccination or following MAP exposure B cell maturation and differentiation has been reduced through the supressed expression of these two genes, which is likely to diminish the memory response and could contribute to MAP persistence.
Although statistically significant differences were seen in this study, only a small number of the vaccinated exposed animals were infected at necropsy (n = 2). To validate the findings of this research, a larger number of vaccinated infected animals should be examined.
Conclusions
In conclusion, B cell responses were shown to be important to vaccine-mediated immune protection. A strong initial B cell response, characterised by MAP-specific IgG1 levels in serum, was seen in vaccinated sheep that cleared infection. Furthermore, this response appears to be toned down or tightly regulated towards the later stages of infection to prevent the dominance of the humoral response that likely marks the progression to clinical disease. The unique insight into the mechanisms behind vaccine immunity provided by this study will allow vaccine development to promote a strong initial humoral response and could possibly contribute to genetic selection for vaccine response in the future.
Methods
Animal trial
Fifty Merino sheep were purchased from a farm participating in the Australian Market Assurance scheme for Paratuberculosis. The wether lambs aged 4 months were sourced from a flock in Armidale, New South Wales (NSW), an area that has no prior history of Johne’s disease (JD). Absence of MAP infection was confirmed through extensive whole flock faecal tests and serum antibody ELISA [48]. On arrival at the university farm in Camden, NSW, sheep were randomly allocated into 4 treatment groups, Gudair™ vaccinated MAP unexposed (n = 5) and exposed (n = 20), Non-vaccinated MAP unexposed (n = 5) and exposed (n = 20). Sheep were managed under conventional Australian sheep farming conditions by grazing in open paddocks on unimproved pasture. During the trial all animals were examined daily and were weighed monthly. Faecal samples were collected from the rectum and blood samples via jugular venepuncture of all animals prior to inoculation and then every 3–4 months; collection order was non-purposive by selecting animals as they entered the handling yards. Blood and faecal samples were collected in sheep handling yards within the university farm land, and undertaken at a time of day when the weather was mild (between 15 and 25 °C) to reduce stress to the animals.
Vaccination with Gudair® was performed in accordance with the manufacturer’s instructions and at 6 weeks post-vaccination a cohort were moved to quarantine paddocks and inoculated with MAP (Telford 9.2). The inoculation consisted of 3 oral doses (within a 4-week period) as described by Begg et al. [48] with a total of 9.25 × 108 viable MAP organisms. A non-vaccinated cohort was similarly exposed to MAP at the same time. Unexposed animals, vaccinated and non-vaccinated, were kept in separate paddocks to their exposed counterparts.
At the conclusion of the trial (52wpe), all animals were euthanised using an intravenous injection of barbiturate (Lethabarb™) at 0.5 ml/kg bodyweight. The disease status of all MAP exposed animals was then categorised based on liquid culture of MAP from gut tissues collected at necropsy, as described previously [49–51]. Animals with positive tissue cultures were classified as infected and those with negative results were classified as uninfected. A smaller subset of animals was used for gene expression examination in gut tissue (Table 1).
Serum antibody levels
MAP specific IgG1 ELISA
ELISA plates (Nunc Maxisorb) were coated with 5 μg/ml MAP 316v, Protoplasmic antigen A (PPA) or heat-killed M. phlei. The ELISA plates were machine washed 5 times (Tecan, Austria) using wash buffer (phosphate buffered saline with 0.05% v/v Tween 20). Diluted serum (1/100) was added in duplicate to each antigen. Plates were incubated at 37 °C for 1 h, washed as described above and anti-IgG1 antibody (AbD Serotec, MCA 2440) was added. After a 1-h incubation at 37 °C, plates were machine-washed 5 times. Goat anti-mouse HRP (Dako P0447) was then added to each well and incubated at 37 °C for 30 min. Plates were washed as described earlier and TMB substrate (Pierce) added prior to incubating at room temperature in the dark for 20 min, the reaction was stopped with 2 M sulphuric acid and plates were read at 450 nm.
A single batch of positive and negative controls were included on each plate to standardise the ELISA. The positive serum controls were sourced from a sheep with high MAP specific serum antibody levels, as identified by the commercial IDEXX ELISA and the IgG1 ELISA. The negative control was serum from a sheep consistently test-negative for MAP-specific antibodies, as determined by the IDEXX ELISA and the IgG1 ELISA.
The MAP 316v antigen-specific IgG1 response was calculated using the following formula:
M. phlei was included as a mycobacterial cross-reactivity control to ensure that responses seen in the IgG1 ELISA were MAP specific. PPA was included in the plate as a second MAP specific antigen and the ratio of PPA and 316 V response (1:0.8) was used to monitor positive control performance. Furthermore, the stronger PPA response was utilised to calculate SP%.
MAP specific IgG ELISA
The commercial IDEXX Pourquier ELISA (Idexx Laboratories, Australia) was used to determine MAP-specific serum IgG antibody levels. The ELISA was performed based on the manufacturer’s instructions.
Faecal MAP detection
A high throughput direct faecal PCR was used to quantify the amount of MAP shed in the faeces of all sheep, as previously described [52].
Gene expression at the site of infection
Gene selection
The genes to be examined were selected from a normalised and statistically analysed data set generated from previous microarray gene expression analysis (Affymetrix GeneChip) on sheep peripheral blood mononuclear cells (PBMC). This data set was generated from two sheep experimental infection trials that compared differential gene expression in animals vaccinated with Gudair™ and experimentally exposed to MAP compared to non-vaccinated MAP-exposed animals. Samples for gene expression analysis were taken at 13 wpe and 18 animals were used (9 vaccinated infected and 9 vaccinated uninfected). The raw data was normalised using the RMA (Robust Multichip Averaging) algorithm and significant differences were determined with ANOVA [53].
The data set was then examined using Ingenuity Pathway Analysis (IPA) software (version 01–01, Qiagen Bioinformatics). IPA was used to search for genes related to B cell functionality, survival, receptor signalling, migration and interaction with other immune cells within the data set. The overlay function was then used to examine the gene expression fold change in the microarray data set in relation to vaccination and disease outcome. Genes for qPCR analysis for the current study (Table 6) were then selected based on a fold change of greater than +/− 1.5 in PBMC.
Table 6.
Selected genes and primers used for gene expression analysis of intestinal tissue
| Gene name | Entrez gene ID | Primer sequence | Tm | Product size (bp) | Gene Function |
|---|---|---|---|---|---|
| LYN | 100,302,103 |
5′-ACGGAGAGTGGTGGAAAGC-3′ 5′-GTGCACGGGGTCATAGTCT-3’ |
59.6 59.1 |
482 | B cell signaling + activation |
| GRB2 | 101,109,893 |
5’-ACTGCTGCTCCTGTTCTTCC-3′ 5′-AAACGCAGAACACAGAAGCG-3’ |
60.0 59.7 |
442 | B cell signal transduction + communication |
| EGR1 | 443,547 |
5’-CCCCGACTATCTGTTTCCACA-3′ 5′-ATGCGGCTGGGTTTGATGA-3’ |
59.5 60.0 |
340 | B cell maturation + activating factor signaling |
| JUN | 443,219 |
5’-CAAGTGCCGGAAAAGGAAGC-3′ 5′-ACAGTCTCGCCTCAAAACGT-3’ |
59.5 59.8 |
314 | Transcription regulator |
| NFIL3 | 100,217,409 |
5’-CACTGTGAGCGCCTTTGTG-3′ 5′-GGGCCCTCCTGTGAATGTT-3’ |
59.7 59.6 |
262 | B Cell survival |
| LCK | 100,216,439 |
5’-CCCAGCTTCTCCACTGCAA-3′ 5′-ACGGAGCTGTTCACCCTTC-3’ |
59.9 59.6 |
270 | B cell activation |
| CD81 | 100,147,790 |
5’-CTGGGCACGTTCTTCACCT-3′ 5′-GCTGCAAGGCCTGGTCATA-3’ |
59.3 60.0 |
479 | B cell development, activation, growth and motility |
| CD84 | 101,102,779 |
5’-CGTGGAACCCTGTCAGCAA-3′ 5′-AGCACAGAAAGCACAGCCA-3’ |
60.2 60.2 |
412 | Found on memory B cells |
| CD40L | 767,628 |
5’-AGCTGGCCGTGAAAAGACA-3′ 5′-AACACCGAAGCACCCGATT-3’ |
59.9 59.9 |
489 | Initiation and maintenance of humoral immune response |
| BAFF (TNFSF13B) | 101,104,901 |
5’-TTGCAGACAGTGACACGCC-3′ 5′-AGGTGTCCCATGGCAAAGG-3’ |
60.9 59.9 |
426 | B Cell survival |
| MIF | 780,466 |
5’-GCAAGCCGGCACAGTACAT-3′ 5′-ATGTAGATCCTGTCCGGGCT-3’ |
61.0 60.1 |
305 | Survival of mature B cells + NF-kB signalling cascade |
| Reference gene | |||||
| Ovine GAPDH | 443,005 |
5’-AGAAACCTGCCAAGTATGATG-3′ 5′-CCTAGAATGCCCTTGAGAGG-3’ |
60.5 62.6 |
76 | |
Tissue sections
At the conclusion of the trial (52 wpe), all animals were necropsied. The gastrointestinal tract was removed and 3–4 cm sections of the mid to terminal ileum were excised. The sections were frozen at − 80 °C prior to RNA extraction.
RNA extraction and quality and quantity assessment
RNA extraction was performed using RNAzol® RT (Merc) per the manufacturer’s instructions. The quality and quantity of purified RNA was assessed using a NanoDrop® ND-1000 UV-Vis Spectrophotometer (Thermo Scientific, Wilmington, DE), using the Nucleic Acid module. The absorbance at 260 nm was used to determine the RNA concentration where an A260 nm reading of 1.0 is equivalent to 40 μg/mL of RNA. Purity was characterised as a A260/A280 ratio between 1.8 and 2 and a A260/A230 ratio between 1.4 to 2.2. To remove contaminating genomic DNA and increase RNA purity, the samples were DNase treated and ethanol precipitated following extraction.
cDNA generation
cDNA was generated from RNA using the iScript™ cDNA Synthesis kit (Bio-Rad) per the manufacturer’s instructions, diluted 1/10 and stored at − 80 °C until required.
Primer selection and validation
Forward and reverse primers (Table 6) were designed specifically for the gene regions of interest using online software Primer 3 [54] and checked for specificity using a BLAST search. As genes were selected based on expression levels in PBMC, gene expression in intestinal tissues was confirmed using cDNA generated from a paratuberculosis infected sheep.
Three additional housekeeping genes were assessed with geNorm analysis in the qBASE plus analysis software (Biogazelle) [55]. This analysis identified the most stable reference; for subsequent analyses one reference gene was used based on the geNorm analysis (Table 5).
qPCR and gene expression level analysis
qPCR was performed using an Mx3000P Real-time PCR system (Stratagene, Agilent) using the SensiMix™ SYBR® kit (Bioline). Assays were prepared in 96 well plates and included duplicates of each sample. Reaction volumes of 25 μl (including 10 μl of target cDNA at a 1/10 dilution) were prepared and amplified under the following conditions: 95 °C for 10 min, then 40 cycles of 95 °C for 20 s, 56 °C for 30 s and 72 °C for 30 s, with fluorescence acquisition at the end of each annealing step. The specificity of the reaction was confirmed using melting curve analysis. Standard curves were performed on each plate for each primer set. Data collected from the quantitative reverse transcription (qRT)-PCR were analysed using qBASE plus analysis software (Biogazelle) utilising a modified Comparative Ct (ΔΔCt) method [56]. Fold changes were determined in comparison to pre-selected baseline group (Table 2) and the biological significance was set at a change of +/− 1.5 fold.
Statistical analysis
Restricted maximum likelihood (REML) in a linear mixed model (Genstat 16th edition; VSN International Ltd., Hemel Hempstead, United Kingdom) was used to analyse the MAP-specific IgG1, IgG serum ELISA results (S/P %) and MAP DNA quantity in the faeces. Sheep were grouped based on treatment coupled with infection status (vaccinated control, vaccinated infected, vaccinated uninfected, non-vaccinated control, non-vaccinated infected or non-vaccinated uninfected), which along with sampling time point was included as a fixed effect in the model. Animal ear tag number was included as a random effect. When the REML analysis was significant, post-hoc tests to determine the significant differences between pairs of predicted means using the Fisher’s Least Significant Difference procedure were performed.
Acknowledgements
The authors would like to thank Anan Waldron, Ann-Michele Whittington, Nicole Carter and Rebecca Maurer for laboratory support. Additionally, we would like to thank Craig Kristo, James Dalton and Nobel Toribio for animal husbandry and sample collection assistance.
Abbreviations
- BAFF
B cell activating factor
- CD40L
CD40 ligand
- CMI
Cell mediated immunity
- ELISA
Enzyme linked immunosorbent assay
- FcR
Fc receptor
- IFNγ
Interferon gamma
- IPA
Ingenuity pathway analysis
- LcK
Lymphocyte specific protein kinase
- MAP
Mycobacterium avium subspecies paratuberculosis
- MIF
Macrophage migration inhibitory factor
- PBMC
Peripheral blood mononuclear cell
- PCR
Polymerase chain reaction
- PPA
Protoplasmic antigen A
- REML
Restricted maximum likelihood
- wpe
Weeks post exposure
Authors’ contributions
Conceptualization, HP, DB, KP, RW, AP, and KdS; Formal Analysis, HP; Investigation, HP; Data Curation, AP; Methodology, DB, HP; Writing-Original Draft Preparation, HP; Writing-Review & Editing, HP, KP, KdS, DB, RW and AP; Supervision, KdS; Funding Acquisition, RW and HP. All authors have read and approved the publication of this manuscript.
Funding
This work was supported by Meat and Livestock Australia and by the Cattle Council of Australia, Sheep Meat Council of Australia, and Wool Producers Australia through Animal Health Australia (Grant no. P.PSH.0576) and an Australian Postgraduate Award and scholarship from Meat and Livestock Australia (HP). The funding body had no role in the design of the study, collection, analysis, and interpretation of data or in the writing of this manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The experimental trial was approved by the University of Sydney Animal Ethics Committee (AEC) (ref no 6064) and conducted in compliance with the Animal Research Act 1985, Animal Research Regulation 2010 and the Australian code for the care and use of animals for scientific purposes 8th edition 2013 (the Code).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Auriol C. Purdie and Kumudika de Silva contributed equally to this work.
Contributor Information
Hannah B. Pooley, Phone: +61 2 9036 7737, Email: hannah.pooley@sydney.edu.au
Douglas J. Begg, Email: doug.begg@outlook.com
Karren M. Plain, Email: karren.plain@sydney.edu.au
Richard J. Whittington, Email: richard.whittington@sydney.edu.au
Auriol C. Purdie, Email: auriol.purdie@sydney.edu.au
Kumudika de Silva, Email: kumi.desilva@sydney.edu.au.
References
- 1.Achkar JM, Prados-Rosales R. Updates on antibody functions in Mycobacterium tuberculosis infection and their relevance for developing a vaccine against tuberculosis. Curr Opin Immunol. 2018;53:30–37. doi: 10.1016/j.coi.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganusov VV, Klinkenberg D, Bakker D, Koets AP. Evaluating contribution of the cellular and humoral immune responses to the control of shedding of Mycobacterium avium spp. paratuberculosis in cattle. Vet Res. 2015;46:62. doi: 10.1186/s13567-015-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stabel JR. Host responses to Mycobacterium avium subsp. paratuberculosis: a complex arsenal. Anim Health Res Rev. 2006;7(1–2):61–70. doi: 10.1017/S1466252307001168. [DOI] [PubMed] [Google Scholar]
- 4.Sweeney RW. Pathogenesis of Paratuberculosis. Vet Clin N Am Food Anim Pract. 2011;27(3):537–546. doi: 10.1016/j.cvfa.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Stabel J. Transitions in immune responses to Mycobacterium paratuberculosis. Veterinary Microbiology. 2000;77(3-4):465–473. doi: 10.1016/s0378-1135(00)00331-x. [DOI] [PubMed] [Google Scholar]
- 6.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon γ in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178(6):2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Silva K, Plain K, Purdie A, Begg D, Whittington R. Defining resilience to mycobacterial disease: characteristics of survivors of ovine paratuberculosis. Vet Immunol Immunopathol. 2018;195:56–64. doi: 10.1016/j.vetimm.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Waters WR, Bannantine JP, Miller JM, Palmer MV, Stabel JR, Jones DE, et al. Early induction of humoral and cellular immune responses during experimental Mycobacterium avium subsp. paratuberculosis infection of calves. Infect Immun. 2003;71(9):5130–5138. doi: 10.1128/IAI.71.9.5130-5138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begg DJ, de Silva K, Carter N, Plain KM, Purdie A, Whittington RJ. Does a Th1 over Th2 dominancy really exist in the early stages of Mycobacterium avium subspecies paratuberculosis infections? Immunobiology. 2011;216(7):840–846. doi: 10.1016/j.imbio.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Eppleston J, Reddacliff L, Windsor P, Links I, Whittington R. Preliminary observations on the prevalence of sheep shedding Mycobacterium avium subsp paratuberculosis after 3 years of a vaccination program for ovine Johne's disease. Aust Vet J. 2005;83(10):637–638. doi: 10.1111/j.1751-0813.2005.tb13279.x. [DOI] [PubMed] [Google Scholar]
- 11.Windsor PA. Understanding the efficacy of vaccination in controlling ovine paratuberculosis. Small Rumin Res. 2013;110(2–3):161–164. [Google Scholar]
- 12.Kaufmann SHE. Future vaccination strategies against tuberculosis: thinking outside the box. Immunity. 2010;33(4):567–577. doi: 10.1016/j.immuni.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Bannantine JP, Hines ME, Bermudez LE, Talaat AM, Sreevatsan S, Stabel JR, et al. Rational framework for evaluating the next generation of vaccines against mycobacterium avium subspecies paratuberculosis. Front Cell Infect Microbiol. 2014;4:126. [DOI] [PMC free article] [PubMed]
- 14.Reddacliff L, Eppleston J, Windsor P, Whittington R, Jones S. Efficacy of a killed vaccine for the control of paratuberculosis in Australian sheep flocks. Vet Microbiol. 2006;115(1–3):77–90. doi: 10.1016/j.vetmic.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Kalis CHJ, Hesselink JW, Barkema HW, Collins MT. Use of long-term vaccination with a killed vaccine to prevent fecal shedding of Mycobacterium avium subsp paratuberculosis in dairy herds. Am J Vet Res. 2001;62(2):270–274. doi: 10.2460/ajvr.2001.62.270. [DOI] [PubMed] [Google Scholar]
- 16.de Silva K, Plain KM, Begg DJ, Purdie AC, Whittington RJ. CD4(+) T-cells, gammadelta T-cells and B-cells are associated with lack of vaccine protection in Mycobacterium avium subspecies paratuberculosis infection. Vaccine. 2015;33(1):149–155. doi: 10.1016/j.vaccine.2014.10.082. [DOI] [PubMed] [Google Scholar]
- 17.Kozakiewicz L, Phuah J, Flynn J, Chan J. The role of B cells and humoral immunity in mycobacterium tuberculosis infection. Adv Exp Med Biol. 2013;783:225–50. [DOI] [PMC free article] [PubMed]
- 18.Coussens PM. Model for immune responses to Mycobacterium avium subspecies paratuberculosis in cattle. Infect Immun. 2004;72(6):3089–3096. doi: 10.1128/IAI.72.6.3089-3096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Achkar JM, Chan J, Casadevall A. B cells and antibodies in the defense against Mycobacterium tuberculosis infection. Immunol Rev. 2015;264(1):167–181. doi: 10.1111/imr.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins KL, Bosio CM, Rhinehart-Jones TR. Importance of B cells, but not specific antibodies, in primary and secondary protective immunity to the intracellular bacterium Francisella tularensis live vaccine strain. Infect Immun. 1999;67(11):6002–6007. doi: 10.1128/iai.67.11.6002-6007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linton PJ, Harbertson J, Bradley LM. A critical role for B cells in the development of memory CD4 cells. J Immunol. 2000;165(10):5558–5565. doi: 10.4049/jimmunol.165.10.5558. [DOI] [PubMed] [Google Scholar]
- 22.Shen H, Whitmire JK, Fan X, Shedlock DJ, Kaech SM, Ahmed R. A specific role for B cells in the generation of CD8 T cell memory by recombinant listeria monocytogenes. J Immunol. 2003;170(3):1443–1451. doi: 10.4049/jimmunol.170.3.1443. [DOI] [PubMed] [Google Scholar]
- 23.Whitmire JK, Asano MS, Kaech SM, Sarkar S, Hannum LG, Shlomchik MJ, et al. Requirement of B cells for generating CD4+ T cell memory. J Immunol. 2009;182(4):1868–1876. doi: 10.4049/jimmunol.0802501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begg DJ, Griffin JF. Vaccination of sheep against M. paratuberculosis: immune parameters and protective efficacy. Vaccine. 2005;23(42):4999–5008. doi: 10.1016/j.vaccine.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 25.Cabral-Marques Otavio, Ramos Rodrigo Nalio, Schimke Lena F., Khan Taj Ali, Amaral Eduardo Pinheiro, Barbosa Bomfim Caio César, Junior Osvaldo Reis, França Tabata Takahashi, Arslanian Christina, Carola Correia Lima Joanna Darck, Weber Cristina Worm, Ferreira Janaíra Fernandes, Tavares Fabiola Scancetti, Sun Jing, D'Imperio Lima Maria Regina, Seelaender Marília, Garcia Calich Vera Lucia, Marzagão Barbuto José Alexandre, Costa-Carvalho Beatriz Tavares, Riemekasten Gabriela, Seminario Gisela, Bezrodnik Liliana, Notarangelo Luigi, Torgerson Troy R., Ochs Hans D., Condino-Neto Antonio. Human CD40 ligand deficiency dysregulates the macrophage transcriptome causing functional defects that are improved by exogenous IFN-γ. Journal of Allergy and Clinical Immunology. 2017;139(3):900-912.e7. doi: 10.1016/j.jaci.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Roodgar M, Ross CT, Tarara R, Lowenstine L, Dandekar S, Smith DG. Gene expression and TB pathogenesis in rhesus macaques: TR4, CD40, CD40L, FAS (CD95), and TNF are host genetic markers in peripheral blood mononuclear cells that are associated with severity of TB lesions. Infect Genet Evol. 2015;36:396–409. doi: 10.1016/j.meegid.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Kai, Zhang Yan, Hu Shizong, Yu Yang, Yang Qianting, Jin Dongdong, Chen Xinchun, Jin Qi, Liu Haiying. Increased Levels of BAFF and APRIL Related to Human Active Pulmonary Tuberculosis. PLoS ONE. 2012;7(6):e38429. doi: 10.1371/journal.pone.0038429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gore Y, Starlets D, Maharshak N, Becker-Herman S, Kaneyuki U, Leng L, et al. Macrophage migration inhibitory factor induces B cell survival by activation of a CD74-CD44 receptor complex. J Biol Chem. 2008;283(5):2784–2792. doi: 10.1074/jbc.M703265200. [DOI] [PubMed] [Google Scholar]
- 29.Lutay Nataliya, Håkansson Gisela, Alaridah Nader, Hallgren Oskar, Westergren-Thorsson Gunilla, Godaly Gabriela. Mycobacteria Bypass Mucosal NF-kB Signalling to Induce an Epithelial Anti-Inflammatory IL-22 and IL-10 Response. PLoS ONE. 2014;9(1):e86466. doi: 10.1371/journal.pone.0086466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raju B, Hoshino Y, Belitskaya-Lévy I, Dawson R, Ress S, Gold JA, et al. Gene expression profiles of bronchoalveolar cells in pulmonary TB. Tuberculosis. 2008;88(1):39–51. doi: 10.1016/j.tube.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Souza CD, Evanson OA, Weiss DJ. Role of cell membrane receptors in the suppression of monocyte anti-microbial activity against Mycobacterium avium subsp. paratuberculosis. Microb Pathog. 2008;44(3):215–223. doi: 10.1016/j.micpath.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Maglione PJ, Xu J, Casadevall A, Chan J. Fcγ receptors regulate immune activation and susceptibility during Mycobacterium tuberculosis infection. J Immunol. 2008;180(5):3329–3338. doi: 10.4049/jimmunol.180.5.3329. [DOI] [PubMed] [Google Scholar]
- 33.Joller N, Weber SS, Müller AJ, Spörri R, Selchow P, Sander P, et al. Antibodies protect against intracellular bacteria by fc receptor-mediated lysosomal targeting. Proc Natl Acad Sci U S A. 2010;107(47):20441–20446. doi: 10.1073/pnas.1013827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolly A, Colavecchia SB, Fernandez B, Fernandez E, Mundo SL. Antibodies induced by Lipoarabinomannan in bovines: characterization and effects on the interaction between Mycobacterium Avium Subsp. Paratuberculosis and macrophages in vitro. Vet Med Int. 2011;2011:258479. doi: 10.4061/2011/258479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jolly A, Lompardía S, Hajos SE, Mundo SL. Evidence of a pro-apoptotic effect of specific antibodies in a bovine macrophage model of infection with Mycobacterium avium subsp. paratuberculosis. Vet Immunol Immunopathol. 2016;169:47–53. doi: 10.1016/j.vetimm.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Armstrong JA, D'Arcy Hart P. Phagosome lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J Exp Med. 1975;142(1):1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins AM, Jackson KJL. A temporal model of human IgE and IgG antibody function. Front Immunol. 2013;4:235. doi: 10.3389/fimmu.2013.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Everman JL, Bermudez LE. Antibodies against invasive phenotype-specific antigens increase Mycobacterium avium subspecies paratuberculosis translocation across a polarized epithelial cell model and enhance killing by bovine macrophages. Front Cell Infect Microbiol. 2015;5:58. doi: 10.3389/fcimb.2015.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avery DT, Kalled SL, Ellyard JI, Ambrose C, Bixler SA, Thien M, et al. BAFF selectively enhances the survival of plasmablasts generated from human memory B cells. J Clin Investig. 2003;112(2):286–297. doi: 10.1172/JCI18025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Awate S, Babiuk LA, Mutwiri G. Mechanisms of action of adjuvants. Front Immunol. 2013;4:114. doi: 10.3389/fimmu.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elgueta R, Benson MJ, De Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229(1):152–172. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Francis DA, Karras JG, Ke X, Sen R, Rothstein TL. Induction of the transcription factors NF-κB, AP-1 and NF-AT during B cell stimulation through the CD40 receptor. Int Immunol. 1995;7(2):151–161. doi: 10.1093/intimm/7.2.151. [DOI] [PubMed] [Google Scholar]
- 43.Schreiber M, Kolbus A, Piu F, Szabowski A, Möhle-Steinlein U, Tian J, et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13(5):607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khalifeh MS, Stabel JR. Clinical disease upregulates expression of CD40 and CD40 ligand on peripheral blood mononuclear cells from cattle naturally infected with Mycobacterium avium subsp. paratuberculosis. Clin Vaccine Immunol. 2013;20(8):1274–1282. doi: 10.1128/CVI.00246-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sefton BM, Taddie JA. Role of tyrosine kinases in lymphocyte activation. Curr Opin Immunol. 1994;6(3):372–379. doi: 10.1016/0952-7915(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 46.Dinkel A, Warnatz K, Ledermann B, Rolink A, Zipfel PF, Bürki K, et al. The transcription factor early growth response 1 (Egr-1) advances differentiation of pre-B and immature B cells. J Exp Med. 1998;188(12):2215–2224. doi: 10.1084/jem.188.12.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karim AF, Chandra P, Chopra A, Siddiqui Z, Bhaskar A, Singh A, et al. Express path analysis identifies a tyrosine kinase Src-centric network regulating divergent host responses to Mycobacterium tuberculosis infection. J Biol Chem. 2011;286(46):40307–40319. doi: 10.1074/jbc.M111.266239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Begg DJ, de Silva K, Di Fiore L, Taylor DL, Bower K, Zhong L, et al. Experimental infection model for Johne's disease using a lyophilised, pure culture, seedstock of Mycobacterium avium subspecies paratuberculosis. Vet Microbiol. 2010;141(3–4):301–311. doi: 10.1016/j.vetmic.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 49.Whittington R, Whittington A, Waldron A. Development and validation of a liquid medium (M7H9C) for routine culture of Mycobacterium avium subsp. paratuberculosis to replace modified Bactec 12B. J Clin. 2013;51(12):3993-4000. [DOI] [PMC free article] [PubMed]
- 50.Plain KM, Waldron AM, Begg DJ, de Silva K, Purdie AC, Whittington RJ. Efficient, validated method for detection of mycobacterial growth in liquid culture media by use of bead beating, magnetic-particle-based nucleic acid isolation, and quantitative PCR. J Clin Microbiol. 2015;53(4):1121–1128. doi: 10.1128/JCM.03521-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whittington RJ, Begg DJ, de Silva K, Purdie AC, Dhand NK, Plain KM. Case definition terminology for paratuberculosis (Johne's disease). BMC Vet Res. 2017;13(1):328. [DOI] [PMC free article] [PubMed]
- 52.High-Throughput Direct Fecal PCR Assay for Detection of Mycobacterium avium subsp. paratuberculosis in Sheep and Cattle, (2014). [DOI] [PMC free article] [PubMed]
- 53.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 54.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 55.Vandesompele Jo, De Preter Katleen, Pattyn Filip, Poppe Bruce, Van Roy Nadine, De Paepe Anne, Speleman Frank. Genome Biology. 2002;3(7):research0034.1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.