Abstract
Traditional anatomical studies of the parahippocampal region (PHR) defined the lateral portion into two subregions, the perirhinal (PRC) and parahippocampal (PHC) cortices. Based on this organization, several models suggested that the PRC and the PHC play different roles in memory through connections with different memory-related brain networks. To identify the key components of the human PHR, we used a well accepted connection-based parcellation method on two independent datasets. Our parcellation divided the PRC and PHC into three subregions, specifically, the rostral PRC, caudal PRC (PRCc), and PHC. The connectivity profile for each subregion showed that the rostral PRC was connected to the anterior temporal (AT) system and the PHC was connected to the posterior medial (PM) system. The transition area (PRCc) integrated the AT-PM systems. These results suggest that the lateral PHR not only contains functionally segregated subregions, but also contains a functionally integrated subregion.
SIGNIFICANCE STATEMENT We redefined the cartography of the human parahippocampal region (PHR) and identified a transition subarea based on distinct anatomical and functional connectivity profiles. This well defined anatomical organization of the PHR is necessary for expanding our understanding and studying the functional relevance of its subregions in recognition memory. We found that the transition subregion [caudal perirhinal cortex (PRCc)] is a functionally integrated subregion that integrates the anterior temporal (AT)–posterior medial (PM) systems. In addition, we found that the core components of the AT and PM systems connect with the PHR in the rostral PRC and parahippocampal cortex (PHC), respectively, rather than connecting with the traditional, larger, and thus less concise PRC and PHC areas. This may lead to new insights into the human memory system and related neurodegenerative diseases.
Keywords: brain connectivity, parahippocampal region
Introduction
The parahippocampal region (PHR) is an interface region between the hippocampus and the neocortex located in the medial temporal lobe (MTL) (Squire and Zola-Morgan, 1991; Lavenex and Amaral, 2000; Squire et al., 2004). The PHR has been implicated in many functions, including long-term memory (for reviews, see Squire et al., 2004; Eichenbaum et al., 2007), working memory (Jeneson and Squire, 2012; Libby et al., 2014), and perception (O'Neil et al., 2009; Barense et al., 2010). Given these important and complex functions of the PHR, identifying its anatomical organization is necessary for expanding our understanding and studying the functional relevance of its subregions.
Based on the traditionally accepted parcellation scheme for the PHR [i.e., entorhinal cortex (ERC), perirhinal cortex (PRC), and parahippocampal cortex (PHC)] (Squire and Zola-Morgan, 1991; Insausti et al., 1998; Burwell, 2000; Ding and Van Hoesen, 2010; Kivisaari et al., 2013), a considerable body of research has focused on searching for the functional segregation of the PRC and PHC (for reviews, see Davachi, 2006; Diana et al., 2007; Eichenbaum et al., 2007; Ranganath, 2010). In particular, the well known memory-guided behavior model proposed by Ranganath and Ritchey (2012) suggested that the PRC and PHC are the core components of two memory-related brain networks, the anterior temporal (AT) and posterior medial (PM) systems, respectively. However, this existing anatomical description of the human PHR is coarse, so functional heterogeneity may exist within the PRC or PHC. First, based on cytoarchitecture and tract-tracing studies, more precise parcellation schemes of the PRC and PHC in monkey brains subdivided the PRC into 35, 36r, and 36c, and divided the PHC into TH and TF (Suzuki and Amaral, 1994a; Lavenex et al., 2002; Suzuki and Amaral, 2003; Saleem et al., 2007). Specifically, area 36c serves as an interface for communication between the 36r and TF in monkeys, as indicated by tracer studies (Lavenex et al., 2002, 2004). Second, although these may only be artifacts, some human fMRI studies found that, along an anterior–posterior axis, the most anterior and posterior portions of the PHR show significant functional differences (Köhler et al., 2005; Diana et al., 2010; Hayes et al., 2010; Libby et al., 2014) and the middle portion seems to have a mixture of functions (Litman et al., 2009; Staresina et al., 2011; Hannula et al., 2013; Liang et al., 2013). Therefore, these clues can be taken to indicate that the actual anatomical partition of the human PHR may be more fine grained than the existing definition and that an integration subregion between the anterior and posterior portions of the PHR may also exist.
Recently, methods based on connection-based parcellation (CBP) techniques have emerged as powerful tools for investigating the organization of the human brain (Cloutman and Lambon Ralph, 2012). Noninvasive CBP techniques have been used to define functionally distinct brain regions that are highly consistent with traditional cytoarchitectonic findings (Johansen-Berg et al., 2004; Beckmann et al., 2009; Fan et al., 2014; Zhang et al., 2014). Conversely, tractography studies that used diffusion imaging have revealed functionally specialized subregions that exhibit a unique complement of connections (Bouhali et al., 2014; Neubert et al., 2014). Therefore, a parcellation strategy based on diffusion MRI is feasible for unraveling the anatomical organization of the PHR and for testing whether a crossroad area exists between the anterior and posterior portions of the PHR.
The goal of this study was to reveal whether a transition area exists and serves as an interface between the anterior–posterior connections. For this purpose, using two independent datasets, we first used an in vivo anatomical CBP technology (Fan et al., 2014; Neubert et al., 2014; Zhang et al., 2014) to identify the fine-grained parcellation of the PHR and then investigated the differences in the connectivity patterns between each subregion using probabilistic fiber tracking and resting-state functional connectivity analysis.
Materials and Methods
Subjects
Two independent groups of healthy subjects were included in this study. Dataset 1 included 20 (10 males, age range, 17–20 years, age, 18.50 ± 0.76 years, mean ± SD) right-handed participants. Each subject was free of any psychiatric or neurological abnormalities. Informed consent was obtained from all subjects as approved by the local medical ethics committee of the University of Electronic Science and Technology of China. Dataset 2 included 10 participants (5 males, age 26–35 years) and was obtained from Human Connectome Project (HCP) dataset (http://www.humanconnectomeproject.org/data/; Van Essen et al., 2013).
Data acquisition
Dataset 1 provided diffusion tensor imaging (DTI), structural MRI, and resting-state fMRI data, which were obtained using a Signa HDx (General Electric) 3.0 tesla MR. The DTI scheme of Dataset 1 produced a collection of 64 images with non-colinear diffusion gradients (b = 1000 s/mm2) and 3 non-diffusion-weighted images (b = 0 s/mm2) using a single-shot echoplanar imaging (SE-EPI) sequence. For each participant, 75 slices were collected with repetition time (TR) = 8500 ms, echo time (TE) = 67.6 ms, field of view (FOV) = 256 mm × 256 mm, acquisition matrix = 128 × 128, flip angle (FA) = 90°, and resolution = 2 mm × 2 mm × 2 mm with no gap. Resting-state fMRI data were obtained using a gradient-echo single-shot EPI (GE-EPI) sequence with the following parameters: TR/TE = 2000/30 ms, FA = 90°, FOV = 240 mm × 240 mm, matrix = 64 × 64, slice thickness = 3.4 mm, 0.6 mm gap, 39 transverse slices, and duration of 8 min. During the fMRI scans, all the subjects were instructed to keep their eyes closed, relax, and move as little as possible. Sagittal 3D T1-weighted images were also acquired with a brain volume (BRAVO) sequence with the following parameters: TR/TE = 1.9/3 ms, inversion time (TI) = 800 ms, FA = 7°, FOV = 256 mm × 256 mm, in-plane resolution = 1 mm × 1 mm, slice thickness = 1 mm with no gap, and 192 sagittal slices.
The methodological details, including the scanner information, diffusion image acquisition, and reconstruction for the HCP dataset are provided in Uğurbil et al. (2013). In brief, the important parameters were TE/TR = 89.5/5520 ms, FOV = 210 mm × 180 mm, FA = 90°, 1.25 mm isotropic spatial resolution, six b = 0, and three diffusion weightings (b = 1000, 2000, and 3000 s/mm2) images.
Data preprocessing
The diffusion and structural MR data were preprocessed using the FMRIB's Diffusion Toolbox (FMRIB Software Library, FSL version 5.0; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL) and the MINC Toolbox (http://www.bic.mni.mcgill.ca/ServicesSoftware/MINC). First, a brain extraction was performed on the non-diffusion-weighted images (b = 0 s/mm2) using the brain extraction tool in FSL. Second, after being converted into the MINC format, the structural MR images were corrected for non-uniformity artifacts using the nonparametric non-uniform intensity normalization (N3) algorithm to improve the accuracy of the subsequent anatomical analyses. Next, the skull-stripped T1-weighted image from each subject was linearly transformed into the previously processed non-diffusion b0 image using the registration program (minctracc) available with MINC tools. This process provided a set of coregistered T1 images in native DTI space. Subsequently, each coregistered T1-weighted image in the diffusion space was first registered to the Montreal Neurological Institute (MNI) template using a linear (lsq9, i.e., 3 rotations, 3 translations, and 3 scales) transformation. Next, the linearly transformed T1-weighted images were nonlinearly warped into the MNI template using the automated nonlinear MRI registration package (MNI_ANIMAL) in MINC tools. Finally, the derived transformation parameters were inverted and used to warp the seed and target masks from MNI space to the native DTI space using a nearest neighbor interpolation.
Preprocessing of the resting-state fMRI data was performed using scripts provided by the 1000 Functional Connectome Project (www.nitrc.org/projects/fcon_1000) using both FSL and AFNI (Automated Functional NeuroImaging; http://afni.nimh.nih.gov/afni). The preprocessing steps consisted of the following: (1) discarding the first 10 volumes in each scan series to allow for signal equilibration, (2) performing slice-timing correction, (3) performing motion correction, (4) performing time-series de-spiking, (5) normalizing the mean-based intensity, (6) performing band-pass temporal filtering (0.005 Hz < f < 0.10 Hz), (7) removing linear and quadratic trends, (8) performing linear and nonlinear spatial normalization of the structural MR images to the MNI152 brain template, (9) coregistering the anatomical volume with the mean functional volume, and (10) performing nuisance signal regression (white matter, CSF, the global signal, and 6 motion parameters).
Finally, 4D residual time series data in individual space were acquired for each subject after the preprocessing. None of the participants had a head motion of >1.5 mm maximum translations in the x, y, and z directions or 1.5° in any angular rotation. To reduce the variability in the EPI time series due to susceptibility artifacts, temporal signal-to-noise ratio (TSNR) (Murphy et al., 2007) maps were calculated for each subject. The averaged TSNR map was then thresholded at 20 to generate a binarized mask in each individual space.
Definition of the PHR boundary
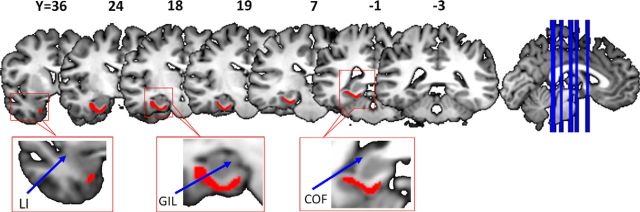
The seeds of the PHR region were defined manually on individual participant T1-weighted images from the DISPLAY software package (MacDonald, 1996) using criteria outlined in previous studies (Insausti et al., 1998; Pruessner et al., 2002; Huntgeburth and Petrides, 2012; Kivisaari et al., 2013; Frankó et al., 2014) (Fig. 1). To avoid overestimating the boundary of the PHR, a conservative boundary was defined. In brief, the most anterior border of the PHR was defined as being 2 mm anterior to the most anterior coronal slice containing gray matter in the limen insulae or the most anterior slice in which the collateral sulcus was visible, whichever was more anterior. From the limen insulae to the end of the gyrus intralimbicus (i.e., the transition from the head to the body of the hippocampus), the medial and lateral borders were separately defined using the medial apex of the parahippocampal gyrus and the shoulder of the lateral bank of the collateral sulcus. After the disappearance of the gyrus intralimbicus in the posterior direction, the border only included the medial bank of the collateral sulcus and extended to the most medial aspect of the parahippocampal gyrus. The most posterior border of the PHR was defined as one slice posterior to the crus of the fornix. For each subject, the PHR mask was linearly registered to the individual fractional anisotropy image. Then, to obtain a mask for a group, the individual PHR masks were nonlinearly registered to the MNI standard template using SPM8.
Figure 1.
Human PHR ROI. Shown is an example of the PHR ROI for a single subject in coronal sections. The numbers are the specific coordinates for each coronal slice. LI, limen insula; COF, crus of the fornix.
Connectivity-based parcellation of the PHR
First, to estimate the connectivity probability, probabilistic fiber tracking was applied by sampling 5000 streamline fibers per voxel within the PHR in native diffusion space using a multiple fiber orientations model (Behrens et al., 2007). The connectivity probability between the ith voxel and jth voxel was defined by the number of fibers passing through the two voxels. Next, the connections were estimated between the voxels in the PHR and all of the remaining voxels in the brain and were downsampled to 5 mm isotropic voxels to construct the native connectivity M-by-N matrix between all the PHR voxels (M) and the brain voxels (N). Finally, based on the above native connectivity matrix, a symmetric cross-correlation M-by-M matrix was calculated to quantify the similarity between the connectivity profiles of each PHR seed.
Many previous methods, such as k-means clustering for the medial frontal cortex (Johansen-Berg et al., 2004) and the PCA/scree test for the occipital lobes (Thiebaut de Schotten et al., 2014), have adopted clustering as a way to parcellate brain regions. In the current study, a spectral clustering algorithm (Von Luxburg, 2007) was applied to parcellate the PHR and no spatial constraint was applied when identifying the clusters. Spectral clustering is able to capture clusters that have complicated shapes (Von Luxburg, 2007), making them suitable for parcellating the structure of complicated brain regions such as the PHR. In addition, the spectral clustering algorithm, which has been used successfully in our previous parcellations of various brain regions (Liu et al., 2013; Fan et al., 2014; Zhang et al., 2014), is less sensitive to spatial distance effects than k-means clustering.
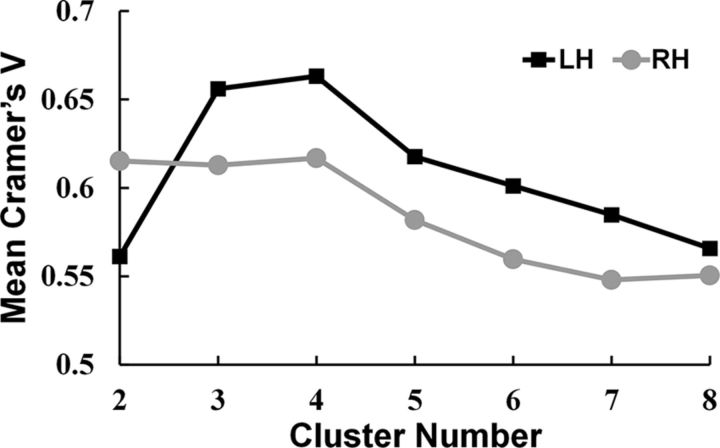
To avoid arbitrarily choosing the number of clusters, we used cross-validation to determine the number of clusters that yielded optimal consistency across the subjects. Specifically, as was done in previous studies (Liu et al., 2013; Fan et al., 2014), a leave-one-out cross-validation method in which each subject's data were excluded from the averaging was performed. For each subject, we checked the consistency between the clustering results of the single and the average across the remaining subjects using Cramer's V (Cramér, 1999). Cramer's V has values in the interval [0, 1], in which higher values indicate better consistency (Cramér, 1999). The intersubject consistency was checked for k = 2, 3, 4, 5, 6, 7, and 8 clusters. Next, the individual segmentation results were transformed into MNI standard space to create population probability maps. Maximum probability maps (MPMs) for each PHR subregion were created in MNI space to retain the quantitative information about intersubject variability.
The main goal of the PHR parcellation was to provide reliable subregions as a tool for neuroimaging studies. To accomplish this, the reproducibility of the connectivity-defined subregions was explored using an independent dataset. The parcellation processing procedures for Dataset 2 were identical to the methods that had been applied to Dataset 1, as described above.
Mapping connectivity patterns
Anatomical connectivity pattern of each PHR subregion.
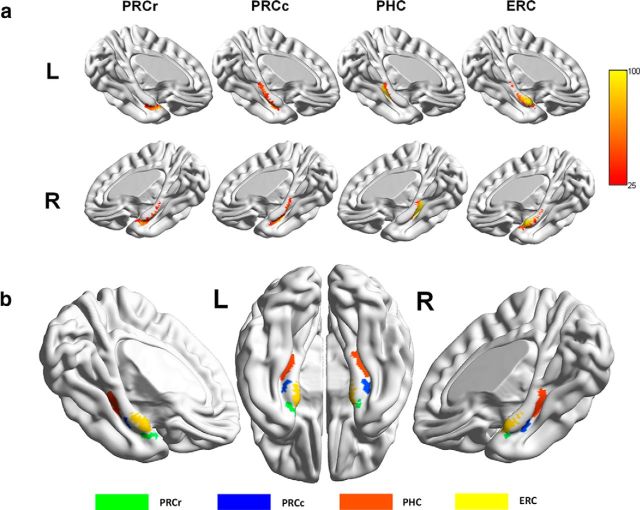
First, each subregion of the PHR was used as a seed area. To avoid an overlap between the different seed regions and possible across-subject averaging, we defined the seed masks of each PHR subregion only if the probability of their voxels in the potential PHR subregion was higher than any other PHR subregion with a probability >50%. According to this procedure, if a voxel was assigned to one of the PHR subregion seed masks, the probability of this voxel's being in this subregion must be >62.5% and the probability of this voxel's belong to other subregion must be <25%. Following this procedure could partially reduce the across-subject averaging issue that could potentially result from performing connectivity analyses on neighboring ROIs (each seed mask of PHR subregion; see Fig. 4b). To prepare for the subsequent connectivity analysis, each seed mask was registered to the T1 space for each individual.
Figure 4.
Probabilities and the seed mask for each PHR subregion. a, Probabilities for each PHR subregion to be classified into one of the four clusters. The color scheme represents the probability that the brains of all the subjects overlap at each voxel. b, Surface view of the location of the seed mask for each PHR subregion.
Probabilistic tractography for each seed region in each hemisphere in individual DTI space was performed by estimating the fiber orientations of each voxel. To reduce calculation error, 100,000 samples for each subject were drawn from the connectivity distribution (starting from the seed voxels in the PHR subregions). To reduce noise and facilitate the qualitative analysis, the connectivity probability value for each subject was thresholded using a connectivity probability at least 3.08 × 10−5 based on a previously published study (Rilling et al., 2008); that is, at least 3.08 of the 100,000 samples generated from each seed voxel were connected. The fiber tracts were then binarized and warped into standard MNI space based on the corresponding estimated transformations. We subsequently averaged the warped fiber tracts across the subjects to obtain population maps, which were then thresholded to display only those voxels that existed in at least 50% of the subjects.
Target areas.
We combined the Dusseldor–Julich histological atlas (JHA; http://www.fz-juelich.de/JuBrain/EN/_node.html) (Zilles and Amunts, 2010), Brainnetome atlas (http://atlas.brainnetome.org/), and Harvard-Oxford cortical structural atlas (HOCSA; http://neuro.imm.dtu.dk/wiki/Harvard-Oxford_Atlas) to define the target areas using the following criteria. First, the target areas of the cerebral cortex were located using the Brainnetome atlas where available. Second, for those regions that have not yet been defined in the Brainnetome atlas, we used the JHA. Third, for those regions that have not yet been defined in the Brainnetome atlas or the JHA, we used the HOCSA. If these three atlases were discrepant from each other, the Brainnetome atlas was used preferentially. In total, 65 target regions were defined for each hemisphere.
Finally, to illustrate the differences in anatomical connectivity for each subregion, we mapped the population threshold maps from the probabilistic tractography for each subregion onto a map of the target regions. To test for the effect of within-subject factors on the “PHR subregions” and “target areas,” multivariate analyses of variance with subsequent univariate ANOVAs and pairwise comparisons were performed to identify the specific differences between each pair of PHR subregional connection probabilities with the ipsilateral target areas. These statistical analyses were performed using SPSS version 22.0).
Functional connectivity pattern of each PHR subregion.
We used the same definitions for the functional connectivity seed regions as those used in the anatomical connectivity. For the functional connectivity analysis, we used the method developed by Libby et al. (2012). First, the functional residual time series data were registered to each individual T1 image using FMRIB's Linear Image Registration Tool (FLIRT) (df = 6) (Jenkinson and Smith, 2001; Jenkinson et al., 2002) to prepare seeds for the time course extraction. Next, a functional connectivity map was computed for each subject space using the correlation coefficient between the mean time series for each seed (in their individual T1 image, unsmoothed) and that of each voxel in the whole brain (in their individual fMRI, 6 mm FWHM Gaussian smoothed) for each person. The correlation coefficients for each functional connectivity map were then normalized using Fisher's z-transform. Finally, to prepare for the subsequent group-level analyses, functional connectivity results maps were registered in a two-step process, first to the individual T1 images (df = 6) and then to the MNI152 brain template (df = 12).
Group level analyses were conducted in two steps. First, to identify brain regions that showed significant correlations with each subregion, each individual's z-values were entered into random-effects one-sample t tests in a voxelwise manner. Second, paired t tests were used to identify the precise regions that differed in their resting-state functional connectivity (RSFC) strengths between every pair of PHR subregions ipsilaterally. All of the resulting t-statistic images were thresholded at p (uncorrected) <0.005, and the size of the cluster, k > 67 voxels (536 mm3), was used to achieve a corrected statistical significance of p < 0.01, as determined by AlphaSim correction.
Because the PHR is an important region of the MTL memory system, we wanted to investigate the connectivity pattern of the PHR subregions (omitting the ERC) with the hippocampus and the ERC, two regions within the MTL memory system. To examine the distribution of the functional connectivity of these PHR subregions along the anterior–posterior axis of the hippocampus and the ERC, we plotted the β-weighted connectivity strengths obtained from a normalized individual-level connectivity analysis for each subregion. The ROI for the hippocampus was extracted from the Harvard–Oxford subcortical atlas thresholded at 75% for each hemisphere and the one for the ERC was extracted from the above potential parcellation result.
Results
Connectivity-based parcellation
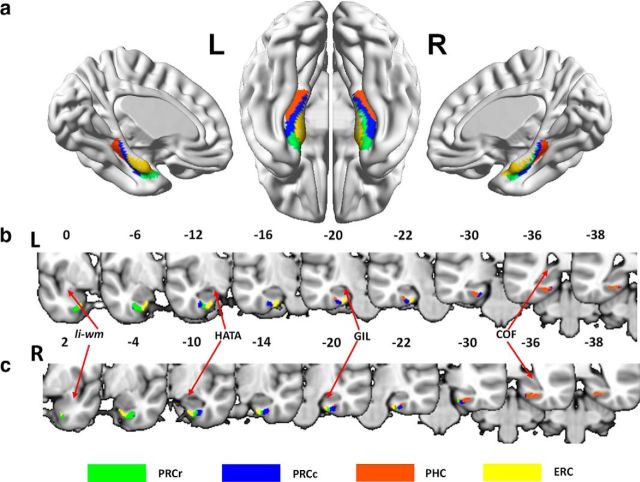
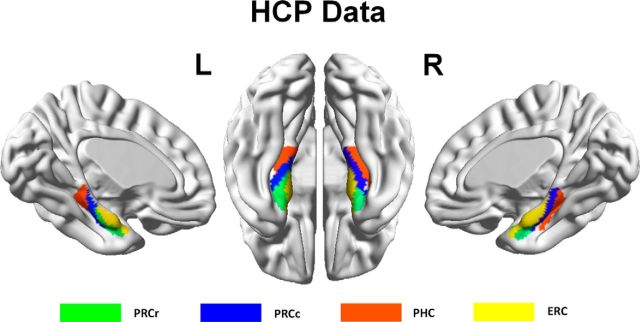
Applying a spectral clustering algorithm (Von Luxburg, 2007) to the features obtained by probabilistic tractography from the in vivo DTI data parcellated the PHR into two to eight subregions. Although no gold standard for selecting the cluster number has yet been determined, we selected the optimal cluster number using a cross-validation method based on the consistency of the clustering across the subjects. We found that a cluster number of four gave the highest consistency for the across-subjects clustering (Fig. 2). The four distinct subregions with different connectivity patterns were identified in individual space for each subject. These results were transformed and combined into standard MNI brain space to create population-based (Fig. 3) and probability-based (Fig. 4a) parcellations of the PHR. The parcellation results for the HCP datasets (Fig. 5) were the same as those obtained using Dataset 1.
Figure 2.
Average Cramer's V as an indication of clustering consistency. Cramer's V has values in the interval [0, 1] in which high values indicate good consistency. The figure shows that the four-cluster solution had the highest Cramer's V for both the left (black) and right (gray) hemispheres.
Figure 3.
PHR parcellation solution. a, Surface view of the PHR parcellation results. b, c, Nine coronal slices of the MNI152 standard brain displayed with the PHR parcellation results from the left and right hemispheres, respectively. Li-wm, Limen insula white matter; HATA, hippocampal-amygdaloidal transitional area; COF, crus of the fornix.
Figure 5.
Replication of the PHR parcellation in the HCP dataset in the left and right hemispheres.
From the MPM of the PHR parcellation results, we were able to identify four separable subregions of the PHR (Fig. 3b,c). Based on cross-species similarities in their topological distribution, these four subregions were named the rostral PRC (PRCr), caudal PRC (PRCc), PHC, and ERC. Compared with traditional cytoarchitectonic maps, the PRCr and PRCc (green and blue) constitute the traditional region PRC (the anterior boundary was defined as 2 mm rostral to the collateral sulcus or its appearance and the posterior boundary was defined as 6 mm caudal to the end of the gyrus intralimbicus (GIL) (Insausti et al., 1998; Ding and Van Hoesen, 2010; Frankó et al., 2014). The anatomical landmark used to identify the PRCr and PRCc was the end slice of the hippocampal–amygdaloidal transitional area. In addition, 2 mm posterior to the GIL, the PRCc shifted from the collateral sulcus to the parahippocampal gyrus. The boundary of the PHC (orange) in our results was the slice of the medial bank of the collateral sulcus from the GIL to 2 mm posterior to the crus of the fornix, a definition that is consistent with the traditional definition of the PHC region (Frankó et al., 2014). The location of the ERC was the slice of the parahippocampal gyrus from the limen insula white matter to 2 mm posterior to the GIL, a definition that is also quite consistent with the previous cytoarchitectonically defined ERC (Insausti et al., 1998; Frankó et al., 2014).
Unlike previous cytoarchitectonic studies (Insausti et al., 1998; Ding and Van Hoesen, 2010), PHR areas 35 and 36 were not identified in the current study. In addition, recent studies using high-resolution fMRI data indicated that the ERC contains two subregions, the anterior–lateral and the posterior–medial (Maass et al., 2015; Navarro Schroder et al., 2015). These differences between our study and the above ones may have been caused by our use of Cramer's V, which could have underestimated the variability. Perhaps the most important explanation for these differences may, however, may have been that the resolution of the current DWI data was not precise enough to segregate it into such small brain regions. The current study results were stable between the individuals within each population and can be used to explain recent fMRI findings about PHR function. We believe that, as the data spatial resolution increases, we will be able to obtain a more fine-grained anatomical structure of the PHR.
Anatomical connectivity patterns of PHR subregions
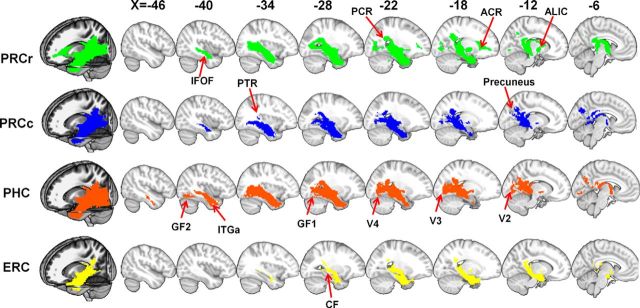
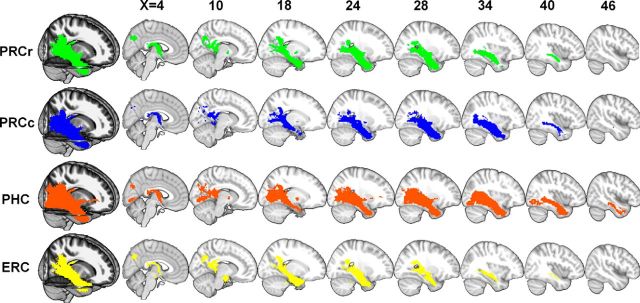
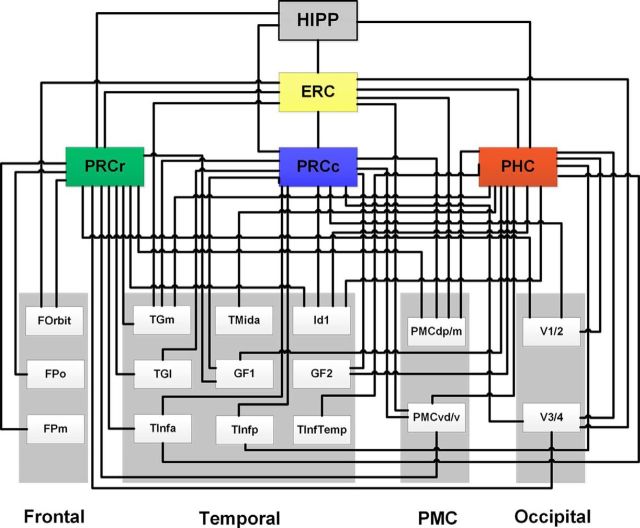
The anatomical connectivity patterns obtained from the average population maps of the probabilistic tractography results were mapped onto the MNI152 template with MRIcron. Each pattern indicates that the population probability of a voxel belonging to the pathway for each subregion was >50% (Figs. 6, 7). The PRCr connected with the frontal pole, orbital frontal cortex, hippocampus, posterior cingulate cortex, anterior temporal regions, and visual regions via the inferior frontooccipital fasciculus (IFOF), posterior thalamic radiation (PTR), posterior corona radiate (PCR), anterior corona radiata, and the anterior limb of the internal capsule. The PRCc connected with the temporal pole, temporal fusiform gyrus, hippocampus, posterior cingulate cortex, precuneus, and visual regions via the IFOF, PTR, PCR, and the splenium of the corpus callosum (SOCC). The PHC connected with the temporal pole, inferior temporal lobe, fusiform region, hippocampus, posterior cingulate cortex, precuneus, and vision regions via the IFOF, PTR, PCR, and SOCC. The ERC connected with the temporal pole, PHR, hippocampus, and posterior cingulate regions by the IFOF and the crus of the fornix. In summary, the connectivity patterns of the PRCr, PRCc and PHC showed an anterior–posterior trend from the orbital frontal region to the visual region and the ERC was primarily connected with the PHR and the hippocampus (Figs. 6, 7). The wiring diagram shows the population probabilistic tractography maps for each PHR subregion (Fig. 8) and indicates the connectivity patterns for each subregion (for details, see Table 1).
Figure 6.
Population maps of the probabilistic tractography results for each subregion from the left hemisphere. Population maps of the probabilistic tractography results from the left PRCr (green), PRCc (blue), PHC (orange), and ERC (yellow) are shown on the ICBM152 template in MNI space with MRIcron. The numbers in the top row number are the sagittal slice numbers from the MNI152 template.
Figure 7.
Population maps of the probabilistic tractography results for each subregion from the right hemisphere.
Figure 8.
Summary of the probabilistic tractography results for each PHR subregion. The wiring diagram illustrates the probabilistic tractography organization for all subregions. These results are integrated in Figures 6 and 7. The target brain regions were obtained from the Dusseldor–Julich, Brainnetome, and Harvard–Oxford atlases (for details, see Table 1).
Table 1.
PHR subregions connected target areas named as provided in the atlases
| Target area | Atlas | Labeling | |
|---|---|---|---|
| Frontal lobe | Frontal orbital cortex | HOCSA | FOrbit |
| Orbital part of frontal pole | Brainnetome | Fpo | |
| Medial part of frontal pole | Brainnetome | FPm | |
| Medial part of temporal pole | Brainnetome | TGm | |
| Lateral part of temporal pole | Brainnetome | TGl | |
| Dysgranular insula Id1 | JHA | Id1 | |
| Temporal lobe | Middle temporal gyrus, anterior part | HOCSA | TMida TInfa |
| Inferior temporal gyrus, anterior part | HOCSA | TInfp | |
| Inferior temporal gyrus, posterior part | HOCSA | TInfTemp | |
| Inferior temporal gyrus, temporooccipital part | HOCSA | GF | |
| Fusiform gyrus | JHA | HIPP | |
| Hippocampus | HOCSA | ||
| Posteromedial | Dorsal posteromedial cortex, posterior part | Brainnetome | PMCdp |
| cortex | Dorsal posteromedial cortex, medial part | Brainnetome | PMCdm |
| Ventral posteromedial cortex, dorsal part | Brainnetome | PMCvd | |
| Ventral posteromedial cortex, ventral part | Brainnetome | PMCvv | |
| Occipital lobe | Visual area hOc1 | JHA | V1 |
| Visual area hOc2 | JHA | V2 | |
| Visual area hOc3 | JHA | V3 | |
| Visual area hOc4 | JHA | V4 |
JHA, Dusseldor-Julich histological atlas; HOCSA, Harvard–Oxford structural atlas.
To quantify the differences in the anatomical connections across the four PHR subregions, the connection probabilities between the four PHR subregions and the 23 target regions were normalized for the size of each of the four subregions. The normalized connection probabilities were entered in pairwise comparisons, which revealed significant differences between these four PHR subregions in their connection patterns (Table 2). Compared with the other three subregions, the PRCr showed higher probabilities of connection with the frontal pole, temporal pole, and orbital frontal regions. The PHC showed higher probabilities of connection with the inferior temporal, fusiform, and the V1 and V2 regions than the other three subregions. Compared with the other three subregions, the PRCc showed higher probabilities of connection with the hippocampus. In the V3 and V4 regions, the PHC showed higher connection probabilities than the PRCr or the ERC, but not a significant difference from the V3 and V4 connections of the PRCc. The PRCc showed higher connection probabilities with the left precuneus than did the PRCr or the ERC, but was not significantly different in this connectivity from the PHC.
Table 2.
Differences in the anatomical connections between each pair of PHR subregions and the target areas
| PHR subregion/target area | Fpo | FPm | Forbit | TGl | TGm | Id1 | TMida | TInfa | TInfp | TInfTemp | GF1 | GF2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Left | ||||||||||||
| F tests (F/P-value) | 7.591/0.000* | 2.958/0.038* | 3.882/0.012* | 2.340/0.080 | 14.697/0.000* | 5.375/0.002* | 5.818/0.001* | 6.300/0.001* | 1.809/0.153 | 6.672/0.000* | 2.464/0.069 | 15.102/0.000* |
| PRCr-PRCc (P) | (+) 0.000* | 0.316 | (+) 0.015* | 0.865 | (+) 0.000* | (+) 0.006* | 0.818 | 0.971 | 0.972 | 0.923 | 0.310 | 0.268 |
| PRCr-PHC (P) | (+) 0.000* | (+) 0.027* | (+) 0.012* | 0.050 | (+) 0.000* | (+) 0.027* | (−) 0.002* | (−) 0.001* | 0.072 | (−) 0.002* | (−) 0.020* | (−) 0.000* |
| PRCr-ERC (P) | (+) 0.000* | (+) 0.009* | 0.811 | 0.756 | (+) 0.000* | (+) 0.000* | 0.634 | 0.684 | 0.823 | 0.391 | 0.953 | 0.503 |
| PRCc-PHC (P) | 0.914 | 0.216 | 0.940 | (+) 0.034* | 0.891 | 0.587 | (−) 0.003* | (−) 0.001* | 0.077 | (−) 0.002* | 0.181 | (−) 0.000* |
| PRCc-ERC (P) | 0.710 | 0.101 | (−) 0.027* | 0.888 | 0.210 | 0.279 | 0.480 | 0.712 | 0.796 | 0.391 | 0.339 | 0.077 |
| PHC-ERC (P) | 0.793 | 0.680 | (−) 0.023* | (+) 0.024* | 0.165 | 0.106 | (+) 0.000* | (+) 0.000* | (+) 0.044* | (+) 0.000* | (+) 0.023* | (+) 0.000* |
| Right | ||||||||||||
| F tests (F/P-value) | 2.834/0.044* | 2.010/0.120 | 4.326/0.007* | 5.803/0.001* | 2.922/0.039* | 4.262/0.008* | 4.498/0.006* | 6.646/0.000* | 9.603/0.000* | 4.763/0.004* | 15.371/0.000* | 6.794/0.001* |
| PRCr-PRCc (P) | (+) 0.018* | 0.091 | 0.088 | 0.713 | 0.603 | 0.425 | 0.942 | 0.344 | 0.053 | 0.927 | 0.358 | 0.165 |
| PRCr-PHC (P) | (+) 0.034* | (+) 0.042* | 0.080 | (−) 0.001* | (+) 0.047* | (−) 0.004* | (−) 0.006* | (−) 0.000* | (−) 0.000* | (−) 0.004* | (−) 0.000* | (−) 0.000* |
| PRCr-ERC (P) | (+) 0.015* | (+) 0.037* | 0.210 | 0.958 | (+) 0.013* | 0.767 | 0.644 | 0.923 | 0.625 | 0.698 | 0.656 | 0.784 |
| PRCc-PHC (P) | 0.790 | 0.717 | 0.962 | (−) 0.003* | 0.139 | (−) 0.036* | (−) 0.007* | (−) 0.006* | (−) 0.021* | (−) 0.005* | (−) 0.000* | (−) 0.021* |
| PRCc-ERC (P) | 0.945 | 0.677 | (−) 0.004* | 0.675 | (+) 0.045* | 0.275 | 0.593 | 0.298 | (+) 0.016* | 0.632 | 0.174 | 0.098 |
| PHC-ERC (P) | 0.737 | 0.957 | (−) 0.003* | (+) 0.001* | 0.592 | (+) 0.002* | (+) 0.001* | (+) 0.000* | (+) 0.000* | (+) 0.001* | (+) 0.000* | (+) 0.000* |
| PHR subregion/target areas | PMCdm | PMCdp | PMCvd | PMCvv | V1 | V2 | V3d | V3v | V4d | V4v | Hipp |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Left | |||||||||||
| F tests (F/P-value) | 1.931/0.132 | 4.576/0.005* | 1.146/0.336 | 2.371/0.077 | 4.232/0.008* | 6.284/0.001* | 1.955/0.128 | 5.771/0.001* | 3.062/0.033* | 5.789/0.001* | 21.764/0.000* |
| PRCr-PRCc (P) | 0.058 | (−) 0.005* | 0.142 | (−) 0.029* | 0.058 | (−) 0.011* | 0.415 | 0.366 | 0.549 | 0.527 | (−) 0.000* |
| PRCr-PHC (P) | 0.840 | (−) 0.045* | 0.835 | 0.074 | (−) 0.013* | (−) 0.001* | 0.344 | (−) 0.004* | 0.201 | (−) 0.045* | (−) 0.003* |
| PRCr-ERC (P) | 0.819 | 0.855 | 0.848 | 0.736 | 0.653 | 0.882 | 0.234 | 0.290 | (+) 0.005* | (+) 0.043* | 0.306 |
| PRCc-PHC (P) | 0.089 | 0.401 | 0.207 | 0.684 | 0.535 | 0.484 | 0.895 | (−) 0.046* | 0.493 | (−) 0.009* | (+) 0.000* |
| PRCc-ERC (P) | (+) 0.034* | (+) 0.003* | 0.098 | 0.063 | (+) 0.020* | (+) 0.007* | (+) 0.047* | 0.052 | (+) 0.026* | 0.158 | (+) 0.000* |
| PHC-ERC (P) | 0.667 | (+) 0.030* | 0.689 | 0.144 | (+) 0.004* | (+) 0.001* | (+) 0.035* | (+) 0.000* | 0.119 | (+) 0.000* | (+) 0.048* |
| Right | |||||||||||
| F tests (F/P-value) | 2.063/0.112 | 0.729/0.538 | 0.996/0.399 | 1.570/0.204 | 10.247/0.000* | 11.886/0.000* | 0.692/0.560 | 12.844/0.000* | 2.534/0.063 | 19.151/0.000* | 17.998/0.000* |
| PRCr-PRCc (P) | 0.153 | 0.247 | 0.327 | 0.337 | 0.584 | 0.189 | 0.820 | 0.367 | 0.872 | 0.535 | (+) 0.018* |
| PRCr-PHC (P) | 0.057 | 0.175 | 0.136 | 0.307 | (−) 0.000* | (−) 0.000* | 0.302 | (−) 0.000* | (−) 0.033* | (−) 0.000* | (+) 0.000* |
| PRCr-ERC (P) | 0.943 | 0.382 | 0.863 | 0.406 | 0.597 | 0.651 | 0.499 | 0.625 | 0.936 | 0.614 | (+) 0.000* |
| PRCc-PHC (P) | 0.624 | 0.840 | 0.606 | 0.951 | (−) 0.000* | (−) 0.001* | 0.209 | (−) 0.000* | (−) 0.023* | (−) 0.000* | (+) 0.011* |
| PRCc-ERC (P) | 0.134 | 0.774 | 0.418 | 0.075 | 0.284 | 0.079 | 0.367 | 0.166 | 0.936 | 0.263 | (+) 0.000* |
| PHC-ERC (P) | (−) 0.048* | 0.625 | 0.187 | 0.066 | (+) 0.000* | (+) 0.000* | 0.720 | (+) 0.000* | (+) 0.028* | (+) 0.000* | 0.068 |
Using univariate ANOVAs and pairwise comparisons, differences in the PHR subregions with each ipsilateral target brain area, as well as the anatomical connections with the different targets among the PHR subregions, are shown. + and − indicate the direction of the pairwise comparison.
*Mean difference was significant at the 0.05 level.
Functional connectivity pattern of the PHR subregions
Whole-brain RSFC of the PHR subregions
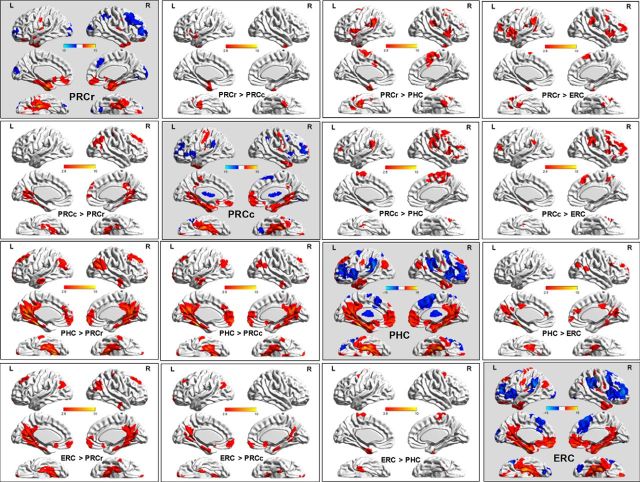
The left whole-brain RSFC pattern for each PHR subregion is displayed in Figure 9. Given the similarity in the functional connectivity patterns of the left (Fig. 9) and the right (Fig. 10) PHRs, we focused on the left PHR subregions. The RSFC results for the cerebellum are not shown in the figures.
Figure 9.
Functional connectivity pattern for each left PHR subregion and each left subregional RSFC variations. Shown are spatial distribution and paired difference maps of the RSFC patterns for the left PHR subregions. Statistical parametric maps are displayed using a statistic thresholded at p (uncorrected) <0.005 and cluster extent k > 67 voxels (536 mm3) was used to achieve a corrected statistical significance of p < 0.01, as determined by AlphaSim correction. The RSFC and the relevant connection differences maps are projected onto a 3D brain surface with intensity scales representing the T-values.
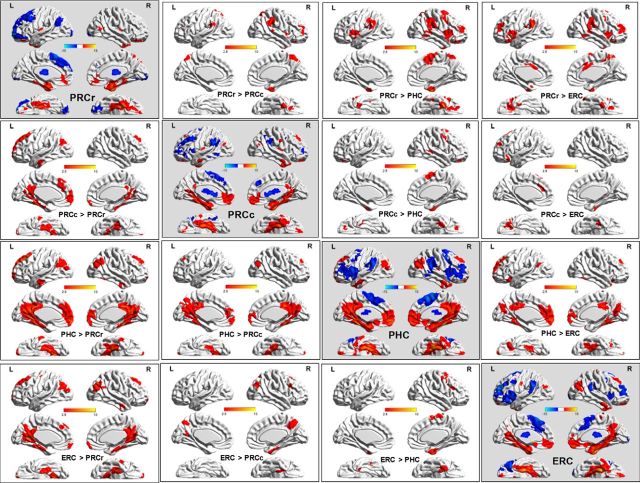
Figure 10.
Functional connectivity pattern for each right PHR subregion and each right subregional RSFC variations.
The first analyses focused on separately characterizing the RSFC patterns for each PHR subregion (see the subfigures on the diagonal in Figs. 9, 10). In brief, the cortex of all the PHR subregions was positively functionally correlated with areas that included the MTL, medial frontal pole, and the lateral superior frontal and angular gyri and all of the subregions were negatively functionally correlated with areas that included BA 44/45/46 and the medial superior frontal and supramarginal gyri. Specially, unlike the three other subregions, the PRCr was positively functionally connected with the lateral orbital frontal cortex and did not connect functionally with the posterior cingulate or the precuneus. The PHC differed from the others in that it positively functionally connected with the dorsal portion of the precuneus and did not connect functionally with the medial orbital frontal cortex. The PRCc not only positively functionally connected with the medial orbital frontal cortex (the PRCr also connected, but the PHC did not), but also functionally connected with the posterior cingulate cortex and ventral precuneus (the PHC also connected, but the PRCr did not).
PHR subregional RSFC variations
In addition, paired t tests were used to reveal significant differences in functional connectivity between each pair of PHR subregions (see the up and down triangles in Figs. 9, 10). In brief, the greatest differences in RSFC strength between the four subregions were in the lateral orbital frontal cortex and the precuneus, which are important component regions in the AT-PM systems. Compared with the other three subregions, the PRCr showed greater RSFC strength with the ipsilateral lateral orbital frontal cortex, which is an important component of the AT system. In contrast, compared with other three subregions, the PHC had greater RSFC strength with the precuneus, which is an important component of the PM system. With respect to the precuneus, the PRCc showed greater RSFC strength than the PRCr, but lower RSFC strength than the PHC.
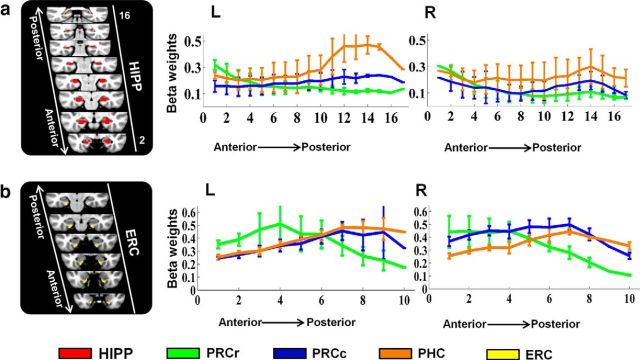
Functional connectivity distribution of the PRCr, PRCc, and PHC in the hippocampus and ERC
The PHR and hippocampus combine to form the MTL memory system. Anatomical studies in rodents (van Strien et al., 2009) and nonhuman primates (Squire and Zola-Morgan, 1991) and functional connectivity studies in humans (Kahn et al., 2008; Libby et al., 2012) have all indicated that each subregion within the MTL system has different pathways. These previous findings motived us to determine whether the patterns in which each PHR subregion connected to the main regions of the MTL-important regions were different. Because the hippocampus is a very important region in the MTL memory system and the ERC is an important gateway between the hippocampus and the other PHR subregions, we focused on discovering the functional connectivity distributions of the PRCr, PRCc, and PHC in the hippocampus and the ERC. The functional connectivity strengths between the PRCr, PRCc, and PHC and the hippocampus are illustrated in Figure 11a. The PRCr showed stronger connectivity with the anterior hippocampus, the PHC showed stronger connectivity with the posterior hippocampus, and the PRCc showed a connectivity strength that was intermediate between the PRCr and PHC in all the hippocampal slices. For the ERC, as illustrated in Figure 11b, the PRCr showed more functional connectivity with the anterior portion of the ERC, whereas the PRCc and PHC showed stronger functional connectivity with the posterior portion of the ERC. This anterior and posterior difference in the functional connectivity pattern of the ERC is consistent with the findings of recent functional subregions and topography in human ERC studies (Maass et al., 2015; Navarro Schroder et al., 2015). Interestingly, the functional connectivity between the PHC and the ERC was less strong than that between the PRCc and the ERC.
Figure 11.
Functional connectivity gradients along the longitudinal hippocampus and the ERC showing the differences between the PRCr, PRCc, and PHC. a, Parameter estimates indexed along the anterior–posterior axis of the hippocampus as red areas in both hemispheres. Middle and right, Mean parameter estimates indexing connectivity extracted from each slice along the anterior–posterior axis of the HIPP for the PRCr, PRCc, and PHC. Error bars indicate SEM. b, Parameter estimates indexing along the anterior–posterior axis of the ERC. Left, Coronal slices through the ERC with anatomical ROIs in the MNI152 template represented as yellow areas in both hemispheres. Middle and right, Mean parameter estimates indexing connectivity extracted from each slice along the anterior–posterior axis of the ERC for the PRCr, PRCc, and PHC.
In summary, we observed that the PRCc was a distinct area in terms of its connectivity profiles. First, the PRCc did not connect functionally to the lateral orbitofrontal area, that is, to the specific area to which the PRCr is functionally connected, or to the dorsal portion of the precuneus cortex; that is, to the specific area to which the PHC is functionally connected (Figs. 9, 10). Second, the PRCc showed a median level of functional connectivity with most slices of the hippocampus, whereas the PRCr and the PHC showed strong functional connectivity with the anterior and the posterior hippocampus, respectively (Fig. 11a). Because a series of studies has indicated that the hippocampus shows functional specialization along its long axis (for a recent review, see Poppenk et al., 2013), the PRCc can logically be regarded as a subregion that is distinct from the PRCr or the PHC. We found that, indeed, the PRCc is a distinct subregion with a connectivity pattern that is distinct from those of either the PRCr or the PHC.
Discussion
The current study, unlike traditional PHR parcellations, consistently identified four major subregions in the PHR in two independent spatial resolution datasets (Figs. 3, 5). Specifically, a widely studied area of the PHR, which is traditionally separated into the PRC and PHC (Eichenbaum et al., 2007; for reviews, see Diana et al., 2007), could be subdivided into the PRCr, PRCc, and PHC. The PRCc was found to be a transition region in the PHR in terms of its connectivity profiles. For the first time, we were able to obtain a well defined parcellation of the human PHR using diffusion MRI. This parcellation scheme should improve our understanding of the anatomical, connectivity, and functional characteristics of the PHR.
Anatomical and connectional organization of the PHR subregions
Our PRCr corresponds to the rostral portion of the PRC as it was anatomically defined using cytoarchitectonic (Insausti et al., 1998; Thangavel et al., 2008), myeloarchitectonic (Sewards, 2011), and modern neuroanatomical techniques (Ding and Van Hoesen, 2010) (Ding and Van Hoesen, 2010) and our PRCc corresponds to the caudal portion of the previously defined PRC. No existing anatomical description corresponds to a rostral–caudal division of the PRC in humans as far as we know, but monkeys have a rostral (36r) and caudal (36c) subdivision of area 36 in the main body of the PRC. In addition, the anterior–posterior topographic relationship that we identified as the PRCr, PRCc, and PHC in the human lateral PHR is consistent with the relationship between areas 36r, 36c, and TF in monkeys (Suzuki and Amaral, 1994a, 2003; Saleem et al., 2007).
The connectivity profiles of the PRCr and PHC are broadly consistent with prior evidence from tracer studies in monkeys indicating the presence of parallel pathways that were distinguished by a greater number of projections to the 36r compared with those of the TF (Suzuki and Amaral, 1994a; Lavenex et al., 2002). In addition, the connectivity profiles for the PRCr and PHC also, respectively, match the traditional PRC and PHC connectivity profiles obtained by human resting-state functional connectivity ROI analyses (Libby et al., 2012).
The location of the ERC in our result resembles that of Brodmann area 28 (Brodmann, 1909), as well as what has often previously been called the EC or ERC (Insausti et al., 1998; Pruessner et al., 2002; Amunts et al., 2005). In the current study, the ERC had a major connection with the hippocampus and with other regions of the PHR, RSC, and posterior cingulate cortex. This finding is consistent with the anatomical pathway of the ERC from tracer studies in nonhuman primates (Suzuki and Amaral, 1994a, 1994b) and rodents (Burwell and Amaral, 1998; van Strien et al., 2009). The review paper by Lavenex and Amaral (2000) and recent human studies (Maass et al., 2015; Navarro Schroder et al., 2015) suggested that the connections of the ERC mean that it is an important funnel for information flow in the neocortex, PHR, and hippocampus. Therefore, the position and the connections of the ERC in our result make it similar to the traditional ERC.
Transition area within the PHR integrating the anterior temporal–posterior medial systems
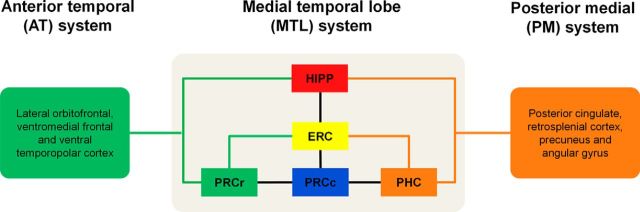
The intermediate area made up of the traditional PRC and PHC is located in the PHR and has always been considered to be merely a relay for transferring information between the neocortex and the hippocampus (Lavenex and Amaral, 2000). Previous studies suggested that the PRC and PHC are structurally and functionally distinct regions and belong to two separate networks (Kahn et al., 2008; Libby et al., 2012; Catani et al., 2013). The model proposed by Ranganath and Ritchey (2012) suggested that the PRC and the PHC are respective core components of the AT and PM systems, both of which are associated with memory-guided behavior. The current findings are similar to those suggested by the AT-PM model, but provide two additional insights. First, the current study defined the core components of the AT systems within the PHR as belonging to the PRCr rather than to the traditional, larger, and thus less concise PRC area. Second, in the PHR we identified a subarea that we termed the PRCc between the PRCr and the PHC that seems to integrate the AT-PM networks.
Compared with the parcellation protocol for the PHR based on tract-tracing studies in animals (Suzuki and Amaral, 1994a; Lavenex et al., 2002; Suzuki and Amaral, 2003), the dichotomized scheme dividing the lateral human PHR into the PRC and PHC is coarse. In the current study, we have parcellated the human PRC into the PRCr and PRCc based on its anatomical connectivity profiles. Therefore, the portion based on the coarse anatomical description of the PRC and PHC organization within the AT-PM model may need to be improved. The AT system takes the PRC as the core component and the connectivity pathway extends to the ventral temporopolar area, lateral orbitofrontal cortex, and anterior hippocampus (Ranganath and Ritchey, 2012). Consistent with this pathway, our results showed that the PRCr was connected with a network that overlapped the AT system. Similarly, the PM system takes the PHC-RSC as the core component and the connectivity pathway extends to include the posterior cingulate, precuneus, lateral parietal cortex, and posterior hippocampus (Ranganath and Ritchey, 2012). Our findings showed that the PHC was connected with a pathway that primarily overlapped with the PM system. Because of the similarity between the PRCr and the PHC and their connected networks and the AT-PM system, we suggest that the PRCr and the PHC may be the core nodes of the AT and PM systems within the PHR, respectively (Fig. 12).
Figure 12.
Schematic model summarizing our findings and hypothesis. In the normal brain, the PRCr and PHC are core components of the AT and PM systems, respectively. Within the MTL system, the PRCc integrates partial information from the AT and PM systems via the ERC to the hippocampus.
In the current study, we observed that the PRCc connected with many of the components of both the AT and PM networks, indicating that it is a transition area between the PRCr and the PHC. This organization has similarities with area 36c, which is in a similar location to the PRCc in humans, in that area 36c serves as an interface for communication between the 36r and the TF in monkeys, as indicated by tracer studies (Lavenex et al., 2002, 2004). Although the current research did not include any task research, some clues can be gained from recent task fMRI studies (Litman et al., 2009; Staresina et al., 2011; Liang et al., 2013), which could be taken to indicate that the middle subarea, the PRCc, may integrate the specific functions of the PRCr and PHC. For example, Staresina et al. (2011) found that the middle parahippocampal gyrus region was activated by both object and scene trials, whereas the anterior and posterior parahippocampal gyrus regions were activated by only object or scene trials, respectively. Other studies found two additional patterns of activation along an anterior–posterior axis in that they identified a consistent phenomenon in which the middle parahippocampal gyrus region responded to a mixture of both visual object and visuospatial information, whereas the anterior and posterior portions responded to either visual object or visuospatial information, respectively (Litman et al., 2009; Liang et al., 2013). Of course, these phenomena may have resulted from group averaging or from smoothing in the fMRI studies. Further studies will be needed to investigate the effects of averaging and smoothing. The parcellation, connectivity pattern, and functional studies described above all suggest that the PRCc is a transition region between the PRCr and PHC, integrating the information from the PRCr and PHC. Finally, we suggest that the PRCc may combine the different types of information from the AT and PM systems (via the PRCr and the PHC), passing them through the posterior portion of the ERC (Fig. 11b) to the hippocampus (Fig. 12).
Conclusion
In the current study, we redefined the cartography of the human PHR and identified a transition subarea based on distinct anatomical and functional connectivity profiles. We found that the core components of the AT and PM systems that connect with the PHR are the PRCr and the PHC and that the transition subregion (the PRCc) is a functionally integrated subregion that integrates the AT-PM systems. This may lead to new insights into the human memory system and related neurodegenerative diseases.
Footnotes
This work was supported by the National Key Basic Research and Development Program (973) (Grants 2011CB707801 and 2012CB720702), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB02030300), the Natural Science Foundation of China (Grants 91132301 and 81270020), and Open Project Funding of National Key Laboratory of Cognitive Neuroscience and Learning-Beijing Normal University (Grant CNLYB1410). We thank Prof. Henry Kennedy for constructive comments on the manuscript, the anonymous reviewers who offered constructive comments and suggestions on the manuscript, and Rhoda E. and Edmund F. Perozzi for editing assistance and discussions.
The authors declare no competing financial interests.
References
- Amunts K, Kedo O, Kindler M, Pieperhoff P, Mohlberg H, Shah NJ, Habel U, Schneider F, Zilles K. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat Embryol (Berl) 2005;210:343–352. doi: 10.1007/s00429-005-0025-5. [DOI] [PubMed] [Google Scholar]
- Barense MD, Henson RN, Lee AC, Graham KS. Medial temporal lobe activity during complex discrimination of faces, objects, and scenes: Effects of viewpoint. Hippocampus. 2010;20:389–401. doi: 10.1002/hipo.20641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann M, Johansen-Berg H, Rushworth MF. Connectivity-based parcellation of human cingulate cortex and its relation to functional specialization. J Neurosci. 2009;29:1175–1190. doi: 10.1523/JNEUROSCI.3328-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? Neuroimage. 2007;34:144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhali F, Thiebaut de Schotten M, Pinel P, Poupon C, Mangin JF, Dehaene S, Cohen L. Anatomical connections of the visual word form area. J Neurosci. 2014;34:15402–15414. doi: 10.1523/JNEUROSCI.4918-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodmann K. Vergleichende Lokalisationslehre der Gro hirnrinde. Berlin: Springer; 1909. [Google Scholar]
- Burwell RD. The parahippocampal region: corticocortical connectivity. Ann N Y Acad Sci. 2000;911:25–42. doi: 10.1111/j.1749-6632.2000.tb06717.x. [DOI] [PubMed] [Google Scholar]
- Burwell RD, Amaral DG. Cortical afferents of the perirhinal, postrhinal, and entorhinal cortices of the rat. J Comp Neurol. 1998;398:179–205. doi: 10.1002/(SICI)1096-9861(19980824)398:2<179::AID-CNE3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Catani M, Dell'acqua F, Thiebaut de Schotten M. A revised limbic system model for memory, emotion and behaviour. Neurosci Biobehav Rev. 2013;37:1724–1737. doi: 10.1016/j.neubiorev.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Cloutman LL, Lambon Ralph MA. Connectivity-based structural and functional parcellation of the human cortex using diffusion imaging and tractography. Front Neuroanat. 2012;6:34. doi: 10.3389/fnana.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramér H. Mathematical methods of statistics. Princeton, NJ: Princeton University; 1999. [Google Scholar]
- Davachi L. Item, context and relational episodic encoding in humans. Curr Opin Neurobiol. 2006;16:693–700. doi: 10.1016/j.conb.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Diana RA, Yonelinas AP, Ranganath C. Imaging recollection and familiarity in the medial temporal lobe: a three-component model. Trends Cogn Sci. 2007;11:379–386. doi: 10.1016/j.tics.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Diana RA, Yonelinas AP, Ranganath C. Medial temporal lobe activity during source retrieval reflects information type, not memory strength. J Cogn Neurosci. 2010;22:1808–1818. doi: 10.1162/jocn.2009.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SL, Van Hoesen GW. Borders, extent, and topography of human perirhinal cortex as revealed using multiple modern neuroanatomical and pathological markers. Hum Brain Mapp. 2010;31:1359–1379. doi: 10.1002/hbm.20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H, Yonelinas AP, Ranganath C. The medial temporal lobe and recognition memory. Annu Rev Neurosci. 2007;30:123–152. doi: 10.1146/annurev.neuro.30.051606.094328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Wang J, Zhang Y, Han W, Yu C, Jiang T. Connectivity-based parcellation of the human temporal pole using diffusion tensor imaging. Cereb Cortex. 2014;24:3365–3378. doi: 10.1093/cercor/bht196. [DOI] [PubMed] [Google Scholar]
- Frankó E, Insausti AM, Artacho-Pérula E, Insausti R, Chavoix C. Identification of the human medial temporal lobe regions on magnetic resonance images. Hum Brain Mapp. 2014;35:248–256. doi: 10.1002/hbm.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannula DE, Libby LA, Yonelinas AP, Ranganath C. Medial temporal lobe contributions to cued retrieval of items and contexts. Neuropsychologia. 2013;51:2322–2332. doi: 10.1016/j.neuropsychologia.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes SM, Baena E, Truong TK, Cabeza R. Neural mechanisms of context effects on face recognition: automatic binding and context shift decrements. J Cogn Neurosci. 2010;22:2541–2554. doi: 10.1162/jocn.2009.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntgeburth SC, Petrides M. Morphological patterns of the collateral sulcus in the human brain. Eur J Neurosci. 2012;35:1295–1311. doi: 10.1111/j.1460-9568.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- Insausti R, Juottonen K, Soininen H, Insausti AM, Partanen K, Vainio P, Laakso MP, Pitkänen A. MR volumetric analysis of the human entorhinal, perirhinal, and temporopolar cortices. AJNR Am J Neuroradiol. 1998;19:659–671. [PMC free article] [PubMed] [Google Scholar]
- Jeneson A, Squire LR. Working memory, long-term memory, and medial temporal lobe function. Learn Mem. 2012;19:15–25. doi: 10.1101/lm.024018.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–156. doi: 10.1016/S1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1006/nimg.2002.1132. [DOI] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TE, Robson MD, Drobnjak I, Rushworth MF, Brady JM, Smith SM, Higham DJ, Matthews PM. Changes in connectivity profiles define functionally distinct regions in human medial frontal cortex. Proc Natl Acad Sci U S A. 2004;101:13335–13340. doi: 10.1073/pnas.0403743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn I, Andrews-Hanna JR, Vincent JL, Snyder AZ, Buckner RL. Distinct cortical anatomy linked to subregions of the medial temporal lobe revealed by intrinsic functional connectivity. J Neurophysiol. 2008;100:129–139. doi: 10.1152/jn.00077.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisaari SL, Probst A, Taylor KI. fMRI: basics and clinical applications. Berlin: Springer; 2013. The perirhinal, entorhinal, and parahippocampal cortices and hippocampus: an overview of functional anatomy and protocol for their segmentation in MR images; pp. 239–267. [Google Scholar]
- Köhler S, Danckert S, Gati JS, Menon RS. Novelty responses to relational and non-relational information in the hippocampus and the parahippocampal region: a comparison based on event-related fMRI. Hippocampus. 2005;15:763–774. doi: 10.1002/hipo.20098. [DOI] [PubMed] [Google Scholar]
- Lavenex P, Amaral DG. Hippocampal-neocortical interaction: a hierarchy of associativity. Hippocampus. 2000;10:420–430. doi: 10.1002/1098-1063(2000)10:4<420::AID-HIPO8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Lavenex P, Suzuki WA, Amaral DG. Perirhinal and parahippocampal cortices of the macaque monkey: projections to the neocortex. J Comp Neurol. 2002;447:394–420. doi: 10.1002/cne.10243. [DOI] [PubMed] [Google Scholar]
- Lavenex P, Suzuki WA, Amaral DG. Perirhinal and parahippocampal cortices of the macaque monkey: Intrinsic projections and interconnections. J Comp Neurol. 2004;472:371–394. doi: 10.1002/cne.20079. [DOI] [PubMed] [Google Scholar]
- Liang JC, Wagner AD, Preston AR. Content representation in the human medial temporal lobe. Cereb Cortex. 2013;23:80–96. doi: 10.1093/cercor/bhr379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby LA, Ekstrom AD, Ragland JD, Ranganath C. Differential connectivity of perirhinal and parahippocampal cortices within human hippocampal subregions revealed by high-resolution functional imaging. J Neurosci. 2012;32:6550–6560. doi: 10.1523/JNEUROSCI.3711-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby LA, Hannula DE, Ranganath C. Medial temporal lobe coding of item and spatial information during relational binding in working memory. J Neurosci. 2014;34:14233–14242. doi: 10.1523/JNEUROSCI.0655-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman L, Awipi T, Davachi L. Category-specificity in the human medial temporal lobe cortex. Hippocampus. 2009;19:308–319. doi: 10.1002/hipo.20515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Qin W, Li W, Fan L, Wang J, Jiang T, Yu C. Connectivity-based parcellation of the human frontal pole with diffusion tensor imaging. J Neurosci. 2013;33:6782–6790. doi: 10.1523/JNEUROSCI.4882-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass A, Berron D, Libby LA, Ranganath C, Duzel E. Functional subregions of the human entorhinal cortex. Elife. 2015:4. doi: 10.7554/eLife.06426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald D. Program for display and segmentation of surfaces and volumes. Montreal, Quebec: McConnell Brain Imaging Center, Montreal Neurological Institute; 1996. [Google Scholar]
- Murphy K, Bodurka J, Bandettini PA. How long to scan? The relationship between fMRI temporal signal to noise ratio and necessary scan duration. Neuroimage. 2007;34:565–574. doi: 10.1016/j.neuroimage.2006.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro Schroder T, Haak KV, Zaragoza Jimenez NI, Beckmann CF, Doeller CF. Functional topography of the human entorhinal cortex. Elife. 2015:4. doi: 10.7554/eLife.06738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert FX, Mars RB, Thomas AG, Sallet J, Rushworth MF. Comparison of human ventral frontal cortex areas for cognitive control and language with areas in monkey frontal cortex. Neuron. 2014;81:700–713. doi: 10.1016/j.neuron.2013.11.012. [DOI] [PubMed] [Google Scholar]
- O'Neil EB, Cate AD, Köhler S. Perirhinal cortex contributes to accuracy in recognition memory and perceptual discriminations. J Neurosci. 2009;29:8329–8334. doi: 10.1523/JNEUROSCI.0374-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenk J, Evensmoen HR, Moscovitch M, Nadel L. Long-axis specialization of the human hippocampus. Trends Cogn Sci. 2013;17:230–240. doi: 10.1016/j.tics.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Pruessner JC, Köhler S, Crane J, Pruessner M, Lord C, Byrne A, Kabani N, Collins DL, Evans AC. Volumetry of temporopolar, perirhinal, entorhinal and parahippocampal cortex from high-resolution MR images: considering the variability of the collateral sulcus. Cereb Cortex. 2002;12:1342–1353. doi: 10.1093/cercor/12.12.1342. [DOI] [PubMed] [Google Scholar]
- Ranganath C. A unified framework for the functional organization of the medial temporal lobes and the phenomenology of episodic memory. Hippocampus. 2010;20:1263–1290. doi: 10.1002/hipo.20852. [DOI] [PubMed] [Google Scholar]
- Ranganath C, Ritchey M. Two cortical systems for memory-guided behaviour. Nat Rev Neurosci. 2012;13:713–726. doi: 10.1038/nrn3338. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TE. The evolution of the arcuate fasciculus revealed with comparative DTI. Nat Neurosci. 2008;11:426–428. doi: 10.1038/nn2072. [DOI] [PubMed] [Google Scholar]
- Saleem KS, Price JL, Hashikawa T. Cytoarchitectonic and chemoarchitectonic subdivisions of the perirhinal and parahippocampal cortices in macaque monkeys. J Comp Neurol. 2007;500:973–1006. doi: 10.1002/cne.21141. [DOI] [PubMed] [Google Scholar]
- Sewards TV. Adolf Hopf's 1954 myeloarchitectonic parcellation of the human temporal lobe: a review and assessment. Brain Res Bull. 2011;86:298–313. doi: 10.1016/j.brainresbull.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Squire LR, Zola-Morgan S. The medial temporal lobe memory system. Science. 1991;253:1380–1386. doi: 10.1126/science.1896849. [DOI] [PubMed] [Google Scholar]
- Squire LR, Stark CE, Clark RE. The medial temporal lobe. Annu Rev Neurosci. 2004;27:279–306. doi: 10.1146/annurev.neuro.27.070203.144130. [DOI] [PubMed] [Google Scholar]
- Staresina BP, Duncan KD, Davachi L. Perirhinal and parahippocampal cortices differentially contribute to later recollection of object- and scene-related event details. J Neurosci. 2011;31:8739–8747. doi: 10.1523/JNEUROSCI.4978-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki WA, Amaral DG. Perirhinal and parahippocampal cortices of the macaque monkey: cortical afferents. J Comp Neurol. 1994a;350:497–533. doi: 10.1002/cne.903500402. [DOI] [PubMed] [Google Scholar]
- Suzuki WA, Amaral DG. Topographic organization of the reciprocal connections between the monkey entorhinal cortex and the perirhinal and parahippocampal cortices. J Neurosci. 1994b;14:1856–1877. doi: 10.1523/JNEUROSCI.14-03-01856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki WA, Amaral DG. Perirhinal and parahippocampal cortices of the macaque monkey: cytoarchitectonic and chemoarchitectonic organization. J Comp Neurol. 2003;463:67–91. doi: 10.1002/cne.10744. [DOI] [PubMed] [Google Scholar]
- Thangavel R, Van Hoesen GW, Zaheer A. Posterior parahippocampal gyrus pathology in Alzheimer's disease. Neuroscience. 2008;154:667–676. doi: 10.1016/j.neuroscience.2008.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut de Schotten M, Urbanski M, Valabregue R, Bayle DJ, Volle E. Subdivision of the occipital lobes: an anatomical and functional MRI connectivity study. Cortex. 2014;56:121–137. doi: 10.1016/j.cortex.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Uğurbil K, Xu J, Auerbach EJ, Moeller S, Vu AT, Duarte-Carvajalino JM, Lenglet C, Wu X, Schmitter S, Van de Moortele PF, Strupp J, Sapiro G, De Martino F, Wang D, Harel N, Garwood M, Chen L, Feinberg DA, Smith SM, Miller KL, et al. WU-Minn HCP Consortium. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage. 2013;80:80–104. doi: 10.1016/j.neuroimage.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Uğurbil K, Consortium WU-MH. The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Strien NM, Cappaert NL, Witter MP. The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nat Rev Neurosci. 2009;10:272–282. doi: 10.1038/nrn2614. [DOI] [PubMed] [Google Scholar]
- Von Luxburg U. A tutorial on spectral clustering. Statistics and Computing. 2007;17:395–416. doi: 10.1007/s11222-007-9033-z. [DOI] [Google Scholar]
- Zhang Y, Fan L, Zhang Y, Wang J, Zhu M, Zhang Y, Yu C, Jiang T. Connectivity-based parcellation of the human posteromedial cortex. Cereb Cortex. 2014;24:719–727. doi: 10.1093/cercor/bhs353. [DOI] [PubMed] [Google Scholar]
- Zilles K, Amunts K. Centenary of Brodmann's map-conception and fate. Nat Rev Neurosci. 2010;11:139–145. doi: 10.1038/nrn2776. [DOI] [PubMed] [Google Scholar]