Abstract
Herein we introduce the Visual Mass-Spec Share (vMS-Share), a new public mass spectrometric (MS) repository and data mining website/resource freely accessible at https://vmsshare.nist.gov. vMS-Share is a web-based application developed for instant visualization of raw MS data with integrated display of metadata optimized for the sharing of proteomics and metabolomics experimental results. Each MS-based identification is linked to a given experiment and the entire experimental data can then be viewed using the link associated with a given peptide and/or small molecule. Interactive and user-friendly visualizations are provided to the user via variety of easily accessible search filters.
Keywords: proteomics, metabolomics, public repository, raw data visualization/mining
INTRODUCTION
Mass spectrometry (MS)-based proteomic and metabolomic studies produce increasingly large amounts of raw experimental data and allied biological findings. To facilitate sharing and dissemination of raw MS data, public web-based repositories have been developed to enable direct access to proteomic and metabolomic researchers and the scientific community at large.(Perez-Riverol et al., 2015, Spicer et al., 2017) These repositories also provide links to corresponding publications to facilitate collaboration between groups in diverse geographical locations.
Mass spectra of peptides and small molecules acquired during mass spectrometric (MS) profiling of biological specimens are the principal output and basis of proteomic and/or metabolomic MS-based analyses.(Fiehn, 2002, Rappsilber and Mann, 2002) Acquired mass spectra are subsequently processed using automated searching algorithms that lead to the identification of proteins and metabolites found in biological specimens. However, researchers still need to examine acquired mass spectra manually to monitor identification processes and validate significant findings. Consequently, the ability to easily visualize, compare, and integrate raw MS data deposited in public repositories using web browser is critical for facile data mining and assessment of their quality and/or reproducibility. The most widely utilized public repositories and freely accessible web applications for visualization/mining of proteomic and metabolomic data are described elsewhere.(Oveland et al., 2015, Spicer et al., 2017)
Herein we describe the Visual Mass-Spec Share (vMS-Share), a new public mass spectrometric (MS) repository and data mining web-application with a user-friendly graphical interface which allows for intuitive analysis and integration of raw MS data.
MATERIAL AND METHODS
Availability and implementation
vMS-Share interface was built using Hypertext Preprocessor (PHP), Structured Query Language (SQL), Hypertext Markup Language (HTML5), JavaScript and Cascading Style Sheet (CSS3). The website is administered by the National Institute of Standards and Technology (NIST; Gaithersburg, MD, USA) and is available at https://vmsshare.nist.gov
Input Pipeline
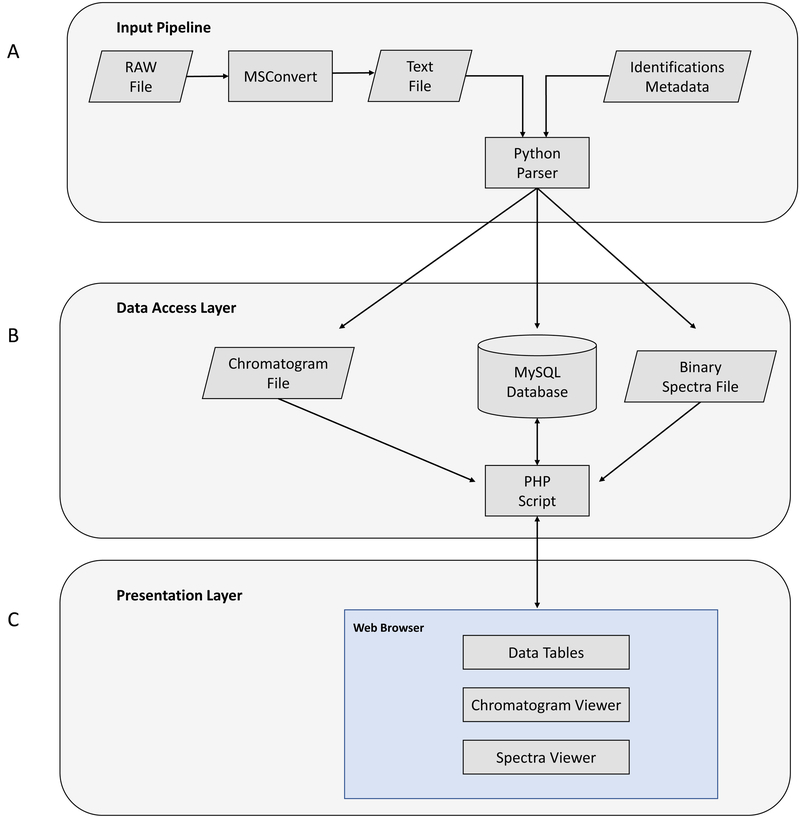
Data input for vMS-Share consists of two files: (i) raw MS data and (ii) experimental meta-data (Fig. 1A). The request for data submission should include a link (e.g., cloud bucket link) from which we can download /access their data.
Figure 1.
(A) Data input pipeline. (B) Data access layer. (C) Data presentation layer.
MSConvert from ProteoWizard tools (http://proteowizard.sourceforge.net/tools.shtml) is used for extraction of raw MS data into text format. A list of data formats that ProteoWizard tools support are available via the ProteoWizard Installation and Data Formats page at http://proteowizard.sourceforge.net/formats/index.html.
Custom Python scripts are used for parsing of the data while MySQL is used as a Back-End Database for storage. Raw MS data for each experiment is parsed and meta-data for each spectrum is entered into MySQL database. During data parsing a Python script also generates files for display of base peak chromatograms as well as file that contains spectral data. Spectral data files are binary encoded to reduce the file size and optimize storage on the web server. File offset pointers for each spectrum are stored in the database for fast search and retrieval of spectral data. Experimental meta-data containing identifications is parsed last, entered into database and linked with raw MS data through scan IDs from raw data files.
Data Access Layer
Our website is running on industry standard open source LAMP stack – the web server runs on Linux operating system that runs Apache web server, MySQL database and PHP scripts for dynamic web page content display (Figure 1 B). Website security considerations include read only privileges for database access as well as SQL injection checks for any search inputs made by the users. Additionally, the website uses SSL certificates for secure HyperText Transfer Protocol (HTTPS).
Presentation Layer
Interactive and searchable data tables were implemented using open source jQuery JavaScript plugin called DataTables (https://datatables.net/). Interactive chromatogram graphing display was implemented using open source JavaScript charting library called dygraphs (http://dygraphs.com/). This library was further modified to create custom identification flags and pop-up labels. Interactive spectra display was created using open source JavaScript plotting library Flot (https://www.flotcharts.org/). This library was also further modified to create dynamic labels for spectrum intensity (Figure 1 C).
RESULTS
In this section, the most important features and the use of the web interface are described. vMS-Share has a framework that allows users to utilize two distinct workflows for raw MS data mining: (i) experiment-based search and/or (ii) study-based search.
Experiment-based search
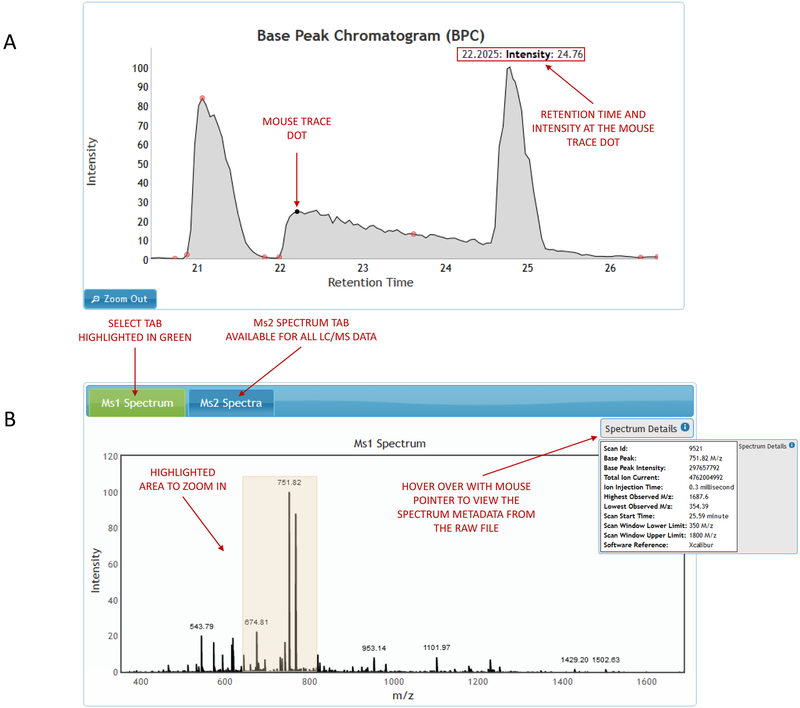
Experiment-based search enables user to pick a data-set of interest and selects an experiment within that data set. The experiment image consists of an interactive chromatogram viewer with flagged identification points coupled with an MS spectrum viewer and a table of selected identifications (Figure 2A). Chromatogram and spectrum viewers offer zoom in/out capabilities.
Figure 2.
(A) Experiment-based search. (B) Viewing a spectrum within the experiment.
Viewing any spectrum within the experiment
Clicking on any retention time point on the chromatogram will bring up a graphical representation of the selected MS1 spectrum and raw file metadata associated with it. In case of MS2 data, the selected MS1 spectrum will also contain a tab for viewing MS2 spectra associated with it (Figure 2B). The MS2 spectra tab contains a table ordered by retention times which are used as links to view each MS2 spectrum and the corresponding raw file metadata.
Viewing identifications within the experiment
Hovering with the mouse pointer above identification flags on the chromatogram viewer brings up a pop-up balloon and associated identification name (Figure 3A). Clicking on the pop-up balloon opens an interactive spectral display of the identification that includes experimental metadata as well as raw file metadata.
Figure 3.
Viewing identifications within the experiment. (A) Chromatogram viewer. (B) Spectral viewer.
Identifications within the selected experiment can also be searched through the table that is displayed on the right side of the chromatogram viewer (Figure 3 A). The identifications table consists of two columns displaying the identification title and the retention time. Searching is enabled for both the identification title and the retention time as well as sorting on any of the two columns. Clicking on the identification title brings up the spectral display and the corresponding experimental and raw file metadata (Figure 3 B).
Study-based search
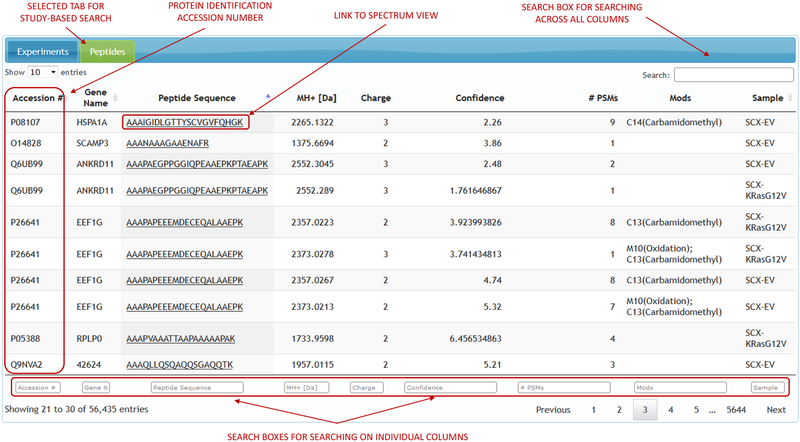
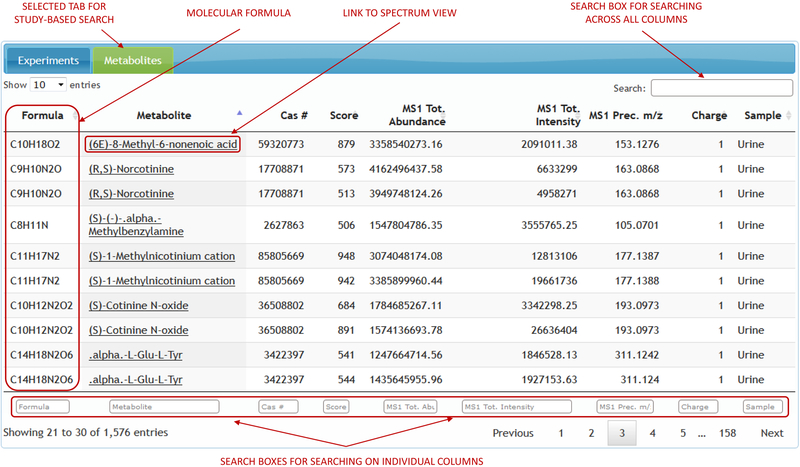
Study-based search approach allows the user to pick a data-set of interest and search for the identifications across all the experiments within that data-set. List of identifications for any given data-set can be searched, filtered and sorted on any single column that the identifications table displays. These search filters vary for different types of ‘omics’. Proteomic data can be searched in relation to peptide sequence, protein accession number, modifications, atomic mass and other attributes (Figure 4). Metabolomic data can be searched on metabolite name, formula, CAS number and other chemical attributes (Figure 5). Clicking on any single identification opens the spectral display with the corresponding experimental and raw file metadata. The spectrum viewer will also have a link pointing to the experiment to which the spectrum belongs to. Clicking on the experiment link brings up the full experiment view.
Figure 4.
Study-based proteomics search.
Figure 5.
Study-based metabolomics search.
DISCUSSION
With the advent of high resolution/accuracy and high-throughput MS technologies, large amounts of proteomic and metabolomic raw data are constantly being generated. A repository housing both proteomic and metabolomic data provides extensive output for the proteome and metabolome annotations. Maintaining these types of repositories is challenging due to the large volume and diversity (i.e., LC-MS, GC-MS, NMR) as well as diverse needs (e.g. visualization, quantitation, statistics) of prospective users. It requires sustained human efforts and constant financial support. Status, features and comparison of contemporary and most widely utilized proteomic and metabolomic public repositories are reviewed elsewhere.(Chen et al., 2015, Spicer et al., 2017) Unlike PRoteomics IDEntifications (PRIDE) database, our application does not require downloading of a third-party software and also permits visualization and data mining of metabolomic raw data. Additionally, vMS-share does not require specific proteome database file formatting (e.g., FASTA).
Upon the completion and testing of the basic infrastructure for the web interface developed at NIST for instant visualization of raw MS data across different analytical platforms (e.g. LC-MS, GC-MS), it was decided that the scope of the web interface could be a foundation for a public repository containing ‘omics’ MS data for both proteins/peptides and small molecules. vMS-Share submissions are labeled as samples and investigations. A sample includes all raw data from a given biological source, irrespective of the number of instrument runs required to collect the raw MS data. An investigation is defined as a pool of related samples and/or experiments (e.g., time point series, healthy vs. diseased series). Each sample has its own list of identifications linked to flagged mass spectra. All studies are uniquely identified by release date and study label, whereas file name and sample name identify corresponding study specimens. Public data may be surfed by investigations or by samples. In the case of pending publication, deposited raw data remains private during review period. As a constantly and dynamically developed public repository and web-based application, vMS-Share is open to proteomic and metabolomic community and scientists at large for suggestions and development of additional computational features.
CONCLUSIONS
The significance of data sharing is widely appreciated in MS-based proteomics and metabolomics. However, visualizing, mining and distribution of raw MS data from proteomic and metabolomic datasets is a challenging task. The vMS-Share mission is to place a prospective user at the center of the interface. It is accomplished by development of user-friendly web application that offers versatile frameworks for data visualization and mining. This enables direct and facile analysis of raw mass spectra and manual evaluation of significant findings. vMS-Share is freely accessible to anyone using a contemporary web browser with no need to install a third-party software whilst preserving data integrity and security. vMS-Share enables dissemination of data not only between different laboratories with an institution but also between universities and/or research institutes at distinct geographical locations.
REFERENCES
- 1.Perez-Riverol Y, Alpi E, Wang R, Hermjakob H, Vizcaino JA (2015) Making proteomics data accessible and reusable: Current state of proteomics databases and repositories. Proteomics 15 (5–6), 930–949. https://onlinelibrary.wiley.com/doi/full/10.1002/pmic.201400302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spicer R, Salek RM, Moreno P, Canueto D, Steinbeck C (2017) Navigating freely-available software tools for metabolomics analysis. Metabolomics 13 (9). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5550549/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiehn O (2002) Metabolomics - the link between genotypes and phenotypes. Plant Mol Biol 48 (1–2), 155–171. https://www.ncbi.nlm.nih.gov/pubmed/11860207 [PubMed] [Google Scholar]
- 4.Rappsilber J, Mann M (2002) What does it mean to identify a protein in proteomics? Trends Biochem Sci 27 (2), 74–78. https://www.sciencedirect.com/science/article/pii/S0968000401020217?via%3Dihub [DOI] [PubMed] [Google Scholar]
- 5.Oveland E, Muth T, Rapp E, Martens L, Berven FS, et al. (2015) Viewing the proteome: How to visualize proteomics data? Proteomics 15 (8), 1341–1355. https://onlinelibrary.wiley.com/doi/full/10.1002/pmic.201400412 [DOI] [PubMed] [Google Scholar]
- 6.Chen T, Zhao J, Ma J, Zhu YP (2015) Web Resources for Mass Spectrometry-based Proteomics. Genom Proteom Bioinf 13 (1), 36–39. https://www.sciencedirect.com/science/article/pii/S1672022915000054?via%3Dihub [DOI] [PMC free article] [PubMed] [Google Scholar]