Abstract
In brain, specific RNA-binding proteins (RBPs) associate with localized mRNAs and function as regulators of protein synthesis at synapses exerting an indirect control on neuronal activity. Thus, the Fragile X Mental Retardation protein (FMRP) regulates expression of the scaffolding postsynaptic density protein PSD95, but the mode of control appears to be different from other FMRP target mRNAs. Here, we show that the fragile X mental retardation-related protein 2 (FXR2P) cooperates with FMRP in binding to the 3′-UTR of mouse PSD95/Dlg4 mRNA. Absence of FXR2P leads to decreased translation of PSD95/Dlg4 mRNA in the hippocampus, implying a role for FXR2P as translation activator. Remarkably, mGluR-dependent increase of PSD95 synthesis is abolished in neurons lacking Fxr2. Together, these findings show a coordinated regulation of PSD95/Dlg4 mRNA by FMRP and FXR2P that ultimately affects its fine-tuning during synaptic activity.
Keywords: FMRP, FXR2P, mRNA translation, PSD95, RNA binding proteins
Introduction
Several forms of synaptic plasticity induce mRNA transport and protein synthesis at synapses resulting in the remodeling of local signaling networks (Xing and Bassell, 2013). Disc, Large Homolog 4 (Drosophila) (DLG4) mRNA encodes the postsynaptic density protein 95 (PSD95), the most abundant scaffold within the postsynaptic density compartment (PSD) (Cheng et al., 2006). PSD95 plays a key role in spine formation (Feyder et al., 2010), as well as LTP and LTD via NMDA and AMPA receptor signaling (Migaud et al., 1998; Stein et al., 2003; Béïque et al., 2006). The absence of PSD95 affects mouse behavioral phenotypes and dendritic spine morphology; furthermore, PSD95 has been linked with the Williams and Angelman syndromes (Feyder et al., 2010; Cao et al., 2013).
Expression of hundreds of mRNAs localized in dendrites and axons (Cajigas et al., 2012) is specifically controlled by neuronal RNA-binding proteins (RBPs) that commonly interact with specific mRNA sequences frequently present in the 5′ and 3′ untranslated regions (UTRs). The 3′-UTR regions modulate the translation and longevity of mRNAs by interacting with RBPs (Gebauer et al., 2012). Loss of expression or mutations in certain RBPs results in diverse neurological disorders characterized by deficits in synaptic structure and function (Kapeli and Yeo, 2012).
Fragile X Mental Retardation Protein (FMRP) governs the fate of many mRNAs in brain as well as in non-neuronal cells (for review, see Pasciuto and Bagni, 2014). Absence or mutations in FMRP cause the Fragile X Syndrome (FXS), the most common form of inherited intellectual disability and a leading cause of monogenic autism (Bagni et al., 2012; Gross et al., 2012). Among the putative FMRP targets identified so far, 240 mRNAs encode postsynaptic proteins, including the PSD95 mRNA (Muddashetty et al., 2007; Zalfa et al., 2007; Darnell et al., 2011). In mice, FMRP controls steady-state and activity-dependent PSD95 levels (Muddashetty et al., 2007; Zalfa et al., 2007).
Here, we have isolated a subset of brain RBPs that bind the 3′-UTR of PSD95 mRNA. Among them, we found FMRP, the members of the ELAV-like family, and FXR2P (fragile X mental retardation-related protein 2). FXR2P associates with PSD95 mRNA in vitro and in vivo, requiring the presence of FMRP for full binding activity. We show that absence of FXR2P impairs PSD95 mRNA translation indicating a role for FXR2P as translational activator. Our results show that the paralogs FMRP and FXR2P control the same mRNA by different mechanisms, shedding new light on the control of the Fragile X-related protein family members on target mRNAs through a cooperative or independent interaction.
Materials and Methods
Animals.
Animal care was conducted according to national and international guidelines (Belgian law of August 14th, 1986, and the following K.B. of November 14th, 1993 and K.B of September 13th, 2004; European Community Council Directive 86/609, OJa L 358, 1, December 12, 1987; National Institutes of Health Guide for the Care and Use of Laboratory Animals, National Research Council, 1996). Four-week-old Fmr1 knock-out (KO) C57BL/6 (Bakker et al., 1994) and Fxr2 KO FVB (Bontekoe et al., 2002) males and wild-type (WT) littermates were used.
Constructs.
The WT and mutated 3′-UTR of PSD95 mRNA (full-length and 5 fragments as described by Zalfa et al., 2007) were transcribed using standard protocols in presence of biotin-16-UTP (Roche).
PSD95 mRNA alignments.
Transcript dataset from FMRP and HuD HITS-CLIP from P11-P25 mouse brain polysomes (Darnell et al., 2011) was downloaded from NCBI GEO (accession GSE45148).
Antibodies.
Antibodies against FMRP (rAMII) were described previously (Napoli et al., 2008); antibodies against FMRP (7G1), FXR2P (1G2), and GAPDH were from Developmental Studies Hybridoma Bank; antibodies against FXR2P and Vinculin were from Sigma-Aldrich; antibodies against HuR, HuD, Tristetraprolin (TTP), phospho-ERK1/2, and ERK1/2 were from Santa Cruz Biotechnology; Alexa-488 anti-rabbit, Alexa-488 anti-goat, and Alexa-568 anti-mouse secondary antibodies were from Invitrogen.
Brain extracts.
Mouse forebrain and cortex or hippocampi were homogenized in 100 mm NaCl, 10 mm MgCl2, 10 mm Tris-HCl, pH 7.4, 1% Triton X-100 (Sigma), 1 tablet/10 ml Roche protease inhibitor, 40 U/ml Rnasin, incubated as in Napoli et al. (2008).
RNA extraction.
Total RNA from cortex and hippocampus was extracted using TRIzol (Invitrogen).
Immunoprecipitation (IP).
Brain extracts were incubated with 2 μg of specific antibodies at 4°C overnight in the absence or presence of a mix of RNase A/T1 (10 μl/ml) (Fermentas). Protein complexes were captured with protein A- or protein G-Dynal beads.
RNA-IP.
RNA-IP was performed as previously described (Napoli et al., 2008). Anti-FMRP and anti-HuD antibodies were incubated with 500–800 μg of brain extract in IP buffer (20 mm Tris-HCl pH 7.4, 150 mm NaCl, 1 mm MgCl2, 0.1% Triton X-100, 0.1 mg/ml heparin) for 1 h at 4°C. The immunocomplexes were captured on Protein A-Sepharose (GE Healthcare). FXR2P-IP was performed as by Guo et al. (2011). Eluted RNA was reverse transcribed, and qPCR was performed using LightCycler 480 SYBR Green I (Roche Applied Science). Primers used were as follows: PSD95 forward, 5′-CATTGCCCTGAAGAACGC-3′; PSD95 reverse, 5′-ATGGATCTTGGCCTCGAA-3′; Cyp46 forward, 5′-CATGAGACTTCTGCCAACCA-3′; Cyp46 reverse, 5′-CTTGGAACCGACAACCTCAT-3′.
RNA pull-down.
In vitro synthesized RNAs were bound to Dynabeads M-270 streptavidin (Invitrogen). The RNAs immobilized to the beads were incubated in binding buffer (200 mm KCl, 10 mm MgCl2, 60 mm HEPES, pH 7.4) with 500 μg of brain protein extracts for 1 h at 4°C or with 15 μl of in vitro-synthesized FXR2P and FMRP protein (TNT System, Promega). Proteins were eluted in Laemmli buffer and separated by SDS-PAGE for immunoblotting. Alternatively, the SDS-PAGE gel was cut, the proteins extracted, digested, and analyzed by LC-MS/MS. The LC-MS/MS analysis was performed as by Klemmer et al. (2009). Briefly, the digested peptides were analyzed on an ABI 4800 proteomics analyzer (AB Sciex). Peptide collision was performed at 2 kV, and MS/MS spectra were each collected from 2500 laser shots. The spectra were annotated through Mascot (MatrixScience) searches against the SwissProt and NCBI databases. “Unique” peptides matching only one protein cluster were considered for protein identification. Only proteins identified with at least two peptides with a confidence interval ≥95% (AB Sciex, percentage) were considered. The “unused” value is defined as the sum of protein scores from all the nonredundant peptide matched to a single protein. Proteins identified with the antisense PSD95 mRNA were considered as background.
Immunohistochemistry.
Primary cortical neurons were fixed and stained as by Napoli et al. (2008). Coronal sections from fixed brains of postnatal 30 d WT mice were permeabilized/blocked at room temperature for 3 h in PBS, 0.3% Triton X-100, 3% BSA, and 5% normal goat or donkey serum, incubated with the primary and secondary antibodies, and mounted on glass coverslips using Fluoroshield (Sigma).
Western blotting.
Western blotting was performed using standard protocols and chemiluminescence (ECL Plus or Advance, GE Healthcare Pharmacia) with Fujifilm LAS-3000 or with the laser scanning Odyssey Infrared Imaging System (LI-COR Biosciences). Quantification was performed using Aida software.
Neuronal cultures.
Primary mouse neurons were prepared as previously described (Napoli et al., 2008) and used after 12–14 d in vitro. Neurons were stimulated with 100 μm DHPG (Sigma-Aldrich) or vehicle for 20 min.
Polysome-mRNA ribonucleoparticle distribution.
Analyses of hippocampal extracts from WT and Fxr2 KO have been performed as by Zalfa et al. (2007).
Statistics.
Comparisons between two groups were performed using the nonparametric Mann–Whitney test unless indicated differently. Significance was accepted with p < 0.05. Error bars indicate SEM.
Results
FMRP-dependent and FMRP-independent complexes associate to the PSD95 3′-UTR
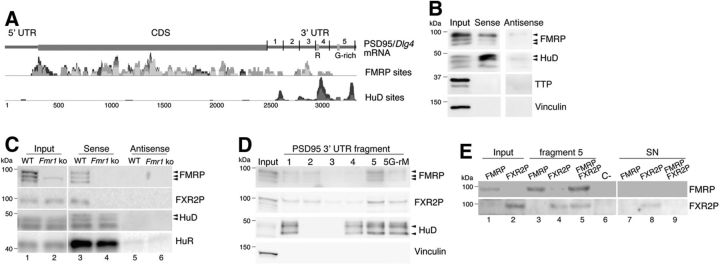
PSD95 mRNA is localized and translated at synapses (Muddashetty et al., 2007; Zalfa et al., 2007). We searched to identify the trans-activating factors that regulate PSD95 mRNA metabolism in an unbiased approach. The 3′-UTR of PSD95 contains numerous cis-acting elements, including the G-rich element involved in the direct binding to FMRP (Zalfa et al., 2007; Muddashetty et al., 2011), and AU-rich elements (AREs) involved in the interaction with the ELAV-like proteins HuD and HuR (Darnell et al., 2011; Mukherjee et al., 2011) (Fig. 1A). According to HITS-CLIP experiments, FMRP interacts with the 3′-UTR of the PSD95 mRNA as well as with its coding sequence (CDS) (Fig. 1A) (Darnell et al., 2011). The absence of the G-rich element in the CLIP dataset (Darnell et al., 2011) might be due to a less favorable UV cross-linking with riboGs (Sugimoto et al., 2012), but it should be noted that there is a cluster of CLIP tags just upstream of the G-rich element (Fig. 1A) (Darnell et al., 2011). First, a biotinylated RNA spanning the entire 3′-UTR of PSD95 mRNA (835 nts) in sense and antisense orientation was used in a RNA pull-down with brain extracts of WT and Fmr1 KO mice. The RBPs FMRP and HuD were principally captured with the sense fragment (Fig. 1B). TTP, an ARE-binding protein, and the protein Vinculin were absent, thus showing the specificity of the assay (Fig. 1B). Second, the PSD95 3′-UTR interactome was identified by mass spectrometry (MS) (LC-MS/MS) in a scaled-up pull-down. Upon subtraction of the proteins pulled down with the antisense probe, 18 copurified proteins were considered to bind specifically the 3′-UTR of PSD95 mRNA (Table 1). All of them displayed the molecular function “RNA binding” (GO: 0003723). Comparison to similar pull-downs in Fmr1 KO extracts showed that the proteins fall into two categories: those that seem to require FMRP for binding and those that are independent of FMRP. Only two proteins were selectively detected in the Fmr1 KO but not in the WT purifications (Table 1). To validate the MS results, the biotin pull-downs were repeated, and the presence or absence of key proteins was verified by Western blotting. The ELAV-like proteins HuD and HuR copurified with the PSD95 3′-UTR from WT and Fmr1 KO extracts (Fig. 1C, bottom two panels), whereas FMRP and the FXR2P were found only in the pull-down from WT, but not from Fmr1 KO extracts (Fig. 1C, top two panels). The antisense RNA of the PSD95 3′-UTR, instead, did not associate with significant amounts of any of the four proteins (Fig. 1C, lanes 5 and 6). Pull-down of WT brain extracts with different fragments of the PSD95 3′-UTR (Zalfa et al., 2007) revealed that FMRP and FXR2P mostly associate to fragment 5 (Fig. 1D). Mutations of the G-rich region (G-rM) in fragment 5 impaired this interaction. HuD interacted with fragments 1, 4, and 5 (Fig. 1D), consistent with HITS-CLIP published data (Fig. 1A) (Darnell et al., 2011). In vitro synthesized FMRP and FXR2P interacted directly with PSD95 fragment 5 (Fig. 1E, lanes 3 and 4). The presence of FMRP increased the efficiency of FXR2P binding (Fig. 1E, lane 5).
Figure 1.
PSD95 3′-UTR mRNA pulls down a specific set of RBPs. A, Scheme of the full-length murine PSD95 mRNA. Fragments 1–5 span the entire 3′-UTR region (Zalfa et al., 2007). FMRP and HuD HITS-CLIP were performed at different developmental stages (Darnell et al., 2011). The reads were cumulatively plotted with gray tones. Box R corresponds to the sequence ATTTCTCATTTATTTTTTCCACTTTTTTTCTTCTCAAAG on human PSD95 mRNA recognized by HuR (Mukherjee et al., 2011); box G-rich is as in Zalfa et al. (2007). B, Protein pull-down from WT mouse brain using the sense or antisense PSD95/Dlg4 3′-UTR. FMRP, HuD, TTP, and Vinculin proteins were detected by Western blotting. C, Pull-down as in B from WT and Fmr1 KO mouse brain. D, Protein pull-down from WT mouse brain using fragments 1–5 and fragment 5 mutated in the G-rich region indicated in A (Zalfa et al., 2007). E, Pull-down of in vitro-translated FMRP and FXR2P using fragment 5. C−, Negative control containing in vitro-translated Luciferase. WT, Wild type; KO, knock-out; CDS, coding sequence; SN, supernatant.
Table 1.
Proteins binding to the PSD-95 3′-UTR
| UniProt accession number | Protein name | Unused value |
% of coverage |
||
|---|---|---|---|---|---|
| WT | Fmr1 KO | WT | Fmr1 KO | ||
| Q61701 | ELAV-like protein 4 (HuD) | 14.23 | 0.72 | 32.5 | 18.2 |
| Q60899 | ELAV-like protein 2 (HuB) | 13.50 | 7.30 | 34.2 | 23.1 |
| Q8R081 | Heterogeneous nuclear ribonucleoprotein L (hnRNP L) | 13.06 | 2.42 | 21.8 | 14.8 |
| Q8VIJ6 | Polypyrimidine tract-binding protein-associated-splicing factor (PSF) | 12.51 | ND | 27.6 | ND |
| P70372 | ELAV-like protein 1 (HuR) | 8.60 | 10.00 | 22.7 | 27.6 |
| P62960 | Y-box-binding protein 1 (YB-1) | 8.00 | ND | 33.5 | ND |
| Q99K48 | Non–POU domain-containing octamer-binding protein (NonO protein) | 4.84 | ND | 6.8 | ND |
| Q60900 | ELAV-like protein 3 (HuC) | 4.67 | 0.31 | 39.5 | 21.8 |
| P35922 | Fragile X mental retardation protein 1 (FMRP) | 3.70 | ND | 24.6 | ND |
| P61979 | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) | 3.05 | ND | 23.8 | ND |
| Q60668 | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0/AUF1) | 2.00 | 0.52 | 14.4 | 9.6 |
| P62631 | Elongation factor 1-alpha 2 (eEF1A-2) | 2.00 | 0.42 | 5.0 | 1.7 |
| Q9Z108 | Double-stranded RNA-binding protein Staufen1 | 2.00 | ND | 11.1 | ND |
| Q64012 | RNA-binding protein Raly | 0.80 | ND | 2.6 | ND |
| Q7TN98 | Cytoplasmic polyadenylation element-binding protein 4 (CPEB4) | 0.41 | ND | 9.3 | ND |
| Q9WVR4 | Fragile X mental retardation-related protein 2 (FXR2P) | 0.28 | ND | 8.9 | ND |
| P62320 | Small nuclear ribonucleoprotein Sm D3 | ND | 0.49 | ND | 15.9 |
| Q9WV02 | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) | ND | 0.48 | ND | 5.7 |
Brain (cortex and hippocampus) proteins from WT and Fmr1 KO mice identified by mass spectrometry using the biotinylated PSD95 3′-UTR. Indicated are the UniProt accession number, the “unused” value (protein score based on the peptide confidence), and the coverage of the protein. ND, Not detected.
Because a mass spectrometry analysis of the reticulocyte lysate excluded the presence of residual amount of FMRP (data not shown), we can conclude that the direct interaction of FXR2P with the fragment 5 was less efficient compared with FMRP as shown by the remaining protein in the supernatant (compare lanes 7 and 8).
Hu proteins associate to PSD95 mRNA in an FMRP-independent manner
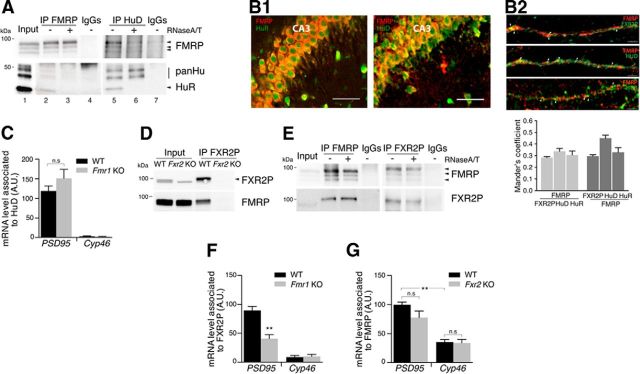
Among the proteins that do not require FMRP for their association with the PSD95 3′-UTR were the ELAV family of RBPs (HuB, HuC, HuD, HuR; see Table 1). These proteins bind to AREs and regulate splicing, translation, and stability of the respective mRNAs (Simone and Keene, 2013). Hu proteins interact and coprecipitate weakly with FMRP, and this association was strongly reduced upon RNase treatment of the extract, further suggesting that Hu proteins and FMRP form distinct RNPs on the same mRNA target (Fig. 2A). The FMRP and HuR/D colocalize in the neuronal cell bodies and dendritic processes in the CA3 as well as in primary neurons (Fig. 2, B1 and B2, respectively). Remarkably, the AREs present in fragment 5 (Fig. 1A) are in close vicinity to the FMRP binding site, and it is therefore conceivable that they sterically and/or functionally compete with FMRP. In support of this notion, RNA-IP of hippocampal HuD showed that the amount of PSD95 mRNA bound to HuD tends to be higher in the absence of FMRP (Fig. 2C). No interaction was found for the control mRNA Cyp46, encoding the endoplasmic reticulum cholesterol hydroxylase.
Figure 2.
Hu proteins and FXR2P bind to PSD95 mRNA. A, Brain extracts were precipitated by specific antibodies against FMRP and HuD in the presence (+) or absence (−) of RNase A/T1 and control IgGs. Coimmunoprecipitated proteins FMRP, HuR, and other Hu members (panHu) are indicated. B1, Representative image of the CA3 region of hippocampus stained with anti-FMRP (red) and anti-HuR or HuD (green). Scale bar, 50 μm. B2, FMRP (red) and FXR2P or HuD or HuR (green) staining on primary neurons at 12 d in vitro. White arrows indicate protein colocalization quantified by the Mander's coefficient (n = 122 for FXR2P; n = 44 for HuD; n = 34 for HuR). Scale bar, 12.5 μm. C, HuD RNA-IP from WT and Fmr1 KO hippocampal extracts. PSD95 and Cyp46 mRNAs were amplified by qRT-PCR. D, FXR2P-IP from WT and Fxr2 KO brain extracts detected by Western blotting. E, Same as in A, FMRP-IP and FXR2P-IP in the presence of RNase A/T1 detected by Western blotting. F, FXR2P RNA-IP from WT and Fmr1 KO hippocampal extracts. PSD95 and Cyp46 mRNAs were amplified by qPCR. G, FMRP-IP from WT and Fxr2 KO hippocampal extracts. PSD95 and Cyp46 mRNAs were amplified by qPCR. mRNA levels were calculated using the formula 2^ − (CtPSD95 − Ctexogenous normalizerBC200) and normalized to the mRNA present in the input and the mock IP. C, n = 7 independent experiments. F, n = 7 independent experiments. G, n = 7 independent experiments. **p < 0.01 (Mann–Whitney nonparametric test). Data are mean ± SEM. n.s, Not statistically significant.
FXR2P interaction with PSD95 3′-UTR is strongly enhanced by FMRP
Among the FMRP-dependent partners of the PSD95 mRNA ribonucleoparticle, we found FXR2P. It has been shown that FXR2P heteromerizes with FMRP in non-neuronal cells (Zhang et al., 1995; Siomi et al., 1996) and in brain (Fig. 2D). However, it is not known whether both proteins bind simultaneously to the same RNA molecule and what is the role that each protein exerts on a given mRNA. We therefore investigated the interaction of the two proteins in targeting and regulating PSD95 mRNA. The Hu proteins were not found to coprecipitate with FXR2P complex (data not shown). The FMRP-FXR2P complex in brain is resistant to RNase treatment (Fig. 2E) (Kanai et al., 2004), and both proteins colocalize in the cell bodies and dendrites of the hippocampus and in primary neurons (Fig. 2B2), consistent with previous findings in cortex (Christie et al., 2009).
To investigate whether the two proteins recognize the PSD95 mRNA collaboratively, we tested whether the association in vivo of PSD95 mRNA with one protein required the presence of the other. FXR2P (Fig. 2F) and FMRP (Fig. 2G) were immunoprecipitated from hippocampal extracts, and coprecipitating mRNAs were analyzed. PSD95 mRNA association to FXR2P was significantly reduced in extracts of Fmr1 KO mice (Fig. 2F). The Cyp46 control mRNA did not precipitate with FXR2P (Fig. 2F). This result indicates that FMRP stabilizes the binding of FXR2P to PSD95 mRNA, confirming the in vitro binding assays (Fig. 1). As expected, FMRP also specifically recognized PSD95 mRNA (Fig. 2G) (Muddashetty et al., 2007; Zalfa et al., 2007) compared with the control Cyp46 mRNA. The association of FMRP with PSD95 mRNA, however, was not affected in the Fxr2 KO (Fig. 2G). This indicates that FMRP is the primary binder that might stabilize the association of FXR2P with PSD95 mRNA.
FXR2P is a positive regulator of PSD95 mRNA translation
FXR2P has been found associated with polyribosomes, predominantly with 60S large ribosomal subunits (Siomi et al., 1996), and its absence might affect the translation of its target mRNAs. We therefore asked whether FXR2P might act as a translational regulator of PSD95 mRNA. In Fxr2 KO hippocampi, PSD95 protein levels were significantly reduced by 30% (Fig. 3A). PSD95 mRNA abundance, in contrast, was not affected by the absence of FXR2P (Fig. 3B), suggesting a change in translational efficiency or protein degradation. We therefore gauged PSD95 mRNA translation efficiency from WT and Fxr2 KO by separating, on a sucrose gradient, nontranslated mRNAs from those actively engaged into polysomes (Fig. 3C1) (Zalfa et al., 2007). In the Fxr2 KO extracts, PSD95 mRNA was shifted toward the lighter fractions containing less efficiently translating polyribosomes (compare fraction 4 with fraction 6; Fig. 3C1,C2) consistent with the observed reduced protein production.
Figure 3.
FXR2P absence impairs PSD95 protein synthesis. A, PSD95 level in cortex and hippocampus of WT and Fxr2 KO mice quantified by Western blotting normalized to GAPDH levels. B, PSD95 mRNA level in cortex and hippocampus of WT and Fxr2 KO mice normalized to control Cyp46 mRNA. mRNA levels were calculated using the formula 2^ − Ct(CtPSD95 − CtCyp46). C1, WT and Fxr2 KO cytoplasmic extracts were fractionated on a sucrose gradient. The panel shows the UV traces and collected fractions. PSD95 and Cyp46 mRNA levels in each fraction were quantified by qPCR versus total RNA. C2, PSD95 and Cyp46 mRNA levels in the heavy polysome (1–4), light polysome (5–7), and ribonucleoparticle (8–12) fractions were quantified over the 12 fractions. *p < 0.05 (Student's t test). **p < 0.01 (Student's t test). Data are mean ± SEM. D, DIV14 primary neurons were treated with vehicle or DHPG and PSD95 expression detected by Western blotting normalized to Vinculin. pERK1/2 expression was normalized to ERK1/2 expression and represented as percentage of the vehicle. A, n = 4 for each genotype. B, n = 4 for WT and n = 3 for Fxr2 KO. C1, n = 4 for WT and n = 3 for Fxr2 KO. C2, n = 4 for WT and n = 3 for Fxr2 KO. D, n = 4 for WT and n = 6 for Fxr2 KO. *p < 0.05 (Mann–Whitney test). **p < 0.01 (Mann–Whitney test). Data are mean ± SEM. E, Model. FMRP and FXR2P cooperate to regulate PSD95 mRNA. In the absence of FXR2P, PSD95 levels are reduced because the translational machinery is not efficiently recruited to the mRNA (C1). Furthermore, no increase is observed upon mGluR activation.
Absence of FXR2P affects activity-driven synthesis of PSD95
In FXS, PSD95 synthesis is affected upon activation of Type I mGluRs (Todd et al., 2003; Muddashetty et al., 2007). To investigate the role of FXR2P, if any, on the activity-dependent PSD95 mRNA translation, we stimulated WT and Fxr2 KO neurons with DHPG (Fig. 3D). Whereas PSD95 levels were increased in WT neurons, no significant changes were observed in the Fxr2 KO neurons, similarly to previous findings in the Fmr1 KO neurons (Todd et al., 2003; Muddashetty et al., 2007). ERK1/2 phosphorylation, in contrast, could still be triggered by DHPG in the Fxr2 KO genotype similarly to WT: a major difference to the Fmr1 KO, where ERK1/2 phosphorylation is already increased in basal conditions (Hou et al., 2006).
Discussion
Here, we defined a specific set of RBPs that interact with the 3′-UTR of PSD95 mRNA, an mRNA involved in synaptic activity (Todd et al., 2003; Muddashetty et al., 2007; Muddashetty et al., 2011). Among others, FMRP (Zalfa et al., 2007), FXR2P, and the members of the ELAV/Hu family were identified. FMRP regulates PSD95 mRNA translation in the cortex (Muddashetty et al., 2011) and its stability in the hippocampus (Zalfa et al., 2007). Hu proteins are implicated in controlling stability and translation of target mRNAs (Abdelmohsen et al., 2010; Simone and Keene, 2013). Specifically, HuD appears to have a stabilizing influence on PSD95 mRNA (Abdelmohsen et al., 2010). We show here that FMRP and HuD belong to the same RNP via RNA interaction (Fig. 2A), indicating that the two proteins may jointly control the stability of their targets by interacting with distinct cis-elements in the 3′-UTR. HuD and FMRP are important regulators of neuronal morphology and maturation, and FMRP- and HuD-associated mRNAs in mouse brain encode proteins with vital roles in neuronal differentiation and cytoskeletal organization (Bolognani et al., 2010; Darnell et al., 2011). In this context, it is likely that both proteins simultaneously regulate PSD95 mRNA stability according to the needs of the synapse.
In addition, the absence of FMRP affects the interaction of certain RNPs associated to PSD95 3′-UTR, in particular its paralog FXR2P (Fig. 2F). Structurally, the FMRP paralogs FXR1P and FXR2P are 60%–90% identical with FMRP (Zhang et al., 1995). However, FXR2P diverges from FXR1P and FMRP in the C-terminal region, as it contains an RG cluster instead of an RGG box (Menon and Mihailescu, 2007), which may explain different RNA-binding properties. For example, the G-quartet-containing structure of the Semaphorin 3F mRNA is strongly recognized by the C termini of FMRP and FXR1P, but less by FXR2P (Menon and Mihailescu, 2007). Here we show that, although FXR2P binds PSD95 mRNA directly (Fig. 1E), it requires FMRP for optimal binding (Figs. 1C,E, 2F).
Surprisingly, the absence of FXR2P leads to a reduction of PSD95 protein levels in hippocampus due to decreased translational efficiency (Fig. 3A,C1,C2). Following mGluR-driven synaptic activation, PSD95 synthesis does not increase (Fig. 3D). Similarly, the absence of FMRP impairs the mGluR-dependent PSD95 protein production (Todd et al., 2003; Schmit et al., 2013). Because PSD95 mRNA de novo transcription is not affected by synaptic activation by Type I mGluRs (Todd et al., 2003), it is tempting to hypothesize that FXR2P increases mRNA translation possibly by recruiting the translation machinery (Fig. 3E). This model is consistent with the initial findings from Dreyfuss and collaborators showing a direct binding of FXR2P to the 60S ribosomal subunit (Siomi et al., 1996) (Fig. 3E). We cannot exclude that FMRP belongs to two distinct complexes on the PSD95 mRNA that could activate and repress mRNA translation: namely, the FMRP/FXR2P and the FMRP/CYFIP1 complexes (Napoli et al., 2008). In non-neuronal cells, the N terminus of FMRP interacts with both FXR2P (Siomi et al., 1996) and CYFIP1 (Schenck et al., 2001), implying that the complexes are mutually exclusive. Indeed, CYFIP1 does not associate with FXR2P in brain (data not shown), arguing against a triple complex and opening the possibility that dynamic switching between FMRP/FXR2P and FMRP/CYFIP1 regulates PSD95 expression. In summary, the FMRP-FXR2P complex might assure a homeostatic as well as activity-driven production of PSD95.
It has been proposed that the same RBP can influence mRNA expression by more than one mechanism probably due to the combinatorial influence of other RBPs (Ule and Darnell, 2006; Gebauer et al., 2012). The fact that both FMRP and FXR2P may exert an overlapping, not identical, control on the same mRNAs would explain the similarities and diversities observed in the phenotypes for Fxr2 and Fmr1 KO mice. Both KO mice show resemblance in certain behavioral paradigms and have immature spines (Spencer et al., 2006). However, both models differ in the outcomes of other behavioral tests as well as in the activation of protein synthesis in response to synaptic plasticity. In the Fmr1 KO mice, hippocampal mGluR-LTD is enhanced and independent of new protein synthesis (Huber et al., 2002), whereas in Fxr2 KO mice, mGluR-LTD is reduced and requires new protein synthesis (Zhang et al., 2009). Furthermore, ERK1/2 phosphorylation is comparable between Fxr2 KO and WT adult brains (Fig. 3D), whereas it is upregulated in the Fmr1 KO (Hou et al., 2006). This is consistent with the view that both FMRP and FXR2P are involved in the mGluR-LTD synaptic activity, but their absence likely has different consequences.
Footnotes
The work is supported by Foundation Lejeune, Associazione Italiana Sindrome X Fragile, FWO-G.0705.11, VIB, EU FP7 SynSys (242167 to C.B., A.B.S., and K.W.L.), and Marie Curie ITN Brain Train (FP7) VIB to C.B. E.F. was recipient of Intra European career development fellowship Marie Curie FP7. We thank Nathalie Leysen, Jonathan Royaert, and Karin Jonckers for excellent technical assistance; Eef Lemmens for the great administrative support; Rob Willemsen (Erasmus MC, The Netherlands) for providing us the Fxr2 KO mice; and Jennifer Darnell for advice on the HITS-CLIP data.
The authors declare no competing financial interests.
References
- Abdelmohsen K, Hutchison ER, Lee EK, Kuwano Y, Kim MM, Masuda K, Srikantan S, Subaran SS, Marasa BS, Mattson MP, Gorospe M. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol Cell Biol. 2010;30:4197–4210. doi: 10.1128/MCB.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Tassone F, Neri G, Hagerman R. Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J Clin Invest. 2012;122:4314–4322. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker CE, Verheij C, Willemsen R, Vanderhelm R, Oerlemans F, Vermey M, Bygrave A, Hoogereen AT, Oostra BA, Reyniers E, De Boulle K, Dhooge R, Cras P, Van Velzen D, Nagels G, Martin JJ, Dedeyn PP, Darby JK, Willems PJ. FMR1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- Béïque JC, Lin DT, Kang MG, Aizawa H, Takamiya K, Huganir RL. Synapse-specific regulation of AMPA receptor function by PSD-95. Proc Natl Acad Sci U S A. 2006;103:19535–19540. doi: 10.1073/pnas.0608492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010;38:117–130. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontekoe CJ, McIlwain KL, Nieuwenhuizen IM, Yuva-Paylor LA, Nellis A, Willemsen R, Fang Z, Kirkpatrick L, Bakker CE, McAninch R, Cheng NC, Merriweather M, Hoogeveen AT, Nelson D, Paylor R, Oostra BA. Knockout mouse model for Fxr2: a model for mental retardation. Hum Mol Genet. 2002;11:487–498. doi: 10.1093/hmg/11.5.487. [DOI] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Rioult-Pedotti MS, Migani P, Yu CJ, Tiwari R, Parang K, Spaller MR, Goebel DJ, Marshall J. Impairment of TrkB-PSD-95 signaling in Angelman syndrome. PLoS Biol. 2013;11:e1001478. doi: 10.1371/journal.pbio.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, Sheng M, Peng J. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J Neurosci. 2009;29:1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyder M, Karlsson RM, Mathur P, Lyman M, Bock R, Momenan R, Munasinghe J, Scattoni ML, Ihne J, Camp M, Graybeal C, Strathdee D, Begg A, Alvarez VA, Kirsch P, Rietschel M, Cichon S, Walter H, Meyer-Lindenberg A, Grant SG, et al. Association of mouse Dlg4 (PSD-95) gene deletion and human DLG4 gene variation with phenotypes relevant to autism spectrum disorders and Williams' syndrome. Am J Psychiatry. 2010;167:1508–1517. doi: 10.1176/appi.ajp.2010.10040484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Preiss T, Hentze MW. From cis-regulatory elements to complex RNPs and back. Cold Spring Harbor Perspect Biol. 2012;4:a012245. doi: 10.1101/cshperspect.a012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Berry-Kravis EM, Bassell GJ. Therapeutic strategies in fragile X syndrome: dysregulated mGluR signaling and beyond. Neuropsychopharmacology. 2012;37:178–195. doi: 10.1038/npp.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhang L, Christopher DM, Teng ZQ, Fausett SR, Liu C, George OL, Klingensmith J, Jin P, Zhao X. RNA-binding protein FXR2 regulates adult hippocampal neurogenesis by reducing Noggin expression. Neuron. 2011;70:924–938. doi: 10.1016/j.neuron.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kapeli K, Yeo GW. Genome-wide approaches to dissect the roles of RNA binding proteins in translational control: implications for neurological diseases. Front Neurosci. 2012;6:144. doi: 10.3389/fnins.2012.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemmer P, Smit AB, Li KW. Proteomics analysis of immunoprecipitated synaptic protein complexes. J Proteomics. 2009;72:82–90. doi: 10.1016/j.jprot.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Menon L, Mihailescu MR. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic Acids Res. 2007;35:5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O'Dell TJ, Grant SG. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelić S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Pasciuto E, Bagni C. SnapShot: FMRP mRNA targets and diseases. Cell. 2014;158:1446–1446.e1. doi: 10.1016/j.cell.2014.08.035. [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit TL, Dowell JA, Maes ME, Wilhelm M. c-Jun N-terminal kinase regulates mGluR-dependent expression of post-synaptic FMRP target proteins. J Neurochem. 2013;127:772–781. doi: 10.1111/jnc.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone LE, Keene JD. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr Opin Genet Dev. 2013;23:35–43. doi: 10.1016/j.gde.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer CM, Serysheva E, Yuva-Paylor LA, Oostra BA, Nelson DL, Paylor R. Exaggerated behavioral phenotypes in Fmr1/Fxr2 double knockout mice reveal a functional genetic interaction between Fragile X-related proteins. Hum Mol Genet. 2006;15:1984–1994. doi: 10.1093/hmg/ddl121. [DOI] [PubMed] [Google Scholar]
- Stein V, House DR, Bredt DS, Nicoll RA. Postsynaptic density-95 mimics and occludes hippocampal long-term potentiation and enhances long-term depression. J Neurosci. 2003;23:5503–5506. doi: 10.1523/JNEUROSCI.23-13-05503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, König J, Hussain S, Zupan B, Curk T, Frye M, Ule J. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, Malter JS. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci U S A. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Xing L, Bassell GJ. mRNA localization: an orchestration of assembly, traffic and synthesis. Traffic. 2013;14:2–14. doi: 10.1111/tra.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, Bagni C. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hou L, Klann E, Nelson DL. Altered hippocampal synaptic plasticity in the FMR1 gene family knockout mouse models. J Neurophysiol. 2009;101:2572–2580. doi: 10.1152/jn.90558.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, O'Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]