Abstract
ArcLight is a genetically encoded fluorescent voltage sensor using the voltage-sensing domain of the voltage-sensing phosphatase from Ciona intestinalis that gives a large but slow-responding optical signal in response to changes in membrane potential (Jin et al., 2012). Fluorescent voltage sensors using the voltage-sensing domain from other species give faster yet weaker optical signals (Baker et al., 2012; Han et al., 2013). Sequence alignment of voltage-sensing phosphatases from different species revealed conserved polar and charged residues at 7 aa intervals in the S1–S3 transmembrane segments of the voltage-sensing domain, suggesting potential coil–coil interactions. The contribution of these residues to the voltage-induced optical signal was tested using a cassette mutagenesis screen by flanking each transmembrane segment with unique restriction sites to allow for the testing of individual mutations in each transmembrane segment, as well as combinations in all four transmembrane segments. Addition of a counter charge in S2 improved the kinetics of the optical response. A double mutation in the S4 domain dramatically reduced the slow component of the optical signal seen in ArcLight. Combining that double S4 mutant with the mutation in the S2 domain yielded a probe with kinetics <10 ms. Optimization of the linker sequence between S4 and the fluorescent protein resulted in a new ArcLight-derived probe, Bongwoori, capable of resolving action potentials in a hippocampal neuron firing at 60 Hz. Additional manipulation of the voltage-sensing domain could potentially lead to fluorescent sensors capable of optically resolving neuronal inhibition and subthreshold synaptic activity.
Keywords: fluorescent protein, omega current, voltage imaging, voltage sensor, voltage-sensing domain, voltage-sensing phosphatase
Introduction
Genetically encoded fluorescent sensors of neuronal activity offer the potential to study the physiology of individual neurons and entire neuronal circuits. The gCAMP series of calcium sensors enable the non-invasive measurement of spiking activity in vivo in transparent preparations, such as Danio rerio embryos (Ahrens et al., 2013). Combined with channel rhodopsin, light is becoming the electrode of choice because it is less detrimental and more flexible than glass pipettes.
Genetically encoded fluorescent voltage sensors (GeFVSs) have lagged behind their calcium counterparts. This is because GeFVSs are transmembrane proteins. A fluorescent transmembrane protein has two unattractive features. The first is that inefficient trafficking to the plasma membrane results in an internal, unresponsive fluorescence that diminishes the signal-to-noise ratio of the probe. The second problem is that membrane expression limits the amount of the probe, which lowers the total light level, again reducing the signal-to-noise ratio.
Poor membrane expression of the original GeFVSs that used voltage-sensing domains (VSDs) from voltage-gated channels (Siegel and Isacoff, 1997; Sakai et al., 2001; Ataka and Pieribone, 2002; Baker et al., 2007) was overcame by using the VSD from the Ciona intestinalis voltage-sensing phosphatase (VSP; Dimitrov et al., 2007). The probe hVoS (hybrid voltage sensor) overcomes the trafficking problem using a farnyselated-GFP to anchor the fluorescence to the plasma membrane and works quite well in slices, but the requirement of a quenching agent may limit its effectiveness in vivo (Wang et al., 2012).
GeFVSs must give a large optical response to yield a detectable signal over stochastically varying fluorescence noise. Improvements have mostly focused on modifications to the fluorescent domain. ArcLight, which contains the VSD from the Ciona VSP (Murata et al., 2005) fused to a variant of Super-ecliptic pHlorin (Ng et al., 2002), gives a 30% change in fluorescence during a 100 mV depolarization (Jin et al., 2012). Arch, a GeFVS based on rhodopsin, gives a large optical signal in response to membrane depolarizations but suffers from relatively low fluorescence (Kralj et al., 2012). The coupling of these rhodopsin-based probes with orange-shifted FPs also optically reports changes in membrane potential (Gong et al., 2014; Hochbaum et al., 2014). The sensor ASAP1 (Accelerated Sensor of Action Potentials 1) uses an extracellular, circularly permuted GFP that improves the speed of the optical signal (St-Pierre et al., 2014). GeFVSs are now capable of reporting neuronal activity in vivo (Cao et al., 2013).
A complimentary approach was done by mutating the VSD of the Ciona VSP. Several GeFVSs using the VSD from the Danio VSP, Zahra I and Zahra II (Baker et al., 2012), and the chicken VSP (Han et al., 2013) have faster kinetics than ArcLight. Alignment of the VSD from the VSP of several species revealed conserved polar and charged amino acids in the S1–S4 transmembrane segments. Two mutations to the S4 transmembrane combined with a mutation to the S2 transmembrane yielded a probe with similar signal size to ArcLight with improved kinetics. This probe, Bongwoori, can optically resolve action potentials in hippocampal neurons firing at 60 Hz.
Materials and Methods
Sequence alignment.
Using the Ciona intestinalis VSP protein sequence as bait, VSP homologs were identified in a BLAST search of the GenBank database (Madden et al., 1996). A broad spectrum of species were included in a multiple sequence alignment using Clustal Omega (McWilliam et al., 2013). The species and accession numbers were as follows: Danio rerio (zebrafish), AAH96934.1; Oryzias latipes (Japanese rice fish), XP_004076268.1; Takifugu rubripes (fugu), XP_003971378.1; Gallus gallus (chicken), XP_417079.2; Taeniopygia guttata (finch), XP_002195057.2; Mus musculus (mouse), AAI07330.1; Ailuropoda melanoleuca (giant panda), XP_002917732.1; Rattus norvegicus (rat), EDM08996.1; Ornithorhynchus anatinus (platypus), XP_001513133.2; Otolemur garnettii (galago), 003801862.1; Cricetulus griseus (hamster), XP_003503371.1; Pongo abelii (orangutan), XP_003778255.1; Macaca mulatta (rhesus), XP_001082960.2; Gorilla gorilla gorilla (gorilla), XP_004062630.1; Homo sapiens (human), AAP45146.1; Sus scrofa (boar), XP_003357879.2; Saimiri boliviensis boliviensis (squirrel monkey), XP_003934442.1; Loxodonta Africana (elephant), XP_003412628.1; Ciona intestinalis (sea squirt), BAD98733.1; Strongylocentrotus purpuratus (sea urchin), XP_003731108.1; Pelodiscus sinensis (turtle), XP_006124971; Xenopus laevis (frog), NP_001267536; Aplysia californica (sea hare), XP_005105026; Odobenus rosmarus divergens (walrus), XP_004399551; Pantholops hodgsonii (antelope), XP_005966581; Latimeria chalumnae (coelacanth), XP_006002818; Crassostrea gigas (oyster), EKC43187; Callithrix jacchus (marmoset), XP_002749061; and Metaseiulus occidentalis (mite), XP_003745380.
Plasmid DNA designs and construction.
The VSD from the Ciona intestinalis VSP was synthesized (Bionicsro) with restriction sites inserted between the four transmembrane domains. The introduction of a BamHI site upstream of transmembrane segment S1 caused a V109I mutation in the N terminus. The ClaI site downstream of S1 in the S1–S2 extracellular loop introduced a P140I mutation. A HindIII site was inserted in the S2–S3 intracellular loop that created an N176L mutation. Creation of a KpnI site in the S3–S4 extracellular loop resulted in an N176L mutation. This construct is called CC1, which contains the wild-type transmembrane domain sequences of Ciona intestinalis VSP gene.
The fluorescent protein (FP) Super Ecliptic pHlorin A227D was fused at position R256, which is 21 aa downstream of the S4 transmembrane segment. The RYR sequence at positions 254–256 allowed the introduction of a silent EcoRV site that was used to insert the FP. The entire construct was flanked by NheI and XhoI sites for cloning into pcDNA3.1hygro+ vector (Invitrogen).
Six additional constructs were synthesized that all had the restriction site architecture described for the CC1 construct with additional point mutations in each transmembrane segment. This allowed the generation of four mutants per synthesis. CC2 contained a D136A mutation in S1, an A154D mutation in S2, an E183N mutation in S3, and an R223Q mutation in S4. CC3 contained a G122A mutation in S1, a D164A mutation in S2, a D186A mutation in S3, and an R217Q/R229I double mutation in S4. CC4 had a D129S mutation in S1, an R168A mutation in S2, a D186S mutation in S3, and an R229I mutation in S4. CC5 contained a D129N mutation in S1, a D164N mutation in S2, a G187A mutation in S3, and a V220T mutation in S4. CC6 had a D136S mutation in S1, an R168H mutation in S2, a T193P mutation in S3, and a V220R mutation in S4. CC7 had a D129A mutation in S1, an A154D mutation in S2, four mutations in S3 consisting of S179T/S180N/K181P/L182W, and an R217Q mutation in S4. Single, double and triple mutants were then generated by transferring the desired transmembrane domain(s) into the CC1 construct via restriction digests. For instance, the triple mutant referred to in the text was generated by transferring the S2 domain from CC2 and the S4 domain from CC3 into CC1.
The construct, Bongwoori, consists of the triple mutant VSD and the optimized linker between the S4 transmembrane domain and the FP Super Ecliptic pHlorin, reported for ArcLight (Jin et al., 2012). This was achieved by a two-step PCR. The first step amplified the VSD from the triple mutant (A154D/R217Q/R229I) and the FP from ArcLight (a kind gift from Lawrence B. Cohen, Yale University School of Medicine, New Haven, CT). The VSD of the triple mutant was amplified using the sense primer RK024 (5′-aagctggctagcaccATGGAGGGATTCGACGGTTCAG-3′) and the reverse primer HP052 (5′-TACTCATGGGATCCCCTTGTTGGTGGGAATAAAATATTCTTGCTAAgat-3′). The FP segment of ArcLight was amplified using the sense primer HP051 (5′-GAATATTTTATTCCCACCAACAAGGGGATCCCATGAGTAAAGGAGAAG-3′) and reverse primer RK027 (5′-ttttctcgAGTTCTAGATCATTTGTATAGTTCATCC-3′). These two PCR products consisted of identical sequences in the linker between S4 and the FP, allowing for a full-length extension when combined in the second step of PCR and amplified with primers RK024 and RK027. All DNA constructs were verified by sequencing using the dye-termination method (Cosmogenetech).
Transient expression of GeFVSs in mammalian cells.
HEK293 cells were maintained in DMEM (High Glucose DMEM; Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS; Invitrogen). HEK293 cells were seeded onto #0 coverslips coated with poly-l-lysine (Sigma) in a 24-well culture dish. Transient transfections in HEK293 cells were accomplished by using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer (Invitrogen).
Primary hippocampal neurons were prepared from C57BL/6/F mice (Koatech) on embryonic day 18 as described previously (Banker and Goslin, 1988). Dissociated neurons were obtained by digesting hippocampi with 10 U/ml papain (Worthington) and 0.001% DNase I (Sigma) for 40 min at 37°C. Papain was inactivated with 10% FBS in HBSS. Neurons were then dissociated by mechanical trituration through Pasteur pipettes (Hilgenberg) with DNase I. Neurons were plated at 1 × 105 cells/ml onto 8–13 mm #0 coverslips (Ted Pella) precoated for 24 h with poly-d-lysine (Sigma) in 0.1 m borate buffer, pH 8.5. Neurons were cultured at 37°C in Neurobasal medium (Gibco) with B-27 supplement (Gibco by Life Technologies), 0.5 mm Glutamax-I (Gibco by Life Technologies) and 10% FBS. The following day, medium was replaced with identical medium without FBS. Transient transfections for neurons were performed at 6–7 d in vitro using the calcium phosphate method (Calphos mammalian transfection kit; Clontech, TaKaRa) as described previously (Jiang and Chen, 2006). Neurons were usually recorded at 7–8 d in vitro (16–24 h after transfection).
Patch clamp.
Coverslips with transfected HEK293 cells were patched at 33°C and chamber perfused with bath solution containing the following (in mm): 150 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2, 5 d-glucose, and 5 HEPES, pH 7.4. We used 3–5 MΩ glass patch pipettes (capillary tubing with 1.5/0.84 mm; World Precision Instruments) that were pulled by a P-97 micropipette puller (Sutter Instruments). The pipette solution consisted of the following (in mm): 120 K-aspartate, 4 NaCl, 4 MgCl2, 1 CaCl2, 10 EGTA, 3 Na2ATP, and 5 HEPES, pH 7.2. Transfected HEK293 cells were recorded in voltage clamp in the whole-cell configuration with a −70 mV holding potential. Electrical activity of cultured hippocampal neurons was recorded in current-clamp mode to evoke action potentials. Patch clamp was performed using an EPC10 amplifier (HEKA).
Arc lamp and wide-field imaging.
During the patch-clamp experiments, the cells were imaged on an IX71 microscope with a 60× 1.35 numerical aperture oil-immersion lens (Olympus). Fluorescence excitation was delivered using a 75 W xenon arc lamp (Cairn Research). The excitation filter was FF02-472/30 (Semrock). The emission filter was FF01-496/LP (Semrock). The dichroic was FF495-Di03 (Semrock). The objective C-mount image was demagnified by an Optem zoom system A45699 (Qioptiq LINOS) and projected onto the e2v CCD39 chip of NeuroCCD-SM 80 × 80 pixel camera (RedShirtImaging). The imaging apparatus was mounted on a Vibraplane Bench Top vibration isolation platform (Kinetic System, Minus K Technology). The mechanical shutter in the incident light patch was mounted on a separate table and did not touch the microscope. Images were recorded at a frame rate of 1 kfps.
Optical signal analysis.
Optical signal recordings were analyzed using Neuroplex (RedShirtImaging). The fluorescent traces for constructs expressed in HEK293 cells were averages of 16 trials. The fluorescent traces for constructs expressed in neuronal cells were from a single trial. The fluorescent traces are the spatial average of the output of all the pixels receiving light from the cell. The offline low-pass temporal filtering used to improve the signal-to-noise ratio is indicated in the figure legends.
The kinetics of the optical response and the voltage sensitivity of the optical response were analyzed using Excel (Microsoft) and Origin 8.6 (OriginLab). For the kinetics, the optical traces were fitted to a double-exponential decay, y = y0 + A1e−(t − t0)/τ1 + A2e−(t − t0)/τ2 (where t is time in milliseconds) and a single exponential decay, y = y0 + A1e−(t − t0)/τ1 (where t is time in milliseconds).
To compare the optical responses that were better fitted to a double-exponential decay to those that could only be fit by a single-exponential decay, a weighted tau was calculated as the sum of τ1 multiplied by the relative amplitude, A1, plus τ2 multiplied by the relative amplitude, A2, as defined by the following formula: τweighted = τ1(A1/(A1 + A2)) + τ2(A2/(A1 + A2)).
The voltage sensitivity was determined by initially fitting individual cell responses to the Boltzmann equation, y = (A1 − A2)/(1 + e(V−V1/2)/dx) + A2, where y is −ΔF/F, and V is membrane potential in millivolts. A1 is the minimum value, and A2 is the maximum value. V1/2 is the membrane potential in millivolts at half-maximal ΔF/F, and dx is the slope. All traces were then normalized such that A1 = 0 and A2 = 1. The trials for each construct were then averaged and refit. The range of the fluorescent voltage response in Table 2 was calculated by extrapolating the V1/2 value to the normalized maximum and minimum fluorescent change using the slope of the Boltzmann fit at V1/2 where, y = (A2 − A1)/4dx.
Table 2.
Range and speed of optical signals
| Construct | V1/2 (mV) | Range (mV) | τon (ms) | τoff (ms) |
|---|---|---|---|---|
| CC1 (wild type) | 96 ± 5 | 55 ± 10 | 17 ± 3 | |
| A154S (S2) | 53 ± 6 | −20 to 120 (140) | 44 ± 6 | 40 ± 16 |
| A154D (S2) | 55 ± 8 | −10 to 120 (130) | 68 ± 15 | 9 ± 1 |
| D164N (S2) | 6 ± 2 | −50 to 60 (110) | 42 ± 4 | 52 ± 5 |
| D164A (S2) | 45 ± 28* | 62 ± 13 | 41 ± 4 | |
| R168A (S2) | 67 ± 5 | −10 to 140 (150) | 39 ± 12 | 19 ± 3 |
| R217Q (S4) | −46 ± 6 | −130 to 30 (160) | 25 ± 3 | 39 ± 5 |
| R229I (S4) | 76 ± 10 | 10 to 140 (130) | 18 ± 1 | 7 ± 1 |
| V220T (S4) | 75 ± 3 | −10 to 160 (170) | 13 ± 2 | 9 ± 2 |
| V220R (S4) | 154 ± 9 | 35 ± 3 | 21 ± 3 | |
| R223Q (S4) | 59 ± 11 | 20 to 100 (80) | 25 ± 2 | 14 ± 1 |
| R217Q/R229I (S4) | −42 ± 6 | −130 to 40 (170) | 9 ± 3 | 10 ± 1 |
| A154D/R217Q/R229I (S2/S4) | −15 ± 5 | −70 to 40 (110) | 8 ± 1 | 9 ± 1 |
| Bongwoori | −3 ± 1 | −60 to 50 (110) | 10 ± 1 | 7 ± 1 |
| ArcLight | −17 ± 3 | −100 to 60 (160) | 34 ± 4 | 29 ± 2 |
The V1/2, range of optical signal, and the weighted time constants at a 100 mV depolarization are listed. The total range is in parentheses. The range is an estimate based on the slope at V1/2. The range was rounded to the nearest 10 mV because it incorporates the error of the inflexion point and the error of the slope (n ≥ 3 for all constructs). No range is listed for CC1, D164A, and V220R because values for the CC1 and V220R constructs are too shifted to positive potentials for a meaningful analysis and the D164A construct did not fit the Boltzmann curve well (Fig. 4B).
Results
Sequence alignment reveals several polar residues conserved throughout the VSD of VSPs
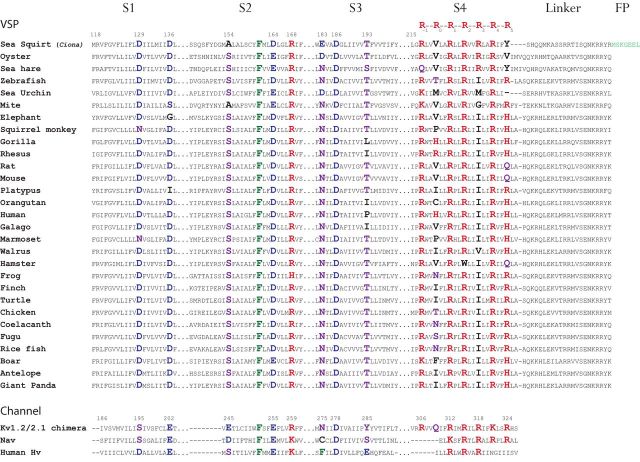
To investigate the potential cause(s) for the different kinetics of VSP-based GeFVSs, the sequences of the VSD transmembrane segments from several VSPs were aligned using Clustal Omega (Sievers et al., 2011; McWilliam et al., 2013). Figure 1 identifies conserved polar amino acids in the transmembrane segments of the VSD. The alignment includes a diverse spectrum of species to limit the bias toward an animal class, i.e., mammals or fish. The alignment of S4 is somewhat arbitrary because the regular repetition of positive charges in S4 can be aligned in different ways. We designate the positive charges in S4 as R−1—R0—R1—R2—R3—R4—R5 to aid comparisons across species. R1–R4 denotes the residues associated with the gating charges (Villalba-Galea et al., 2013). The conserved polar amino acids in S1–S3 are also shown in the VSDs of a chimeric, voltage-gated potassium channel Kv 1.2/2.1 (Tao et al., 2010), a voltage-gated proton channel (GenBank accession number XP_005254006.1), and a voltage-gated sodium channel (Jeong et al., 2000).
Figure 1.
Alignment of the VSD transmembrane segments from several VSP homologs. Conserved polar residues are in large font. Acidic residues are in blue. Basic residues are in red, and polar residues are in purple. A highly conserved phenylalanine in S2 is in green. The linker sequence was added after alignment to show the position of the FP Super Ecliptic pHlorin A227D (light green). The numbers for the VSP sequences are based on the Ciona protein. The numbers for the Kv sequence are based on the Kv1.2/2.1 chimera crystal structure.
The S1–S3 transmembrane domains exhibit a highly conserved structural motif separated by 7 aa, suggesting potential coil–coil interactions between the α-helices of the VSD. For instance, the S1 transmembrane domain in a vast majority of VSP proteins has two aspartic acids, D129 and D136 (the position of the VSP residues refers to the Ciona sequence denoted in Fig. 1), that are 7 aa apart, which implies that these amino acids reside on the same side of the α-helix. This conservation can also be seen in the S1 transmembrane domain of other voltage-sensing proteins except that the voltage-gated potassium channel (the Kv1.2/2.1 chimera in Fig. 1) and the voltage-gated sodium channel have a polar serine residue at the position homologous to D129 as opposed to a potentially charged aspartic acid.
The S2 transmembrane domain exhibits an even starker conservation to other voltage-sensing proteins with a similar 7 aa theme. In the center of the S2 domain resides an invariable phenylalanine (F161). Three amino acids downstream is a conserved negative residue (D/E164). Seven amino acids downstream of F161 is a conserved basic residue (R168). Interestingly, 7 aa upstream of F161 is a highly conserved polar residue, S154. One notable exception is the Ciona VSP protein that has an alanine (A154) at that position, which could account for the difference in speed observed for Ciona-based sensors versus zebrafish-based sensors. Again, this conservation in the S2 transmembrane domain is also seen in voltage-gated ion channels (Fig. 1). Note that, in the voltage-gated potassium and sodium channels, there is a potentially charged acidic residue 7 aa upstream of the conserved phenylalanine in S2, whereas a majority of the VSP homologs have a polar serine. This is the reverse situation of the conservation in S1 in which the voltage-gated ion channels have a polar serine residue (S195, Kv1.2/2.1 chimera) and the VSP homologs have an aspartic acid (D129).
The S3 domain exhibits conserved polar and/or charged residues 7 aa apart as well. However, the pattern in S3 seems to be antiparallel to that found in the S2 domain. For instance, S3 has an invariable aspartic acid (D186) in all of the VSP homolog sequences. Three amino acids upstream is a highly conserved asparagine (N183). Four amino acids upstream of N183 is a fairly well conserved serine residue (S179); however, this region of the S3 α-helix may be intracellular because there exists a high proportion of charged and polar residues. This is supported by the recently reported crystal structure of the VSD from Ciona VSP (Li et al., 2014). Also, 7 aa downstream of D186 is another fairly well conserved polar residue, T193. This distribution of conserved polar and charged residues in the VSD suggests interhelical interactions (all of the charged residues tend to reside on the same side of S1–S3), which may be good targets for modulating the voltage response of GeFVSs.
Additional evidence suggesting that these conserved residues have a role in sensing voltage changes comes from the crystal structures of Kv1.2/2.1 chimera (Tao et al., 2010) and the VSD from Ciona VSP (Li et al., 2014). Using the Cn3D program v4.3 (Wang et al., 2000) to select amino acids in the crystal structure of Kv1.2/2.1, residues in neighboring helices within 5 Å were identified. For instance, to identify residues in the S2–S4 transmembrane domains that potentially interact with D129 (Ciona S1 domain), residues within 5 Å of S195 (homologous residue in Kv1.2/2.1) were selected. The results are listed in Table 1. Surprisingly, many of these conserved amino acids in the S1–S3 transmembrane domains are within 3 Å of a positive residue from the S4 transmembrane domain (Table 1). Both D129 and D136 in the S1 domain of Ciona may interact with different arginines in the S4 domain. However, only D129 is within 5 Å of conserved residues in the S2 domain (A154 and F161). A154 in the S2 domain of Ciona, which is a serine for most of the VSP sequences, is within 3 Å of the S4 domain and 4 Å of the conserved T193 in the S3 domain. D164 is the only residue to be within 5 Å of the other three transmembrane domains. The recently reported structure of the Ciona VSP also shows a close proximity of many of these conserved polar amino acids to the arginine residues in S4 (Table 1, bold font). The conservation and close proximity of these conserved polar residues to the S4 domain suggest potential interactions affecting the speed, size, and voltage sensitivity of the optical signal from Ciona-based GeFVSs.
Table 1.
Potential inter-transmembrane interactions in the VSD of the Ciona VSP
| Residue | 3 Å | 4 Å | 5 Å |
|---|---|---|---|
| D129/S195 (S1) | R2226 (S4) | L157 (S2) | A154 (S2) |
| R3318 (S4) | C248 (S2) | F161 (S2) | |
| F252 (S2) | E245 (S2) | ||
| D136/E202 (S1) | K142 (S1–S2) | Y150 (S2) | D151 (S2) |
| Q147 (S2) | G230 (S1–S2) | I229 (S1–S2) | |
| R2315 (S4) | Y231 (S1–S2) | ||
| F241 (S2) | |||
| R1312 (S4) | |||
| A154/E245 (S2) | R3318 (S4) | L132 (S1) | D129 (S1) |
| Y231 (S1–S2) | T197 (S3) | ||
| Y285 (S3) | R2226 (S4) | ||
| R2315 (S4) | S195 (S1) | ||
| S198 (S1) | |||
| D164/E255 (S2) | S188 (S1) | I113 (S0) | |
| F172 (N-terminal) | |||
| K4321 (S4) | D278 (S3) | ||
| R168/R259 (S2) | I113 (S0) | E183 (S3) | |
| F177 (S2–S3) | G187 (S3) | ||
| D186 (S3) | I190 (S3) | ||
| W169 (N-terminal) | R3229 (S4) | ||
| F172 (N-terminal) | R4232 (S4) | ||
| R5324 (S4) | F269 (S3) | ||
| N275 (S3) | |||
| D278 (S3) | |||
| E183/N275 (S3) | R4232 (S4) | R5324 (S4) | R168 (S2) |
| F256 (S2) | |||
| R259 (S2) | |||
| D186/D278 (S3) | R5324 (S4) | R168 (S2) | F252 (S2) |
| R3229 (S4) | E225 (S2) | ||
| R4232 (S4) | F256 (S2) | ||
| K4321 (S4) | R259 (S2) | ||
| T193/Y285 (S3) | T246 (S2) | L225 (S4) | S158 (S2) |
| R2226 (S4) | A222 (S4) | ||
| Y231 (S1–S2) | Q233 (S1–S2) | ||
| F242 (S1–S2) | |||
| E245 (S2) | |||
| I249 (S2) |
The conserved polar and charged residues in the S1–S3 transmembrane segments identified in Figure 1 were mapped onto the crystal structures of the Ciona VSD upstate crystal structure (bold, 4G7V; Li et al., 2014) and the Kv1.2/2.1 chimera crystal structure (black, 2R9R; Long et al., 2007). Amino acids within 5 Å are listed. The bold numbers correspond to the Ciona VSP sequence. The black numbers correspond to the Kv1.2/2.1 chimera crystal structure. The secondary structure of the residue is in parentheses. For example, (S1–S2) indicates the extracellular loop between S1 and S2. The putative position of the charged residue in S4 (R−1-X-X-R0-X-X-R1-X-X-R2-X-X-R3-X-X-R4-X-X-R5) is depicted as in Figure 1.
The size, speed, and voltage response of a GeFVS can be modulated by mutating the conserved residues in the S1, S2, and S4 transmembrane domains
With the potential coil–coil interactions in the VSD (Fig. 1) and the close proximity of the homologous residues in both the Kv1.2/2.1 crystal structure and the Ciona VSD structure (Table 1), we used cassette mutagenesis to enable the characterization of individual mutations, as well as combinations of these mutations from different transmembrane segments. A construct using the VSD from Ciona VSP was synthesized containing restriction sites that flank each transmembrane domain (CC1 in Fig. 2). In addition, the Super Ecliptic pHlorin A227D (SE A227D) FP that gives signals of ∼30% ΔF/F per 100 mV depolarization was used as the optical reporter because this FP gives a very good signal-to-noise ratio (Jin et al., 2012).
Figure 2.
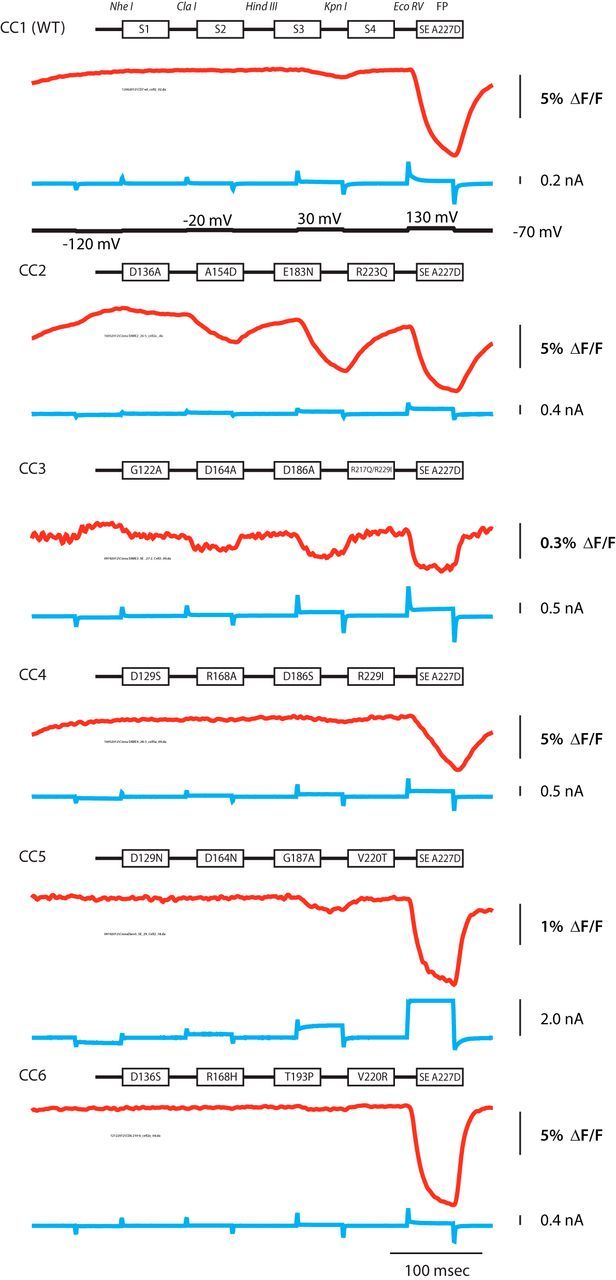
Cassette mutagenesis strategy for combinatorial screening of the VSD. Each transmembrane segment was flanked with unique restriction sites. CC1 contains the wild-type transmembrane segments denoted as S1–S4. CC2 consists of the quadruple mutant D136A (S1), A154D (S2), E183N (S3), and R223Q (S4). CC3 is a quintuple mutant consisting of two mutations in S4 in addition to the single mutations in S1–S3. The red trace is a representative fluorescent signal from voltage-clamped HEK293 cells (average of 16 trials). The holding potential was −70 mV. The pulse protocol is given in the black trace and was identical for all constructs. The current is shown in the blue trace. CC5 exhibited a voltage-dependent current not seen in the other constructs.
Seven total Ciona-based constructs were synthesized. CC1 has the wild-type Ciona VSD transmembrane segments fused to SE A227D. The other six constructs, CC2–CC7, consist of four mutations, one in each transmembrane domain (CC3 has two mutations in the S4 domain for a total of five mutations). Many of these mutations change the Ciona to the corresponding Danio amino acid. Single mutants were then generated by cutting and pasting the appropriate transmembrane domain into CC1. Figure 2 shows the schematic of the quadruple mutants and compares the optical traces of these constructs. HEK293 cells expressing the appropriate construct were voltage clamped at −70 mV and given a series of depolarizations while being imaged at 1 kHz. Representative traces from each construct are shown. Unfortunately, CC7, which attempted to fuse the SE A227D protein to the S4 α-helix, did not express in HEK293 cells (data not shown). Although the only rationale for generating quadruple mutants was to reduce the number of constructs synthesized, Figure 2 shows clearly the potential of these mutations because they are capable of affecting the size, speed, and voltage dependence of the optical responses. For instance, the fluorescence trace from a cell expressing CC2 shows a clear shift in the optical signal toward negative potentials compared with CC1. CC3 shows a much reduced optical signal (0.5% ΔF/F). CC5 and CC6 have a faster τoff compared with wild type, whereas CC4 shows a much slower τon. CC5 also has a voltage-dependent current. Because there is no pore in the VSP, this current is most likely attributable to an omega current through the VSD (Tombola et al., 2005, 2007). Whatever the mechanism, an ideal sensor should have minimal effect on the physiology of the cell. Thus, it was important to determine the mutation(s) responsible for the unwanted current.
A mutation in the S1 transmembrane domain results in an omega current
Figure 3 compares the optical and electrical traces of HEK293 cells expressing the CC5 quadruple mutant (D129N/D164N/G187A/V220T) to cells expressing each individual mutation found in the CC5 construct (i.e., D129N only). Only the D129N mutation in the S1 transmembrane domain yielded a current during a 200 mV depolarization. The D129N mutation also has a reduced optical response at 150 and 200 mV depolarizations compared with the CC5 construct, which may be in part attributable to a shift in the voltage response toward more positive potentials. The need for a stronger depolarization to illicit this current suggests that at least one other mutation in the S2–S4 transmembrane domains shifted the voltage dependence of the CC5 current to more negative potentials. Indeed, the D164N mutation in S2 and the V220T mutation in S4 are both capable of shifting the voltage dependence of these optical signals toward more negative potentials (Fig. 3).
Figure 3.
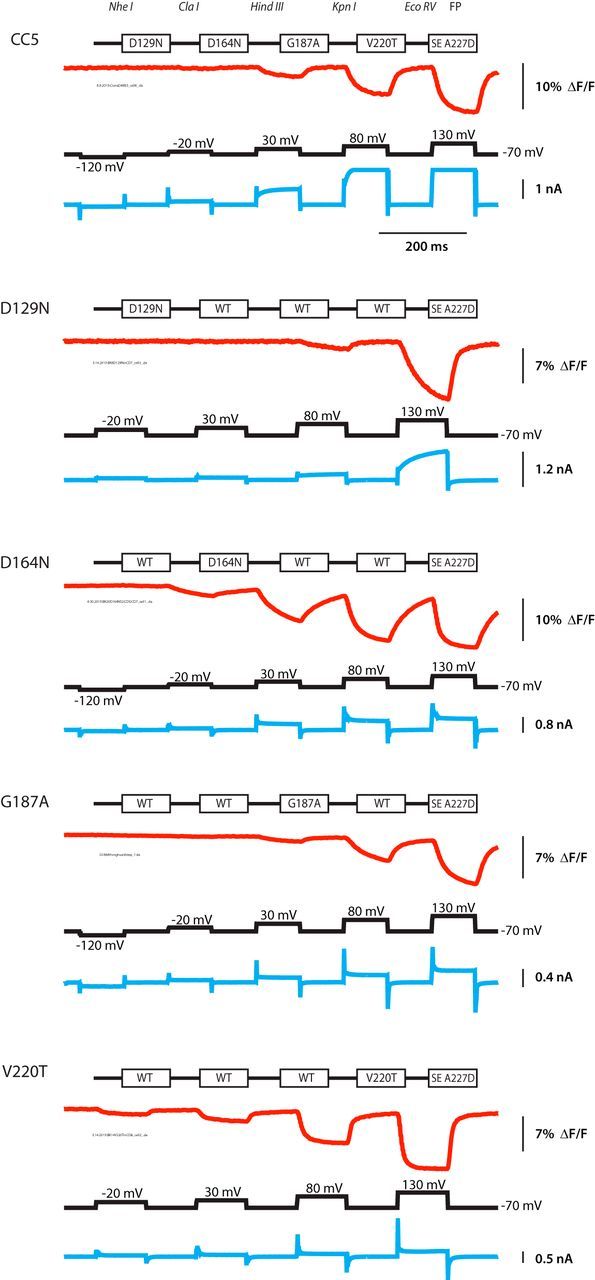
The D129N mutation in S1 results in a voltage-dependent current. The individual mutants were subcloned into the wild-type construct to determine whether the current was attributable to a single mutation. The voltage-pulse protocol is depicted in black and was varied because of the different voltage sensitivities. The current is in blue. Only constructs with the D129N mutation exhibited a voltage-dependent current.
It is interesting to note that the four individual mutations of CC5 had a semicumulative effect on the voltage sensitivity of the optical signal. The quadruple mutant CC5 has a V1/2 = 91 ± 1 mV (n = 4, SEM) which is very similar to the wild-type V1/2 of 96 ± 5 mV (n = 4). The D164N mutation in S2 results in a dramatic shift toward more negative potentials (V1/2 = 6 ± 2 mV, n = 3). The V220T mutation in S4 slightly shifts the optical response to more negative potentials (V1/2 = 75 ± 3 mV, n = 3). However, the D129N mutation requires a very strong depolarization to observe a change in the fluorescence. Figure 3 also shows that the omega current seems to follow the voltage sensitivity of the optical signal.
Mutations of the conserved polar and charged amino acids in the S2 transmembrane domain affected the voltage dependence and improved the speed of the optical signal
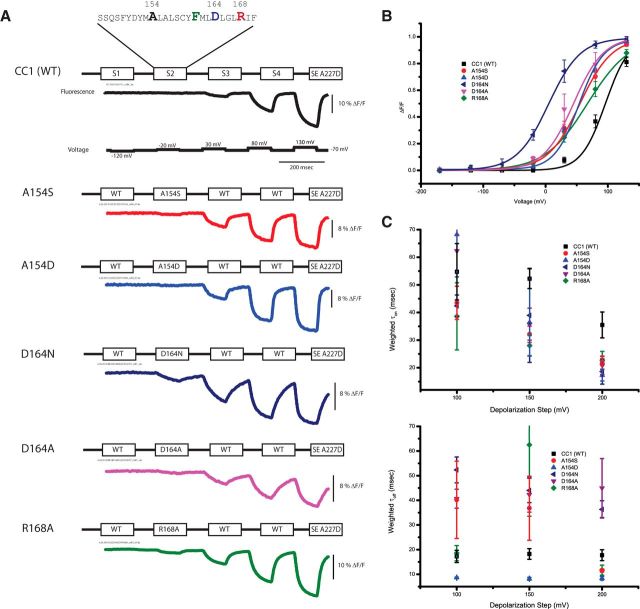
The conserved helical motif [S,E,D]-X6-F-X2-[D,E]-X3-[R,K,H], where X is primarily any hydrophobic residue, suggests that the S2 α-helix has four complete turns, with the S154–F161–D164–R168 residues residing on the same side of the helix. Figure 4 compares the traces of several S2 mutants with that of the CC1 wild-type Ciona construct. The Boltzman fit to the CC1 construct is tentative because its V1/2 ≥ 90 mV but provides a reference point for the mutations to the VSD. All of the S2 mutations tested shifted the voltage response to more negative potentials, with D164N having the largest effect.
Figure 4.
Mutations in the S2 domain affect the speed and voltage sensitivity of the optical signal. A, Representative fluorescent traces of HEK293 cells expressing either the wild-type construct or an S2 mutant. The voltage protocol is the same for all constructs. The color coding is maintained for all panels. B, The optical signal was normalized and fitted to a Boltzman equation. All S2 mutations shifted the voltage response to more negative potentials. C, The weighted on and off taus of the optical signals for the 100, 150, and 200 mV depolarization steps. The optical signals were fitted to a single- or double-exponential decay. Because not all constructs were fit to a double exponential, the weighted taus (τ1 × % amplitude + τ2 × % amplitude) are plotted for comparison. The τon becomes faster as the depolarization strength increases, whereas τoff remains fairly constant over the different voltages. Error bars are SEM.
An important parameter rarely discussed for GeFVSs is the slope of the Boltzman fit to the voltage response. Although the V1/2 reflects a shift in the voltage response, the slope indicates the relative voltage range in which the probe gives an optical response. Depending on the desired measurements, a probe with a 5% signal over a 20 mV range may be more useful than a probe with a 30% signal over a 200 mV range. Comparison of the slopes for the S2 mutations in Figure 4B shows that the R168A mutation reduces the slope, thereby increasing the range of the optical response to 150 mV (from −10 to 140 mV; Table 2). The A154D mutant exhibits a shift in the V1/2 (55 ± 8 mV), and the slope of the response is steeper (range is 130 mV from −10 to 120 mV; Table 2).
The time constants of the S2 mutants proved to be a complicated issue. Some of the constructs could be fit with a single-exponential decay (CC1), but others were best fit with a double-exponential decay (D164N). Interestingly, some constructs exhibited a double-exponential decay for smaller depolarization steps but were better fit by a single-exponential decay at larger depolarizations (A154S τoff). To compare the single-exponential decays to the double-exponential decays, Figure 4C plots the weighted τon and τoff values comparing the S2 mutants with wild type. The τon gets faster as the depolarization step increases in magnitude. This was attributable to two factors. One factor was an increase in the percentage of the fast component, whereas the other factor was an increase in the speed of the slow component. For instance, the 100 mV depolarization step for A154S exhibited a fast τon of 8 ± 1 ms, which accounted for 30 ± 4% of the amplitude (n = 4). At 150 mV depolarization, the τon was statistically unchanged at 9 ± 1 ms but accounted for 44 ± 8% of the amplitude. The 200 mV depolarization also had a similar fast τon of 8.0 ± 1.0 ms consisting of 58 ± 5% of the amplitude. The slow component also increased in speed with 59 ± 10 ms for a 100 mV depolarization, 51 ± 3 ms for a 150 mV depolarization, and 41 ± 1 ms for a 200 mV depolarization step. The fast component of the A154D τon was slightly slower than A154S but exhibited the same sort of pattern (12 ± 1 ms at 100 mV, 33 ± 2%; 12 ± 1 ms at 150 mV, 49 ± 10%; 8 ± 1 ms at 200 mV, 68 ± 3%). The slow component of A154D τon also improved in speed during stronger depolarizations (96 ± 22 ms, 100 mV; 65 ± 20 ms, 150 mV; 32 ± 3 ms, 200 mV).
The A154 position also exhibited interesting off kinetics. Again, the fast component percentage increased for A154S as the depolarization got stronger until only a single exponential could be observed at 200 mV (8 ± 1 ms at 100 mV, 58 ± 2%; 12 ± 1 ms at 150 mV, 80 ± 9%; 12 ± 1 ms at 200 mV, 100%). The off kinetics of the A154D construct could only be fit by a single exponential at all depolarizations (9 ± 1 ms at 100 mV, 8 ± 1 ms at 150 mV, and 8 ± 1 ms at 200 mV). The fast, single-exponential off rate and the steeper slope for the A154D mutant made it a viable candidate for additional probe development.
Mutations to the S4 transmembrane domain had the most dramatic contributions to the speed and voltage sensitivity of the optical response
A major difference between Ciona-based GeFVSs and Danio-based sensors that may contribute to the different kinetics of the optical response is the S4 transmembrane domain (Fig. 1). Although it is difficult to correctly align the S4 domain because of the distribution of positively charged amino acids (usually arginine), the alignment in Figure 1 suggests two distinct patterns in the S4 domain for the VSPs of different species. The first pattern has an uninterrupted distribution of four arginine residues every third amino acid (R1-X-X-R2-X-X-R3-X-X-R4). In Ciona, these four arginine residues have been shown to be responsible for gating currents (Villalba-Galea et al., 2013). However, this pattern is present in a small minority of the S4 domains in VSP homologs. The Danio VSP and the majority of other species has the pattern R1-X-X-R2-X-X-I3-X-X-R4-X-X-R5 in which the arginine repeat is interrupted by an isoleucine at position R3. Six amino acids upstream of the R1 residue is another highly conserved arginine residue (R217) that is found in all but one of the sequences aligned in Figure 1. This residue is not part of the gating current (or sensing current), but the R217Q mutant significantly shifts the voltage dependence to more negative potentials (Dimitrov et al., 2007). We thereby designate this position as R−1 (Fig. 1). The homologous mutation in zebrafish also shifts the voltage dependence in the same manner as Ciona, which is another justification for the alignment in Figure 1 (Hossain et al., 2008, Baker et al., 2012). Figure 5 shows just how drastic a change the R217Q mutant causes, giving an optical signal that will now respond to hyperpolarizations [R217Q V1/2 = −46 ± 6 mV, n = 6 vs CC1 (wild type) V1/2 = 95 ± 6 mV, n = 4]. The R217Q mutation also changes the kinetics of the optical response. R217Q has a faster τon (100 mV depolarization, τ1 = 6 ± 1 ms, 48 ± 5%; τ2 = 42 ± 5 ms, n = 5) but a slower τoff (100 mV depolarization τ1 = 39 ± 5 ms, 100%) compared with wild type (Fig. 5C).
Figure 5.
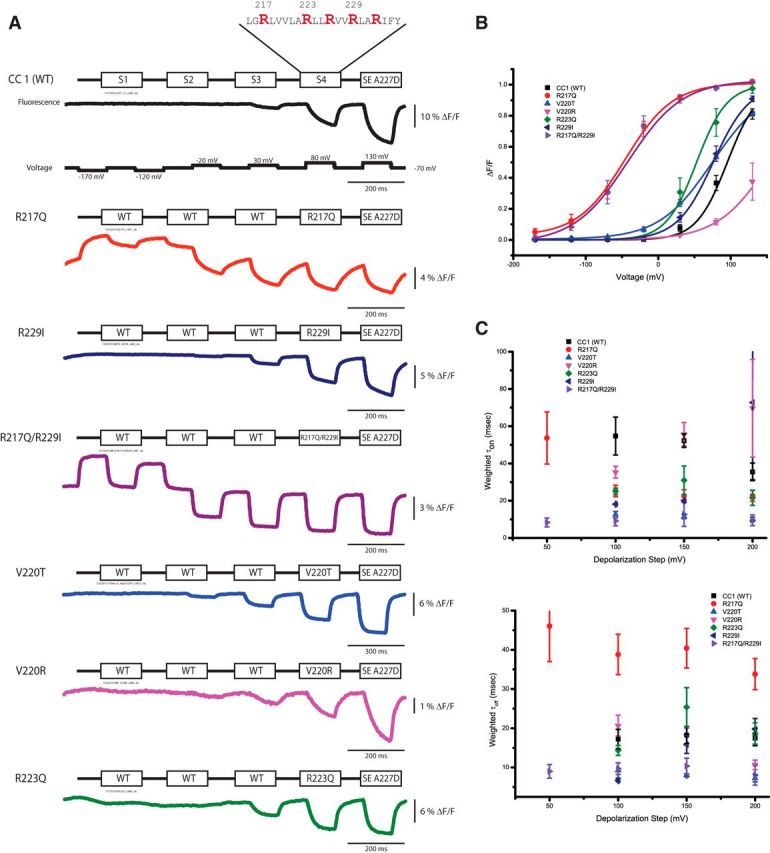
Mutations in the S4 domain affect the speed, voltage sensitivity, and size of the optical signal. A, Representative fluorescent traces of HEK293 cells expressing either the wild-type construct or an S4 mutant. The voltage protocol is the same for all constructs, but the pulse length for V220T was longer. The color coding is maintained for all panels. B, The optical signal was normalized and fitted to a Boltzman equation. Most S4 mutations shifted the voltage response to more negative potentials, with the exception of the V220R mutant, which appears to shift the voltage response to more positive potentials. C, The weighted on and off taus of the optical signals for the 50, 100, 150, and 200 mV depolarization steps. The optical signals were fitted to a single- or double-exponential decay. Like the S2 mutants, not all S4 constructs could be fit to a double exponential. The weighted taus (τ1 × % amplitude + τ2 × % amplitude) are plotted for comparison. Unlike the S2 mutants, the τon for the S4 mutants remains relatively constant as the depolarization strength increases, as does the τoff. Error bars are SEM.
The R229I mutation converts the Ciona S4 distribution of positive charges to the pattern found in most other VSPs, including zebrafish. Changing the R3 position to isoleucine has a very modest effect on the voltage sensitivity (V1/2 = 76 ± 10 mV, n = 4) but creates a slow component in the τon that increases in amplitude as the depolarization step increases. This increase in the contribution of the slow component causes the weighted τ to become larger as the depolarization step increases. The V220T is another S4 mutation that makes the Ciona-based construct more like zebrafish. This mutation at the R0 position also has a modest shift in the voltage sensitivity (V1/2 = 75 ± 3 mV, n = 5) but significantly increases the range of the voltage response to 170 mV (from −10 to 160 mV; Table 2).
The double mutant R217Q/R229I shows a marked increase in the speed of the optical response. For the τon, this is attributable to an increase in the speed of the fast component, an increase in the overall contribution of the fast component, and an increase in the speed of the slow component (τon for 100 mV depolarization is 6 ± 1 ms, 86 ± 10%, n = 3). Unlike the CC5 mutations that exhibited a cumulative voltage sensitivity, the R229I mutation did not significantly alter the voltage response of the optical signal for the R217Q mutation (Fig. 5B). Another important feature of the R217Q/R229I mutant is that the τon and τoff do not vary in response to stronger depolarizations. The same effect was noticed for the zebrafish-based sensors Zahra I and Zahra II (Baker et al., 2012). Remarkably, the R217Q/R229I construct does not exhibit the double-exponential characteristic of the R229I individual mutation. Although the size of the R217Q/R229I double mutant is nearly threefold lower than the CC1 construct, the shifted voltage sensitivity and the rapid optical response made the R217Q/R229I double mutant a viable candidate for additional development.
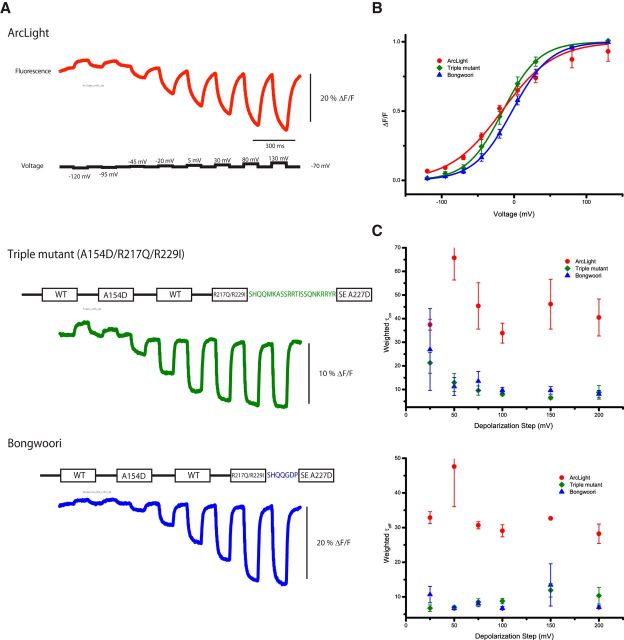
A triple mutant with an optimized linker length between the S4 domain and the FP yields a fast probe with a large optical signal
With some combinations of mutations showing an accumulative effect, we combined the A154D mutation in S2 with the R217Q/R229I double mutation in S4. Figure 6 compares the optical response of this triple mutant to that of ArcLight in HEK293 cells. The signal strength of the optical response for the A154D/R217Q/R229I triple mutant is less than half of the signal exhibited by ArcLight, but the slope of the voltage sensitivity has increased as has the speed of the optical response. The triple-mutant construct and ArcLight have very similar V1/2 values (triple mutant, −15 ± 5 mV; ArcLight, −17 ± 3 mV). Replacing the linker sequence between the S4 domain and the FP in the triple mutant with that optimized for ArcLight (Jin et al., 2012) shifted the voltage response to more positive potentials (V1/2 = −3 ± 1 mV) without a significant change in the slope of the response of the triple mutant (compare green and blue traces in Fig. 6B). This optimized linker length shown in blue in Figure 6A also doubled the signal strength, making it comparable with ArcLight. This ArcLight-derivative called Bongwoori has improved speed with weighted taus three to four times faster than ArcLight. However, the fast component of the on response for a 100 mV depolarization is virtually the same (Bongwoori, 8 ± 1 ms; ArcLight, 7 ± 1 ms), but the amplitude of the fast component for Bongwoori accounts for 91 ± 6% of the total exponential decay, whereas the fast component for ArcLight is only 22 ± 8% (n = 4 for both constructs). Bongwoori also has an improved off rate that could only be fit to a single exponential for the 100 mV depolarization step (τoff = 7 ± 1 ms, 100%).
Figure 6.
A combination of S2 and S4 mutations results in a significantly faster optical signal. A, Representative optical traces from HEK293 cells expressing ArcLight, a triple mutant consisting of A154D(S2)/R217Q(S4)/R229I(S4) and Bongwoori. The linker sequence between S4 and the FP is shown in green for the triple mutant and blue for Bongwoori. The Bongwoori linker site is identical to that of ArcLight. B. The voltage-sensitivity of the normalized optical signal. Bongwoori has a steeper and a positively shifted voltage response. C, The speed of the optical signal is significantly increased in the triple mutant and Bongwoori.
Resolution of high-frequency action potentials in dissociated, hippocampal neurons
A true test of the speed of the optical response for a GeFVS is to image action potentials in neurons and not high-frequency pulse protocols in HEK293 cells. To compare the ability to optically report action potentials between Bongwoori and ArcLight, dissociated, hippocampal neurons were transfected with the appropriate construct and submitted to current clamp to elicit action potentials. Figure 7 shows a low-pass-filtered trace and the unfiltered trace from a single trial for both ArcLight and Bongwoori. As can be seen even in the unfiltered trace, Bongwoori is capable of resolving action potentials firing at 60 Hz. This improved resolution over ArcLight is primarily attributable to its improved speed and the shifted voltage sensitivity. Bongwoori has a V1/2 near 0 mV, which is better suited to resolve action potentials from subthreshold depolarizations than ArcLight, which has a V1/2 near −20 mV. Bongwoori also has a steeper slope so the range of voltage that results in a fluorescence change is reduced (Fig. 6B). Therefore, Bongwoori exhibits less of a fluorescent change for subthreshold depolarizations but a similar fluorescence change after threshold depolarization. These factors make Bongwoori better at resolving action potentials and ArcLight better at reporting subthreshold activity.
Figure 7.
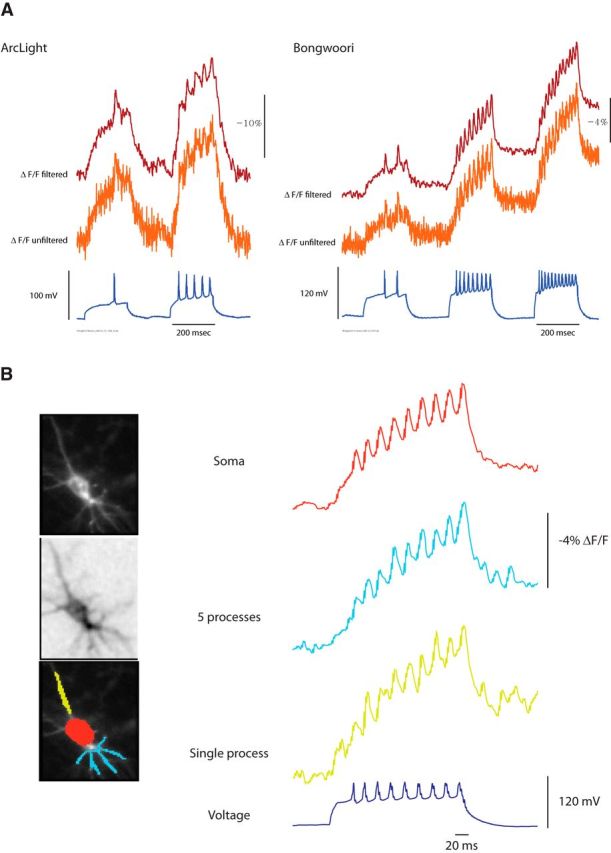
Optical resolution of action potentials from hippocampal neurons. A, Hippocampal neurons expressing either ArcLight or Bongwoori under current clamp were induced to fire action potentials. The orange trace is from a single trial, unfiltered acquisition at 1 kHz. The red trace has been filtered using a low-pass Gaussian 100 filter. Bongwoori has a positively shifted voltage response compared with ArcLight, resulting in a lower fluorescent change during the subthreshold depolarization. B, The resting light image, the frame subtraction image, and the optical changes observed in the soma (red), five lower processes (light blue), and a single process (yellow) filtered using a low-pass Gaussian 100 filter. The signal-to-noise ratio in the soma is better than the process traces and is sufficient for resolving action potentials at 60 Hz (dark blue).
The resolution of high-frequency action potentials was also seen in the processes of the hippocampal neuron. Figure 7B compares the optical traces from the soma and different processes to the current-clamped induced action potentials. The soma has the highest expression and therefore the most light, which results in a higher signal-to-noise ratio. The processes have less membrane for expression of the voltage sensor, resulting in less light, which lowers the signal-to-noise ratio. Even with the lower signal-to-noise ratio, action potentials could be observed optically in a single process.
One negative aspect of Bongwoori is the change in baseline fluorescence during the current-clamp experiment. There are two potential reasons for this. The change in baseline may well be a result of the acidification of the cell as it fires multiple action potentials. The FP for Bongwoori is a mutated version of a pH-sensing FP (Ng et al., 2002; Jin et al., 2012). Increasing the buffering capacity of the internal and external solutions during current clamp reduced but did not eliminate the baseline change (data not shown). The second reason for the increase in slope is most likely the contribution of the slow component during high-frequency firing of the neuron. Although the action potential is rapid (2–3 ms), the neuron experiences a long-term depolarization of at least 50 mV for 200 ms (Fig. 7A, voltage trace in blue), which is why it is important to reduce the contribution of the slow component of GeFVSs. Regardless of the cause for the baseline change, Bongwoori can resolve action potentials at high frequencies even after the baseline fluorescence has changed (Fig. 7A, second and third pulse of the current clamp).
Discussion
Mutagenesis of the conserved polar amino acids in the VSD enabled the development of a novel GeFVS, Bongwoori, capable of resolving action potentials in a neuron firing at 60 Hz. Using the alignment of the VSD from many voltage-gated phosphatases to guide the mutagenesis of the transmembrane domains, we have generated several Ciona-based GeFVSs that exhibit a variety of kinetic responses and altered voltage dependencies. Fortunately, in some instances, mutations in one transmembrane domain can function independently from mutations in the other transmembrane domains. This cumulative effect was seen in the additive contribution of individual mutations to the voltage dependence of the optical signal (Fig. 3, CC5). In contrast, the speed of the optical response was not a sum of the parts, which may result from individual mutations generating a rate-limiting hindrance to the change in conformation.
In addition to identifying desirable mutations, we have identified D129 in S1 as a residue not to be tampered with. Mutagenesis of this amino acid to a polar, noncharged asparagine shifted the voltage response to more positive potentials and created a voltage-gated pore, which is extremely undesirable. D129 is also one turn of the α-helix away from I126, which has been reported to be part of a hydrophobic core that also affects the voltage sensitivity of the VSP (Lacroix and Bezanilla, 2012). In the S4 downstate, D129 clearly interacts with R1223 and is very close to the hydrophobic barrier created by F161 in S2 (Li et al., 2014). In the S4 upstate, D129 appears to interact with R2226 (R1 and R2 refer to the position in S4; Fig. 1), which could potentially account for the omega current once S4 moves.
Many GeFVSs of membrane potential are able to optically report action potentials (Barnett et al., 2012; Jin et al., 2012; Kralj et al., 2012; Han et al., 2013; St-Pierre et al., 2014), but the ability to resolve action potentials at high frequencies in neurons remains an issue. This is attributable to several factors: (1) the speed of the optical response (both on and off); (2) the strength of the optical signal; (3) the V1/2 and the slope of the optical response as a function of voltage; and (4) efficient membrane targeting of the probe (which vastly improves the signal-to-noise ratio). Optimizing one of these characteristics can overcome deficiencies in other areas. The rhodopsin-based probes exhibit poor membrane expression but give large signals (Kralj et al., 2012; Gong et al., 2014; Hochbaum et al., 2014). ArcLight exhibits relatively slow kinetics but is still able to optically resolve an action potential attributable to a large fluorescent change (Jin et al., 2012).
In vivo, without electrodes, the speed and size of the optical signal are the two parameters important for resolving action potentials from subthreshold depolarizations. Most GeFVSs published (including Bongwoori) exhibit kinetics slower than an action potential. The consequences are twofold. Not only will the fluorescent signal lag behind the voltage change of an action potential, but the signal size will be diminished since the membrane voltage returned to baseline before the probe reached its maximal response. The slower speed and diminished signal are potential confounds to the optical resolution of action potentials from subthreshold depolarizations. If the probe gives a smaller signal in vivo than what was observed in neuronal culture, then the resolution of action potentials becomes even more difficult. Shifting the V1/2 toward more positive potentials would bias the optical signal toward action potentials. Increasing the slope of the optical response would improve the signal size and thus the signal-to-noise ratio of the current probes (as long as the V1/2 is in a useful position).
The strategy of using probes that are tuned to specific voltage ranges, one that would only respond to action potentials for instance, requires multiple probes for measuring different types of neuronal activity. Although a single probe with a large, fast, linear fluorescent response over a large voltage range could detect inhibition, synaptic activity, and action potentials in a single cell, the signals from a population of cells expressing that probe would be difficult to interpret. A pixel would encounter light from different cells potentially experiencing different voltage changes. Even in the ideal case of specific cell-type expression, neuronal activity will probably not be synchronized. The fluorescent output would therefore be a summation of those activities resulting in the loss of information. Probes tuned to specific voltage ranges would have the advantage of maintaining that information while populations of neurons are being imaged.
By focusing on the VSD of the Ciona VSP, we were able to tune the voltage sensitivity of GeFVSs. The V1/2 for the wild-type Ciona VSP is nearly +100 mV. The R217Q mutation in S4 drastically shifts the V1/2 to −70 mV for VSFP2.1 (Dimitrov et al., 2007) and to −30 mV for ArcLight (Jin et al., 2012) but also results in a much slower optical signal (Fig. 5). In this report, we identified several mutations that shift the voltage sensitivity of the optical probe. Mutagenesis of D129 in S1 shifted the voltage response to more positive potentials. Mutagenesis of the conserved polar residues in S2 (A154, D164, and R168) all shifted the voltage response of the optical signal to more negative potentials. Mutations to the S4 domain also shifted the voltage response. Curiously, most mutations that shift the voltage response to more negative potentials slow the speed of the optical response. The exceptions to this observation are the A154D and V220T mutants. Unfortunately, the V220T mutant dramatically reduced the slope of the voltage dependence. The V220T construct gives an optical signal over a range of 170 mV. The only mutant that increased the slope of the voltage dependence was the R223Q mutation (range of 80 mV; Table 2), which also slowed the speed of the optical response.
Several mutations to the VSD exhibited positive qualities that could be combined without losing the desired qualities of the individual mutations. For instance, the R217Q drastically shifts the voltage dependence of the optical signal to more physiological potentials. The R229I mutant yields a faster optical signal but only slightly changes the voltage sensitivity. Both the R217Q and the R229I mutations individually exhibit kinetics that are better fitted by double exponentials (Fig. 5). This is not true of the R217/R229I double mutant. This double mutant has reduced the on and off taus to below 10 ms yet maintains the voltage dependence of the R217Q mutant. The main drawback of this probe is that the signal strength is only ∼5% ΔF/F per 100 mV. Combining the R217Q/R219I mutation with the A154D mutation in S2 doubled the signal size to ∼10% ΔF/F per 100 mV while maintaining the speed and voltage dependence of the R217Q/R229I mutant. The A154D mutant introduces a potential counter charge in S2 that is not present in the wild-type, CC1 construct, which may account for the change in the signal size.
Additional improvement in the signal size came from reducing the linker length between the S4 transmembrane domain and the FP to the composition that was optimized for ArcLight (Jin et al., 2012). This probe, Bongwoori, has a signal size comparable with ArcLight that is just slightly slower than the triple mutant (A154D/R217Q/R229I) but has a better V1/2 (triple mutant, −16 ± 5 mV; ArcLight, −17 ± 3 mV; Bongwoori, −3 ± 1 mV) for resolving action potentials from subthreshold depolarizations (Figs. 6, 7). For ArcLight, we observed optical signals up to 18% ΔF/F for an action potential in hippocampal neurons, but almost 90% of that optical signal came from the subthreshold depolarization with only 10% of the signal resulting from the spike of the action potential. Bongwoori exhibits optical signals up to 5% ΔF/F for an action potential, but 50% of that signal is attributable to spiking. The speed and voltage sensitivity of this probe enables the resolution of action potentials at a frequency of 60 Hz in a single trial without filtering the optical signal (Fig. 7). It is unclear why shortening the linker between S4 and the FP shifted the voltage response to more positive potentials, but hopefully this parameter contributing to the voltage response of the probe can also be exploited to better resolve different components of neuronal activity.
To date, improvements in genetically encoded fluorescent voltage probes have mostly centered on the FP. Here, we have shown that the VSD is capable of improving the speed, size, and voltage dependence of these probes. Being able to adjust the voltage sensitivity of a probe will be critical for in vivo analyses of neuronal circuits. The ultimate goal is to replace cumbersome, electrical pipettes with light. Optically differentiating an action potential from a subthreshold event will be difficult unless the probe is tuned to only respond to action potentials. Identifying neuronal inhibition will be extremely difficult because most current GeFVSs give an opposite fluorescent signal under hyperpolarizing conditions. One neuron may increase its fluorescence but the decrease in neighboring cells could potentially mask that fluorescence change. Additional manipulation of the VSD should eventually lead to probes that are tuned to optically resolve action potentials, synaptic activity, and potentially inhibition.
Footnotes
This work was supported by the World Class Institute (WCI) program of the National Research Foundation of Korea funded by Ministry of Education, Science, and Technology of Korea Grant WCI 2009-003 and Korea Institute of Science and Technology Institutional Program Project 2E24210. We thank D. Choi and L. Cohen for critical review of this manuscript. We thank Ji Hee Kim for translational assistance.
References
- Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J. 2002;82:509–516. doi: 10.1016/S0006-3495(02)75415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BJ, Lee H, Pieribone VA, Cohen LB, Isacoff EY, Knopfel T, Kosmidis EK. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J Neurosci Methods. 2007;161:32–38. doi: 10.1016/j.jneumeth.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Baker BJ, Jin L, Han Z, Cohen LB, Popovic M, Platisa J, Pieribone V. Genetically encoded fluorescent voltage sensors using the voltage-sensing domain of Nematostella and Danio phosphatases exhibit fast kinetics. J Neurosci Methods. 2012;208:190–196. doi: 10.1016/j.jneumeth.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G, Goslin K. Developments in neuronal cell culture. Nature. 1988;336:185–186. doi: 10.1038/336185a0. [DOI] [PubMed] [Google Scholar]
- Barnett L, Platisa J, Popovic M, Pieribone VA, Hughes T. A fluorescent, genetically-encoded voltage probe capable of resolving action potentials. PLoS One. 2012;7:e43454. doi: 10.1371/journal.pone.0043454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Platisa J, Pieribone VA, Raccuglia D, Kunst M, Nitabach MN. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D, He Y, Mutoh H, Baker BJ, Cohen L, Akemann W, Knöpfel T. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS One. 2007;2:e440. doi: 10.1371/journal.pone.0000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Wagner MJ, Zhong Li J, Schnitzer MJ. Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors. Nat Commun. 2014;5:3674. doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Jin L, Platisa J, Cohen LB, Baker BJ, Pieribone VA. Fluorescent protein voltage probes derived from ArcLight that respond to membrane voltage changes with fast kinetics. PLoS One. 2013;8:e81295. doi: 10.1371/journal.pone.0081295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, Kapoor V, Zou P, Kralj JM, Maclaurin D, Smedemark-Margulies N, Saulnier JL, Boulting GL, Straub C, Cho YK, Melkonian M, Wong GK, Harrison DJ, Murthy VN, Sabatini BL, Boyden ES, Campbell RE, Cohen AE. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MI, Iwasaki H, Okochi Y, Chahine M, Higashijima S, Nagayama K, Okamura Y. Enzyme domain affects the movement of the voltage sensor in ascidian and zebrafish voltage-sensing phosphatases. J Biol Chem. 2008;283:18248–18259. doi: 10.1074/jbc.M706184200. [DOI] [PubMed] [Google Scholar]
- Jeong SY, Goto J, Hashida H, Suzuki T, Ogata K, Masuda N, Hirai M, Isahara K, Uchiyama Y, Kanazawa I. Identification of a novel human voltage-gated sodium channel alpha subunit gene, SCN12A. Biochem Biophys Res Commun. 2000;267:262–270. doi: 10.1006/bbrc.1999.1916. [DOI] [PubMed] [Google Scholar]
- Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- Jin L, Han Z, Platisa J, Wooltorton JR, Cohen LB, Pieribone VA. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix JJ, Bezanilla F. Tuning the voltage-sensor motion with a single residue. Biophys J. 2012;103:L23–L25. doi: 10.1016/j.bpj.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wanderling S, Paduch M, Medovoy D, Singharoy A, McGreevy R, Villalba-Galea CA, Hulse RE, Roux B, Schulten K, Kossiakoff A, Perozo E. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, Mackinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Madden TL, Tatusov RL, Zhang J. Applications of network BLAST server. Methods Enzymol. 1996;266:131–141. doi: 10.1016/S0076-6879(96)66011-X. [DOI] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenböck G. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36:463–474. doi: 10.1016/S0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Sakai R, Repunte-Canonigo V, Raj CD, Knöpfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur J Neurosci. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–741. doi: 10.1016/S0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, Lin MZ. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F, Pathak MM, Isacoff EY. Voltage-sensing arginines in a potassium channel permeate and occlude cation-selective pores. Neuron. 2005;45:379–388. doi: 10.1016/j.neuron.2004.12.047. [DOI] [PubMed] [Google Scholar]
- Tombola F, Pathak MM, Gorostiza P, Isacoff EY. The twisted ion-permeation pathway of a resting voltage-sensing domain. Nature. 2007;445:546–549. doi: 10.1038/nature05396. [DOI] [PubMed] [Google Scholar]
- Villalba-Galea CA, Frezza L, Sandtner W, Bezanilla F. Sensing charges of the Ciona intestinalis voltage-sensing phosphatase. J Gen Physiol. 2013;142:543–555. doi: 10.1085/jgp.201310993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, McMahon S, Zhang Z, Jackson MB. Hybrid voltage sensor imaging of electrical activity from neurons in hippocampal slices from transgenic mice. J Neurophysiol. 2012;108:3147–3160. doi: 10.1152/jn.00722.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. doi: 10.1016/S0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]