Abstract
DNA 5-hydroxylmethylcytosine (5hmC) catalyzed by ten-eleven translocation methylcytosine dioxygenase (TET) occurs abundantly in neurons of mammals. However, the in vivo causal link between TET dysregulation and nociceptive modulation has not been established. Here, we found that spinal TET1 and TET3 were significantly increased in the model of formalin-induced acute inflammatory pain, which was accompanied with the augment of genome-wide 5hmC content in spinal cord. Knockdown of spinal TET1 or TET3 alleviated the formalin-induced nociceptive behavior and overexpression of spinal TET1 or TET3 in naive mice produced pain-like behavior as evidenced by decreased thermal pain threshold. Furthermore, we found that TET1 or TET3 regulated the nociceptive behavior by targeting microRNA-365-3p (miR-365-3p). Formalin increased 5hmC in the miR-365-3p promoter, which was inhibited by knockdown of TET1 or TET3 and mimicked by overexpression of TET1 or TET3 in naive mice. Nociceptive behavior induced by formalin or overexpression of spinal TET1 or TET3 could be prevented by downregulation of miR-365-3p, and mimicked by overexpression of spinal miR-365-3p. Finally, we demonstrated that a potassium channel, voltage-gated eag-related subfamily H member 2 (Kcnh2), validated as a target of miR-365-3p, played a critical role in nociceptive modulation by spinal TET or miR-365-3p. Together, we concluded that TET-mediated hydroxymethylation of miR-365-3p regulates nociceptive behavior via Kcnh2.
SIGNIFICANCE STATEMENT Mounting evidence indicates that epigenetic modifications in the nociceptive pathway contribute to pain processes and analgesia response. Here, we found that the increase of 5hmC content mediated by TET1 or TET3 in miR-365-3p promoter in the spinal cord is involved in nociceptive modulation through targeting a potassium channel, Kcnh2. Our study reveals a new epigenetic mechanism underlying nociceptive information processing, which may be a novel target for development of antinociceptive drugs.
Keywords: hydroxymethylation, Kcnh2, miRNA, nociceptive behavior
Introduction
DNA methylation, an epigenetic mechanism by which a relative stable and long-term gene is silenced, plays an important role in the transcription regulation underlying such neurophysiological and neuropathological processes as neuronal development, plasticity, and CNS diseases (Barbier et al., 2015). In mammals, methylations at the fifth position of cyosine (5mC) in CpG are catalyzed by de novo DNA methyltransferase3a and 3b (DNMT3a, 3b) and subsequently maintained by DNMT1 (Okano et al., 1999; Goll and Bestor, 2005). Increasing evidence has shown that 5mC is implicated in the nociceptive processing via modulating pronociceptive or antinociceptive gene expression in nociceptive pathways, including those in the dorsal root ganglion (DRG), spinal cord, and several pain matrix brain regions (Tajerian et al., 2011; Viet et al., 2011). However, recent accumulating data indicate that 5mC in the nociceptive gene is reversible (Tochiki et al., 2012; Qi et al., 2013; Pan et al., 2014; Pollema-Mays et al., 2014), which suggests that DNA demethylation may modulate the nociceptive process.
Although 5mC as well as DNMT enzymes have been well characterized (Tajerian et al., 2011; Viet et al., 2011), the complementary process of DNA demethylation has, until recently, remained elusive. A major breakthrough came in 2009, when ten-eleven translocation methylcytosine dioxygenase (TET) 1 was verified to be able to convert 5mC to 5-hydroxymethylcytosine (5hmC; Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Soon after, two other TET family members, TET2 and TET3, were also shown to have 5mC hydroxylase activity (Ito et al., 2010; Ko et al., 2010). At present, all three TET proteins have been found to successively oxidize 5mC to 5hmC as well as to 5-formalcytosine and 5-carboxylcytosine (He et al., 2011; Ito et al., 2011), with 5hmC being distributed in a cell- or tissue-dependent manner (Branco et al., 2012). Recent genome-wide profiling has further revealed that 5hmC majorly distributes in individual gene promoters, and this feature contributes to gene transcription by enrolling DNA-binding proteins or changing the remolding of the chromatin structure (Desplats et al., 2011). Consequently, 5hmC-mediated epigenetic modification serves as a critical intermediate step in active demethylation pathways, and is essential in a range of biological processes, such as embryonic development, stem cell function, and cancer formation (Abdolmaleky et al., 2006; Wang et al., 2008). Notably, 5hmC is relatively enriched in CNS tissues (Kriaucionis and Heintz, 2009; Münzel et al., 2010; Li and Liu, 2011; Szulwach et al., 2011), suggesting that its importance in functional regulation in the CNS. Consistently, emerging evidence has shown that TET proteins are implicated in CNS-related diseases or neuronal activity, such as inflammation (Taylor et al., 2014), drug addiction (Feng et al., 2015), Alzheimer's disease (Coppieters et al., 2014), and learning and memory (Kaas et al., 2013; Rudenko et al., 2013; Zhang et al., 2013). However, the role of TET-mediated regulation of 5hmC and subsequent active DNA demethylation in nociceptive modulation has not been explored.
Recent reports have shown a strong connection between miRNA modulation and the nociceptive response. microRNA-365-3p (miR-365-3p) is abundant and highly conserved across species, and modulates various diseases, such as cancer (Guo et al., 2013), adipose metabolism (Mori et al., 2014), and atherosclerosis (Kim et al., 2014). Moreover, miR-365-3p is upregulated not only in the blood of schizophrenia patients (Liu et al., 2013) but also in the primary microglia of the brain cortex of an amyotrophic lateral sclerosis mouse model (Parisi et al., 2013). Furthermore, the overexpression of miR-365-3p significantly downregulates the IL-6 level in cortex microglia (Parisi et al., 2013). Although limited evidence suggests a potential modulatory function of miR-365-3p in CNS diseases, it is unclear whether and how miR-365-3p participates in the nociceptive response.
Here, we investigated the epigenetic mechanism of TET and 5hmC in the nociceptive response. We found increased expression of TET1, TET3, and 5hmC, but not TET2, in the spinal cord of mice in a formalin-induced acute inflammation pain model. Furthermore, spinal TET1 or TET3 modulated the nociception by regulating the 5hmC level of the miR-365-3p promoter, and subsequent expression of miR-365-3p and Kcnh2, a validated target of miR-365-3p. Together, our study reveals a new epigenetic mechanism underlying nociceptive processing that TET-mediated hydroxymethylation of miR-365-3p regulates nociceptive behaviors via Kcnh2.
Materials and Methods
Animals and tissue.
Adult male Shanghai populations of Kunming mice (20–25 g) were used in this study. The nociceptive model was induced by intraplantar injection of 5% formalin in PBS buffer (20 μl; F8775, Sigma-Aldrich) into the dorsal surface of the mouse hindpaw as described previously (Kynast et al., 2013). The time spent licking the formalin-injected paw was recorded at 5 min intervals up to 60 min, starting immediately after formalin injection. Thermal nociceptive behavior was assessed in a blinded fashion by measuring each mouse's paw-withdrawal latency in response to thermal stimulus (Pan et al., 2014). All animal procedures were approved by the Animal Care Committee of Xuzhou Medical College. The lumbar (L3–L5) segments of the spinal cord were removed, and the ipsilateral dorsal spinal cord was separated rapidly and snap-frozen in liquid nitrogen and stored at −80°C.
Climbing test.
A metal wire-mesh with 0.5 mm diameter and 5-mm-wide grid was placed vertically 30 cm above the table. Each mouse started from the bottom of the mesh with its head facing downward. After the mouse was released, the time required to climb all the way to the top was recorded (Zhang et al., 2014).
DNA dot blot assay.
Genomic DNA was extracted using QIAamp DNA Mini Kit (51306; Qiangen). Dot blot analysis was performed as detailed previously (Wang et al., 2012). Briefly, DNA was denatured in TE buffer for 10 min at 95°C and immediately chilled on ice for 5 min. One-hundred nanograms of each DNA sample were transferred onto positively charged nylon membrane, baked for 2 h at 80°C crosslinked with UV254 for 10 min, blocked with 5% nonfat milk for 1.5 h at 25°C, and incubated with the antibody specific to 5hmC (1:10,000; 39999, Active Motif) at 4°C overnight, followed by incubation with a peroxidase-conjugated rabbit IgG antibody. The signal was visualized by using ECL (345818, Millipore). Equal DNA loading was verified by staining the membranes with 0.2% methylene blue. The intensity of each dot was normalized to total DNA.
5hmC immunoprecipitation.
According to the manufacturer's instructions (17–295, Millipore) with partial alterations, genomic DNA was sonicated to an average fragment size of 100–1000 bp, then precipitated and pre-cleared at 4°C for 60 min with G agarose beads. As input control, a small volume (10%) of sonicated DNA was used; the remainder was immunoprecipitated with monoclonal anti-5mC (C15200081, Diagenode), anti-5hmC antibody (C15200200, Diagenode), or negative control antibody IgG (PP64, Millipore) at 4°C overnight. The complexes were captured using agarose beads for 2 h at 4°C with rotation, centrifuged, and washed beads several times. Eluted DNA using immunoprecipitation (IP) elution buffer at room temperature, and then centrifuged and transferred supernatant to clean tubes. DNA was then purified using a QIAquick PCR purification kit (28106, Qiagen). Input was diluted 1/100 and used for (semi-) quantitative PCR (qPCR) analyses. 5hmC immunoprecipitation (hMeIP) 365F (5′-TTAGTTTTGGCTCTCTGTGAG-3′) and hMeIP 365R (5′-ATGATAGCGGAATGGAAACA-3′) set primers were used to amplify immunoprecipitated DNA and input DNA in quantitative analysis for 5hmC or 5mC in the miR-365-3p promoter.
5hmC and 5mC sequencing.
Genomic DNA was sonicated to 200–900 bp using a Covaris instrument, and 800 ng of the fragmented sample was end-repaired, A-tailed, and ligated to single-end Illumina's genomic adapters with Genomic DNA Sample Kit (FC-102-1002, Illumina). Then, 300–1000 bp of ligated DNA fragments were selected after agarose gel purification. Genomic DNA IP was performed by IP with 1 μl monoclonal anti-5mC (C15200081, Diagenode), anti-5hmC antibody (C15200200, Diagenode), or control IgG antibody (PP64, Millipore) in 400 μl IP buffer (0.5% BSA in PBS). Next, 25 ng 5mC- or 5hmC-captured DNA or input DNA was used for preparation of sequencing libraries following the instructions of Illumina's seq sample prep kit (111257047, Rev A). Briefly, 14 cycles of PCR were performed on 5 μl immunoprecipitated DNA using the single-end Illumina PCR primers. PCR productions were purified with Qiagen MinElute columns, after which a final size selection (300–1000 bp) was performed with electrophoresis in 2% agarose. PCR-amplified DNA libraries were quality controlled by Agilent 2100 Bioanalyzer and finally diluted in elution buffer to 5 ng/μl. Then, 1 μl was used in real-time qPCR (RT-qPCR) reactions to confirm the enrichment for the hydroxymethylated region. The library was denatured with 0.1 m NaOH to generate single-stranded DNA molecules, loaded onto channels of the flow cell at 8 pm concentration, and amplified in situ using the TruSeq Rapid PE Cluster Kit (PE-402-4001, Illumina to generate the sequencing cluster. Then, 100 sequencing cycles was performed using the TruSeq SBS Kit v5 (FC-104-5001, Illumina) protocol on the Illumina HiSeq 2000. Image analysis and base calling were performed using Off-Line Basecaller software (OLB v1.8). After passing through the Solexa CHASTITY quality filter, the clean reads were aligned to the mouse genome (UCSC MM10) using BOWTIE software (v2.1.0). A hydroxymethylation score for any region in the genome was defined as the number of fragments per kilobase (Maunakea et al., 2010). Two biological replicates were performed for each condition.
RNA, miRNA isolation, and RT-qPCR.
Spinal samples were homogenized in Trizol (15596-026, Invitrogen). miRNA was reversely transcribed using specific primers including: 365RT (5′-TTAACTGGATACGAAGGGTCCGAACACCGGTCGTATCCAGTTAAATAAGGAT-3′); 335RT (5′-TTAACTGGATACGAAGGGTCCGAACACCGGTCGTATCCAGTTAAACATTTTTC-3′); and 92RT (5′-TTAACTGGATACGAAGGGTCCGAACACCGGTCGTATCCAGTTAAGGAGGCC-3′). The transcription of other genes was performed with oligo (dT)18. The cDNA products were used as templates to detect miRNA or gene content in RT-qPCR, respectively using forward primer (365f: 5′-TGCGGTAATGCCCCT AAAAA-3′; 335f: 5′-TGCGGTCAAGAGCAATAACGAAA-3′; and 92f: 5′-TGCGGTATTGCACTCGTCCCGG-3′), and universal reverse primer (Mr: 5′-TACGAA GGGTCCGAACAC-3′); or using Tet1 primer pairs (T1f: 5′-GAGCCTGTTCCT CGATGTGG-3′ or T1r: 5′-CAAACCCACCTGAGGCTGTT-3′), Tet2 primer pairs (T2f: 5′-TGTTGTTGTCAGGGTGAGAATC-3′; T2r: 5′-TCTTGCTTCTGGCAAACTTACA-3′), Tet3 primer pairs (T3f: 5′-CCGGATTGAGAAGGTCATCTAC-3′ or T3r: 5′-CCGGATTGAGAAGGTCATCTAC-3′), and Kcnh2 primer pairs (Kf1: 5′-GTGCTGCCTGAGTATAAGCTG-3′ or Kr1: 5′-CCGAGTACGGTGTGAAGA CT-3′). SYBR Green method was used to perform RT-qPCR with the SYBR Premix Ex TaqII kit (RR820A, Takara). Each reaction was run in triplicate. The ΔΔCt method was used to analyze the expression as previously described (Pan et al., 2014). U6 RNA (Uf: 5′-CTCGCTTCGGCAGCACATATACT-3′ or Ur: 5′-ACGCTTCACGAATTTGCGTGTC-3′) was used as an internal control of miRNAs; Gapdh (Gf: 5′-GGTGAAGGTCGGTGTGAACG-3′ or Gr: 5′-CTCGCTCCTGGAAGATGGTG-3′) was used as an internal control of Tet1, Tet2, Tet3, and Kcnh2.
Plasmid construction.
Tet overexpression: three Tet1 or four Tet3 inserts (100 ng/insert) prepared by PCR using primers for Tet1 (f11: 5′-AGGTTTAAACTACGGATGGAAGCTGCACCCTGTGACTGTGATGG-3′, r11: 5′-CATCTGCTTGTAAAGTGGAGCTAA-3′; f12: 5′-AGAATTGGCTACAGTATTAGCTC-3′, r12: 5′-GCTGTGGGAAGAGACTGGTGAGG-3′; and f13: 5′-CCGAGGTGGCTGCTCATCCTCAC-3′, r13: 5′-GTAAGCTTAGGCCTGTCATTAGAC CCAACGATTGTAGGGTC-3′), and Tet3 (f31: 5′-AGGTTTAAACTACGGATGGAGTTCCCTACCTGCGATTGTGT-3′, r31: 5′-GAGGGGCAGCACGTGCAGTTGCT-3′; f32: 5′-CAGATCCCTGAGGACGAGCAACT-3′, r32: 5′-GTGGTTGTCTGTGGCAACAGCCT-3′; f33: 5′-CGAGCTGCCAGGTCAGGCTGTT-3′, r33: 5′-GCTCAAGGGCATGCCAAAGGCTT-3′; and f34: 5′-TAGACAAAGCCTGGCAAGCCTTTG-3′, r34: 5′-GTAAGCTTAGGCCTGTCACTAGATCCAGCGGCTGTAGGGGCCA-3′), and 50 ng PWPXL vector digested by BamHI (R0136L, NEB) were, respectively, assembled to a Tet1- or Tet3-expressing vector, namely Lenti-T1 and Lenti-T3, using Gibson DNA assembly reactions kit (E5510S, NEB). All constructs were confirmed by DNA sequencing.
Lentivirus package and infection.
The constructed core plasmid (16 μg) and two envelope plasmids, PSPAX2 (12 μg) and PMD2G (4.8 μg), were cotransfected into HEK293T cells in a 6-well plate according to manufacturer instructions of Lipofectamine 2000 (11668-027, Invitrogen). The supernatant was collected at 48 h after transfection, and concentrated by using a Centricon Plus-70 filter unit (UFC910096, Millipore). Lentivirus with titers >108 TU/ml was used in the experiment. The assays of lentivirus in vitro and in vivo infection were performed according to the previous description (Pan et al., 2014). Briefly, 20 μl lentivirus and 1.5 μl polybrene (1.4 μg/μl; H9268, Sigma-Aldrich) were added in a 24-well plate containing 1 × 105 HEK293T cells and 400 μl DMEM without FBS; after 24 h, the transfection medium was replaced with 500 μl fresh complete medium containing 10% FBS; cells were collected at 48 h after culture. Each time, 1 μl lentivirus was injected intrathecally in vivo.
SiRNA, mimics, inhibitor, and agonist delivery.
TET1 siRNA (sense: 5′-UUGUGUCACGGCCAUCUGCdTdT-3′; antisense: 5′-GCAGAUGGCCGUGACACAAdTdT-3′) and TET3 siRNA (sense: 5′-UUGCUCCGAGCAUAUUUGCdTdT-3′; antisense: 5′-GCAAAUAUGCUCGGAGCAAdTdT-3′) were designed and validated in vitro and in vivo. Five microliters of 20 μm mimics (365S: 5′-UAAUGCCCCUAAAAAUCCUUAU-3′; 365R: 5′-AUAAGGAUUUUUAGGGGCAUUA-3′), siRNA or inhibitor (miR-365-3p inhibitor: 5′-AUAAGGAUUUUUAGGGGCAUUA-3′), or scrambled siRNA (sense: 5′-UUCUCCGAACGUGUCACGUdTdT-3′, antisense: 5′-ACGUGACACGUUCGGAGAAdTdT-3′), or KCNH2 agonist NS1643 (0.02 mg/4 μl; ab141825, Abcam) was intrathecally injected at the L3–L5 level of the mouse spinal cord.
Construction of methylated or hydroxymethylated promoter reporter.
The defined region of miR-365-3p promoter was amplified from mouse genomic DNA using a primer set, including forward primer (5′-CGCTCGAGAGGACGGAGCCCAGGCAG-3′) and reverse primer (5′-CGACGCGTAACCAATCAGCTGGCGGGA-3′). The PCR products and pGL6 (5:1) were mixed, and digested with XhoI (R0146S, NEB) and HindIII (R3104S, NEB), then ligated with T4 ligase. The ligation product is named pGL6-365. pGL6-365 or pGL6 was methylated using methyltransferase (M.SssI; M0226S, NEB) at 37°C for 1 h, and then purified with Wizard Plus SV Minipreps DNA Purification System (A1330, Promega). The methylated pGL6-365 or pGL6 was hydroxymethylated by the use of recombinant TET1 protein (31363, Active Motif) in 50 mm HEPES, pH8.0, 50 μm Fe(NH4)2(SO4)2, 2 mm ascorbate, and 1 mm α-ketoglutarate for 3 h at 37°C. pRL-TK plasmid was used as an internal control (Promega).
miR-365-3p target construction.
The 3′-untranslated region (3′-UTR) sequence of miR-365-3p targeting Kcnh2 was synthesized [wild-type (wt)-Kf: 5′-P-TCGATGTGGTCCCCCCACTGCCCTGGGGGCATTAGCTGGTCTAACTGCCCGGAGGC-3′, and wt-Kr: 5′-P-GGCCGCCTCCGGGCAGTTAGACCAGCTAATGCCCCCAGGGCAGTGGGGGGACCACA-3′; mutation type (mut)-Kf: 5′-P-TCGATGTGGTCCCCCCACTGCCCTGGGAATACCAGCTGGTCTAACTGCCCGGAGGC-3′, mut-Kr: 5′-P-GGCCGCCTCCGGGCAGTTAGACCAGCTGGTATTCCCAGGGCAGTGGGGGGACCACA-3′]. The annealed forward and reverse oligos (wt-Kf/wt-Kr and mut-Kf/mut-Kr) were, respectively, inserted into a dual-reporter psiCHECK2 plasmid digested by Xhol and NotI, and named as wild-type CHK-wt-Kcnh2 and mutant-type CHK-mut-Kcnh2.
Luciferase reporter assay.
HEK293T cells were cultured in DMEM with10% FBS. HEK293T cells were seeded at 1 × 105 cells per 24-well. Two-hundred nanograms per well of the methylated and hydroxymethylated pGL6-365 or empty vector or control pRL-TK plasmid, were transfected into HEK293T cells using Lipofectamine 2000 (11668-027, Invitrogen) in a 24-well plate. Identification of miR-365-3p targets was performed by transfecting CHK-wt-Kcnh2 or CHK-mut-Kcnh2 plasmids (50 ng) and miR-365-3p mimics (80 ng) or inhibitor (50 ng) into HEK293T cells. Cell lysates were prepared and subjected to luciferase assays using the Dual-luciferase reporter kit (E1910, Promega) at 48 h after transfection according to the manufacturer's instruction.
Immunofluorescence and fluorescence in situ hybridization.
The procedure was performed as previous described (Pan et al., 2014). Briefly, spinal cords were rapidly dissected from perfused mice with 4% PFA, fixed with 4% PFA, then cryoprotected in 30% sucrose. For double-labeling immunofluorescence (IF) analyses, the spinal sections were incubated overnight at 4°C with a mixture of NeuN antibody (MAB377, Millipore) and TET1 (1:50; sc-163443, Santa Cruz Biotechnology) or TET3 (1:50; sc-139186, Santa Cruz Biotechnology) or 5hmC antibody (1:2000; 39999, Active Motif). The sections were then incubated with fluorescent-conjugated secondary antibody (AB10113, AB10081, AB10053; Shanghai Sangon Biotech) at 37°C for 1 h. For fluorescence in situ hybridization (FISH) and IF costaining, miR-365-3p probe with double-digoxin modification was hybridized to spinal slices as instructed in the FISH kit (Guangzhou Exon), and then incubated with KCNH2 antibody (1:100; sc20130, Santa Cruz Biotechnology), and finally incubated with AlexaFluor (Cell Signaling Technology). After the sections were rinsed in 0.01 m PBS, coverslips were applied.
Western blot analysis.
Protein (20–50 μg) was separated with 10% SDS-PAGE gel, transferred onto a nitrocellulose membrane, and incubated with antibody against TET1 (1:100; 61443, Active Motif), TET2 (1:100; sc-136926, Santa Cruz Biotechnology), TET3 (1:100; sc139186, Santa Cruz Biotechnology), and KCNH2 antibody (1:1000; BS7461, Bioworld) or control β-Actin (1:1000; TA-09, ZSGB-Bio). Blots were washed and incubated in peroxidase-conjugated goat anti-rabbit IgG secondary antibody (1:1000; VA001, Vicmed). Protein blots were visualized using chemiluminescence detection (72091, Sigma-Aldrich).
Statistical analysis.
Data are presented as mean ± SEM and were analyzed using GraphPad Prism v5.00. The data for DNA dot blot was analyzed with one-way ANOVA. The data for behavioral tests, luciferase reporter assay, and 5hmC level analysis after Tet knockdown or overexpression were analyzed with two-way ANOVA with two repeated factors followed by Tukey multiple-comparison tests. Unpaired comparisons were assessed by two-tailed Student's t test for 5hmC, protein, image, luciferase unit, or RT-qPCR analysis as appropriate.
Results
Formalin-induced acute inflammation pain increases spinal 5hmC levels
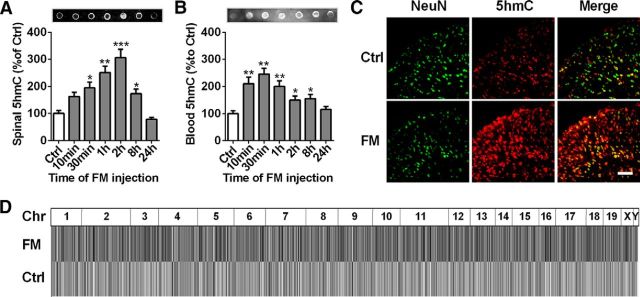
The abundance of 5hmC in the CNS suggests its potential roles in the regulation and dysregulation of neural functions. To explore the relationship between 5hmC-mediated epigenetic modulation and the regulation of spinal nociceptive processing, we first examined the alteration of 5hmC content in mouse spinal cord in a formalin-induced acute inflammatory pain model. Dot blot assay revealed a marked increase in spinal 5hmC from 30 min to 2 h after formalin injection, which then gradually returned to basal levels at 24 h after formalin injection (Fig. 1A). High correlation of epigenetic marks between spinal cord and blood may be useful for diagnostic and therapeutic application in pain. Therefore, we want to know whether modification of 5hmC would be altered in whole blood similarly to spinal cord underlying the formalin-induced acute inflammatory pain status. We found that the blood 5hmC significantly increased from 10 to 30 min, then gradually decreased to a basal level from 1 to 24 h after formalin injection (Fig. 1B), suggesting that the gain of 5hmC in both mouse spinal cord and blood is a novel epigenetic feature in nociceptive processing.
Figure 1.
Genomic 5hmC analysis in spinal cord or blood in a formalin-induced acute inflammation pain model. A, B, Time course of dot blot assay of spinal 5hmC (A) or blood 5hmC (B) in formalin (FM)-induced acute inflammatory pain mice. Spinal or blood DNA samples were collected from 10 min to 24 h after FM injection and then dot-blotted using a 5hmC-specific antibody. *p < 0.05 versus control (Ctrl) group, **p < 0.01 versus control group, ***p < 0.001, versus control group; n = 6. C, Combined 5hmC (red) and NeuN (a neuronal marker; green) immunofluorescence staining in spinal cord at 2 h after formalin or saline (Sal) injection. Scale bar, 25 μm. D, Genomic 5hmC profiles in formalin-induced acute inflammatory pain. The read densities of each chromosome (Chr) in heatmap have been equally scaled and then normalized through the total number of mapped reads per sample.
To further explore the regulatory mechanism underlying the role of 5hmC in the nociceptive process, we investigated the genomic characteristics of spinal 5hmC change at 2 h after formalin injection. First, we used immunofluorescence staining experiments to further confirm a significant increase in global 5hmC in the spinal cord of acute pain mice (Fig. 1C) compared with that of the control group. Additionally, immunofluorescence costaining showed ∼94.6% overlap between the 5hmC signal and NeuN (a neuron marker) in the control group, and 82% overlap in the formalin group (Fig. 1C), suggesting that 5hmC may occur in mouse spinal neurons. Then, we performed a genome-wide evaluation of spinal 5hmC distribution at 2 h after saline and formalin injection by a genome DNA IP sequencing method using anti-5hmC antibody, which was used for the analysis of the differential 5hmC (hMeDIP). Before sequencing, the specificity of anti-5hmC and anti-5mC antibody was tested by the use of negative control IgG antibody. The results showed that DNAs were not pulled down by IgG, but by the anti-5hmC and anti-5mC antibody in DNA detection using gel staining and NanoDrop 2000 spectrophotometer (data not shown), respectively, indicating that the anti-5hmC and anti-5mC antibodies are specific for DNA immunoprecipitation. In the following sequencing, ∼40.1 or 36.2 million reads were obtained from the spinal cord of the saline-treated group and the formalin-injected group, respectively (Table 1). When raw reads were aligned to the mouse reference genome (mm10), multiple reads without mapping were filtered out, leaving 20.9 and 19.2 million uniquely mapped reads, respectively, for subsequent analysis (Table 1). The genome-scaled 5hmC read density of formalin-injected mice exhibited significant differences from those of control animals, as determined by an IGV Browser (IGV 1.4.2; Fig. 1D). These data indicate that 5hmC modification may be involved in the nociceptive response.
Table 1.
Summary of sequencing data
*The mean data in a duplicate experiment.
Spinal TET1 and TET3 contribute to formalin-induced nociceptive response
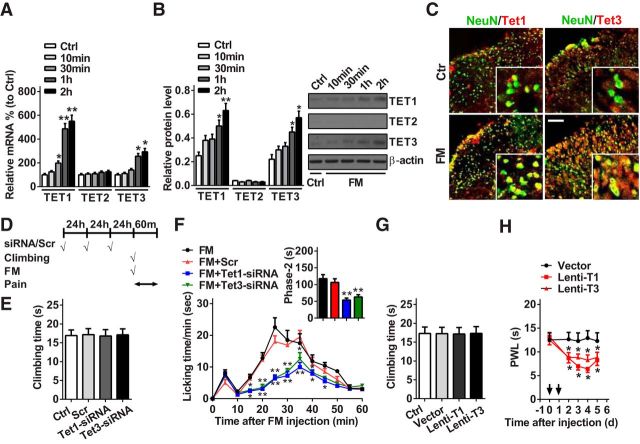
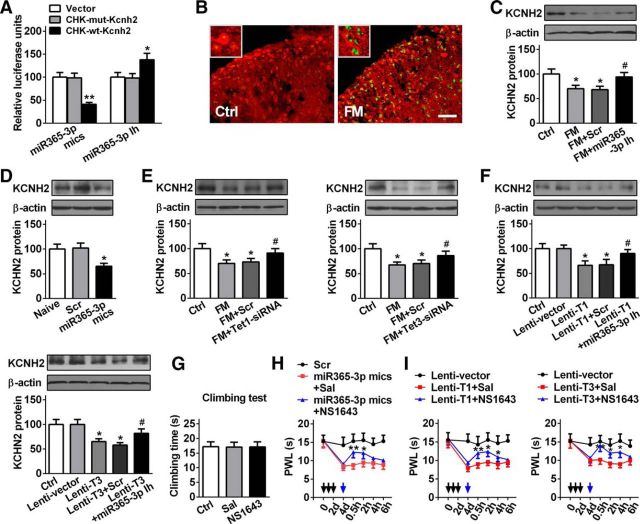
As TET proteins convert 5mC to 5hmC, spinal Tet expression and its role in pain behavioral regulation were investigated in the formalin-induced pain model. We found that Tet1 and Tet3, but not Tet2, in mRNA and protein expression were gradually increased from 10 min to 2 h after formalin injection. Compared with the control group, Tet1 and Tet3 mRNAs were increased by 450% and 193%, respectively, at 2 h after formalin injection; correspondingly, TET1 and TET3 proteins were increased by 152% and 160%, respectively (Fig. 2A,B). These results suggest that increased TET1 and TET3, but not TET2, may be involved in the nociceptive response induced by formalin. Double-immunofluorescence staining revealed that the increased TET1 and TET3 after formalin injection were expressed in spinal neurons (Fig. 2C). Then, we explored the potential effects of manipulation of spinal TET1 or TET3 expression on the nociceptive behavior of mice in formalin test. The two modulation tools of siRNA (Tet1-siRNA or Tet3-siRNA) for TET1 or TET3 downregulation and lentivirus (Lenti-T1 or Lenti-T3) for TET1 or TET3 overexpression were used in the experiments. The transfection efficiency of siRNA or lentivirus was validated in the spinal cord of naive mice. Before the measure of nociceptive behavior, a vertical climbing test was performed to determine whether the modulation tools could affect the motor behavior. The results showed that the knockdown of TET1 or TET3 (Fig. 2D,E) or the overexpression of TET1 or TET3 (Fig. 2G) could not influence the motor function of the mice. However, knockdown of TET1 or TET3 via intrathecal injection of Tet1-siRNA or Tet3-siRNA for 3 consecutive days before formalin injection alleviated the formalin-induced second phase of the nociceptive response compared with negative control-treated mice (Fig. 2D, F). In contrast, overexpression of TET1 or TET3 via intrathecal injection of Lenti-T1 or Lenti-T3, but not empty vector, for 2 consecutive days significantly produced pain-like behavior as evidenced by a decrease of thermal pain threshold (Fig. 2H). These findings suggest that spinal TET1 and TET3 contribute to acute nociceptive modulation.
Figure 2.
Spinal TET1 and TET3 regulate acute nociceptive behavior. A–C, Quantitative analysis of spinal Tet1, Tet2, and Tet3 expression in mRNA using RT-qPCR (A) and protein using Western blotting (B) at 10 min, 30 min, 1 h, and 2 h, or immunofluorescent staining (C) at 2 h after formalin injection. Scale bar, 25 μm. *p < 0.05 versus control group, **p < 0.01 versus control group; n = 6. D, The strategy of formalin injection, motor and pain test. Scrambled (Scr) indicates a negative control. E, Time to climb a vertical wire mesh at 72 h after intrathecal injection of Tet1- and Tet3-siRNA for 3 consecutive days in naive mice. No significance versus Ctrl or Scr group; n = 8. F, Intrathecal injection of Tet1- and Tet3-siRNA for 3 consecutive days reversed formalin-induced acute pain in phase 2. *p < 0.05 versus FM+Scr group, **p < 0.01 versus FM+Scr group; n = 8. G, Time of climbing test at 96 h after intrathecal injection of Lenti-T1 and Lenti-T3 for 2 consecutive days in naive mice. No significance versus Ctrl or vector group; n = 8. H, Intrathecal injection of Lenti-T1 and Lenti-T3 for 2 consecutive days produced thermal hyperalgesia (paw withdrawal latency; PWL) in naive mice. Black arrow indicates Lenti-T1 and Lenti-T3 or Lenti-vector injection. *p < 0.05 versus Lenti-vector group; n = 8.
TET1 and TET3 regulate the nociceptive behavior by targeting miR-365-3p
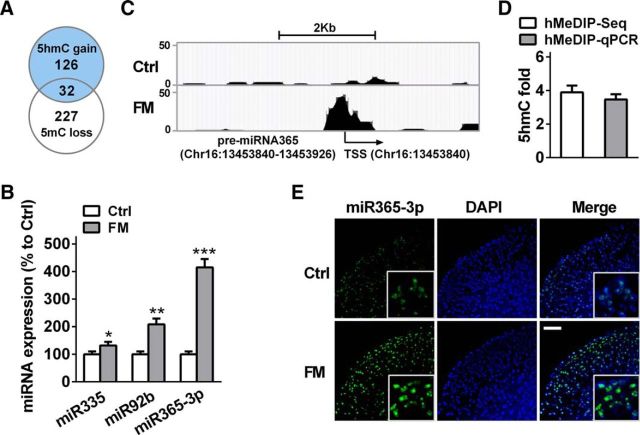
Recent emerging evidence has shown a strong connection between miRNA and nociceptive modulation, but the functional regulatory mechanism in miRNA expression itself remains elusive. In the present study, we further investigated whether 5hmC was involved in the regulation of miRNA expression under nociceptive status. As the enrichment of CpG dinucleotide in the gene promoter plays an important role in transcription efficiency (Pan et al., 2014), we analyzed the spinal 5hmC level in the CpG dinucleotide of global miRNA promoters (−500 bp ∼ +2000 bp) after formalin injection. In total, we identified 735 miRNAs with differential 5hmC promoters (data not shown). The cross-analysis of 126 miRNAs with >1.5-fold 5hmC gain (hMeDIP-Seq score >20) and 277 with >1.5-fold 5mC loss showed 34 crossed miRNAs (Fig. 3A; Table 2). Among them, the 5hmC level in the promoters of 3 miRNAs—miR-335, miR-92b, and miR-365-3p—were increased by 281.6, 291.6, and 287.9%, respectively, exhibiting relatively marked change compared with the control group (Table 2). RT-qPCR analysis revealed that the expressions of three miRNAs were upregulated in the spinal cord of formalin mice (Fig. 3B). However, compared with miR-335 and miR-92b, miR-365-3p exhibited significant alteration not only in 5hmC and 5mC levels (Table 2), but also in transcription expression (Fig. 3B) after formalin treatment. Furthermore, the visual 5hmC profile also showed an increase, ranging from −110 to +60 bp in the promoter of pre-miR-365-3p (Chr16:13453840-13453926) in the formalin group versus the control group (Fig. 3C). These results were further verified with hMeDIP-qPCR (Fig. 3D). Additionally, FISH confirmed an increase in spinal miR-365-3p expression at 2 h after formalin injection (Fig. 3E). Therefore, we chose to further investigate the functional regulatory role of miR-365-3p in the nociceptive process mediated by TET1 and TET3.
Figure 3.
Acute inflammatory pain leads to the alteration of spinal 5hmC and 5mC level in miR-365-3p promoter and its expression. A, Overlap of 5hmC gain and 5mC loss in genome-wide promoters in the spinal cord of mice with formalin-induced acute pain. The hMeDIP score (fragment-counts/kb) >20. B, The expression of three miRNAs [miR-335 (miR335), miR-92b (miR92b), and miR-365-3p (miR365–3p)] was analyzed using RT-qPCR. *p < 0.05 versus control group, **p < 0.01 versus control group, ***p < 0.001 versus control group; n = 6. C, The hMeDIP CpG island region using Genome browser shot of USUC in spinal pre-miR-365-3p promoter in FM group versus control group. TSS represents the transcript start site. D, Validation for hMeDIP-Seq results in the miR-365-3p promoter using qPCR. E, miR-365-3p FISH (green) and cell nuclear immunofluorescence staining (DAPI; blue) in spinal cord at 2 h after formalin or saline injection. Scale bar, 25 μm.
Table 2.
The cross analysis of 5hmC and 5mC in miRNA promoter
| miRNA information |
5hmC |
5mC |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Chr | TSS | TTS | Fold | FM | Ctrl | Fold | FM | Ctrl |
| miR1983 | chr13 | 21897049 | 21896917 | 3.3466 | 81 | 24 | −1.6772 | 19 | 11 |
| miR365-3p | chr16 | 13453839 | 13453926 | 3.8788 | 60 | 16 | −4.8028 | 31 | 7 |
| miR92b | chr3 | 89227198 | 89227115 | 3.9162 | 57 | 15 | −2.846 | 28 | 10 |
| miR1895 | chr3 | 134240583 | 134240504 | 2.0823 | 54 | 26 | −2.5348 | 33 | 13 |
| miR499 | chr2 | 155622879 | 155622958 | 3.6747 | 51 | 14 | −1.9821 | 23 | 11 |
| miR615 | chr15 | 103014909 | 103015001 | 1.5311 | 48 | 31 | −1.8678 | 12 | 7 |
| miR335 | chr6 | 30741298 | 30741396 | 3.8156 | 44 | 11 | −1.8562 | 37 | 20 |
| miR1942 | chr15 | 76215673 | 76215610 | 2.3384 | 44 | 19 | −1.8296 | 21 | 11 |
| miR149 | chr1 | 92850377 | 92850443 | 1.8373 | 44 | 24 | −1.8677 | 24 | 13 |
| miR3962 | chr7 | 144932135 | 144932115 | 1.908 | 43 | 22 | −3.7355 | 12 | 3 |
| miR3110 | chr8 | 126721800 | 126721780 | 1.9905 | 41 | 21 | −2.1345 | 17 | 8 |
| miR26a-1 | chr9 | 119031795 | 119031885 | 1.8373 | 41 | 22 | −1.7343 | 23 | 13 |
| miR1247 | chr12 | 110278129 | 110278047 | 2.8708 | 40 | 14 | −1.7788 | 9 | 5 |
| miR130b | chr16 | 17124142 | 17124060 | 2.2967 | 40 | 17 | −4.696 | 38 | 8 |
| let7b | chr15 | 85707318 | 85707403 | 2.2967 | 40 | 17 | −2.1346 | 14 | 7 |
| miR3572 | chr7 | 3655961 | 3656047 | 1.8373 | 35 | 19 | −2.846 | 14 | 5 |
| miR5126 | chr1 | 84695838 | 84695915 | 3.8584 | 33 | 9 | −2.7749 | 23 | 8 |
| miR487b | chr12 | 109727332 | 109727414 | 2.756 | 33 | 12 | −1.8296 | 21 | 11 |
| miR491 | chr4 | 88122039 | 88122125 | 2.0415 | 32 | 16 | −1.5247 | 35 | 23 |
| miR421 | chr15 | 103195810 | 103195830 | 1.9394 | 30 | 16 | −1.6009 | 5 | 3 |
| miR137 | chr3 | 118433856 | 118433929 | 3.3072 | 29 | 9 | −2.1345 | 17 | 8 |
| miR100 | chr9 | 41531424 | 41531504 | 2.3623 | 29 | 12 | −2.1346 | 31 | 15 |
| miR208b | chr14 | 54975776 | 54975699 | 2.2311 | 27 | 12 | −2.7749 | 45 | 16 |
| miR3473c | chr1 | 191998634 | 191998553 | 2.2311 | 27 | 12 | −1.7788 | 9 | 5 |
| miR1298 | chrX | 147064904 | 147065002 | 3.2154 | 22 | 7 | −100 | 7 | 0 |
| miR34c | chr9 | 51103110 | 51103033 | 2.5723 | 22 | 9 | −1.5247 | 17 | 11 |
| miR669k | chr2 | 10475299 | 10475426 | 1.8373 | 22 | 12 | −2.1346 | 14 | 7 |
| miR762 | chr7 | 127708486 | 127708562 | 1.8373 | 22 | 12 | −2.846 | 28 | 10 |
| miR127 | chr12 | 109592845 | 109592915 | 1.6077 | 22 | 14 | −1.7076 | 28 | 16 |
| miR1951 | chr2 | 115638724 | 115638813 | 1.6077 | 22 | 14 | −3.9133 | 19 | 5 |
| miR463 | chrX | 66799297 | 66799222 | 1.9905 | 21 | 10 | −1.6009 | 21 | 13 |
| miR150 | chr7 | 45121756 | 45121821 | 1.7061 | 21 | 12 | −3.2019 | 21 | 7 |
| miR490 | chr6 | 36421741 | 36421825 | 1.7061 | 21 | 12 | −2.0011 | 26 | 13 |
| miR5101 | chr12 | 75909102 | 75909185 | 1.7061 | 21 | 12 | −8.5377 | 14 | 2 |
TSS, Transcription start site; TTS, transcription terminal site; Fold, FM versus Ctrl.
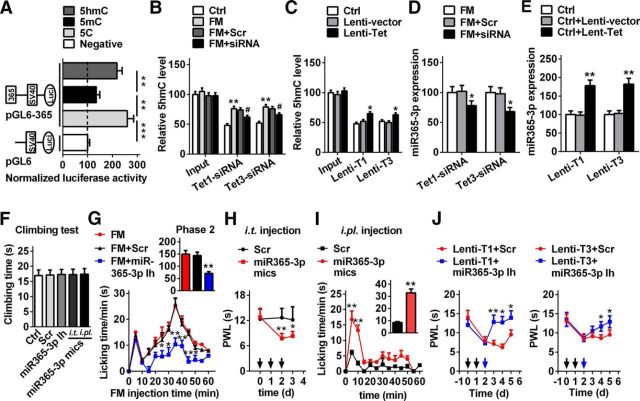
To determine whether TET proteins catalyzed 5hmC in the miR-365-3p promoter, we performed two strategies in vivo and in vitro to verify the hypothesis. First, in vitro, we evaluated whether the hydroxymethylated promoter of miR-365-3p could increase the efficiency of transcription. We cloned a 1817 bp segment of pre-miR-365-3p promoter (TTS: −1817 to 0 bp) into the pGL6 firefly luciferase reporter (pGL6-365) and examined its ability to drive luciferase expression in HEK293T cells according to a previous method (Sunahori et al., 2009). We found that this promoter region produced high luciferase activities compared with the negative control (Fig. 4A), indicating that the cloned region contains the regulatory element, and can control miR-365-3p expression. However, the pGL6-365 in vitro methylated by methyltransferase M.SssI before being introduced into cells abolished the enhanced luciferase activity (Fig. 4A). Further, the hydroxymethylated pGL6-365, which was converted in vitro from the methylated pGL6-365 via TET1 protein catalysis, reversed the activities of the methylated pGL6-365 group (Fig. 4A). These results suggest a direct positive role of DNA hydroxymethylation of the pGL6-365–3p promoter in transcriptional efficiency. Then, we analyzed the 5hmC change in the miR-365-3p promoter after TET1 or TET3 knockdown in formalin mice or overexpression in naive mice. We found that the 5hmC level was decreased by 18.4 and 16.5%, respectively, after the injection of Tet1- or Tet3-siRNA in the spinal cord of formalin mice (Fig. 4B), accompanied by a decrease in miR-365-3p expression (Fig. 4D). Overexpression of TET1 or TET3 increased the level of 5hmC by 31.3% and 21.2%, respectively (Fig. 4C), subsequently resulting in augmentation of the miR-365-3p expression in naive mice (Fig. 4E). Together, these results suggested that TET1 and TET3 regulate miR-365-3p expression by converting 5mC to 5hmC in the miR-365-3p promoter in the spinal cord of nociceptive pain mice.
Figure 4.
DNA hydroxymethylation mediated by TET1 and TET3 in the miR-365-3p promoter regulates spinal miR-365-3p expression and acute pain behavior. A, The activities of the methylated or demethylated promoter of the cloned miR-365-3p promoter encompassing TSS of pre-miR-365-3p were detected by firefly luciferase reporter assays in HEK293T cells. The pGL6 plasmid (empty vector) was used as the negative control; pGL6-365, plasmid with miR-365-3p promoter. 5C, Unmethylated group; 5mC, methylated group; 5hmC, hydroxyl-methylated group. Values of luciferase activities for each plasmid were normalized for transfection efficiency by cotransfection with pRL-TK plasmid. **p < 0.01 versus adjacent groups, ***p < 0.001 versus negative group; n = 6. B, Tet1- and Tet3-siRNA reversed the increase in spinal 5hmC of miR-365-3p caused by formalin injection. The relative iv5hmC level was measured using hMeDIP-qPCR. Input represents PCR products from genomic DNA without hMeDIP. **p < 0.01 versus control group, #p < 0.05 versus FM+Scr; n = 6. C, TET1 and TET3 overexpression mediated by lentivirus increased the 5hmC level of the miR-365-3p promoter in naive mice. *p < 0.05 versus the Lenti-vector group; n = 6. D, Downregulation of spinal TET1 and TET3 induced by siRNA decreased the expression of miR-365-3p. *p < 0.05 versus the FM+ Tet1-siRNA or Tet3-siRNA group; n = 6. E, Upregulation of spinal TET1 and TET3 induced by lentivirus increased the expression of miR-365-3p. **p < 0.01 versus the Lenti-vector group; n = 6. F, Time of climbing test at 72 h after intrathecal (i.t.) injection of miR-365-3p inhibitor (Ih), miR-365-3p mimics (mics) or Scr for 3 consecutive days, or at 5 min after intraplantar (i.pl.) injection of miR-365-3p mimics or Scr in naive mice. No significance versus Ctrl or Scr group; n = 6. G, Intrathecal injection of miR-365-3p inhibitor for 3 consecutive days alleviated formalin-induced acute pain in phase 2. *p < 0.05 versus FM+ Scr group, **p < 0.01 versus FM+Scr group; n = 6. H, Intrathecal injection of miR-365-3p mimics for 3 consecutive days induced the production of thermal hyperalgesia in naive mice. Black arrow indicates miR-365-3p mimics or Scr injection. *p < 0.05 versus Scr group, **p < 0.01 versus Scr group; n = 8. I, Intraplantar injection of miR-365-3p mimics produced pain-like behavior. Five microliters of miR-365-3p mimics or Scr (20 μm) was injected. **p < 0.01 versus Scr group; n = 8. J, Intrathecal injection of miR-365-3p inhibitor markedly inhibited thermal hyperalgesia induced by Lenti-T1 and Lenti-T3 in naive mice. Black arrow indicates Lenti-T1 and Lenti-T3 injection; blue arrow indicates miR-365-3p inhibitor or Scr injection. *p < 0.05 versus Lenti-T1 or Lenti-T3+Scr, **p < 0.01, versus Lenti-T1+Scr; n = 8.
To further investigate whether miR-365-3p contributed to the modulation of nociceptive behavior, we assessed the changes of motor and nociceptive behavior after manipulating spinal miR-365-3p. The results showed that the motor function could not be affected by the knockdown or overexpression of miR-365-3p in spinal cord, or overexpression of miR-365-3p in plantar surface of hindpaw in naive mice (Fig. 4F). However, downregulation of spinal miR-365-3p relieved the second phase nociceptive response induced by formalin; no effect was found in the scrambled group (Fig. 4G). In contrast, overexpression of miR-365-3p via intrathecal injection of mimics, but not control scrambled miRNAs, for 3 consecutive days, significantly induced thermal hyperalgesia (Fig. 4H). Interestingly, a formalin-induced nociceptive response could be mimicked by miR-365-3p overexpression in peripheral tissue via intraplantar injection of miR-365-3p mimics, but not control scrambled miRNAs, and this formalin pain-like nocifensive behavior could last for ∼15 min (Fig. 4I). These results suggest that miR-365-3p is involved in acute nociceptive regulation. Finally, to determine the role of miR-365-3p in mediation of nociceptive modulation by TET1 and TET3 at the behavior level, we post-treated the animals with inhibitor to knockdown miR-365-3p after intrathecal injection of Lenti-T1 or T3, and then observed the behavioral response. We found that knockdown of miR-365-3p markedly alleviated the thermal hyperalgesia induced by TET1 or TET3 overexpression in naive mice (Fig. 4J), suggesting that miR-365-3p acts as a mediator to modulate the nociceptive response induced by spinal TET proteins.
miR-365-3p targeting Kcnh2 regulates nociceptive response
To identify potential target genes of miR-365-3p, we performed an in silico target prediction by three independent programs: miRanda, PicTar, and Targetscan in TarMir. A total of 54 genes were predicted as potential targets of miR-365-3p (data not shown). Some of these target genes (e.g., Csk, Lin7c, and Skg1) have been reported to contribute to the regulation of pain behavior (Monastyrskaya et al., 2013; Peng et al., 2013; Deiteren et al., 2015). As Kcnh2 mRNA constitutes a conserved miR-365-3p binding site within its 3′-UTR (bp 220–227), and is involved in modulation of neuronal plasticity through a deactivating K+ current in CNS (Huffaker et al., 2009), we chose to evaluate the potential role of Kcnh2 as a target of miR-365-3p in nociception modulation. To validate the in silico prediction, we cloned a fragment of the Kcnh2 3′-UTR containing the miR-365-3p seed region into psiCHECK luciferase vector and tested the effects of miR-365-3p on the activities of the reporter gene in HEK293T cells. We found that cotransfection of miR-365-3p mimics with the reporter CHK-wt-Kcnh2 decreased luciferase activities by 58.5% compared with CHK-mut-Kcnh2. Contrarily, miR-365-3p inhibitor increased luciferase activities by 40.8% in CHK-wt-Kcnh2 but not in CHK-mut-Kcnh2 (Fig. 5A). Furthermore, we investigated the regulatory role of miR-365-3p in Kcnh2 expression in vivo. FISH-immunofluorescent costaining revealed that miR-365-3p and KCNH2 were coexpressed in spinal cells, and the increase in miR-365-3p expression was accompanied by a decrease of KCNH2-positive expression in the formalin group compared with the control group (Fig. 5B). The decreased expression of KCNH2 protein was reversed by pre-injection of miR-365-3p inhibitor, but not by scrambled control in formalin mice (Fig. 5C). Additionally, spinal KCNH2 expression was significantly reduced in naive mice by intrathecal injection of miR-365-3p mimics (Fig. 5D). The in vitro and in vivo findings suggest that Kcnh2 is a direct target of miR-365-3p. Based on the consideration that TET1 and TET3 regulate the expression of spinal miR-365-3p, we further wanted to know whether manipulating TET1 or TET3 would influence the expression of KCNH2 under nociception condition. We found that knockdown of TET1 or TET3 via pre-injection of siRNA, but not Scr, reversed the decrease of KCNH2 protein in formalin mice (Fig. 5E). In contrast, overexpression of TET1 or TET3 decreased the level of KCNH2 protein by 34.2 and 35.1%, respectively, in naive mice, which could be reversed by knockdown of miR-365-3p via intrathecal injection of miR-365-3p inhibitor (Fig. 5F). These results suggest that TET1 or TET3 regulates expression of KCNH2 through miR-365-3p.
Figure 5.
miR-365-3p targeting Kcnh2 regulates acute inflammatory pain. A, Luciferase assay confirming Kcnh2 as the target of miR-365-3p was analyzed using a mimic and an inhibitor of miR-365-3p in HEK293T cells. A mutation was generated in the Kcnh2-3′-UTR sequence in the complementary site for the seed region of miR-365-3p as indicated in CHK-mut-Kcnh2 plasmid. *p < 0.05 versus CHK-mut-Kcnh2, **p < 0.01 versus CHK-mut-Kcnh2; n = 5. B, FISH (miR-365-3p; green) and immunofluorescent (KCNH2; red) costaining in spinal cord at 2 h after formalin injection. Scale bar, 25 μm. C, The increased spinal KCNH2 in FM mice was reversed by intrathecal injection of the miR-365-3p inhibitor. KCNH2 was measured at 2 h after formalin injection (74 h after injection of miR-365-3p inhibitor). *p < 0.05 versus Ctrl group, #p < 0.05 versus FM+Scr; n = 5. D, Spinal KCNH2 expression was decreased by intrathecal injection of miR-365-3p mimics in naive mice. KCNH2 was measured at 2 d after intrathecal injection of miR-365-3p mimics. *p < 0.05 versus Scr groups; n = 5. E, Intrathecal injection of Tet1-siRNA (left) and Tet3-siRNA (right) reversed the increase of spinal KCNH2 in formalin mice. KCNH2 was measured at 2 h after formalin injection (74 h after Tet1- or Tet3-siRNA injection). *p < 0.05 versus Ctrl group, #p < 0.05 versus FM+Scr; n = 5. F, TET1 (top right) and TET3 (bottom left) overexpression mediated by lentivirus decreased KCNH2 expression in naive mice, which was reversed by the miR-365-3p inhibitor. KCNH2 was measured at 2 d after miR-365-3p inhibitor injection (96 h after Lenti-T1 or Tet3 injection). *p < 0.05 versus lentivector, #p < 0.05 versus Lenti-T1 or Lenti-T3+Scr; n = 5. G, Climbing time at 0.5 h after single intrathecal injection of NS1643 (KCNH2 agonist) in naive mice. No significance versus Ctrl or saline group; n = 6. H, Intrathecal injection of NS1643 inhibited thermal hyperalgesia induced by miR-365-3p mimics in naive mice. Black arrows indicate injection of miR-365-3p mimics or Scr. Blue arrow indicates NS1643 or saline injection. *p < 0.05 versus miR-365-3p mimics+Sal, **p < 0.01 versus miR-365-3p mimics+Sal; n = 6. I, Intrathecal injection of NS1643 markedly alleviated the thermal hyperalgesia induced by Lenti-T1 (left) and Lenti-T3 (right) in naive mice. Black arrows indicate Lenti-T1 and Lenti-T3 or Lenti-vector injection. Blue arrow indicates NS1643 or saline injection. *p < 0.05 versus Lenti-T1 or Lenti-T3+Sal, **p < 0.01 versus Lenti-T1 or Lenti-T3+Sal; n = 6.
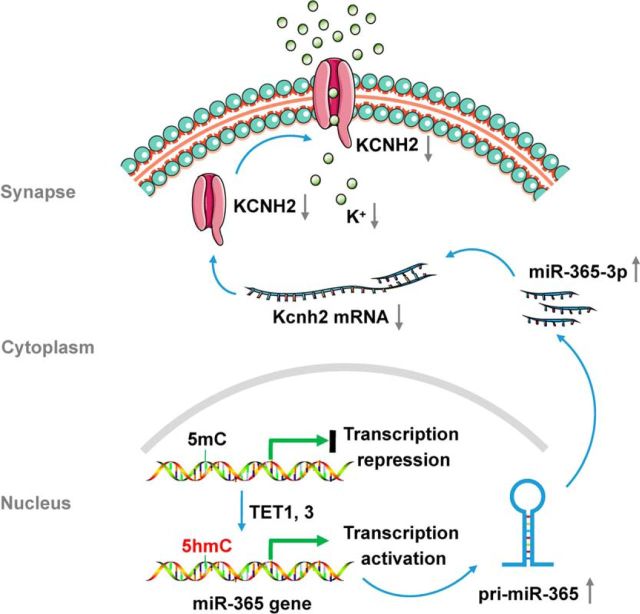
To gain insight into the role of KCNH2 in mediating nociceptive modulation by miR-365-3p at the behavioral level, we treated animals with the KCNH2 agonist NS1641 to activate the KCNH2 channel after intrathecal injection of miR-365-3p mimics, then measured the behavioral response. In the climbing behavior test, we found no difference between KCNH2 agonist (NS1643) group and saline group in the climbing time (Fig. 5G). However, the activation of KCNH2 significantly alleviated the thermal hyperalgesia induced by overexpression of spinal miR-365-3p in naive mice, indicating a direct mediatory role of KCNH2 in the nociceptive response induced by miR-365-3p (Fig. 5H). Similarly, activating KCNH2 significantly inhibited the thermal hyperalgesia induced by Tet1 or Tet3 overexpression in naive mice (Fig. 5I). Together, these results suggest that spinal TET1 and TET3 regulates the nociceptive response directly via miR-365-3p targeting Kcnh2 (Fig. 6).
Figure 6.
The schematic of TET1 or TET3 regulating the nociceptive response via miR-365-3p targeting Kcnh2.
Discussion
The dysfunction of pain-related gene expression is the most prominent contributor to the different nociceptive pathways (e.g., DRG, spinal cord, and pain-related brain regions) underlying the different types of pain (e.g., inflammatory pain and neuropathic pain; Tochiki et al., 2012; Pan et al., 2014). Recently, epigenetic mechanisms have been identified as essential constituents of the nociceptive pathway (Zhang et al., 2011; Cao et al., 2015). Accumulating evidence has implicated DNA methylation in nociceptive modulation. Emerging facts have shown that a potent demethylation mechanism is embedded in the pain process (Qi et al., 2013; Pan et al., 2014), which prompted us to determine whether TET proteins, as active demethylation enzymes, and 5hmC are involved in the epigenetic regulation of the nociceptive response. In the present study, we demonstrated that formalin-induced acute inflammatory pain increased the expression of spinal TET1 and TET3 proteins; furthermore, knockdown of TET1 and TET3 significantly alleviated the formalin-induced nociceptive response.
Previous studies have confirmed that TET proteins are associated with neurological and psychiatric disorders, such as Alzheimer's disease (Coppieters et al., 2014), learning and memory (Kaas et al., 2013; Rudenko et al., 2013), drug addiction (Feng et al., 2015), and inflammation (Taylor et al., 2014) by altering 5hmC patterns and gene expression level in response to neuronal activity. Mice lacking Tet1 exhibit the abnormal hippocampal long-term depression and the impaired memory extinction (Rudenko et al., 2013). TET1 overexpression in the dorsal hippocampus specifically impairs long-term memory formation (Kaas et al., 2013). TET2 has been detected in some CNS regions (Santiago et al., 2014). However, whether TET2 is involved in the neurophysiological or the neuropathological process remains unclear. The absence of TET2 in the modulation of CNS diseases may be merely associated with the tumor suppressor function of TET2 found in hematopoietic stem cells from animals and humans (Saint-Martin et al., 2009; Wakita et al., 2013; Chen et al., 2015). This may be why Tet2 was less expressed in the spinal cord of mice with formalin-induced pain. Previous reports revealed an important role of TET3 in neural progenitor cell maintenance, terminal differentiation, and neuronal development. Knock-out of Tet3 in embryonic stem cells leads to the impairment of neuronal differentiation (Xu et al., 2012; Li et al., 2015). In contrast, overexpression of TET3 rescues miR-15b-induced impaired proliferation of cortical neural progenitor cells (Lv et al., 2014). The autistic cerebellum exhibits a significant increase in TET1 and TET3, suggesting that the disease may be cooperatively caused by TET1 and TET3 (James et al., 2014). In the present study, our data confirmed an important mechanism of TET1 and TET3 underlying nociceptive modulation. These results further provide a new insight into the role of TET proteins in the modulation of CNS diseases.
5hmC is a newly discovered base in DNA which is converted from 5mC by TET proteins. Functionally, 5hmC is responsible for enhancing transcriptional efficiency. Recent studies have confirmed its extensive distribution in various tissues. Interestingly, the enrichment of 5hmC is nearly 10-fold more in neuronal cells than in some peripheral tissues or embryonic stem cells (Ficz et al., 2011; Taylor et al., 2014), suggesting that 5hmC is a stable epigenetic modification and may provide an epigenetic signature to a particular neuronal function or dysfunction in CNS. Consistently, global 5hmC is markedly gained in multiple CNS tissues under the progress of CNS development and diseases, such as the brain neurons of mouse embryonic differentiation (Hahn et al., 2013), aging mouse hippocampus (Chen et al., 2012), and the spinal cord of amyotrophic lateral sclerosis disease (Figueroa-Romero et al., 2012). Likewise, the loss of global 5hmC appears in CNS development processes and some diseases, such as terminally differentiated Purkinje neurons (Kriaucionis and Heintz, 2009) or in the striatum and cortex of Huntington's disease (Wang et al., 2013). These studies reveal a critical role of TET-mediated 5hmC modification under physiological and pathological conditions in the CNS, and an abnormal 5hmC level is a novel epigenetic feature in CNS diseases. In agreement with this viewpoint, we here observed that the nociceptive response is associated with a marked increase of 5hmC content in the spinal cord and blood.
MiRNAs are 20–25 nucleotide noncoding RNAs that regulate gene expression post-transcriptionally by binding the 3′-UTRs of mRNAs (Bartel, 2004). They have been implicated in various biological effects, including neurodegeneration (Eacker et al., 2009). Recent reports from different pain models have shown a strong connection between miRNA modulation and pain pathways from primary afferent nociceptors, DRG, spinal cord, and brain areas associated with pain perception. Manipulation of miRNA expression prevents and reverses persistent inflammatory, neuropathic, and cancer pain behavior (Favereaux et al., 2011; Imai et al., 2011). Although increasing molecular and functional evidence supports the alteration of miRNA expression in different CNS tissue under different pain conditions, little is known about how the miRNA gene expression itself is regulated in these processes. In our previous study, we reported, for the first time, the 5mC-mediated epigenetic modification of miR-219 expression in vivo, and its functional significance in chronic inflammatory pain induced by complete Freund's adjuvant (Pan et al., 2014). However, whether DNA hydroxymethylation causes dysregulation of miRNA in the nociceptive response still needs to be determined.
As miR-365-3p exhibited a significant difference in 5hmC and 5mC content of promoter, and in its expression after formalin treatment, miR-365-3p was chosen as an experimental target in this study. In fact, miR-365-3p was first discovered to express differentially in primary breast cancer (Yan et al., 2008). Furthermore, it has been demonstrated to be involved in the pathological process of various diseases, such as adipose metabolism (Sun et al., 2011; Feuermann et al., 2013; Mori et al., 2014), atherosclerosis lesion (Kim et al., 2014), and cancer formation (Guo et al., 2013; Seviour et al., 2015; Tao et al., 2015). However, to our best knowledge, the functional regulation of miR-365-3p in CNS-related diseases is still not reported. Here, we provide the first epigenetic evidence that formalin-mediated pain induced a significant increase in 5hmC and a decrease in 5mC in the miR-365-3p promoter, resulting in the enhancement of miR-365-3p transcription and the subsequent increase of miR-365-3p expression. Moreover, knockdown of TET1 or TET3 markedly reversed the increased 5hmC in miR-365-3p, accompanied by a decrease in miR-365-3p expression and alleviation of nociceptive behavior. Therefore, miR-365-3p hydroxymethylation mediated by TET1 and TET3 and its functional significance in nociceptive modulation elucidate a novel mechanism of miR-365-3p in the induction and maintenance of nociceptive behavior, which expands our knowledge about the functional role of miR-365-3p in CNS diseases.
Voltage-gated potassium (Kv) channels are membrane proteins that allow the rapid and selective flow of K+ ions across the cell membrane, generating electrical signals in neuron, that further play an essential role in controlling neuronal excitability. Currently, at least six Kv channel proteins—including Kv1.2 (Fan et al., 2014), Kv2.2 (Thibault et al., 2012), Kv3.4, Kv4.3 (Chien et al., 2007; Cao et al., 2010), Kv7, and Kv9.1 (Costigan et al., 2010; Tsantoulas et al., 2012)—have been identified in association with regulation of nociceptive signals. These Kv proteins are significantly decreased in spinal cord or DRG in inflammatory pain (Xu et al., 2010), neuropathic pain (Cao et al., 2010; Thibault et al., 2012), and a metastatic bone-cancer pain model (Zheng et al., 2013), whereas rescuing the reduction of Kv1.2 in the injured L5 DRG attenuates the pain hypersensitivity induced by spinal nerve ligation (Fan et al., 2014). In contrast, lentivirus- or siRNA-mediated knockdown of spinal Kv2.2 (Thibault et al., 2012) or Kv9.1 (Tsantoulas et al., 2012) induces a distinct nociceptive response.
Kcnh2 (Kv11.1 or Erg1a) is isolated from the hippocampal cDNA library (Warmke and Ganetzky, 1994) and encodes the α-subunits that form rapidly activating delayed rectifier K+ channels (Sanguinetti et al., 1995). KCNH2 has been found to be implicated in the neuropathology of CNS-related diseases (Kongsamut et al., 2002; Winterer and Weinberger, 2004). Overexpression of Kcnh2 isoform (3.1) in primary cortical neurons causes a dramatic alteration in KCNH2 channel physiology via induction of a rapidly deactivating K+ current and a high-frequency, non-adapting firing pattern, suggesting that KCNH2 is a potential new Kv channel for the treatment of CNS diseases (Huffaker et al., 2009). Although KCNH2 have been confirmed as a link to the modulation of CNS disease, whether KCNH2 has a direct connection with the nociceptive response, and its functional regulatory mechanisms are still unknown. We observed that Kcnh2 is a new regulatory target of miR-365-3p, and further demonstrated that the dysfunction of KCNH2 is critical in the generation of the nociceptive response. Based on the potent regulatory role of KCNH2 in the excitability of neurons and transport of ions (Huffaker et al., 2009), we propose that KCNH2 directly mediates the role of miR-365-3p in nociception via negative regulation of the polarization of spinal neurons.
In summary, the present study demonstrated that the increase of 5hmC content mediated by TET1 or TET3 in the miR-365-3p promoter at the spinal cord level was involved in nociceptive modulation through targeting the potassium channel KCNH2. Our findings reveal a new epigenetic mechanism underlying nociceptive information processing, indicating a possible novel target for the development of antinociceptive drugs.
Footnotes
The work was supported by grants from the National Natural Science Foundation of China (81070888, 81230025 to J.-L.C., 81271231 to Z.P., 31500855 to L.-J.Z., 81200859 to H.-L.D.); Key project of the Natural Science Foundation of Jiangsu Education Department (11KJA320001 to J.-L.C.; 15KJA320004 to Z.P.); the Natural Science Foundation of Jiangsu Province (BK2011196 to Z.P.); and a Project Funded by the Qing Lan Project of Jiangsu, by the Six Talent Summit Project of Jiangsu, by the 333 High-level Personnel Training Project of Jiangsu, China, and by the Jiangsu Provincial Special Program of Medical Science (BL2014029). We thank Dr Lydia Wai Tai (Department of Anesthesiology, University of Hong Kong, China) and Sarah E. Toombs Smith (University of Texas Medical Branch, Galveston, TX) for their proofreading of the paper.
The authors declare no competing financial interests.
References
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, Sun H, Schuebel K, Zhou Z, Yuan Q, Vendruscolo LF, Goldman D, Heilig M. DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J Neurosci. 2015;35:6153–6164. doi: 10.1523/JNEUROSCI.4571-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nri3380. [DOI] [PubMed] [Google Scholar]
- Cao DY, Bai G, Ji Y, Traub RJ. Epigenetic upregulation of metabotropic glutamate receptor 2 in the spinal cord attenuates oestrogen-induced visceral hypersensitivity. Gut. 2015;64:1913–1920. doi: 10.1136/gutjnl-2014-307748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XH, Byun HS, Chen SR, Cai YQ, Pan HL. Reduction in voltage-gated K+ channel activity in primary sensory neurons in painful diabetic neuropathy: role of brain-derived neurotrophic factor. J Neurochem. 2010;114:1460–1475. doi: 10.1111/j.1471-4159.2010.06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Schneider RK, Breyfogle LJ, Rosen EA, Poveromo L, Elf S, Ko A, Brumme K, Levine R, Ebert BL, Mullally A. Distinct effects of concomitant Jak2V617F expression and Tet2 loss in mice promote disease progression in myeloproliferative neoplasms. Blood. 2015;125:327–335. doi: 10.1182/blood-2014-04-567024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Dzitoyeva S, Manev H. Effect of aging on 5-hydroxymethylcytosine in the mouse hippocampus. Restor Neurol Neurosci. 2012;30:237–245. doi: 10.3233/RNN-2012-110223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien LY, Cheng JK, Chu D, Cheng CF, Tsaur ML. Reduced expression of A-type potassium channels in primary sensory neurons induces mechanical hypersensitivity. J Neurosci. 2007;27:9855–9865. doi: 10.1523/JNEUROSCI.0604-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer's disease human brain. Neurobiol Aging. 2014;35:1334–1344. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, Wu T, Kiselycznyk C, Poddar M, Lu Y, Diatchenko L, Smith S, Cobos EJ, Zaykin D, Allchorne A, Gershon E, Livneh J, Shen PH, Nikolajsen L, Karppinen J, et al. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 2010;133:2519–2527. doi: 10.1093/brain/awq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiteren A, van der Linden L, de Wit A, Ceuleers H, Buckinx R, Timmermans JP, Moreels TG, Pelckmans PA, De Man JG, De Winter BY. P2X3 receptors mediate visceral hypersensitivity during acute chemically-induced colitis and in the post-inflammatory phase via different mechanisms of sensitization. PloS One. 2015;10:e0123810. doi: 10.1371/journal.pone.0123810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats P, Spencer B, Coffee E, Patel P, Michael S, Patrick C, Adame A, Rockenstein E, Masliah E. Alpha-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem. 2011;286:9031–9037. doi: 10.1074/jbc.C110.212589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Guan X, Wang W, Zhao JY, Zhang H, Tiwari V, Hoffman PN, Li M, Tao YX. Impaired neuropathic pain and preserved acute pain in rats overexpressing voltage-gated potassium channel subunit Kv1.2 in primary afferent neurons. Mol Pain. 2014;10:8. doi: 10.1186/1744-8069-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favereaux A, Thoumine O, Bouali-Benazzouz R, Roques V, Papon MA, Salam SA, Drutel G, Léger C, Calas A, Nagy F, Landry M. Bidirectional integrative regulation of Cav1.2 calcium channel by microRNA miR-103: role in pain. EMBO J. 2011;30:3830–3841. doi: 10.1038/emboj.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, Le T, Ferguson D, Cahill ME, Li Y, Koo JW, Ribeiro E, Labonte B, Laitman BM, Estey D, Stockman V, Kennedy P, Couroussé T, Mensah I, Turecki G, et al. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat Neurosci. 2015;18:536–544. doi: 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuermann Y, Kang K, Gavrilova O, Haetscher N, Jang SJ, Yoo KH, Jiang C, Gonzalez FJ, Robinson GW, Hennighausen L. MiR-193b and miR-365-1 are not required for the development and function of brown fat in the mouse. RNA Biol. 2013;10:1807–1814. doi: 10.4161/rna.27239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Figueroa-Romero C, Hur J, Bender DE, Delaney CE, Cataldo MD, Smith AL, Yung R, Ruden DM, Callaghan BC, Feldman EL. Identification of epigenetically altered genes in sporadic amyotrophic lateral sclerosis. PloS One. 2012;7:e52672. doi: 10.1371/journal.pone.0052672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Ann Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Guo SL, Ye H, Teng Y, Wang YL, Yang G, Li XB, Zhang C, Yang X, Yang ZZ, Yang X. Akt-p53-miR-365-cyclin D1/cdc25A axis contributes to gastric tumorigenesis induced by PTEN deficiency. Nat Commun. 2013;4:2544. doi: 10.1038/ncomms3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker SJ, Chen J, Nicodemus KK, Sambataro F, Yang F, Mattay V, Lipska BK, Hyde TM, Song J, Rujescu D, Giegling I, Mayilyan K, Proust MJ, Soghoyan A, Caforio G, Callicott JH, Bertolino A, Meyer-Lindenberg A, Chang J, Ji Y, et al. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med. 2009;15:509–518. doi: 10.1038/nm.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Saeki M, Yanase M, Horiuchi H, Abe M, Narita M, Kuzumaki N, Suzuki T, Narita M. Change in microRNAs associated with neuronal adaptive responses in the nucleus accumbens under neuropathic pain. J Neurosci. 2011;31:15294–15299. doi: 10.1523/JNEUROSCI.0921-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SJ, Shpyleva S, Melnyk S, Pavliv O, Pogribny IP. Elevated 5-hydroxymethylcytosine in the engrailed-2 (EN-2) promoter is associated with increased gene expression and decreased MeCP2 binding in autism cerebellum. Transl Psychiatry. 2014;4:e460. doi: 10.1038/tp.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Ham O, Lee SY, Choi E, Lee CY, Park JH, Lee J, Seo HH, Seung M, Choi E, Min PK, Hwang KC. MicroRNA-365 inhibits the proliferation of vascular smooth muscle cells by targeting cyclin D1. J Cell Biochem. 2014;115:1752–1761. doi: 10.1002/jcb.24841. [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsamut S, Kang J, Chen XL, Roehr J, Rampe D. A comparison of the receptor binding and HERG channel affinities for a series of antipsychotic drugs. Eur J Pharmacol. 2002;450:37–41. doi: 10.1016/S0014-2999(02)02074-5. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynast KL, Russe OQ, Möser CV, Geisslinger G, Niederberger E. Modulation of central nervous system-specific microRNA-124a alters the inflammatory response in the formalin test in mice. Pain. 2013;154:368–376. doi: 10.1016/j.pain.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Li T, Yang D, Li J, Tang Y, Yang J, Le W. Critical role of Tet3 in neural progenitor cell maintenance and terminal differentiation. Mol Neurobiol. 2015;51:142–154. doi: 10.1007/s12035-014-8734-5. [DOI] [PubMed] [Google Scholar]
- Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids. 2011;2011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yuan YB, Guan LL, Wei H, Cheng Z, Han X, Yang L, Pu CC, Yang FD, Lu Z, Deng H, Zhao JP, Yu X. MiRNA-365 and miRNA-520c-3p respond to risperidone treatment in first-episode schizophrenia after a 1 year remission. Chin Med J. 2013;126:2676–2680. [PubMed] [Google Scholar]
- Lv X, Jiang H, Liu Y, Lei X, Jiao J. MicroRNA-15b promotes neurogenesis and inhibits neural progenitor proliferation by directly repressing TET3 during early neocortical development. EMBO Rep. 2014;15:1305–1314. doi: 10.15252/embr.201438923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrskaya K, Sánchez-Freire V, Hashemi Gheinani A, Klumpp DJ, Babiychuk EB, Draeger A, Burkhard FC. miR-199a-5p regulates urothelial permeability and may play a role in bladder pain syndrome. Am J Pathol. 2013;182:431–448. doi: 10.1016/j.ajpath.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Mori MA, Thomou T, Boucher J, Lee KY, Lallukka S, Kim JK, Torriani M, Yki-Järvinen H, Grinspoon SK, Cypess AM, Kahn CR. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J Clin Invest. 2014;124:3339–3351. doi: 10.1172/JCI73468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzel M, Globisch D, Brückl T, Wagner M, Welzmiller V, Michalakis S, Müller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pan Z, Zhu LJ, Li YQ, Hao LY, Yin C, Yang JX, Guo Y, Zhang S, Hua L, Xue ZY, Zhang H, Cao JL. Epigenetic modification of spinal miR-219 expression regulates chronic inflammation pain by targeting CaMKIIγ. J Neurosci. 2014;34:9476–9483. doi: 10.1523/JNEUROSCI.5346-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi C, Arisi I, D'Ambrosi N, Storti AE, Brandi R, D'Onofrio M, Volonté C. Dysregulated microRNAs in amyotrophic lateral sclerosis microglia modulate genes linked to neuroinflammation. Cell Death Dis. 2013;4:e959. doi: 10.1038/cddis.2013.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HY, Chen GD, Lai CY, Hsieh MC, Lin TB. Spinal serum-inducible and glucocorticoid-inducible kinase 1 mediates neuropathic pain via kalirin and downstream PSD-95-dependent NR2B phosphorylation in rats. J Neurosci. 2013;33:5227–5240. doi: 10.1523/JNEUROSCI.4452-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollema-Mays SL, Centeno MV, Apkarian AV, Martina M. Expression of DNA methyltransferases in adult dorsal root ganglia is cell-type specific and up regulated in a rodent model of neuropathic pain. Front Cell Neurosci. 2014;8:217. doi: 10.3389/fncel.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Zhou Y, Xiao Y, Tao J, Gu J, Jiang X, Xu GY. Promoter demethylation of cystathionine-beta-synthetase gene contributes to inflammatory pain in rats. Pain. 2013;154:34–45. doi: 10.1016/j.pain.2012.07.031. [DOI] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Martin C, Leroy G, Delhommeau F, Panelatti G, Dupont S, James C, Plo I, Bordessoule D, Chomienne C, Delannoy A, Devidas A, Gardembas-Pain M, Isnard F, Plumelle Y, Bernard O, Vainchenker W, Najman A, Bellanné-Chantelot C French Group of Familial Myeloproliferative Disorders. Analysis of the ten-eleven translocation 2 (TET2) gene in familial myeloproliferative neoplasms. Blood. 2009;114:1628–1632. doi: 10.1182/blood-2009-01-197525. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- Santiago M, Antunes C, Guedes M, Sousa N, Marques CJ. TET enzymes and DNA hydroxymethylation in neural development and function: how critical are they? Genomics. 2014;104:334–340. doi: 10.1016/j.ygeno.2014.08.018. [DOI] [PubMed] [Google Scholar]
- Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T, Zhang F, Hill SM, Liu W, Maiti SN, Cooper L, Azencot R, Lopez-Berestein G, Rodriguez-Aguayo C, Roopaimoole R, Pecot C, Sood AK, Mukherjee S, Gray JW, Mills GB, Ram PT. Functional proteomics identifies miRNAs to target a p27/Myc/phospho-Rb signature in breast and ovarian cancer. Oncogene. 2015 doi: 10.1038/onc.2014.469. Advance online publication. Retrieved Feb. 2, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunahori K, Juang YT, Tsokos GC. Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac alpha promoter determines CREB binding and activity. J Immunol. 2009;182:1500–1508. doi: 10.4049/jimmunol.182.3.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajerian M, Alvarado S, Millecamps M, Dashwood T, Anderson KM, Haglund L, Ouellet J, Szyf M, Stone LS. DNA methylation of SPARC and chronic low back pain. Mol Pain. 2011;7:65. doi: 10.1186/1744-8069-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Ji J, Li X, Ding N, Wu H, Liu Y, Wang XW, Calvisi DF, Song G, Chen X. Distinct anti-oncogenic effect of various microRNAs in different mouse models of liver cancer. Oncotarget. 2015;6:6977–6988. doi: 10.18632/oncotarget.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SE, Smeriglio P, Dhulipala L, Rath M, Bhutani N. A global increase in 5-hydroxymethylcytosine levels marks osteoarthritic chondrocytes. Arthritis Rheumatol. 2014;66:90–100. doi: 10.1002/art.38200. [DOI] [PubMed] [Google Scholar]
- Thibault K, Calvino B, Dubacq S, Roualle-de-Rouville M, Sordoillet V, Rivals I, Pezet S. Cortical effect of oxaliplatin associated with sustained neuropathic pain: exacerbation of cortical activity and down-regulation of potassium channel expression in somatosensory cortex. Pain. 2012;153:1636–1647. doi: 10.1016/j.pain.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Tochiki KK, Cunningham J, Hunt SP, Géranton SM. The expression of spinal methyl-CpG-binding protein 2, DNA methyltransferases and histone deacetylases is modulated in persistent pain states. Mol Pain. 2012;8:14. doi: 10.1186/1744-8069-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsantoulas C, Zhu L, Shaifta Y, Grist J, Ward JP, Raouf R, Michael GJ, McMahon SB. Sensory neuron downregulation of the Kv9.1 potassium channel subunit mediates neuropathic pain following nerve injury. J Neurosci. 2012;32:17502–17513. doi: 10.1523/JNEUROSCI.3561-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viet CT, Ye Y, Dang D, Lam DK, Achdjian S, Zhang J, Schmidt BL. Re-expression of the methylated EDNRB gene in oral squamous cell carcinoma attenuates cancer-induced pain. Pain. 2011;152:2323–2332. doi: 10.1016/j.pain.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita S, Yamaguchi H, Omori I, Terada K, Ueda T, Manabe E, Kurosawa S, Iida S, Ibaraki T, Sato Y, Todoroki T, Hirakawa T, Ryotokuji T, Arai K, Kitano T, Mitamura Y, Kosaka F, Dan K, Inokuchi K. Mutations of the epigenetics-modifying gene (DNMT3a, TET2, IDH1/2) at diagnosis may induce FLT3-ITD at relapse in de novo acute myeloid leukemia. Leukemia. 2013;27:1044–1052. doi: 10.1038/leu.2012.317. [DOI] [PubMed] [Google Scholar]
- Wang F, Yang Y, Lin X, Wang JQ, Wu YS, Xie W, Wang D, Zhu S, Liao YQ, Sun Q, Yang YG, Luo HR, Guo C, Han C, Tang TS. Genome-wide loss of 5-hmC is a novel epigenetic feature of Huntington's disease. Hum Mol Genet. 2013;22:3641–3653. doi: 10.1093/hmg/ddt214. [DOI] [PubMed] [Google Scholar]
- Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer's disease. PloS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Pan Q, Lin L, Szulwach KE, Song CX, He C, Wu H, Warren ST, Jin P, Duan R, Li X. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet. 2012;21:5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci U S A. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterer G, Weinberger DR. Genes, dopamine and cortical signal-to-noise ratio in schizophrenia. Trends Neurosci. 2004;27:683–690. doi: 10.1016/j.tins.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Xu W, Wu Y, Bi Y, Tan L, Gan Y, Wang K. Activation of voltage-gated KCNQ/Kv7 channels by anticonvulsant retigabine attenuates mechanical allodynia of inflammatory temporomandibular joint in rats. Mol Pain. 2010;6:49. doi: 10.1186/1744-8069-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, Diao J, Wu F, He HH, Cui Q, Clark E, Ma C, Barbara A, Veenstra GJ, Xu G, Kaiser UB, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chung SK, Chow BK. The knockout of secretin in cerebellar Purkinje cells impairs mouse motor coordination and motor learning. Neuropsychopharmacology. 2014;39:1460–1468. doi: 10.1038/npp.2013.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP, Liu W, Xu ZM, Yang L, Ding YQ, Tang F, Meissner A, Ding C, Shi Y, Xu GL. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Cai YQ, Zou F, Bie B, Pan ZZ. Epigenetic suppression of GAD65 expression mediates persistent pain. Nat Med. 2011;17:1448–1455. doi: 10.1038/nm.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Fang D, Liu M, Cai J, Wan Y, Han JS, Xing GG. Suppression of KCNQ/M (Kv7) potassium channels in dorsal root ganglion neurons contributes to the development of bone cancer pain in a rat model. Pain. 2013;154:434–448. doi: 10.1016/j.pain.2012.12.005. [DOI] [PubMed] [Google Scholar]