Abstract
Rapid and reliable assessment of mitochondrial bioenergetics is a vital tool in drug discovery studies aimed at reversing or improving mitochondrial dysfunction. Induced pluripotent stem cell (iPSC)-derived neural stem cells (NSCs) carry and replicate the donor disease pathology and can be an ideal cellular model for phenotypic screening of compounds. Herein we describe the use of Seahorse XFe96 analyzer to assess mitochondrial functions in iPSC-derived NSCs for drug screening.
Keywords: iPSCs, Mitochondrial functions, LRRK-2, Parkinson’s disease, Drug screening, Kinase inhibitors
1. Introduction
Mitochondria play a critical role in cellular metabolism, reactive oxygen species (ROS) production, and apoptosis. Thus, the assessment of mitochondrial functions provides valuable insight into the physiology and bioenergetics of the cells. Mitochondrial dysfunction has been linked to the pathogenesis of various neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and others [1]. Drugs targeting or improving mitochondrial functions could therefore be of interest for developing novel therapies. Investigation of these drugs requires rapid and reliable assessment of their effect on mitochondrial function. Current assays available for mitochondrial function can be broadly classified as either extracellular flux analyzers or complementary functional analyzers [2]. The extracellular flux analyzers enable a noninvasive quantification of mitochondrial respiration, whereas the complementary functional analyzers, such as ATP (adenosine triphosphate) assays, mitochondrial membrane potential assays, and ROS assay kits, measure specific functions of the organelle [3]. Although rapid, the complementary functional analysis methods are not extremely reliable due to the presence of artifacts and low sensitivity. Furthermore, these methods are insufficient in delineating underlying mechanisms of mitochondria-associated metabolism. In contrast, extracellular flux analyzers provide realtime measurements of changes in mitochondrial respiration and bioenergetics. The development of 96-well plate systems, such as the Seahorse XFe96 analyzer, has made it possible to screen compounds for their effect on mitochondrial energetics in real time and short span [4–6].
The XFe96 analyzer has been widely used to measure the changes in mitochondrial respiration and glycolysis in different cell types such as: neurons [7, 8], astrocytes [9, 10], iPSCs [11–13], lymphocytes [14], and cancer cells [15]. We describe a detailed methodology of using induced pluripotent stem cell (iPSC)-derived neural stem cells (NSCs) for drug screening with the XFe96 analyzer. The NSCs were generated and propagated using the NN1 media (see Chap. 1) with basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF). The dopaminergic (DA) neurons were generated from the NSCs as described in Chap. 7. NSCs or their differentiated progeny, DA neurons, astrocytes, and oligodendrocytes offer an authentic and reliable system for screening drugs that target mitochondrial dysfunction in the central nervous system (CNS). The protocol described can be used to investigate acute versus chronic effects of the compounds on oxygen consumption rate (OCR) and on extracellular acidification rates (ECAR) of the cells. Furthermore the OCR and ECAR values can be used to calculate additional parameters of mitochondrial bioenergetics.
2. Materials
2.1. Equipment
Seahorse XFe96 analyzer (Agilent, Seahorse Bioscience, North Billerica, MA, USA).
Seahorse XFe96 cell culture microplates (Agilent, Seahorse Bioscience, North Billerica, MA, USA).
Seahorse XFe96 sensor cartridges (Agilent, Seahorse Bioscience, North Billerica, MA, USA).
Hemocytometer (Hausser Scientific, Horsham, PA, USA) or automated cell counter (Countess, Invitrogen, Carlsbad, CA, USA).
Cell culture incubator (Nuaire, Plymouth, MN, USA).
37 °C non-CO2 incubator (Fisher Scientific, Pittsburgh, PA, USA).
Phase contrast microscope (Zeiss, Oberkochen, Germany).
pH meter (Corning/Fisher Scientific, Pittsburgh, PA, USA).
Centrifuge tubes (15–50 mL) (Corning, Oneonta NY, USA).
Syringe filters (Corning, Oneonta NY, USA).
60 mL syringes (BD, Franklin Lakes, NJ, USA).
Centrifuge (Eppendorf, Hamburg, Germany).
Multichannel pipettes (20–200 μL) (Eppendorf).
Reservoir basins (Fisher Scientific, Pittsburgh, PA, USA).
Pipette aids (10–1000 μL) (Eppendorf, Hamburg, Germany).
Pipette tips (10–1000 μL) (Accuflow, E&K Scientific, Santa Clara, CA, USA).
Water bath (Fisher Scientific, Pittsburgh, PA, USA).
2.2. Reagents
XF calibrant (Agilent, Seahorse Bioscience, North Billerica, MA, USA).
- Seahorse XF Cell Mito Stress Test Kit (Agilent, Seahorse Bio- science, North Billerica, MA, USA):
- Oligomycin.
- Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP).
- Rotenone.
NN1 media (Neoneuron, San Antonio, TX, USA).
Fetal bovine serum (FBS, GE, Logan, Utah, USA).
Accutase (Gibco, Life Technologies, NY, USA).
Trypsin neutralizer (Gibco, Life Technologies, NY, USA).
Poly-L-ornithine hydrobromide (Sigma-Aldrich, St. Louis, MO, USA).
Sterile phosphate buffered solution (PBS, Gibco, Life Technologies, NY, USA).
Double-distilled water (Gibco, Life Technologies, NY, USA).
- Artificial cerebrospinal fluid solution (ACSF):
- Sodium chloride (NaCl) (Sigma-Aldrich, St. Louis, MO, USA).
- Potassium chloride (KCl) (Sigma-Aldrich, St. Louis, MO, USA).
- Calcium chloride (CaCl2) (Sigma-Aldrich, St. Louis, MO, USA).
- Monopotassium phosphate (KH2PO4) (Sigma-Aldrich, St. Louis, MO, USA).
- Magnesium chloride (MgCl2) (Sigma-Aldrich, St. Louis, MO, USA).
- HEPES (Sigma-Aldrich, St. Louis, MO, USA).
- Glucose (Sigma-Aldrich, St. Louis, MO, USA).
- Sodium pyruvate (Sigma-Aldrich, St. Louis, MO, USA).
- Sodium hydroxide (NaOH) (Sigma-Aldrich, St. Louis, MO, USA).
- Hydrochloric acid (HCl) (Sigma-Aldrich, St. Louis, MO, USA).
3. Methods
3.1. Preparation of Reagents
ACSF: To prepare 100 mL of ACSF, dissolve the reagents in 90 mL of double-distilled (D.D, 18 MΩ cm at 25 °C) (see Note 1) water to attain the following final concentrations. 120 mM NaCl, 3.5 mM KCl, 1.3 mM CaCl2, 0.4 mM KH2PO4, 1 mM MgCl2, 5 mM HEPES, 10 mM glucose, and 10 mM sodium pyruvate. Adjust the pH to 7.4 using either 1 N NaOH or 1 N HCL. Increase the volume to 100 mL, and recheck the pH. Filter sterilize and store at 4 °C (see Notes 2 and 3).
Poly-L-ornithine hydrobromide (PLO): Dissolve poly-L-ornithine hydrobromide (PLO) in D.D water under sterile conditions to prepare a 10X stock solution of 0.16 mg/mL. The stock solution can be aliquoted and stored at −20 °C for future use (see Note 1).
Culture media: To 50 mL of NN1 media, add 500 μL fetal bovine serum (final concentration 1%) to prepare culture media for plating NSCs. Filter sterilize and store at 4 °C.
3.2. Coating XFe96 Cell Culture Microplates
Under sterile cell culture conditions, prepare working solution of PLO (16 μg/mL) by diluting 1 mL of 10× stock solution (0.16 mg/mL) in 9 mL of D.D water.
Add 25–30 μL of PLO working solution to each well, and incubate in cell culture incubator for a minimum of 2 h.
After incubation, wash the plate twice with sterile PBS and plate the cells. The plates can be prepared a day in advance and stored at 4 °C under sterile conditions.
3.3. Plating Cells in Seahorse XFe96 Analyzer Cell Culture Microplates
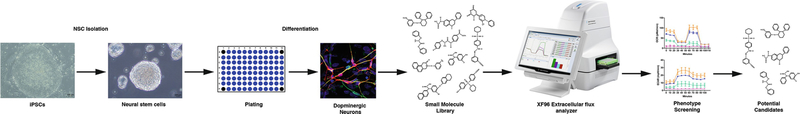
(For the isolation and culture of iPSC-derived NSCs, please refer to Chap. 1. For the differentiation of NSCs to DA neurons, please refer to Chap. 7). In this protocol the cells should be plated in the XFe96 cell culture microplates a day before the actual assay. The workflow of the assay is outlined in Fig. 1.
Fig. 1.
Workflow for screening mitoeffective compounds using iPSC-derived dopaminergic neurons and XFe96 analyzer. Generate iPSCs from patient somatic cells and isolate NSCs. Culture and differentiate the NSCs into dopaminergic neurons in cell culture plate of XFe96 analyzer. Treat (acute or chronic) the dopaminergic neurons with small molecule library. Measure changes in OCR using XFe96 analyzer. Evaluate the effect of the test compounds on different components of mitochondrial respiration. Select potential compounds for further testing from the pool based on their efficacy on improving mitochondrial respiration
Prewarm the Accutase, trypsin neutralizer, and culture media (NN1 + 1% FBS) in 37 °C water bath.
Collect all the neurospheres from the T-75 flask into a 50 mL tube, and centrifuge at 1000 rpm (200 × g) for 5 min.
Carefully remove the supernatant by using vacuum suction without disturbing the pellet.
Resuspend the pellet in 1 mL Accutase, and incubate for 3 min at 37 °C.
Inhibit the action of Accutase by adding 1 mL of trypsin neutralizer, and gently triturate 8–10 times to dissociate the cells into single-cell suspension. The trituration should be gentle enough to dissociate the cells without damaging the cellular integrity (see Note 4).
Add 8 mL of culture media, and centrifuge at 1000 rpm (200 × g) for 5 min.
Remove the supernatant, and resuspend the cells in 3–5 mL of culture media.
Count the cells using either hemocytometer or automate cell counter, and prepare a cell suspension of 500,000 cells/mL (see Note 5).
Using a multichannel pipette, add 100 μL of cell suspension (50,000 cells/well) to all wells except A1, A12, H1, and H12 (Fig. 2). Add only media to A1, A12, H1, and H12 wells, as they will serve as blanks during the assay for background correction.
Incubate the cells for 1 h at 37 °C in CO2 incubator.
To study long-term (24-h) effect of compounds on mitochondrial functions, prepare the compounds in 2× concentration in culture media and add to wells (100 μL/well). For control wells add 100 μL of culture media (see Note 6).
Incubate the cells at 37 °C in CO2 incubator.
The following day check the morphology of the cells under a phase contrast microscope. If the cell density and plating was done properly, all the cells should attach and appear as a monolayer. If large numbers of vacant spaces are visible between the cells, then the plating density should be increased until a monolayer is observed.
Fig. 2.
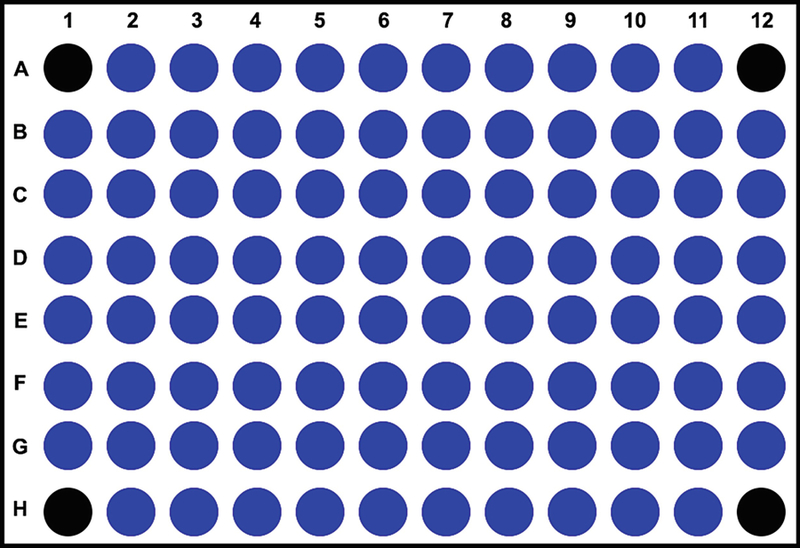
Cell culture plate design of XFe96 analyzer. The protocol for XFe96 analyzer necessitates leaving four wells (depicted in black color: A1, A12, H1, and H12) blank for the measurement of background during the assay. The remaining 92 wells are used for plating cells and screening compounds
3.4. Hydrating Seahorse XFe96 Analyzer Sensor Cartridge
Hydrate the Seahorse XFe96 analyzer sensor cartridge at least for 12–24 h before the start of the assay;
Place the cartridge container label side up, and open the packaging.
Carefully lift the cartridge sensor along with the hydration plate, and place it on a level surface. The sensor cartridges come with two delivery guides that are helpful for loading compounds into the port.
Carefully place the sensor cartridges along with lid and delivery guides upside down. The probes of the sensor cartridge should not touch any kind of surface.
Add 200 μL of calibrant solution to each well of the hydration plate.
Place the sensor cartridge back on to the hydration plate making sure all the probes are in the calibrant solution.
Wrap the sensor cartridge in plastic, and place it overnight at 37 °C in a non-CO2 incubator.
Turn on the XFe96 analyzer, and let it warm overnight.
3.5. Preparation of Cell Culture Microplate and Sensor Cartridge Plate for the Assay
The following steps are performed on the day of the assay:
Prewarm the ACSF in a 37 °C water bath.
Check the cells under a phase contrast microscope for their morphology. The cells should appear as a monolayer with clear media without any floating cells. If large vacant spaces are visible between the cells, then the plating number should be adjusted until a monolayer is formed.
Using a multichannel pipette, carefully remove 180 μL of the media leaving 20 μL in each well (including blanks). Care should be taken to not disturb the cells.
Wash the cells twice with 200 μL of ACSF.
After the second wash, add 160 μL of ACSF to all the wells.
Check the morphology of the cells under a phase contrast microscope to make sure the monolayer is intact in all wells and no cells are lost (see Note 7).
Place the cell culture microplate in a non-CO2 incubator at 37 °C for the ACSF to equilibrate.
Meanwhile prepare stock solutions of the compounds for the assay in ACSF.
- For mitochondrial modulators in the Mito Stress Kit, prepare the stock solution according to manufacturer’s instructions in ACSF (assay media):
- Oligomycin: add 630 μL of ACSF and mix well, final concentration of 100 μM.
- FCCP: add 720 μL of ACSF and mix well, final concentration of 100 μM.
- Rotenone: add 540 μL of ACSF and mix well, final concentration of 50 μM (see Note 8).
- Prepare 10× concentration working solutions of the mitochondrial modulators:
- Oligomycin: add 600 μL of stock solution to 2.4 mL of ACSF, concentration of 20 μM (10×).
- FCCP: add 600 μL of stock solution to 2.4 mL of ACSF, concentration of 20 μM (10×).
- Rotenone: add 300 μL of stock solution to 2.7 mL of ACSF, concentration of 5 μM (10×).
Prepare the test compounds for drug screening (acute response) at 10X concentration in ACSF. Calculate the required volume for each drug by multiplying the number of replicates with 20 μL/well.
Load the compounds using delivery guide into the ports of the sensor cartridge. Do not remove the hydration plate during this process. The probes should always remain hydrated during the entire procedure.
- For experiments investigating chronic response to compounds on mitochondrial respiration, load the compounds in the following order:
Port Compound Volume (μL) Concentration (μM)
Port Well A Oligomycin 20 20 2 B FCCP 22 20 2 C Rotenone 25 5 0.5 - For experiments investigating acute response to compounds on mitochondrial respiration, load the compounds in the following order:
Port Compound Volume (μL) Concentration (μM)
Port Well A Test Compound 20 10× × B Oligomycin 20 20 2 C FCCP 22 20 2 D Rotenone 25 5 0.5 Place the pipette tip through the holes in the delivery guide into each port, and carefully dispense the solution.
Ensure all the ports are filled with their designated compounds.
3.6. Real-Time Measurement of Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR)
In this protocol, the XFe96 analyzer should be turned on the evening before the day of the assay:
Initiate the seahorse wave software program.
Select the Mito Stress template.
Enter the information regarding the experiment, i.e., cell type, cell number, assay media treatment, date, etc.
Select the wells and label them accordingly (blanks, control, and treatment).
Label the injection ports with their respective compounds.
-
Enter the measurement settings:
Calibrate.
Equilibrate.
Baseline:
Mix: 3 min.
Wait: 1 min.
Measure: 3 min.
Loop 3 times.
Injection 1: Oligomycin.
Mix: 3 min.
Wait: 1 min.
Measure: 3 min.
Loop 3 times.
Injection 2: FCCP.
Mix: 3 min.
Wait: 1 min.
Measure: 3 min.
Loop 3 times.
Injection 3: Rotenone.
Mix: 3 min.
Wait: 1 min.
Measure: 3 min.
Loop 3 times.
-
For experiments investigating the acute response of the investigational compound, the following settings can be used:
Calibrate.
Equilibrate.
Baseline:
Mix: 3 min.
Wait: 1 min.
Measure: 3 min.
Loop 3 times.
Injection 1: Test compound.
Mix: 3 min.
Wait: 1 min.
Measure: 3 min.
Loop 3 times.
Injection 2: Oligomycin.
Mix: 3 min.
Wait: 1 min.
Measure: 3 min.
Loop 3 times.
Injection 3: FCCP.
Mix: 3 min.
Wait: 1 min.
Measure: 3 min.
Loop 3 times.
Injection 4: Rotenone.
Mix: 3 min.
Wait: 1 min.
Measure: 3 min.
Loop 3 times.
Proceed to next step.wload the sensor cartridge along with the hydration plate into the slot provided in the machine. Make sure the sensor cartridges are placed in proper orientation and in alignment with the slot of the machine.
Begin the calibration process.
Once the calibration is completed, the software will prompt to load the cell culture microplate. Replace the hydration plate with the cell culture microplate. Make sure the cell culture microplate is in proper orientation and in alignment with the slot of the machine.
Start the assay.
After completion of the assay, save the results file and export results to an excel document.
Save the plate for further analysis, such as protein estimation.
3.7. Data Analysis
The excel document provides detailed information regarding assay configuration, raw values, rate, and operation log.
The raw (plate) values provide information of each measurement and can be used to plot the changes in OCR or ECAR in response to the compounds used in the test against time.
- In this protocol using the OCR values from the assay results, the following additional parameters can be calculated (see Note 9):
- Non-mitochondrial respiration: third OCR measurement after rotenone injection.
- Basal respiration: (third baseline OCR measurement) – non-mitochondrial respiration.
- Maximal respiration: (Highest OCR measurement after the injection of FCCP)—(non-mitochondrial respiration).
- ATP-linked respiration: (third baseline OCR measurement)—(third measurement following oligomycin injection).
- Proton leak: (third measurement following oligomycin injection)—non-mitochondrial respiration.
- Spare respiratory capacity: Maximal respiration—basal respiration.
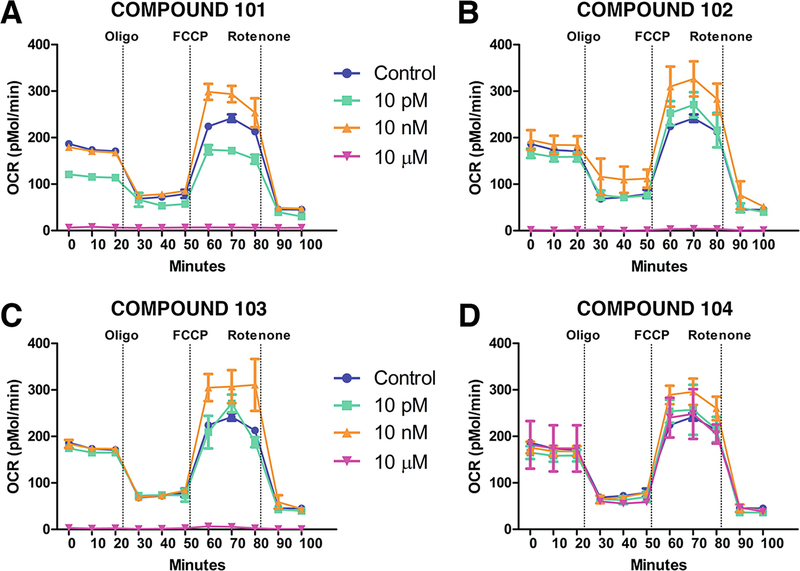
The data can be presented as absolute OCR (Mmol/min) (Fig. 3) and ECAR (pH/min) values or normalized to the cell number or protein content of the wells.
Fig. 3.
Measurement of changes in OCR in response to treatment with different compounds. Representative plots demonstrating the changes in OCR in response to treatment with different test compounds. Each compound was tested in three different concentrations to determine the differential response of drug concentration on cellular OCR
Acknowledgment
The authors thank the members of the Daadi laboratory for their helpful support and suggestions. This work was supported by the Worth Family Fund, the Perry & Ruby Stevens Charitable Foundation, the Robert J., Jr. and Helen C. Kleberg Foundation, the NIH primate center base grant (Office of Research Infrastructure Programs/OD P51 OD011133), and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1 TR001120.
Disclosures: Dr. Marcel M. Daadi is founder of the biotech company NeoNeuron.
Footnotes
Notes
Prepare all solutions and reagents in double-distilled water or ultrapure water (18 MΩ cm at 25 °C).
In contrast to other protocols that use XF assay medium, we used ACSF in our assays. We have compared the results from XF assay medium and ACSF and observed that NSCs survived and responded better in ACSF compared to XF assay media. Furthermore, ACSF is recommended for the assays with primary rat cortical neurons.
Adjust the pH of ACSF accurately to 7.4. The assay is sensitive to changes in pH, and this may affect the outcome of the tests.
During the dissociation of neurospheres to generate single-cell suspension, the trituration should be done gently to reduce damage to the integrity of the cells but should also be firmly enough to dissociate larger clumps as they may affect the plating density and thus the results of the assay.
A cell density titration should be performed to determine the cell number for optimal detection of response in the assay. Usually a cell density of 30,000–50,000 cells/well should be sufficient to generate a discernable response in the assay. How- ever, it may change according to the experiment parameters (cell type, treatment).
It is recommended to have at least 4–6 replicates per group to obtain accurate results in the assay.
Care should be taken during the rinsing process as loss of cells results in high variability between and within groups.
Rotenone is toxic and known to cause Parkinson’s disease. Personal protective equipment should be worn all the time while working with rotenone and other toxic compounds.
The calculations are based on the experimental setup described in the current protocol. Changes in the number of measurements may require modification to the equations for the calculation of the parameters.
References
- 1.Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443(7113):787–795 [DOI] [PubMed] [Google Scholar]
- 2.Zhang J et al. (2012) Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat Protoc 7(6):1068–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand MD, Nicholls DG (2011) Assessing mitochondrial dysfunction in cells. Biochem J 435(2):297–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R et al. (2015) The acute extracellular flux (XF) assay to assess compound effects on mitochondrial function. J Biomol Screen 20 (3):422–429 [DOI] [PubMed] [Google Scholar]
- 5.Koopman M et al. (2016) A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nat Protoc 11 (10):1798–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozsvari B et al. (2017) Mitoriboscins: Mitochondrial-based therapeutics targeting cancer stem cells (CSCs), bacteria and pathogenic yeast. Oncotarget 8(40):67457–67472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribeiro SM, Gimenez-Cassina A, Danial NN (2015) Measurement of mitochondrial oxygen consumption rates in mouse primary neurons and astrocytes. Methods Mol Biol 1241:59–69 [DOI] [PubMed] [Google Scholar]
- 8.Clerc P, Polster BM (2012) Investigation of mitochondrial dysfunction by sequential microplate-based respiration measurements from intact and permeabilized neurons. PLoS One 7(4):e34465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang T, Cadenas E (2014) Astrocytic metabolic and inflammatory changes as a function of age. Aging Cell 13(6):1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy Choudhury G et al. (2015) Methylene blue protects astrocytes against glucose oxygen deprivation by improving cellular respiration. PLoS One 10(4):e0123096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park SJ et al. (2017) Metabolome profiling of partial and fully reprogrammed induced pluripotent stem cells. Stem Cells Dev 26 (10):734–742 [DOI] [PubMed] [Google Scholar]
- 12.Panopoulos AD et al. (2012) The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res 22(1):168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rana P et al. (2012) Characterization of human-induced pluripotent stem cell-derived cardiomyocytes: bioenergetics and utilization in safety screening. Toxicol Sci 130 (1):117–131 [DOI] [PubMed] [Google Scholar]
- 14.van der Windt GJ, Chang CH, Pearce EL (2016) Measuring bioenergetics in T cells using a seahorse extracellular flux analyzer. Curr Protoc Immunol 113:3 16B 1–3 16B 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong Z et al. (2016) Focused screening of mitochondrial metabolism reveals a crucial role for a tumor suppressor Hbp1 in ovarian reserve. Cell Death Differ 23(10):1602–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]