Abstract
Neuropeptide S (NPS) has generated substantial interest due to its anxiolytic and fear-attenuating effects in rodents, while a corresponding receptor polymorphism associated with increased NPS receptor (NPSR1) surface expression and efficacy has been implicated in an increased risk of panic disorder in humans. To gain insight into this paradox, we examined the NPS system in rats and mice bred for high anxiety-related behavior (HAB) versus low anxiety-related behavior, and, thereafter, determined the effect of central NPS administration on anxiety- and fear-related behavior. The HAB phenotype was accompanied by lower basal NPS receptor (Npsr1) expression, which we could confirm via in vitro dual luciferase promoter assays. Assessment of shorter Npsr1 promoter constructs containing a sequence mutation that introduces a glucocorticoid receptor transcription factor binding site, confirmed via oligonucleotide pull-down assays, revealed increased HAB promoter activity—an effect that was prevented by dexamethasone. Analogous to the human NPSR1 risk isoform, functional analysis of a synonymous single nucleotide polymorphism in the coding region of HAB rodents revealed that it caused a higher cAMP response to NPS stimulation. Assessment of the behavioral consequence of these differences revealed that intracerebroventricular NPS reversed the hyperanxiety of HAB rodents as well as the impaired cued-fear extinction in HAB rats and the enhanced fear expression in HAB mice, respectively. These results suggest that alterations in the NPS system, conserved across rodents and humans, contribute to innate anxiety and fear, and that HAB rodents are particularly suited to resolve the apparent discrepancy between the preclinical and clinical findings to date.
Keywords: anxiety, basolateral amygdala, fear, paraventricular nucleus, promoter fragmentation
Introduction
Anxiety disorders are among the most common psychiatric illnesses, with a lifetime prevalence of ∼30% (Kessler and Wang, 2008). While a number of pharmacotherapies are available, the lack of truly novel-acting compounds has led to a focus on the development of non-GABAergic compounds (Cryan and Slattery, 2007). Neuropeptides represent such potential targets due to their distinct synthesis and release sites, and multiple behavioral functions (Landgraf et al., 2007; Slattery and Neumann, 2010a).
Neuropeptide S (NPS) together with its G-protein-coupled receptor, NPS receptor 1 (NPSR1), represent such a candidate system. NPS is synthesized in discrete clusters of brainstem nuclei, including the peri-locus ceruleus area (hereafter termed LC), whereas its receptor exhibits a widespread distribution pattern (Xu et al., 2007; Leonard and Ring, 2011). In rodent studies, central and nasal administration of NPS elicits potent anxiolytic and arousal effects, as well as facilitating the extinction of conditioned fear (Jüngling et al., 2008; Leonard et al., 2008; Meis et al., 2008; Ionescu et al., 2012; Lukas and Neumann, 2012; Wegener et al., 2012).
In humans, Npsr1 is located in a chromosomal region linked to panic disorder, and a corresponding single nucleotide polymorphism (SNP; Asn107Ile-rs324981) has been associated with increased risk of the overinterpretation of fear (Okamura et al., 2007; Donner et al., 2010; Raczka et al., 2010; Domschke et al., 2011). However, while the preclinical findings to date suggest that increasing NPS levels within the brain reduces anxiety and fear responses, the human Ile107 receptor variant exhibits increased surface receptor expression and a 10-fold higher NPS-induced signaling response than the Asn107 variant (Reinscheid et al., 2005; Bernier et al., 2006).
To investigate the discrepancy between rodent and human literature, and to test the efficacy of NPS as a potential anxiolytic therapeutic, its behavioral effects have to be confirmed in psychopathological animal models (Landgraf et al., 2007). Wistar rats or CD-1 mice selectively bred for high anxiety-related behavior (HAB; rHABs and mHABs, respectively) versus low anxiety-related behavior (LAB; rLABs and lHABs, respectively) represent such models (Neumann et al., 2010; Sartori et al., 2011a). In addition to their high innate anxiety, HAB mice display enhanced expression, and HAB rats display impaired extinction of conditioned fear responses, respectively (Muigg et al., 2008; Sartori et al., 2011b; Yen et al., 2012). While these behaviors can be attenuated in rHABs with traditional anxiolytics, mHABs do not respond to such treatment (Sah et al., 2012). These inborn differences make these rodents attractive models to further assess the genetic underpinnings of extremes in anxiety-related behavior and the anxiolytic potential of novel drugs (Czibere et al., 2011; Neumann et al., 2011).
Therefore, to address the putative discrepancy between the preclinical and clinical findings regarding the NPS system and anxiety, we tested the hypothesis that selective breeding for anxiety leads to genetic, expressional, and functional differences in the brain NPS–NPSR1 system. We next determined whether NPS administration could exert its anxiolytic and fear extinction effects in these animals with genetically predisposed psychopathologies and limited efficacy of traditional anxiolytics.
Materials and Methods
Animals
Adult male rHAB and rLAB Wistar rats (280–350 g), mHAB and mLAB CD-1 mice (30–35 g) and weight-matched nonselected Wistar rats (rNAB; Charles River) and CD1 mice (mNAB; Munich breeding) were used in these studies. Rats and mice were housed in groups of four, until 1 week before tissue harvesting or undergoing surgical procedures, when they were single housed. Animals were maintained on a 12 h light/dark cycle (lights on: rats, 6:00 A.M.; mice, 8:00 A.M.) in a temperature-controlled colony (21–23°C, 55% humidity). The animals had free access to food and water. All experimental procedures were performed in the morning (8:30–11:30 A.M.). The primary testing for the selection of experimental HAB and LAB rats and mice was performed on the elevated plus-maze (EPM) at the ages of 9 and 7 weeks, respectively, as previously described (Krömer et al., 2005; Neumann et al., 2010). All experiments were conducted with the approval of the local governments of the Oberpfalz and Oberbayern.
Measurement of Npsr1 and Nps mRNA levels, and NPSR1 protein levels
Rats and mice were killed under basal conditions and brief 20 s isoflurane anesthesia. Brains were snap frozen in N-methylbutane stored at −80°C, and LC, paraventricular nucleus (PVN), and amygdala tissue samples were collected. The 3 × 200 μm PVN-targeted, 3 × 200 μm amygdala-targeted, and 5 × 200 μm LC-targeted sections were mounted on slides with the aid of histological staining and atlases (Paxinos and Watson, 1998; Paxinos and Franklin, 2001). Tissue punchers with diameters of 1.8 mm (rats) and 0.8 mm (mice; Fine Science Tools) were used to harvest PVN, amygdala, and LC tissues.
Quantitative PCR
Total rat and mouse RNA was extracted from PVN, amygdala, and LC tissue, and cDNA was prepared and the real-time quantitative PCR (qPCR) for Nps and Npsr1 performed as previously described (Elfving et al., 2008; Bunck et al., 2009). Briefly, real-time qPCR was performed on cDNA using a QuantiFast SYBR Green Kit (Qiagen) based on the manufacturer's instruction on a LightCycler (Roche Diagnostics) or an Mx3000P (Stratagene). Standard curves were generated, and each experiment was performed in duplicate. Relative transcript concentrations were calculated using the 2(−ΔΔCt) method (Livak and Schmittgen, 2001). For a list of all primers used, see Tables 1 and 2.
Table 1.
List of primers used for mRNA expression studies in the mHABs and mLABs
| Gene | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|
| Housekeeping genes | ||
| Rpl13a | (+) | CACTCTGGAGGAGAAACGGAAGG |
| Rpl13a | (−) | GCAGGCATGAGGCAAACAGTC |
| B2 mg | (+) | CTATATCCTGGCTCACACTG |
| B2 mg | (−) | CATCATGATGCTTGATCACA |
| Target genes | ||
| Nps | (+) | TGGTGTTATCCGGTCCTCTC |
| Nps | (−) | GGACCTTTTCATCGATGTCT |
| Npsr1 | (+) | CTCTTCACTGAGGTGGGCTC |
| Npsr1 | (−) | CCAGTGCTTCAGTGAACGTC |
Table 2.
List of primers used for mRNA expression studies in the rHABs and rLABs
| Gene | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|
| Target and housekeeping genes for rat Nps measurements | ||
| Polr2b | (+) | GAAGCCAGGTTAAGAAATCTC |
| Polr2b | (−) | GACACTCATTCAGCTCACAC |
| Gapdh | (+) | TGGAGTCTACTGGCGTCTT |
| Gapdh | (−) | TGTCATATTTCTCGTGGTTCA |
| Actb | (+) | GGCACCACCATGTACCCAGGC |
| Actb | (−) | CGATGGAGGGGCCGGACTCA |
| Nps | (+) | ATCTTAGCTCTGTCGCTGTC |
| Nps | (−) | CGACGTCTTCTCCAAAATTG |
| Target and housekeeping genes for rat Npsr1 measurements | ||
| 18srRNA | (+) | ACGGACCAGAGCGAAAGCAT |
| 18srRNA | (−) | TGTCAATCCTGTCCGTGTCC |
| Actb | (+) | TGTCACCAACTGGGACGATA |
| Actb | (−) | GGGGTGTTGAAGGTCTCAAA |
| CycA | (+) | AGCACTGGGGAGAAAGGATT |
| CycA | (−) | AGCCACTCAGTCTTGGCAGT |
| Gapdh | (+) | TCACCACCATGGAGAAGGC |
| Gapdh | (−) | GCTAAGCAGTTGGTGGTGCA |
| Hmbs | (+) | TCCTGGCTTTACCATTGGAG |
| Hmbs | (−) | TGAATTCCAGGTGAGGGAAC |
| Hprt1 | (+) | GCAGACTTTGCTTTCCTTGG |
| Hprt1 | (−) | CGAGAGGTCCTTTTCACCAG |
| Rpl13a | (+) | ACAAGAAAAAGCGGATGGTG |
| Rpl13a | (−) | TTCCGGTAATGGATCTTTGC |
| Ywhaz | (+) | TTGAGCAGAAGACGGAAGGT |
| Ywhaz | (−) | GAAGCATTGGGGATCAAGAA |
| Npsr1 | (+) | CTGTTCTCCATCCCCACACT |
| Npsr1 | (−) | GCAGTTGGAAATCACCGTCT |
Western blotting
Total protein was extracted from PVN-containing tissue punches (separate cohorts from those used for qPCR). Briefly, tissue was homogenized in RIPA buffer (Sigma) supplemented with 0.1 m PMSF, 0.1 m sodium orthovanadate, and protease inhibitor, and incubated on ice for 30 min. After centrifugation (15 min, 14,000 × g, 4°C), the supernatant was collected. Protein concentration was measured using the Pierce BCA Protein Assay Kit. Sixty micrograms of rat proteins and 15 μg of mouse proteins were separated on a 10% SDS-polyacrylamide gel. After transfer onto a nitrocellulose membrane (Bio-Rad), nonspecific binding was blocked in Tris-buffered saline/0.1% Tween-20 (TBST), pH 7.4, supplemented with 5% nonfat milk powder (Sigma) for 1 h at room temperature. The semi-quantitative assessment of NPSR1 protein was performed using an anti-NPSR1 antibody purchased from Abcam (ab92425) or an anti-NPSR1 (Ab2; Leonard et al., 2011), which was provided by Dr. Robert Ring (Pfizer, New York, NY). Membranes containing rat proteins were incubated in Ab2 (1:500) and those containing mouse proteins were incubated in Ab2 (1:500) or ab92425 (1:500) overnight at 4°C with gentle shaking. Thereafter, the membranes were washed three times for 5 min each in TBST, incubated in a 1:1000 dilution of HRP-conjugated anti-rabbit antibody (New England Biolabs) in 2.5% milk in TBST for 30 min at room temperature, washed three times, and visualized by chemiluminescence (Western Lighting, PerkinElmer). β-Tubulin (1:1000) was used as a loading control (New England Biolabs).
Genotyping of NPSR1 knock-out mice
Genomic DNA was prepared from hypothalamic punches of NPSR1 wild-type (WT; NPSR1+/+) and NPSR1 knock-out (KO; NPSR1−/−) mouse brains (provided by Dr. Chiara Ruzza, University of Ferrara, Ferrara, Italy). DNA was incubated overnight at 55°C and 1000 rpm with horizontal shaking conditions in 200 μl of proteinase K lysis buffer containing 50 mm KCl, 10 mm Tris-HCl, pH 8.3, 1 mm MgCl2, 0.45% Nonidet P-40, 0.45% Tween 20, 0.1 mg/ml gelatin, and 0.5 mg/ml proteinase K. Thereafter, samples were incubated for 10 min at 95°C and centrifuged for 10 min at 14,000 g, 4°C. Two microliters of genomic DNA was added to a PCR mix containing DreamTaq PCR Master Mix (Thermo Scientific) and primers in a final concentration of 0.2 μm. The following three oligonucleotide primers were used: the forward primer was specific to the endogenous NPSR locus [5′-CCTTATCCTCAAACCACGAAGTAT-3′]; the second was a common reverse primer [5′-GTGGGTACATGAGAAGGTTAGGAG-3′]; and the third was a forward primer [5′-AAATGCCTGCTCTTTACTGAAGG-3′] specific to the targeting plasmid. The reaction mix was placed in a thermal cycler and incubated for 5 min at 95°C. The PCR proceeded for 40 cycles as follows: 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. Additionally, samples were incubated at 72°C for 5 min and was finally stored at 12°C. Amplification products were separated by agarose gel electrophoresis at 140 V for 1 h and then stained with RotiStain (Roth), and images were captured using ChemiDoc XRS system (Bio-Rad).
Nps and Npsr1 sequencing and bioinformatic analysis
Genomic DNA was extracted from HAB and LAB rodents (NucleoSpin Tissue Kit), and sequencing primers (Tables 3, 4, 5, 6; Sigma-Aldrich) were designed using Primer-BLAST (National Center for Biotechnology Information, Bethesda, MD). Sequencing and bioinformatic analyses were performed as previously described (Bunck et al., 2009). The Nps and Npsr1 DNA fragments (Frags) were amplified using Taq-polymerase (Fermentas). Cleaned PCR products were used for sequencing by the Big Dye Terminator kit version 3.1 (Applied Biosystems), and sequences were resolved by capillary electrophoresis on a 3730 DNA analyzer (Applied Biosystems). Rat and mouse Nps and Npsr1 DNA sequences were analyzed using BioEdit version 7.0.2 (Hall, 1999). Transcription factor binding sites in the Npsr1 promoter were predicted using the Transcription Element Search System (TESS; Schug, 2008). Genomic coordinates are based on genome builds RGSC3.4 (EnsEMBL release 68) for rats and Mm10 (GRCm38, EnsEMBL release 68) for mice.
Table 3.
List of primers used for sequencing of the rat Npsr1 gene with location and exons/promoter/downstream enhancing region the primers hybridized to
| Sequential order | Location | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|---|
| 1 | Intron 1 | (+) | GGTGAGCAATAGCCAGAAGC |
| 1 | Intron 2 | (−) | CAGAATTTAAAGCCAGGGCA |
| 2 | Intron 2 | (+) | GCGCAAGTGACTGTGTCATC |
| 2 | Intron 3 | (−) | CTTCTCTCCCGCTGGTACTG |
| 3 | Intron 3 | (+) | TCCATGCCTCACTTTTCCTC |
| 3 | Intron 4 | (−) | AGCTAGGGAGAAAGGCGTGT |
| 4 | Intron 4 | (+) | AGCCCAGATCTGCTTCCAGT |
| 4 | Intron 5 | (−) | ATGGCGTGAGGATCAGGTAG |
| 5 | Intron 5 | (+) | GTCCTAGTGACTCCCAGCCA |
| 5 | Intron 6 | (+) | GTTCCCACAAGGAGTTTGGA |
| 6 | Intron 6 | (−) | TGGCACCTTCAGTATGAGCA |
| 6 | Intron 7 | (+) | CCAGATACCCCTATTTCCAGC |
| 7 | Intron 7 | (−) | AGCCGCCACTAATCCATCTT |
| 7 | Intron 8 | (+) | ACACTCCTTCCCTGCATGTC |
| 8 | Intron 8 | (+) | AAGAGGGATGCTTCTGGTGA |
| 8 | Intron 9 | (−) | GAGCATTGGGAGCACAACTT |
| 9 | Intron 9 | (+) | TGGAGGAAGAGGTCCAGTTG |
| 9 | Exon 10 | (−) | ATGGTGAAGGTCTGGGTGAG |
| 10 | Exon 10 | (+) | CTCTCCAAGCCTGAATTCATC |
| 10 | Exon 10 | (−) | CTAACATCTTCTCCTCCACATG |
| 11 | Exon 10 | (+) | GGAGGACAACAAAGGTTAGAC |
| 11 | Exon 10 | (−) | ATAAGACCAGCACTTCCTTG |
| 12 | Exon 10 | (+) | AAATAGTGATAGACCCTGGC |
| 12 | Exon 10 | (−) | ACATGTTAACGACTGAACGA |
| 13 | Exon 10 | (+) | CCCACAGCCCTATGACGCACG |
| 13 | Exon 10 | (−) | TGCTAGCTAGGACACCCGCCA |
| 14 | Exon 10 | (+) | GCTGACGGCTCGTTCAGTCG |
| 14 | Exon 10 | (−) | AGGGGATGGTGTCGGCATGTG |
| 15 | Exon 10 | (+) | ACAGGACTGGTGCTGAAATCGC |
| 15 | Exon 10 | (−) | ACTTCAACATCCTCTGCTACACTGC |
| 16 | Exon 10 | (+) | GTCCTATGATGCTGGATGAATCATGC |
| 16 | Exon 10 | (−) | CCTGAAAGGAGAGGATCTTTCGCCA |
| 17 | Exon 10 | (+) | GGTGCCCACCTTCCACACCAAGATG |
| 17 | DER | (−) | GGCCATCAGACGTGTGGCTTCC |
| 18 | DER | (+) | CCAGCTTCATAGAGACAGCTCTGC |
| 18 | DER | (−) | ACCCCCATTCTCCCACCCCAC |
| 19 | DER | (+) | TCATTATCCACAACAGGGCTGGACC |
| 19 | DER | (−) | ATGGCCTGCAAGGCTAAGGCG |
| 20 | DER | (+) | CAGCACTTGGGAGACAGAGA |
| 20 | DER | (−) | TGCTGAGCTAAATGTCAAAG |
| −1 | Promoter | (+) | AGACAAACACAGACCCCTGC |
| −1 | Intron 1 | (−) | GAGTTCAGTTAGCCAGGGCA |
| −2 | Promoter | (+) | TGTCATGTCGAAACCCTTCA |
| −2 | Promoter | (−) | CAGCTGAGATCGCTTTTGTCT |
| −3 | Promoter | (+) | GTCAGCAGCTTCTGTGCATC |
| −3 | Promoter | (−) | AAGGGGTATGTCCCAGGAAG |
| −4 | Promoter | (+) | TGCACCCATTTTTAGTTCCC |
| −4 | Promoter | (−) | AGGGGTATGTCCCAGGAAGT |
| −5 | Promoter | (+) | GATCCTACTTTGGGCCTGTCG |
| −5 | Promoter | (−) | GAAGATGCTCAACCACATTATTAGC |
| −6 | Promoter | (+) | GTCCACCCCTGAGAGTTCCAG |
| −6 | Promoter | (−) | CAGGGCATCAAGTGAGGGCATC |
| −7 | Promoter | (+) | GGTGTGGATTTGTGAGGGAGGT |
| −7 | Promoter | (−) | GTTCAGTCAGGGAAGATGCATC |
DER, Downstream enhancing region.
Table 4.
List of primers used for sequencing of the mouse Npsr1 gene with location and exons/promoter/downstream enhancing region
| Sequential order | Location | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|---|
| −7 | Promoter | (+) | GCAGAGGAGACCACACTGGCG |
| −7 | Promoter | (−) | GCCTGACGACAAGGAAGATCCACG |
| −6 | Promoter | (+) | TTGTCATCTCCTGTCTGTGCCCCT |
| −6 | Promoter | (−) | CGCCAGTGTGGTCTCCTCTGC |
| −5 | Promoter | (+) | TGCAGCGTAATGAACACCCCCA |
| −5 | Promoter | (−) | GTAGGCCAACCTTTGCTTTACTGCC |
| −4 | Promoter | (+) | CTGTATGTGCAAATGTGTGTC |
| −4 | Promoter | (−) | GGAGAGCAGAATGTCATGAG |
| −3 | Promoter | (+) | AAGCCCTCATCTCTAACCTG |
| −3 | Promoter | (−) | TCATGGTTTCCCCTCCTCCA |
| −2 | Promoter | (+) | GGGCAAACAAACACTATTGATC |
| −2 | Promoter | (−) | ACATCCCCTAAATACCACTGAGT |
| −1 | Promoter | (+) | CACCTACAAACTTTTCCATC |
| −1 | Promoter | (−) | AATCTCCACATTTCCCTGAG |
| 1 | Promoter | (+) | GGGCAGGTCTGTGGGATGGTG |
| 1 | Intron 2 | (−) | GCCTCCCTAGCAGCAGCTAAGACT |
| 2 | Intron 2 | (+) | CCTGGGCATTTGCTGGGCGG |
| 2 | Intron 3 | (−) | TGTGAGGACACTGAAGGTGGCA |
| 3 | Intron 3 | (+) | AGCAAGCCCTCTCCTGGGACC |
| 3 | Intron 4 | (−) | AAGGAGTGTCTGATTGTGCAGGAC |
| 4 | Intron 4 | (+) | CTGCTTCCAGCAGGGAGGGC |
| 4 | Intron 5 | (−) | TGGGGTGAGGATCAGGCAGCA |
| 5 | Intron 5 | (+) | AGGTAGGTGGGCCTGCACCC |
| 5 | Intron 6 | (−) | AAGCAGGGTCCAGCCCGTGG |
| 6 | Intron 6 | (+) | CAAGCAGAGCTGTCAAGGATGGT |
| 6 | Intron 7 | (−) | GCTTTCAGGGAGGCCGAGTGG |
| 7 | Intron 7 | (+) | TGGGCATTTGCATTGGGTTGC |
| 7 | Intron 8 | (−) | TGGCTCTTGCAGCAGTCAAACAC |
| 8 | Intron 8 | (+) | TGTTAGCACACCCAAGGCCAC |
| 8 | Intron 9 | (−) | GGAAGTGTACGGAGGTTCGCAGC |
| 9 | Intron 9 | (+) | ACTGTCCACTAGGCTGTGATGGC |
| 9 | Exon 10 | (−) | TGCAGGTGCTGGGCTAACGG |
| 10 | Exon 10 | (+) | TGCCACCTGCAATTCACGCAC |
| 10 | Exon 10 | (−) | TGTGCCTGCATGGTGTCCTTGT |
| 11 | Exon 10 | (+) | AGCAAGAGCAAACTCCCAAGCA |
| 11 | Exon 10 | (−) | GCATCATAGGGCTGTGGGTGG |
| 12 | Exon 10 | (+) | GGCACCTCTGGCACCTCTGC |
| 12 | Exon 10 | (−) | CCACCATGACCTTAAGCAGGCAGC |
| 13 | Exon 10 | (+) | TGGCTGACTGCTGGTTGAGTCG |
| 13 | Exon 10 | (−) | CAAGGGCCTGGGCCTCCTGT |
| 14 | Exon 10 | (+) | AGCAAGCAGAAGCATTGAGTGGC |
| 14 | Exon 10 | (−) | GTGGTGCCCAGAGACACAGCA |
| 15 | Exon 10 | (+) | GCCATCTATGCAGAACTTGCTCTAG |
| 15 | Exon 10 | (−) | AACACATTTGCCCGATCAGCCT |
| 16 | Exon 10 | (+) | AGGTGCCTACCTTCCACACCAAG |
| 16 | DER | (−) | GGCTGTCAAATGTGCAGCTTCCCT |
DER, Downstream enhancing region.
Table 5.
List of primers used for sequencing of the rat Nps gene with location and exons/introns/promoter/downstream enhancing region the primers hybridized to
| Sequential order | Location | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|---|
| 1 | Promoter | (+) | CCCCTGGCCACCCATGTCAC |
| 1 | Exon2, Intron 2 | (−) | AGCCGTGAAGCCCTTACCTTGGA |
| 2 | Promoter | (+) | GCAGGCTCAGACAGCGAGCG |
| 2 | Intron 2 | (−) | GAAACAGCCATTTCCATGTGCAGG |
| 3 | Intron 2 | (+) | TCAGGATGGTGGAGTGCCCAA |
| 3 | Intron 2 | (−) | GCTCATGGCATAGGAGCAAGGACA |
| 4 | Intron 2 | (+) | AAATGATTGCCTTTCTTCGGGGGT |
| 4 | Intron 2 | (−) | ACACCACCTTGTGGCCCAGGA |
| 5 | Intron 2 | (+) | TCCAAGTGGCAACTCCTGCAAGC |
| 5 | Intron 2 | (−) | AGGCAGCACCATCGCTCACC |
| 6 | Intron 2 | (+) | TGTCCCTAAAGGTTTGCTCACCGC |
| 6 | Intron 2 | (−) | ACTGCCCATTTTAAGTCTTGAGCCACC |
| 7 | Intron 2 | (+) | AGTGGCCTCTGGGAAGAGTGG |
| 7 | Intron 2 | (−) | GCCCTGGCTGAGTGAATGACTGG |
| 8 | Intron 2 | (+) | TGCACATCTTCTTTCCTCCAGAGCCA |
| 8 | Intron 2 | (−) | GCCTCCGATGGGAGCTGCTG |
| 9 | Intron 2 | (+) | TCCCAACCCCCAAACAGAGCG |
| 9 | Intron 2 | (−) | ACCGGGCCAAAGGAACCTGC |
| 10 | Intron 2 | (+) | TGGCTCTGGCGCTTGGCTTC |
| 10 | Intron 2 | (−) | AGCCCTAGGTTTAGCCCCCAGC |
| 11 | Intron 2 | (+) | CGGCCTGCCCATGCACACTTA |
| 11 | Exon 3 | (−) | GCCTGGCTGGGCAGGTACTC |
| 12 | Intron 2 | (+) | GCTGTGTTTCAGTGATGTTTCTCCCCA |
| 12 | DER | (−) | GGCGGAAGTTTGAGACAGGTTTGC |
| 13 | Exon 3 | (+) | ACGGAGTCGGCTCAGGGGTG |
| 13 | DER | (−) | CGCTGGCGATCCCTTGCTGC |
| 14 | DER | (+) | ACGACGCGTGGGCGTTTCTAC |
| 14 | DER | (−) | TGACCTGGCAGGGACAGCGA |
| 15 | DER | (+) | CCTGGGTCTGTTTCTCCCCCTC |
| 15 | DER | (−) | CTGGAAGCTGGTGCCAAGGATAC |
| −6 | Promoter | (+) | GGAGCTGCAGGCAAAGCCTCA |
| −6 | Promoter | (−) | ACCCAAACCAAGGTTCCTCACCA |
| −5 | Promoter | (+) | GATACAGCAAACAGGAGGGA |
| −5 | Promoter | (−) | TCTCCAAAGAACAGAGCTCC |
| −4 | Promoter | (+) | CAAGAAGAAGGGAAGTGATGTGGCA |
| −4 | Promoter | (−) | AGGACAAGGAGGTGACCCAGCT |
| −3 | Promoter | (+) | CCCAGGCTTCCAGCTTGGCA |
| −3 | Promoter | (−) | CGGCAGAGGAAAACGTCAGAGGG |
| −2 | Promoter | (+) | CGGATCCTTGTGCTTGCATGGC |
| −2 | Promoter | (−) | GGCCAGGGGCCTCCAAAGGA |
| −1 | Promoter | (+) | CAGCCCTGTCAGCCTGCATCA |
| −1 | Intron 1 | (−) | AGGACCTTGGGTGGGATCTCACAC |
DER, Downstream enhancing region.
Table 6.
List of primers used for sequencing of the mouse Nps gene with location and exons/promoter/downstream enhancing region
| Sequential order | Location | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|---|
| −4 | Promoter | (+) | CCAGGCTTCCAGCTTGGCAC |
| −4 | Promoter | (−) | GCTGCTATTGCTGCTGTTTCTGAAG |
| −3 | Promoter | (+) | GGGTATCTTTGCCCTCCAAAAGGTG |
| −3 | Promoter | (−) | GGCAATCTGTTGTCACTGGTCCCTG |
| −2 | Promoter | (+) | TCCCTGCTCAACACCCCAAACC |
| −2 | Promoter | (−) | ACTGGTTGGCCTGGCTGTGG |
| −1 | Promoter | (+) | GAGGCTCCTGGCCACCCATG |
| −1 | Intron 2 | (−) | GGGCCCTCCACCATCCTGATCA |
| 1 | Promoter | (+) | TGGCAAGCTCTGAGTGAAGTCAACC |
| 1 | Intron 2 | (−) | TTTGGGCCCTCCACCATCCTGA |
| 2 | Promoter, Exon 1 | (+) | CCCATCTGCGCAGGTCTCGG |
| 2 | Intron 2 | (−) | TCCACTGTGCGGGTTTTTGGT |
| 3 | Promoter, Exon 1 | (+) | CATCTGCGCAGGTCTCGG |
| 3 | Intron 2 | (−) | CCAGAGTTACCTACTGTCACATAC |
| 4 | Intron 2 | (+) | AGCCGGTGGTAGCCCTACACT |
| 4 | Exon 3 | (−) | ACTCTGAGCCCGTTAGGAGAAGGG |
| 5 | Exon 3 | (+) | CCTTTCGCAACGGAGTCGGCT |
| 5 | Exon 3 | (−) | CGAGCCCTTGCTGCAGGTACC |
| 6 | Exon 3 | (+) | GTGCCACCAAGTGCAGTGGC |
| 6 | DER | (−) | GCTGGTGACCAAGGACAGGGT |
DER, Downstream enhancing region.
Copy number variation
The isolated genomic DNA was also used to measure the number of gene copies of Nps and Npsr1 using qPCR. The corresponding primers are listed in Table 7. No cytosine–phosphodiester–guanine (CpG) islands, as assessed using CpG island searcher (Takai and Jones, 2002, 2003), were identified.
Table 7.
List of primers used for the measurement of copy number variations of Nps and Npsr1 sequence
| Species, gene | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|
| Rat, Npsr1 | (+) | GCTGCTGCTGCCCTGGCTAA |
| Rat, Npsr1 | (−) | GCCCTCTGTGAGGTTGGCCG |
| Rat, Nps | (+) | AGCTCTGTCGCTGTCCGTGGT |
| Rat, Nps | (−) | AGCCGTGAAGCCCTTACCTTGG |
| Rat, Gapdh | (+) | CGTGTGTAGCGGGCTGCTGT |
| Rat, Gapdh | (−) | CCAGGCGTCCGATACGGCCA |
| Mouse, Npsr1 | (+) | CAGCTGCTGCCCCGGCTAAC |
| Mouse, Npsr1 | (−) | GGTTGGCTGGCATGGCTCAGG |
| Mouse, Nps | (+) | ACGTGCTTTGGTGTTATCCGGTCC |
| Mouse, Nps | (−) | TTGGGCCCTCCACCATCCTGA |
| Mouse, Gapdh | (+) | TCCCCCTATCAGTTCGGAGC |
| Mouse, Gapdh | (−) | AGTAGCTGGGCCTCTCTCAT |
Allele-specific transcription assay
PVN and amygdala tissues were punched as described above from F1 offspring of crossed HAB versus LAB rats or mice, respectively. Both crosses (i.e., HAB or LAB father) were performed to rule out imprinting-based effects. Total RNA was isolated, and the converted cDNA was used to quantify HAB and LAB alleles by qPCR, as described above. The primers were designed to differentiate between the HAB and LAB alleles by incorporating the SNPs between the rat (A(227016)G) and mouse (A(156453)G; rs37572071) lines (Table 8) at the 3′ end of the respective primers.
Table 8.
List of primers used for allele specific quantification of HAB and LAB alleles in F1 offspring of rats and mice
| Species, gene, specificity | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|
| Rat, Npsr1, common | (+) | ACTGTGGCCAGACGACTCCT |
| Rat, Npsr1, rHAB-specific | (−) | CCTCGGTTGTAGCTGCAGCATATC |
| Rat, Npsr1, rLAB-specific | (−) | CCTCGGTTGTAGCTGCAGCATACT |
| Mouse, Npsr1, common | (+) | GTGCTGTTCTCCACGTGCAG |
| Mouse, Npsr1, mLAB-specific | (−) | TCAGGGGCCATGAAGTCTCGT |
| Mouse, Npsr1, mHAB-specific | (−) | TCAGGGGCCATGAAGTCTCGC |
Dual luciferase assays
Npsr1 promoter constructs.
Homologous Npsr1 sequences of HAB and LAB animals were cloned into pGL3 basic luciferase vectors and transfected into mouse neuro-2a cells using Exgen 500 in vitro reagent (Thermo Scientific; Table 9, primer list). The Npsr1 promoter constructs were cotransfected with a pRK5-Gaussia-KDEL vector, to normalize transfection efficiency, and an SV40-pGL3 vector was used as a positive control (a gift from Dr. Theo Rein and Dr. Ulrike Schmidt, Max Planck Institute of Psychiatry, Munich, Germany). Selected constructs were stimulated with 1 and 10 μm/5 μl dexamethasone (DEX; Ratiopharm) for 24 h before carrying out the dual reporter assay (Schülke et al., 2010).
Table 9.
List of primers used for cloning rHAB/rLAB and mHAB/mLAB Npsr1 promoter fragments into luciferase vector
| Species, gene, designation | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|
| Rat, Npsr1, Frag E | (+) | ATCGGTACCGATGGTGAGGGCTGTGCTGG |
| Rat, Npsr1, Frag D | (+) | ATCGGTACCCGATCTCAGCTGAAACAAACTCATAACTC |
| Rat, Npsr1, Frag C | (+) | ATCGGTACCCTCTAGGAATGCACACTTACTCAGCTCTG |
| Rat, Npsr1, Frag B | (+) | ATCGGTACCCCCACTCTAGGGCCTTTCATCTAGG |
| Rat, Npsr1, Frag A | (+) | ATCGGTACCCTGGGTCCTCCAGTCTCTTGAGG |
| Rat, Npsr1, common | (−) | CAGGCTAGCGGCTCAGGCAGGGTCAAGTCTTA |
| Mouse, Npsr1, Frag S | (+) | ATCGGTACCGTGATACCAGCTGAAACAAACACATAACTAC |
| Mouse, Npsr1, Frag R | (+) | ATCGGTACCGAGACAAACACAGACTCCTACCTC |
| Mouse, Npsr1, Frag Q | (+) | ATCGGTACCGCAAAGGTTGGCCTACATGGCTC |
| Mouse, Npsr1, Frag P | (+) | ATCGGTACCGGATTGTCATCTCCTGTCTGTGCC |
| Mouse, Npsr1, common | (−) | CAGGCTAGCGGCTCAGGCAGGGTCAGGTC |
| –, –, RVprimer3 | (+) | CTAGCAAAATAGGCTGTCCC |
| –, –, GLprimer2 | (−) | CTTTATGTTTTTGGCGTCTTCCA |
Npsr1 cDNA constructs.
Both rat and mouse HAB/LAB Npsr1 cDNA were amplified with primers (Table 10) carrying additional FLAG tag sequence using Phusion DNA polymerase (New England Biolabs). Finally, the HAB and LAB Npsr1 cDNAs were cloned into the XhoI/BamHI sites of pcDNA3.1/Zeo(−) (Life Technologies GmbH). Previous studies have shown that NPSR1 signals via the cAMP pathway (Reinscheid et al., 2005); thus, we used a cell line stably expressing a cAMP response element (CRE) cloned upstream of a luciferase reporter (HEK293-CRE-luc; Nordemann et al., 2013) to test the functional properties of HAB and LAB Npsr1 pcDNA constructs. The HEK293-CRE-luc cells were seeded at 2.5 × 104 cells/well in a 96-well plate with DMEM containing 10% FBS, 1% sodium pyruvate, and hygromycin B (200 μg/ml; Sigma-Aldrich). Twenty-two hours later, cells were replaced in media containing no hygromycin B, and then transfection was performed using 30 ng of HAB/LAB Npsr1 pcDNA constructs and 70 ng Gaussia vector per well using lipofectamine 2000 (Life Technologies). The cells were stimulated with 1 nmol/5 μl NPS (Bachem) 40 h after transfection, and then the dual luciferase assay was performed at 48 h after transfection.
Table 10.
List of primers used for cDNA amplification of rat and mouse Npsr1 cDNA sequences with additional FLAG tag
| Species, gene, designation | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|
| Rat, Npsr1, cDNA | (+) | CATGCTCGAGGCCACCATGCCGGCCAACCTCACAGAGGGCA |
| Rat, Npsr1, cDNA with FLAG | (−) | CATGGGATCCTTACTTGTCGTCATCGTCTTTGTAGTCGATGAATTCAGGCTTGGAGA |
| Mouse, Npsr1, cDNA | (+) | CATGCTCGAGGCCACCATGCCAGCCAACCTCACAGAGGGCA |
| Mouse, Npsr1, cDNA with FLAG | (−) | CATGGGATCCTTACTTGTCGTCATCGTCTTTGTAGTC GATGAATTCCGGCTTGGAGA |
| –, –, T7 | (+) | TAATACGACTCACTATAGGG |
| –, –, Bgh | (−) | TAGAAGGCACAGTCGAGG |
Oligonucleotide pull-down assay
To further determine the functionality of the SNP in the promoter region (rHAB, −388; mHAB, −506), we performed oligonucleotide pull-down assays to assess glucocorticoid receptor (GR) binding.
5′-biotinylated forward oligonucleotides, containing either the HAB or LAB sequence variant with putative GR binding properties, and their respective complementary nonbiotinylated strands (Table 11) were resuspended to a final concentration of 20 μm, and then 5 μl each of forward and reverse primer and 5 μl of 10× NEB buffer 2 were diluted up to 35 μl with distilled water. The reaction mix was incubated for 4 min at 95°C and 10 min at 70°C, and was allowed to cool at room temperature for several hours to allow annealing of the oligonucleotides. Streptavidin beads (GE Healthcare Europe GmbH) were washed with 1 ml of ice-cold PBS and incubated with the annealed 5′-biotinylated oligonucleotides overnight. Nuclear extracts were prepared from total brain tissue from a Wistar rat subjected to restraint stress (by placing it in a Plexiglas restrainer for 30 min and killing it an hour later) using the NE-PER nuclear and cytoplasmic extraction reagent (Piercenet). The extract was precleared using streptavidin beads overnight at 4°C, and the protein concentration was estimated using the Pierce BCA Protein Assay Kit (Thermo Scientific). Then, 300 μg of precleared nuclear proteins was incubated with the biotinylated double-stranded streptavidin Sepharose complex for 4 h at room temperature on a rotating shaker. Afterward, the entire complex was washed twice with RIPA buffer to remove unbound proteins, heated at 95°C in sample buffer, and then separated on 10% SDS-PAGE gels. The separated proteins were blotted on a nitrocellulose membrane, blocked for nonspecific binding using 5% milk-TBST for 1 h, and incubated overnight with a polyclonal GR antibody (sc-1004X, Santa Cruz Biotechnology) at 1:200 dilution in 2.5% milk in TBST. The next day, the blot was washed three times for 5 min with TBST and then incubated for 1 h at room temperature in an anti-rabbit HRP-conjugated secondary antibody (1:1000) dilution in 2.5% milk/TBST. Subsequently, the blot was washed three times for 5 min each in TBST, and proteins were visualized by chemiluminescence (Western Lighting, PerkinElmer). Finally, the blot was also subjected to colloidal silver staining (Harper and Speicher, 2001) to serve as the protein loading control.
Table 11.
List of primers used for oligonucleotide pull-down assay
| Species, gene, designation | Orientation | Primer sequence (5′ → 3′) |
|---|---|---|
| Mouse, Npsr1, mLAB-506 specific | (+) | ACATAACTGACAGATT |
| Mouse, Npsr1, mLAB-506 specific | (−) | AATCTGTCAGTTATGT |
| Mouse, Npsr1, mHAB-506 specific | (+) | ACATAACTCTGACAGATT |
| Mouse, Npsr1, mHAB-506 specific | (−) | AATCTGTCAGAGTTATGT |
| Rat, Npsr1, rHAB-388 specific | (+) | TGGAAACTAGAAGAGAGA |
| Rat, Npsr1, rHAB-388 specific | (−) | TCTCTCTTCTAGTTTCCA |
| Rat, Npsr1, rLAB-388 specific | (+) | TGGAAACTGGAAGAGAGA |
| Rat, Npsr1, rLAB-388 specific | (−) | TCTCTCTTCCAGTTTCCA |
Surgical and infusion procedures
Rats and mice were fixed in a stereotaxic apparatus and implanted with an indwelling guide cannula using isoflurane anesthesia under semi-sterile conditions, as previously described (Slattery and Neumann, 2010b; Kessler et al., 2011). A guide cannula (rat: 21 gauge, 12 mm long; mice: 23 gauge, 8 mm long) was implanted 2 or 1.5 mm above the right lateral ventricle [rat: anteroposterior (AP), −1.0 mm from bregma; mediolaterial (ML), +1.6 mm; dorsoventral (DV), +1.8 mm; Paxinos and Watson, 1998; mice: AP, −0.3 mm; ML, +1.1 mm; DV, +1.6 mm; Franklin and Paxinos, 2007] and anchored to two stainless steel screws using dental acrylic. Animals were allowed to recover for at least 7 d before undergoing behavioral testing and injected with either vehicle (rats, 5 μl; mice, 2 μl of Ringer's solution), NPS (0.1 or 1 nmol) or the NPSR1-antagonist (NPSR1-A) D-Cys(tBu)5-NPS (10 nmol; Guerrini et al., 2009; rats, 45 min before the anxiety tests or 20 min before fear extinction training; mice, 25 min before behavioral tests). Cannula placements were verified as previously described (Muigg et al., 2008), leading to the exclusion of three HAB mice from further analysis.
Elevated plus-maze
The EPM was used to assess anxiety-related behavior in both rats and mice, as previously described (Krömer et al., 2005; Slattery and Neumann, 2010b). The 5 min test was performed on a plus-shaped maze, which was elevated (rats, 70 cm; mice, 37 cm) from the floor and consisted of two closed arms (rats: 50 × 10 × 40 cm; 25–30 lux; mice: 30 × 5 × 15 cm; 10 lux) and two open arms (rats: 50 × 10 cm; 90–100 lux; mice: 30 × 5 cm; 300 lux) connected by a central neutral zone (rats: 10 × 10 cm; mice: 5 × 5 cm). A camera above the maze enabled the assessment of behavior. The test started by placing the animal in the neutral zone facing a closed arm. During the test, the percentage of time spent on the open arms, as indicative of anxiety-related behavior, and the number of closed arm entries (rats) or total distance traveled (mice; Anymaze software, Stoelting), as indicative of locomotor behavior, were determined by an observer blind to treatment.
Light-dark box
This test was used to assess anxiety-related behavior in nonselected Wistar rats to confirm the anxiolytic effect of NPS. Briefly, the experimental setup consisted of one lit (40 × 50 cm, 350 lux) and one dark (40 × 30 cm, 70 lux) compartment connected via a small opening (7.5 × 7.5 cm), enabling transition between the compartments. The floors in each compartment were divided into squares (10 × 10 cm). Rats were placed in the dark compartment, and the time spent in the light compartment and the number of line crosses were assessed during the 5 min test. The time spent in the light compartment was used to assess anxiety-related behavior, and the number of line crosses as an indicator of locomotor activity (Slattery and Neumann, 2010b).
Auditory fear conditioning
The auditory fear-conditioning experiment was performed according to previous protocols for mice (Sartori et al., 2011b) and rats (Muigg et al., 2008) involving 120 s of stimulus-free habituation, and consolidation periods before and after the last stimulus presentations in all experimental sessions.
Mice acquired conditioned fear by the pairing of five audible cues [conditioned stimulus (CS); white noise, 80 dB, 2 min] and coterminating mild, short, scrambled foot shocks [unconditioned stimulus (US); 0.7 mA, 2 s] in conditioning chambers (TSE). Twenty-four hours later, three CSs in the absence of the US separated by 5 s were presented to mice for a fear expression test in a novel context (single mouse cage, illuminated with red light to 10 lux).
Rats also received five-tone CSs (white noise, 80 dB, 30 s) that each coterminated with a US (0.7 mA scrambled foot shock, 2 s) in the conditioning context (26 × 30 × 32 cm chamber cleaned with water and illuminated to 300 lux; Coulbourn Instruments). Twenty-four hours later, rats were exposed to 30 CSs in the absence of the US, each separated by an intertrial interval of 5 s for extinction training of cued conditioned fear in a novel context (standard rat cage swiped with ethanol and illuminated with red light to 10 lux). On the third day of the experiment, only three CSs were presented to rats in the extinction context.
The time remaining in freezing behavior during acquisition, fear expression test, and fear extinction training was manually assessed by an observer blind to treatment. Freezing scores during fear expression and during extinction, respectively, were binned into three-block averages. The percentage change in freezing behavior between the first-trial block of the extinction training and the extinction retrieval session was used as an indicator of fear reduction induced by the training and/or drug treatment. Animals showing >25% freezing to the context prior to the first CS presentation were excluded from the analysis.
Statistical analyses
Data were analyzed using either ANOVA (one-way, two-way, or two-way with repeated measures) followed by Fisher's least significant difference post hoc test or an unpaired t test. Statistical significance was set to p < 0.05 and performed using SPSS version 19.0.
Results
HAB rodents display lower Npsr1 mRNA expression and numerous polymorphisms compared with LAB rodents
Initial evidence for a role of the NPSR1 system in the phenotype of the breeding lines was provided by the demonstration of lower expression levels within the PVN and amygdala of HAB rodents compared with their LAB counterparts. rHABs displayed lower Npsr1 mRNA expression than rLABs only in the PVN (p < 0.001; Fig. 1A), and mHABs had lower expression only in the basolateral amygdala compared with mLABs (p < 0.05; Fig. 1B).
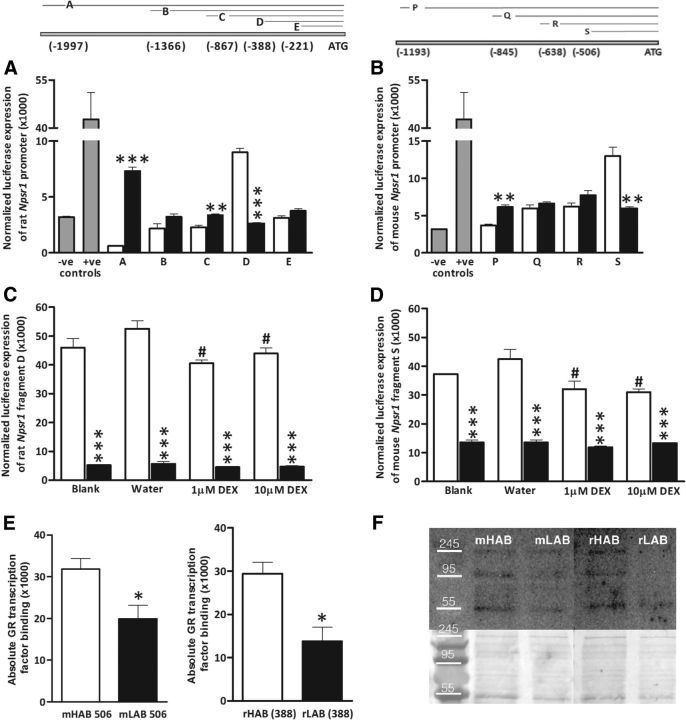
Figure 1.
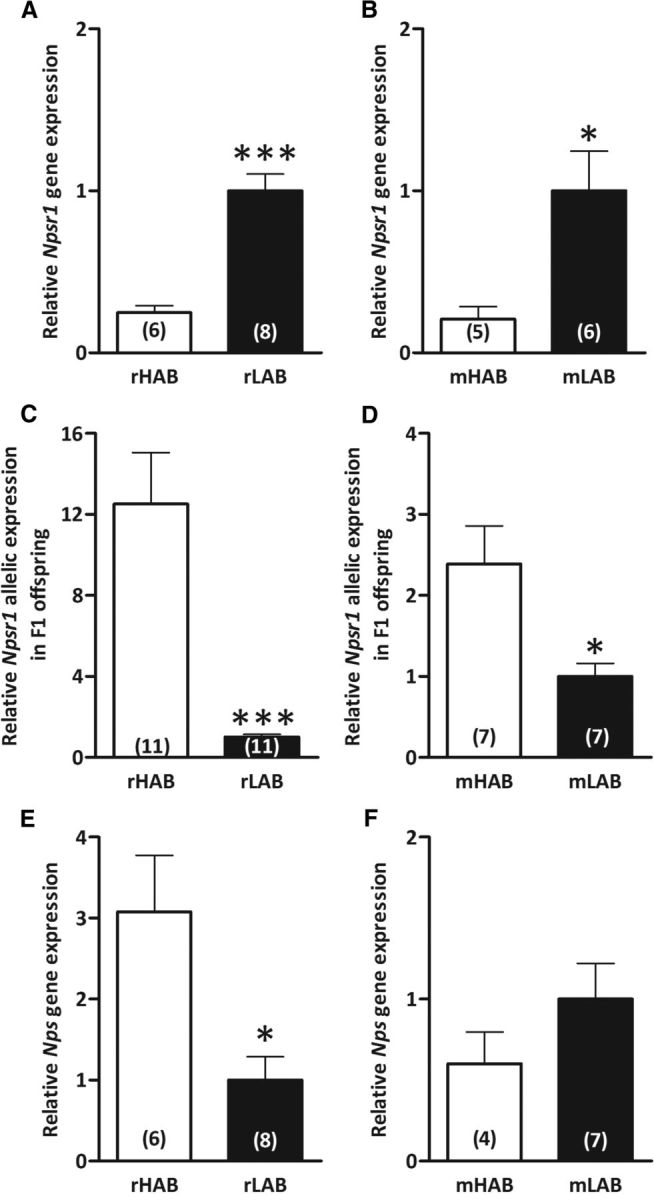
Npsr1 and Nps mRNA expression levels in rats and mice selectively bred for high (rHABs and mHABs) versus low (rLABs and mLABs) anxiety-related behavior, respectively. A, B, Basal Npsr1 mRNA expression in the hypothalamic PVN in rHABs versus rLABs (A), and in the amygdala of mHABs versus mLABs (B). C, D, Allele-specific quantification of Npsr1 alleles using A(227,016)G as a marker to distinguish rHAB- and rLAB-specific alleles in the PVN of F1 offspring (C), and using (A(156,453)G; rs37572071) to distinguish mHAB- and mLAB-specific alleles in the amygdala of F1 offspring (D). E, F, Basal Nps mRNA expression in the locus coeruleus area of rHABs versus rLABs (E) and mHABs versus mLABs (F). Data are shown as the mean ± SEM, and numbers in parentheses indicate group size. *p < 0.05; ***p < 0.001.
Sequencing of the Npsr1 gene demonstrated numerous insertions, deletions, and SNPs between both breeding lines (Tables 12, 13). In both rats and mice, the LAB sequence was identical to that of the reference strain (BN/SsNHsdMCW and C57BL/6J). TESS analysis revealed a nuclear factor-1 binding site within 500 bp of the DNA region in rLABs, while it was altered to a GR binding site in the corresponding rHAB position. In the homologous region of the mouse gene, adjacent to an activator protein-1 (AP-1) binding site, an insertion in the mHAB sequence also led to the introduction of a GR binding site. Intriguingly, within the coding region, synonymous SNPs [A(227016)G and A(156453)G; rs37572071] in exons 8 and 4 of rHABs and mHABs, respectively, were observed (Tables 12, 13).
Table 12.
Npsr1 polymorphisms in rHAB versus rLAB: SNPs, deletions, and insertions
| Variation type | HAB | LAB | Location in Npsr1 gene | Relative position |
|---|---|---|---|---|
| SNP | C | G | Promoter | −1926 |
| SNP | T | C | Promoter | −1856 |
| SNP | C | A | Promoter | −1813 |
| SNP | C | T | Promoter | −1593 |
| Insertion | T | Promoter | −1366 | |
| Deletion | C | Promoter | −1300 | |
| SNP | A | G | Promoter | −908 |
| SNP | G | A | Promoter | −867 |
| SNP | A | G | Promoter | −388 |
| SNP | G | C | Promoter | −221 |
| Deletion | A | Intron 4 | 168,623 | |
| SNP | T | A | Intron 4 | 198,121 |
| SNP | C | T | Intron 5 | 198,323 |
| Deletion | CTT | Intron 5 | 198,338 ∼198,340 | |
| SNP | G | A | Exon 8 | 227,016 |
| SNP | C | T | Intron 8 | 227,154 |
| SNP | C | T | Exon 10 | 231,601 |
| Deletion | C | Exon 10 | 231,993 | |
| Insertion | AGAGAGAGAGAG | Exon 10 | 232,152 | |
| SNP | C | T | Exon 10 | 232,218 |
| SNP | C | T | Exon 10 | 232,505 |
| Insertion | TGTCTCTCTCT | DER | 234,193 | |
| SNP | A | G | DER | 234,331 |
| SNP | A | G | DER | 234,985 |
| SNP | C | T | DER | 235,041 |
| SNP | G | A | DER | 235,223 |
| SNP | T | C | DER | 235,279 |
Positions are relative to the ATG start codon. DER, Downstream enhancing region.
Table 13.
Npsr1 polymorphisms in mHAB versus mLAB: SNPs, deletions and insertions
| Variation type | HAB | LAB | Location in Npsr1 gene | Relative position | SNP identifier |
|---|---|---|---|---|---|
| SNP | G | A | Promoter | −2251 | rs50949943 |
| SNP | C | T | Promoter | −2248 | rs48292984 |
| SNP | C | G | Promoter | −2123 | rs47083749 |
| SNP | T | C | Promoter | −2112 | rs49887483 |
| SNP | T | A | Promoter | −2104 | rs47000117 |
| SNP | T | C | Promoter | −2046 | rs48022291 |
| SNP | G | A | Promoter | −1942 | rs46860992 |
| SNP | T | C | Promoter | −1863 | rs51840884 |
| SNP | T | A | Promoter | −1842 | rs45839541 |
| SNP | C | T | Promoter | −1775 | rs52096988 |
| SNP | G | A | Promoter | −1667 | |
| Deletion | AA (GA) × 18 | Promoter | Approximately −1608 to −1571 | ||
| SNP | G | A | Promoter | −1516 | |
| SNP | T | C | Promoter | −1469 | |
| SNP | G | A | Promoter | −1418 | |
| SNP | A | G | Promoter | −1376 | |
| SNP | A | T | Promoter | −1315 | |
| SNP | C | T | Promoter | −1236 | rs48864073 |
| Deletion | T | Promoter | −1227 | ||
| SNP | A | T | Promoter | −1226 | rs36643873 |
| SNP | A | T | Promoter | −1132 | |
| SNP | T | G | Promoter | −1072 | rs51941766 |
| SNP | G | A | Promoter | −1032 | |
| SNP | C | T | Promoter | −860 | rs45719875 |
| SNP | G | A | Promoter | −816 | rs37067240 |
| SNP | C | T | Promoter | −731 | rs50871983 |
| SNP | C | T | Promoter | −714 | rs48580633 |
| SNP | T | G | Promoter | −713 | rs47842102 |
| SNP | C | G | Promoter | −674 | rs46047101 |
| Deletion | T | Promoter | −648 | ||
| SNP | T | A | Promoter | −637 | rs45879530 |
| SNP | T | C | Promoter | −612 | rs51858460 |
| SNP | T | C | Promoter | −591 | rs46930781 |
| SNP | C | A | Promoter | −557 | rs50633535 |
| Insertion | C | Promoter | −479 | ||
| Insertion | T | Promoter | −478 | ||
| SNP | A | T | Intron 1 | −82 | rs48722200 |
| Insertion | A | Intron 3 | 156,233 | ||
| SNP | G | A | Exon 4 | 156,453 | rs37572071 |
| SNP | C | T | Intron 7 | 205,648 | |
| Deletion | C | Intron 7 | 205,718 | ||
| SNP | T | C | Intron 8 | 215,240 | |
| SNP | G | C | Intron 9 | 215,243 | |
| SNP | G | A | Intron 9 | 215,244 | |
| SNP | A | G | Intron 9 | 215,245 | |
| SNP | G | A | Exon 10 | 216,508 | rs49543460 |
| SNP | G | A | Exon 10 | 217,782 | rs49030747 |
| SNP | A | G | DER | 218,543 |
Positions are relative to the ATG start codon. DER, Downstream enhancing region.
cis-Acting polymorphisms drive higher HAB allelic expression
As the differences in expression could be due to cis–trans factor interactions, we assessed HAB and LAB allelic expression in cross-mated F1 offspring. Higher HAB allelic expression, regardless of the maternal line, was observed in the F1 generation (rats: p < 0.001; Fig. 1C; mice: p < 0.05; Fig. 1D), which is in apparent contrast to the expression levels reported above.
HAB and LAB rodents differ in Nps mRNA expression and sequence
We could show higher Nps mRNA expression in the LC of rHABs compared with rLABs (p < 0.05; Fig. 1E), but no polymorphisms were observed between the rat lines. In contrast, while no mRNA expression differences between mHABs and mLABs were observed (Fig. 1F), numerous polymorphisms in the Nps gene were found (Table 14). In total, 35 SNPs and one insertion were found within the mLAB Nps sequence, whereas the mHAB Nps sequence was identical to the reference mouse strain (C57BL/6J). The gene-coding locus of mLAB carried four SNPs, leading to amino acid changes at positions leucine(5)isoleucine, valine(10)isoleucine, and arginine(54)glycine; and a synonymous mutation coding for threonine at position 65 in the amino acid sequence—all before the mature 20 aa peptide (Table 14).
Table 14.
Nps polymorphisms in mHAB versus mLAB: SNPs, deletions, and insertions
| Variation type | HAB | LAB | Location in Nps gene | Relative position | SNP identifier |
|---|---|---|---|---|---|
| SNP | A | T | Promoter | −1031 | |
| SNP | T | C | Promoter | −1030 | |
| Insertion | GTGT | Promoter | −995 | ||
| SNP | T | C | Promoter | −924 | rs49048062 |
| SNP | A | G | Promoter | −920 | rs42460586 |
| SNP | T | C | Promoter | −871 | |
| SNP | C | T | Promoter | −868 | |
| SNP | T | G | Promoter | −867 | |
| SNP | A | G | Promoter | −819 | rs50307957 |
| SNP | C | T | Promoter | −788 | rs50890340 |
| SNP | G | A | Promoter | −712 | rs49326925 |
| SNP | A | C | Promoter | −621 | rs52014995 |
| SNP | T | C | Promoter | −316 | |
| SNP | C | T | Promoter | −163 | |
| SNP | C | T | Promoter | −13 | rs33467230 |
| SNP | T | A | Exon 2 | 125 | |
| SNP | G | A | Exon 2 | 140 | |
| SNP | G | A | Intron 2 | 250 | |
| SNP | G | A | Intron 2 | 273 | |
| SNP | A | T | Intron 2 | 380 | |
| SNP | A | C | Intron 2 | 460 | |
| SNP | C | T | Intron 2 | 502 | |
| SNP | A | G | Exon 3 | 3624 | rs33470378 |
| SNP | A | G | Exon 3 | 3659 | rs33470381 |
| SNP | C | T | Exon 3 | 3937 | rs33471194 |
| SNP | A | G | Exon 3 | 4022 | rs33471197 |
| SNP | T | C | Exon 3 | 4127 | rs33471203 |
| SNP | A | G | Exon 3 | 4155 | rs33471946 |
| SNP | C | A | Exon 3 | 4195 | rs50157889 |
| SNP | C | G | Exon 3 | 4196 | rs33466004 |
| SNP | C | T | Exon 3 | 4215 | rs47207120 |
| SNP | A | G | Exon 3 | 4217 | rs46716508 |
| SNP | G | A | Exon 3 | 4240 | rs49462104 |
| SNP | T | C | Exon 3 | 4264 | rs51623072 |
| SNP | T | A | DER | 4321 | rs33466010 |
| SNP | G | A | DER | 4369 |
Positions are relative to the ATG start codon. DER, Downstream enhancing region.
HAB and LAB Npsr1 promoter assays help explain the mRNA expression differences
In both rats and mice, an equal number of gene copies of Npsr1 and Nps were observed in HAB and LAB (Nps: rHABs, 1.1 ± 0.13; rLABs, 1.0 ± 0.06; mHABs, 0.83 ± 0.12; mLABs, 1.0 ± 0.26; Npsr1: rHABs, 1.1 ± 0.09; rLABs, 1.0 ± 0.05; mHABs, 0.94 ± 0.21; mLABs, 1.0 ± 0.31), and no CpG islands were detected in the promoter region of either gene. Thus, to further assess the discrepancy between lower basal HAB Npsr1 mRNA and higher HAB-specific allelic expression in F1 offspring, we performed dual-luciferase assays. When rat promoter constructs of ∼2000 bp upstream of the ATG translation start site were assessed, rHAB activity was approximately half of the corresponding rLAB promoter (p < 0.001; Fig. 2A). We also assessed the activity of promoter–deletion constructs to deduce the contribution of individual SNPs. No difference was observed between rHAB and rLAB Frag B (−1366 to start codon), while a higher activity in rLABs than rHABs was seen in Frag C (−867 to start codon; p < 0.01). These fragments comprise numerous SNPs, making their interpretation difficult, but they do reveal that the SNPs alter promoter activity. However, Frag D (−388 to start codon; containing only one SNP, which introduces a putative GR binding site) had a nearly twofold higher rHAB rat promoter activity in comparison with that in rLABs (p < 0.001). Subsequent deletion of the G (−388)A SNP led to Frag E (−221 to start codon) with the C (−221)G SNP, where promoter activity did not differ between rHABs and rLABs (Fig. 2A). Similarly, using homologous mouse Npsr1 promoter constructs, we observed that the activity of the full-length mHAB construct was lower than the corresponding mLAB construct [Frag P (−1193 to start codon); p < 0.01]. As the putative promoter length decreased, no difference in the corresponding promoter activity between mHABs and mLABs was observed ≥Frag Q (−845 to start codon) and R (−634 to start codon)]; although, again, interpretation is difficult due to these fragments containing numerous SNPs. However, Frag S (−506 to start codon), which harbors the putative GR binding site and only one SNP, displayed higher mHAB promoter activity than the corresponding mLAB construct (p < 0.01; Fig. 2B). Moreover, while single CpG bases may influence promoter activity, as shown for human NPSR1 (Reinius et al., 2013), these data suggest that the SNPs in Npsr1 promoter regions are sufficient to explain the observed differences in mRNA expression.
Figure 2.
Dual luciferase assays reveal differences in HAB (white columns) and LAB (black columns) rodent Npsr1 promoter fragments, and pull-down assays show preferential GR binding to the HAB promoter. Fragments displayed in the upper pictogram, with fragments A–E in rats (left), and P–S in mice (right). A–D, Dual luciferase assays of rHAB versus rLAB Npsr1 promoter constructs in pGL3 basic vector (A); mHAB versus mLAB Npsr1 promoter constructs in pGL3 basic vector (B); rHAB versus rLAB fragment D treated with 1, 10 μm DEX, and water as a control for 24 h (C); and mHAB versus mLAB fragment S carrying the putative GR binding site treated with 1, 10 μm DEX, and water as control for 24 h before the assay (D). E, F, Semi-quantitative Western blots for GR transcription factor binding to mHAB 506 versus mLAB 506 and rHAB 388 versus rLAB 388 (E), and representative images with left and right lower images showing corresponding colloidal silver staining (F). Data are shown as the mean ± SEM (n = 3 for each group and represent independent assays performed in triplicate; except n = 4 for Fig. 2E); −ve and +ve controls represent pGL3 basic and SV40-pGL3 vectors, respectively. *p < 0.05, **p < 0.01, ***p < 0.001, compared with HAB; #p < 0.05, when compared with water.
DEX administration reduces the activity of the HAB Npsr1 promoter
We next assessed the effect of DEX administration on HAB and LAB Npsr1 promoter activity to determine whether the predicted GR binding site close to the ATG start codon in the HAB rodents was functional. In keeping with our in silico data, the high activity of HAB Fragments D (F(3,16) = 3.37; p = 0.045) and S (F(3,16) = 4.12; p = 0.023), which was recapitulated here (p < 0.001 for all HAB vs LAB comparisons), could be reduced by DEX administration (p < 0.05 vs water; Fig. 2C,D).
Oligonucleotide pull-down assay reveals functional significance of the HAB NPSR1 SNP
We could further show that the GR has significantly higher binding to the mHAB (−506) and rHAB (−388) Npsr1 promoters compared with the corresponding LAB regions (both p < 0.05; Fig. 2E,F). In addition to the expected 95 kDa band size, there were degradation bands at ∼55 and >245 kDa, which represent different antibody-specific immunoprecipitates (i.e., different GR isoforms or binding to their interaction partners).
Synonymous SNP in the HAB rat and mouse NPSR1 coding region causes increased functional activity
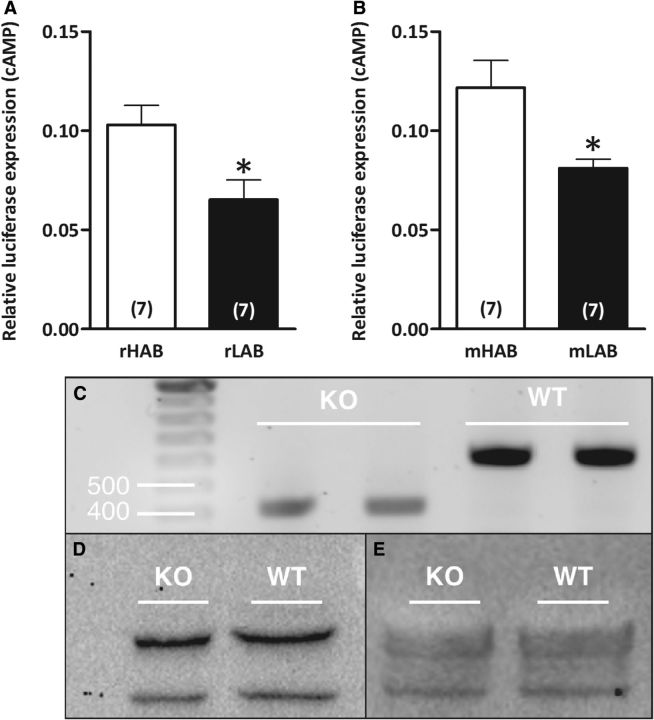
Previous studies have shown that synonymous SNPs are not always silent (Duan et al., 2003; Hunt et al., 2009) and that the nonsynonymous SNP in the coding region of NPSR1 leads to increased NPS efficacy and cell surface expression (Bernier et al., 2006). Therefore, we tested the functional impact of the synonymous SNPs in the HAB NPSR1 coding regions by transiently transfecting the HAB versus LAB NPSR1 constructs into cells expressing a luciferase gene downstream of CRE. This assay revealed higher NPS-dependent luciferase expression in cells expressing the HAB NPSR1 protein in both rats and mice compared with transient expression of respective LAB NPSR1 proteins (both p < 0.05; Fig. 3A,B), which is akin to the human Ile107 risk isoform. However, we could not determine NPSR1 protein levels owing to the nonspecific nature of the two presently available antibodies, as revealed in genotype-confirmed WT and NPSR1 KO mice (Fig. 3C–E). Thus, despite an ∼200 bp deletion in the coding region in NPSR1 KO animals, as revealed by Western blot, a 43 kDa band (expected NPSR1 size; ab92425 antibody) or 72 kDa band (Ab2; Leonard and Ring, 2011) were observed in both WT and KO animals. Thus, both the presence of a respective band and the lack of difference in its weight provide evidence for the unspecificity of the currently available NPSR1 antibodies.
Figure 3.
Functional differences in HAB and LAB NPSR1 variants. A, B, HEK-CRE-luc cells were transfected with rHAB or rLAB Npsr1 cDNA constructs carrying A(227,016)G (A) and mHAB or mLAB Npsr1 cDNA constructs carrying A(156,453)G (B); rs37572071 along with Gaussia vector and stimulated with 1 nmol/5 μl NPS at 40 h after transfection until assay at 48 h. C, NPSR1 genotyping using hypothalamic tissue from WT and NPSR1 KO mice. D, E, Semi-quantitative Western blots for total NPSR1 protein of WT and NPSR1 KO mice in hypothalamic paraventricular nucleus tissue with Abcam antibody ab92425 (43 kDa; D) and with Ab2 (72 kDa; Leonard and Ring, 2011; E). β-Tubulin served as a loading control for both Western blots. Data are shown as the mean ± SEM, and numbers in parentheses indicate the group size. **p < 0.01. The luciferase assay was performed independently, and each n was an average of three replicates.
Central NPS administration rescues the anxiety-related behavior and fear deficits in HAB rodents
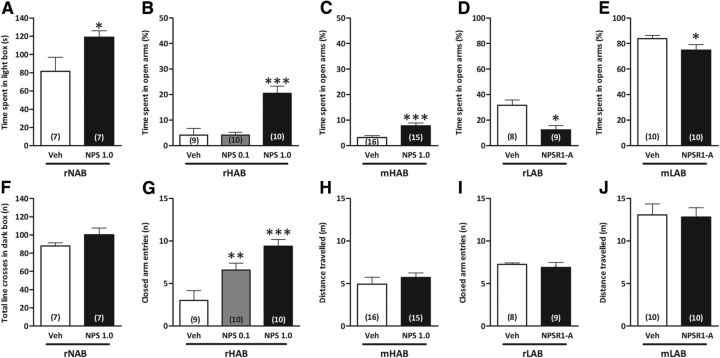
We next determined whether these genetic and functional differences resulted in behavioral consequences of NPS and NPSR1-A administration in HAB and LAB rodents with genetic predisposition to extremes in anxiety. Initially, we confirmed the anxiolytic effect of intracerebroventricular NPS (1 nmol) administration in rNAB rats, as reflected by an increased amount of time spent in the light compartment of the light-dark box (LDB; p < 0.05; Fig. 4A), whereas the locomotor activity was not altered by NPS (Fig. 4F). The anxiolytic effect of intracerebroventricular NPS administration has also previously been shown in nonselected CD-1 mice (Ionescu et al., 2012).
Figure 4.
Central NPS and NPSR1-A administration attenuate the anxiety phenotypes of HAB and LAB rodents. A–C, Intracerebroventricular NPS administration exerted an anxiolytic effect in rNABs in the LDB (A); in rHABs in a dose-dependent manner, with 1 nmol, but not 0.1 nmol, increasing the time an rHAB spent on the open arms of the EPM (B); and in mHABs compared with vehicle treatment (C). D, E, Contrastingly, intracerebroventricular administration of D-Cys(tBu)5-NPS (10 nmol), an NPSR1-A, increased the anxiety of rLABs (D) and mLABs (E) on the EPM. F–H, Intracerebroventricular NPS administration differentially affected locomotor activity with unchanged number of line crosses of rNABs in the LDB (F), increased closed arm entries of rHABs on the EPM (G), and unchanged distances that mHABs traveled on the EPM (H). I, J, D-Cys(tBu)5-NPS did not affect locomotor activity in either rLABs (I) or mLABs (J). Data represent the mean ± SEM. Numbers in parentheses indicate group size. *p < 0.05, **p < 0.01, ***p < 0.001, compared with vehicle group. Veh, Vehicle.
In rHABs, intracerebroventricular NPS administration dose-dependently altered the percentage of time spent in the open arms of the EPM (F(2,26) = 17.7; p < 0.001) with 1 nmol (p < 0.001), but not 0.1 nmol, leading to an anxiolytic effect (Fig. 4B); whereas both doses (F(2,26) = 12.0; p < 0.001) increased the number of closed arm entries (0.1 nmol, p < 0.01; 1 nmol, p < 0.001; Fig. 4G). Central NPS administration also significantly increased the percentage of time spent in the open arms of the EPM in mHABs (p < 0.001; Fig. 4C) without altering locomotor activity (Fig. 4H).
On the other hand, blockade of the endogenous NPS system by intracerebroventricular administration of NPSR1-A (10 nmol) decreased the percentage of time rLABs and mLABs spent on the open arms of the EPM (p < 0.05; Fig. 4D,E), although it did not affect locomotion (Fig. 4I,J).
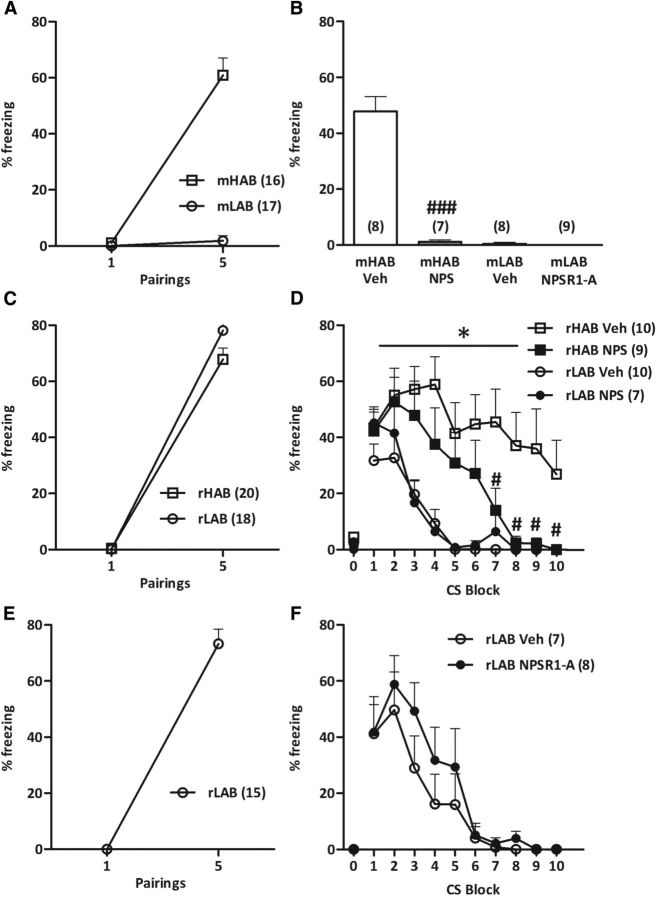
We next tested whether intracerebroventricular NPS could reduce the enhanced fear expression and/or facilitate the impaired fear extinction previously described in mHABs (Sartori et al., 2011b; Yen et al., 2012) and rHABs (Muigg et al., 2008), respectively.
Conditioned freezing responses increased in mHABs (pairing: F(1,15) = 93.0; p < 0.001), but not mLABs (pairing: F(1,16) = 1.036; p = 0.324; Fig. 5A) across the five CS–US pairings, which is in line with our previous study (Yen et al., 2012). Twenty-four hours later, intracerebroventricular NPS completely abolished fear expression in mHABs (p < 0.001; Fig. 5B), while the NPSR1-A did not alter freezing levels in mLABs (p = 0.307; Fig. 5B).
Figure 5.
Central NPS administration reverses the cued-fear conditioning deficits of HAB rodents. A, C, E, As a result of acquisition, mHABs (A), rHABs, and rLABs (C, E), but not mLABs (A), showed an increased freezing response to the presentation of the conditioned stimulus (tone). B, D, Twenty-four hours after fear conditioning, intracerebroventricular administration of NPS (1 nmol) 20 min before extinction training completely abolished the expression of cue-conditioned fear in mHABs (B) and facilitated the impaired extinction of conditioned fear in rHABs (D). B, F, Intracerebroventricular administration of an NPSR1-A (10 nmol) did not alter cued fear expression in mLABs (B) or cued fear extinction in rLABs (F). D, F, Neither rHABs nor rLABs showed any freezing to the context on day 2 before the first CS presentation. Data represent the mean ± SEM; numbers in parentheses indicate group size. *p < 0.05, compared with vehicle-treated LAB animals; #p < 0.05 and ###p < 0.001, compared with respective vehicle-treated HAB group. Veh, Vehicle.
In the rat study, rHABs and rLABs acquired cue-conditioned fear to the same extent (pairing: F(1,35) = 495; p < 0.001; Fig. 5C; and rLAB pairing: F(1,14) = 202; p < 0.001; Fig. 5E). Importantly, neither rHABs nor rLABs showed any freezing to the context on day 2 before the first CS presentation (Fig. 5D,F). While there was no difference in fear expression between rHABs and rLABs (F(1,32) = 1.17; p = 0.288), vehicle-administered rHABs exhibited a retarded fear extinction compared with vehicle-administered rLABs (group: F(3,29) = 5.961; p = 0.003; CS: F(9,261) = 18.4; p < 0.001; Fig. 5D), confirming our previous results (Muigg et al., 2008). However, intracerebroventricular NPS administration significantly accelerated fear extinction in rHABs, but not in rLABs (Fig. 5D), which was also evident in the extinction retrieval test, as evidenced by a greater reduction in freezing between the first block of extinction training and the extinction retrieval session (line × treatment: F(1,29) = 10.4; p = 0.003; data not shown). NPSR1-A administration did not affect the decrease in freezing levels displayed by rLABs during fear extinction training (drug: F(1,13) = 0.669; p = 0.428; CS: F(9,117) = 19.5; p < 0.001; drug × CS interaction: F(9,117) = 0.667; p = 0.738; Fig. 5E), and between the first block of extinction training and the extinction retrieval test (p = 0.320; data not shown).
Discussion
In this study, we demonstrate that selective breeding of rats and mice for extremes in anxiety-like behavior leads to numerous differences in the Npsr1 gene sequences between HAB and LAB rodents, which, in turn, cause alterations in transcriptional and functional activity. SNPs were observed in the Npsr1 promoter region in HAB rodents that causes transcriptional inhibition. However, analogous to the effect of the Ile107 SNP in the human NPSR1, a SNP in the exonal region of the rHAB and mHAB receptor sequence resulted in increased NPSR1 signal transduction upon agonist exposure. This increase in NPSR1 activity counteracts the effect of the SNPs in the promoter region and rescue NPSR1 function. Indeed, we could demonstrate that NPS administration elicits anxiolytic and fear-attenuating effects in HAB rodents, supporting this hypothesis. Together, these results suggest that attenuated endogenous NPS system activity mediates, at least in part, the anxiety- and fear-related phenotype of HAB rodents.
Given that the NPS system has been implicated in both anxiety and fear regulation, we speculated that genetic differences may contribute to the behavioral phenotypes of HAB and LAB rats and mice. Indeed, we initially demonstrated that Npsr1 mRNA expression within the PVN (rHABs) and basolateral amygdala (mHABs), regions implicated in local NPS-mediated effects (Smith et al., 2006; Jüngling et al., 2008; Meis et al., 2008), were lower compared with their respective LAB line. Moreover, numerous polymorphisms were also found in the Npsr1 gene sequence of HABs, while, in contrast, the LAB lines did not differ from their respective reference genomes. While no difference in Nps mRNA expression was observed between the mouse lines, we could demonstrate a threefold higher expression level in rHABs compared with rLABs in the LC. In contrast, although no Nps sequence differences were observed between the rat lines, polymorphisms in the Nps gene leading to 3 aa substitutions within the putative signal peptide sequence of mLABs were identified (Table 11). However, it remains to be determined whether these differences in Nps expression and sequence play a role in the rHAB and mLAB phenotypes, respectively.
To discern whether the Npsr1 polymorphisms underlie the differential mRNA expression between the breeding lines, we next performed dual luciferase assays. These assays recapitulated the in vivo findings, as whole HAB rat and mouse constructs had lower promoter activity in vitro than their respective LAB constructs, which must be driven by the sequence variation. In contrast, as the putative promoter length was decreased, there was a twofold higher promoter activity in rHABs and mHABs, which is analogous to the higher HAB-specific allelic expression in the F1 offspring. This indicates the importance of the whole promoter, and the cross talk between cis-acting polymorphisms and trans-acting factors. Intriguingly, TESS analyses revealed the introduction of a plausible GR binding site in the HAB sequence compared with a nuclear factor (NF)-1 or AP-1 site in the corresponding LAB promoters. A previous study (Mori et al., 1997) showed that GR causes the suppression of downstream interleukin-5 gene expression by interfering with activities of AP-1 or NF-κB. Moreover, the presence of AP-1 and NF-1 has been shown to enhance chromatin accessibility and subsequent GR binding, which in turn helps to recruit other coactivators in the absence of glucocorticoids (Belikov et al., 2004; Biddie et al., 2011). Thus, in HAB rodents it is possible that the adjacent NF-1 or AP-1 site enhances GR binding, and that this would, in turn, recruit other coactivators, leading to enhanced HAB-specific expression of Npsr1.
In contrast, DEX stimulation led to GR activation, which interferes with the activity of basal transcription factors like AP-1 and NF-κB, leading to trans-repression (Newton and Holden, 2007). Our finding that DEX administration decreased the promoter activity of Npsr1 in HAB rodents only suggests that such a cis–trans interaction is present in this line. Further, using oligonucleotide pull-down assays, we could confirm that the GR binds to this corresponding region of the HAB Npsr1 promoters. Although synonymous mutations, such as those observed in both rHABs and mHABs, do not lead to an altered amino acid sequence, they have previously been shown to affect mRNA splicing, stability, protein structure, synthesis, and folding (Cartegni and Krainer, 2002; Cartegni et al., 2002). It has been shown that guanine nucleotides are more frequently observed than adenine nucleotides in the third position of synonymous substitutions (Hunt et al., 2009), and that this may impact both the incorporation rate of amino acids into newly synthesized proteins as well as its subsequent translocation. Such strong codon usage bias has been shown to affect many aspects of gene synthesis in the human dopamine D2 receptor (Duan et al., 2003). Indeed, we found that both the rHAB and mHAB NPSR1 protein products resulted in greater NPS-induced cAMP response, similar to the human NPSR1 Ile107 isoform (Reinscheid et al., 2005). As we could not confirm NPSR1 protein levels, owing to the nonspecific nature of currently available antibodies, two possible explanations for this finding exist. Thus, the synonymous SNP may lead to higher surface receptor expression in HAB rodents, as observed for the human Ile107 isoform (Bernier et al., 2006), or it may directly affect NPS–NPSR1 signaling. Future experiments are required to determine which of the two mechanisms is responsible for the HAB NPSR1 phenotype. The findings demonstrating that selective breeding for anxiety leads to alterations in both the Nps and Npsr1 genes and/or expression are intriguing, given the apparent discrepancy between recent preclinical findings of anxiolytic NPS effects and those from human studies. Thus, HAB rodents provide a suitable model to study the effect of NPS administration in animals displaying a similar NPSR1 functional phenotype to that of humans with the Ile107 allele. We initially recapitulated and confirmed the anxiolytic effect of intracerebroventricular NPS administration in nonselected Wistar rats (Lukas and Neumann, 2012) and CD-1 mice bred for intermediate anxiety (Ionescu et al., 2012) that has been reported in a number of rat and mouse strains (Xu et al., 2004; Leonard et al., 2008; Rizzi et al., 2008; Wegener et al., 2012). Importantly, we could extend this by demonstrating that even hyperanxiety levels driven by a (seemingly) rigid genetic predisposition and resistance to traditional anxiolytic drugs in mHABs (Sah et al., 2012) are reversed by intracerebroventricular administration of NPS in HAB rats and mice. Although a possible link between anxiety and locomotion has been discussed (Escorihuela et al., 1999), and has been repeatedly described after intracerebroventricular NPS administration (Xu et al., 2007; Ruzza et al., 2012), we could dissociate the anxiolytic and arousal effects of NPS in both HAB rats and mice. Moreover, intracerebroventricular administration of a NPSR1-A (Guerrini et al., 2009) specifically increased anxiety in LAB rats and mice, suggesting an involvement of the endogenous NPS system in the determination of their low innate anxiety levels. This is particularly likely as NPSR1-A administration has not been shown to have an effect on anxiety alone in rodents.
As the Ile107 NPSR1 is associated with panic and startle responses in humans (Domschke et al., 2012), we finally determined whether intracerebroventricular NPS administration could reduce conditioned fear expression and facilitate the impaired fear extinction, respectively, in HAB rodents, as shown before in nonselected mice (Jüngling et al., 2008; Meis et al., 2008). Indeed, intracerebroventricular NPS administration was able to reduce the exaggerated fear expression seen in mHABs and to accelerate fear extinction in rHABs. In contrast to the anxiogenic effect observed on the EPM, NPSR1-A administration did not affect fear extinction or fear expression in LAB rats and mice. The fact that NPS administration can rescue the behavioral phenotypes of HAB rodents, coupled with our molecular findings, suggests that a lack of endogenous NPS release may underlie their high anxiety- and fear-like phenotypes. Therefore, we propose that a similar situation may exist in Ile107 NPSR1 carriers, given the similar increased NPS-induced signaling in mice, rats, and humans.
In summary, we could reveal genetic differences within the NPS system of rats and mice bred for extremes in anxiety-related behavior, which result in expressional and functional differences. Moreover, we have shown that synthetic NPS is an efficient anxiolytic agent, even in rats and mice with a strong genetic predisposition to high anxiety, and that it can reverse their deficits in fear extinction. Our findings suggest that differences in the brain NPS system underlie, at least partly, the HAB versus LAB behavioral phenotypes, which seems all the more probable given that functional differences have evolved in both rat and mouse lines, and mirror those in humans with the NPSR1 Ile107 allele. Finally, these findings provide evidence to explain the apparent discrepancy between the preclinical and clinical observations to date, and suggest that NPS administration may be particularly effective in anxious patients with the high-risk variant.
Footnotes
This work was supported by the Austrian Science Fund FWF P25375 and SFB F4410 to N.S., European Research Council, Bundesministerium für Bildung und Forschung, Elitenetwork of Bavaria (to I.D.N.) and the Deutsche Forschungsgemeinschaft (to I.D.N. and D.A.S.). We thank Professor A. Buschauer (University of Regensburg) for the cell line, and Drs. U. Schmidt and T. Rein for the vectors. We also thank Dr. D. Beiderbeck, R. Maloumby, and M. Nussbaumer for their excellent technical assistance.
The authors declare no competing financial interests.
References
- Belikov S, Astrand C, Holmqvist PH, Wrange O. Chromatin-mediated restriction of nuclear factor 1/CTF binding in a repressed and hormone-activated promoter in vivo. Mol Cell Biol. 2004;24:3036–3047. doi: 10.1128/MCB.24.7.3036-3047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier V, Stocco R, Bogusky MJ, Joyce JG, Parachoniak C, Grenier K, Arget M, Mathieu MC, O'Neill GP, Slipetz D, Crackower MA, Tan CM, Therien AG. Structure-function relationships in the neuropeptide S receptor: molecular consequences of the asthma-associated mutation N107I. J Biol Chem. 2006;281:24704–24712. doi: 10.1074/jbc.M603691200. [DOI] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, Vinson C, Stamatoyannopoulos JA, Hager GL. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunck M, Czibere L, Horvath C, Graf C, Frank E, Kessler MS, Murgatroyd C, Müller-Myhsok B, Gonik M, Weber P, Pütz B, Muigg P, Panhuysen M, Singewald N, Bettecken T, Deussing JM, Holsboer F, Spengler D, Landgraf R. A hypomorphic vasopressin allele prevents anxiety-related behavior. PLoS One. 2009;4:e5129. doi: 10.1371/journal.pone.0005129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Slattery DA. Animal models of mood disorders: recent developments. Curr Opin Psychiatry. 2007;20:1–7. doi: 10.1097/YCO.0b013e3280117733. [DOI] [PubMed] [Google Scholar]
- Czibere L, Baur LA, Wittmann A, Gemmeke K, Steiner A, Weber P, Pütz B, Ahmad N, Bunck M, Graf C, Widner R, Kühne C, Panhuysen M, Hambsch B, Rieder G, Reinheckel T, Peters C, Holsboer F, Landgraf R, Deussing JM. Profiling trait anxiety: transcriptome analysis reveals cathepsin B (Ctsb) as a novel candidate gene for emotionality in mice. PLoS One. 2011;6:e23604. doi: 10.1371/journal.pone.0023604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domschke K, Reif A, Weber H, Richter J, Hohoff C, Ohrmann P, Pedersen A, Bauer J, Suslow T, Kugel H, Heindel W, Baumann C, Klauke B, Jacob C, Maier W, Fritze J, Bandelow B, Krakowitzky P, Rothermundt M, Erhardt A, et al. Neuropeptide S receptor gene—converging evidence for a role in panic disorder. Mol Psychiatry. 2011;16:938–948. doi: 10.1038/mp.2010.81. [DOI] [PubMed] [Google Scholar]
- Domschke K, Klauke B, Winter B, Gajewska A, Herrmann MJ, Warrings B, Muhlberger A, Wosnitza K, Dlugos A, Naunin S, Nienhaus K, Fobker M, Jacob C, Arolt V, Pauli P, Reif A, Zwanzger P, Deckert J. Modification of caffeine effects on the affect-modulated startle by neuropeptide S receptor gene variation. Psychopharmacology (Berl) 2012;222:533–541. doi: 10.1007/s00213-012-2678-0. [DOI] [PubMed] [Google Scholar]
- Donner J, Haapakoski R, Ezer S, Melén E, Pirkola S, Gratacòs M, Zucchelli M, Anedda F, Johansson LE, Söderhäll C, Orsmark-Pietras C, Suvisaari J, Martín-Santos R, Torrens M, Silander K, Terwilliger JD, Wickman M, Pershagen G, Lönnqvist J, Peltonen L, et al. Assessment of the neuropeptide S system in anxiety disorders. Biol Psychiatry. 2010;68:474–483. doi: 10.1016/j.biopsych.2010.05.039. [DOI] [PubMed] [Google Scholar]
- Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- Elfving B, Bonefeld BE, Rosenberg R, Wegener G. Differential expression of synaptic vesicle proteins after repeated electroconvulsive seizures in rat frontal cortex and hippocampus. Synapse. 2008;62:662–670. doi: 10.1002/syn.20538. [DOI] [PubMed] [Google Scholar]
- Escorihuela RM, Fernández-Teruel A, Gil L, Aguilar R, Tobeña A, Driscoll P. Inbred Roman high- and low-avoidance rats: differences in anxiety, novelty-seeking, and shuttlebox behaviors. Physiol Behav. 1999;67:19–26. doi: 10.1016/S0031-9384(99)00064-5. [DOI] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. Ed 3. San Diego: Academic; 2007. [Google Scholar]
- Guerrini R, Camarda V, Trapella C, Caló G, Rizzi A, Ruzza C, Fiorini S, Marzola E, Reinscheid RK, Regoli D, Salvadori S. Further studies at neuropeptide s position 5: discovery of novel neuropeptide S receptor antagonists. J Med Chem. 2009;52:4068–4071. doi: 10.1021/jm900604g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Harper S, Speicher DW. Detection of proteins on blot membranes. Curr Protoc Protein Sci. 2001 doi: 10.1002/0471140864.ps1008s00. chapter10:unit 10.8. [DOI] [PubMed] [Google Scholar]
- Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol. 2009;578:23–39. doi: 10.1007/978-1-60327-411-1_2. [DOI] [PubMed] [Google Scholar]
- Ionescu IA, Dine J, Yen YC, Buell DR, Herrmann L, Holsboer F, Eder M, Landgraf R, Schmidt U. Intranasally administered neuropeptide S (NPS) exerts anxiolytic effects following internalization into NPS receptor-expressing neurons. Neuropsychopharmacology. 2012;37:1323–1337. doi: 10.1038/npp.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüngling K, Seidenbecher T, Sosulina L, Lesting J, Sangha S, Clark SD, Okamura N, Duangdao DM, Xu YL, Reinscheid RK, Pape HC. Neuropeptide S-mediated control of fear expression and extinction: role of intercalated GABAergic neurons in the amygdala. Neuron. 2008;59:298–310. doi: 10.1016/j.neuron.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MS, Bosch OJ, Bunck M, Landgraf R, Neumann ID. Maternal care differs in mice bred for high vs. low trait anxiety: impact of brain vasopressin and cross-fostering. Soc Neurosci. 2011;6:156–168. doi: 10.1080/17470919.2010.495567. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Wang PS. The descriptive epidemiology of commonly occurring mental disorders in the United States. Annu Rev Public Health. 2008;29:115–129. doi: 10.1146/annurev.publhealth.29.020907.090847. [DOI] [PubMed] [Google Scholar]
- Krömer SA, Kessler MS, Milfay D, Birg IN, Bunck M, Czibere L, Panhuysen M, Pütz B, Deussing JM, Holsboer F, Landgraf R, Turck CW. Identification of glyoxalase-I as a protein marker in a mouse model of extremes in trait anxiety. J Neurosci. 2005;25:4375–4384. doi: 10.1523/JNEUROSCI.0115-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf R, Kessler MS, Bunck M, Murgatroyd C, Spengler D, Zimbelmann M, Nussbaumer M, Czibere L, Turck CW, Singewald N, Rujescu D, Frank E. Candidate genes of anxiety-related behavior in HAB/LAB rats and mice: focus on vasopressin and glyoxalase-I. Neurosci Biobehav Rev. 2007;31:89–102. doi: 10.1016/j.neubiorev.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Leonard SK, Ring RH. Immunohistochemical localization of the neuropeptide S receptor in the rat central nervous system. Neuroscience. 2011;172:153–163. doi: 10.1016/j.neuroscience.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Leonard SK, Dwyer JM, Sukoff Rizzo SJ, Platt B, Logue SF, Neal SJ, Malberg JE, Beyer CE, Schechter LE, Rosenzweig-Lipson S, Ring RH. Pharmacology of neuropeptide S in mice: therapeutic relevance to anxiety disorders. Psychopharmacology. 2008;197:601–611. doi: 10.1007/s00213-008-1080-4. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lukas M, Neumann ID. Nasal application of neuropeptide S reduces anxiety and prolongs memory in rats: social versus non-social effects. Neuropharmacology. 2012;62:398–405. doi: 10.1016/j.neuropharm.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Meis S, Bergado-Acosta JR, Yanagawa Y, Obata K, Stork O, Munsch T. Identification of a neuropeptide S responsive circuitry shaping amygdala activity via the endopiriform nucleus. PLoS One. 2008;3:e2695. doi: 10.1371/journal.pone.0002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori A, Kaminuma O, Suko M, Inoue S, Ohmura T, Hoshino A, Asakura Y, Miyazawa K, Yokota T, Okumura Y, Ito K, Okudaira H. Two distinct pathways of interleukin-5 synthesis in allergen-specific human T-cell clones are suppressed by glucocorticoids. Blood. 1997;89:2891–2900. [PubMed] [Google Scholar]
- Muigg P, Hetzenauer A, Hauer G, Hauschild M, Gaburro S, Frank E, Landgraf R, Singewald N. Impaired extinction of learned fear in rats selectively bred for high anxiety—evidence of altered neuronal processing in prefrontal-amygdala pathways. Eur J Neurosci. 2008;28:2299–2309. doi: 10.1111/j.1460-9568.2008.06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann ID, Veenema AH, Beiderbeck DI. Aggression and anxiety: social context and neurobiological links. Front Behav Neurosci. 2010;4:12. doi: 10.3389/fnbeh.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann ID, Wegener G, Homberg JR, Cohen H, Slattery DA, Zohar J, Olivier JD, Mathé AA. Animal models of depression and anxiety: what do they tell us about human condition? Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1357–1375. doi: 10.1016/j.pnpbp.2010.11.028. [DOI] [PubMed] [Google Scholar]
- Newton R, Holden NS. Separating transrepression and transactivation: a distressing divorce for the glucocorticoid receptor? Mol Pharmacol. 2007;72:799–809. doi: 10.1124/mol.107.038794. [DOI] [PubMed] [Google Scholar]
- Nordemann U, Wifling D, Schnell D, Bernhardt G, Stark H, Seifert R, Buschauer A. Luciferase reporter gene assay on human, murine and rat histamine H4 receptor orthologs: correlations and discrepancies between distal and proximal readouts. PLoS One. 2013;8:e73961. doi: 10.1371/journal.pone.0073961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura N, Hashimoto K, Iyo M, Shimizu E, Dempfle A, Friedel S, Reinscheid RK. Gender-specific association of a functional coding polymorphism in the Neuropeptide S receptor gene with panic disorder but not with schizophrenia or attention-deficit/hyperactivity disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:1444–1448. doi: 10.1016/j.pnpbp.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotoxic coordinates. Ed 2. San Diego: Academic; 2001. [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotactic coordinates. San Diego: Academic; 1998. [Google Scholar]
- Raczka KA, Gartmann N, Mechias ML, Reif A, Buchel C, Deckert J, Kalisch R. A neuropeptide S receptor variant associated with overinterpretation of fear reactions: a potential neurogenetic basis for catastrophizing. Mol Psychiatry. 2010;15:1045, 1067–1074. doi: 10.1038/mp.2010.79. [DOI] [PubMed] [Google Scholar]
- Reinius LE, Gref A, Sääf A, Acevedo N, Joerink M, Kupczyk M, D'Amato M, Bergström A, Melén E, Scheynius A, Dahlén SE, Pershagen G, Söderhäll C, Kere J. DNA methylation in the Neuropeptide S Receptor 1 (NPSR1) promoter in relation to asthma and environmental factors. PLoS One. 2013;8:e53877. doi: 10.1371/journal.pone.0053877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinscheid RK, Xu YL, Okamura N, Zeng J, Chung S, Pai R, Wang Z, Civelli O. Pharmacological characterization of human and murine neuropeptide s receptor variants. J Pharmacol Exp Ther. 2005;315:1338–1345. doi: 10.1124/jpet.105.093427. [DOI] [PubMed] [Google Scholar]
- Rizzi A, Vergura R, Marzola G, Ruzza C, Guerrini R, Salvadori S, Regoli D, Calo G. Neuropeptide S is a stimulatory anxiolytic agent: a behavioural study in mice. Br J Pharmacol. 2008;154:471–479. doi: 10.1038/bjp.2008.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzza C, Pulga A, Rizzi A, Marzola G, Guerrini R, Calo' G. Behavioural phenotypic characterization of CD-1 mice lacking the neuropeptide S receptor. Neuropharmacology. 2012;62:1999–2009. doi: 10.1016/j.neuropharm.2011.12.036. [DOI] [PubMed] [Google Scholar]
- Sah A, Schmuckermair C, Sartori SB, Gaburro S, Kandasamy M, Irschick R, Klimaschewski L, Landgraf R, Aigner L, Singewald N. Anxiety- rather than depression-like behavior is associated with adult neurogenesis in a female mouse model of higher trait anxiety- and comorbid depression-like behavior. Transl Psychiatry. 2012;2:e171. doi: 10.1038/tp.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori SB, Landgraf R, Singewald N. The clinical implications of mouse models of enhanced anxiety. Future Neurol. 2011a;6:531–571. doi: 10.2217/fnl.11.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori SB, Hauschild M, Bunck M, Gaburro S, Landgraf R, Singewald N. Enhanced fear expression in a psychopathological mouse model of trait anxiety: pharmacological interventions. PLoS One. 2011b;6:e16849. doi: 10.1371/journal.pone.0016849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics chapter. 2008;2 doi: 10.1002/0471250953.bi0206s21. unit 2.6. [DOI] [PubMed] [Google Scholar]
- Schülke JP, Wochnik GM, Lang-Rollin I, Gassen NC, Knapp RT, Berning B, Yassouridis A, Rein T. Differential impact of tetratricopeptide repeat proteins on the steroid hormone receptors. PloS One. 2010;5:e11717. doi: 10.1371/journal.pone.0011717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery DA, Neumann ID. Oxytocin and major depressive disorder: experimental and clinical evidence for links to aetiology and possible treatment. Pharmaceuticals. 2010a;3:702–724. doi: 10.3390/ph3030702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery DA, Neumann ID. Chronic ICV oxytocin attenuates the pathological high anxiety state of selectively bred Wistar rats. Neuropharmacology. 2010b;58:56–61. doi: 10.1016/j.neuropharm.2009.06.038. [DOI] [PubMed] [Google Scholar]
- Smith KL, Patterson M, Dhillo WS, Patel SR, Semjonous NM, Gardiner JV, Ghatei MA, Bloom SR. Neuropeptide S stimulates the hypothalamo-pituitary-adrenal axis and inhibits food intake. Endocrinology. 2006;147:3510–3518. doi: 10.1210/en.2005-1280. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- Wegener G, Finger BC, Elfving B, Keller K, Liebenberg N, Fischer CW, Singewald N, Slattery DA, Neumann ID, Mathé AA. Neuropeptide S alters anxiety, but not depression-like behaviour in Flinders Sensitive Line rats: a genetic animal model of depression. Int J Neuropsychopharmacol. 2012;15:375–387. doi: 10.1017/S1461145711000678. [DOI] [PubMed] [Google Scholar]
- Xu YL, Reinscheid RK, Huitron-Resendiz S, Clark SD, Wang Z, Lin SH, Brucher FA, Zeng J, Ly NK, Henriksen SJ, de Lecea L, Civelli O. Neuropeptide S: a neuropeptide promoting arousal and anxiolytic-like effects. Neuron. 2004;43:487–497. doi: 10.1016/j.neuron.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Xu YL, Gall CM, Jackson VR, Civelli O, Reinscheid RK. Distribution of neuropeptide S receptor mRNA and neurochemical characteristics of neuropeptide S-expressing neurons in the rat brain. J Comp Neurol. 2007;500:84–102. doi: 10.1002/cne.21159. [DOI] [PubMed] [Google Scholar]
- Yen YC, Mauch CP, Dahlhoff M, Micale V, Bunck M, Sartori SB, Singewald N, Landgraf R, Wotjak CT. Increased levels of conditioned fear and avoidance behavior coincide with changes in phosphorylation of the protein kinase B (AKT) within the amygdala in a mouse model of extremes in trait anxiety. Neurobiol Learn Mem. 2012;98:56–65. doi: 10.1016/j.nlm.2012.04.009. [DOI] [PubMed] [Google Scholar]