Abstract
Background
Non-small cell lung carcinomas (NSCLC) are prevalent, lethal cancers with especially grim prospects due to late-stage detection and chemoresistance. Circular RNAs (circRNAs) are non-coding RNAs that participate in tumor development. However, the role of circRNAs in NSCLC is not well known. This study investigated the role of one circRNA – circPTPRA– in NSCLC and characterized its molecular mechanism of action.
Methods
circPTPRA expression was analyzed in human NSCLC tumors and matched healthy lung tissue. We performed functional characterization in NSCLC cell lines and a mouse xenograft model of NSCLC to elucidate the molecular role of circPTPRA in epithelial-mesenchymal transitioning (EMT). We also assessed the regulatory action of circPTPRA on the microRNA miR-96-5p and its target the tumor suppressor Ras association domain-containing protein 8 (RASSF8).
Findings
circPTPRA was significantly downregulated in NSCLC tumors relative to matched healthy lung tissue. Lower circPTPRA levels correlated with metastasis and inferior survival outcomes in NSCLC patients. circPTPRA suppressed EMT in NSCLC cell lines and reduced metastasis in the murine xenograft model by sequestering miR-96-5p and upregulating RASSF8. Correlation analyses in patient-derived NSCLC tumor specimens supported the involvement of the circPTPRA/miR-96-5p/RASSF8/E-cadherin axis dysregulation in NSCLC tumor progression.
Interpretation
circPTPRA suppresses EMT and metastasis of NSCLC cell lines by sponging miR-96-5p, which upregulates the downstream tumor suppressor RASSF8. The circPTPRA/miR-96-5p/RASSF8/E-cadherin axis can be leveraged as a potential treatment avenue in NSCLC.
Fund
The Key research and development projects of Anhui Province (201904a0720079), the Natural Science Foundation of Anhui Province (1908085MH240), the Graduate Innovation Program of Bengbu Medical College (Byycx1843), the National Natural Science Foundation of Tibet (XZ2017ZR-ZY033) and the Science and Technology Project of Shannan (SNKJYFJF2017-3) and Academic Subsidy Project for Top Talents in Universities of Anhui in 2019 (gxbjZD16)
Keywords: Lung cancer; NSCLC; circRNA; Epithelial-mesenchymal transition, EMT; miR-96-5p
Research in context.
Evidence before this study
Lung cancer ranks first among all malignancies in cancer-related deaths globally. Approximately 70% of lung cancer cases are diagnosed at an advanced stage of the disease. Consequently, earlier methods of detection and more efficacious therapeutic options are urgently needed to address the significant mortality associated with lung cancer. The involvement of long non-coding RNAs on tumor development is increasingly appreciated, but the role of circular RNAs (circRNAs) in non-small cell lung carcinomas (NSCLC) is much less well-established.
Added value of this study
We identified the long non-coding circRNA circPTPRA as differentially downregulated in NSCLC tumors versus matched adjacent healthy lung tissue. Downregulation of circPTPRA was associated with metastatic disease and shorter survival in NSCLC patients. Functional studies in NSCLC cell lines and a mouse xenograft model of NSCLC revealed that circPTPRA suppressed EMT in NSCLC cell lines and reduced metastasis in the murine xenograft model by sequestering miR-96-5p and upregulating RASSF8. Correlation analyses in patient-derived NSCLC tumor specimens supported the involvement of the circPTPRA/miR-96-5p/RASSF8/E-cadherin axis dysregulation in NSCLC tumor progression.
Implications of all the available evidence
Our study presents a novel body of experimental data in support of circPTPRA as a tumor-suppressive circRNA in NSCLC. circPTPRA mediates its EMT-suppressive effects by sponging miR-96-5p, which upregulates the downstream tumor suppressor RASSF8. Furthermore, our results advocate the circPTPRA/miR-96-5p/RASSF8/E-cadherin axis as a potential research avenue for new drug development strategies in NSCLC.
Alt-text: Unlabelled Box
1. Introduction
Lung cancer ranks first among all malignancies in cancer-related deaths globally and in China [1,2]. In 2018, the estimated worldwide death toll from lung cancer was 154,050, accounting for 25.3% of all cancer-related deaths [1]. Approximately 70% of lung cancer cases are diagnosed at an advanced stage of the disease, since the symptoms are non-specific and there are no effectual diagnostic tests [3,4]. This results in poorer patient prospects due to more extensive tumor burdens and metastases and frequent treatment failures due to the development of chemo- and radio resistance [5,6]. Consequently, earlier methods of detection and more efficacious therapeutic options are urgently needed to address the significant mortality associated with lung cancer.
The majority of lung cancer cases, constituting approximately 80 to 85%, are non-small cell lung carcinomas (NSCLC) [7]. Although NSCLC tumors often possess mutations in genes encoding proteins, such as to KRAS [8] and receptor tyrosine kinases (RTKs) [9], the involvement of non-coding RNAs (ncRNAs), such as circular RNAs (circRNAs), long non-coding RNAs (lncRNAs), and microRNAs (miRNAs), on tumor development is increasingly appreciated [10,11]. In particular, miRNAs [12] and lncRNAs [13] have been extensively studied in NSCLC, where they may be up- or downregulated, and act as biomarkers or predictors of treatment response. On the other hand, the participation of circRNAs in NSCLC is much less well-established. circRNAs form a closed loop because their 5′ and 3 ends are covalently linked [14,15]. Advances in the detection of circRNAs using high-throughput RNA sequencing (RNA-seq) [16] is revealing their roles in the pathogenesis of cancer [17,18], but their participation in NSCLC is still in the early stages of discovery. For instance, one study has reported that circRNA_100876 is overexpressed in NSCLC and could serve as a prognostic indictor since its expression correlates with tumor staging, lymph node metastasis, and overall survival [19].
circPTPRA (circ_PTPRA_4–5, hsa_circRNA_102984, hsa_circ_0006117, circRNA.8325) is transcribed from the protein tyrosine phosphatase receptor type A (PTPRA) gene (ENSG00000132670). Interestingly, circPTPRA was found to be downregulated in bladder carcinomas [20,21]; however, its role in NSCLC is not fully elucidated. To shed light on its potential role in NSCLC, we undertook an investigation on the relation of circPTPRA to the clinicopathological characteristics of NSCLC and on its underlying molecular pathways and functions. We report that lowered circPTPRA transcripts in NSCLC play a crucial role in tumor progression and could constitute a predictive biomarker and potential treatment strategy for the disease.
2. Materials and methods
2.1. Ethics statement
This study was reviewed and approved by the Ethics Review Committee at Bengbu Medical College and was performed in accordance with Declaration of Helsinki. Every patient provided their informed consent in writing prior to their participation in the study. Animal protocols, housing, and care were approved by the same Ethics Review Committee and conducted according to the guidelines set forth in the National Institutes of Health's (NIH) “Guide for the Care and Use of Laboratory Animals” (8th edition).
2.2. Collection of patient-derived lung tissue
For the initial microarray and circPTPRA quantitative reverse-transcription PCR (qPCR) experiments, a set of NSCLC tumors and matched neighboring healthy lung tissue was obtained from NSCLC patients (n = 34) who underwent lobectomy between January 2004 and December 2010 at our hospital.
For all further experiments, a larger set of NSCLC tumor tissue and matched neighboring healthy lung tissue was obtained from 80 NSCLC patients who underwent lobectomy during the same time period at our hospital. The international tumor-node-metastasis (TNM) classification from the Union Internationale Contre le Cancer (UICC) (UICC; 1974, 2nd edition) was employed to stage lung tumors [22]. De-identified patient information has been outlined in Supplementary Table S1. Every patient had frequent follow-up visits post-surgery and was monitored for signs of cancer relapse to determine overall survival (OS) and disease-free survival (DFS). DFS times were censored at the date of death from non-NSCLC causes or at the date of last follow-up. Tumor and healthy lung tissue samples were flash frozen and stored in liquid nitrogen until required for quantification of circRNA transcripts and for immunohistochemistry (IHC).
2.3. circRNA microarray
The initial set of NSCLC specimens and matched non-tumor tissues (n = 34) were employed for the initial microarray analysis. This microarray analysis was performed by Kangcheng Biotech (Shanghai, China). The microarray results are presented in Supplementary File 1.
2.4. Quantitative real-time PCR (qPCR) analysis
TRIzol™ (Invitrogen) was employed to purify total RNA from NSCLC specimens and cell lines. The SYBR Premix Ex Taq II kit (Takara Bio, Beijing, China) was utilized to perform qPCR on a 7500 Fast Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). Transcripts were normalized to GAPDH for mRNAs or to small nucleolar RNA U6 for circRNAs and miRNAs. Primers were as follows: (i) circPTPRA, forward 5′- ACA CAC ACA CAC ACA CAC AC, reverse 5′-CTG CTC ACA AGA CCT ACC CA, (ii) PTPRA, forward 5′-CAA CAA TGC TAC CAC AGT, reverse, 5′-AAG AGA AGT TAG TGA AGA AGT T, (iii) miR-96-5p, forward 5′-TTT GGC ACT AGC ACA TTT TTG CT, reverse primer provided with kit; (iv) Ras association domain-containing protein 8 (RASSF8), forward 5′-AAG TAT GGG TGG ATG GAG TTC AG, reverse 5′-ATG AGG TGC TAA GTG TCT TTC AG; (v) GAPDH, forward 5′-TGA AGG TCG GAG TCA ACG GAT TTG GT, reverse 5′-CAT GTG GGC CAT GAG GTC CAC CAC, and (vi) U6, forward 5′- GCT TCG GCA GCA CAT ATA CTA AAA T, reverse primer provided with kit. Relative quantification was calculated with the comparative CT method (DDCT) method.
2.5. Animal care and xenograft model
Animals for this study were procured from Charles River Laboratories (Beijing, China). Xenograft mouse models of NSCLC were generated in nude BALB/c mice (aged 4 weeks) via tail vein injection of 0.5 × 106 NSCLC cells. Mice were euthanized six weeks post-injection, and their lungs were excised and fixed in phosphate buffered formaldehyde. Lungs were embedded in paraffin, and serial sections were used to count metastatic lung lesions.
2.6. Cell lines and culture conditions
The NSCLC cell lines (H23, H1755, and H522) and a non-cancerous lung cell line (BEAS-2B) were procured from American Type Culture Collection (ATCC). Lines were validated three months prior to the start of this study by morphology and growth kinetics and were cultured for no longer than two months. All cell-lines were grown in RPMI-1640 (Invitrogen, Thermo Fisher Scientific, Waltham, MA) with fetal bovine serum (FBS, 10%; HyClone™, Thermo Fisher Scientific).
2.7. Transient and stable transfection of cell lines
Vectors to overexpress (OE) or knockdown (KD) circPTPRA (circPTPRA-OE and circPTPRA-KD, respectively), as well as RASSF8-OE, were procured from OBiO Technology (Shanghai, China). Plasmids expressing circPTPRA, a short hairpin RNA (shRNA) against circPTPRA (circPTPRA-shRNA), and a small-interfering RNA against linear circPTPRA (si-circPTPRA) were purchased from GenePharma (Suzhou, China). miR-96-5p mimics were from RiboBio (Guangzhou, China). Lentiviral plasmids of miR-96-5p inhibitor (HmiR-AN0852-AM03) and scrambled control (CmiR-AN0001-AM03) were purchased from GeneCopoeia (Rockville, MD).
Transient transfection was performed on NSCLC cells plated into six-well culture plates at a confluence of 50 to 60%. One day later, cultures underwent transfection of miR-96-5p mimics or inhibitor using Lipofectamine® 2000 (Invitrogen). For stable transfections, lentiviruses were generated and used to infect cells using Lenti-Pac™ HIV Expression Packaging Kit (GeneCopoeia). Selection of cells with OE circPTPRA or negative control plasmid was performed in puromycin [23]. Doubly-transfected cells that additionally underwent transfection of miR-96-5p mimics, miR-96-5p inhibitor, and respective controls were selected in hygromycin. Cells that were used to generate the in vivo metastatic murine model by tail vein injection were generated from cells with stable miR-96-5p inhibitor transfection or circPTPRA-shRNA transection.
2.8. Scratch and Transwell assays
The scratch assay was performed on confluent NSCLC cultures in six-well culture plates using a P200 pipette tip. Images were taken at time points 0- and 24-h post-scratch. The Transwell assay was conducted with Boyden chambers comprising 24-well Transwell plates (BD Biosciences, San Jose, CA) with membranes possessing 8 mm pores. Cells (5 × 104) were plated to the top chamber in 500 μl serum-free media, which did not have an extracellular matrix coat. RPMI-1640 media supplemented with FBS (20%) was included in the bottom chamber. Roughly one day later, fixation and staining were performed of cells that had adhered to the membrane's bottom surface. Cell counts were made following microscopic examination. Scratch and Transwell assays were performed with two technical replicates and three independent biological replicates.
2.9. Western blot (WB) analysis
Cells were lysed in lysis buffer, and an equal amount of protein was loaded into each well of a sodium dodecyl sulfate (SDS)-PAGE (12%) gel. Proteins were resolved by an applied potential and then blotted to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Burlington, MA). Blots were rinsed, blocked for 1 h, and probed at 4 °C with primary antibodies against: (i) PTPRA (1:1000; Proteintech, Rosemont, IL), (ii) RASSF8(1:1000; Proteintech), (iii) N-cadherin (1:1000; Cell Signaling Technologies, Danvers, MA), (iv) E-cadherin (1:1000; Cell Signaling Technologies), (v) ALIX (1:1000; Proteintech), (vi) CD63 (1:1000; Abcam, Cambridge, UK), (vii) heat shock protein 70 (HSP70, 1:2000; Proteintech), (viii) tumor susceptibility gene 101 TSG101 (1:1000; Proteintech), (ix) GAPDH (1:5000; Proteintech), and (x) vimentin (1:1000; Cell Signaling Technologies).
The following day, blots were rinsed and probed for 1 h at room temperature (RT) with horseradish peroxidase (HRP)-tagged secondary antibody. Following additional rinse step, blots were developed with a Pierce™ ECL (enhanced chemiluminescence) Western Blotting Substrate (Thermo Fisher Scientific) and detected bands were quantified using Quantity One® 1-D Analysis Software (Bio-Rad, USA).
2.10. Inhibition of transcription
Actinomycin D (2 mg/ml) was added to inhibit transcription versus negative control vehicle (DMSO; Sigma-Aldrich, St Louis, MO) [24]. RNase R (3 U/μg; Epicentre Technologies) was added to total RNA (2 μg) samples at 37 °C for 15 min. circPTPRA and PTPRA levels were quantified in actinomycin D- or RNase R-treated samples by qPCR.
2.11. Northern blot analysis
A NorthernMax™ Kit (Ambion®, Thermo Fisher Scientific) was utilized for Northern blot analysis. RNase R was used to digest an aliquot of total RNA (10 μg), which was loaded and resolved on an agarose gel (2%), followed by capillary transfer to an Amersham Hybond-N+ membrane (GE Healthcare, Chicago, IL). Following an overnight hybridization at 58 °C, blots were extensively rinsed with a one-time 0.2 × saline‑sodium citrate (SSC) buffer, 0.1% SDS rinse (RT, 10 min), and two-times 0.2 × SSC, 0.1% SDS rinses (58 °C, 3 min). Finally, blots were developed with a Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher Scientific). Biotin-conjugated probes were obtained from Takara Bio (Beijing, China) and had sequences as follows: (i) circPTPRA, 5′-biotin-AGT TGA CCA CCC CTG TAG GGA CTA CAG CCC TGC TTA AAA GCA AAT CAG TCC CTG AAA CTG and (ii) GAPDH, 5′-biotin-TAT CCA CTT TAC CAG AGT TAA AAG CAG CCC TGG TGA CCA GGC GCC CAA TAC GAC CAA A.
2.12. RNA fluorescence in situ hybridization (RNA-FISH) analysis
Cells were rinsed in phosphate buffer saline (PBS) and fixation was achieved in paraformaldehyde (PFA, 4% in PBS). Cells were then deposited onto sterilized glass slides and dehydrated using serial ethanol solutions (70, 80, and 100% ethanol). Probes were then hybridized at 37 °C in a humidified chamber away from light. The following day, slides were rinsed two times in 2 × SCC/formamide (50%) for 5 min, treated with Alexa Fluor™ 488 Tyramide SuperBoost™ Kits (Thermo Fisher Scientific) for 30 min, and treated with DAPI. A FV1000 confocal microscope was used for image acquisition (Olympus, Tokyo, Japan). RNA probes for circPTPRA (biotin-labeled) and miR-96-5p [digoxigenin (DIG)-labeled] were from Takara Bio (Beijing, China) and had sequences as follows: (i) circPTPRA 5′-biotin-CCC CUG UAG GGA CUA CAG CCC UGC UUA AAA GCA AAU CAG UCC, and (ii) miR-96-5p, 5′-DIG-AGC AAA AAU GUG CUA GUG CCA AA.
2.13. Fluorescence immunocytochemistry (ICC)
NSCLC cells were cultured adhered to Corning® confocal glass-bottom dishes (Corning, Corning, NY). After media was removed, they were washed 2 times in PBS and fixation was achieved in PFA (4% in PBS) on ice for 15 min. Following rinse steps (PBS, 3 times), permeabilization was performed in Triton X-100 (0.1% in PBS, i.e., PBS-T) on ice for 10 min, followed by more rinse steps (PBS, 2 times), and blocking in PBS-T with bovine serum albumin (BSA, 5%) at 37 °C for 30 min. Cells underwent additional rinse steps in PBS-T, probing with primary antibodies at 37 °C for 1 h, rinse steps, secondary antibody treatment (37 °C, 30 min), rinse steps, DAPI nuclear counterstain, and final rinse steps. A FV1000 confocal microscope was used for image acquisition (Olympus) and relative mean fluorescence intensities were quantified by Image J (https://imagej.nih.gov/ij/).
2.14. Preparation of cytoplasmic and nuclear extracts
An Ambion®PARIS™ Kit (Thermo Fisher Scientific) was employed to prepare cytoplasmic and nuclear extracts from H23 cells.
2.15. RNA immunoprecipitation (RIP) assay
A Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore) was utilized for the RIP assay employing antibodies against: (i) FLAG tag (catalog no. 66008–3-Ig, Proteintech) and (ii) IgG control (catalog no. 30000–0-AP, Proteintech).
2.16. Luciferase reporter assays
The luciferase reporter plasmid (Luc) was procured from OBiO Technology (Shanghai, China). The circPTPRA or RASSF8 sequence was inserted downstream of its luciferase gene promoter (Luc-circPTPRA or Luc-RASSF8). Site directed mutagenesis was employed to introduce mutations into the miR-96-5p binding sites on circPTPRA or RASSF8 (Luc-circPTPRA-mutant or Luc-RASSF8-mutant).
All luciferase reporter assays were conducted in H23 cells (1000 per well) that were seeded onto 96-well culture plates and cultured for 24 h. For the initial validation assay, H23 cells underwent co-transfection with a mixture of Luc-circPTPRA or empty Luc (50 ng), a siRNA against circPTPRA (si-circPTPRA, 100 pmol), and a Renilla luciferase standardization control (pRL-CMV, 5 ng). For the circPTPRA-miR-96-5p assay, H23 cells underwent co-transfection with a mixture of Luc-circPTPRA or Luc-circPTPRA-mutant (50 ng), miR-96-5p mimics (5 pmol), and a Renilla luciferase standardization control (pRL-CMV, 5 ng). For the RASSF8-miR-96-5p assay, H23 cells underwent co-transfection with a mixture of Luc-RASSF8 or Luc-RASSF8-mutant (50 ng), miR-96-5p mimics (5 pmol), and a Renilla luciferase standardization control (pRL-CMV, 5 ng). The luciferase signal was quantified using a Dual-Luciferase® Reporter Assay System (Promega, Madison, WI) after 48 h.
2.17. Bioinformatic evaluation of circPTPRA-miR-96-5p interaction
Predictive analysis of an interaction between hsa_circRNA_102984 (circPTPRA) and hsa-miR-96-5p was produced by Arraystar miRNA target prediction package [25].
2.18. Immunohistochemistry (IHC) analysis
Tissue sections were probed with primary antibodies against: (i) E-cadherin (1:200; Cell Signaling Technology) and (ii) RASSF8 (1:100; Proteintech). A Dako REAL™ EnVision™ Detection System, (Dako, Agilent Technologies) was then used to detect protein distribution. Sections were semi-quantified according to a scoring system that took into account the total immunoreactive area and the intensity of immunoreactivity with minor modifications [23]. For each E-cadherin and RASSF8: (i) percent of immunoreactive cells (i.e., total immunoreactive area) was scored by 1 < 10%, 2 = 10–50%, and 3 > 50%, and (ii) intensity of immunoreactivity was scored by 0 = negative, 1 = weak, 2 = moderate, and 3 = strong. The combined score from area and intensity of immunoreactivity were used to compute a staining index, which ranged from a score of 0 to 9. In instances tissues were heterogeneously stained, the staining index was measured for each area, and a weighted average was calculated [26]. IHC analysis and semi-quantification was performed by two independent blinded investigators.
2.19. Statistical tests and analyses
Statistical tests and analyses were performed with Prism 6 (GraphPad, La Jolla). Data were expressed as means ± standard errors of the mean (SEM). χ2 test was used to assess the correlation of circPTPRA levels in patient-derived tissue samples to clinicopathological characteristics. Unpaired Student's t-test was used to assess comparison across groups. Kaplan-Meier survival analysis was used to evaluate OS and DFS, with application of the log-rank test for comparison across groups. A Spearman correlation coefficient was used to analyze correlations. A P-value of <0.05 was taken to be statistically significant for all tests.
3. Results
3.1. circPTPRA downregulated in NSCLC; correlated with clinicopathological features in NSCLC patients
In our initial microarray analysis of NSCLC tumor specimens (n = 34 patients), hsa_circRNA_102984 was found to be the most significantly downregulated circRNA in NSCLC specimens relative to adjacent healthy lung tissue (Supplementary File 1). Therefore, hsa_circRNA_102984 was selected for further analysis by qPCR. By qPCR, hsa_circRNA_102984 was downregulated in patient-derived NSCLC tumor specimens versus adjacent healthy lung tissue (tumor:healthy lung expression ratio (mean ± SEM) = 0.17 ± 0.036, p = .039 [unpaired Student's t-test]). We searched the Genome Reference Consortium Human Build 37 (GRCh37/hg19), which revealed that hsa_circRNA_102984 situated on chromosome 20 (chr20 (+ strand): 2,944,918–2,945,848) resided within the protein tyrosine phosphatase receptor type A gene (PTPRA; ENSG00000132670; chr20 (+ strand): 2,844,830-3,019,722; GRCh37/hg19). Consequently, hsa_circRNA_102984 has been designated circPTPRA in this study.
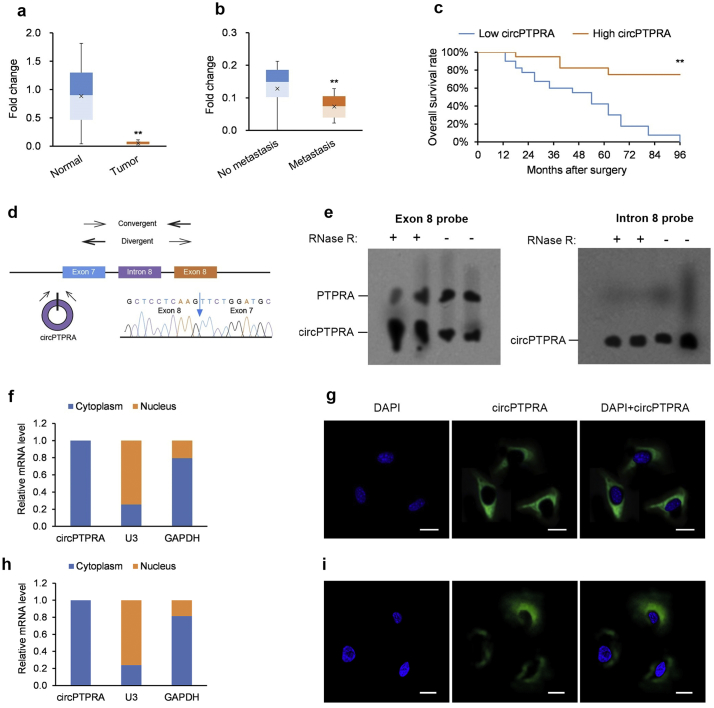
A larger set of NSCLC tumor specimens and their paired healthy lung tissue (n = 80 patients) were collected to determine the relationship between circPTPRA levels to clinicopathological characteristics (see Supplementary Table S1 for patient details). circPTPRA levels were significantly lower in NSCLC tumor versus paired healthy specimens (Fig. 1a) and were also significantly lower in lymph node metastatic disease versus non-metastatic disease (Fig. 1b). Patients with low (below median) circPTPRA levels displayed lower OS rates (Fig. 1c).
Fig. 1.
circPTPRA downregulation in NSCLC; correlates with clinicopathological features in NSCLC patients.
(a) Fold-change in circPTPRA transcripts in NSCLC tumors versus matched neighboring healthy lung tissue quantified by qPCR (n = 80 specimens); *p < .05, **p < .01 [paired Student's t-test]. (b) Fold-change in circPTPRA transcripts in lymph node metastatic versus non-metastatic NSCLC tumors quantified by qPCR (n = 48 non-metastatic, n = 32 metastatic specimens); *p < .05, **p < .01 [unpaired Student's t-test]. (c) Kaplan-Meier univariate analysis of overall survival in NSCLC patients with high (above median) versus low (below median) circPTPRA levels; *p < .05, **p < .01 [log-rank test]. (d) Schematic of convergent and divergent primers used to confirm circPTPRA as a circRNA. (e) Left panel: Northern blot of PTPRA and circPTPRA transcripts using an exon-8 hybridization probe with RNase R. Right panel: Northern blot of circPTPRA transcripts using an intron-8 hybridization probe without RNase R with GAPDH Northern blot used as a positive control. (f, g) Cytoplasmic distribution of circPTPRA in (f) H23 cells and (g) H1755 cells shown in typical RNA-FISH images; scale bars = 10 μm. Quantitation of nuclear and cytoplasmic fractions demonstrate circPTPRA's cytoplasmic localization.
All in vitro experiments: n = 3 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard errors of the mean (SEMs). Abbreviations: gDNA = genomic DNA, qPCR = quantitative real-time PCR, RNA-FISH = RNA fluorescence in situ hybridization, RT-PCR = reverse transcription PCR.
We extended our analysis from patient-derived NSCLC tumor specimens to NSCLC cell lines; consistently, circPTPRA levels were lower in the NSCLC cell lines versus the non-cancerous lung cell line BEAS-2B (Supplementary Fig. S1a). We performed qPCR of circPTPRA from H23 cells using divergent primers followed by Sanger sequencing of the amplified product from 5'exon-8 to 3'exon-7 (Fig. 1d) [27]. This enabled the design of hybridization probes that bound only to circPTPRA (intron-8) and to both circPTPRA and PTPRA (exon-8) for Northern blot (Fig. 1e). Furthermore, the putative circular species was resistant to breakdown by the exonuclease RNase R, verifying its circular nature, and was stable in H23 cultures (half-life >24 h) (Supplementary Figs. S1b and S1c). Moreover, circPTPRA's localization is largely cytoplasmic, as determined by fractionation experiments and RNA fluorescence in situ hybridization (RNA-FISH) (Fig. 1f and g).
3.2. circPTPRA fosters an invasive phenotype in NSCLC cell lines
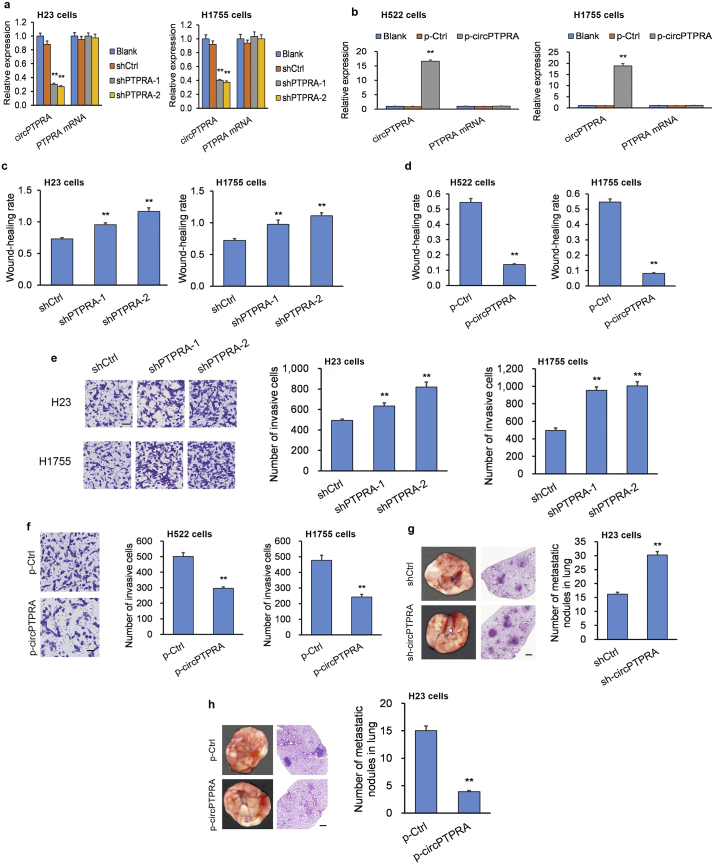
Of the three NSCLC cell lines, circPTPRA expression was lowest in the H23 and H1755 cell lines, which were selected for subsequent circPTPRA-knockdown (circPTPRA-KD) experiments (Supplementary Fig. S1a). Short hairpin RNAs (shRNAs) targeting the back-spliced section of circPTPRA were generated to efficiently knockdown circPTPRA expression in H23 cells and H1755 cells (Fig. 2a). Moreover, circPTPRA expression was highest in the H522 and H1755 cell lines (Supplementary Fig. S1a), which were selected for the subsequent circPTPRA-overexpression (circPTPRA-OE) experiments. Transfection of H522 and H1755 cells with a circPTPRA-OE plasmid resulted in a robust increase in circPTPRA levels (Fig. 2b).
Fig. 2.
circPTPRA suppresses the invasive phenotype in NSCLC cell lines.
(a) Quantification of circPTPRA and PTPRA transcript levels in circPTPRA-KD H23 cells and circPTPRA-KD H1755 cells assessed by qPCR. *p < .05, **p < .01 vs. Blank [unpaired Student's t-test]. (b) Quantification of circPTPRA and PTPRA transcript levels in circPTPRA-OE H522 cells and circPTPRA-OE H1755 cells assessed by qPCR. *p < .05, **p < .01 vs. Blank [unpaired Student's t-test]. (c) circPTPRA-KD H23 cells and circPTPRA-KD H1755 cells exhibited attenuated migration compared to cells transfected with a scrambled shRNA control in a scratch assay. *p < .05, **p < .01 vs. shCtrl [unpaired Student's t-test]. (d) circPTPRA-OE H522 cells and circPTPRA-OE H1755 cells exhibited enhanced migration compared to cells transfected with an empty plasmid control in a scratch assay. *p < .05, **p < .01 vs. p-Ctrl [unpaired Student's t-test]. (e) circPTPRA-KD H23 cells and circPTPRA-KD H1755 cells exhibited attenuated invasion compared to cells transfected with a scrambled shRNA control in a Transwell assay; scale bar = 100 μm. *p < .05, **p < .01 vs. shCtrl [unpaired Student's t-test]. (f) circPTPRA-OE H522 cells and circPTPRA-OE H1755 cells exhibited attenuated invasion compared to cells transfected with an empty plasmid control in a Transwell assay; scale bar = 100 μm. *p < .05, **p < .01 vs. p-Ctrl [unpaired Student's t-test]. (g and h) In vivo xenografts into nude BALB/c mice generated by tail vein injection of stably-transfected H23 cells were sacrificed 6 weeks later to assess lung lesions (n = 9 mice per group). Injection of (g) circPTPRA-KD H23 cells or scrambled shRNA control H23 cells or (h) circPTPRA-OE H23 cells or empty plasmid control H23 cells. *p < .05, **p < .01 vs. shCtrl or p-Ctrl [unpaired Student's t-test]. Left panels: Typical lung images with metastatic lesions; Middle panels: H&E lung tissue sections of metastatic lesions (100× magnification; scale bar = 100 μm); Right panel: Quantification of metastatic lesions.
All in vitro experiments: n = 3 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard errors of the mean (SEMs). Abbreviations: circPTPRA-KD = NSCLC cells with shRNA-mediated circPTPRA knockdown, circPTPRA-OE = NSCLC cells with circPTPRA overexpression plasmid, H&E = hematoxylin and eosin, qPCR = quantitative real-time PCR.
Migration and invasion were heightened in circPTPRA-KD H23 cells and circPTPRA-KD H1755 cells (Fig. 2c and e; Supplementary Fig. S2a), while they were markedly diminished in circPTPRA-OE H522 cells and circPTPRA-OE H1755 cells (Fig. 2d and f; Supplementary Fig. S2b). To substantiate the in vitro experiments, we generated an in vivo mouse xenograft model by tail-vein injection of stably-transfected H23 cells. We sacrificed mice six weeks later to assess lung tumor lesions. Mice injected with circPTPRA-KD H23 cells had more lung lesions compared to mice injected with H23 cells stably transfected with a scrambled shRNA control (Fig. 2g). Conversely, mice injected with circPTPRA-OE H23 cells had fewer lung lesions compared to mice injected with H23 cells transfected with an empty plasmid control (Fig. 2h).
3.3. circPTPRA suppresses EMT in NSCLC cell lines
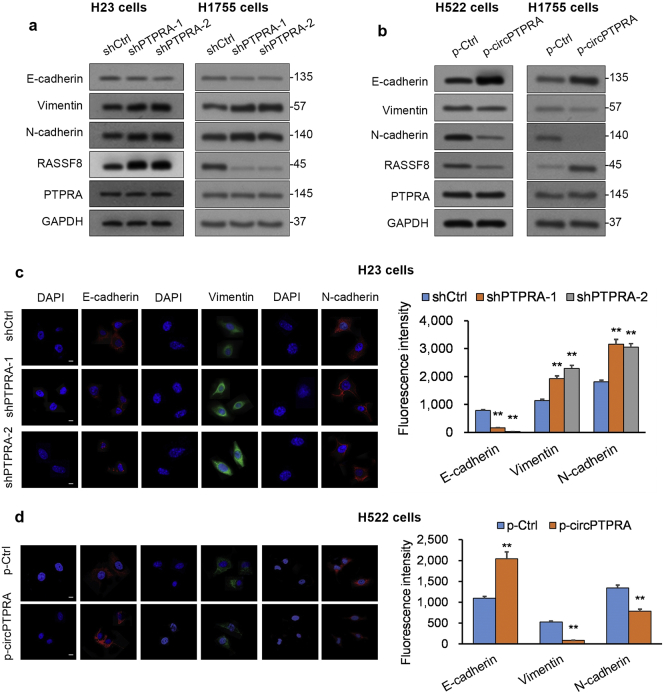
The epithelial-mesenchymal transition (EMT) is a mechanism associated with loss of differentiation in epithelial cells, which gain a more migration-prone mesenchymal phenotype [28]. In tumors, EMT is associated with a more invasive phenotype [28]. Since circPTPRA levels were decreased in NSCLC tumors from patients with metastatic disease compared to those from patients with non-metastatic disease (Fig. 1b), we evaluated the effect of circPTPRA on EMT in NSCLC cell lines. Western blot (WB) analysis revealed that circPTPRA-KD H23 cells and circPTPRA-KD H1755 cells exhibited lower E-cadherin protein levels (epithelial markers) and higher N-cadherin and vimentin protein levels (mesenchymal markers) compared to NSCLC cells transfected with scrambled shRNA (Fig. 3a). Conversely, circPTPRA-OE H522 cells and circPTPRA-OE H1755 cells exhibited lower E-cadherin protein levels and higher N-cadherin and vimentin protein levels compared to NSCLC cells transfected with empty vector (Fig. 3b). Notably, circPTPRA KD or OE did not impact PTPRA levels in these cell lines (Fig. 3a, b). Fluorescence immunocytochemistry (ICC) analysis in circPTPRA-KD H23 cells and circPTPRA-OE H522 cells corroborated our WB results (Fig. 3c and d). Cumulatively, circPTPRA suppresses EMT in NSCLC cells in a PTPRA-independent manner.
Fig. 3.
circPTPRA suppresses EMT in NSCLC cell lines.
(a and b) WB analysis of E-cadherin (epithelial marker), N-cadherin and vimentin (mesenchymal markers), RASSF8, and PTPRA in (a) circPTPRA-KD H23 cells and circPTPRA-KD H1755 cells as well as (b) circPTPRA-OE H522 cells and circPTPRA-OE H1755 cells. GAPDH served as loading control. (c and d) Fluorescent ICC analysis of E-cadherin, N-cadherin, and vimentin in (c) circPTPRA-KD H23 cells and (d) circPTPRA-OE H522 cells; scale bar = 5 μm. Left panels: Typical images from fluorescent ICC analysis; Right panels: Quantification of mean fluorescence intensity using Image J. *p < .05, **p < .01 vs. shCtrl or p-Ctrl [unpaired Student's t-test].
All in vitro experiments: n = 3 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard errors of the mean (SEMs). Abbreviations: circPTPRA-KD = NSCLC cells with shRNA-mediated circPTPRA knockdown, circPTPRA-OE = NSCLC cells with circPTPRA overexpression plasmid, ICC = immunocytochemistry, RASSF8 = Ras association domain-containing protein 8, WB = Western blot.
3.4. circPTPRA sequesters miR-96-5p in NSCLC cell lines
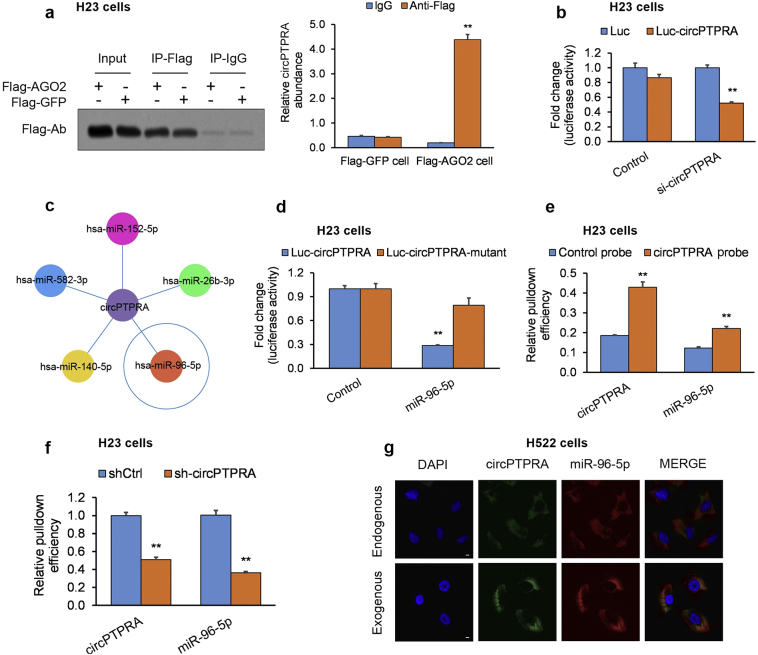
Since several circRNAs persist within the cytoplasm and sequester miRNAs [29], we hypothesized that circPTPRA's anti-malignant effects may be an effect of sponging tumorigenic miRNA(s) in NSCLC cells. circPTPRA possesses a putative Argonaute 2 (AGO2) binding site; therefore, we determined whether the AGO2 site was occupied in circPTPRA by RNA immunoprecipitation (RIP). H23 cells that had undergone stable transfection with either AGO2-FLAG or a non-specific protein control GFP-FLAG, were subjected to RIP with anti-FLAG and anti-IgG antibodies. Endogenous circPTPRA was enriched in the AGO2-FLAG IP fraction in comparison to the GFP-FLAG and IgG control fractions (Fig. 4a). AGO2 occupancy on circPTPRA implies circPTPRA can be integrated into an RNA-induced silencing complex (RISC complex).
Fig. 4.
circPTPRA sequesters miR-96-5p in NSCLC cell lines.
(a) RIP assay in NSCLC cells expressing AGO2-FLAG or GFP-FLAG; endogenous circPTPRA was enriched in the AGO2-FLAG IP fraction in comparison to the GFP-FLAG and IgG control fractions; *p < .05, **p < .01 vs. matching IgG group [unpaired Student's t-test]. (b) A luciferase reporter assay plasmid was generated by cloning the whole circPTPRA sequence downstream of the luciferase gene promoter (circPTPRA-FLWT). H23 cells underwent co-transfection of circPTPRA-FLWT with circPTPRA-shRNA or scrambled control shRNA; *p < .05, **p < .01 vs. matching Luc group [unpaired Student's t-test]. (c) Schematic of predicted miRNA targets of circPTPRA. (d) H23 cells underwent co-transfection of circPTPRA-FLWT or circPTPRA-FLMUT with miR-96-5p mimics; *p < .05, **p < .01 vs. matching Control group [unpaired Student's t-test]. (e) Biotin-tagged circPTPRA or control probes were used in pull-down assay of miR-96-5pin NSCLC cells; percent of RNA that pulled-down (%input) was expressed relative to10-fold diluted input RNA, and standardized relative to RNA pulled-down by the control probe; *p < .05, **p < .01 vs. matching Control probe group [unpaired Student's t-test]. (f) Biotin-tagged circPTPRA or control probes were used in pull-down assay of miR-96-5p in circPTPRA-KD or scrambled shRNA control NSCLC cells; % input standardized relative to RNA pulled-down in control NSCLC cells; *p < .05, **p < .01 vs. matching shCtrl group [unpaired Student's t-test]. (g) Typical fluorescent ICC images of circPTPRA co-localized withmiR-96-5pin circPTPRA-OE or empty plasmid control NSCLC cells; scale bar = 5 μm.
All in vitro experiments: n = 3 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard errors of the mean (SEMs). Abbreviations: circPTPRA-KD = NSCLC cells with shRNA-mediated circPTPRA knockdown, circPTPRA-OE = NSCLC cells with circPTPRA overexpression plasmid, ICC = immunocytochemistry, miRNA = micro RNA, RIP = RNA immunoprecipitation assay, shRNA = short hairpin RNA.
Next, we utilized a luciferase reporter assay to screen miRNA binding to circPTPRA in H23 cells. The sequence for circPTPRA was cloned downstream of the luciferase gene promoter (Luc-circPTPRA), which was co-transfected with si-circPTPRA, causing a drop in the luciferase signal and confirming specificity (Fig. 4b). Arraystar software [25] predicted that the miR-96-5p possessed a binding location within the circPTPRA sequence (Fig. 4c, Supplementary Fig. S3). Next, H23 cells underwent co-transfection with Luc-circPTPRA or Luc-circPTPRA-mutant (with eliminated miR-96-5p binding sites) and miR-96-5p mimics. miR-96-5p mimics decreased the Luc-circPTPRA luciferase signal by over 70% in comparison to a control miRNA mimics. However, co-transfection of Luc-circPTPRA-mutant with miR-96-5p mimics did not significantly impact luciferase signal intensity (Fig. 4d). The findings suggest that circPTPRA can bind to and sequester miR-96-5p.
To test the relevance in situ in H23 cells, we conducted an RNA pull-down using a biotin-tagged circPTPRA probe and found that only miR-96-5p was enriched in the biotin IP fraction in comparison to control IgG (Fig. 4e). In circPTPRA-KD H23 cells, miR-96-5p was not enriched to an appreciable extent (Fig. 4f). When circPTPRA-OE H522 cells were analyzed by RNA-FISH, exogenous circPTPRA localized with endogenous miR-96-5p (Fig. 4g). Cumulatively, the findings support a direct interaction between circPTPRA and miR-96-5p, which was selected for further in-depth examination.
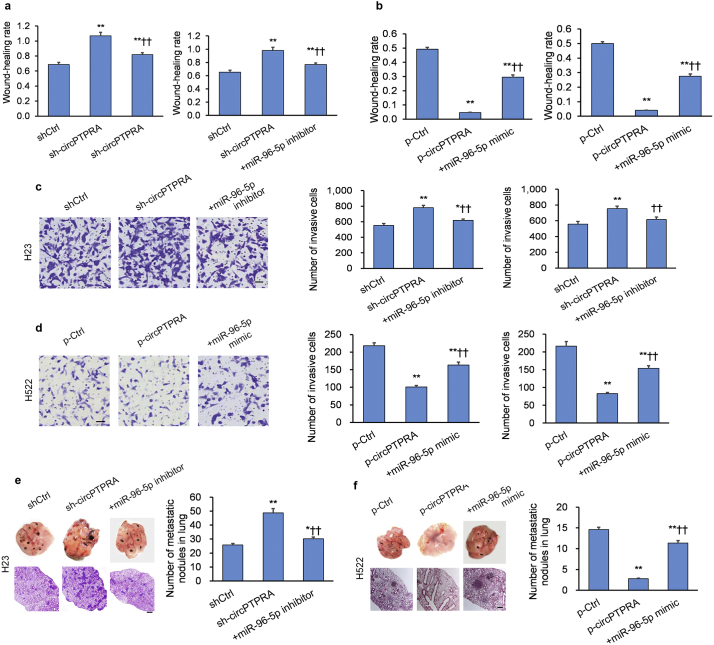
3.5. circPTPRA suppresses EMT in NSCLC cell lines by sponging miR-96-5p
Since circPTPRA can bind to and sequester miR-96-5p, we questioned whether this was one mechanism through which circPTPRA mediated its anti-malignant effects in NSCLC. Therefore, we transfected NSCLC cells with either circPTPRA-KD or circPTPRA-KD + miR-96-5p inhibitor to determine whether the miR-96-5p inhibitor would re-establish the original NSCLC phenotype. Indeed, migration and invasion capacity were reinstated in circPTPRA-KD NSCLC cells with miR-96-5p inhibitor (Fig. 5a and c; Supplementary Figs. S4a and S4c). We also transfected NSCLC cells with either circPTPRA-OE or circPTPRA-OE + miR-96-5p mimics to determine whether the miR-96-5p mimics would re-establish the original NSCLC phenotype. Migration and invasion capacity were reinstated in circPTPRA-OE NSCLC cells with miR-96-5p mimics (Fig. 5b and d; Supplementary Figs. S4b and S4d).
Fig. 5.
circPTPRA suppresses EMT in NSCLC cell lines by sequestering miR-96-5p.
(a and c) circPTPRA-KD H23 cells and circPTPRA-KD H1755 cells exhibit heightened (a) migration and (c) invasiveness in a scratch and Transwell assay, respectively; co-transfection with miR-96-5p inhibitor reversed the phenotype; scale bar = 100 μm. *p < .05, **p < .01 vs. shCtrl, †p < .05, ††p < .01 vs. sh-circPTPRA [unpaired Student's t-test]. (b and d) circPTPRA-OE H522 cells and circPTPRA-OE H1755 cells exhibit diminished (b) migration and (d) invasiveness in a scratch and Transwell assay, respectively; co-transfection with miR-96-5p mimics reversed the phenotype; scale bar = 100 μm. *p < .05, **p < .01 vs. p-Ctrl, ††p < .01 vs. p-circPTPRA [unpaired Student's t-test]. (e and f) In vivo xenografts into nude BALB/c mice generated by tail-vein injection of stably-transfected H23 cells were sacrificed 6 weeks later to assess lung lesions (n = 9 mice per group). (e) Injection of circPTPRA-KD H23 cells promotes metastasis, which is reversed by co-transfection of miR-96-5p inhibitor, while (f) injection of circPTPRA-OE H23 cells suppresses metastasis, which is reversed by co-transfection of miR-96-5p mimics; *p < .05, **p < .01 vs. p-Ctrl, †p < .05, ††p < .01 vs. sh-circPTPRA or p-circPTPRA [unpaired Student's t-test]. Left panels: Typical lung images with metastatic lesions; middle panels: H&E lung tissue sections of metastatic lesions (100× magnification; scale bar = 100 μm); lower right panel: Quantification of metastatic lesions.
All in vitro experiments: n = 3 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard errors of the mean (SEMs). Abbreviations: circPTPRA-KD = NSCLC cells with shRNA-mediated circPTPRA knockdown, circPTPRA-OE = NSCLC cells with circPTPRA overexpression plasmid, H&E = hematoxylin and eosin, ICC = immunocytochemistry, WB = Western blot.
To validate the in vitro experiments, we conducted experiments in our in vivo mouse xenograft model by tail-vein injection of stably-transfected H23 cells. Injection of circPTPRA-KD H23 cells co-transfected with miR-96-5p inhibitor reversed the effect on tumor metastasis of circPTPRA knockdown (Fig. 5e). Conversely, injection of circPTPRA-OE H23 cells co-transfected with miR-96-5p mimics reversed the effect on tumor metastasis of circPTPRA overexpression (Fig. 5f).
Since these findings advocate that circPTPRA stimulates EMT in NSCLC cell lines via miR-96-5p sequestration, we sought to determine whether circPTPRA resulted in upregulation of miR-96-5p gene targets. We queried miRanda [30] to identify miR-96-5p gene targets and discovered a highly-conserved miR-96-5p binding site on the 3′ untranslated region (3′-UTR) of Ras association domain-containing protein 8 (RASSF8) (Supplementary Fig. S5a), a putative lung tumor suppressor that blocks EMT [31]. In order to validate that RASSF8 is a miR-96-5p target, H23 cells underwent co-transfection with Luc-RASSF8 or Luc-RASSF8-mutant (with eliminated miR-96-5p binding sites) and miR-96-5p mimics. miR-96-5p mimics decreased the Luc-RASSF8 luciferase signal in comparison to a control miRNA mimics (Supplementary Fig. S5b). However, co-transfection of Luc-c RASSF8-mutant with miR-96-5p mimics did not significantly impact luciferase signal intensity (Supplementary Fig. S5b). Moreover, H23 cells transfected with miR-96-5p mimics expressed significantly lower RASSF8 mRNA and protein levels compared to cell lines transfected with a control mimics (Supplementary Figs. S5c and S5d), experimentally corroborating that RASSF8 is a miR-96-5p target.
Since these findings advocate that RASSF8 is a miR-96-5p target, we sought to determine whether miR-96-5p inhibition rescues RASSF8 and EMT marker expression in circPTPRA-silenced NSCLC cells. Indeed, the addition of miR-96-5p inhibitor in circPTPRA-KD NSCLC cells reversed the circPTPRA KD-mediated decrease in RASSF8 expression, reversed the circPTPRA KD-mediated decrease in the downstream RASSF8 target E-cadherin [31], and reversed the circPTPRA KD-mediated increases in N-cadherin and vimentin (Supplementary Figs. 6a and 6c). On the other hand, addition of miR-96-5p mimics in circPTPRA-OE NSCLC cells reversed the circPTPRA OE-mediated increase in RASSF8, reversed the circPTPRA OE-mediated increase in E-cadherin, and reversed circPTPRA OE-mediated decreases in N-cadherin and vimentin (Supplementary Figs. 6b and 6d). Overall, the findings support that circPTPRA stimulates EMT in NSCLC cell lines by sequestering miR-96-5p.
3.6. RASSF8 overexpression rescues invasive NSCLC phenotype produced by circPTPRA knockdown
We questioned whether RASSF8 downregulation is the primary mechanism by which circPTPRA mediates its anti-malignant effects in NSCLC cells. Therefore, we sought to determine whether overexpression of RASSF8 would rescue the invasive phenotype produced by circPTPRA knockdown. Therefore, we transfected the H23 and H1755 cell lines with either circPTPRA-KD or circPTPRA-KD + RASSF8-OE to determine whether RASSF8 overexpression would re-establish the original NSCLC phenotype (Supplementary Fig. 7a). Indeed, migration and invasion capacity were reinstated in circPTPRA-KD NSCLC cells with RASSF8-OE (Supplementary Fig. 7b and 7c). To validate the in vitro experiments, we conducted experiments in our in vivo mouse xenograft model by tail-vein injection of stably-transfected H23 cells. Injection of circPTPRA-KD H23 cells with RASSF8-OE reversed the effect on tumor metastasis of circPTPRA knockdown (Supplementary Fig. 7d). The addition of RASSF8-OE in circPTPRA-KD H23 and H1755 cells reversed the circPTPRA KD-mediated decrease in RASSF8 expression, reversed the circPTPRA KD-mediated decrease in the downstream RASSF8 target E-cadherin, and reversed the circPTPRA KD-mediated increases in N-cadherin and vimentin (Supplementary Fig. 7e). Overall, the findings support that RASSF8 downregulation is the primary mechanism by which circPTPRA mediates its anti-malignant effects in NSCLC cells.
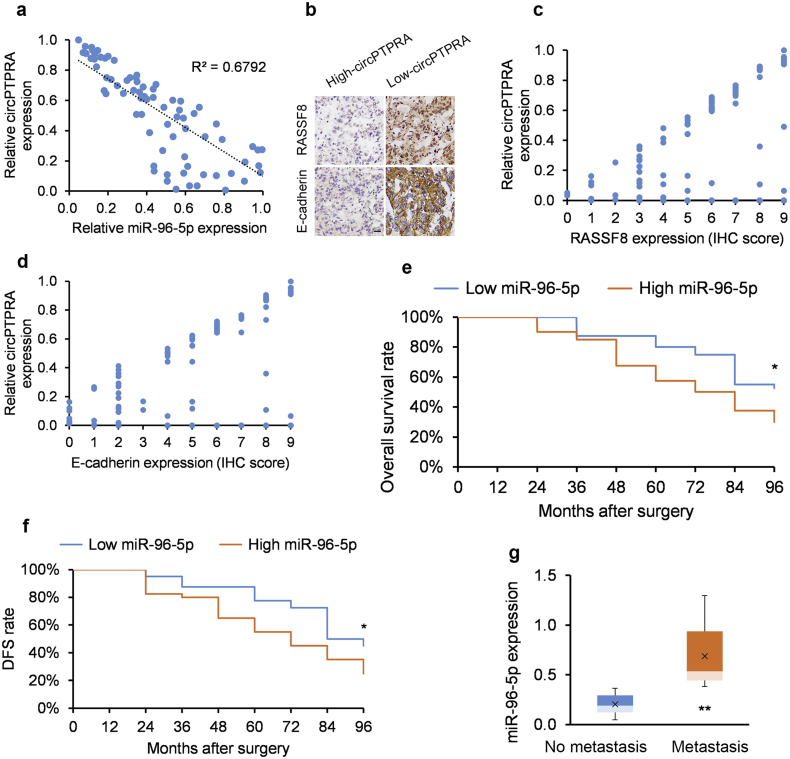
3.7. circPTPRA expression clinically correlates with miR-96-5p/RASSF8/E-cadherin axis expression in human NSCLC tumors
Since our functional studies support that circPTPRA stimulates EMT in NSCLC cell lines via the miR-96-5p/RASSF8/E-cadherin axis, we next analyzed this axis in clinical samples. circPTPRA and miR-96-5p levels correlated inversely in patient-derived NSCLC tumor samples (Fig. 6a). Furthermore, circPTPRA levels positively correlated with IHC-measured RASSF8 and E-cadherin expression in our study group (Figs. 6b-d). The findings support circPTPRA's role in suppressing EMT and metastasis in NSCLC tumors. Accordingly, low (below median) miR-96-5p levels were associated with improved OS and DFS (Fig. 6e and f). Moreover, metastatic NSCLC tumors displayed higher miR-96-5p levels (Fig. 6g). These clinical correlates support our contention that circPTPRA sponging of miR-96-5p suppresses EMT and NSCLC tumor progression.
Fig. 6.
circPTPRA suppresses EMT in NSCLC cell lines via regulating the miR-96-5p/RASSF8/E-cadherin axis.
(a) circPTPRA and miR-96-5p levels correlate inversely (R = −0.8241, R2 = 0.6792 [Spearman correlation]) in patient-derived NSCLC tumors (n = 80). (b) Typical IHC images of RASSF8 and E-cadherin protein levels in patient-derived NSCLC tumors stratified by circPTPRA expression; scale bar = 50 μm. (c and d) circPTPRA positively correlate with (c) RASSF8 transcript levels and (d) E-cadherin transcript levels in patient-derived NSCLC tumors (n = 80). (e and f) Kaplan-Meier univariate analysis of (e) overall survival and (f) disease-free survival (DFS) in NSCLC patients with high (above median) versus low (below median) miR-96-5p tumor levels (n = 40 in each group). *p < .05, **p < .01 [log rank test]. (g) miR-96-5p levels in NSCLC lymph node metastasis (n = 32) and non-metastatic NSCLC tumors (n = 48); *p < .05, **p < .01 [unpaired Student's t-test].
Data presented as means with error bars representing standard errors of the mean (SEMs). Abbreviations: DFS = disease-free survival, EMT = epithelial-mesenchymal transition, RASSF8 = Ras association domain-containing protein 8.
4. Discussion
Herein, we identified the long non-coding circular RNA circPTPRA as differentially downregulated in NSCLC tumors versus matched adjacent healthy lung tissue. Downregulation of circPTPRA was associated with metastatic disease and shorter survival in NSCLC patients. Functional studies in NSCLC cell lines and a mouse xenograft model of NSCLC revealed that circPTPRA sequesters the oncogenic miRNA miR-96-5p, thereby suppressing EMT in NSCLC cell lines via the miR-96-5p/RASSF8/E-cadherin axis. Since the RASSF8/E-cadherin pathway is critical in regulating cell–cell adhesion in NSCLC cells [31], our study provides the first evidence on circPTPRA's role in suppressing NSCLC tumor progression.
Studies of circRNAs indicate they have important regulatory functions and are not just idle by-products of mRNA splicing [32]. For instance, aberrant regulation of circRNAs has been noted in gliomas, lung carcinomas, renal cell carcinomas, and other human malignancies [33] [[34], [35], [36]].Our work adds to this list by supporting circPTPRA's role in suppressing NSCLC tumor progression. Our in vitro experiments revealed that elevated circPTPRA levels decreased NSCLC cell migration and invasiveness and reduced their expression of the EMT markers N-cadherin and vimentin. Our in vivo experiments in mouse xenografts also demonstrated the metastatic-suppressive potential of circPTPRA. In conjunction with the lower circPTPRA levels observed in more aggressive clinical NSCLC samples, our in vitro and in vivo results consistently advocate for circPTPRA's role in suppressing NSCLC EMT and tumor progression.
circRNAs are known to possess roles in sponging miRNA [29]. Specific miRNAs, however, may only be sequestered by the subset of circRNAs that possess appropriate miRNA binding sites. Most putative interactions have been suggested by in silico analysis, as opposed to bench experiments. This study provides a solid body of experimental evidence supporting the sponging action of circPTPRA upon miR-96-5p in H23 cells. Specifically, we demonstrated that: (i) endogenous circPTPRA was enriched in the AGO2-FLAG IP fraction, implying that circPTPRA can be integrated into a RISC complex; (ii) luciferase reporter assays confirmed circPTPRA's binding to miR-96-5p; (iii) RNA pull-down assays revealed that miR-96-5p was enriched in the circPTPRA biotin IP fraction; and (iv) RNA-FISH assays demonstrated that exogenous circPTPRA localizes with endogenous miR-96-5p.
miR-96-5p is well-established as an oncogenic miRNA species in several human cancers. For example, miR-96-5p directly suppresses Forkhead Box Protein O1 (FOXO1) and Forkhead Box Protein O3 (FOXO3), key tumor suppressor proteins in prostate and breast cancer [37,38] as well as ovarian cancer [39], respectively. miR-96-5p also functions as an oncogenic miRNA in bladder cancer cells through targeting the tumor suppressor Cyclin Dependent Kinase Inhibitor 1A (CDKN1A) [40]. Herein, we add the tumor suppressor RASSF8 [31] to the list of miR-96-5p gene targets in NSCLC cells. The impact of circPTPRA KD on EMT properties in NSCLC cells could be reversed by co-transfection of a miR-96-5p inhibitor or RASSF8 OE. Conversely, the effect of circPTPRA OE could be rescued by co-transfection with miR-96-5p mimics. These in vitro findings were corroborated by correlation analyses in patient-derived NSCLC tumors. These combined findings suggest that circPTPRA primarily mediates its suppressive effect on NSCLC cell EMT via miR-96-5p sequestration and consequent RASSF8 upregulation.
In conclusion, our study presents a novel body of experimental data in support of circPTPRA as a tumor-suppressive circRNA in NSCLC. circPTPRA mediates its EMT-suppressive effects by sponging miR-96-5p, which upregulates the downstream tumor suppressor RASSF8. Furthermore, our results advocate the circPTPRA/miR-96-5p/ RASSF8/E-cadherin axis as a potential research avenue for new drug development strategies in NSCLC.
Acknowledgments
Acknowledgments
This work was funded by the Key research and development projects of Anhui Province(201904a0720079),the Natural Science Foundation of Anhui Province (1908085MH240), the Graduate Innovation Program of Bengbu Medical College (Byycx1843), the National Natural Science Foundation of Tibet (XZ2017ZR-ZY033), the Science and Technology Project of Shannan (SNKJYFJF2017-3) and Academic Subsidy Project for Top Talents in Universities of Anhui in 2019 (gxbjZD16). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceived and designed the study: WL, JHZ. ZQQ, YQC, JG, and XJL.
Performed the literature search and data extraction: SLW, YYZ, and CLZ.
Analyzed the data: YRJ, YBS, and XJW.
Drafted the manuscript: WL, YYZ, and QL.
Conflicts of interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.05.032.
Contributor Information
Jihong Zhou, Email: zhoujihongww@126.com.
Wei Li, Email: bbmcliwei@126.com.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2018. 2018;vol. 68(1):7–30. [DOI] [PubMed]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]; Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66(2):115-32. [DOI] [PubMed]
- 3.Smith R.A., Manassaram-Baptiste D., Brooks D., Cokkinides V., Doroshenk M., Saslow D. Cancer screening in the United States, 2014: a review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2014;64(1):30–51. doi: 10.3322/caac.21212. [DOI] [PubMed] [Google Scholar]; Smith RA, Manassaram-Baptiste D, Brooks D, Cokkinides V, Doroshenk M, Saslow D, et al. Cancer screening in the United States, 2014: a review of current American Cancer Society guidelines and current issues in cancer screening. CA: a cancer journal for clinicians. 2014;64(1):30-51. [DOI] [PubMed]
- 4.Yang J., Liu H., Wang H., Sun Y. Down-regulation of microRNA-181b is a potential prognostic marker of non-small cell lung cancer. Pathol. Res. Pract. 2013;209(8):490–494. doi: 10.1016/j.prp.2013.04.018. [DOI] [PubMed] [Google Scholar]; Yang J, Liu H, Wang H, Sun Y. Down-regulation of microRNA-181b is a potential prognostic marker of non-small cell lung cancer. Pathology-Research and Practice. 2013;209(8):490-4. [DOI] [PubMed]
- 5.Tan C.-S., Gilligan D., Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16(9) doi: 10.1016/S1470-2045(15)00246-6. e447-e59. [DOI] [PubMed] [Google Scholar]; Tan C-S, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. The Lancet Oncology. 2015;16(9):e447-e59. [DOI] [PubMed]
- 6.Willers H., Azzoli C.G., Santivasi W.L., Xia F. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer J. (Sudbury, Mass) 2013;19(3):200. doi: 10.1097/PPO.0b013e318292e4e3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Willers H, Azzoli CG, Santivasi WL, Xia F. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer journal (Sudbury, Mass). 2013;19(3):200. [DOI] [PMC free article] [PubMed]
- 7.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]; Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63(1):11-30. [DOI] [PubMed]
- 8.Tomasini P., Walia P., Labbe C., Jao K., Leighl N.B. Targeting the KRAS pathway in non-small cell lung cancer. Oncologist. 2016 doi: 10.1634/theoncologist.2015-0084. [theoncologist. 2015-0084] [DOI] [PMC free article] [PubMed] [Google Scholar]; .Tomasini P, Walia P, Labbe C, Jao K, Leighl NB. Targeting the KRAS pathway in non-small cell lung cancer. The oncologist. 2016:theoncologist. 2015-0084. [DOI] [PMC free article] [PubMed]
- 9.Sullivan I., Planchard D. Next-generation EGFR tyrosine kinase inhibitors for treating EGFR-mutant lung cancer beyond first line. Front Med. 2017;3:76. doi: 10.3389/fmed.2016.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sullivan I, Planchard D. Next-generation EGFR tyrosine kinase inhibitors for treating EGFR-mutant lung cancer beyond first line. Frontiers in medicine. 2017;3:76. [DOI] [PMC free article] [PubMed]
- 10.Anastasiadou E., Jacob L.S., Slack F.J. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]; Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nature Reviews Cancer. 2018;18(1):5. [DOI] [PMC free article] [PubMed]
- 11.Liu B., Ye B., Yang L., Zhu X., Huang G., Zhu P. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18(5):499. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]; Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu P, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nature immunology. 2017;18(5):499. [DOI] [PubMed]
- 12.Del Vescovo V., Denti M.A. microRNA: Cancer. Springer; 2015. microRNA and lung cancer; pp. 153–177. [DOI] [PubMed] [Google Scholar]; Del Vescovo V, Denti MA. microRNA and lung cancer. microRNA: Cancer: Springer; 2015. p. 153-77. [DOI] [PubMed]
- 13.Wei M.-M., Zhou G.-B. Long non-coding RNAs and their roles in non-small-cell lung cancer. Genomics Proteomics Bioinformatics. 2016;14(5):280–288. doi: 10.1016/j.gpb.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wei M-M, Zhou G-B. Long non-coding RNAs and their roles in non-small-cell lung cancer. Genomics, proteomics & bioinformatics. 2016;14(5):280-8. [DOI] [PMC free article] [PubMed]
- 14.Haque S., Harries L. Circular RNAs (circRNAs) in health and disease. Genes. 2017;8(12):353. doi: 10.3390/genes8120353. [DOI] [PMC free article] [PubMed] [Google Scholar]; Haque S, Harries L. Circular RNAs (circRNAs) in health and disease. Genes. 2017;8(12):353. [DOI] [PMC free article] [PubMed]
- 15.Holdt L.M., Kohlmaier A., Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75(6):1071–1098. doi: 10.1007/s00018-017-2688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cellular and Molecular Life Sciences. 2018;75(6):1071-98. [DOI] [PMC free article] [PubMed]
- 16.Sorek R., Cossart P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet. 2010;11(1):9. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]; Sorek R, Cossart P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nature Reviews Genetics. 2010;11(1):9. [DOI] [PubMed]
- 17.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014 doi: 10.1101/gad.251926.114. [gad. 251926.114] [DOI] [PMC free article] [PubMed] [Google Scholar]; Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes & development. 2014:gad. 251926.114. [DOI] [PMC free article] [PubMed]
- 18.Luo Y.-H., Zhu X.-Z., Huang K.-W., Zhang Q., Fan Y.-X., Yan P.-W. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. doi: 10.1016/j.biopha.2017.12.015. [DOI] [PubMed] [Google Scholar]; Luo Y-H, Zhu X-Z, Huang K-W, Zhang Q, Fan Y-X, Yan P-W, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomedicine & Pharmacotherapy. 2017;96:892-8. [DOI] [PubMed]
- 19.Yao J.-T., Zhao S.-H., Liu Q.-P., Lv M.-Q., Zhou D.-X., Liao Z.-J. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol. Res. Pract. 2017;213(5):453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]; Yao J-T, Zhao S-H, Liu Q-P, Lv M-Q, Zhou D-X, Liao Z-J, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathology-Research and Practice. 2017;213(5):453-6. [DOI] [PubMed]
- 20.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017 doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO reports. 2017:e201643581. [DOI] [PMC free article] [PubMed]
- 21.Zhong Z., Lv M., Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Scientific reports. 2016;6:30919. [DOI] [PMC free article] [PubMed]
- 22.Yoh Watanabe M. TNM classification for lung cancer. Ann Thorac Cardiovasc Surg. 2003;9(6) [PubMed] [Google Scholar]; Yoh Watanabe M. TNM classification for lung cancer. Ann Thorac Cardiovasc Surg. 2003;9(6). [PubMed]
- 23.Li X., Zhang J., Jiang L., Wang F., Liu L., Liao Y. Overexpression of maelstrom promotes bladder urothelial carcinoma cell aggressiveness by epigenetically downregulating MTSS1 through DNMT3B. Oncogene. 2016;35(49):6281. doi: 10.1038/onc.2016.165. [DOI] [PubMed] [Google Scholar]; Li X, Zhang J, Jiang L, Wang F, Liu L, Liao Y, et al. Overexpression of maelstrom promotes bladder urothelial carcinoma cell aggressiveness by epigenetically downregulating MTSS1 through DNMT3B. Oncogene. 2016;35(49):6281. [DOI] [PubMed]
- 24.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H. Recognition of RNA N 6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N 6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nature cell biology. 2018;20(3):285. [DOI] [PMC free article] [PubMed]
- 25.Shi Y., Shang J. Long Non-Coding RNAs. Springer; 2016. Long noncoding RNA expression profiling using Arraystar LncRNA microarrays; pp. 43–61. [DOI] [PubMed] [Google Scholar]; Shi Y, Shang J. Long noncoding RNA expression profiling using Arraystar LncRNA microarrays. Long Non-Coding RNAs: Springer; 2016. p. 43-61. [DOI] [PubMed]
- 26.Lao X., Feng Q., He G., Ji M., Zhu D., Xu P. Immature colon carcinoma transcript-1 (ICT1) expression correlates with unfavorable prognosis and survival in patients with colorectal cancer. Ann Surg Oncol. 2016;23(12):3924–3933. doi: 10.1245/s10434-016-5305-1. [DOI] [PubMed] [Google Scholar]; Lao X, Feng Q, He G, Ji M, Zhu D, Xu P, et al. Immature colon carcinoma transcript-1 (ICT1) expression correlates with unfavorable prognosis and survival in patients with colorectal cancer. Annals of surgical oncology. 2016;23(12):3924-33. [DOI] [PubMed]
- 27.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]; Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nature structural & molecular biology. 2015;22(3):256. [DOI] [PubMed]
- 28.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat Rev Cancer. 2018;18(2):128. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]; Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nature Reviews Cancer. 2018;18(2):128. [DOI] [PubMed]
- 29.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]; Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384. [DOI] [PubMed]
- 30.Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]; Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome biology. 2010;11(8):R90. [DOI] [PMC free article] [PubMed]
- 31.Lock F.E., Underhill-Day N., Dunwell T., Matallanas D., Cooper W., Hesson L. The RASSF8 candidate tumor suppressor inhibits cell growth and regulates the Wnt and NF-κB signaling pathways. Oncogene. 2010;29(30):4307. doi: 10.1038/onc.2010.192. [DOI] [PubMed] [Google Scholar]; Lock FE, Underhill-Day N, Dunwell T, Matallanas D, Cooper W, Hesson L, et al. The RASSF8 candidate tumor suppressor inhibits cell growth and regulates the Wnt and NF-κB signaling pathways. Oncogene. 2010;29(30):4307. [DOI] [PubMed]
- 32.Chen L.-L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]; Chen L-L. The biogenesis and emerging roles of circular RNAs. Nature reviews Molecular cell biology. 2016;17(4):205. [DOI] [PubMed]
- 33.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI. 2018;110(3) doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI: Journal of the National Cancer Institute. 2018;110(3). [DOI] [PMC free article] [PubMed]
- 34.Wang K., Sun Y., Tao W., Fei X., Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. doi: 10.1016/j.canlet.2016.12.036. [DOI] [PubMed] [Google Scholar]; Wang K, Sun Y, Tao W, Fei X, Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer letters. 2017;394:1-12. [DOI] [PubMed]
- 35.Qiu M., Xia W., Chen R., Wang S., Xu Y., Ma Z. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018 doi: 10.1158/0008-5472.CAN-17-2808. [canres. 2808.017] [DOI] [PubMed] [Google Scholar]; Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer research. 2018:canres. 2808.017. [DOI] [PubMed]
- 36.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]; Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer research. 2013;73(18):5609-12. [DOI] [PubMed]
- 37.Yu J.-J., Wu Y.-X., Zhao F.-J., Xia S.-J. miR-96 promotes cell proliferation and clonogenicity by down-regulating of FOXO1 in prostate cancer cells. Med Oncol. 2014;31(4):910. doi: 10.1007/s12032-014-0910-y. [DOI] [PubMed] [Google Scholar]; Yu J-J, Wu Y-X, Zhao F-J, Xia S-J. miR-96 promotes cell proliferation and clonogenicity by down-regulating of FOXO1 in prostate cancer cells. Medical oncology. 2014;31(4):910. [DOI] [PubMed]
- 38.Shi Y., Zhao Y., Shao N., Ye R., Lin Y., Zhang N. Overexpression of microRNA-96-5p inhibits autophagy and apoptosis and enhances the proliferation, migration and invasiveness of human breast cancer cells. Oncol Lett. 2017;13(6):4402–4412. doi: 10.3892/ol.2017.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shi Y, Zhao Y, Shao N, Ye R, Lin Y, Zhang N, et al. Overexpression of microRNA-96-5p inhibits autophagy and apoptosis and enhances the proliferation, migration and invasiveness of human breast cancer cells. Oncology letters. 2017;13(6):4402-12. [DOI] [PMC free article] [PubMed]
- 39.Xu X., Dong Z., Li Y., Yang Y., Yuan Z., Qu X. The upregulation of signal transducer and activator of transcription 5-dependent microRNA-182 and microRNA-96 promotes ovarian cancer cell proliferation by targeting forkhead box O3 upon leptin stimulation. Int J Biochem Cell Biol. 2013;45(3):536–545. doi: 10.1016/j.biocel.2012.12.010. [DOI] [PubMed] [Google Scholar]; Xu X, Dong Z, Li Y, Yang Y, Yuan Z, Qu X, et al. The upregulation of signal transducer and activator of transcription 5-dependent microRNA-182 and microRNA-96 promotes ovarian cancer cell proliferation by targeting forkhead box O3 upon leptin stimulation. The international journal of biochemistry & cell biology. 2013;45(3):536-45. [DOI] [PubMed]
- 40.Wu Z., Liu K., Wang Y., Xu Z., Meng J., Gu S. Upregulation of microRNA-96 and its oncogenic functions by targeting CDKN1A in bladder cancer. Cancer Cell Int. 2015;15(1):107. doi: 10.1186/s12935-015-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu Z, Liu K, Wang Y, Xu Z, Meng J, Gu S. Upregulation of microRNA-96 and its oncogenic functions by targeting CDKN1A in bladder cancer. Cancer cell international. 2015;15(1):107. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2