Abstract
γ-Secretase is an intramembrane-cleaving protease that produces amyloid-β peptide 42 (Aβ42), which is the toxic and aggregation-prone species of Aβ that causes Alzheimer's disease. Here, we used the substituted cysteine accessibility method to analyze the structure of transmembrane domains (TMDs) 4 and 5 of human presenilin 1 (PS1), a catalytic subunit of γ-secretase. We revealed that TMD4 and TMD5 face the intramembranous hydrophilic milieu together with TMD1, TMD6, TMD7, and TMD9 of PS1 to form the catalytic pore structure. Notably, we found a correlation in the distance between the cytosolic sides of TMD4/TMD7 and Aβ42 production levels, suggesting that allosteric conformational changes of the cytosolic side of TMD4 affect Aβ42-generating γ-secretase activity. Our results provide new insights into the relationship between the structure and activity of human PS1.
SIGNIFICANCE STATEMENT Modulation of γ-secretase activity to reduce toxic amyloid-β peptide species is one plausible therapeutic approaches for Alzheimer's disease. However, precise mechanistic information of γ-secretase still remains unclear. Here we identified the conformational changes in transmembrane domains of presenilin 1 that affect the proteolytic activity of the γ-secretase. Our results highlight the importance of understanding the structural dynamics of presenilin 1 in drug development against Alzheimer's disease.
Keywords: allosteric change, amyloid, enzyme, membrane protein, protease, secretase
Introduction
γ-Secretase is an intramembrane-cleaving protease that cleaves amyloid precursor protein (APP) to generate amyloid-β peptide (Aβ) with C-terminal heterogeneity (Tomita and Iwatsubo, 2013; Tomita, 2014). Aβ42 is a minor but toxic and aggregation-prone Aβ species that is predominantly deposited in the brains of patients with Alzheimer's disease (AD; Iwatsubo et al., 1994; Holtzman et al., 2011). Thus, understanding the molecular mechanism by which γ-secretase produces Aβ42 is important for the development of therapeutic agents against AD. γ-Secretase comprises four transmembrane proteins: presenilin (PS), anterior pharynx-defective 1, Nicastrin, and presenilin enhancer 2 (Pen-2; Takasugi et al., 2003). PS is an aspartic protease subunit that shows proteolytic activity and autoproteolysis to generate N- and C-terminal fragments (NTF and CTF, respectively) on formation of the full complex. Recently, the crystal structure of Methanoculleus marisnigri JR1 (mmPSH), which is an archaeon PS homolog, was resolved (Li et al., 2013). However, unlike PS, mmPSH can catalyze the proteolysis of substrates without additional cofactors, and, furthermore, it is difficult to resolve the crystal structure of multimeric membrane proteins. We previously analyzed the structure of membrane-embedded, proteolytically active PS1 using the substituted cysteine accessibility method (SCAM) and cross-linking experiments (Sato et al., 2006, 2008; Takagi et al., 2010). SCAM enables identification of the hydrophilic environment by the accessibility of sulfhydryl reagents to cysteine residues (Cys) introduced at a desired position. Cross-linking experiments reveal the arrangement of the transmembrane domains (TMDs). We also performed competition experiments using γ-secretase inhibitors to identify residues that are critical to proteolytic activity. Using a combination of these approaches, we found that PS1 harbors a hydrophilic “catalytic pore” structure formed by TMD1, TMD6, TMD7, and TMD9 in the membrane. In this study, we analyzed the structure of PS1 TMD4 and TMD5, which are thought to play important roles in the binding of PS1 with Pen-2 and stabilization of the complex, respectively (Kim and Sisodia, 2005; Watanabe et al., 2005, 2010). We found that both TMD4 and TMD5 face the hydrophilic pore in the membrane and are involved in forming the catalytic site structure of γ-secretase. In particular, the cytoplasmic side of TMD4 forms a flexible transmembrane structure located in proximity to the catalytic site. We found that alterations in the distance between the cytosolic sides of TMD4 and TMD7 correlate with Aβ42 secretion, suggesting that TMD4 conformation is a critical factor regulating Aβ42-generating activity of γ-secretase.
Materials and Methods
The polyclonal antibodies PS1NT, G1Nr3, and G1Nr5 were raised against the N terminus of the recombinant human PS1 protein (Leem et al., 2002; Sato et al., 2008). Anti-M5 antibody was against the hydrophilic loop (HL) region, namely, amino acids 299–313 of human PS1 (Honda et al., 1999). Pen-2 was detected by anti-PNT3 antibody (Isoo et al., 2007). The anti-human Aβ 82E1 antibody (1:2500 dilution; catalog #10323; Immuno-Biological Laboratories), the anti-Xpress antibody (1:2500 dilution; catalog #R910-25; Thermo Fisher Scientific) and the anti-cleaved Notch1 V1744 antibody (1:500 dilution; catalog #4147; Cell Signaling Technology) were purchased from the indicated companies. l-685,458 [(1S-Benzyl-4R-[1-(1S-carbamoyl-2-phenylethylcarbamoyl)-1S-3-methyl-butylcarbamoyl]-2R-hydroxy-5-phenylpentyl) carbamic acid tert-butyl ester (Shearman et al., 2000)] and peptide 15 (pep15; Das et al., 2003) were purchased from Bachem and BEX, respectively. N-[N-(3,5-difluorophenacetyl)]-L-alanyl]-S-phenylglycine-tert-butyl ester (DAPT) was synthesized as described previously (Dovey et al., 2001; Kan et al., 2004; Morohashi et al., 2004). Methanethiosulfonate (MTS) cross-linkers (Sato et al., 2006) were dissolved in dimethylsulfoxide at 8 mm. A catalyst was made from copper sulfate in distilled water and phenanthroline in ethanol at 10 and 50 mm, respectively. These compounds were used at a final concentration of 100 and 300 μm, respectively. cDNAs encoding APP carrying Swedish mutation (APPNL), NotchΔE, PS1, Cysless PS1 [PS1/Cys(−)], and His-Xpress-tagged PS1 were described previously (Kopan et al., 1996; Watanabe et al., 2005; Sato et al., 2006; Isoo et al., 2007). Single- or double-Cys mutant (mt) PS1 cDNAs were generated using a long PCR-based protocol. Maintenance of embryonic fibroblasts derived from Psen1/Psen2 double knock-out (DKO) mice of either sex (Herreman et al., 2000) or from Psenen knock-out (P2KO) mice of either sex (Bammens et al., 2011), retroviral infections (Kitamura et al., 2003), and generation of stable infectants were performed as described previously (Watanabe et al., 2005; Sato et al., 2006, 2008; Watanabe et al., 2010). Microsome preparation and immunoblot analysis were performed as described previously (Tomita et al., 1997, 1999; Takahashi et al., 2003). For the measurement of secreted Aβ, recombinant retroviruses encoding each Cys mt PS1 were transiently infected into DKO cells stably expressing APPNL (Watanabe et al., 2005). After 24 h of incubation, conditioned media were collected and subjected to two-site ELISAs (i.e., BNT77/BA27 and BNT77/BC05 for Aβ40 and Aβ42, respectively; Asami-Odaka et al., 1995). Biotinylation and competition experiments using N-biotinoylaminoethyl methanethiosulfonate (MTSEA-biotin) in intact cells or microsome fractions have been described previously (Sato et al., 2006, 2008). Cross-linking experiments were performed as described previously except that microsomes were incubated with MTS cross-linkers (200 μm) or CuSO4 (100 μm) and 1,10-phenanthroline (300 μm) as a catalyst for 30 min at 37°C (Kobashi, 1968; Sato et al., 2006; Takagi et al., 2010; Takeo et al., 2012). For cross-linking experiments, microsomes were incubated with a catalyst for 10 min at 37°C. Aβ42-lowering compound GSM-1 (30 μm; Ohki et al., 2011) was preincubated for 30 min at 4°C before incubation for cross-linking. For the competition assay, sodium 2-sulfonatoethyl methanethiosulfonate (MTSES) or 2-(trimethylammonium)ethyl methanethiosulfonate bromide (MSTET) was preincubated with intact cells at 2 mm for 30 min or microsomes at 2 mm for 5 min at 4°C and washed once before biotinylation. Inhibitors were preincubated with microsome aliquots for 30 min at 4°C before incubation for biotinylation at concentrations that completely abolish the proteolytic activity of γ-secretase (l-685,458, 1 μm; pep15, 1 μm; DAPT, 10 μm; Morohashi et al., 2006; Sato et al., 2006).
Results
SCAM analysis of TMD4 and TMD5 of PS1
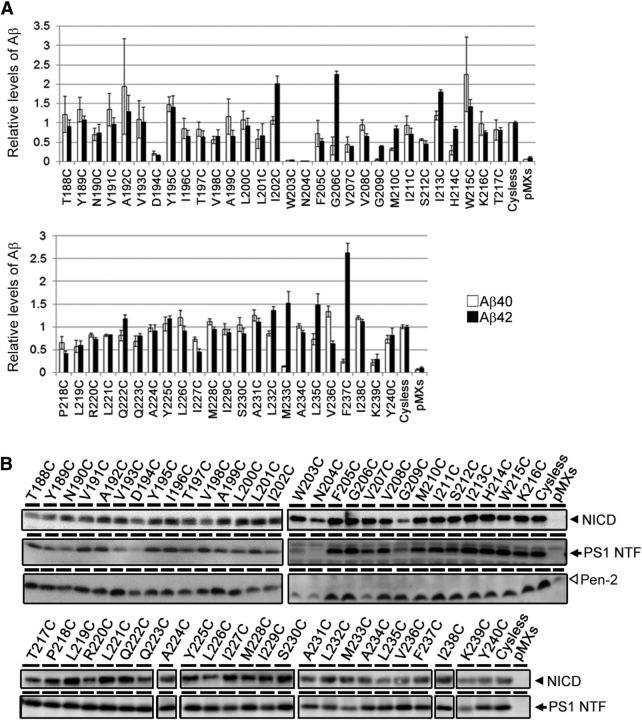
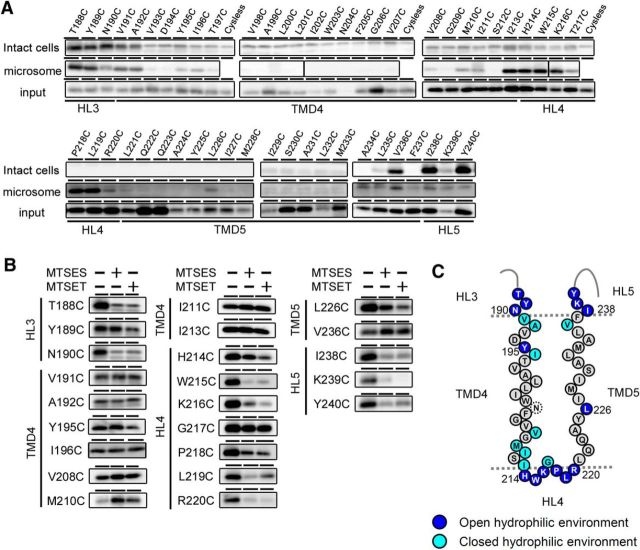
We previously demonstrated that PS1 forms an intramembranous hydrophilic pore-like structure in the area around its catalytic aspartates (Sato et al., 2006). To examine whether TMD4 and TMD5 of PS1 are embedded in the hydrophobic membrane or face this hydrophilic environment, we constructed single-Cys mt PS1s mutated at all 53 amino acid positions (i.e., T188 to Y240; Fig. 1). Some single-Cys mt PS1s failed to recover Aβ production (i.e., total Aβ for W203C and N204C; Aβ40 for G209C and M233C; Aβ42 for D194C; Fig. 2A). In addition, consistent with previous findings that TMD4 is critical for Pen-2 interaction (Kim and Sisodia, 2005; Watanabe et al., 2005), levels of Pen-2 and PS1 NTF were low in some single-Cys mt PS1s (i.e., D194C, W203C, N204C, and V207C for Pen-2; N190C, V193C, D194C, W203C, N204C, and G209C for PS1 NTF). These data suggest the possibility that substitution at these amino acid residues affected the metabolism and stability of γ-secretase complex. However, all single-Cys mt PS1s retained their γ-secretase activities to generate Notch intracellular domain (NICD; Fig. 2B), suggesting that these mutants harbor γ-secretase activity at minimal level. We then tested the reactivities of these mt PS1s to MTSEA-biotin. SCAM analysis of intact cells revealed that T188C, Y189C, N190C, V191C, A192C, V236C, I238C, K239C, and Y240C mt PS1s all reacted with MTSEA-biotin from the extracellular side (Fig. 3A). In addition to these residues, SCAM analysis using microsome fractions demonstrated that Y195C, I196C, V208C, M210C, I211C, I213C, H214C, W215C, K216C, G217C, P218C, L219C, R220C, and L226C mt PS1s were labeled with MTSEA-biotin. These results confirmed that TMD4 and TMD5 span the lipid bilayer with a type I and II orientation, respectively. To clarify whether the residues face hydrophilic environments inside or outside of the membrane, we used charged MTS reagents (i.e., MTSES and MTSET, which have a negatively charged and positively charged group, respectively), which are not able to access closed hydrophilic environments such as inside the membrane, as labeling competitors in the SCAM analysis (Sato et al., 2006, 2008; Takagi et al., 2010). The labeling of T188C, Y189C, N190C, H214C, W215C, K216C, P218C, L219C, R220C, L226C, I238C, K239C, and Y240C by MTSEA-biotin was decreased by preincubation with the charged MTS reagents, suggesting that these positions in the HL face an open hydrophilic environment (Fig. 3B). These data indicated that TMD4 and TMD5 comprise 23 (V191 to I213) and 17 (L221 to F237) residues, respectively (Fig. 3C). Notably, several residues on the cytosolic side of TMD4 face the intramembranous hydrophilic environment.
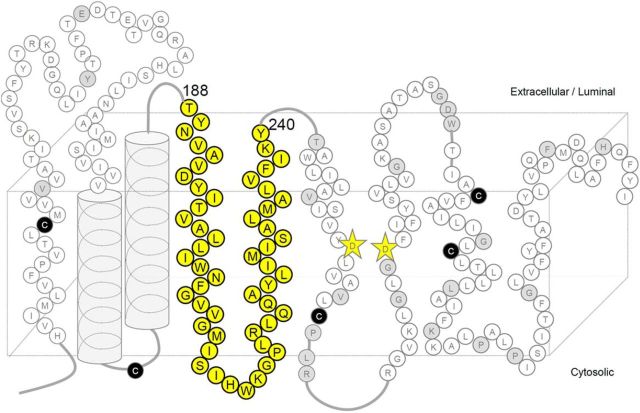
Figure 1.
Locations of the Cys mutations of PS1 used in this study. Schematic diagram of human PS1. Endogenous Cys that were replaced with Ser in PS1/Cys(−) are shown as black circles. Amino acid residues substituted to Cys in this and previous studies are shown as circles with their single-letter character representing the original amino acids. Residues analyzed in this study are shown as yellow circles. Residues in which Cys substitution resulted in a loss of γ-secretase activity are shown as gray circles. Catalytic aspartates are shown as yellow stars.
Figure 2.
γ-Secretase activity of single-Cys mt PS1 used in this study. A, Sandwich ELISA of secreted Aβ from APPNL-stable DKO cells transiently transfected with single-Cys mt PS1 (n = 3–12, means ± SEs). The levels of secreted Aβ in the conditioned media were normalized by that of cells transfected with PS1/Cys(−). Amounts of Aβ40 production are shown as white bars, and amounts of Aβ42 production are shown as black bars. B, NICD generation from NotchΔE-stable DKO cells transiently transfected with single-Cys mt PS1 was detected by immunoblot analysis using an anti-cleaved Notch1 antibody (top panels). The levels of expressed PS1 NTF and Pen-2 were detected by immunoblot analysis using anti-PS1 NTF and Pen-2 antibodies (bottom panels).
Figure 3.
SCAM analysis of single-Cys mt PS1s mutated around TMD4 and TMD5. A, Biotinylation experiment using MTSEA-biotin in intact cells (top panels) and microsomes (middle panels). Amounts of PS1 NTF in the input fraction are shown in the bottom panels. Putative domains are indicated below the panels. B, Labeling competition of single-Cys mt PS1s that were labeled by MTSEA-biotin in A was performed after preincubation with negatively charged MTSES or positively charged MTSET. Putative domains are indicated at the left of the panels. C, Summary of the biotinylation experiments using MTSEA-biotin and competition experiments using charged MTS reagents. All charged reagents were accessible to the Cys-substituted amino acids shown as dark blue circles (open hydrophilic environment). Residues in which labeling were not competed by MTSES or MTSET are shown as light blue circles (closed hydrophilic environment). Residues that were not labeled by MTSEA-biotin are shown as black letters in gray circles. N204C (dotted circle) was not endoproteolyzed.
Reactivity of substituted Cys in the presence of γ-secretase inhibitors
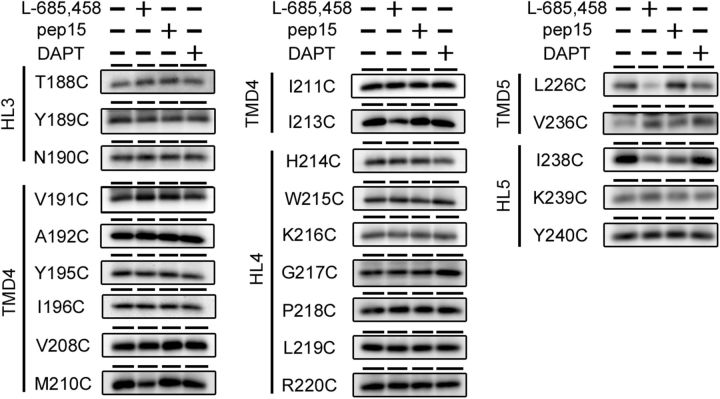
Next, we examined the effects of γ-secretase inhibitors (GSIs) on the water accessibility of residues in TMD4 and TMD5. We used three types of inhibitors, namely, l-685,458, pep15, and DAPT. l-685,458 is a transition-state-analog-type GSI that binds to catalytic aspartates (Li et al., 2000). pep15 is a helical-peptide-type GSI and targets the initial substrate-binding site of γ-secretase near its active site (Das et al., 2003; Kornilova et al., 2005). DAPT is a dipeptide-type GSI that binds to a domain distinct from the catalytic site or the substrate-binding site in PS1 CTF (Morohashi et al., 2006). Using these GSIs as competitors in the SCAM analyses, we were able to identify the GSI binding sites and/or the conformational changes on γ-secretase inhibition (Sato et al., 2006, 2008; Takagi et al., 2010; Ohki et al., 2011). The labeling of M210C, I213C, L226C, and I238C decreased with l-685,458 treatment, suggesting that these residues are involved in formation of the catalytic site (Fig. 4). In addition, labeling of I238C also decreased with pep15 treatment. Notably, the amount of labeling of V236C increased with all inhibitors, similarly to that of G78C (Takagi et al., 2010). These data suggested that hydrophilic environment around V236 and I238 is allosterically altered in the inhibitory conformation of γ-secretase induced by the GSI binding.
Figure 4.
Labeling competition by GSIs. Labeling of single-Cys mt PS1s by MTSEA-biotin was performed after preincubation with l-685,458, pep15, or DAPT. Putative domains are indicated at the left of the panels.
Topological mapping of TMD4 and TMD5 using cross-linking experiments
To understand the topological features of these functional residues in TMD4 and TMD5, we performed cross-linking experiments using double-Cys mt PS1 harboring a pair of Cys mutations: one Cys was located within TMD4 or TMD5, and the other Cys was chosen from the residues that face the catalytic pore in the CTF (L383 in the GxGD motif, I387 in TMD7, and L435 in the PAL motif). In addition, we analyzed the I238C/D450C double mutant, because the water accessibilities of both I238C and D450C were decreased with pep15 (Sato et al., 2008). All double-Cys mt PS1s retained their γ-secretase activity (Fig. 5A). We also used M2M, M3M, M4M, M6M, M8M, M11M, M14M, and M17M, which are sulfhydryl-to-sulfhydryl cross-linking reagents with spacer arms 5.2, 6.5, 7.8, 10.4, 13.0, 16.9, 20.8, and 24.7 Å long, respectively (Loo and Clarke, 2001). In addition, we used a complex of copper ions and 1,10-phenanthroline (Cu-Phe) as a catalyst for the oxidation-reduction reaction to form a disulfide bond between Cys that are able to collide with each other (Kobashi, 1968). Most active mutants were cross-linked to the NTF and CTF of PS1, except for V191C/I387C or V208C/L383C (Fig. 5B–D). These results show that the hydrophilic interface of TMD4 and TMD5 face the catalytic pore. In particular, I213C/L383C, L226C/L383C, and L226C/I387C are cross-linked directly by the catalysis (Cu-Phe; Fig. 5B,C), indicating that TMD4 and TMD5 are located close to TMD7 around the catalytic site. In contrast, I213C/L435C, L226C/L435C, and V236C/L435C are cross-linked with cross-linkers that are longer than M8M (Fig. 5C,D), suggesting that these residues are located in proximity to the catalytic aspartate but not the PAL motif. Notably, I213 at the membrane boundary of TMD4 is closer to L383 and I387 than M210, suggesting that the cytosolic side of TMD4 bends into the catalytic pore as part of the loop region. In addition, I238C and D450C are also cross-linked (Fig. 5D), suggesting the possibility that the luminal side of TMD5 is involved in the formation of the substrate binding site.
Figure 5.
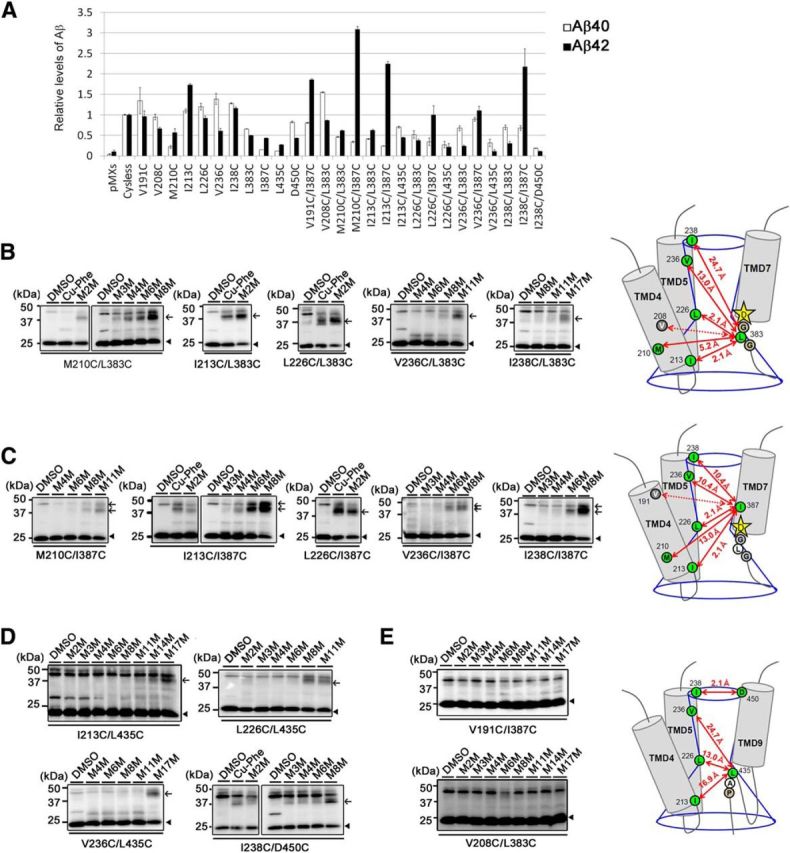
Cross-linking experiments using catalysts or MTS cross-linkers. A, Sandwich ELISA of secreted Aβ from APPNL-stable DKO cells transiently transfected with double-Cys mt PS1 (n = 3, mean ± SEs). Cross-linking experiments of double-Cys mt PS1 with Cys mutations in TMD4 or TMD5 and L383C (B), I387C (C), L435C, or D450C (D). Immunoblot analysis was performed using an anti-G1Nr5 antibody. PS1 NTFs and cross-linked products (NTF–CTF heterodimers) are shown by black arrowheads and black arrows, respectively. Double-Cys mt PS1s that failed to be cross-linked were shown in E. The predicted maximum lengths between two residues are indicated as red lines in the schematic illustrations on the right. Residues mutated to Cys are shown by circles. Positions of cross-linked cysteines were indicated by green circles. Predicted structure of the catalytic pore and distances between cross-linked residues were indicated by blue lines and red arrows, respectively. Positions of catalytic aspartates and conserved motifs were shown by stars and white circles, respectively.
Conformational changes in PS1 are associated with altered Aβ42 production
In the cytoplasmic region of TMD4, several point mutations associated with familial AD (FAD) have been identified (i.e., G206S, G206D, G206A, G206V, G209R, G209E, G209V, S212Y, I213L, I213F, and I213T; Cruts et al., 2012). The tripeptide Trp-Asn-Phe (WNF) sequence in proximity to the above glycines of TMD4 binds to Pen-2, which is an active γ-secretase component (Kim and Sisodia, 2005; Watanabe et al., 2005). Therefore, we speculate that the glycines listed above give flexibility to the cytosolic side of TMD4, which might alter the catalytic structure of γ-secretase to affect its activity, including Aβ42 production. To test this hypothesis, we first examined the distance between I213 and L383 in P2KO fibroblasts derived from Pen-2 KO mouse that completely lack γ-secretase activity (Bammens et al., 2011). We overexpressed N-terminally 6×His-tagged Cys mt PS1, which can be distinguished from endogenous PS1 (Fig. 6A). As expected, I213C/L383C was retained as a holoprotein because of lack of the functional γ-secretase complex. We reported previously that cross-linking reagents generated a faster migrating band from the proteolytically active PS1 holoprotein, in a similar manner to that from PS1 NTF and CTF (Takeo et al., 2012). However, no cross-linked band was observed in P2KO cell membranes (Fig. 6B). In contrast, a faster migrating product of His-tagged I213C/L383C was detected in Pen-2-expressing P2KO cells. These data indicated that the cytoplasmic side of TMD4 and TMD7 are located in proximity on assembly and activation of the γ-secretase complex. These results are consistent with our previous results that binding of the γ-secretase subunits induces the TMDs to come into close proximity, facing the catalytic pore (Takeo et al., 2012).
Figure 6.
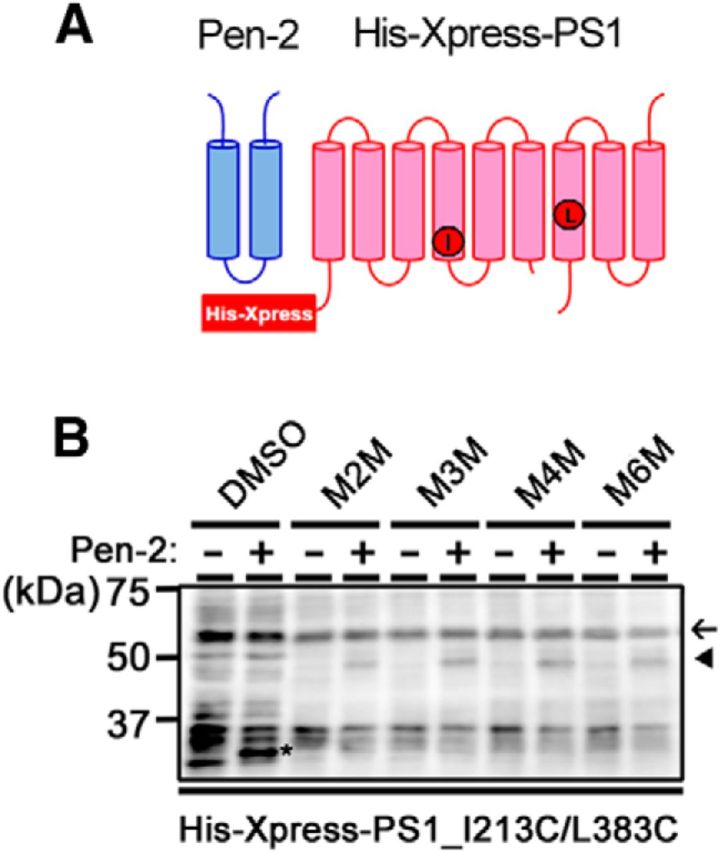
Conformational changes in TMD4 are associated with γ-secretase complex assembly. A, Schematic diagram of Pen-2 and His-Xpress-PS1 used in this study. Positions of I213 and L383 were shown as red circles. B, Cross-linking experiments of double-Cys mt PS1 with I213C and L383C mutations, in P2KO cells with or without Pen-2 overexpression. Western blotting was performed using the anti-Xpress antibody. His-Xpress-PS1 NTF (asterisk) appeared only in the Pen-2-expressing cells. His-Xpress-PS1 holoprotein, NTFs and cross-linked products (NTF–CTF heterodimers) are shown by black arrow, asterisk, and arrowhead, respectively. Note that the cross-linked product also appeared only in the Pen-2-expressing cells.
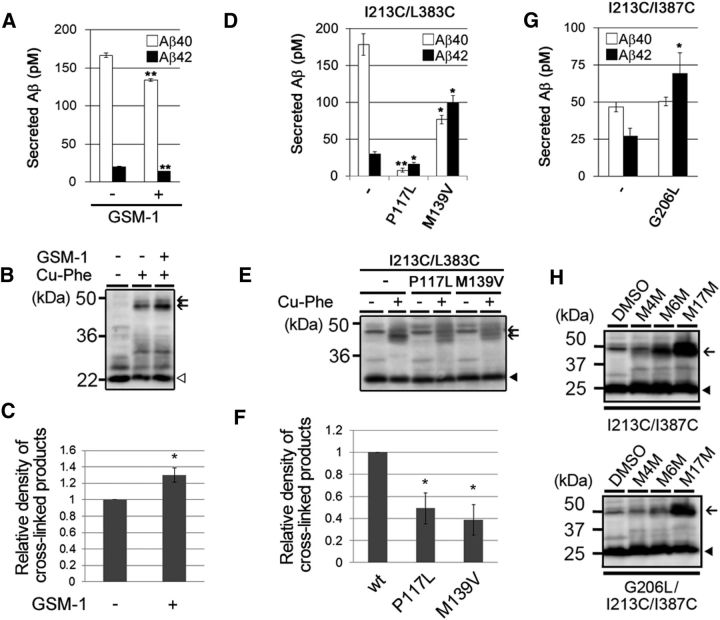
Next, we tested whether the conformational changes in TMD4 and TMD7 correlate with Aβ42 production. We performed cross-linking experiments using I213C/L383C PS1 incubated with GSM-1, which is a γ-secretase modulator that reduces Aβ42 production (Page et al., 2010; Ohki et al., 2011). We confirmed that Aβ42 production ratio from DKO cells expressing I213C/L383C PS1 was lowered by GSM-1 treatment (10.7% for DMSO and 9.4% for 30 μm GSM-1). Notably, preincubation of I213C/L383C PS1 with GSM-1 at an Aβ42-lowering concentration increased the amount of cross-linked PS1 NTF and CTF (Fig. 7A–C). Considering our previous results that GSM-1 directly bound to the TMD1 of PS1 (Ohki et al., 2011), the distance between I213 and L383 appears to be allosterically shortened by GSM-1. Next, we examined the effect of FAD mutations that elevate the Aβ42 production ratio. Because both I213 and L383 are located on the cytoplasmic side of PS1, we chose FAD-linked mutations located on the extracellular side, namely, P117L in HL1 (Wisniewski et al., 1998) and M139V in TMD2 (Clark et al., 1995). These FAD mutations increased the Aβ42 production ratio (14.3% for wild type, 69.3% for P117L, and 56.3% for M139V) and reduced the amount of cross-linked PS1 NTF and CTF (Fig. 7D–F), suggesting that the distance between TMD4 and TMD7 correlates with Aβ42 production. Finally, to understand the importance of glycines in the distance between TMD4 and TMD7, we generated G206L/I213C/I387C mt PS1 and performed cross-linking experiments. As expected, the G206L mutation caused an increased Aβ42 (42.3% for I213C/I387C and 54.4% for G206L/I213C/I387C; Fig. 7G). Moreover, I213C and I387C did not cross-link with either M4M or M6M in the G206L mutant background (Fig. 7H; also compare with Fig. 5C), indicating that the G206L mutation altered the conformation of the cytosolic side of TMD4 to increase Aβ42 production. Together, our results demonstrate that the proximity between the cytoplasmic sides of TMD4 and TMD7 correlates with the degree of activation of the trimming activity of the γ-secretase complex (Fig. 8A).
Figure 7.
Conformational changes in TMD4 associated with altered Aβ42 ratio. A, Sandwich ELISA of secreted Aβ from APPNL-stable DKO cells expressing I213C/L383C mt PS1/Cys(−) after preincubation with 30 μm GSM-1 (n = 3, means ± SEs; **p < 0.01 by Student's t test). B, Cross-linking experiments of I213C/L383C mt PS1 with a catalyst (Cu-Phe) after preincubation with GSM-1 (30 μm). Immunoblot analysis was performed using an anti-M5 antibody. PS1 CTFs and cross-linked products (NTF–CTF heterodimers) are indicated by the white arrowhead and black arrows, respectively. Densitometric analysis of the results are shown in C (n = 3, means ± SEs; *p < 0.05 by Student's t test). D, Sandwich ELISA of secreted Aβ from APPNL-stable DKO cells expressing I213C/L383C mt PS1 with an FAD-linked mutation (I213C/L383C/P117L or I213C/L383C/M139V; n = 3, means ± SEs; *p < 0.05, **p < 0.01 by Student's t test). E, Cross-linking experiments of I213C/L383C mt PS1 with or without FAD-linked mutations and a catalyst (Cu-Phe). Immunoblot analysis was performed using an anti-G1Nr5 antibody. PS1 NTFs and cross-linked products are indicated by a black arrowhead and black arrows, respectively. Densitometric analysis of the results are shown in F (n = 3, means ± SEs; *p < 0.05 by Student's t test). G, Sandwich ELISA of secreted Aβ from APPNL-stable DKO cells expressing I213C/I387C or G206L/I213C/I387C mt PS1 (n = 3, means ± SEs; *p < 0.05 by Student's t test). H, Cross-linking experiments of I213C/L383C or G206L/I213C/I387C mt PS1 with MTS cross-linkers. Immunoblot analysis was performed using an anti-G1Nr5 antibody. PS1 NTFs, and cross-linked products are indicated by black arrowheads and black arrows, respectively.
Figure 8.
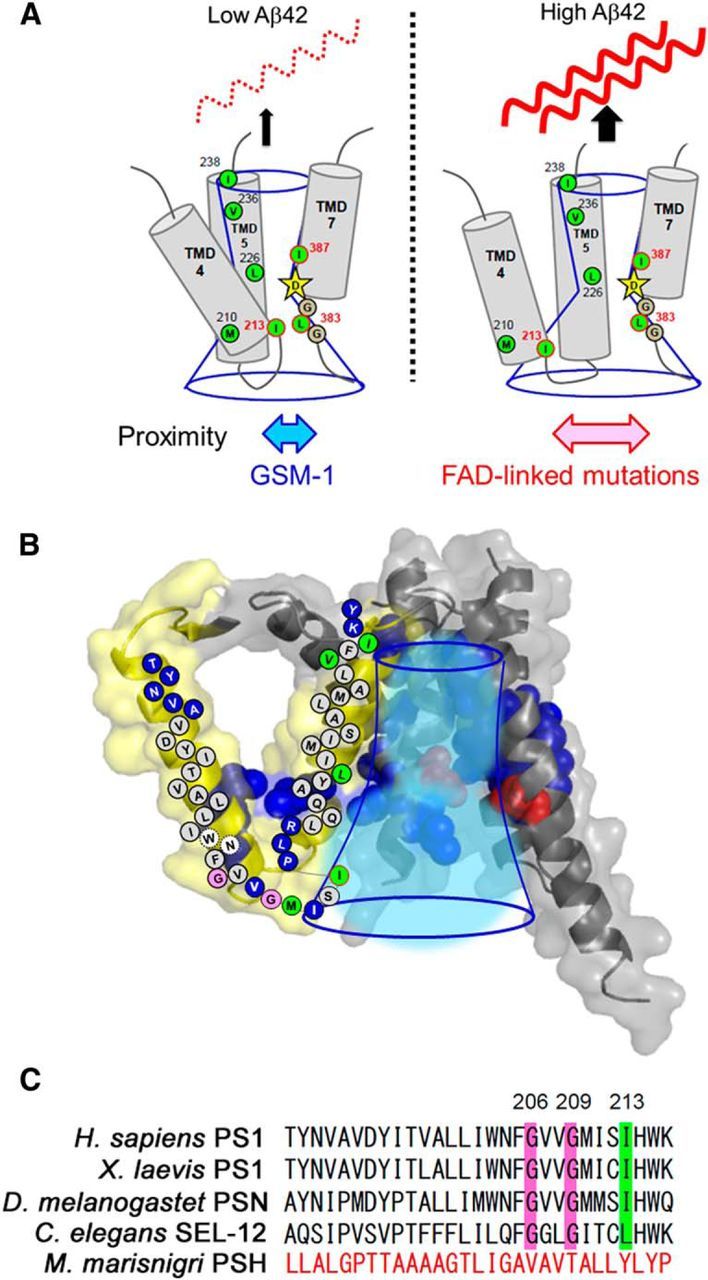
Structure and function of TMD4 and TMD5 of PS1. A, Summary of SCAM analyses and cross-linking experiments. The cytosolic residues of TMD4, TMD5, and L383, which face the catalytic pore, are shown as green circles. Catalytic aspartate on TMD7 is shown by stars. Conserved glycines in the catalytic motif are denoted as gray circles. GSM-1 treatment reduced the Aβ42 production and the distance between I213 and L383 in the catalytic site (left). In contrast, FAD-linked mutation increased the Aβ42 levels and the distance (right), suggesting that the proximity between the cytosolic sides of TMD4 and TMD7 correlates with Aβ42-generating activity. B, Schematic diagram of TMD4 and TMD5 (shown in yellow) in the PS1 model (Li et al., 2013). Residues facing the hydrophilic environment are shown as blue spheres and circles. Catalytic aspartates are shown as red spheres. Conserved glycines and the residues involved in the binding with l-685,458 are shown as pink and green circles, respectively. The catalytic pore is indicated by blue lines. C, Alignment of primary sequences of PS1 or its homolog TMD4 of Homo sapiens, Xenopus laevis, Drosophila melanogaster, Caenorhabditis elegans, and M. marisnigri. Positions of conserved glycines and isoleucine are indicated in pink and green, respectively.
Discussion
In this study, we found that TMD4 and TMD5 face the catalytic pore with TMD1, TMD6, TMD7, and TMD9 by SCAM. The main issue of this method is that several mutants with different activities (Fig. 2) were used. However, recent studies have shown that overall conformation revealed by SCAM almost corresponds to the structure resolved by x-ray crystallographic analysis [e.g., LacY (Abramson et al., 2003), P-glycoprotein (Aller et al., 2009), and YidC (Shimokawa-Chiba et al., 2015)]. Moreover, conformational changes and fluctuations of the membrane proteins in the lipid bilayer of living cells can be analyzed only by SCAM (Kaback et al., 2011). Thus, combination with biochemical and biophysical techniques is critical to the interpretation of the results by SCAM. TMD4 has a WNF sequence that is responsible for its interaction with Pen-2 (Kim and Sisodia, 2005; Watanabe et al., 2005) and two highly conserved glycine residues on its cytoplasmic side (i.e., G206 and G209). Importantly, the WNF sequence and glycine residues give different structural characteristics to the luminal and cytosolic sides of TMD4, respectively. First, SCAM analysis revealed that the luminal side of TMD4 is completely embedded and/or tightly packed in the hydrophobic environment, whereas the cytosolic side is located in the hydrophilic milieu in the membrane and forms part of the catalytic site. Intriguingly, we and others have reported that proximal and central regions of the luminal side of TMD4 are cooperatively involved in the PS1–Pen-2 interaction independently of the primary sequence of the proximal region (Kim and Sisodia, 2005; Watanabe et al., 2005). Thus, the α-helical nature of the luminal side of TMD4 is required for surface presentation of the side chains of the WNF sequence that is indispensable for Pen-2 binding. In contrast, the highly conserved M210 and I213 residues on the cytosolic side of TMD4 face the hydrophilic pore and are part of the catalytic site structure. Intriguingly, G206 and G209 were also highly conserved among species. In TMD environment, glycine is known as a helix-breaking residue, which generates a kink in the TMD because of access to a much greater φ and ψ torsional space (Dong et al., 2012). In addition, x-ray crystallographic analyses of membrane proteins revealed that some glycine residues in the TMD participate in helix–helix interaction (Javadpour et al., 1999). Our SCAM analysis results demonstrate that the consecutive residues (I213 to R220) after the glycines face the hydrophilic environment in the membrane, suggesting that the cytoplasmic region of TMD4 forms a loop-like structure. Moreover, from results of cross-linking experiment, we hypothesize that the cytosolic side of PS1 TMD4 including these residues is involved in the regulation of the velocity of the trimming of Aβ42 to Aβ38 and/or the dissociation of Aβ42. Supporting this notion, GSM-1 increased the velocity of the cleavage of Aβ42 and decreased the velocity of the dissociation of Aβ42 from PS1/γ-secretase. In contrast, FAD-associated PS1 mutants reduced the velocity of the cleavage and increased the velocity of the dissociation (Okochi et al., 2013). Although I213C mutants caused a high Aβ42 production ratio compared with PS1/Cys(−) (Fig. 5), either GSM or FAD mutation affected the Aβ42 ratio, suggesting that conformational changes related to modulation of Aβ42 production was still allowed in this mutant. However, it still remains unclear whether this allosteric change is necessary and sufficient for the regulation of γ-secretase activity in wild-type PS1. Fine cryoelectron microscopic analysis using recombinant γ-secretase complex containing wild-type PS1 with GSMs might provide a molecular evidence for functional aspects of this conformational change (Lu et al., 2014; Bai et al., 2015a,b; Elad et al., 2015). Nevertheless, our results suggest that the regulation of the distance between the cytosolic side of TMD4 and TMD7 would be a novel strategy for the development of drugs against AD that reduce Aβ42 production. Notably, several FAD-associated mutations were found in G206 and G209, including amino acid substitutions to alanine. Therefore, glycine residues in the cytoplasmic region of TMD4 break the helical structure and give flexibility to the catalytic site, which affects γ-secretase activity.
A structural model of human PS1 has been reported, based on the crystal structure of mmPSH, a PS homolog from M. marisnigri JR1 (Li et al., 2013). In addition, recently, an atomic structure of PS1 in human γ-secretase at 3.4 Å resolution also has been revealed by cryoelectron microscopy single-particle analysis (Bai et al., 2015b). Notably, overall structure of PS1 in the γ-secretase complex was quite similar to that of mmPSH. However, the distance between the residues in TMD4 and TMD7 measured in our experiments is different from those indicated by these atomic structures (Fig. 8B). Distances between I213 and L383 in the PS1 model based on crystal structure of mmPSH, atomic structure of PS1, and our SCAM analysis were 14.8, 12.8, and 2.1 Å, respectively. The distance measured by SCAM analysis is shorter than those calculated in the atomic structures. However, in these structures, the distance between two catalytic aspartates is longer than those of other activated aspartate proteases such as pepsin, suggesting that these structures reflects the inactive conformation of the γ-secretase. Consistent with this, classification of cryoelectron microscopy images implicated the dynamic conformational changes in TMD4 along with Pen-2 (Bai et al., 2015a). Thus, it would be important to compare the structures of the TMD4 of PS1 in the enzymatically active conformation.
In TMD5, L226 and the luminal side of TMD5 face the catalytic pore. Consistent with this, TMD5 in the mmPSH-based PS1 model and the atomic structure of PS1 is located near TMD7 (Li et al., 2013; Bai et al., 2015b). Unexpectedly, L226 faces the open hydrophilic environment in the middle of TMD5, whereas both L383 and I387 are located in the closed hydrophilic environment (Sato et al., 2006). Notably, L226 locates at the inner core of TMD2–TMD5, which is implicated as the FAD-linked mutational hotspot (Bai et al., 2015b), suggesting the critical role of TMD5 in the γ-secretase-mediated intramembrane proteolysis. However, the functional effect of this structural character of TMD5 still remains unknown. It is intriguing that an internal water-retention site was found very near the catalytic site in the rhomboid protease GlpG, which ensures catalytic efficiency (Zhou et al., 2012). Simulation analysis revealed that this water is spontaneously supplied by bulk exchange. Thus, one possibility is that L226 is involved in the formation of an internal water-retention site in PS1, and conformational changes during catalysis would allow MTSEA-biotin to gain access to L226. Additional possibility is that TMD5 participates the substrate binding during the endoproteolysis. The atomic structure of the γ-secretase revealed that TMD5 is involved in the formation of the cavity, which contains unknown rod-shaped density connected from the extracellular side (Bai et al., 2015a,b). This cavity was not observed in some conformation revealed by cryoelectron microscopy (Bai et al., 2015a). Notably, the extracellular side of TMD5 is located near TMD2 and TMD6, both of which are considerably flexible and are known as substrate binding sites (Watanabe et al., 2010). In addition, the accessibility of V236 at the luminal side of TMD5 was increased on preincubation with GSIs, indicating that TMD5 is exposed to the extracellular side in the inhibitory conformation. These results support our notion that TMD5 harbors a critical role in the intramembrane cleavage, and the conformation of TMD5 was allosterically changed during the catalytic process.
Together, using chemical biology and biochemical structural analyses, we have identified the structure and function of TMD4 and TMD5 of PS1 in γ-secretase-mediated cleavage. High-resolution structure of the human γ-secretase complex revealed suggested that the folding of PS1 was similar to that of mmPSH (Li et al., 2013; Lu et al., 2014; Bai et al., 2015b). However, the resolution of the structural information on the intramembrane region is not sufficient for identification of the exact location of the loop region in the catalytic site of this structure. Also, dynamic conformational flexibility of this enzyme during catalysis has been implicated (Bai et al., 2015a; Elad et al., 2015). Importantly, our SCAM and cross-linking approaches are able to annotate the structure of cellular membrane-embedded, proteolytically active PS1, although the target residue is limited. Thus, in combination of SCAM, additional detailed analyses of the structure of PS1 at atomic resolution, as well as molecular dynamics simulations are required toward understanding the functional aspect of the flexibility of the cytoplasmic region of these TMDs and toward development of novel GSMs as effective therapeutics against AD.
Footnotes
This work was supported by grants-in-aid for Scientific Research (A) from the Japan Society for the Promotion of Science, Scientific Research on Innovative Areas for Brain Environment from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by grants from Takeda Science Foundation, the Cell Science Research Foundation, the Tokyo Biochemical Research Foundation, Daiichi Sankyo Foundation of Life Science, Ono Medical Research Foundation, Nagase Science Technology Foundation, and by a donation from Chuichi Imai. S.T.-N. was a research fellow of the Japan Society for the Promotion of Science. We thank Drs. Bart De Strooper (Catholic University of Leuven, Leuven, Belgium), T. Kitamura (University of Tokyo, Tokyo, Japan), R. Kopan (Cincinnati Children's Hospital Medical Center, Cincinnati, Ohio), G. Thinakaran (University of Chicago, Chicago, Illinois), Yigong Shi (Tsinghua University, Beijing, China), S. Yokoshima, T. Fukuyama (Nagoya University, Nagoya, Japan), and A. Takashima (National Center for Geriatrics and Gerontology, Obu, Japan) for valuable reagents, and Takeda Pharmaceutical Company for the Aβ ELISA kit. We are also grateful to our laboratory members for helpful discussions and technical assistance.
The authors declare no competing financial interests.
References
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami-Odaka A, Ishibashi Y, Kikuchi T, Kitada C, Suzuki N. Long amyloid beta-protein secreted from wild-type human neuroblastoma IMR-32 cells. Biochemistry. 1995;34:10272–10278. doi: 10.1021/bi00032a022. [DOI] [PubMed] [Google Scholar]
- Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife. 2015a doi: 10.7554/eLife.11182. doi: 10.7554/eLife.11182. Advance online publication. Retrieved December 28, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Yan C, Yang G, Lu P, Ma D, Sun L, Zhou R, Scheres SH, Shi Y. An atomic structure of human gamma-secretase. Nature. 2015b;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammens L, Chávez-Gutiérrez L, Tolia A, Zwijsen A, De Strooper B. Functional and topological analysis of Pen-2, the fourth subunit of the gamma-secretase complex. J Biol Chem. 2011;286:12271–12282. doi: 10.1074/jbc.M110.216978. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Clark RF, Hutton M, Fuldner M, Froelich S, Karran E, Talbot C, Crook R, Lendon C, Prihar G, He C, Korenblat K, Martinez A, Wragg M, Busfield F, Behrens MI, Myers A, Norton J, Morris J, Mehta N, Pearson C, et al. The structure of the presenilin 1 (S182) gene and identification of six novel mutations in early onset AD families. Nat Genet. 1995;11:219–222. doi: 10.1038/ng1095-219. [DOI] [PubMed] [Google Scholar]
- Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33:1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Berezovska O, Diehl TS, Genet C, Buldyrev I, Tsai JY, Hyman BT, Wolfe MS. Designed helical peptides inhibit an intramembrane protease. J Am Chem Soc. 2003;125:11794–11795. doi: 10.1021/ja037131v. [DOI] [PubMed] [Google Scholar]
- Dong H, Sharma M, Zhou HX, Cross TA. Glycines: role in alpha-helical membrane protein structures and a potential indicator of native conformation. Biochemistry. 2012;51:4779–4789. doi: 10.1021/bi300090x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- Elad N, De Strooper B, Lismont S, Hagen W, Veugelen S, Arimon M, Horré K, Berezovska O, Sachse C, Chávez-Gutiérrez L. The dynamic conformational landscape of gamma-secretase. J Cell Sci. 2015;128:589–598. doi: 10.1242/jcs.164384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herreman A, Serneels L, Annaert W, Collen D, Schoonjans L, De Strooper B. Total inactivation of gamma-secretase activity in presenilin-deficient embryonic stem cells. Nat Cell Biol. 2000;2:461–462. doi: 10.1038/35017105. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Morris JC, Goate AM. Alzheimer's disease: the challenge of the second century. Sci Transl Med. 2011;3:77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T, Yasutake K, Nihonmatsu N, Mercken M, Takahashi H, Murayama O, Murayama M, Sato K, Omori A, Tsubuki S, Saido TC, Takashima A. Dual roles of proteasome in the metabolism of presenilin 1. J Neurochem. 1999;72:255–261. doi: 10.1046/j.1471-4159.1999.0720255.x. [DOI] [PubMed] [Google Scholar]
- Isoo N, Sato C, Miyashita H, Shinohara M, Takasugi N, Morohashi Y, Tsuji S, Tomita T, Iwatsubo T. Abeta42 overproduction associated with structural changes in the catalytic pore of gamma-secretase: common effects of Pen-2 N-terminal elongation and fenofibrate. J Biol Chem. 2007;282:12388–12396. doi: 10.1074/jbc.M611549200. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- Javadpour MM, Eilers M, Groesbeek M, Smith SO. Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys J. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback HR, Smirnova I, Kasho V, Nie Y, Zhou Y. The alternating access transport mechanism in LacY. J Membr Biol. 2011;239:85–93. doi: 10.1007/s00232-010-9327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan T, Tominari Y, Rikimaru K, Morohashi Y, Natsugari H, Tomita T, Iwatsubo T, Fukuyama T. Parallel synthesis of DAPT derivatives and their gamma-secretase-inhibitory activity. Bioorg Med Chem Lett. 2004;14:1983–1985. doi: 10.1016/j.bmcl.2004.01.067. [DOI] [PubMed] [Google Scholar]
- Kim SH, Sisodia SS. Evidence that the “NF” motif in transmembrane domain 4 of presenilin 1 is critical for binding with PEN-2. J Biol Chem. 2005;280:41953–41966. doi: 10.1074/jbc.M509070200. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genornics. Exp Hematol. 2003;31:1007–1014. doi: 10.1016/S0301-472X(03)00260-1. [DOI] [PubMed] [Google Scholar]
- Kobashi K. Catalytic oxidation of sulfhydryl groups by o-phenanthroline copper complex. Biochim Biophys Acta. 1968;158:239–245. doi: 10.1016/0304-4165(68)90136-0. [DOI] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci U S A. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornilova AY, Bihel F, Das C, Wolfe MS. The initial substrate-binding site of gamma-secretase is located on presenilin near the active site. Proc Natl Acad Sci U S A. 2005;102:3230–3235. doi: 10.1073/pnas.0407640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem JY, Saura CA, Pietrzik C, Christianson J, Wanamaker C, King LT, Veselits ML, Tomita T, Gasparini L, Iwatsubo T, Xu H, Green WN, Koo EH, Thinakaran G. A role for presenilin 1 in regulating the delivery of amyloid precursor protein to the cell surface. Neurobiol Dis. 2002;11:64–82. doi: 10.1006/nbdi.2002.0546. [DOI] [PubMed] [Google Scholar]
- Li X, Dang S, Yan C, Gong X, Wang J, Shi Y. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493:56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
- Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio-Mower J, Harrison T, Lellis C, Nadin A, Neduvelil JG, Register RB, Sardana MK, Shearman MS, Smith AL, Shi XP, Yin KC, Shafer JA, Gardell SJ. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Determining the dimensions of the drug-binding domain of human P-glycoprotein using thiol cross-linking compounds as molecular rulers. J Biol Chem. 2001;276:36877–36880. doi: 10.1074/jbc.C100467200. [DOI] [PubMed] [Google Scholar]
- Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SH, Shi Y. Three-dimensional structure of human gamma-secretase. Nature. 2014;512:166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi Y, Tominari Y, Watanabe N, Kan T, Fuwa H, Natsugari H, Fukuyama T, Tomita T, Iwatsubo T. Presenilin-1 is a molecular target of a dipeptidic gamma-secretase inhibitor, DAPT. Neurobiol Aging. 2004;25:S566–S567. doi: 10.1074/jbc.M513012200. [DOI] [PubMed] [Google Scholar]
- Morohashi Y, Kan T, Tominari Y, Fuwa H, Okamura Y, Watanabe N, Sato C, Natsugari H, Fukuyama T, Iwatsubo T, Tomita T. C-terminal fragment of presenilin is the molecular target of a dipeptidic gamma-secretase-specific inhibitor DAPT (N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester) J Biol Chem. 2006;281:14670–14676. doi: 10.1074/jbc.M513012200. [DOI] [PubMed] [Google Scholar]
- Ohki Y, Higo T, Uemura K, Shimada N, Osawa S, Berezovska O, Yokoshima S, Fukuyama T, Tomita T, Iwatsubo T. Phenylpiperidine-type gamma-secretase modulators target the transmembrane domain 1 of presenilin 1. EMBO J. 2011;30:4815–4824. doi: 10.1038/emboj.2011.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okochi M, Tagami S, Yanagida K, Takami M, Kodama TS, Mori K, Nakayama T, Ihara Y, Takeda M. gamma-secretase modulators and presenilin 1 mutants act differently on presenilin/gamma-secretase function to cleave Abeta42 and Abeta43. Cell Rep. 2013;3:42–51. doi: 10.1016/j.celrep.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Page RM, Gutsmiedl A, Fukumori A, Winkler E, Haass C, Steiner H. Beta-amyloid precursor protein mutants respond to gamma-secretase modulators. J Biol Chem. 2010;285:17798–17810. doi: 10.1074/jbc.M110.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Morohashi Y, Tomita T, Iwatsubo T. Structure of the catalytic pore of gamma-secretase probed by the accessibility of substituted cysteines. J Neurosci. 2006;26:12081–12088. doi: 10.1523/JNEUROSCI.3614-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Takagi S, Tomita T, Iwatsubo T. The C-terminal PAL motif and transmembrane domain 9 of presenilin 1 are involved in the formation of the catalytic pore of the gamma-secretase. J Neurosci. 2008;28:6264–6271. doi: 10.1523/JNEUROSCI.1163-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman MS, Beher D, Clarke EE, Lewis HD, Harrison T, Hunt P, Nadin A, Smith AL, Stevenson G, Castro JL. L-685,458, an aspartyl protease transition state mimic, is a potent inhibitor of amyloid beta-protein precursor gamma-secretase activity. Biochemistry. 2000;39:8698–8704. doi: 10.1021/bi0005456. [DOI] [PubMed] [Google Scholar]
- Shimokawa-Chiba N, Kumazaki K, Tsukazaki T, Nureki O, Ito K, Chiba S. Hydrophilic microenvironment required for the channel-independent insertase function of YidC protein. Proc Natl Acad Sci U S A. 2015;112:5063–5068. doi: 10.1073/pnas.1423817112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Tominaga A, Sato C, Tomita T, Iwatsubo T. Participation of transmembrane domain 1 of presenilin 1 in the catalytic pore structure of the gamma-secretase. J Neurosci. 2010;30:15943–15950. doi: 10.1523/JNEUROSCI.3318-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Hayashi I, Tominari Y, Rikimaru K, Morohashi Y, Kan T, Natsugari H, Fukuyama T, Tomita T, Iwatsubo T. Sulindac sulfide is a noncompetitive gamma-secretase inhibitor that preferentially reduces Abeta 42 generation. J Biol Chem. 2003;278:18664–18670. doi: 10.1074/jbc.M301619200. [DOI] [PubMed] [Google Scholar]
- Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T. The role of presenilin cofactors in the gamma-secretase complex. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- Takeo K, Watanabe N, Tomita T, Iwatsubo T. Contribution of the gamma-secretase subunits to the formation of catalytic pore of presenilin 1 protein. J Biol Chem. 2012;287:25834–25843. doi: 10.1074/jbc.M111.336347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T. Molecular mechanism of intramembrane proteolysis by gamma-secretase. J Biochem. 2014;156:195–201. doi: 10.1093/jb/mvu049. [DOI] [PubMed] [Google Scholar]
- Tomita T, Iwatsubo T. Structural biology of presenilins and signal peptide peptidases. J Biol Chem. 2013;288:14673–14680. doi: 10.1074/jbc.R113.463281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T, Maruyama K, Saido TC, Kume H, Shinozaki K, Tokuhiro S, Capell A, Walter J, Grünberg J, Haass C, Iwatsubo T, Obata K. The presenilin 2 mutation (N141I) linked to familial Alzheimer disease (Volga German families) increases the secretion of amyloid beta protein ending at the 42nd (or 43rd) residue. Proc Natl Acad Sci U S A. 1997;94:2025–2030. doi: 10.1073/pnas.94.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T, Takikawa R, Koyama A, Morohashi Y, Takasugi N, Saido TC, Maruyama K, Iwatsubo T. C terminus of presenilin is required for overproduction of amyloidogenic Abeta42 through stabilization and endoproteolysis of presenilin. J Neurosci. 1999;19:10627–10634. doi: 10.1523/JNEUROSCI.19-24-10627.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Tomita T, Sato C, Kitamura T, Morohashi Y, Iwatsubo T. Pen-2 is incorporated into the gamma-secretase complex through binding to transmembrane domain 4 of presenilin 1. J Biol Chem. 2005;280:41967–41975. doi: 10.1074/jbc.M509066200. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Takagi S, Tominaga A, Tomita T, Iwatsubo T. Functional analysis of the transmembrane domains of presenilin 1 participation of transmembrane domains 2 and 6 in the formation of initial substrate-binding site of gamma-secretase. J Biol Chem. 2010;285:19738–19746. doi: 10.1074/jbc.M110.101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski T, Dowjat WK, Buxbaum JD, Khorkova O, Efthimiopoulos S, Kulczycki J, Lojkowska W, Wegiel J, Wisniewski HM, Frangione B. A novel Polish presenilin-1 mutation (P117L) is associated with familial Alzheimer's disease and leads to death as early as the age of 28 years. Neuroreport. 1998;9:217–221. doi: 10.1097/00001756-199801260-00008. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Moin SM, Urban S, Zhang Y. An internal water-retention site in the rhomboid intramembrane protease GlpG ensures catalytic efficiency. Structure. 2012;20:1255–1263. doi: 10.1016/j.str.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]