Abstract
B cell adaptor for PI3-kinase (BCAP) is a signaling adaptor that activates the phosphoinositide 3-kinase (PI3K) pathway downstream of B cell receptor signaling in B cells and Toll-like receptor (TLR) signaling in macrophages. BCAP binds to the regulatory p85 subunit of class I PI3K, and is a large, multidomain protein. We used proteomic analysis to identify other BCAP-interacting proteins in macrophages and found that BCAP specifically associated with the caspase-1 pseudosubstrate inhibitor Flightless-1 and its binding partner leucine-rich repeat flightless-interacting protein 2 (LRRFIP2). Because these proteins inhibit the NLRP3 inflammasome, we investigated the role of BCAP in inflammasome function. Independent of its effects on TLR priming, BCAP inhibited NLRP3- and NLRC4-induced caspase-1 activation, cell death, and IL-1β release from macrophages. Accordingly, caspase-1–dependent clearance of a Yersinia pseudotuberculosis mutant was enhanced in BCAP-deficient mice. Mechanistically, BCAP delayed the recruitment and activation of pro-caspase-1 within the NLRP3-ASC pre-inflammasome through its association with Flightless-1. Thus, BCAP is a multifunctional signaling adaptor that inhibits key pathogen-sensing pathways in macrophages.
Introduction
B cell adapter for PI3-kinase (BCAP) is a signaling adapter expressed in several immune lineages, including macrophages, B cells, and NK cells. First identified in B cells, BCAP is phosphorylated on four YxxM tyrosines after B cell receptor (BCR) ligation, which promotes the binding and activation of class I phosphoinositide 3-kinase (PI3K) (1). In B cells, BCR-induced BCAP tyrosine phosphorylation depends upon ITAM-mediated activation of Syk and Btk, and is important for B cell proliferation and survival (2, 3). In NK cells, BCAP is phosphorylated and activates PI3K after signaling by the NK cell activating receptor NKR-P1C and lack of BCAP alters NK cell development (4). In T cells, BCAP participates in both TCR and IL-1R PI3K activation(5, 6). Thus, BCAP acts as a signaling adapter linking ITAM signaling and PI3K activation in B cells, T cells and NK cells (4–7).
In addition to its function downstream of ITAM-containing receptor complexes in lymphocytes, BCAP signals downstream of Toll-like receptors (TLR) in macrophages (8, 9). TLR ligation leads to activation of several signaling pathways, including the NF-κB, MAPK and PI3K pathways. BCAP is specifically required for optimal TLR-induced PI3K pathway activation in macrophages (8, 9). As PI3K is a potent inhibitory signal that keeps TLR responses in check, BCAP-deficient macrophages display increased TLR-induced TNF, IL-6 and IL-12 p40 production compared to wild-type (WT) macrophages, which depends upon the 4 YxxM tyrosines in BCAP (8). TLRs do not contain ITAM sequences, nor are they known to associate with ITAM-containing adapters, thus BCAP activation in macrophages is fundamentally distinct from that in lymphocytes. BCAP is a large protein of ~800 amino acids with several protein-protein interaction domains, including 3 proline-rich sequences, a coiled-coil domain, ankyrin repeats, and a homodimerization domain in addition to the YxxM tyrosines (1, 10). Additionally, the N-terminal domain of BCAP has been defined as a TIR domain important for limiting TLR responses (9, 11, 12). Because BCAP is a large multi-domain signaling adapter, we reasoned that BCAP may interact with other signaling pathways in addition to the TLR pathway in macrophages.
Inflammasomes are multiprotein cytoplasmic complexes that concentrate and activate the cysteine protease caspase-1, resulting in the processing and release of IL-1β and IL-18 and the caspase-1-dependent cell death known as pyroptosis (13, 14). Inflammasome activation contributes to clearance of a variety of pathogens through the recruitment and activation of immune cells and the depletion of an intracellular niche for pathogen replication. However, aberrant or prolonged inflammasome activation can be pathogenic, as is seen in sterile inflammatory disorders such as gout and pseudogout, and in autoinflammatory diseases caused by autosomal dominant activating mutations in inflammasome components, such as cryopyrin-associated periodic syndromes (CAPS) with mutations in NLRP3 and Familial Mediterranean Fever with mutations in MEFV (15, 16).
Structural studies of inflammasome components have revealed a nucleation dependent polymerization mechanism for inflammasome assembly (17, 18). Sensor proteins such as NLRP3 recruit the adaptor protein ASC, which in turn recruits and activates pro-caspase-1 through proximity-induced autoproteolysis. ASC polymerizes in a prion-like manner that allows for the assembly of a single inflammasome focus containing NLR, ASC and caspase-1, which acts as a platform for IL-1β and IL-18 processing. Therefore, the sequential nature of inflammasome assembly provides opportunities for host and pathogen inhibition. Inflammasomes serve to restrict a variety of pathogens, as is underscored by the numerous inflammasome inhibitors encoded by bacteria and viruses (13, 19–21). Poxviruses and other virus families encode direct competitive inhibitors of caspase-1 proteolytic activity, as do pathogenic Yersinia species. Enteropathogenic Escherichia coli and viruses such as KSHV and Myxoma virus encode proteins that interfere with earlier events in the assembly of the inflammasome complex (19, 21). Therefore, pathogens have multiple strategies to evade inflammasome-mediated immune responses, which highlights the importance of inflammasome activation for proper pathogen control. Whereas many pathogen-derived proteins that interfere with inflammasome activation are defined, relatively few host proteins that dampen this process to control potentially pathogenic inflammation are described. Similar to the pathogen-encoded inflammasome inhibitors, host inflammasome inhibitors either directly inhibit the proteolytic activity of caspase-1 or interfere with inflammasome assembly (22–24).
One host inflammasome inhibitor is Flightless-1(24, 25), a ubiquitously expressed protein with an N-terminal leucine rich repeat (LRR) domain and C-terminal gelsolin repeats, which regulates actin remodeling and cell migration (26, 27). Flightless-1 also contains a caspase-1 pseudosubstrate site, which inhibits caspase-1 activity (24). Here we show that BCAP interacted with Flightless-1 and its binding partner LRRFIP2 in macrophages. Therefore, we investigated whether BCAP inhibited inflammasome activation through its association with Flightless-1.
Results
BCAP interacts with the endogenous caspase-1 inhibitor Flightless-1.
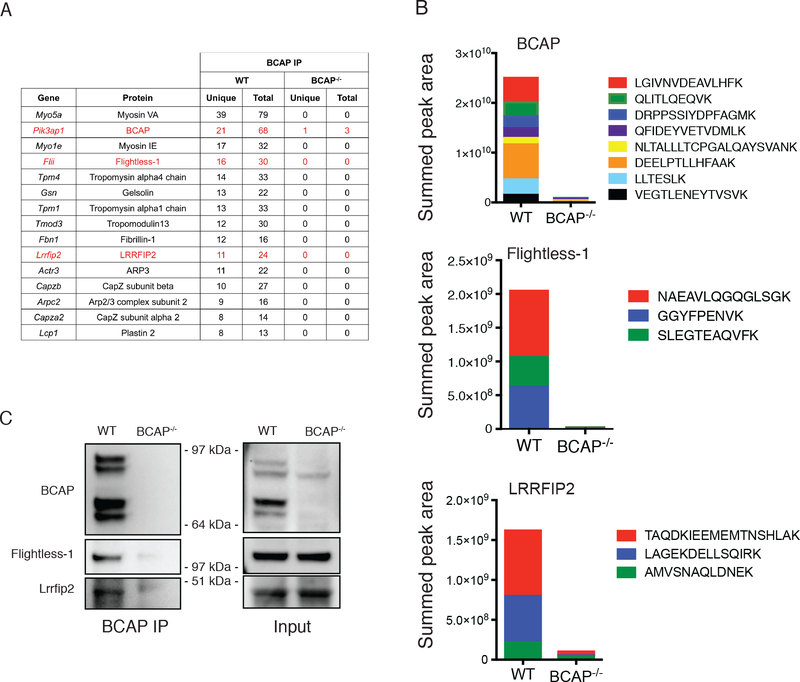
To identify novel pathways that may be regulated by BCAP in macrophages, we performed an unbiased screen for BCAP-interacting proteins using immunoprecipitation coupled with tandem mass spectrometry. We immunoprecipitated BCAP using a specific monoclonal antibody from lysates of WT macrophages and BCAP−/- macrophages. Identified proteins were filtered using the following criteria: at least 5 unique peptides identified in all three anti-BCAP immunoprecipitates from WT macrophages, 0 or 1 unique peptides in negative control immunoprecipitate from BCAP−/- macrophages, and low non-specific interactions as determined by the CRAPome database (28) (<10 average spectral counts). This analysis yielded 14 proteins, many of which were proteins that bind to and/or regulate the actin cytoskeleton, including several myosins, gelsolin, and components of the Arp2/3 complex (Figure 1A).
Figure 1. BCAP interacts with Flightless-1 and LRRFIP2 in macrophages.
(A and B) Mass spectrometry analysis of proteins associated with BCAP in lysates from WT or BCAP−/- BMDMs immunoprecipitated for BCAP. Tabulated data (A) are the average numbers of unique and total peptides from each protein. Mean peak area quantification (B) for each BCAP, Flightless-1 and LRRFIP2 peptide isolated are representative of 3 independent experiments. The intensity of each peptide was summed to generate an overall protein quantification score (peak area). (C) Western blot analysis of the indicated proteins in WT or BCAP−/- macrophages lysates immunoprecipitated for BCAP. Blots (upper) are representative of 3 independent experiments. Quantified band intensity values (lower) of Flightless-1 and LRRFIP2 are means ± SEM pooled from all experiments. ***P<0.001 by two-sided unpaired t-test.
We also noted in our list of specific BCAP-interactors the presence of two proteins that have defined functions in inhibiting NLRP3 inflammasomes, Flightless-1 (encoded by Flii) and leucine-rich repeat Flightless-1-interacting protein 2 (LRRFIP2, encoded by Lrrfip2) (Figure 1A). Flightless-1 is a ubiquitously expressed protein that contains both a gelsolin domain, which participates in actin capping, and a leucine rich repeat domain, for protein-protein interactions. LRRFIP2 is predominantly expressed in myeloid cells, and binds to Flightless-1 in macrophages where together they inhibit NLRP3 inflammasome activation through the ability of Flightless-1 to directly inhibit caspase-1 activity (24, 25). To quantitatively assess the presence of BCAP, Flightless-1 and LRRFIP2 in our immunoprecipitates, we performed precursor area quantification on selected peptides that we found to be abundant and to exhibit reproducible elution profiles. We quantified these peptides derived from each protein in WT and BCAP−/- macrophages, showing a strong enrichment in immunoprecipitates from WT macrophages (Figure 1B). We then confirmed the interaction of BCAP, Flightless-1 and LRRFIP2 in macrophages by co-immunoprecipitation and Western blot (Figure 1C). Together, these results demonstrated BCAP interacts with the NLRP3 inflammasome inhibitors Flightless-1 and LRRFIP2 in macrophages.
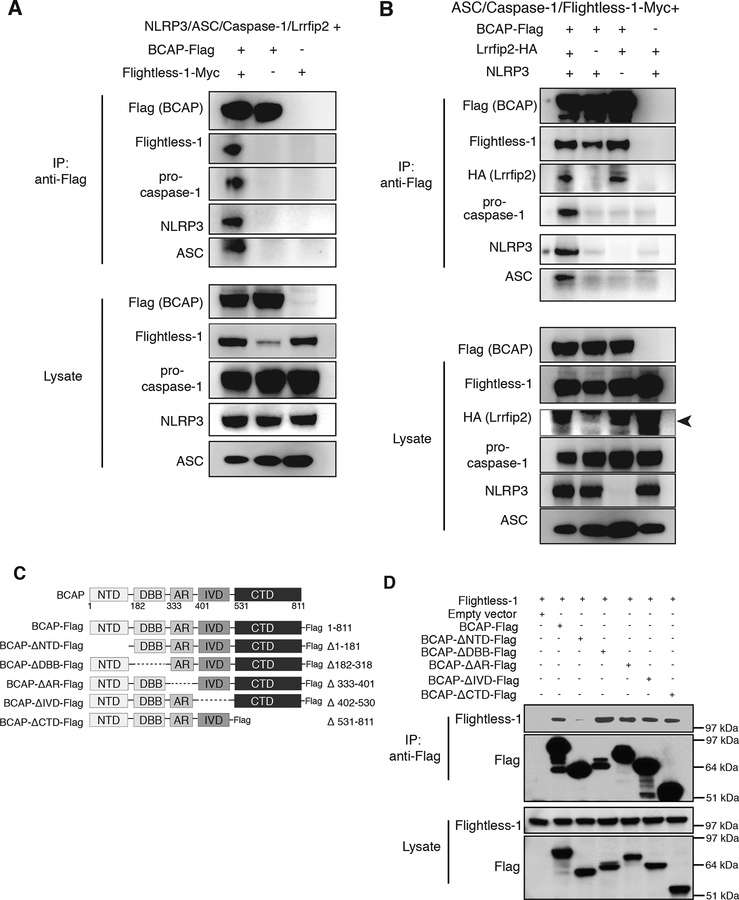
BCAP interacts with the NLRP3 inflammasome in the presence of Flightless-1 and LRRFIP2
Flightless-1 binds to pro-caspase-1 in the forming NLRP3 inflammasome, which is mediated by LRRFIP2 (25). To directly test whether BCAP is also a part of this complex, we co-transfected NLRP3, ASC and a pro-caspase-1 catalytic site mutant into 293T cells along with BCAP, Flightless-1 and LRRFIP2. Immunoprecipitation of BCAP using a C-terminal Flag-tag also precipitated Flightless-1 (Figure 2A) as we found in macrophages. BCAP also co-immunoprecipitated with pro-caspase-1, which was in a larger complex containing NLRP3 and ASC (Figure 2A). We next determined if this association required the presence of LRRFIP2. The ability of BCAP and Flightless-1 to co-immunoprecipitate did not require LRRFIP2, however, pro-caspase-1 association with this complex required LRRFIP2 expression (Figure 2B). The interaction of BCAP and Flightless-1 with pro-caspase-1 also required NLRP3 expression, unlike the association of BCAP and Flightless-1 (Figure 2B). Thus, BCAP interacts with the NLRP3 inflammasome in the presence of both Flightless-1 and LRRFIP2.
Figure 2. BCAP interacts with NLRP3 inflammasome components through its N-terminal domain.
(A and B) Western blot analysis of the indicated proteins in lysates from 293T cells co-transfected with plasmids encoding BCAP-Flag, Flightless-1, LRRFIP2-HA, NLRP3, ASC, and pro-caspase-1-RFP as indicated and immunoprecipitated for Flag. Black arrowhead in (B) indicates LRRFIP2 band. (C) Flag-tagged BCAP deletion mutants used in (D). (D) Western blot analysis of 293T cells co-transfected with plasmids encoding BCAP-Flag deletion mutants and Flightless-1. Lysates were immunoprecipitated for Flag and immunoblotted with anti-Flag and anti-Flightless-1 antibodies. Data are representative of 3 independent experiments.
We next investigated which domain of BCAP was required for its ability to bind to Flightless-1. BCAP has a variety of sequence motifs, including an N-terminal TIR domain, ankyrin repeats, a “DBB” domain shared with the drosophila protein Dof and the mammalian protein BANK1, and proline rich regions and coiled coil sequences in its C-terminal region, in addition to four tyrosines present in YxxM motifs required for p85 PI3K binding (1) (Figure 2C). To examine which BCAP region binds to Flightless-1, we constructed a panel of Flag-tagged BCAP deletion mutants and examined their ability to immunoprecipitate Flightless-1 when transfected into 293T cells. As shown in Figure 2D, only the BCAP mutant lacking the N-terminal domain was unable to co-immunoprecipitate Flightless-1. BCAP’s four YxxM tyrosines, which are required for inhibition of TLR responses but are not located in the N-terminal domain (8), were not necessary for BCAP to co-immunoprecipitate Flightless-1 (Figure S1). Thus, BCAP uses its N-terminal domain, but does not require sequences for PI3K binding/activation, for Flightless-1 association.
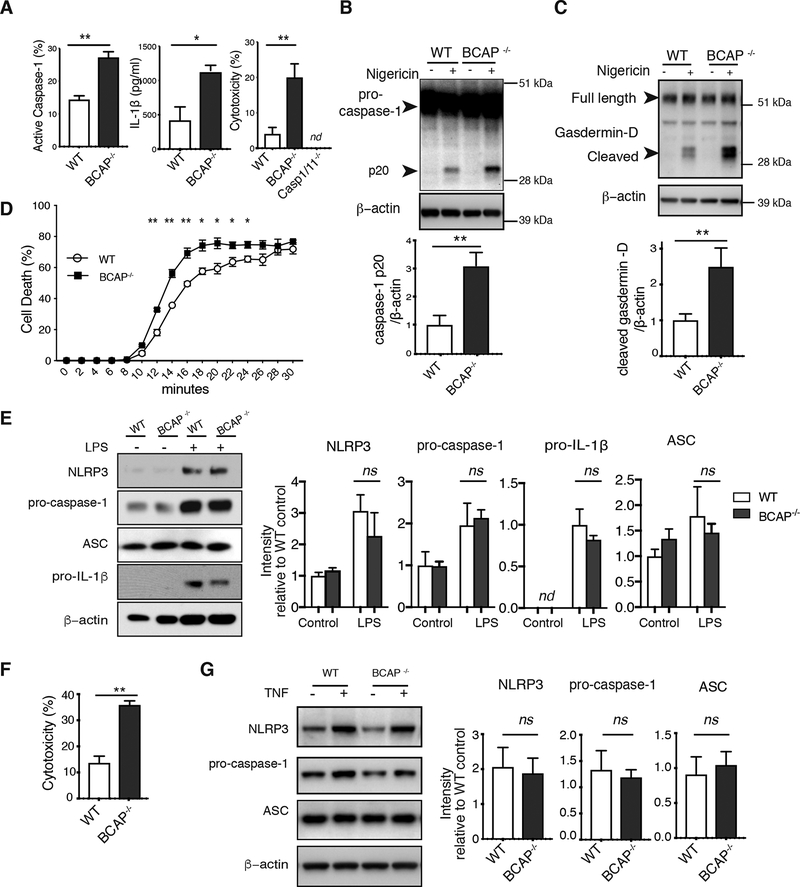
BCAP inhibits NLRP3 inflammasome activation independent of LPS priming
As we found that BCAP interacts with Flightless-1 and LRRFIP2, which together inhibit NLRP3 inflammasomes, we reasoned that BCAP may promote Flightless-1 function. We LPS-primed WT and BCAP-deficient macrophages and assessed NLRP3 inflammasome activity after treatment with the NLRP3-activating Streptomyces hygroscopicus toxin Nigericin. BCAP−/- macrophages had increased measures of inflammasome activation, including increased caspase-1 activation, cell death, and IL-1β secretion, compared to WT macrophages (Figure 3A). This Nigericin-induced cell death was due to pyroptosis as we detected no death of Casp1/11−/− macrophages. In addition to increased active caspase-1 as measured by fluorescence microscopy, BCAP−/- macrophages also had increased caspase-1 (Figure 3B) and Gasdermin D (Figure 3C) cleavage compared to WT macrophages. To assess the kinetics of NLRP3 inflammasome activation in the absence of BCAP, we performed a time course of cell death using live cell fluorescent microscopy to assess uptake of the membrane-impermeant fluorescent probe SYTOX green. BCAP-deficient macrophages had significantly increased cell death compared to WT macrophages as early as 12 minutes after Nigericin treatment, which was sustained until close to maximal cell death at ~30 minutes (Figure 3D). Thus, the absence of BCAP led to increased NLRP3 inflammasome function in macrophages, suggesting that in WT macrophages BCAP serves to inhibit inflammasome activation.
Figure 3. Increased NLRP3 inflammasome activation in the absence of BCAP.
(A) Immunofluorescence staining for active caspase-1 (left), ELISA analysis of IL-1β secretion (middle) and LDH assay of cell death (right) in BMDMs primed with LPS for 4 hours then treated with nigericin for 30 min. Data are means + SEM pooled from 3 experiments. (B and C) Western Blot analysis for pro-caspase-1 (B) and gasdermin-D (C) cleavage in cell lysates of LPS-primed nigericin-treated BMDMs. Blots (upper) are representative of 3 independent experiments and band intensity values (lower) are means + SEM pooled from experiments. (D) Live cell imaging of cell death in LPS-primed nigericin-treated BMDMs. Data are means ± SEM of 3 independent experiments. (E) Western Blot analysis of indicated proteins in lysates from cells treated for 16 hours with LPS. Blots (left) are representative of 3 experiments and quantified band intensity values are means + SEM pooled from all experiments. (F) LDH assay of cell death in BMDMs primed with TNF for 6 hrs and treated with nigericin for 30 minutes. Data are means ± SEM of 3 independent experiments. (G) Western Blot analysis of indicated proteins from lysates of BMDMs primed with TNF for 6 hrs. Blots (left) are representative of 3 independent experiments and quantified band intensity values (right) are means + SEM pooled from all experiments. ns, p>0.05, *p<0.05, **p <0.01, ***p<0.001 by two-sided unpaired t-test. nd=not detected.
LPS priming of macrophages increases NLRP3 inflammasome activation by inducing expression of NLRP3 and pro-IL-1β. Because we have previously shown that BCAP inhibits LPS-induced TNF, IL-6 and IL-12 p40 production in macrophages due to a reduction in PI3K activation (8), we directly examined expression of components of the NLRP3 inflammasome to determine whether the increased NLRP3 inflammasome activation in BCAP−/- macrophages was due to increased LPS-priming. We found no difference between WT and BCAP−/- macrophages in NLRP3, ASC, pro-caspase-1 or pro-IL-1β protein by Western blot at concentrations of LPS used for priming in our experiments at either 4 or 16 hours (Figure 3E, Figure S2A). Though we did not find increased NLRP3 or pro-IL-1β protein in BCAP−/- macrophages, we did find increased LPS-induced TNF secretion consistent with our previous work (Figure S2B). qPCR analysis also showed no difference in induction of mRNA encoding NLRP3 and pro-IL-1β in response to LPS between WT and BCAP−/- macrophages (Figure S2C).
Though differences in LPS priming of NLRP3 inflammasome components measured did not explain the increased NLRP3 activation in BCAP−/- macrophages, it was possible that BCAP−/- macrophages differentially secreted a soluble factor that could affect NLRP3 inflammasome function. We first measured IL-10 production and found no difference in IL-10 secretion between LPS-treated WT and BCAP−/- macrophages, consistent with our previous work (Figure S2B) (8). Next, we took a broader approach, assessing whether supernatants from LPS-primed BCAP−/- macrophages increased NLRP3 inflammasome function when transferred onto WT macrophages. Macrophages were LPS-primed for 4 hours, washed, and then cultured for an additional 2 hours in supernatants from either LPS-primed WT or BCAP−/- macrophages before treatment with Nigericin. Culturing WT macrophages with supernatants from LPS-primed BCAP−/- macrophages did not increase Nigericin-induced cell death or IL-1β release (Figure S2D). Conversely, culturing BCAP−/- macrophages with supernatants from LPS-primed WT macrophages did not decrease these measures (Figure S2D). We conclude that the difference in NLRP3 inflammasome function between LPS-primed WT and BCAP−/- macrophages is not due to differential secretion of a soluble factor and is therefore cell-intrinsic.
We and others previously found that BCAP does not inhibit TNF signaling in macrophages (8, 9), thus we also examined NLRP3 inflammasome activation after TNF priming. BCAP-deficiency also increased Nigericin-mediated cytotoxicity in macrophages after TNF priming without affecting NLRP3, pro-caspase-1 or ASC expression (Figure 3F, G). Thus, we conclude that the effect of BCAP on NLRP3-induced caspase-1 activation is independent of priming, and therefore BCAP must inhibit inflammasome activation through a distinct cell-intrinsic mechanism, likely through interactions with Flightless-1.
BCAP inhibits NLRP3 inflammasome activation during Yersinia pseudotuberculosis infection in vitro and in vivo
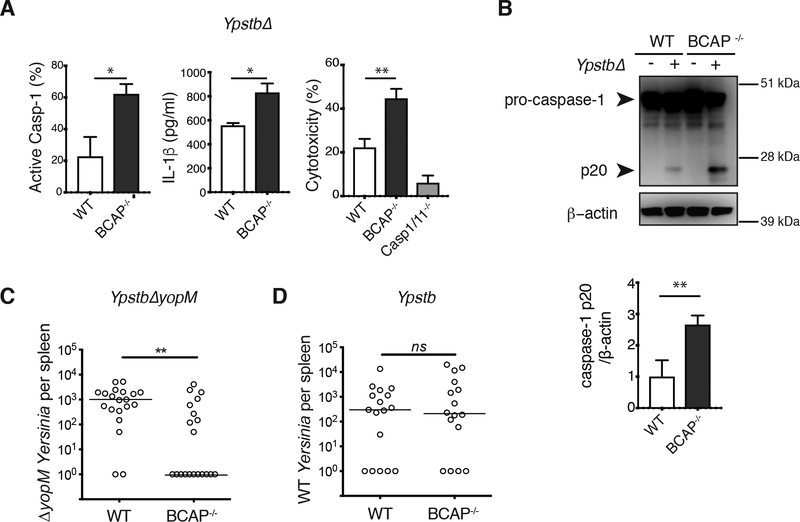
To ask whether BCAP also inhibits inflammasome activation during bacterial infection, we infected LPS-primed WT and BCAP−/- macrophages with a Yersinia pseudotuberculosis (Ypstb) mutant that efficiently activates the NLRP3 inflammasome. This YpstbΔ mutant has no Yop effectors to translocate into the host cytoplasm that interfere with inflammasome activation (29–31). Similar to when we activated the NLRP3 inflammasome with Nigericin, during infection with YpstbΔ BCAP−/- macrophages had increased active caspase-1, IL-1β secretion and caspase-1/11-dependent cell death compared to WT macrophages (Figure 4A, Fig. S3). Caspase-1 cleavage, as measured by Western blot, was also increased during YpstbΔ infection in BCAP−/- macrophages compared to WT macrophages (Figure 4B). Therefore, BCAP inhibited inflammasome activation not only during toxin-induced NLRP3 inflammasome activation, but also during bacterial infection with YpstbΔ.
Figure 4. Increased Yersinia-mediated inflammasome activation in the absence of BCAP.
(A) Immunofluorescence staining for active caspase-1 (left), ELISA analysis of IL-1β secretion (middle) and LDH assay of cell death (right) in BMDMs primed with LPS overnight, then infected with YpstbΔ for 90 min. Data are means + SEM pooled from 4 independent experiments. (B) Western blot analysis of pro-caspase-1 cleavage in lysates of BMDMs primed with LPS overnight, then infected with YpstbΔ for 90 min. Blots (upper) are representative of 3 independent experiments and quantified band intensity values (lower) are means + SEM pooled from all experiments. (C and D) Bacterial burden in the spleens of WT and BCAP−/- mice on day 4 after infection with YpstbΔYopM (C) or Ypstb (D). Data and median values (line) of at least 16 mice are from 2 independent experiments. **p <0.01, ***p<0.001 by two-sided unpaired t-test (A and B) and Mann-Whitney test (C and D).
To examine whether BCAP inhibits inflammasome activation in vivo, we examined the clearance of a Ypstb mutant solely lacking YopM, a Ypstb caspase-1 inhibitor. Clearance of YpstbΔYopM is attenuated compared to wild-type Ypstb, and this attenuation depends upon caspase-1 (29). We infected WT and BCAP−/- mice i.p. with YpstbΔYopM and measured CFU in the spleen 4 days after infection. BCAP−/- mice had significantly increased clearance of YpstbΔYopM, with ~2 logs fewer bacteria in the spleen than WT mice (Figure 4C). 11/20 BCAP−/- mice had completely cleared the infection by day 4, whereas only 2/20 WT mice had done so. To exclude that increased TLR signaling, rather than increased caspase-1 activation, in BCAP−/- mice led to the improved bacterial clearance, we infected WT and BCAP−/- mice with wild-type Ypstb that express YopM, clearance of which is caspase-1-independent (29). We observed no difference in bacterial clearance between WT and BCAP−/- mice infected with wild-type Ypstb (Figure 4D). Together, these results suggest that during YpstbΔYopM infection increased caspase-1 activation, rather than enhanced TLR signaling, is responsible for enhanced clearance of caspase-1-dependent YpstbΔYopM bacteria by BCAP−/- mice compared to WT mice.
BCAP inhibits the NAIP5/NLRC4 and NAIP2/NLRC4 inflammasomes
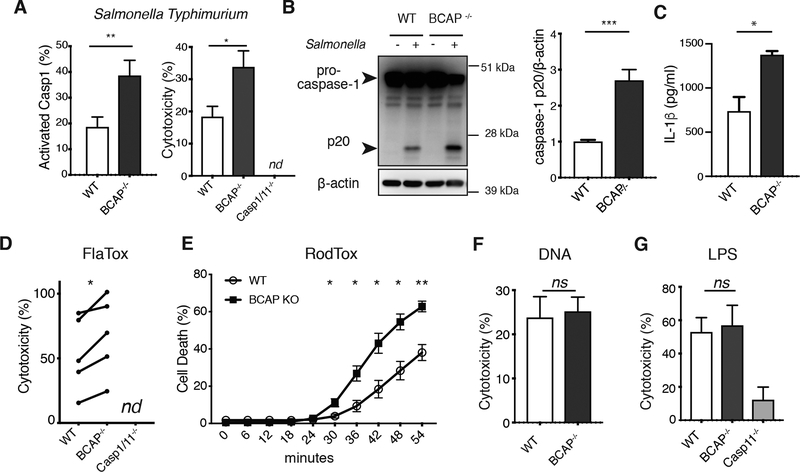
We next examined whether BCAP plays a role in inhibition of inflammasomes in addition to NLRP3. First, we assessed inflammasome activation after infection with Salmonella Typhimurium, which is principally sensed by NLRC4 inflammasomes and does not require LPS-priming (32). We observed increased caspase-1 activation in S. Typhimurium-infected unprimed BCAP−/- macrophages compared to unprimed WT macrophages as measured by fluorescence microscopy, cell death and pro-caspase-1 cleavage (Figure 5A and B). Cell lysis was not detected during infection of Casp1/11−/− macrophages with S. Typhimurium, which together with the observed cleavage of pro-caspase-1 indicates pyroptotic cell death during this infection. We also assessed IL-1β release from S. Typhimurium infected macrophages that had been LPS-primed to induce high amounts of pro-IL-1β protein. LPS-primed BCAP−/- macrophages secreted significantly more IL-1β than LPS-primed WT macrophages (Figure 5C), consistent with increased NLRC4 inflammasome activation.
Figure 5. BCAP inhibits NLRC4 inflammasomes.
(A) Immunofluorescence staining for active caspase-1 (left) and LDH assay of cell death (right) in unprimed BMDMs infected with S. Typhimurium for 30 minutes. Data are means + SEM pooled from 3 independent experiments. (B) Western blot analysis of pro-caspase-1 cleavage in lysates of BMDMs infected with S. Typhimurium for 30 minutes. Blots (left) are representative of 3 independent experiments and quantified band intensity values are means + SEM pooled from all experiments. (C) ELISA analysis of IL-1β release from LPS-primed BMDMs infected with S. Typhimurium for 30 minutes. Data are means + SEM pooled from 3 independent experiments. (D) LDH assay of cell death in unprimed BMDMs treated with FlaTox for 20 minutes. Data are individual means from 5 independent experiments (connected with lines). (E) Live cell imaging to measure cell death in BMDMs treated with RodTox. Data are means ± SEM of 3 independent experiments. (F) LDH assay of cell death in unprimed BMDMs transfected with calf thymus DNA for 3 hours. Data are means + SEM pooled from 3 independent experiments. (G) LDH assay of cell death in unprimed BMDMs treated with LPS and Cholera Toxin B for 3.5 hours. Data are means ± SEM of 3 independent experiments. *p<0.05, **p <0.01, ***p<0.001 by two-sided unpaired (A-C and E-G) and paired t-test (D).
Additionally, we observed increased cell death in unprimed BCAP−/- macrophages upon administration of FlaTox (Figure 5D), a direct NAIP5/NLRC4 activator that consists of Legionella pneumophila flagellin fused with the amino-terminal domain of Bacillus anthracis lethal factor (LFn), which facilitates cytosolic delivery in combination with the anthrax protective antigen (PA) channel in the absence of TLR activation (33). Because components of the FlaTox we used were generated in E. coli and could have LPS contamination (33), we also used a mammalian-expressed NLRC4 activating agent RodTox, composed of the inner rod protein (PrgJ) from S. Typhimurium SPI-1 type III secretion system fused to LFn combined with mammalian-expressed PA, which requires both NAIP2 and NLRC4 for caspase-1 activation (34). Kinetic analysis of cell death with mammalian-expressed RodTox showed significantly increased cell death of BCAP−/- macrophages compared to WT macrophages at all time points where cell death was detected (Figure 5E). BCAP deficiency did not affect NLRC4 expression in macrophages (Figure S4). Together, these data strongly support a role for BCAP in suppressing caspase-1 activation downstream of NLRC4.
BCAP plays a role in inhibiting both NLRP3 and NLRC4 inflammasomes, which suggests that BCAP may play a general role in the inhibition of inflammasome function. We therefore examined if BCAP inhibits the DNA-sensing AIM2 inflammasome (35). We transfected unprimed WT and BCAP−/- macrophages with calf thymus DNA and measured cell death. Unexpectedly, cell lysis of WT and BCAP−/- macrophages was similar after DNA transfection (Figure 5F). Therefore, BCAP does not inhibit all caspase-1 activating inflammasomes.
In addition to caspase-1, caspase-11 is also an inflammasome effector caspase that induces pyroptosis (36). We therefore investigated whether BCAP inhibits the non-canonical caspase-11 inflammasome that senses cytosolic LPS. To activate caspase-11, we delivered LPS to the cytoplasm of macrophages using the noncatalytic B-subunit of cholera toxin (37). Under these conditions, caspase-11-mediated cell lysis of LPS-primed WT and BCAP−/- macrophages was similar (Figure 5G). These data demonstrate that BCAP does not inhibit non-canonical caspase-11 inflammasomes. Thus, BCAP inhibits NLRP3 and NLRC4 inflammasomes, but not AIM2 inflammasomes or non-canonical caspase-11 inflammasomes.
BCAP inhibits the entry of pro-caspase-1 into inflammasome foci
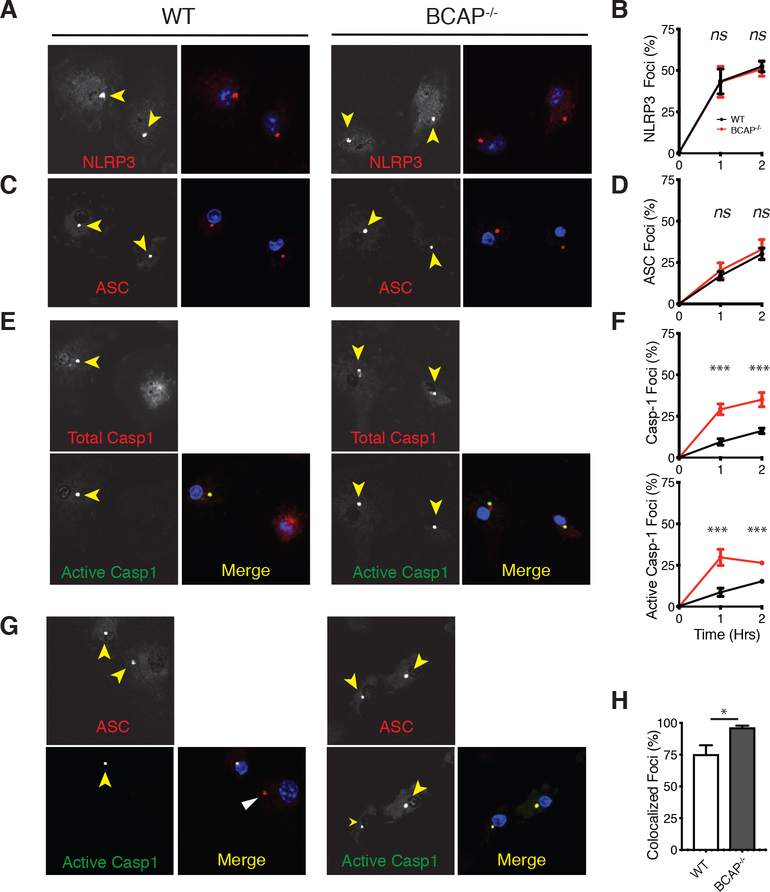
To determine mechanistically how BCAP inhibits caspase-1 activation, we examined the formation of NLRP3/ASC/caspase-1 inflammasome foci during infection with YpstbΔ. Upon detection of YpstbΔ infection, NLRP3 undergoes a conformational change that results in self-oligomerization into a single focus. This conformational change leads to NLRP3 recruitment of the adaptor protein ASC to the pre-inflammasome focus. ASC then oligomerizes and recruits pro-caspase-1, resulting in increased local concentrations of pro-caspase-1 and enzymatic activation. We used immunofluorescence to assess whether BCAP-deficiency affects the kinetics of NLRP3 inflammasome formation (29). The kinetics of formation of NLRP3 and ASC foci between WT and BCAP−/- macrophages during infection with YpstbΔ were similar (Figure 6A–D), reinforcing our finding that differences between WT and BCAP−/- macrophages are not due to differences in NLRP3 or ASC expression after LPS-priming. In contrast to NLRP3 and ASC foci formation, there was a significant increase in the recruitment of total and active caspase-1 to inflammasome foci in BCAP−/- macrophages compared to WT macrophages at both one and two hours after infection (Figure 6E, F).
Figure 6. BCAP delays caspase-1 recruitment into NLRP3 inflammasome foci.
(A to F) Immunofluorescence analysis of NLRP3, ASC or caspase-1 foci formation in BMDMs primed overnight with LPS and then infected with YpstbΔ. Yellow arrows indicate foci. Images (A, C, and E) are representative of 3 independent experiments and quantification of the percent of cells with foci of the indicated protein (B, D, and F) are means ± SEM from all experiments. (G and H) Immunofluorescence analysis of active caspase-1 and ASC in BMDMs primed overnight with LPS and then infected with YpstbΔ for 90 min. White arrow indicates pre-inflammasome focus containing ASC, but not active caspase-1. Images (G) are representative of 3 independent experiments and quantification of the percent of cells with ASC foci with active caspase-1 (H) are means ± SEM from all experiments. *p<0.05, **p <0.01, ***p<0.001 by two-sided unpaired t-test.
These data show that in the absence of BCAP, pro-caspase-1 is more quickly recruited to and activated within NLRP3 inflammasome foci. To assess this more directly, we co-stained for ASC and active caspase-1 after YpstbΔ infection. ~70% of the WT macrophages with ASC foci also had active caspase-1 in the same focus at one hour after infection, while >95% of BCAP−/- macrophages with ASC foci had active caspase-1 in the same focus (Figure 6G, H). Thus, in the absence of BCAP, entry and activation of caspase-1 into the NLRP3 inflammasome occurred more rapidly showing that BCAP acts to delay the recruitment of pro-caspase-1 to the NLRP3 inflammasome, where it becomes activated by autoproteolysis.
BCAP is required for Flightless-1 inhibition of NLRP3 inflammasome activation
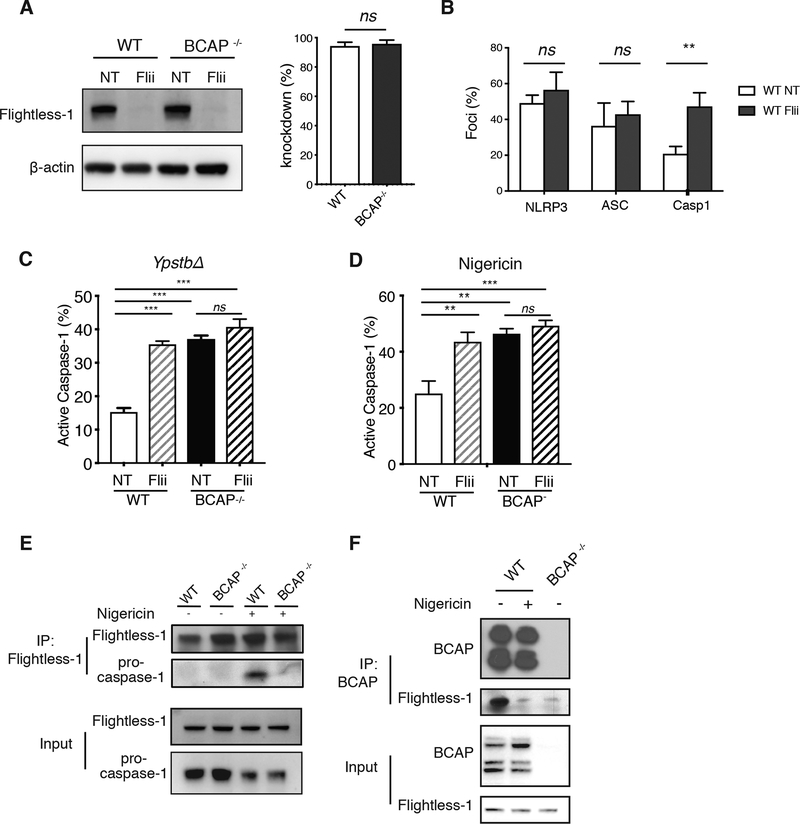
Our proteomics data suggested that BCAP may inhibit NLRP3 inflammasome function by associating with Flightless-1. Flightless-1 directly inhibits caspase-1 proteolytic activity through its pseudosubstrate site, but whether it affects recruitment of NLRP3 and ASC into forming NLRP3 inflammasomes has not been addressed. Thus, we knocked down Flightless-1 in macrophages and assessed formation of NLRP3/ASC/caspase-1 foci. shRNA-mediated knockdown (KD) of Flightless-1 efficiently reduced Flightless-1 protein in both WT and BCAP−/- macrophages compared to the non-target (NT) control (Figure 7A, Figure S5A). Similar to the effect of BCAP deficiency in macrophages, Flightless-1 KD caused increased recruitment of active caspase-1, but not NLRP3 or ASC, to foci after YpstbΔ infection (Figure 7B, Figure S5B). These data suggest that BCAP and Flightless-1 both impact inflammasome maturation at the stage of pro-caspase-1 recruitment and activation.
Figure 7. BCAP inhibition of NLRP3 inflammasome activation requires Flightless-1.
(A) Western blot analysis of Flightless-1 in BMDMs transduced with lentiviruses encoding shRNA #1 (Flii) or non-targeting shRNA (NT). Blots (left) are representative of 3 independent experiments and quantified knockdown efficiency values (right) are means + SEM from 3 independent experiments. (B) Immunofluorescence analysis of NLRP3, ASC, and active caspase-1 in WT BMDM transduced with NT or Flii-specific shRNA #1 and primed with LPS overnight, then infected with YpstbΔ. Data are means + SEM from 3 independent experiments. (C and D) Immunofluorescence analysis of active caspase-1 in WT and BCAP−/- BMDM transduced with NT shRNA or Flii-specific shRNA #1 and were either LPS-primed overnight and infected with YpstbΔ for 60 min (C) or LPS-primed for 4 h and treated with nigericin for 30 min (D). Data are means + SEM of 3 independent experiments. (E and F) Western blot analysis of proteins immunoprecipitated with antibodies to Flightless-1 (E) or BCAP (F) in lysates of WT or BCAP−/- BMDM primed with LPS for 4 h and then treated with nigericin for 15 min. Blots are representative of 3 independent experiments. ns, not significant, *p<0.05, **p <0.01, ***p<0.001 by two-sided unpaired t-test (A and B), or One-way ANOVA with Tukey’s post-test (C and D).
To determine if BCAP requires Flightless-1 to inhibit inflammasome activation, we knocked down Flightless-1 in both WT and BCAP−/- macrophages and infected with YpstbΔ to assess NLRP3 inflammasome activation (Figure 7C). Whereas Flightless-1 KD in WT macrophages significantly increased YpstbΔ-induced caspase-1 activation, Flightless-1 KD had no effect in BCAP−/- macrophages, proving that the interaction between BCAP and Flightless-1 is required to inhibit inflammasome activation during YpstbΔ infection. We found a similar result with Nigericin-induced caspase-1 activation using two distinct shRNAs; Flightless-1 KD significantly increased caspase-1 activation in WT macrophages, but not in BCAP−/- macrophages (Figure 7D, Figure S5C).
We next assessed what role BCAP serves in Flightless-1 inhibition of NLRP3 inflammasomes. Flightless-1 was not constitutively associated with pro-caspase-1 in macrophages in the LPS-primed state, but was inducibly associated with pro-caspase-1 after activation of NLRP3 with Nigericin, confirming the data of Jin et al. (25). Not only was BCAP required for Flightless-1 inhibition of NLRP3 inflammasomes, but BCAP was required for Flightless-1 to associate with pro-caspase-1 (Figure 7E). These data reinforced our findings that BCAP was required for Flightless-1 function in inflammasome inhibition (Figure 7C and D).
We initially found that BCAP and Flightless-1 were associated in resting macrophages (Figure 1). We asked if BCAP remained associated with Flightless-1 after LPS-priming and NLPR3 inflammasome activation. BCAP and Flightless-1 were associated in LPS-primed macrophages. However, Nigericin treatment caused the dissociation of BCAP and Flightless-1 (Figure 7F), at a time when Flightless-1 is bound to pro-caspase-1 (Figure 7E). Our data suggest a model in which BCAP facilitates the delivery of Flightless-1 and LRRFIP2 to the forming NLRP3 inflammasome, and then BCAP is subsequently released while Flightless-1 remains associated with pro-caspase-1 (Figure S6).
Discussion
Inflammasome activation is a potent inflammatory defense required for clearance of pathogens, yet excessive inflammasome activation leads to pathological inflammation and autoinflammatory syndromes. Therefore, tight regulation of this pathway is required to resist infection yet prevent excess collateral damage. In this study, we showed that the macrophage signaling adaptor protein BCAP dampened NLRP3 and NLRC4 inflammasome activation through interaction with the endogenous caspase-1 inhibitor Flightless-1. Together with our previous work showing BCAP can inhibit TLR signaling in macrophages by activating PI3K, we have identified BCAP as a multifunctional signaling adapter tuning macrophage innate immunity.
Together, BCAP and Flightless-1 inhibit pro-caspase-1 recruitment to NLRP3 pre-inflammasomes to limit inflammasome activation. BCAP-deficient macrophages have similar formation of NLRP3 and ASC containing pre-inflammasome foci as WT macrophages, yet have increased total and active caspase-1 containing foci and increased measures of caspase-1 activation similar to cells with Flightless-1 knockdown (24, 25). We confirmed that Flightless-1 was not complexed with pro-caspase-1 in resting macrophages, but that early after activation of NLRP3 inflammasomes, Flightless-1 interacted with pro-caspase-1 (25). Importantly, we found that the ability of Flightless-1 to bind to pro-caspase-1, and inhibit its function, required BCAP. Thus, we propose that BCAP facilitates binding of Flightless-1 to pro-caspase-1, which delays pro-caspase-1 entry into the forming NLRP3 inflammasome focus (Figure S6). Whereas BCAP and Flightless-1 interacted without co-expression of NLRP3, ASC and pro-caspase-1, the ability of BCAP and Flightless-1 to form a complex with pro-caspase-1 required NLRP3 and ASC, as well as the Flightless-1 binding partner LRRFIP2. These findings support a model in which NLRP3 inflammasome activation signals for BCAP to facilitate Flightless-1-pro-caspase-1 interactions. Our data suggests that this complex is then targeted to NLRP3-containing inflammasome foci by LRRFIP2, and BCAP is released. Identification of the signals that lead to BCAP release from the Flightless-1/pro-caspase-1 complex is an important area of future work.
We found here that BCAP does not inhibit formation of all inflammasomes. Of those we tested, BCAP inhibited NLRP3, NAIP5/NLRC4 and NAIP2/NLRC4 inflammasomes, but not AIM2 inflammasomes or non-canonical caspase-11 inflammasomes. We found the BCAP-facilitated complex between Flightless-1 and pro-caspase-1 required the expression of the Flightless-1 binding partner LRRFIP2, which is required for Flightless-1 to associate with and inhibit only the NLRP3 inflammasome (25). Given that BCAP required both Flightless-1 and LRRFIP2 to associate with NLRP3 inflammasomes, but that LRRFIP2 only associates with NLRP3 (25), we hypothesize that there may be an analogous protein to LRFIP2 that allows BCAP and Flightless-1 to inhibit NLRC4 inflammasomes. This adapter would likely bind to NLRC4 and Flightless-1 similar to how LRRFIP2 binds to NLRP3 and Flightless-1.
In contrast to NLRP3 and NLRC4 inflammasomes, BCAP did not inhibit AIM2 inflammasome function. This may suggest there are not adapter proteins in macrophages to bridge AIM2 to Flightless-1. Similarly, BCAP did not inhibit non-canonical caspase-11 inflammasomes. Though Flightless-1 was previously identified as a protein that interacts with caspase-11, Flightless-1 did not have the capacity to inhibit caspase 11 activity (24). This may explain why BCAP-deficiency has no effect on non-canonical inflammasome-mediated cell death. Thus, we speculate that the reasons why BCAP inhibits certain inflammasomes, but not others, may depend upon Flightless-1 specificity for caspase-1 over caspase-11, as well as the availability of Flightless-1 binding partners to assist in recruitment of the Flightless-1/BCAP/pro-caspase-1 complex. The discovery of the protein or proteins that bridge Flightless-1 to NLRC4, allowing for BCAP inhibition, is under investigation.
We and others first defined a role for BCAP in limiting pathogen sensing through the TLR family of pattern recognition receptors (8, 9). BCAP inhibits TLR-induced inflammatory cytokine production, specifically TNF, IL-6 and IL-12 p40 production, through its ability to bind the p85 subunit of class I PI3K and thereby activate PI3K catalytic activity (8). Therefore, upon finding that NLPR3 inflammasome function was increased in BCAP-deficient macrophages compared to WT cells, we examined whether this was due to increased LPS-dependent priming, which transcriptionally induces NLRP3 and pro-IL-1β. We showed that high dose LPS-induced similar expression of NLRP3 and pro-IL-1β mRNA and protein in WT and BCAP-deficient macrophages, even under conditions where BCAP-deficient cells produced more TNF than WT macrophages. Additionally, WT and BCAP-deficient macrophages did not differ in expression of ASC, pro-caspase-1, Flightless-1 or LRRFIP2. Consistent with our previous findings, LPS-induced IL-10 production was also similar between WT and BCAP-deficient macrophages. Therefore, BCAP inhibits some LPS-induced responses through its ability to activate PI3K (8, 9). However, BCAP is not able to inhibit expression of all LPS-induced genes, including Nlrp3 and Il1b.
Furthermore, supporting that the ability of BCAP to inhibit NLRP3 inflammasomes is not due to effects on LPS priming, BCAP-deficient macrophages also showed increased NLRP3 inflammasome activation after TNF priming, which is a signaling pathway not inhibited by BCAP (8, 9). Additionally, we showed that the kinetics NLRP3 and ASC foci formation are not different between WT and BCAP−/- macrophages, supporting our conclusion that increased NLRP3 inflammasome activation in BCAP−/- macrophages was not due to increased NLRP3 expression. We also demonstrated that soluble factors produced in response to LPS priming are not responsible for the enhanced NLRP3 inflammasome activation of BCAP−/- macrophages. Our finding that BCAP inhibited NAIP5/NLRC4 and NAIP2/NLRC4 inflammasomes induced by S. Typhimurium infection or by direct cytosolic delivery of the NAIP5/NLRC4 activator FlaTox (33) and the NAIP2/NLRC4 activator RodTox (34) in the absence of LPS-priming also supports a direct role for BCAP in inhibiting inflammasome activation. Furthermore, distinguishing the role of BCAP in limiting activation of the TLR pathway through PI3K and the NLRP3 and NLRC4 inflammasomes through Flightless-1, is our finding that mutation of the four YxxM tyrosines in BCAP required for PI3K p85 binding had no effect on BCAP binding to Flightless-1. The N-terminal domain of BCAP that contains a TIR domain is both required for Flightless-1 interaction and for inhibition of TLR signaling (9), supporting the key role of this BCAP domain in limiting innate immune signaling.
Flightless-1 not only binds and inhibits pro-caspase-1 activation, but it also regulates actin remodeling through its gelsolin domains (26). Though we have not directly assessed how BCAP- and Flightless-1−mediated actin cytoskeletal rearrangements may limit inflammasome activity, this is an intriguing possibility, given the emerging information on how actin polymerization may regulate and be sensed by various inflammasomes (38–42). We found that in addition to Flightless-1 and LRRFIP2, BCAP and Flightless-1 also were associated with other actin binding proteins in macrophages, including members of the ARP2/3 complex and several myosins. Thus, the ability of BCAP and Flightless-1 to allow for sensing of aberrant actin dynamics will be the focus of future studies.
Our results reveal a role for BCAP in the inhibition of NLRP3 and NLRC4 inflammasome function, as BCAP and Flightless-1 work cooperatively to delay pro-caspase-1 recruitment and activation to forming inflammasomes containing NLRP3 and ASC. This delay in entry of pro-caspase-1 into NLRP3 inflammasomes may allow pro-caspase-1 to be directed to sites within the cell where it can be activated and cleave substrates distinct from the prototypical caspase-1 targets, pro-IL-1β, pro-IL-18 and gasdermin D. These targets may include subunits of the NADPH oxidase on phagosomal membranes (43) or metabolic proteins (44), pathways targeted by caspase-1 to regulate the key macrophage cellular processes, phagosomal acidification and glycolysis, respectively (45). It is interesting to speculate that pro-caspase-1 association with Flightless-1 and the actin cytoskeleton may facilitate some of these alternative functions of caspase-1 in macrophages.
Our data show that BCAP and Flightless-1 are a critical hub for limiting inflammasome activity in macrophages. Inhibition of inflammasome activation is a novel function for BCAP, which had previously only been shown to stimulate PI3K-dependent signaling pathways in hematopoietic cells, which include the TLR pathway in macrophages, ITAM-dependent pathways in B cells, NK cells and T cells, and the IL-1R pathway in T cells. Our work highlights how inhibition is intrinsically integrated into innate sensing pathways to produce the appropriate amount of inflammation, key to both protective and pathogenic immune responses.
Materials and Methods
Mice
Wild-type C57BL/6J mice were bred at the Benaroya Research Institute or purchased from Jackson Laboratories. BCAP−/- mice lacking the Pik3ap1 gene were back-crossed to C57BL/6J mice for 9 generations (3). Caspase 1/11−/- mice (46) were provided by R. Flavell, Yale University, New Haven, CT. Caspase 11−/- mice (47) were provided by M. Bevan, University of Washington, Seattle, WA. All experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at Benaroya Research Institute and the University of Washington.
Cell Culture
Bone marrow-derived macrophages were generated from bone marrow from femurs and/or tibias of mice by culture for 7 days at 37°C in 5% CO2 in DMEM (Invitrogen) supplemented with 10% FCS, 5 mM HEPES, 0.2 mg/ml L-glutamine, 0.05 mM 2-ME, 50 mg/ml gentamicin sulfate, and 10,000 U/ml penicillin and streptomycin with 30% L cell-conditioned medium or 10% CMG12–14 supernatant as a source of M-CSF. Macrophages were then harvested using PBS containing 1 mM EDTA, suspended in antibiotic free DMEM supplemented with 5% FCS. Macrophages were activated with 100 ng/ml Salmonella minnesota R595 LPS (List Biologicals) 4 hrs prior to nigericin treatment or 16 hrs prior to bacterial infection unless otherwise noted.
Mass spectrometry-based proteomics
Bone marrow derived macrophages (108 cells/condition) were lysed with 1 ml of radioimmunoprecipitation assay buffer (RIPA; 150 mM NaCl, 50 mM Tris [pH 7.4], 1 mM EDTA (pH 8.0), 1% Triton X-100, 0.25% sodium deoxycholate, protease and phosphatase inhibitors). Immunoprecipitation was performed by incubation of lysates with 2 μg anti-BCAP antibody (4L8E6)(4) overnight (4°C) followed by subsequent enrichment using protein A agarose beads (Calbiochem). The resulting protein complexes were washed several times in lysis buffer, twice in ammonium bicarbonate buffer (50 mM), and subsequently were reduced, alkylated and digested into peptides using Trypsin (V5280, Promega). Peptides were loaded onto a 3-cm self-packed C18 capillary pre-column (Reprosil 5 μM, Dr. Maisch). After a 10-min rinse (0.1% Formic Acid), the pre-column was connected to a 25-cm self-packed C18 (Reliasil 3 μM, Orochem) analytical capillary column (inner diameter, 50-μm; outer diameter, 360-μm) with an integrated electrospray tip (~1-μm orifice). Online peptide separation followed by mass spectrometric analyses was performed on a 2D-nanoLC system (nanoAcquity UPLC system, Waters Corp.). Peptides were eluted using a 150-min gradient with solvent A (H2O/Formic Acid, 99.9:1 (v/v)) and B (Acetonitrile/Formic Acid, 99.9:1 (v/v)): 10 min from 0% to 10% B, 105 min from 10% to 40% B, 15 min from 40% to 80% B, and 20 minutes with 100% A. Eluted peptides were directly electrosprayed into a Orbitrap QExactive mass spectrometer (Thermo Fisher Scientific) equipped with a high energy collision cell (HCD).
The mass spectrometer was operated in a data-dependent mode to automatically switch between MS and MS/MS acquisitions. Each full scan (from m/z 300–1500) was acquired in the Orbitrap analyzer (resolution = 70,000), followed by MS/MS analyses on the top twenty most intense precursor ions that had charge states greater than one. The HCD MS/MS scans were acquired using the Orbitrap system (resolution = 17,500) at normalized collision energy of 28%. The precursor isolation width was set at 2 m/z for each MS/MS scan and the maximum ion accumulation times were as follows: MS (100ms), MS/MS (100ms). MS/MS data files were searched using the Comet algorithm (48) and the data was further processed using the Institute for System’s Biology’s Trans-Proteomic Pipeline (49). Static modification of cysteine (carbamidomethylation; 57.02 Da) was used in the search. For precursor area quantification of LRRFIP2, PIK3AP1 and FLII peptides, the data files were imported into the Skyline software package (50) whereupon the retention time windows for each peptide was determined based upon co-elution the MS-MS identification. For each peptide, the relative intensity of the precursor ions was quantified using this retention time window.
Bacterial growth, in vitro infection, and inflammasome activation
Yersinia pseudotuberculosis YPIII serogroup O:3 (Ypstb) and its mutant YpstbΔ (yopJ-, yopO-, yopE-, yopH-, yopK-, yopM-) (31) were grown overnight at 25°C in LB, then diluted 1:40 into LB with 20 mM MgCl2 and 20 mM Na2C2O4, grown at 25°C for 1 hr and then 37°C for 2 hrs. Bacteria were then washed in PBS, added to a MOI of 10, and spun onto macrophages for 3 minutes at 150 g. NLRP3 was induced with 5 μM nigericin (Sigma-Aldrich). Salmonella Typhimurium was grown in LB overnight at 37°C, diluted 1:40, and grown 3 hrs in LB containing 0.3 M NaCl. NAIP5/NLRC4 was induced with FlaTox, consisting of 5 μg/ml LFn-FlaA and 10 μg/ml Protective Antigen (List Biologicals). Recombinant LFn-FlaA was expressed and purified as previously described (33). NAIP2/NLRC4 was induced by RodTox, consisting of mammalian-expressed 20 ng/ml LFn-PrgJ and 4 μg/ml Protective Antigen as previously described (34). To activate the AIM2 inflammasome, macrophages were transfected with calf thymus DNA (Sigma-Aldrich) complexed with Lipofectamine 2000 (Invitrogen). To induce caspase-11 activation, 2 μg/ml LPS (Salmonella minnesota R595, List Biologicals) was mixed with 20 μg/ml Cholera Toxin B subunit (Sigma-Aldrich) in OPTI-MEM (Gibco), added to macrophages primed overnight with LPS after removal of culture media, and spun at 1000 g for 5 min (37). Cytotox 96 kit (Promega) was used to determine LDH release. Experiments measuring IL-1β release, caspase-1 activation, or inflammasome assembly, were performed in the presence of 5 mM glycine to reduce cell lysis (51). IL-1β in supernatants was quantified by ELISA (R&D Systems).
Real-time cell death analysis
BMDMs were seeded on 96-well (RodTox stimulation) or 24-well tissue culture plates (Nigericin stimulation). Prior to live-cell imaging, media was replaced with fresh macrophage media containing 1:5000 Sytox Green dye (Life Technologies) and Hoechst 33342 nucleic acid stain (Molecular Probe). Cells were incubated at 37°C for 20 minutes before adding 5 μM nigericin or 20 ng/ml RodTox. Following the stimuli treatment, cells were immediately visualized for Sytox green uptake (cell death) and Hoechst nuclei staining (total cell number) using CYTATION 3 Imaging reader (BioTek). Images were taken every 2 (nigericin) or 6 (RodTox) minutes for up to 30 (nigericin) or 54 minutes (RodTox). Cell death percentage was calculated as number of Sytox green positive cells divided by the Hoechst positive cell number.
Fluorescence microscopy
Caspase-1 activation and active caspase-1 foci formation was determined by FAM-YVAD-FMK (Immunochemistry Technologies) staining of macrophages infected on glass coverslips. DNA was stained with Hoescht 33342. Cells were washed with PBS and subsequently fixed with Cytofix (BD Biosciences). Immunofluorescent staining was performed post-fixation in PermWash (BD Biosciences) with antibodies specific to NLRP3 (sc-66846, Santa Cruz), ASC (ADI-905-173-100, Enzo), or pro-caspase-1 (sc-514, Santa Cruz) with Alexa fluor-conjugated secondary antibodies (Invitrogen). Coverslips were mounted with ProLong (Molecular Probes) and examined by confocal microscope (Leica SP8X) at the W. M. Keck Microscopy Center. Caspase-1 activation or foci formation was enumerated by counting the fraction of positive cells in four separate fields for each coverslip. Caspase-1 and ASC foci colocalization were enumerated by counting the fraction of positive cells in eight separate fields from duplicate coverslips.
Plasmid construction and transfection
Recombinant vectors encoding mouse Flightless-1 with a C-terminal myc tag and LRRFIP2 with a C-terminal HA tag were constructed by PCR-based amplification of cDNA from mouse bone marrow derived macrophages. Truncated mutants of BCAP were generated by PCR-based mutation and amplification of wild-type BCAP expression vector (3) using Q5® Site-Directed Mutagenesis Kit (E0554S, NEB). Expression plasmids encoding mouse ASC (pUNO1-mASC, Invivogen), NLRP3 (puno1ha-mnalp3, Invivogen), RFP-caspase-1 (C285A) (43), LRRFIP2, BCAP and Flightless-1 were transfected into 293T cells with TransIT®-LT1 transfection reagent (MIR 2300, Mirus) according to the manufacturer’s instructions.
Immunoprecipitation and Immunoblot analysis
Macrophages were lysed at the indicated times in lysis buffer containing 1% Triton X-100, protease inhibitors (Sigma P8340) and sodium orthovanadate (1 mM; Sigma). For immunoprecipitation, macrophages or 293T cells were lysed using lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.25% sodium deoxycholate, 1 mM PMSF, 1 mM sodium orthovanadate and protease inhibitor (Sigma)). Whole cell lysates were incubated with 1.5 μg anti-Flag (2 hours at 4 °C), anti-BCAP or anti-Flightless-1 antibodies (overnight at 4 °C) followed by protein A-agarose (30 minutes 4°C), extensively washed with lysis buffer, and then eluted with boiling in 1x LDS sample buffer (Invitrogen). Lysates or immunoprecipitates were separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membrane, and detected by the indicated antibodies and Immobilon chemiluminescence system (Millipore). The ImageJ software was used to quantify immunoblot bands. For immunoprecipitation blots, the relative intensity was calculated by dividing the intensity of the immunoprecipitated protein by the intensity of same protein in lysates. Relative intensity was calculated by normalizing to WT. For all other experiments, the intensity of the protein of interest was first normalized to β-actin expression and then relative intensity was calculated by normalizing to WT BMDMs. Antibodies used were to Flightless-1 (sc-55583, Santa Cruz), LRRFIP2 (sc-100025, Santa Cruz), caspase-1 p20 (AG-20B-0042B-C100, Adipogen), human caspase-1 (sc-622, Santa Cruz), NLRP3 (15101S, Cell Signaling), ASC (sc-22514-R, Santa Cruz), IL-1β (AF-401-NA, R&D Systems), BCAP (4LE86)(4), Flag M2 (F3165, Sigma Aldrich), c-Myc (sc-40, Santa Cruz).
Lentivirus-Mediated shRNA Knockdown
293T cells were cotransfected with packaging plasmid (psPAX2), envelope plasmid (VSVg) together with a plasmid expressing short hairpin RNA (shRNA) specific for mouse Fli1 (TRCN0000324345, shRNA#1 or TRCN000032413, shRNA#2, Sigma Aldrich) or non-target control shRNA (Sigma Aldrich). We transfected 293T cells in 10-cm dish and harvested the virus-containing culture medium 48 hours after changing the transfection medium to DMEM containing 10% FCS. The collected medium was filtered through 0.45-μm filters. The filtered virus were diluted in DMEM containing 10% FCS and used to infect bone marrow cells after one day of culture. 24 hours after infection, the medium was replaced with fresh macrophage medium containing 5 μg/ml puromycin. After 72 hours of puromycin selection, the macrophages were replated for further experiments.
In Vivo infection
Yersinia pseudotuberculosis YPIII yopM- (29) or Yersinia pseudotuberculosis YPIII cultures were grown overnight in LB broth and enumerated by Coulter counter (Multisizer 4, Beckman Coulter). Yersinia were diluted in PBS for intraperitoneal delivery of 2000 CFU (Yersinia pseudotuberculosis YPIII yopM-) or 1000 cfu (Yersinia pseudotuberculosis YPIII) in 200 μl to 6–8 wk mice. Mice were sacrificed on day 4 and spleens were homogenized and plated for CFU.
Statistical Analysis
Statistics were calculated using GraphPad Prism and tests used for individual experiments are listed in each figure legend.
Supplementary Material
Acknowledgments:
We thank members of the Cookson and Hamerman labs and Drs. Dan Campbell, Adam Lacy-Hulbert and Naeha Subramanian for helpful comments on the manuscript, Dr. C. LaRock for thoughtful discussions, Dr. Caroline Stefani for assistance with live cell imaging, Dr. Wendy Loomis for bacteria and helpful discussions, and Drs. Kerry Campbell, Tomohiro Kurosaki, Adam Lacy-Hulbert, Lynda Stuart and Russell Vance for mice and reagents. We also acknowledge G. Martin and NIH award S10OD016240 for support of the W.M. Keck Microscopy Center at the University of Washington.
Funding: This work was supported by the NIH (R01AI113325, R01AI073441, and R01AI124693 to J.A.H, R01AI057141 to B.T.C, R00HL103768 to R.G.J, T32GM007270–38 to S.J.C, and T32CA009537 and T32AI106677 to J.M.D.).
Footnotes
Publisher's Disclaimer: “This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
Supplementary Materials
Figure S1. BCAP YxxM tyrosines are not required for association with Flightless-1.
Figure S2. No difference in expression of NLRP3 inflammasome components or soluble factors induced by LPS between WT and BCAP−/- macrophages.
Figure S3. BCAP−/- macrophages have increased caspase-1 activation after YpstbΔ infection.
Figure S4. No difference in NLRC4 or AIM2 expression between WT and BCAP−/- macrophages.
Figure S5. BCAP inhibition of inflammasome activation requires Flightless-1.
Figure S6. Model of NLRP3 inflammasome inhibition by BCAP and Flightless-1.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials
References and Notes
- 1.Okada T, Maeda A, Iwamatsu A, Gotoh K, Kurosaki T, BCAP: the tyrosine kinase substrate that connects B cell receptor to phosphoinositide 3-kinase activation, Immunity 13, 817–827 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Yamazaki T, Takeda K, Gotoh K, Takeshima H, Akira S, Kurosaki T, Essential immunoregulatory role for BCAP in B cell development and function, J. Exp. Med 195, 535–545 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamazaki T, Kurosaki T, Contribution of BCAP to maintenance of mature B cells through c-Rel, 4, 780–786 (2003). [DOI] [PubMed] [Google Scholar]
- 4.MacFarlane AW, Yamazaki T, Fang M, Sigal LJ, Kurosaki T, Campbell KS, Enhanced NK-cell development and function in BCAP-deficient mice, Blood 112, 131–140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh MD, Ni M, Sullivan JM, Hamerman JA, Campbell DJ, B cell adaptor for PI3-kinase (BCAP) modulates CD8+ effector and memory T cell differentiation, Journal of Experimental Medicine 215, 2429–2443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deason K, Troutman TD, Jain A, Challa DK, Mandraju R, Brewer T, Ward ES, Pasare C, BCAP links IL-1R to the PI3K-mTOR pathway and regulates pathogenic Th17 cell differentiation, Journal of Experimental Medicine 215, 2413–2428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada S, Zhang H, Hatano M, Tokuhisa T, A Physiologic Role of Bcl-xL Induced in Activated Macrophages. [PubMed]
- 8.Ni M, MacFarlane AW, Toft M, Lowell CA, Campbell KS, Hamerman JA, B-cell adaptor for PI3K (BCAP) negatively regulates Toll-like receptor signaling through activation of PI3K, Proc. Natl. Acad. Sci. U.S.A. 109, 267–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Troutman TD, Hu W, Fulenchek S, Yamazaki T, Kurosaki T, Bazan JF, Pasare C, Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt, Proc. Natl. Acad. Sci. U.S.A. 109, 273–278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battersby A, Csiszár A, Leptin M, Wilson R, Isolation of proteins that interact with the signal transduction molecule Dof and identification of a functional domain conserved between Dof and vertebrate BCAP, J. Mol. Biol 329, 479–493 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Guven-Maiorov E, Keskin O, Gursoy A, Nussinov R, A Structural View of Negative Regulation of the Toll-like Receptor-Mediated Inflammatory Pathway, Biophys. J 109, 1214–1226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halabi S, Sekine E, Verstak B, Gay NJ, Moncrieffe MC, Structure of the Toll/Interleukin-1 Receptor (TIR) Domain of the B-cell Adaptor That Links Phosphoinositide Metabolism with the Negative Regulation of the Toll-like Receptor (TLR) Signalosome, J. Biol. Chem 292, 652–660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Moltke J, Ayres JS, Kofoed EM, Chavarría-Smith J, Vance RE, Recognition of bacteria by inflammasomes, Annu. Rev. Immunol 31, 73–106 (2013). [DOI] [PubMed] [Google Scholar]
- 14.LaRock CN, Cookson BT, Burning down the house: cellular actions during pyroptosis, PLoS Pathog 9, e1003793 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rock KL, Latz E, Ontiveros F, Kono H, The sterile inflammatory response, Annu. Rev. Immunol 28, 321–342 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broderick L, De Nardo D, Franklin BS, Hoffman HM, Latz E, The inflammasomes and autoinflammatory syndromes, Annu Rev Pathol 10, 395–424 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Hauenstein AV, Zhang L, Wu H, The hierarchical structural architecture of inflammasomes, supramolecular inflammatory machines, Curr. Opin. Struct. Biol 31, 75–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schröder GF, Fitzgerald KA, Wu H, Egelman EH, Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes, Cell 156, 1193–1206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taxman DJ, Huang MTH, Ting JP-Y, Inflammasome inhibition as a pathogenic stealth mechanism, Cell Host Microbe 8, 7–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupfer C, Malik A, Kanneganti T-D, Inflammasome control of viral infection, Curr Opin Virol 12, 38–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamkanfi M, Dixit VM, Modulation of inflammasome pathways by bacterial and viral pathogens, J. Immunol 187, 597–602 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Le HT, Harton JA, Pyrin- and CARD-only Proteins as Regulators of NLR Functions, Front Immunol 4, 275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorfleutner A, Chu L, Stehlik C, Inhibiting the inflammasome: one domain at a time, Immunol. Rev 265, 205–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Yin HL, Yuan J, Flightless-I regulates proinflammatory caspases by selectively modulating intracellular localization and caspase activity, J. Cell Biol. 181, 321–333 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin J, Yu Q, Han C, Hu X, Xu S, Wang Q, Wang J, Li N, Cao X, LRRFIP2 negatively regulates NLRP3 inflammasome activation in macrophages by promoting Flightless-I-mediated caspase-1 inhibition, Nat Commun 4, 2075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopecki Z, Cowin AJ, Flightless I: an actin-remodelling protein and an important negative regulator of wound repair, Int. J. Biochem. Cell Biol. 40, 1415–1419 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Kopecki Z, Ludwig RJ, Cowin AJ, Cytoskeletal Regulation of Inflammation and Its Impact on Skin Blistering Disease Epidermolysis Bullosa Acquisita, Int J Mol Sci 17 (2016), doi: 10.3390/ijms17071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellacheruvu D, Wright Z, Couzens AL, Lambert J-P, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin Z-Y, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJR, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras A-C, Nesvizhskii AI, The CRAPome: a contaminant repository for affinity purification-mass spectrometry data, Nat. Methods 10, 730–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaRock CN, Cookson BT, The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing, Cell Host Microbe 12, 799–805 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brodsky IE, Palm NW, Sadanand S, Ryndak MB, Sutterwala FS, Flavell RA, Bliska JB, Medzhitov R, A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system, Cell Host Microbe 7, 376–387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viboud GI, So SSK, Ryndak MB, Bliska JB, Proinflammatory signalling stimulated by the type III translocation factor YopB is counteracted by multiple effectors in epithelial cells infected with Yersinia pseudotuberculosis, Mol. Microbiol 47, 1305–1315 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Miao EA, Rajan JV, Salmonella and Caspase-1: A complex Interplay of Detection and Evasion, Front Microbiol 2, 85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, Vance RE, Rapid induction of inflammatory lipid mediators by the inflammasome in vivo, Nature 490, 107–111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauch I, Tenthorey JL, Nichols RD, Al Moussawi K, Kang JJ, Kang C, Kazmierczak BI, Vance RE, NAIP proteins are required for cytosolic detection of specific bacterial ligands in vivo, Journal of Experimental Medicine 213, 657–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schattgen SA, Fitzgerald KA, The PYHIN protein family as mediators of host defenses, Immunol. Rev 243, 109–118 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Jørgensen I, Miao EA, Pyroptotic cell death defends against intracellular pathogens, Immunol. Rev 265, 130–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA, Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock, Science 341, 1250–1253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Man SM, Ekpenyong A, Tourlomousis P, Achouri S, Cammarota E, Hughes K, Rizzo A, Ng G, Wright JA, Cicuta P, Guck JR, Bryant CE, Actin polymerization as a key innate immune effector mechanism to control Salmonella infection, Proc. Natl. Acad. Sci. U.S.A. 111, 17588–17593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim ML, Chae JJ, Park YH, De Nardo D, Stirzaker RA, Ko H-J, Tye H, Cengia L, DiRago L, Metcalf D, Roberts AW, Kastner DL, Lew AM, Lyras D, Kile BT, Croker BA, Masters SL, Aberrant actin depolymerization triggers the pyrin inflammasome and autoinflammatory disease that is dependent on IL-18, not IL-1β, Journal of Experimental Medicine 212, 927–938 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong Y-N, Peng X, Xi JJ, Chen S, Wang F, Shao F, Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome, Nature 513, 237–241 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Richards N, Schaner P, Diaz A, Stuckey J, Shelden E, Wadhwa A, Gumucio DL, Interaction between pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis, J. Biol. Chem 276, 39320–39329 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Burger D, Fickentscher C, de Moerloose P, Brandt KJ, F-actin dampens NLRP3 inflammasome activity via Flightless-I and LRRFIP2, Sci Rep 6, srep29834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokolovska A, Becker CE, Ip WKE, Rathinam VAK, Brudner M, Paquette N, Tanne A, Vanaja SK, Moore KJ, Fitzgerald KA, Lacy-Hulbert A, Stuart LM, Activation of caspase-1 by the NLRP3 inflammasome regulates the NADPH oxidase NOX2 to control phagosome function, Nat. Immunol 14, 543–553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao W, Yeretssian G, Doiron K, Hussain SN, Saleh M, The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock, J. Biol. Chem 282, 36321–36329 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Rathinam VAK, Fitzgerald KA, Inflammasome Complexes: Emerging Mechanisms and Effector Functions, Cell 165, 792–800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuida K, Lippke J, Ku G, Harding M, Livingston D, Su M, Flavell R, Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme, Science 267, 2000–2003 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM, Non-canonical inflammasome activation targets caspase-11, Nature 479, 117–121 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Eng JK, Jahan TA, Hoopmann MR, Comet: an open-source MS/MS sequence database search tool, Proteomics 13, 22–24 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Keller A, Eng J, Zhang N, Li X-J, Aebersold R, A uniform proteomics MS/MS analysis platform utilizing open XML file formats, Mol Syst Biol 1, 2005.0017–E8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ, Skyline: an open source document editor for creating and analyzing targeted proteomics experiments, Bioinformatics 26, 966–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fink SL, Cookson BT, Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages, Cell. Microbiol 8, 1812–1825 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.