Abstract
Background
This study aimed to use a network pharmacology approach to establish the effects of plumbagin on pancreatic cancer (PC) and to predict core targets and biological functions, pathways, and mechanisms of action.
Material/Methods
Genes associated with the pathogenesis of PC were obtained from a database of gene-disease associations (DisGeNET). Putative genes associated with plumbagin were identified from the databases of drug target identification (PharmMapper), target prediction of bioactive components (SwissTargetPrediction), and comprehensive drug target information (DrugBank). PC targets of plumbagin were harvested by using a functional enrichment analysis tool (FunRich). The data of function-related protein-protein interactions (PPIs) with a confidence score >0.9 were obtained by using functional protein association networks (STRING). The network interactions of plumbagin and PC targets and function-related proteins were constructed through complex network analysis and visualization (Cytoscape). The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analysis were used to identify the effects of plumbagin.
Results
The most important biotargets for plumbagin in PC were identified as TP53, MAPK1, BCL2, and IL6. A total of 1,731 annotations and 121 enriched pathways for plumbagin and PC were identified by KEGG and GO analysis. The top 10 signaling pathways of plumbagin and PC were screened, followed by identification of biological components and functions.
Conclusions
Network pharmacology established the effects of plumbagin on PC, predicted core targets, biological functions, pathways, and mechanisms of action. Further studies are needed to validate these predictive biotargets in PC.
MeSH Keywords: Health Care Evaluation Mechanisms, Medical Oncology, Pharmacology
Background
Pancreatic cancer (PC) is an aggressive malignancy with high patient mortality that accounts for approximately 3% of all cases of cancer and has a 5-year mortality of 90% [1,2]. In the USA, PC is the second leading cause of cancer-related death, just behind lung cancer, and the incidence has increased recently [3]. The pathogenesis of PC is characterized by a tumor that is difficult to diagnose in its early stage, resulting in patients presenting with late-stage PC at initial diagnosis. Because early detection of PC can be difficult, and most patients are initially diagnosed with metastatic PC, treatment includes radiation therapy and chemotherapy, including gemcitabine, but these treatments may nor improve prognosis and have unwanted side effects [4–6]. Therefore, it is necessary to develop alternative treatment approaches for PC.
Plumbagin (5-hydroxy-2-methyl-1,4-naphthene) is a compound that is isolated from Plumbago zeylanica, and has been shown to have pharmacological activity as a traditional Chinese medicine. Although there have been previous reports on the toxic effects of plumbagin, there have also been reports of its anti-neoplastic and anti-proliferative effects on tumor cells in vitro and in vivo, and its role in the inhibition of cell migration in breast cancer, liver cancer, and lung cancer [7–9]. Currently, few studies have investigated the effects of plumbagin on PC or the biotargets and molecular mechanisms involved by the effects of plumbagin. Network pharmacology is an emerging and novel method for identifying the systemic mechanisms of therapeutic compounds in disease [10–11]. Systems pharmacology may be a promising strategy for understanding the pharmacological targets and mechanisms of plumbagin in PC. Therefore, this study aimed to use a network pharmacology approach to establish the effects of plumbagin on pancreatic cancer (PC) and to predict core targets and biological functions, pathways, and mechanisms of action. A schematic diagram of the study design is shown in Figure 1.
Figure. 1.
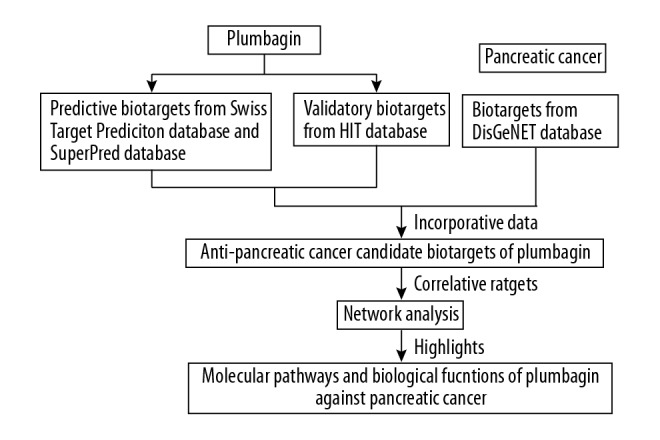
The schematic flowchart was designed in the current study through conducting a network pharmacology strategy.
Material and Methods
Identification of candidate targets of plumbagin and pancreatic cancer (PC)
The study used databases of drug classification and target prediction (SuperPred) and SwissTargetPrediction to predict the putative targets of plumbagin, followed by identification of known targets of plumbagin through the herb identification target (HIT) database. Also, the DisGeNET database (http://www.disgenet.org/) was used to collect the PC-associated gene targets. A Venn diagram of the plumbagin-PC network targets was plotted and visualized using FunRich software. The overlapping targets were harvested as correlative targets between plumbagin and PC.
Cluster analysis and the protein-protein interaction (PPI) network of plumbagin and PC
The identifiable targets from plumbagin and PC were introduced into the STRING database of functional protein-association networks to obtain the PPI network and the function-related proteins of plumbagin in PC. Also, the ClusterOne algorithm setting of Cytoscape software was used to cluster the proteins in the network, and the biological function and pathway enrichment analysis of each cluster were implemented.
Biological functions and enrichment pathway analysis of identifiable targets
NetworkAnalyzer was used to calculate network topological parameters, such as the median degree of freedom and the maximum degree of freedom from the PPI network targets for plumbagin in PC. Core targets were screened according to the degree values of topological data, indicating the upper limit of the screening range as the maximum degree value and the lower limit as the median degree. Also, the gene/protein functional annotation online platform, KOBAS, was used to integrate the biological function and pathway enrichment of the core targets, followed by being imported in the OmicShare cloud platform. The biological processes and the signaling pathways involved in all core targets of plumbagin in PC were visualized, and high-level Venn diagrams were plotted according to the P-values.
Results
Construction of protein-protein interaction (PPI) network and clustering analysis of plumbagin and pancreatic cancer (PC)
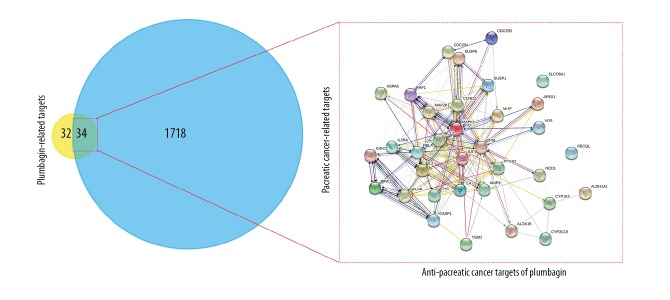
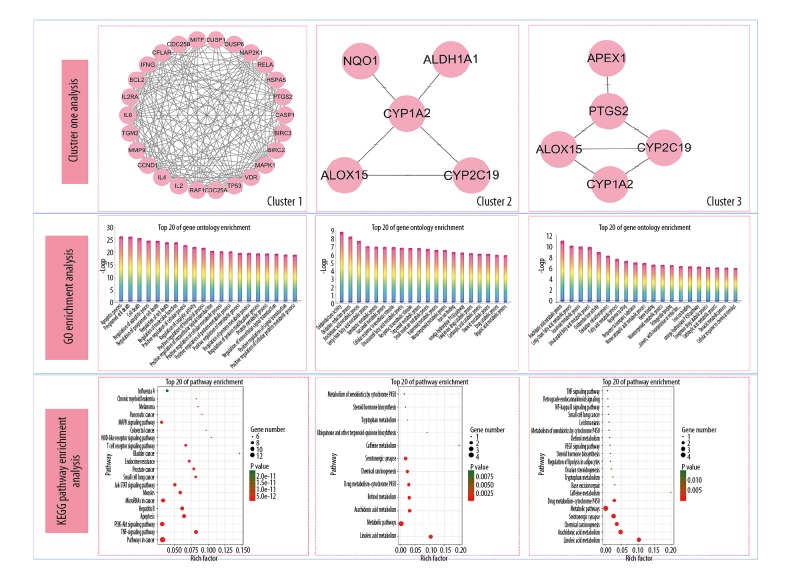
In total, there were 1,752 cancer target genes, and 66 putative plumbagin target genes. The data from FunRich showed 34 interactive targets of plumbagin in PC. After removing the duplicates, the data from the STRING database identified 34 node proteins and 180 edges between the interactive targets (Figure 2). Also, cluster analysis from the ClusterONE algorithm resulted in three groups of protein-protein function-related PPI networks of plumbagin in PC (Figure 3).
Figure 2.
A protein-protein interaction (PPI) network of plumbagin in pancreatic cancer (PC) targets was constructed for visualization of interactive targets.
Figure 3.
Candidate core targets of plumbagin in pancreatic cancer (PC), with the most important targets identified as TP53, MAPK1, BCL2, and IL6.
Screening of core targets of plumbagin and PC
NetworkAnalyzer was used to determine network topology parameters, including the freedom of degree, the shortest path, and the central median value of the PPI network for plumbagin in PC. As shown in Table 1, the smaller of shortest path length and the larger vector value indicated the more important proteins. After calculation, the median degree of freedom was 10.5, and the maximum degree of freedom was 28. Therefore, the screening condition range of core targets was set as 21–28, and the four most important proteins, TP53, MAPK1, BCL2, and IL6 were identified.
Table 1.
Details of corn targets of plumbagin against pancreatic cancer.
| Targets | Degree | Average shortest path length | Betweenness centrality | Closeness centrality | Clustering coefficient |
|---|---|---|---|---|---|
| TP53 | 28 | 1.09677419 | 0.23638787 | 0.91176471 | 0.37830688 |
| MAPK1 | 23 | 1.25806452 | 0.07097866 | 0.79487179 | 0.52173913 |
| BCL2 | 22 | 1.29032258 | 0.04904667 | 0.775 | 0.56277056 |
| IL6 | 22 | 1.29032258 | 0.06234832 | 0.775 | 0.53679654 |
| PTGS2 | 18 | 1.41935484 | 0.0731364 | 0.70454545 | 0.58823529 |
| CCND1 | 18 | 1.41935484 | 0.02421563 | 0.70454545 | 0.63398693 |
| RAF1 | 17 | 1.48387097 | 0.03205762 | 0.67391304 | 0.57352941 |
| IL4 | 17 | 1.4516129 | 0.02307854 | 0.68888889 | 0.68382353 |
| RELA | 17 | 1.4516129 | 0.02055342 | 0.68888889 | 0.64705882 |
| IL2 | 16 | 1.48387097 | 0.00672515 | 0.67391304 | 0.8 |
| MAP2K1 | 16 | 1.48387097 | 0.02639246 | 0.67391304 | 0.625 |
| CFLAR | 16 | 1.48387097 | 0.01300061 | 0.67391304 | 0.71666667 |
| MMP9 | 15 | 1.51612903 | 0.02254651 | 0.65957447 | 0.6952381 |
| IFNG | 13 | 1.58064516 | 0.00349326 | 0.63265306 | 0.85897436 |
| MITF | 12 | 1.61290323 | 0.00517425 | 0.62 | 0.81818182 |
| CASP1 | 12 | 1.61290323 | 0.00429753 | 0.62 | 0.8030303 |
| BIRC2 | 9 | 1.74193548 | 8.82E-04 | 0.57407407 | 0.88888889 |
| BIRC3 | 9 | 1.74193548 | 8.82E-04 | 0.57407407 | 0.88888889 |
| IL2RA | 8 | 1.83870968 | 3.80E-04 | 0.54385965 | 0.92857143 |
| DUSP1 | 8 | 1.74193548 | 0.00618055 | 0.57407407 | 0.82142857 |
| CDC25A | 7 | 1.80645161 | 7.89E-04 | 0.55357143 | 0.9047619 |
| HSPA5 | 7 | 1.80645161 | 1.65E-04 | 0.55357143 | 0.95238095 |
| CYP1A2 | 6 | 1.83870968 | 0.0153646 | 0.54385965 | 0.33333333 |
| CDC25B | 6 | 1.83870968 | 3.58E-04 | 0.54385965 | 0.93333333 |
| ALOX15 | 5 | 1.87096774 | 0.00552057 | 0.53448276 | 0.6 |
| VDR | 5 | 1.87096774 | 0 | 0.53448276 | 1 |
| TGM2 | 5 | 1.90322581 | 1.65E-04 | 0.52542373 | 0.9 |
| DUSP6 | 4 | 2.09677419 | 0 | 0.47692308 | 1 |
| CYP2C19 | 3 | 2.22580645 | 5.38E-04 | 0.44927536 | 0.66666667 |
| NQO1 | 3 | 1.96774194 | 7.17E-04 | 0.50819672 | 0.66666667 |
| APEX1 | 3 | 1.93548387 | 0 | 0.51666667 | 1 |
| ALDH1A1 | 2 | 2 | 0 | 0.5 | 1 |
Biological functions and pathway enrichment assays of core targets of plumbagin and PC
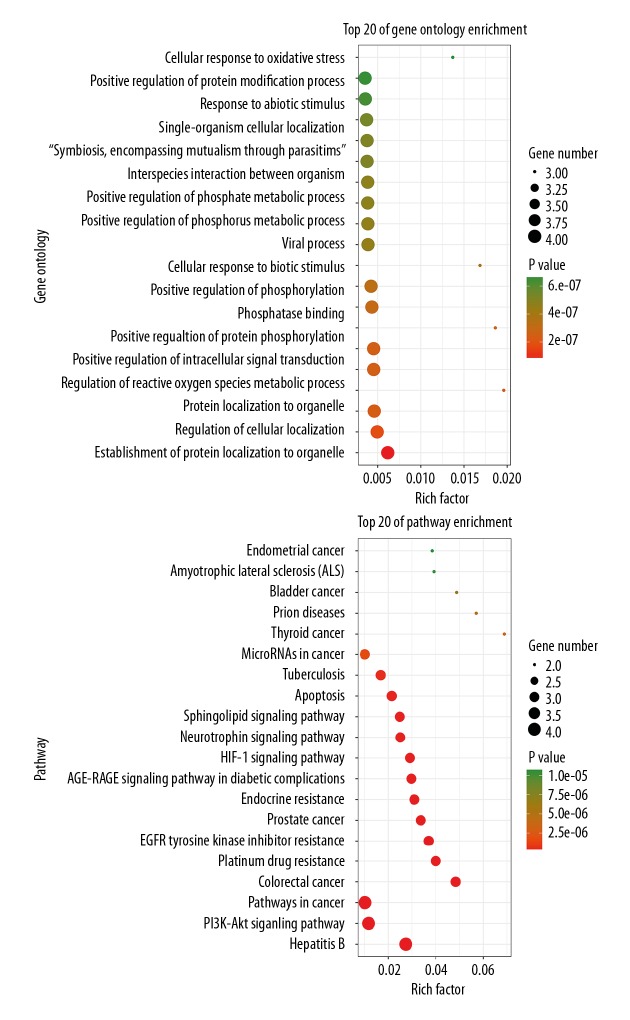
The biological functions and signaling pathways from all core targets were enriched and analyzed by using KOBAS software. A total of 1,731 Gene Ontology (GO) annotations and 121 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were obtained. The inclusive data were uploaded to the OmicShare cloud platform to produce the Venn diagrams of the top 20 biological functions and molecular pathways of the core targets (Figure 4). The plumbagin effects on PC were related to 1731 GO annotations, including the establishment of protein localization to organelles, regulation of cell localization, protein localization to organelles, regulation of reactive oxygen species (ROS) metabolic processes, and positive regulation of intracellular signal transduction. As shown in the KEGG analysis, the molecular pathways of plumbagin in PC were mainly enriched in 121 signaling pathways, including hepatitis B, PI3K-Akt signaling, pathways in cancer, colorectal cancer, platinum-based drug resistance, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) resistance, prostate cancer, endocrine resistance, the advanced glycation endproducts (AGE)-receptor for advanced glycation endproducts (RAGE) signaling pathway, and the hypoxia-inducible factor 1 (HIF-1) signaling pathway.
Figure 4.
Biological functions and molecular pathways from all core targets of plumbagin in pancreatic cancer (PC), with the top 20 biological processes and signaling pathways screened and classified.
Discussion
In this study, the bioinformatics findings from web-based databases obtained core targets and identified the biological functions, pathways, and mechanisms of plumbagin in pancreatic cancer (PC). These findings supported that plumbagin might have specific therapeutic effects in PC. The important predictive biotargets for plumbagin in PC were identified as TP53, MAPK1, BCL2, and IL6. It is known that p53 plays a key role in regulating downstream genes involved in cell cycle arrest, DNA repair, and programmed cell death, and p53 dysfunction leads to genomic instability, uncontrolled cell growth, and unregulated cell cycle inhibition [12]. Therefore, p53 mutation is a common feature of malignant tumors. Mutant somatic TP53 can induce resistance to chemotherapeutic agents or targeted drugs and may be associated with resistance to treatment in patients with PC [13].
MAPK is a transduction protein that controls several cellular processes, including cell proliferation and survival [14]. Studies have shown that overexpression of MAPK1 is found in primary PC and is associated with poor prognosis following chemotherapy [15]. The anti-apoptotic effector, BCL2, is associated with chemotherapy resistance and poor prognosis in patients with hematological malignancies [16]. BCL2 is downstream of the p53 signaling pathway and is regulated by the p53 gene and plays different roles in cellular events, including in DNA damage and hypoxia [17]. Abnormal expression of BCL2 is associated with the progression of PC [18]. Also, IL6 is a regulatory cytokine, which is associated with the progression of cancer and other diseases, including hypertension and cardiovascular disease [19,20]. IL-6 can induce the migration of PC cells in vitro, and it may be an important effector in the development and progression of PC, to function as a potential therapeutic target [21,22]. In this study, the core targets that were identified included TP53, MAPK1, BCL2, and IL6 in plumbagin in PC, which might be potential therapeutic biomolecules for the treatment of PC.
Conclusions
The findings from this study showed that the pharmacological activities of plumbagin in pancreatic cancer (PC) might be associated with the regulation of four main targets that include TP53, MAPK1, BCL2, and IL6. The possible biological mechanisms involved include the modulation of the PI3K/Akt signaling pathway, the advanced glycation endproducts (AGE)-receptor for advanced glycation endproducts (RAGE) signaling pathway, and the hypoxia-inducible factor 1 (HIF-1) signaling pathway, which exert crucial roles in cell survival, apoptosis, and metabolism.
Footnotes
Source of support: This study was funded by the National Natural Science Foundation of China (No. 81560134)
Conflict of interest
None.
References
- 1.Conroy T, Bachet J-B, Ayav A, et al. Current standards and new innovative approaches for treatment of pancreatic cancer. Eur J Cancer. 2016;57:10–22. doi: 10.1016/j.ejca.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 2.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 3.Sanabria Mateos R, Conlon KC. Pancreatic cancer. Surgery (Oxford) 2016;34:282–91. [Google Scholar]
- 4.Zhang Y, Zhang XX, Yuan RY, et al. Cordycepin induces apoptosis in human pancreatic cancer cells via the mitochondrial-mediated intrinsic pathway and suppresses tumor growth in vivo. Onco Targets Ther. 2018;11:4479–90. doi: 10.2147/OTT.S164670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong W, Chen BZ, Lee AKY, et al. Chinese herbal medicine effectively prolongs the overall survival of pancreatic cancer patients: A case series. Integr Cancer Ther. 2019;18 doi: 10.1177/1534735419828836. 1534735419828836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao J, Wang G, Wu J, et al. Arsenic trioxide inhibits Skp2 expression to increase chemosensitivity to gemcitabine in pancreatic cancer cells. Am J Transl Res. 2019;11:991–97. [PMC free article] [PubMed] [Google Scholar]
- 7.Roy A, Bharadvaja N. Biotechnological approaches for the production of pharmaceutically important compound: Plumbagin. Curr Pharm Biotechnol. 2018;19:372–81. doi: 10.2174/1389201019666180629143842. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Cai Y, He C, et al. Anticancer properties and pharmaceutical applications of plumbagin: A review. Am J Chin Med. 2017;45:423–41. doi: 10.1142/S0192415X17500264. [DOI] [PubMed] [Google Scholar]
- 9.De U, Son JY, Jeon Y, et al. Plumbagin from a tropical pitcher plant (Nepenthes alata Blanco) induces apoptotic cell death via a p53-dependent pathway in MCF-7 human breast cancer cells. Food Chem Toxicol. 2019;123:492–500. doi: 10.1016/j.fct.2018.11.040. [DOI] [PubMed] [Google Scholar]
- 10.Su M, Guo C, Liu M, et al. Therapeutic targets of vitamin C on liver injury and associated biological mechanisms: A study of network pharmacology. Int Immunopharmacol. 2019;66:383–87. doi: 10.1016/j.intimp.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 11.Li R, Ma X, Song Y, et al. Anti-colorectal cancer targets of resveratrol and biological molecular mechanism: Analyses of network pharmacology, human and experimental data. J Cell Biochem. :2019. doi: 10.1002/jcb.28404. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Vousden KH, Lu X. Live or let die: The cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 13.Abrams SL, Lertpiriyapong K, Yang LV, et al. Introduction of WT-TP53 into pancreatic cancer cells alters sensitivity to chemotherapeutic drugs, targeted therapeutics and nutraceuticals. Adv Biol Regul. 2018;69:16–34. doi: 10.1016/j.jbior.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Roskoski R., Jr ERK1/2 MAP kinases: Structure, function, and regulation. Pharmacol Res. 2012;66:105–43. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Furukawa T, Kanai N, Shiwaku HO, et al. AURKA is one of the downstream targets of MAPK1/ERK2 in pancreatic cancer. Oncogene. 2006;25:4831–39. doi: 10.1038/sj.onc.1209494. [DOI] [PubMed] [Google Scholar]
- 16.Deng X, Gao F, May WS. Protein phosphatase 2A inactivates Bcl2’s antiapoptotic function by dephosphorylation and up-regulation of Bcl2-p53 binding. Blood. 2009;113:422–28. doi: 10.1182/blood-2008-06-165134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raffo AJ, Kim AL, Fine RL. Formation of nuclear Bax/p53 complexes is associated with chemotherapy induced apoptosis. Oncogene. 2000;19:6216–28. doi: 10.1038/sj.onc.1203995. [DOI] [PubMed] [Google Scholar]
- 18.Bold RJ, Virudachalam S, McConkey DJ. BCL2 expression correlates with metastatic potential in pancreatic cancer cell lines. Cancer. 2001;92:1122–29. doi: 10.1002/1097-0142(20010901)92:5<1122::aid-cncr1429>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 19.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta. 2011;1813:878–88. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 20.Kumari N, Dwarakanath BS, Das A, Bhatt AN. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 2016;37:11553–72. doi: 10.1007/s13277-016-5098-7. [DOI] [PubMed] [Google Scholar]
- 21.Holmer R, Goumas FA, Waetzig GH, et al. Interleukin-6: a villain in the drama of pancreatic cancer development and progression. Hepatobiliary Pancreat Dis Int. 2014;13:371–80. doi: 10.1016/s1499-3872(14)60259-9. [DOI] [PubMed] [Google Scholar]
- 22.Razidlo GL, Burton KM, McNiven MA. Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42. J Biol Chem. 2018;293:11143–53. doi: 10.1074/jbc.RA118.003276. [DOI] [PMC free article] [PubMed] [Google Scholar]