ABSTRACT
This study analyzed the microbiological quality of drinking and source water from three First Nations communities in Manitoba, Canada that vary with respect to the source, storage and distribution of drinking water. Community A relies on an aquifer and Community B on a lake as source water to their water treatment plants. Community C does not have a water treatment plant and uses well water. Quantification of free residual chlorine and fecal bacterial (E. coli and coliforms), as well as detection of antibiotic resistance genes (sul, ampC, tet(A), mecA, vanA, blaSHV, blaTEM, blaCTX-M, blaOXA-1, blaCYM-2, blaKPC, blaOXA-48, blaNDM, blaVIM, blaGES and blaIMP) was carried out. While water treatment plants were found to be working properly, as post-treatment water did not contain E. coli or coliforms, once water entered the distribution system, a decline in the chlorine concentration with a concomitant increase in bacterial counts was observed. In particular, water samples from cisterns not only contained high number of E. coli and coliforms, but were also found to contain antibiotic resistance genes. This work shows that proper maintenance of the distribution and storage systems in First Nations communities is essential in order to provide access to clean and safe drinking water.
Keywords: E. coli, total coliforms, antibiotic resistance genes, drinking water, First Nations
Maintaining proper water distribution system is essential to ensure safe drinking water to communities.
INTRODUCTION
Access to clean drinking water is one of the most important determinants of human health (World Health Organization 2011). Consumption of polluted drinking water can result in waterborne infectious diseases, such as gastroenteritis, which has been shown to affect thousands of people, and sometimes lead to death (Mac Kenzie et al. 1994; Leclerc, Schwartzbrod and Dei-Cas 2002). In urban areas of Canada, people have access to clean and plentiful drinking water because of advanced water treatment technologies, sufficient water operator knowledge and access to land management decision-making. However, many residents living on First Nation reserves lack drinking water security, in part because of unacceptable levels of fecal bacteria in treated water and drinking water supplies (Patrick 2011).
Chlorination inactivates bacteria and is commonly used to treat water (Cutler and Miller 2005). In order to prevent bacterial regrowth when water is distributed from the treatment plant to the homes, the free residual chlorine concentration in the water distribution systems must remain ≥0.2 mg/L (LeChevallier, Welch and Smith 1996). The U.S. Environmental Protection Agency (EPA) Surface Water Treatment Rule requires a minimum disinfectant residual of 0.2 mg/L for water entering the distribution system (EPA 2002). Free chlorine concentrations in most Canadian drinking water distribution systems range from 0.04 to 2.0 mg/L (Health Canada 2017). In a recent study from a fly-in community in Manitoba, water samples obtained from the water truck and in homes with cisterns all showed free residual chlorine concentrations <0.2 mg/L and unacceptable levels of E. coli (1 to 2100 CFUs/100 mL) (Farenhorst et al. 2017). In this community that relies on a lake as their source water to the water treatment plant (WTP), the water in the cisterns also contained antibiotic resistance genes (ARGs) (Fernando et al. 2016).
ARGs are present in low amounts in various natural environments (Martínez 2008). While human activity is an important selection pressure, some bacterial species are intrinsically resistant to antibiotics as seen in instances where ARGs have been reported in environments not impacted by human activity (D'Costa et al. 2006; Bhullar et al. 2012). Surface waters such as lakes and rivers can be an important reservoirs of ARGs (Marti, Variatza and Balcazar 2014). ARGs can enter surface waters through point-sources such as the discharge of municipal and aquacultural wastes, and through non-point sources such as urban and agricultural runoff (Marti, Variatza and Balcazar 2014). Because lake and river water is used as source water for drinking WTPs, ARGs can enter drinking water distribution systems when WTP does not remove them completely (Guo et al. 2014; Su et al. 2018).
Given that we detected fecal bacteria and ARGs in the tap water of cistern homes in a fly-in First Nation community in Manitoba, Canada (Fernando et al. 2016; Farenhorst et al. 2017), the purpose of this study was to examine such water quality parameters in a broader range of First Nation households and communities in Manitoba, Canada.
MATERIALS AND METHODS
Water distribution systems and sample collection
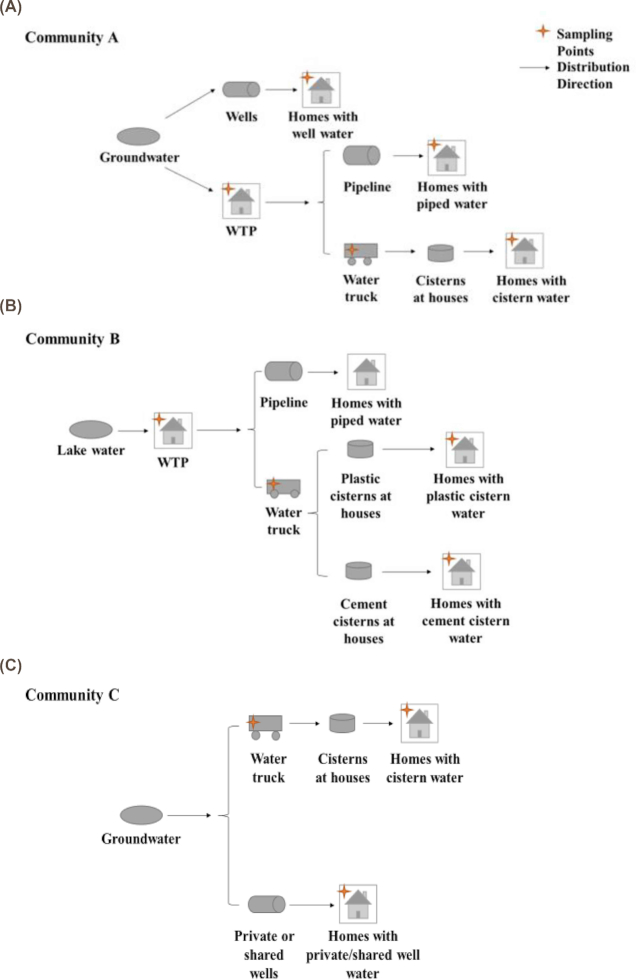
A total of 92 water samples were collected across the water distribution systems of three First Nation reserves (Table 1) with two sampling rounds per community. These communities are distinct from our previously published work (Fernando et al. 2016). All samples were collected between July and November 2016. The sampling points along the drinking water distribution system in each community are illustrated in Figure 1. Community A utilizes an aquifer for its source water that is processed through a membrane reverse-osmosis water treatment system and disinfected using chlorine. There are more than 300 homes in community A with the majority of homes receiving well water, and the remaining homes using piped water or aboveground polyethylene cisterns stored in insulated shelters. Community B utilizes lake water that is processed through a conventional water treatment system equipped with a reverse osmosis system and disinfected using chlorine. There are more than 200 homes in community B with about 30% of the homes receiving piped water and 70% of the homes using underground cisterns made of concrete or polyethylene. In both communities A and B, a water truck is filled at the WTP to deliver water to the cisterns. Community C has more than 100 homes and no drinking WTP. Most homes (66%) have individual wells (not chlorinated), while 28% of homes use public wells (2–3 households per well, not chlorinated) and 6% of homes use underground cisterns made of concrete or polyethylene. The wells in Community C are drilled wells. The water truck is filled up at a public well and the water is chlorinated in the truck prior to filling the cistern at homes. A typical household cistern in these communities holds 5500–7500 litres of water.
Table 1.
The description of water samples collected in this study.
| Community | Sample size and description |
|---|---|
| Community A | A-WTP: finished water at WTP |
| A-Tr1 and A-Tr2: water samples collected from water truck 1 and water truck 2 | |
| A-P1 to A-P6: water samples collected from six pipeline homes, A-P1 to A-P6 are listed in the order of closest to further away from the WTP | |
| A-C1 to A-C5: water samples collected from five cistern (aboveground) homes | |
| A-W1 to A-W6: water samples collected from six well homes | |
| Community B | B-WTP: finished water at WTP |
| B-Tr water: water samples collected from water truck 1 and water truck 2 | |
| B-P1 to B-P6: water samples collected from six pipeline homes, B-P1 to B-P6 are listed in the order of closest to further away from the WTP | |
| B-C1 to B-C7: water samples collected from seven cistern (underground) homes | |
| Community C | C-Tr: water samples collected from the water truck |
| C-SW1 to C-SW5: water samples collected from five shared well water | |
| C-PW1 to C-PW5: water samples collected from five private well water | |
| C-C1 to C-C5: water samples collected from five cistern (underground) homes |
Figure 1.
The drinking water distribution systems in community A (A), community B (B) and community C (C). The black star represents the sampling location in each community.
Sampling was done following standard method SM 9060A for sample bottle pre-treatment and SM 9060B for sample preservation and storage as described (Rice et al. 2012). Water samples were transported in coolers with icepacks to the University of Manitoba on the same day as the samples were collected and immediately processed upon arrival for bacterial count. DNA extraction was carried out the following day. Communities were located between 200 and 400 km from the University of Manitoba each sampling round took 1 or 2 days. During sample collection, a Hatch Chlorine Pocket Colorimeter II (VWR, Mississauga, ON, Canada) was used to determine free residual chlorine according to the adapted USEPA DPD Method 8021 (Hach 2002).
E. coli and coliform quantification
Samples were processed in duplicate following SM 9222 for standard membrane filter procedure (Rice et al. 2012). Briefly, 100 mL of water sample or negative control (autoclaved water) was filtered through sterile polyethersulfone membranes (0.45-µm pore size, 47-nm diameter; Pall Corporation, Mississauga, ON, Canada). Filter papers were then placed on agar plates containing Brilliance E. coli/coliform medium (Fisher Scientific, Ottawa, ON, Canada) and incubated at 37°C for 24 h. Some samples required dilution because of high bacterial counts. Brilliance agar can distinguish E. coli and coliforms from other bacteria based on β-D-glucuronidase production. E. coli colonies appear purple E. coli colonies; whereas coliforms result in pink colonies (Wohlsen 2011). E. coli counts in water samples were calculated as CFU/100 mL = number of purple colonies/volume of filtered sample (100 mL) × dilution factor. Total coliform bacteria count in water were calculated as CFU/mL = number of purple + pink colonies/volume of filtered sample (100 mL) × dilution factor.
DNA extraction
DNA extraction was conducted for samples collected in the second sampling round. For DNA extraction, 300–500 mL of water sample was filtered through sterile polyethersulfone membranes (0.22-µm pore size; 47-mm diameter; Pall Corporation, Mississauga, ON, Canada). DNA was extracted by using the DNeasy PowerWater Kit (QIAGEN, Germantown, MD, USA) following the manufacturer's instructions. An unused filter membrane was used as the extraction control. DNA concentrations were quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and the quality of DNA was tested on a 1% agarose gel.
ARGs quantification
Detection of ARGs was carried out for water samples from the second sampling round. Absolute quantitative PCR (qPCR) was used to quantify the abundance of seven different ARGs. ampC, vanA, tet(A), mecA, sul1, sul2 and sul3. The ampC, vanA, tet(A) and mecA genes were quantified for all water samples and sul1, sul2 and sul3 genes were only quantified for cistern samples in community B (we were unable to carry out the detection for sul genes communities A and C due to the lack of adequate DNA). All primers used in this study, except sul primers, have been standardized previously (Fernando et al. 2016). Prior to their use, the efficiency of sul primers was determined using methods previously described (Fernando, Zhanel and Kumar 2013). Using positive control samples, we were able to detect as low as 2 copies/μL of target DNA for each of the reactions. The primers are listed in Table 1 (Supporting Information). Bacterial strains used as positive controls have been described previously (Fernando et al. 2016). Primers were designed using Custom Primers—OligoPerfect™ Designer at ThermoFisher Scientific (parameters chosen for primers were: 18–22 bp for primer length; 55°C to 65°C for annealing temperature; 30% to 40% for GC content; ∼ 120 bp for amplification length). The qPCR reaction was conducted on the StepOnePlus real-time PCR system (Life Technologies Inc., Burlington, ON, Canada) following a previously described method (Alexander et al. 2015; Fernando et al. 2016). Reactions were carried out in triplicate. The calculation was carried out as follows: Copy No./ng DNA/100 mL = (10(Ct-Y_inter)/slope)/1 ng/µL of DNA/(Vfilteration/100 mL).
Multiplex PCR detection of β-lactamase and carbapenemase genes
Multiplex PCR was carried out using the method described previously (Fernando et al. 2016). Primers used for PCR detection are listed in Table 1 (Supporting Information). The presence of five different β-lactamase genes: blaSHV, blaTEM, blaCTX-M, blaOXA-1 and blaCMY-2 were determined using a multiplex PCR for cistern samples in community B. The specific primers are listed in Table 1 (Supporting Information). The PCR reaction was conducted using Q5 high-fidelity DNA polymerase (New England Biolabs, Whitby, ON, Canada) with final concentrations of the primers were 0.27 µM. The run parameters for amplification cycle were: 1 cycle of initial denaturation at 95°C for 15 min, 40 cycles of denaturation at 94°C for 30 s, annealing at 63.5°C for 90 s, extension at 72°C for 90 s and final extension at 72°C for 7 min.
Four different carbapenemase-encoding genes, blaKPC, blaNDM, blaGES-5 and blaOXA-48, were determined using multiplex PCRs for cistern samples in community B. The blaIMP/blaVIM detections were carried out in separate reactions.
Statistical analysis
Analysis of variance (ANOVA) was performed using GLIMMIX in SAS 9.4. (SAS Institute Inc., Cary, NC, USA) to test for the effect of the type of water distribution on free residual chlorine concentrations in tap water of homes (i.e. piped versus cisterns versus wells for community A; piped versus cisterns for community B; cisterns versus private wells versus shared wells for community C). Distribution of free chlorine concentrations was considered as log normal. The Tukey multiple comparison procedure was used to compare least square means for communities A and C. The multiple comparison procedure was set as default to compare least square means for community B. For all statistical analyses, significance was determined at α = 0.05.
RESULTS
Chlorine concentrations and fecal bacteria counts
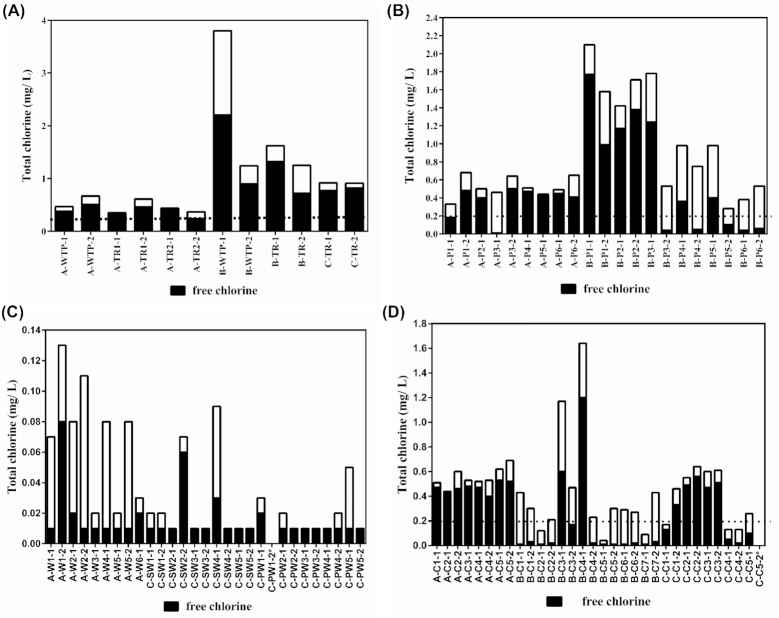
For communities A and B (Fig. 2a), the treated water at the WTP had free residual chlorine concentrations within the 0.4 to 2.0 mg/L range typically reported in Canada (Health Canada 2009). For community A, homes with piped (Fig. 2b) and cistern (Fig. 2c) water always had free residual chlorine concentrations >0.2 mg/L, except for A-P1-1 and A-P3-1 (Fig. 2b). Well water is typically not chlorinated and thus tap water in homes with well water had free residual chlorine concentrations <0.2 mg/L (Fig. 2d). For community A, free residual chlorine concentration in tap water of homes with wells was always significantly less than in tap water of pipeline homes (Table 2, Supporting Information). For both sampling rounds in community B, homes with cisterns almost always had free residual chlorine concentrations of <0.2 mg/L (Fig. 2c). In contrast, only pipeline homes located further away from the WTP showed such low concentrations when (Fig. 2b). Thus, homes with cisterns showed significantly less free residual chlorine concentration than homes with piped water (Table 2, Supporting Information). In community C, the tap water in some homes with cisterns had free residual chlorine concentrations <0.2 mg/L (Fig. 2c). However, free residual chlorine concentration in tap water of cistern homes was significantly greater than in tap water of homes with wells. This is not unexpected, since well water was not chlorinated.
Figure 2.
Total (black + white bar) and free residual (black bar) chlorine concentrations in water samples collected from communities A and B from the WTP and water trucks (A); communities A and B from the pipelines (B); water samples collected in communities A and C from the wells (C) and communities A, B and C from the cisterns (D). Dashed line is the minimum recommended free residual chlorine concentration. Description of the samples is provided in Table 1.
No E. coli/coliforms bacteria were detected in any of the water samples collected in communities A or C, except for C-C4 that tested positive for total coliforms and E. coli during both sampling rounds (Fig. 3). Not surprisingly, this cistern sample from community C also had the lowest free residual chlorine concentrations among the cistern homes (Fig. 2c). In community B, no E. coli/coliforms bacteria were detected in water samples from the treatment plant, water trucks and homes with piped water. However, six out of seven homes with cisterns tested positive for total coliforms and E. coli in both sampling rounds (Fig. 3). There was a large variation across the six homes in the counts of E. coli/coliforms bacteria, for example E. coli and total coliforms were particularly abundant in samples B-C4 and B-C6 and relatively small in B-C7 and C-C4.
Figure 3.
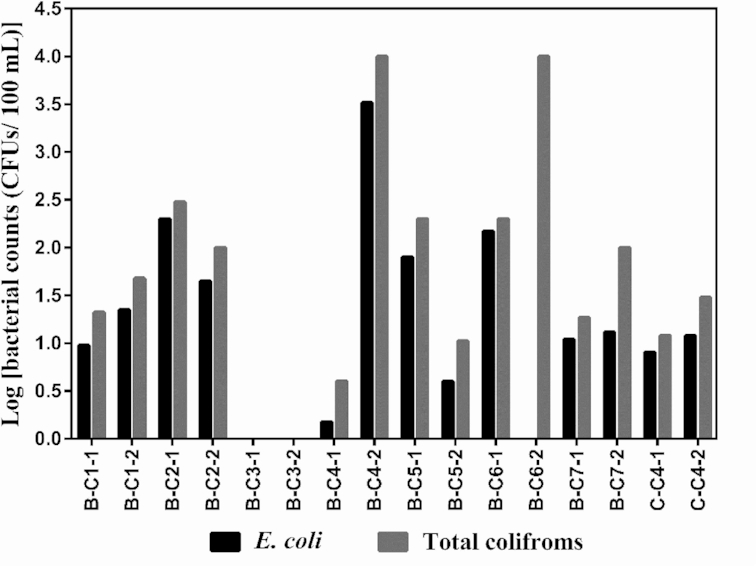
The CFUs of E. coli (black) and total coliforms (grey) detected in cistern samples from communities B and C. Description of the samples is provided in Table 1.
Detection of ARGs
No ARGs were detected in any of the water samples collected in communities A or C, except for A-W1 in community A that tested positive by qPCR for the ampC gene (Fig. 4). In community B, no ARGs were detected in water samples from the treatment plant, water trucks and piped water. However, water samples from homes with cistern were observed positive for several ARGs: the sul1gene was detected in B-C5 and B-C6 (Fig. 4). The sample B-C5 was also positive for the mecA gene (Fig. 4). The genes, sul2, sul3, ampC and vanA, were not detected in any water samples in community B.
Figure 4.
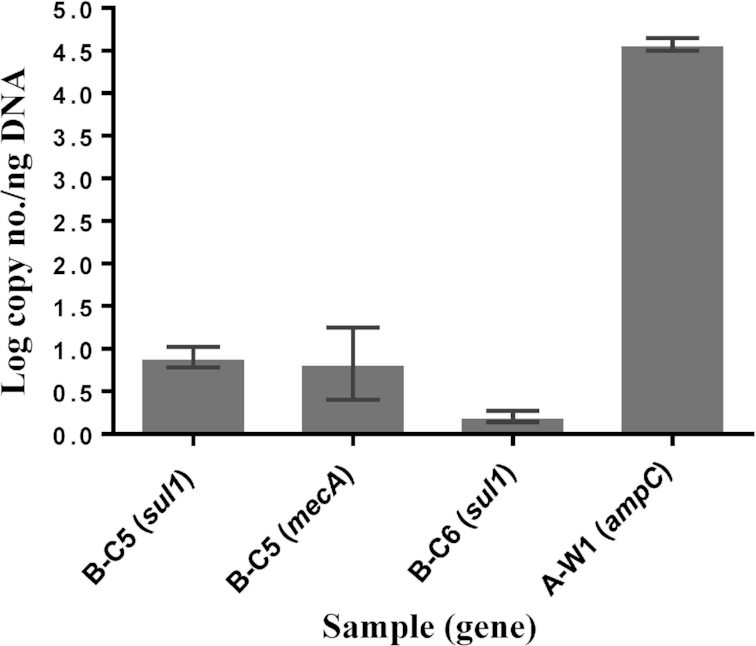
Quantification of ARGs in water samples from communities A and B. No ARGs were detected in water samples from community C. Log copy number/ng of total DNA was determined using using a standard curve created using control bacterial strains. Description of the samples is provided in Table 1.
Among β-lactamase genes tested, blaSHV gene was detected in three cistern samples (B-C4, B-C5 and B-C7) (Table 2). Both blaCTX-M and blaOXA-1 gene were detected in two cistern water samples: they were B-C2 and B-C6, and B-C3 and B-C5, respectively. The blaTEM gene was found in one cistern (B-C6). blaCYM-2 was not detected in water samples in community B. As for the carbapenemase genes, blaKPC and blaNDM were the most frequently detected genes: both were detected in five cistern water samples (B-C3, B-C4, B-C5, B-C6 and B-C7). blaOXA-48 was detected in three cistern water samples (B-C2, B-C4 and B-C5). blaVIM, blaGES and blaIMP gene were not detected in water samples in community B.
Table 2.
The detection of ARGs in cistern water samples in community B. blaSHV, blaTEM, blaCTX-M, blaOXA-1, blaCYM-2, blaKPC, blaOXA-48, blaNDM and blaGES were detected by multiplex PCR. blaVIM and blaIMP were detected by separate PCRs. Shaded boxes represent the presence of genes.
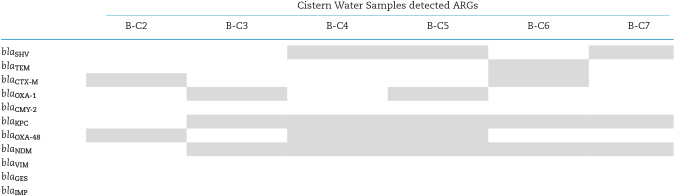
|
In summary, the sample B-C5 was positive for 7 ARGs, followed by B-C6 in which 5 ARGs were detected. The sample B-C4 was positive for four ARGs (Table 2). Both B-C3 and B-C7 were positive for three ARGs. The sample B-C2 was positive for two ARGs. B-C1 was the only cistern water sample in community B for which no ARGs were detected.
DISCUSSION
Chlorine concentrations and E. coli/coliforms counts
The objective of this study was to investigate the microbiological quality of drinking water in three First Nation reserves with community A relying on an aquifer and community B relying on a lake as source water to their WTPs, and community C having no WTP and relying on well water from an aquifer for further water distribution. These types of water distribution systems are representative of what is typical First Nation reserves in Manitoba and other provinces in Canada. Treated water at the waste WTP in communities A and B contained adequate levels of free residual chlorine and was free of fecal bacteria, suggesting that the water treatment process in both communities is functioning well. For the pipeline water in communities A and B, samples were always negative for fecal bacteria although some of water samples had free residual chlorine concentrations less than the minimum recommended level, particularly pipeline homes that were furthest away from the waste WTP in community B. The relatively low free residual chlorine concentration in pipeline water further away from the WTP could result from the decay of chlorine which is affected by a) water quality parameters, such as dissolved organic carbon concentrations and inorganic substances (Warton et al. 2006), and b) pipeline system parameters, such as pipe age, pressure variation and flow conditions (LeChevallier, Cawthon and Lee 1988). In large water distribution systems, maintaining the appropriate level so free residual chlorine is often not possible without the process of re-chlorination within the distribution system (https://www.ontario.ca/page/procedure-disinfection-drinking-water-ontario#section-4). None of the First Nation communities in this study have the ability to carry out re-chlorination within their distribution systems, which at least partly explains the relatively low free residual chlorine levels in households further from the WTP. However, since these communities are quite small in size, setting up re-chlorination stations may not be an option. The results from both communities suggest that the drinking water distribution system of WTP and the pipeline provides safe drinking water (free of fecal bacterial contamination) in First Nation reserves. For well water in communities A and C, no fecal bacteria were detected in any sample, despite water not being chlorinated. This observation indicates that groundwater wells can provide water free of fecal bacteria, however, since the deep well water can contain high levels of salts and minerals (Kundu, Mandal and Hazra 2009; Edmunds, Ahmed and Whitehead 2015), safety of the water needs to be further evaluated.
No E. coli and total coliforms were detected in the tap water of homes that had aboveground polyethylene cisterns stored in insulated shelters (all located in community A). Fecal contamination in water was detected in homes with underground cisterns, only in one home in community C (this home has a cracked underground concrete cistern) but in almost all of the homes samples in community B. In community B, underground cisterns are constructed of either concrete or polyethylene materials) and fecal bacterial levels in tap water of cistern homes ranged from 0 to 3000 CFUs/100 mL for E. coli and from 0 to 10 000 CFUs/100 mL for total coliforms (Fig. 3). This relatively large variation across homes might be due to two reasons. First, some cisterns have been used for a longer time and were cracked. For example, higher fecal bacterial counts were detected in samples B-C6 than B-C5. The cistern in home B-C6 had been used for 27 years and this house is on the top priority list to have its cistern replaced because it shows signs of cracking. The cistern in home B-C5 had been used for only 4 years and cracking was not observed. Second, the timing of cistern cleaning varied. For instance, high fecal bacterial counts were detected in the sample B-C4, while no bacteria were detected in the sample B-C3. Cistern B-C4 had not been cleaned in the year of sampling (2016), but cistern B-C3 had been cleaned in July just prior to the sampling.
In this study, the presence of fecal bacteria was detected but the source of contamination remains unclear. However, our study did observe high number of fecal bacteria in samples from homes that had concrete underground cisterns samples, compared to those with above ground cisterns. Underground concrete cisterns have an increased risk of drinking water becoming contaminated when cracked, as well as that the concrete rough surface favors the formation of biofilms (Characklis, McFeters and Marshall 1990). Overall, the results underscore the importance of regular cleaning of cisterns to reduce the microbial contamination. Results also suggest that aboveground polyethylene cisterns may offer a safer option for water storage than underground concrete cisterns.
Detection of ARGs
The presence of ARGs was investigated by using qPCR and multiplex PCR methods. For qPCR method, seven ARGs: sul1, sul2, sul3 (sulfonamides resistance), tet(A) (tetracycline resistance), ampC (β-lactam resistance), mecA (methicillin resistance) and vanA (vancomycin resistance) were examined for water samples. The genes sul2, sul3, tet(A) and vanA were not detected in any water samples across the three communities. This is despite them being commonly detected in various environments (Suhartono, Savin and Gbur 2016; Young et al. 2016; Zhang, Lin and Yu 2016). Three genes, ampC, mecA and sul1 were detected in a few water samples (Fig. 4). Three of these samples came from cisterns, and one from the well. The ampC gene belongs to β-lactamase group, whose corresponding antibiotics, β-lactam, are commonly used in the treatment of infections in both humans and animals (van Hoek et al. 2011; Roberts, Schwarz and Aarts 2012). The mecA gene is an indication of the presence of methicillin-resistant Staphylococcus aureus, although it was also reported in non-staphylococcal pathogens (Kassem, Esseili and Sigler 2008). Sulfonamides are one of the earliest antibiotics introduced in the world and widely used for treating human infections (van Hoek et al. 2011; Roberts, Schwarz and Aarts 2012). These genes had been commonly detected in various environments such as drinking water, surface water and wastewater (Schwartz et al. 2003; Alexander et al. 2015; Suhartono, Savin and Gbur 2016). However, even though only a fraction of the samples tested via qPCR were positive for the ARGs, the numbers detected are alarmingly high. For example ampC numbers detected in the sample A-W1 is equivalent to those reported in the influent of wasteWTPs (Alexander et al. 2015). Further, compared to our previous study (Fernando et al. 2016), the numbers observed in this study are up to 1000-fold higher.
For multiplex PCR method, five β-lactamase genes and six carbapenemase genes were detected in cistern samples from community B (Table 2). β-lactamase and carbapenemase genes have been detected in many aquatic environments around the world. For example, blaOXA-1 and blaTEM-1 genes were detected in a WTP in China (Zhang, Lin and Yu 2016). The blaCTX-M gene was detected in water samples collected from an Indian river (Bajaj et al. 2015), and the OXA-48 gene was found in river water in Algeria (Tafoukt et al. 2017). In this study, blaSHV, blaCTX-M, blaOXA-1, blaKPC, blaNDM and blaOXA-48 were the more frequently detected genes in cistern water samples (at least in two out of seven water samples) (Table 2). Among carbapenemase genes, blaKPC and blaNDM, were most common our samples, whereas in our previous study (Fernando et al. 2016), these two genes were detected at a much lower frequency (blaNDM in 2/27 and blaKPC in 3/27 samples). Further, in the same study we did not detect observe blaOXA-1 in any of the samples, whereas in the present study, we detected blaOXA-1 in two of the samples.β-lactams and carbapenems are commonly used to treat infections in human and animals (van Hoek et al. 2011; Roberts, Schwarz and Aarts 2012), with carbapenems often being used as the last resort for treating antibiotic-resistant infections. Although, ARGs have been shown to be present in various environmental bacterial species (D'Costa et al. 2006), their presence in pathogenic bacteria can pose a significant risk to the health of individuals exposed to such bacteria (Yong et al. 2009; Robledo, Aquino and Vazquez 2011; Ben-David et al. 2012). Therefore, even though the number of samples that tested positive for carbapenemase and β-lactamase is much lower than our previous study (Fernando et al. 2016), the prominence of blaNDM and blaKPC is concerning. Hence the detection of ARGs in cistern water in community B could further suggest that the water in these homes is unsafe to drink and that steps must be taken to replace the underground cisterns with alternate options for supplying safe drinking water to homes. It is worth mentioning that in multiplex PCR experiment, the presence of ARGs was detected but not their expression.
Overall, based on the results of the cistern samples in community B, the connection between the detection of fecal bacteria and the detection of ARGs is not clear. For example, the water from B-C4 and B-C6 had higher bacterial counts than B-C5, but more ARGs were detected in B-C5 than B-C4 and B-C6. Similarly, B-C3 was negative for fecal bacteria but positive for ARGs. This observation indicates that ARGs may be present in non-coliform organisms.
Both this study and the one by Fernando et al. (Fernando et al. 2016) detected ampC, mecA, β-lactamase genes and carbapenemase genes in drinking water of taps in homes, suggesting that the presence of ARGs in drinking water sources of First Nation reserves might not be an isolated case. Although ampC, mecA, sul1, β-lactamase genes and carbapenemase genes were detected, the selection pressure for the presence of these genes in organisms is unclear because of the absence of public data on the usage of antibiotics in different geographical regions of Canada.
CONCLUSIONS
This study investigated drinking water quality in three First Nation communities in Manitoba, Canada by determining free residual chlorine concentrations, and screening for the presence of E. coli and total coliforms, as well as ARGs. Regardless of water source to the WTP as ground or surface water, our findings suggest that families in homes connected via pipes from the waste WTP had access to water free of fecal bacteria and ARGs. Families that had groundwater wells or aboveground polyethylene cisterns also tend to have access to safe drinking water. In contrast, families with underground cisterns are likely to be frequently exposed to fecal bacteria and ARGs, particularly if these cisterns are cracked and/or not recently (<7 months) cleaned. This trend was consistent in both sampling rounds. Therefore, our study strongly suggests that underground concrete cisterns may pose a significant risk to the residents due to high degree of microbial contamination. Further, since concrete cisterns are quite commonly used in various developing countries, our findings are likely to be applicable beyond the communities described in this study.
FUNDING
This work is supported by the Natural Science and Engineering Research Council of Canada's (NSERC) Collaborative Research and Training Experience (CREATE) Program, and by the Canadian Institutes of Health Research (CIHR).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the three First Nation reserves for their ongoing partnership in this research. The authors also wish to thank Wendy Ross, Rob Ellis, Ross McQueen and Edward Topp (Agriculture and Agri-Food Canada) for their contributions to this research partnership.
Conflict of interest. None declared.
REFERENCES
- Alexander J, Bollmann A, Seitz W et al.. Microbiological characterization of aquatic microbiomes targeting taxonomical marker genes and antibiotic resistance genes of opportunistic bacteria. Sci Total Environ. 2015;512–513:316–25. [DOI] [PubMed] [Google Scholar]
- Bajaj P, Singh NS, Kanaujia PK et al.. Distribution and molecular characterization of genes encoding CTX-M and AmpC beta-lactamases in Escherichia coli isolated from an Indian urban aquatic environment. Sci Total Environ. 2015;505:350–6. [DOI] [PubMed] [Google Scholar]
- Ben-David D, Kordevani R, Keller N et al.. Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin Microbiol Infect. 2012;18:54–60. [DOI] [PubMed] [Google Scholar]
- Bhullar K, Waglechner N, Pawlowski A et al.. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One. 2012;7:e34953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Characklis WG, McFeters GA, Marshall KC. Biofilms. In: Characklis WG, Marshall KC(eds). Wiley Series in Ecological and Applied Microbiology. New York: Wiley, 1990, 796. [Google Scholar]
- Cutler D, Miller G. The role of public health improvements in health advances: the twentieth century United States. Demography. 2005;42:1–22. [DOI] [PubMed] [Google Scholar]
- D'Costa VM, McGrann KM, Hughes DW et al.. Sampling the antibiotic resistome. Science. 2006;311:374–7. [DOI] [PubMed] [Google Scholar]
- Edmunds WM, Ahmed KM, Whitehead PG. A review of arsenic and its impacts in groundwater of the Ganges–Brahmaputra–Meghna delta, Bangladesh. Environ Sci Process Impacts. 2015;17:1032–46. [DOI] [PubMed] [Google Scholar]
- EPA US. National Primary Drinking Water Regulations.Document No. 40 CFR Part 141.2002, 436–8. [Google Scholar]
- Farenhorst A, Li R, Jahan M et al.. Bacteria in drinking water sources of a First Nation reserve in Canada. Sci Total Environ. 2017;575:813–9. [DOI] [PubMed] [Google Scholar]
- Fernando D, Zhanel GG, Kumar A. Antibiotic resistance and expression of resistance-nodulation-division pumps and outer membrane porins in Acinetobacter species isolated from Canadian hospitals. Can J Infect Dis Med Microbiol. 2013;24:17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando DM, Tun HM, Poole J et al.. Detection of antibiotic resistance genes in source and drinking water samples from a First Nations community in Canada. Appl Environ Microbiol. 2016;82:4767–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Li J, Yang F et al.. Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci Total Environ. 2014;493:626–31. [DOI] [PubMed] [Google Scholar]
- Hach C. Water Analysis Handbook. Colorado: Loveland, 2002. [Google Scholar]
- Health Canada. Guidelines for Canadian Drinking Water Quality: Guideline Technical Document. Ottawa, Ontario: Health Canada, 2009. [Google Scholar]
- Health Canada. Guidelines for Canadian Drinking Water Quality—Summary Table, Health Canada; Ottawa, Ontario, 2017. [Google Scholar]
- Kassem II, Esseili MA, Sigler V. Occurrence of mecA in nonstaphylococcal pathogens in surface waters. J Clin Microbiol. 2008;46:3868–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu MC, Mandal B, Hazra GC. Nitrate and fluoride contamination in groundwater of an intensively managed agroecosystem: a functional relationship. Sci Total Environ. 2009;407:2771–82. [DOI] [PubMed] [Google Scholar]
- LeChevallier MW, Cawthon CD, Lee RG. Factors promoting survival of bacteria in chlorinated water supplies. Appl Environ Microbiol. 1988;54:649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeChevallier MW, Welch NJ, Smith DB. Full-scale studies of factors related to coliform regrowth in drinking water. Appl Environ Microbiol. 1996;62:2201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc H, Schwartzbrod L, Dei-Cas E. Microbial agents associated with waterborne diseases. Crit Rev Microbiol. 2002;28:371–409. [DOI] [PubMed] [Google Scholar]
- Mac Kenzie WR, Hoxie NJ, Proctor ME et al.. A massive outbreak in Milwaukee of cryptosporidium infection transmitted through the public water supply. New Engl J Med. 1994;331:161–7. [DOI] [PubMed] [Google Scholar]
- Marti E, Variatza E, Balcazar JL. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014;22:36–41. [DOI] [PubMed] [Google Scholar]
- Martínez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–7. [DOI] [PubMed] [Google Scholar]
- Patrick RJ. Uneven access to safe drinking water for First Nations in Canada: connecting health and place through source water protection. Health Place. 2011;17:386–9. [DOI] [PubMed] [Google Scholar]
- Rice EW, Baird RB, Eaton AB et al.. Standard Methods for the Examination of Water and Wastewater, 22nd Edition American Public Health Association, American Water Works Association, Water Environment Federation, 2012. [Google Scholar]
- Roberts MC, Schwarz S, Aarts HJM. Erratum: acquired antibiotic resistance genes: an overview. Front Microbiol. 2012;3:384–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo IE, Aquino EE, Vazquez GJ. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob Agents Chemother. 2011;55:2968–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T, Kohnen W, Jansen B et al.. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol Ecol. 2003;43:325–35. [DOI] [PubMed] [Google Scholar]
- Suhartono S, Savin M, Gbur EE. Genetic redundancy and persistence of plasmid-mediated trimethoprim/sulfamethoxazole resistant effluent and stream water Escherichia coli. Water Res. 2016;103:197–204. [DOI] [PubMed] [Google Scholar]
- Su HC, Liu YS, Pan CG et al.. Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: From drinking water source to tap water. Sci Total Environ. 2018;616–617:453–61. [DOI] [PubMed] [Google Scholar]
- Tafoukt R, Touati A, Leangapichart T et al.. Characterization of OXA-48-like-producing Enterobacteriaceae isolated from river water in Algeria. Water Res. 2017;120:185–9. [DOI] [PubMed] [Google Scholar]
- van Hoek AHAM, Mevius D, Guerra B et al.. Acquired antibiotic resistance genes: an overview. Front Microbiol. 2011;2:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warton B, Heitz A, Joll C et al.. A new method for calculation of the chlorine demand of natural and treated waters. Water Res. 2006;40:2877–84. [DOI] [PubMed] [Google Scholar]
- Wohlsen TD. Comparative evaluation of chromogenic agar CM1046 and mFC agar for detection of E. coli and thermotolerant coliform bacteria from water samples. Lett Appl Microbiol. 2011;53:155–60. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Guidelines for Drinking-Water Quality. Geneva: World Health Organization, 2011. [Google Scholar]
- Yong D, Toleman MA, Giske CG et al.. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S, Nayak B, Sun S et al.. Vancomycin-Resistant Enterococci and Bacterial Community Structure following a Sewage Spill into an Aquatic Environment. Appl Environ Microbiol. 2016;82:5653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Lin W, Yu X. Effects of full-scale advanced water treatment on antibiotic resistance genes in the Yangtze Delta area in China. FEMS Microbiol Ecol. 2016;92:fiw065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.