SUMMARY
Producing healthy, high-oleic oils and eliminating trans-fatty acids from foods are two goals that can be addressed by reducing activity of the oleate desaturase, FAD2, in oilseeds. However, it is essential to understand the consequences of reducing FAD2 activity on the metabolism, cell biology and physiology of oilseed crop plants. Here, we translate knowledge from studies of fad2 mutants in Arabidopsis (Arabidopsis thaliana) to investigate the limits of non-GMO approaches to maximize oleic acid in the seed oil of canola (Brassica napus), a species that expresses three active FAD2 isozymes. A series of hypomorphic and null mutations in the FAD2.A5 isoform were characterized in yeast (Saccharomyes cerevisiae). Then, four of these were combined with null mutations in the other two isozymes, FAD2.C5 and FAD2.C1. The resulting mutant lines contained 71–87% oleic acid in their seed oil, compared with 62% in wild-type controls. All the mutant lines grew well in a greenhouse, but in field experiments we observed a clear demarcation in plant performance. Mutant lines containing less than 80% oleate in the seed oil were indistinguishable from wild-type controls in growth parameters and seed oil content. By contrast, lines with more than 80% oleate in the seed oil had significantly lower seedling establishment and vigor, delayed flowering and reduced plant height at maturity. These lines also had 7–11% reductions in seed oil content. Our results extend understanding of the B. napus FAD2 isozymes and define the practical limit to increasing oil oleate content in this crop species.
Keywords: FAD2, fatty acids, high-oleic oil, seed lipid metabolism, seed oil, triacylglycerol
INTRODUCTION
Vegetable oils are major components of human diets, but the fatty-acid compositions of oils from many crop species are not optimal for human nutrition. For example, while high-oleic (18:1) oils have recognized health benefits (Gillingham et al., 2011), many oils contain 30–60% linoleic (18:2) and linolenic (18:3) fatty acids that are synthesized from 18:1 by desaturase enzymes (Wallis et al., 2002; Bates et al., 2013). High 18:2 + 18:3 in the oil reduces the shelf life of food products made from the oil because these polyunsaturated fatty acids are substrates for oxidation and free-radical reactions that produce off flavors and rancidity. Industrial processing by partial hydrogenation has traditionally been used to lower the polyunsaturated content of soybean and other oils, but this results in the production of trans-fats that are unhealthy and no longer acceptable in foods (Steinhart et al., 2003; Micha and Mozaffarian, 2009).
The gateway enzyme for synthesis of polyunsaturated fatty acids in seeds is the oleoyl-phosphatidylcholine Δ12 desaturase of the endoplasmic reticulum, and in Arabidopsis (Arabidopsis thaliana) this enzyme is encoded by the FAD2 gene (Ohlrogge and Browse, 1995; Wallis et al., 2002). Our previous characterization of Arabidopsis fad2 mutants illuminated the biochemistry of 18:1 desaturation (Miquel and Browse, 1992), indicated that FAD2 is constitutively expressed in all tissues of the plant (Okuley et al., 1994), and established that 18:1 desaturation by FAD2 is essential for low-temperature survival (Miquel et al., 1993; Miquel and Browse, 1994). A T-DNA allele of fad2, fad2-5 allowed us to clone FAD2 by a gene-tagging approach (Okuley et al., 1994).
Reducing 18:1 desaturation in oilseed crops by mutation or molecular-genetic silencing of FAD2 homologs can provide high-oleic oil, precluding the need for partial hydrogenation with its attendant production of trans-fats. Soybean (Glycine max) contains seed-specific FAD2 isogenes, FAD2-1A and FAD2-1B (Heppard et al., 1996), and reducing expression of these by gene silencing has led to the successful release of high-oleic soybean varieties marketed by Dupont-Pioneer (as Plenish™) and Monsanto (as Vistive Gold™; Kinney, 1996; Wilson, 2012) that contain 72–77% 18:1 in the oil.
In Brassica species, the FAD2 genes are expressed in all plant tissues. Canola (Brassica napus) is an allotetraploid and contains four FAD2 isogenes. Using the standardized B. napus genome nomenclature (Chalhoub et al., 2014), these are designated FAD2.A1, FAD2.A5, FAD2.C1 and FAD2.C5, where the nomenclature indicates the subgenome (A or C) and the chromosome location of the gene. In B. napus, the FAD2.A1 isoform contains deletion and insertion events that preclude it from encoding an active enzyme (Wells et al., 2014). (Wells et al., use BnaA.FAD2.b, BnaA.FAD2.a, BnaC.FAD2.b and BnaC.FAD2.a for FAD2.A1, FAD2.A5, FAD2.C1 and FAD2.C5, respectively.) The FAD2.A5, .C5 and .C1 isoforms encode closely related proteins (> 90% sequence identity). Mutations in these genes demonstrate that they contribute to polyunsaturated fatty-acid synthesis in all organs and tissues of the plant. In leaves, the changes in fatty-acid composition are relatively subtle, because the chloroplast localized desaturase encoded by the fad6 genes provides considerable desaturation (Miquel and Browse, 1992; Table S1a). However, in roots, polyunsaturated fatty acids (18:2 + 18:3) are reduced from 48% of the total fatty acids in wild-type to less than 20% in the Bnfad2.a5/c5 double-mutant (Table S1b), and these changes are comparable to those seen in the Arabidopsis fad2 mutants (Lemieux et al., 1990).
Conventional and mutant breeding programs have produced commercial canola varieties that have 72–80% 18:1 in the oil (and about 12% 18:2 + 18:3), and these are successfully grown around the world. By combining mutations at all three BnFAD2 loci, Wells et al. (2014) produced lines with up to 84% 18:1 (6% 18:2 + 18:3), while Peng et al. (2010) using RNA-interference generated lines with up to 85% 18:1. These lines apparently grew well under greenhouse conditions. Several patents (e.g. U.S. patent 6 414 223; Kodali et al., 2002) also describe the production of mutant canola lines containing up to 85% oleate in the seed oil, but detailed field performance and yield data are not available. Based on our characterization of Arabidopsis fad2 mutants, we hypothesize that plant growth and performance of fad2 null lines will be poor at low temperatures. In particular, cellular functions may be compromised by reduced membrane unsaturation in vegetative tissues at low temperatures (Miquel et al., 1993). Here, we describe genetic and biochemical experiments that use information obtained from the Arabidopsis fad2 alleles to inform the consideration of 41 mutations generated in the three functional FAD2 genes of canola. Five selected mutations were brought together in various combinations to produce lines ranging from 60 to 87% 18:1 in their seed oils. Field evaluation of growth parameters and seed oil content among these lines indicate that seed oil levels of 18:1 above 80% are associated with poor seedling establishment, reduced plant vigor and reduced seed oil content in field-grown plants.
RESULTS
Characterization of Arabidopsis fad2 alleles
To extend our previous characterization of Arabidopsis fad2 mutants, we analyzed the fatty-acid compositions of mature seeds from six allelic fad2 lines grown at 22°C (Table 1). The fad2-2, fad2-3 and fad2-4 seeds all contained approximately 65% 18:1 and less than 5% 18:2 + 18:3, compared with approximately 50% 18:2 + 18:3 in wild-type seeds. On this basis, these lines are considered null, or near null, at the fad2 locus. The small amounts of polyunsaturated fatty acids in seed samples of these mutants are likely products of the chloroplast 16:1/18:1 desaturase encoded by the FAD6 gene (Browse et al., 1989). The fad2-1 and fad2-6 seeds contained less than 56% 18:1 and more than 12% 18:2 + 18:3, suggesting that these alleles encode proteins with a low level of desaturase activity. The fad2-5 allele contains a T-DNA insert in the 5’-untranslated region of FAD2 and expresses a reduced level of wild-type FAD2 mRNA (Okuley et al., 1994).
Table 1.
Fatty-acid compositions of seeds from wild-type and six allelic fad2 mutants of Arabidopsis
| Allele | Fatty acids (percent in total) | |||||
|---|---|---|---|---|---|---|
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:1 | |
| WT | 8.2 | 2.5 | 14.6 | 28.2 | 21.9 | 19.5 |
| fad2-1 | 6.9 | 2.8 | 55.7 | 4.2 | 8.2 | 19.2 |
| fad2-2 | 6.3 | 2.8 | 65.3 | 0 | 2.5 | 19.8 |
| fad2-3 | 7.2 | 3.0 | 65.0 | 0 | 2.9 | 21.2 |
| fad2-4 | 6.2 | 2.3 | 64.6 | 0.6 | 3.3 | 19.2 |
| fad2-5a | 8.5 | 4.0 | 37.7 | 8.1 | 11.3 | 26.0 |
| fad2-6 | 6.9 | 2.8 | 53.1 | 5.3 | 8.5 | 20.9 |
Data for this T-DNA allele are from Okuley et al. (1994).
WT, wild-type.
While wild-type Arabidopsis grows and completes its life-cycle at 6°C, the three null fad2 alleles stop growing and eventually die at this temperature (Miquel et al., 1993). By contrast, fad2-1 plants survive and grow like wild-type at 6°C, presumably because the fad2-1 encoded protein is stable and active at low temperatures (Miquel et al., 1993).
To further explore the molecular genetics and biochemistry of these Arabidopsis fad2 isoforms, we cloned full-length fad2 cDNAs from fad2-1, fad2-2, fad2-3, fad2-4 and fad2-6 plants. Sequencing of the fad2-3 cDNA revealed a G→A mutation that encodes an in-frame stop codon following residue 286 of the FAD2 protein (Figure 1). The predicted mutant protein lacks the third histidine motif (Figure 1) that is essential for binding iron at the active site of the enzyme. This is consistent with fad2-3 being a null allele. The similarity of seed fatty-acid composition (Table 1) and plant phenotype (Miquel et al., 1993) of fad2-2 (S136F) and fad2-4 (P280L) with fad2-3 suggests that these alleles are also null. Both fad2-1 (A104T) and fad2-6 (T148I) support some desaturation in planta (Table 1), and data indicate that fad2-1 is a temperature-sensitive allele (Miquel et al., 1993).
Figure 1.
Five mutant alleles of Arabidopsis FAD2. The positions of mutations listed in Table 2 are shown in red. Conserved sequences that coordinate two Fe atoms at the active site are in bold. Four putative transmembrane sequences are underlined in green.
We expressed the fad2-1, fad2-2, fad2-3, fad2-4 and fad2-6 cDNAs in yeast (Saccharomyes cerevisiae) under control of the strong, constitutive alcohol dehydrogenase (ADH) promoter. Because yeast only synthesizes saturated and monounsaturated fatty acids, including 18:1, it is well suited for the heterologous expression and characterization of eukaryotic 18:1 desaturases (Kajiwara et al., 1996; Peyou-Ndi et al., 2000). We measured the fatty-acid composition of transgenic yeast strains by gas chromatography after 48 h growth at 30 and 20°C, and calculated the percentage conversion of endogenous 18:1–18:2. Consistent with the data in Table 1, yeast expressing fad2-2 and fad2-3 cDNAs did not show any synthesis of 18:2 (Table 2). Yeast expressing the fad2-1 cDNA showed only 2% conversion of 18:1 to 18:2 when grown at 30°C, but the extent of conversion was 10-fold higher in yeast grown at 20°C. By contrast, yeast expressing fad2-6 showed only a small increase in 18:2 synthesis when the temperature was lowered from 30 to 20°C (Table 2). These results indicate that fad2-1 is a temperature-sensitive allele and provides a biochemical rationale for the survival of fad2-1 plants at low temperatures (Miquel et al., 1993).
Table 2.
18:2 synthesis in yeast expressing wild-type or mutant Arabidopsis FAD2 proteins
| Strain | Mutation | 18:2 synthesis | |
|---|---|---|---|
| 30°C | 20°C | ||
| Empty vector | 0 | 0 | |
| WT | – | 32.1 ± 4.5 | 58.5 ± 1.5 |
| fad2-1 | A104T | 2.1 ± 0.1 | 40.3 ± 1.3 |
| fad2-2 | S136F | 0 | 0 |
| fad2-3 | W287stop | 0 | 0 |
| fad2-4 | P280L | 0 | 0 |
| fad 2-6 | T148I | 6.1 ± 1.0 | 35.1 ± 0.8 |
Yeast expressing each protein were cultured for 48 h at either 30 or 20°C.
Data are 18:2 as percentage of 18:1 and 18:2; mean ± SE from three separate experiments.
WT, wild-type.
Transcript abundance of BnFAD2 genes in canola seeds
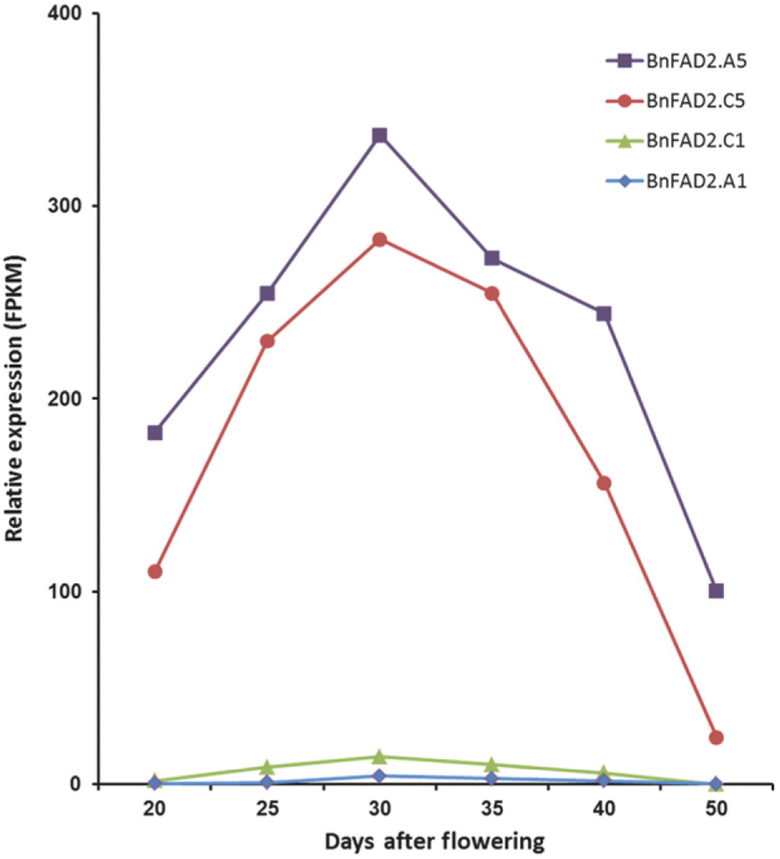
Transcript profiles of developing B. napus seeds were analyzed using established methods (Troncoso-Ponce et al., 2011) to determine expression of the four BnFAD2 genes at six time points over the course of seed development. The data (Figure 2) indicate that transcripts encoding FAD2.A5 and FAD2.C5 dominate during seed development, while transcripts for FAD2.C1 are approximately 10-fold lower in abundance. Almost no sequences attributable to the FAD2.A1 gene were detected, consistent with the prediction that this gene is non-functional. The patterns of expression of the FAD2.A5, .C5 and .C1 isoforms in seeds were similar, with transcript abundance (measured as fragments per kilobase of transcript per million mapped reads; FPKM) doubling between 14–20 days after flowering (DAF) and 26–30 DAF before falling to lower levels as seeds approach maturity (Figure 2).
Figure 2.
Expression of BnFAD2 genes during seed filling in Brassica napus. Data are from RNAseq analyses on seed harvested at six stages of development.
Identification of mutations in BnFAD2 genes
We determined FAD2.A5, .C5 and .C1 genomic DNA sequences from 9600 plants in a B. napus mutant M2 population generated from ethyl methanesulfonate treatment. One line, designated high-oleate 103 (HIOL103) was determined to have a G→A transition that converts the codon for tryptophan 101 of the FAD2.C5 protein to a stop codon (W101 stop). The HIOL109 line contains a W190 stop mutation in the FAD2.C1 protein. Each of these mutations is predicted to prevent translation of a functional protein and we consider the mutations null.
Because results from B. napus and Arabidopsis indicate that completely eliminating FAD2 activity may compromise plant growth and survival at low temperatures, we identified a collection of 39 mutations in the FAD2.A5 gene with the aim of identifying a range of hypomorphic mutations that could help us define the upper limit of 18:1 content in seed oil that can be achieved without compromising plant performance and oil yield under field conditions. Six of the 39 mutations were silent DNA changes (no amino acid change) and 13 encoded conserved changes in the protein sequence (Table S2). Another five mutations, including four that change an amino acid in one of the three histidine-rich sequences (histidine boxes) that bind iron at the active site, and one encoding S136F equivalent to the Arabidopsis fad2-2 null allele were considered likely null. The remaining 15 mutations encode non-conserved amino acid substitutions in residues that are conserved among FAD2 sequences from different species (Table S2).
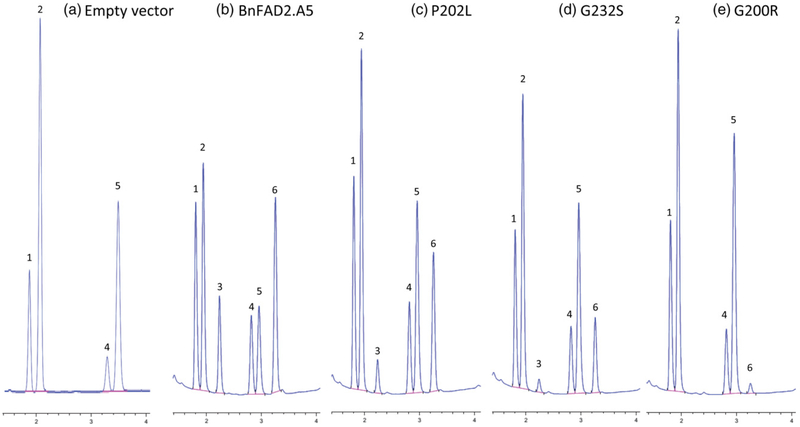
To test our prediction that these remaining mutations would include a series of FAD2 proteins spanning a range of desaturase enzyme activity, we chose to determine the enzyme activities of these mutant fad2 proteins using our yeast expression system. Each mutation was incorporated into an otherwise wild-type FAD2 cDNA that was transformed into yeast under control of the ADH promoter. Similar to other eukaryotic Δ12 desaturases (Kajiwara et al., 1996; Peyou-Ndi et al., 2000), the wild-type BnFAD2.5 enzyme allowed for the synthesis of both 18:2 and 16:2 from their monounsaturated precursors (Figure 3). When grown at 30°C, five of the yeast strains showed no detectable conversion of 18:1 to 18:2, while the remaining 10 strains exhibited between 5% and 65% conversion of 18:1 to 18:2 compared with 77% conversion for the wild-type FAD2 enzyme (Figure 3; Table 3). When grown at 20°C, several of the tested strains showed higher conversion of 18:1 and 18:2; for example, conversion by the G232D mutant was more than sixfold higher at 20°C than at 30°C.
Figure 3.
Oleate desaturation by wild-type and mutant BnFAD2.A5 desaturases expressed in yeast. Fatty acid methyl esters were prepared from yeast lipids and analyzed using gas chromatography. (a) Yeast transformed with empty vector; (b) wild-type BnFAD2.A5; (c) P202L; (d) G232S; (e) G200R. Fatty acids are: 1, 16:0; 2, 16:1; 3, 16:2; 4, 18.0; 5, 18:1; 6, 18:2.
Table 3.
18:2 synthesis in yeast expressing wild-type or mutant Brassica FAD2.A5 proteins
| Mutation | 18:2 synthesis | |
|---|---|---|
| 30°C | 20°C | |
| Empty | 0 | 0 |
| vector | ||
| WT | 66.6 ± 0.4 | 67.8 ± 1.3 |
| G257R | 0 | 0 |
| P159L | 0 | 0 |
| G123D | 0 | 0 |
| E152K | 0 | 0 |
| T297I | 0 | 0 |
| S199L | 0.19 ± 0.15 | 1.84 ± 0.78 |
| G200R | 3.22 ± 0.13 | 8.68 ± 0.54 |
| P216L | 3.57 ± 0.27 | 21.3 ± 3.9 |
| G232D | 5.11 ± 0.69 | 33.1 ± 1.1 |
| S150F | 8.56 ± 0.50 | 40.4 ± 5.4 |
| G248R | 20.7 ± 1.5 | 64.9 ± 1.3 |
| P191L | 24.3 ± 2.4 | 54.5 ± 2.0 |
| G232S | 36.8 ± 6.3 | 65.9 ± 1.7 |
| P202L | 40.7 ± 2.6 | 64.0 ± 3.4 |
| E287K | 57.7 ± 1.7 | 76.9 ± 2.6 |
Yeast expressing each protein was cultured for 48 h at either 30 or 20°C.
Data are 18:2 as percentage of 18:1 + 18:2; mean ± SE for three separate experiments.
WT, wild-type.
Production of Brassica napus lines with 60–87% 18:1 in seeds
Based on the information summarized in Table S2 and the results of yeast assays (Table 3), we selected four B. napus lines carrying mutations in the FAD2.A5 gene (summarized in Table 4). These lines were each crossed to plants containing null mutations in both the FAD2.C5 and FAD2.C1 genes (W101 stop and W190 stop, respectively; Table 4). Following selfing of the resulting F1 plants, allele-specific oligonucleotides were employed to identify F2 plants homozygous for one, two or all three of the mutations, as well as plants homozygous wild-type at all three loci. Plants representing the 19 possible genotypes were grown to maturity in a greenhouse, and the fatty-acid composition of seed samples was determined by gas chromatography. Table 5 shows part of the seed analysis, and full data on the fatty-acid compositions are included in Table S3. Across the 19 lines, seed 18:1 content ranged from 61.1 to 87.1%, while content of polyunsaturated fatty acids (18:2 + 18:3) varied inversely from 30 to 3.6%. The highest 18:1 (and lowest 18:2 + 18:3) was achieved by the combination of the fad2.a5-1 (S199L) mutation (HIOL112) with the null mutations in .C5 and .C1 genes. The fad2.a5-2 (G200R) mutation (HIOL113) was only slightly less effective in blocking 18:1 desaturation. Plants carrying the fad2.a5-3 (G232S) mutation (HIOL116) produced seeds containing 77.2% 18:1 and 13.9% 18:2 + 18:3. The fad2.a5-4 (P202L) mutation (HIOL114) had a substantially smaller effect on raising seed 18:1 content (Table 5). Under greenhouse conditions, plants of all the mutant lines showed good seedling establishment and growth. No differences in plant height, flowering time or yield were observed.
Table 4.
Summary information for six Brassica napus lines containing mutations in FAD2 genes
| Gene | Mutation | Allele | Abbreviation | Line |
|---|---|---|---|---|
| FAD2.A5 | S199L | fad2.a5-1 | a5-1 | HIOL 112 |
| FAD2.A5 | G200R | fad2.a5-2 | a5-2 | HIOL 113 |
| FAD2.A5 | G232S | fad2.a5-3 | a5-3 | HIOL 116 |
| FAD2.A5 | P202L | fad2.a5-4 | a5-4 | HIOL 114 |
| FAD2.C5 | W101stop | fad2.c5-1 | c5 | HIOL 103 |
| FAD2.C1 | W190stop | fad2.c1-1 | c1 | HIOL 109 |
Table 5.
Distribution of 18-carbon unsaturated fatty acids in seeds of Brassica napus lines with mutations in FAD2 genes grown in a greenhouse
| Genotype | Percent of total fatty acids | ||
|---|---|---|---|
| 18:1 | 18:2 | 18:3 | |
| Wild-type | 61.8 | 18.9 | 9.2 |
| a5-1 | 72.6 | 10.1 | 8.2 |
| a5-2 | 73.1 | 9.1 | 8.6 |
| a5-3 | 66.8 | 14.7 | 9.3 |
| a5-4 | 61.0 | 19.3 | 10.7 |
| c5 | 69.0 | 12.7 | 8.5 |
| c1 | 63.3 | 18.4 | 9.1 |
| c5/c1 | 70.8 | 11.3 | 8.6 |
| a5-1/c5 | 85.5 | 1.8 | 3.5 |
| a5-1/c1 | 74.0 | 8.5 | 7.9 |
| a5-1/c5/c1 | 87.1 | 1.3 | 2.3 |
| a5-2/c5 | 85.6 | 1.7 | 3.7 |
| a5-2/c1 | 75.0 | 7.9 | 8.1 |
| a5-2/c5/c1 | 86.5 | 1.3 | 2.7 |
| a5-3/c5 | 75.4 | 7.8 | 7.8 |
| a5-3/c1 | 74.3 | 8.5 | 8.3 |
| a5-3/c5/c1 | 77.2 | 6.6 | 7.3 |
| a5-4/c5 | 68.8 | 12.5 | 9.6 |
| a5-4/c1 | 62.4 | 18.3 | 10.0 |
| a5-4/c5/c1 | 70.6 | 11.5 | 9.0 |
Assessment of plant performance and oil composition in the field
Because the fad2.a5 P202L mutation had only a small effect on seed oil composition (Table 5), we did not further investigate the HIOL114 lines. For the other three fad2.a5 mutations, we excluded sublines that contained only one (homozygous) mutation and selected F3 sublines homozygous for two or three of the mutations, along with segregants that were wild-type at all three loci. This provided a set of 10 sublines that in the greenhouse had produced seeds with 62–87% 18:1, and these were grown in the field for further evaluation.
The field experiment had a randomized, complete-block design with four replicates at each of three locations to the west and south of Ghent, Belgium (see Experimental procedures for details). Each plot was assessed when the plants were at the 2–3 leaf stage (Establishment) and at the 5–6 leaf stage (Vigor) using a nine-point scale, in which 1 indicates very poor, 5 normal and 9 very strong. Days from the beginning and to the end of flowering, and the height of plants at maturity were also recorded. Finally, the protein and oil contents of seeds were measured. Analysis of variance (ANOVA) was used to calculate least significant difference for each parameter (P<0.05). The mean values of the seven parameters for each line are shown in Table 6, along with 18:1 contents of the greenhouse-grown seed used for these trials. Asterisks indicate values that differ significantly from the wild-type control. Analysis of the fatty-acid compositions of seed harvested in this field experiment (Table S4) shows 18:1 content for each line that is similar to, but slightly lower than, the value recorded in Table 5 for the greenhouse-grown seed used for planting this experiment.
Table 6.
Growth and physiology of Brassica napus lines with mutations in FAD2 genes during field trials
| Genotype | Seed 18:1 (%) | Establishment | Vigor | Days to flowering | End of flowering | Plant height (cm) | Protein (%) | Oil (%) |
|---|---|---|---|---|---|---|---|---|
| Wild-type | 61.8 | 4.9 | 6.8 | 46.6 | 67.9 | 122 | 29.2 | 41.6 |
| c5/c1 | 70.8 | 4.8 | 6.5 | 46.8 | 68.1 | 124 | 28.6 | 42.7 |
| a5-1/c5 | 85.5 | 4.1* | 2.8* | 47.7* | 71.8* | 98* | 34.4* | 36.8* |
| a5-1/c1 | 74.0 | 5.0 | 6.6 | 46.7 | 68.3 | 120 | 29.3 | 42.0 |
| a5-1/c5/c1 | 87.1 | 3.9* | 2.2* | 47.9* | 71.8* | 93* | 33.8* | 38.2* |
| a5-2/c5 | 85.6 | 4.3* | 2.8* | 47.5* | 70.6* | 97* | 32.3* | 38.6* |
| a5-2/c1 | 75.0 | 4.8 | 5.7 | 46.6 | 67.8 | 121 | 28.4 | 42.3 |
| a5-2/c5/c1 | 86.5 | 4.1* | 2.3* | 47.8* | 71.7* | 90* | 32.9* | 38.1* |
| a5-3/c5 | 75.4 | 4.8 | 6.5 | 46.5 | 67.8 | 117 | 29.5 | 40.6 |
| a5-3/c1 | 74.3 | 4.9 | 6.5 | 47.4 | 68.7 | 123 | 30.3 | 39.9 |
| a5-3/c5/c1 | 77.2 | 5.0 | 6.7 | 46.9 | 67.9 | 123 | 29.2 | 41.0 |
Asterisks indicate values that are significantly different (P < 0.05) from wild-type, based on ANOVA analysis.
The results in Table 6 show that all the mutant lines that contain less than 80% 18:1 in their seed oil were indistinguishable from wild-type in the parameters for growth, physiology and seed composition that we measured. By contrast all the mutant lines with more than 80% 18:1 in the seed oil had significantly lower seedling establishment and vigor, delayed flowering and reduced plant height at maturity. These lines also had 7–11% reductions in seed oil content relative to wild-type, with concomitant increases in seed protein. It is unlikely that these phenotypes are caused by mutations in genes other than FAD2. All the mutant lines were backcrossed to wild-type at least three times before being used in the experiments described here. Furthermore, crosses of the Bnfad2.c5 Bnfad2.c1 double-mutant with both Bnfad2.a5-1 and Bnfad2.a5-2 produced triple-mutant progeny with similar defects in growth and oil yield in the field.
DISCUSSION
The imperative of eliminating trans-fats from human diets (Micha and Mozaffarian, 2009) and the recognized health benefits of high-oleic vegetable oils (Gillingham et al., 2011) have provided the impetus to develop cultivars of major oilseed crops with maximum levels of 18:1 in the oil. High-oleic sunflower oil contains up to 83% 18:1 (Flagella et al., 2002), and this represents a level that has not yet been achieved in available soybean and canola varieties. Transgenic approaches that effectively silence the seed-specific FAD2-1 genes of soybean were first developed 20 years ago (Kinney, 1996). However, high-oleic soy oil was not of interest to the food industry until restrictions on trans-fats, and thus partial hydrogenation, led to a 22% reduction in market share for soy oil in the USA (United Soybean Board, 2013). Although the high-oleic Plenish™ and Vistive Gold™ varieties developed by Dupont-Pioneer and Monsanto have been tested by the food industry over the last 10 years (Waltz, 2010), regulatory barriers to genetically modified crops, especially in some non-US markets, have limited commercial development. The identification and combining of mutations in the soy FAD2-1A and FAD2-1B genes has also led to the production of high-oleic lines, with up to 86% 18:1 and approximately 4% 18:2 + 18:3. Preliminary trials indicate that these non-GMO lines grow and yield well. Further development and evaluation of these lines are in progress (Pham et al., 2011).
By contrast, breeding efforts with canola (B. napus) have had less success, producing commercial lines with up to 80% 18:1 and a minimum of 12% 18:2 + 18:3 (Wells et al., 2014). A key question is whether it is possible to produce canola lines with greater than 80% 18:1 that perform well during growth under normal field conditions. Here, we used information from our work on Arabidopsis desaturases, and an extensive collection of sequenced mutations in the B. napus FAD2.A5, FAD2.C5 and FAD2.C1 genes to explore the biochemical basis and physiological limits of high-oleic low-polyunsaturated traits in canola.
Arabidopsis plants that have null mutations at the FAD2 locus, fad2-2 and fad2-3 grow poorly, especially at low temperatures (Miquel et al., 1993), and this suggested the likelihood that B. napus lines that are null at all three FAD2 loci will similarly be compromised in growth and development under field conditions. Whereas fad2-2 plants stopped growing and eventually died when exposed to 6°C growth temperature, plants of the Arabidopsis fad2-1 line grew as well as wild-type controls, because the fad2-1 mutation is leaky, particularly at low temperatures (Miquel et al., 1993). When we expressed a cDNA of the fad2-1 coding sequence in yeast, the mutant protein, FAD2 A104T did indeed provide for the synthesis of 18:2 in yeast lipids (Table 2). These results indicated that identifying a set of leaky alleles in one or more of the three isogenes for FAD2 in canola would allow us to produce the range of multiple-mutant lines needed to investigate the relationship between seed fatty-acid composition and the physiology of plants grown under greenhouse and field conditions.
We first identified B. napus lines with null mutations in the FAD2.C5 and FAD2.C1 genes (Table 4). Next, we used information about essential and conserved residues in FAD2 and other desaturase proteins, chemical properties of amino acids, and data from mutant Arabidopsis alleles to assess 39 different mutations in the B. napus FAD2.A5 sequence that were identified by sequencing mutant populations. Fifteen mutations were identified as good candidates to provide low 18:1 desaturation, and a cDNA corresponding to each of the mutant alleles was expressed in yeast. Five of the mutant constructs did not support any detectable synthesis of 18:2, indicating that these alleles are probably null. The remaining 10 mutant constructs supported 18:2 synthesis to different extents (Table 3). Based on data from yeast grown at 30°C, we identified two mutations that provided less than 5% the desaturase activity of the control wild-type construct (S199L and G200R) and two that provided 55–60% of wild-type activity (G232S and P202L). After at least three cycles of backcrossing to wild-type, the B. napus lines containing these mutations (HIOL112, HIOL113, HIOL116 and HIOL114, respectively) were each crossed to a line (HIOL103/109) with null mutations in the FAD2.C5 and FAD2.C1 genes to allow the selection of 19 F2 plant lines containing mutations in one, two or all three of the FAD2 genes. Under greenhouse conditions, these lines grew well and produced seed with 18:1 varying from 61 to 87% of total fatty acids (Table 5). However, when 10 of these lines producing high-oleic oils (seed 18:1 ranging from 71 to 87%) were grown in field trials, the four with 18:1 higher than 80% grew poorly and produced seed with reduced oil content (Table 6). Lines with up to 77% 18:1 were indistinguishable from wild-type during growth in these field trials and produced seed with wild-type oil content.
Our results indicate that conventional and mutation breeding approaches have reached the limit of increased 18:1 levels in canola. More targeted approaches, such as seed-specific expression of RNAi constructs (Peng et al., 2010) targeting the three B. napus FAD2 genes, allow for the maintenance of wild-type levels of polyunsaturated fatty acids in non-seed tissues. However, all current methods for seed-specific knockdown of FAD2 expression require the use of transgenic approaches, and objections to genetically modified crops is a substantial barrier to commercialization. In addition, results from Arabidopsis fad2 mutants suggest that normal seed development and germination require some minimal content of polyunsaturated (18:2 + 18:3) fatty acids (Miquel and Browse, 1994), so more research and new approaches are needed to increase 18:1 content of B. napus seed oil above 80%.
EXPERIMENTAL PROCEDURES
Arabidopsis mutants
Seeds of Arabidopsis (A. thaliana) ecotype Col-0 (wild-type) and the fad2 mutants were sown directly on soil. The sown seeds were incubated at 5°C for 48 h, then cultivated at 22°C with 16 or 24 h light at 100–150 μE m−2. Seeds from mature plants were collected for analysis.
Fatty acid and lipid analysis
The overall fatty-acid composition of leaves was determined by heating samples at 80°C in 1 mL of 2.5% (v/v) H2SO4 in methanol for 1 h in screw-capped tubes. After the addition of 1.5 mL of 0.9% NaCl solution and 1 mL of hexane, fatty acids were extracted into the hexane phase by shaking and the tubes were centrifuged at low speed. Samples (1 μL of the hexane phase were separated by gas chromatography on a 15 m × 0.53 mm Carbowax column, and quantified using a flame ionization detector. The gas chromatograph was programmed for an initial temperature of 150°C for 3 min followed by an increase of 15°C per min to 210°C; this final temperature was maintained for a further 12 min.
For the analysis of canola seed samples, about 0.8 g of seed (ground or whole) was weighed and added to 2.0 mL of sodium methoxide solution, 1.0 mL of petroleum ether and a metal crushing rod in a scintillation vial, and crushed in an Eberbach shaker to extract the oil. The fatty acids of the o-acyl lipids were converted to their methyl ester derivatives using sodium methoxide and extracted into the hexane phase. These methyl esters were analyzed by capillary gas-liquid chromatography on a Varian Model 430, with a flame ionization detector and a Zebron ZB-Wax column (15 m × 0.32 mm ID × 0.50 μm film thickness).
Yeast expression
The Arabidopsis FAD2 gene does not have an intron within the open reading frame. We therefore used gene-specific primers to amplify each mutant allele from genomic DNA prepared from leaf tissue of the corresponding mutant. These were each ligated into the episomal yeast expression vector pMK195 (Peyou-Ndi et al., 2000). Directional cloning of the cDNA into this vector provided for expression of the FAD2 proteins under the control of the constitutive ADH promoter. The resulting construct was introduced into S. cerevisiae strain YRP685 (MATa, leu2, lys2, his4, trp1, ura3) using the lithium acetate procedure (Peyou-Ndi et al., 2000). Transformed cells were grown in a complete minimal media supplemented with 2% glucose but lacking uracil, as pMK195 encodes ura prototrophy. For fatty-acid analyses, cells were grown for 48 h in selective media in the presence of glucose. One milliliter of the culture was centrifuged and the cells were resuspended in 1 mL of 2.5% (v/v) H2SO4 in methanol, and then derivatized, extracted and analyzed by gas chromatography as described above. In these experiments, novel fatty acids were identified by comparison of their retention times with authentic 16:2 and 18:2 fatty acids (NuChek-Prep, Elysian, MN, USA).
For analyses of the Brassica FAD2.A5 mutant alleles, a wild-type sequence was first cloned into the pMK195 vector. Then site-directed mutagenesis using overlap polymerase chain reaction (Bok and Keller, 2012) was employed to introduce mutations corresponding to the 15 mutant alleles listed in Table 3. These were each expressed in yeast and analyzed for 18:2 synthesis as described above.
Identification of Brassica FAD2 genes
A TBLASTN homology search using the Arabidopsis FAD2 gene sequence (At3g12120) was used as the query in a search of databases of Brassica rapa sequences and of Brassica oleracea sequences. The BLAST analyses resulted in the identification of two FAD2 gene homologs for B. rapa (BrFAD2.1, BrFAD2.2), and two FAD2 gene homologs for B. oleracea (BoFAD2.1, BoFAD2.2). cDNAs corresponding to these sequences were predicted using Fgenesh software (Salamov and Solovyev, 2000). A BLAST homology search of a database containing B. napus mRNA sequences using the B. rapa BrFAD2 gene sequences resulted in the identification of the cDNA sequences of B. napus BnFAD2.A5 and BnFAD2.A1. Similarly, a BLAST homology search of the in-house database containing B. napus mRNA sequences using the B. oleracea BoFAD2 gene sequences as a query resulted in the identification of the cDNA sequences of BnFAD2.C5 and BnFAD2.C1. The B. napus BnFAD2.A5, BnFAD2.C5 and BnFAD2.C1 genes encode FAD2 proteins of 384 amino acids (Figure S1).
Expression analysis of Brassica napus FAD2 genes
The relative gene expression levels of B. napus FAD2 genes were determined through analysis of Illumina mRNAseq-derived transcriptome databases obtained for six different seed developmental stages according to the methods of Troncoso-Ponce et al. (2011). Gene expression levels were calculated taking into account a normalization step for the sequencing depth per database (target reads per million reads in the database) and for the target gene length [fragments per kilobase per million reads in the database (RPKM); Mortazavi et al., 2008].
Generation and isolation of mutant Brassica napus fad2 alleles
Mutations in the BnFAD2 genes were generated and identified as follows: 30 000 seeds from an elite spring oilseed rape breeding line were pre-imbibed for 2 h on wet filter paper in deionized water. Half of the seeds were exposed to 0.8% ethyl methanesulfonate (EMS) and half to 1% EMS (Sigma: M0880), and incubated for 4 h. The mutagenized seeds (M1 seeds) were rinsed three times and dried in a fume hood overnight. M1 plants were grown in soil and selfed to generate M2 seeds. M2 seeds were harvested for each individual M1 plant. Two times 4800 M2 plants derived from different M1 plants were grown, and DNA samples were prepared from leaf samples of each individual M2 plant. The DNA samples were screened for the presence of point mutations in the FAD2 genes by direct sequencing, and analyzing the sequences for the presence of the point mutations using the NovoSNP software (http://www.molgen.ua.ac.be/bioinfo/novosnp/. All of the mutant lines selected for detailed investigation were backcrossed to wild-type at least three times before being used in the experiments reported.
Oil composition in seeds from Brassica napus combining BnFAD2.A5, BnFAD2.C5 and BnFAD2.C1 alleles grown in the greenhouse
Brassica plants comprising mutant BnFAD2.A5. BnFAD2.C5 and BnFAD2.C1 alleles were crossed. Following selfing of the resulting progeny, seeds from plants homozygous for FAD2.A5, FAD2.C5 or FAD2.C1 mutations, and combinations thereof, and wild-type seg-regants were identified by sequencing of the three loci. The fatty-acid composition of the seed oil of these lines was determined by gas chromatography, as described above.
Measurements of plant growth and seed composition of mutant lines in the field
Fatty-acid composition and plant performance parameters were determined from plants grown in the field. Brassica lines with mutant BnFAD2.A5, BnFAD2.C5 or BnFAD2.C1 alleles, and combinations thereof, and wild-type segregants were grown at three different sites, west and south of Ghent, Belgium at 1, Astene; 2, Kruishoutem; and 3, Maarkedal. The sites had different soil types (1, sandy; 2, sand + low loam; 3, sand + high loam) but similar seasonal weather patterns. At the time of planting (22–23 April 2014), night-time minimum temperatures were close to the longterm averages (7–8°C). At each site, plants of each line were grown in four, replicate 2 × 1.5-m plots with six rows of plants in each plot. During most of the 4-month growing season, average diurnal maximum and minimum temperatures were in the ranges 17–22°C and 10–14°C, respectively. Rainfall over the 4-month growing season at the sites was approximately: 1, 240 mm; 2, 200 mm; 3, 280 mm. Harvest dates were: 1, 28 August; 2, 1 September; 3, 21 August.
Fatty-acid composition in the seed oil was determined as described above for the seed from each plot. The following plant performance parameters were determined: Establishment at the 2–3 leaf stage on a scale 1–9, where 1 =very thin, 5 = average, 9 = very thick; Vigor at the 5–6 leaf stage on a scale 1–9, where 1 = poor, 5 = average, 9 = vigorous; Flowering Start: the stage (in days after seeding) at which 10% is in flower; Flowering End: the stage (in days after seeding) at which 10% remains in flower; Plant Height at the stage of flowering end in cm; Oil content in the seed (as percent of whole seed); protein content in the seed (as percent of whole seed). Seed quality parameters were obtained through gas chromatography analysis. For the statistical analysis an ANOVA test was performed to identify significant differences between the mutant lines and the wild-type segregants.
Supplementary Material
Figure S1. Amino acid sequences of three isozymes of FAD2 from Brassica napus.
Table S1 Fatty-acid compositions of leaves (a) and roots (b) from Brassica napus wild-type and two Bnfad2 double-mutants
Table S2 Summary information for 39 mutations in the Brassica napus FAD2.A5 gene identified in this study
Table S3 Fatty-acid compositions of seeds of Brassica napus lines with mutations in FAD2 genes grown in a greenhouse
Table S4 Distribution of 18-carbon unsaturated fatty acids in seeds of Brassica napus lines with mutations in FAD2 genes grown in the field
ACKNOWLEDGEMENTS
This work was supported by BASF Agricultural Solutions by Agriculture and Food Research Initiative (AFRI) competitive award number 2018-67013-27459 from the USDA National Institute of Food and Agriculture, and by the Agricultural Research Center at Washington State University. The authors are grateful to Jo Dierickx, Liesbeth Vercruysse and Lieven Paermentier for growing and molecular characterization of the plants, and to their colleagues at BASF Agricultural Solutions and WSU for helpful discussion.
Footnotes
CONFLICT OF INTEREST
The authors declare no competing financial interests.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Bates PD, Stymne S and Ohlrogge J (2013) Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol 16, 358–364. [DOI] [PubMed] [Google Scholar]
- Bok JW and Keller NP (2012) Fast and easy method for construction of plasmid vectors using modified quick-change mutagenesis. Methods Mol. Biol 944, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, Kunst L, Anderson S, Hugly S and Somerville C (1989) A mutant of Arabidopsis deficient in the chloroplast 16:1/18:1 desaturase. Plant Physiol. 90, 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S et al. (2014) Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Flagella Z, Rotunno T, Tarantino E, Di Caterina R and De Caro A (2002) Changes in seed yield and oil fatty acid composition of high oleic sunflower (Helianthus annuus L.) hybrids in relation to the sowing date and the water regime. Eur. J. Agron 17, 221–230. [Google Scholar]
- Gillingham LG, Harris-Janz S and Jones PJ (2011) Dietary monounsaturated fatty acids are protective against metabolic syndrome and cardiovascular disease risk factors. Lipids 46, 209–228. [DOI] [PubMed] [Google Scholar]
- Heppard EP, Kinney AJ, Stecca KL and Miao GH (1996) Developmental and growth temperature regulation of two different microsomal ω-6 desaturase genes in soybeans. Plant Physiol. 110, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara S, Shirai A, Fujii T, Toguri T, Nakamura K and Ohtaguchi K (1996) Polyunsaturated fatty acid biosynthesis in Saccharomyces cerevisiae: expression of ethanol tolerance and the FAD2 gene from Arabidopsis thaliana. Appl. Environ. Microbiol 12, 4309–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney AJ (1996) Genetic engineering of soybean oils for food applications. J. Food Lipids 3, 273–292. [Google Scholar]
- Kodali DR, Fan Z and DeBonte LR (2002) Plants, seeds and oils having an elevated total monounsaturated fatty acid content. US Patent 6(414), 223. [Google Scholar]
- Lemieux B, Miquel M, Somerville C and Browse J (1990) Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor. Appl. Genet 80, 234–240. [DOI] [PubMed] [Google Scholar]
- Micha R and Mozaffarian D (2009) Trans fatty acids: effects on metabolic syndrome, heart disease and diabetes. Nat. Rev. Endocrinol 5, 335–344. [DOI] [PubMed] [Google Scholar]
- Miquel M and Browse J (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J. Biol. Chem 267, 1502–1509. [PubMed] [Google Scholar]
- Miquel M and Browse J (1994) High-oleate oilseeds fail to develop at low temperature. Plant Physiol. 106, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel M, James D Jr, Donner H and Browse J (1993) Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc. Natl Acad. Sci. USA 90, 6208–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L and Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB and Browse JA (1995) Lipid biosynthesis. Plant Cell 7, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E and Browse J (1994) The Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell6, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q, Hu Y, Wei R, Zhang Y, Guan C, Ruan Y and Liu C (2010) Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds. Plant Cell Rep. 29, 317–325. [DOI] [PubMed] [Google Scholar]
- Peyou-Ndi MM, Watts JL and Browse J (2000) Identification and characterization of an animal Δ12 fatty acid desaturase gene by heterologous expression in Saccharomyces cerevisiae. Arch. Biochem. Biophys 376, 399–408. [DOI] [PubMed] [Google Scholar]
- Pham AT, Lee JD, Shannon JG and Bilyeu KD (2011) A novel FAD2-1 A allele in a soybean plant introduction offers an alternate means to produce soybean seed oil with 85% oleic acid content. Theor. Appl. Genet 123, 793–802. [DOI] [PubMed] [Google Scholar]
- Salamov AA and Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhart H, Rickert R and Winkler K (2003) Trans fatty acids (TFA): analysis, occurrence, intake and clinical relevance. Eur. J. Med. Res 8, 358–362. [PubMed] [Google Scholar]
- Troncoso-Ponce MA, Kilaru A, Cao X, Durrett TP, Fan J, Jensen JK, Thrower NA, Pauly M, Wilkerson C and Ohlrogge JB (2011) Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 68, 1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Soybean Board (2013) FY 2013 Action Plan, pp. 49–56. Available at www.unitedsoybean.org
- Wallis JG, Watts JL and Browse J (2002) Polyunsaturated fatty acid synthesis: what will they think of next? Trends Biochem. Sci 27, 467–473. [DOI] [PubMed] [Google Scholar]
- Waltz E (2010) Food firms test fry Pioneer’s trans fat-free soybean oil. Nat. Biotech 28, 1129. [DOI] [PubMed] [Google Scholar]
- Wells R, Trick M, Soumpourou E, Clissold L, Morgan C, Werner P, Gibbard G, Clarke M, Jennaway R and Bancroft I (2014) The control of seed oil polyunsaturate content in the polyploid crop species Brassica napus. Mol. Breed 33, 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RF (2012) The role of genomics and biotechnology in achieving global food security for high-oleic vegetable oil. J. Oleo Sci 61, 357–367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Amino acid sequences of three isozymes of FAD2 from Brassica napus.
Table S1 Fatty-acid compositions of leaves (a) and roots (b) from Brassica napus wild-type and two Bnfad2 double-mutants
Table S2 Summary information for 39 mutations in the Brassica napus FAD2.A5 gene identified in this study
Table S3 Fatty-acid compositions of seeds of Brassica napus lines with mutations in FAD2 genes grown in a greenhouse
Table S4 Distribution of 18-carbon unsaturated fatty acids in seeds of Brassica napus lines with mutations in FAD2 genes grown in the field