Editorial
Blum’s laboratory has dedicated work to develop an accurate genetic test to predict true liability/risk for addiction and Reward Deficiency Syndrome (RDS) behaviours [1,2]. Genius Health LLC., scientists, in conjunction with their Genomic Testing Centre GTC), have successfully developed the first Genetic Addiction Risk Score (GARS™). The actual association to determine risk using a clinical outcome, termed the Addiction Severity IndexMedia Version (ASI-MV), was accomplished with the Institute of Behavioural Genetics, University of Colorado, Boulder [3–8] and Dominion Diagnostics. The commercial test has just received a Notice of Allowance (application number 14/796.989) on 7–26-18 from the United States Patent and Trademark Office.
To develop this patented Genetic Addiction Risk Score (GARS™), we first selected ten reward candidate genes including Dopamine receptors (DRD1, 2, 3, 4); Dopamine Transporter (DAT1); serotonin transporter, COMT, MAO, GABA, Mu opiate receptor and a number of SNPs and point mutations that influence the net release of dopamine at the brain reward site. The variants or SNPs, including point-mutations, were chosen to reflect a hypo dopaminergic trait. This was based on thousands of association studies providing clear evidence of specific risk alleles for all addictions (Figure 1).
Figure 1:
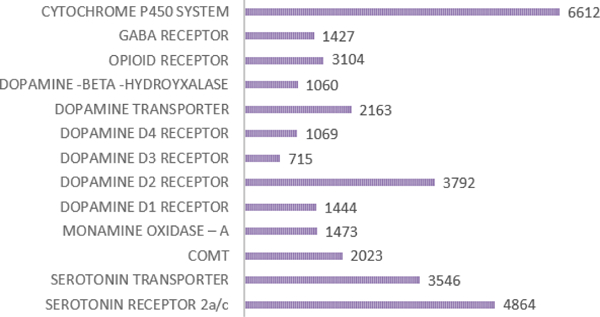
Illustrates the number of studies per genetic risk allele produced in 2014 and is reproduced with permission from Blum et. al [7].
After a preliminary selection phase reviewing many reward gene polymorphisms and risk alleles, we selected the following 10 reward genes. This list was compiled from PubMed in 2014 involving 25,620 various reported articles on the GARS gene panel.
(Table 1) illustrates the reward genes that have been extensively researched and include but are not limited to D1-D4 receptors; Dopamine Transporter (DAT1); Serotonin Transporter (5-HTTLPR); Mu Opiate Receptor (OPRM1); GABA Receptor (GABRB3); Catechol-O-Methyl Transferase (COMT) val158met; and MAO-A gene promoter VNTR.
Table 1:
Proposed Genetic Addiction Risk Score (GARS™) Panel of Reward Genes.
| Dopamine D1 Receptor Gene |
| Dopamine D2 Receptor Gene |
| Dopamine D3 Receptor Gene |
| Dopamine D4 Receptor Gene |
| Serotonin Transporter Gene |
| Dopamine Transporter Gene |
| Mu-opiate Receptor Gene |
| GABA - B3 Receptor Gene |
| Monoamine Oxidase A Gene |
| Catechol-O- Methyl transferase Gene |
| Cytochrome P450 Gene (optional PGX) |
In terms of validation, we partnered with the developers of the Addiction Severity Index- Media Version (ASI-MV), a test mandated in at least 13 states, for both alcohol and drug severity risk scores.8 We contacted seven very diverse treatment centres across the United States resulting in a total of 393 subjects that were genotyped using the selected GARS™ panel. All the data was genotyped and analysed at the Institute for Behavioural Genetics (IBG) at the University of Colorado Boulder. The results indicate a significant association between a summed score of all GARS panel risk alleles (variant forms) and both the ASI-MV alcohol (p<0.004) and drug (P<0.05) severity indices in a total of 273 subjects. Further analysis revealed a clear path to predicting additive behavioural risk. Clearly, carriers of any four risk alleles had a significant prediction of drug severity risk, and carriers of any seven had a significant prediction of alcohol severity risk.
Furthermore, the higher the number of risk alleles, the stronger the prediction of alcohol or drug use severity. Results also demonstrate that family problems, psychological issues and medicalization significantly correlate with risk as well. One important caveat was that if we changed any specific SNP within the GARS panel, the significance was lost. This strongly suggests the importance of the combined GARS™ panel, with any deviation producing false results as may occur with other commercial tests with little to no validation research. These results are further substantiated by other studies including the work of Blum et al [2] revealing that carriers of the DRD2 A1 allele at birth have a 74% chance to become addicted to any one of the RDS behaviours.
The GARS test is indeed a cluster analysis linking these polymorphisms synergistically with an overall expression of DNA predictability to many addictive behaviours. In a recent paper published in the journal Science, the researchers measured the amount of genetic overlap across the disorders using Genome-Wide Association Studies (GWAS) of 265,218 patients and 784,643 controls. They examined the relationships between brain disorders and 17 physical or cognitive measures, such as years of education, from 1,191,588 individuals. The dataset ultimately included all GWAS consortia studying common brain disorders with sufficient sample sizes identified by the team. Their results demonstrated that psychiatric disorders share many genetic variants, while neurological disorders (such as Parkinson’s or Alzheimer’s) appear more distinct.
The results indicate that psychiatric disorders likely have important similarities at a molecular level, which current diagnostic categories do not reflect [9]. This suggests that using GARS may have predictive relevance for addictive behaviours inclusive of RDS, not as a diagnostic, but as a test to identify predisposition of high addiction risk for Substance Use Disorder (SUD).
It is well-known that DNA polymorphisms, in by itself, is impacted by the environment or epigenetics. In fact, it is the known relationship between the two elements whereby the mathematical equation P=G =E represents the resultant phenotype. We believe that this novel tool could help psychiatrists and other clinical professionals identify people at risk for not only substance use disorder but a remarkable array of Reward Deficiency Syndrome (RDS) behaviours [1].
References
- 1.Blum K, Sheridan PJ, Wood RC, Braverman ER, Chen TJ, et al. (1996) Comings DE. The D2 dopamine receptor gene as a determinant of reward deficiency syndrome. J R Soc Med 89: 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum K, Wood RC, Braverman ER, Chen TJ, Sheridan PJ (1995) The D2 dopamine receptor gene as a predictor of compulsive disease: Bayes’ theorem. Funct Neurol 10: 37–44. [PubMed] [Google Scholar]
- 3.Blum K, Madigan MA, Fried L, Braverman ER, Giordano J, et al. (2017) Coupling genetic addiction risk score (GARS) and pro dopamine regulation (KB220) to combat substance use disorder (SUD). Glob J Addict Rehabil Med 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blum K, Chen ALC, Thanos PK, Febo M, Demetrovics Z, et al. (2018) Genetic addiction risk score (GARS)™, a predictor of vulnerability to opioid dependence. Front Biosci (Elite Ed).10: 175–196. [DOI] [PubMed] [Google Scholar]
- 5.Blum K, Downs BW, Dushaj K, Li M, Braverman ER, et al. (2016) The benefits of customized DNA directed nutrition to balance the brain reward circuitry and reduce addictive behaviours. Precis Med (Bangalore) 1: 18–33. [PMC free article] [PubMed] [Google Scholar]
- 6.Blum K, Oscar-Berman M, Demetrovics Z, Barh D, Gold MS (2014) Genetic Addiction Risk Score (GARS): molecular Neurogenetic evidence for predisposition to Reward Deficiency Syndrome (RDS). Mol Neurobiol 50: 765–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blum K, Badgaiyan RD, Agan G, Fratantonio J, Simpatico T, et al. (2015) Molecular Genetic Testing in Reward Deficiency Syndrome (RDS): Facts and Fiction. J Reward Defic Syndr 1: 65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler SF, McNaughton EC, Black RA (2015) Tapentadol abuse potential: a postmarketing evaluation using a sample of individuals evaluated for substance abuse treatment. Pain Med 16: 119–130. [DOI] [PubMed] [Google Scholar]
- 9.Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, et al. (2018) Analysis of shared heritability in common disorders of the brain. Science 360: eaap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]


