Abstract
Entrainment to environmental light/dark (LD) cycles is a central function of circadian clocks. In Drosophila, entrainment is achieved by Cryptochrome (CRY) and input from the visual system. During activation by brief light pulses, CRY triggers the degradation of TIMELESS and subsequent shift in circadian phase. This is less important for LD entrainment, leading to questions regarding light input circuits and mechanisms from the visual system. Recent studies show that different subsets of brain pacemaker clock neurons, the morning (M) and evening (E) oscillators, have distinct functions in light entrainment. However, the role of CRY in M and E oscillators for entrainment to LD cycles is unknown. Here, we address this question by selectively expressing CRY in different subsets of clock neurons in a cry-null (cry0) mutant background. We were able to rescue the light entrainment deficits of cry0 mutants by expressing CRY in E oscillators but not in any other clock neurons. Par domain protein 1 molecular oscillations in the E, but not M, cells of cry0 mutants still responded to the LD phase delay. This residual light response was stemming from the visual system because it disappeared when all external photoreceptors were ablated genetically. We concluded that the E oscillators are the targets of light input via CRY and the visual system and are required for normal light entrainment.
Keywords: circadian clock, clock neurons, Cryptochrome, Drosophila melanogaster, light entrainment
Introduction
Light is the most important environmental time cue [zeitgeber time (ZT)] for circadian clocks in most organisms. The light-resetting mechanism in the clock of the model organism Drosophila melanogaster is primarily explained by the rapid degradation of one clock protein, TIMELESS (TIM), by light (Lee et al., 1996; Myers et al., 1996; Zeng et al., 1996). Although the Drosophila visual system, which includes the compound eyes, ocelli, and Hofbauer–Buchner (H–B) eyelets, mediates light information to the clock, these pathways do not cause a rapid degradation of TIM (Stanewsky et al., 1998; Yang et al., 1998; Emery et al., 2000). In contrast, light-dependent TIM degradation is mainly caused by the blue light-sensitive protein Cryptochrome (CRY), which is expressed in pacemaker clock neurons in the brain and can interact directly with TIM in these cells (Ceriani et al., 1999; Klarsfeld et al., 2004; Benito et al., 2008). After exposure to light, CRY is activated and undergoes a conformational change at the C terminus, leading to its binding to TIM (Busza et al., 2004; Dissel et al., 2004; Vaidya et al., 2013). After this, the TIM/CRY complex is ubiquitinated by JETLAG (JET) and undergoes degradation via the proteasomal pathway (Koh et al., 2006; Peschel et al., 2006).
There are ∼150 neurons expressing clock genes in the Drosophila brain, provisionally called “clock neurons” (Muraro et al., 2013). They are divided into nine groups: (1) dorsal neuron (DN)1a; (2) DN1p; (3) DN2; (4) DN3; (5) lateral posterior neuron; (6) lateral neuron (LN)d; (7) fifth small type (s)-LNv; (8) large type (l)-LNv; and (9) s-LNv (Helfrich-Förster, 2003, 2007). s-LNv and l-LNv neurons contain pigment-dispersing factor (PDF), a neuropeptide that is involved in intercellular communication between the clock neurons (Shafer et al., 2008; Yoshii et al., 2009b). Furthermore, the s-LNv group is important for anticipatory morning activity and the pacemaker center that maintains free-running rhythms in constant darkness (DD) (Stoleru et al., 2005; Picot et al., 2007). In contrast, LNd and fifth s-LNv neurons are important for anticipatory evening activity. Therefore, neurons in the former group are called morning (M) cells and the latter are named evening (E) cells.
CRY is expressed in most of the important clock neurons (Yoshii et al., 2008), but several clock neurons that do not contain CRY or express only very low levels are nevertheless synchronized by light. There are additional differences in the expression levels of CRY between different clock neurons (Yoshii et al., 2008). These facts have raised the question of whether separate clock neurons have different functions in CRY-dependent light entrainment. Furthermore, several studies have shown that the responses to short light pulses are different in M and E oscillators (Shang et al., 2008; Tang et al., 2010; Lamba et al., 2014). Nevertheless, little is known about the role of M and E cells, or CRY, for entrainment to regular light/dark (LD) cycles. To address this issue, in the present study, we generated fly lines that express CRY in subsets of different neurons and tested their entrainability to LD cycles.
Materials and Methods
Fly strains.
w1118 (w) flies were used as a control strain in this study. This line was used originally to create the cry01 mutant; therefore, both have the same genetic background except for the replacement of the cry locus by a mini-white containing P-element transgene in the mutant (Dolezelova et al., 2007). To test the effect of eye color, CantonS flies were outcrossed into the w background for six generations to create red-eyed flies, named w+, that have the w genetic background. To rescue cry expression in a subset of clock neurons, cry01 mutants were crossed with the following transgenic flies: uas–cry (Emery et al., 1998), tim(uas)–gal4 (Blau and Young, 1999), gmr–gal4 (Freeman, 1996), cry–gal4 #39 (Klarsfeld et al., 2004), Pdf–gal4 (Renn et al., 1999), npf–gal4 (Wu et al., 2003), trpA1SH–gal4 (Hamada et al., 2008), c929–gal4 (Taghert et al., 2001), mai179–gal4 (Grima et al., 2004), r6–gal4 (Helfrich-Förster et al., 2007), Pdf–gal8096A (Stoleru et al., 2004), Clk4.1M–gal4 (Zhang et al., 2010), R78G01–gal4, and R54D11–gal4 (Pfeiffer et al., 2008). Clk4.1M–gal4, R78G01–gal4, and R54D11–gal4 transgenes were inserted on the third chromosome in which the cry gene is located; therefore, cry01 strains containing the gal4 transgenes were generated by meiotic recombination. w;uas–GFP S65t (#1522) flies were obtained from the Bloomington Drosophila Stock Center. Flies were reared under 12/12 h LD cycles on Drosophila medium (0.7% agar, 8.0% glucose, 3.3% yeast, 4.0% cornmeal, 2.5% wheat embryo, and 0.25% propionic acid) at 25°C.
Determination of s-tim and ls-tim alleles.
To determine s-tim and ls-tim polymorphisms of each fly stain, PCR was performed using the following primer set: sense, TAGGTATCGCCCTCCAAG; and antisense, TAGGCAGCTCCACAATCA (Schlichting et al., 2014). The resulting PCR products were subjected to DNA sequencing. CantonS, Pdf–gal8096A, and trpA1SH–gal4 strains possess the ls-tim allele. w1118, cry01, uas–cry, tim(uas)–gal4, cry–gal4 #39, Pdf–gal4, npf–gal4, c929–gal4, mai179–gal4, r6–gal4, R78G01–gal4, and R54D11–gal4 strains possess the s-tim allele.
Activity recording and data analysis.
Three- to 6-d-old male flies were used to record locomotor activity rhythms. Flies were confined in recording tubes containing agar/sugar food (2% agar and 4% sucrose) for the Drosophila Activity Monitor (DAM2; Trikinetics). The monitors were placed in an incubator (CN-40A; Mitsubishi Electric) at a constant temperature of 20°C. White LEDs were set above the monitors in the incubator, and lights on and off were controlled by an LC4 light controller (Trikinetics). The light intensity used in all experiments was 100 lux (3.2 μW/cm−2). To ensure that all strains were entrained completely under 16/8 h LD, the activity rhythms of all strains were first recorded for 7 d under 16/8 h LD. Subsequently, the flies were subjected to an 8 h phase delay of LD by a prolongation of the light phase as shown in Figure 3.
Figure 3.
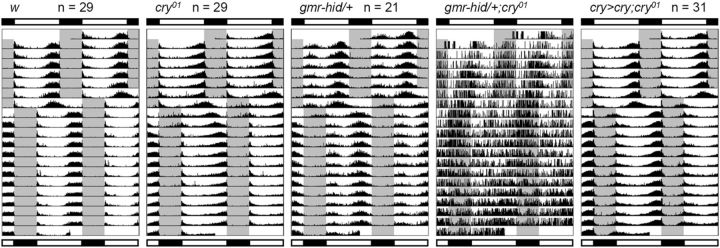
Activity rhythms under 16/8 h LD before and after an 8 h LD phase shift. The gray area in the actograms indicates the dark phase. Bars above and below the actograms indicate light conditions before and after the LD phase shift, respectively. Actograms show mean activity rhythms calculated from the number of flies indicated next to the strain names. Because gmr–hid/+;cry01 flies were not able to synchronize to LD cycles, the actogram is of a representative individual fly. cry>cry;cry01 indicates w;cry–gal4 #39/uas–cry;cry01.
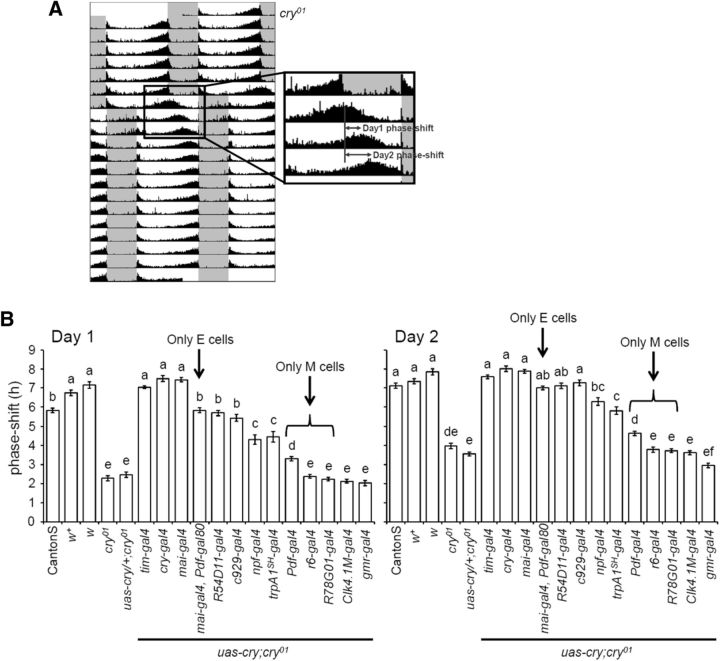
For visual inspection, raw data were displayed as actograms using ActogramJ (http://actogramj.neurofly.de/; Schmid et al., 2011). To calculate the magnitude of the phase shift, daily activity profiles of individual flies were plotted, and the phase of the evening peak of each fly was determined manually, as described previously (Rieger et al., 2003). The phase angles between the evening peak on the day of the LD phase shift and the evening peak on day 1 (or day 2) after the LD phase shift were calculated for individual flies as shown in Figure 5A, and the mean phase angles were calculated for each strain. For statistical analysis, Tukey's multiple comparison test was used after testing for a normal distribution with the Kolmogorov–Smirnov test. Statistics were calculated using EZR software, which is based on R (Kanda, 2013).
Figure 5.
The magnitude of the phase shifts after an 8 h LD phase delay. A, Determination of the phase angle between the evening activity peak before and after the LD phase shift. The phase of the evening activity peak was determined on the day when the light phase was prolonged to induce a shift in the LD cycle. The next day was regarded as day 1, when the activity phase shift occurred. B, The magnitude of the phase shifts (mean ± SEM) on day 1 (left) and day 2 (right) after an 8 h LD phase delay. The control strains (CantonS, w+, and w) were phase shifted by 6–7 h in response to an 8 h LD shift on the first day, suggesting that these flies almost finished re-entrainment within 1 d. Lowercase letters above the bars indicate significant differences revealed by Tukey's multiple comparison test (p < 0.01).
Immunohistochemistry and confocal imaging.
Whole flies were fixed in 4% paraformaldehyde in PBS with 0.1% Triton X-100 for 2.5 h at room temperature (RT). Fixed flies were washed three times in PBS, and then their brains were dissected. After washing three times with PBS containing 0.5% Triton X-100 (PBS-T), brains were blocked in PBS-T containing 5% normal donkey or goat serum for 1 h at RT and subsequently incubated in primary antibodies at 4°C for 48 h. After washing six times in PBS-T, the brains were incubated with secondary antibodies at RT for 3 h. Then brains were washed six times in PBS-T and mounted in Vectashield mounting medium (Vector Laboratories). The primary antibodies used were chicken anti-GFP (1:2000; ab13970; Abcam), mouse anti-PDF (1:1000; Developmental Studies Hybridoma Bank; Cyran et al., 2005), guinea pig anti-vrille (VRI; 1:3000; Glossop et al., 2003), rat anti-TIM (1:1000; Yoshii et al., 2008), rabbit anti-CRY (1:1000; Yoshii et al., 2008), rabbit anti-ion transport peptide (ITP; 1;10,000; Hermann-Luibl et al., 2014), and rabbit anti-Par domain protein 1 (PDP1; 1:3000; Cyran et al., 2003). We used the following fluorescence-conjugated secondary antibodies at a 1:500 concentration: Alexa Fluor antibodies (Life Technologies) of 488 nm (goat anti-chicken and goat anti-mouse), 555 nm (goat anti-rabbit), 635 nm (goat anti-mouse and goat anti-guinea pig), and goat anti-rabbit or anti-rat IgG Cy3 (Millipore).
Staining was visualized using laser scanning confocal microscopes (Fluoview300 from Olympus; and TCS SPE from Leica). For quantification of PDP1 immunostaining, the confocal microscope settings were kept constant throughout the experiments. For each time point, nine hemispheres from nine different brains were analyzed. Measurement of staining intensity was performed using NIH ImageJ (W. S. Rasband, National Institutes of Health, Bethesda, MD), as described previously (Yoshii et al., 2009a).
Results
CRY-dependent TIM degradation by a light pulse
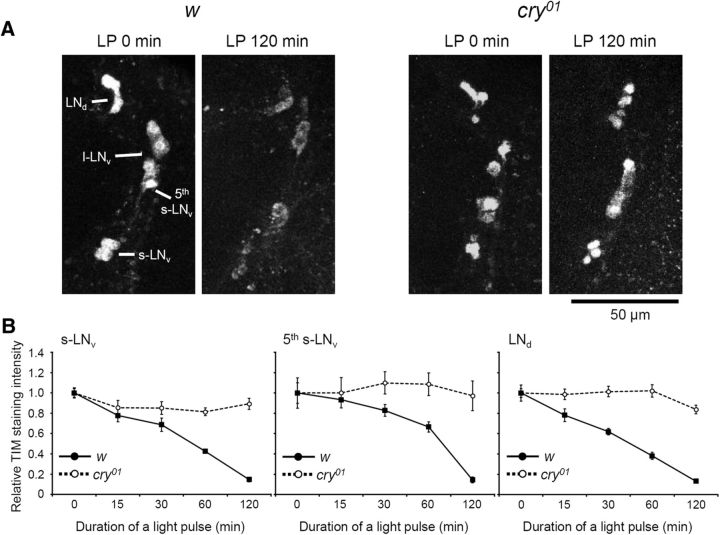
Previous studies using whole-head extracts and S2 cells have shown that TIM is degraded rapidly by light in a CRY-dependent manner (Busza et al., 2004; Peschel et al., 2009). To test the speed of TIM degradation in the brain clock neurons under light, w control flies and cry01 mutants, which were entrained to 12/12 h LD, were subjected to a light pulse of different durations, starting at different ZTs, before collecting the flies at ZT 21. At ZT 21, TIM is normally at its peak level, whereas the tim mRNA level is already declined (So and Rosbash, 1997). Thus, we expected to observe relatively clear TIM degradation kinetics at this time point, unaffected by its mRNA synthesis. Our immunohistochemical analysis revealed that TIM required a 120 min light pulse to decline to a level almost undetectable by immunohistochemistry in all LN groups of the control flies, including the CRY-negative LNds (Fig. 1A,B). These data fit to TIM degradation kinetics from head extracts (Busza et al., 2004) but not to in vitro studies (Ozturk et al., 2014), in which TIM is more rapidly degraded by light. It is worth mentioning also that much shorter light pulses (although depending on light intensity) are sufficient for causing TIM degradation (Vinayak et al., 2013), albeit complete depletion is not reached until a few hours after the light pulse. Even after a 120 min light pulse, there was still some residual TIM detectable, especially in the cytoplasm (Fig. 1A). Therefore, light-dependent TIM degradation may be more efficient in the nucleus than in the cytoplasm.
Figure 1.
TIM degradation kinetics after light pulses (LP). Flies were exposed to light pulses of different durations that started at different ZTs until ZT 21, when the flies were collected. A, TIM immunostaining in LN cells in w control brains (left 2 panels) and cry01 mutant brains (right 2 panels) at ZT 21. PDF costaining was also performed to distinguish the groups of LN cells (images not shown). B, TIM degradation kinetics in three LN groups in response to different light pulses. To normalize data, TIM staining intensity with no light pulse was set to 1.0. The mean ± SEM staining intensity was calculated from 10 hemispheres of 10 different brains. TIM staining was almost completely abolished after a 120 min light pulse in w control brains (solid lines), whereas it remained at a high level in cry01 mutant brains (dashed lines).
In cry01 mutants, TIM levels did not change even after a 120 min light pulse, clearly indicating that TIM degradation by the light pulse was indeed mediated by CRY. The same TIM kinetics were also observed in the DN groups, including CRY-negative DN1p and DN2 neurons (data not shown). Thus, CRY-dependent TIM degradation also occurs in CRY-negative clock neurons, as shown in a previous study (Yoshii et al., 2008). For unknown reasons, TIM staining in the DN groups in cry01 mutants was very weak even without a light pulse. Therefore, quantification was not performed in these cells. Possible reasons for this weak TIM staining in the DN groups are as follows: (1) a phase shift of the TIM peak in the mutants away from ZT21, when the staining was performed, or (2) a novel role of CRY in modulating TIM stability in the dark. However, both hypotheses require additional investigation to be validated.
Characterization of gal4 lines
Before generating fly lines that express CRY only in subsets of clock neurons, we screened the Janelia gal4 stocks (Pfeiffer et al., 2008) for strains suitable for our purpose. We found two gal4 lines that showed rather specific GFP expression in the clock neurons when crossed with uas–gfp flies. In particular, GFP expression of R78G01–gal4/uas–gfp flies was observed in the l-LNv and few additional non-clock cells in the brain (Fig. 2A). R54D11–gal4/uas–gfp flies showed GFP signal in the fifth s-LNv, one ITP-positive LNd (CRY-positive neuron; Johard et al., 2009), one l-LNv, very weakly in two s-LNv neurons, and very few non-clock cells in the brain (Fig. 2B). We also investigated GFP expression in trpA1SH–gal4/uas–gfp flies showing GFP signal in the fifth s-LNv, three CRY-positive LNd, and one DN1a (Fig. 2C; Hamada et al., 2008). The c929–gal4 line had been used previously as an l-LNv-specific driver (Taghert et al., 2001). Here, we found that it also shows expression in the fifth s-LNv and one ITP (and CRY)-positive LNd, in addition to all l-LNv neurons (Fig. 2D). We used these lines together with well characterized clock-neuron-specific gal4 drivers to generate the cry-rescue flies (Table 1).
Figure 2.
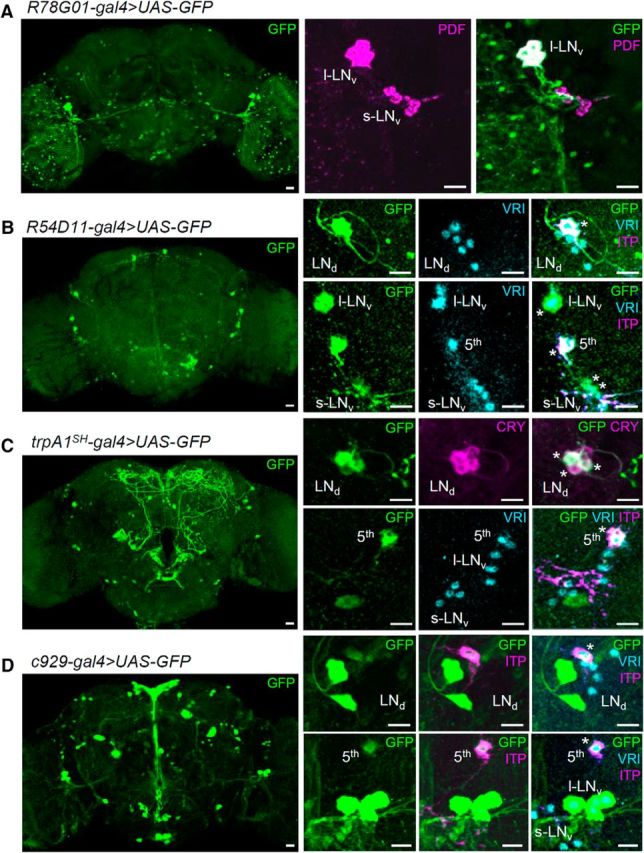
Characterization of gal4 strains. gal4 lines were crossed with uas–GFP–S65t to visualize the gal4 expression patterns. Flies were entrained to 12/12 h LD for 5 d and were dissected at ZT 19. Brains were triple stained with anti-GFP (green), anti-VRI (cyan), and anti-PDF (magenta in A) or anti-ITP (magenta in bottom rows of B–D). The ITP-positive LNd and fifth s-LNv are CRY-positive neurons (Johard et al., 2009). trpA1SH–gal4/uas–GFP flies were double stained with anti-GFP (green) and anti-dCRY (magenta) to determine whether GFP-positive LNd were also CRY positive (C, top row). Scale bars, 10 μm.
Table 1.
Expression patterns of different gal4 divers in clock neurons
| Gal4 driver | Expression sites in clock neurons |
|---|---|
| tim(uas)gal4 | All clock neurons |
| cry–gal4 #39 | All cry-positive neurons (+ 3 CRY-negative LNd) |
| mai179–gal4 | Three CRY-positive LNd, fifth s-LNv, ∼2 l-LNv, s-LNv, *DN1a |
| mai179–gal4/Pdf–gal80 | Three CRY-positive LNd, fifth s-LNv, *DN1a |
| R54D11–gal4 | One CRY-positive LNd, fifth s-LNv, *one l-LNv, *one to two s-LNv |
| c929–gal4 | One CRY-positive LNd, fifth s-LNv, l-LNv |
| npf–gal4 | Three LNd (one CRY-positive and two CRY-negative), fifth s-LNv, one to two l-LNv |
| trpA1SH–gal4 | Three LNd (CRY-positive), fifth s-LNv, one DN1a |
| Pdf–gal4 | l-LNv, s-LNv |
| r6–gal4 | s-LNv |
| R78G01–gal4 | l-LNv |
| Clk4.1M–gal4 | Six CRY-positive DN1p |
| gmr–gal4 | Eyes |
*Indicates very weak expression.
Re-entrainment experiments to a phase-shifted LD cycle
To investigate entrainability to light, flies were subjected to an 8 h phase delay of the given LD cycle. We chose a 16/8 h LD cycle for this experiment because it is easier to discriminate the actual phase of the evening activity peak from the startle response caused by lights off (Rieger et al., 2012). In 16/8 LD cycles, the evening activity of w control flies peaked before lights off, whereas cry01 mutants and eyeless grm–hid/+ mutants showed activity peaks later and earlier than the control flies, respectively (Fig. 3). After an 8 h phase delay of LD, the activity rhythms of w control flies and grm–hid/+ mutants immediately phase shifted and established a steady phase within almost 1 d (Fig. 3). In contrast, cry01 mutants needed several days to completely synchronize to the new phase of LD. This slow re-entrainment was rescued by cry expression in CRY-positive neurons in cry–gal4/uas–cry;cry01 flies (Fig. 3). When both the eyes and CRY were absent in grm–hid/+;cry01 mutants, the flies were not able to synchronize to LD cycles, in agreement with previous studies (Helfrich-Förster et al., 2001; Klarsfeld et al., 2004).
cry rescue in the fifth s-LNv and LNd neurons
To investigate the role of CRY in different clock neurons, we then generated 11 cry-rescue strains by crossing different clock-specific gal4 drivers with a uas–cry line in the cry01 background and subjected them to the 8 h phase delay. Average actograms of all strains are shown in Figure 4. To evaluate the speed of the re-entrainment, we determined the phase delays accomplished by individual flies on days 1 and 2 after the shift (Fig. 5A). The evening activity of w control flies phase shifted by almost 7 h on day 1 after the 8 h phase shift of LD (Figs. 3, 5B), meaning that w flies almost completed re-entrainment to the new LD cycle within 1 d. w+ flies, which have the same genetic background as w control flies but have red eyes, similarly showed fast re-entrainment, proving that eye color does not matter in this assay. CantonS wild-type flies were slightly slower than w control flies in the re-entrainment. The difference was >1 h and statistically significant. Thus, the genetic background including the tim allele (Peschel et al., 2006; w: s-tim; CantonS: ls-tim) slightly affected the speed of the re-entrainment. cry01 mutants phase shifted by ∼2 h on both day 1 and day 2, suggesting that the phase shift mediated by visual system-dependent light entrainment is limited to ∼2 h/d. In addition, rescuing cry in the eyes with a gmr–gal4 driver did not improve the cry01 phase shift.
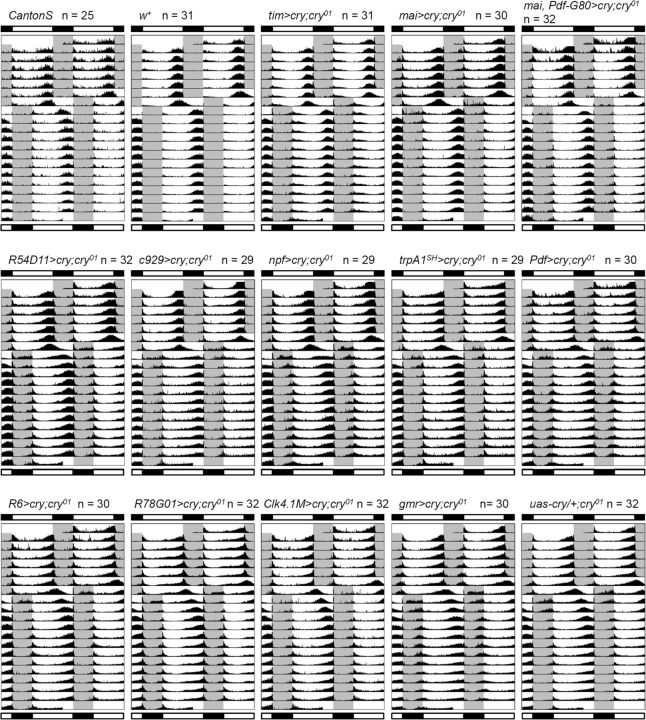
Figure 4.
Activity rhythms of other strains, which were not included in Figure 3, under 16/8 h LD before and after an 8 h LD phase shift. The gray area in the actograms indicates the dark phase. Bars above and below the actograms indicate light conditions before and after the LD phase shift, respectively. Actograms show mean activity rhythms calculated from the number of flies indicated next to the strain names.
When all clock neurons, all CRY-positive clock neurons, or the majority of the CRY-positive LN groups expressed cry by tim–gal4, cry–gal4, or mai–gal4, respectively, the flies were able to re-entrain at the same speed as the w and w+ flies. mai–gal4 is expressed in the LN groups (except for the three CRY-negative LNd and one to two l-LNv) and very weakly in the DN1a neurons (Picot et al., 2007; Rieger et al., 2009). Thus, this suggests that the DN groups are not major players in CRY-dependent fast light entrainment. The Clk4.1M–gal4 is a specific driver for CRY-positive DN1p neurons (Zhang et al., 2010), and the phase shift of cry-rescued flies with this line was not significantly different from cry01 mutants or uas–cry/+;cry01 flies, further confirming that CRY in the DN groups does not contribute to light re-entrainment to phase delays.
The r6–gal4 and R78G01–gal4 are specific drivers for the s-LNv and l-LNv, respectively. The phase shifts of cry-rescued flies with these drivers were not statistically different from cry01 mutants (Fig. 5B). However, when cry was rescued in both s-LNv and l-LNv neurons with the Pdf–gal4 driver, the flies were able to phase shift by ∼3 h/d, and this was statistically significant from the phase shift in cry01 mutants. In contrast, mai–gal4/Pdf–gal80 is a specific line for three CRY-positive LNd and the fifth s-LNv. cry-rescued flies with this driver achieved ∼6 h of phase shift. Other gal4 drivers that are expressed in both the fifth s-LNv and LNd neurons (R54D11–gal4, c929–gal4, trpA1SH–gal4, and npf–gal4) also showed considerably improved phase shifts. Together, these data show that CRY expression in the fifth s-LNv and LNd neurons is particularly important for fast light entrainment. However, because cry-rescued flies with the mai–gal4 driver shifted at a significantly faster rate than the mai–gal4/Pdf–gal80 cry-rescued flies (at least on day 1), CRY expression in the PDF neurons (s-LNv and l-LNv) seems to additionally support the phenotypic rescue. We should also note that gal4 expression in the fifth s-LNv and LNd neurons in c929–gal4, trpA1SH–gal4 and npf–gal4 strains is relatively weak compared with the tim–gal4, cry–gal4, and mai–gal4 strains. In addition, the R54D11–gal4, c929–gal4, trpA1SH–gal4 strains show gal4 expression only in one CRY-positive LNd. Therefore, the minor differences in behavior between the E cell-specific cry-rescue strains may be attributable to the level of CRY expression and/or the number of CRY-positive E cells.
Phase shifts in clock neurons via CRY and the eyes
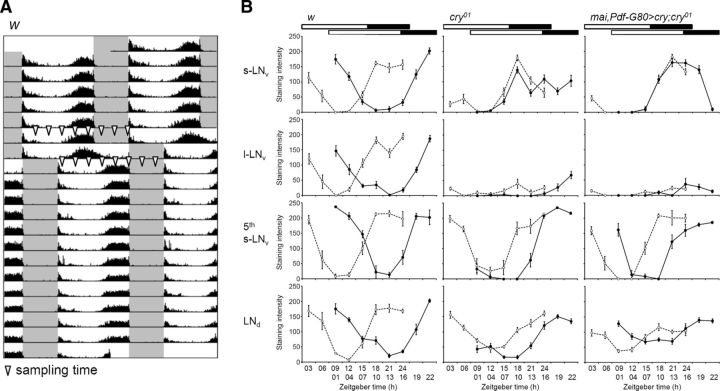
Next, we investigated the cycling of a core clock protein in the clock neurons under the phase shift condition. The PDP1 clock protein was chosen, because the cycling of PERIOD (PER) and TIM are thought to be influenced directly by light-mediated TIM degradation, but here we intended to show contributions from other pathways to the protein cycling, in particular pacemaker neurons. Three strains, w control flies, cry01 mutants, and mai–gal4/Pdf–gal80>cry;cry01 flies, were entrained to 16/8 h LD for 7 d and subjected to an 8 h phase shift of the LD cycle using the same behavioral protocol that was performed in the previous experiments shown in Figure 3. The flies were collected at 3 h intervals for immunostaining before and after day 1 of the phase shift (Fig. 5A).
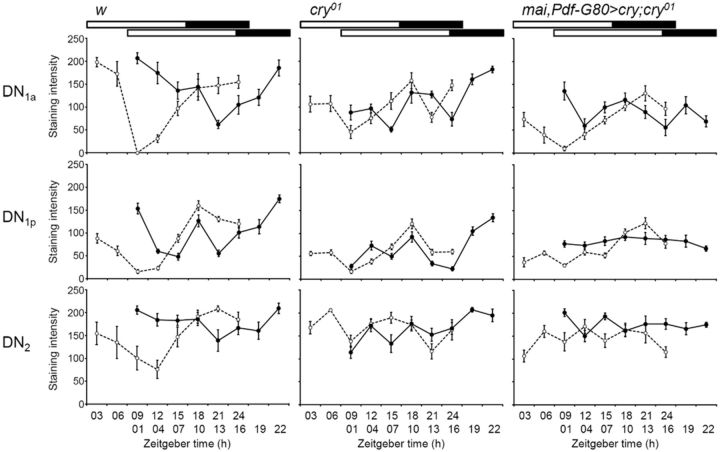
In w control flies, all LN groups displayed large phase shifts in PDP1 cycling after an 8 h phase shift of LD (Fig. 6). PDP1 began to increase at approximately ZT 12 and reached peak levels at ZT 18 before the LD phase shift in all LN groups, and these were phase shifted by 9–15 h after the LD shift. Thus, PDP1 cycling was surprisingly shifted by >8 h in terms of the onset of the PDP1 increase and the phase of the peaks. Among the DN groups, the DN1a showed similar phase shifts to the LN groups (Fig. 7). Only a subset of the DN1p neurons phase shifted. The other DN groups seemed to be insensitive to the LD phase shift and showed no difference in the phase at which PDP1 peaked before or after the shift. The phase shift of the DN2 neurons was hard to judge because these neurons seemed to resynchronize slowly.
Figure 6.
PDP1 cycling in LN clock neurons before and after an 8 h LD phase shift. A, Sampling schedule for PDP1 staining. The arrowheads on the actogram indicate when flies were sampled for immunostaining. B, Mean ± SEM PDP1 staining intensity was calculated from nine hemispheres of nine different brains for each time point in the course of a single experiment. The open circles with dashed lines and filled circles with solid lines indicate the PDP1 cycling before and after the LD phase shift, respectively. PDF costaining was performed on all brains to distinguish different LN groups (images not shown). The bars above the graphs indicate light conditions.
Figure 7.
PDP1 cycling in DN clock neurons before and after an 8 h LD phase shift. Mean ± SEM PDP1 staining intensity was calculated from nine hemispheres of nine different brains for each time point in the course of a single experiment. The open circles with dashed lines and filled circles with normal lines indicate PDP1 cycling before and after the LD phase shift, respectively. The bars above the graphs indicate the light conditions.
In cry01 mutants, the l-LNv neurons did not show any strong PDP1 cycling before or after the phase shift; therefore, we could not reliably determine any peak (Fig. 6). The same was true for the DN neurons, although the DN1p showed some similarities to the controls (Fig. 7). Prominent PDP1 oscillations were present in the s-LNv, fifth s-LNv, and LNd neurons of cry01 mutants (Fig. 6). In the s-LNv neurons, PDP1 showed a narrower peak than in the controls, but, most importantly, PDP1 cycling did not follow the phase delay of the LD cycle (Fig. 6). In contrast, PDP1 cycling in the fifth s-LNv and LNd of cry01 mutants was reasonably phase shifted as judged from an increase in PDP1 levels. We could not determine unequivocally the phase of the PDP1 peak before the shift, because PDP1 seemed to reach its maximal level at ZT 24, when our time series was stopped. Importantly, PDP1 of the fifth s-LNv and LNd reached its peak levels later in cry01 mutants than in the controls, and this coincided with the late E activity peak of the mutants (Fig. 3), further providing evidence for the proposed role of the E cells in controlling this peak. The response of the molecular clock in the fifth s-LNv and LNd neurons of cry01 mutants to the phase-delayed LD cycles shows that visual system-dependent light input pathways, in addition to CRY, entrain the E cells. The magnitude of the shifts matches the behavioral data that revealed a phase delay of 2 h/d of the E peak in cry01 mutants (Fig. 5B).
When cry expression was rescued in the E cells in mai–gal4/Pdf–gal80>cry;cry01 flies, the wild-type phase of the PDP1 peak (at approximately ZT 18–ZT 21) was restored in the fifth s-LNv and LNd neurons; in addition, these neurons also responded with a phase delay to the shifted LD cycles in a manner very similar to the wild-type neurons (Fig. 6). The increase in PDP1 levels after the phase shift occurred earlier in the rescued flies than in the controls, but the PDP1 peaks were reached at the same time (at approximately ZT 19 to ZT 22) in both fly groups, suggesting that CRY expression in the E cells was sufficient for almost normal phase delays of the E cells. Once again, this corresponds to the delay of E activity shown in the behavioral experiments (Fig. 5B). Interestingly, CRY rescue in the E cells had little effect on PDP1 cycling in the s-LNv (Fig. 6), which did not show any apparent phase delay in response to the LD shift. In addition, there was no effect on PDP1 cycling in the l-LNv (Fig. 6) or DN (Fig. 7). Thus, cry rescue in the E cells increased the speed of the phase shift in the E cells themselves but not in other clock neurons. Nevertheless, we emphasize that there must be crosstalk between the different clock neurons, because the shape and phase of the PDP1 oscillations in s-LNv neurons appeared more normal after CRY rescue in the E cells (Fig. 6).
Discussion
The Drosophila circadian clock is extremely light sensitive (Helfrich-Förster et al., 2002; Hirsh et al., 2010). Short-term exposure to a weak light pulse is sufficient for TIM degradation and a large phase shift in activity rhythms (Vinayak et al., 2013). This rapid response of the Drosophila clock to light is attributed to TIM degradation by the CRY-dependent pathway in the clock neurons (Stanewsky et al., 1998; Emery et al., 2000; Busza et al., 2004). In the present study, we demonstrated that the fifth s-LNv and LNd neurons, known as the E cells, play an important role in the CRY-dependent fast light entrainment. In the absence of CRY, molecular cycling in the E cells can respond to an 8 h phase shift of LD, whereas cycling in the M cells (l-LNv and s-LNv neurons) cannot (Fig. 6). Thus, visual input pathways entrain the E cells but not the M cells, at least not during a short period. The rescue of cry in the E cells alone increases the speed of the phase shift of activity rhythms and PDP1 cycling in these cells. Together, both the visual system and CRY initially reset the E cells, enabling extremely sensitive light entrainment in the Drosophila clock.
Circadian network for light entrainment
To compound a hierarchical structure in the clock network, some clock neurons would dominate others to drive a coherent circadian rhythm in behavior (Yoshii et al., 2012). It has been shown that the M cells are master pacemakers and maintain the molecular cycling in some of the LNd neurons in DD (Stoleru et al., 2005; Yao and Shafer, 2014). In contrast, the E cells are more important under constant light conditions. Functional E cells are able to drive free-running rhythms in constant light, but functional M cells are not (Picot et al., 2007; Stoleru et al., 2007; Rieger et al., 2009). This fact, together with our present results, suggests that the E cells are a specialized group of clock cells for light entrainment, and they may have a dominant power to control activity rhythms under light.
However, there are lines of evidence showing that the PDF neurons are also important for light resetting. The group of larger PDF neurons, the so-called l-LNv, is less important for generating activity rhythms (Grima et al., 2004; Shafer and Taghert, 2009). However, flies lacking l-LNv neurons are relatively insensitive to a light pulse at dawn but not at dusk (Shang et al., 2008). Double mutants of cry and Pdf (or Pdf-receptor), which lack an evening activity peak but show a large morning peak under LD (Cusumano et al., 2009; Zhang et al., 2009), show damping PER oscillations in the fifth s-LNv and LNd neurons and no behavioral evidence of light re-entrainment in response to an 8 h LD phase shift (Im et al., 2011). Thus, the PDF neurons and PDF itself are also important for the light response of the clock.
To explain all the results of the present and previous studies, one can speculate that light input from the visual system may pass through the PDF neurons before arriving in the E cells. Therefore, light entrainment in cry/Pdf double mutants may be defective because of the lack of cry and the main neurotransmitter of the PDF neurons. Although all neuronal connections between the visual system and the clock neurons in the adult fly have not been identified, it is known that the neuronal projections from the H–B eyelets terminate very close to where the PDF neurons send projections into the accessory medulla (Helfrich-Förster et al., 2002, 2007; Malpel et al., 2002). The larval precursor of the H–B eyelet, the Bolwig organ, which constitutes the larval visual system, signals to the larval LNv via acetylcholine (Wegener et al., 2004; Keene et al., 2011; Yao et al., 2012). This leads to a reset of the DN1 clock neurons via PDF signaling (Klarsfeld et al., 2011). Therefore, the same mechanism may also exist in the adult fly, with the PDF neurons receiving light information from the visual system and then signaling to the fifth s-LNv and LNd cells via PDF. Indeed, both the fifth s-LNv and the LNd were shown to be receptive to PDF, and there is evidence for a functional neuronal connection between the PDF neurons and the LNd (Shafer et al., 2008; Yao et al., 2012; Yao and Shafer, 2014). Guo et al. (2014) reported that temporal activation of PDF neurons by the temperature-sensitive TrpA1 channel mimics a phase response curve (PRC) in response to light pulses and that this activation of PDF neurons promotes TIM degradation in all clock neurons (Guo et al., 2014). This system is CRY independent but PDF-receptor dependent. Thus, it is possible that the PDF neurons are activated by visual input pathways and cause degradation of TIM in the E cells, resetting their molecular clocks.
One remaining question is how the M cells and PDF can contribute to the LD entrainment in the absence of CRY, although PDP1 cycling in the s-LNv and l-LNv was insensitive to an LD phase shift in cry01 and mai–gal4/Pdf–gal80>cry;cry01 flies (Fig. 6). To address this, we have to ask whether the molecular clock in the M cells is required for LD entrainment of the E cells and of activity rhythms in future work.
PRC or LD phase shift
Two recent reports also proposed a non-cell-autonomous mechanism involved in light-dependent phase resetting of the clock (Tang et al., 2010; Lamba et al., 2014). Tang et al. (2010) revealed that the TIM degradation in M cells is insensitive to a light pulse at ZT 15, which is the phase delay zone in the PRC to light, whereas the E cells are still sensitive (Tang et al., 2010). However, CRY expression in the M cells is necessary for the phase delay, because knockdown of cry in the s-LNv neurons attenuates the amplitude of the phase delay. Lamba et al. (2014) further examined this using two genetic methods: (1) rescue of wild-type jet under a jet mutant background in a manner similar to the protocol performed for cry rescue in the present study; and (2) the knockdown of jet by RNAi (Lamba et al., 2014). JET is an E3 ubiquitin ligase that promotes degradation of TIM together with CRY (Koh et al., 2006; Peschel et al., 2009). Interestingly, when there is sufficient jet expression in the M cells, TIM degradation occurs in both the M and E cells (Lamba et al., 2014). This suggests that an interaction between the M and E cells is important for phase resetting. This is in good agreement with our data, showing TIM degradation in CRY-negative clock neurons after light exposure (Fig. 1).
Both studies showed that activity phase shifts are mediated by network interactions between M and E cells. Our data match this, because the rescue of cry in both the M and E cells completely restored wild-type entrainment. However, the cry rescue in fifth s-LNv and LNd cells consistently showed a significant improvement in the speed of re-entrainment (Fig. 5B), suggesting the importance of the E cells. One difference between our study and the above mentioned studies is the way in which we evaluated the light response of the clock. In the LD phase-shift experiment, a long-term light exposure covering the delay and advance zones of the PRC strongly resets the clock in day 1 of the LD shift. In contrast, the PRC experiment reveals finer parameters of the clock with respect to a short light response, e.g., the direction of the phase shift, the magnitude of the shift, and the sensitive phase. Therefore, it is not easy to compare our data with the PRC studies.
A longer light pulse usually causes a larger phase shift (Kistenpfennig et al., 2012). When flies receive the same number of photons, a dimmer yet longer light pulse is more effective than a brighter yet shorter pulse (Vinayak et al., 2013). Among the clock neurons, TIM degradation in the fifth s-LNv neuron is especially sensitive to a long light pulse (Vinayak et al., 2013), which may imply that the role of the E cells in light-resetting mechanisms emerges only under a long-term light exposure.
Footnotes
This work was funded by Japan Society for the Promotion of Science (JSPS) Science Research Funds Grants 23870021, 25840121, and 23657056 and German Research Foundation Grant Fo-207/12-1 (Collaborative Research Center Grant SFB 1047 “Insect Timing” INST 93/784-1). C.K. was supported by JSPS Fellowship PE12050. The Division of Instrumental Analysis, Okayama University, supported us in the use of a laser scanning confocal microscope (Fluoview300). We thank J. Hirsh for comments on this manuscript, J. C. Hall, M. W. Young, M. Rosbash, R. Stanewsky, P. Emery, P. E. Hardin, F. Rouyer, A. Klarsfeld, P. H. Taghert, M. Freeman, P. A. Garrity, P. Shen, and the Bloomington Drosophila Stock Center for providing fly lines. We are also grateful to T. Todo, J. Giebultwicz, J. Blau, P. E. Hardin, H. Dircksen, and the Developmental Studies Hybridoma Bank for providing antibodies.
The authors declare no competing financial interests.
References
- Benito J, Houl JH, Roman GW, Hardin PE. The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J Biol Rhythms. 2008;23:296–307. doi: 10.1177/0748730408318588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/S0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Más P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- Cusumano P, Klarsfeld A, Chélot E, Picot M, Richier B, Rouyer F. PDF-modulated visual inputs and cryptochrome define diurnal behavior in Drosophila. Nat Neurosci. 2009;12:1431–1437. doi: 10.1038/nn.2429. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/S0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci. 2005;25:5430–5437. doi: 10.1523/JNEUROSCI.0263-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel S, Codd V, Fedic R, Garner KJ, Costa R, Kyriacou CP, Rosato E. A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci. 2004;7:834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- Dolezelova E, Dolezel D, Hall JC. Rhythm defects caused by newly engineered null mutations in Drosophila's cryptochrome gene. Genetics. 2007;177:329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/S0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Förster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/S0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/S0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/S0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, Rosbash M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. Elife. 2014;3:e02780. doi: 10.7554/eLife.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Tech. 2003;62:94–102. doi: 10.1002/jemt.10357. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/S0896-6273(01)00277-X. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Edwards T, Yasuyama K, Wisotzki B, Schneuwly S, Stanewsky R, Meinertzhagen IA, Hofbauer A. The extraretinal eyelet of Drosophila: development, ultrastructure, and putative circadian function. J Neurosci. 2002;22:9255–9266. doi: 10.1523/JNEUROSCI.22-21-09255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C, Shafer OT, Wülbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- Hermann-Luibl C, Yoshii T, Senthilan PR, Dircksen H, Helfrich-Förster C. The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. J Neurosci. 2014;34:9522–9536. doi: 10.1523/JNEUROSCI.0111-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh J, Riemensperger T, Coulom H, Iché M, Coupar J, Birman S. Roles of dopamine in circadian rhythmicity and extreme light sensitivity of circadian entrainment. Curr Biol. 2010;20:209–214. doi: 10.1016/j.cub.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Li W, Taghert PH. PDFR and CRY signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila. PLoS One. 2011;6:e18974. doi: 10.1371/journal.pone.0018974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard HA, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Förster C, Nässel DR. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol. 2009;516:59–73. doi: 10.1002/cne.22099. [DOI] [PubMed] [Google Scholar]
- Kanda Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Mazzoni EO, Zhen J, Younger MA, Yamaguchi S, Blau J, Desplan C, Sprecher SG. Distinct visual pathways mediate Drosophila larval light avoidance and circadian clock entrainment. J Neurosci. 2011;31:6527–6534. doi: 10.1523/JNEUROSCI.6165-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistenpfennig C, Hirsh J, Yoshii T, Helfrich-Förster C. Phase-shifting the fruit fly clock without cryptochrome. J Biol Rhythms. 2012;27:117–125. doi: 10.1177/0748730411434390. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A, Malpel S, Michard-Vanhée C, Picot M, Chélot E, Rouyer F. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarsfeld A, Picot M, Vias C, Chélot E, Rouyer F. Identifying specific light inputs for each subgroup of brain clock neurons in Drosophila larvae. J Neurosci. 2011;31:17406–17415. doi: 10.1523/JNEUROSCI.5159-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba P, Bilodeau-Wentworth D, Emery P, Zhang Y. Morning and evening oscillators cooperate to reset circadian behavior in response to light input. Cell Rep. 2014;7:601–608. doi: 10.1016/j.celrep.2014.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- Malpel S, Klarsfeld A, Rouyer F. Larval optic nerve and adult extra-retinal photoreceptors sequentially associate with clock neurons during Drosophila brain development. Development. 2002;129:1443–1453. doi: 10.1242/dev.129.6.1443. [DOI] [PubMed] [Google Scholar]
- Muraro NI, Pírez N, Ceriani MF. The circadian system: plasticity at many levels. Neuroscience. 2013;247:280–293. doi: 10.1016/j.neuroscience.2013.05.036. [DOI] [PubMed] [Google Scholar]
- Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- Ozturk N, Selby CP, Zhong D, Sancar A. Mechanism of photosignaling by Drosophila cryptochrome: role of the redox status of the flavin chromophore. J Biol Chem. 2014;289:4634–4642. doi: 10.1074/jbc.M113.542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N, Veleri S, Stanewsky R. Veela defines a molecular link between Cryptochrome and Timeless in the light-input pathway to Drosophila's circadian clock. Proc Natl Acad Sci U S A. 2006;103:17313–17318. doi: 10.1073/pnas.0606675103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19:241–247. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot M, Cusumano P, Klarsfeld A, Ueda R, Rouyer F. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 2007;5:e315. doi: 10.1371/journal.pbio.0050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/S0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rieger D, Stanewsky R, Helfrich-Förster C. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J Biol Rhythms. 2003;18:377–391. doi: 10.1177/0748730403256997. [DOI] [PubMed] [Google Scholar]
- Rieger D, Wülbeck C, Rouyer F, Helfrich-Förster C. Period gene expression in four neurons is sufficient for rhythmic activity of Drosophila melanogaster under dim light conditions. J Biol Rhythms. 2009;24:271–282. doi: 10.1177/0748730409338508. [DOI] [PubMed] [Google Scholar]
- Rieger D, Peschel N, Dusik V, Glotz S, Helfrich-Förster C. The ability to entrain to long photoperiods differs between three D. melanogaster wildtype strains and is modified by twilight simulation. J Biol Rhythms. 2012;27:37–47. doi: 10.1177/0748730411420246. [DOI] [PubMed] [Google Scholar]
- Schlichting M, Grebler R, Peschel N, Yoshii T, Helfrich-Förster C. Moonlight detection by Drosophila's endogenous clock depends on multiple photopigments in the compound eyes. J Biol Rhythms. 2014;29:75–86. doi: 10.1177/0748730413520428. [DOI] [PubMed] [Google Scholar]
- Schmid B, Helfrich-Förster C, Yoshii T. A new ImageJ plug-in “ActogramJ” for chronobiological analyses. J Biol Rhythms. 2011;26:464–467. doi: 10.1177/0748730411414264. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Taghert PH. RNA-interference knockdown of Drosophila pigment dispersing factor in neuronal subsets: the anatomical basis of a neuropeptide's circadian functions. PLoS One. 2009;4:e8298. doi: 10.1371/journal.pone.0008298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/S0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, Fernández MP, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Taghert PH, Hewes RS, Park JH, O'Brien MA, Han M, Peck ME. Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J Neurosci. 2001;21:6673–6686. doi: 10.1523/JNEUROSCI.21-17-06673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CH, Hinteregger E, Shang Y, Rosbash M. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron. 2010;66:378–385. doi: 10.1016/j.neuron.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya AT, Top D, Manahan CC, Tokuda JM, Zhang S, Pollack L, Young MW, Crane BR. Flavin reduction activates Drosophila cryptochrome. Proc Natl Acad Sci U S A. 2013;110:20455–20460. doi: 10.1073/pnas.1313336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak P, Coupar J, Hughes SE, Fozdar P, Kilby J, Garren E, Yoshii T, Hirsh J. Exquisite light sensitivity of Drosophila melanogaster cryptochrome. PLoS Genet. 2013;9:e1003615. doi: 10.1371/journal.pgen.1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener C, Hamasaka Y, Nässel DR. Acetylcholine increases intracellular Ca2+ via nicotinic receptors in cultured PDF-containing clock neurons of Drosophila. J Neurophysiol. 2004;91:912–923. doi: 10.1152/jn.00678.2003. [DOI] [PubMed] [Google Scholar]
- Wu Q, Wen T, Lee G, Park JH, Cai HN, Shen P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron. 2003;39:147–161. doi: 10.1016/S0896-6273(03)00396-9. [DOI] [PubMed] [Google Scholar]
- Yang Z, Emerson M, Su HS, Sehgal A. Response of the timeless protein to light correlates with behavioral entrainment and suggests a nonvisual pathway for circadian photoreception. Neuron. 1998;21:215–223. doi: 10.1016/S0896-6273(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Yao Z, Shafer OT. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science. 2014;343:1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Macara AM, Lelito KR, Minosyan TY, Shafer OT. Analysis of functional neuronal connectivity in the Drosophila brain. J Neurophysiol. 2012;108:684–696. doi: 10.1152/jn.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila's clock neurons. J Comp Neurol. 2008;508:952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Vanin S, Costa R, Helfrich-Förster C. Synergic entrainment of Drosophila's circadian clock by light and temperature. J Biol Rhythms. 2009a;24:452–464. doi: 10.1177/0748730409348551. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Förster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila's clock. J Neurosci. 2009b;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Rieger D, Helfrich-Förster C. Two clocks in the brain: an update of the morning and evening oscillator model in Drosophila. Prog Brain Res. 2012;199:59–82. doi: 10.1016/B978-0-444-59427-3.00027-7. [DOI] [PubMed] [Google Scholar]
- Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lear BC, Seluzicki A, Allada R. The CRYPTOCHROME photoreceptor gates PDF neuropeptide signaling to set circadian network hierarchy in Drosophila. Curr Biol. 2009;19:2050–2055. doi: 10.1016/j.cub.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]