Abstract
Myelinating Schwann cells in the vertebrate peripheral nervous system rely on Brg1 (Smarca4) for terminal differentiation. Brg1 serves as central ATP-hydrolyzing subunit of the chromatin remodelling BAF complexes and is recruited during myelination as part of these complexes by the transcription factor Sox10 in Schwann cells. Here, we analyzed the role of Brg1 during development of myelinating oligodendrocytes in the CNS of the mouse. Following Brg1 deletion in oligodendrocyte precursors, these cells showed normal survival, proliferation, and migration. A mild but significant reduction in the number of oligodendrocytes with myelin gene expression in the absence of Brg1 points to a contribution to oligodendroglial differentiation but also shows that the role of Brg1 is much less prominent than during Schwann cell differentiation. Additionally, we failed to obtain evidence for a genetic interaction between Brg1 and Sox10 comparable with the one in Schwann cells. This argues that similarities exist between the regulatory networks and mechanisms in both types of myelinating glia but that the exact mode of action and the relevance of functional interactions differ, pointing to a surprising degree of variability in the control of myelination.
Keywords: chromatin, myelination, oligodendrocyte, Schwann cell, Sox, transcription
Introduction
The generation of myelin-forming oligodendrocytes from neuroepithelial precursor cells in the ventricular zone of the developing CNS is a multistep process that begins with specification of oligodendrocyte precursor cells (OPCs) and ends with cell-cycle exit and initiation of the terminal differentiation and myelination program (Rowitch, 2004; Wegner and Stolt, 2005; Richardson et al., 2006).
It requires the concerted action of numerous transcription factors that cooperate and influence each other in a regulatory network where microRNAs serve as additional modulators (Emery, 2010). Participating transcription factors include several bHLH proteins such as Olig1, Olig2, Mash1/Ascl, and Hes5, Sox proteins such as Sox5, Sox6, Sox9, Sox10, and Sox17, zinc finger proteins such as Zfp488 and Zfp191, the homeodomain proteins Nkx2.2 and Nkx6.2, and the Ntd80-domain containing Myrf (Wegner, 2008; Emery, 2010).
Network activity is also coupled to the exact state of the chromatin as the existing chromatin landscape defines the possible changes in gene expression and as network-induced chromatin changes help to implement the differentiation process (Li et al., 2009; Emery, 2010; Liu and Casaccia, 2010). This argues for a role of chromatin-modifying factors, such as histone deacetylases and acetylases, and chromatin remodelling complexes in oligodendroglial development (Marin-Husstege et al., 2002; Cunliffe and Casaccia-Bonnefil, 2006).
Recently, Brg1-containing chromatin remodelling complexes were shown to be involved in the differentiation of myelin-forming Schwann cells in the peripheral nervous system (PNS) (Weider et al., 2012; Limpert et al., 2013). Brg1 (Brahma-related gene product 1, also known as Smarca4) is one of two alternatively used ATP-hydrolyzing subunits in BAF chromatin remodelling complexes (Ho and Crabtree, 2010; Hargreaves and Crabtree, 2011). The other is the related Brm (Brahma, also known as Smarca2). Brg1-containing chromatin remodelling complexes were shown to be recruited in a Sox10-dependent fashion via their Baf60a subunit to target genes that are then required for initiation and execution of the myelination program (Weider et al., 2012; Marathe et al., 2013). These findings also offered an explanation of how to link the regulatory network via a specific and centrally positioned transcription factor to the chromatin remodelling machinery.
Considering the analogy between Schwann cells and oligodendrocytes and the central role of Sox10 in the regulatory network of both types of myelinating glia, it seemed plausible to assume that Brg1-containing chromatin remodelling complexes would be similarly relevant for oligodendroglial differentiation and linked to the corresponding regulatory network via Sox10.
Materials and Methods
Generation of mice, tissue preparation, immunochemistry, and in situ hybridization.
For conditional Brg1 deletion in the oligodendrocyte lineage, the Brg1fl allele (Sumi-Ichinose et al., 1997) was combined with a Cnp1Cre allele (Lappe-Siefke et al., 2003) or a Ng2::Cre BAC transgene (Zhu et al., 2008) (see Fig. 1A) to generate Cnp1Cre/+ Brg1fl/fl and Ng2::Cre Brg1fl/fl mice. Use of the Brn4::Cre transgene (bcre-32 line) (Ahn et al., 2001) additionally allowed early Brg1 deletion throughout the CNS. Controls included Brg1fl/fl mice without Cre allele as well as Cnp1Cre/+ mice, Ng2::Cre mice, and Brn4::Cre mice with wild-type Brg1 alleles, and showed no distinguishable differences among each other or to wild-type mice. Some mice additionally carried the Rosa26stopfloxEYFP allele (Srinivas et al., 2001), the Sox10fl allele (Finzsch et al., 2010), or the Sox8lacZ allele (Sock et al., 2001). Genotyping was performed by PCR. All mice were on a mixed C3H × C57BL/6J background. They were kept under standard housing conditions with 12:12 h light-dark cycles and continuous access to food and water in accordance with animal welfare laws. Experiments were approved by the responsible local committees and government bodies.
Figure 1.
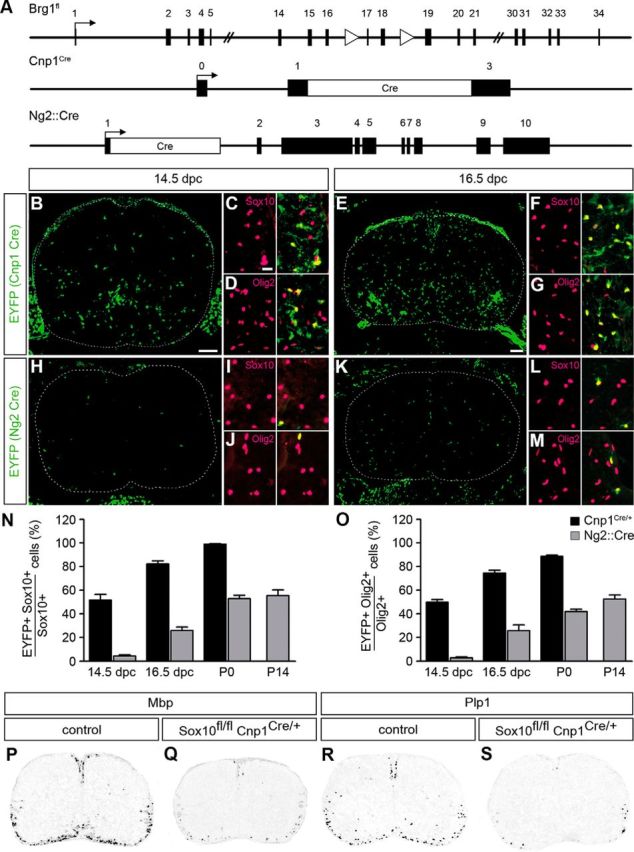
In vivo gene deletion strategy. A, Schematically represented are the floxed Brg1 allele (Brg1fl) and the Cre alleles (Cnp1Cre and Ng2::Cre) used for the study. Exons are numbered. Arrows indicate transcription start sites. Triangles represent loxP sites. The Cre reading frame is in white. B–M, Using the Rosa26stopfloxEYFP reporter, timing and specificity of deletion were determined for the Cnp1Cre and Ng2::Cre alleles by IHC on transverse spinal cord sections (forelimb level) at 14.5 dpc and 16.5 dpc. Cre-induced EYFP expression was detected by an antibody against GFP (green), cells of the oligodendrocyte lineage by antibodies against Sox10 (C, F, I, L), and Olig2 (D, G, J, M) (magenta). C, D, F, G, I, J, L, M, Magnifications. B, E, H, K, Overviews. The stippled line indicates the spinal cord perimeter. Scale bars: B, E, H, K, 100 μm; C, D, F, G, I, J, L, M, 25 μm. N, O, From these and comparable stainings, deletion rates at the Rosa26 locus in cells of the oligodendrocyte lineage (identified by Sox10 staining in N and by Olig2 staining in O) were quantified for the Cnp1Cre (black bars) and Ng2::Cre (gray bars) alleles at 14.5 dpc, 16.5 dpc, P0, and P14. At least nine separate sections from the forelimb region of three independent specimens were counted for each age and genotype. Data are mean ± SEM for biological replicates. No EYFP-positive cells were detected in the absence of Cre alleles. P–S, ISH was performed with probes for Mbp (P, Q) and Plp1 (R, S) on spinal cord sections (forelimb level) of Sox10fl/fl CnpCre/+ embryos and control (Sox10fl/fl Cnp1+/+) littermates at P0 to show that the Cnp1-driven Cre recombinase is sufficiently active early enough to prevent Sox10 from inducing myelin gene expression. Spinal cords were placed on a white background.
Both male and female embryos and pups were used for the study. Embryos were recovered by Cesarean section from 12.5 d post coitum (dpc) to 18.5 dpc from staged pregnancies. Pups were collected at the day of birth (P0) or after 2 weeks (P14). Spinal cord tissue was immediately isolated from killed animals and underwent fixation in 4% PFA before transfer to 30% sucrose and freezing in Tissue Freezing Medium (Leica). For immunohistochemistry (IHC), 10 μm cryotome sections at forelimb level were used (Stolt et al., 2003). The following primary antibodies were applied, some of which were also used for immunocytochemistry (ICC) on OLN93 cells and on rat primary oligodendroglia kept under proliferating or differentiating conditions: guinea pig anti-Sox10 antiserum (1:1000 dilution for IHC) (Maka et al., 2005), guinea pig anti-Lmx1b antiserum (1:10,000 dilution for IHC, gift from C. Birchmeier and T. Müller, MDC, Berlin), guinea pig anti-Glast antiserum (1:200 dilution for IHC, Millipore Bioscience Research Reagents), goat anti-Brg1 antiserum N15 (1:50 dilution for ICC, Santa Cruz Biotechnology), rabbit anti-Brg1 antiserum H88 (1:50 dilution for IHC and 1:200 dilution for ICC, Santa Cruz Biotechnology), rabbit anti-Brm antiserum (1:50 dilution for IHC and 1:200 dilution for ICC, Abcam), rabbit anti-Olig2 antiserum (1:1000 dilution for IHC, Millipore), rabbit anti-Myrf antiserum (1:1000 dilution for IHC) (Hornig et al., 2013), rabbit anti-Gfap (1:1000 dilution for IHC, Invitrogen), rabbit anti-cleaved caspase 3 antiserum (1:200 dilution for IHC, Cell Signaling Technology), rabbit anti-Pdgfra antiserum (1:300 dilution for IHC and 1:500 dilution for ICC, Santa Cruz Biotechnology), rat anti-Pdgfra antiserum (1:100 dilution for ICC, BD Biosciences PharMingen), rabbit anti-Ng2 antiserum (1:500 dilution for ICC, Millipore), rat anti-Ng2 monoclonal (1:100 for IHC, gift from J. Trotter, University of Mainz, Mainz, Germany), rabbit anti-GFP antiserum (1:1000 dilution for IHC, Molecular Probes), rat anti-Gfp antiserum (1:1000 dilution for IHC, Gerbu Biotechnik), rat anti-Mbp antiserum (1:250 dilution for IHC and 1:750 dilution for ICC, Serotec), mouse anti-glutamine synthetase (GlnS) monoclonal (1:500 for IHC, BD Transduction Laboratories), and mouse anti-Nkx2.2 monoclonal (1:5000 dilution for IHC, Hybridoma Bank). For anti-Nkx2.2 antibodies, signal intensity was enhanced by using the TSA-Plus Fluorescence system (PerkinElmer). For coimmunochemistry of Brg1 or Brm antibody, treatment was followed by incubation with goat anti-rabbit IgG Fab fragment (1:100, Dianova) before addition of antibodies directed against Gfap, Pdgfra, Myrf, or Olig2. Secondary antibodies were coupled to Cy3 or Alexa-488 fluorescent dyes (Dianova). The 10 μm sections were also used for TUNEL according to the manufacturer's protocol (Millipore Bioscience Research Reagents). Samples were documented with a Leica DMI 6000B inverted microscope (Leica) equipped with a DFC 360FX camera (Leica).
For ISH, 10 μm cryotome sections from the forelimb level of mouse embryos were used with DIG-labeled antisense riboprobes specific for Mbp and Plp1 as described previously (Stolt et al., 2002). Samples were analyzed and documented with a Leica MZFLIII stereomicroscope equipped with an Axiocam (Zeiss).
Plasmids.
Plasmids for expression of Brg1 and Brm in eukaryotic cells were as described previously (Khavari et al., 1993; Muchardt and Yaniv, 1993). For gene-specific knockdown, shRNA coding sequences were inserted into the pSuper vector according to the manufacturer's instruction (Oligoengine). Targeted sequences included the following: 5′-CCAAAGCAACCATCGAACT-3′ for rat Brg1 (Weider et al., 2012) and 5′-CCCAGCTGAGAAGTTGTCACCAAAT-3′ for rat Brm. shRNAs were validated for their ability to suppress Brg1 or Brm induction in transfected cells (data not shown). For generation of glutathione-S-transferase (GST) fusion proteins with Sox10 or Olig2, sequences corresponding to dimerization and high-mobility-group domain (amino acids 66–182) of rat Sox10 (Weider et al., 2012) or full-length mouse Olig2 were inserted in frame into pGEX-KG.
Cell culture, preparation of protein extracts, and Western blotting.
Primary oligodendroglia were obtained from newborn rat brain after preparation of mixed glial cultures and shake-off (McCarthy and de Vellis, 1980). They were grown in Sato proliferation medium in the presence of bFGF and PDGF-AA or kept in differentiation medium after replacement of the aforementioned mitogens by 1% FCS as described previously (Hoffmann et al., 2014). Cells were harvested in the undifferentiated state or after 1 or 4 d in differentiation medium and used for preparation of protein extracts (Kuhlbrodt et al., 1998) or total RNA using Trizol reagent (Life Sciences). For some experiments, proliferating primary oligodendroglia were transfected with shRNA and GFP expression plasmids using Xfect reagent (Clontech) and placed for 4 d in differentiation medium before fixation and staining with anti-Mbp antibodies.
Protein extracts from oligodendroglial cultures and spinal cord tissue were size-fractionated on polyacrylamide-SDS gels, blotted onto nitrocellulose membranes, and analyzed by Western blotting using the antisera against Brg1, Brm (1:200 dilution each), and Sox10 (1:1000 dilution) as well as rabbit antiserum against Gapdh (1:5000 dilution, Santa Cruz Biotechnology), protein A coupled to HRP (Bio-Rad), and Luminol reagent for detection.
Additionally, rat oligodendroglial OLN93 and CG4 cells were kept in culture as described previously (Sock et al., 1997; Hornig et al., 2013). OLN93 cells were transfected using SuperFect reagent (QIAGEN). Fixation and staining with antibodies directed against Brg1 or Brm followed 48 h after transfection. CG4 cells were used to prepare extracts that were used in pulldown assays with GST-Sox10 and GST-Olig2 fusions and analyzed by Western blotting as described previously (Weider et al., 2012).
Quantitative PCR (qPCR) analysis.
RNA samples from mouse spinal cord, primary rat oligodendroglial cultures, and Schwann cells were reverse transcribed and used to analyze expression levels by qPCR on a CFX96 Real Time PCR System (Bio-Rad). The following primer pairs were used: 5′-CCTGCAGTCTGTCTTCACCA-3′ and 5′-GGTCCTCCTCCTGCTCCTC-3′ for mouse and rat Brg1, 5′-GGCAGAAAATTGCCAAAGAA-3′ and 5′-CCACTTGCTTCTGACTGTTCG-3′ for mouse and rat Brm, 5′-TCAGTCTCGGCTGTCCAGCC-3′ and 5′-GTCCTCGTGAAGAGCCCAAC-3′ for mouse Sox10. Transcript levels were normalized to Rpl8.
Chromatin immunoprecipitation (ChIP).
Chromatin was prepared after cross-linking of endogenous proteins to DNA and shearing from primary rat oligodendroglial cultures kept under proliferating conditions or 4 d under differentiating conditions and from brain tissue of newborn mice as previously described (Weider et al., 2012). Immunoprecipitation was overnight at 4°C using antibodies directed against Brg1, Brm, Olig2, and Sox10 or control immunoglobulins in the presence of protein A/G Sepharose CL-4B beads (Life Technologies) pretreated with BSA and salmon sperm DNA. Antibodies were the same as for immunochemistry. For genomic detection of Brm, an additional rat antibody (Millipore, clone 1H7A10) was used. Both anti-Brm antibodies yielded very similar results so that only the ones obtained with the rat antibody are shown.
After washing, crosslink reversal, proteinase K treatment, phenol/chloroform extraction, and ethanol precipitation, the amount of DNA from input and precipitated chromatin was quantified by qPCR using the Bio-Rad CFX96 Real Time PCR system (Weider et al., 2012). The following rat-specific primer pairs were used for ChIP on cultured cells: (1) for the Cnp1 gene 5′-ATGGTGGTGGGTACCAAGAG-3′ and 5′-AGCCCTAGCCCTGAGAAGAC-3′ to amplify positions 6311 to 6616 (Cnp1 enh), and 5′-CTTGGCGAAGAAGATGGAAG-3′ and 5′-AGAGGGTGGGTGAAGGAACT-3′ to amplify positions 5481 to 5744 (Cnp1 neg); (2) for the Myrf gene 5′-CACGTGGCTGACGGGATTTC-3′ and 5′-CCACAGCTGTGGCTGCTGGC-3′ to amplify positions 7620 to 8432 (Myrf enh) and 5′-GGGTCTGGTATTCGTAGGTC-3′ and 5′-GTGTTCTCTGTACCTCTTGG-3′ to amplify positions −3973 to −4152 (Myrf neg); (3) for the Zfp191 gene 5′-TGCTTGTGGACACTCTGAGAA-3′ and 5′-ATAGGGCAAGGCTTGAGGAT-3′ to amplify positions −824 to −1123 (Zfp191 enh) and 5′-AATGCGGCCATTGTCATC-3′ and 5′-CGGCCTCTGGACCCTTAC-3′ to amplify positions 89 to −181 (Zfp191 neg). The following mouse-specific primer pairs were used for ChIP on brain tissue: (1) for the Cnp1 gene 5′-CCTACCCTCACCCTGTGTTG-3′ and 5′-CCTTGCTGAGAGGTCTTTGG-3′ (Cnp1 enh) to amplify positions 6718 to 7110, and 5′-GATCAGGAGCTGCAGTTGTG-3′ and 5′-GGCCTTGACCTCCATCTTCT-3′ to amplify positions 5501 to 5767 (Cnp1 neg); (2) for the Myrf gene 5′-CACGTGGCTGACGGGATTTC-3′ and 5′-CCACAGCTGTGGCTGCTGGC-3′ (Myrf enh) to amplify positions 7856 to 8025 and 5′-GTAAGTGGGTCTCTGTGTGC-3′ and 5′-GTGGGTTCAGAATCTGCATAG-3′ to amplify −9392 to −9581 (Myrf neg); (3) for the Zfp191 gene 5′-CCTGGTTAGCACAGTACCCA-3′ and 5′-ATTCACTACGGGAGCAGAGG-3′ to amplify positions −1336 to −1631 (Zfp191 enh) and 5′-AGCTCGTGGAATGTGCAATC-3′ and 5′-CGGGCCGGTTTTGTTCTATT-3′ to amplify positions 88 to −202 (Zfp191 neg). For localization of amplified fragments, see also Figure 6A.
Figure 6.
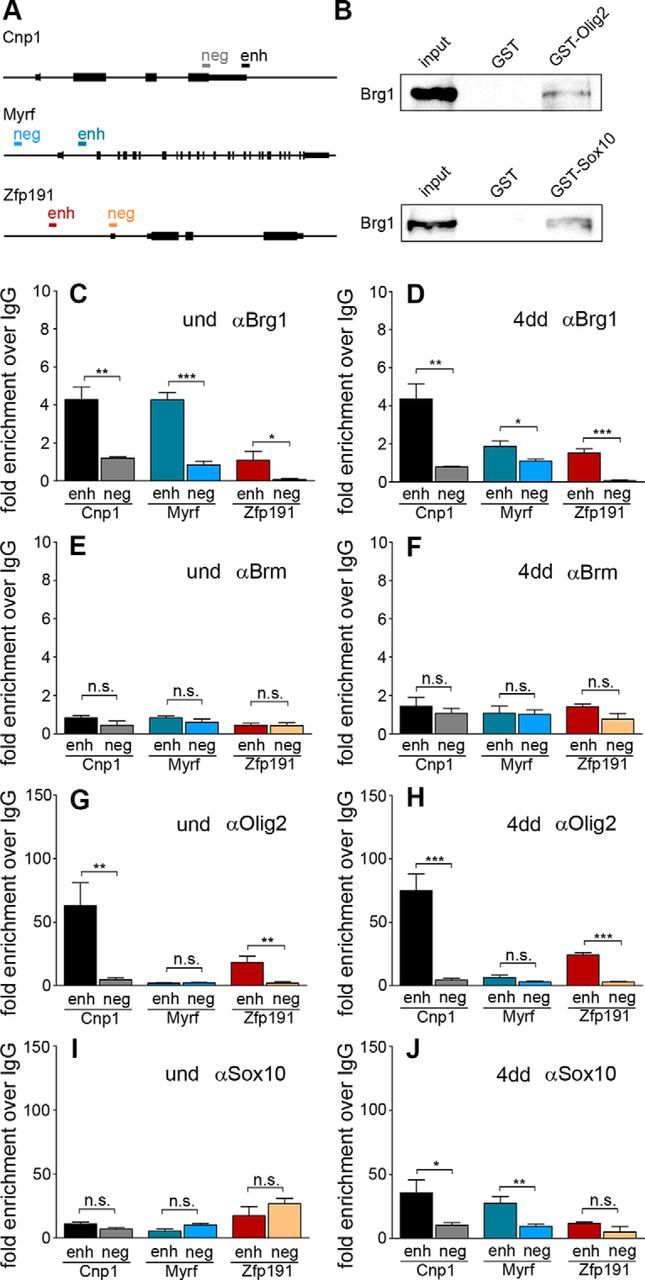
Relation of Brg1 and Brm with oligodendroglial transcription factors. A, The Cnp1, Myrf, and Zfp191 genes are schematically depicted as well as the position of specific enhancer (enh) and negative control (neg) regions probed in ChIP experiments. B, The conserved dimerization and high-mobility group domains of Sox10 and full-length Olig2 were fused to GST (GST-Sox10, GST-Olig2) and used in pulldown assays to precipitate Brg1 from oligodendroglial CG4 cell extracts. GST alone served as control. Brg1 in input and precipitated fractions was detected by Western blot using rabbit anti-Brg1 antibody. Results were similar with extracts from undifferentiated and 4 d differentiated CG4 cells. Shown are the ones obtained with undifferentiated cells. C–J, ChIP was performed on the Cnp1, Myrf, and Zfp191 genes with antibodies directed against Brg1 (C, D), Brm (E, F), Olig2 (G, H), Sox10 (I, J), and control IgGs (C–J) on chromatin prepared from primary oligodendrocyte cultures kept under proliferating conditions (und) or differentiated for 4 d (4dd). qPCR was then used to detect specific regions in immunoprecipitated chromatin, and relative enrichments were determined in the immunoprecipitate obtained with the specific antibody over the control IgG. Experiments were performed at least two times with each PCR in triplicate (Student's t test). n.s., Not significant. *p ≤ 0.05. **p ≤ 0.01. ***p ≤ 0.001.
Results
Both Cnp1Cre and Ng2::Cre allow specific gene deletion in oligodendrocyte precursors during embryonic development
To delete Brg1 in cells of the oligodendroglial lineage, we used the same floxed allele (Sumi-Ichinose et al., 1997) previously used to study its role in Schwann cells and combined it with Cre alleles that exhibit oligodendroglial expression (Fig. 1A). One such Cre is the Cnp1Cre allele that was generated by inserting Cre sequences into the Cnp1 locus (Lappe-Siefke et al., 2003). The resulting Brg1fl/fl Cnp1Cre/+ mice are henceforth referred to as Brg1ΔCnp mice. The other Cre is the Ng2::Cre in which the Cre was integrated into the Ng2 (Cspg4) genomic region in the context of a BAC transgene (Zhu et al., 2008). These Brg1fl/fl Ng2::Cre mice are named Brg1ΔNg2 mice.
In the majority of Cnp1Cre/+ mice, Cre expression started in oligodendrocytes as they began terminal differentiation. However, we observed that a small percentage of Cnp1Cre/+ mice exhibited higher and earlier Cre expression and transmitted this trait to their progeny. These mice were exclusively used in the study and combined with Brg1fl and/or Rosa26stopfloxEYFP alleles. In Cnp1Cre/+ Rosa26stopfloxEYFP/+ mice, Cre-dependent EYFP activation was not yet detectable in the spinal cord at 12.5 dpc (data not shown) but clearly visible at 14.5 dpc (Fig. 1B). It had further increased at 16.5 dpc (Fig. 1E). Cell-type specificity of Cre expression was preserved in these Cnp1Cre expressors as most EYFP-expressing cells were positive for the oligodendroglial markers Olig2 and Sox10 (Fig. 1C,D,F,G). Only few ventral neurons were additionally labeled (Fig. 1E; data not shown). Quantification revealed that ∼50%-52% of all oligodendroglial cells had undergone Cre-dependent recombination of the Rosa26stopfloxEYFP allele at 14.5 dpc (Fig. 1N,O). Recombination increased to 75%–82% at 16.5 dpc and was near complete at the day of birth (Fig. 1N,O). Considering that oligodendroglial cells in the spinal cord are mostly OPCs at 14.5 dpc and 16.5 dpc, use of these Cnp1Cre carrying mice allowed efficient gene ablation in OPCs.
The same characterization was also performed on mice carrying the Ng2::Cre transgene. Again, Cre-dependent EYFP expression was detectable in Ng2::Cre Rosa26stopfloxEYFP/+ mice from 14.5 dpc onwards (Fig. 1H,K) and mostly restricted to oligodendroglial cells (Fig. 1I,J,L,M). However, deletion rates were much lower and reached only 3%–4% at 14.5 dpc, 26%–27% at 16.5 dpc, and approximately half the cells at P0 and P14 (Fig. 1N,O) in accord with the literature (Zhu et al., 2008). The Ng2::Cre transgene thus targets the same cells as the Cnp1Cre allele, but with lower efficiency.
As another test for the deletion efficiency of the Cnp1Cre allele, we generated Sox10fl/fl Cnp1Cre/+ mice. If Cnp1Cre-dependent deletion is efficient, oligodendroglial differentiation should be stalled as observed after Brn4::Cre-dependent Sox10 deletion in neuroepithelial precursor cells of the CNS (Hornig et al., 2013). In line with this assumption, expression of the two oligodendroglial differentiation markers Mbp and Plp1 was dramatically reduced relative to the wild-type control (Fig. 1P–S) and almost comparable with expression in the constitutive or Brn4::Cre-dependent conditional Sox10 knock-out (Britsch et al., 2001; Hornig et al., 2013).
Brg1 is expressed at all stages of oligodendrocyte development
Immunohistochemical analysis of the perinatal spinal cord revealed that Brg1 is expressed in oligodendroglial cells. Both Sox10 and Olig2 as markers of this lineage were coexpressed with Brg1 (Fig. 2A,B). Brg1 was found in OPCs as evident by costaining with Pdgfra and Ng2 (Fig. 2C,D) as well as in maturing Myrf- and Mbp-positive oligodendrocytes (Fig. 2E,F). There was no obvious difference between OPCs and maturing oligodendrocytes regarding intensity of the immunoreactivity. Brg1 was additionally present in neurons as evident from costaining with the panneuronal marker NeuN (data not shown) or the dorsal interneuron marker Lmx1b (Fig. 2G), and in astrocytes labeled with Gfap, GlnS, or Glast (Fig. 2H–J).
Figure 2.
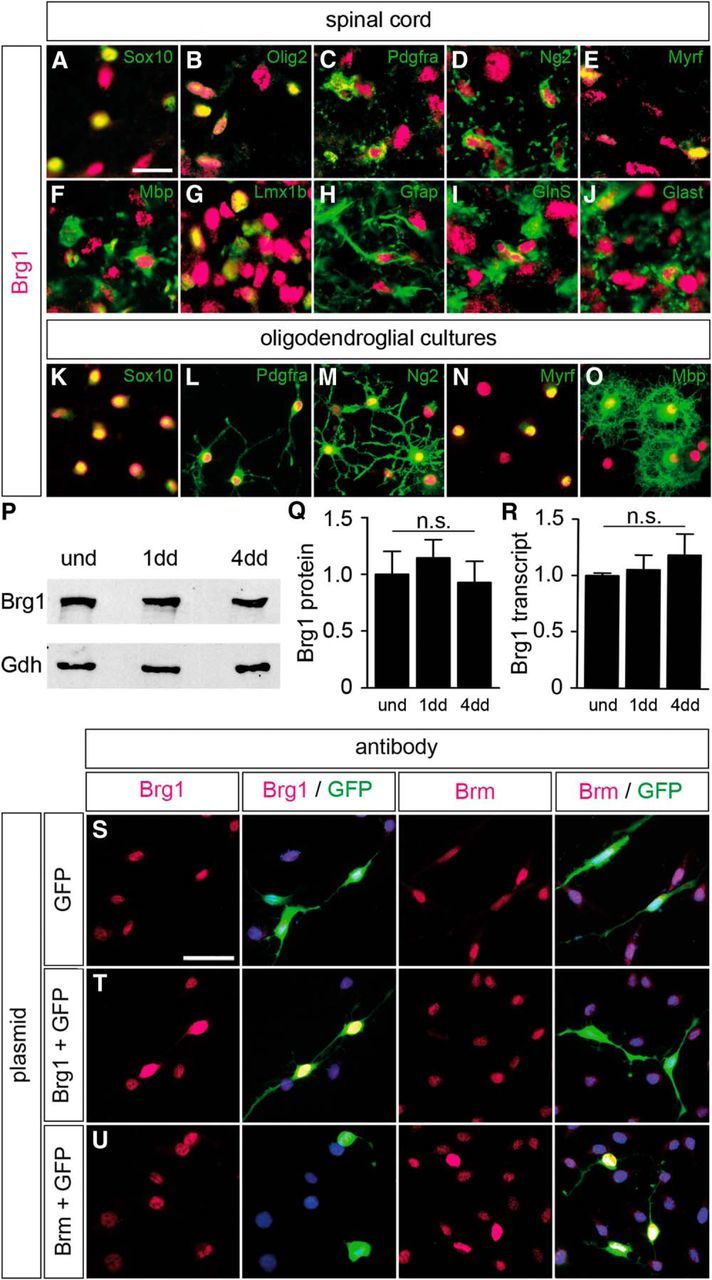
Brg1 expression in the oligodendrocyte lineage. A–J, Co-IHC was performed on spinal cord sections (forelimb level) at P0 to determine the occurrence of Brg1 (magenta) in cells of the oligodendrocyte lineage (Sox10 in A, Olig2 in B, Pdgfra in C, Ng2 in D, Myrf in E, and Mbp in F), in neurons (Lmx1b in G), and in astrocytes (Gfap in H, GlnS in I, and Glast in J) (all in green). Pictures were taken from either gray (G, I, J) or white matter (A–F, H). Scale bar, 25 μm. K–O, Stainings with antibodies directed against Brg1 (magenta) and various oligodendroglial markers (Sox10 in K, Pdgfra in L, Ng2 in M, Myrf in N, and Mbp in O) (all in green) were also performed on primary oligodendroglial cultures kept under proliferating (K–M) or differentiating (N, O) conditions. Scale bar, 25 μm. P, Q, By Western blot, amounts of Brg1 protein were comparable in oligodendroglial cultures kept under proliferating conditions (und) or differentiating conditions for 1 (1dd) and 4 (4dd) days when normalized to Gapdh (Gdh) levels. P, Representative Western blot. Q, Quantification from three independent biological replicates with the amounts in proliferating cultures set to 1. R, Amounts of Brg1 transcripts did not significantly differ between oligodendroglial cultures kept under proliferating conditions or differentiating conditions for 1 and 4 d. After normalization to Rpl8, amounts in the proliferating cultures were set to 1. Experiments were repeated at least three times with material from three independent cultures for each condition, and a Student's t test was applied. n.s., Not significant. S–U, To determine antibody specificity, immunohistochemistry was performed on OLN93 cells transfected with expression plasmids for Brg1 or Brm in the presence of GFP (indicated on the right) using antibodies directed against either of the two paralogs (in magenta, indicated on top). Transfected cells were identified by GFP expression (green). Scale bar, 50 μm.
Brg1 was also detected in nuclei of cultured Sox10-positive oligodendroglial cells (Fig. 2K). As judged from staining intensities, amounts appeared comparable between Pdgfra- and Ng2-positive OPCs on the one hand and Myrf- or Mbp-positive oligodendrocytes on the other (Fig. 2L–O). Similar expression levels were confirmed by Western blot on extracts prepared from oligodendroglial cultures kept in proliferating or in differentiating conditions for 1 or 4 d (Fig. 2P,Q). qRT-PCR yielded similar results for transcript levels with two different primer pairs for Brg1 (Fig. 2R; and data not shown). We conclude that Brg1 is widely expressed in most cell types of the perinatal spinal cord. This includes cells of the oligodendrocyte lineage from OPCs to maturing oligodendrocytes.
To confirm specificity of the anti-Brg1 antibody, we transfected oligodendroglial OLN93 cells with expression plasmids for Brg1 or Brm in the presence of GFP for detection of transfected cells (Fig. 2S–U). Because of the presence of endogenous Brg1, all nuclei exhibited a weak staining with the anti-Brg1 antibody. However, only nuclei of Brg1-transfected cells, but not Brm-transfected cells, exhibited a strongly increased staining intensity. Likewise, anti-Brm antibodies stained all nuclei weakly. This time, enhanced nuclear staining was only observed for Brm-transfected cells, but not for Brg1-transfected cells. These results support the conclusion that both anti-Brg1 and anti-Brm antibodies are specific and do not cross-react with the other paralog to a significant extent. In agreement, both antibodies were reported by the manufacturers to be raised against unique epitopes.
Brg1 is effectively deleted in oligodendroglial cells by Cnp1Cre and partially by Ng2::Cre
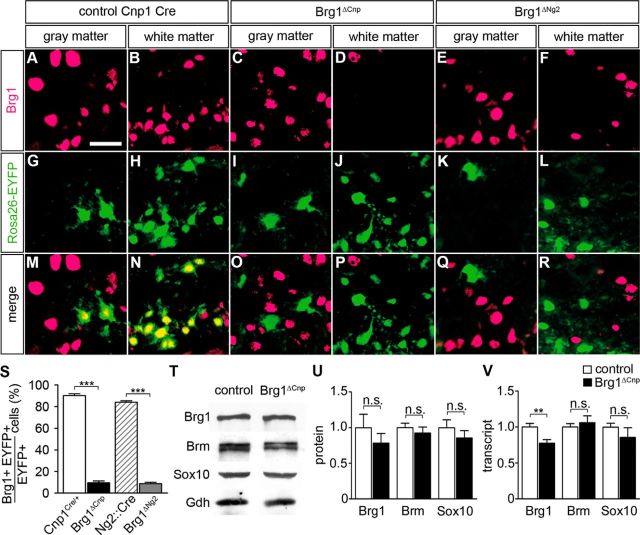
Using the Rosa26stopfloxEYFP allele, we analyzed the rate of oligodendroglial Brg1 deletion in Brg1ΔCnp and Brg1ΔNg2 mice at the time of birth. Mice with Cre and Rosa26stopfloxEYFP alleles served as controls. Although Brg1 was detected in nearly all cells that had experienced Cre activity and were EYFP-positive in gray and white matter of control spinal cord tissue, there was hardly any Brg1 expression left in EYFP-positive cells of Brg1ΔCnp mice (compare Fig. 3A,B,G,H,M,N with Fig. 3 C,D,I,J,O,P).
Figure 3.
Efficiency of Brg1 deletion in vivo. A–R, Co-IHC was performed with antibodies directed against Brg1 (magenta) and GFP (green) on transverse spinal cord sections (forelimb level) of control embryos (Rosa26+/stopfloxEYFP Cnp1+/Cre in A, B, G, H, M, N) and their Brg1ΔCnp Rosa26+/stopfloxEYFP (C, D, I, J, O, P) and Brg1ΔNg2 Rosa26+/stopfloxEYFP (E, F, K, L, Q, R) littermates at P0. Scale bar, 25 μm. S, From these staining, Brg1 occurrence was quantified in Cre-expressing cells for the following genotypes: Rosa26+/stopfloxEYFP Cnp1+/Cre (white bar), Brg1ΔCnp Rosa26+/stopfloxEYFP (black bar), Rosa26+/stopfloxEYFP Ng2::Cre (hatched bar), and Brg1ΔNg2 Rosa26+/stopfloxEYFP (gray bar). At least nine separate sections from the forelimb region of three independent specimens were counted for each age and genotype. Data are presented as mean ± SEM for biological replicates. T, U, Amounts of Brg1, Brm, and Sox10 proteins were compared in extracts from spinal cord tissue of control and Brg1ΔCnp embryos at P0 by Western blotting. Normalization was to Gapdh (Gdh). T, Representative Western blot. U, Quantification from three independent biological replicates. V, Quantitative RT-PCR was used to determine the amounts of Brg1, Brm, and Sox10 transcripts in spinal cord tissue from control (white bars) and Brg1ΔCnp (black bars) embryos at P0. After normalization to Rpl8, amounts in the control were set to 1. Experiments were repeated three times with material from three independent spinal cord preparations for each genotype. Differences to control were statistically significant where indicated (Student's t test). n.s., Not significant. **p ≤ 0.01. ***p ≤ 0.001.
Cre recombinase activity was clearly less in Brg1ΔNg2 mice as evidenced by the lower number of EYFP-positive cells. However, those cells that had experienced Cre activity and had undergone recombination at the Rosa26 locus were again overwhelmingly Brg1-deficient (Fig. 3E,F,K,L,Q,R). Quantification confirmed the efficient deletion with only 10 ± 2% of all EYFP-positive cells being Brg1-positive in Brg1ΔCnp and 9 ± 1% in Brg1ΔNg2 mice compared with 90 ± 2% in Cnp1+/Cre Rosa26+/stopfloxEYFP and 84 ± 1% in Ng2::Cre Rosa26+/stopfloxEYFP control mice (Fig. 3S). Considering the previously determined percentage of oligodendroglial cells that undergo recombination of the Rosa26stopfloxEYFP allele, we can therefore conclude that the vast majority of oligodendrocytes are Brg1-deficient in the spinal cord of Brg1ΔCnp mice at birth, whereas one-third to one-half have lost Brg1 expression in Brg1ΔNg2 mice. Deletion in Brg1ΔCnp mice led to small reductions in protein and transcript amounts for Brg1 as expected for a deletion regimen that affects Brg1 expression only in oligodendroglial cells while sparing most other remaining CNS cells (Fig. 3T–V). Reductions reached only significance for transcripts, but not for the protein. Amounts of the related Brm and the oligodendroglial transcription factor Sox10 remained unchanged both on protein and transcript levels (Fig. 3T–V). This argues that there is no compensatory upregulation of Brm in the absence of Brg1. Likewise, maintenance of Sox10 expression appears Brg1-independent.
Brg1 deficiency in oligodendrocytes leads to a mild differentiation defect
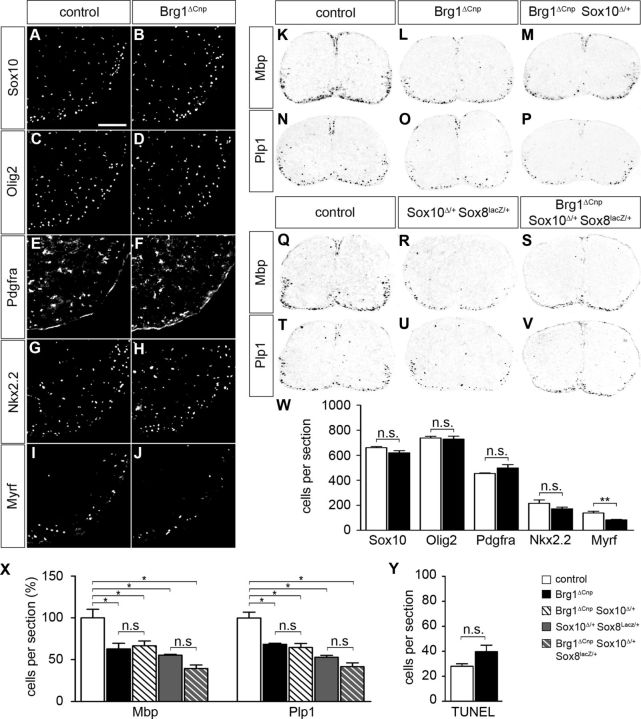
Using IHC and ISH, we characterized the oligodendroglial phenotype in Brg1ΔCnp mice immediately after birth. On P0, differences were not apparent in the number or distribution of Sox10- or Olig2-positive cells between the mutant and the wild-type control (Fig. 4A–D and Fig. 4W, respectively). The slightly higher number of Olig2-positive cells compared with Sox10-positive cells could be because Olig2 also recognizes a subset of astrocytes. Alternatively, the antibody against Olig2 might have been slightly more sensitive than the one against Sox10. This argues that the number of oligodendroglial cells in the spinal cord of mutant mice is comparable with the wild-type after birth. In agreement, apoptosis was not increased in the spinal cord of newborn Brg1ΔCnp mice or among the cells that had undergone recombination as judged by TUNEL (Fig. 4Y; and data not shown). There was also no substantial change in the number of Pdgfra-positive OPC (Fig. 4E,F and Fig. 4W, respectively) or in the number of Nkx2.2-positive promyelinating oligodendrocytes (Fig. 4G,H and Fig. 4W, respectively). In contrast, the number of Myrf-positive myelinating oligodendrocytes was reduced to 60% (Fig. 4I,J and Fig. 4W, respectively). This reduction was statistically significant and points to a mild influence of Brg1 on terminal differentiation of oligodendrocytes. This assumption was confirmed by ISH with probes for myelin gene transcripts. Mbp- and Plp1-positive cells were reduced by ∼37% and 31%, respectively (Fig. 4K,L,N,O and Fig. 4X).
Figure 4.
Perinatal consequences of Brg1 deletion in Brg1ΔCnp mice. A–J, IHC was performed on transverse spinal cord sections (forelimb region) of newborn control mice (A, C, E, G, I) and Brg1ΔCnp littermates (B, D, F, H, J) with antibodies directed against Sox10 (A, B), Olig2 (C, D), Pdgfra (E, F), Nkx2.2 (G, H), and Myrf (I, J). Ventral horn region is shown placed on a black background. Scale bar, 100 μm. K–V, Myelin gene expression was analyzed by ISH on transverse spinal cord sections (forelimb region) of newborn control (K, N, Q, T), Brg1ΔCnp (L, O), Brg1ΔCnp Sox10Δ/+ (M, P), Sox10Δ/+ Sox8lacZ/+ (R, U), and Brg1ΔCnp Sox10Δ/+ Sox8lacZ/+ (S, V) mice with antisense probes for Mbp (K–M, Q–S) and Plp1 (N–P, T–V). Spinal cords were placed on a white background. W, X, From these immunohistochemical stainings and ISH, cells were quantified that expressed the various oligodendroglial markers in control (white bars), Brg1ΔCnp (black bars), Brg1ΔCnp Sox10Δ/+ (hatched white bars), Sox10Δ/+ Sox8lacZ/+ (gray bars), and Brg1ΔCnp Sox10Δ/+ Sox8lacZ/+ (hatched gray bars) mice. Y, Additionally, cells undergoing apoptosis were identified by TUNEL and counted in control and Brg1ΔCnp mice. At least nine separate sections from the forelimb region of three independent specimens were counted for each genotype in W–Y. Data are shown in absolute (W, Y) or relative numbers (X) and are presented as mean ± SEM. Differences to control were statistically significant for Myrf, Mbp, and Plp1 as indicated (Student's t test). n.s., Not significant. *p ≤ 0.05. **p ≤ 0.01.
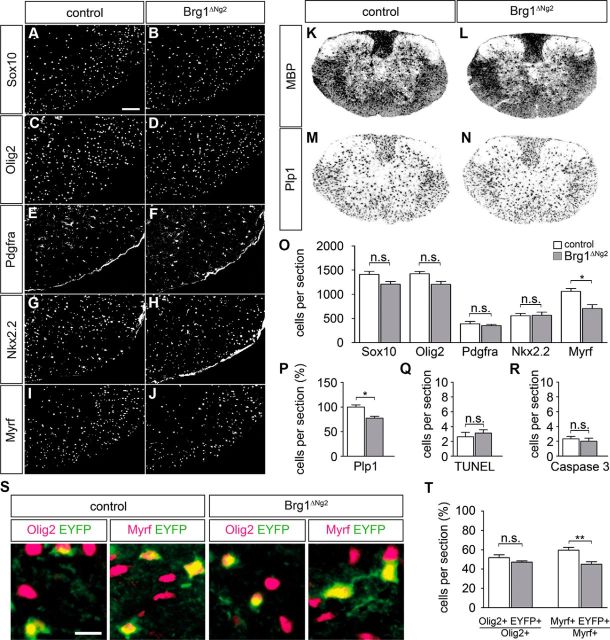
Considering the low deletion rate of the Ng2::Cre recombinase, Brg1ΔNg2 mice could not be expected to exhibit a phenotype at birth. Therefore, analysis was primarily performed on these mice at P14. In contrast, Brg1ΔCnp mice could not be studied at this later time point as pups died during the first day after birth. Although the reasons for the early postnatal death have not been studied in detail, this is most likely due to Cnp1Cre activity and Brg1 deletion in tissues outside the CNS. Results for Brg1ΔNg2 mice at P14 were comparable with those for Brg1ΔCnp mice at P0 with similar numbers of Sox10-, Olig2-, Pdgfra-, and Nkx2.2-positive cells (Fig. 5A–H and Fig. 5O, respectively), and similar rates of apoptosis between control and mutant mice (Fig. 5Q,R). Again, we detected a statistically significant reduction in the number of Myrf- and Plp1-positive cells in the order of 23%–33% relative to the wild-type (Fig. 5I,J,M–P). The Mbp-specific signal from ISH could no longer be quantified at this age but looked only slightly reduced in the Brg1ΔNg2 mutant relative to controls (Fig. 5K,L).
Figure 5.
Postnatal consequences of Brg1 deletion in Brg1ΔNg2 mice. A–J, IHC was performed on transverse spinal cord sections (forelimb region) of 2-week-old control mice (A, C, E, G, I) and Brg1ΔNg2 littermates (B, D, F, H, J) with antibodies directed against Sox10 (A, B), Olig2 (C, D), Pdgfra (E, F), Nkx2.2 (G, H), and Myrf (I, J). Ventral horn region is shown placed on a black background. Scale bar, 100 μm. K–N, Myelin gene expression was analyzed on transverse spinal cord sections (forelimb region) of control (K, M), and Brg1ΔNg2 (L, N) mice at 2 weeks with antisense probes for Mbp (K, L) and Plp1 (M, N). Spinal cords were placed on a white background. O, P, Cells were quantified that expressed various oligodendroglial markers in control (white bars) and Brg1ΔNg2 (gray bars) mice. Q, R, Additionally, cells undergoing apoptosis were identified by TUNEL (Q) and activated caspase-3 staining (R), and counted in control and Brg1ΔNg2 mice. At least nine separate sections from the forelimb region of three independent specimens were counted for each genotype in O–R. Data are shown in absolute (O, Q, R) or relative numbers (P, T) and are presented as mean ± SEM. Differences to control were statistically significant for Myrf as indicated (Student's t test). n.s., Not significant. *p ≤ 0.05. S, Co-IHC was performed with antibodies directed against GFP (green) and the oligodendroglial markers Olig2 and Myrf (all magenta) on transverse spinal cord sections of 2-week-old control (Rosa26+/stopfloxEYFP Ng2::Cre) and Brg1ΔNg2 Rosa26+/stopfloxEYFP mice. Scale bar, 25 μm. T, Quantifications from these stainings revealed that the percentage of GFP-labeled among all Olig2-positive cells did not vary between control and mutant, whereas the percentage of GFP-labeled among Myrf-positive cells was reduced in Brg1ΔNg2 Rosa26+/stopfloxEYFP mice (Student's t test). n.s., Not significant. **p ≤ 0.01.
Considering that recombination in Brg1ΔNg2 mice was restricted to approximately half the oligodendroglial cells at P14, we also asked whether a potentially stronger oligodendroglial differentiation defect would be masked by the normal development of oligodendrocytes that escaped Cre-mediated recombination. We crossed the Rosa26stopfloxEYFP allele into Brg1ΔNg2 mice and compared oligodendroglial cells with those in Ng2::Cre Rosa26stopfloxEYFP/+ controls. These comparisons showed that EYFP-positive cells are not underrepresented among Sox10- or Olig2-positive oligodendroglial cells relative to control mice (Fig. 5S,T; and data not shown). It also confirmed that a substantial number of EYFP-positive cells still expressed the differentiation marker Myrf in the Brg1ΔNg2 mutant at P14, although these cells were underrepresented (Fig. 5S,T). These results on Brg1ΔNg2 and Brg1ΔCnp mice confirm that Brg1 positively acts on the ability of oligodendrocytes to undergo terminal differentiation. However, they also show that Brg1 is not essential as significant levels of oligodendroglial differentiation will occur in the absence of Brg1 in both mouse mutants.
Genetic interaction between Sox10 and Brg1 cannot be detected during oligodendroglial differentiation
To analyze a potential genetic interaction between Brg1 and Sox10 during oligodendroglial differentiation, Brg1ΔCnp mice were generated that additionally carried a Sox10fl allele and thus were heterozygous for Sox10 in oligodendroglial cells. These mice exhibited the same mild oligodendroglial differentiation defect as Brg1ΔCnp mice. The number of Mbp- and Plp1-positive cells were reduced in P0 spinal cords relative to the wild-type, but not any more than in Brg1ΔCnp carrying two intact Sox10 alleles (Fig. 4M,P; for quantification, see Fig. 4X). To account for the possibility that genetic interaction between Sox10 and Brg1 is masked by partial redundancy between Sox10 and the related Sox8 in differentiating oligodendrocytes (Stolt et al., 2004), we also compared myelin gene expression in Brg1ΔCnp mice that carried a heterozygous Sox8 deletion on top of a heterozygous loss of Sox10 in oligodendrocytes (i.e., Brg1ΔCnp Sox10 Δ/+ Sox8lacZ/+). Reduction of Mbp and Plp1 expression in P0 spinal cords was not any higher than already observed in Cnp1Cre/+ Sox10 fl/+ Sox8lacZ/+ littermates (Fig. 4Q–V; for quantification, see Fig. 4X). Thus, there is no indication for a genetic interaction between Brg1 and Sox10 (or more generally speaking between Brg1 and SoxE proteins) in differentiating oligodendrocytes.
Brg1 is already associated with the regulatory regions of myelination-associated genes in OPC
Brg1 had previously been shown to be associated in differentiating oligodendrocytes with regulatory regions of several differentiation genes (Yu et al., 2013). Considering that we also detected Brg1 in OPC, we studied by ChIP whether Brg1 was already present on these regulatory regions before the onset of the differentiation process. We focused on regions from the Cnp1, Myrf, and Zfp191 genes (Fig. 6A) that had previously been shown to be bound by Brg1 in differentiating oligodendrocytes (Hornig et al., 2013; Yu et al., 2013). All three regulatory regions were indeed not only bound by Brg1 in cultures of differentiating oligodendrocytes but were also enriched in chromatin prepared from cultured primary OPCs and precipitated with anti-Brg1 antibodies, relative to precipitates of the same material with control IgG (Fig. 6C,D). Control fragments from the three genomic loci (again chosen according to previous publications) did not exhibit a comparable enrichment. The related Brm was not detected on these regulatory regions in OPCs or oligodendrocyte cultures by ChIP using two different anti-Brm antibodies (Fig. 6E,F; and data not shown). Brg1 binding to these regions furthermore overlapped substantially with binding of Olig2 and Sox10, particularly in differentiating oligodendrocytes, but also in cultured OPCs (Fig. 6G–J), and correlated with the ability of both proteins to interact with oligodendroglial Brg1 in GST-pulldown assays (Fig. 6B). These findings are compatible with a model in which Brg1 is already bound to these regions before the actual differentiation process starts. Both Olig2 and Sox10 may participate in recruiting Brg1.
Brg1 is coexpressed with Brm at all times in oligodendroglial cells
As Brg1 can be replaced by Brm as ATP-hydrolyzing subunit in BAF chromatin remodelling complexes, we analyzed Brm in more detail in oligodendroglial cells. Using antibodies directed against Brm on spinal cord sections, we detected Brm in virtually all Sox10- or Olig2-positive cells (Fig. 7A,B) independent of whether these cells expressed Pdgfra and Ng2 as OPC markers or Myrf and Mbp as markers of the differentiating state (Fig. 7C–F). Similar to Brg1, Brm expression was not restricted to the oligodendrocyte lineage but also occurred in neurons and astrocytes (Fig. 7G–J).
Figure 7.
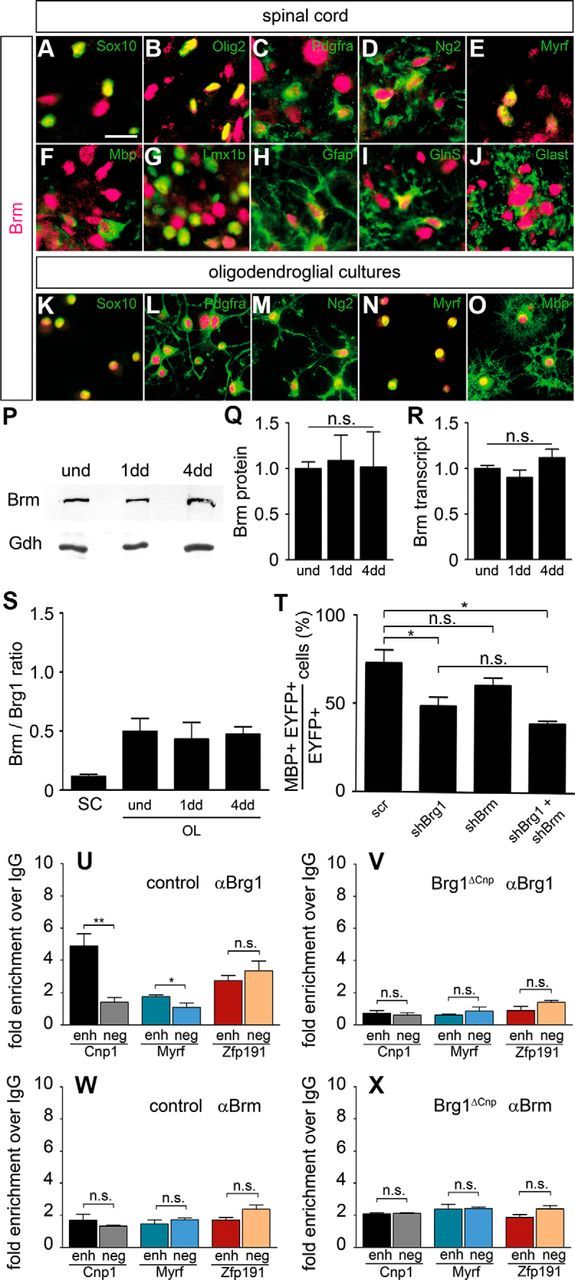
Expression and function of Brm in the oligodendrocyte lineage. A–J, Co-IHC was performed on spinal cord sections (forelimb level) at P0 to determine the occurrence of Brm (magenta) in cells of the oligodendrocyte lineage (Sox10 in A, Olig2 in B, Pdgfra in C, Ng2 in D, Myrf in E, and Mbp in F), in neurons (Lmx1b in G), and in astrocytes (Gfap in H, GlnS in I, and Glast in J) (all in green). Pictures were taken from either gray (G, I, J) or white matter (A–F, H). Scale bar, 25 μm. K–O, Stainings with antibodies directed against Brm (magenta) and various oligodendroglial markers (Sox10 in K, Pdgfra in L, Ng2 in M, Myrf in N, and Mbp in O) (all in green) were also performed on primary oligodendroglial cultures kept under proliferating (K–M) or differentiating (N, O) conditions. Scale bar, 25 μm. P, Q, By Western blot, Brm protein amounts were comparable in oligodendroglial cultures kept under proliferating conditions (und) or differentiating conditions for 1 (1dd) and 4 (4dd) days when normalized to Gapdh (Gdh) levels. P, A representative Western blot. Q, Quantification from three independent biological replicates with protein amounts in proliferating conditions set to 1. R, Brm transcript amounts did not significantly differ between oligodendroglial cultures kept under proliferating conditions or differentiating conditions for 1 and 4 d. After normalization to Rpl8, amounts in the proliferating cultures were set to 1. Experiments were repeated at least three times with material from three independent cultures for each condition, and a Student's t test was applied. n.s., Not significant. S, Transcript amounts were determined for Brm and Brg1 by qPCR in three independent experiments in Schwann cells as well as in oligodendroglial cultures kept under proliferating or differentiating conditions. After normalization to Rpl8, amounts were used to determine a Brm to Brg1 ratio. Although this ratio bears no information about the absolute Brm and Brg1 levels in these cells, it allows to conclude that oligodendrocytes contain more Brm relative to Brg1 than Schwann cells. T, Mbp expression was determined by immunocytochemistry on primary OPCs transfected with scrambled (scr), Brg1-specific (shBrg1), and Brm-specific (shBrm) shRNA or combinations thereof. A total of 100 cells were counted for each transfection in three separate experiments. The percentage of Mbp-expressing cells among all transfectants was then determined in three independent experiments (Student's t test). ns, Not significant. *p ≤ 0.05. U–X, ChIP was performed on the Cnp1, Myrf, and Zfp191 genes with antibodies directed against Brg1 (U, V), Brm (W, X), and control IgGs (U–X) on chromatin prepared from brains of wild-type and Brg1ΔCnp mice at P0. qPCR was then used to detect specific regions in immunoprecipitated chromatin, and relative enrichments were determined in the immunoprecipitate obtained with the specific antibody over the control IgG. Experiments were performed three times with each PCR in triplicate (Student's t test). n.s., Not significant. *p ≤ 0.05. **p ≤ 0.01.
Confirming these results, antibodies against Brm also stained the nuclei in cultured oligodendroglial cells. Virtually all Sox10-expressing cells were again positive for Brm, independent of whether proliferating or differentiating cultures were analyzed (Fig. 7K; and data not shown). Among oligodendroglial cells, Pdgfra- or Ng2-positive OPC were equally stained as Myrf- or Mbp-positive myelinating oligodendrocytes (Fig. 7L–O). This was corroborated by similar signal intensities in Western blots and RT-PCR experiments on material from proliferating compared with differentiating oligodendroglial cultures (Fig. 7P–R). Oligodendroglial cells thus coexpress Brg1 and its paralog Brm at all stages of development. Results from comparative qPCR on reverse-transcribed RNA from cultured Schwann cells and oligodendroglia furthermore suggest that the amounts of Brm relative to Brg1 are substantially higher in OPC and differentiating oligodendrocytes than in Schwann cells, at least on a transcript level (Fig. 7S).
However, only shRNA-dependent knockdown of Brg1, but not of Brm, led to a significant reduction of Mbp expression in transfected oligodendroglial cells kept for 4 d in culture under differentiating conditions (Fig. 7T). Additionally, there was no further significant decrease of Mbp expression when both Brg1 and Brm were knocked down. The absence of Brm-dependent effects correlates with our inability to detect Brm on regulatory regions of myelin genes in cultured oligodendrocytes (Fig. 6E,F). We also failed to detect Brm on these regulatory regions when ChIP was performed on brain of wild-type mice at P0 (Fig. 7W). In contrast, Brg1 was present on the Cnp1 and Myrf regions (Fig. 7U). Failure to detect a specific enrichment for the Zfp191 regulatory region may be due to the fact that Zfp191 expression is not restricted to oligodendrocytes but also found in neurons (Howng et al., 2010). Intriguingly, Brm could not even be identified on the Cnp1 and Myrf regulatory regions when ChIP was performed on brain of Brg1ΔCnp mice (Fig. 7X) in which Brg1 occupancy had been lost (Fig. 7V). Despite the presence of substantial amounts, Brm therefore appears unable to functionally replace Brg1 in oligodendrocytes.
Brg1 is required for Sox10 induction in OPC
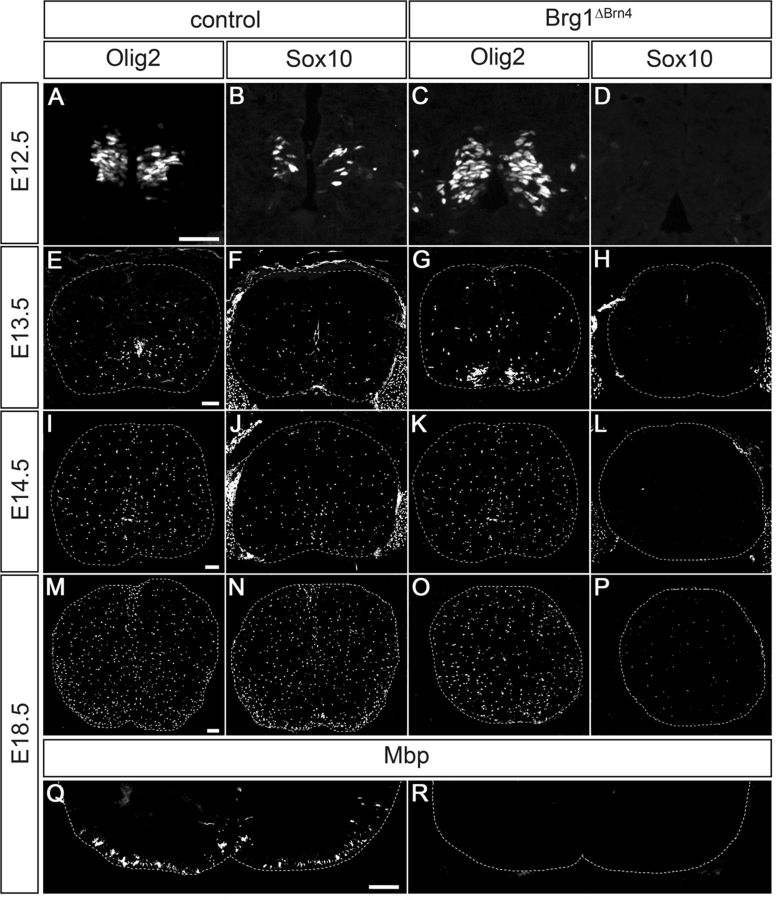
Considering the mild disruptions of oligodendrocyte development after Cnp1Cre- and Ng2::Cre-dependent Brg1 deletion, we were interested to study the consequences of an earlier deletion. For that purpose, we combined the Brg1fl allele with a Brn4::Cre transgene that efficiently deletes throughout the spinal cord ventricular zone at early times (Thein et al., 2010). The resulting Brg1ΔBrn4 mice were analyzed during different times of embryonic development (Fig. 8). Although Brg1 deletion in the spinal cord has been reported to result in several defects (Matsumoto et al., 2006), the progenitors of motor neurons (pMN) domain in the ventral ventricular zone appeared fairly normal at 12.5 dpc in Brg1ΔBrn4 mice (compare Fig. 8A with Fig. 8C) and gave rise to emigrating Olig2-positive cells that in the following days colonized the mantle zone and correspond to OPCs (compare Fig. 8E,I,M with Fig. 8G,K,O). Intriguingly, the majority of these cells failed to turn on Sox10 expression (Fig. 8D,H,L,P), which is normally induced in OPCs immediately upon specification (Fig. 8B) and remains expressed from then on (Fig. 8F,J,N). This is in stark contrast to the situation in Brg1ΔCnp and Brg1ΔNg2 mice where Sox10 expression is unaltered (Fig. 3T–V; and data not shown). A few Sox10-positive cells eventually appeared at 18.5 dpc in Brg1ΔBrn4 spinal cords (Fig. 8P). Myelin gene expression could not be detected at this age (compare Fig. 8Q with Fig. 8R; and data not shown). These data argue that Brg1 is required for the induction of Sox10 expression during or shortly after OPC specification, whereas maintenance of Sox10 expression at later times is independent of Brg1.
Figure 8.
Consequences of Brg1 deletion in Brg1ΔBrn4 mice. IHC was performed on transverse spinal cord sections (forelimb region) of control mice (A, B, E, F, I, J, M, N, Q) and Brg1ΔBrn4 littermates (C, D, G, H, K, L, O, P, R) at 12.5 dpc (A–D), 13.5 dpc (E–H), 14.5 dpc (I–L), and 18.5 dpc (M–R) with antibodies directed against Olig2 (A, C, E, G, I, K, M, O), Sox10 (B, D, F, H, J, L, N, P), and Mbp (Q, R). Sox10 and Mbp expression is strongly reduced in Brg1ΔBrn4 mice. M–P, Spinal cords were placed on a black background. Scale bar (each valid for the whole row): A, 50 μm; E, I, M, Q, 100 μm.
Discussion
Chromatin remodelling is an essential activity in many developmental processes, and oligodendroglial development is no exception (Li et al., 2009; Liu and Casaccia, 2010). Our analysis of mice with Brg1-deficient oligodendrocytes argues that part of this remodelling activity is performed by Brg1-containing BAF complexes and that these complexes are active during terminal differentiation and the onset of myelin gene expression. In this respect, there are similarities to Schwann cells where Brg1 is essential during the initial phases of terminal differentiation and myelination (Weider et al., 2012).
Apart from these similarities between oligodendrocytes and Schwann cells, there are also substantial differences in the role of Brg1 between both cell types. First and foremost, this concerns the severity of the mutant phenotype and the actual impact of Brg1 on terminal differentiation. Whereas Brg1 deletion in immature Schwann cells arrests development at this stage (Weider et al., 2012), deletion in OPCs only reduces the number of terminally differentiating oligodendrocytes by one-third, arguing that Brg1 is not essential and thus less important for oligodendrocyte than for Schwann cell differentiation. We do not think that the mild effect on oligodendroglial development is due to trivial reasons, such as inefficient and late Brg1 deletion. Analyses indicate that our Cnp1Cre-dependent deletion occurred in the vast majority of OPCs at an early enough time to ensure Brg1 absence before the onset of terminal differentiation and myelination. Additionally, we see that many cells that have undergone recombination still express terminal differentiation markers, such as Myrf, Mbp, and Plp1 in both Brg1ΔCnp and Brg1ΔNg2 mice, again arguing that differentiation is not blocked, but rather attenuated.
One of the reasons for this mild impact of Brg1 deficiency on oligodendroglial development and differentiation may be the presence of Brm in these cells. Brm can replace Brg1 in BAF complexes (Ho and Crabtree, 2010; Hargreaves and Crabtree, 2011). Considering that Brm and Brg1 are largely coexpressed during oligodendroglial development and that expression of Brm relative to Brg1 appears higher in oligodendrocytes than in Schwann cells, partial compensation of the loss of Brg1 by Brm is a possibility in oligodendrocytes. However, our results from knockdown and ChIP experiments in oligodendroglial cultures and CNS tissue have yielded no indication that this may indeed be the case.
Alternatively, other chromatin remodelling complexes may be coexpressed with BAF complexes and may have a greater impact on oligodendroglial differentiation and thereby reduce the importance of BAF complexes. During Schwann cell differentiation, for instance, both Brg1-containing BAF and Chd4-containing NuRD complexes are expressed (Hung et al., 2012; Weider et al., 2012). The much stronger myelination defect in Brg1-deficient Schwann cells than in Chd4-deficient Schwann cells clearly argues for a dominant role of the BAF complex (Hung et al., 2012; Weider et al., 2012). In differentiating oligodendrocytes, the roles may be reversed.
The other main difference between the role of Brg1 in oligodendrocytes and Schwann cells concerns the functional link to Sox10. Although there is genetic and biochemical indication that Sox10 is essential for Brg1 recruitment in Schwann cells to genes that need to be activated as a precondition for terminal differentiation (Weider et al., 2012) or during the process itself (Marathe et al., 2013), we found no evidence for a similarly tight relationship between both proteins in oligodendrocytes. This is despite the fact that Sox10 seems capable of interacting with Brg1 and of recruiting Brg1-containing BAF complexes to chromatin in oligodendrocytes as suggested by GST pulldown and ChIP experiments.
However, it has to be considered that Sox10 is the sole SoxE protein in Schwann cells, whereas it is coexpressed with Sox9 and Sox8 in oligodendrocytes (Stolt et al., 2003, 2005). Although the exact relationship between these closely related proteins has yet to be determined, there is evidence for partial redundancy between them (Stolt et al., 2005; Finzsch et al., 2008). Considering that Sox9 is expressed during oligodendroglial development into the early phases of differentiation and that Sox8 is expressed throughout (Stolt et al., 2003, 2005), especially Sox8 may be able to recruit Brg1-containing BAF complexes in the absence of Sox10. Redundancy among SoxE proteins may thus mask the link between Sox10 and Brg1. However, even the analysis of mice with combined deletions of Brg1, Sox8, and Sox10 yielded no hints at genetic interaction, arguing that SoxE redundancy is not the main reason for the different relationship between Sox10 and Brg1 in oligodendrocytes compared with Schwann cells.
In contrast to Schwann cells, oligodendroglial cells do not only rely on Sox10 for lineage progression and differentiation, but also on the bHLH proteins Olig2 and Olig1 (Meijer et al., 2012). Their presence is not dramatically altered by the absence of Sox10 in OPCs and differentiating oligodendrocytes. Our GST-pulldown experiments and ChIP experiments furthermore indicate that Olig2 also physically interacts with Brg1 in oligodendrocytes and may also trigger chromatin remodelling events by recruiting Brg1-containing BAF complexes. This could easily influence the relationship between Sox10 and Brg1.
Such a link between Olig2 and chromatin remodelling activities has also been reported in a recent study (Yu et al., 2013). Yu et al. (2013) reported that Olig2 works as a prepatterning factor in oligodendroglial cells and recruits Brg1-containing chromatin complexes to a large set of genes at the beginning of the differentiation process. To what extent Sox10 or other SoxE proteins are involved in this recruitment process has not been analyzed. It is also unclear to us why the authors did not see widespread occurrence of Brg1 in all CNS cell types but instead reported a selective enrichment in differentiating oligodendrocytes relative to OPCs, astrocytes, or neurons.
When the authors investigated the consequences of Brg1 deletion on oligodendroglial development in the mouse, they reported a complete block of differentiation. Such a severe phenotype is again at odds with our findings, as is the conclusion of an absolutely essential function of Brg1 during oligodendroglial differentiation.
Yu et al. (2013) used the same floxed Brg1 allele that we used. What differs are the Cre lines that were used. Whereas we used Ng2::Cre and Cnp1Cre lines, Yu et al. (2013) used an Olig1Cre previously generated as an Olig1 null allele by replacing Olig1 coding sequences by Cre sequences (Xin et al., 2005). This Cre line deletes already in neuroepithelial cells of the pMN domain and thus before specification of most oligodendroglial cells. In those OPCs that are not pMN-derived, Cre deletion occurs immediately after specification (Xin et al., 2005). Therefore, one of the differences between the two strategies is the timing of deletion, with Olig1Cre being earlier. If the main function of Brg1 during oligodendrocyte development is at the onset of terminal differentiation, this difference should not matter as Cnp1Cre- and Ng2::Cre-dependent deletion occurs early enough to ensure timely Brg1 absence.
However, if this assumption is wrong and the different timing is relevant for the observed phenotypic differences, it may be postulated that Brg1 must already be present and active in OPC at the time of specification or shortly thereafter as a prerequisite for the later effect on terminal differentiation. In support of such an assumption, we show in this study that an early deletion of Brg1 throughout the ventricular zone of the spinal cord interferes with Sox10 induction in those cells that arise as newly specified OPCs from the pMN domain. Even at the end of embryogenesis, most OPCs still do not express Sox10. Strong reduction of Sox10 expression has also been observed following Olig1Cre-dependent deletion of Brg1 (Yu et al., 2013).
The most important role of Brg1 during oligodendrocyte development therefore seems to be its early requirement for Sox10 induction during OPC specification. If Sox10 cannot be induced properly, terminal differentiation will fail as a consequence (Stolt et al., 2002; Hornig et al., 2013). Further modulatory roles for Brg1-containing BAF complexes during the differentiation process itself are suggested by the presence of Brg1 on regulatory regions of many terminal differentiation genes, and there is good reason to assume that Olig2 and/or Sox10 would be involved in the recruitment. However, this later activity of Brg1 and the BAF remodelling complex appears to be not essential.
We find it intriguing that Brg1 has a less prominent role in oligodendroglial than in Schwann cell differentiation and that it seems instead required for Sox10 induction during specification. The functional link between Brg1 and Sox10 during terminal differentiation is less well developed in oligodendrocytes than in Schwann cells. This supports the notion that substantial differences exist between the regulatory networks that coordinate development in these two types of myelinating glia.
Footnotes
The present work was performed by M.B. in fulfillment of the requirements for obtaining the degree Dr. rer. biol. hum. It was supported by Deutsche Forschungsgemeinschaft Grant We1326/8-2 to M. Wegner. We thank D. Metzger for supplying the floxed Brg1 allele; C. Birchmeier, T. Müller, and J. Trotter for the gift of antibodies; T. Archer and C. Muchardt for plasmids; J. Bader for experimental help; and S. Reiprich for critical reading of the manuscript.
The authors declare no competing financial interests.
References
- Ahn K, Mishina Y, Hanks MC, Behringer RR, Crenshaw EB., 3rd BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal-ventral patterning of the limb. Development. 2001;128:4449–4461. doi: 10.1242/dev.128.22.4449. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Emery B. Transcriptional and post-transcriptional control of CNS myelination. Curr Opin Neurobiol. 2010;20:601–607. doi: 10.1016/j.conb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Finzsch M, Stolt CC, Lommes P, Wegner M. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor α expression. Development. 2008;135:637–646. doi: 10.1242/dev.010454. [DOI] [PubMed] [Google Scholar]
- Finzsch M, Schreiner S, Kichko T, Reeh P, Tamm ER, Bösl MR, Meijer D, Wegner M. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J Cell Biol. 2010;189:701–712. doi: 10.1083/jcb.200912142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann SA, Hos D, Küspert M, Lang RA, Lovell-Badge R, Wegner M, Reiprich S. Stem cell factor Sox2 and its close relative Sox3 have differentiation functions in oligodendrocytes. Development. 2014;141:39–50. doi: 10.1242/dev.098418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig J, Fröb F, Vogl MR, Hermans-Borgmeyer I, Tamm ER, Wegner M. The transcription factors Sox10 and Myrf define an essential regulatory network module in differentiating oligodendrocytes. PLoS Genet. 2013;9:e1003644. doi: 10.1371/journal.pgen.1003644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howng SY, Avila RL, Emery B, Traka M, Lin W, Watkins T, Cook S, Bronson R, Davisson M, Barres BA, Popko B. ZFP191 is required by oligodendrocytes for CNS myelination. Genes Dev. 2010;24:301–311. doi: 10.1101/gad.1864510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H, Kohnken R, Svaren J. The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J Neurosci. 2012;32:1517–1527. doi: 10.1523/JNEUROSCI.2895-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Li H, He Y, Richardson WD, Casaccia P. Two-tier transcriptional control of oligodendrocyte differentiation. Curr Opin Neurobiol. 2009;19:479–485. doi: 10.1016/j.conb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpert AS, Bai S, Narayan M, Wu J, Yoon SO, Carter BD, Lu QR. NF-kappaB forms a complex with the chromatin remodeler BRG1 to regulate Schwann cell differentiation. J Neurosci. 2013;33:2388–2397. doi: 10.1523/JNEUROSCI.3223-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maka M, Stolt CC, Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev Biol. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Marathe HG, Mehta G, Zhang X, Datar I, Mehrotra A, Yeung KC, de la Serna IL. SWI/SNF enzymes promote SOX10-mediated activation of myelin gene expression. PLoS One. 2013;8:e69037. doi: 10.1371/journal.pone.0069037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer DH, Kane MF, Mehta S, Liu H, Harrington E, Taylor CM, Stiles CD, Rowitch DH. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat Rev Neurosci. 2012;13:819–831. doi: 10.1038/nrn3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Sock E, Leger H, Kuhlbrodt K, Schreiber J, Enderich J, Richter-Landsberg C, Wegner M. Expression of Krox proteins during differentiation of the O2-A progenitor cell line CG-4. J Neurochem. 1997;68:1911–1919. doi: 10.1046/j.1471-4159.1997.68051911.x. [DOI] [PubMed] [Google Scholar]
- Sock E, Schmidt K, Hermanns-Borgmeyer I, Bösl MR, Wegner M. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol Cell Biol. 2001;21:6951–6959. doi: 10.1128/MCB.21.20.6951-6959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Schmitt S, Lommes P, Sock E, Wegner M. Impact of transcription factor Sox8 on oligodendrocyte specification in the mouse embryonic spinal cord. Dev Biol. 2005;281:323–331. doi: 10.1016/j.ydbio.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein DC, Thalhammer JM, Hartwig AC, Crenshaw EB, 3rd, Lefebvre V, Wegner M, Sock E. The closely related transcription factors Sox4 and Sox11 function as survival factors during spinal cord development. J Neurochem. 2010;115:131–141. doi: 10.1111/j.1471-4159.2010.06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a soxist's view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Weider M, Küspert M, Bischof M, Vogl MR, Hornig J, Loy K, Kosian T, Müller J, Hillgärtner S, Tamm ER, Metzger D, Wegner M. Chromatin-remodeling factor Brg1 is required for Schwann cell differentiation and myelination. Dev Cell. 2012;23:193–201. doi: 10.1016/j.devcel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, Chan JR, Wu J, Lu QR. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135:145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]