Abstract
Nascent RNA may form a three-stranded structure with DNA, called an R-loop, which has been linked to fundamental biological processes such as transcription, replication and genome instability. Here, we provide a detailed protocol for a newly developed strategy, named R-ChIP, for robust capture of R-loops genome-wide. Distinct from R-loop-mapping methods based on the monoclonal antibody S9.6, which recognizes RNA–DNA hybrid structures, R-ChIP involves expression of an exogenous catalytically inactive RNASEH1 in cells to bind RNA–DNA hybrids but not resolve them. This is followed by chromatin immunoprecipitation (ChIP) of the tagged RNASEH1 and construction of a strand-specific library for deep sequencing. It takes ~3 weeks to establish a stable cell line expressing the mutant enzyme and 5 more days to proceed with the R-ChIP protocol. In principle, R-ChIP is applicable to both cell lines and animals, as long as the catalytically inactive RNASEH1 can be expressed to study the dynamics of R-loop formation and resolution, as well as its impact on the functionality of the genome. In our recent studies with R-ChIP, we showed an intimate spatiotemporal relationship between R-loops and RNA polymerase II pausing/pause release, as well as linking augmented R-loop formation to DNA damage response induced by driver mutations of key splicing factors associated with myelodysplastic syndrome (MDS).
Introduction
Genomes are the underlying templates for many biological processes in the nucleus that are temporally and spatially coordinated, including transcription, replication and epigenetic DNA and histone modification. In addition to many well-documented regulatory proteins, functional RNAs and specific nucleic acid structures, such as R-loops, have also been increasingly recognized as having important regulatory functions in the genome.
R-loops form when nascent RNA exiting from the exit channel within RNA polymerase II anneals back to template DNA, leaving non-template DNA single stranded. Although initially considered to be rare co-transcriptional by-products, emerging evidence suggests that R-loops are more widely distributed across the genome1–3. A number of specific genomic regions are thought to have high R-loop-forming propensity, including promoter and terminator regions of numerous genes2,4, enhancers4,5 and telomere and centromere regions6,7. Previous biochemical and genomics studies have demonstrated that R-loop-forming regions are tightly associated with GC-rich, especially G-rich (known as GC-skew), sequences on the non-template DNA8,9. Such DNA segments have the potential to form a G-quadruplex, a secondary DNA structure containing guanine tetrads that are thought to promote R-loop formation by disrupting the normal helical structure of DNA, allowing a newly transcribed RNA to anneal back to template DNA4,10–12. Importantly, we and others have recently shown that the presence of a free RNA end or a DNA nick greatly enhances R-loop formation by facilitating the invasion of RNA into double-stranded DNA (dsDNA)4,13.
It has been increasingly recognized that R-loops are highly dynamic, implying that R-loops are regulated during their formation and/or resolution2,4,14. R-loop formation is promoted by active transcription15, RNA polymerase pausing4 and, intriguingly, head-on transcription–replication collisions16, and is counteracted by various RNA-binding proteins involved in RNA splicing17, nuclear export18,19 and degradation5, regulators of DNA conformation20 and replication21, and chromatin modifiers22. As for the resolution of existing R-loop structures, RNase H endonucleases and various helicases, e.g., DHX9 (refs. 23,24) and SETX25, are known to either cleave the RNA moiety in the RNA–DNA hybrid or unwind the hybrid to resolve the R-loop. Despite the rapid accumulation of knowledge on R-loop biology, our understanding of the regulatory pathway for R-loop formation and resolution has remained incomplete. As R-loops appear to play critical roles in DNA replication26,27, transcriptional control28,29, DNA damage response30 and chromosome segregation7, excessive numbers of R-loops have been shown to interfere with both transcription31 and replication32, leading to elevated genome instability and other functional defects in gene expression, which have been linked to various forms of cancer and neurodegenerative disease33–35.
Given its important impact on both physiological and pathological processes, R-loop biology is of broad interest to researchers in multiple fields. Key to the study of R-loop formation and regulation is the development of robust methodologies for the accurate and comprehensive analysis of R-loops in the genome. Traditionally, R-loops have been analyzed by electron microscopy10, dot-blot hybridization and immunostaining7,36. Although useful for visualizing and quantifying overall R-loop levels, these approaches do not provide detailed information on R-loop formation and dynamics at individual genomic loci. Immunoprecipitation (IP) with the S9.6 monoclonal antibody specific for RNA–DNA hybrids, coupled with deep-sequencing technology, has enabled genome-wide mapping of R-loops in vitro, which has greatly advanced our knowledge of R-loop biology2,3,8,37,38. However, various limitations of these S9.6-based approaches have also been noted in recent studies (see below).
To precisely probe R-loops in vivo, we have taken advantage of the catalytically inactive form of RNASEH1, a conserved enzyme from bacteria to humans that performs R-loop resolution39, to develop the R-ChIP technology, and have revealed the functional relationship between R-loops and transcription pausing4. On the basis of its successful application in both adherent (HEK293T) and suspension (K562) cells, we have further performed R-ChIP to reveal excessive R-loop formation as a key disease mechanism in MDS33. Here, we provide a detailed R-ChIP protocol as a complementary R-loop-mapping approach to those based on the use of S9.6 to facilitate its application in the field of R-loop biology.
Overview of the R-ChIP procedure
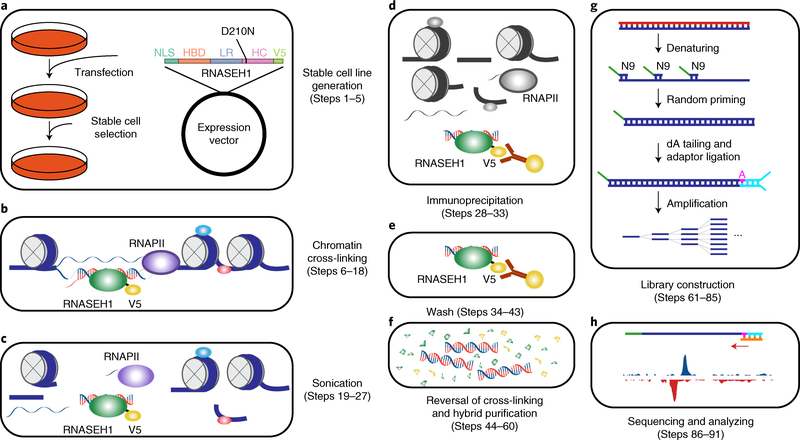
The whole procedure is summarized in Fig. 1, which consists of five stages: establishment of stable cells expressing a mutant RNASEH1 tagged with a V5 sequence at the C terminus (Steps 1–5), chromatin cross-linking and fragmentation (Steps 6–27), ChIP (Steps 28–60), library construction (Steps 61–85) and sequencing and data analysis (Steps 86–91).
Fig. 1 |. Overview of the R-ChIP workflow.
a, Cells are transfected with an expression vector containing a mutant RNASEH1 (D210N) gene with a V5 tag at its N terminus, followed by drug selection to obtain a stable cell line expressing the mutant protein. b, Cells from a are first cross-linked to stabilize DNA–protein interactions. RNA and DNA molecules are colored in red and blue, respectively. Proteins are shown as ellipses. c, Nuclei are isolated and sheared by sonication. d, RNASEH1 mutant associated with RNA–DNA hybrids is immunoprecipitated with V5 antibody-conjugated beads. e, Beads are washed to remove nonspecifically bound chromatin fragments. f, RNA–DNA hybrids are recovered by reversal of cross-linking and purification. g, RNA–DNA hybrids are denatured, and the resultant ssDNA is converted into dsDNA by extension with a primer containing a 9-nt random sequence at the 3′ end. dA is added to the 3′ end of the extension product, followed by adaptor ligation (shown in cyan) to only one end, as the other end is blocked by the 5′ overhang generated by the N9 primer. PCR amplification is then performed for library construction. h, Single-end sequencing is performed and the results are analyzed. dA, deoxyadenosine triphosphate; HBD, RNA–DNA hybrid binding domain; HC, RNA–DNA hybrid catalytic domain; LR, linker region; NLS, nuclear localization signal; RNAPII, RNA polymerase II; ssDNA, single-stranded DNA; V5, V5 tag.
Generation of stable cell lines is the most time-consuming step; it involves the introduction of a construct expressing a mutant form (D210N in the catalytic domain) of RNASEH1 into the cell of interest by transfection or viral transduction followed by antibiotic selection4. A stable cell line expressing the WKKD mutant (W43A, K59A, K60A and D210N in both the catalytic and binding domains) can be constructed in parallel for comparison, although this type of control may not be needed in every experiment. For cross-linking and fragmentation, cells are harvested and fixed with formaldehyde, followed by cell lysis, nuclei extraction and sonication. Fragmented chromatin is then subjected to ChIP with antibody-conjugated beads, multiple rounds of washes, reversal of crosslinking, RNase A and proteinase K treatment and DNA recovery. If the quality of the recovered DNA passes the qPCR assessment for signal enrichment on certain known R-loop sites, both immunoprecipitated (IPed) and the corresponding input DNA are further processed for strand-specific library construction. A primer with a 9-nt random sequence at its 3′ end is used to anneal to the DNA moiety of the RNA–DNA hybrid, followed by extension to generate dsDNA. Next, ‘dA tailing’ is performed at the 3′ end of the dsDNA, followed by adaptor ligation. The ligated product is amplified by PCR with high-fidelity DNA polymerase and a barcode-containing primer. DNA fragments in the size range of 140–350 bp are gel-selected and purified for quantification by a Qubit fluorometer, followed by deep sequencing on a standard Illumina sequencer.
Although the R-ChIP procedure presented above is related to many aspects of standard ChIP-seq, it has several unique features: (i) the establishment of a set of mutant RNASEH1-expressing cell lines, in which the cell line expressing the WKKD mutant provides a critical negative control for R-loop peak calling; (ii) construction of the library by using a unidirectional strategy for adaptor ligation and signal amplification, thus preserving the strand information of the sequencing reads; and (iii) performance of a bioinformatics analysis of the resulting sequencing data by first confirming the strand specificity and then performing peak calling by using the background information from input or WKKD mutant control DNA.
Comparison with other sequencing-based methods
Early efforts used indirect footprint approaches to locate R-loop-forming regions. In principle, single-stranded DNA (ssDNA) in the R-loop structure can be modified at cytosine residues by bisulfite under non-denaturing conditions, and several early studies thus combined bisulfite conversion and Sanger sequencing to map R-loops at specific genomic loci of interest17,28,40.
DNA/RNA immunoprecipitation sequencing (DRIP-seq) is the first and most widely used technique developed for genome-wide capture of R-loops by using fragmented chromatin for IP with S9.6 antibody, followed by sequencing of the recovered R-loop-containing DNA fragments8. Similar approaches have been applied to multiple biological systems, including yeast3, plants38 and mammals41. To further improve the resolution and robustness of DRIP-seq, a number of strategies have been developed to sequence RNA2,41 or template DNA38 in S9.6-captured material. Other efforts to increase the resolution, specificity or sensitivity of DRIP-seq-based approaches have also been made through combining DRIP with bisulfite footprinting to identify R-loop-associated ssDNA (bisDRIP-seq)42, or via S1 nuclease digestion to remove non-template ssDNA in R-loop regions before sonication in order to prevent its re-annealing back to template DNA during IP (S1-DRIP-seq)3. It remains unclear to what extent sonication may disrupt fragile R-loops, especially with unfixed cells43,44.
DRIP-seq and its derivatives rely on the specificity of the S9.6 antibody, which has been recently questioned44–46. S9.6 predominantly recognizes RNA–DNA hybrids in a sequence-independent manner. However, it also binds dsRNA with a lower affinity than RNA–DNA hybrids47, leading to substantial false-positive signals44,46. This is because nascent RNA transcripts, part of which may be folded into dsRNA, are known to extensively associate with transcribing DNA in the nucleus48,49, and thus, such chromatin-tethered RNAs would give rise to false-positive signals. A key control in both DRIP-seq and DRIPc-seq is the treatment of the same sample before IP with purified bacterial RNase H, to digest RNA within R-loops2,8. However, caution must be taken in interpreting data from such ‘control’. Owing to biased restriction digestion of DNA or insufficient RNA fragmentation, DRIP-seq and DRIPc-seq may capture not only R-loops, but also their associated DNA and RNA fragments43. In DRIPc-seq, for example, a portion of an RNA may be engaged in R-loop formation, whereas other parts of the RNA remain as unengaged RNA that contains both single- and double-stranded regions. Once the anchor in the R-loop is removed by RNase H treatment, all associated RNA would be lost. Therefore, it would be difficult to conclude that all RNase H-sensitive signals correspond exactly to the R-loop formation regions. The S9.6 antibody may also capture any dsRNA or dsRNA-containing RNA anchored to DNA via triplex formation50–55 or other unknown mechanisms. This is supported by a recent report showing that DRIPc-seq detects a substantial fraction of RNase H-resistant signals that are instead sensitive to a dsRNA-specific endoribonuclease (RNase III) in yeast46.
In contrast to S9.6, which has only a fivefold higher affinity for RNA–DNA hybrids than for dsRNA47, RNASEH1 encoded in mammalian genomes has an affinity for RNA–DNA hybrids 25-fold and 100-fold higher than for dsRNA and dsDNA, respectively56. Therefore, in theory, it can be used as an alternative way to capture R-loops, which may help address various controversies related to R-loops detected by S9.6-based methods43,44. Indeed, DRIVE-seq was initially developed on the basis of such a principle by using purified catalytically inactive but binding-competent RNASEH1; however, DRIVE-seq appears quite inefficient in recovering RNA–DNA hybrids in vitro, as compared with DRIP-seq8. The low efficiency in the in vitro capture experiment might be due to suboptimal experimental conditions, or alternatively, to the possibility that RNASEH1 might be more efficient in targeting R-loops inside cells through functional interactions with various cofactors57; therefore, such mutant RNASEH1 can be used for robustly capturing R-loops in vivo instead of in vitro58–60. On the basis of this reasoning, we developed R-ChIP by using binding competent but catalytically inactive RNASEH1 under in vivo conditions. By cell fixation followed by sonication, as in a standard ChIP protocol, R-ChIP preserves R-loop confirmations formed in vivo, provides signals with high resolution and demonstrates a striking strand specificity of the signal4,33.
A thorough comparison of R-ChIP and S9.6-based methods, including DRIP-seq, DRIPc-seq and RDIP-seq, has been performed as reported4. R-ChIP-mapped R-loops showed high resolution and accordance with all known sequence features of R-loop-forming regions, suggesting improved accuracy and specificity. Although both R-ChIP and S9.6-based methods revealed enrichment of R-loops at transcription start sites (TSSs), the major discrepancy is highly enriched signals in both gene bodies and transcription termination sites (TTSs) detected by S9.6-based methods, but not by R-ChIP. The theoretical basis for such discrepancy is currently unknown. A formal possibility is that R-ChIP captures the action sites of RNASEH1 in the genome, but not in other regions, such as gene bodies and gene ends.
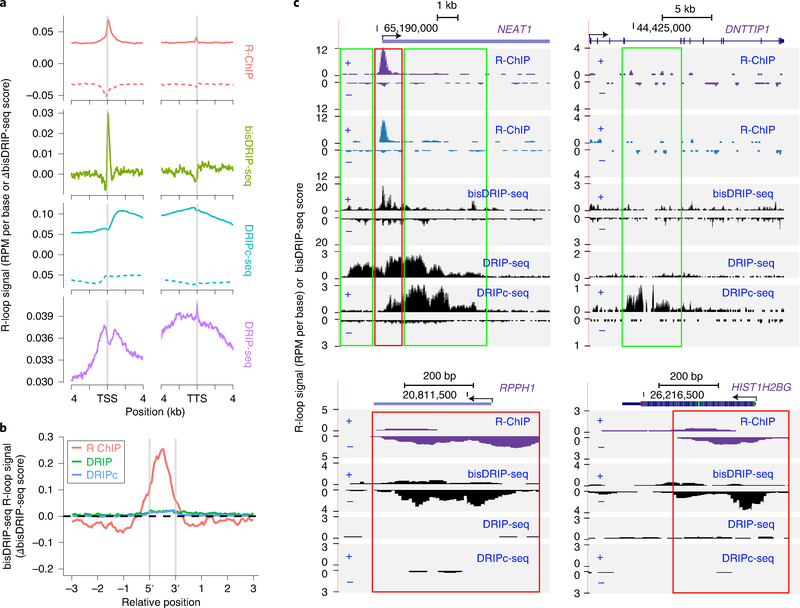
Because of the major discrepancy between the existing R-loop mapping methods, it is important to develop technologies independent of S9.6 or RNASEH1. The problem is that, currently, there is no ‘gold standard’ for detecting R-loops. The recently developed bisDRIP-seq technology combines S9.6 capture with bisulfite sequencing42, which is appealing on the basis of the principle of its experimental design. Probably because of the noisy nature of the data, the authors had mainly used ensemble data for analysis after combining multiple independently generated libraries. We used the same ensemble bisDRIP-seq data to compare with the signals captured by R-ChIP versus those detected by DRIP-seq and DRIPc-seq. On the basis of the meta-gene analysis, both R-ChIP and bisDRIP-seq detected Rloops at TSSs, but few in gene bodies or ends, which is in contrast to strong signals detected by both DRIP-seq and DRIPc-seq (Fig. 2a). A high consistency between R-ChIP and bisDRIP-seq is further illustrated by compiling bisDRIP-seq signals on mapped genomic regions with different methods, showing that the bisDRIP-seq signals are highly enriched at the peak regions detected by R-ChIP, but not by DRIPc-seq and DRIP-seq (Fig. 2b). On specific gene examples, such as NEAT1, a broadly expressed long noncoding RNA, R-loop signals were detected by all four methods, but the signals detected by both R-ChIP and bisDRIP-seq were narrowly enriched near its TSS, whereas much broader signals were detected by both DRIP-seq and DRIPc-seq (Fig. 2c, top left). For DNTTIP1, signals in a broad region of the gene body were detected by DRIPc-seq and less so by DRIP-seq, but no specific signal was detected by either R-ChIP or bisDRIP-seq (Fig. 2c, top right). By contrast, for RPPH1 and HIST1H2BG, the two gene loci previously documented by bisDRIP-seq, R-ChIP, but not DRIPc-seq or DRIP-seq, showed the same signals as bisDRIP-seq (Fig. 2c, bottom panels). Together, these comparisons suggest a high degree of agreement between R-ChIP and bisDRIP-seq.
Fig. 2 |. Comparison of R-ChIP and S9.6-based methods.
a, Meta-gene analysis of signals detected by R-ChIP4, bisDRIP-seq42, DRIPc-seq2 and DRIP-seq15 at the ±4-kb region of TSS and TTS, respectively. Note that y-axis units are different for individual methods. R-loop signals are measured in terms of reads per million (RPM) per base by R-ChIP, DRIPc-seq and DRIP-seq, or the difference between template and non-template strand bisDRIP-seq scores by bisDRIP-seq42. bisDRIP-seq signals above or below 0 indicate the existence of R-loops on the forward or reverse strand. For R-ChIP and DRIPc-seq, solid and dotted lines represent potential R-loop signals from the forward and reverse strands, respectively. b, bisDRIP-seq R-loop signals at peak regions defined by R-ChIP, DRIPc-seq (DRIPc) and DRIP-seq (DRIP) data. c, Signals detected by R-ChIP (purple for K562 cells and blue for HEK293T cells), bisDRIP-seq, DRIPc-seq and DRIP-seq at representative genomic loci. Regions with consistent signal profiles between R-ChIP and bisDRIP-seq are highlighted by red rectangles, whereas those detected only by DRIP-seq or DRIPc-seq are highlighted by green rectangles. TSS, transcription start site; TTS, transcription termination site.
Despite various theoretical considerations and comparisons among the existing data, we believe that it is premature to suggest which method is more accurate than another at this point, which clearly requires further investigation. It is entirely possible that different approaches may query distinct aspects of R-loops. Here, we focus on describing the advantages, potential limitations and future improvements of R-ChIP in detail in order to enable future users to explore this alternative R-loop mapping method.
Advantages of R-ChIP
High specificity for capturing R-loops in vivo
RNASEH1 is a conserved endonuclease that specifically recognizes RNA–DNA hybrids and has been previously used to detect R-loops at specific genomic loci58–60. By comparing the sequencing data from R-ChIP performed with RNASEH1 containing a point mutation in the catalytic site (D210N) with those from R-ChIP performed with RNASEH1 carrying additional mutations in its nucleic acid binding domain (WKKD), we further confirmed that almost all captured R-loop sites are RNASEH1 binding domain dependent4, indicating a high specificity of this strategy. Furthermore, unlike the majority of S9.6-based methods, the R-ChIP procedure includes a cell-fixation step, by which the native state of the R-loop configuration is stabilized during the IP step.
R-loop mapping with high resolution
For DRIP-seq and most of its derivatives, genomic DNA is fragmented by restriction digestion, which limits their resolution (around a few kilobases for DRIP-seq and DRIPc-seq)2,43. By contrast, the resolution of R-ChIP depends on the fragment size after sonication, as in standard ChIP-seq, resulting in an average size of mapped R-loop peaks ~200–300 bp, comparable with the R-loop size range directly visualized by electron microscopy10. Such high resolution greatly helps to locate actual R-loop-forming sites and to reveal critical sequence features associated with R-loop formation relative to surrounding epigenetic modification events, as we demonstrated4,33.
Strand information
We have taken advantage of the hybrid composition of R-loops by selectively amplifying the DNA moiety in RNA–DNA hybrids, thus preserving the strand information of the sequencing reads. We noted that sequencing either the RNA or the DNA strand of the RNA–DNA hybrid has been recently implemented in DRIPc-seq2 and ssDRIP-seq38. Such strand specificity is critical for judging library quality, especially in various genomic regions where DNA is transcribed in both directions. This strand specificity also helps reveal a precise spatial relationship of an R-loop with certain strand-specific features (e.g., G-quadruplexes), most of which reside at the 5′ end of R-loops on the non-template strand.
Limitations of R-ChIP
Expression of an exogenous RNASEH1
R-ChIP requires engineered expression of a catalytically dead RNASEH1 in the cells of interest. Establishing such a cell line is time consuming and limits the application of R-ChIP for certain systems, especially when working with tissues or animals. In addition, although we did not notice any phenotypical defects, such as cell cycle arrest and cell death, for the cell lines we have studied, it is conceivable that the mutant RNASEH1 may compete with endogenous enzymes or other proteins for binding to RNA–DNA hybrids. One particular concern is the potential stabilization of R-loops by exogenously expressed RNASEH1. However, this may provide an opportunity for capturing some labile R-loops.
Potentially incomplete R-loop map
Although RNASEH1 is supposed to specifically target R-loops in vivo, we cannot rule out the possibility that this enzyme binds only a subset of R-loops in the genome. To date, a number of proteins involved in the regulation of R-loop dynamics have been reported21,23,25, some of which may have functions complementary to those of RNASEH1 in resolving R-loops. Second, as for all ChIP-based methods, it is also possible that certain R-loop-forming regions may not be fully accessible to exogenous RNASEH1 due to binding competition with other factors or the formation of certain DNA/chromatin structures in vivo. Future efforts in developing alternative R-loop mapping technologies may help test these possibilities and address this and other potential limitations of R-ChIP.
Lack of differentiation between R-loops and other types of RNA–DNA hybrids
RNA–DNA hybrids are known to accompany transcription, as well as DNA replication and repair. As both RNASEH1 and S9.6 recognize RNA–DNA hybrids instead of the whole R-loop structure, none of the R-loop-mapping methods to date can differentiate R-loops from other types of RNA–DNA hybrids. Interestingly, we noted during re-analysis of the bisDRIP-seq data that bisDRIP-seq detects numerous signals in intergenic regions that do not correspond to annotated transcription regions, and more importantly, those bisDRIP-seq signals lack strand specificity, implying that such signals might reflect RNA–DNA hybrids associated with DNA replication, an intriguing possibility to be further investigated in future studies.
Future improvements
So far, the R-ChIP protocol has been applied to several human cell lines to generate genome-wide R-loop maps. We expect to see its potential applications to other cell types or tissues from transgenic animals. To avoid the potential side effects of a constitutively expressed mutant RNASEH1, it may be necessary to express the mutant in an inducible system, so that its expression can be transiently induced before performing R-ChIP. The induced level of RNASEH1 can also be titrated in order to achieve efficient R-loop capture without eliciting any molecular interference due to overexpression.
The resolution of R-ChIP may be further increased by including a nuclease digestion step, as shown for ChIP-exo61. During deep sequencing, we currently use single-read sequencing to read 40 nt from one end of the library, and the sequencing reads must be computationally extended to the size of the averaged DNA fragments in the library. Paired-end sequencing is clearly advantageous in reading the sequences that cover the actual regions bound by the mutant RNASEH1. It may also be important to test a variation of R-ChIP by sequencing recovered RNA instead of DNA, which may further improve the resolution of the R-ChIP technology.
It is unclear to what extent R-ChIP has missed real R-loop signals. Similar to recent coupling between DRIP-seq and bisulfite sequencing42, it may be interesting to couple R-ChIP with bisulfite sequencing at two levels. The first would be to perform bisulfite sequencing on captured R-loops, which will score information on both strands (the template strand by R-ChIP and the non-template strand by bisulfite sequencing) for comparison. Second, it may be important to perform genome-scale bisulfite sequencing to identify all exposed single-stranded regions in an undenatured condition. The results can then be compared with R-ChIP-captured R-loops, which may help differentiate between transcription-induced RNA–DNA hybrids and those associated with DNA replication.
Like ChIP-seq62, R-ChIP can be further developed to a single-cell level and/or to generate quantitative information in the future, which will be critical for studying the functional relationship between R-loop formation and dynamics and gene expression.
Experimental design
Establishment of stable cell lines expressing an exogenous V5-tagged RNASEH1
To generate a stable cell line expressing the mutant RNASEH1 (D210N), we previously cloned the mutant RNASEH1-coding region into the pPyCAG expression vector. We also replaced the N-terminal mitochondrial localization signal with a nuclear localization signal sequence and fused a V5 tag at the C terminus4. Next, we transfected HEK293T or K562 cells with the vector and selected for stable cell lines expressing the mutant RNASEH1 with hygromycin B. No clonal selection was performed, which could be done if preferred. Because the vector contains a strong CAG promoter, the expression level of an exogenous RNASEH1 is much higher than that of its endogenous counterpart33. However, on the basis of cell counting, cell cycle profiling and assay for apoptosis, we detected no obvious cellular defects in any of our selected stable cell lines in comparison with parental cells. Other expression vectors, such as lentiviral- or retroviral-based gene expression systems, might be optimal for certain cell types that are difficult to transfect.
When seeding cells for R-ChIP, we usually estimate the initial cell number and growth rate in order to reach 70–80% cell confluence on the day of the experiment. One day before harvesting, we change the medium once to maintain active cell growth. For a single R-ChIP experiment, we typically use 5 × 106–107 cells to obtain sufficient IPed DNA for library construction. To obtain statistically meaningful results, we normally generate libraries from two to three replicates of R-ChIP, each with a control library generated from conresponding input DNA, and use comparable numbers of sequencing reads from these libraries for data analysis.
Preparation of V5 antibody-conjugated beads
Conjugation of the antibody targeting the V5 tag with magnetic beads is performed before harvesting the cells (see the ‘Reagents’ section for further information on antibody bead preparation). The conjugated antibody beads have been tested for IP efficiency and then for R-ChIP-enriched signals on several known promoters by qPCR. We previously tested two types of magnetic beads from Thermo Fisher, Pierce Protein A/G beads and Dynabeads Protein A/Protein G, and found both suitable for generating highly reproducible results. Conjugation was usually performed overnight at 4 °C. We found that 2.5–3 μg of V5 antibody per IP is typically enough to pull down 5–10 ng of chromatin fragments for library preparation, although such yield may vary due to cell-type differences and antibody efficiency, and thus, the antibody amount and cell number may need to be optimized in a specific experimental setting.
Cross-linking and cell lysis
Fresh 1% (vol/vol) formaldehyde is recommended for cross-linking the cells. As RNASEH1 directly binds to RNA–DNA hybrids, 10 min of fixation at room temperature (20–25 °C) is sufficient for the formation of the protein–chromatin complex for HEK293T (adherent) and K562 (suspension) cells. The best fixation timing must be empircally determined for individual cell types.
Sonication
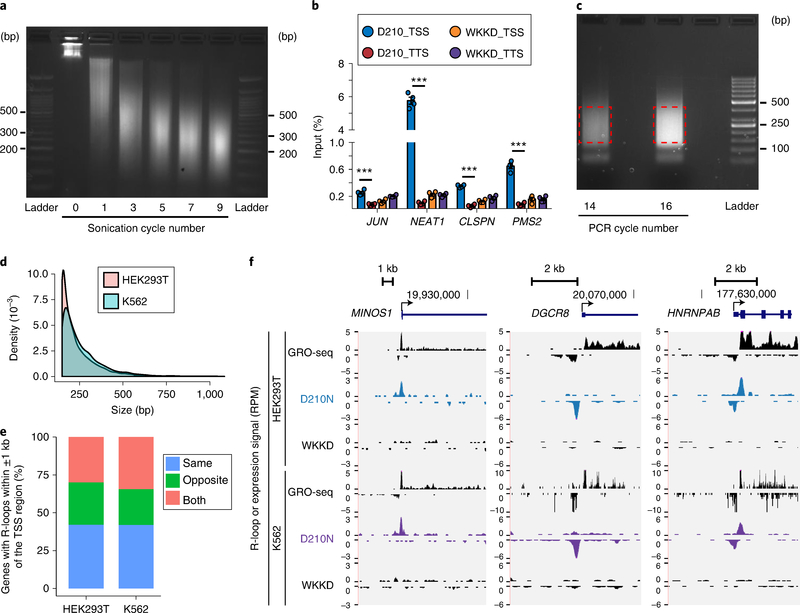
As in regular ChIP experiments, sonication is one of the most critical steps for R-ChIP. To obtain good enrichment and resolution, sufficient chromatin shearing is required, yet overheating during sonication could result in reversal of cross-linking and disruption of protein complexes. We use a probe sonicator for shearing the chromatin. Using 500 μl of nuclear lysate in a 1.5-ml LoBind tube, we immerse the bottom part of the tube in ice water and sonicate the lysate for 10 s, followed by recovery for 1 min on ice per sonication cycle. According to our experience, seven to eight rounds of sonication are sufficient to shear chromatin into 100- to 600-bp fragments in our sonication setting (see also Fig. 3a). As many variables, such as cell type and number, and different sonication systems, may markedly affect the sonication efficiency, a pilot test to find the best sonication conditions is strongly recommended.
Fig. 3 |. Expected results of R-ChIP.
a, Size distribution of chromatin fragments before and after different numbers of sonication cycles is assayed on a 2% (wt/vol) agarose gel. As denoted, seven cycles of sonication enable the majority of DNA to be sufficiently fragmented. b, The R-ChIP-qPCR signals relative to input for several genomic loci on K562 cells are shown as mean ± s.e.m. (n = 4, P < 0.001, two-sided t test). c, Amplification of an R-ChIP library by different numbers of PCR cycles. d, Size distribution of R-ChIP peaks in HEK293T and K562 cells. e, Out of all genes with R-loops detected within ±1 kb of the TSS region (false-discovery rate <0.01; signal enrichment >5-fold), the percentages of three groups of genes are shown, for which R-loops are identified in the same, opposite or both directions of transcriptional activity. f, Snapshots of three representative R-loop-containing genomic loci corresponding to the three gene groups in e. R-ChIP (D210N) and control (WKKD), as well as GRO-seq signals, are displayed for HEK293T (ref. 4) and K562 (ref. 68) cell lines.
ChIP
Thorough wash of magnetic beads greatly reduces background noise. When processing a large number of samples, we usually divide them into a few groups; samples used for direct comparison are put in the same group (four to six samples per group), such that long incubation time with high-salt buffer and experimental variations can be minimized for samples in the same group.
Quality assessment of IPed RNA–DNA hybrids
To ensure ChIP enrichment quality and reproducibility, we suggest preparing two to three biological replicates and making aliquots of a small amount of purified DNA for a qPCR test before library construction. In addition, in order to more specifically assess signal enrichment, purified ChIPed DNA from cells expressing the RNASEH1 (WKKD) mutant may be included for comparison, although this type of control may not be needed in every experiment. The results are calculated and shown as the percentage of input (Fig. 3b). The relative enrichment can be evaluated as the fold change by comparing the signals from R-loop-positive regions, such as some TSS regions of highly expressed genes with high GC content and GC skew, with those from control regions, such as intergenic regions close to the selected positive regions.
Library construction and sequencing
We recommend measuring the concentration of the recovered DNA and using a similar amount of DNA from each sample for library construction. Input samples usually yield a large amount of DNA (4–8 μg). We usually use 5–10 ng for library construction. A primer with a 9-nt random sequence at its 3′ end for random seeding on the DNA moiety of RNA–DNA hybrids is used to generate dsDNA with a 5′ overhang. Phi29 DNA polymerase is chosen for primer extension because of its exceptional processivity and strand displacement activity. Other DNA polymerases with similar features, for example, DNA polymerase I, large (Klenow) fragment, may be used for primer extension as well. We recommend running a test PCR to choose the lowest number of PCR cycles at which bright smear signals can be seen in a 2% (wt/vol) agarose gel (see also Fig. 3c; 16 cycles is chosen in this case). To ensure high complexity and avoid excessive PCR duplication, it is essential to start with a sufficient amount of ChIPed DNA for library construction and keep the number of PCR cycles as low as possible (normally ≤18).
We previously sequenced R-ChIP libraries by using the single-end mode, and the read length is 40 nt on a standard Illumina sequencer (i.e., HiSeq 2500). At least 15–20 million uniquely mapped reads for each replicate of human cell lines (30–40 million after combining two replicates) are minimally required for peak calling and downstream data analysis. We also suggest obtaining similar or more sequencing reads for the corresponding input DNA in order to provide sufficient coverage of background signals for enrichment analysis.
Data analysis
R-ChIP is designed to sequence the 5′ end of the template strand DNA (Fig. 1h), thus making the data analysis different from that of typical ChIP-seq. Uniquely mapped reads after removing PCR duplicates should first be separated on the basis of their strand information. The reads also must be extended to the average size of gel-isolated DNA fragments to better pinpoint the location of individual R-loop regions, as the original mapped reads cannot represent the actual length of each recovered fragment. We use MACS v.2 software63 to identify R-loop-forming regions by separately using sequencing reads from the Watson or Crick strand in comparison with either the WKKD or the input library. If each biological replicate is sequenced with sufficient depth, we recommend calling robust peaks by irreproducible discovery rate methodology, developed by the ENCODE consortium64,65, or by counting common peaks of all replicates. Alternatively, if a high reproducibility (e.g., R ≥ 0.8) is seen, we usually combine data from all replicates to obtain a conservative set of Rloop peaks by using a stringent cutoff, e.g., fold change ≥5 and a q value ≤0.01. Usually, the average peak size is ~200 bp (Fig. 3d). Note that R-loop signals in the antisense direction with regard to the annotated genes are prevalent at promoter regions (Fig. 3e), possibly due to divergent transcription. The strand specificity offered by R-ChIP technology could faithfully assign these R-loops to where they originated, which is further supported by corresponding nascent RNA transcription from global nuclear run-on sequencing (GRO-seq) data (Fig. 3f).
Materials
Biological materials
HEK293T cells (a gift from S. Dowdy, University of California, San Diego)
K562 cells (ATCC, cat. no. CCL-243) ! CAUTION The cell lines used should be regularly checked to ensure that they are authentic and are not infected with mycoplasma.
Reagents
DMEM (Corning, cat. no. 10–013-CV)
RPMI1640 (Corning, cat. no. 10–040-CV)
Opti-MEM I reduced-serum medium (Thermo Fisher, cat. no. 31985–070)
FBS (Omega, cat. no. FB-11)
Sodium pyruvate (Thermo Fisher, cat. no. 11360–070)
Penicillin–streptomycin (Thermo Fisher, cat. no. 15140–122)
Expression vector pPyCAG (a gift from the J.C. Izpisua Belmonte lab)
RNASEH1 expression vectors (wild type (WT; Addgene, plasmid no. 111906), D210N (Addgene, plasmid no. 111904) and WKKD (Addgene, plasmid no. 111905)) were deposited in Addgene. Refer to the following link for more information: https://www.addgene.org/Xiang-Dong_Fu/)
Lipofectamine 2000 (Thermo Fisher, cat. no. 11668–030)
Hygromycin B (Thermo Fisher, cat. no. 10687010)
The antibody against the V5 tag (Santa Cruz, cat. no. sc-83849-R) originally used in our experiments for IP, R-ChIP and western blot has been discontinued. Alternative antibodies used by our colleagues include anti-V5 tag antibody from Abcam (cat. no. ab15828) and monoclonal antibody - V5-tag (D3H8Q) rabbit mAb from Cell Signaling (cat. no. 13202)
Pierce protein A/G beads (Thermo Fisher, cat. no. PI88802)
Glycogen (Thermo Fisher, cat. no. FERR0561)
Sodium chloride (NaCl; Sigma-Aldrich, cat. no. s9888)
EDTA (0.5 M, pH 8.0; Thermo Fisher, cat. no. 15575–020)
Tris-Cl buffer (pH 8.0; Lonza, cat. no. 51238)
Triton X-100 (Sigma-Aldrich, cat. no. X100–500ML)
Molecular-grade H2O (Corning, cat. no. 46–000-CM)
BSA (Gemini, cat. no. 700–100P)
PBS (Thermo Fisher, cat. no. 14190–144)
Formaldehyde (37% (vol/vol); Sigma-Aldrich, cat. no. 252549–100ML) ! CAUTION Formaldehyde is toxic if swallowed, upon contact with the skin or if inhaled. Wear a lab coat, goggles and gloves and work in a chemical hood. All formaldehyde waste must be kept and disposed of according to local and institutional regulations.
Glycine (Sigma-Aldrich, cat. no. G7126–500G)
SigmaFast Protease Inhibitor Cocktail Tablets (Sigma-Aldrich, cat. no. S8830–2TAB)
RiboLock RNase inhibitor (Thermo Fisher, cat. no. FEREO0382)
Igepal CA-630, for molecular biology (Sigma-Aldrich, cat. no. I8896–50ML)
SDS (Affymetrix, cat. no. 151–21-3)
Sodium deoxycholate (Sigma-Aldrich, cat. no. D6750–25G)
Lithium chloride (LiCl; Sigma-Aldrich, cat. no. L9650)
RNase A (Thermo Fisher, cat. no. EN0531)
Proteinase K (New England Biolabs, cat. no. P8107S)
Phase-lock tubes (5Prime, cat. no. 2302820)
Phenol, equilibrated, molecular biology grade (Sigma-Aldrich, cat. no. P4557–400ML) ! CAUTION Phenol is toxic if swallowed, upon contact with the skin or if inhaled. Wear a lab coat, goggles and gloves and work in a chemical hood. All phenol waste must be kept and disposed of according to local and institutional regulations.
UltraPure phenol:chloroform:isoamyl alcohol (25:24:1 (vol/vol); Thermo Fisher, cat. no. 15593031) ! CAUTION Phenol:chloroform:isoamyl alcohol is toxic if swallowed, upon contact with the skin or if inhaled. Wear a lab coat, goggles and gloves and work in a chemical hood. All waste must be kept and disposed of according to local and institutional regulations.
Ethanol, pure (Koptec, cat. no. 64–17-5)
Sodium acetate (pH 5.2; Sigma-Aldrich, cat. no. S2889)
FastStart Universal SYBR Green Master (ROX) (2×; Roche, cat. no. 4913850001)
phi29 DNA polymerase (New England Biolabs, cat. no. M0269S)
dNTP mix (New England Biolabs, cat. no. N0447S)
PureLink PCR Micro Kit (Thermo Fisher, cat. no. K310050)
Klenow fragment (3′→5′ exo-; New England Biolabs, cat. no. M0212S)
Deoxyadenosine triphosphate (dATP; New England Biolabs, cat. no. N0447S)
T4 DNA ligase (New England Biolabs, cat. no. M0202S)
Phusion High-Fidelity DNA polymerase (New England Biolabs, cat. no. M0530S)
Agarose (Seakem LE Agarose; Lonza, cat. no. 50004)
PureLink Quick Gel Extraction Kit (Thermo Fisher, cat. no. K210012)
Loading buffer (6×), orange (New England Biolabs, cat. no. B7022S)
DNA ladder, 50-bp (Thermo Fisher, cat. no. SM0371)
Ethidium bromide solution (10,000×, 10 mg/ml; Bio-Rad, cat. no. 161–0433)
Primers (Table 1 and synthesized by IDT)
10× NEB buffer 2 (New England Biolabs, cat. no. B7002S)
DNA purification kit (GeneJET PCR Purification Kit, Thermo Fisher, cat. no. K0702)
TAE buffer (AccuGENE 10× TAE Buffer, Lonza, cat. no. 50841)
Table 1 |.
Primer and oligo sequences
| Primer name | Purpose | Sequence (5′–3′) | Reference |
|---|---|---|---|
| JUN-TSS forward | For checking R-ChIP enrichment (Step 60) | GGGTGACATCATGGGCTATT | |
| JUN-TSS reverse | For checking R-ChIP enrichment (Step 60) | TCGGACTATACTGCCGACCT | |
| JUN-TTS forward | For checking R-ChIP enrichment (Step 60) | AAATAAGCAGGCTGGGGAAT | |
| JUN-TTS reverse | For checking R-ChIP enrichment (Step 60) | CAATCAAGCATGGGGATAGG | |
| NEAT1-TSS forward | For checking R-ChIP enrichment (Step 60) | TAGTTGTGGGGGAGGAAGTG | |
| NEAT1-TSS reverse | For checking R-ChIP enrichment (Step 60) | ACCCTGCGGATATTTTCCAT | |
| NEAT1-TTS forward | For checking R-ChIP enrichment (Step 60) | AGAGGGAGGGAGAGCTGAAG | |
| NEAT1-TTS reverse | For checking R-ChIP enrichment (Step 60) | GCATGAAGTCAGACCAGCAA | |
| CLSPN-TSS forward | For checking R-ChIP enrichment (Step 60) | GGCTGAGGGAATCAGAGACA | 4 |
| CLSPN-TSS reverse | For checking R-ChIP enrichment (Step 60) | GGGCGTGTGCATAAACTCA | |
| CLSPN-TTS forward | For checking R-ChIP enrichment (Step 60) | GGCACACAGCTTGGAATGTA | |
| CLSPN-TTS reverse | For checking R-ChIP enrichment (Step 60) | CCCCAGCAACTCTGAGACAG | |
| PMS2-TSS forward | For checking R-ChIP enrichment (Step 60) | AGCTGAGAGCTCGAGGTGAG | 4 |
| PMS2-TSS reverse | For checking R-ChIP enrichment (Step 60) | GAGATCGCTGCAACACTGAG | |
| PMS2-TTS forward | For checking R-ChIP enrichment (Step 60) | GCCAGACGTTGAGGAAGAAG | |
| PMS2-TTS reverse | For checking R-ChIP enrichment (Step 60) | ATCAACCCTTCCACTGCTTG | |
| N9 random primera | For primer extension (Step 61) | 5′-/invddt/CAAGCAGAAGACGGCAT ACGAGNN NNNNNNN-3′ |
4 |
| Oligo Aa | For making an adaptor (Step 69) | 5′-/Phos/GATCGGAAGAGCGTC GTGTAGGGAAAGAGTGT-3′ | 4 |
| Oligo Ba | For making an adaptor (Step 69) | 5′-AGACGTGTGCTCTTCCGATCT-3′ | 4 |
| PCR primera | For PCR amplification (Step 74) | 5′-CAAGCAGAAGACGGCATACGAG −3′ | |
| Barcode primera | For PCR amplification (Step 74) | 5′-AATGATACGGCGACCACCGAGATCTACACNNNNNACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ |
NNNNN, flexible barcode sequence; NNNNNNNNN, random sequence.
Oligonucleotides may need to be re-designed based on the specific sequencing platform. You may find the nxCode website (http://hannonlab.cshl.edu/nxCode/nxCode/main.html) useful for barcode sequence design. All oligonucleotides can be dissolved in H2O at a stock concentration of 10 μM and stored for at least 1 year at −80 °C.
Equipment
CO2 incubator (Thermo Fisher, model no. 3110). Cells should be maintained at 37 °C with 5% CO2
Culture dishes (Nest, cat. no. 704001)
Cell scraper (Sigma-Aldrich, cat. no. SIAL0008–150EA)
Rotating platform (BenchRocker 2D Rocker; Benchmark Scientific, model no. BR2000)
Rotating device with a thermoblock (ThermoMixer C; Eppendorf, cat. no. 5382000023)
−80 °C Freezer (FORMA 900 series, Thermo Fisher, model no. 989)
−20 °C Freezer (Panasonic, model no. BZ10145190)
Single-channel manual pipette (0.5–10 μl; Rainin, cat. no. 17014388)
Single-channel manual pipette (20–200 μl; Rainin, cat. no. 17014391)
Single-channel manual pipette (100–1,000 μl; Rainin, cat. no. 17014382)
Electric dispensing pipette (Fisherbrand Electric Pipet Controller; Thermo Fisher, cat. no. 14–955-202)
Refrigerated centrifuge (Eppendorf, model no. 5804R)
Refrigerated microcentrifuge (Eppendorf, model no. 5425R)
DNA/RNA LoBind microcentrifuge tubes (1.5 ml; Eppendorf, cat. no. 022431021)
Probe sonicator (Sonifier Cell Disruptor; Branson, model no. 185) ! CAUTION Ultrasonic waves have an adverse effect on human hearing. Wearing ear protection is strongly recommended during operation.
Magnetic separator (DynaMag-2 magnet; Thermo Fisher, cat. no. 12321D)
Tube roller and rotator (Labnet, cat. no. H5500)
Vortex mixer (Vortex-Genie 2; Scientific Industries, model no. G560/SI-0236)
Mini centrifuge (MyFuge12; Benchmark Scientific, model no. C1012)
Laboratory water bath (Precision, cat. no. 51221058)
Optical adhesive film (MicroAmp; Thermo Fisher, cat. no. 4360954)
Fast optical 96-well reaction plate (MicroAmp; Thermo Fisher, cat. no. 4346906)
Real-time PCR system (StepOnePlus; Thermo Fisher, cat. no. 4376600)
96-Well thermal cycler (SimpliAmp; Thermo Fisher, model. no. A24811)
Qubit fluorometer (Thermo Fisher, model no. Q32857)
PCR tubes (Axygen; Corning, cat. no. PCR-02-C)
Agarose electrophoresis cell (wide mini-sub cell GT cell; Bio-Rad, model no. 1704469edu)
Transilluminator (Dark Reader; Clare Chemical, model. no. DR22A)
0.22-μm Filter unit (Olympus, cat. no. 25–244)
T75 tissue culture flasks (Corning, cat. no. 430641U)
Sequencing system (Illumina, model no. HiSeq 2500)
Hemocytometer (Cynmar, cat. no. 012–00150)
Software
FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
Cutadapt (http://cutadapt.readthedocs.io/en/stable/index.html)
Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)
SAMtools (http://samtools.sourceforge.net/)
bedtools (http://bedtools.readthedocs.io/en/latest/)
Picard Tools (http://broadinstitute.github.io/picard)
MACS v.2.0 (https://github.com/taoliu/MACS)
UCSC Genome Browser (https://genome.ucsc.edu/)
Reagent setup
Bead wash buffer
To prepare 50 ml of bead wash buffer, add 1 ml of 1 M Tris-Cl, pH 8.0, 1.5 ml of 5 M NaCl, 0.2 ml of 0.5 M EDTA, pH 8.0, and 5 ml of 10% (vol/vol) Triton X-100 to 42.3 ml of molecular-grade H2O and filter-sterilize using a 0.22-μm filter unit. Store at 4 °C for up to 6 months.
Antibody-binding buffer
To prepare 50 ml of antibody-binding buffer, dissolve 0.25 g of BSA in 45 ml of PBS and adjust the volume to 50 ml. Filter-sterilize using a 0.22-μm filter unit. This stock solution is stable at 4 °C for up to 3 months.
Bead blocking buffer
To prepare1 ml of bead blocking buffer, add 10 μl of 20 mg/ml glycogen and 40 μl of antibody-binding buffer to 1 ml of PBS. Prepare fresh before use.
1.375 M glycine
To prepare 250 ml of 1.375 M glycine, dissolve 25.8 g of glycine in 220 ml of molecular-grade H2O and adjust the volume to 250 ml. Filter-sterilize using a 0.22-μm filter unit. Store at 4 °C for up to 12 months.
Cell lysis buffer
To prepare 50 ml of cell lysis buffer, add 0.5 ml of 1 M Tris-Cl, pH 8.0, 0.1 ml of 5 M NaCl and 2.5 ml of 10% (vol/vol) Igepal CA-630 to 46.9 ml of molecular-grade H2O and filter-sterilize using a 0.22-μm filter unit. Store at 4 °C for up to 6 months.
20% (wt/vol) SDS solution
To prepare 50 ml of 20% (wt/vol) SDS solution, dissolve 10 g of SDS in 40 ml of molecular-grade H2O. Heat at 60 °C in a laboratory water bath to facilitate dissolving. Adjust the volume to 50 ml. This stock solution is stable at room temperature for up to 6 months. ! CAUTION SDS is a strong denaturant and irritant. Wear a mask when weighing SDS and clean the area after weighing, as the powder disperses easily.
Nuclear lysis buffer
To prepare 50 ml of nuclear lysis buffer, add 2.5 ml of 1 M Tris-Cl, pH 8.0, 1 ml of 0.5 M EDTA, pH 8.0, and 2.5 ml of 20% (wt/vol) SDS to 44 ml of molecular-grade H2O; then filter-sterilize using a 0.22-μm filter unit. Store at room temperature for up to 6 months.
TE buffer
To prepare 50 ml of TE buffer, add 0.5 ml of 1 M Tris-Cl, pH 8.0, and 0.1 ml of 0.5 M EDTA, pH 8.0, to 49.4 ml of molecular-grade H2O and filter-sterilize using a 0.22-μm filter unit. Store at 4 °C for up to 6 months.
5 M NaCl
To prepare 100 ml of 5 M NaCl, dissolve 29.22 g of NaCl in 90 ml of molecular-grade H2O and adjust the volume to 100 ml. Filter-sterilize using a 0.22-μm filter unit and store at room temperature for up to 12 months.
2.5 M LiCl
To prepare 100 ml of 2.5 M LiCl, dissolve 10.6 g of LiCl in 90 ml of molecular-grade H2O and adjust the volume to 100 ml. Filter-sterilize using a 0.22-μm filter unit and store at room temperature for up to 12 months.
10% (wt/vol) Sodium deoxycholate solution
To prepare 100 ml of 10% (wt/vol) sodium deoxycholate solution, dissolve 10 g of sodium deoxycholate in 90 ml of molecular-grade H2O and heat at 60 °C in a laboratory water bath to facilitate dissolving. Adjust the volume to 100 ml. Filter-sterilize, using a 0.22-μm filter unit, and store at room temperature for up to 12 months. ▲ CRITICAL This solution should be protected from light.
Wash buffer I
To prepare 50 ml of wash buffer I, add 1 ml of 1 M Tris-Cl, pH 8.0, 0.2 ml of 0.5 M EDTA, pH 8.0, 5 ml of 10% (vol/vol) Triton X-100, 0.25 ml of 20% (wt/vol) SDS and 1.5 ml of 5 M NaCl to 42.05 ml of molecular-grade H2O; then filter-sterilize using a 0.22-μm filter unit. Store at 4 °C for up to 6 months.
Wash buffer II
To prepare 50 ml of wash buffer II, add 1 ml of 1 M Tris-Cl, pH 8.0, 0.2 ml of 0.5 M EDTA, pH 8.0, 5 ml of 10% (vol/vol) Triton X-100, 0.25 ml of 20% (wt/vol) SDS and 5 ml of 5 M NaCl to 38.55 ml of molecular-grade H2O; then filter-sterilize using a 0.22-μm filter unit. Store at 4 °C for up to 6 months.
Wash buffer III
To prepare 50 ml of wash buffer III, add 0.5 ml of 1 M Tris-Cl, pH 8.0, 0.1 ml of 0.5 M EDTA, pH 8.0, 5 ml of 10% (vol/vol) Igepal CA-630, 5 ml of 2.5 M LiCl and 5 ml of 10% (wt/vol) sodium deoxycholate to 34.4 ml of molecular-grade H2O; then filter-sterilize using a 0.22-μm filter unit. Store at 4 °C for up to 6 months.
Elution buffer
To prepare 50 ml of elution buffer, add 0.5 ml of 1 M Tris-Cl, pH 8.0, 0.1 ml of 0.5 M EDTA, pH 8.0, and 2.5 ml of 20% (wt/vol) SDS to 46.9 ml of molecular-grade H2O and filter-sterilize using a 0.22-μm filter unit. Store at room temperature for up to 6 months.
1× Annealing buffer
To prepare 50 ml of 1× annealing buffer, add 0.5 ml of 1 M Tris-Cl, pH 8.0, 0.5 ml of 5 M NaCl and 0.1 ml of 0.5 M EDTA to 48.9 ml of molecular-grade H2O; then filter-sterilize using a 0.22-μm filter unit. Store at room temperature for up to 12 months.
70% (vol/vol) Ethanol
To prepare 50 ml of 70% (vol/vol) ethanol, add 15 ml of pure ethanol to 35 ml of molecular-grade H2O.
Procedure
Establishment of a stable cell line expressing the catalytically inactive RNASEH1 ● Timing 3–4 weeks
-
1
Transfect cells with the pPyCAG-RNASEH1 (D210N) vector by using Lipofectamine 2000 according to the manufacturer’s instructions. Briefly, seed 2–3 × 105 cells in one well of a six-well plate. When cells reach 60–70% confluence, replace the medium with 1.5 ml of Opti-MEM I. Next, prepare a transfection mixture, including 4 μl of Lipofectamine 2000 and 1.5 μg of pPyCAG-RNASEH1 (D210N) vector in 0.5 ml of Opti-MEM I according to the manufacturer’s instructions. Add the mixture to the well and culture for 6 h in a CO2 incubator. It is recommended to prepare two to three biological replicates for transfection.
▲ CRITICAL STEP Usually K562 cells have a lower transfection efficiency than HEK293T cells, so it may take longer for stable K562 cell lines to become established.
▲ CRITICAL STEP A stable cell line expressing RNASEH1 (WKKD) can be established in parallel to serve as a control for R-ChIP experiments; however, this is not always required, as input chromatin can be used as a control as well.
-
2
Replace the medium with fresh culture medium. 2 d after transfection, add hygromycin B to a final concentration of 100–200 μg/ml.
▲CRITICAL STEP Sensitivity to hygromycin B may vary depending on the cell type. It is therefore recommended to first test the cell line with different concentrations of hygromycin B and to choose the lowest concentration, at which all cells die within 3–5 d, for later selection steps.
-
3
Replace the medium with fresh medium supplemented with hygromycin B at the optimal concentration determined in Step 2 every 2–3 d until new clones repopulate. The selection process may take ~2 weeks. Passage and allow the cells to grow for one to two more passages before Step 4.
-
4
Prepare lysates of stable cells for western blot to examine the expression level of exogenous RNASEH1 (ref. 4).
▲CRITICAL STEP A single strong band at ~35 kDa by western blot using the V5 antibody would suggest a successful selection of stable cells, as shown in our previous publication4. We recommend freezing a few vials of the newly established stable cells at −80 °C or in liquid nitrogen for long-term (up to 1 year) storage. Considering the variation in gene expression profiles between individual cells, we did not select for single-cell clones, expecting that the R-ChIP signals would represent the average R-loop signals of a cell population at individual genomic loci.
? TROUBLESHOOTING
-
5
Grow adherent cells, such as HEK293T cells, in 10-cm culture dishes, or grow suspension cells, such as K562 cells, in T75 tissue culture flasks. For R-ChIP experiments, seed cells at 20–30% confluence and prepare to harvest the cells at 70–80% confluence, at which point enough cells can be collected for one experiment from one plate or flask (the final cell number is usually 1–1.5 × 107 per plate or flask). When cells are ready for harvest, proceed to Step 12.
Bead preparation ● Timing ~3 h hands-on plus overnight incubation
-
6
The day before harvesting the cells, wash protein A/G beads in a 1.5-ml LoBind tube with 1 ml of bead wash buffer by gentle pipetting or vortexing for 15 s (25 μl of slurry per sample, maximal 100 μl of slurry for four samples in one tube).
▲CRITICAL STEP Do not centrifuge at high speed, or dry or freeze the magnetic beads, as these operations will result in aggregation of beads, thus reducing their binding ability. Wash up to 100 μl of protein A/G beads per tube (for four R-ChIP experiments).
-
7
Place the tube into a magnetic separator for 15–20 s, then remove and discard the supernatant. Repeat Steps 6 and 7 twice for a total of three washes.
-
8
Add 1 ml of bead blocking buffer and incubate the beads for 1 h at room temperature on a rotating platform.
-
9
Wash the beads with 1 ml of antibody-binding buffer, as described in Steps 6 and 7, for three times.
-
10
Resuspend the beads with 1 ml of antibody-binding buffer on ice, add an appropriate amount of anti-V5-tag antibody to the buffer and mix well (2.5 μg of antibody per 25 μl of beads; refer to Step 6).
-
11
Incubate the antibody–bead mixture overnight at 4 °C on a rotating platform until use (4 h of incubation is the minimum recommended if shortening the experimental time is preferred).
Cross-linking and harvesting the cells ● Timing 1–2 h
-
12
For adherent cells (e.g., HEK293T), aspirate and discard the medium from the culture dish and wash the cells twice with 10 ml of cold PBS on ice. Then add 10 ml of PBS to the culture dish. For suspension cells (e.g., K562), count the cells with a hemocytometer and transfer 0.5–1.5 × 107 cells to a 50-ml conical tube, spin at 300g for 5 min at 4 °C and aspirate the supernatant. Wash the cells twice with 10 ml of cold PBS and then resuspend the cells in 10 ml of PBS by pipetting gently.
-
13
Add 270 μl of 37% (vol/vol) formaldehyde directly to the culture dish (HEK293T cells) or a conical tube (K562 cells) from Step 12 to obtain a final concentration of 1% (vol/vol) formaldehyde. Incubate the cells for 10 min at room temperature on a rotating platform.
▲CRITICAL STEP To prevent chromatin overfixation, do not fix the cells with a concentration of formaldehyde higher than 1% (vol/vol) and do not cross-link the cells for more than 20 min.
-
14
To quench the formaldehyde, add 1 ml of 1.375 M glycine to the cells from Step 13 to a final concentration of 0.125 M and incubate the mixture on a rotating platform for 15 min at room temperature.
-
15
Aspirate and discard the supernatant for adherent cells. For suspension cells, spin down fixed cells at 600g for 5 min at 4 °C before aspirating and discarding the supernatant.
-
16
Wash fixed adherent cells three times with 10 ml of cold PBS and add 1 ml of cold PBS to the culture dish. For suspension cells, wash the cells with 10 ml of cold PBS and spin at 600g for 5 min at 4 °C. Aspirate the supernatant and repeat for a total of three washes. Aspirate the supernatant and resuspend the cells in 1 ml of cold PBS.
▲CRITICAL STEP After completing this step, handle the samples on ice and use ice-cold buffers.
-
17
For adherent cells, scrape the cells off the plate, using a cell scraper, and transfer them to a 1.5-ml LoBind tube. For suspension cells, directly transfer cold PBS with cells to a new 1.5-ml LoBind tube.
-
18
Centrifuge at 600g for 5 min at 4 °C. Aspirate and discard the supernatant.
∎PAUSE POINT The cell pellet can be stored for 1–2 weeks at −80 °C.
Nuclei isolation ● Timing 1–1.5 h
-
19
Prepare the cell lysis buffer with 1× protease inhibitor cocktail (10 μl of 100× protease inhibitor cocktail per ml of cell lysis buffer, final concentration) and RiboLock RNase inhibitor (1 μl/ml final concentration) before use. 2 ml of cell lysis buffer is needed for one sample.
-
20
Resuspend the cell pellet from Step 18 in 1 ml of cell lysis buffer and mix well by pipetting several times.
-
21
Incubate the cell suspension for 15 min on ice with gentle inversion of the tube every 2–3 min.
-
22
Spin at 700g for 5 min at 4 °C and discard the supernatant.
-
23
Resuspend the pellet with 1 ml of cell lysis buffer and repeat Step 22.
-
24
Prepare an appropriate volume of nuclear lysis buffer (0.5 ml of buffer per sample) with 1× protease inhibitor cocktail and RiboLock RNase inhibitor (refer to Step 19).
-
25
Resuspend the pellet from Step 23 with 0.5 ml of nuclear lysis buffer by gentle pipetting or vortexing and then incubate the nuclear suspension for 10 min on ice.
Sonication and mixing of sheared chromatin with beads ● Timing 3–4 h hands-on plus overnight incubation
-
26
Sonicate the nuclear suspension for seven cycles. In each cycle, sonicate for 10 s, followed by resting for 1 min on ice. Keep the remaining samples on ice. We usually process six to eight samples at the same time.
-
27
To evaluate the chromatin shearing efficiency, add 10 μl of the nuclear suspension from Step 26 to a new 1.5-ml tube. Add 90 μl of TE buffer and 1 μl of RNase A and incubate on a ThermoMixer for 1 h at 37 °C with agitation (alternating between rotating at 1,200 r.p.m. for 30 s and resting for 2 min). Add 1 μl of proteinase K to the tube and continue incubating with the same agitation settings for 2 h at 65 °C. Extract the DNA by following Steps 50–57 or by using a DNA purification kit. Resolve the extracted DNA (50–100 ng) on a 2% (wt/vol) agarose gel in 1× TAE buffer by electrophoresis. Properly sheared chromatin fractions should have a size range between 100 and 600 bp.
▲CRITICAL STEP Sonication is one of the most critical steps for generating a high-quality R-ChIP library. Optimizing the sonication conditions by pilot experiments is highly recommended when working with a new cell type.
? TROUBLESHOOTING
∎PAUSE POINT Sheared chromatin can be stored for up to 1 week at −80 °C, but immediate processing is preferable.
-
28
Keep the tubes for 30–60 min on ice until the SDS has precipitated and the chromatin suspension becomes cloudy.
-
29
Spin at 16,000g for 10 min at 4 °C and then transfer the supernatant containing sheared chromatin to a new 1.5-ml LoBind tube.
▲CRITICAL STEP Steps 28 and 29 serve to reduce the concentration of SDS in the buffer, which may facilitate antibody binding to the exogenously expressed V5-tagged RNASEH1.
-
30
Add the following reagents to bring the volume to 1.3 ml: 644 μl of TE buffer, 130 μl of 10% (vol/vol) Triton X-100, 13 μl of 10% (wt/vol) sodium deoxycholate, 13 μl of 100× proteinase inhibitor cocktail and 0.5 μl of RiboLock RNase inhibitor. Mix well by gentle vortexing.
-
31
Add 65 μl of the chromatin suspension to a new 1.5-ml tube as the input chromatin fraction and store it overnight at −20 °C for Step 44. Take 50 μl of the chromatin suspension as the input for testing the IP efficiency at Step 43 by western blot; this can be stored for 1 week at −80 °C.
-
32
Transfer the buffer containing antibody-conjugated beads from Step 11 to 1.5-ml tubes, adding 25 μl of the original bead slurry per tube. Place the bead-containing tubes in a magnetic separator and wait until all the beads attach to the side of the tube wall close to the magnetic separator (15–20 s). Remove and discard the buffer. Add 1.2 ml of chromatin suspension from Step 31 to the tube containing the antibody-conjugated beads.
-
33
Incubate the tube on a tube roller overnight at 4 °C.
Washing the beads and elution of the IPed DNA ● Timing 3–3.5 h
-
34
Prepare aliquots of an appropriate volume of wash buffers I, II and III with 0.1× protease inhibitor cocktail before use.
▲CRITICAL STEP To reduce experimental variation, we recommend handling up to six to eight samples for washing at a time. If there are more samples to process, divide them into several groups. Process samples used for direct comparison in one group at a time, following Steps 35–40.
-
35
To wash the beads, place the tube in a magnetic separator and wait until all the beads attach to the side of the tube wall close to the magnetic separator. Aspirate the supernatant carefully. Add 1 ml of wash buffer I and remove the tube from the magnetic separator. Mix the beads and buffer well by gentle pipetting several times and then continue to incubate the tube on a tube roller for 3 min at 4 °C.
-
36
Place the tube in a magnetic separator and remove the wash buffer once the beads have attached to the side of the tube. Repeat Steps 35 and 36 three times.
-
37
Wash the beads three times as described in Steps 35 and 36, using wash buffer II (1 ml per tube each time).
-
38
Wash the beads once as described in Steps 35 and 36, using 1 ml of wash buffer III.
-
39
Wash the beads once as described in Steps 35 and 36, using 1 ml of TE buffer.
-
40
Spin at 1,400g for 1 min at 4 °C and aspirate residual TE buffer.
-
41
Add 170 μl of elution buffer and pipette briefly to resuspend the beads.
-
42
Incubate the beads for 30 min at 65 °C on a ThermoMixer. Vortex cycling should be set to alternate between rotating at 1,200 r.p.m. for 30 s and resting for 2 min.
Reversal of cross-linking ● Timing overnight
-
43
43 Spin the tube from Step 42 in a microcentrifuge at the maximum speed for 30 s at room temperature and transfer the supernatant to a new tube. Aliquot 20 μl of the supernatant as the IPed protein for western blot analysis, together with the input chromatin collected at Step 31.
▲CRITICAL STEP This is an important checkpoint for evaluating antibody quality and IP efficiency4.
? TROUBLESHOOTING
-
44
Add 85 μl of elution buffer to the 65 μl of input chromatin collected at Step 31.
-
45
Reverse cross-link the IPed and input chromatin on a ThermoMixer overnight at 65 °C (alternating agitation at 1,200 r.p.m. for 30 s with resting for 2 min).
RNase A and proteinase K treatment ● Timing ~4 h
-
46
Add 150 μl of TE buffer and 6 μl of 10 mg/ml RNase A to each tube.
-
47
Incubate the tubes on a ThermoMixer for 2 h at 37 °C (alternating between agitation at 1,200 r.p.m. for 30 s and resting for 2 min).
-
48
Add 7 μl of proteinase K and 1.5 μl of glycogen to each tube.
-
49
Continue to incubate the mixture on a ThermoMixer for 2 h at 65 °C (alternating between agitation at 1,200 r.p.m. for 30 s and resting for 2 min).
Recovery of the IPed DNA ● Timing 3–3.5 h
-
50
Add 300 μl of phenol, mix well by pipetting 20 times and transfer the mixture to a new phase-lock gel tube.
-
51
Spin at 16,000g for 5 min at 4 °C.
-
52
Transfer the upper aqueous solution to a new 1.5-ml tube and repeat Steps 50 and 51.
-
53
Transfer the upper aqueous solution to a new 1.5-ml tube, and add 300 μl of phenol:chloroform: isoamyl alcohol. Mix by pipetting 20 times and repeat Step 51.
-
54
Transfer the upper aqueous solution to a new tube. Add 750 μl of 100% ethanol and 30 μl of sodium acetate to precipitate DNA for 30 min at −80 °C.
-
55
Spin at 16,000g for 15 min at 4 °C. Remove the supernatant.
▲CRITICAL STEP After centrifugation, there should be a tiny white precipitate at the bottom of the tube, which is the glycogen and recovered DNA.
-
56
Wash the pellet twice with 1 ml of 70% (vol/vol) cold ethanol by gentle vortexing and centrifuge at 16,000g for 5 min at 4 °C.
-
57
Remove the supernatant, dry the pellet for 5 min at room temperature and dissolve the pellet in 25 μl of TE buffer.
-
58
Measure the concentration of the IPed DNA with a Qubit fluorometer. The concentration of the recovered DNA is expected to be between 0.1 and 0.7 ng/μl.
∎PAUSE POINT The recovered DNA can be stored for up to 1 week at −80 °C, but immediate processing for qPCR examination, followed by library construction the next day, is recommended.
qPCR ● Timing ~3 h
-
59
Take 10 μl of the IPed DNA and dilute it fourfold with molecular-grade H2O.
-
60Perform a qPCR on several typical R-loop-forming regions, such as the TSS regions of JUN, NEAT1, CLSPN and PMS2 (Fig. 3b)4. Perform qPCR with 1 μl of IPed DNA in a 10-μl reaction mixture with FastStart Universal SYBR Green Master in one well of a 96-well plate, using the qPCR program listed below. For each DNA sample, perform qPCR in three or four technical replicates. The results are calculated as percentage of input, and relative enrichment is evaluated by comparing R-ChIP signals at the expected R-loop-forming region at TSS with those of the terminator region of the same gene as the control (see Table 1 for primer sequences used for Fig. 3b). If the qPCR results indicate successful R-ChIP enrichment of select genes, proceed to Step 61.
Step Denature Anneal and extend Melt curve Hold 1 95 °C, 10 min 2–41 95 °C, 15 s 60 °C, 45 s 42 60 °C→95 °C, 2 °C/s 43 25 °C ▲CRITICAL STEP qPCR is a fast way to evaluate the quality of R-ChIP experiments, which may save time and reagents if this quality control step is performed. It also helps to identify potential problems encountered during the R-ChIP procedure.
? TROUBLESHOOTING
Primer extension and DNA cleanup ● Timing 1–1.5 h
-
61Prepare the primer extension mixture in a 0.2-ml PCR tube on ice as follows:
Component Amount(μl) Final concentration Recovered DNA (Step 58) 11 — 10× phi29 DNA polymerase buffer 2 1× 1 mg/ml BSA 4 0.2 mg/ml 3 mM dNTP mix 1 0.15 mM 20 μM N9 random primer (Table 1) 1 1 μM Total 19 — -
62
Incubate the mixture in a thermal cycler for 5 min at 95 °C, followed by 5 min at 25 °C.
-
63
Add 1 μl of 10 U/μl phi29 DNA polymerase and incubate in a thermal cycler for 20 min at 30 °C, followed by 10 min at 65 °C.
-
64
Purify the extension product with the PureLink PCR Micro Kit and elute with 20 μl of elution buffer, provided with the kit. Proceed directly to Step 65.
dA tailing and cleanup ● Timing 1–1.5 h
-
65Prepare the reaction mixture in a 0.2-ml PCR tube on ice as follows:
Component Amount (μl) Final concentration DNA (Step 64) 20 — 10× NEB buffer 2 3 1× 1 mM dATP 6 0.2 mM Klenow fragment (3′→5′ exo-) 1 0.16 U/μl Total 30 — -
66
Incubate the reaction mixture on a thermal cycler for 30 min at 37 °C.
-
67
Purify the extension product with a PureLink PCR Micro Kit and elute with 13 μl of elution buffer, provided by the kit.
Adaptor preparation and ligation ● Timing ~3 h
-
68
To prepare the adaptor for ligation, separately dissolve oligo A and oligo B (see Table 1 for their sequences) in 1× annealing buffer, making the final concentration 40 μM.
-
69
Mix the oligo A and oligo B solutions in a 1:1 molar ratio (30 μl of each oligo) in a 0.2-ml PCR tube to anneal the adaptors. Place the mixture on a thermal cycler with the following program: 2 min at 95 °C, ramping down at the rate of 0.1 °C/s from 95 to 25 °C.
-
70
Prepare aliquots of annealed adaptors (final concentration is ~20 μM) in several 1.5-ml tubes.
∎PAUSE POINT The annealed adaptors can be stored for at least 1 year at −20 °C.
-
71
For adaptor ligation, prepare 2 μM adaptors by diluting the stock solution tenfold with molecular-grade H2O.
-
72Prepare the ligation mixture as follows:
Component Amount (μl) Final concentration DNA (Step 67) 13 — 10× Ligation buffer (provided with T4 DNA ligase) 2 1× 2 μM adaptor 1 0.1 μM T4 DNA ligase 4 80 U/μl Total 20 — -
73
Incubate the reaction mixture for 2 h at room temperature; then proceed to PCR amplification in Step 74.
∎PAUSE POINT The ligation products can be stored for a few weeks at –20 °C.
PCR and library recovery ● Timing ~5 h
▲CRITICAL We recommend running a test PCR to determine the lowest number of PCR cycles required for obtaining a sufficient amount of library DNA for sequencing. We usually test 14 and 16 cycles by preparing two tubes of the PCR mixture for each sample and running two PCR programs separately. If the PCR product is too weak, as assessed by gel electrophoresis, a final PCR with 18 cycles can be done using the remaining sample.
-
74Prepare the following PCR mix for a test PCR (10 μl). The remaining ligation product can be kept on ice:
Component Amount (μl) Final concentration DNA ligation product (Step 73) 0.5 — 5× Phusion HF buffer (provided with Phusion High-Fidelity DNA
polymerase)2 1× 10 μM barcode primer (Table 1) 0.2 0.2 μM 10 μM PCR primer (Table 1) 0.2 0.2 μM 10 mM dNTP mix 0.5 0.5 mM Phusion High-Fidelity DNA polymerase 0.2 0.04 U/μl Molecular-grade H2O 6.4 – Total 10 -
75Amplify the DNA using the following PCR program:
Step Denature Anneal Extend Hold 1 98 °C, 30 s 2 (14 or 16 cycles) 98 °C, 10 s 65 °C, 30 s 72 °C, 30 s 3 72 °C, 5 min 4 4 °C -
76
Add 2 μl of 6× loading buffer to the tube and resolve all PCR mixtures on a 2% (wt/vol) agarose gel in 1× TAE buffer for ~30–40 min; then choose an optimal PCR cycle number for the final library amplification.
-
77Prepare the following PCR reaction mix (100 μl):
-
78
Amplify the ligation product using the same program as in Step 75 and the optimal cycle number determined in Steps 74–76.
-
79
Transfer the PCR product to a new 1.5-ml tube, add 1 μl of 20 mg/ml glycogen and 10 μl of 3 M sodium acetate, pH 5.2, and mix well by pipetting. Add 250 μl of ethanol and invert the tube several times.
-
80
Precipitate the PCR products for 30 min at −80 °C.
-
81
Spin at 16,000g for 15 min at 80 °C; then remove and discard the supernatant.
-
82
Dry the pellet for 5 min at room temperature. Dissolve the DNA pellet in 20 μl of 1× loading buffer and resolve it together with the 50-bp DNA ladder on a 2% (wt/vol) agarose gel containing 1 μg/ml ethidium bromide in 1× TAE buffer for ~30–40 min.
-
83
Use a new blade to cut out the gel chunk that contains the DNA library in the size range of 140–350 bp and transfer the gel slice to a new 1.5-ml tube.
-
84
Extract the library using a PureLink Quick Gel Extraction Kit and elute it in 15 μl of molecular-grade H2O.
-
85
Measure the concentration of R-ChIP libraries using a Qubit fluorometer. It is expected that 5–15 ng/μl PCR product will be recovered for each sample.
? TROUBLESHOOTING
Sequencing and basic data analysis ● Timing ~30 h
-
86
Sequence the R-ChIP library with Illumina read 1 sequencing primer on the HiSeq 2500 platform. Multiplexing of libraries can be performed if individual libraries are generated by PCR using different barcoding primers (Table 1).
-
87
Remove adaptor sequences via the Cutadapt software. Assess the quality of raw sequencing data using FastQC. When necessary, filter out low base-calling-quality reads and reads with excessive amounts of ambiguous bases.
> cutadapt –a CTCGTATGCCGTCTTCTGCTTG –m 15 –o D210N.flt.fq D210N.raw.fq
> fastqc R-ChIP.flt.fq
-
88
Align the filtered reads to pre-built Bowtie 2 indexes using the default local mode of Bowtie 2 (ref. 66). Keep only uniquely mapped reads with high mapping quality (≥30). Remove potential PCR duplicates using SAMtools67 or Picard.
> bowtie2 -p 12 --local -x $idx -U D210N.flt.fq | samtools view -bS - | samtools sort -m 4G -@ 8 | samtools rmdup -s - /dev/stdout | samtools view -F4 -bh -q 30 > D210N.flt.bam
-
89
Separate the resultant reads (in .bam format) into two files according to the strand (Watson or Crick) to which they are mapped.
> samtools view -f 0×10 -b D210N.flt.bam > D210N.strand1.flt.bam
> samtools view -F 0×10 -b D210N.flt.bam > D210N.strand2.flt.bam
-
90
Process the sequencing data for D210N and WKKD or input with the same pipeline (Steps 87–89). Call narrow peaks (in narrowPeak format) separately for strand-specific reads with the MACS v.2 software63, by taking WKKD or the input library as a control, and extending the reads to the average size of gel-isolated DNA fragments (150 bp).
> macs2 callpeak -t D210N.strand1.flt.bam -c input.strand1.flt.bam -f BAM -n strand1 -g hs -q 0.01 --nomodel --extsize 150
> macs2 callpeak -t D210N.strand2.flt.bam -c input.strand2.flt.bam -f BAM -n strand2 -g hs -q 0.01 --nomodel --extsize 150
-
91
Visualization. Convert the .bam format files resulting from Step 89 into bigwig files via genomeCoverageBed (from bedtools) and bedGraphToBigWig (from the UCSC Genome Browser utilities). Usually, the sequencing coverage is normalized to reads per million. Upload the bigwig files as custom tracks of the UCSC Genome Browser for visualization.
> scale=ècho “scale=5;1000000/$(samtools view D210N.flt.bam | wc –l)” | bc`
> bedtools genomecov –ibam D210N.strand1.flt.bam –g $chromsizes –bg –scale $scale –fs 150 | sort –k 1,1 –k 2,2n > D210N.strand1.bdg
> bedGraphToBigWig D210N.strand1.bdg $chromsizes D210N.strand1.bw
> bedtools genomecov –ibam D210N.strand2.flt.bam –g $chromsizes –bg –scale $scale –fs 150 | sort –k 1,1 –k 2,2n
> D210N.strand2.bdg > bedGraphToBigWig D210N.strand2.bdg $chromsizes D210N.strand2.bw
Troubleshooting
Troubleshooting advice can be found in Table 2.
Table 2 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 4 | Low expression level of exogenous RNASEH1 | Poor effect of hygromycin B | Always include a well of untransfected cells as a control for selection. At an effective concentration of hygromycin B, untransfected cells will be killed witdin 5 d |
| 27 | Low chromatin-shearing quality | Inefficient cell lysis | Incubate the cells in nuclear lysis buffer for a longer time and avoid the formation of cell clumps |
| Insufficient sonication | Increase the cycle number of sonication and always check the sonication efficiency before IP. We also recommend optimization of the sonication conditions for each new experimental setting (different number of cells or different volume of nuclear lysate) or when working with a new cell type | ||
| 43 | Low IP quality | Poor antibody quality | Increase the amount of antibody per sample or switch to another ChIP-grade antibody if the current one has a specificity issue |
| Excessive washing of the chromatin–bead complex | Reduce the incubation time of the chromatin–bead complex with wash buffer II | ||
| 60 | Low R-ChIP enrichment as assessed by qPCR(≤2-fold enrichment) | Low cell quality | Harvest the cells when they are in the active proliferation state and have reached 70–80% confluence |
| Insufficient cross-linking | Use fresh formaldehyde and increase the cross-linking time, but do not use a formaldehyde concentration higher than 1% (vol/vol) | ||
| Poor sonication quality | Optimize the sonication conditions. Avoid overheating of chromatin fractions during sonication | ||
| Insufficient washing of beads | Increase the wash time with wash buffers, especially high- salt wash buffer II | ||
| 85 | Low library yield | Insufficient cell number for the experiment | Prepare more cells before harvesting |
| Sample lost through library construction | Before using IPed DNA, perform Steps 61–85 with an aliquot of input DNA to test the recovery efficiency of the PureLink PCR Micro Kit used, by measuring the concentration of recovered DNA and adaptor ligation efficiency by estimating the PCR product amount by gel electrophoresis | ||
| Poor adaptor quality and ligation efficiency | Check the adaptor purity by size on a PAGE gel after annealing and increase the adaptor concentration during ligation | ||
| Insufficient PCR cycle number | Always run a test PCR to determine the most appropriate cycle number |
Timing
Steps 1–5, establishment of a stable cell line expressing RNASEH1: 3–4 weeks
Steps 6–11, bead preparation: ~3 h hands-on plus overnight incubation
Steps 12–18, cross-linking and harvesting the cells: 1–2 h
Steps 19–25, nuclei isolation: 1–1.5 h
Steps 26–32, sonication and mixing of sheared chromatin with beads: 3–4 h hands-on
Step 33, incubation of sheared chromatin with beads: overnight
Steps 34–42, washing the beads and elution of IPed DNA: 3–3.5 h
Steps 43–45, reversal of cross-linking: overnight
Steps 46–49, RNase A and proteinase K treatment: ~4 h
Steps 50–58, recovery of IPed DNA: 3–3.5 h
Steps 59 and 60, qPCR: ~3 h
Steps 61–64, primer extension and cleanup: 1–1.5 h
Steps 65–67, dA tailing: 1–1.5 h
Steps 68–73, adaptor preparation and ligation: ~3 h
Steps 74–85, PCR and library recovery: ~5 h
Step 86, sequencing: ~1 d
Steps 87–91, basic data analysis: 6 h
Anticipated results
Fragmentation by sonication
We consider a sonication result acceptable when the size of the sheared chromatin fraction is between 100 and 600 bp, equivalent to DNA fragments that cover one to three nucleosomes (Fig. 3a). A larger fragment size due to insufficient sonication will adversely affect the resolution and enrichment of R-ChIP signals.
IP and bead washing
The quality of the antibody substantially affects the specificity and sensitivity of R-ChIP signals. We recommend testing the IP efficiency of the antibody by western blot. After IP with the anti-V5-tag antibody, we usually obtain a single band at ~35 kDa, and the IP efficiency is ~10–30% of input4. In our experience, a well-executed R-ChIP experiment with ~107 cells usually generates an amount of final IPed DNA ranging from 0.1 to 0.7 ng/μl in a total of 25–30 μl of elution buffer. A much higher concentration of recovered DNA may indicate insufficient washing of the beads to reduce nonspecifically bound chromatin, leading to a low signal-to-noise ratio in qPCR and sequencing.
Signal enrichment after ChIP
On the basis of our previous data, we choose several typical TSS regions with strong R-loop signals observed in multiple cell types as positive genomic loci for the qPCR test4. To determine if there is an adequate signal enrichment, R-ChIP qPCR results are first converted to percentage of input; then if there is four- to eightfold enrichment for the majority of these loci in comparison with the corresponding control regions, such as the downstream terminator region of the same gene or certain distal intergenic regions, we then consider the R-ChIP efficiency acceptable and proceed to library construction steps. As a control, no enrichment should be observed for the WKKD mutant (Fig. 3b).
Library construction
The majority of the PCR products should be within the range of 140–350 bp. After subtracting the length of flanking primer sequences (31 + 67 bp), the final products should range from ~40 to ~250 bp in size, which largely determines the R-ChIP resolution. A larger library size may indicate insufficient chromatin fragmentation by sonication, which will result in less accurate localization of R-loop sites.
Evaluation of sequencing data
R-loops are highly dynamic and associated with transcription activity, which varies among different cell types and growth conditions. Therefore, it is reasonable to observe differentially formed R-loop regions and varying peak intensity of the same R-loop sites when comparing R-ChIP replicates. In addition, the total called peak number largely depends on sequencing depth. We usually detect several thousands to 14,000 peaks, given 10–30 million uniquely mapped reads per sample, and the correlation coefficient of signal intensity between biological replicates is normally between 0.8 and 0.9. As R-ChIP also provides the strand information for the called R-loop peaks, it is expected that the direction of R-ChIP peaks will match the direction of transcription when comparing R-ChIP data with RNA-seq or GRO-seq data.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
R-ChIP data for HEK293T and K562 cell lines are available at the NCBI GEO repository under the accession number GSE97072. All public datasets used in this study are indicated in the corresponding figure legends.
Acknowledgements
The authors are grateful to members of the Fu lab for cooperation, reagent sharing and insightful discussion that improved this work. We thank T. Hishida of the J.C. Izpisua Belmonte lab (Gene Expression Laboratories, Salk Institute for Biological Studies) for providing the pPyCAG plasmid and W. Li for critical comments on the manuscript. This work was supported by NIH grants (GM049369, GM052872, HG004659 and DK098808) to X.-D.F. and Start-up funds of Wuhan University (1304/413100052 and 1304/413100072) to L.C. We thank S. Dowdy (University of California, San Diego) for providing HEK293T cells
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41596-019-0154-6.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Chen, L. et al. Mol. Cell 68, 745–757 (2017): https://doi.org/10.1016/j.molcel.2017.10.008
Chen, L. et al. Mol. Cell 69, 412–425.e6 (2018): https://doi.org/10.1016/j.molcel.2017.12.029
References
- 1.Aguilera A & Garcia-Muse T R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Sanz LA et al. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahba L, Costantino L, Tan FJ, Zimmer A & Koshland D S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 30, 1327–1338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L et al. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell 68, 745–757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pefanis E et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graf M et al. Telomere length determines TERRA and R-loop regulation through the cell cycle. Cell 170, 72–85 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Kabeche L, Nguyen HD, Buisson R & Zou L A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 359, 108–114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginno PA, Lott PL, Christensen HC, Korf I & Chedin F R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugimoto N et al. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34, 11211–11216 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Duquette ML, Handa P, Vincent JA, Taylor AF & Maizels N Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 18, 1618–1629 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy D, Yu K & Lieber MR Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol. Cell. Biol 28, 50–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy D & Lieber MR G clustering is important for the initiation of transcription-induced R-loops in vitro,whereas high G density without clustering is sufficient thereafter. Mol. Cell. Biol 29, 3124–3133 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy D, Zhang Z, Lu Z, Hsieh CL & Lieber MR Competition between the RNA transcript and thenontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol. Cell. Biol 30, 146–159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chedin F Nascent connections: R-loops and chromatin patterning. Trends Genet. 32, 828–838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stork CT et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 5, e17548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamperl S, Bocek MJ, Saldivar JC, Swigut T & Cimprich KA Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170, 774–786 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X & Manley JL Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Benitez F, Gaillard H & Aguilera A Physical proximity of chromatin to nuclear pores prevents harmful R loop accumulation contributing to maintain genome stability. Proc. Natl. Acad. Sci. USA 114, 10942–10947 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huertas P & Aguilera A Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12, 711–721 (2003). [DOI] [PubMed] [Google Scholar]
- 20.El Hage A, French SL, Beyer AL & Tollervey D Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 24, 1546–1558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang EY & Stirling PC Replication fork protection factors controlling R-loop bypass and suppression. Genes (Basel) 8, 33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taneja N et al. SNF2 family protein Fft3 suppresses nucleosome turnover to promote epigenetic inheritance and proper replication. Mol. Cell 66, 50–62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty P & Grosse F Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst.) 10, 654–665 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Cristini A, Groh M, Kristiansen MS & Gromak N RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 23, 1891–1905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skourti-Stathaki K, Proudfoot NJ & Gromak N Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itoh T & Tomizawa J Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. USA 77, 2450–2454 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu B & Clayton DA RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. EMBO J. 15, 3135–3143 (1996). [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ & Dean C R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340, 619–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boque-Sastre R et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. USA 112, 5785–5790 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohle C et al. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell 167, 1001–1013 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Belotserkovskii BP, Soo Shin JH & Hanawalt PC Strong transcription blockage mediated by R-loop formation within a G-rich homopurine-homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Res. 45, 6589–6599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan W et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 25, 2041–2056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L et al. The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol. Cell 69, 412–425.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costantino L & Koshland D The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol 34, 39–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richard P & Manley JL R loops and links to human disease. J. Mol. Biol 429, 3168–3180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powell WT et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc. Natl. Acad. Sci. USA 110, 13938–13943 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nadel J et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin 8, 46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu W et al. The R-loop is a common chromatin feature of the Arabidopsis genome. Nat. Plants 3, 704–714 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Cerritelli SM & Crouch RJ Cloning, expression, and mapping of ribonucleases H of human and mouse related to bacterial RNase HI. Genomics 53, 300–307 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Yu K, Chedin F, Hsieh CL, Wilson TE & Lieber MR R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol 4, 442–451 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Chen PB, Chen HV, Acharya D, Rando OJ & Fazzio TG R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol 22, 999–1007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dumelie JG & Jaffrey SR Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. Elife 6, e28306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halasz L et al. RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Res. 27, 1063–1073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanoosthuyse V Strengths and weaknesses of the current strategies to map and characterize R-loops. Noncoding RNA 4, E9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konig F, Schubert T & Langst G The monoclonal S9.6 antibody exhibits highly variable binding affinities towards different R-loop sequences. PLoS ONE 12, e0178875 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartono SR et al. The affinity of the S9.6 antibody for double-stranded RNAs impacts the accurate mapping of R-loops in fission yeast. J. Mol. Biol 430, 272–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips DD et al. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit 26, 376–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X et al. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol 35, 940–950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bell JC et al. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. Elife 7, e27024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitz KM, Mayer C, Postepska A & Grummt I Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24, 2264–2269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bacolla A, Wang G & Vasquez KM New perspectives on DNA and RNA triplexes as effectors of biological activity. PLoS Genet. 11, e1005696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mondal T et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun 6, 7743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Leary VB et al. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 11, 474–485 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Postepska-Igielska A et al. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell 60, 626–636 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Syed J & Sugiyama H RNA-DNA triplex formation by long noncoding RNAs. Cell Chem. Biol 23, 1325–1333 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Nowotny M et al. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 27, 1172–1181 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen HD et al. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell 65, 832–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhatia V et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Legros P, Malapert A, Niinuma S, Bernard P & Vanoosthuyse V RNA processing factors Swd2.2 and Sen1 antagonize RNA Pol III-dependent transcription and the localization of condensin at Pol III genes. PLoS Genet. 10, e1004794 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tresini M et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature 523, 53–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rhee HS & Pugh BF Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rotem A et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol 33, 1165–1172 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Q, Brown JB, Huang H & Bickel PJ Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat 5, 1752–1779 (2011). [Google Scholar]
- 65.Landt SG et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Core LJ et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet 46, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
R-ChIP data for HEK293T and K562 cell lines are available at the NCBI GEO repository under the accession number GSE97072. All public datasets used in this study are indicated in the corresponding figure legends.