Abstract
Pancreatic endocrine islets are vital for glucose homeostasis. However, the islet developmental trajectory and its regulatory network are not well understood. To define the features of these specification and differentiation processes, we isolated individual islet cells from TgBAC(neurod1:EGFP) transgenic zebrafish and analyzed islet developmental dynamics across four different embryonic stages using a single-cell RNA-seq strategy. We identified proliferative endocrine progenitors, which could be further categorized by different cell cycle phases with the G1/S subpopulation displaying a distinct differentiation potential. We identified endocrine precursors, a heterogeneous intermediate-state population consisting of lineage-primed alpha, beta and delta cells that were characterized by the expression of lineage-specific transcription factors and relatively low expression of terminally differentiation markers. The terminally differentiated alpha, beta, and delta cells displayed stage-dependent differentiation states, which were related to their functional maturation. Our data unveiled distinct states, events and molecular features during the islet developmental transition, and provided resources to comprehensively understand the lineage hierarchy of islet development at the single-cell level.
Keywords: single-cell RNA-seq, zebrafish, pancreas, islet, precursor cell, progenitor cell
Introduction
The islets of Langerhans mainly consist of alpha, beta, delta, and epsilon cells, which are pivotal for glucose homeostasis (Tehrani and Lin, 2011). Islet dysfunction contributes to diabetes mellitus, the global prevalence of which in 2014 was estimated to be 9% (Prince et al., 2017). To gain a better understanding of diabetic pathogenesis, the developmental mechanism regulating islet biology has been intensively investigated, though still far from being fully elucidated.
Zebrafish (Danio rerio) represents an ideal model organism for the study of embryonic development, especially for dissecting the formation of internal organs such as the pancreas, due to its unique advantages, including the transparency of early embryos as well as rapid embryonic development outside of the mother. Zebrafish primary islets are observed during early embryonic development, and the existence of large primary islets and elevated free glucose level following its inhibition on 3 days post fertilization (dpf) indicated that the early forming primary islets are vital for glucose homeostasis (Prince et al., 2017). Comparing with pancreas development in mammals, where endocrine cells and exocrine cells could derive from both dorsal bud and ventral bud, the pancreatic dorsal bud in zebrafish only gives rise to endocrine cells, offering a unique advantage to study endocrine cell fate specification without the ‘interference’ from exocrine cells. Many transcription factors driving islet development have been reported. Unlike the key role of Ngn3 in regulating mouse pancreatic endocrine cell differentiation, its homolog in zebrafish is not expressed in pancreas and no endocrine defects were detected in the zebrafish ngn3 mutant (Flasse et al., 2013). Instead, the basic helix-loop-helix (bHLH) transcription factor Neurod1, which works together with Ascl1b, has been shown to be a key transcription factor for the initiation of zebrafish islet specification (Flasse et al., 2013). After the initiation stage, the expression of endocrine islet markers, including neurod1, pax6b, isl1, and nkx2.2a, persists throughout islet formation (Pauls et al., 2007; Verbruggen et al., 2010; Wilfinger et al., 2013; Dalgin and Prince, 2015). At the onset of cell fate determination, the expression of lineage-specific transcription factors, including arxa for alpha cells, pdx1 and mnx1 for beta cells, and cdx4 for delta cells, is elevated (Kinkel et al., 2008; Dalgin et al., 2011; Kimmel et al., 2011, 2015; Djiotsa et al., 2012). Despite research on critical transcription factors for islet development, however, the lineage hierarchy and gene regulatory network of islet specification are still poorly understood. The presence of progenitors and precursors, which represent the gene regulatory network of each cell type, remains elusive. In search of potential intermediate states during endocrine lineage specification, genome-wide high-throughput and unbiased analyses of the developmental process are required.
Single-cell RNA-seq is a powerful tool that has been used to reveal the heterogeneity of adult pancreatic islet cells, cell-subtype-specific expression patterns and alterations in gene expression associated with type 2 diabetes (Baron et al., 2016; Li et al., 2016; Muraro et al., 2016; Segerstolpe et al., 2016; Wang et al., 2016; Xin et al., 2016a, b; Lawlor et al., 2017). In addition, several distinct states and hallmark features during beta cell maturation have been captured via single-cell transcriptome analysis (Zeng et al., 2017; Qiu et al., 2017a). Bulk RNA-seq analyses of pancreatic endocrine cells isolated from embryonic and adult zebrafish revealed many novel markers in different cell types (Tarifeño-Saldivia et al., 2017). By contrast, reports focusing on deciphering early endocrine pancreas development at the single-cell level are very limited and so far only available for the mouse model with few detailed analysis (Xin et al., 2016a; Stanescu et al., 2017; Yu et al., 2018), and the zebrafish islet specification process at the single-cell level remains to be unveiled in details (Farrell et al., 2018; Wagner et al., 2018), partially due to the difficulty in isolating enough tissue-specific single cells from tiny zebrafish embryos.
To define transcriptomic dynamics and the lineage hierarchy of the pancreatic islet specification at the single-cell level, we performed single-cell RNA-seq on the developing islets at four important embryonic stages in zebrafish by isolating GFP+ cells from the islets manually dissected from a TgBAC(neurod1:EGFP) transgenic fish (Obholzer et al., 2008). Five clusters were identified based on 413 single-cell transcriptomes, including newly identified candidate proliferative progenitor and precursor in addition to the differentiated alpha cells, beta cells and delta cells. Candidate progenitors were further categorized by cell cycle states into G1/S, G2/M or quiescent subpopulations with distinct differentiation potentials. Precursors were further classified into lineage-primed alpha, beta, and delta precursor cells, which could be specified into corresponding terminally differentiated cell types by elevating metabolic and secretory activities. We also defined stage-dependent features of the three types of terminally differentiated cells. Finally, we analyzed the conservation of genes specific to the major cell types across species, and explored the developmental process in islets using pseudo-time analysis. Overall, our study characterized the developing populations and timeline during islet specification and our results provided a resource for understanding endocrine pancreas development at the single-cell level.
Results
The transcriptomic landscape of primary islet formation during zebrafish embryogenesis
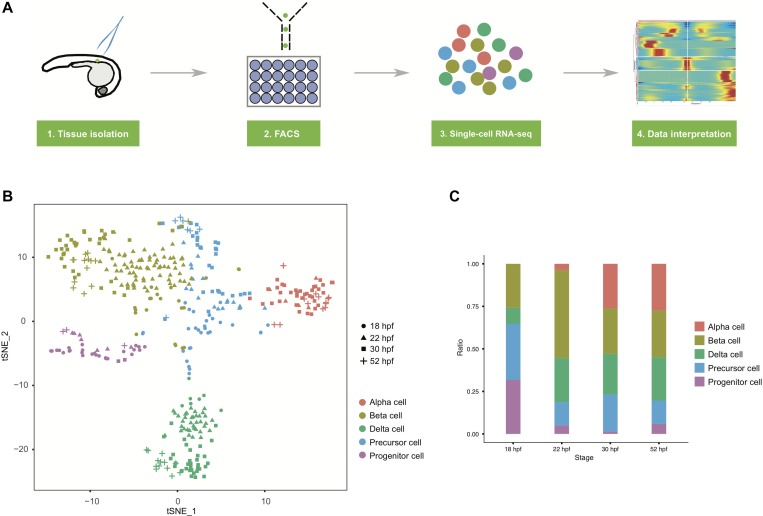
To analyze the developmental process during zebrafish primary islet formation at the single-cell level, we performed single-cell RNA-seq of the developing endocrine islet, represented by GFP+ cells isolated from islet dissections of TgBAC(neurod1:EGFP) transgenic fish at four important embryonic stages, namely, 18 hours post fertilization (hpf), 22 hpf, 30 hpf, and 52 hpf (Figure 1A and Supplementary Figure S1A). Pancreatic insulin expression was first detected at 15 hpf (Biemar et al., 2001), and nascent islet emerges as these cells converged to the midline around 19 hpf (Kinkel et al., 2008). Major endocrine cell types appeared by 22 hpf (Biemar et al., 2001), and the principal islet was established by 30 hpf. At 52 hpf, the pancreatic dorsal and ventral buds merged into a single structure (Field et al., 2003). To avoid contamination of the neurod1+ neuronal system, which also displays GFP expression, pancreatic islets were carefully dissected manually from embryos under a fluorescent microscope and digested into a single-cell suspension (Supplementary Figure S1A). GFP+ single islet cells were then sorted into 96-well plates by fluorescence-activated cell sorting (FACS) (Supplementary Figure S1B). Single-cell RNA sequencing was performed according to STRT-seq steps with slight modifications on reverse transcription and amplification primers (Islam et al., 2011, 2014; Li et al., 2017; Zhong et al., 2018).
Figure 1.
Overview of single-cell RNA-seq of the zebrafish embryonic primary islet. (A) Workflow of single-cell RNA-seq of the primary islet. (B) Distribution of all cells (n = 413) as shown with the first two dimensions of t-SNE analysis. The cell type is color-coded, with embryonic stages represented by different shapes. (C) Cell type ratio at different developmental stages. See also Supplementary Figure S1 and Table S1.
After the quality control, 413 high-quality single-cell transcriptomes, including 85 for 18 hpf, 139 for 22 hpf, 138 for 30 hpf and 51 for 52 hpf, were obtained for the downstream analyses. The median numbers of detected genes of different stages (transcripts per million [TPM] > 0) were 4109, 3618, 3890, and 3106, respectively (Supplementary Figure S1C and Table S1).
The entire single-cell dataset was unbiasedly projected onto the two dimensions of the t-distributed stochastic neighbor embedding (t-SNE) plot. Overall, five clusters were identified, including newly postulated islet precursors and proliferative progenitors, in addition to three expected cell types (alpha, beta, and delta cells) (Figure 1B). Interestingly, only four out of these five islet cell types were detected at 18 hpf, except for the alpha cell population, which was not present until 22 hpf. On the other hand, the ratio of the progenitor cells clearly decreased across the developmental timeline with the most dramatic reduction occurring after 18 hpf. These results reflect the rapid dynamics of islet cell determination and development (Figure 1C and Supplementary Table S1).
Characterization of potential proliferative islet progenitors
Endocrine progenitor specification from the endoderm is the initial step for lineage development of the pancreatic islet. In mice, Ngn3 expression marks the initiation of endocrine specification (Gu et al., 2002). However, zebrafish ngn3 was not expressed in the pancreas and the ngn3 mutant showed no defects in endocrine islet development (Flasse et al., 2013). Although the functional equivalents of mammalian Ngn3 were shown to be neurod1 and ascl1b in zebrafish, the existence and characteristics of pancreatic endocrine progenitors remained elusive due to the wide expression of neurod1 in all islet cell types (Flasse et al., 2013).
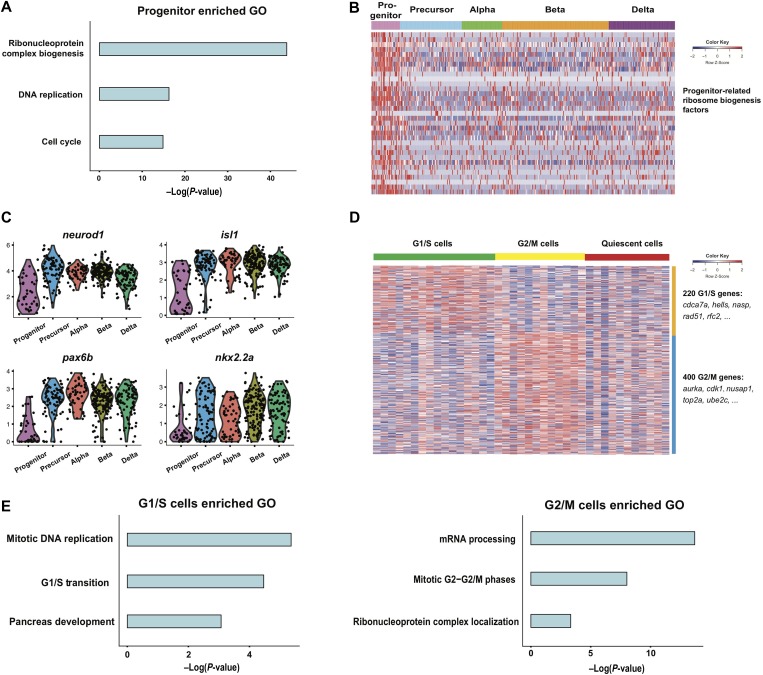
To define the identity of the potential progenitors from our single-cell RNA-seq data, differentially expressed genes for each of the five clusters were evaluated. Gene ontology (GO) analysis results showed that differentially expressed genes from the potential progenitor cluster were highly enriched in terms including ‘ribonucleoprotein complex biogenesis’, ‘DNA replication’, and ‘cell cycle’, which led us to define this cluster as potential proliferative progenitors (Figure 2A and Supplementary Table S2). Recent studies showed that certain populations of ribosome biogenesis factors contributed to the identity of stem or progenitor cells by selecting specific mRNA species to be translated (Brombin et al., 2015). Our data demonstrated that these particular ribosome biogenesis factors were specifically enriched in our progenitor cluster, which further confirmed the progenitor identity for this cluster (Figure 2B and Supplementary Table S2).
Figure 2.
Analyses of proliferative progenitors. (A) GO analysis results of differentially expressed genes (power >0.4) in the progenitor cluster. (B) Heat map displaying the expression of ribosome biogenesis factors related to the stem cell or progenitor cell identity. (C) Violin plot showing the relatively low expression level of known endocrine markers neurod1, nkx2.2a, pax6b, and isl1 in the progenitor population. (D) Heat map showing further classification of the progenitors according to the cell cycle state. (E) Enriched GO terms of G1/S cells in comparison with G2/M and quiescent progenitor cells, and enriched GO terms of G2/M cells in comparison with G1/S and quiescent progenitor cells. See also Supplementary Figure S2 and Table S2.
Transcription factors are important in regulating the stepwise endocrine lineage specification and determination, though limited factors have been identified, and the dynamic expression of known important transcription factors has not been systematically investigated, especially at the single-cell level. We first evaluated the expression profile of known endocrine markers and regulators, including neurod1, isl1, pax6b, and nkx2.2a in our single-cell RNA-seq data and found that their mRNA levels were relatively lower in the progenitor population compared with the remaining four clusters, where their expression levels were comparable and remained relatively constant, indicating that the expression of these four transcription factors was first initiated in progenitors to specify the endocrine identity of the progenitor cells. Thereafter, their expression was upregulated as well as maintained throughout islet development to direct and ensure the differentiation of the progenitors into distinct terminal cell lineages (Figure 2C). We similarly analyzed the expression profiles of 516 transcription factors (those with average expression >10 TPM) (Zhang et al., 2012, 2015), and found fev, insm1a, pou2f2a, and tox3 to display similar patterns as the above-mentioned known endocrine markers, suggesting their potential role in regulating endocrine development but also possibly functioning as endocrine initiators (Supplementary Figure S2A). The expression of tox3 in zebrafish endocrine islet was confirmed by RT-PCR of single-cell RNA-seq libraries of pancreatic islets isolated from TgBAC(neurod1:EGFP) transgenic embryos (Supplementary Figure S2C). Expression of these genes in zebrafish pancreatic islet was also suggested previously by the bulk RNA-seq analyses (Tarifeño-Saldivia et al., 2017), though their function in zebrafish pancreas development has not been reported and requires further investigation. In mammals, Fev loss-of-function mice displayed a reduced production and secretion of insulin as well as impaired glucose tolerance (Ohta et al., 2011), and Insm1 has been reported to be essential for mouse endocrine islet development (Gierl et al., 2006). But the function of Pou2f2 and Tox3 in pancreas has not been studied in mammals. In addition, we found that 12 transcription factors were specifically enriched in the progenitor population (Supplementary Table S2), indicating the unique feature of the progenitors. The expression of 11 out of the 12 transcription factors in zebrafish endocrine islet was confirmed by RT-PCR of single-cell RNA-seq libraries of pancreatic islets isolated from TgBAC(neurod1:EGFP) transgenic embryos (Supplementary Figure S2C). Specific expression of hoxb8a in the pancreas of zebrafish embryos was further confirmed by whole mount in situ hybridization (Supplementary Figure S2D).
Next, we analyzed the cell cycle phase of the progenitors based on the expression levels of G1/S and G2/M marker genes. Progenitors could be easily classified into G1/S cells (enriched in G1/S gene expression), G2/M cells (enriched in G2/M gene expression) and the quiescent population (low expression of G1/S and G2/M genes) (Figure 2D and Supplementary Table S2). To decipher more significant features of the cells at different phases, we further compared the expression pattern of G1/S and G2/M cells by GO analysis. Besides cell cycle-related activities such as ‘mitotic DNA replication’ and ‘G1/S transition’, the G1/S cells also showed a potential for ‘pancreas development’ (Figure 2E). On the other hand, G2/M cells were enriched in terms related to the capacity of cell proliferation rather than differentiation, as indicated by ‘mRNA processing’ and ‘ribonucleoprotein complex localization’, suggesting that the decision of the progenitor cells towards differentiation may take place during the G1/S phase (Figure 2E). Interestingly, the cell cycle phase of the mouse pancreatic progenitors has also been reported to be correlated with different differentiation potentials (Bechard et al., 2016). Our results indicate that the mechanism of cell proliferation and differentiation that couples during pancreatic endocrine development may well be conserved throughout vertebrate evolution.
Epigenetic regulators, including DNA methyltransferase, histone deacetylase, and histone demethylase, have been shown to direct pancreatic endocrine differentiation and maintain beta cell identity (Haumaitre et al., 2008; Xie et al., 2015; Yu et al., 2018). By profiling the expression of 115 epigenetic factors, we found that certain epigenetic regulators were specifically enriched in progenitors, including DNA methyltransferase Dnmt3b and Dnmt3bb.2, histone-modifying factors such as lysine methyltransferase Setdb1b and Ehmt1b, arginine methyltransferase Carm1 and Prmt5, and acetyltransferase Hat1 and Pcbp4 (Supplementary Figure S2B). The expression of dnmt3bb.2, ehmt1b, hat1, and setdb1b in endocrine islet was confirmed by RT-PCR of single-cell RNA-seq libraries of pancreatic islets isolated from TgBAC(neurod1:EGFP) transgenic embryos (Supplementary Figure S2C). To our knowledge, the functions of these epigenetic factors for the pancreas development have not been reported. Overall, all the above results suggested the proliferative as well as differentiation potentials of the progenitor population.
Lineage priming and specification of candidate islet precursors
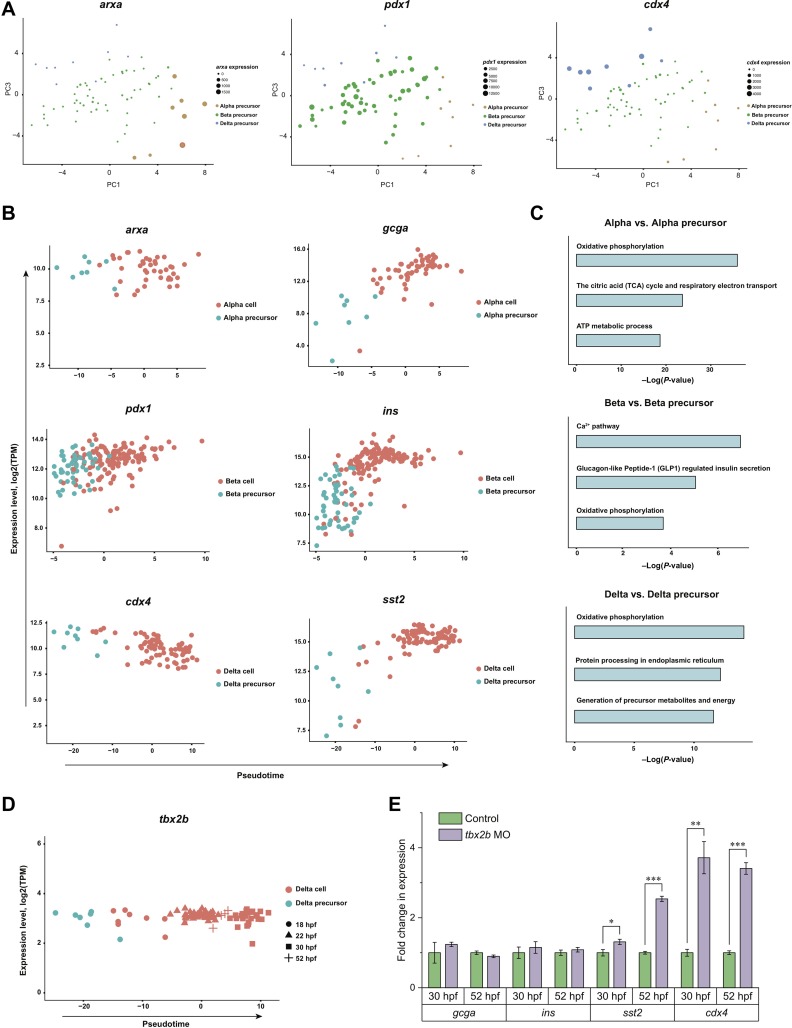
The existence of precursors during islet specification has not yet been reported, mainly due to technical difficulties. Single-cell transcriptome analysis gave us a unique opportunity to investigate this issue. Next, we focused on the analysis of the cluster in the center of the t-SNE map that was adjacent to all clusters representing the terminally differentiated cells (Figure 1B). We projected the cells of this cluster on the principal component analysis (PCA) map and profiled the expression of three lineage-specific transcription factors, representing the alpha (arxa), beta (pdx1), or delta (cdx4) cell type, respectively (Figure 3A). Three distinct subpopulations could be revealed by the mutually exclusive expression of these three transcription factors. Interestingly, when compared with their corresponding terminally differentiated cells, these three subpopulations all exhibited relatively lower expression of corresponding hormone markers (gcga, ins, or sst2), though there were no significant differences regarding the expression levels of the corresponding transcription factors, indicating that these cells were at an intermediate state of differentiation and their differentiation potential has been specified towards a defined lineage (Figure 3B). Based on these observations, we postulated this middle cluster as lineage-primed precursors. In searching for differentially expressed genes for these three types of precursors, we found interesting patterns for beta precursor cells, where two groups, one composed of 18 hpf cells and the other composed of all the remaining cells (22, 30, and 52 hpf), could be distinguished by distinct mRNA expression profiles (Supplementary Figure S3A and Table S3). While the beta precursor cells from a relatively early developmental stage (18 hpf) were enriched in GO terms such as ‘metabolism of RNA’, ‘cell cycle’, and ‘chromosome organization’, displaying a clear potential for cell proliferation, all the remaining cells at later stages (22 hpf, 30 hpf, and 52 hpf) were undergoing metabolic activities reflecting physiological properties of the beta cell lineage, including ‘glycolysis’, ‘oxidative phosphorylation’, and ‘type B pancreatic cell development’, indicating a developmental stage-related stepwise maturation process of beta cell precursors (Supplementary Figure S3B).
Figure 3.
Analyses of lineage-primed precursors. (A) PCA plot displaying the distribution of alpha, beta, and delta precursors. Cell types are indicated by colors, and the dot size is according to log2 TPM values. (B) The lineage specification of alpha (first row), beta (second row), and delta (third row) precursors and corresponding terminally differentiated cells. The x-axis indicated the pseudo-time value and the y-axis represented the expression level (log2(TPM)) of respective markers. (C) GO analysis showing the enriched terms of terminally differentiated alpha, beta and delta cells in comparison with respective precursors (from the top down). (D) The tbx2b expression level across delta lineage specification. (E) The fold change of gcga, ins, sst2, and cdx4 expression in 30 and 52 hpf zebrafish embryos between tbx2b morpholino-injected and uninjected groups as identified by qRT-PCR. The expression level was normalized to the housekeeping gene gapdh. Statistical significance was calculated by one-way analysis of variance (ANOVA). See also Supplementary Figure S3 and Table S3.
To further dissect the lineage priming and specification process from precursors to their corresponding terminally differentiated cells, we profiled the precursor as well as the terminal cells on the PCA-based pseudo-time. The results showed a clear path of transcriptional increase for each of the terminal markers following differentiation and maturation from precursors to terminal cells across the pseudo-time (Figure 3B). GO analysis results revealed that terminal differentiation was achieved by an upregulation of metabolic and secretory activities including ‘oxidative phosphorylation’, ‘calcium pathway’ and so on (Figure 3C and Supplementary Table S3).
In addition to well-established lineage-specific transcription factors, we profiled all the transcription factors expressed (with average expression >10 TPM) in the precursors and terminal cells and identified tbx2b as a potential new delta cell lineage specification factor (Figure 3D and Supplementary Figure S3C). By knocking down tbx2b expression using a morpholino disrupting its mRNA splicing, we observed an increase in the expression of the delta cell transcription factor cdx4 and terminal marker sst2, without affecting the expression level of the alpha cell marker gcga and beta cell marker ins (Figure 3E and Supplementary Figure S3D). Specific upregulation of sst2 expression following tbx2b knockdown was further confirmed by in situ hybridization (Supplementary Figure S3E). These results indicate that Tbx2b may serve as the delta cell lineage-specific factor, acting as an inhibitory regulator in a cdx4-dependent way. Taken together, our results provide evidence for the existence of heterogeneous and lineage-primed pancreatic precursors for alpha, beta, and delta cells, and their specification mechanisms towards terminal differentiation in zebrafish.
Terminally differentiated islet cells
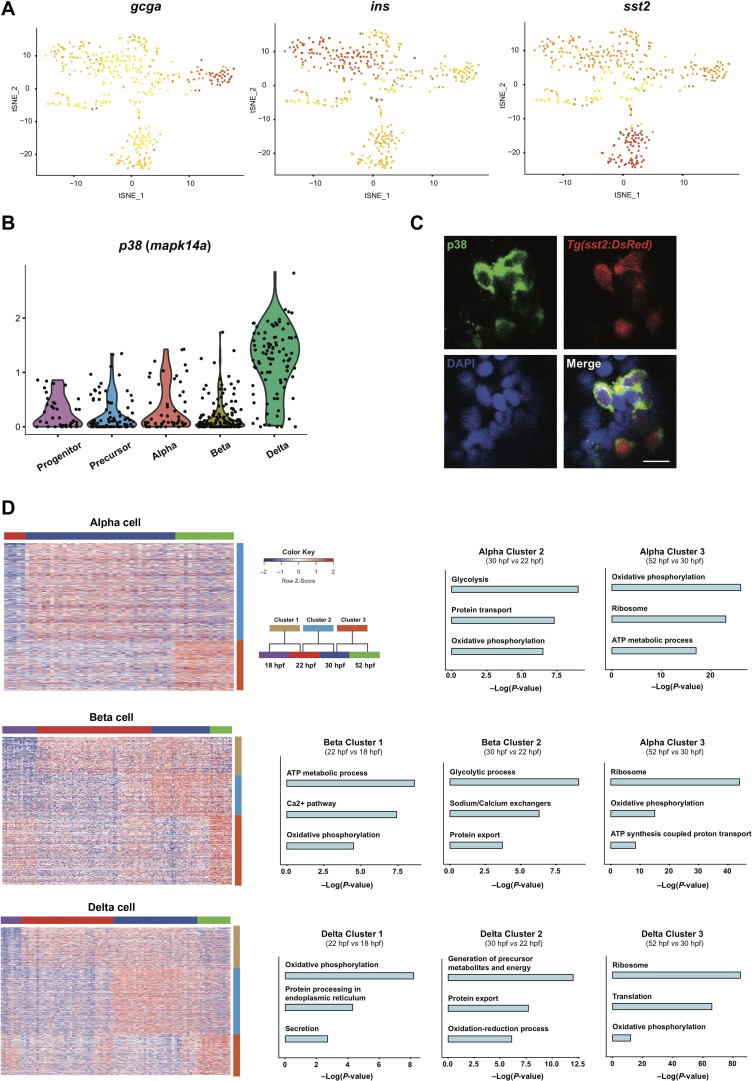
Next, we focused on the analysis of terminally differentiated alpha, beta, and delta cells, which were characterized by the relatively high expression of gcga, ins, and sst2, respectively (Figure 4A). The specifically expressed genes for each cell type were revealed by heat map profiling, and their shared GO terms showed upregulation of metabolic and secretory activities related to the functional maturation of terminally differentiated cells, as expected (Supplementary Figure S4A, B and Table S4). In addition, we identified mapk14a/p38 as a delta cell-specific marker and confirmed its expression pattern by immunofluorescent staining (Figure 4B and C).
Figure 4.
Analyses of terminally differentiated cells. (A) t-SNE representation of cells showing the expression of endocrine hormones gcga, ins, sst2. Color scale is according to log2RPKM values with the gradient from yellow to red corresponding to minimum (zero) and maximum expression, respectively. (B) The relative expression of p38/mapk14a across different cell types. (C) Immunofluorescence staining result of p38 (green) in a 6 dpf Tg(sst2:DsRed) zebrafish embryo. The bar represents 10 μm. (D) Heat map showing stage-specific genes for alpha, beta, and delta cells and their related GO terms. Clusters 1–3 indicated the enriched genes of the consecutive stage comparison of 22 hpf vs. 18 hpf, 30 hpf vs. 22 hpf, 52 hpf vs. 30 hpf, respectively. See also Supplementary Figure S4 and Table S4.
By profiling the transcriptional dynamics of each cell type from 18 hpf to 52 hpf, we revealed the functional maturation process of terminally differentiated islet cells. The comparison between consecutive stages showed that the differentiated cells undergo maturation by upregulating and maintaining ATP metabolic and secretory gene expression (Figure 4D and Supplementary Table S4). These results suggested that the terminally differentiated cells may undergo a functional maturation process with a gradual increase in metabolic and physiological gene expression after emergence and reaching a peak between 30 hpf to 52 hpf, which coincided with the increased energy demand of developing zebrafish embryos.
Pseudo-time and conservation analysis
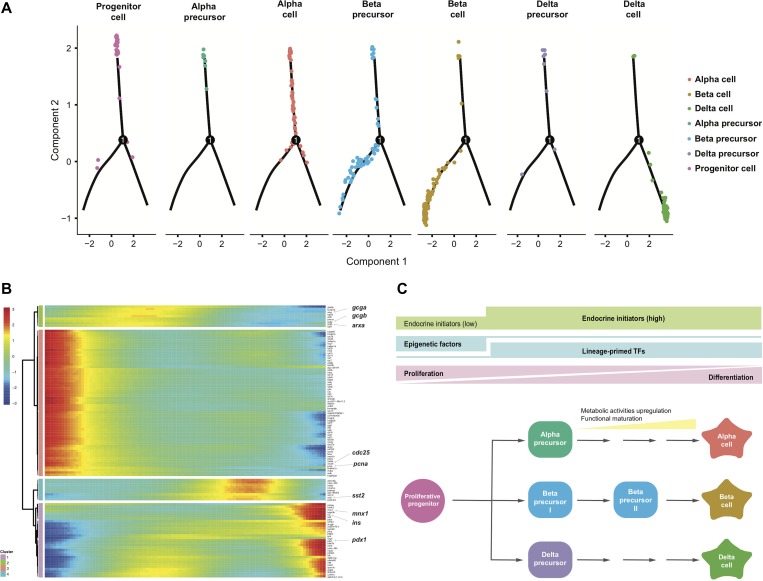
The cell state transition of endocrine islets was further characterized by a series of transcriptional dynamics. Monocle 2 (Trapnell et al., 2014; Qiu et al., 2017b, c) has emerged as a powerful tool for reconstructing the lineage differentiation trajectory and its governing gene regulatory network. Islet cells were aligned on the developmental trajectory termed as pseudo-time, and two branches were reconstructed using Monocle 2 (Figure 5A and Supplementary Figure S5A). This pseudo-time analysis was performed with the whole set of sequenced 413 cells as listed in Supplementary Table S1, the same as those applied for Figure 1B. The overall result of this pseudo-time analysis is shown in Supplementary Figure S5A, and this result was then split into seven subpanels according to particular cell types (Figure 5A), for better presentation and evaluation. In silico development started with proliferative progenitors, followed by alpha precursors and alpha terminal cells, then diverged into two fates, the precursor and terminal beta cells, and the delta cells, respectively (Figure 5A). Furthermore, branched heat map revealed four clusters of genes (not cells) showing distinct dynamics of expression patterns across pseudo-time of islet development, while the genes in Cluster 1 showed lowest expression at the beginning and their expression gradually increased following the process of islet development and reached highest level at the terminal stage, the genes in Cluster 3 showed the opposite pattern, with highest expression at initial stage of development and lowest expression at the terminal differentiation stage (Figure 5B). The associated regulatory program was first characterized by the high expression of proliferation-associated genes like pcna and cdc25 in Cluster 3, coincident with the proliferative property of the progenitor cell. Thereafter, alpha cell-associated genes including arxa and gcga in Cluster 2 were upregulated, then followed by the increased expression of genes specific for beta lineage including pdx1 in Cluster 1 and delta cell-related genes such as sst2 in Cluster 4, indicating the successive appearance of alpha, beta and delta cell lineages following the progenitor cell. Finally, the expression of beta cell marker genes including pdx1, ins, and mnx1 in Cluster 1 reached to the highest level, suggesting that the beta cells may experience a longer path towards terminal differentiation (Figure 5B and Supplementary Table S5). Overall, the pseudo-time analysis results, together with all the above-mentioned other analyses, led to a framework of regulatory events governing pancreatic islet development (Figure 5C).
Figure 5.
Pseudo-time analysis and summary of primary islet formation. (A) Monocle 2 trajectory of primary islet specification by aligning islet cells on the developmental trajectory termed as pseudo-time. Each subpanel corresponds to previously identified cell types as shown in Figure 1B. (B) Branched heat map showing the dynamically changed genes (q-value < 0.00001) across pseudo-time. Here the four clusters represented four groups of genes (not cells) showing distinct dynamics of expression patterns across pseudo-time of zebrafish islet development. (C) Summary on the process of primary islet proliferation and differentiation implied from our single-cell RNA-seq data. See also Supplementary Figure S5 and Table S5.
Although many transcription factors were conserved between zebrafish and mammals, a comparative analysis of all the protein-coding genes at the transcriptomic level is lacking. Our data showed that the transcriptome of the proliferative progenitors was more conserved across species, compared with all the other clusters, especially with nearly 90% of protein-coding orthologs found in mice and humans (Supplementary Figure S5B) (Athanasiadis et al., 2017; Carmona et al., 2017). This pattern potentially reflects the conservation of the transcriptome and molecular mechanisms regulating the development of endocrine proliferative progenitors and other cell types.
Gene duplication is a general event during the evolution of eukaryotic organisms, which is important for functional divergence. We analyzed gene duplication events before and after zebrafish speciation for differentially expressed genes and found that the majority of the paralogs were duplicated before ray-finned fish speciation (Supplementary Figure S5C) (Athanasiadis et al., 2017; Carmona et al., 2017). The percentage of functionally conserved genes (i.e. all the paralog genes were expressed in the same cell type) in the paralogs post-ray-finned fish speciation varied according to cell types, with terminally differentiated cells >50% and the progenitor cells to be the lowest, suggesting that the terminal cells may share conserved mechanisms while the progenitors adapted differential strategies (Supplementary Figure S5D).
Discussion
Although bulk RNA-seq analysis of pancreatic endocrine cells isolated from embryonic or adult zebrafish has been reported, which identified many novel markers in different cell types (Tarifeño-Saldivia et al., 2017), our work analyzed zebrafish islet development with different embryonic stages at the single-cell transcriptome level and revealed new and important cell types such as potential proliferative progenitors as well as lineage-primed precursors, which showed the unique power of single-cell RNA-seq to explore important intermediate states of cell differentiation during developmental processes. Furthermore, we identified various properties of these cells, including cell cycle status, enriched novel transcription factors, and epigenetic factors as well as the dynamic expression of these factors. Overall, our single-cell RNA-seq analysis of the islet revealed a detailed differentiation program during zebrafish embryo development and provided a potential lineage hierarchy for understanding the principal islet formation in details at the single-cell level.
Progenitors displayed a distinct proliferative potential as revealed by specific enrichment of cell cycle-related biological activities. This population could be further divided into G1/S and G2/M cells with distinct differentiation potentials, and precursors were primed for alpha, beta, and delta lineages by expressing specific transcription factors. We further described their molecular characteristics by applying several different comparison and analysis methods. For the proliferative progenitors, we identified a group of new transcription factors showing expression patterns similar to known important endocrine regulators as well as a group of epigenetic factors specifically enriched in progenitors, whose functions have not yet been revealed in endocrine development.
We also demonstrated that lineage-primed precursors formed by increasing the expression of lineage-specific transcription factors and differentiated into terminal cells by maintaining the level of these transcription factors while gradually enhancing metabolic and physiological functions. In particular, we identified tbx2b and p38 to be new delta cell-specific markers and further provided evidence demonstrating tbx2b to be a potential new negative regulator of delta cell lineage determination by a morpholino-based knockdown experiment. Finally, we revealed the presence of a stage-dependent maturation process for the terminal differentiation of the three different functional islet cells, i.e. alpha, beta, and delta cells.
Intensive analyses have focused on dissecting the functions of signaling pathways and transcription factors regulating the emergence of terminally differentiated islet cells. However, comprehensive and precise regulatory mechanisms for the differentiation program have remained elusive, partially due to the lack of characterization of intermediate developmental steps and cell populations. The potential islet lineage hierarchy, based on our data, provided a framework to understand the specific and dynamic functions of genes at the single-cell level with higher resolution.
The classic concept of discrete developmental hierarchy has been widely accepted in developmental biology, as has been used to describe the processes of hematopoietic as well as pancreatic development (Shih et al., 2013; Zhang et al., 2018). Single-cell RNA-seq analysis offered a powerful tool to analyze more detailed developmental processes at the single-base resolution and single-cell level, revealing the increasing number of subpopulations of developmental intermediates (Tritschler et al., 2017; Zhang et al., 2018). In our study, the identification of beta lineage precursor I and beta lineage precursor II indicated the potential stepwise specification of islet development. We believe that with the accumulation of more and more single-cell RNA-seq results and more detailed analyses, a continuous proliferation and differentiation landscape of the endocrine islet will gradually be completely elucidated.
Efficiently generating endocrine islet cells from stem cell cultures relies on enough knowledge about the developmental mechanisms of islets. Although progenitors transiently expressing Ngn3 were identified in mouse pancreas, the detailed mechanisms of further lineage specification/determination towards each specific islet cell type remain unclear. We propose that single-cell RNA-seq studies are necessary and would benefit in vitro differentiation methodologies. Our data showed that the beta cell lineage fate seemed to be determined at the precursor stage by upregulating and maintaining pdx1 expression, and other hormones such as gcga and sst2 could not be highly co-expressed once fate was determined. According to our data, it may not be inconceivable to maintain the expression level of beta lineage factors during a late stage of the in vitro differentiation protocol. By further uncovering the similarities and differences between in vivo and in vitro differentiation programs and developmental paths, the timing for the initiation of key transcription factors could be revealed, which might help to improve the production of functional islet cells through directed differentiation of embryonic stem (ES) cells or induced pluripotent stem (iPS) cells.
Materials and methods
Zebrafish care and use
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Peking University and conducted according to the rules of the Committee on Animal Care of Beijing, China. The IACUC reference number at Peking University was LSC-ZhangB-1.
Zebrafish were maintained at 28°C in a circulating system with a 14 h/10 h light/dark cycle. The in-tank breeding method was used to obtain embryos. Embryos were maintained in the incubator at 28°C. Larvae were anesthetized with 0.16 mg/ml Tricaine (MS-222, Sigma-Aldrich Inc.) before the dissection.
Isolation of zebrafish endocrine islets by manual dissection and single-cell preparation by FACS
The endocrine pancreas was isolated from TgBAC(neurod1:EGFP) transgenic fish (Obholzer et al., 2008) by manual dissection of the embryos into DPBS (ThermoFisher Scientific, 14040133) using tweezers (Dumont, RS-5015) under a fluorescent dissecting microscope (Leica, M165FC). Thereafter, the tissue was transferred to Accutase solution (Sigma, A6964) and digested for 3–5 min at 37°C, followed by up and down pipetting (3 min for the 18 hpf and 22 hpf embryos, 5 min for the 30 hpf and 52 hpf embryos). The dissociated sample was collected by centrifugation at 500× g for 5 min at 4°C, followed by resuspension in 350 μl DPBS. Single cells from the digested sample were sorted by the BD FACS Aria SORP (Special Order Research Product) and collected directly into the 96-well plate (Axygen, PCR-96-LP-AB-C) containing lysis buffer (2.5 μl per well). The lysis buffer consisted of 2 mM dNTP mixture (TaKaRa, 4019), 0.8 U/μl Recombinant RNase Inhibitor (TaKaRa, 2313B), 0.38% Triton-X 100 (Sigma, T8787) and 300 nM RT primer.
Single cell RNA-seq library preparation and sequencing
We followed STRT-seq steps (Islam et al., 2011, 2014) to perform single-cell transcriptome amplification with a few modifications in the RT and amplification primers (Li et al., 2017; Zhong et al., 2018). After amplification for 20 cycles, cDNAs labeled with different barcodes were pooled together and purified using the DNA Clean and Concentration Kit (ZYMO, D5044) to remove primer dimers and free primers. Thereafter, we performed a second amplification using biotin-labeled primers containing the Illumina read2 primer sequence and indexes for four cycles. The cDNAs were then fragmented by sonication with the Covaris S220 (ThermoFisher Scientific, 4465653) and the 5′ end of the first strand cDNA was enriched using C1 streptavidin beads (Invitrogen, 65002). A library was constructed using the KAPA Hyper Prep Kit from Illumina (KAPA, KK8505) according to the manufacturer’s instructions. 0.5 G of 150 bp pair-end reads were obtained for each single cell by sequencing on the Illumina Hiseq4000 as performed by Novogene.
Processing of single-cell RNA-seq data
We first segregated the raw reads on the basis of the cell-specific barcode information in read2 of the pair-ended reads. Thereafter, sequences in read1 were trimmed with customized scripts to remove the PCR primer sequence (AAGCAGTGGTATCAACGCAGAGT), poly(A) tail sequence, sequences with low-quality bases (N > 10%) or sequences contaminated with adapters. The stripped read1 sequences were then aligned to the Zv9 zebrafish reference genome (Ensembl) using TopHat (Version 2.0.12; Kim et al., 2013). Uniquely mapped reads were counted using HTseq-count from the HTSeq package (Anders et al., 2015). For each single gene, we discarded duplicated transcripts with identical UMIs. Finally, the transcript number for each gene in each cell was quantified by the number of different UMIs of that gene.
Nonlinear dimensional reduction (t-SNE) analysis
To analyze single-cell data, we ran Seurat (Version 2.2) on the TPM expression value (Butler et al., 2018). Initially, genes with expression in at least three cells were selected, whereas cells with an expressed gene number below 2000 were filtered. The highly variable genes were calculated using the FindVariableGenes function (x.low.cutoff = 0.5, x.high.cutoff = 7, y.cutoff = 0.7). Batch effects were mitigated by performing the function ScaleData. Thereafter, the function RunPCA was applied on the scaled data for PCA.
Identification of cell types
To determine statistically significant principal components, the JackStraw function (num.replicate = 100) was performed, and PCs from 1 to 13 were chosen for clustering. We then applied the FindClusters function (reduction.type = ‘pca’, resolution = 1) to cluster cells. Terminally differentiated populations (alpha, beta, and delta cells) were identified by the marker gene expression of gcga, ins, and sst2, respectively.
Identification of cell type-specific genes
To identify markers in each single cluster against all other cells, the FindAllMarkers function (min.pct = 0.25, thresh.use = 0.25, test.use = ‘roc’) was used, and genes with a power >0.4 were chosen. Seurat also provides the FindMarkers function (min.pct = 0.25, thresh.use = 0.25, test.use = ‘roc’) for differential expression, which can be used to test one cluster against all other clusters, and genes with a power >0.4 were selected. The GO enrichment analysis was performed using Metascape (http://metascape.org) (Tripathi et al., 2015).
Cell cycle analysis
A gene set of 220 G1/S related genes and 400 G2/M related genes (Supplementary Table S2) were applied to define the proliferative progenitors and to group them into three categories, including G1/S cells, G2/M cells, and quiescent cells.
Pseudo-time analysis by PCA
Pseudo-time analysis was based on PCA. PCA was performed on lineage-primed precursors and corresponding terminally differentiated cells (high expression of both hormonal markers and transcriptional factors) using variable genes, and the PC1 value was used as the pseudo-time value.
Constructing the lineage hierarchy in the islet specification
The lineage hierarchy reconstruction was performed using Monocle 2 (Trapnell et al., 2014; Qiu et al., 2017b, c). Differentially expressed genes identified by Seurat were chosen to order cells. To facilitate easy visualization and interpretation, ‘DDRTree’ was selected for dimension reduction. The beginning of the trajectory was defined according to the stage and cell type previously identified using the root_state argument in the orderCells function.
Quantification and statistical analysis
To identify marker genes, we performed the roc test in the Seurat package and chose genes with a power >0.4 as marker genes. To identify genes dynamically changed across pseudo-time, we performed the differentialGeneTest function in the Monocle 2 package and selected genes with a q-value <0.00001. ANOVA was used in Figure 3E.
Supplementary Material
Acknowledgements
We sincerely thank the Zebrafish International Resource Center (ZIRC) for providing TgBAC(neurod1:EGFP). We thank Yuying Gao and Yan Shen for lab management; Yingdi Jia and Jingliang Chen for zebrafish husbandry. We sincerely thank Shujing Wang, Zhonglin Fu and Xuefang Zhang from the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with FACS, and members of the Zhang and Tang laboratories for helpful advice and discussions. We also thank Xiaochen Li and Xuemei Hao from the Core Facilities at School of Life Sciences, Peking University in Beijing, China, for assistance with confocal microscopy. We thank Xiaohui Xue from Wuhan University for technical assistance.
Funding
This work was partially supported by grants from the National Key Basic Research Program of China (2015CB942800), the National Key Research and Development Program of China, Stem Cell and Translational Research (2016YFA0100500), and the National Natural Science Foundation of China (NSFC) (31871458, 31671500, 31730060, and 81371264).
Conflict of interest
none declared.
References
- Anders S., Pyl P.T., and Huber W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis E.I., Botthof J.G., Andres H., et al. (2017). Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat. Commun. 8, 2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M., Veres A., Wolock S.L., et al. (2016). A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechard M.E., Bankaitis E.D., Hipkens S.B., et al. (2016). Precommitment low-level Neurog3 expression defines a long-lived mitotic endocrine-biased progenitor pool that drives production of endocrine-committed cells. Genes Dev. 30, 1852–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemar F., Argenton F., Schmidtke R., et al. (2001). Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev. Biol. 230, 189–203. [DOI] [PubMed] [Google Scholar]
- Brombin A., Joly J.S., and Jamen F. (2015). New tricks for an old dog: ribosome biogenesis contributes to stem cell homeostasis. Curr. Opin. Genet. Dev. 34, 61–70. [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., et al. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona S.J., Teichmann S.A., Ferreira L., et al. (2017). Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 27, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgin G., and Prince V.E. (2015). Differential levels of Neurod establish zebrafish endocrine pancreas cell fates. Dev. Biol. 402, 81–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgin G., Ward A.B., Hao L.T., et al. (2011). Zebrafish mnx1 controls cell fate choice in the developing endocrine pancreas. Development 138, 4597–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djiotsa J., Verbruggen V., Giacomotto J., et al. (2012). Pax4 is not essential for β-cell differentiation in zebrafish embryos but modulates α-cell generation by repressing arx gene expression. BMC Dev. Biol. 12, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell J.A., Wang Y., Riesenfeld S.J., et al. (2018). Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H.A., Dong P.D., Beis D., et al. (2003). Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev. Biol. 261, 197–208. [DOI] [PubMed] [Google Scholar]
- Flasse L.C., Pirson J.L., Stern D.G., et al. (2013). Ascl1b and Neurod1, instead of Neurog3, control pancreatic endocrine cell fate in zebrafish. BMC Biol. 11, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl M.S., Karoulias N., Wende H., et al. (2006). The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic β cells and intestinal endocrine cells. Genes Dev. 20, 2465–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J., and Melton D.A. (2002). Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447–2457. [DOI] [PubMed] [Google Scholar]
- Haumaitre C., Lenoir O., and Scharfmann R. (2008). Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol. Cell. Biol. 28, 6373–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Kjallquist U., Moliner A., et al. (2011). Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 21, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Zeisel A., Joost S., et al. (2014). Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., et al. (2013). Tophat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel R.A., Dobler S., Schmitner N., et al. (2015). Diabetic pdx1-mutant zebrafish show conserved responses to nutrient overload and anti-glycemic treatment. Sci. Rep. 5, 14241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel R.A., Onder L., Wilfinger A., et al. (2011). Requirement for Pdx1 in specification of latent endocrine progenitors in zebrafish. BMC Biol. 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkel M.D., Eames S.C., Alonzo M.R., et al. (2008). Cdx4 is required in the endoderm to localize the pancreas and limit β-cell number. Development 135, 919–929. [DOI] [PubMed] [Google Scholar]
- Lawlor N., George J., Bolisetty M., et al. (2017). Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 27, 208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Dong J., Yan L., et al. (2017). Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20, 858–873. [DOI] [PubMed] [Google Scholar]
- Li J., Klughammer J., Farlik M., et al. (2016). Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 17, 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro M.J., Dharmadhikari G., Grun D., et al. (2016). A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obholzer N., Wolfson S., Trapani J.G., et al. (2008). Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J. Neurosci. 28, 2110–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y., Kosaka Y., Kishimoto N., et al. (2011). Convergence of the insulin and serotonin programs in the pancreatic β-cell. Diabetes 60, 3208–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls S., Zecchin E., Tiso N., et al. (2007). Function and regulation of zebrafish nkx2.2a during development of pancreatic islet and ducts. Dev. Biol. 304, 875–890. [DOI] [PubMed] [Google Scholar]
- Prince V.E., Anderson R.M., and Dalgin G. (2017). Zebrafish pancreas development and regeneration: Fishing for diabetes therapies. Curr. Top. Dev. Biol. 124, 235–276. [DOI] [PubMed] [Google Scholar]
- Qiu X., Hill A., Packer J., et al. (2017. b). Single-cell mRNA quantification and differential analysis with census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., et al. (2017. c). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W.-L., Zhang Y.-W., Feng Y., et al. (2017. a). Deciphering pancreatic islet β cell and α cell maturation pathways and characteristic features at the single-cell level. Cell Metab. 25, 1194–1205. [DOI] [PubMed] [Google Scholar]
- Segerstolpe A., Palasantza A., Eliasson P., et al. (2016). Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H.P., Wang A., and Sander M. (2013). Pancreas organogenesis: from lineage determination to morphogenesis. Annu. Rev. Cell Dev. Biol. 29, 81–105. [DOI] [PubMed] [Google Scholar]
- Stanescu D.E., Yu R., Won K.J., et al. (2017). Single cell transcriptomic profiling of mouse pancreatic progenitors. Physiol. Genomics 49, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarifeño-Saldivia E., Lavergne A., Bernard A., et al. (2017). Transcriptome analysis of pancreatic cells across distant species highlights novel important regulator genes. BMC Biol. 15, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani Z., and Lin S. (2011). Endocrine pancreas development in zebrafish. Cell Cycle 10, 3466–3472. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., et al. (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S., Pohl M.O., Zhou Y., et al. (2015). Meta-and orthogonal integration of influenza ‘OMICs’ data defines a role for UBR4 in virus budding. Cell Host Microbe 18, 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler S., Theis F.J., Lickert H., et al. (2017). Systematic single-cell analysis provides new insights into heterogeneity and plasticity of the pancreas. Mol. Metab. 6, 974–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen V., Ek O., Georlette D., et al. (2010). The Pax6b homeodomain is dispensable for pancreatic endocrine cell differentiation in zebrafish. J. Biol. Chem. 285, 13863–13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D.E., Weinreb C., Collins Z.M., et al. (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.J., Schug J., Won K.J., et al. (2016). Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65, 3028–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfinger A., Arkhipova V., and Meyer D. (2013). Cell type and tissue specific function of islet genes in zebrafish pancreas development. Dev. Biol. 378, 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R., Carrano A.C., and Sander M. (2015). A systems view of epigenetic networks regulating pancreas development and β-cell function. Wiley Interdiscip. Rev. Syst. Biol. Med. 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y., Kim J., Ni M., et al. (2016. a). Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc. Natl Acad. Sci. USA 113, 3293–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y., Kim J., Okamoto H., et al. (2016. b). RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 24, 608–615. [DOI] [PubMed] [Google Scholar]
- Yu X.-X., Qiu W.-L., Yang L., et al. (2018). Dynamics of chromatin marks and the role of JMJD3 during pancreatic endocrine cell fate commitment. Development 145, pii: dev163162. [DOI] [PubMed] [Google Scholar]
- Zeng C., Mulas F., Sui Y., et al. (2017). Pseudotemporal ordering of single cells reveals metabolic control of postnatal β cell proliferation. Cell Metab. 25, 1160–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.-M., Chen H., Liu W., et al. (2012). Animal TFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 40, D144–D149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao S., Xia J., et al. (2018). Hematopoietic hierarchy–an updated roadmap. Trends Cell Biol. 28, 976–986. [DOI] [PubMed] [Google Scholar]
- Zhang H.-M., Liu T., Liu C.J., et al. (2015). Animal TFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 43, D76–D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhang S., Fan X., et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.