Abstract
Studies conducted in the last years have demonstrated that human milk represents a continuous supply of beneficial bacteria to the infant gut, which contribute to the maturation of the digestive and immune functions in the developing infant. Nevertheless, the origin of bacterial populations in milk is not fully understood yet and they have been proposed to originate from maternal skin, infant’s mouth, and (or) endogenously, from the maternal digestive tract through a mechanism involving immune cells. Understanding the composition, functions and assembly of the human milk microbiota has important implications not only for the infant gut microbiota establishment, but also for the mammary health since dysbiosis in the milk bacteria may lead to mastitis. Besides, host, microbial, medical and environmental factors may affect the composition of the human milk microbiome, with implications for the mother-infant health. Application of both culture-dependent and -independent techniques to assess the milk microbiome faces some practical limitations but, together, have allowed providing novel and complementary views on its origin, composition and functioning as summarized in this minireview. In the next future, the application of the ultimate advances in next-generation sequencing and omics approaches, including culturomics, will allow a detailed and comprehensive understanding of the composition and functions of these microbial communities, including their interactions with other milk components, expanding the opportunities to design novel microbiome-based modulation strategies for this ecosystem.
Keywords: microbiome, human milk, breastfeeding, infant nutrition, microbiome modulation
Human Milk Contains a Microbiome
Human milk is the gold standard for infant nutrition as it contains multiple biologically active components, including immune factors, lipids, oligosaccharides, microRNAs and hormones among others (Hennet and Borsig, 2016), whose concentration vary with maternal and environmental factors, getting adapted to the variable needs of the baby (Hunt et al., 2011; Andreas et al., 2015; Feng et al., 2016; Gomez-Gallego et al., 2016; Kumar et al., 2016; Ruiz et al., 2017). Globally, its complex and dynamic composition promotes a healthy infant growth and development, which has prompted international organizations to recommend exclusive breastfeeding during the first 6 months of life (World Health Organization, 2003).
Microorganisms have emerged as important bioactive components of human milk. Its occurrence in milk was first recognized the second half of the past century, while studying the potential transmission of infections through breastfeeding (Rantasalo and Kauppinen, 1959; Foster and Harris, 1960; Kenny, 1977; Eidelman and Szilagyi, 1979). Later, several studies described the presence of viable commensal, mutualistic or potentially probiotic bacteria in healthy human milk (Fernández et al., 2013), leading to an increasing interest in human milk microbiota and their effects in maternal-infant health. Indeed, among the microorganisms occurring in milk, some strains belonging to the species Lactobacillus salivarius, Lactobacillus fermentum, Lactobacillus gasseri, Bifidobacterium breve, Bifidobacterium adolescentis, and Bifidobacterium longum subsp. infantis have demonstrated potential to promote mother and infant health, including the prevention or treatment of lactational mastitis, the promotion of a normal gut bacterial colonization in preterm neonates, or the amelioration of diarrhea in IBS patients (Arroyo et al., 2010; Chiu et al., 2014; Maldonado-Lobón et al., 2015; Moles et al., 2015; Fernández et al., 2016; Shin et al., 2018).
The scientific interest in human milk bacteria is evolving in parallel to the unveiled roles of human microbiomes in health and disease, and to the advances in the methodologies available for their study. Nowadays, research interests in this field are focused on (a) the roles that milk bacterial communities play on infant health and development, including their influence on the acquisition of the gut microbiota in early life; (b) the roles that they may play on maternal health, including breast health; (c) the maternal, infant, environmental and medical factors that shape and modulate their compositions, both under physiological conditions and when dysbiosis occurs; and (d) their origin, in order to confirm or refuse the existence of an endogenous oral-entero-mammary route allowing the selective translocation of some bacteria from the maternal digestive tract to the mammary gland.
The ever-growing asset of-omic tools is opening new avenues to understand the benefits of breastfeeding from microbiological, metabolic and immunological perspectives, and to establish new microbiota-based strategies to promote maternal-infant health, including the development of procedures for the transfer of the milk microbiota to neonates or infants that are not breastfed or the development of human milk-derived probiotics (Fernández et al., 2018). This review summarizes the main advances on our comprehension of the human milk microbiota gained through the refinement of tools available for microbial ecology studies, identifying opportunities to address existing knowledge gaps.
Culture-Dependent Based Studies
Microbiology studies have traditionally relied on culturing, isolating and characterizing, phenotypically and (or) genotypically, the bacterial isolates, though these approaches fail to obtain information on non-culturable bacteria. Additionally, most microbial studies on milk were performed on samples from women whose infants were suffering from potential milk-borne infections and thus, microbial growth media and conditions have been biased toward detecting pathogens including viral particles (cytomegalovirus, Zika, HIV, etc.), and pathogenic microbial species typically associated with mammary infections (e.g., Staphylococcus aureus) or infant infections (e.g., group B Streptococcus) (Chen et al., 2016). Overall, culturing conditions for such pathogenic bacteria require the utilization of standard clinical cultivation media and relatively short incubation periods (16–18 h) at 37°C under standard atmospheric incubations. These conditions have allowed isolating dominant and rapidly growing culturable microorganisms mainly including Gram-positive species (Staphylococcus, Streptococcus, Corynebacterium, and Propionibacterium). Other bacterial groups present in milk from healthy lactating mothers are more fastidious to grow under routine laboratory conditions as they require either specific atmospheric incubations (e.g., anaerobic conditions) or specific nutrients or ingredients in the cultivation media (for instance, reducing agents for growing anaerobic species), and thus may have passed unnoticed in early microbiological studies not specifically designed to recover such species. For instance, lactic acid bacteria (Lactobacillus, Lactococcus, Leuconostoc, Weissella...) and bifidobacteria have only been isolated from milk following the utilization of specific growth media and longer anaerobic incubations (Martín et al., 2003, 2009; Abrahamsson et al., 2009; Solís et al., 2010; Arboleya et al., 2011; Murphy et al., 2017). Globally, over 200 different bacterial species representing approximately 50 different genera have been isolated from milk, including new bacterial species, such as Streptococcus lactarius (Martín et al., 2011).
Culture-dependent approaches have been pioneering in demonstrating that healthy human milk harbors a microbial community, representing a source of commensal microorganisms for the neonate. Albeit their limitations to access unculturable bacteria, culturing enables preserving the strains facilitating further studies and exploitation of potential biotechnological applications (Lara-Villoslada et al., 2007; Arboleya et al., 2011; Langa et al., 2012; Cárdenas et al., 2014, 2015). Specifically, the activities most commonly studied on milk isolates include virulence and antibiotic resistance (Jiménez et al., 2008a; Kozak et al., 2015), but also bacteriocin production (Heikkilä and Saris, 2003; Olivares et al., 2006; Lara-Villoslada et al., 2007; Kozak et al., 2015; Sharma et al., 2017) and probiotic traits (Fernández et al., 2013; Reis et al., 2016). Indeed, bacteria from milk of healthy women fulfill the criteria recommended for human probiotics: human origin, a history of safe prolonged intake, adaptation to mucosal and dairy substrates (Lara-Villoslada et al., 2007; Fernández et al., 2013), and to the human gut (Jeurink et al., 2013). Thus, some Lactobacillus and Bifidobacterium species (L. gasseri, L. salivarius, L. rhamnosus, L. plantarum, L. fermentum, L. reuteri, B. breve, B. longum…) from milk have awakened strong interest as potential probiotic bacteria, enjoying the GRAS (Generally Recognized As Safe) and the QPS (Qualified Presumption of Safety) status conceded by the Food and Drug Administration and the European Food Safety Authority, respectively.
Bacterial isolation coupled to phenotypical and genotypical characterization, including whole genome sequencing (WGS) analyses, are essential steps to characterize and evaluate the safety and the probiotic potential of bacterial isolates from human origin, including those isolated from milk. Generally, the in vitro assessment of probiotic properties in human milk isolates has included their antimicrobial activity against pathogenic microorganisms and the compounds that may be responsible for such activity (organic acids, bacteriocins, hydrogen peroxide), the ability to survive when exposed to conditions similar to those found in the human gastrointestinal tract, their adherence to human intestinal cells and mucin, their ability to degrade mucin or to produce biogenic amines or their susceptibility to antibiotics (Martín et al., 2005, 2006, 2010). The safety and health-promoting properties of some strains have also been confirmed in vivo, including the use of animal models (Olivares et al., 2006) and human clinical trials (Jiménez et al., 2008b; Arroyo et al., 2010; Maldonado et al., 2010, 2012; Gil-Campos et al., 2012; Espinosa-Martos et al., 2016; Fernández et al., 2016). Finally, a few strains isolated from this fluid have been analyzed to date through WGS, including representatives from the genera Lactobacillus, Bifidobacterium, and Streptococcus (Jiménez et al., 2010a,b, 2012; Martín et al., 2012, 2013).
In the framework of recent research conducted in the field of the gut microbiota, the “culturomics” approach, which is based on the utilization of a wide range of culturing and incubation conditions, is emerging as a promising strategy to aid in the discovery and description of novel microorganisms associated to the human host, including pathogenic and commensal species (Bilen et al., 2018). Such approach has not been applied to the human milk ecosystem yet, although it can be anticipated that it will lead to significant advances in our knowledge of this human ecosystem and to the discovery of novel health-promoting bacterial strains.
Culture-Independent Based Studies
Since cultivable microorganisms represent a fraction of the communities inhabiting a specific ecological niche, culture-independent molecular techniques, i.e., quantitative PCR, denaturing (or temperature) gradient gel electrophoresis, and Next generation sequencing (NGS) approaches, from metataxonomics (16SrRNA amplicon analysis) to metagenomics (total DNA sequencing), have allowed valuable complementary assessment of the human milk microbiota, including information on microorganisms which remain unculturable to date (Jeurink et al., 2013; McGuire and McGuire, 2015; Mira and Rodriguez, 2017). Nonetheless, such techniques introduce other limitations and biases as they may over- or underestimate some groups because of differences in cell structures affecting DNA extraction efficiencies (McGuire and McGuire, 2015; Gomez-Gallego et al., 2016; Mira and Rodriguez, 2017). For instance, it has been well established that differences in the storage and manipulation of the samples and in the procedures for DNA extraction and storage strongly affect the yield and quality of extracted DNA, but may also lead to the underrepresentation of those microorganisms more recalcitrant to cellular lysis (Gram-positive bacteria or eukaryotic species), strongly biasing the results obtained from microbiome analysis of complex microbial communities (Wesolowska-Andersen et al., 2014), and precluding the comparison of results from different studies. While strong efforts have attempted to standardize methodologies so as to allow comparison of results from different studies in the frame of the human gut microbiome, such standardization has not been implemented in human milk microbiome studies, yet.
Besides, culture-independent techniques cannot differentiate between live and dead microorganisms, as DNA from dead organisms may persist in the environment, affecting the biological significance of the conclusions from sequencing-based analysis of microbial communities. Several methodologies have been implemented to incorporate a viability assessment in DNA-based analysis (Emerson et al., 2017). Among them, the use of propidium monoazide (PMA) should be highlighted. PMA is a dye which penetrates damaged membranes and, thus, only binds to extracellular DNA or DNA from damaged cells. Upon light exposure, PMA irreparably damage the DNA and, consequently, further DNA extraction and amplification procedures will only affect live cells. Nevertheless, such methodologies have not been widely implemented in microbiome-based surveys because of their current drawbacks; as an example, their efficacy to differentiate live and dead cells may depend on the particular composition of the community (Li R. et al., 2017). Finally, other limitations are inherent to the specific 16S rRNA region(s) targeted, sequencing platform used and/or the specific bioinformatic analysis, particularly in terms of the criteria followed to filter sequencing errors, sequencing clustering methods, and reference databases employed for taxonomic assignation or functional annotation (Fouhy et al., 2016). These facts highlight the urgent need to standardize methodologies for the sequencing-based assessment of microbial populations within the human body, including specific procedures adapted to the particular characteristics of different human body ecosystems (e.g., samples with a low biomass or with a high proportion of Gram-positive bacteria, such as human milk).
Anyhow, culture-independent studies confirmed the presence in milk of DNA from bacterial groups previously isolated from this ecosystem, including Staphylococcus, Streptococcus, Corynebacterium, Propionibacterium, lactic acid bacteria, and Bifidobacterium (Martín et al., 2007; Collado et al., 2009; Hunt et al., 2011; Cabrera-Rubio et al., 2012, 2016; Jost et al., 2013, 2014; Ward et al., 2013; Jiménez et al., 2015; Fitzstevens et al., 2017); but also reported the presence of DNA from microbial groups which had not been previously recovered through culturing healthy human milk samples, such as Clostridium and Bacteroides (Delgado et al., 2008; Collado et al., 2009). Overall, these techniques offered novel tools to study the milk ecosystem and defined some changes in total or specific microbial groups representation and in microbial activities of interest, e.g., virulence or bacteriocin production (Obermajer et al., 2015) linked to specific maternal/infant or environmental characteristics (Table 1). For instance, qPCR-based studies associated antibiotic administration to reduced lactobacilli and bifidobacterial presence (Soto et al., 2014) and identified reduction in bifidobacterial presence in association to preterm delivery-(Khodayar-Pardo et al., 2014).
Table 1.
Summary including the main conclusions and experimental design of culture-independent studies reported to date on human milk microbiota.
| Study group | Methodology | Factor study | Main conclusions | References |
|---|---|---|---|---|
| Healthy women (Spain), (n = 65) | qPCR and 28S rRNA sequencing | – | Detection of fungi species in a high proportion of samples from healthy lactating women | Boix-Amorós et al., 2017 |
| Healthy women (Spain), (n = 32) | qPCR using 16S rRNA for target populations | Preterm vs. Term deliveries Longitudinal study |
Prematurity was associated with reduced Bifidobacterium levels C-section was associated with reduced Streptococcus in transitional milk | Khodayar-Pardo et al., 2014 |
| Healthy women (Germany and Austria) (n = 160) | qPCR using 16S rRNA for target populations | Antibiotherapy effect | Reduced lactobacilli or bifidobacteria positive samples following antibiotherapy. | Soto et al., 2014 |
| Healthy women (United States), Longitudinal study (n = 16) | 16S rRNA sequencing (V1–V2 regions/454 platform) | Longitudinal study in healthy lactating women | High inter-individual variability. Define a 9 core microbial genera including: Staphylococcus, Streptococcus, Serratia, Pseudomonas, Corynebacterium, Ralstonia, Propionibacterium, Sphingomonas, Bradyrhizobiaceae | Hunt et al., 2011 |
| Healthy women (Finland), Longitudinal study (n = 18) | 16S rRNA sequencing (V3–V4 regions/454 platform) | Longitudinal study and Maternal BMI | Mature milk contains a higher representation of oral inhabitants Maternal BMI and weight gain during pregnancy positively associated with staphylococci and lactobacilli; but negatively associated with bifidobacteria Elective C-section impacts the milk microbiota structure |
Cabrera-Rubio et al., 2012 |
| Healthy women (Spain), (n = 10) | 16S rRNA sequencing (V1–V3 regions/454 platform) | Delivery mode | C-section is associated with higher abundance of Staphylococcus and lower of Streptococcus; and lower overall diversity | Cabrera-Rubio et al., 2016 |
| Healthy women (Canada), (n = 39), | 16S rRNA sequencing (V6 regions/Illumina Miseq platform) | Term vs. preterm, vaginal vs. C-section | High inter-individual variation, though gestation, mode of delivery or infant gender were not associated with specific milk microbiota differences | Urbaniak et al., 2016 |
| Healthy women (Mexico) (n = 10) | 16S rRNA sequencing (V4 regions/Illumina Hiseq platform) | Pre-pregnancy BMI, Comparison to infant saliva microbiota |
Dominant genera display statistically significant differences in their representation in breast milk and infant saliva Higher abundance and diversity of Streptococcus than in other studies Pre-pregnancy BMI correlated with lower Streptococcus abundance and higher microbial diversity |
Davé et al., 2016 |
| Healthy women (Spain), (n = 21) | 16S rRNA sequencing (V1–V4 regions/454 platform) | Longitudinal study from colostrum to mature milk | Define a core of 7 genera: Finegoldia, Streptococcus, Corynebacterium, Staphylococcus, Acinetobacter, Peptoniphilus, and Pseudomonas No differences in the representation of the most abundant genera at different time points |
Boix-Amorós et al., 2016 |
| Healthy lactating women (n = 8) vs. women receiving chemotherapy (n = 1) | 16S rRNA sequencing (V6 region/Ion Torrent platform) + metabolomics | Chemotherapy | Chemotherapy associated with decreased bacterial diversity in human milk including shifts in Bifidobacterium, Eubacterium, Staphylococcus, Cloacibacterium, Acinetobacter, Xanthomonadaceae, and Stenotrophomonas Docosahexaenoic acid and inositol were also reduced |
Urbaniak et al., 2014b |
| Healthy women (n = 79) (China, South Africa, Finland, and Spain) | 16S rRNA sequencing (V4 region/Illumina Miseq platform) + metabolomics | Geographical and delivery type-associated variation | Variation in microbial and metabolic profiles, geographically and in association with type of delivery Proteobacteria, Actinobacteria and Bacilli show statistical significant correlations with some of the milk metabolites evaluated, including human milk oligosaccharides, lactate, polyamines and riboflavin |
Gómez-Gallego et al., 2018 |
| Same cohort as in Gómez-Gallego et al., 2018 | 16S rRNA sequencing (V4 region/Illumina Miseq platform) + metabolomics | Geographical variation | Geographical variation in breastmilk microbiota composition, and association with fatty acid profiles Expand previous universal core microbiome definitions, including 23 groups, and taxa specifically associated with certain geographical locations |
Kumar et al., 2016 |
| Healthy women (n = 41) (Centro African Republic) | 16S rRNA sequencing (V1–V3 region/Illumina Miseq platform) | Lifestyle variation: hunter-gatherers vs. horticulturalists | Most abundant taxa very different from those reported in cohorts, including groups not previously detected in milk e.g., Rhizobium Seasonal and lifestyle differences related to variation in breastmilk microbiome composition, likely associated with variation in environmental and dietary exposures |
Meehan et al., 2018 |
| Healthy women (n = 33) (Taiwan) | 16S rRNA sequencing (V3–V4 region/Illumina Miseq platform) | Longitudinal study | Describes the interindividual variability in the prevalence and relative abundance of commensal and opportunistic pathogenic bacteria in breastmilk from healthy women | Chen et al., 2018 |
| Healthy women (n = 29), (Italy) | 16S rRNA sequencing (V2 and V3 region/Ion Torrent platform) | Delivery type | Define variations in colostrum based on delivery type, with an overall greater abundance of environmental bacteria in samples from mothers that underwent C-section | Toscano et al., 2017 |
| Healthy lactating women (n = 10) vs. mastitis (n = 10), (Spain) | Shotgun sequencing (454 platform) | Mastitis | Detect the presence or archaea, viruses, fungi and protozoa Mastitis relates to reduced microbiota diversity |
Jiménez et al., 2015 |
| Healthy women (n = 10) (Canada) | Shotgun metagenomics (Illumina platform) | na | Identify functional differences in breastmilk microbiome as compared to the gut microbiome of formula fed and breast fed infants Identify presumptive immunomodulatory DNA motifs |
Ward et al., 2013 |
| Healthy women (n = 18) vs. women with mastitis (n = 32) | 16S rRNA sequencing (V2–V3 region/Ion Torrent platform) | Mastitis | Milk samples of mastitis women have lower microbial diversity, increased abundance of opportunistic pathogens and depletion of commensal obligate anaerobes. | Patel et al., 2017 |
| Healthy women (n = 393) (Canada) | 16S rRNA sequencing (V4 region/Illumina MiSeq platform) | na | Breastfeeding practices, maternal BMI and parity are significantly associated with milk microbiota coposition | Moossavi et al., 2019 |
| Healthy women (n = 394) (Ethiopia, Gambia, United States, Ghana, Kenya, Peru, Spain, Sweden) | 16S rRNA sequencing (V1–V3 region/Illumina Miseq platform) | Different geographical settings and relationship to infant fecal microbiotas | Geographical variation in total and core breastmilk microbiota, and a high percentage of taxa (70–88%) are encountered in both breast milk and infant fecal samples Positive correlations between the milk microbiota and the infant fecal microbiota |
Lackey et al., 2019 |
High-Throughput Culture-Independent Based Studies
The description of commensal bacteria in milk was coincidental in time with the advent of NGS approaches, which permit a deeper resolution of microbial communities and revealed that the complexity of milk microbiota is much more extensive than previously anticipated (Jeurink et al., 2013; McGuire and McGuire, 2017). Most NGS-based studies conducted on milk have focused on metataxonomics to define bacterial populations presence and their variation with selected factors (Table 1). Briefly, such studies revealed the existence of large inter-individual variations (Hunt et al., 2011; Cabrera-Rubio et al., 2012), some of which have been associated with geographical and lifestyles differences, with a general higher microbial diversity in samples from developing and (or) rural locations (Kumar et al., 2016; Sakwinska et al., 2016; Drago et al., 2017; Li S.-W. et al., 2017; Lackey et al., 2019), in agreement with observations within the gut microbiota ecosystem. Other specific factors frequently associated with overall variation in the milk microbiome structure include delivery mode (Cabrera-Rubio et al., 2012, 2016; Khodayar-Pardo et al., 2014; Toscano et al., 2017) and time from birth (Cabrera-Rubio et al., 2012, 2016; Khodayar-Pardo et al., 2014; Toscano et al., 2017 (Ma et al., 2015; Yu et al., 2016; Patel et al., 2017), though some inconsistencies have been reported among studies. For instance, differences in the milk microbiome structure of women who gave birth vaginally and those delivering by C-section have been frequently reported, though the specific taxa driving such differences seem to be study- and cohort-dependent: a higher abundance of lactobacilli species has been associated with C-section in South African, Finnish, Chinese and Taiwanese cohorts, whereas such difference has not been detected in Spanish women (Khodayar-Pardo et al., 2014; Kumar et al., 2016), some authors have even reported a higher lactobacilli representation following vaginal delivery in a Taiwanese population (Chen et al., 2018), and other studies found no differences in milk microbiomes depending on delivery type (Sakwinska et al., 2016; Urbaniak et al., 2016). Remarkably, particular cohort-dependent factors that might be driving variation in the human milk microbiomes have not been thoroughly explored, but might contribute to explain these apparent discordances. Similarly, differences between colostrum, transition and mature milk microbiotas have been reported by some authors, who reported an increased abundance of typical oral inhabitants in transition and mature milk, though such differences have not been consistently observed in different studies (Cabrera-Rubio et al., 2012; Chen et al., 2018).
Other factors less explored but described to affect the milk microbiome are summarized in Table 1 and include environmental exposure to disinfection agents (Bever et al., 2018), chemotherapy (Urbaniak et al., 2014b), maternal nutrient intakes (Boix-Amorós et al., 2016; Williams et al., 2017a), and the structure of human social networks, including cooperative breeding (Meehan et al., 2018). Correlation and network analyses have also related variation in the milk microbiome with variation in other milk components including immune cells, polyamines and fatty acids, though such observations have been reported in limited studies and their potential implications to monitor or improve maternal-infant health have not been thoroughly explored (Gómez-Gallego et al., 2017; Williams et al., 2017b). Despite of the large inter-individual and inter-population variations, most studies agree to identify the genera Staphylococcus, Streptococcus, and Propionibacterium as core members of the milk microbiota (Hunt et al., 2011; Jiménez et al., 2015), as they appear to be universally present in healthy milk irrespectively of maternal and infant characteristics studied to date. The presence of potentially beneficial bacteria including lactobacilli and bifidobacterial species, is also commonly reported in human milk, though these are not consistently detected across all samples and populations studied, and usually represent a minority of the bacterial populations detected (Chen et al., 2018).
Intriguingly, NGS-based studies have also identified in milk DNA from strictly anaerobic gut-associated microbes (Bacteroides, Blautia, Clostridium, Collinsella, Coprococcus, Eubacterium, Faecalibacterium, Roseburia, Ruminococcus,...), which are either non-culturable or hard to culture in the laboratory, and that have not been recovered through milk culturing to date (Cabrera-Rubio et al., 2012; Jost et al., 2013, 2014; Jiménez et al., 2015; Gomez-Gallego et al., 2016). Confirming their presence within healthy human milk would require its isolation through culturomic approaches or their detection through other techniques, including the specific labeling of target microorganisms (either through qPCR or flow-cytometry based approaches), or target specific RNA-based analysis, which would also confirm its functionality within the ecosystem and may provide additional proofs on the possible role of these bacteria for human health. Traditionally bacterial cells in milk were considered contaminants originating from the infant’s oral cavity or the mother’s skin. However, the detection of such anaerobic species typical from gut environments has suggested that breastfeeding could be an ingenious way to provide the newborn infant a set of commensal microbes to initially colonize the gut (McGuire and McGuire, 2015). Indeed, some works have proposed that selected bacteria in the maternal gut could reach the mammary gland through an endogenous route, involving complex interactions between bacteria, epithelial and immune cells (Martín et al., 2004). Although the mechanisms governing this process have not been elucidated yet, studies with pregnant and lactating mice models have offered a plausible scientific basis (Rodríguez, 2014; de Andrés et al., 2017; Mira and Rodriguez, 2017). Additionally, oral administration of some lactobacilli strains to lactating women led to their presence in milk supporting the existence of an endogenous gut-mammary gland connection during lactation (Abrahamsson et al., 2009; Arroyo et al., 2010).
Finally, metataxonomic studies have frequently revealed the presence in milk of sequences belonging to a group of soil- and water-associated bacterial genera, including Acinetobacter, Bradyrhizobium, Methylobacterium, Microbacterium, Novosphingobium, Pseudomonas, Ralstonia, Sphingopyxis, Sphingobium, Sphingomonas, Stenotrophomonas, and Xanthomonas, which have not been detected through milk culturing (Hunt et al., 2011; Cabrera-Rubio et al., 2012; Urbaniak et al., 2014a; Li S.-W. et al., 2017). In some works, the DNA sequences from such microorganisms were so frequent and abundant that were even consider part of the “core microbiome” of human milk (Hunt et al., 2011). However, contaminating DNA in PCR and DNA extraction reagents frequently belongs to these groups and may lead to artifacts when assessing low abundance microbiomes, such as human milk (Grahn et al., 2003; Mühl et al., 2010; Laurence et al., 2014; Salter et al., 2014; Kim et al., 2017; Perez-Muñoz et al., 2017). Thus, caution should be taken when analyzing the human milk microbiome using these methodologies.
Past Pitfalls and New Opportunities
The constant refinement of culture-independent techniques has boosted our understanding of the milk microbial ecosystem although there are still limitations to unravel the study of this community. First, the analysis of low biomass microbiomes faces some challenges due to the risk of DNA contamination. Recommendations to reduce the impact of contaminants in sequence-based low-biomass microbiota studies have already been provided (Salter et al., 2014), including the utilization of DNA extractions procedures demonstrated to introduce the lowest DNA background possible. Besides, reporting sequencing of negative controls or describing identification and removal of contaminant sequences during the bioinformatics analysis of the sequencing reads (Davis et al., 2018; Karstens et al., 2018; Zinter et al., 2019), should be considered in milk microbiome studies.
Secondly, in view of the high inter-individual, inter-populations and inter-study variations reported to date, milk microbiome research needs an urgent standardization of samples collection and processing as previously proposed for other human microbiomes (Costea et al., 2017). Standardization should cover aseptic techniques, cautions to prevent sample contamination from skin or milk extraction devices, the convenience of using preservation protocols (Lackey et al., 2017); and DNA processing and sequencing analysis, which would enormously facilitate comparison of results from different studies (Costea et al., 2017). Besides, multiple maternal and environmental factors affect the milk microbiome, which together with the reduced number of samples analyzed in most available studies strongly hampers drawing biologically and universally valid significant conclusions. Defining appropriate inclusion/exclusion criteria and metadata collection is necessary to ensure the biological data obtained will answer the specific question under investigation. Furthermore, factors like maternal diet, known to affect the gut microbiota composition and some milk components, have not been considered in milk microbiomes studies and thus may need to be evaluated in further studies (Dingess et al., 2017; Bzikowska-Jura et al., 2018).
Most studies on milk microbiome to date have exclusively identified bacterial population shifts, but have not provided any functional insights into these communities. Results from gut microbiome studies have demonstrated that microbial ecosystems are more conserved at functional than at taxonomic levels, reflecting the existence of redundant functions among community members and suggesting that some species might be interchangeable without affecting the functional attributes of the community (Moya and Ferrer, 2016). Thus, functions may be better biomarkers for health-disease states than taxonomical composition though they have been scarcely studied on milk. A pooled milk sample resulting from 10 donors was analyzed through shotgun-sequencing, identifying abundant and prevalent functions in the milk microbiota, as compared to the infant gut microbiome and identified putative immunomodulatory motifs in microbiota-derived DNA sequences, providing basis for further mechanistic studies (Ward et al., 2013). Additionally, shotgun metagenomics enhance the resolution of taxonomic assignment and has demonstrated vertical transmission of strains from the mother to the infant (Asnicar et al., 2017); and identified novel functions of interest from unculturable bacteria in complex communities (Berini et al., 2017), though this potential has not been harnessed within the milk microbiota.
Although milk microbiome studies have focused on bacteria, milk may also vehiculate yeasts or viruses (Daudi et al., 2012; Dupont-Rouzeyrol et al., 2016; Mutschlechner et al., 2016). Since shotgun metagenomics sequences the whole DNA present at a given environment, including eukaryote, prokaryote and viral DNA, such approaches, together with 18S rRNAmetagenomic sequencing, have shed light on the existence of fungal species in milk samples from healthy women, opening new avenues to study other milk microbiota components that have been overlooked (Jiménez et al., 2015; Boix-Amorós et al., 2016). Similarly, a milk virome analyses recently revealed the dominance of bacteriophages as opposed to eukaryotic viruses (Pannaraj et al., 2018), raising the possibility of modulating milk bacterial populations by bacteriophages present within the community (Jiménez et al., 2015; Duranti et al., 2017).
It is worth highlighting that DNA-based studies provide information on the community’s composition and their metabolic potential but do not demonstrate the activities they perform in situ. To partly overcome such limitations, some studies have proposed the utilization of propidium monoazide (PMA), a DNA binding dye that enters only dead and membrane-compromised cells, thus preventing detection of damaged cells by qPCR or NGS approaches (Tantikachornkiat et al., 2016), although such strategies have been never reported on milk microbiota studies. Besides, a combination of flow-cytometry and NGS approaches has been proposed to specifically identify bacteria coated with host antibodies (Simón-Soro et al., 2015). In fact, bacterial interactions with the host immune system appear to differ between body sites and, understanding how the maternal immune system interacts with milk bacteria may help elucidate the mechanisms allowing bacterial translocation through an enteromammary pathway (Rodríguez, 2014). Other NGS-based methodologies that have not yet been applied to milk microbiota studies, include RNA-based (metatranscriptomics) or single-cell methodologies, which may offer novel information about the communities that are metabolically active at a given point and their functionality (Gosalbes et al., 2012; Yao et al., 2017).
Finally, little attention has been paid to the associations of milk microbiota with other milk components (Roncada et al., 2013; Kumar et al., 2016; Bardanzellu et al., 2017; Gómez-Gallego et al., 2017, 2018; Figure 1) although the combination of multiple “-omic” approaches, at DNA, RNA, protein and metabolite levels, will undoubtedly facilitate our comprehension on the inter-relationships among milk components, and ultimately, with the host. Besides, further expansion of culturing conditions, coupled to anaerobic maintenance of samples from collection to processing, will facilitate the isolation and recovery of microorganisms currently recalcitrant to grow under laboratory conditions (Stewart, 2012; Browne et al., 2016), enabling further mechanistic and functional studies. In conclusion, the constant refinement in culture-independent techniques has shed important information on the composition and modulation of the milk microbiome, although there are still important knowledge gaps on its functions and modulation. Emerging approaches offer novel opportunities to address studies in this field contributing to identify the mechanisms governing the milk microbiota assembly and its impact on maternal-infant health, paving the way to design novel microbiota-based strategies to promote maternal-infant health.
FIGURE 1.
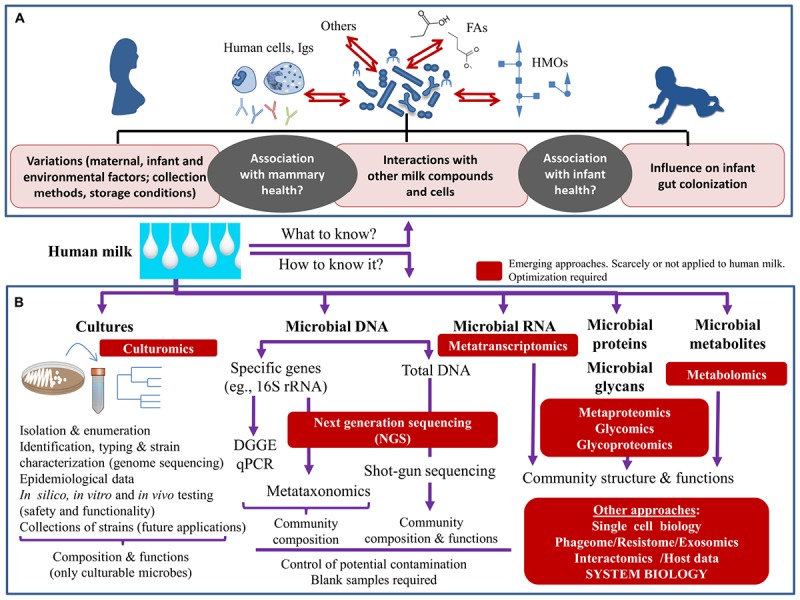
Schematic overview of what we need to know about the human milk microbiome (A) and the main tools that can be applied to achieve their study (B).
Author Contributions
LR and JR conceived this review article. All authors wrote and approved the final version of the manuscript.
Conflict of Interest Statement
CG-C is now employed by Probisearch SL, but did not at the time of the research being undertaken. The company had no role in the study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. LR is a postdoctoral researcher supported by the Juan de la Cierva Postdoctoral Trainee Program of the Spanish Ministry of Economy and Competitiveness (MINECO; IJCI-2015-23196). Our research in this field has been supported by the grant AGL2016-75476-R (Ministry of Economy and Competitiveness, Spain).
References
- Abrahamsson T. R., Sinkiewicz G., Jakobsson T., Fredrikson M., Björkstén B. (2009). Probiotic lactobacilli in breast milk and infant stool in relation to oral intake during the first year of life. J. Pediatr. Gastroenterol. Nutr. 49 349–354. 10.1097/MPG.0b013e31818f091b [DOI] [PubMed] [Google Scholar]
- Andreas N. J., Kampmann B., Mehring Le-Doare K. (2015). Human breast milk: a review on its composition and bioactivity. Early Hum. Dev. 91 629–635. 10.1016/J.EARLHUMDEV.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Arboleya S., Ruas-Madiedo P., Margolles A., Solís G., Salminen S., de Los Reyes-Gavilán C. G., et al. (2011). Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int. J. Food Microbiol. 149 28–36. 10.1016/j.ijfoodmicro.2010.10.036 [DOI] [PubMed] [Google Scholar]
- Arroyo R., Martín V., Maldonado A., Jiménez E., Fernández L., Rodríguez J. M. (2010). Treatment of infectious mastitis during lactation: antibiotics versus oral administration of Lactobacilli isolated from breast milk. Clin. Infect. Dis. 50 1551–1558. 10.1086/652763 [DOI] [PubMed] [Google Scholar]
- Asnicar F., Manara S., Zolfo M., Truong D. T., Scholz M., Armanini F., et al. (2017). Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2:e00164-16. 10.1128/mSystems.00164-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardanzellu F., Fanos V., Reali A. (2017). “Omics” in human colostrum and mature milk: looking to old data with new eyes. Nutrients 9:E843. 10.3390/nu9080843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berini F., Casciello C., Marcone G. L., Marinelli F. (2017). Metagenomics: novel enzymes from non-culturable microbes. FEMS Microbiol. Lett. 364:fnx211 10.1093/femsle/fnx211 [DOI] [PubMed] [Google Scholar]
- Bever C. S., Rand A. A., Nording M., Taft D., Kalanetra K. M., Mills D. A., et al. (2018). Effects of triclosan in breast milk on the infant fecal microbiome. Chemosphere 203 467–473. 10.1016/J.CHEMOSPHERE.2018.03.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilen M., Dufour J.-C., Lagier J.-C., Cadoret F., Daoud Z., Dubourg G., et al. (2018). The contribution of culturomics to the repertoire of isolated human bacterial and archaeal species. Microbiome 6:94. 10.1186/s40168-018-0485-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boix-Amorós A., Collado M. C., Mira A. (2016). Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol. 7:492 10.3389/fmicb.2016.00492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boix-Amorós A., Martinez-Costa C., Querol A., Collado M. C., Mira A. (2017). Multiple approaches detect the presence of fungi in human breastmilk samples from healthy mothers. Sci. Rep. 7:13016. 10.1038/s41598-017-13270-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne H. P., Forster S. C., Anonye B. O., Kumar N., Neville B. A., Stares M. D., et al. (2016). Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature 533 543–546. 10.1038/nature17645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzikowska-Jura A., Czerwonogrodzka-Senczyna A., Olêdzka G., Szostak-Wêgierek D., Weker H., Wesołowska A., et al. (2018). Maternal nutrition and body composition during breastfeeding: association with human milk composition. Nutrients 10:1379. 10.3390/nu10101379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Rubio R., Collado M. C., Laitinen K., Salminen S., Isolauri E., Mira A. (2012). The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 96 544–551. 10.3945/ajcn.112.037382 [DOI] [PubMed] [Google Scholar]
- Cabrera-Rubio R., Mira-Pascual L., Mira A., Collado M. C. (2016). Impact of mode of delivery on the milk microbiota composition of healthy women. J. Dev. Orig. Health Dis. 7 54–60. 10.1017/S2040174415001397 [DOI] [PubMed] [Google Scholar]
- Cárdenas N., Laiño J. E., Delgado S., Jiménez E., Juárez del Valle M., Savoy de Giori G., et al. (2015). Relationships between the genome and some phenotypical properties of Lactobacillus fermentum CECT 5716, a probiotic strain isolated from human milk. Appl. Microbiol. Biotechnol. 99 4343–4353. 10.1007/s00253-015-6429-0 [DOI] [PubMed] [Google Scholar]
- Cárdenas N., Martín V., Delgado S., Rodríguez J. M., Fernández L. (2014). Characterization of Lactobacillus gastricus strains isolated from human milk. Int. Dairy J. 39 167–177. 10.1016/j.idairyj.2014.06.008 [DOI] [Google Scholar]
- Chen P.-W., Lin Y.-L., Huang M.-S. (2018). Profiles of commensal and opportunistic bacteria in human milk from healthy donors in Taiwan. J. Food Drug Anal. 26 1235–1244. 10.1016/J.JFDA.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Pan W. G., Xian W. Y., Cheng H., Zheng J. X., Hu Q. H., et al. (2016). Identification of infantile diarrhea caused by breast milk-tTransmitted Staphylococcus aureus infection. Curr Microbiol. 73 498–502. 10.1007/s00284-016-1088-7 [DOI] [PubMed] [Google Scholar]
- Chiu Y.-H., Tsai J.-J., Lin S.-L., Choritosvakin C., Lin M.-Y. (2014). Characterisation of bifidobacteria with immunomodulatory properties isolated from human breast milk. J. Funct. Foods 7 700–708. 10.1016/j.jff.2013.12.015 [DOI] [Google Scholar]
- Collado M. C., Delgado S., Maldonado A., Rodríguez J. M. (2009). Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett. Appl. Microbiol. 48 523–528. 10.1111/j.1472-765X.2009.02567.x [DOI] [PubMed] [Google Scholar]
- Costea P. I., Zeller G., Sunagawa S., Pelletier E., Alberti A., Levenez F., et al. (2017). Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 35:1069. 10.1038/nbt.3960 [DOI] [PubMed] [Google Scholar]
- Daudi N., Shouval D., Stein-Zamir C., Ackerman Z. (2012). Breastmilk hepatitis a virus rna in nursing mothers with acute hepatitis a virus infection. Breastfeed. Med. 7 313–315. 10.1089/bfm.2011.0084 [DOI] [PubMed] [Google Scholar]
- Davé V., Street K., Francis S., Bradman A., Riley L., Eskenazi B., et al. (2016). Bacterial microbiome of breast milk and child saliva from low-income Mexican-American women and children. Pediatr. Res. 79 846–854. 10.1038/pr.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N. M., Proctor D. M., Holmes S. P., Relman D. A., Callahan B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. 10.1186/s40168-018-0605-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Andrés J., Jiménez E., Chico-Calero I., Fresno M., Fernández L., Rodríguez J. M. (2017). Physiological translocation of lactic acid bacteria during pregnancy contributes to the composition of the milk microbiota in mice. Nutrients 10:E14. 10.3390/nu10010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S., Arroyo R., Martín R., Rodríguez J. M. (2008). PCR-DGGE assessment of the bacterial diversity of breast milk in women with lactational infectious mastitis. BMC Infect. Dis. 8:51. 10.1186/1471-2334-8-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingess K. A., Valentine C. J., Ollberding N. J., Davidson B. S., Woo J. G., Summer S., et al. (2017). Branched-chain fatty acid composition of human milk and the impact of maternal diet: the global exploration of human milk (GEHM) study. Am. J. Clin. Nutr. 105 177–184. 10.3945/ajcn.116.132464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drago L., Toscano M., De Grandi R., Grossi E., Padovani E. M., Peroni D. G. (2017). Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus burundi. ISME J. 11 875–884. 10.1038/ismej.2016.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont-Rouzeyrol M., Biron A., O’Connor O., Huguon E., Descloux E. (2016). Infectious zika viral particles in breastmilk. Lancet 387:1051. 10.1016/S0140-6736(16)00624-3 [DOI] [PubMed] [Google Scholar]
- Duranti S., Lugli G. A., Mancabelli L., Armanini F., Turroni F., James K., et al. (2017). Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5:66. 10.1186/s40168-017-0282-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidelman A. I., Szilagyi G. (1979). Patterns of bacterial colonization of human milk. Obstet. Gynecol. 53 550–552. [PubMed] [Google Scholar]
- Emerson J. B., Adams R. I., Román C. M. B., Brooks B., Coil D. A., Dahlhausen K., et al. (2017). Schrödinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5:86. 10.1186/s40168-017-0285-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Martos I., Jiménez E., de Andrés J., Rodríguez-Alcalá L. M., Tavárez S., Manzano S., et al. (2016). Milk and blood biomarkers associated to the clinical efficacy of a probiotic for the treatment of infectious mastitis. Benef. Microbes 7 305–318. 10.3920/BM2015.0134 [DOI] [PubMed] [Google Scholar]
- Feng P., Gao M., Burgher A., Hui Zhou T., Pramuk K. (2016). A nine-country study of the protein content and amino acid composition of mature human milk. Food Nutr. Res. 60:31042. 10.3402/fnr.v60.31042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L., Cárdenas N., Arroyo R., Manzano S., Jiménez E., Martín V., et al. (2016). Prevention of infectious mastitis by oral administration of Lactobacillus salivarius PS2 during late pregnancy. Clin. Infect. Dis. 62 568–573. 10.1093/cid/civ974 [DOI] [PubMed] [Google Scholar]
- Fernández L., Langa S., Martín V., Maldonado A., Jiménez E., Martín R., et al. (2013). The human milk microbiota: origin and potential roles in health and disease. Pharmacol. Res. 69 1–10. 10.1016/j.phrs.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Fernández L., Ruiz L., Jara J., Orgaz B., Rodríguez J. M. (2018). Strategies for the preservation, restoration and modulation of the human milk microbiota. implications for human milk banks and neonatal intensive care units. Front Microbiol. 9:2676. 10.3389/fmicb.2018.02676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzstevens J. L., Smith K. C., Hagadorn J. I., Caimano M. J., Matson A. P., Brownell E. A. (2017). Systematic review of the human milk microbiota. Nutr. Clin. Pract. 32 354–364. 10.1177/0884533616670150 [DOI] [PubMed] [Google Scholar]
- Foster W. D., Harris R. E. (1960). The incidence of staphylococcus pyogenes in normal human breast milk. J. Obstet. Gynaecol. Br. Emp. 67 463–464. 10.1111/j.1471-0528.1960.tb07027.x [DOI] [PubMed] [Google Scholar]
- Fouhy F., Clooney A. G., Stanton C., Claesson M., Cotter P. D. (2016). 16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 16:123. 10.1186/s12866-016-0738-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Campos M., López M. Á., Rodriguez-Benitez M. V., Romero J., Linares M. D., Maldonado J., et al. (2012). Lactobacillus fermentum CECT 5716 is safe and well tolerated in infants of 1–6 months of age: a randomized controlled trial. Pharmacol. Res. 65 231–238. 10.1016/j.phrs.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Gomez-Gallego C., Garcia-Mantrana I., Salminen S., Collado M. C. (2016). The human milk microbiome and factors influencing its composition and activity. Semin. Fetal Neonatal Med. 21 400–405. 10.1016/j.siny.2016.05.003 [DOI] [PubMed] [Google Scholar]
- Gómez-Gallego C., Kumar H., García-Mantrana I., du Toit E., Suomela J.-P., Linderborg K. M., et al. (2017). Breast milk polyamines and microbiota interactions: impact of mode of delivery and geographical location. Ann. Nutr. Metab. 70 184–190. 10.1159/000457134 [DOI] [PubMed] [Google Scholar]
- Gómez-Gallego C., Morales J., Monleón D., du Toit E., Kumar H., Linderborg K., et al. (2018). Human breast milk NMR metabolomic profile across specific geographical locations and its association with the milk microbiota. Nutrients 10:1355. 10.3390/nu10101355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes M. J., Abellan J. J., Durban A., Perez-Cobas A. E., Latorre A., Moya A. (2012). Metagenomics of human microbiome: beyond 16s rDNA. Clin. Microbiol. Infect. 18 47–49. 10.1111/j.1469-0691.2012.03865.x [DOI] [PubMed] [Google Scholar]
- Grahn N., Olofsson M., Ellnebo-Svedlund K., Monstein H. J., Jonasson J. (2003). Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol. Lett. 219 87–91. 10.1016/s0378-1097(02)01190-4 [DOI] [PubMed] [Google Scholar]
- Heikkilä M. P., Saris P. E. J. (2003). Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J. Appl. Microbiol. 95 471–478. 10.1046/j.1365-2672.2003.02002.x [DOI] [PubMed] [Google Scholar]
- Hennet T., Borsig L. (2016). Breastfed at tiffany’s. Trends Biochem. Sci. 41 508–518. 10.1016/j.tibs.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Hunt K. M., Foster J. A., Forney L. J., Schütte U. M. E., Beck D. L., Abdo Z., et al. (2011). Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One 6:e21313. 10.1371/journal.pone.0021313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeurink P. V., van Bergenhenegouwen J., Jiménez E., Knippels L. M. J., Fernández L., Garssen J., et al. (2013). Human milk: a source of more life than we imagine. Benef. Microbes 4 17–30. 10.3920/BM2012.0040 [DOI] [PubMed] [Google Scholar]
- Jiménez E., de Andrés J., Manrique M., Pareja-Tobes P., Tobes R., Martínez-Blanch J. F., et al. (2015). Metagenomic analysis of milk of healthy and mastitis-suffering women. J. Hum. Lact. 31 406–415. 10.1177/0890334415585078 [DOI] [PubMed] [Google Scholar]
- Jiménez E., Delgado S., Fernández L., García N., Albújar M., Gómez A., et al. (2008a). Assessment of the bacterial diversity of human colostrum and screening of staphylococcal and enterococcal populations for potential virulence factors. Res. Microbiol. 159 595–601. 10.1016/j.resmic.2008.09.001 [DOI] [PubMed] [Google Scholar]
- Jiménez E., Fernández L., Maldonado A., Martín R., Olivares M., Xaus J., et al. (2008b). Oral administration of Lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. Appl. Environ. Microbiol. 74 4650–4655. 10.1128/AEM.02599-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E., Langa S., Martín V., Arroyo R., Martín R., Fernández L., et al. (2010a). Complete genome sequence of Lactobacillus fermentum CECT 5716, a probiotic strain isolated from human milk. J. Bacteriol. 192:4800. 10.1128/JB.00702-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E., Martín R., Maldonado A., Martín V., Gómez de Segura A., Fernández L., et al. (2010b). Complete genome sequence of Lactobacillus salivarius CECT 5713, a probiotic strain isolated from human milk and infant feces. J. Bacteriol. 192 5266–5267. 10.1128/JB.00703-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E., Villar-Tajadura M. A., Marín M., Fontecha J., Requena T., Arroyo R., et al. (2012). Complete genome sequence of bifidobacterium breve CECT 7263, a strain isolated from human milk. J. Bacteriol. 194 3762–3763. 10.1128/JB.00691-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost T., Lacroix C., Braegger C., Chassard C. (2013). Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br. J. Nutr. 110 1253–1262. 10.1017/S0007114513000597 [DOI] [PubMed] [Google Scholar]
- Jost T., Lacroix C., Braegger C. P., Rochat F., Chassard C. (2014). Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 16 2891–2904. 10.1111/1462-2920.12238 [DOI] [PubMed] [Google Scholar]
- Karstens L., Asquith M., Davin S., Fair D., Gregory W. T., Wolfe A. J., et al. (2018). Controlling for contaminants in low biomass 16S rRNA gene sequencing experiments. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny J. F. (1977). Recurrent group B streptococcal disease in an infant associated with the ingestion of infected mother’s milk. J. Pediatr. 91 158–159. 10.1016/s0022-3476(77)80473-3 [DOI] [PubMed] [Google Scholar]
- Khodayar-Pardo P., Mira-Pascual L., Collado M. C., Martínez-Costa C. (2014). Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J. Perinatol. 34 599–605. 10.1038/jp.2014.47 [DOI] [PubMed] [Google Scholar]
- Kim D., Hofstaedter C. E., Zhao C., Mattei L., Tanes C., Clarke E., et al. (2017). Optimizing methods and dodging pitfalls in microbiome research. Microbiome 5:52. 10.1186/s40168-017-0267-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak K., Charbonneau D., Sanozky-Dawes R., Klaenhammer T. (2015). Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. Gut Microbes 6:341. 10.1080/19490976.2015.1103425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H., du Toit E., Kulkarni A., Aakko J., Linderborg K. M., Zhang Y., et al. (2016). Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front. Microbiol. 7:1619. 10.3389/fmicb.2016.01619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey K. A., Williams J. E., Meehan C. L., Zachek J. A., Benda E. D., Price W. J., et al. (2019). What’s Normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: the inspire study. Front. Nutr. 6:45. 10.3389/fnut.2019.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey K. A., Williams J. E., Price W. J., Carrothers J. M., Brooker S. L., Shafii B., et al. (2017). Comparison of commercially-available preservatives for maintaining the integrity of bacterial DNA in human milk. J. Microbiol. Methods 141 73–81. 10.1016/J.MIMET.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Langa S., Maldonado-Barragán A., Delgado S., Martín R., Martín V., Jiménez E., et al. (2012). Characterization of Lactobacillus salivarius CECT 5713, a strain isolated from human milk: from genotype to phenotype. Appl. Microbiol. Biotechnol. 94 1279–1287. 10.1007/s00253-012-4032-1 [DOI] [PubMed] [Google Scholar]
- Lara-Villoslada F., Sierra S., Martín R., Delgado S., Rodríguez J. M., Olivares M., et al. (2007). Safety assessment of two probiotic strains, Lactobacillus coryniformis CECT5711 and Lactobacillus gasseri CECT5714. J. Appl. Microbiol. 103 175–184. 10.1111/j.1365-2672.2006.03225.x [DOI] [PubMed] [Google Scholar]
- Laurence M., Hatzis C., Brash D. E. (2014). Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PLoS One 9:e97876. 10.1371/journal.pone.0097876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Tun H. M., Jahan M., Zhang Z., Kumar A., Dilantha Fernando W. G., et al. (2017). Comparison of DNA-, PMA-, and RNA-based 16S rRNA Illumina sequencing for detection of live bacteria in water. Sci. Rep. 7:5752. 10.1038/s41598-017-02516-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-W., Watanabe K., Hsu C.-C., Chao S.-H., Yang Z.-H., Lin Y.-J., et al. (2017). Bacterial composition and diversity in breast milk samples from mothers living in taiwan and mainland china. Front. Microbiol. 8:965. 10.3389/fmicb.2017.00965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Guan Q., Ye C., Zhang C., Foster J. A., Forney L. J. (2015). Network analysis suggests a potentially ‘evil” alliance of opportunistic pathogens inhibited by a cooperative network in human milk bacterial communities. Sci. Rep. 5:8275. 10.1038/SREP08275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado J., Cañabate F., Sempere L., Vela F., Sánchez A. R., Narbona E., et al. (2012). Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants. J. Pediatr. Gastroenterol. Nutr. 54 55–61. 10.1097/MPG.0b013e3182333f18 [DOI] [PubMed] [Google Scholar]
- Maldonado J., Lara-Villoslada F., Sierra S., Sempere L., Gómez M., Rodriguez J. M., et al. (2010). Safety and tolerance of the human milk probiotic strain Lactobacillus salivarius CECT5713 in 6-month-old children. Nutrition 26 1082–1087. 10.1016/j.nut.2009.08.023 [DOI] [PubMed] [Google Scholar]
- Maldonado-Lobón J. A., Díaz-López M. A., Carputo R., Duarte P., Díaz-Ropero M. P., Valero A. D., et al. (2015). Lactobacillus fermentum CECT 5716 reduces Staphylococcus load in the breastmilk of lactating mothers suffering breast pain: a randomized controlled trial. Breastfeed. Med. 10 425–435. 10.1089/bfm.2015.0070 [DOI] [PubMed] [Google Scholar]
- Martín R., Heilig G. H. J., Zoetendal E. G., Smidt H., Rodríguez J. M. (2007). Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J. Appl. Microbiol. 103 2638–2644. 10.1111/j.1365-2672.2007.03497.x [DOI] [PubMed] [Google Scholar]
- Martín R., Jiménez E., Heilig H., Fernández L., Marín M. L., Zoetendal E. G., et al. (2009). Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl. Environ. Microbiol. 75 965–969. 10.1128/AEM.02063-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín R., Jiménez E., Olivares M., Marín M. L., Fernández L., Xaus J., et al. (2006). Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair. Int. J. Food Microbiol. 15 35–43. 10.1016/j.ijfoodmicro.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Martín R., Langa S., Reviriego C., Jiménez E., Marín M., Olivares M., et al. (2004). The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci. Technol. 15 121–127. 10.1016/j.tifs.2003.09.010 [DOI] [Google Scholar]
- Martín R., Langa S., Reviriego C., Jimínez E., Marín M. L., Xaus J., et al. (2003). Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 143 754–758. 10.1016/j.jpeds.2003.09.028 [DOI] [PubMed] [Google Scholar]
- Martín R., Olivares M., Marín M. L., Fernández L., Xaus J., Rodríguez J. M. (2005). Probiotic potential of 3 Lactobacilli strains isolated from breast milk. J. Hum. Lact. 21 8–17. 10.1177/0890334404272393 [DOI] [PubMed] [Google Scholar]
- Martín V., Cárdenas N., Jiménez E., Maldonado A., Rodríguez J. M., Fernández L. (2013). Genome sequence of Lactobacillus gastricus PS3, a strain isolated from human milk. Genome Announc. 11:e00489-13. 10.1128/genomeA.00489-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Maldonado A., Fernández L., Rodríguez J. M., Connor R. I. (2010). Inhibition of human immunodeficiency virus type 1 by lactic acid bacteria from human breastmilk. Breastfeed. Med. 5 153–158. 10.1089/bfm.2010.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Maldonado-Barragán A., Jiménez E., Ruas-Madiedo P., Fernández L., Rodríguez J. M. (2012). Complete genome sequence of Streptococcus salivarius PS4, a strain isolated from human milk. J. Bacteriol. 194 4466–4467. 10.1128/JB.00896-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Mañes-Lázaro R., Rodríguez J. M., Maldonado-Barragán A. (2011). Streptococcus lactarius sp. nov., isolated from breast milk of healthy women. Int. J. Syst. Evol. Microbiol. 61 1048–1052. 10.1099/ijs.0.021642-0 [DOI] [PubMed] [Google Scholar]
- McGuire M. K., McGuire M. A. (2015). Human milk: mother nature’s prototypical probiotic food? Adv. Nutr. 6 112–123. 10.3945/an.114.007435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire M. K., McGuire M. A. (2017). Got bacteria? The astounding, yet not-so-surprising, microbiome of human milk. Curr. Opin. Biotechnol. 44 63–68. 10.1016/J.COPBIO.2016.11.013 [DOI] [PubMed] [Google Scholar]
- Meehan C. L., Lackey K. A., Hagen E. H., Williams J. E., Roulette J., Helfrecht C., et al. (2018). Social networks, cooperative breeding, and the human milk microbiome. Am. J. Hum. Biol. 30:e23131. 10.1002/ajhb.23131 [DOI] [PubMed] [Google Scholar]
- Mira A., Rodriguez J. (2017). “The origin of human milk bacteria,” in Prebiotics and Probiotics in Human Milk: Origins and Functions of Milk-Borne Oligosaccharides and Bacteria eds McGuire M., McGuire M., Bode L. (Cambridge, MA: Academic Press; ) 349–364. 10.1016/b978-0-12-802725-7.00013-0 [DOI] [Google Scholar]
- Moles L., Escribano E., Andrés J., De Montes M. T., Rodríguez J. M., Jiménez E., et al. (2015). Administration of Bifidobacterium breve PS12929 and Lactobacillus salivarius PS12934, two strains isolated from human milk, to very low and extremely low birth weight preterm infants: a pilot study. J. Immunol. Res. 2015:538171. 10.1155/2015/538171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moossavi S., Atakora F., Miliku K., Sepehri S., Robertson B., Duan Q. L.et al. (2019). Integrated analysis of human milk microbiota with oligosaccharides and fatty acids in the CHILD cohort. Front. Nutr. 6:58. 10.3389/fnut.2019.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A., Ferrer M. (2016). Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 24 402–413. 10.1016/J.TIM.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Mühl H., Kochem A.-J., Disqué C., Sakka S. G. (2010). Activity and DNA contamination of commercial polymerase chain reaction reagents for the universal 16SrDNA real-time polymerase chain reaction detection of bacterial pathogens in blood. Diagn. Microbiol. Infect. Dis. 66 41–49. 10.1016/j.diagmicrobio.2008.07.011 [DOI] [PubMed] [Google Scholar]
- Murphy K., Curley D., O’Callaghan T. F., O’Shea C.-A., Dempsey E. M., O’Toole P. W., et al. (2017). The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study. Sci. Rep. 7:40597. 10.1038/srep40597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschlechner W., Karall D., Hartmann C., Streiter B., Baumgartner-Sigl S., Orth-Höller D., et al. (2016). Mammary candidiasis: molecular-based detection of Candida species in human milk samples. Eur. J. Clin. Microbiol. Infect. Dis. 35 1309–1313. 10.1007/s10096-016-2666-0 [DOI] [PubMed] [Google Scholar]
- Obermajer T., Lipoglav L., Tompa G., Treven P. (2015). Colostrum of healthy slovenian mothers: microbiota composition and bacteriocin gene prevalence. PLoS One 10:e0123324. 10.1371/journal.pone.0123324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares M., Diaz-Ropero M. P., Martin R., Rodriguez J. M., Xaus J. (2006). Antimicrobial potential of four Lactobacillus strains isolated from breast milk. J. Appl. Microbiol. 101 72–79. 10.1111/j.1365-2672.2006.02981.x [DOI] [PubMed] [Google Scholar]
- Pannaraj P. S., Ly M., Cerini C., Saavedra M., Aldrovandi G. M., Saboory A. A., et al. (2018). Shared and distinct features of human milk and infant stool viromes. Front. Microbiol. 9:1162. 10.3389/fmicb.2018.01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. H., Vaidya Y. H., Patel R. J., Pandit R. J., Joshi C. G., Kunjadiya A. P. (2017). Culture independent assessment of human milk microbial community in lactational mastitis. Sci. Rep. 7:7804. 10.1038/S41598-017-08451-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Muñoz M. E., Arrieta M.-C., Ramer-Tait A. E., Walter J. (2017). A critical assessment of the sterile womb and in utero colonization hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5:48. 10.1186/s40168-017-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantasalo I., Kauppinen M. A. (1959). The occurrence of Staphylococcus aureus in mother’s milk. Ann. Chir. Gynaecol. Fenn. 48 246–258. [PubMed] [Google Scholar]
- Reis N. A., Saraiva M. A. F., Duarte E. A. A., de Carvalho E. A., Vieira B. B., Evangelista-Barreto N. S. (2016). Probiotic properties of lactic acid bacteria isolated from human milk. J. Appl. Microbiol. 121 811–820. 10.1111/jam.13173 [DOI] [PubMed] [Google Scholar]
- Rodríguez J. M. (2014). The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 5 779–784. 10.3945/an.114.007229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncada P., Stipetic L. H., Bonizzi L., Burchmore R. J. S., Kennedy M. W. (2013). Proteomics as a tool to explore human milk in health and disease. J. Proteomics 88 47–57. 10.1016/J.JPROT.2013.04.008 [DOI] [PubMed] [Google Scholar]
- Ruiz L., Espinosa-Martos I., García-Carral C., Manzano S., McGuire M. K., Meehan C. L., et al. (2017). What’s normal? Immune profiling of human milk from healthy women living in different geographical and socioeconomic settings. Front. Immunol. 8:696. 10.3389/fimmu.2017.00696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakwinska O., Moine D., Delley M., Combremont S., Rezzonico E., Descombes P., et al. (2016). Microbiota in breast milk of chinese lactating mothers. PLoS One 11:e0160856. 10.1371/journal.pone.0160856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter S. J., Cox M. J., Turek E. M., Calus S. T., Cookson W. O., Moffatt M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:87. 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C., Singh B. P., Thakur N., Gulati S., Gupta S., Mishra S. K., et al. (2017). Antibacterial effects of Lactobacillus isolates of curd and human milk origin against food-borne and human pathogens. 3 Biotech 7:31. 10.1007/s13205-016-0591-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. P., Choi Y. M., Kim W. H., Hong S. P., Park J. M., Kim J., et al. (2018). A double blind, placebo-controlled, randomized clinical trial that breast milk derived-Lactobacillus gasseri BNR17 mitigated diarrhea-dominant irritable bowel syndrome. J. Clin. Biochem. Nutr. 62 179–186. 10.3164/jcbn.17-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Soro Á, D’Auria G., Collado M. C., Džunková M., Culshaw S., Mira A. (2015). Revealing microbial recognition by specific antibodies. BMC Microbiol. 15:132. 10.1186/s12866-015-0456-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís G., de Los Reyes-Gavilan C. G., Fernández N., Margolles A., Gueimonde M. (2010). Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 16 307–310. 10.1016/j.anaerobe.2010.02.004 [DOI] [PubMed] [Google Scholar]
- Soto A., Martín V., Jiménez E., Mader I., Rodríguez J. M., Fernández L. (2014). Lactobacilli and bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors. J. Pediatr. Gastroenterol. Nutr. 59 78–88. 10.1097/MPG.0000000000000347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194 4151–4160. 10.1128/JB.00345-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantikachornkiat M., Sakakibara S., Neuner M., Durall D. M. (2016). The use of propidium monoazide in conjunction with qPCR and Illumina sequencing to identify and quantify live yeasts and bacteria. Int. J. Food Microbiol. 234 53–59. 10.1016/J.IJFOODMICRO.2016.06.031 [DOI] [PubMed] [Google Scholar]
- Toscano M., De Grandi R., Peroni D. G., Grossi E., Facchin V., Comberiati P., et al. (2017). Impact of delivery mode on the colostrum microbiota composition. BMC Microbiol. 17:205. 10.1186/s12866-017-1109-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., Angelini M., Gloor G. B., Reid G. (2016). Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome 4:1. 10.1186/s40168-015-0145-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., Cummins J., Brackstone M., Macklaim J. M., Gloor G. B., Baban C. K., et al. (2014a). Microbiota of human breast tissue. Appl. Environ. Microbiol. 80 3007–3014. 10.1128/AEM.00242-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., McMillan A., Angelini M., Gloor G. B., Sumarah M., Burton J. P., et al. (2014b). Effect of chemotherapy on the microbiota and metabolome of human milk, a case report. Microbiome 2:24. 10.1186/2049-2618-2-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T. L., Hosid S., Ioshikhes I., Altosaar I. (2013). Human milk metagenome: a functional capacity analysis. BMC Microbiol. 13:116. 10.1186/1471-2180-13-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesolowska-Andersen A., Bahl M. I., Carvalho V., Kristiansen K., Sicheritz-Pontén T., Gupta R., et al. (2014). Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2:19. 10.1186/2049-2618-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. E., Carrothers J. M., Lackey K. A., Beatty N. F., York M. A., Brooker S. L., et al. (2017a). Human milk microbial community structure is relatively stable and related to variations in macronutrient and micronutrient intakes in healthy lactating women. J. Nutr. 147 1739–1748. 10.3945/jn.117.248864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. E., Price W. J., Shafii B., Yahvah K. M., Bode L., McGuire M. A., et al. (2017b). Relationships among microbial communities, maternal cells, oligosaccharides, and macronutrients in human milk. J. Hum. Lact. 33 540–551. 10.1177/0890334417709433 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2003). Global Strategy for Infant and Young Child Feeding. Geneva: WHO. [Google Scholar]
- Yao G., Yu J., Hou Q., Hui W., Liu W., Kwok L.-Y., et al. (2017). A perspective study of koumiss microbiome by metagenomics analysis based on single-cell amplification technique. Front. Microbiol. 8:165. 10.3389/fmicb.2017.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Deng H., Ma J., Huang S., Yang J., Huang Y., et al. (2016). Clinical metagenomic analysis of bacterial communities in breast abscesses of granulomatous mastitis. Int. J. Infect. Dis. 53 30–33. 10.1016/J.IJID.2016.10.015 [DOI] [PubMed] [Google Scholar]
- Zinter M. S., Mayday M. Y., Ryckman K. K., Jelliffe-Pawlowski L. L., DeRisi J. L. (2019). Towards precision quantification of contamination in metagenomic sequencing experiments. Microbiome 7:62. 10.1186/s40168-019-0678-6 [DOI] [PMC free article] [PubMed] [Google Scholar]


