Abstract
Fibroblast growth factors (Fgfs) play important roles in developmental processes of the inner ear, including the ontogeny of the statoacoustic ganglia (SAG) and hair cells. However, the detailed genetic mechanism(s) underlying Fgf/Fgfr-dependent otic neural development remains elusive. Using conditional genetic approaches and inhibitory small molecules, we have revealed that Fgfr-PI3K/Akt signaling is mainly responsible for zebrafish SAG development and have determined that Sox9a and Atoh1a act downstream of Fgfr-Akt signaling to specify and/or maintain the otic neuron fate during the early segmentation stage. Sox9a and Atoh1a coregulate numerous downstream factors identified through our ChIP-seq analyses, including Tlx2 and Eya2. Fgfr-Erk1/2 signaling contributes to ultricular hair cell development during a critical period between 9 and 15 hours postfertilization. Our work reveals that a genetic network of the previously known sensory determinant Atoh1 and the neural crest determinant Sox9 plays critical roles in SAG development. These newly uncovered roles for Atoh1and Sox9 in zebrafish otic development may be relevant to study in other species.
Keywords: Fgf signaling, Fgfr-Erk1/2, Fgfr-PI3K/Akt, hair cells, statoacoustic ganglia
Introduction
Zebrafish otic neurogenesis, referring to otic neuronal specification and differentiation (Maier et al., 2014), occurs during a relatively long developmental period. After initial specification and differentiation of otic neuroblasts at the early to midsegmentation stages, migration of neuroblasts begins at the 22 hours postfertilization (hpf) and ends at approximately the 42 hpf stage in zebrafish (Haddon and Lewis, 1996; Whitfield et al., 2002). After further cell divisions, delaminated neuroblasts differentiate into postmitotic statoacoustic (VIIIth) ganglia (SAG), which send peripheral projections to the sensory patches in the ear and central connections to their CNS targets. Fgf signaling has been suggested to regulate otic neurogenesis in many species (Adamska et al., 2001; Alsina et al., 2004; Millimaki et al., 2007; Vemaraju et al., 2012), but how the signaling functions to direct SAG development remains elusive.
Many transcription factors that are dependent on Fgf signaling to be activated or maintained in the ear are otic neurogenic factors (Neurogenin, Neurod, Hmx3, etc.) (Adamska et al., 2000; Ota and Ito, 2006). Fgf-dependent sox9a is expressed in the pre-otic region in zebrafish (Yan et al., 2005; Esain et al., 2010). In embryos carrying a homozygous deletion that removes dlx3b, dlx4b, and sox9a (Dfb380), all otic sensory lineages are lost, yet expression of otic neuroblast markers remains in residual otic cells (Liu et al., 2003; Hans et al., 2013). Because the additional knockdown of sox9b function in the Dfb380 or Dfb380;fgf8a−/− double mutant shows a complete loss of the residual otic cells (Liu et al., 2003), the role of Sox9, particularly Sox9a (it is upstream of sox9b), in otic neurogenesis is implicated. Sox9 is involved in neural differentiation within the CNS (Scott et al., 2010; Martini et al., 2013), but whether it also promotes neurogenic development in the peripheral nervous system is unknown. Zebrafish atoh1a marks the prospective maculae from otic specification stages and indicates hair cell differentiation (Chen et al., 2002; Adolf et al., 2004; Millimaki et al., 2007), the process called otic sensorigenesis (Maier et al., 2014). It is worth noting that Atoh1 is expressed at low levels in SAG/hair cell precursors of mouse otocyst (Matei et al., 2005), implicating additional involvement of Atoh1 in otic neurogenesis.
In the present study, we examined how Fgf signaling and candidate neurogenic factors such as Sox9a and Atoh1a are involved in zebrafish otic neurogenesis. Using conditional approaches to investigate the requirement of Fgf signaling, we discovered that inactivation of Fgfr-PI3K/Akt signaling abolishes the formation of the majority of SAG neurons, whereas Fgfr-Erk1/2 signaling partially contributes to otic sensory (hair cell) development. Downstream of Fgfr-PI3K/Akt signaling, Sox9a and Atoh1a bind to their direct/common targets such as eya2 and tlx2 to allow the otic ganglion development to proceed.
Materials and Methods
Zebrafish stains and maintenance.
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Peking University. The reference from IACUC of Peking University is LSC-LiuD-01. Wild-type Tübingen strain and all transgenic zebrafish lines were kept and bred under standard conditions (Westerfield, 2007). One-cell stage embryos were collected and incubated at 28°C in E2 medium as described previously. We used the following transgenic lines: Tg(hsp70:fgf8) (Hans et al., 2007), Tg(hsp70:dnfgfr1) (Lee et al., 2005), and TgBAC(neurod:EGFP) (Obholzer et al., 2008). Heat-shock-inducible transgenes were activated by incubating embryos at 38.5°C for 40 min as indicated in the Results. We could not tell the sex of zebrafish embryos or larvae at 10–72 h after fertilization.
Antisense morpholino oligonucleotides and mRNA misexpression.
For morpholino oligonucleotide (MO) knockdowns, embryos were injected at the one- to two-cell stage with translation-blocking MOs obtained from Gene Tools Inc. MO injections were as follows: 10 ng of fgf3-MO, 5′-CAGTAACAACAAGAGCAGAATTATA-3′(Léger and Brand, 2002); 7.5 ng of fgf8a-MO, 5′-GAGTCTCATGTTTATAGCCTCAGTA-3′(Maroon et al., 2002); 8 ng of sox9a-MO, 5′-TCAGGTAGGGGTCGAGGAGATTCAT-3′(Liu et al., 2003); 6 ng of sox9b-MO, 5′-TGCAGAGAGAGAGTGTTTGAGTGTG-3′(Dutton et al., 2009);4 ng of atoh1a-MO, 5′-ATCCATTCTGTTGGTTTGTGCTTTT-3′ (Millimaki et al., 2007); 6 ng of atoh1b-MO, 5′-TCATTGCTTGTGTAGAAATGCATAT-3′(Millimaki et al., 2007); 15 ng of tlx2-MO, 5′-CGCGGTTCTATATGTTACGCACAGC-3′, 6 ng of eya2-MO, 5′-CGTCTGTCCGAAGCTGCCATGAGC-3′.
For mRNA injections, 80–100 pg of sox9a mRNA, 60–80 pg of atoh1a mRNA, and 40–60 pg of eya2 or tlx2 mRNA was individually injected into one-cell-stage embryos and incubated at 28°C until the desired stages.
In situ hybridization and immunostaining.
In situ hybridization and immunostaining were performed as described previously (Westerfield, 2007). The following probes and antibodies were used: neurod (Korzh et al., 1998), atoh1a (Millimaki et al., 2007), atoh1b (Millimaki et al., 2007), sox9a (Yan et al., 2002), tlx2 (Langenau et al., 2002), eya2 (Lupo et al., 2011), anti-GFP (1:500; rabbit or mouse; Invitrogen), anti-HuC/D (1:500; mouse; Invitrogen), and anti-MyoVI (1:200, rabbit; Proteus Biosciences).
Chemical inhibition.
Embryos were treated with 50 μm SU5402 (Calbiochem), 2.5 μm wortmannin (Calbiochem), 25 μm PD98059 (Calbiochem), 10 μm U73122 (Calbiochem) in 10 μm DMSO or with 10 μm DMSO alone in E2 medium.
Western blotting.
Embryos were homogenized in lysis buffer with proteinase inhibitors, run on a 12% SDS-PAGE gel, blotted onto a PVDF membrane (Millipore), and then incubated with primary antibodies (β-actin 1:1000 and c-Myc 1:500; Santa Cruz Biotechnology) at 4°C overnight. Subsequently, the membranes were rinsed three times with TBST and then incubated with anti-rabbit or anti-mouse IRDye (Odyssey) for 2 h at room temperature and visualized using an Odyssey Infrared Imaging System.
Quantitative RT-PCR.
Total RNA was extracted from embryos using TRIzol (Invitrogen). SuperScript II reverse transcriptase (Invitrogen) was used to synthesize cDNA from 0.5 μg of total RNA. Quantitative RT-PCR (qRT-PCR) was performed with an ABI-7500 using 2×FastStart SYBR Green Master Mix (Invitrogen). Total RNA from each sample was normalized to β-actin. In each experiment, three pools of embryos were run in duplicate. Primer pairs are listed in Table 1.
Table 1.
Primer pairs used for RT-PCR
| Gene | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|
| atoh1a | CAACGCCCTGTCCGACTTAC | GGGAGATCGGTCCGTTTCTAA |
| atoh1b | TTCGGGAGGAGTTTTCCATCT | TCCACGGCAGAGGAGTACTCA |
| b-actin | TTTTCCCCTCCATTGTTGGA | CCTCTCTTGCTCTGAGCCTCAT |
| bnc1 | TCTGAGCAAGCTGAAGGTTCATC | GTCCCATAAAGCATCAGACTACAGATATC |
| col2a1a | AAGTCCATCAACGGGCAGATT | ATTCAGGGTGGCAGAGTTTCAG |
| cx43 | GCAGCACCATCTCCAATTCA | GCCAATGGCTGCAACTCAT |
| dacha | GGCTGGACCTGCCATTTATG | TTCAGGTGGTTCATCTGGTTCA |
| dlc | AGGCCCCCTCTGTGATGAGT | TAGACCGCCCCAACCTTCTT |
| eya2 | ATCACCACGGCGATACCCTAT | GACTCGTGGGAGCCTGGTACT |
| fn1 | GAGCTGCATCGCCTACTCTCA | GTAGCCTTCGTCGTGCCTTTT |
| hes6 | GCATGCATGAAGTGCACACAT | TCGTTCAGAGGCATACATTCGA |
| irx2a | GGTGGTTCATTCCTCTGTGGCG | CATATTACTCCTGACTGATCTGTGG |
| jag1b | CACCCCCTCCACCCTCTAAA | CCTTGAGGCATGTTCTCCTTGT |
| neo1 | CGCGGCTATAAGATCGGTTATG | TGGCATCGAGGTTATCAATGC |
| notch1b | ACTTCGCAAACAATCCGGG | TTCTTTTATCTTTGCCGTGATGAC |
| sox9a | CGCGCAGGAAACTGGCCGAC | CGCTTTTCCACCTCGTTCAGTAATCTCC |
| sox9b | TGGAGACTGCTGAACGAGGGCGA | GCTGGTATTTGTAGTCTGGATGATCTTTCTTATGCT |
| spry1 | GGACTCAGCCCAAATCTCTGAA | CAGATGTACAGATGCGCAGAGTCT |
| tbx1 | TCCCATTTTTGCGATGTTGC | GCACGGCTCAAACTGGCTT |
| tlx2 | GACCAGTCGAGTAGTTGCATGCT | AGGCCGGATACATGCTGTTG |
| unc5b | CATGCAGATGGCAGCGATTA | ATGCATCCTCGGGCTCACT |
| zgc77614 | CTCTCACTGGAGCAGTTGTTGAAG | AGCACAGGCTGGACTGGTACA |
ChIP analysis and ChIP-Seq.
ChIP assays were performed as described previously (Wardle et al., 2006). Thirty nanograms of pull-down DNA from ChIP was amplified and sequenced using the Illumina platform (HiSeq 2000). ChIP-seq reads were then mapped to the zebrafish genome assembly (Zv9 genome assembly, July 2010) using BWA (Li and Durbin, 2009) and improper mapped reads were filtered out by Samtools (Li et al., 2009). The regions significantly enriched in ChIP-seq reads from sample and comparison controls were called by MACS (Zhang et al., 2008). Potential target genes in each ChIP-seq experiment were identified by HOMER (Heinz et al., 2010) and annotated with gtf file of zebrafish genome assembly (ZV9).
For the ChIP-PCR assay, ∼1/50 of the precipitated DNA was used for PCR; an equivalent amount of starting material was used as Input DNA. PCR was performed in 2×PCR buffer (GenStar), 10 μm forward and reverse primers, and one unit of Hot Start Taq polymerase (GenStar). Primer pairs used for the PCR are listed in Tables 2 and 3.
Table 2.
Primer pairs used to verify Atoh1a targets in ChIP-PCR assay
| Gene | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|
| bnc1 | CACGGCTGAAACAGGGGAT | CAAAGGCAGAGAGTGGGAAAAC |
| dlc | CAGAATGTGTCCTAAAAGTGTGAGCAAATGA | GTGTGATCCAGTGGCAGAAGGCG |
| eya2 | TCAACACTCGCACTATAGGTGAATTGTAT | CTTATGTCGCCAAAGCAGAATGAAC |
| fn1 | TGAGGGTTCCAGCGAGAGATTATCC | GAGGACATCTGATCTCCGGTGTCAG |
| irx2a | GGTGGTTCATTCCTCTGTGGCG | CATATTACTCCTGACTGATCTGTGGTCATTCT |
| jag1b | CGCGCAGTGTTTCAACCTGG | GATTTCTCACCTTGGCATGATGTG |
| neo1 | GATGGGTAGATGGTTTTATAGGCG | ATTCATCCATTGATTCATTAGTCTGC |
| notch1b | ACTTCGCAAACAATCCGGG | TTCTTTTATCTTTGCCGTGATGAC |
| spry1 | GACTCAGCCCAAATCTCTGAACGCTCCCTTT | GTTCTCCGCCGAACACAAACACAATCCG |
| tlx2 | CAACCAATGTAGGCAGAAGGGCA | CCAGGCAGCGTTTTTCCAATCC |
| zgc77614 | TGGGTAAATAAGAAACAAGCAACA | GCAGAATATCCAAAATCTGAAGTGAT |
| hes6 | CCTTTTCTATTCATACACT | TCAGAATCACAGAGGGAG |
Table 3.
Primer pairs used to verify Sox9a targets in ChIP-PCR assay
| Gene | Forward primer 5′-3′ | Reverse primer 5′-3′ |
|---|---|---|
| bnc1 | TATTAATTTCATGTTCAGCTCAACC | TGTATTGTCTTCATTCCATTGTAAATAC |
| cx43 | ACCCTGAGCACACTGCCA | TCAACTCTGCCCACACATTC |
| dacha | GAGAGAGATTTTACTGTTTAGTTTGGCA | TTTTCTTAGGTGGGGCTTGATG |
| eya2 | GCTGTTAGACTGTTGCACTTTTTT | GTGTTTCTTACACCAGGGGTTC |
| fn1 | CTGAAGGGAGTCTTGGCGGAGGTT | ATCCAGAGAAGGTTCAGGCTGTGGTAAA |
| neo1 | TTTGCCTAGTCTGAATGTATGTTTGCTTATG | GCCTTCACTGACAGAGACATGAAGATGT |
| spry1 | CACCAACCCAGACGATATTTCACT | TTTTCGGTATTTTGCGCTTCAT |
| tbx1 | GGACAGTGACTTTGGCCTTG | CTTTGCTTCATTATTTCTCCTTTG |
| tlx2 | CTGCTTCTTTAGTTTTTCAGTAGA | CTGTAAAATGATGGTATCACAAATA |
| unc5b | AAATGGGCTCAGTGGGGGGTGG | TGTTTGTGGGTGTCTACAGTGTTTCTGGGTA |
| zgc77614 | TTTATTCCCCTTGATGATATTGTGCTTT | TGCCATGAGATTTCTGAATTCTACATGC |
| col2a1a | CCTGGTGGTAAGTGAGACCATTAGA | GGTGTAAAATGTGGCCTCCTGTAG |
Imaging and data analysis.
Live embryos and embryos processed for ISH were imaged under a Zeiss Imager A1 microscope equipped with differential interference contrast optics. Live embryos were first anesthetized in tricaine before imaging. Immunostained embryos were imaged under a Zeiss Imager A1 fluorescence microscope or a Leica SP5 confocal microscope using 20× or 40× objectives. Embryos were prepared on glass-bottomed dishes in 1.5% LMP-agarose. For quantitative results, only one side of each embryo was included. Area measurements were performed on reconstructed confocal z-stacks using software equipped in Leica SP5. One-way ANOVA test was used to compare control and experimental groups. *p < 0.05 is considered significant.
Results
Fgfr-PI3K/Akt signaling regulates otic neurogenesis at 10–14 hpf
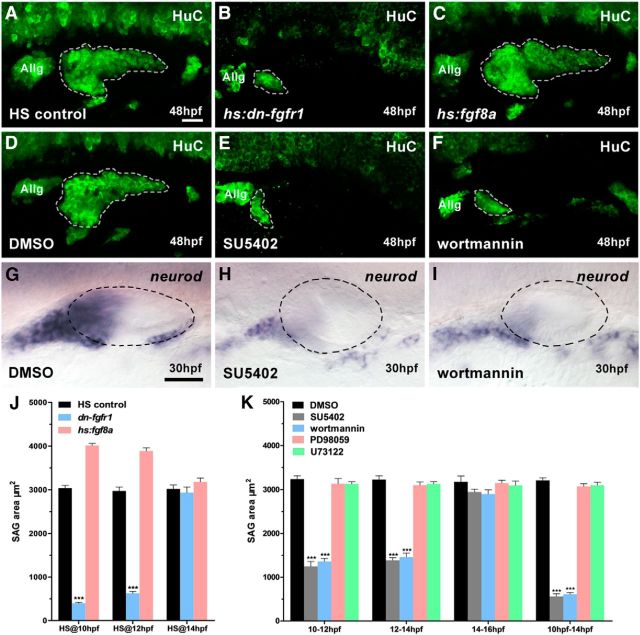
To assess zebrafish SAG development, we measured the otic HuC immunostaining area (SAG area) during different developmental stages. At 26 hpf, the SAG area of wild-type embryos was nearly 15% and 35% larger than that in fgf3 and fgf8a morphants, respectively (Table 4). The SAG area progressively increased in control embryos over the course of development from 26 to 48 hpf, whereas in fgf3 and fgf8a morphants, the SAG area failed to expand (Table 4), suggesting a defect in the initial SAG development due to a low level of Fgf signaling. Fgf8 is sufficient for otic induction (Yao et al., 2014), but when both Fgf3 and Fgf8a functions are knocked down or abolished in zebrafish, no ear forms (Phillips et al., 2001; Léger and Brand, 2002; Maroon et al., 2002; Liu et al., 2003), making it hard to judge the role of these Fgf ligands at later stages of otic neurogenesis. We instead used two heat-inducible transgenic lines to test when Fgf signaling is critical for the otic neurogenesis: heat-activated dominant-negative Fgfr1 (hs:dn-fgfr1) and heat-activated Fgf8 (hs:fgf8a). In a series of tests with heat shock treatment at different developmental stages, we found that the activation of heat-activated dominant-negative Fgfr1 (hs:dn-fgfr1) at 10 and 12 hpf led to a drastic reduction in the SAG area (Fig. 1A,B,J). Conversely, an increase of Fgf8a at 10 and 12 hpf slightly enlarged the SAG population (Fig. 1A,C,J). In both sets of experiments, however, heat activation of transgenes at 14 hpf did not show any significant change of SAG area (Fig. 1J). Therefore, it appeared that Fgf signaling is involved in the otic neurogenesis before14 hpf.
Table 4.
SAG populations in morphants
| Genotype | Stage |
|||
|---|---|---|---|---|
| 26 hpf | 36 hpf | 42 hpf | 48 hpf | |
| Wild-type | 1221 ± 196 μm2 (n = 16) | 2048 ± 388 μm2 (n = 12) | 2968 ± 258 μm2 (n = 15) | 3273 ± 381 μm2 (n = 11) |
| fgf3 MO | 1034 ± 95 μm2 (n = 13) | 1018 ± 104 μm2 (n = 9) | 1171 ± 68 μm2 (n = 14) | 1150 ± 51 μm2 (n = 10) |
| fgf8 MO | 790 ± 52 μm2 (n = 14) | 840 ± 79 μm2 (n = 13) | 1020 ± 82 μm2 (n = 11) | 973 ± 102 μm2 (n = 13) |
| sox9a MO | 736 ± 109 μm2 (n = 19) | 1010 ± 127 μm2 (n = 19) | 1019 ± 60 μm2 (n = 18) | 1033 ± 48 μm2 (n = 20) |
| sox9b MO | 787 ± 72 μm2 (n = 22) | 785 ± 171 μm2 (n = 15) | 828 ± 71 μm2 (n = 21) | 1095 ± 228 μm2 (n = 13) |
| atoh1a MO | 789 ± 76 μm2 (n = 13) | 1064 ± 131 μm2 (n = 17) | 1216 ± 277 μm2 (n = 13) | 1292 ± 324 μm2 (n = 11) |
| atoh1b MO | 969 ± 117 μm2 (n = 20) | 2032 ± 204 μm2 (n = 18) | 2355 ± 395 μm2 (n = 11) | 2410 ± 247 μm2 (n = 13) |
Listed is a summary of SAG areas at 26, 36, 42, and 48 hpf in both wild-type embryos and fgf3, fgf8a, sox9a, sox9b, atoh1a, or atoh1b morphants.
Figure 1.
SAG development is impaired when Fgf-PI3K/Akt signaling is blocked. A–C, Heat-shocked controls (A), hs:dn-fgfr1 embryos (B), and hs:fgf8a embryos (C) are immunostained by HuC antibody at 48 hpf. The 40 min heat shock starts at 10 hpf. D–F, HuC antibody staining in DMSO- (D), SU5402- (E), and wortmannin (F)-treated embryos at 48 hpf. The chemical treatment period is 10–14 hpf. G–I, Expression of neurod in DMSO- (G), SU5402- (H), and wortmannin (I)-treated embryos at 30 hpf. The chemical treatment period is 10–14 hpf. J, Summary of SAG size after manipulating Fgf signaling at different heat shock time point (n = 10; 48 hpf). K, Summary of size of SAG populations after blocking the Fgf signaling (n = 10; 48 hpf). Allg, Anterior lateral line ganglion. White dotted lines indicate otic neurogenic region; black dotted lines indicate otic region. A–I, lateral views with anterior to left and dorsal up. Results in J and K are presented as mean ± SEM. ***p < 0.001, one-way ANOVA. Scale bar, 30 μm.
To further determine when Fgf signaling is directly involved in SAG development, SU5402 was used to block Fgfr signaling at 10–12, 12–14, and 14–16 hpf. A reduction in the SAG area was observed only in the 10–12 and 12–14 hpf groups (Fig. 1K). If embryos were treated for 4 h (10–14 hpf), then SAG reduction was comparable to that of hs:dn-fgfr1embryos heat shocked at 10 and 12 hpf (Fig. 1A,B,D,E,J,K). Together, these experiments have defined the critical time when otic neurogenesis is sensitive to Fgf signaling to be between 10 and 14 hpf.
To determine which signaling pathways were downstream of Fgf in SAG development, we also used PD98059 to block Fgfr-Erk1/2 signaling, wortmannin to block Fgfr-PI3K/Akt signaling, and U73122 to block phospholipase C, the noncanonical intracellular Fgfr signaling active in the otic region between 10 and 14 hpf (Millimaki et al., 2007; Boeldt et al., 2014; Penumarti and Abdel-Rahman, 2014; Tocharus et al., 2014). We found that only wortmannin reduced SAG area, comparable to that of SU5402 treatment (Fig. 1D–F,K). Furthermore, SU5402 and wortmannin treatments between 10 and14 hpf drastically reduced the expression of SAG marker neurod at 30 hpf, yet the ear sizes of treated and untreated embryos were comparable (Fig. 1G–I), suggesting that the otic induction is largely unaffected by the chemical treatments during this period. Together, these experiments demonstrated that the critical timing of Fgf/Fgfr-Akt-dependent SAG development is between 10 and 14 hpf.
Sox9a and Atoh1a are Fgfr-PI3K/Akt signaling dependent to direct otic neurogenesis
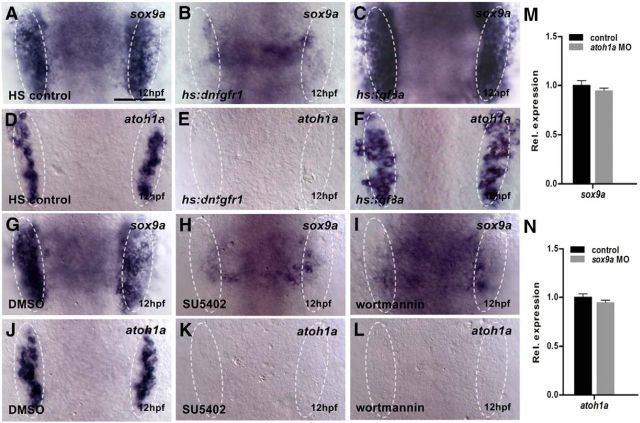
The onset of zebrafish sox9a at the end of gastrulation in the pre-otic region suggested that it was a good candidate to regulate otic neurogenesis. We first analyzed whether sox9a expression was regulated by Fgf signaling at 12 hpf. Activation of hs:dnfgfr1 and hs:fgf8a heat shocked at 10 hpf resulted in dramatically changed expression of sox9a in the otic region (Fig. 2A–C). The reduced expression of sox9a was also evident in SU5402- or wortmannin-treated embryos treated between 10 and 12 hpf (Fig. 2G–I). When Sox9a function was knocked down by MO, neurod expression and SAG area were reduced (Fig. 3A,B,D,E, Table 4), suggesting that Sox9a may mediate the Fgf-dependent otic neurogenesis.
Figure 2.
The sox9a and atoh1a are two parallel targets downstream of Fgfr-PI3K/Akt signaling. A–C, Expression of sox9a in heat-shocked controls (A), hs:dn-fgfr1 embryos (B), and hs:fgf8a embryos (C) at 12 hpf. The 40 min heat shock starts at 10 hpf. D–F, Expression of atoh1a in heat-shocked controls (D), hs:dn-fgfr1 embryos (E), and hs:fgf8a embryos (F) at 12 hpf. The 40 min heat shock starts at 10 hpf. G–I, Expression of sox9a in DMSO- (G), SU5402- (H), and wortmannin (I)-treated embryos at 12 hpf. The chemical treatment period is 10–12 hpf. J–L, Expression of atoh1a in DMSO- (J), SU5402- (K), or wortmannin (L)-treated embryos at 12 hpf. The chemical treatment period is 10–12 hpf. M–N, Analyzing the expression profiles of sox9a (M) or atoh1a (N) genes by qRT-PCR at 14 hpf. White dotted lines indicate the otic region. A–L, Dorsal views with anterior to top. Results in M and N are presented as mean ± SEM (one-way ANOVA test). Scale bar, 30 μm.
Figure 3.
sox9a and atoh1a both work in otic neurogenesis. A–C, Expression of neurod in wild-type embryos (A), sox9a morphants (B), and atoh1a morphants (C) at 30 hpf. D–F, HuC antibody staining in wild-type embryos (D), sox9a morphants (E), and atoh1a morphants (F) at 48 hpf. G–J, sox9a (I) and atoh1a (J) mRNA can individually and partially restore SAG areas of SU5402-treated (10–14 hpf) neurod:EGFP embryo (H) at 30 hpf. K, Summary of the restored SAG areas in mRNA-injected embryos. The chemical treatment period is 10–14 hpf (n = 10; 48 hpf). Allg, Anterior lateral line ganglion; Pllg, posterior lateral line ganglion. White dotted lines indicate the otic neurogenic region; black dotted lines indicate the otic region. A–J, Lateral views with anterior to left and dorsal up. Results in K are presented as mean ± SEM. **p < 0.01, ***p < 0.001 (one-way ANOVA). Scale bar, 30 μm.
Fgfrs are required to activate atoh1a (Millimaki et al., 2007; Sweet et al., 2011). In hs:dnfgfr1 transgenic embryos heat shocked at 10 hpf, atoh1a expression was almost lost in the otic region, whereas in hs:fgf8a embryos, it was significantly increased (Fig. 2D–F). When embryos were treated with SU5402 and wortmannin between 10 and 12 hpf, atoh1a expression was completely blocked (Fig. 2J–L). In embryos injected with atoh1a MOs, otic neurod expression and SAG area showed significant decreases (Fig. 3A,C,D,F, Table 4), suggestive of a role of Atoh1a in Fgf-dependent otic neurogenesis.
We next tested whether we could rescue impaired otic neurogenesis in neurod::EGFP transgenic and wild-type embryos treated with SU5402 (10–14 hpf) by mRNA misexpression of sox9a and atoh1a and found partially restored SAG area in both cases (Fig. 3G–K). Because of early onset of both genes in the pre-otic region, it is possible that Sox9a and Atoh1a regulate each other at the transcriptional level. However, qRT-PCR assays failed to detect any changes in corresponding expression after MO injection for atoh1a or sox9a (Fig. 2M,N), indicating that they act independently to mediate Fgfr-PI3K/Akt-dependent otic neurogenesis.
Identification of direct targets of Sox9a and Atoh1a
To determine whether Sox9a and Atoh1a shared common downstream targets, we performed ChIP-seq analysis. We injected Myc-tagged versions of atoh1a or sox9a plasmid DNA (Fig. 4A) individually into one-cell-stage embryos and collected crude protein from 10 to 14 hpf embryos. Sox9a-Myc or Atoh1a-Myc fusion protein could be detected by Western blot analysis (Fig. 4B). We validated the effectiveness of our ChIP assay by detecting the presence of known Sox9 and Atoh1 target genes such as col2a1a and hes6 (Bell et al., 1997; Scheffer et al., 2007; Yokoi et al., 2009), respectively, in embryo (14 hpf) extract by ChIP-PCR assay (Fig. 4C). The enriched Sox9a- and Atoh1a-binding chromatin DNA fragments collected from injected embryos of at least 10 separate experiments were then sequenced. A total of 2112 (Sox9a group) and 4580 (Atoh1a group) peaks were mapped to 1217 and 3106 candidate genes, respectively, with a vast majority of binding sites in either intergenic regions (59.34% Sox9a and 49.63% Atoh1a) or intronic regions (34.92% and 33.78%, respectively; Fig. 4D). Based on the distance of peaks from the transcription starting site to 50 kb upstream of the last intron of each candidate gene (Fig. 4E) and their otic expressions, we narrowed the list to 18 candidate targets of Sox9a and 21 targets of Atoh1a and found that 15 targets were common to both datasets (Table 5). ChIP-PCR assay verified 11 of 21 genes for Atoh1a and 11 of 18 genes for Sox9a as reproducible targets (Fig. 4F, Table 5).
Figure 4.

Identifying direct targets of Sox9a and Atoh1a. A, Diagram of the recombinant plasmid pN3-X-Myc. B, Western blot analysis using a Myc antibody detects Sox9a-Myc and Atoh1a-Myc fusion proteins in embryos at 14 hpf. C, ChIP-PCR analysis detects known targets of Sox9a (col2a1a) and Atoh1a (hes6). D, Distribution of all sorts of Sox9a (left) or Atoh1a (right) genomic DNA-binding sites as revealed by ChIP-Seq analyses. E, Peaks of Sox9a (gray) or Atoh1a (dark gray) genomic DNA binding areas is largely mapped near Ensembl transcriptional starting sites (TSS). F, ChIP-PCR analysis confirms selected potential targets for both Sox9a and Atoh1a. G, qRT-PCR assay confirms selected targets for both Sox9a and Atoh1a at 16–22 hpf. The col2a1a and hes6 bars serve as positive controls. Results in G are presented as mean ± SEM. *p < 0.05 (one-way ANOVA).
Table 5.
A summary of targets of Sox9a and Atoh1a
| Sox9a targets (gene/location) | Atoh1a targets (gene/location) | |
|---|---|---|
| Verified, targets of both | bnc1/-8217bp | bnc1/-1331bp |
| eya2/ 1st intron | eya2/-39376bp | |
| fn1/-14566bp | fn1/-26089bp | |
| neo1/-1010bp | neo1/1st intron | |
| spry1/-3886bp | spry1/ 5′UTR | |
| tlx2/1st intron | tlx2/-4239bp | |
| zgc77614/-4658bp | zgc77614/-187bp5 | |
| Verified, targets unique to one | dacha/ 2nd intron | dlc/-2774bp |
| cx43/-16069bp | irx2a/-8618bp | |
| tbx1/1st intron | jag1b/1st intron | |
| unc5b/1st intron | notch1b/-7642bp | |
| Not verified | cdh2/-4914bp | cdh2/-32746bp |
| irx1a/-29014bp | dacha/1st intron | |
| notch1b/2nd intron | irx1a/-36793bp | |
| prdm1a/-1668bp | irx5a/1st intron | |
| robo1/1st intron | pax2b/-18427bp | |
| spry2/-31106bp | pou3f3b/-27665bp | |
| spry4/4th intron | prdm1a/4th intron | |
| spry2/-16082bp | ||
| spry4/-27068bp | ||
| unc5b/1st intron |
All of these genes are relevant to the otic development. By performing ChIP-PCR assay, we verified 11 of 21 genes for Atoh1a and 11 of 18 genes for Sox9a. Among verified genes, seven are common to both sets.
On the Atoh1a list, dlc, jag1b and notch1b are related Notch signaling, but Sox9a target genes did not fit into this category. The transcription factor-encoding genes eya2, irx2a, and tlx2 and structural protein encoding genes such as bnc1 and fn1 were also identified. On the Sox9a list, we found nucleotide-binding protein-coding genes such as dacha, tbx1, and unc5b that were previously shown to be involved in the inner ear development. In addition, Fgf signaling related spry1 and neural development factor gene neo1 were also listed as Sox9a-targeted genes. Among all verified genes, seven were common to both sets (Fig. 4F, Table 5). Furthermore, our qRT-PCR assay confirmed that these genes were substantially reduced in sox9a or atoh1a morphants (Fig. 4G). Together, these results suggest that we have successfully identified a number of novel players that are directly regulated by Sox9a, Atoh1a, or both.
Eya2 and Tlx2 are involved in otic neurogenesis
We selected tlx2 and eya2 for further study as potential dual targets of Atoh1a and Sox9a. Our ChIP-PCR assay revealed binding sites for Atoh1a (−4239 bp) and Sox9a (+1543 bp) on the tlx2 locus and binding sites for Atoh1a (−39376 bp) and Sox9a (+22707 bp) on the eya2 locus (Table 5). Whole-mount in situ hybridization revealed that both genes are expressed early and specifically in ear neurogenic region. Expression of tlx2 was downregulated in both sox9a and atoh1a morphants at 24 hpf (Fig. 5A–C). Notably, eya2 expression was also reduced in the morphants at 14 hpf, when it was first detectable in the developing otic placode at both anterior and posterior portions (Fig. 5E–G). In sox9a and atoh1a double morphants, expression of tlx2 and eya2 were further reduced (Fig. 5A–H). SAG area was obviously decreased in eya2 or tlx2 morphants at 30 hpf, but in double morphants, no further reduction in SAG area was readily evident (Fig. 5I–L). The reduction in SAG areas was apparently not as pronounced as those for sox9a and atoh1a morphants or for SU5402-treated embryos (Fig. 3G,H,K). When either tlx2 or eya2 mRNA was injected into two-cell-stage embryos that were previously injected with either sox9a or atoh1a MO at the one-cell stage, the area of the SAG was partially restored (Fig. 5M). Together, these experiments suggest a model in which Fgfr-PI3K/Akt signaling regulates otic neurogenesis through Sox9a and Atoh1a, at least partially by joint regulation of their common downstream targets, Tlx2 and Eya2.
Figure 5.
tlx2 and eya2 contribute to otic neurogenesis downstream of Sox9a and Atoh1a. A–D, Expression of tlx2 in wild-type embryos (A), sox9a morphants (B), atoh1a morphants (C), and double morphants (D) at 24 hpf. E–H, Expression of eya2 in wild-type embryos (E), sox9a morphants (F), atoh1a morphants (G), and double morphants (H) at 14 hpf. I–L, Otic neurons are reduced in wild-type embryos (I), tlx2 morphants (J), eya2 morphants (K), and tlx2/eya2 double morphants (L) at 30 hpf. M, Summary of the restored SAG areas in mRNA-injected embryos (n = 10; 48 hpf). Allg, Anterior lateral line ganglion. White dotted lines indicate the otic neurogenic region; black dotted lines indicate the otic region. A–L, Lateral views with anterior to left and dorsal up. Results in M are presented as mean ± SEM. **p < 0.01, ***p < 0.001 (one-way ANOVA test). Scale bar, 30 μm.
Fgfr-Erk1/2 signaling regulates otic sensory development
Blocking Fgfr-Erk1/2 signaling did not show any influence on sox9a expression, but slightly reduced atoh1a and diminished atoh1b expression (Fig. 6A–F). Because Atoh1b plays an important role in otic sensorigenesis, we sought to test whether Fgfr-Erk1/2 signaling is responsible for hair cell development. We treated embryos with SU5402 or PD98059 from 10 to 14 hpf and observed >50% reduction of ultricular hair cells in both cases (Fig. 6G–I,K), supporting the idea that Fgfr-Erk1/2 signaling is a primary pathway for otic sensorigenesis. Such a reduction was not found in wortmannin-treated embryos (Fig. 6J,K), indicating that Fgfr-PI3K/Akt signaling does not play a role in the otic sensory development.
Figure 6.
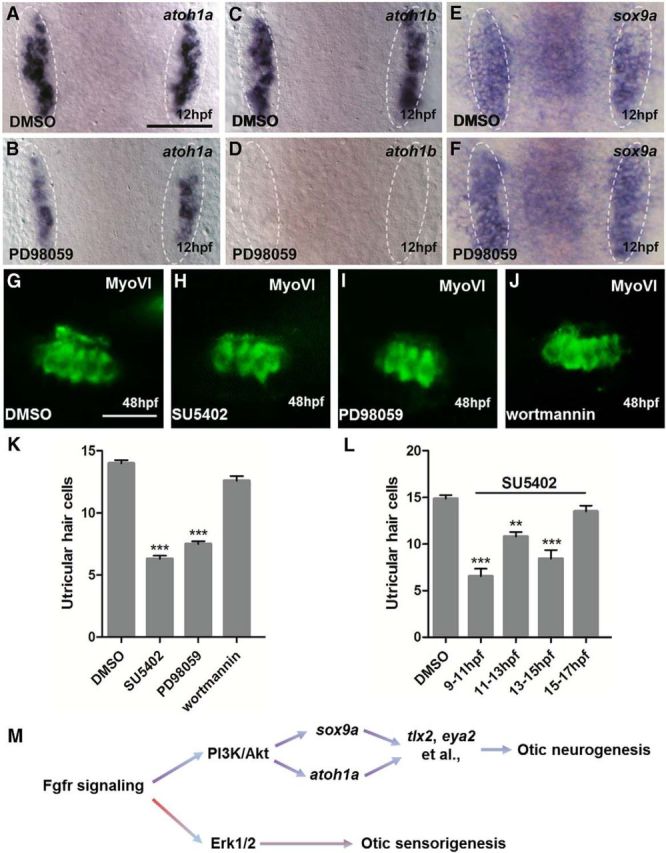
Fgfr-Erk1/2 signaling contributes to otic sensorigenesis. A–F, Expression of atoh1a, atoh1b, and sox9a in embryos treated by DMSO (A,C,E) and PD98059 (B,D,F) at 12 hpf. The chemical treatment period is 10–12 hpf. G–J, Utricular hair cells in 48 hpf embryos treated with DMSO (G), SU5402 (H), PD98059 (I), or Wortmannin (J). K, Utricular hair cell number is severely reduced by blocking Fgfr-Erk1/2 signaling starting from 10–14 hpf (n = 10; 48 hpf). L, Blocking Fgfr signaling using 100 μm SU5402 at different stages indicates the number of utricular hair cells is drastically reduced upon 9–11 hpf and 13–15 hpf treatments (n = 10; 48 hpf). M, Model of zebrafish otic neurogenesis and sensorigenesis is proposed. White dotted lines indicate the otic region. A–F, Dorsal views with anterior to top. G–J, Lateral views with anterior to left and dorsal up. Results in K and L are presented as mean ± SEM. **p < 0.01, ***p < 0.001 (one-way ANOVA test). Scale bar, 30 μm.
To determine when Fgf signaling is needed for otic sensorigenesis, we performed SU5402 treatment for distinct windows of development. Treatment at 9–11, 11–13, and 13–15 hpf, but not at 15–17 hpf, resulted in significant reductions of hair cells in the utricle, although the drop in hair cell counts was less pronounced in the 11–13 hpf treatment group (Fig. 6L). These experiments support the notion that Fgfr-Erk1/2 signaling controls part of otic sensory development starting as early as gastrulation ends.
Discussion
In the present study, we investigated when and how Fgfr signaling regulates zebrafish otic neurogenesis. We bypassed the early critical period for Fgf-dependent otic induction using conditional approaches and found both Atoh1a and Sox9a to be involved primarily in Fgfr-PI3K/Akt signaling-mediated otic neurogenesis between 10 and 14 hpf. Using ChIP-seq, we identified potential genetic targets of Atoh1a and Sox9a and, among their shared targets, we show that eya2 and tlx2 also participate in SAG development. We have also discovered that blocking Fgfr-Erk1/2 signaling between 9 and 15 hpf leads to reduced ultricular hair cells. Therefore, we have confirmed the predicted role of Sox9a and demonstrated a direct involvement of Atoh1a in otic neurogenesis. We propose that zebrafish otic neurogenesis and sensorigenesis require distinct Fgf-dependent gene networks (Fig. 6M).
In zebrafish, Fgf signaling participates in otic neurogenesis (Vemaraju et al., 2012). By blocking Fgf-Fgfr signaling at selected developmental stages, we have found that Fgf-Fgfr signaling activates and/or maintains sox9a and atoh1a via Fgfr-PI3K/Akt signaling at 10–14 hpf (Figs. 1, 2). When Fgfr signaling is blocked between 10 and 14 hpf, overall ear development is normal but the SAG area is drastically reduced. Otic neuroblasts develop normally when Fgf signaling is blocked after 14 hpf. It is therefore reasonable to claim that 10–14 hpf is the critical time of SAG development in zebrafish. However, the starting point of the otic neurogenesis remains unclear, because residual SAG area remains even when Fgf signaling is blocked at 10 hpf. It is possible that otic neurogenesis overlaps with Fgf-dependent specification of the otic placode at earlier stages, precluding analysis, or that the ground neuronal state of the caudal preplacodal region, which gives rise to the otic and epibranchial fates, is preset by Fgf-independent factors such as Foxi1 (Hans et al., 2013) before 10 hpf.
It is intriguing that Fgf signaling is partitioned so that Fgfr-PI3K/Akt signaling directs SAG whereas Fgfr-Erk1/2 signals play a role in HC development. Active PI3K/Akt signaling can be detected in developing chick inner ear in organotypic cultures (Aburto et al., 2012). Consistently, we observed the colocalization of p-AKT and sox9a mRNA in many pre-otic cells at 12 hpf (our unpublished observation), suggesting active PI3K/Akt signaling during otic neurogenesis. In chick organ culture studies, PI3K/Akt signaling, triggered by Igf-1, is found to affect the survival of neuronal progenitors that actively divide during inner ear development (Aburto et al., 2012), whereas Fgfr-Erk1/2 signaling is thought to be crucial for HC survival of mammalian cochlea (Battaglia et al., 2003). However, we observed normal SAG proliferation or cell death when Fgf signaling (both wortmanin and PD98059 treatments) was blocked during otic development (our unpublished observation). Our experiments indicate that a combination of downstream neurogenic factors of Fgfr-PI3K/Akt signaling are responsible for otic neurogenesis, but neither sox9a nor atoh1a is Fgfr-Erk1/2 signaling dependent (Fig. 6). The in vivo experiments conducted in our study may partially explain the discrepancy.
Perhaps the most important finding of our study was that Atoh1a participates in otic neurogenesis. Atoh1a and Atoh1b function in otic hair cell fate determination (Millimaki et al., 2007) and hair cell damage regeneration (Lewis et al., 2012; Kraft et al., 2013). In the developing otic region, the initial expression of atoh1a is genetically downstream of Atoh1b and the maintenance of atoh1b expression requires Atoh1a (Millimaki et al., 2007). Therefore, the reduction of ultricular hair cells by blocking Fgfr-Erk1/2 signaling at 10–12 hpf is most likely due to temporarily reduced atoh1b expression, which affects the atoh1a expression level (Fig. 6). In fact, a slight reduction of SAG area was also evident in atoh1b morphants (Table 4), reinforcing the idea that atoh1b is genetically upstream of atoh1a. Interestingly, in Atoh1-null mice, fewer afferent fibers from SAG were found to innervate sensory epithelia from embryonic day 12.5 to 18.5 (Fritzsch et al., 2005). Because SAG afferent neurons send a single axon toward their targets, it is likely that correspondingly SAG cell body number was also reduced in these mice (Fritzsch et al., 2005; Matei et al., 2005). Because of the presence of both atoh1a and atoh1b, a duplicated pair of mammalian Atoh1 ortholog resulting from an evolutionary genome-wide duplication event (Adolf et al., 2004), we were able to distinguish additional roles of Atoh1 in both otic neurogenesis and sensorigenesis. Because PD98059-treated animals (during 9–15 hpf) still develop significant number of ultricular hair cells (Fig. 6), even with much lower level of atoh1b, further investigation of the role of Atoh1b and perhaps other factors(s) in the otic sensorigenesis is desirable. Nevertheless, it appears that, during vertebrate evolution, one of the duplicated Atoh1 encoding genes (atoh1a) retains its role in otic neurogenesis, whereas the other (atoh1b) drives sensory epithelia differentiation in zebrafish.
It is already known that Sox9 is involved in altering the potential of CNS stem cells from neurogenic to gliogenic states and that Sox9 may be a direct target of Fgf signaling critical for neural crest development (Scott et al., 2010; Martínez-Morales et al., 2011; Yardley and Garcia-Castro, 2012). Because zebrafish sox9b initially labels the neural crest whereas sox9a marks mainly otic cells at the early segmentation stage (Yan et al., 2005), we focused our attention on Sox9a to illustrate its role in otic neurogenesis and indeed confirmed our speculation that it is important in defining otic neuronal fate (Figs. 2, 3). Because the SAG area of sox9b morphants is similarly reduced compared with that of sox9a morphants (Table 4) and onset of sox9b in the otic region depends on sox9a, we speculate that both Sox9s are required sequentially and continuously in otic SAG development.
Using a ChIP-PCR assay, we have verified 11 targets of Atoh1a and 11 targets of Sox9a that are all expressed in the otic placode or vesicle. The low number of verified genes might be because we used cell extract of whole embryos misexpressing Sox9a-Myc or Atoh1a-Myc fusion proteins. Among the verified genes, irx2a and tbx1 were already known to be involved in the neurogenesis (Choy et al., 2010; Radosevic et al., 2011), indicative of the relevance of our ChIP-seq analysis. Sox9a and Atoh1a share seven targets, which may indicate their essential roles in SAG development. Targets unique to Atoh1a are Notch signaling components such as dlc, jag1b, and notch1b. Whether Atoh1a triggers Notch lateral inhibition in Sox9a+ cells to direct neurogenic development may warrant further investigation.
In addition, we have verified that eya2 and tlx2, the common targets of Sox9a and Atoh1a, are two novel neurogenic genes in the ear (Figs. 4, 5). Some members of Eya family serve as transcriptional cofactors both in Drosophila and vertebrates and are related to inner ear developmental defects (Schönberger et al., 2005; Wang et al., 2008). Our study adds Eya2 to Eya family members that participate in zebrafish otic neurogenesis and suggest that its role lies downstream of Sox9a and Atoh1a. Among four zebrafish tlx genes, only tlx2 is expressed in the otic vesicle (Andermann and Weinberg, 2001; Langenau et al., 2002). Tlx2 has been shown previously to regulate the neural crest development and enteric neuron specification (Borghini et al., 2006; Borghini et al., 2007), so our study potentially expands its role to the ear. Importantly, whereas tlx2 and eya2 show effects on SAG development, they are milder than those seen for atoh1a or sox9a loss, suggesting that other Atoh1a/Sox9a targets are also important. Therefore, understanding the individual roles/functions for other genes identified from our ChIP-seq analysis will broaden our view of the complicated process of neurogenesis in the ear.
In summary, between 10 and 14 hpf during zebrafish embryogenesis, Fgfr-PI3K/Akt signaling sequentially activates and maintains sox9a/b and atoh1a functions in newborn otic cells and both Sox9 and Atoh1a turn on their target genes, including tlx2 and eya2, in some pre-otic cells, allowing a portion of them to eventually become SAG.
Footnotes
This work was supported by the National Basic Research Program of Chinese Ministry of Science and Technology (973 Grant 2012CB944503 and partially funded by 973 Grant 2011CBA01102), the Peking-Tsinghua Center for Life Sciences (D.L.). D.L. was partially supported by the PKU School of Life Science (Project 111) for academic travel expenses. Y.W. is a recipient of the PKU President Graduate Scholarship. We thank A. Zhu, J. Zhang, and J. Peng for critical comments on the manuscript; David W. Raible and Louis L. Tao for proofreading, and Ping Huang for excellent management of the Liu laboratory.
The authors declare no competing financial interests.
References
- Aburto MR, Magariños M, Leon Y, Varela-Nieto I, Sanchez-Calderon H. AKT signaling mediates IGF-I survival actions on otic neural progenitors. PLoS One. 2012;7:e30790. doi: 10.1371/journal.pone.0030790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska M, Léger S, Brand M, Hadrys T, Braun T, Bober E. Inner ear and lateral line expression of a zebrafish Nkx5-1 gene and its downregulation in the ears of FGF8 mutant, ace. Mech Dev. 2000;97:161–165. doi: 10.1016/S0925-4773(00)00414-7. [DOI] [PubMed] [Google Scholar]
- Adamska M, Herbrand H, Adamski M, Krüger M, Braun T, Bober E. FGFs control the patterning of the inner ear but are not able to induce the full ear program. Mech Dev. 2001;109:303–313. doi: 10.1016/S0925-4773(01)00550-0. [DOI] [PubMed] [Google Scholar]
- Adolf B, Bellipanni G, Huber V, Bally-Cuif L. atoh1.2 and beta3.1 are two new bHLH-encoding genes expressed in selective precursor cells of the zebrafish anterior hindbrain. Gene Expr Patterns. 2004;5:35–41. doi: 10.1016/j.modgep.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Alsina B, Abelló G, Ulloa E, Henrique D, Pujades C, Giraldez F. FGF signaling is required for determination of otic neuroblasts in the chick embryo. Dev Biol. 2004;267:119–134. doi: 10.1016/j.ydbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Andermann P, Weinberg ES. Expression of zTlxA, a Hox11-like gene, in early differentiating embryonic neurons and cranial sensory ganglia of the zebrafish embryo. Dev Dyn. 2001;222:595–610. doi: 10.1002/dvdy.1239. [DOI] [PubMed] [Google Scholar]
- Battaglia A, Pak K, Brors D, Bodmer D, Frangos JA, Ryan AF. Involvement of ras activation in toxic hair cell damage of the mammalian cochlea. Neuroscience. 2003;122:1025–1035. doi: 10.1016/j.neuroscience.2003.08.041. [DOI] [PubMed] [Google Scholar]
- Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, Cheah KS. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- Boeldt DS, Grummer MA, Magness RR, Bird IM. Altered VEGF-Stimulated Ca2+ Signaling In Part Underlies Pregnancy-Adapted eNOS Activity in UAEC. J Endocrinol. 2014;223:1–11. doi: 10.1530/JOE-14-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghini S, Bachetti T, Fava M, Di Duca M, Cargnin F, Fornasari D, Ravazzolo R, Ceccherini I. The TLX2 homeobox gene is a transcriptional target of PHOX2B in neural-crest-derived cells. Biochem J. 2006;395:355–361. doi: 10.1042/BJ20051386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghini S, Di Duca M, Santamaria G, Vargiolu M, Bachetti T, Cargnin F, Pini Prato A, De Giorgio R, Lerone M, Stanghellini V, Jasonni V, Fornasari D, Ravazzolo R, Ceccherini I. Transcriptional regulation of TLX2 and impaired intestinal innervation: possible role of the PHOX2A and PHOX2B genes. Eur J Hum Genet. 2007;15:848–855. doi: 10.1038/sj.ejhg.5201852. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- Choy SW, Cheng CW, Lee ST, Li VW, Hui MN, Hui CC, Liu D, Cheng SH. A cascade of irx1a and irx2a controls shh expression during retinogenesis. Dev Dyn. 2010;239:3204–3214. doi: 10.1002/dvdy.22462. [DOI] [PubMed] [Google Scholar]
- Dutton K, Abbas L, Spencer J, Brannon C, Mowbray C, Nikaido M, Kelsh RN, Whitfield TT. A zebrafish model for Waardenburg syndrome type IV reveals diverse roles for Sox10 in the otic vesicle. Dis Model Mech. 2009;2:68–83. doi: 10.1242/dmm.001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esain V, Postlethwait JH, Charnay P, Ghislain J. FGF-receptor signalling controls neural cell diversity in the zebrafish hindbrain by regulating olig2 and sox9. Development. 2010;137:33–42. doi: 10.1242/dev.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Matei VA, Nichols DH, Bermingham N, Jones K, Beisel KW, Wang VY. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn. 2005;233:570–583. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddon C, Lewis J. Early ear development in the embryo of the zebrafish, Danio rerio. J Comp Neurol. 1996;365:113–128. doi: 10.1002/(SICI)1096-9861(19960129)365:1<113::AID-CNE9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hans S, Christison J, Liu D, Westerfield M. Fgf-dependent otic induction requires competence provided by Foxi1 and Dlx3b. BMC Dev Biol. 2007;7:5. doi: 10.1186/1471-213X-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Irmscher A, Brand M. Zebrafish Foxi1 provides a neuronal ground state during inner ear induction preceding the Dlx3b/4b-regulated sensory lineage. Development. 2013;140:1936–1945. doi: 10.1242/dev.087718. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzh V, Sleptsova I, Liao J, He J, Gong Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev Dyn. 1998;213:92–104. doi: 10.1002/(SICI)1097-0177(199809)213:1<92::AID-AJA9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Kraft S, Hsu C, Brough DE, Staecker H. Atoh1 induces auditory hair cell recovery in mice after ototoxic injury. Laryngoscope. 2013;123:992–999. doi: 10.1002/lary.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Palomero T, Kanki JP, Ferrando AA, Zhou Y, Zon LI, Look AT. Molecular cloning and developmental expression of Tlx (Hox11) genes in zebrafish (Danio rerio) Mech Dev. 2002;117:243–248. doi: 10.1016/S0925-4773(02)00187-9. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Léger S, Brand M. Fgf8 and Fgf3 are required for zebrafish ear placode induction, maintenance and inner ear patterning. Mech Dev. 2002;119:91–108. doi: 10.1016/S0925-4773(02)00343-X. [DOI] [PubMed] [Google Scholar]
- Lewis RM, Hume CR, Stone JS. Atoh1 expression and function during auditory hair cell regeneration in post-hatch chickens. Hear Res. 2012;289:74–85. doi: 10.1016/j.heares.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Chu H, Maves L, Yan YL, Morcos PA, Postlethwait JH, Westerfield M. Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development. 2003;130:2213–2224. doi: 10.1242/dev.00445. [DOI] [PubMed] [Google Scholar]
- Lupo G, Gestri G, O'Brien M, Denton RM, Chandraratna RA, Ley SV, Harris WA, Wilson SW. Retinoic acid receptor signaling regulates choroid fissure closure through independent mechanisms in the ventral optic cup and periocular mesenchyme. Proc Natl Acad Sci U S A. 2011;108:8698–8703. doi: 10.1073/pnas.1103802108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier EC, Saxena A, Alsina B, Bronner ME, Whitfield TT. Sensational placodes: neurogenesis in the otic and olfactory systems. Dev Biol. 2014;389:50–67. doi: 10.1016/j.ydbio.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroon H, Walshe J, Mahmood R, Kiefer P, Dickson C, Mason I. Fgf3 and Fgf8 are required together for formation of the otic placode and vesicle. Development. 2002;129:2099–2108. doi: 10.1242/dev.129.9.2099. [DOI] [PubMed] [Google Scholar]
- Martínez-Morales PL, Diez del Corral R, Olivera-Martínez I, Quiroga AC, Das RM, Barbas JA, Storey KG, Morales AV. FGF and retinoic acid activity gradients control the timing of neural crest cell emigration in the trunk. J Cell Biol. 2011;194:489–503. doi: 10.1083/jcb.201011077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini S, Bernoth K, Main H, Ortega GD, Lendahl U, Just U, Schwanbeck R. A critical role for Sox9 in notch-induced astrogliogenesis and stem cell maintenance. Stem Cells. 2013;31:741–751. doi: 10.1002/stem.1320. [DOI] [PubMed] [Google Scholar]
- Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, Morris K, Feng F, Jones K, Lee J, Fritzsch B. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millimaki BB, Sweet EM, Dhason MS, Riley BB. Zebrafish atoh1 genes: classic proneural activity in the inner ear and regulation by Fgf and Notch. Development. 2007;134:295–305. doi: 10.1242/dev.02734. [DOI] [PubMed] [Google Scholar]
- Obholzer N, Wolfson S, Trapani JG, Mo W, Nechiporuk A, Busch-Nentwich E, Seiler C, Sidi S, Söllner C, Duncan RN, Söllner C, Duncan RN, Boehland A, Nicolson T. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J Neurosci. 2008;28:2110–2118. doi: 10.1523/JNEUROSCI.5230-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Ito K. BMP and FGF-2 regulate neurogenin-2 expression and the differentiation of sensory neurons and glia. Dev Dyn. 2006;235:646–655. doi: 10.1002/dvdy.20673. [DOI] [PubMed] [Google Scholar]
- Penumarti A, Abdel-Rahman AA. Neuronal nitric oxide synthase dependent elevation in adiponectin in the rostral ventrolateral medulla underlies GPR18-mediated hypotension in conscious rats. J Pharmacol Exp Ther. 2014;351:44–53. doi: 10.1124/jpet.114.216036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BT, Bolding K, Riley BB. Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev Biol. 2001;235:351–365. doi: 10.1006/dbio.2001.0297. [DOI] [PubMed] [Google Scholar]
- Radosevic M, Robert-Moreno A, Coolen M, Bally-Cuif L, Alsina B. Her9 represses neurogenic fate downstream of Tbx1 and retinoic acid signaling in the inner ear. Development. 2011;138:397–408. doi: 10.1242/dev.056093. [DOI] [PubMed] [Google Scholar]
- Scheffer D, Sage C, Plazas PV, Huang M, Wedemeyer C, Zhang DS, Chen ZY, Elgoyhen AB, Corey DP, Pingault V. The alpha1 subunit of nicotinic acetylcholine receptors in the inner ear: transcriptional regulation by ATOH1 and co-expression with the gamma subunit in hair cells. J Neurochem. 2007;103:2651–2664. doi: 10.1111/j.1471-4159.2007.04980.x. [DOI] [PubMed] [Google Scholar]
- Schönberger J, Wang L, Shin JT, Kim SD, Depreux FF, Zhu H, Zon L, Pizard A, Kim JB, Macrae CA, Mungall AJ, Seidman JG, Seidman CE. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat Genet. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, Briscoe J. SOX9 induces and maintains neural stem cells. Nat Neurosci. 2010;13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- Sweet EM, Vemaraju S, Riley BB. Sox2 and Fgf interact with Atoh1 to promote sensory competence throughout the zebrafish inner ear. Dev Biol. 2011;358:113–121. doi: 10.1016/j.ydbio.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocharus C, Puriboriboon Y, Junmanee T, Tocharus J, Ekthuwapranee K, Govitrapong P. Melatonin enhances adult rat hippocampal progenitor cell proliferation via ERK signaling pathway through melatonin receptor. Neuroscience. 2014;275:314–321. doi: 10.1016/j.neuroscience.2014.06.026. [DOI] [PubMed] [Google Scholar]
- Vemaraju S, Kantarci H, Padanad MS, Riley BB. A spatial and temporal gradient of Fgf differentially regulates distinct stages of neural development in the zebrafish inner ear. PLoS Genet. 2012;8:e1003068. doi: 10.1371/journal.pgen.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sewell WF, Kim SD, Shin JT, MacRae CA, Zon LI, Seidman JG, Seidman CE. Eya4 regulation of Na+/K+-ATPase is required for sensory system development in zebrafish. Development. 2008;135:3425–3434. doi: 10.1242/dev.012237. [DOI] [PubMed] [Google Scholar]
- Wardle FC, Odom DT, Bell GW, Yuan B, Danford TW, Wiellette EL, Herbolsheimer E, Sive HL, Young RA, Smith JC. Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol. 2006;7:R71. doi: 10.1186/gb-2006-7-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. Ed 5. Eugene, OR: University of Oregon; 2007. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio) [Google Scholar]
- Whitfield TT, Riley BB, Chiang MY, Phillips B. Development of the zebrafish inner ear. Dev Dyn. 2002;223:427–458. doi: 10.1002/dvdy.10073. [DOI] [PubMed] [Google Scholar]
- Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, Chung BC, Westerfield M, Haffter P, Hopkins N, Kimmel C, Postlethwait JH, Nissen R. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129:5065–5079. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
- Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- Yao D, Zhao F, Wu Y, Wang J, Dong W, Zhao J, Zhu Z, Liu D. Dissecting the differentiation process of the pre-placodal ectoderm in zebrafish. Dev Dyn. 2014;243:1338–1351. doi: 10.1002/dvdy.24160. [DOI] [PubMed] [Google Scholar]
- Yardley N, García-Castro MI. FGF signaling transforms non-neural ectoderm into neural crest. Dev Biol. 2012;372:166–177. doi: 10.1016/j.ydbio.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi H, Yan YL, Miller MR, BreMiller RA, Catchen JM, Johnson EA, Postlethwait JH. Expression profiling of zebrafish sox9 mutants reveals that Sox9 is required for retinal differentiation. Dev Biol. 2009;329:1–15. doi: 10.1016/j.ydbio.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]