Abstract
In order to survive the exposure to acid pH, Escherichia coli activates molecular circuits leading from acid tolerance to extreme acid resistance (AR). The activation of the different circuits involves several global and specific regulators affecting the expression of membrane, periplasmic and cytosolic proteins acting at different levels to dampen the harmful consequences of the uncontrolled entry of protons intracellularly. Many genes coding for the structural components of the AR circuits (protecting from pH ≤ 2.5) and their specific transcriptional regulators cluster in a genomic region named AFI (acid fitness island) and respond in the same way to global regulators (such as RpoS and H-NS) as well as to anaerobiosis, alkaline, cold and respiratory stresses, in addition to the acid stress. Notably some genes coding for structural components of AR, though similarly regulated, are non-AFI localised. Amongst these the gadBC operon, coding for the major structural components of the glutamate-based AR system, and the ybaS gene, coding for a glutaminase required for the glutamine-based AR system. The yhiM gene, a non-AFI gene, appears to belong to this group. We mapped the transcription start of the 1.1 kb monocistronic yhiM transcript: it is an adenine residue located 22 nt upstream a GTG start codon. By real-time PCR we show that GadE and GadX equally affect the expression of yhiM under oxidative growth conditions. While YhiM is partially involved in the RpoS-dependent AR, we failed to detect a significant involvement in the glutamate- or glutamine-dependent AR at pH ≤ 2.5. However, when grown in EG at pH 5.0, the yhiM mutant displays impaired GABA export, whereas when YhiM is overexpressed, an increases of GABA export in EG medium in the pH range 2.5–5.5 is observed. Our data suggest that YhiM is a GABA transporter with a physiological role more relevant at mildly acidic pH, but not a key component of AR at pH < 2.5.
Keywords: inner membrane proteins, acid resistance, Escherichia coli, GadX, GadE, anaerobiosis
1. Introduction
The ability to sense and respond to a mild-to-harsh acid stress (from pH 6.0 down to pH 2.0) is essential for neutralophilic bacteria, such as Escherichia coli, because inorganic and organic acid stresses encountered in the external environment, but also in host districts such as the gastric and distal gut compartments as well as the macrophage phagolysosome, can lead to bacterial death [1],[2]. Several studies have provided insight into the environmental and regulatory factors affecting the expression of the genes belonging to the most potent acid resistance (AR) system in the enteric bacterium Escherichia coli K12, namely the glutamate-dependent AR (GDAR or AR2) system (For reviews: [1],[3]). In order to function the E. coli GDAR system requires glutamate supplementation in the pH 2.5 minimal medium used for acid challenge [4], the intracellular proton-consuming activity of at least one of the two isoforms of glutamate decarboxylase (GadA or GadB) and the antiport activity of GadC, an inner membrane protein which exchanges exogenous glutamate for γ-aminobutyrate (GABA), the decarboxylation product [5],[6].
Acid stress, starvation, hyperosmolar stress, anaerobiosis, cold and respiratory stress were shown to cause significant up-regulation of most of GDAR genes (Figure 1A and references therein). The activation of these genes is essentially controlled via two circuits which function under different growth conditions: the cross-talking phosphorelay EvgSA and PhoQP circuits become operative when exponentially growing cells are cultivated in minimal medium at mildly acidic pH and under a low Mg2+ concentration, respectively, whereas the GadXYW circuit plays a major role when cells grown in rich medium enter into the stationary phase [7],[8],[9]. Both circuits converge on the activation of the regulator GadE, the role of which is essential to fully develop GDAR [10], as also reported for pathogenic E. coli [3]. The genes involved in GDAR encode in addition to the two paralogs of glutamate decarboxylase (gadA and gadB) and the glutamate/GABA antiporter (gadC), also two periplasmic chaperons (hdeA and hdeB), and specific transcriptional regulators belonging either to the AraC-family (gadX, gadW, ydeO) or to the LuxR-family (gadE and yhiF). Though less extensively investigated, the protein products of the slp, yhiD and hdeD genes also contribute to AR [11]. With the notable exception of ydeO and gadBC, all the above listed genes are clustered at approximately 3.66 Mb on the E. coli K12 chromosome, in the acid fitness island (AFI) [12]. The gene ybaS (glsA) coding for a glutaminase is also distantly located from AFI and gadBC, and plays a key role in the glutamine-dependent AR system (AR2_Q), which also requires GadC for the entry of glutamine and the export of glutamate or GABA [1],[13],[14],[15]. Besides the activating circuits, the TorSR phosphorelay system was shown to negatively affect GDAR genes expression under alkaline conditions [16]. As part of the complex network controlling the expression of these AR genes, RpoS, the general stress sigma factor of the RNA polymerase, and H-NS, a global transcriptional regulator acting mostly as gene silencer, are both playing an important role as they positively and negatively influence the expression of the above genes, respectively [17],[18].
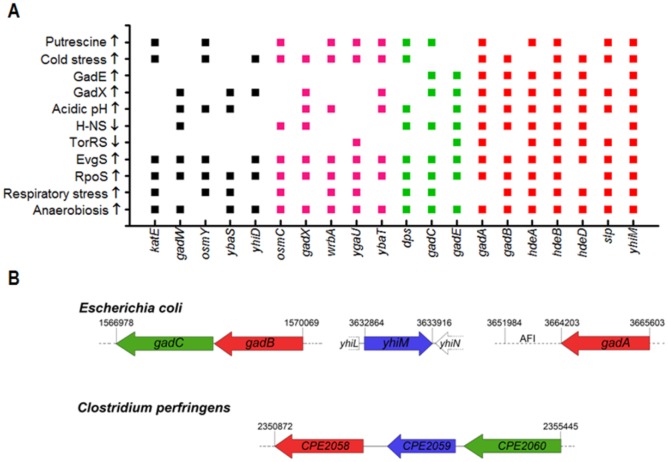
Figure 1. Preliminary observations suggesting that yhiM might belong to the AR circuit.
(A) In the graph the genes found in 5–6 (black squares), 7 (magenta squares), 8 (green squares), 9–11 (red squares) out of the 11 transcriptomic studies taken under consideration [16],[17],[19]–[23],[25],[26],[28],[29],[30] are listed for each group in alphabetical order. The environmental stimuli and the regulators analysed in each transcriptomic study are listed on the Y axis and arrows show up-regulation (↑) or down-regulation (↓) of the reported genes. (B) Schematic representation of the genomic regions containing the gadB, gadC, gadA and yhiM genes in E. coli K12 MG1655 and its homologues in Clostridium perfringens str.13. Orthologs and paralogs are depicted in the same colour for both microorganisms. The 5′ end of the E. coli AFI is indicated.
Notably, in several transcriptomic studies in addition to ybaS, yhiM, another non-AFI gene, is frequently found in the list of genes regulated alike many AR genes (Figure 1A). In particular yhiM is activated under acidic, cold and respiratory stresses as well as in anaerobic conditions, repressed by H-NS and TorSR, in addition to being RpoS-dependent and activated by EvgAS, GadE and GadX [16],[18],[19]–[26]. In this report we analyse in further detail the transcriptional regulation of the yhiM gene in the E. coli strains MC4100 and MG1655, a widespreadly used strain and a reference K12 strain for acid resistance studies, respectively. Respect to what was suggested by other authors using a less-characterized E. coli strain UCB [27], in MC4100 and MG1655 we observed only a partial involvement of the inner membrane protein YhiM in the RpoS-dependent AR system, but did not observe a clear involvement in the glutamate- or glutamine-dependent AR at pH 2.5. On the other hand, we observed that YhiM affects GABA export at pH 5.0 in minimal medium.
2. Materials and Methods
2.1. Bacterial strains, plasmids and growth media
The bacterial strains and the plasmids used in this study are listed in Table 1. Bacterial growth was monitored by determining the optical density at 600 nm (OD600) with a diode array spectrophotometer (HP8452; Agilent Technologies). The media used were: LB [31], LB-MES (LB buffered at pH 5.5 with 100 mM morpholineethanesulfonic acid [MES]) or brought to pH 2.5, LB-MOPS (LB buffered at pH 7.4 with 100 mM 3-[N-Morpholino]propanesulfonic acid [MOPS]) and EG, minimal medium E [32] supplemented with 0.4% glucose, at the desired pH. Antibiotics were added at the following concentrations: ampicillin, 100 µg/ml; streptomycin, 20 µg/ml; kanamycin, 25 µg/ml. The ΔyhiM and ΔgadC deletion mutants from the E. coli K12 strains MG1655 and MC4100 were generated using the procedure described by Datsenko and Wanner [33] with the oligonucleotides pairs ΔyhiM_for-ΔyhiM_rev and ΔgadC_for-ΔgadC_rev, respectively (Table 2). The replacement of the yhiM (or gadC) ORF with a KanR cassette, following a double crossing-over on the target gene, was checked by comparing the size of the PCR products obtained by amplifying the genomic DNA of the wild type and the mutant strains with pairs of primers annealing upstream and downstream of the deleted ORFs yhiM_for/yhiM (Bgl)_rev, for the yhiM mutant; α/β_out/gadC_rev, for the gadC mutant (Table 2).The insertion of the KanR cassette was checked with oligo k1 (specific for the cassette) and yhiM (Bgl)_rev, for the yhiM mutant or gadC_rev, for the gadC mutant (Table 2). Both mutants did not show any growth defect in LB and EG media at neutral and mildly acidic pH, when growth was carried out at 37 °C.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or Plasmid | Relevant Genotype | Reference or source |
| MC4100 | F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC ptsF25 rbsR | [34] |
| MG1655 | F− λ− rph−1 | GSC |
| JM109 | (F' traD36 proA+ proB+ lacIq lacZΔM15) recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) | [31] |
| MC4100gadX | MC4100 gadX::KanR | [9] |
| MC4100gadE | MC4100 gadE::KanR | [35] |
| MG1655gadX | MG1655 gadX::KanR | [9] |
| MG1655gadE | MG1655 gadE::KanR | [35] |
| MG1655yhiM | MG1655 yhiM::KanR | This work |
| MC4100yhiM | MC4100 yhiM::KanR | This work |
| MG1655gadC | MG1655 gadC::KanR | [36] |
| MC4100gadC | MC4100 gadC::KanR | This work |
| pBs | (pBluescriptSK) multicopy phagemid vector; ColE1 replicon, lacZ bla | Stratagene |
| pCRIIyhiM | 1050-bp fragment encompassing the entire yhiM coding sequence ligated in pCRII-TOPO | This work |
| pQE60 | Expression vector; ColE1 replicon, PT5-lacO RBSII | QIAGEN |
| pQEyhiM-6×His | 1050-bp fragment encompassing the entire yhiM coding sequence ligated to the NcoI-BglII sites of pQE60 | This work |
Table 2. Oligonucleotides used in this study.
| Primer | Sequence | Restriction site | |
| ΔyhiM_for | 5′-ATTATCATTAATGCATATTTCAATATTAGCAGGGATACC gtgtaggctggagctgcttc-3′ |
||
| ΔyhiM_rev | 5′-CCGCTTCTAATATTGAAACGATTGAGAACAACGTAAAGC attccggggatccgtcgacc-3′ |
||
| ΔgadC_for | 5′-TACCGTTTTAGGGGGATAATATGGCTACATCAG TACAGACgtgtaggctggagctgcttc-3′ |
||
| ΔgadC_rev | 5′-TTAGTGTTTCTTGTCATTCATCACAATATAGTGTGGTGAA attccggggatccgtcgacc-3′ | ||
| yhiM_revRT | 5′-TAACAGTGCCCAACCCCATATCAT-3′ | ||
| yhiM_for | 5′-ggataccatgGTGAACATATATATCGGGTGG-3′ | NcoI | |
| yhiM (Bgl)_rev | 5′-ggttagatctTTTTTTAGCAGAACCCGCTTC-3′ | BglII | |
| gadC_rev | 5′-GCACATATGGATATCTGCTCCC-3′ | ||
| α/β_out | 5′-CGAAACTGCAGGGTATTGC-3′ | ||
| rRNA16S_for | 5′-CGTTACCCGCAGAAGAAGC-3′ | ||
| rRNA16S_rev | 5′-GTGGACTACCAGGGTATCTAATCC-3′ | ||
| k1 | 5′-CAGTCATAGCCGAATAGCCT-3′ | ||
Restriction sites are underlined. Nucleotides not matching the target sequence are in lower-case.
2.2. RNA analysis
RNA was isolated from cells grown under the specified conditions, using either a modified hot-phenol extraction method [5] or the RNeasy mini kit (QIAGEN). RNA concentration and quality were assessed by determining (in 0.1 N NaOH) the optical density at 260 nm (OD260) and the 260 nm/280 nm ratio, respectively. Electrophoresis was carried out essentially as previously described [5] loading 5 µg of total RNA per sample. To detect yhiM-specific mRNA, northern blots were hybridized with a 1050-bp yhiM probe obtained by PCR amplification using plasmid pCRIIyhiM (see below) as template and the oligonucleotide pair yhiM_for and yhiM (Bgl)_rev (Table 2). The yhiM-specific probe was labelled and hybridized with the DIG High Prime Labeling and Detection Starter Kit II (Roche), according to manufacturer instructions.
Primer extension analysis was performed with the oligonucleotide yhiM_revRT (Table 2), which anneals 286 nt downstream the putative GTG start codon, and using total RNA (7.0 µg) extracted from either E. coli MC4100 or E. coli MG1655 grown to the stationary phase in LB-MES, pH 5.5. The entire procedure was essentially as previously described [5].
2.3. RT-qPCR
Quantitative real time RT-PCR (RT-qPCR) reactions were performed in two steps. Reverse transcription of DNase-treated RNAs (3 µg) was carried out using 10 U Transcriptor Reverse Transcriptase (Roche) and oligonucleotides rRNA16S_rev, and yhiM_revRT (Table 2). Real Time PCR was performed on a Chromo4 Real Time PCR Instrument (BioRad Laboratories) using the LightCycler FastStart DNA Master SYBR Green I (Roche) and the following pairs of oligonuleotides (Table 2): yhiM_for-yhiM_revRT for amplification of yhiM-specific mRNA, and rRNA16S_for-rRNA16S_rev for amplification of the 16S rRNA (internal control).
2.4. Cloning of yhiM
A 1050-bp DNA fragment encompassing the entire yhiM open reading frame was generated by PCR using Fast Start High Fidelity Enzyme (Roche) and the E. coli MG1655 genomic DNA as template, with the oligonucleotides yhiM_for and yhiM (Bgl)_rev (Table 2). The amplicon was subcloned into pCRII-TOPO, using the TOPO TA Cloning System (Invitrogen) and sequenced on both strands. The NcoI-BglII fragment from pCRIIyhiM was then ligated to the corresponding sites of the expression vector pQE60, linearized with the same restriction enzymes. In the newly generated plasmid construct, named pQEyhiM-6×His, the yhiM gene is cloned in frame at its 3′ end with the nucleotide sequence coding for a 6×His tag carried by pQE60. This plasmid pQEyhiM-6×His was used to transform competent cells of E. coli JM109/pREP4.
To assay YhiM-directed GABA export, single colonies of the E. coli strains JM109/pREP4/pQE60 and JM109/pREP4/pQEyhiM-6×His were inoculated in 3 ml of LB, containing ampicillin and kanamycin, grown to the late exponential phase and then induced by addition of 0.2 mM IPTG. Growth was prolonged overnight, after which cultures were centrifuged at 3500 rpm for 10 min and the pellet resuspended in 1 ml EG, pH 7.0. The washing and resuspension steps were repeated one more time. Aliquots of 25 µl were transferred in 475 µl of EG medium, at different pH (2.5, 3.5, 4.5 and 5.5), containing 1 mM sodium glutamate. Incubation was carried out for 90 min at 37 °C and the reactions were stopped and neutralized by adding 4 µl of 5 N NaOH. Following a centrifugation step to pellet the bacteria, 50 µl of the reaction mixtures were assayed with the Gabase assay, as previously described [37]. Cell viability was checked by plating out the bacteria before and after the challenge at the indicated pH.
2.5. Cell fractionation
Cytoplasmic and membrane fractions from E. coli wild-type strain MC4100 and its mutant derivative MC4100ΔyhiM were obtained as described by Capitani et al. [38] with some minor modifications. Briefly, after 24-hours growth in LB, the bacterial pellet from a 0.5 L culture was resuspended in 25 ml of 50 mM Tris-HCl, pH 7.5, containing 1 mM DTT and a protease inhibitor cocktail (Complete, Roche). The resuspension was then divided into two 13-ml aliquots. One of the aliquots was brought to pH 5.6 by dropwise addition of 8 N HCl. The two samples were then sonicated and if necessary the pH was adjusted to 7.4 and 5.6, respectively. An additional slow speed centrifugation step was carried out to remove cell debris and unbroken cells, thus generating the cell extract. The cytoplasmic and membrane fractions from each cell extract were separated by ultracentrifugation at 50,000 rpm (Beckman model L8-70) for 1 h at 10 °C. The pellet (corresponding to the membrane fraction) was resuspended in 2 ml of 100 mM Tris-HCl, pH 8.0, containing 150 mM NaCl, 5 mM EDTA and 0.5% lauroyl sarcosine.
2.6. AR assays
Oxidative and fermentative AR systems were assayed essentially as described by Lin et al. [4]. Briefly, 24-hours cultures in either LBG, pH 5.0 (for fermentative AR) or LB-MES, pH 5.5 (for oxidative AR) were acid challenged by 1:1000 dilution in rich medium at pH 2.5 or minimal medium EG at pH ≤ 2.5, supplemented when required with the relevant amino acids to test for the fermentative amino acid-dependent AR systems.
2.7. Gad activity assay and GABA measurements
Glutamate decarboxylase activity assays and GABA measurements were performed as previously described [37].
3. Results and Discussion
3.1. Transcriptional regulation of yhiM
As mentioned in the Introduction, in several transcriptomic studies the E. coli yhiM gene, a non-AFI gene, is frequently found in the list of genes regulated alike many AR genes (Figure 1A). In the E. coli K12 reference strain MG1655 the yhiM gene (b3491) is located in between the divergently oriented yhiL and yhiN genes (Figure 1B), thus suggesting that its transcript is monocistronic. The genomic context in E. coli is rather different from that found in Clostridium perfringens str.13, where the gene CPE2059 coding for a 351 amino acid-long protein homologous to E. coli YhiM, is located in between the genes CPE2060 and CPE2058, coding for the homologs of E. coli GadC and GadB, respectively (Figure 1B and [12]). A similar gene assembly is also present in other strains of C. perfringens for which the genome sequence is available (data not shown).
These preliminary observations, i.e. co-regulation with AR genes (Figure 1A) and co-localization with the genes coding for GadB and GadC in C. perfringens (Figure 1B), prompted us to further investigate both the transcriptional regulation of E. coli yhiM and the biological role played by its protein product, in light of a possible involvement in AR and specifically in GDAR. As a further reason of interest, yhiM codes for an integral inner membrane protein belonging to a protein family, DUF2776, the function of which is not known.
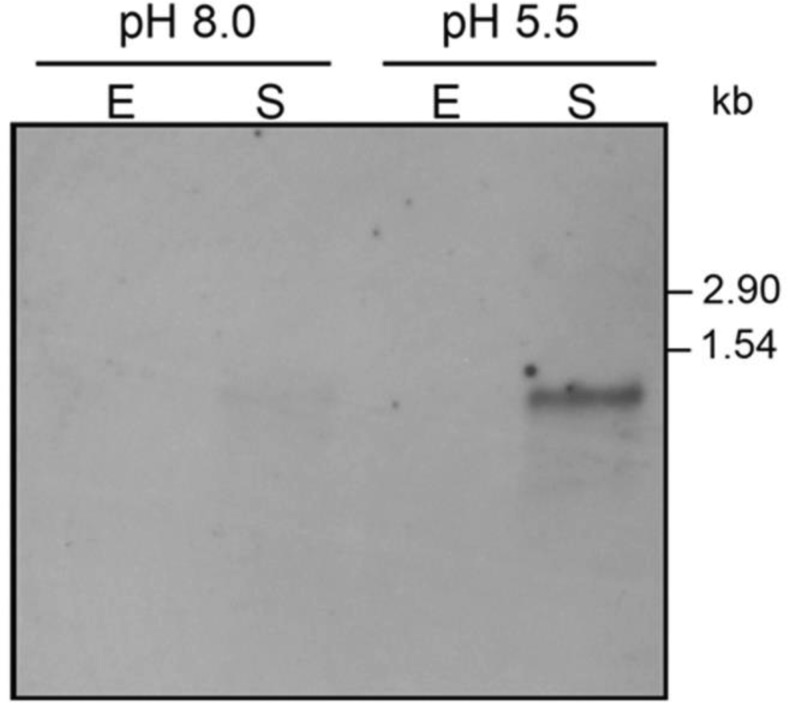
To study yhiM transcriptional regulation, total RNA was extracted from E. coli K12 MG1655 cells grown to the exponential or stationary phases either in LB-MOPS medium (pH 7.4) or in LB-MES medium (pH 5.5). Northern blots of total RNA (5 µg/sample), resolved on 1.2% agarose gels, were hybridised with a DIG-labelled yhiM-specific probe. Figure 2 shows that the yhiM probe detects a single mRNA species with an apparent length of 1.1 kb, as deduced from its electrophoretic mobility. The yhiM mRNA is detectable only in the stationary phase samples, particularly in that obtained from bacteria grown at pH 5.5 under oxidative conditions, i.e. LB-MES pH 5.5. Similar results were obtained when RNA extracted from E. coli MC4100 was analysed by Northern blot (data not shown).
Figure 2. Northern blot analysis of yhiM-specific transcript.
Total RNA was extracted from E. coli strain MG1655 grown to the exponential (E, OD600 ∼ 0.6) and stationary (S, OD600 ∼ 2) phases, in LB-MOPS (pH 8.0) or LB-MES (pH 5.5).
The yhiM transcription was then analysed quantitatively by real time PCR in E. coli MG1655 and in its isogenic gadX and gadE derivatives grown in different media and conditions (Table 3). We found that the yhiM transcript is 9 folds more abundant at pH 7.4 in exponential phase under anaerobic conditions and 12.5 folds more abundant in the stationary phase of growth at pH 5.5 with respect to the exponential phase under aerobic conditions. In the gadX and gadE mutants, analysed under the most inducing condition (i.e. stationary phase in LB-MES pH 5.5), yhiM expression is still higher than in the stationary phase at pH 7.4, but nevertheless reduced by half with respect to that observed in the wild-type strain at pH 5.5 (Table 3, last three columns).
Table 3. Expression fold change of the yhiM-specific transcript in E. coli MG1655 in different conditions and mutant backgrounds as assessed by quantitative real time PCR.
| Strain | MG1655 | ΔgadX | ΔgadE | |||
| Medium | LB-MOPS pH 7.4 | LB-MES pH 5.5 | ||||
| Condition | E-O2 | E | S | S | ||
| Expr. fold | 9 | 1 | 1.5 | 12.5 | 5.7 | 6 |
| SEM | 3 | 0.08 | 1.5 | 1.1 | 1.2 | |
| P value | 0.0001 | 0.01 | 0.002 | 0.002 | 0.006 | |
Total RNA was extracted from E. coli K12 strain MG1655 and its ΔgadX and ΔgadE derivatives grown to the exponential aerobic (E), exponential anerobic (E-O2) and stationary (S) phases, in the indicated medium.
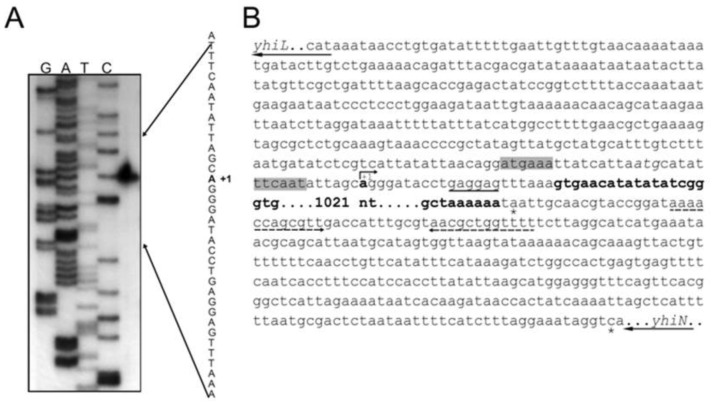
In order to localize more precisely the –10 and –35 promoter elements (also in light of more in depth transcription regulation studies) and to drive the design of primers correctly placed for mutant construction, primer extension analysis was conducted to identify the 5′-terminus of the yhiM mRNA using total RNA extracted from E. coli MC4100 grown to the stationary phase in LB-MES, pH 5.5. Results reported in Figure 3A show that transcription of yhiM originates from an adenine (A) residue located 22 nt upstream a GTG codon, which functions as a translation start in E. coli [40]. An identical result was obtained when total RNA from E. coli MG1655 was used instead (data not shown). At compatible distance from the 5′ end of the yhiM transcript, we identified –35 (ATGAAA) and –10 (TTCAAT) promoter elements, optimally spaced by 17 nt (Figure 3B). Both –10 and –35 sequences match 4 out of 6 nucleotides of the recognition sites for σ70-dependent RNA polymerase. Moreover, the sequence GAGGAG-N6, immediately upstream the GTG start codon, is fully compatible with the Shine-Dalgarno sequence for ribosome binding (Figure 3B). The 1127-nt length of the yhiM transcript, deduced from the distance between the experimentally determined transcription start site and a putative stem-loop structure centred 35 nt downstream the yhiM stop codon (Figure 3B), perfectly agrees with the predicted length of the transcript as deduced by Northern blot analysis (Figure 2).
Figure 3. Sequence analysis and mapping of the yhiM transcriptional start site.
(A) Mapping of the 5′' end of the yhiM transcript by primer extension analysis. RNA was extracted from E. coli MC4100 stationary phase cells grown at pH 5.5 and reverse transcribed after priming with the oligonucleotide yhiM_revRT (Table 2), labelled at its 5′-end. Lanes G, A, T, and C are sequencing ladders of pCRIIyhiM with the same oligonucleotide used for the primer extension reaction. Sequencing reactions were run in parallel with the cDNA transcript (right lane). (B) Nucleotide sequence of the yhiL-yhiN genome region in E. coli MG1655. The bent arrow indicates the yhiM transcriptional start site at the residue in bold (+1). The sequences for the –10 and –35 putative promoter elements are on gray background. The potential Shine-Dalgarno sequence for ribosome binding is underlined. The yhiM ORF is in bold. The asterisks indicate the yhiM and yhiN stop codons. The dashed arrows indicate the inverted repeats probably generating a stem-loop like structure. The black arrows at the 5′ and 3′ ends of the sequence show the start and stop triplets as well as the direction of transcription of the yhiL and yhiN genes, respectively.
3.2. YhiM does not affect GadA/B and GadC cellular localization and is not a key component of GDAR
E. coli YhiM, starting from the methionine residue coded by the GTG alternative start codon, is a 350 amino acid-long protein with a calculated molecular mass of 37.66 kDa and a theoretical pI 8.2. A BLAST [41] search, using E. coli YhiM (Swiss-Prot: P37630) as query sequence, retrieved as best scoring sequences (>45% identity) those from members of a protein family (pfam10951: DUF2776) for which a biochemical function has not been assigned yet. The Clustal Omega alignment of E. coli YhiM and its homologues from Bordetella avium, Laribacter hongkongensis, C. perfringens, Bacteroides tethaiotaomicron and Parabacteroides distasonis is provided in the Supplementary Figure 1. Based on several topology prediction programs (www.expasy.org), E. coli YhiM consists of 10 transmembrane (TM) spanning domains, with both the N-terminal and C-terminal ends on the cytoplasmic side. The amino acids predicted in TM domains are indicated by black lines above the alignment in Supplementary Figure 1. Overall the alignment shows that the degree of identity at the level of the predicted TM domains (35% on average) is that mostly contributing to the overall identity.
In order to answer the question on the role played by YhiM, we first investigated a possible involvement in the physical and/or functional interaction with GadA/B or GadC. In fact, one possibility is that YhiM may provide the membrane anchoring site for GadB when it is recruited to the membrane district upon acidification of the cytoplasm [38]. Indeed, it was shown that in the cell extracts from the E. coli strain JM109 overexpressing GadB, the enzyme at neutral pH is mainly localized in the cytoplasmic fraction, whereas at mildly acidic pH (pH 5.6) more than 55% of the protein is detected and assayed in the detergent-solubilized membrane fraction [38]. To date the chemical nature (protein or lipid) of the anchoring partner is still not known. Experiments carried out in our laboratory suggest that recruitment of GadB to the membrane does not occur via GadC, the cognate membrane antiporter (data not shown) in GDAR. In order to investigate whether YhiM could be involved in the pH-dependent recruitment of glutamate decarboxylase to the membrane, both the cytosolic and membrane fractions from the E. coli strain MC4100 and its ΔyhiM derivative were assayed for Gad activity both at pH 7.4 and pH 5.6. The results are shown in Table 4 and provide evidence that, as in the overexpressing strain [38], in the wild-type strain at acidic pH native glutamate decarboxylase (consisting of a mixture of GadA and GadB; [37]) partitions between the cytosolic and membrane fractions and that in the absence of yhiM no major changes are observed. According to these results, we can exclude that YhiM is the protein partner recruiting GadA/B to the membrane and also exclude that the yhiM mutation affects the GadA/B activity in the cell. In fact the total enzyme units in wild type MC4100 (300 Units/OD600) are very close to those measured in the ΔyhiM mutant (330 Units/OD600).
Table 4. Partition of Glutamate decarboxylase (GadA/B) activity* in the cytosolic (Cyto) and membrane (Mem) fractions from stationary phase cellular extract brought to pH 7.4 or 5.6.
| pH 7.4 | pH 5.6 | |||
|
Cyto |
Mem |
Cyto |
Mem |
|
| MC4100 | 265 ± 21 | 75 ± 6 | 146 ± 11 | 124 ± 9 |
| MC4100ΔyhiM | 275 ± 23 | 56 ± 4 | 190 ± 8 | 143 ± 5 |
* the activity is reported in Units/mg.
Being a membrane protein, an alternative role played by YhiM could be that of participating to GDAR either by physically interacting with GadC or by functionally overlapping its function. Therefore we analysed the effect of yhiM deletion on both the GadC localization in the cell membrane and the involvement in the GDAR phenotype.
Immunoblot analysis of total cell extracts and membrane fractions from the E. coli wild-type strain MC4100 and its ΔyhiM derivative, using anti-GadC antibodies, showed that GadC expression and localization in the cell membrane are not altered in the absence of YhiM (data not shown).
Also GDAR was not affected in the absence of a functional yhiM gene. Using the standard conditions to assay the RpoS-, glutamate- and glutamine-dependent AR phenotypes, we analysed both the ΔyhiM and the ΔgadC strains and compared their AR phenotype with that of wild-type MG1655. The results (Figure 4) are in disagreement with the data provided by Nguyen and Sparks-Thissen [27] as we failed to observe any major involvement of yhiM in acid resistance, with only the RpoS-dependent circuit being slightly affected. On the contrary the gadC mutation is sufficient to give rise to an acid sensitive phenotype, as expected from previous studies [5],[6].
Figure 4. Participation of E. coli yhiM to fermentative and oxidative AR systems.
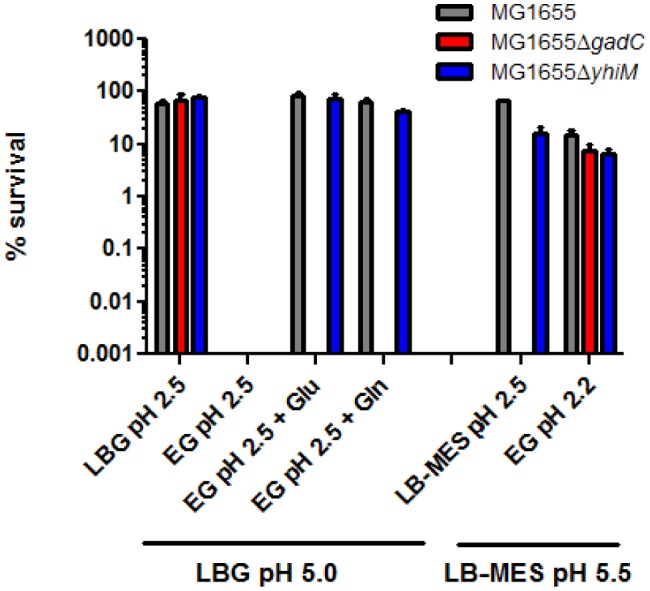
Wild type MG1655 and the isogenic gadC and yhiM mutants were tested for different AR phenotypes. G glutamate- and glutamine-dependent AR were assayed by diluting 1:1000 24-hours cultures grown in LBG pH 5.0 in the indicated media. The oxidative AR system activity was assayed by diluting 1:1000 24-hours cultures grown in LB-MES pH 5.5 in the indicated media.
It has been reported that MG1655 survives extremely well in pH 2.5 spent EG medium, whereas it succumbs in fresh EG at pH 2.5, in the absence of glutamate. A likely explanation for this observation is that glutamate secretion, which is proposed to originate by metabolic overflow [42], would provide sufficient substrate in the spent medium to support GDAR, even in the absence of glutamate supplementation. We therefore assayed GDAR in spent EG medium, acidified to pH 2.5, obtained from either E. coli MG1655 or MG1655ΔyhiM grown for 14–15 hours. Wild type MG1655 survived well in both acidified spent challenge media (data not shown). This suggests that the ΔyhiM strain is not impaired in the ability to export glutamate (or any other molecule) required to protect against acid challenge in spent EG medium.
3.3. YhiM participates to GABA export in E. coli at acidic pH
It has been reported that yhiM is amongst the genes significantly up-regulated in an E. coli cydAB mutant, in which the terminal oxidase cytochrome bd-I, typically induced under microaerobic conditions, is missing [19]. Based on experimental evidence, the authors suggest that GadC might substitute for terminal oxidase-mediated (cytochrome bd-I and bo') proton translocation, thus contributing to proton motive force (PMF) maintenance. Therefore the possibility exists that yhiM codes for a membrane protein involved in GABA export at pH ≥ 5, where gadC is expected to be inactive [43]. In addition to this the localization of the E. coli yhiM homolog in C. perfringens in between the gadB and gadC genes (Figure 1A) suggests that YhiM might be involved in GABA transport. In order to test this possibility, we measured the amount of GABA exported by wild type E. coli MC4100 and by its ΔyhiM and ΔgadC isogenic derivatives grown in minimal medium EG, pH 5.0, supplemented with 10 mM glutamate. Aliquots of the growth medium were withdrawn at time intervals and assayed for GABA content. During growth the ΔyhiM strain exports in the extracellular medium significantly less GABA (50% on average) than the wild-type and ΔgadC strains, with the latter mutant strain showing no effect on GABA export under the conditions used for the assay (Figure 5A), in line with a role of GadC more restricted to pH values ≤ 3 [43]. Though less pronounced, similar results were obtained when growth was carried out in rich LBG medium (LB, pH 5.0, containing 0.4% glucose) or using E. coli MG1655 and its yhiM and gadC deletion mutants (data not shown).
Figure 5. GABA export in the E. coli yhiM mutant and overexpressing strains.
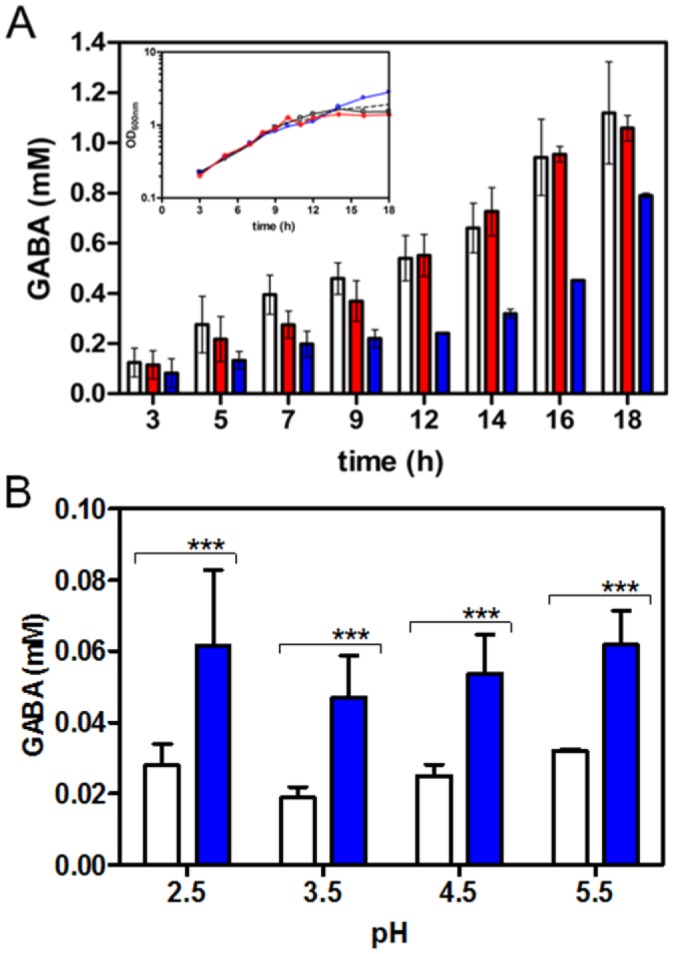
(A) The E. coli strains MC4100 (white) and its knockout mutants MC4100yhiM (blue) and MC4100gadC (red) were grown in minimal medium EG, pH 5.0, supplemented with 10 mM sodium glutamate. At the indicated time intervals 1-ml aliquots were collected, the OD600 was measured and GABA in the spent medium quantified. The values are means ± standard deviation of at least 3 independent cultures. Inset: the growth profiles (OD600) of MC4100 (black), MC4100yhiM (blue) and MC4100gadC (red) are reported. (B) Cells from IPTG-induced cultures of the E. coli strains JM109/pREP4/pQE60 (white) and JM109/pREP4/pQEyhiM-6×His (blue) were incubated at 37 °C for 90 min in EG medium, at the indicated pH, supplemented with 1 mM sodium glutamate. The GABA exported was assayed as previously described [37]. The values are means ± standard deviation of 3 independent experiments (in duplicate) with p-value of 0.0005 (***).
In order to obtain further evidence on the involvement of YhiM in GABA export, we compared the extracellular GABA produced by the E. coli strain JM109/pREP4/pQEyhiM-6×His with that produced by the strain JM109/pREP4 carrying the empty expression vector pQE60. The ability to export GABA was assayed by incubating at 37 °C for 90 min a fixed amount of cells (4–5 × 107) in 0.5 ml of EG medium, at different pHs, containing 1 mM glutamate. The results shown in Figure 4B confirm that YhiM is involved in GABA export and that its activity is not significantly affected by the pH of the medium. Together these results suggest that YhiM contributes to GABA export in E. coli.
4. Conclusions
In the present work we have analysed in further detail the transcriptional regulation of yhiM and its function in E. coli. Our data suggest that YhiM is dispensable for GDAR and thus its function does not overlap that of GadC, which remains a key structural component of AR under both fermentative and oxidative conditions. However, while the GABA export activity of GadC is restricted to extremely acidic pH conditions [43], as confirmed by the unaffected ability to export GABA in minimal medium at pH 5.0 (Figure 5A) of the otherwise acid-sensitive gadC mutant (Figure 4). As a matter of fact, the yhiM mutant is not impaired in GDAR (or in glutamine-dependent AR), though it exports less GABA than the parental strain in minimal medium EG at pH 5.0. Thus, we conclude that YhiM has an involvement in GABA export, though we noticed some differences among the strains under analysis (i.e. between MC4100 and MG1655). Heterogeniety at the strain level, might also provide an explanation on why we could not obtain results in agreement with those obtained by other authors, using a different E. coli strain and a different approach for mutagenesis (tn10 insertion) [27].
While yhiM expression and function are not strictly related to AR, they might be linked to other physiological functions which also the GDAR genes participate to. Amongst these functions there are the adaptations of E. coli cells to respiratory stress and anaerobiosis, conditions in which many GDAR genes as well as yhiM are induced (Figure 1A and Table 3). The role played by YhiM is still elusive in the E. coli physiology and will requires further work to be clearly assessed, however the high similarity (> 60%) in amino acid sequence amongst YhiM homologs in bacteria from different phyla (See Supplementary Figure 1) reinforces the hypothesis that this protein might play an important role in bacterial cell physiology. As a matter of fact, a recent report suggests an involvement of YhiM at high temperatures and low osmolarity [44].
Footnotes
Conflict of Interest: All authors declare no conflicts of interest in this paper.
References
- 1.Lund P, Tramonti A, De Biase D. Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol Rev. 2014;38:1091–1125. doi: 10.1111/1574-6976.12076. [DOI] [PubMed] [Google Scholar]
- 2.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 3.De Biase D, Lund PA. The Escherichia coli Acid Stress Response and Its Significance for Pathogenesis. Adv Appl Microbiol. 2015;92:49–88. doi: 10.1016/bs.aambs.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Lin J, Smith MP, Chapin KC, et al. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Biase D, Tramonti A, Bossa F, et al. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol. 1999;32:1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- 6.Castanie-Cornet MP, Penfound TA, Smith D, et al. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eguchi Y, Ishii E, Hata K, et al. Regulation of acid resistance by connectors of two-component signal transduction systems in Escherichia coli. J Bacteriol. 2011;193:1222–1228. doi: 10.1128/JB.01124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton NA, Johnson MD, Antczak P, et al. Novel aspects of the acid response network of E. coli K-12 are revealed by a study of transcriptional dynamics. J Mol Biol. 2010;401:726–742. doi: 10.1016/j.jmb.2010.06.054. [DOI] [PubMed] [Google Scholar]
- 9.Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol. 2008;70:965–982. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- 10.Sayed AK, Foster JW. A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol Microbiol. 2009;71:1435–1450. doi: 10.1111/j.1365-2958.2009.06614.x. [DOI] [PubMed] [Google Scholar]
- 11.Mates AK, Sayed AK, Foster JW. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J Bacteriol. 2007;189:2759–2768. doi: 10.1128/JB.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Biase D, Pennacchietti E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol Microbiol. 2012;86:770–768. doi: 10.1111/mmi.12020. [DOI] [PubMed] [Google Scholar]
- 13.Tsai MF, McCarthy P, Miller C. Substrate selectivity in glutamate-dependent acid resistance in enteric bacteria. Proc Natl Acad Sci USA. 2013;110:5898–5902. doi: 10.1073/pnas.1301444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma D, Lu P, Shi Y. Substrate selectivity of the acid-activated glutamate/gamma-aminobutyric acid (GABA) antiporter GadC from Escherichia coli. J Biol Chem. 2013;288:15148–15153. doi: 10.1074/jbc.M113.474502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu P, Ma D, Chen Y, et al. L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 2013;23:635–644. doi: 10.1038/cr.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bordi C, Theraulaz L, Mejean V, et al. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol Microbiol. 2003;48:211–223. doi: 10.1046/j.1365-2958.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- 17.Hommais F, Krin E, Laurent-Winter C, et al. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 18.Weber H, Polen T, Heuveling J, et al. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepherd M, Sanguinetti G, Cook GM, et al. Compensations for diminished terminal oxidase activity in Escherichia coli: cytochrome bd-II-mediated respiration and glutamate metabolism. J Biol Chem. 2010;285:18464–18472. doi: 10.1074/jbc.M110.118448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes ET, Wilks JC, Sanfilippo P, et al. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 2006;6:89. doi: 10.1186/1471-2180-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tucker DL, Tucker N, Ma Z, et al. Genes of the GadX-GadW regulon in Escherichia coli. J Bacteriol. 2003;185:3190–3201. doi: 10.1128/JB.185.10.3190-3201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tucker DL, Tucker N, Conway T. Gene expression profiling of the pH response in Escherichia coli. J Bacteriol. 2002;184:6551–6558. doi: 10.1128/JB.184.23.6551-6558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eguchi Y, Oshima T, Mori H, et al. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology. 2003;149:2819–2828. doi: 10.1099/mic.0.26460-0. [DOI] [PubMed] [Google Scholar]
- 24.Masuda N, Church GM. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J Bacteriol. 2002;184:6225–6234. doi: 10.1128/JB.184.22.6225-6234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kailasan-Vanaja S, Bergholz TM, Whittam TS. Characterization of the Escherichia coli O157:H7 Sakai GadE regulon. J Bacteriol. 2009;191:1868–1877. doi: 10.1128/JB.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito A, May T, Kawata K, et al. Significance of rpoS during maturation of Escherichia coli biofilms. Biotechnol Bioeng. 2008;99:1462–1471. doi: 10.1002/bit.21695. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen TM, Sparks-Thissen RL. The inner membrane protein, YhiM, is necessary for Escherichia coli (E. coli) survival in acidic conditions. Arch Microbiol. 2012;194:637–641. doi: 10.1007/s00203-012-0798-x. [DOI] [PubMed] [Google Scholar]
- 28.White-Ziegler CA, Davis TR. Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. J Bacteriol. 2009;191:1106–1110. doi: 10.1128/JB.00599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White-Ziegler CA, Um S, Perez NM, et al. Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology. 2008;154:148–166. doi: 10.1099/mic.0.2007/012021-0. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida M, Kashiwagi K, Shigemasa A, et al. A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J Biol Chem. 2004;279:46008–46013. doi: 10.1074/jbc.M404393200. [DOI] [PubMed] [Google Scholar]
- 31.Ausubel FM, Brent R, Kingston RE, et al. Current protocols in molecular biology. New York, NY: John Wiley and Sons; 1987. [Google Scholar]
- 32.Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casadaban MJ, Chou J, Cohen SN. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tramonti A, De Canio M, Delany I, et al. Mechanisms of transcription activation exerted by GadX and GadW at the gadA and gadBC gene promoters of the glutamate-based acid resistance system in Escherichia coli. J Bacteriol. 2006;188:8118–8127. doi: 10.1128/JB.01044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Occhialini A, Jimenez de Bagues MP, Saadeh B, et al. The glutamic acid decarboxylase system of the new species Brucella microti contributes to its acid resistance and to oral infection of mice. J Infect Dis. 2012;206:1424–1432. doi: 10.1093/infdis/jis522. [DOI] [PubMed] [Google Scholar]
- 37.De Biase D, Tramonti A, John RA, et al. Isolation, overexpression, and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr Purif. 1996;8:430–438. doi: 10.1006/prep.1996.0121. [DOI] [PubMed] [Google Scholar]
- 38.Capitani G, De Biase D, Aurizi C, et al. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. Embo J. 2003;22:4027–4037. doi: 10.1093/emboj/cdg403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin J, Lee IS, Frey J, et al. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riley M, Abe T, Arnaud MB, et al. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burkovski A, Kramer R. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl Microbiol Biotechnol. 2002;58:265–274. doi: 10.1007/s00253-001-0869-4. [DOI] [PubMed] [Google Scholar]
- 43.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186:6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson MA, Mann MD, Evans MA, et al. The inner membrane protein YhiM is necessary for Escherichia coli growth at high temperatures and low osmolarity. Arch Microbiol. 2016 doi: 10.1007/s00203-016-1288-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.