Abstract
G protein-coupled receptors (GPCRs) are targets for ~35% of approved drugs but only ~15% of the ~800 human GPCRs are currently such targets. GPCRomics, the use of unbiased, hypothesis-generating methods (e.g., RNA-sequencing [RNA-seq]), with tissues and cell types to identify and quantify GPCR expression, has led to the discovery of previously unrecognized GPCRs that contribute to functional responses and pathophysiology and that may be therapeutic targets. The combination of GPCR expression data with validation studies (e.g., signaling and functional activities) provides opportunities for the discovery of disease-relevant GPCR targets and therapeutics. Here, we review insights from GPCRomic approaches, gaps in knowledge and future directions by which GPCRomics can advance GPCR biology and the discovery of new GPCR-targeted drugs.
Keywords: gene expression, RNA-sequencing, G protein-coupled receptors (GPCRs), cancer associated fibroblasts, G proteins, cyclic AMP
An overview of past and current efforts on GPCRs: Progress and concerns
G protein-coupled receptors (GPCRs, also termed 7-transmembrane receptors based on their structure and topology in cell membranes) are the largest family of signaling receptors in eukaryotes, including ~800 GPCRs in humans. GPCRs modulate cell and tissue physiology by regulating signaling pathways via 4 major classes of heterotrimeric (αβγ) GTP binding proteins (G proteins, see Glossary), Gs, Gi/o, Gq/11 and G12/13 and/or the scaffolding protein β-arrestin. G proteins regulate the activity of enzymes and channels that alter cellular levels of second messengers and, in turn, functional activities of cells via pathways that include protein kinases and altered phosphorylation/activity of proteins, ion channel activity, low molecular weight GTP binding proteins and changes in gene expression. Through these diverse signaling mechanisms, GPCRs regulate a wide range of cellular functions, which can include, for example, general features of cell biology (e.g., metabolism, growth, death, migration) as well as cell-specific responses (e.g., muscle contraction/relaxation, glandular/epithelial secretion, alterations in neuronal activity, etc.).
GPCRs have been a major research focus in pharmacology [1–3]. Initial efforts assessed functional activities (e.g., muscle contraction/relaxation, metabolic responses, etc.) in organisms and tissues. Subsequent biochemical studies with tissues, cells and subcellular fractions led to the discovery of cyclic AMP (cAMP) and calcium as GPCR second messengers and set the stage for direct analyses of GPCRs by radioligand binding and other techniques. The latter approaches facilitated GPCR purification, molecular cloning, biophysical analyses and structural studies (e.g., X-ray crystallography and cryo-electron microscopy) which have transformed understanding of GPCRs from hypothesized concepts (e.g., the description of α- and β-adrenoceptors [4]) to atomic level data at a few angstroms resolution (e.g., [5–11]).
GPCRs are also highly useful targets for drugs, indeed the most frequently targeted gene family: ~35% of approved drugs target GPCRs but only ~15% of the ~800 human GPCRs are currently targeted. Hence, new opportunities likely exist for GPCRs as drug targets [1–3]. Factors contributing to this “limited” targeting of GPCRs include the >100 orphan GPCRs that lack “tool compounds” to aid in defining their functional roles and, incomplete knowledge of signaling mechanisms of many GPCRs and of the identity of GPCRs expressed in healthy and diseased cells.
The application of new methodologies has provided molecular insights that have aided in the discovery of new GPCR-targeted drugs and new ideas (e.g., inverse agonism, signaling bias, GPCR oligomerization, allosteric modulation and compartmentalized GPCR signaling including from intracellular sites [10–17]). Such experiments are often conducted on well-studied GPCRs that are transfected into heterologous cells at supra-physiological levels prior to assessment of GPCR signaling, localization, regulation and action in response to agonists, antagonists and/or allosteric modulators. But do such studies overlook or misconstrue important features of GPCRs? For example, are conclusions based on experiments with well-studied GPCRs applicable to all, or even most, GPCRs? Does transfection reproduce the endogenous localization and/or alter the stoichiometry of GPCR signaling components? Do mutations and partners used to facilitate structural studies change properties of GPCRs? More generally, has sufficient attention been given to experimental artifacts (for example, the impact of mutations on receptor biology) and the “observer effect” whereby the act of measurement and methods used to assess a phenomenon can change its properties? Such questions have led us and others to assess GPCR expression and action in native cells and tissues ([3,18–32]) whereby unbiased, hypothesis-generating approaches allow one to pose questions regarding the expression of GPCRs in normal and diseased (human) cells and tissues. We term this approach GPCRomics (Figure 1, Key Figure).
Figure 1, Key Figure.
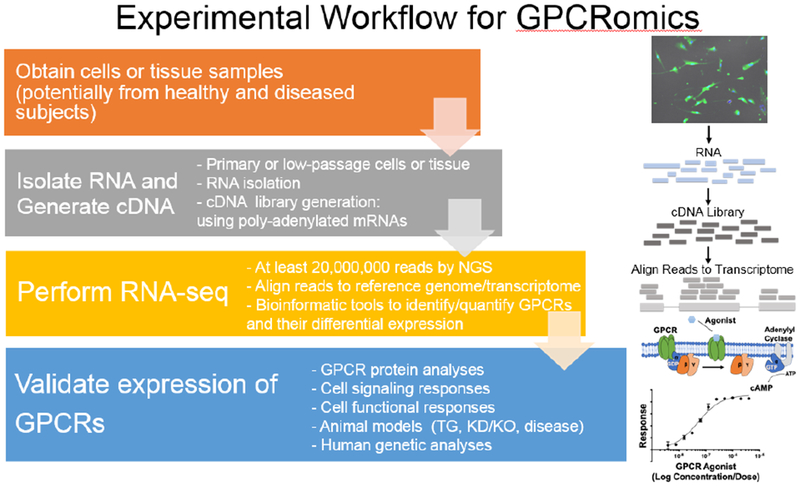
Evaluation of GPCR mRNA expression in biological samples, such as primary cells or tissues, relies on isolation of high-quality RNA, i.e. with minimal protein and DNA contamination and minimal degradation. High-throughput methods, such as RNA-seq, then identify and quantify GPCRs. This GPCR expression data forms the basis for further studies to validate the expression of GPCRs via independent mRNA and protein analyses, cell signaling and functional assays, use of animal models and human genetic analyses. Abbreviations used: NGS: next-generation sequencing, TG = transgenic; KD/KO = Knockdown/Knockout
GPCRomics: Its utility for pharmacology and drug discovery
The suffix “-ome” describes the totality of a family of entities and “-omics” defines a field of study or methodology that characterizes an -ome. Many omics areas have been proposed, e.g., genomics, transcriptomics, epigenomics, proteomics, lipidomics, glycomics and metabolomics. GPCRomics thus seeks to define and study the GPCRome. A key goal of GPCRomics is to “discover” (i.e., reveal the existence of) and characterize endogenous GPCRs (endoGPCRs) that are functionally important in health and disease. This in turn could help to elucidate potential signaling events and functional activities that endoGPCRs regulate, how states of altered physiology (e.g., age, environmental stress, hormonal status) impact on endoGPCR expression and action and whether endoGPCR expression and action change in diseased tissues and cells. In addition, GPCRomic studies can identify “new” (previously unrecognized) GPCRs that can be targets for drug discovery efforts and in turn, may yield new, efficacious and safe therapeutics, especially for conditions that lack such agents.
One seeks to identify, quantify and assess responses of GPCRs expressed in tissues or cells of experimental animal or humans. Our laboratory has focused on the assessment of individual cell types following their isolation or growth in low-passage tissue culture [3]. For drug discovery, human cells are the optimal system, especially cells from patients who are matched with healthy subjects for age, sex, ethnicity and other variables (e.g., smoking history, physical training, drug usage, etc.). GPCRomic data that compare cells from healthy and diseased subjects can reveal disease-related GPCRs that may be therapeutic targets. Culture of the cells ex vivo can facilitate screening and assessment of small molecules or biologies as potential therapeutics, thus facilitating drug discovery.
Methods for conducting and analyzing GPCRomic studies
Based on the low abundance of most GPCRs and limited availability of protein analytical methods to identify GPCRs, the currently favored GPCRomic approach is to assess mRNA expression and then undertake studies to validate the mRNA data. Several methods can be used, including real-time qPCR analyses (with GPCR probes), hybridization-based DNA microarrays (that assess a cell’s or tissue’s transcriptome), GPCR-targeted microarrays (which have DNA probes for individual GPCRs) and RNA-sequencing (RNA-seq) [3]. Because the DNA probes on whole transcriptome arrays are typically not optimized to detect individual GPCRs, those arrays are unable to detect certain GPCRs (false negatives) and in addition, yield false positives, thus limiting the utility of hybridization-based transcriptome microarrays for GPCRomic studies [3]. Commercial qPCR-based (e.g., TaqMan®) GPCR arrays can also be used to quantify GPCR expression but are limited to the analysis of ~350 GPCRs and certain house-keeping genes (e.g., [3]).
RNA-seq is currently preferred for GPCRomic studies due to its increased sensitivity and breadth of detection. A typical RNA-seq workflow (Figure 1) begins with the isolation of minimally degraded RNA [33] and its conversion (using standard methods, e.g., Truseq mRNA kits and protocols [Illumina, Inc., San Diego, CA, USA]) to complementary DNA (cDNA) libraries for sequencing. Sequencing of >20 million single 75bp reads per sample is generally sufficient to quantify expression of >20,000 coding genes (including GPCRs) and their splice variants. While RNA-seq yields more comprehensive data than the qPCR-based assay mentioned above, a challenge in the use of RNA-seq is that it requires more extensive data analysis.
Analysis of RNA-seq data involves multiple steps which include the use of various bioinformatic tools. We briefly review these steps here and in Figure 2. Raw data is first obtained as ‘FASTQ’ files that are checked for sequencing quality (e.g., using FASTQC [35]) to identify low quality reads and non-mRNA contaminants; various tools can remove these artifacts [33] (Figure 2). The FASTQ files are then input into Kallisto [36], an ‘alignment-free’, fast and computationally cheap method [33] that estimates transcript expression. The gene expression from this transcript-level data is determined using tximport [37], which yields gene-level counts that can be used for differential expression (DE) analysis. By inputting gene expression as counts into edgeR [38] or similar tools (e.g., DEseq2 [39]), one can calculate the fold-change between groups and statistical significance (False Discovery Rates [FDRs]). DE analysis can also be performed using bootstraps from Kallisto as input for the ‘Sleuth’ tool [40]. Further, one can analyze genes with DE using tools to explore enrichment of pathways involved in other aspects of GPCR biology (e.g., [41–43]).since RNA-seq provides data on such components, including GPCR ligands, G proteins and post-G protein effectors. Using tools such as those above and other methods for transcript expression [33], one can also assess if GPCRs genes undergo alternative splicing (e.g., [45]). This is a particularly interesting avenue since even though approximately 50% of GPCRs have 2 or more exons [44],alterative splicing of GPCRs remains largely unexplored.
Figure 2: RNA-sequencing and data analysis for GPCRomics.
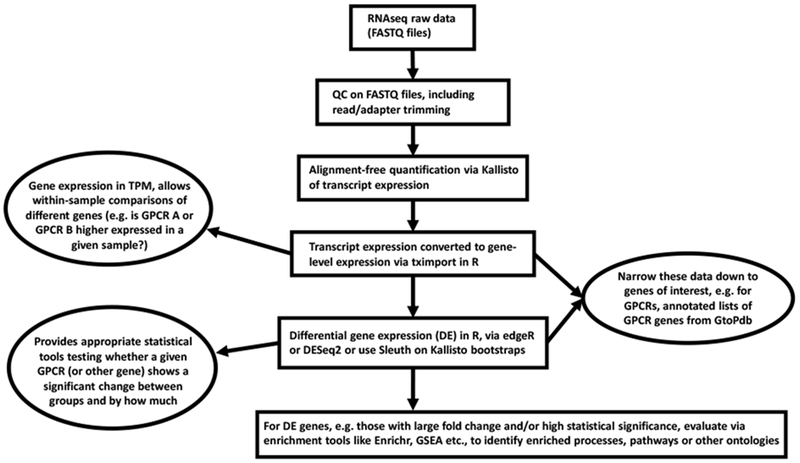
The schematic describes a workflow and tools for analyzing RNA-seq raw-data that yields gene (GPCR) expression data together with output that includes analyses of differential expression (DE), enrichment of GPCRs and mRNAs involved in GPCR-regulated signaling and response.
It is important to note here that a challenge in analyzing and interpreting GPCR expression data is how best to identify and classify GPCR genes. The Guide to Pharmacology Database (GtoPdb)i [46] provides the most comprehensive, expert-curated annotation of GPCRs and includes information on signaling, ligands, etc. Annotated GPCR genes obtained from this source can be used to query gene (GPCR) expression data to yield GPCRomic results, including the number and level of expression of GPCRs detected, that have DE (e.g., comparing GPCR expression in normal and diseased cells/tissue) and that couple to various G proteins and signaling mechanisms. One can pool results for expression of GPCRs known to link to a particular G protein and calculate potential changes in G protein signaling pathways, for example, alteration in the predicted activation of Gs versus Gi.
What does one learn from GPCRomic analyses?
GPCRomic analyses define the identity and level of expression of GPCRs in cells and tissues. One can decrease variables introduced by the heterogeneous cell types in tissues by focusing on individual cell types (e.g., [1,18,19,21,26,27,47–52]). Such analyses have yielded several observations, which include:
-
1)
Many/most cell types express at least 100 different endoGPCRs and individual cell types in a tissue have different GPCR profiles.
-
2)
These endoGPCRs include GPCRs from each of the 5 major GPCR families (Class A [Rhodopsin-like], Class B [Secretin-like], Class C [Glutamate-like], Adhesion and Frizzled); in some cases, small numbers of olfactory and taste receptors are also detected, often at low magnitudes of expression.
-
3)
G protein linkage is unknown or uncertain for many of the prominently expressed endoGPCRs.
-
4)
Many highly expressed GPCRs (including numerous orphan GPCRs) were not previously known to be expressed by the cells being studied. This is a key observation that derives from GPCRomic studies since the use of unbiased techniques can reveal evidence for functional and/or disease roles for newly recognized GPCRs (e.g., [3, 18–20]).
-
5)
Exposure of cells to perturbations such as growth factors, differentiation, hypoxia, etc. can alter GPCR expression, both in terms of the GPCRs that are expressed and their level of expression (e.g. [49–52]).
-
6)
Cells from subjects with diseases can have altered GPCR expression profiles, in terms of the identity of expressed receptors and their magnitude of expression (e.g., ]18,23,25,28,32,51]).
However, it is important to keep in mind that GPCRs identified by their mRNA expression may not be functional, hence one must test their ability to regulate cellular signaling and functional activity using cell biological, biochemical and physiological approaches.
Information regarding druggable GPCRs [1] can help guide subsequent studies and may identify GPCRs for which approved drugs might be repurposed for new therapeutic indications. By comparing the expression of individual GPCRs and the GPCR repertoire in cells from individuals of different age, sex, exercise conditioning and environmental exposures, one can define interindividual variability, age- and sex-related differences and physiological adaptations resulting in altered GPCR repertoires. Cells can be perturbed ex vivo, for example by changes in oxygen, nutrients, exposure to drugs, etc., in order to assess for cell-autonomous changes in GPCR expression and function. An example is the treatment of human lung fibroblasts with transforming growth factor-β (TGF-β) which enhances the fibrotic state of the cells and produces many changes in gene expression, including of prostanoid receptors [52]. Other studies have revealed culture time-dependent changes in GPCR expression and response, for example in human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs), findings that have implications in the use of hiPSC-CMs for disease modeling and drug testing [49].
From a drug discovery perspective, as mentioned above, one can use GPCRomics to test for disease-related changes in cellular GPCR expression in order to determine if particular GPCRs contribute to pathobiology and may be therapeutic targets. At least 3 patterns of changes in GPCR expression and action may influence disease processes and guide drug discovery efforts. These patterns involve GPCRs with:
-
1)
Low expression in normal cells but with an increase (e.g., ≥ 2-fold) or perhaps unique expression in a disease state.
-
2)
High expression (e.g., >10 tpm by RNA-seq) in both normal and diseased cells.
-
3)
Decreased expression (e.g., by >50%) in a disease setting.
Limited studies have evaluated individual GPCRs based on these patterns. We envision therapeutic challenges with patterns 2 and 3, since drug actions will occur on both normal and diseased cells. Accordingly, we have focused on pattern 1 and recently conducted studies with pancreatic ductal adenocarcinoma (PDAC) cancer associated fibroblasts (PCAFs). We compared GPCR expression by PCAFs with that of the PCAF precursor cells: pancreatic stellate cells (PSCs) and pancreatic fibroblasts (PFs) [32,51]. We discovered that PCAFs express 37 and 39 GPCRs with at least 2-fold increased expression compared to PFs and PSCs, respectively. Of note, GPR68, a proton (H+) sensing GPCR, has a prominently increased expression in PCAFs and in PDAC tumors (in which the pH in the tumor microenvironment is typically ~6.5). Further, we found that GPR68 contributes to PDAC-PCAF cell interaction via induction of GPR68 expression in PSCs promoted by PDAC cell-derived tumor necrosis factor (TNF)-a. PCAFs expressing GPR68 respond to low pH by increasing production and secretion of interleukin-6 (IL-6) which then promotes PDAC cell proliferation. GPR68 can couple to Gs and Gq [53] but its promotion of IL-6 production in PCAFs appears to occur via a cAMP/Protein Kinase A/CREB (cAMP response element-binding) protein-dependent mechanism, thus implicating Gs signaling as the mechanism for the increase in IL-6 production and secretion from PCAFs [51].
Data from GPCRomic analyses need to be validated by further studies (Figure 1), including independent qPCR and assessment of protein expression, such as by immunological (e.g., immunoblotting, flow cytometry, immunohistochemistry, etc.) or proteomic (e.g., mass spectrometry) techniques. Evidence for protein expression can also be obtained by assessment of signaling pathways and functional activity. For disease-related changes in GPCR expression, one seeks to define GPCR-mediated responses that contribute to the disease phenotype. Such studies may be aided by using short interfering (si) RNA/ short hairpin (sh) RNA, CRISPR/Cas9 and/or GPCR-targeted drugs. For validation of a potential target from GPCRomic findings, evidence of functional activity is key (the sine qua non) for validation of a GPCR prior to initiating drug discovery efforts. Functional and signaling responses may also be useful as readouts in drug screening programs.
Limitations of current GPCRomic approaches
RNA-seq and other methods that assess GPCR expression have certain limitations. Most studies to date have assessed tissues or mixed populations of cells. However, recent evidence, especially from assessment of mRNA expression in single cells, i.e., single cell gene expression, indicates that sub-populations of cells differentially express many mRNAs, including GPCRs with different expression profiles and magnitude of expression, including in individual neurons [54,55], vascular and immune cells [56,57) and with cell-specific changes in disease models. Such studies typically isolate single cells using microfluidic techniques. Further, RNA expression methods provide “snapshot” data unless multiple samples are collected to determine the temporal dynamics of mRNA expression [26]. It is conceivable that changes in GPCR expression occur in response to perturbations (e.g., drug treatments) as a result of alterations in mRNA formation and/or degradation, neither of which is assessed by “bulk” measurements of a cell’s or tissue’s GPCR mRNA expression. Moreover, although recent studies have implied a closer relationship between mRNA and protein than was previously thought [58–61], RNA expression does not necessarily reflect protein expression in all cases. Unbiased methods to assess cellular GPCR protein expression are currently limited, in part because of the paucity of well-validated, sensitive and specific GPCR antibodies (e.g., [62,63]). Other methods, such as proteomic/phosphoroteomic (mass spectrometry) [e.g., 64-66] and translatomic (differential centrifugation to isolate polysomes) [67] approaches have not been extensively used to assess GPCR expression. The generally lower expression of GPCR proteins compared to many other cellular proteins further makes the detection of GPCR proteins a challenge. It can also be difficult to draw conclusions about the physiological actions of GPCRs from GPCRomic data. For example, a GPCR might have constitutive (inherent) activity but occupancy by an endogenous agonist is usually required to elicit functional, especially maximal, responses. While studies with cells ex vivo generally require activation by an agonist, this may not be possible when assessing orphan GPCRs following their identification by GPCRomics analyses.
A further challenge in GPCRomic studies relates to the cells being investigated. In some cases, one can readily isolate and assess primary cells (e.g., circulating blood cells). For other cell types, tissues must be disrupted, cells isolated and then grown in tissue culture (optimally for a limited number of passages) prior to mRNA isolation. Such experiments typically involve exposure of cells to conditions not identical to those in vivo and may introduce experimental artifacts and the observer effect noted above. This is a particular concern with human cell lines derived from tumors, immortalized by viral infection or introduction of certain genes--manipulations that may alter cellular gene (including GPCR) expression (e.g., [68]).
Concluding remarks
GPCRomics can help identify a role for individual and groups of GPCRs in health and disease and reveal “new” GPCRs as drug targets, thus providing a basis for drug discovery efforts or repurposing of approved drugs. It may identify previously unrecognized GPCRs in diseases, including in types of cancer that are major contributors to morbidity and mortality but for which GPCRs have been largely ignored as targets (e.g., [18,23,25,32,69–75]). However, numerous questions exist in the field (see Outstanding Questions) and will likely be addressed in future studies. A focus will be on assessing GPCR expression in single cells, which, as noted above, may identify sub-populations of cell types with distinct GPCR profiles, perhaps cell- and disease-specific functional splice variants and interactions of GPCRs with G proteins and other partners [45,76–80]. Studies are also needed to identify the mechanisms that mediate changes in GPCR mRNA expression in physiological and disease settings. Such effects may occur via altered transcription or post-transcriptional events. Relatively little is known regarding the transcription factors, nuclear events and epigenetic mechanisms that regulate GPCR gene expression and is a potentially fruitful area for research efforts (e.g., [81–83]). Understanding is also limited regarding mechanisms that alter GPCR protein expression as related to translation, post-translational modification, and protein degradation and will likely be a focus of future studies.
Further, drug discovery and approved therapeutics have expanded from an earlier focus on small molecules to a broader palate of biological entities, including peptides, proteins, antibodies/nanobodies, anti-sense oligonucleotides, aptamers and genes (gene therapy). GPCRomics may identify GPCRs amenable to such approaches and may help guide appropriate hiPSC-derived models for drug screening and perhaps, cell therapy. Newly recognized GPCRs may also be useful as biomarkers that can be analyzed on circulating (tumor, fetal) cells, exosomes in the blood or urine or as cell surface proteins detectable with antibodies or other probes, perhaps via imaging methods [32,84–89]. Such techniques may further facilitate personalized/precision medicine approaches for GPCR therapeutics: identification of individual and groups of patients who express one or more GPCRs that can be treated with appropriate GPCR-targeted agents and whose response is monitored by biomarker analyses. While such ideas are currently hypothetical, we opine that GPCRomics will likely aid in their actualization. In sum, we believe that GPCRomics--an unbiased, hypothesis-generating approach--is a powerful tool for revealing aspects of GPCR biology and can be a gateway for drug discovery and ultimately, clinical/translational pharmacology and therapeutics.
Highlights.
GPCRomic analysis, currently based on mRNA studies (in particular, the use of RNAseq) is a hypothesis-generating approach that can identify and quantify previously unrecognized GPCRs.
GPCRomic studies reveal that various cell types typically express >100 of the ~360 known human endoGPCRs, including numerous orphan GPCRs.
Previously unrecognized (“new”) GPCRs may be physiologically important, contribute to pathophysiology and will likely expand the utility of GPCRs as therapeutic targets in multiple disease settings.
GPCRomic analyses may reveal increased GPCR mRNA expression in such disease settings and thereby new GPCRs as therapeutic targets.
The therapeutic application of GPCRomic discoveries will benefit from new approaches, such as gene editing, nanobodies, aptamers and gene therapy.
Outstanding Questions Box.
Can more precise ways than those that currently exist be developed to assess and predict GPCR protein expression in cells?
Can new, non-invasive approaches be discovered that can define and quantify cellular GPCR signaling, in particular in single cells?
What are the mechanisms that mediate alterations in GPCR expression and functionality?
Can GPCRomic studies identify previously unrecognized GPCRs that can be targets for drug discovery efforts, especially for conditions that lack safe and effective therapeutics?
How can one optimally utilize results from GPCRomics to accelerate drug discovery?
Acknowledgements
The authors are grateful to the prior investigators in our laboratory and collaborating investigators in other laboratories who have participated in our GPCRomic studies. Recent efforts on this topic were supported by research and training grants from the National Institute of Health (HL 098062, GM007752, CA121938, HL007444, CA189477, AG053568) with additional research support from the Department of Defense (W81XWH-14–1-0372), Bristol Myers Squibb, an ASPET David Lehr Award and the Padres Pedal the Cause PTC2017 award.
GLOSSARY
- Allosteric modulation
alteration of the effects of a primary ligand by indirectly influencing (modulating) the primary ligand’s effects on a receptor (or other protein)
- α- and β-adrenoceptors
also called α- and β-adrenergic receptors, these GPCRs are expressed on many cell types and are activated endogenously by norepinephrine (noradrenaline) and epinephrine (adrenaline). The receptors consist of the multiple subtypes (α-1, α-2 and their sub-types; β-1,β-2, β-3), and have important functions in the nervous system, heart, blood vessels, kidney, lung, fat and other tissues.
- Biomarkers
a biological marker indicative of a disease state or lack thereof. Used in medicine to screen or monitor patients for a disease.
- Bootstraps
iterative estimates that assess uncertainty in abundance of transcript quantification in the analysis of RNA-seq data.
- endoGPCRs
GPCRs endogenously expressed by a cell or tissue.
- Ex vivo
experiments done on tissues or cells from an organism with minimal changes made to the natural conditions the tissues/cells would be exposed to in a live organism. This is in contrast to in vitro experiments whereby cells derived from organisms have been adapted in the laboratory to conditions outside of their natural biological context.
- False Discovery Rate (FDR)
a statistical approach that attempts to define the rate of rejected null hypotheses that are incorrect rejections in a setting of multiple comparisons.
- FASTQ files
large text files with sequenced reads from RNA-seq along (or other NGS platforms) with quality scores that indicate the confidence of accurate identification of individual bases.
- GPCRomics
the comprehensive study of GPCR expression and function in a cell, cell type or tissue (i.e., the GPCRome). As shown in Figure 1, a typical GPCRomics workflow involves several steps: mRNA isolation, cDNA generation, RNA-sequencing and associated data analysis, and validation studies of GPCRs that are identified.
- Inverse agonism:
a drug action that decreases a response below the level of activity observed in the absence of the drug.
- Kallisto
an ‘alignment-free’, fast and computationally cheap method that estimates transcript expression (in transcripts per million, tpm- the number of times a particular transcript occurs per million transcripts sequenced) and counts (estimated number of times a transcript occurs).
- Orphan GPCR
a GPCR whose physiological agonist is unknown.
- Prostanoid receptors.
a family of GPCRs that selectively binds and responds to prostaglandins, which are arachidonic acid-derived molecules.
- Radioligand binding
a method to detect and quantify GPCRs (and other types of receptors) by the use of radiolabeled molecules (“radioligands”) that bind to the receptors.
- RNA-sequencing (RNA-seq)
a technique that utilizes next-generation sequencing (NGS) to identify and quantify the mRNA (prepared as complementary DNA [cDNA]) of a biologic sample.
- Signaling bias
(also termed functional selectivity): activation of a receptor by a drug/compound that selectively activates a particular signal transduction pathway relative to other drugs/compounds that bind to the same receptor and activate multiple pathways.
- Single cell gene expression
sequencing the RNA of an individual cell shows the variability between individual cells within a given population. This is in contrast to non-single cell analyses, which give the average gene expression for an entire population.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Resources
i) The Guide to Pharmacology Database (GtoPdb): http://www.guidetopharmacology.org/
References
- 1.Sriram K and Insel PA (2018) G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol. Pharmacol. 93, 251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milligan G (2018) G protein-coupled receptors not currently in the spotlight: free fatty acid receptor 2 and GPR35. Br J Pharmacol 175, 2543–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Insel PA et al. (2015) G Protein-Coupled Receptor (GPCR) Expression in Native Cells: “Novel” endoGPCRs as Physiologic Regulators and Therapeutic Targets. Mol. Pharmacol. 88, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahlquist RP (1948) A Study of the Adrenotropic Receptors. Am. J. Physiol. Content 153, 586–600 [DOI] [PubMed] [Google Scholar]
- 5.Wu F et al. (2017) Structure and Function of Peptide-Binding G Protein-Coupled Receptors. J. Mol. Biol. 429, 2726–2745 [DOI] [PubMed] [Google Scholar]
- 6.Hilger D et al. (2018) Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 25, 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vass M et al. (2018) Chemical Diversity in the G Protein-Coupled Receptor Superfamily. Trends Pharmacol. Sci. 39, 494–512 [DOI] [PubMed] [Google Scholar]
- 8.Thal DM et al. (2018) Recent advances in the determination of G protein-coupled receptor structures. Curr. Opin. Struct. Biol. 51, 28–34 [DOI] [PubMed] [Google Scholar]
- 9.Manglik A and Kruse AC (2017) Structural Basis for G Protein-Coupled Receptor Activation. Biochemistry 56, 5628–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thal DM et al. (2018) Structural insights into G-protein-coupled receptor allostery. Nature 559, 45–53 [DOI] [PubMed] [Google Scholar]
- 11.Munk C et al. (2019) An online resource for GPCR structure determination and analysis. Nat. Methods 16, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wootten D et al. (2018) Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 19, 638–653 [DOI] [PubMed] [Google Scholar]
- 13.Smith JS et al. (2018) Biased signalling: from simple switches to allosteric microprocessors. Nat. Rev. Drug Discov. 17, 243–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milligan G et al. (2019) GPCR homo-oligomerization. Curr. Opin. Cell Biol. 57, 40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurevich VV and Gurevich EV (2018) GPCRs and Signal Transducers: Interaction Stoichiometry. Trends Pharmacol. Sci. 39, 672–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eichel K and von Zastrow M (2018) Subcellular Organization of GPCR Signaling. Trends Pharmacol. Sci. 39, 200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg ZY and Puthenveedu MA (2018) Regulation of G protein-coupled receptor signaling by plasma membrane organization and endocytosis. Traffic DOI: 10.1111/tra.12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiga A et al. (2016) Transcriptome analysis of G protein-coupled receptors in distinct genetic subgroups of acute myeloid leukemia: identification of potential disease-specific targets. Blood Cancer J. 6, e431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koyama H et al. (2016) Comprehensive Profiling of GPCR Expression in Ghrelin-Producing Cells. Endocrinology 157, 692–704 [DOI] [PubMed] [Google Scholar]
- 20.Amisten S (2016) Chapter 5 - Quantification of the mRNA expression of G protein-coupled receptors in human adipose tissue In G Protein-Coupled Receptors 132 (Shukla K, A. B. T.-M. in C. B., ed), pp. 73–105, Academic Press; [DOI] [PubMed] [Google Scholar]
- 21.Flegel C et al. (2016) Characterization of non-olfactory GPCRs in human sperm with a focus on GPR18. Sci. Rep. 6, 32255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang M et al. (2016) Gene expression profiling reveals different molecular patterns in G-protein coupled receptor signaling pathways between early- and late-onset preeclampsia. Placenta 40, 52–59 [DOI] [PubMed] [Google Scholar]
- 23.Balenga N et al. (2017) Orphan Adhesion GPCR GPR64/ADGRG2 Is Overexpressed in Parathyroid Tumors and Attenuates Calcium-Sensing Receptor-Mediated Signaling. J. Bone Miner. Res. 32, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amisten S et al. (2017) A comparative analysis of human and mouse islet G-protein coupled receptor expression. Sci. Rep. 7, 46600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhat RR et al. (2018) GPCRs profiling and identification of GPR110 as a potential new target in HER2+ breast cancer. Breast Cancer Res. Treat. 170, 279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egerod KL et al. (2018) Profiling of G protein-coupled receptors in vagal afferents reveals novel gut-to-brain sensing mechanisms. Mol. Metab. 12, 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts MJ et al. (2018) The inhibition of human lung fibroblast proliferation and differentiation by Gs-coupled receptors is not predicted by the magnitude of cAMP response. Respir. Res. 19, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kübler E and Albrecht H (2018) Large set data mining reveals overexpressed GPCRs in prostate and breast cancer: potential for active targeting with engineered anti-cancer nanomedicines. Oncotarget 9, 24882–24897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehrlich AT et al. (2018) Expression map of 78 brain-expressed mouse orphan GPCRs provides a translational resource for neuropsychiatric research. Commun. Biol. 1, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flegel C et al. (2015) RNA-Seq Analysis of Human Trigeminal and Dorsal Root Ganglia with a Focus on Chemoreceptors. PLoS One 10, e0128951–e0128951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray P et al. (2018) Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 159, 11325–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Insel PA et al. (2018) GPCRomics: GPCR Expression in Cancer Cells and Tumors Identifies New, Potential Biomarkers and Therapeutic Targets. Front. Pharmacol. DOI: 10.3389/fphar.2018.00431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conesa A et al. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sriram K et al. High content detection of GPCR mRNA expression: Which methods work best? (manuscript submitted).
- 35.Andrews S (2010) FastQC A Quality Control tool for High Throughput Sequence Data, Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc [Google Scholar]
- 36.Bray NL et al. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525. [DOI] [PubMed] [Google Scholar]
- 37.Soneson C et al. (2016) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD et al. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI et al. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. DOI: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pimentel H et al. (2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687. [DOI] [PubMed] [Google Scholar]
- 41.Ogata H et al. (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 27, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D et al. (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markovic D and Challiss RAJ (2009) Alternative splicing of G protein-coupled receptors: physiology and pathophysiology. Cell. Mol. Life Sci. 66, 3337–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piltonen M et al. (2018) Alternative Splicing of the Delta-Opioid Receptor Gene Suggests Existence of New Functional Isoforms. Mol. Neurobiol. DOI: 10.1007/s12035-018-1253-z [DOI] [PubMed] [Google Scholar]
- 46.Harding SD et al. (2018) The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 46, D1091–D1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snead AN and Insel PA (2012) Defining the cellular repertoire of GPCRs identifies a profibrotic role for the most highly expressed receptor, protease-activated receptor 1, in cardiac fibroblasts. FASEB J. 26, 4540–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker CS et al. (2015) A second trigeminal CGRP receptor: function and expression of the AMY 1 receptor. Ann. Clin. Transl. Neurol. 2, 595–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung G et al. (2016) Time-dependent evolution of functional vs . remodeling signaling in induced pluripotent stem cell-derived cardiomyocytes and induced maturation with biomechanical stimulation. FASEB J. 30, 1464–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klepac K et al. (2016) The Gq signalling pathway inhibits brown and beige adipose tissue. Nat. Commun. 7, 10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiley SZ et al. (2018) GPR68, a proton-sensing GPCR, mediates interaction of cancer-associated fibroblasts and cancer cells. FASEB J. 32, 1170–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subhendu M, Sheng W, Michkov A, Sriram K, Sun R, Dvorkin A, I.P. J.L. Prostaglandin E2 inhibits pro-fibrotic function of human pulmonary fibroblasts by disrupting Ca2+-signaling. Revis. Manuscr. Submitt. [DOI] [PMC free article] [PubMed]
- 53.Huang X-P et al. (2015) Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 527, 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spaethling JM et al. (2016) Single-cell transcriptomics and functional target validation of brown adipocytes show their complex roles in metabolic homeostasis. FASEB J. 30, 81–92 [DOI] [PubMed] [Google Scholar]
- 55.Spaethling JM et al. (2014) Serotonergic neuron regulation informed by in vivo single-cell transcriptomics. FASEB J. 28, 771–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tischner D et al. (2017) Single-cell profiling reveals GPCR heterogeneity and functional patterning during neuroinflammation. JCI insight 2, 95063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaur H et al. (2017) Single-cell profiling reveals heterogeneity and functional patterning of GPCR expression in the vascular system. Nat. Commun. 8, 15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li JJ et al. (2014) System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ DOI: 10.7717/peerj.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Csárdi G et al. (2015) Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast. PLOS Genet. 11, e 1005206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koussounadis A et al. (2015) Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. DOI: 10.1038/srep10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edfors F et al. (2016) Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. DOI: 10.15252/msb.20167144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beermann S et al. (2012) Commercially available antibodies against human and murine histamine H4-receptor lack specificity. Naunyn. Schmiedebergs. Arch. Pharmacol. 385, 125–135 [DOI] [PubMed] [Google Scholar]
- 63.Marchalant Y et al. (2014) Validating Antibodies to the Cannabinoid CB2 Receptor. J. Histochem. Cytochem. 62, 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu JJ et al. (2018) In vivo brain GPCR signaling elucidated by phosphoproteomics. Science 360, eaao4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao K and Sun J (2018) Elucidating structural and molecular mechanisms of β-arrestin-biased agonism at GPCRs via MS-based proteomics. Cell. Signal. 41, 56–64 [DOI] [PubMed] [Google Scholar]
- 66.Mincarelli L et al. (2018) Defining Cell Identity with Single-Cell Omics. Proteomics DOI: 10.1002/pmic.201700312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tráfier A et al. (2018) Investigation methods to explore G protein-coupled receptor-regulated translatome. C. R. Biol. 341, 65–74 [DOI] [PubMed] [Google Scholar]
- 68.Atwood BK et al. (2011) Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics DOI: 10.1186/1471-2164-12-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aust G et al. (2016) Adhesion GPCRs in Tumorigenesis. pp. 369–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nohata N et al. (2017) Onco-GPCR signaling and dysregulated expression of microRNAs in human cancer. J. Hum. Genet. 62, 87–96 [DOI] [PubMed] [Google Scholar]
- 71.Moody TW et al. (2018) Neuropeptide G Protein-Coupled Receptors as Oncotargets. Front. Endocrinol. (Lausanne). DOI: 10.3389/fendo.2018.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma X et al. (2018) Application of Nanoparticles for Targeting G Protein-Coupled Receptors. Int. J. Mol. Sci. DOI: 10.3390/ijms19072006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng C-M et al. (2018) Frizzled Receptors as Potential Therapeutic Targets in Human Cancers. Int. J. Mol. Sci. DOI: 10.3390/ijms19051543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nieto Gutierrez A and McDonald PH (2018) GPCRs: Emerging anticancer drug targets. Cell. Signal. 41, 65–74 [DOI] [PubMed] [Google Scholar]
- 75.Nugent A and Proia RL (2017) The role of G protein-coupled receptors in lymphoid malignancies. Cell. Signal. 39, 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Masuho I et al. (2015) Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci. Signal. 8, ral23–ral23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flock T et al. (2017) Selectivity determinants of GPCR–G-protein binding. Nature 545, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sokolina K et al. (2017) Systematic protein-protein interaction mapping for clinically relevant human GPCRs. Mol. Syst. Biol. 13, 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Atanes P et al. (2018) Defining G protein-coupled receptor peptide ligand expressomes and signalomes in human and mouse islets. Cell. Mol. Life Sci. 75, 3039–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Crépieux P et al. (2017) A Comprehensive View of the β-Arrestinome. Front. Endocrinol. (Lausanne). 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nag JK and Bar-Shavit R (2018) Transcriptional Landscape of PARs in Epithelial Malignancies. Int. J. Mol. Sci. 19, 3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Itcho K et al. (2018) Aberrant G protein-receptor expression is associated with DNA methylation in aldosterone-producing adenoma. Mol. Cell. Endocrinol. 461, 100–104 [DOI] [PubMed] [Google Scholar]
- 83.Law IKM et al. (2017) Role of G protein-coupled receptors-microRNA interactions in gastrointestinal pathophysiology. Am. J. Physiol. Gastrointest. Liver Physiol. 313, G361–G372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cazorla-Vázquez S and Engel FB (2018) Adhesion GPCRs in Kidney Development and Disease. Front. Cell Dev. Biol. DOI: 10.3389/fcell.2018.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang X et al. (2018) GPRC5A: An Emerging Biomarker in Human Cancer. Biomed Res. Int. 2018, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matters G and Harms J (2018) Utilizing Peptide Ligand GPCRs to Image and Treat Pancreatic Cancer. Biomedicines DOI: 10.3390/biomedicines6020065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang H et al. (2018) Lighting up the brain: genetically encoded fluorescent sensors for imaging neurotransmitters and neuromodulators. Curr. Opin. Neurobiol. 50, 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Medapati MR et al. (2017) Chapter 10 - Characterization of GPCRs in extracellular vesicle (EV) In G Protein-Coupled Receptors Part A 142 (Shukla AKBT-M in C. B., ed), pp. 119–132, Academic Press; [DOI] [PubMed] [Google Scholar]
- 89.El Buri A et al. (2018) The sphingosine 1-phosphate receptor 2 is shed in exosomes from breast cancer cells and is N-terminally processed to a short constitutively active form that promotes extracellular signal regulated kinase activation and DNA synthesis in fibroblasts. Oncotarget 9, 29453–29467 [DOI] [PMC free article] [PubMed] [Google Scholar]


