Abstract
Background
Metastasis is the primary cause of tumor death in renal cell carcinoma (RCC). Improved diagnostic markers of metastasis are critically needed for RCC. MicoRNAs are demonstrated to be stable and significant biomarkers for several malignancies. In this study, we aimed to explore the metastasis related microRNAs and its mechanism in RCC.
Methods
The relationship between microRNAs expression and prognosis and metastasis of RCC patients were explored by data mining through expression profiles from The Cancer Genome Atlas (TCGA). A total of 80 RCC tissues and adjacent normal kidney tissues were obtained from Department of Urology, Peking University First Hospital. Expression of microRNA-200b (miR-200b) in RCC tissues and cell lines were determined by bioinformatic data mining and quantitative real-time PCR (qRT-PCR). The effects of miR-200b on cell proliferation, migration and invasion were determined by cell counting kit-8 and colony formation assay, wound healing assay and Boyden chamber assay. Mouse cell-derived xenograft and patient-derived xenograft model were also performed to evaluate the effects of miR-200b on tumor growth and metastasis in vivo. The molecular mechanism of miR-200b function was investigated using bioinformatic target predication and high-throughput cDNA sequencing (RNA-seq) and validated by luciferase reporter assay, qRT-PCR, Western blot and immunostaining in vitro and in vivo.
Findings
Our findings indicates that miR-200b is frequently downregulated and have potential utility as a biomarker of metastasis and prognosis in RCC. Interestingly, ectopic expression of miR-200b in the Caki-1 and OSRC-2 cell lines suppresses cell migration and invasion in vitro as well as tumor metastases in vivo. However, miR-200b has no effect on cell proliferation in vitro and tumor growth in vivo. In addition, bioinformatics target predication and RNA-seq results reveals that Laminin subunit alpha 4 (LAMA4) is one target of miR-200b and significantly inhibited by miR-200b in vitro and in vivo.
Interpretation
These results demonstrate a previously undescribed role of miR-200b as a suppressor of tumor metastasis in RCC by directly destabilizing LAMA4 mRNA.
Keywords: Renal cell carcinoma, MicroRNA-200b, LAMA4, Metastasis
Abbreviations: RCC, renal cell carcinoma; LAMA4, Laminin subunit alpha 4; PBS, phosphate-buffered saline; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; RQ, relative quantification; PDX, patient derived xenograft; CDX, cell-derived xenograft; EMT, epithelial-mesenchymal transition
Research in context.
Evidence before this study
MicroRNA-200b (miR-200b), as a member of the miR-200 family, is a well-known small RNA which suppresses cancer cell metastasis, affecting many target genes such as transcription factors, stem cell properties' markers, cellular kinases, and key elements of epithelial–mesenchymal transition (EMT) process. Laminin subunit alpha 4 (LAMA4) is part of the Laminin family, which functions as part of a larger family of extracellular matrix glycoproteins. LAMA4 was demonstrated to promote angiogenesis and tumor metastasis. However, to the best of our knowledge, the association between miR-200b and LAMA4 in renal cell carcinoma (RCC) and the implications of this association in the metastasis of RCC has not been investigated.
Added value of this study
Our findings revealed for the first time a potential tumor suppressive role for miR-200b in RCC progression and it may serve as a biomarker or even a therapeutic target for RCC. Additionally, we provided evidence that LAMA4 is a critical target of miR-200b. Using patient derived xenograft (PDX) model and in vivo RNAi transfection technology, tumor invasion of RCC was significantly suppressed and metastasis was effectively blocked. The mechanism of miR-200b-LAMA4 axis on metastasis in RCC was verified by rescue experiments in vitro and in vivo. The downstream of LAMA4-mediated metastasis of RCC was found to be integrin α5β1/ILK/FAK/ERK pathway.
Implications of all the available evidence
Dysregulation of miR-200b-LAMA4 axis may serve as early metastatic biomarker in RCC. Moreover, targeting metastasis in RCC through epigenetically regulates LAMA4 expression could be achieved by miR-200b.
Alt-text: Unlabelled Box
1. Introduction
Renal cell carcinoma (RCC), which accounts for approximately 3% of adult malignancies and approximately 90% of kidney cancers, is the third most common urological cancer after prostate and bladder cancer, but has the highest mortality rate at about 25% [1,2]. Currently, the best curative therapy for RCC is surgical resection, but 20–40% patients will develop recurrence or metastasis after nephrectomy [3]. No adjuvant therapy is available in the clinic due to both chemotherapy- and radiotherapy- resistant RCC [3]. Moreover, available biomarkers for early detection and follow-up of this disease are sparse, which results in a poor prognosis. Therefore, the discovery of new biomarkers that enable the prediction of early metastasis after nephrectomy is warranted.
MicroRNAs (miRNA), which are a group of small non-coding RNAs that have been demonstrated to regulate gene expression at the post-transcriptional level, play important roles in numerous cellular processes including development, proliferation, and apoptosis [4]. MiR-200b is a microRNA that is part of the miR-200 family, the dysregulation of which has indicated its possible involvement in the development and progression of many malignant tumors, such as gastric carcinoma, pancreatic cancer and lung cancer [[5], [6], [7]]. Moreover, studies have suggested that circulating miR-200b is associated with aggressive tumor progression and could be recognized as a reliable marker that predicts the prognosis and survival of patients with ovarian cancer [8] and colorectal cancer [9]. Additionally, other studies have found that miR-200b is strongly deregulated in clear cell renal cell cancer [10,11].
Laminin subunit alpha 4 (LAMA4) is part of the Laminin family, which functions as part of a larger family of extracellular matrix glycoproteins. LAMA4, which is expressed in tissues of mesenchymal origin, endothelial basement membranes, and certain epithelial basement membranes [12], seems to promote the migration of various cell types including cancer cells of kidney [12,13]. Laminins are the major noncollagenous constituent of basement membranes, are expressed in renal cell carcinoma and have a de-adhesive function, which indicates that they may play a role in migration and invasiveness of renal carcinoma cells in vivo [13,14]. In addition, the function of LAMA4, a constituent of laminin-8, 9 and 14, was demonstrated to promote angiogenesis [12,15], which is important for wound healing and tumor metastasis. Moreover, strong LAMA4 expression was shown to predict metastasis and poor survival in RCC [14].
In this study, we conducted a systematic study to identify metastatic markers of miRNAs for RCC using bioinformatics algorithms. Low expression of miR-200b was demonstrated to be associated with metastasis and poor prognosis in RCC patients. We questioned the tumor suppressive role of miR-200b and wondered the key target gene of miR-200b. Extracellular matrix (ECM) dysregulation contributes to neoplastic progression through directing cell growth, survival, migration and differentiation and modulate vascular development and immune function [16]. Combinating RNA sequencing assay with bioinformatics algorithms, we firstly showed that LAMA4, among numbers of ECM genes, was the key target of miR-200b. Then we explored the association between miR-200b and LAMA4 in RCC and the implications of this association in the metastasis of RCC.
2. Materials and methods
2.1. Bioinformatics data mining
Using the LinkedOmics website, we screened the miRNAs that are significantly negatively correlated with overall survival and M stage of patients with RCC in The Cancer Genome Atlas (TCGA) dataset. Then, we downloaded TCGA KIRC RNA-Seq gene expression data and the clinical data from the UCSC Xena database (http://xena.ucsc.edu/). In all, 70 normal kidney and 241 ccRCC tissues were determined to express hsa-miR-200b (MIMAT0000318) and LAMA4 mRNA. All 241 ccRCC tumors had corresponding clinical data that were used to perform the clinical correlation and survival analysis. Predictions of potential targets of miR-200b were performed by computational algorithms based on ‘seed regions’ between miRNAs and target genes. miRanda (http://miRdb.org/miRDB/index.html), TargetScan (http://www.targetscan.org), miRGen v.3 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=miRgenv3%2Findex), and PicTar (https://pictar.mdc-berlin.de/) were used in this study.
2.2. Cell lines and tansfection
The ccRCC cell lines Caki-1, 786-O, ACHN, Caki-2, and OSRC-2 and the normal renal tubular epithelial line HK-2 were purchased from American Type Culture Collection (ATCC, Manassas, VA). HK-2 cells were cultured in DMEM/F12 medium with 10% fetal calf serum (HyClone Laboratories Inc., Logan, UT), and the other cells were cultured in RPMI-1640 (HyClone, Logan, UT) medium supplemented with 10% Gibco™ FBS (Life Technologies, Grand Island, NY). All cells were cultured at 37 °C in a standard humidified incubator containing 5% CO2 and 95% O2. For overexpression and downregulation of miR-200b, chemically synthesized miR-200b mimics, inhibitors and control oligoribonucleotides (Genepharma Co., Ltd.,Shanghai, China) were transiently transfected into RCC cells using Lipofectamine 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA).
For establishing constant express or against miR-200b cell lines, miR-200b sequence and tough decoy (TUD) RNAs (5′-GGATCCgacggcgctaggatcatcaactcatcattaccaatctggcagtattacaagtattctggtcacagaatacaactcatcattaccaatctggcagtattacaagatgatcctagcgccgtcttttttCTCGAG-3′) were cloned into lentiviral shuttle vector pLenti6 or pLenti6-U6 (Invitrogen). For rescuing LAMA4, knockdown of RNAs against LAMA4 (NM_001105206.2), 5′-ccggGCCTAAAGCAAGTCAGAATAActcgagTTATTCTGACTTGCTTTAGGCtttttg-3′ was cloned into lentiviral vector backbone pLKO.1-puro (addgene #8453), which was validated [17]. Lentiviral constructs were transfected with the ViraPower Packaging Mix (Invitrogen) into 293FT cells to generate lentivirus. Cells infected with virus are selected by 5 μg/mL blasticidin (Invitrogen) and/or 2 μg/mL puromycin (Invitrogen). Recombinant human laminin alpha 4 (rLAMA4) was obtained from R&D company (Cat: 7340-A4-050).
2.3. Patient samples
Written informed consent was also obtained from all patients. In all, 80 paired normal and cancer specimens with pathologically confirmed RCC were included in this study. All patients in this study underwent radical nephrectomy at Peking University First Hospital (PUFH) between 2012 and 2013. After resection, the samples were immediately snap-frozen in liquid nitrogen and stored at −196 °C for further studies. Specimen collection for research purposes was approved by Clinical Research Ethics Board of PUFH. Written informed consent was also obtained from all the patients.
2.4. RNA extraction and quantitative real time-polymerase chain reaction (qRT-PCR)
Total RNA of the tissues and cultured cells was extracted by the TRIzol reagent (Qiagen, Hilden, Germany) for both miRNA and mRNA analyses. To quantify mature miRNA, 1 μg total RNA was added to a poly (A) tail by poly (A) polymerase (NEB, Beverly, MA, USA), which was followed by reverse transcription with an oligo (dT) adapter primer [18]. For protein-coding genes' mRNA quantification, oligo (dT) primers was used to generalize cDNAs from 4 μg total RNA. For the analysis of mature miR-200b and protein-coding genes in cDNAs, qRT-PCR was performed using the SYBR Green PCR Master Mix (Transgen, Beijing, China) on an ABI7500 PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. U6 and GAPDH were selected as the internal control. The relative expression (RQ) was calculated as the fold change relative to internal reference, which was based on the following equation: RQ = 2−ΔΔCt, ΔΔCt = (meanCtcancer − meanCtreference) − (meanCtnormal − meanCtreference) [19]. The primers used are listed in Table S1.
2.5. In vitro cell spreading, motility and invasion assays
Wound-healing assay was performed to determine cell spreading ability, while Boyden chamber assay with Matrigel (1:3 dilution in PBS, Product #354234, Corning Inc., NY, USA) was used to evaluate cell motility and invasion abilities. These assays were performed as previously described [20].
2.6. In vitro cell proliferation and colony formation assay
The cell proliferation assay was performed using a Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) and flow cytometry antibody staining for human Ki-67 (Biolegend, San Diego, CA, USA) according to the manufacturer's instructions. For the colony formation assay, 500 Caki-1, OSRC-2 or 786-O cells were incubated in 6-well plates for 7 days, and then the cells were stained with 0.5% crystal violet. The colonies were imaged and counted under a microscope (OLYMPUS IX71).
2.7. Flow cytometry
Cell apoptosis was assayed by staining with Annexin V-FITC and PI (KeyGEN) following manufacturer's instructions and detected by a flow cytometer (FACSCalibur, Becton Dickinson). Cells without staining, Annexin V-FITC or PI single staining and Annexin V-FITC and PI double staining were tested to adjust voltage and compensation before collecting data. Caki-1 miR-200b cells treated with PBS or rLAMA4 for 48 h, then stained with flow cytometry antibodies against human ITGA1, ITGA5, ITGA9 and ITGB1 (Biolegend) and detected by a flow cytometer (FACSCalibur). The detailed information for flow cytometry antibodies were listed in Table S2.
2.8. In vivo tumor growth and metastasis assays
RCC cell-derived xenograft (CDX) and patient-derived xenograft (PDX) models were established as previously described [21]. Briefly, five-week-old male BALB/C nude mice or NOD/SCID mice (Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were subcutaneously injected in the right flank with 1.5 × 106 cancer cells, derived from cultured RCC cells or human RCC tissues, in 0.2 mL phosphate-buffered saline (PBS). Once cancer cells or tissues developed into palpable tumors, caliper measurements were performed every three or four days, and the tumor volume (V) was calculated using the formula V = (L × W2)/2, where L is the length and W is the width of the tumor. When the tumors reached an average volume of 100–150 mm3, the mice were randomly divided into three groups (n = 6). In vivo treatment with miRNA mimics in the CDX and PDX models was performed as previously described [21]. Then, 20 nM chemically modified mi-Ribo™ hsa-miR-200b mimics or mi-Ribo™ hsa-miR-200b control (Ribobio Co., Guangzhou, China) in 50 μL PBS mixed with 50 μL in vivo transfection reagent (Entran-ster™-in vivo, Engreen, Beijing, China) was locally injected into the tumor mass once every 3 days for 3 weeks.
Growth curves were plotted using the average tumor volume within each experimental group at the established time points. The tumor volumes in the mice were recorded until death occurred from the tumor. The dissected tumors were collected and prepared for RNA extraction, hematoxylin-eosin (HE) staining and immunohistochemistry (IHC).
Quantification of the RCC cell-derived lung metastatic colonies was performed by examining the murine lungs using a TCS 4D laser scanning confocal microscope (Leica, Heidelberg, Germany). Animal experiments were conducted in accordance with the NIH Guidelines for the Care and Use of Laboratory Animals with the approval of the Review Board of PUFH, Beijing. Mice were maintained under pathogen-free conditions with regulated temperature and humidity levels. Mice were randomly assigned to cages in groups of 5 and fed ad libitum under controlled light/dark cycles.
2.9. Quantitative detection of human tumor cell metastasis
The detection of RCC metastasis in the murine lungs was performed as previously described [22]. Briefly, genomic DNA was extracted from murine lung tissues using an EasyPure Genomic DNA Kit (Transgen Biotech, Beijing, China). Quantitative real-time PCR was used to measure human Alu-sequences specific for the most conserved region in human DNA. The primers for the Alu-sequences and the PCR conditions used were previously described [22]. The level of human Alu-sequences was normalized to the amount of mouse/human GAPDH genomic DNA that was amplified using mouse/human GAPDH primers. The primers for Alu and mouse/human GAPDH were listed in Table S1.
2.10. Luciferase reporter assay
The 3′-UTRs of LAMA4 carrying the putative miR-200b binding sites and the mutant binding sites were amplified by PCR using XbaI/EcoRI restriction sites and were inserted immediately downstream of the firefly luciferase cDNA in the pGL3-control vector (Promega, Madison, WI, USA) to construct pGL3-LAMA4 WT and pGL3-LAMA4 MUT. All the constructs were verified by sequencing. For the luciferase reporter assay, 300 ng pGL3 constructs (pGL3-LAMA4 WT or pGL3-LAMA4 MUT) together with 26 ng pRL-TK plasmid that expressed Renilla luciferase were co-transfected with 60 pmol miR-200b mimics or miR-200b mimics-control (GenePharma) using Lipofectamine 2000 (Invitrogen). Luciferase activity was detected using the dual luciferase assay system (Promega) on the Thermo Scientific™ Varioskan™ Flash Multimode Reader (Thermo Fisher Scientific, Inc., Waltham, MA), according to the manufacturer's instructions 24 h after transfection. Normalized Firefly luciferase activitiy to Renilla luciferase activity for each sample was calculated. The experiments were performed in quadruple and repeated at least three times. The primers used are listed in Table S1.
2.11. Western blot analysis
Total protein was extracted from the cultured cells and specimens using RIPA buffer (ShineGene Molecular Biotech, Inc., Shanghai, China) according to the manufacturer's instructions. SDS-PAGE and Western blots were carried out following standard protocols. The immunoreactive bands were visualized by Immobilon™ Western Kit (Millipore, Billerica, MA) using a SYNGENE G: BOX imaging system (Frederick). The primary antibodies and the secondary HRP-conjugated antibodies were described in Table S2.
2.12. Immunohistochemistry
Paraffin-embedded tissues were cut into 5-μm-thick consecutive sections and were then deparaffinized in xylene and rehydrated in graded ethanol solutions. Antigen retrieval was performed with 0.01 M citrate buffer (pH 6.0) in a pressure cooker (95–99 °C for 20 min). Sections were cooled and immersed in a 0.3% hydrogen peroxide solution for 15 min to block endogenous peroxidase activity. After blocking with 10% sheep serum albumin, the sections were incubated with different primary antibodies at 4 °C overnight. Next, the sections were incubated with secondary antibody (1:1000, sc-2005, Santa Cruz, CA) at room temperature for 20 min. All the sections were stained for 3 min using a DAB kit (ZLI-9018, ZSGB-BIO). For the negative controls, the primary antibody was replaced with PBS. Slides were stained with primary antibodies against LAMA4 (1:100, ab205568, Abcam, UK) and PCNA (1:100, ab29, Abcam, UK). Two pathologists, who were not informed about the patients' clinical data, examined all slides independently.
2.13. High-throughput cDNA sequencing (RNA-Seq)
The RNA-Seq experiments were performed by Novogene (Beijing, China). The RNA-seq library was prepared for sequencing using standard Illumina protocols. Briefly, total RNAs from Caki-1 control and miR-200b cells were isolated using TRIzol reagent (Invitrogen) and treated with RNase-free DNase I (New England Biolabs, MA, USA), to remove any contaminating genomic DNA. RNA extraction was performed using Dynabeads oligo(dT) (Invitrogen Dynal). Double-stranded complementary DNAs were synthesized from 1 μg of total RNA using Superscript II reverse transcriptase (Invitrogen) and random hexamer primers. Escherichia coli RNase H (New England Biolabs) were added to remove RNA complementary to the cDNA. The cDNA was then fragmented by nebulization, and the standard Illumina protocol was followed thereafter to create the mRNA-seq library. The libraries were sequenced on an Illumina HiSeq 2000 platform. Sequencing reads were aligned to the human genome (hg19) using the TopHat program (v2.1.1) set to the default parameters. Total read counts for each protein-coding gene were extracted using HTSeq (version 0.6.0) and then loaded into R package DESeq2 to calculate the differentially expressed genes with a cut-off fold change of ≥1.5 and an FDR < 0.05. Gene expression was calculated using the RPKM (reads per kilobase transcriptome per million reads) method.
2.14. Statistical analysis
All data in the figures are presented as the mean ± SD from at least three independent experiments. All data were analyzed using the SPSS 20.0 statistical software (IBM, Chicago, IL, USA). The statistical differences between two groups were determined using a double-sided Student's t-test, unless otherwise specified. Survival curves for patients and mice were plotted according to the Kaplan-Meier method, and the Tarone-Ware test was used for statistical significance. Statistical significance was defined as a two-tailed p < .05.
2.15. Data access
The RNA-sequencing data was uploaded to the public Database: the Genome Sequence Archive in Beijing Institute of Genomics (BIG) Data Center, BIG, Chinese Academy of Sciences, under accession number #PRJCA001428.
3. Results
3.1. MiR-200b is associated with clinical overall survival and M stage properties in a public database of RCC
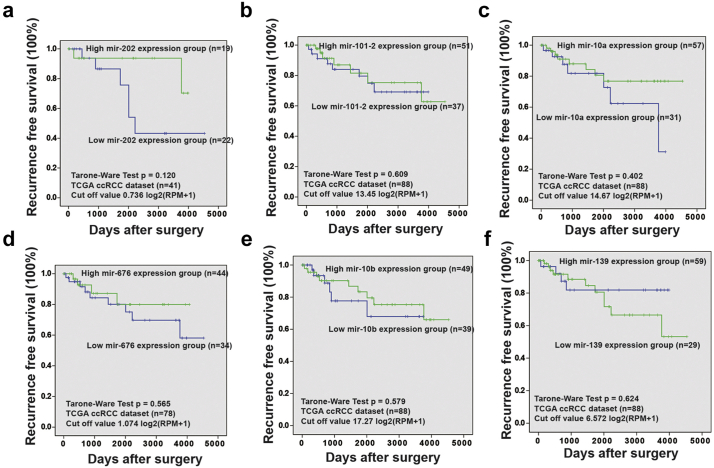
Through an analysis of TCGA database of RCC using the LinkedOmics website [23], we verified the numbers of microRNAs that are relevant to clinical survival and the relationships between their expression in primary tumors and metastatic tissues. As described in Fig. 1a, 215 miRNAs (out of 713 total) were negatively correlated with overall survival (OS) in 254 RCC cases (Fig. S1a). Moreover, 34 of these miRNAs were significantly associated with OS (p < .05) (Fig. S1b). Furthermore, after combination with pathologic M stage (Wilcox Test) data, 26/34 miRNAs were revalidated and showed a negative correlation, and only 7 miRNAs including miR-200b, miR-202, miR-101-2, miR-10a, miR-676, miR-10b, and miR-139 were significantly associated with M stage (p < .05, Mann-Whitney Test) out of 26 negatively correlated miRNA candidates (Fig. 1b and Fig. S1c). Moreover, through a bioinformatics analysis of TCGA KIRC data from UCSC, we found that the expression of miR-200b, miR-10a, miR-10b and miR-139 was significantly lower in 241 ccRCC tissues compared with 70 normal kidney tissues (Fig. 1c and Fig. S2). Additionally, low miR-200b expression in ccRCC patients was related to inferior recurrence-free survival (Fig. 1d) but was not related to the expression of miR-202, miR-101-2, miR-10a, miR-676, miR-10b, or hsa-miR-139 (Fig. S3). TCGA data analysis suggests that low miR-200b expression might be a candidate indicator of poor prognosis in RCC patients after surgery.
Fig. 1.
Downregulation of miR-200b was associated with metastasis and poor prognosis in RCC. (a) Study flow for seeking microRNAs that related with overall survival (OS) and metastasis of RCC patients by in silico analysis using LinkedOmics website. (b) Relative miR-200b expression between M0 and M1 stage from TCGA KIRC dataset. The boxes span the interquartile range. Horizontal lines in the boxes represent median value of miR-200b expression. Whiskers extend to the highest and lowest values. (c) Relative miR-200b expression in normal kidney tissues and ccRCC tissues from TCGA KIRC dataset. RPM, reads per million. (d)Kaplan-Meier curves of recurrence free survival (RFS) time between high and low miR-200b group using Tarone-Ware test in 88 ccRCC patients. Cutoff value was the mean values of miR-200b expression in carcinoma tissues. (e) Relative miR-200b expression in 80 RCC tissues and matched adjacent normal tissues. (f, g) Relative expression data of miR-200b in 80 cases were further analyzed. The expression levels of miR-200b were negatively correlated with T stages (f) and G grades (g). Horizontal lines in (c) and (e–g) represent the mean value of miR-200b expression in TCGA KIRC dataset (c), and mean values of miR-200b expression in 80 RCC patients from PUFH (e-g).
Fig. S1.
Analyzing the TCGA database of RCC using LinkedOmics website. (a) Volcano plot of association results between microRNAs (n = 713) and overall survival in TCGA RCC dataset. (b) Heat plot of 215 microRNAs which are negatively correlated with overall survival in TCGA RCC dataset. (c) Comparison analysis of relative expression of 6 microRNAs (miR-202, miR-101-2, miR-10a, miR-676, miR-10b and miR-139) in primary carcinoma tissues between non-metastatic RCC (M0) and metastatic RCC (M1) in the TCGA database of RCC. All the boxes span the interquartile range. Horizontal lines in the boxes represent median value of miRNAs' expression. Whiskers extend to the highest and lowest values.
Fig. S2.
Analyzing the TCGA database of RCC using UCSC Xena website. Comparison analysis of relative miR-202 (a), miR-101-2 (b), miR-10a (c), miR-676 (d), miR-10b (e) and miR-139 (f) expression between tumor tissues (T) and normal kidney tissues (N) in the TCGA database of RCC. RPM, reads per million.
Fig. S3.
Kaplan-Meier curves of 6 microRNAs in TCGA ccRCC dataset. Kaplan-Meier curves of recurrence free survival (RFS) time between high and low miR-202 (a), miR-101-2 (b), miR-10a (c), miR-676 (d), miR-10b (e) and miR-139 (f) group using Tarone-Ware test in TCGA ccRCC dataset. Cutoff value was the mean values of the corresponding microRNA expression in carcinoma tissues.
3.2. Low expression of miR-200b is associated with metastasis
Metastasis is the primary cause of death in patients with RCC. To explore the relationship between miR-200b and RCC metastasis in clinical specimen, the relative expression of miR-200b normalized to U6 was measured in our 80 paired RCC and adjacent normal kidney tissues. As shown in Fig. 1e, miR-200b expression was significantly decreased in the RCC tissues (median value 0.0013; range 0.000009–4.59) comparing with the matched adjacent normal kidney tissues (median value 0.008; range 0.00002–22.32). The relative expression data of miR-200b in 80 cases was further analyzed. The expression level of miR-200b was negatively correlated with T stage (Fig. 1f) and tumor grade (Fig. 1g). The correlation between the clinicopathological characteristics and the levels of miR-200b; the data are summarized in Table S3. No significant correlation was observed between miR-200b expression levels and gender, age at surgery or histological subtype. However, the expression of miR-200b in the RCC tissues with local tumor invasion was significantly lower than that in RCC tissues without local tumor invasion. Furthermore, the expression of miR-200b was markedly decreased in high-grade RCC.
3.3. miR-200b impedes cell spreading and migration but not growth in RCC
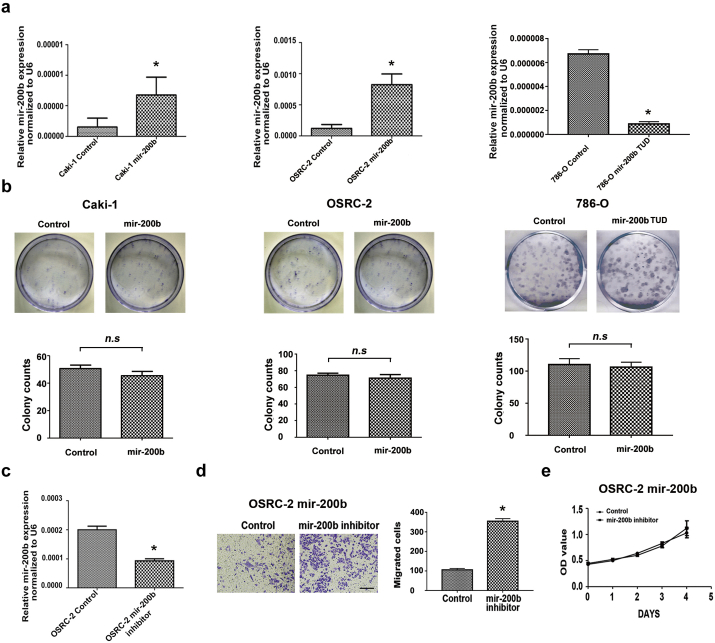
To evaluate whether miR-200b has a functional role in RCC cell metastasis, we first examined the expression of miR-200b in renal proximal tubule epithelial cells and RCC cell lines with different metastatic properties using qRT-PCR. As indicated in Fig. 2a, compared with HK-2, potential metastatic OSRC cell line showed lower level of miR-200b. Moreover, metastasis-derived cell lines (Caki-1 and ACHN) exhibited significant lower levels of miR-200b than primary-derived cell lines (786-O and Caki-2), respectively. Next, we infected the cell lines Caki-1 and OSRC-2 with miR-200b and knock down miR-200b in 786-O cells using lentivirus system. The effects of miR-200b on cell abilities of spreading and migration were measured in vitro. As shown in Fig. S4a the expression levels of miR-200b were increased about 3.1 and 5.7 folds in miR-200b infected Caki-1 and OSRC-2 cells, respectively, and reduced to 13% in 786-OmiR-200b TUD cells. Consistent with the increase of miR-200b expression, the migration ability of Caki-1 and OSRC-2cells infected with miR-200b into the wound was attenuated (58.9% and 56.2%, respectively) compared with the control cells, revealing by wound-healing assay (Fig. 2b). Boyden chamber assays shown that while Caki-1 and OSRC-2 cells was infected with miR-200b mimics, the number of migratory cells was significantly decreased (56.3% and 72.9%, respectively) compared with the controls (Fig. 2c). On the contrast, miR-200b TUD expression in 786-O enhanced cell migration about 2 folds (Fig. 2d). The above results suggested that miR-200b plays a suppressive role in cell spreading and migration in vitro. However, according to CCK-8 assays, miR-200b has no influence on proliferation and colony formation as shown in Fig. 2e and Fig. S4b. Furthermore, miR-200b inhibitor, which was validated by qRT-PCR (Fig. S4c), was used to rescue the migration and proliferation behaviors, in OSRC-2miR-200b cells. The miR-200b inhibitor enhanced OSRC-2-miR-200b migration by almost 40%, which was demonstrated by Boyden chamber assays (Fig. S4d). However, according to cell proliferation assays including Ki-67 staining and CCK-8 assay, no significant difference was observed in cell growth between the miR-200b expression changed group and control group (Fig. 2f–g and Fig. S4e). Additionally, Annexin V-FITC /PI staining was used to testing cell apoptosis in each groups. There is still no significant difference between them (Fig. 2h). These data suggest that miR-200b is specifically highly correlated with a metastatic phenotype, but not with proliferation in RCC cell lines.
Fig. 2.
Ectopic expression of miR-200b impedes cell migration and invasion but not growth inRCC cells in vitro. (a) qRT-PCR analysis of relative miR-200b expression in renal proximal tubule epithelial cells and RCC cell lines. (b) Wound-healing assay. Left panel showed the representative images of wound of Caki-1 and OSRC-2 cells transfected with Control and miR-200b at 0 h and 24 h. Scale bar, 100 μm. Relative cell migratory area (%) of Caki-1 and OSRC-2 cells were statistically analyzed (right panel). (c) Boyden chamber assays were performed with matrigel showed the cell migration and invasion abilities through matrigel of miR-200b over-expressed cells compared with the control cells after 16–18 h' culture. Scale bar, 100 μm. (d) Migration and invasion abilities of 786-O cells were analyzed by Boyden chamber invasive assays between miR-200b TUD cells and control cells. Scale bar, 100 μm. (e) CCK-8 assays were performed to evaluate proliferation ability of miR-200b over-expressed and TUD cells comparing with control cells. (f) Illustration of flow cytometric analysis of Ki-67 staining to evaluate cell proliferation abilitiy between miR-200b over-expressed group and control group. (g) Statistical analysis of positive percentage of Ki-67 staining in miR-200b over-expressed and TUD cells comparing with control cells. (h) Apoptosis detection assay with Annexin V-FITC and PI dual staining. Left panel showed the representative results of apoptosis rate in miR-200b over-expressed and TUD cells comparing with control cells. Apoptosis rate of Caki-1, OSRC-2 and 786-O cells were statistically analyzed (right panel). Data were presented as mean ± SD from at least three independent experiments. *, p < .05. n.s, not significant.
Fig. S4.
Transfection efficiency validation and cell function assays of miR-200b. (a) qRT-PCR validated the over-expression of miR-200b mimics infected Caki-1 and OSRC-2 cells, and inhibition of miR-200b expression in stable 786-O miR-200b TUD cells. (b) Colony formation ability of Caki-1, OSRC-2 and 786-O cells were evaluated by colony formation assays between miR-200b up or down-regulated group and control group. (c) qRT-PCR validated the knock down of miR-200b in OSRC-2 cells by miR-200b inhibitor. (d) Boyden chamber assays with matrigel evaluated cell migration and invasion abilities between miR-200b inhibition group and control group in OSRC-2-miR-200b after 16–18 h' culture. Scale bar, 100 μm. (e) Proliferation ability of renal cancer cells were evaluated by CCK-8 assays between miR-200b inhibitor group and control group in OSRC-2-miR-200b cells. Data were presented as mean ± SD from at least three independent experiments. *, p < .05. n.s, Not significant.
3.4. LAMA4 is a direct target of miR-200b and is highly expressed in RCC cells
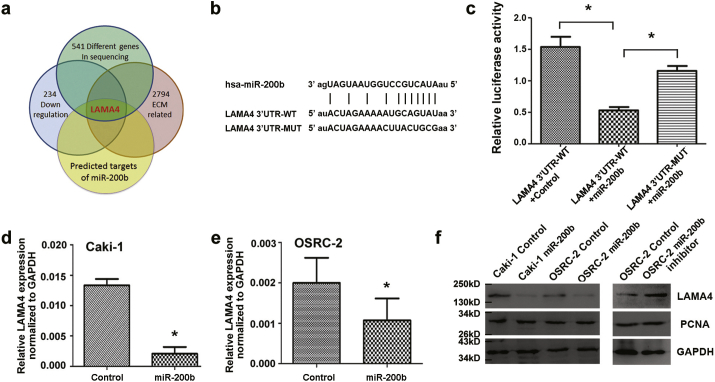
To understand the underlying mechanisms that are involved in the suppression of tumor metastasis by miR-200b in RCC, we first performed a RNA sequencing assay using Caki-1miR-200b and control cells. In all, 307 genes were significantly upregulated and 234 genes were significantly downregulated (Fig. S5a); among the downregulated genes, 102 genes were related to the extracellular matrix (ECM). Since ECM remodeling could alter renal carcinoma cell metastasis [24], we were interested in the top 10 genes that were significantly downregulated, which were annotated with functions associated with tumor metastasis (e.g., LSM1, LAMA4, SEPP1, NR4A2, WFS1, ZEB2, LDB2, ACOX1, CALM1, and EXO1) (Fig. S5b). The in silico analysis using miRGen and the intersection of miRanda, PicTar and TargetScan provided a list of predicted candidate miR-200b target genes. VASH2 and LAMA4 of full scores were found to be directly interacted with miR-200b in malignant events of RCC (Table S4). Through intersection of above independent lists of genes, LAMA4 was the ideal target of miR-200b (Fig. 3a). Furthermore, when TCGA data were analyzed, LAMA4 among the 10 most downregulated ECM-related genes by miR-200b was found to be a unique target, which highlights its relationship with clinical properties such as tumor/normal expression and OS (Fig. S5c–e, and Fig. S6, Fig. S7).
Fig. S5.
Relative LAMA4 mRNA expression and clinical significance in TCGA KIRC dataset. (a) Total different genes were found by RNA sequencing assay in Caki-1-miR-200b cells compared with the control cells. (b) Top 10 significant genes of 102 genes related to extracellular matrix (ECM) were shown. (c) Relative LAMA4 mRNA expression in TCGA KIRC dataset. (d) Kaplan-Meier curves of overall survival time between high and low LAMA4 group using Log-Rank test in TCGA KIRC dataset. Cutoff value was the median value of LAMA4 expression in carcinoma tissues. (e) Relative LAMA4 mRNA expression in 16 human cancer types from STRING profiles. All the boxes in (c, e) span the interquartile range. Horizontal lines in the boxes represent median value of LAMA4 expression. Whiskers extend to the highest and lowest values.
Fig. 3.
MiR-200b targets LAMA4 directly. (a) Venn diagram describing the process of predicting target genes of miR-200b that related with metastasis. (b) Sequence alignment of human miR-200b seed sequence with the 3′-UTR of LAMA4. The mutated sequence in the matched binding sites for the gene that was used for creating firefly luciferase reporter constructs is shown in the bottom of the gene set. (c) Luciferase reporter assay demonstrates that miR-200b inhibited the transcription of wild-type, but not the mutant 3′-UTRs of LAMA4. (d, e) The expression of endogenous LAMA4 was inhibited in the pool of miR-200b infected Caki-1 and OSRC-2 cells, compared with the control, at mRNA level as detected by qRT-PCR. The mRNA expression of LAMA4 was normalized to GAPDH mRNA. (f) The expression of LAMA4 but not PCNA in miR-200b infected renal cancer cell lines was inhibited compared with control counterparts at protein level as detected by Western blot. Data were presented as mean ± SD from at least three independent experiments. *, p < .05.
Fig. S6.
Relative expression of top 10 genes (except of LAMA4) which down regulated significantly by miR-200b in TCGA KIRC dataset. Relative CALM1 (a), EXO1 (b), WFS1 (c), ZEB2 (d), SEPP1 (e), NR4A2 (f), LSM1 (g), LDB2 (h) and ACOX1 (i) mRNA expression in TCGA KIRC dataset. All the boxes span the interquartile range. Horizontal lines in the boxes represent median value of LAMA4 expression. Whiskers extend to the highest and lowest values.
Fig. S7.
Kaplan-Meier curves of top 10 genes (except of LAMA4) which down regulated significantly by miR-200b in TCGA KIRC dataset. Kaplan-Meier curves of overall survival time between high and low CALM1 (a), EXO1 (b), WFS1 (c), ZEB2 (d), SEPP1 (e), NR4A2 (f), LSM1 (g), LDB2 (h) and ACOX1 (i) mRNA expression group using Log-Rank test in TCGA KIRC dataset. Cutoff value was the mean value of genes expression in carcinoma tissues.
Next, we performed a luciferase reporter assay with vectors containing 3′-UTRs that flanked the putative binding sites of miR-200b in the gene. Mutations were generated in the putative binding sites, which were used as controls (Fig. 3b). As shown in Fig. 3c, a statistically significant inhibition of luciferase activity was observed in the wild-type 3′-UTR of the gene compared with the respective mutant construct or with the co-transfection of pcDNA3.0. The inhibition rate for the wild-type 3′-UTR of LAMA4 is 54.3% compared with the corresponding rate of the mutant 3′UTR. Hence, the miR-200b binding-sites in the 3′-UTR of LAMA4 are responsible for the inhibition of the reporter activity, which suggests that miR-200b directly modulates the expression of this gene through its 3′-UTR. Furthermore, qRT–PCR analysis confirmed that the expression of LAMA4 was decreased by approximately 82.1% and 50.1% in miR-200b infected Caki-1 and OSRC-2 cells, respectively, compared with the controls (Fig. 3d–e). These results were validated at the protein level by Western blot for miR-200b in Caki-1 and OSRC-2 cells (Fig. 3f, left).
To further assess whether miR-200b is involved in cell motility in RCC via the targeting of LAMA4, we inhibited the expression of miR-200b with a synthesized miR-200b inhibitor in OSRC-2 cells. As shown in Fig. 3f (right), the protein expression of LAMA4 was increased when miR-200b was inhibited with the miR-200b inhibitor compared with the negative control (scramble oligo). These data demonstrate that ectopic expression of miR-200b downregulates the endogenous expression of LAMA4 at both the mRNA and protein levels.
3.5. Expression of LAMA4 is increased in RCC patients and miR-200b expression is inversely correlated with LAMA4 mRNA levels in RCC tissues
We determined that the expression of LAMA4 was increased in clinical samples. First, we detected its expression in 80 paired RCC and normal tissues. Fig. 4a shows that, relative to the expression of GAPDH, LAMA4 was significantly upregulated in RCC tissues (median value 0.016; range 0.00001–0.44) compared with the matched adjacent normal tissues (median value 0.0057; range 0.00002–0.06). The relative expression data of LAMA4 in 80 cases was further analyzed. The expression levels of LAMA4 were positively correlated with T stage (Fig. 4b) and grade (Fig. 4c).
Fig. 4.
Expression of LAMA4 increases in RCC patients and miR-200b expression inversely correlates with LAMA4 mRNA levels in RCC tissues. (a) Relative LAMA4 mRNA expression in 80 RCC tissues and matched adjacent normal tissues by qRT-PCR. (b, c) Relative expression data of LAMA4 in 80 cases were further analyzed. The relationship between LAMA4 expression and T stages (b) and G grades (c). (d) Western blot results showed protein expressions of LAMA4 in 12 RCC specimens. (e) The linear regression and correlation between miR-200b-3p and LAMA4 mRNA levels in all 80 RCC tissues. Expression status is shown as the T (tumor)/N (normal) ratio in a log scale. (f) The linear regression and correlation between miR-200b-3p and LAMA4 mRNA levels in TCGA 241 ccRCC tissues. Expression status is shown as log2 (RPM + 1) from RNA-seq results, RPM, reads per million. Horizontal lines in (a–c) represent the mean values of LAMA4 expression relative to endogenous reference GAPDH for each series of samples.
Consistent with the results at the mRNA level, the LAMA4 protein was also increased in clinical RCC specimens (Fig. 4d). We then analyzed the expression of miR-200b and the mRNA level of LAMA4 in the same set of 80 primary RCC tissues by qRT–PCR and found a statistically significant inverse correlation between miR-200b and LAMA4 mRNA levels (r = −0.6425, p < .0001, Pearson correlation test) (Fig. 4e). Either, we interested in the relationship between LAMA4 and other miR-200b family, such as miR-200a, miR-200c, miR141 and etc. So the correlation of them were tested by qRT-PCR as well, none of them has a significant relationship with LAMA4 in our 80 primary RCC tissues (Fig. S8a–c). Moreover, TCGA data indicated a similar relationship between miR-200b and LAMA4 (Fig. 4f), but no assess to anyone of miR-200a, miR-200c or miR141 (Fig. S8d–f). These results support the premise that miR-200b downregulation increases the levels of LAMA4 in RCC tissues.
Fig. S8.
Correlation between LAMA4 and other miR-200b family (miR-200a, miR-200c, miR141). The linear regression and correlation between LAMA4 and other miR-200b family (miR-200a (a), miR-200c (b), miR141 (c)) mRNA levels in all 80 RCC tissues. The linear regression and correlation between LAMA4 and other miR-200b family (miR-200a (d), miR-200c (e), miR141 (f)) mRNA levels in TCGA KIRC dataset.
3.6. LAMA4 is a critical role in tumor metastasis which induced by silencing miR-200b in cell-derived xenografts nude mice
In order to understand the function of miR-200b in vivo, CDX was performed to compare the growth ability between miR-200b group and control group. As shown in Fig. 5a, the similar growth curves of xenografts formed by Caki-1 control and miR-200b cells was insignificant. The xenografts were shown in Fig. 5b (Left), and comparison of the tumor weights between these two groups were insignificant (Fig. 5b, Right). Either, it is presented no difference of growth ability between xenografts derived from OSCR-2 control and OSRC-2 miR-200b cells (Fig. 5c, d). Moreover, no significant difference was observed among CDXs derived from 786-O control, 786-O miR-200b TUD and 786-O miR-200b TUD + shLAMA4 cells (Fig. 5e, f). It is suggested that miR-200b had no effect on RCC cells' proliferation in vivo.
Fig. 5.
LAMA4 is a critical role in cell metastasis which induced by silencing miR-200b in cell-derived xenografts (CDXs). (a) Growth curves of mice subcutaneous xenografts derived from Caki-1 Control and Caki-1 miR-200b cells. (b) Image of 10 xenografts formed by Caki-1 Control and Caki-1 miR-200b cells (Left panel). Tumor weights were statistical analyzed (Right panel). (c) OSRC-2 subcutaneous CDXs growth curves. (d) Left panel shown the OSRC-2 subcutaneous CDXs of control and miR-200b group. Statistically analysis of tumor weights of OSRC-2 subcutaneous CDXs (Right panel). (e) Growth curves of 786-O subcutaneous CDXs of control, miR-200b TUD and miR-200b TUD + shLAMA4 group. (f) The image of 786-O subcutaneous CDXs (Left panel). Statistically analysis of tumor weights of 786-O subcutaneous CDXs (Right panel). (g) Left panel shown the mice lung metastatic lesions derived from three groups: Control cells, miR-200b TUD cells and miR-200b TUD + shLAMA4 cells by tail vein injection. Black circles present the position of metastatic nodules. The middle panel is to enlarge the metastatic nodules clearly (arrows). The right panel shown the morphology of lung metastasis by HE staining. Scale bar, 100 μm. (h) QRT-PCR determined relative Alu-sequence expression of mice lung metastases from 786-O CDXs formed by 786-O control, 786-O miR-200b TUD and 786-O miR-200b TUD + shLAMA4 cells, respectively. *, p < .05. n.s, not significant.
The effect of miR-200b-LAMA4 axis on tumor metastasis was evaluated by mice lung metastasis model. As shown in Fig. 5g, lung metastatic lesions of 786-O TUD cells were much more than control group, however, the metastatic colonies significantly decreased while we knockdown LAMA4 in 786-O TUD cells. Statistical analysis of qRT-PCR of human-specific Alu-sequences indicated that downregulation of miR-200b increased the risk of lung metastasis significantly. Interestingly, rescue experiments by shLAMA4 significantly reduced lung metastasis (Fig. 5h). Therefore, LAMA4 is a critical role in tumor metastasis mediated by downregulation of miR-200b.
3.7. Extrinsic injection of miR-200b in RCC PDXs decreases the expression of LAMA4 and prolongs animal survival
To further demonstrate the contribution of miR-200b to RCC treatment, a patient-derived xenograft (PDX) model was established to determine the effects of miR-200b. Because of hard obtained infection patient's tissues, firstly, we detected the effects of ectopic expression of miR-200b on the growth and metastasis of cell-derived xenografts (CDXs). The intratumor injection of miR-200b mimics could significantly increase the expression level of miR-200b (Fig. 6A). However, miR-200b did not affect the growth of CDXs (Fig. 6b), and the expression of PCNA was not different within the tumor tissues (Fig. 6c). Nevertheless, the quantification of human-specific Alu-sequences, which was measured by qRT-PCR, showed that transfection of cells with miR-200b in vivo could suppress lung metastasis in mice with tail vein transplanted tumor cells (Fig. 6d). The number of metastatic colonies was captured by fluorescence microscopy after their isolation from the murine lung tissues (Fig. 6e, Left) and were counted by Image J software (Fig. 6e, Right). These in vivo data indicate that at least half of RCC metastasis and its effects could be significantly arrested by miR-200b mimics.
Fig. 6.
Extrinsic injection of miR-200b mimics in RCC PDXs decreases the expression of LAMA4 and prolongs animal survival. (a) qRT-PCR analysis of miR-200b expression in Caki-1 CDX tumor tissues one week after intratumoral injection of miR-200b mimics or control RNA. (b) Growth curves of Caki-1 CDXs formed by Caki-1 cells injected with miR-200b mimics (n = 6) or control RNA (n = 6). (c) Representative picture of IHC staining of PCNA positive cells of tissues from Caki-1 CDXs. Scale bar, 200 μm. (d) Relative Alu-sequence expression of mice lung metastases from Caki-1 CDXs formed by Caki-1 cells injected with miR-200b mimics or control RNA by qRT-PCR. (e) Representative micrographs and quantitative data of metastatic colonies in mice lung. DiI-positive lung metastatic colonies were photographed and counted under a laser confocal microscope. (f) The hematoxylin and eosin (HE) staining of PDX tissues in nude mice was similar to that of the tumor tissues of the patient with RCC. (g) qRT-PCR analysis of miR-200b expression in PDX tumor tissues one week after intratumoral injection of control RNA, PBS and miR-200b mimics. (h) Growth curves of PDX injected with control RNA (n = 6), PBS (n = 6) and miR-200b-3p mimics (n = 6). (i) Relative Alu-sequence expression of mice lung metastases from PDXs injected with control RNA, PBS and miR-200b mimics by qRT-PCR. (j) Kaplan-Meier curves of overall survival time between control RNA, PBS and miR-200b group using Tarone-Ware test. (k) Representative pictures of IHC staining of PCNA and LAMA4 positive cells of tissues from PDXs. Scale bar, 100 μm. Data were presented as mean ± SD from at least three independent experiments. *, p < .05.
Next, we performed this injection technology with PDX model. As shown in Fig. 6f, the HE staining of the PDXs in nude mice was similar to that of the tumors derived from patients, which indicates that the PDX is homologous with the tumors of patients. Furthermore, the expression level of miR-200b was dramatically increased by intratumor injection compared with the control and PBS injection (Fig. 6g). Although miR-200b has no influence on tumor growth as shown in Fig. 6h, according to Alu-PCR, the intratumor injection of miR-200b mimics decreased the extent of lung metastases in the PDX model compared with the control and PBS (Fig. 6i). In addition, the transfection of miR-200b mimics significantly increased the survival of tumor-bearing nude mice (Fig. 6j). Furthermore, PCNA expression was not significantly different between the miR-200b mimics treatment and control groups (Fig. 6k, top). However, LAMA4 was significantly reduced by miR-200b mimics in tumor specimens isolated from PDXs (Fig. 6k, bottom). Taken together, we found that LAMA4 plays an important role in the contribution of miR-200b to RCC metastasis.
3.8. LAMA4 increases integrin α5β1 and induces migration by ILK/FAK/ERK pathway
LAMA4, an member of extracellular matrix glycoproteins and component of the laminin complex, promoted tumor cell migration and was identified as a ligand for integrin α6β1 and melanoma cell adhesion molecule [[25], [26], [27]]. To explore the molecular mechanism of LAMA4-mediated metastasis in RCC, we analyzed the relationship between LAMA4 and ECM related genes in TCGA ccRCC dataset. Firstly, we found that numbers of integrin family genes' mRNA expression were positively correlated with LAMA4 mRNA expression (Fig. 7a). The top four relevant genes of LAMA4 were ITGB1, ITGA5, ITGA1 and ITGA9, which code α or β proteins of integrins (Fig. 7b). To investigate the influence of LAMA4 on expression of integrins, rLAMA4 was added into the culture medium of Caki-1 miR-200b cells. qRT-PCR results suggested that ITGB1 and ITGA5 mRNA expression were significantly upregulated by rLAMA4 (Fig. 7c). Flow cytometry results showed that percentage of ITGA5 and ITGB1 positive cells were significantly increased by exposure of rLAMA4 (Fig. 7d). To investigate the role of miR-200b-LAMA4 axis on metastasis in renal cell carcinoma, we conducted rescue experiments of LAMA4. Compared with control group, rLAMA4 promoted migration ability of Caki-1 miR-200b cells, however, this effect was abolished by ATN-161, an integrin α5β1 inhibitor (Fig. 7e, left). In order to knockdown LAMA4 in 786–0 miR-200b TUD stably, we constructed shRNA and selected with puromycin (6 μg/mL). Transfection efficiency were validated by quantitative real-time PCR (Fig. S9a) and western blot (Fig. S9b). Moreover, in 786-O miR-200b TUD cells, downregulation of LAMA4 and/or inhibition of integrin α5β1 suppressed cell migrated ability (Fig. 7e, right). However, it is no significant effect of LAMA4 and integrin α5β1 on cell proliferation (Fig. 7f). As shown in Fig. 7g, integrin α5β1 was significantly downregulated by miR-200b mediated decrease of LAMA4. The integrin α5β1 targets, ILK, FAK and ERK were also found lowly phosphorylated in Caki-1 miR-200b cells, compared with control cells. Remarkably, addition of rLAMA4 caused a considerable increase in integrin α5β1 expression and ILK/FAK/ERK pathway activation. Furthermore, in the presence of rLAMA4, a clear reduction of ILK, FAK and ERK phosphorylation could be detected after blockage of integrin α5β1. We also studied the effect of ILK and ERK inhibition on rLAMA4 activated ILK/FAK/ERK pathway. Phosphorylation of ILK, FAK and ERK were markedly reduced upon ILK inhibition, while ERK phosphorylation decreased after ERK inhibition. These results confirmed that LAMA4 increases integrin α5β1 expression and induces migration by activating ILK/FAK/ERK pathway.
Fig. 7.
Effects of LAMA4 on ECM related genes expression and the mechanism of miR-200b-LAMA4 axis on metastasis in RCC. (a) Analysis of the correlation between LAMA4 and numbers of integrin family genes expression in TCGA ccRCC dataset. (b) The linear regression and correlation between LAMA4 and ITGB1, ITGA5, ITGA1 or ITGA9 mRNA levels in TCGA ccRCC dataset. (c, d) Changes of ITGB1, ITGA5, ITGA1 and ITGA9 expression affected by rLAMA4 in Caki-1 miR-200b cells using qRT-PCR (c) and flow cytometry (d). (e) Using Boyden chamber invasive assays, the migration and invasion abilities of Caki-1 miR-200b cells treated with rLAMA and ATN-161 were shown in Left panel. And the effect of LAMA4 knockdown and/or ATN-161 on migration and invasion abilities of 786-O miR-200b TUD cells measured by Boyden chamber invasive assays (right panel). (f) CCK-8 assays was performed to evaluate the role of rLAMA and ATN-161 in cell proliferation of Caki-1 miR-200b cells (left panel). The right panel shown the cell proliferation abilities of 786-O miR-200b TUD cells treated with LAMA4 knockdown and/or ATN-161. (g) Western blot shown the expression of ITGA5, ITGB1, ILK, FAK and ERK after overexpression of miR-200b and treatment with rLAMA4, ATN-161, Cpd-22 and SCH772984 in Caki-1 cells. (h) Diagram of miR-200b-LAMA4 axis on metastasis in RCC. Step 1. Oncology signals induced downregulation of pri-miR-200b; Step 2. Pre-miR-200b was exported and matured in cytoplasm; Step 3. Loss of miR-200b in RCC contributes to upregulation of LAMA4; Step 4. LAMA4 was secreted into extracellular matrix; Step 5. LAMA4 stimulates expression of integrin α5β1; Step 6. ILK/FAK/ERK pathway was activated. Step 7. ERK induces expression of the oncogenes. Data were presented as mean ± SD from at least three independent experiments. *, p < .05.
Fig. S9.
Validation of LAMA4 knock down stably in 786-O miR-200b TUD cells. qRT-PCR (a) and Western blot (b) assays shown the downregualtion of LAMA4 expression by shLAMA4.
4. Discussion
MiR-200b is located on chromosome 1p36.33, the loss of which is observed in 14% clear-cell RCCs [28]. A recent study demonstrated that miR-200b is an epithelial-mesenchymal transition (EMT)-related microRNA [29]. EMT is involved in the initial steps of invasion and metastasis and is associated with a loss of epithelial characteristics and an acquisition of mesenchymal markers, which is prevented by the targeting of ZEB1 and ZEB2 expression by miR-200b [[30], [31], [32]]. In this study, we found a significantly low expression level of miR-200b in clinical RCC specimens in comparison with adjacent normal tissues. Yoshino H and colleagues [29] drew a similar conclusion in a previous study. Moreover, the downregulation of miR-200b has also been reported in several other cancers [5,6,[33], [34], [35], [36], [37], [38]]. Significant relationships between miRNA expression and some clinicopathological parameters, including tumor stage and tumor grade, have also been reported. However, in those studies, the follow-up period was too short to evaluate the relationship between miRNA expression and patient prognosis. In our study, the overexpression of miR-200b not only impeded cell spreading and migration in Caki-1 and OSRC-2 cells but also inhibited the metastasis of patient- derived xenografts and cell-derived xenografts in nude mice. These data clearly demonstrate that miR-200b plays a suppressor role in cell migration and metastasis of RCC.
The role of miR-200b in cancer cell growth seems paradoxical under the environment of different types of cancer. Evidences have suggested that miR-200b behave as a tumor suppressor in various cancer [39,40] by inhibiting cell proliferation. On the contrary, miR-200b have been demonstrated as a promoter of cell proliferation in several other malignancies, such as acute lymphoblastic leukemia [41] and colorectal cancer [42]. However, the effect of miR-200b on RCC cells remains unknown. That lower expression of miR-200b in RCC specimen correlated with higher tumor stage in our study, suggested that miR-200b could possibly execute antiproliferative function in RCC. Against this possibility is the observation that overexpression or downregulation of miR-200b didn't significantly affect RCC cell proliferation in vitro and in vivo. Consistent with these results, regardless of the expression of LAMA4, RCC tumor growth curve of xenograft remained unchanged.It has been suggested that the overexpression of LAMA4 is correlated with malignancy and invasive capacity in breast cancer[17]. Interestingly, LAMA4 can serve as a potential new target antigen for cancer immunotherapy [43]. Strong LAMA4 expression was also shown to predict poor survival in RCC [14]. In this study, we have shown for the first time that LAMA4 is a direct functional target for the microRNA miR-200b. The binding of miR-200b to the LAMA4 3′-UTR results in the downregulation of endogenous LAMA4 expression in renal cancer cells. Moreover, the LAMA4 3′-UTR contains specific conserved binding sites for miR-200b, as shown in luciferase reporter assays. MiR-200b is markedly downregulated in renal cancer, which possibly accounts for LAMA4 overexpression in tumors. This has been supported by the negative correlation between tumor-specific changes in the mRNA levels of miR-200b and LAMA4 and the overexpression of LAMA4 protein in RCC samples. These results provide strong evidence for a novel mechanism of LAMA4 regulation.
LAMA4 was found strongly upregulated on tumor blood vessels in RCC, compared to adjacent non-malignant tissue14. In addition, according to the public profiles on the STRING website, LAMA4 is obviously much more highly expressed in renal cancer than in other cancers (Fig. S6e). It is suggested that LAMA4 co-distributed and interacted with integrins αvβ3, α3β1, and α6β1, thus increased blood vessel development [44,45]. In this study, we found that co-expression of LAMA4 with integrin α5β1, and LAMA4 increased integrin α5β1 expression and induced RCC cell migration by activating ILK/FAK/ERK pathway.
These findings, along with the roles of LAMA4 in tumor progression, signify the involvement of the miR-200b-LAMA4 regulation chain in RCC progression, and demonstrate that miR-200b-LAMA4 axis may serve as a prognostic marker and a therapeutic target in RCC.
Taken together, we document that downregulation of miR-200b is closely associated with metastatic features and poor prognosis in patients with RCC. In vitro and in vivo models suggest that miR-200b-LAMA4 axis plays a functional role in RCC progression. Further, mechanism exploration demonstrate that LAMA4 induces integrin α5β1 expression, and promotes cell migration by ILK/FAK/ERK pathway. Our findings revealed for the first time a potential tumor suppressive role for miR-200b in RCC progression and it may serve as a biomarker or even a therapeutic target for RCC through epigenetically regulates LAMA4 expression.
The following are the supplementary data related to this article.
Primers for qRT-PCR and vector construct
Antibodies for flow cytometry and Western blot
Relationship between mir-200b-3p expression and pathological features in RCC patients
Venn - Screening the targets of miR-200b from three miRNA database websites (miRanda, TargetScan and miRTarBase) using TCGA KIRC dataset analyzing.
Authors' contributions
WZ, LZ and XL supervised this study. YL, BG, WZ, SH, YZ, BS, HH and XZ conceived the experiments and analyzed the data. BW analyzed the data from the public database. YL, BG, ZZ and JL performed experiments and YL, BG and LZ wrote the manuscript. All authors were involved in writing the paper and all approved the submitted manuscript.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Funding
This study was funded by the National Natural Science Foundation of China (81502578, 81672546, 81872083, 81872025 and 81460360), the Beijing Natural Science Foundation (7182030), the National Key R&D Program of China (No. 2016YFC0902601), the Clinical Features Research of Capital (No. Z151100004015173) and the Capital Health Research and Development of Special (2016-1-4077). Natural Science Foundation of Xinjiang Uygur Autonomous Region (2015211C126).
Acknowledgements
The authors thank the staff at the Department of Urology, Peking University First Hospital, Beijing, China, and the Institute of Urology, Peking University, Beijing 100034, China, for technical support.
Contributor Information
Xuesong Li, Email: pineneedle@sina.com.
Liqun Zhou, Email: zhoulqmail@sina.com.
Wei Zhao, Email: linelong@126.com.
References
- 1.Siegel R.L., Miller K.D., Ahmedin J. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]; Siegel RL, Miller KD, Ahmedin J. Cancer statistics, 2018. CA: a cancer journal for clinicians 2018; 68(1): 7-30. [DOI] [PubMed]
- 2.American Cancer Society Survival rates for kidney cancer. 2017. https://www.cancer.org/cancer/kidney-cancer/detection-diagnosis-staging/survival-rates.html (accessed May 1 2019); American Cancer Society. Survival Rates for Kidney Cancer. 2017. https://www.cancer.org/cancer/kidney-cancer/detection-diagnosis-staging/survival-rates.html (accessed May 1 2019).
- 3.Ljungberg B., Bensalah K., Canfield S. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67(5):913–924. doi: 10.1016/j.eururo.2015.01.005. [DOI] [PubMed] [Google Scholar]; Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. European urology 2015; 67(5): 913-24. [DOI] [PubMed]
- 4.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]; Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11(9): 597-610. [DOI] [PubMed]
- 5.Shinozaki A., Sakatani T., Ushiku T. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 2010;70(11):4719–4727. doi: 10.1158/0008-5472.CAN-09-4620. [DOI] [PubMed] [Google Scholar]; Shinozaki A, Sakatani T, Ushiku T, et al. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer research 2010; 70(11): 4719-27. [DOI] [PubMed]
- 6.Li A., Omura N., Hong S.M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70(13):5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li A, Omura N, Hong SM, et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer research 2010; 70(13): 5226-37. [DOI] [PMC free article] [PubMed]
- 7.Tang Q., Li M., Chen L., Bi F., Xia H. miR-200b/c targets the expression of RhoE and inhibits the proliferation and invasion of non-small cell lung cancer cells. Int J Oncol. 2018;53(4):1732–1742. doi: 10.3892/ijo.2018.4493. [DOI] [PubMed] [Google Scholar]; Tang Q, Li M, Chen L, Bi F, Xia H. miR-200b/c targets the expression of RhoE and inhibits the proliferation and invasion of non-small cell lung cancer cells. International journal of oncology 2018; 53(4): 1732-42. [DOI] [PubMed]
- 8.Zuberi M., Mir R., Das J. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol. 2015;17(10):779–787. doi: 10.1007/s12094-015-1303-1. [DOI] [PubMed] [Google Scholar]; Zuberi M, Mir R, Das J, et al. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico 2015; 17(10): 779-87. [DOI] [PubMed]
- 9.Yuan Z., Baker K., Redman M.W. Dynamic plasma microRNAs are biomarkers for prognosis and early detection of recurrence in colorectal cancer. Br J Cancer. 2017;117(8):1202–1210. doi: 10.1038/bjc.2017.266. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yuan Z, Baker K, Redman MW, et al. Dynamic plasma microRNAs are biomarkers for prognosis and early detection of recurrence in colorectal cancer. Br J Cancer 2017; 117(8): 1202-10. [DOI] [PMC free article] [PubMed]
- 10.Duns G., van den Berg A., van Dijk M.C. The entire miR-200 seed family is strongly deregulated in clear cell renal cell cancer compared to the proximal tubular epithelial cells of the kidney. Genes Chromosomes Cancer. 2013;52(2):165–173. doi: 10.1002/gcc.22016. [DOI] [PubMed] [Google Scholar]; Duns G, van den Berg A, van Dijk MC, et al. The entire miR-200 seed family is strongly deregulated in clear cell renal cell cancer compared to the proximal tubular epithelial cells of the kidney. Genes, chromosomes & cancer 2013; 52(2): 165-73. [DOI] [PubMed]
- 11.Silva-Santos R.M., Costa-Pinheiro P., Luis A. MicroRNA profile: a promising ancillary tool for accurate renal cell tumour diagnosis. Br J Cancer. 2013;109(10):2646–2653. doi: 10.1038/bjc.2013.552. [DOI] [PMC free article] [PubMed] [Google Scholar]; Silva-Santos RM, Costa-Pinheiro P, Luis A, et al. MicroRNA profile: a promising ancillary tool for accurate renal cell tumour diagnosis. Br J Cancer 2013; 109(10): 2646-53. [DOI] [PMC free article] [PubMed]
- 12.Shan N., Zhang X., Xiao X. Laminin α4 (LAMA4) expression promotes trophoblast cell invasion, migration, and angiogenesis, and is lowered in preeclamptic placentas. Placenta. 2015;36(8):809–820. doi: 10.1016/j.placenta.2015.04.008. [DOI] [PubMed] [Google Scholar]; Shan N, Zhang X, Xiao X, et al. Laminin α4 (LAMA4) expression promotes trophoblast cell invasion, migration, and angiogenesis, and is lowered in preeclamptic placentas. Placenta 2015; 36(8): 809-20. [DOI] [PubMed]
- 13.Vainionpää N., Lehto V.-P., Tryggvason K., Virtanen I. Alpha4 chain laminins are widely expressed in renal cell carcinomas and have a de-adhesive function. Lab Invest. 2007;87:780. doi: 10.1038/labinvest.3700592. [DOI] [PubMed] [Google Scholar]; Vainionpää N, Lehto V-P, Tryggvason K, Virtanen I. Alpha4 chain laminins are widely expressed in renal cell carcinomas and have a de-adhesive function. Laboratory Investigation 2007; 87: 780. [DOI] [PubMed]
- 14.Wragg J.W., Finnity J.P., Anderson J.A. MCAM and LAMA4 are highly enriched in tumor blood vessels of renal cell carcinoma and predict patient outcome. Cancer Res. 2016;76(8):2314–2326. doi: 10.1158/0008-5472.CAN-15-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wragg JW, Finnity JP, Anderson JA, et al. MCAM and LAMA4 Are Highly Enriched in Tumor Blood Vessels of Renal Cell Carcinoma and Predict Patient Outcome. Cancer research 2016; 76(8): 2314-26. [DOI] [PMC free article] [PubMed]
- 15.Stenzel D., Franco C.A., Estrach S. Endothelial basement membrane limits tip cell formation by inducing Dll4/Notch signalling in vivo. EMBO Rep. 2011;12(11):1135–1143. doi: 10.1038/embor.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stenzel D, Franco CA, Estrach S, et al. Endothelial basement membrane limits tip cell formation by inducing Dll4/Notch signalling in vivo. EMBO Rep 2011; 12(11): 1135-43. [DOI] [PMC free article] [PubMed]
- 16.Pickup M.W., Mouw J.K., Weaver V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15(12):1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO reports 2014; 15(12): 1243-53. [DOI] [PMC free article] [PubMed]
- 17.Ross J.B., Huh D., Noble L.B., Tavazoie S.F. Identification of molecular determinants of primary and metastatic tumour re-initiation in breast cancer. Nat Cell Biol. 2015;17:651. doi: 10.1038/ncb3148. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ross JB, Huh D, Noble LB, Tavazoie SF. Identification of molecular determinants of primary and metastatic tumour re-initiation in breast cancer. Nature cell biology 2015; 17: 651. [DOI] [PMC free article] [PubMed]
- 18.Shi R., Chiang V.L. Facile means for quantifying microRNA expression by real-time PCR. BioTechniques. 2005;39(4):519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]; Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. BioTechniques 2005; 39(4): 519-25. [DOI] [PubMed]
- 19.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9) doi: 10.1093/nar/29.9.e45. [e45] [DOI] [PMC free article] [PubMed] [Google Scholar]; Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research 2001; 29(9): e45. [DOI] [PMC free article] [PubMed]
- 20.Lan L., Han H., Zuo H. Upregulation of myosin Va by Snail is involved in cancer cell migration and metastasis. Int J Cancer. 2010;126(1):53–64. doi: 10.1002/ijc.24641. [DOI] [PubMed] [Google Scholar]; Lan L, Han H, Zuo H, et al. Upregulation of myosin Va by Snail is involved in cancer cell migration and metastasis. International journal of cancer 2010; 126(1): 53-64. [DOI] [PubMed]
- 21.Su B., Zhao W., Shi B. Let-7d suppresses growth, metastasis, and tumor macrophage infiltration in renal cell carcinoma by targeting COL3A1 and CCL7. Mol Cancer. 2014;13:206. doi: 10.1186/1476-4598-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]; Su B, Zhao W, Shi B, et al. Let-7d suppresses growth, metastasis, and tumor macrophage infiltration in renal cell carcinoma by targeting COL3A1 and CCL7. Molecular cancer 2014; 13: 206. [DOI] [PMC free article] [PubMed]
- 22.Zijlstra A., Mellor R., Panzarella G. A quantitative analysis of rate-limiting steps in the metastatic cascade using human-specific real-time polymerase chain reaction. Cancer Res. 2002;62(23):7083–7092. [PubMed] [Google Scholar]; Zijlstra A, Mellor R, Panzarella G, et al. A quantitative analysis of rate-limiting steps in the metastatic cascade using human-specific real-time polymerase chain reaction. Cancer research 2002; 62(23): 7083-92. [PubMed]
- 23.Vasaikar S.V., Straub P., Wang J., Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic acids research 2018; 46(D1): D956-D63. [DOI] [PMC free article] [PubMed]
- 24.Cox T.R., Erler J.T. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech. 2011;4(2):165–178. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cox TR, Erler JT. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Disease Models & Mechanisms 2011; 4(2): 165-78. [DOI] [PMC free article] [PubMed]
- 25.Ishikawa T., Wondimu Z., Oikawa Y. Laminins 411 and 421 differentially promote tumor cell migration via α6β1 integrin and MCAM (CD146) Matrix Biol. 2014;38:69–83. doi: 10.1016/j.matbio.2014.06.002. [DOI] [PubMed] [Google Scholar]; Ishikawa T, Wondimu Z, Oikawa Y, et al. Laminins 411 and 421 differentially promote tumor cell migration via α6β1 integrin and MCAM (CD146). Matrix Biology 2014; 38: 69-83. [DOI] [PubMed]
- 26.Flanagan K., Fitzgerald K., Baker J. Laminin-411 is a vascular ligand for MCAM and facilitates TH17 cell entry into the CNS. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0040443. [DOI] [PMC free article] [PubMed] [Google Scholar]; Flanagan K, Fitzgerald K, Baker J, et al. Laminin-411 is a vascular ligand for MCAM and facilitates TH17 cell entry into the CNS. PloS one 2012; 7(7): e40443. [DOI] [PMC free article] [PubMed]
- 27.Ishikawa T., Wondimu Z., Oikawa Y., Ingerpuu S., Virtanen I., Patarroyo M. Monoclonal antibodies to human laminin α4 chain globular domain inhibit tumor cell adhesion and migration on laminins 411 and 421, and binding of α6β1 integrin and MCAM to α4-laminins. Matrix Biol. 2014;36:5–14. doi: 10.1016/j.matbio.2014.03.003. [DOI] [PubMed] [Google Scholar]; Ishikawa T, Wondimu Z, Oikawa Y, Ingerpuu S, Virtanen I, Patarroyo M. Monoclonal antibodies to human laminin α4 chain globular domain inhibit tumor cell adhesion and migration on laminins 411 and 421, and binding of α6β1 integrin and MCAM to α4-laminins. Matrix Biology 2014; 36: 5-14. [DOI] [PubMed]
- 28.Girgis A.H., Iakovlev V.V., Beheshti B. Multilevel whole-genome analysis reveals candidate biomarkers in clear cell renal cell carcinoma. Cancer Res. 2012;72(20):5273–5284. doi: 10.1158/0008-5472.CAN-12-0656. [DOI] [PubMed] [Google Scholar]; Girgis AH, Iakovlev VV, Beheshti B, et al. Multilevel whole-genome analysis reveals candidate biomarkers in clear cell renal cell carcinoma. Cancer research 2012; 72(20): 5273-84. [DOI] [PubMed]
- 29.Yoshino H., Enokida H., Itesako T. Epithelial-mesenchymal transition-related microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. J Hum Genet. 2013;58(8):508–516. doi: 10.1038/jhg.2013.31. [DOI] [PubMed] [Google Scholar]; Yoshino H, Enokida H, Itesako T, et al. Epithelial-mesenchymal transition-related microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. Journal of human genetics 2013; 58(8): 508-16. [DOI] [PubMed]
- 30.Huber M.A., Kraut N., Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17(5):548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]; Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Current opinion in cell biology 2005; 17(5): 548-58. [DOI] [PubMed]
- 31.Xu J., Lamouille S., Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell research 2009; 19(2): 156-72. [DOI] [PMC free article] [PubMed]
- 32.Kim T., Veronese A., Pichiorri F. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208(5):875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kim T, Veronese A, Pichiorri F, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. The Journal of experimental medicine 2011; 208(5): 875-83. [DOI] [PMC free article] [PubMed]
- 33.Leskela S., Leandro-Garcia L.J., Mendiola M. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr Relat Cancer. 2011;18(1):85–95. doi: 10.1677/ERC-10-0148. [DOI] [PubMed] [Google Scholar]; Leskela S, Leandro-Garcia LJ, Mendiola M, et al. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocrine-related cancer 2011; 18(1): 85-95. [DOI] [PubMed]
- 34.He M., Liu Y., Deng X. Down-regulation of miR-200b-3p by low p73 contributes to the androgen-independence of prostate cancer cells. Prostate. 2013;73(10):1048–1056. doi: 10.1002/pros.22652. [DOI] [PubMed] [Google Scholar]; He M, Liu Y, Deng X, et al. Down-regulation of miR-200b-3p by low p73 contributes to the androgen-independence of prostate cancer cells. The Prostate 2013; 73(10): 1048-56. [DOI] [PubMed]
- 35.Pacurari M., Addison J.B., Bondalapati N. The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS-2B cells. Int J Oncol. 2013;43(2):548–560. doi: 10.3892/ijo.2013.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pacurari M, Addison JB, Bondalapati N, et al. The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS-2B cells. International journal of oncology 2013; 43(2): 548-60. [DOI] [PMC free article] [PubMed]
- 36.Gregory P.A., Bert A.G., Paterson E.L. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]; Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology 2008; 10(5): 593-601. [DOI] [PubMed]
- 37.Ladeiro Y., Couchy G., Balabaud C. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology (Baltimore, Md) 2008;47(6):1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]; .Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology (Baltimore, Md) 2008; 47(6): 1955-63. [DOI] [PubMed]
- 38.Yao Y., Hu J., Shen Z. MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med. 2015;19(4):760–769. doi: 10.1111/jcmm.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yao Y, Hu J, Shen Z, et al. MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. Journal of cellular and molecular medicine 2015; 19(4): 760-9. [DOI] [PMC free article] [PubMed]
- 39.Tang H., Deng M., Tang Y. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19(20):5602–5612. doi: 10.1158/1078-0432.CCR-13-1326. [DOI] [PubMed] [Google Scholar]; Tang H, Deng M, Tang Y, et al. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clinical cancer research: an official journal of the American Association for Cancer Research 2013; 19(20): 5602-12. [DOI] [PubMed]
- 40.Zhang H.F., Alshareef A., Wu C. miR-200b induces cell cycle arrest and represses cell growth in esophageal squamous cell carcinoma. Carcinogenesis. 2016;37(9):858–869. doi: 10.1093/carcin/bgw079. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang HF, Alshareef A, Wu C, et al. miR-200b induces cell cycle arrest and represses cell growth in esophageal squamous cell carcinoma. Carcinogenesis 2016; 37(9): 858-69. [DOI] [PMC free article] [PubMed]
- 41.Ning F., Zhou Q., Chen X. miR-200b promotes cell proliferation and invasion in t-cell acute lymphoblastic leukemia through NOTCH1. J Biol Regul Homeost Agents. 2018;32(6):1467–1471. [PubMed] [Google Scholar]; Ning F, Zhou Q, Chen X. miR-200b promotes cell proliferation and invasion in t-cell acute Lymphoblastic leukemia through NOTCH1. Journal of biological regulators and homeostatic agents 2018; 32(6): 1467-71. [PubMed]
- 42.Zhang Z., Xing T., Chen Y., Xiao J. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-beta1 exposure. Biomed Pharmacother. 2018;106:1135–1143. doi: 10.1016/j.biopha.2018.07.042. [DOI] [PubMed] [Google Scholar]; Zhang Z, Xing T, Chen Y, Xiao J. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-beta1 exposure. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 2018; 106: 1135-43. [DOI] [PubMed]
- 43.Lee B.S., Fujita M., Khazenzon N.M. Polycefin, a new prototype of a multifunctional nanoconjugate based on poly(beta-l-malic acid) for drug delivery. Bioconjug Chem. 2006;17(2):317–326. doi: 10.1021/bc0502457. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee BS, Fujita M, Khazenzon NM, et al. Polycefin, a new prototype of a multifunctional nanoconjugate based on poly(beta-L-malic acid) for drug delivery. Bioconjugate chemistry 2006; 17(2): 317-26. [DOI] [PMC free article] [PubMed]
- 44.Gonzalez A.M., Gonzales M., Herron G.S. Complex interactions between the laminin alpha 4 subunit and integrins regulate endothelial cell behavior in vitro and angiogenesis in vivo. Proc Natl Acad Sci U S A. 2002;99(25):16075–16080. doi: 10.1073/pnas.252649399. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gonzalez AM, Gonzales M, Herron GS, et al. Complex interactions between the laminin alpha 4 subunit and integrins regulate endothelial cell behavior in vitro and angiogenesis in vivo. Proceedings of the National Academy of Sciences of the United States of America 2002; 99(25): 16075-80. [DOI] [PMC free article] [PubMed]
- 45.Lian J., Dai X., Li X., He F. Identification of an active site on the laminin alpha4 chain globular domain that binds to alphavbeta3 integrin and promotes angiogenesis. Biochem Biophys Res Commun. 2006;347(1):248–253. doi: 10.1016/j.bbrc.2006.06.069. [DOI] [PubMed] [Google Scholar]; Lian J, Dai X, Li X, He F. Identification of an active site on the laminin alpha4 chain globular domain that binds to alphavbeta3 integrin and promotes angiogenesis. Biochemical and biophysical research communications 2006; 347(1): 248-53. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers for qRT-PCR and vector construct
Antibodies for flow cytometry and Western blot
Relationship between mir-200b-3p expression and pathological features in RCC patients
Venn - Screening the targets of miR-200b from three miRNA database websites (miRanda, TargetScan and miRTarBase) using TCGA KIRC dataset analyzing.