Abstract
This study aimed to investigate the effects of isoleucine (Ile) on the synthesis and secretion of digestive enzymes and cellular signalling in the pancreatic tissue of dairy goats. The pancreatic tissues were incubated in buffer containing 0, 0.40, 0.80, and 1.60 mM Ile. High levels of Ile significantly increased the buffer release and total concentration of ɑ-amylase in the tissues (P < 0.001). The total trypsin and chymotrypsin concentrations in each of the Ile groups were significantly higher than those in the control group (P < 0.05); however, lipase was not affected. High levels of Ile significantly increased ɑ-amylase mRNA expression (P < 0.001) but had no effect on the mRNA expression of trypsin, chymotrypsin, or lipase. Ile did not affect S6K1 phosphorylation levels. High levels of Ile significantly increased the expression of the γ isoform of 4EBP1 (P < 0.001), which indicated that the phosphorylation of 4EBP1 was significantly increased. The phosphorylation level of eEF2 gradually decreased with the addition of Ile (P < 0.001). These results suggested that high doses of Ile can regulate the excretion of enzymes, especially ɑ-amylase, in the pancreatic tissues of dairy goats by modulating mTOR signalling, and this regulation is independent of the mTOR-S6K1 pathway.
1. Introduction
The specificity of ruminal digestion is different from that of monogastric animals. Studies on the digestive functions of ruminants have thus mostly concentrated on the rumen, whereas relatively few studies exist on the metabolism of nutrients that bypass the rumen. The digestion of rumen bypass nutrients depends on the digestive enzymes secreted by the intestine and other organs, including amylase, oligosaccharides, polysaccharide-hydrolysing enzymes, trypsin, chymotrypsin, and lipase, among others, of which pancreatic exocrine digestive enzymes (ɑ-amylase, trypsin, and lipase) predominate. Recent studies have shown that inadequate secretion of pancreatic ɑ-amylase might limit intestinal starch utilization in ruminants [1, 2]. The energy utilization efficiency of starch degraded in the small intestine is 42% higher compared with its fermentation products in the rumen, which can help to avoid rumen acidosis [3]. Therefore, improving the exocrine function of the pancreas is conducive to improving the digestion and utilization of rumen bypass nutrients. Proteins and amino acids play important roles in animal energy balance, protein synthesis, function, and substance transformation, and functional amino acids (leucine, isoleucine, phenylalanine, etc.) can be used as signalling molecules to control metabolic function in animals [4, 5].
An increasing number of scholars have begun to focus on the study of branched-chain amino acids. Branched-chain amino acids are a group of neutral, aliphatic amino acids with a branched carbon chain structure and include leucine, isoleucine (Ile) and valine [6]. Leucine is a ketogenic amino acid, Ile is both glucogenic and ketogenic, and proline is a glucogenic amino acid [6]. Animals cannot synthesize branched-chain amino acids and the intake of these amino acids occurs only through food. Branched-chain amino acids mainly have physiological functions including repairing muscles, promoting gluconeogenesis, oxidizing and supplying energy for the body, enhancing immunity, and promoting hormone synthesis and secretion [7, 8]. Branched-chain amino acids are functional amino acids. Our research group has devoted many years to the study of functional amino acids and pancreatic exocrine function in ruminants, especially leucine [9–11]; however, Ile, a similar branched-chain and functional amino acid, has been rarely investigated. Like leucine, Ile not only is a substrate for protein synthesis but also plays an important role in intracellular protein synthesis signalling pathways [7]. A study in monogastric animals showed that Ile increased milk protein and milk fat production rates in sows [8]. Previous studies in fish showed that Ile increased the growth rate, feed intake, protein synthesis, and protein deposition in juvenile Jian carps and that the activities of trypsin, chymotrypsin, lipase, and amylase were improved [12]. Ile also has an important regulatory role in protein synthesis. Few studies on Ile in ruminants exist and even fewer studies regarding the effects of Ile on pancreatic functions have been done.
Mammalian target of rapamycin (mTOR) is a highly conserved protein factor that can regulate the growth and metabolism of animals through a mechanism involving the regulation of protein synthesis that is relatively clear [13, 14]. mTOR plays a very important role in the initial stage of translation. Amino acids can regulate protein synthesis by affecting the phosphorylation levels of upstream and downstream target protein molecules in the mTOR pathway [15]. The best-characterized downstream effectors of mTOR include two signaling pathways that act in parallel to control mRNA translation: the 70-kDa ribosomal protein S6 kinase 1 (S6K1) pathway and the eukaryotic translation initiation factor 4E binding protein 1 (4EBP1) pathway [16]. mTOR activated by amino acids, in turn, catalyses the phosphorylation of ribosomal protein S6K1 and 4EBP1 [17]. Another potentially important signaling protein in the control of translation is eukaryotic elongation factor 2 (eEF2), which is indirectly regulated by mTOR through S6K1 [18]. In mammalian epithelial cell culture assays [19, 20], the level of mTOR phosphorylation was approximately 100% higher in the 3.5 mM essential amino acid group compared to the nonessential amino acid group (P < 0.001). With the removal of Ile alone, the level of mTOR phosphorylation was decreased by 47% (P < 0.05). Under energy or amino acid depletion conditions, intracellular mTORC1 signalling is strongly inhibited, while the resupplementation of amino acids to starved cells can significantly stimulate mTORC1 activity [8]. However, the mechanisms by which amino acids act as mTORC1 signalling factors remain unclear.
The aim of this study was to investigate the molecular mechanisms and signalling pathways associated with Ile and the regulation of pancreatic exocrine function.
2. Materials and Methods
All procedures used in this study complied with an animal care protocol that was approved by the Northwest A&F University Animal Care and Use Committee.
2.1. Pancreatic Tissue Preparation
Pancreatic tissue was obtained from three different, 1-year-old, healthy Guanzhong dairy goats. A total mixed ration (Table 1) composed of alfalfa hay (17.5% of dry matter (DM)), corn silage (27.5% of DM), and concentrate (55.0% of DM) was prepared for the goats. Goats were fed twice daily at 0700 and 1900 h on an ad libitum basis (allowing for 5-10% orts) with free access to fresh water. Three goats were slaughtered over 3 days, one goat per day, to provide fresh pancreatic tissues for culturing.
Table 1.
Ingredients and chemical composition of the experimental diet.
| Item | % (DM) |
|---|---|
| Ingredient | |
| Alfalfa hay | 17.50 |
| Corn silage | 27.50 |
| Corn | 40.00 |
| Soybean meal | 13.00 |
| Calcium phosphate | 0.25 |
| Limestone | 0.75 |
| NaCl | 0.50 |
| Vitamin-mineral premix1 | 0.50 |
| Nutrient composition | |
| DM | 50.58 |
| ADF | 18.88 |
| NDF | 34.38 |
| CP | 16.40 |
| Starch | 27.66 |
1Vitamin-mineral premix (per kg): 600 mg of Mn, 950 mg of Zn, 430 mg of Fe, 650 mg of Cu, 30 mg of Se, 45 mg of I, 20 mg of Co, 450 mg of nicotinic acid, 800 mg of vitamin E, 45,000 IU of vitamin D, and 120,000 IU of vitamin A.
Approximately 10 g of pancreas tissue was quickly excised after animal slaughtering, placed in cold normal saline (0.9% NaCl), and immediately sent to the laboratory. The tissue was transferred to KRB buffer (25 mM HEPES, pH 7.4, 118 mM NaCl, 4.7 mM KCl, 1.2 mM MgSO4, 2.5 mM CaCl2, and 1.2 mM KH2PO4) [21] and cut into pieces 2 × 2 mm in size. After moisture removal with absorbent papers, tissue pieces were weighed and then transferred to a culture bottle containing 3 mL of KRB solution. The culture bottle was placed in a cell incubator (95% O2 /5% CO2) with the bottle cap left open for incubation. After incubation, the culture bottle was placed on ice and the buffer was collected and stored frozen. The buffer was used to determine the amount of enzymes released from the pancreatic tissue. The pancreatic tissue was frozen and then used to determine the enzyme activities (ɑ-amylase, trypsin, and lipase), as well as the mRNA abundance and activity and the degree of phosphorylation of the mTOR signalling pathway factors (4EBP1, S6K1, and eEF2).
2.2. Treatments and Experimental Design
The Ile solutions containing Ile at 0 mg/mL, 2.62 mg/mL, 5.24 mg/mL, and 10.48 mg/mL were prepared in KRB buffer and were added to cell culture medium to achieve final Ile concentrations of 0 mM, 0.40 mM, 0.80 mM, and 1.60 mM, respectively. The incubation time was 180 min and three replicates were included for each treatment. After incubation for 180 min, tissues were harvested by scraping in the presence of ice-cold lysis buffer containing 1% (v:v) of protease and phosphatase inhibitors cocktail (Roche, China). The culture medium was also collected for further analysis of enzyme activities. In total, each Ile treatment included three replicates from three goats (n = 3).
2.3. Sample Analysis
2.3.1. Chemical Composition of Diet
Feed samples were collected, dried at 55°C for 72 h, and then ground through a 1 mm screen. These samples were analysed using the procedures described by AOAC (1999) for dry matter (DM, ID 930.5) and crude protein (CP, N×6.25; ID 984.13) [22]. The neutral detergent fibre (NDF) and acid detergent fibre (ADF) contents were determined according to the method of Van Soest et al. [23] using sodium sulphite and a heat-stable α-amylase (Ankom A200I fibre analyser, Ankom Technology, Macedon, NY). The starch content in the bag was determined using an enzymatic method (α-amylase and amyloglucosidase) with a commercial starch analysis kit (Megazyme, Megazyme International Ireland Ltd., Bray, Ireland).
2.3.2. In Vitro Enzyme Release
To measure the activity of the digestive enzymes in the tissue segments, the homogenate supernatant was collected after homogenizing. The activity of α-amylase, trypsin, and lipase in the supernatant and culture medium were determined using commercial kits (Nanjing Jiancheng Bioengineering Institute, China). One unit of enzyme activity was defined as 1 μmol of the product released per minute at 39°C.
2.3.3. Quantification of Amylase, Trypsin, and Lipase mRNA Levels
RNA extraction was performed according to the method described by Wathes et al. [24]. Frozen pancreatic tissue samples were homogenized with 1 mL of Trizol (Invitrogen, USA) and the supernatants were collected by centrifugation. RNA was precipitated by adding chloroform, followed by isopropanol. The RNA pellet was rinsed with 75% ethanol and dissolved in RNAse-free water. Total RNA was measured using 260 nm spectrometry.
Reverse transcription was performed using the RNA PCR Kit 3.0 (TAKaRa Biotechnology (Dalian) Co. Ltd., China). Solutions included 2 μL of MgCl2, 3.75 μL of RNAse-free water, 1 μL of 10× RT buffer, 0.25 μL of RNAse inhibitor, 1 μL of dNTP mix, 0.5 μL of random primers, 0.5 μL of AMV reverse transcriptase, and 1 μL of the RNA sample. The reaction conditions were as follows: 42°C for 30 min, 95°C for 5 min, 5°C for 5 min, and storage at −20°C.
For quantitative real-time PCR, 25 μL of the reaction system containing SYBR Green (TAKaRa Biotechnology (Dalian) Co. Ltd., China) was used. The reaction solution contained 12.5 μL of 2× SYBR premix Ex Taq II, 2 μL of sample cDNA, 8.5 μL of dH2O, 1 μL of 10 μmol/L forward primer, and 1 μL of 10 μmol/L reverse primer. The reaction conditions were as follows: predenaturation at 95°C for 30 s, 40 cycles of denaturation at 95°C for 5 s, annealing at 63°C for 30 s (different temperatures for different primers), and final extension at 72°C for 10 min. Primers for ɑ-amylase, trypsin, chymotrypsin, lipase, and β-actin are listed in Table 2 [4]. mRNA expression was determined using the 2-ΔΔCT method [25] with β-actin used as the housekeeping gene [26].
Table 2.
Real-time PCR primers.
| Gene name | Reference sequence | Primer sequence (5′-3′) | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| Amylase | NM_001035016 | F: GAAATGGCCGTGTGACAGAATTTA | 142 | 64.3 |
| R: ACAAAGACAAGTGCCCTGTCAGAA | ||||
| Trypsin | NM_001113727 | F: TGTCTGCGGCTCACTGCTAC | 119 | 62.7 |
| R: GCTGGGATGGACGATACTCTTG | ||||
| Chymotrypsin | NM_001098965.1 | F: ATGTTGGGCATCACGGTCTT | 172 | 60.0 |
| R: TGTGCCTCCACGTGTTATCC | ||||
| Lipase | NM_001205820 | F: GTGGAAGCAAATGATGGACAAG | 81 | 61.8 |
| R: TGGGTTGAGGGTGAGCAGA | ||||
| β-Actin | AF481159 | F: ACCACTGGCATTGTCATGGACTCT | 152 | 60.0 |
| R: TCCTTGATGTCACGGACGATTTCC |
2.3.4. Protein Immunoblot Analysis
Western blot analysis was performed according to the methods described by Crozeir et al. [27]. The frozen tissue samples were ground to powders with a mortar (under a liquid nitrogen environment) and then homogenized using 2 mL of cold lysis buffer (pH = 7.4). The lysis buffer contained the following: 50 mM TrisHCl, 25 mM sodium fluoride, 5 mM ethylenediaminetetraacetic acid, 50 mM β -glycerophosphate, 10 mM sodium pyrophosphate, 0.2 mM sodium vanadate, 1 mM benzylsulfonyl fluoride, 0.2% polyethylene glycol octyl phenyl ether (v/v), 1 mM dithiothreitol, 10 μg/mL aprotinin, 5% β-mercaptoethanol (v/v), and 10 μg/mL leupeptin. Upon homogenization, the solution was immediately centrifuged (20000 × g, 4°C, 15 min) to collect the supernatant, which was then boiled for 15 min and centrifuged a second time (10000 × g, 4°C, 30 min). The new supernatant was collected for use in immunoblot analyses to determine 4EBP1, S6K1, and eEF2 expression. Prior to western blot analysis, the sample was boiled for 5 min and then cooled on ice. The western blot procedure was as follows: first, protein separation was conducted based on the protein molecular weight using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) with a polyacrylamide gel concentration of 15%. Then, the protein was transferred to a nitrocellulose membrane and the band of interest was incubated with the corresponding primary antibody for 24 h at 4°C. Next, the membrane was rinsed three times with phosphate buffered saline (PBS) and incubated with secondary antibody for 1 h at room temperature. Finally, the ChemiDOC XRS+ imaging system (Bio-Rad, Germany) was used to develop the protein of interest through enhanced chemiluminescence reactions. The primary antibodies used were rabbit anti-goat polyclonal serum antibodies (4EBP1, Calbiochem, Germany; S6K1 and eEF2, Abcam, UK) and the secondary antibodies were goat anti-rabbit horseradish peroxidase conjugates (Beijing Synthetic Technology Co., Ltd., China).
2.4. Calculations and Statistical Analysis
Enzyme activities, mRNA expression, and 4EBP1, S6K1 and eEF2 phosphorylation were analysed using the GLM procedure for the one-way analysis of variance (ANOVA) model with SAS software. Protein expression content was calculated as the ratio of the band intensity of β-actin. Differences of P < 0.05 were considered significant and data are presented as the means ± standard errors of the means (SEM).
3. Results
As shown in Table 3, high levels of Ile (0.8 mM and 1.6 mM) significantly increased the tissue concentration of ɑ-amylase (P < 0.001), the buffer release concentration (P < 0.001), and the total concentration (P < 0.001). The total concentrations of trypsin and chymotrypsin in each of the Ile groups were significantly higher than those in the control group (P < 0.05), while lipase was not affected by Ile (P > 0.05).
Table 3.
The effects of isoleucine on the activities of enzymes in vitro.
| Item | Level of isoleucine (mM1) | SEM2 | P-value | |||
|---|---|---|---|---|---|---|
| 0 | 0.40 | 0.80 | 1.60 | |||
| Tissue concentration (U/g) | ||||||
| ɑ-amylase | 814b | 923b | 4207a | 3631a | 244.7 | < 0.001 |
| Trypsin | 4.45 | 5.13 | 4.87 | 4.91 | 0.445 | 0.148 |
| Chymotrypsin | 3.51 | 4.12 | 4.32 | 4.28 | 0.629 | 0.093 |
| Lipase | 587 | 634 | 608 | 616 | 88.6 | 0.347 |
| Release (U/g tissue) | ||||||
| ɑ-amylase | 325b | 308b | 876a | 804a | 48.3 | < 0.001 |
| Trypsin | 1.92 | 2.14 | 2.06 | 2.26 | 0.318 | 0.271 |
| Chymotrypsin | 1.22 | 1.56 | 1.48 | 1.49 | 0.475 | 0.248 |
| Lipase | 189 | 195 | 213 | 204 | 49.7 | 0.418 |
| Total activity (U/g tissue) | ||||||
| ɑ-amylase | 1139b | 1231b | 5083a | 4435a | 138.1 | < 0.001 |
| Trypsin | 6.37b | 7.27a | 6.93a | 7.17a | 0.582 | 0.014 |
| Chymotrypsin | 4.73b | 5.68a | 5.80a | 5.77a | 0.536 | 0.028 |
| Lipase | 776 | 819 | 821 | 820 | 72.3 | 0.303 |
1One-way ANOVA. Differences were considered significant at P < 0.05.
2Pooled standard error of the means, n=3.
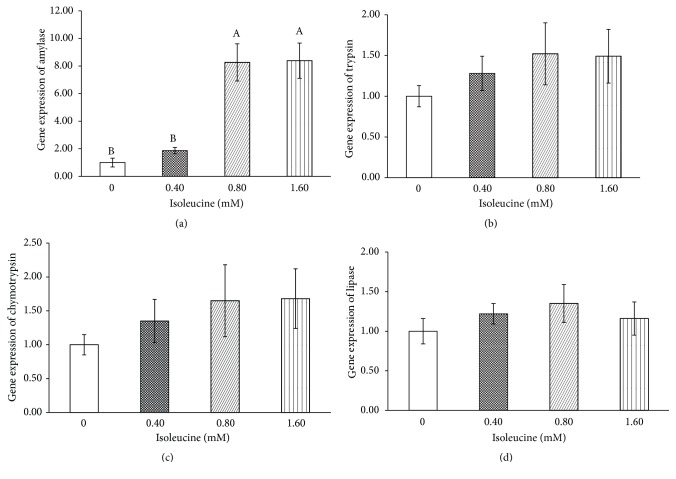
As shown in Figure 1, high levels of Ile (0.8 mM and 1.6 mM) significantly increased ɑ-amylase mRNA expression (P < 0.001), but had no effect on the mRNA expression of trypsin, chymotrypsin, or lipase (P > 0.05).
Figure 1.
The effects of isoleucine on pancreatic amylase (a), trypsin (b), chymotrypsin (c), and lipase (d) mRNA levels in vitro. The values are the means and pooled standard errors of the means (SEM). The enzyme mRNA levels were normalized to β-actin and the values were compared to the control group, which was set to 1.00. Different letters represent significantly different values (P < 0.05, n = 3).
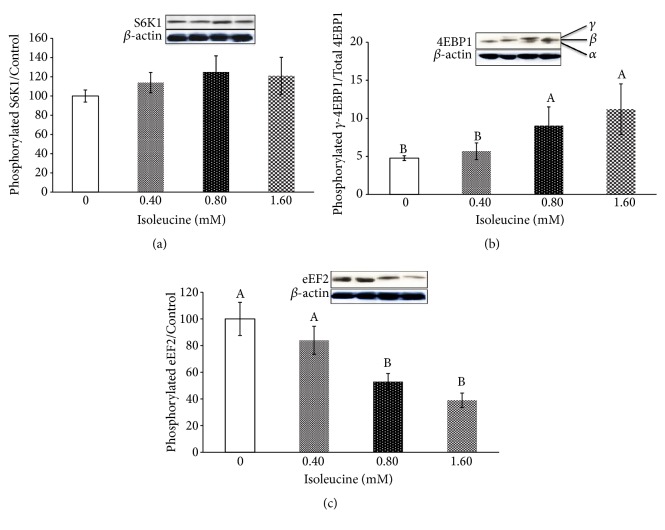
As shown in Figure 2, Ile did not affect the level of S6K1 phosphorylation (P > 0.05). Detection of the 4EBP1 isoforms (ɑ, β, and γ) indicated that high levels of Ile (0.8 mM and 1.6 mM) significantly increased the expression of the γ isoform of 4EBP1 (P < 0.001), which means that the phosphorylation of 4EBP1 was significantly increased. The level of eEF2 phosphorylation gradually decreased with the addition of Ile and was most pronounced in the 0.8 mM and 1.6 mM groups (P < 0.001).
Figure 2.
The effects of isoleucine on the ratio of phosphorylated to total mTOR signalling pathway factors in pancreas tissue. Panel (a) represents S6K1; Panel (b) represents 4EBP1 (ɑ, β, and γ-forms are denoted); Panel (c) represents eEF2. The data are the means and pooled standard errors of the means (SEM). Different letters represent significantly different values (P < 0.05, n = 3).
4. Discussion
In this study, Ile significantly increased the total ɑ-amylase concentration in pancreatic tissues. Compared to the control group, the total ɑ-amylase concentrations in the 0.8 mM and 1.6 mM groups were increased by 346.3% and 289.4%, respectively. A study on juvenile Jian carp noted that Ile significantly increased the trypsin, chymotrypsin, and ɑ-amylase activities in the hepatopancreas and intestines [12], which was similar to the results of a study in mice by Lyman and Wilcox [28]. A previous study conducted by our research team [29] also showed that short-term duodenal infusion of Ile could increase the ɑ-amylase concentration (U/mg) and the secretion rate (U/h) by up to 84.6% and 78.6%, respectively, and long-term infusion could increase the same parameters by 122.7% and 133.8%, respectively. The reason why Ile has different degrees of stimulatory effects on the synthesis and secretion of ɑ-amylase may be because when nutrient infusion is performed in animals, pancreatic enzyme secretion is affected by various factors, including nutrients, hormones, nerves, and gastrointestinal motility, as well as the interactions and antagonistic effects among these factors [2]. By contrast, with in vitro incubation of pancreatic tissues, other impacting factors in in vivo studies can be ruled out and the effects of nutrients on pancreatic enzyme secretion can be studied in isolation, producing a more direct and obvious effect. Low-dose Ile has no effect on pancreatic enzyme secretion. The reason for this remains to be further investigated as few studies regarding the effects of Ile on pancreatic exocrine functions in ruminants exist and no current studies regarding the effects of Ile on enzyme secretion in in vitro pancreatic tissues or acinar cells in ruminants are known. The effects of Ile on pancreatic enzyme mRNA expression may, to a certain extent, explain that low-dose Ile had no effect on ɑ-amylase secretion in the pancreas. High levels of Ile (0.8 mM and 1.6 mM) promoted ɑ-amylase mRNA expression and the effect was extremely significant.
Amino acids not only serve as substrates for protein synthesis but also regulate protein synthesis by modulating translation initiation and translation rates [30]. Previous studies have shown that Ile can increase mTOR phosphorylation and the synthesis rate of casein fragments in mammary gland tissues [19, 20]. Arriola Apelo et al. also showed that Ile can stimulate the mTOR signalling pathway in mammary gland tissues [31]. In this study, Ile significantly increased the level of 4EBP1 phosphorylation. 4EBP1 acts as a downstream signalling factor of mTOR and is directly regulated by mTOR [14]. Once being phosphorylated by mTOR, 4EBP1 releases eukaryotic initiation factor 4E (eIF4E) from the inactive eIF4E·4EBP1 complex to form the active eIF4G·eIF4E complex that binds to mRNA and initiates translation [32]. Therefore, Ile can regulate protein synthesis by modulating the mTOR signalling pathway. However, since Ile does not affect the level of S6K1 phosphorylation, the results of this study indicate that the regulation of protein synthesis by Ile does not depend on the mTOR-S6K1 pathway.
eEF2 plays a crucial role during the elongation stage of protein synthesis, in which the ribosome moves by the equivalent of one codon relative to the mRNA, and the peptidyl-tRNA migrates from the ribosomal A site into the P site, following formation of the new peptide bond [16], while phosphorylated eEF2 is inactive during protein translation [18]. Studies have shown that amino acid deficiency increases the degree of eEF2 phosphorylation [33]. By studying the effects of different amino acids on the protein synthesis pathway in mammary gland tissues, Arriola Apelo et al. [31] showed that Ile can linearly reduce the level of eEF2 phosphorylation and thus increase its activity. A study by Proud et al. showed that eEF2 can be regulated by mTOR [34]. In this study, as the level of Ile increased, the degree of eEF2 phosphorylation gradually decreased, with a particularly significant effect under high-doses of Ile, which was consistent with the above findings.
5. Conclusions
High doses of Ile can regulate the excretion of enzymes, especially ɑ-amylase, in dairy goat pancreatic tissue by modulating mTOR signalling and this regulation is independent of the mTOR-S6K1 pathway.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (nos. 2018YFD0501600 and 2017YFD0500500) and the National Natural Science Foundation of China (nos. 31672451 and 31472122).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Huntington G. B. Starch utilization by ruminants: from basics to the bunk. Journal of Animal Science. 1997;75(3):852–867. doi: 10.2527/1997.753852x. [DOI] [PubMed] [Google Scholar]
- 2.Harmon D. L., Yamka R. M., Elam N. A. Factors affecting intestinal starch digestion in ruminants: A review. Canadian Journal of Animal Science. 2004;84(3):309–318. doi: 10.4141/A03-077. [DOI] [Google Scholar]
- 3.Harmon D. L. Understanding starch utilization in the small intestine of cattle. Asian-Australasian Journal of Animal Sciences. 2009;22(7):915–922. doi: 10.5713/ajas.2009.r.08. [DOI] [Google Scholar]
- 4.Wu G. Functional amino acids in nutrition and health. Amino Acids. 2013;45(3):407–411. doi: 10.1007/s00726-013-1500-6. [DOI] [PubMed] [Google Scholar]
- 5.Wu G., Bazer F. W., Dai Z., Li D., Wang J., Wu Z. Amino acid nutrition in animals: Protein synthesis and beyond. Annual Review of Animal Biosciences. 2014;2:387–417. doi: 10.1146/annurev-animal-022513-114113. [DOI] [PubMed] [Google Scholar]
- 6.Brosnan J. T., Brosnan M. E. Branched-chain amino acids: enzyme and substrate regulation. Journal of Nutrition. 2006;136(1):207S–211S. doi: 10.1093/jn/136.1.207S. [DOI] [PubMed] [Google Scholar]
- 7.Nair K. S., Short K. R. Hormonal and signaling role of branched-chain amino acids. Journal of Nutrition. 2005;135(6):1547S–1552S. doi: 10.1093/jn/135.6.1547S. [DOI] [PubMed] [Google Scholar]
- 8.Richert B. T., Goodband R. D., Tokach M. D., Nelssen J. L. Increasing valine, isoleucine, and total branched-chain amino acids for lactating sows. Journal of Animal Science. 1997;75(8):2117–2128. doi: 10.2527/1997.7582117x. [DOI] [PubMed] [Google Scholar]
- 9.Yu Z. P., Xu M., Liu K., Yao J. H., Yu H. X., Wang F. Leucine markedly regulates pancreatic exocrine secretion in goats. Journal of Animal Physiology and Animal Nutrition. 2014;98(1):169–177. doi: 10.1111/jpn.12069. [DOI] [PubMed] [Google Scholar]
- 10.Yu Z. P., Xu M., Wang F., et al. Effect of duodenal infusion of leucine and phenylalanine on intestinal enzyme activities and starch digestibility in goats. Livestock Science. 2014;162(1):134–140. doi: 10.1016/j.livsci.2014.01.023. [DOI] [Google Scholar]
- 11.Liu K., Liu Y., Liu S. M., et al. Relationships between leucine and the pancreatic exocrine function for improving starch digestibility in ruminants. Journal of Dairy Science. 2015;98(4):2576–2582. doi: 10.3168/jds.2014-8404. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J., Liu Y., Jiang J., et al. Effects of dietary isoleucine on growth, the digestion and absorption capacity and gene expression in hepatopancreas and intestine of juvenile Jian carp (Cyprinus carpio var. Jian) Aquaculture. 2012;368-369:127–128. doi: 10.1016/j.aquaculture.2012.09.019. [DOI] [Google Scholar]
- 13.Schalm S. S., Blenis J. Identification of a conserved motif required for mTOR signaling. Current Biology. 2002;12(8):632–639. doi: 10.1016/S0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- 14.Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Current Biology. 2003;13(10):797–806. doi: 10.1016/S0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 15.Burgos S. A., Dai M., Cant J. P. Nutrient availability and lactogenic hormones regulate mammary protein synthesis through the mammalian target of rapamycin signaling pathway. Journal of Dairy Science. 2010;93(1):153–161. doi: 10.3168/jds.2009-2444. [DOI] [PubMed] [Google Scholar]
- 16.Fingar D. C., Richardson C. J., Tee A. R., Cheatham L., Tsou C., Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Molecular and Cellular Biology. 2004;24(1):200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Proud C. G. The mTOR pathway in the control of protein synthesis. Physiology Journal. 2006;21(5):362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 18.Wang X., Li W., Williams M., Terada N., Alessi D. R., Proud C. G. Regulation of elongation factor 2 kinase by p90RSK1 and p70 S6 kinase. EMBO Journal. 2001;20(16):4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appuhamy J. A., Bell A. L., Nayananjalie W. A., Escobar J., Hanigan M. D. Essential amino acids regulate both initiation and elongation of mRNA translation independent of insulin in MAC-T cells and bovine mammary tissue slices. Journal of Nutrition. 2011;141(6):1209–1215. doi: 10.3945/jn.110.136143. [DOI] [PubMed] [Google Scholar]
- 20.Appuhamy J. A., Knoebel N. A., Nayananjalie W. A., Escobar J., Hanigan M. D. Isoleucine and leucine independently regulate mtor signaling and protein synthesis in MAC-T cells and bovine mammary tissue slices. Journal of Nutrition. 2012;142(3):484–491. doi: 10.3945/jn.111.152595. [DOI] [PubMed] [Google Scholar]
- 21.Umbreit W. W., Burris R. H., Stauffer J. F. Manometric Techniques. 1964;10 [Google Scholar]
- 22.AOAC. Official Methods of Analysis. 16th. Washington, DC, USA: Association of Analytical Chemists; 1999. [Google Scholar]
- 23.van Soest P. J., Robertson J. B., Lewis B. A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. Journal of Dairy Science. 1991;74(10):3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- 24.Wathes D. C., Cheng Z., Chowdhury W., et al. Negative energy balance alters global gene expression and immune responses in the uterus of postpartum dairy cows. Physiological Genomics. 2009;39(1):1–13. doi: 10.1152/physiolgenomics.00064.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Frota I. M., Leitao C. C., Costa J. J., et al. Effects of BMP-7 and FSH on the development of goat preantral follicles and levels of mRNA for FSH-R, BMP-7 and BMP receptors after in-vitro culture. Animal Reproduction. 2011;8(1-2):25–31. [Google Scholar]
- 27.Crozier S. J., Sans M. D., Lang C. H., D'Alecy L. G., Ernst S. A., Williams J. A. CCK-induced pancreatic growth is not limited by mitogenic capacity in mice. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2008;294(5):G1148–G1157. doi: 10.1152/ajpgi.00426.2007. [DOI] [PubMed] [Google Scholar]
- 28.Lyman R. L., Wilcox S. S. Effect of acute amino acid deficiencies on carcass composition and pancreatic function in the force-fed rat. Journal of Nutrition. 1963;79(1):28–36. doi: 10.1093/jn/79.1.28. [DOI] [PubMed] [Google Scholar]
- 29.Liu K., Shen J., Cao Y., Cai C., Yao J. Duodenal infusions of isoleucine influence pancreatic exocrine function in dairy heifers. Archives of Animal Nutrition. 2018;72(1):31–41. doi: 10.1080/1745039X.2017.1396144. [DOI] [PubMed] [Google Scholar]
- 30.Kimball S. R., Jefferson L. S. Control of protein synthesis by amino acid availability. Current Opinion in Clinical Nutrition & Metabolic Care. 2002;5(1):63–67. doi: 10.1097/00075197-200201000-00012. [DOI] [PubMed] [Google Scholar]
- 31.Arriola Apelo S. I., Singer L. M., Lin X. Y., McGilliard M. L., St-Pierre N. R., Hanigan M. D. Isoleucine, leucine, methionine, and threonine effects on mammalian target of rapamycin signaling in mammary tissue. Journal of Dairy Science. 2014;97(2):1047–1056. doi: 10.3168/jds.2013-7348. [DOI] [PubMed] [Google Scholar]
- 32.Kimball S. R., Jefferson L. S. Regulation of global and specific mRNA translation by oral administration of branched-chain amino acids. Biochemical and Biophysical Research Communications. 2004;313(2):423–427. doi: 10.1016/j.bbrc.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Campbell L. E., Miller C. M., Proud C. G. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochemical Journal. 1998;334(1):261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proud C. G. Amino acids and mTOR signalling in anabolic function. Biochemical Society Transactions. 2007;35(5):1187–1190. doi: 10.1042/BST0351187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.