Abstract
Brain inflammation is associated with various types of neurodegenerative diseases, including Alzheimer disease (AD). Quantifying inflammation with PET is a challenging and invasive procedure, especially in frail patients, because it requires blood sampling from an arterial catheter. A widely used alternative to arterial sampling is a supervised clustering algorithm (SVCA), which identifies the voxels with minimal specific binding in the PET images, thus extracting a reference region for noninvasive kinetic modeling. Methods: We tested this algorithm on a large population of subjects injected with the translocator protein radioligand 11C-PBR28 and compared the kinetic modeling results obtained with the gold standard of arterial input function (VT/fp) with those obtained by SVCA (distribution volume ratio [DVR] with Logan plot). The study comprised 57 participants (21 healthy controls, 11 mild cognitive impairment patients, and 25 AD patients). Results: We found that VT/fp was greater in AD patients than in controls in the inferior parietal, combined middle and inferior temporal, and entorhinal cortices. SVCA-DVR identified increased binding in the same regions and in an additional one, the parahippocampal region. We noticed however that the average amplitude of the reference curve obtained from subjects with genetic high-affinity binding for 11C-PBR28 was significantly larger than that from subjects with moderate affinity. This suggests that the reference curve extracted by SVCA was contaminated by specific binding. Conclusion: SVCA allows the noninvasive quantification of inflammatory biomarker translocator protein measured with 11C-PBR28 but without the need of arterial sampling. Although the reference curves were contaminated with specific binding, the decreased variance of the outcome measure, SVCA DVR, allowed for an apparent greater sensitivity to detect regional abnormalities in brains of patients with AD.
Keywords: PET, 11C-PBR28, Alzheimer, brain, clustering, inflammation
The need of arterial sampling to quantify the binding potential of PET tracers in the brain is probably the main obstacle to the widespread use of translocator protein (TSPO) radioligands in research protocols, let alone in clinical practice. In expert hands, placing an arterial catheter in the radial artery takes only a few minutes, is well tolerated by patients, and carries little risk, but it is a logistically challenging and costly procedure. It requires trained personnel and special techniques to analyze the plasma samples, such as high-performance liquid chromatography. In addition, plasma measurements of parent concentrations and plasma free fraction are noisy and prone to errors (1). At least 5 approaches have been proposed as noninvasive alternatives to arterial sampling: image-derived input function, population-derived input function, simultaneous estimation of the input function, pseudo reference region, and supervised clustering algorithm (SVCA), the approach examined in this paper. Image-derived input function from small brain vessels is seldom trustworthy: not only are partial-volume effects challenging to correct without scaling with blood samples, but PET images cannot distinguish the photons emitted by the parent compound from those of its radiometabolites. Template curves such as those used for population-derived input functions may not faithfully capture individual variability, and robust estimates are obtained only when at least one blood sample is used to properly scale the template curve. Although the simultaneous estimation of the input function needs at least one blood sample, together with the time–activity curves of several brain regions (2), Schain et al. estimated BPND (binding potential) noninvasively for 11C-PBR28 using a template input curve and demonstrated that it was able to distinguish between healthy controls and patients with Alzheimer disease (AD) (3). However, a template curve still has to be previously generated from subjects scanned with arterial sampling, and the shape of the curve obtained from a population of healthy subjects may not necessarily reflect that of pathologic conditions (3).
A relative method of measurement, by comparing the uptake of the target region to that of a region devoid of receptors, would obviate arterial sampling and arguably reduce the variability of the estimates, since the input function and the free fraction are common to both regions. However, since TSPO is expressed ubiquitously in the brain, a proper reference region does not exist. Nevertheless, Lyoo et al. successfully quantified 11C-PBR28 in a clinical protocol using a pseudo reference region (4). A pseudo reference region is one that has a certain amount of specific binding but is not affected by the course of the disease. Using this approach, Lyoo et al. reanalyzed a large database of 57 subjects, comprising healthy controls, patients with mild cognitive impairment (MCI), and patients with AD (5). By using the SUV ratio (SUVR) over the cerebellum, they were able to replicate the findings obtained with full kinetic modeling. They showed that TSPO binding was greater in AD patients than in either healthy or MCI subjects, and also identified 1 additional region, suggesting that this approach may have greater sensitivity (4). Of course, an important limitation of this method is that it requires previous knowledge that the designated pseudo reference region is not affected by the disease under study. In the study of Lyoo et al., the use of the cerebellum as a pseudo reference region was justified by several lines of evidence: during AD, the cerebellum is relatively spared from neurodegeneration and the morphology of its microglia differs from that of the activated microglia in the neocortex. In addition, similar values were observed for arterial input function (VT/fp) and SUV in the cerebellum among the healthy controls, MCI patients. and AD patients (4).
A few years ago, Turkheimer et al. described an SVCA (6). SVCA uses predefined kinetic classes to segment the tissue and to automatically extract reference curves, defined as the average curve of all voxels where the specific binding component is minimal. This algorithm, initially validated for 11C-(R)-PK11195, would allow the noninvasive quantification of TSPO across a variety of diseases, independently of the availability of a reference region. Indeed, it has been used in different clinical conditions associated with microglial activation (7–10). However, 11C-(R)-PK11195 is a radioligand with low specific binding, and there seems to be an inverse relationship between the affinity of the tracer and the successful implementation of SVCA (11). Recently García-Lorenzo et al. (12) showed that SVCA can also be applied to 18F-DPA-714, a TSPO tracer with intermediate affinity for TSPO (∼1.5-fold that of 11C-(R)-PK11195 (13)). 11C-PBR28 is one of the most widely used TSPO tracers worldwide and has high affinity for TSPO (∼5- to 6-fold that of 11C-(R)-PK11195 (13)).
The aim of this work was to test the suitability of SVCA to quantify 11C-PBR28 noninvasively. We attempt to replicate the results of the same large database analyzed by Lyoo et al., which comprised healthy controls, MCI patients, and AD patients. This also allowed a direct comparison with the pseudo reference approach.
MATERIALS AND METHODS
Subjects
All data from the previous study were included and reanalyzed for the current study (4). We included 57 participants (21 healthy controls, 11 MCI patients, and 25 AD patients). All MCI and AD patients were positive for amyloid-β after screening with 11C-Pittsburgh compound B PET scans. Therefore, AD patients met the criteria for probable AD dementia with evidence of an AD pathophysiologic process (14), and MCI patients met the criteria for MCI due to a high or intermediate likelihood of developing AD (15). Individual TSPO binding affinity was determined using the leukocyte binding assay (16). All participants were included in our previous study (4), and image data were reanalyzed for the current study.
This study was approved by the Combined Neuroscience Institutional Review Board of the National Institutes of Health Intramural Research Program. All subjects or their surrogate provided written informed consent to participate.
Acquisition of PET and MR Images
PET images were acquired in an Advance PET scanner (GE Healthcare). Before the emission scan, a 68Ge transmission scan was acquired for 8 min for later attenuation correction. After the intravenous injection of 678.2 ± 35.5 MBq of 11C-PBR28 for 1 min, dynamic PET data were acquired for 90 min, and arterial blood was sampled 23 times with time intervals ranging from 0.25 to 15 min during the emission scan. In 27 time frames with increasing scan duration from 0.5 to 5 min, 3-dimensional dynamic PET images were reconstructed with a filtered backprojection algorithm in a 128 × 128 × 35 matrix with 2 × 2 × 4.25 mm voxels. Sampled arterial blood was corrected for metabolite fraction measured by reverse-phase chromatography, and a plasma input function was obtained. The fP of radiotracer was measured by ultrafiltration and normalized to standard plasma (17).
T1-weighted MR images were acquired in a 3-T Achieva scanner (Philips) using a turbo field echo sequence (repetition time, 8.1 ms; echo time, 3.7 ms; flip angle, 8; matrix, 181 × 256 × 256; voxel size, 1 × 0.983 × 0.983 mm).
Image Processing Steps
FreeSurfer 5.1 (Massachusetts General Hospital, Harvard Medical School; http://surfer.nmr.mgh.harvard.edu) was used for creation of participant-specific volume of interest (VOI). In brief, T1-weighted MR images were processed with inhomogeneity correction, skull-stripping, and segmentation into gray and white matter based on the intensity gradient and connectivity of voxels. After tessellation into trigones, 3-dimensional gray and white matter surfaces were created. Cerebral cortex was segmented into smaller cortical areas with the probabilistic labeling algorithm by inflating the white matter surface and overlaying the curvature information on the inflated surface (18,19). Subcortical structures were also segmented using the probabilistic registration technique (20). Composite VOI mask images including 112 cortical and subcortical regions were created. Finally, we created participant-specific composite VOI mask images for 12 cortical (prefrontal, sensorimotor, inferior parietal, superior temporal, middle and inferior temporal, precuneus, anterior cingulate, posterior cingulate, occipital, entorhinal, parahippocampal cortices, and hippocampus) and 3 subcortical regions (striatum, thalamus, and cerebellar cortex) by merging the anatomically related regions.
Statistical parametric mapping 12 (SPM12; Wellcome Department of Cognitive Neurology) and in-house programs implemented in MATLAB R2015b (MathWorks) were used to process the PET images. Except for the first 3 time frames, dynamic PET images were realigned to correct head motion during the scan time. For the VOI analysis, the mean PET images were coregistered to individual T1-weighted MR images, and then all time frames of realigned dynamic PET images were coregistered using the transformation matrix coregistering mean PET to MR images. By overlaying the composite VOI masks, regional time–activity curves were obtained.
For the SVCA procedure, 4 types of tissue mask images (normal gray matter, normal white matter, sinus, and pathologic gray matter) were created in the MR space. To create the white matter mask least affected by the activity from the surrounding gray matter, we first extracted binarized gray matter and cerebrospinal fluid masks from the composite VOI mask. After smoothing these masks using the gaussian kernel with a 7-mm full width at half maximum, we chose the white matter voxels affected by less than 1% of gray matter and cerebrospinal fluid activity and created a white matter mask image in the MR space. Pathologic gray matter masks were created with the voxels for the inferior parietal and middle and inferior temporal cortices in which 11C-PBR28 binding was significantly increased in AD patients (5). Sinus masks were manually drawn on the PET images coregistered to MR images with reference to the first 3 time frames of the dynamic PET images.
SVCA Procedure
We used a modification of an optimized SVCA method with 4 kinetic classes (10). Three kinetic classes (normal gray and white matter and blood) were obtained from 21 controls, and 1 (pathologic gray matter) from the 25 AD patients. First, the means and SD of activities for each time frame were calculated within the whole brain masks. The activities of coregistered dynamic PET images were then standardized by subtracting the means and dividing by the SD. By overlaying the masks for each tissue type on the standardized PET images, standardized time–activity curves for each kinetic class were obtained in each individual. Finally, 4 kinetic classes were established after averaging time–activity curves. Additionally, 2 sets of kinetic classes were separately established for each TSPO genotype.
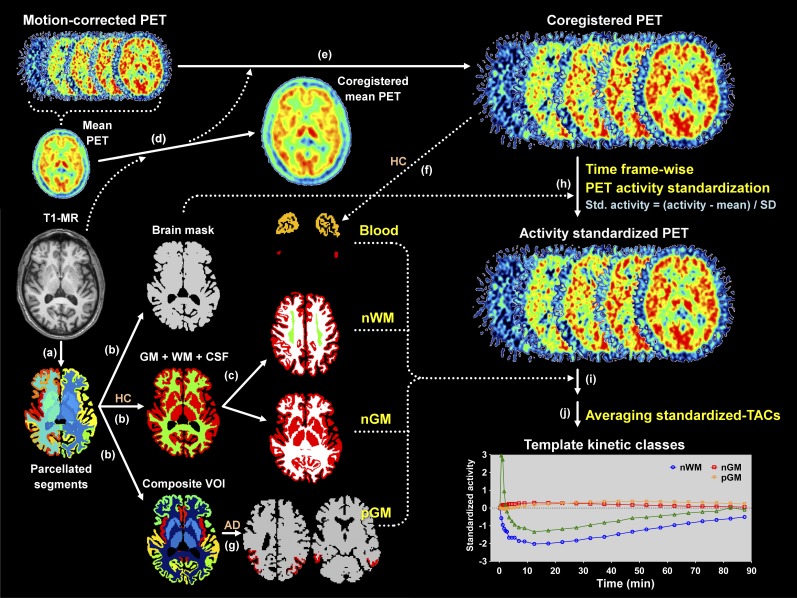
For each voxel in the activity-standardized PET images, the nonnegative least squares algorithm was used to find 4 coefficients for each kinetic class that would minimize the difference between the estimated time–activity curve and the standardized time–activity curve (6). Thereby, each coefficient was mapped. Finally, the SVCA reference time–activity curve was established using the coefficient map for normal gray matter within the gray matter mask to reduce the contamination from the noisy white matter time–activity curve (6,10). The whole procedure is summarized in Figure 1.
FIGURE 1.
Processing steps for acquiring 4 template kinetic classes. (a) By using FreeSurfer, T1-weighted MR images were segmented into 112 regions. (b) After merging regions in parcellated segments, masks for whole brain, gray and white matter, and composite VOI mask images were created. (c) White matter VOI masks least affected by activity of surrounding structures were created for each control. (d and e) PET images were coregistered to T1-MR image using mean PET image. (f) Sinus masks for each control were created. (g) In AD patients, masks for pathologic gray matter (pGM) were created. (h) Activity of PET images was standardized using whole brain masks. (i) Standardized time–activity curves for normal white matter (nWM), normal gray matter (nGM), and blood were obtained from 21 controls. Likewise, standardized time–activity curve for pGM was obtained from 25 AD patients. (j) By averaging standardized time–activity curves (TACs), template time–activity curves for 4 kinetic classes were established. HC = healthy control.
Kinetic Analysis
11C-PBR28 binding values were quantified as distribution volume ratio (DVR) using a Logan reference plot, with k2′ set at 0.13 min−1 (21). The SVCA-derived Logan DVR values were compared with the gold standard of total distribution volume, obtained with a 2-tissue-compartment model, and corrected for plasma-free fraction (VT/fP).
Statistical Analysis
For direct comparison with the pseudo reference region approach, we replicated the same statistical analyses performed by Lyoo et al. (4). SVCA-derived Logan DVR values of 11C-PBR28 were compared among the 3 groups using factorial ANOVA with TSPO genotype as a fixed factor, to correct for affinity differences related to the rs6971 single-nucleotide polymorphism (22). Age was entered as a covariate. Bonferroni adjustment for multiple comparisons was used for comparisons between groups. SPSS (SPSS Inc.) was used for the statistical analysis.
The coefficient of variation was calculated as SD/mean × 100 and used to determine the variability of binding values. The correlation between the binding values and the severity of dementia was explored in 2 stages: first by obtaining the standardized residuals of the Clinical Dementia Rating scale (sum-of-boxes scores and the DVR values in combined middle and inferior temporal cortex after adjusting for age and TSPO genotype) and second by calculating the Pearson correlation between the 2 standardized residuals.
RESULTS
The SVCA-derived DVR values of the present study were greater in AD patients than controls in the inferior parietal, combined middle and inferior temporal, parahippocampal, and entorhinal cortices. Compared with MCI patients, SVCA-DVR values were greater in AD patients in the combined temporal region and in the entorhinal cortex, although the entorhinal cortex did not survive correction for multiple comparisons (Table 1). The results of the present study closely replicated those of Kreisl et al. (5) and are virtually identical to those of Lyoo et al. (4). Indeed, Kreisl et al. found that VT/fp was greater in AD patients than controls in the inferior parietal, combined middle and inferior temporal, and entorhinal cortices, and greater than MCI subjects in the combined middle and inferior temporal and entorhinal cortices (5). With the pseudo reference region approach, Lyoo et al. found an additional significant result in the parahippocampal region (4), the same additional region found in the present study.
TABLE 1.
Level of Statistical Significance by Region with Different Quantification Methods
| Region |
P (AD vs. HC) |
P (AD vs. MCI) |
||||
| VT/fp | SUVR | SVCA-DVR | VT/fp | SUVR | SVCA-DVR | |
| Inferior parietal | 0.028 | <0.0005 | 0.001 | NS | NS | NS |
| Middle and inferior temporal | 0.023 | <0.0005 | <0.0005 | 0.043 | 0.010 | 0.001 |
| Precuneus | NS | 0.048* | NS | NS | NS | NS |
| Entorhinal | 0.048 | 0.009 | 0.001 | 0.048 | NS | 0.025* |
| Parahippocampal | NS | 0.006 | 0.003 | NS | NS | NS |
Did not survive regionwise correction for multiple comparisons.
NS = not significant.
P values were obtained from univariate ANOVA. Diagnosis and genotype were used as fixed factors.
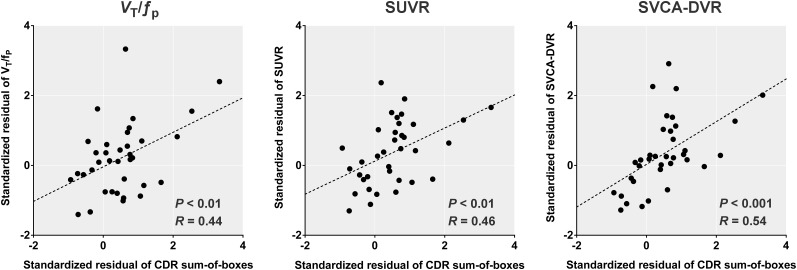
SVCA-DVR values in the combined middle and inferior temporal cortex were positively correlated with the Clinical Dementia Rating sum-of-boxes scores. The level of significance was greater for SVCA-DVR (P < 0.001) than that of either VT/fp (P < 0.01) or SUVR (P < 0.01) (Fig. 2). Notably, simple SUVs did not show any significant group difference and were not significantly correlated to VT/fp values (data not shown).
FIGURE 2.
Correlation between binding values of combined middle and inferior temporal cortex and sum of boxes of clinical dementia rating (CDR) score. SVCA-derived DVR values show correlation similar to, and even slightly stronger than, those with VT/fp and SUVR, as reported by Lyoo et al. (4).
The variability of SVCA-DVR was much lower than that of VT/fp and similar to that of SUVR values (Table 2) (4). Similar to SUVR results, the SVCA-DVR values in mixed-affinity binder (MAB) were paradoxically greater than those of high-affinity binder (HAB) in all diagnostic groups and all regions (mean SVCA-DVR among all regions [HAB vs. MAB]: 1.00 vs. 1.04 in healthy controls, 1.00 vs. 1.07 in MCI patients, and 1.01 vs. 1.08 in AD patients) (Table 3).
TABLE 2.
Coefficients of Variation for VT/fp, SUVR Obtained with Pseudo Reference Region, and Logan-DVR Obtained with SVCA
| Parameter | Coefficient of variation (%) |
||
| VT/fp | SUVR | SVCA-DVR | |
| HAB | 13%–27% | 1%–9% | 2%–7% |
| MAB | 16%–36% | 4%–13% | 3%–11% |
TABLE 3.
Regional 11C-PBR28 Binding Values in Patients with AD, Individuals with MCI, and Healthy Controls, Stratified by TSPO Genotype
| Parameter | Patient | Inferior parietal |
Middle and inferior temporal |
Precuneus |
Occipital |
Hippocampus |
Entorhinal |
Parahippocampal |
| VT (mL·cm−3) | ||||||||
| HAB | AD | 4.7 ± 1.0 | 4.8 ± 1.1 | 4.5 ± 0.9 | 4.3 ± 0.9 | 4.4 ± 1.1 | 4.8 ± 1.1 | 4.0 ± 1.0 |
| MCI | 3.6 ± 1.1 | 3.8 ± 1.0 | 3.8 ± 0.9 | 3.6 ± 1.0 | 3.7 ± 0.9 | 3.8 ± 1.0 | 3.3 ± 0.8 | |
| HC | 4.4 ± 1.1 | 4.3 ± 1.1 | 4.3 ± 1.0 | 4.3 ± 1.0 | 4.3 ± 1.2 | 4.5 ± 1.1 | 4.0 ± 1.1 | |
| MAB | AD | 3.0 ± 1.1 | 3.1 ± 1.1 | 3.0 ± 1.1 | 2.8 ± 0.9 | 2.9 ± 1.1 | 3.2 ± 1.1 | 2.6 ± 0.9 |
| MCI | 3.0 ± 0.6 | 3.0 ± 0.6 | 3.1 ± 0.7 | 2.8 ± 0.5 | 3.0 ± 0.6 | 3.1 ± 0.6 | 2.6 ± 0.5 | |
| HC | 2.6 ± 0.6 | 2.6 ± 0.6 | 2.6 ± 0.6 | 2.6 ± 0.6 | 2.6 ± 0.5 | 2.7 ± 0.7 | 2.3 ± 0.5 | |
| VT/fP (mL·cm−3) | ||||||||
| HAB | AD | 130.9 ± 28.0 | 135.1 ± 26.5 | 125.1 ± 24.6 | 119.9 ± 26.8 | 123.6 ± 25.3 | 134.9 ± 24.1 | 112.9 ± 24.5 |
| MCI | 98.0 ± 20.5 | 102.9 ± 18.5 | 104.8 ± 17.6 | 97.3 ± 17.8 | 100.0 ± 13.1 | 103.9 ± 14.2 | 89.9 ± 14.4 | |
| HC | 104.2 ± 25.2 | 103.7 ± 28.1 | 104.6 ± 28.0 | 103.0 ± 25.9 | 102.8 ± 27.6 | 109.7 ± 29.0 | 94.1 ± 22.3 | |
| MAB | AD | 79.4 ± 25.8 | 81.2 ± 29.2 | 78.8 ± 28.2 | 74.4 ± 22.5 | 76.3 ± 29.5 | 85.7 ± 29.5 | 68.4 ± 24.4 |
| MCI | 68.9 ± 13.7 | 68.7 ± 13.2 | 70.7 ± 13.9 | 66.1 ± 10.3 | 69.1 ± 15.6 | 71.6 ± 17.5 | 60.1 ± 11.5 | |
| HC | 65.9 ± 20.5 | 66.7 ± 19.0 | 65.8 ± 19.3 | 66.9 ± 20.8 | 65.2 ± 17.2 | 69.1 ± 21.6 | 59.3 ± 16.2 | |
| SUVR | ||||||||
| HAB | AD | 1.056 ± 0.041 | 1.081 ± 0.068 | 1.054 ± 0.071 | 1.022 ± 0.043 | 0.996 ± 0.057 | 1.009 ± 0.091 | 0.920 ± 0.056 |
| MCI | 0.939 ± 0.011 | 0.980 ± 0.051 | 1.019 ± 0.072 | 0.948 ± 0.035 | 0.934 ± 0.084 | 0.921 ± 0.063 | 0.853 ± 0.059 | |
| HC | 0.985 ± 0.045 | 0.975 ± 0.071 | 1.006 ± 0.050 | 0.997 ± 0.023 | 0.948 ± 0.024 | 0.946 ± 0.054 | 0.877 ± 0.026 | |
| MAB | AD | 1.117 ± 0.101 | 1.148 ± 0.060 | 1.111 ± 0.115 | 1.068 ± 0.053 | 1.062 ± 0.092 | 1.154 ± 0.060 | 0.993 ± 0.053 |
| MCI | 1.084 ± 0.122 | 1.100 ± 0.104 | 1.121 ± 0.147 | 1.047 ± 0.062 | 1.085 ± 0.075 | 1.118 ± 0.066 | 0.976 ± 0.039 | |
| HC | 1.011 ± 0.061 | 1.046 ± 0.067 | 1.024 ± 0.055 | 1.029 ± 0.053 | 1.027 ± 0.053 | 1.070 ± 0.081 | 0.945 ± 0.058 | |
| SVCA-DVR | ||||||||
| HAB | AD | 1.046 ± 0.037 | 1.060 ± 0.041 | 1.070 ± 0.043 | 1.062 ± 0.042 | 0.980 ± 0.049 | 0.973 ± 0.064 | 0.914 ± 0.052 |
| MCI | 0.981 ± 0.031 | 0.993 ± 0.037 | 1.084 ± 0.053 | 1.009 ± 0.021 | 0.933 ± 0.045 | 0.903 ± 0.037 | 0.864 ± 0.033 | |
| HC | 1.008 ± 0.023 | 0.980 ± 0.042 | 1.061 ± 0.026 | 1.042 ± 0.034 | 0.938 ± 0.048 | 0.909 ± 0.042 | 0.873 ± 0.044 | |
| MAB | AD | 1.112 ± 0.074 | 1.135 ± 0.053 | 1.138 ± 0.099 | 1.099 ± 0.043 | 1.035 ± 0.088 | 1.102 ± 0.065 | 0.982 ± 0.058 |
| MCI | 1.079 ± 0.097 | 1.080 ± 0.082 | 1.151 ± 0.131 | 1.076 ± 0.064 | 1.053 ± 0.081 | 1.044 ± 0.054 | 0.957 ± 0.048 | |
| HC | 1.026 ± 0.026 | 1.041 ± 0.037 | 1.066 ± 0.046 | 1.062 ± 0.029 | 1.011 ± 0.048 | 1.018 ± 0.065 | 0.935 ± 0.044 |
HC = healthy control; VT = total distribution volume; VT/fP = total distribution volume/free fraction of radioligand.
Data are presented as mean ± SD.
DISCUSSION
In this study we showed that a noninvasive SVCA could accurately replicate the quantitative results obtained with arterial sampling in a large clinical study involving healthy controls, MCI patients, and AD patients. SVCA appears to be more sensitive than modeling obtained with arterial sampling, as it identified an additional region (parahippocampal gyrus) that differed between controls and AD patients. This same additional region was found in this same population of subjects also using the pseudo reference region approach (4). Compared with full kinetic modeling, SVCA shares with the pseudo reference region approach the important advantage of not relying on blood data. Indeed, measuring plasma concentrations of radioactivity, separating the parent from its radiometabolites, and measuring the free fraction are complex procedures (1) that introduce an element of variability that is avoided when using DVR or SUVR. For instance, a recent test–retest study of 11C-PBR28 SUVR values in AD yielded low variability and high intraclass correlation coefficients and compared favorably with published values of 11C-PBR28 variability (23).
In our study, the coefficient of variation was 13%–36% for VT/fp and only 2%–11% for DVR (Table 2). However, the obvious advantage of SVCA over SUVR is that a suitable pseudo reference region does not need to be identified beforehand and may not even exist. Thus, this technique can potentially be applied to any population and any disease.
By design, SVCA selects reference tissue voxels that are assumed to be without specific binding (6). Of course, no voxel is actually entirely devoid of TSPO, because this protein is ubiquitously, if heterogeneously, distributed in the brain. For instance, TSPO is highly expressed in the vascular endothelium (24). SVCA was initially validated, and then repeatedly used (7–9), to quantify 11C-(R)-PK11195, a tracer with a low specific binding. Recently, Rizzo et al. analyzed 3 different TSPO tracers (11C-(R)-PK11195, 18F-DPA-714, and 11C-PBR28) and argued that there may be an inverse relationship between binding affinity and suitability for clustering. An increase in affinity would also be associated with a lower tissue contrast. Specifically, tracer affinity would modulate the contrast between tissue kinetics by acting on both parenchymal and vascular binding. High-affinity compounds would display a higher vascular binding, and thus the tissue contrast would be lower and the clustering more difficult (11).
In this study, the tracer with the highest affinity, 11C-PBR28, displayed the highest similarity of tissue kinetics between the gray and white matter classes (as defined by the trigonometric angle between the kinetic vectors of the classes), 18F-DPA-714 had intermediate similarity values, and 11C-(R)-PK11195 had the lowest similarity. SVCA was subsequently validated for 18F-DPA-714 (12), although the nonspecific curves of HABs and MABs were not compared, as we did in the present study. Despite having the highest class similarity, in this study we showed that 11C-PBR28 can be amenable to quantification with SVCA and we were able to accurately replicate the results obtained with standard kinetic modeling.
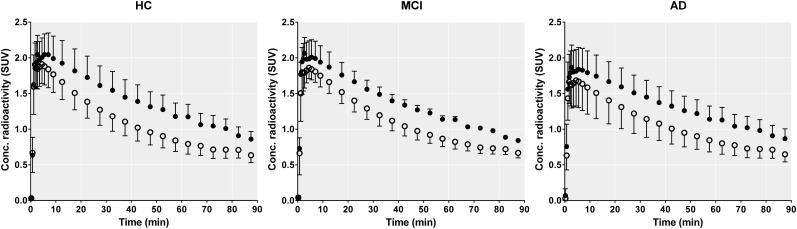
To further probe the suitability of 11C-PBR28 for cluster analysis, we compared the nonspecific curves in HABs and MABs. If SVCA could perfectly extract the nonspecific curve from 11C-PBR28 scans, this curve would have the same amplitude in both populations, since the TSPO polymorphism affects only the specific component.
However, as shown in Figure 3, the nonspecific curves of HABs are significantly higher than those of MABs. This is of course due to the contamination of the reference curves by the different amount of specific binding. Notably, the difference between HABs and MABs is evident even in the healthy brain, where the level of TSPO expression is low (25). This could be explained by the presence of TSPO in cells other than microglia or astrocytes, in particular endothelial cells (24,25). Recently, Matheson et al. warned against hasty implementation of reference models and ratio methods (26), as both SUVR and DVR showed little to no association with 11C-PBR28-VT. Specifically, they showed that almost all variability between brain regions can be attributed to a single underlying dimension of variance, and the ratio between a target region and a highly correlated reference region leaves minimal residual differences between individuals (26). Although they used only a small number of healthy subjects, whose restricted range of VT values may have affected the correlation, and they did not correct the VT values for the plasma free fraction, we do share their concern. A truly nonspecific reference curve should not be affected by genotype, but it did affect our SVCA-derived analyses, strongly suggesting that the analysis is biased by specific binding contaminating the reference curve. Despite this bias, the current results with SVCA-DVR perfectly replicated those from a pseudo reference region (4). Both methods showed apparently greater sensitivity than the gold standard method using VT/fP presumably because of the decreased variance in these 2 bloodless methods compared with VT/fP.
FIGURE 3.
Comparison of SVCA-extracted reference curves between HAB (black dots) and MAB (white dots). Errors bars are SD. In all populations, area under curve was significantly higher for HAB (all P < 0.005), which suggests that SVCA reference curve is not completely free of specific uptake and amount of this uptake depends on genotype status. HC = healthy control.
In summary, using a noninvasive SVCA algorithm, we were able to accurately replicate the findings of a clinical protocol on a large population of healthy controls, MCI patients, and AD patients. In addition, we were able to identify an additional significant region, likely because SVCA analyses do not require delicate and error-prone blood analyses and thus are more sensitive. Although the current results with SVCA-DVR perfectly replicated those from a pseudo reference region, the extracted reference curves were biased to some extent because they are not entirely free of specific binding. The large population and effect size of this study may have facilitated its replication, despite the bias present in the extracted reference curves. It would be plausible that in a study with less statistical power (e.g., fewer patients) a biased SVCA would produce falsely negative results. Therefore, the impact of this bias should be investigated thoroughly before envisioning a widespread application of this technique.
CONCLUSION
SVCA allows the noninvasive quantification of inflammatory biomarker translocator protein measured with 11C-PBR28 but without the need of arterial sampling. Although the reference curves were contaminated with specific binding, the decreased variance of the outcome measure, SVCA DVR, allowed for an apparent greater sensitivity to detect regional abnormalities in brains of patients with AD.
DISCLOSURE
Dr. Lyoo was financially supported by the Basic Science Research Program through the National Research Foundation of Korea grant funded by the Ministry of Science, ICT & Future Planning (2017R1A2B2006694). No other potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Tonietto M, Rizzo G, Veronese M, et al. Plasma radiometabolite correction in dynamic PET studies: Insights on the available modeling approaches. J Cereb Blood Flow Metab. 2016;36:326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanotti-Fregonara P, Chen K, Liow JS, Fujita M, Innis RB. Image-derived input function for brain PET studies: many challenges and few opportunities. J Cereb Blood Flow Metab. 2011;31:1986–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schain M, Zanderigo F, Ogden RT, Kreisl WC. Non-invasive estimation of [11C]PBR28 binding potential. Neuroimage. 2018;169:278–285. [DOI] [PubMed] [Google Scholar]
- 4.Lyoo CH, Ikawa M, Liow JS, et al. Cerebellum can serve as a pseudo-reference region in Alzheimer Disease to detect neuroinflammation measured with PET radioligand binding to translocator protein. J Nucl Med. 2015;56:701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreisl WC, Lyoo CH, McGwier M, et al. In vivo radioligand binding to translocator protein correlates with severity of Alzheimer’s disease. Brain. 2013;136:2228–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turkheimer FE, Edison P, Pavese N, et al. Reference and target region modeling of [11C]-(R)-PK11195 brain studies. J Nucl Med. 2007;48:158–167. [PubMed] [Google Scholar]
- 7.van der Doef TF, de Witte LD, Sutterland AL, et al. In vivo (R)-[11C]PK11195 PET imaging of 18kDa translocator protein in recent onset psychosis. NPJ Schizophr. 2016;2:16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuitemaker A, Kropholler MA, Boellaard R, et al. Microglial activation in Alzheimer’s disease: an (R)-[11C]PK11195 positron emission tomography study. Neurobiol Aging. 2013;34:128–136. [DOI] [PubMed] [Google Scholar]
- 9.Schuitemaker A, van der Doef TF, Boellaard R, et al. Microglial activation in healthy aging. Neurobiol Aging. 2012;33:1067–1072. [DOI] [PubMed] [Google Scholar]
- 10.Yaqub M, van Berckel BN, Schuitemaker A, et al. Optimization of supervised cluster analysis for extracting reference tissue input curves in (R)-[11C]PK11195 brain PET studies. J Cereb Blood Flow Metab. 2012;32:1600–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzo G, Veronese M, Tonietto M, et al. Generalization of endothelial modelling of TSPO PET imaging: considerations on tracer affinities. J Cereb Blood Flow Metab. January 1, 2017. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Lorenzo D, Lavisse S, Leroy C, et al. Validation of an automatic reference region extraction for the quantification of [18F]DPA-714 in dynamic brain PET studies. J Cereb Blood Flow Metab. 2018;38:333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ory D, Celen S, Verbruggen A, Bormans G. PET radioligands for in vivo visualization of neuroinflammation. Curr Pharm Des. 2014;20:5897–5913. [DOI] [PubMed] [Google Scholar]
- 14.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreisl WC, Jenko KJ, Hines CS, et al. A genetic polymorphism for translocator protein 18 kDa affects both in vitro and in vivo radioligand binding in human brain to this putative biomarker of neuroinflammation. J Cereb Blood Flow Metab. 2013;33:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abi-Dargham A, Gandelman M, Zoghbi SS, et al. Reproducibility of SPECT measurement of benzodiazepine receptors in human brain with iodine-123-iomazenil. J Nucl Med. 1995;36:167–175. [PubMed] [Google Scholar]
- 18.Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. [DOI] [PubMed] [Google Scholar]
- 19.Fischl B, van der Kouwe A, Destrieux C, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. [DOI] [PubMed] [Google Scholar]
- 20.Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. [DOI] [PubMed] [Google Scholar]
- 21.Fujita M, Imaizumi M, Zoghbi SS, et al. Kinetic analysis in healthy humans of a novel positron emission tomography radioligand to image the peripheral benzodiazepine receptor, a potential biomarker for inflammation. Neuroimage. 2008;40:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen DR, Yeo AJ, Gunn RN, et al. An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J Cereb Blood Flow Metab. 2012;32:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair A, Veronese M, Xu X, et al. Test-retest analysis of a non-invasive method of quantifying [11C]-PBR28 binding in Alzheimer’s disease. EJNMMI Res. 2016;6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizzo G, Veronese M, Tonietto M, Zanotti-Fregonara P, Turkheimer FE, Bertoldo A. Kinetic modeling without accounting for the vascular component impairs the quantification of [11C]PBR28 brain PET data. J Cereb Blood Flow Metab. 2014;34:1060–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betlazar C, Harrison-Brown M, Middleton RJ, Banati R, Liu GJ. Cellular sources and regional variations in the expression of the neuroinflammatory marker translocator protein (TSPO) in the normal brain. Int J Mol Sci. 2018;19:E2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matheson GJ, Plaven-Sigray P, Forsberg A, Varrone A, Farde L, Cervenka S. Assessment of simplified ratio-based approaches for quantification of PET [11C]PBR28 data. EJNMMI Res. 2017;7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]