Abstract
Osteoarthritis (OA) is a common and troublesome disease among the elderly, and is characterized by extracellular matrix (ECM) degradation. The function of the long non-coding RNA X-inactive-specific transcript (XIST) and its working mechanism in ECM degradation remains unclear. In the present study, XIST was revealed to be upregulated in OA specimens and in articular chondrocytes (ACs) derived from OA tissue (AC/OA) and interleukin-1β (IL-1β)-treated ACs. Loss-of-function experiments demonstrated that downregulation of XIST suppressed the degradation of the ECM in AC/OA and AC/IL-1β-5.0 cells. Furthermore, XIST, matrix metalloproteinase 13 (MMP-13) and ADAM metallopeptidase with thrombospondin type 1 motif 5 (ADAMTS5) were identified as targets of microRNA (miR)-1277-5p, and the reciprocal inhibitive effect between XIST and miR-1277-5p was elucidated. Furthermore, the role of XIST in ECM degradation was confirmed to be functioning as a competing endogenous RNA (ceRNA) of miR-1277-5p. Finally, the protective effect of the downregulation of XIST on ECM degradation was verified in an OA rat model. In conclusion, the present study suggests that XIST promotes MMP-13 and ADAMTS5 expression, indicating ECM degradation, by functioning as a ceRNA of miR-1277-5p in OA. The present study proposed a novel potential target with a new working mechanism in molecular treating of OA.
Keywords: long non-coding RNA X-inactive-specific transcript, miR-1277-5p, matrix metalloproteinase -13/ADAM metallopeptidase with thrombospondin type 1 motif 5, competing endogenous RNA, osteoarthritis
Introduction
Osteoarthritis (OA) is a common disease in elderly individuals worldwide, and is characterized by articular cartilage degradation, which leads to joint pain, particularly in the knee joint (1). Although OA is primarily associated with aging, diverse etiologies also contribute to the progression of this disease. Various reasons, including congenital factors, biomechanical imbalance, variation of gene expression in chondrocytes and secondary acquired chondral or osteochondral injuries, are involved in its pathological changes (2). During the development of OA, imbalanced anabolism and catabolism of the extracellular matrix (ECM) facilitates its degradation and aggravates the lesion of cartilage overlying the bone at the joint surface (3). During the progression of ECM degradation, catabolic factors such as matrix metalloproteinase 13 (MMP-13) and ADAM metallopeptidase with thrombospondin type 1 motif 5 (ADAMTS5)-mediated loss of type II collagen/aggrecan contribute significantly to this process (4). Verma and Dalal (5) revealed that ADAMTS-5 was responsible for aggrecan degradation in a human model of OA. Chen et al (6) reported that the degradation and destruction of type II collagen caused by MMP-13 was the core factor in the occurrence and development of OA. Therefore, identifying the factors that can simultaneously regulate MMP-13 and ADAMTS5 is of particular importance.
Long non-coding RNAs (lncRNAs) are non-protein-coding transcripts that are longer than 200 nucleotides and are involved in the regulation of various molecular and cellular functions (7,8). As an lncRNA, the X-inactive-specific transcript (XIST) is widely reported to be an oncogene in numerous types of cancer, including colon, breast, bladder and prostate cancer (9-12). Fu et al (13) analyzed the differentially expressed lncRNAs in 6 pairs of OA tissue specimens using an lncRNA array assay and demonstrated that XIST was upregulated in OA tissue compared with normal cartilage (fold-change, 32.71). To date, the role that XIST serves in OA and its detailed molecular mechanism remains unclear.
Generally, lncRNAs function through chromatin regulation, histone modification, chromatin remodeling, genomic imprinting and as competing endogenous RNAs (ceRNAs) in post-transcriptional regulation (14-19). The ceRNA theory was first proposed by Salmena et al (20) in 2011 and hypothesized that all types of RNA transcripts could communicate with each other through a new 'language' mediated by microRNA (miR/miRNA) response elements. Gu et al (21) reported that XIST contributed to neuronal apoptosis by functioning as a ceRNA of the phosphatase and tensin homolog gene in rat spinal cord injury. Zhang and Xia (22) found that XIST inhibited the proliferation and mobility of osteosarcoma cells by competitively binding to miR-21-5p and leading to the upregulation of the programmed cell death 4 gene.
The present study focused on the facilitative role of XIST in ECM degradation. Furthermore, XIST was revealed to promote MMP-13/ADAMTS5 expression by functioning as a ceRNA of miR-1277-5p.
Materials and methods
Patients and tissue samples
The present study and the experimental protocols within were performed in compliance with ethical guidelines and approved by the Institute Research Medical Ethics Committee of the Central Hospital Affiliated to Shenyang Medical College, Shenyang, China (approval no. 2016JULY15-7). Written informed consent was obtained from the patients whose tissues were used in this study. Samples of cartilage from 40 patients with OA undergoing total knee arthroplasty (10 males and 30 females with an average age of 63 years), and non-OA cartilage from 20 patients undergoing total hip arthroplasty (diagnosis, femoral neck fracture; average age, 62 years) were collected at the Central Hospital Affiliated to Shenyang Medical College between January 2017 and January 2018. All the samples were obtained at the time of surgery and immediately sent to the central laboratory of Dalian Medical University, (Dalian, China) for further testing.
Articular chondrocyte (AC) isolation, culture and IL-1β treatment
Primary ACs were isolated from the collected cartilage tissues as described previously (23,24). The isolated ACs were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 mg/ml streptomycin in an incubator at 37°C and a humidified atmosphere containing 5% CO2. ACs isolated from non-OA and OA tissues were labeled AC and AC/OA, respectively. Interleukin-1β (IL-1β) treatment was used to simulate an OA chondrocyte model, according to a previous study (25). ACs treated with 1.0 and 5.0 ng/ml ectogenic recombinant human IL-1β (R&D Systems, Inc.) were labeled AC/IL-1β-1.0 and AC/IL-1β-5.0, respectively.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the tissue specimens and the ACs using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and RT reactions were performed at 44°C for 1 h then 92°C for 10 min using a Takara RNA PCR kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. PCR reactions containing SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.) were performed according to the manufacturer's protocol. GAPDH and U6 were used as the reference genes. PCR amplification conditions are as follows: 95°C for 5 min, 38 cycles of 95°C for 5 sec and 61°C for 30 sec. The primers used in this study (Guangzhou RiboBio, Co., Ltd.) are listed in Table I. The results were quantified using the 2-∆∆Cq method (26).
Table I.
Oligonucleotide and primer sequences used in this research.
| A, Primers | |
|
| |
| Target | Sequences (5′-3′) |
|
| |
| XIST forward primer | AGGGAGCAGTTTGCCCTACT |
| XIST reverse primer | CACATGCAGCGTGGTATCTT |
| MMP-13 forward primer | TAAGGAGCATGGCGACTTCT |
| MMP-13 reverse primer | GGTCCTTGGAGTGGTCAAGA |
| ADAMTS5 forward primer | GTGTCACATGAATGATGCCC |
| ADAMTS5 reverse primer | CGACCCTCAAGAACTTTTGC |
| GAPDH forward primer | CTCTGCTCCTCCTGTTCGAC |
| GAPDH reverse primer | GCGCCCAATACGACCAAATC |
| miR-1277-5p forward primer | ACACTCCAGCTGGGAAATATATATATATATGT |
| miR-1277-5p reverse primer | TGGTGTCGTGGAGTCG |
| U6 forward primer | CTCGCTTCGGCAGCACA |
| U6 reverse primer | AACGCTTCACGAATTTGCGT |
|
| |
| B, Oligonucleotides | |
|
| |
| Target | Sequences (5′-3′) |
|
| |
| XIST shRNA | GGCAATTTTTAATATTTAA |
| NC shRNA | GCTAGCGGTATTGGCCAA |
| miR-1277-5p mimics | AAATATATATATATATGTACGTAT |
| Mimic control | ATATGCATGCATTAGAATTGATTA |
| miR-1277-5p inhibitor | ATACGTACATATATATATATTTT |
| Inhibitor control | TATACGTACGTATATTGATTAAT |
XIST, X-inactive-specific transcript; MMP-13, matrix metalloproteinase 13; ADAMTS5, ADAM metallopeptidase with thrombospondin type 1 motif 5; miR, microRNA; NC, negative control.
Western blot analysis
Total protein was extracted using a radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich; Merck KGaA) and quantified using a bicinchoninic acid protein assay kit (Santa Cruz Biotechnology, Inc.). The samples (100 µg) were separated by 10% SDS-PAGE, transferred onto a polyvinylidene difluoride membrane, and then blocked for 1 h at room temperature by using of 5% bovine serum albumin (MP Biomedicals, LLC). The membranes were incubated with primary antibodies at 4°C overnight and subsequently with secondary antibodies [goat anti-rabbit immunoglobulin G horseradish peroxidase (HRP)-conjugated; cat. no. ab205718; dilution, 1:2,000; Abcam) at room temperature for 1 h. The primary antibodies used in the present study were the following: Anti-MMP-13 (cat. no. ab39012; 1:3,000 dilution), anti-ADAMTS5 (cat. no. ab231595; 1:250 dilution) and anti-GAPDH (cat. no. ab9485; 1:500 dilution; all Abcam). Following washing with 0.1% TBST 3 times, an ECL Western Blotting Substrate kit (cat. no. ab65623; Abcam) was used for chemiluminescence imaging and bands were analyzed with ImageJ software version 2 (National Institutes of Health).
Oligonucleotide transfection
XIST smart silencer short hairpin RNA (shRNA), negative control (NC) shRNA, miR-1277-5p mimics, mimic control, miR-1277-5p inhibitor and inhibitor control were chemically synthesized by Guangzhou RiboBio Co., Ltd. The plasmids wt-pcDNA-XIST and mut-pcDNA-XIST were purchased from Shanghai GenePharma Co., Ltd. When the cells reached 80% confluence, the oligonucleotides (100 nM) were transfected into the cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The ACs were harvested 48 h after transfection for further experiments. The sequences of the above oligonucleotides were also listed in Table I.
Immunofluorescence analysis
An immunofluorescence assay was performed as reported previously (24). Briefly, AC/OA and AC/IL-1β-5.0 cells (5×105/ml) were seeded onto glass coverslips (0.8×0.8 cm) and cultured until they reached 50-60% confluence. The culture medium was discarded, and the coverslips were washed twice with PBS, fixed with 4% paraformaldehyde for 15 min at room temperature, blocked with 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA) for 1 h at room temperature and incubated with primary antibodies (anti-MMP13: Dilution 1:500; Abcam; cat. no. ab39012; and anti-ADAMTS5: Dilution 1:100; Abcam; cat. no. ab231595) at 4°C overnight. The next day, following incubation with fluorescent Alexa Fluor® 555 secondary antibody (dilution 1:500; cat. no. A-21428; Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h and DAPI (Cell Signaling Technology, Inc.) for 5 min at room temperature, the coverslips were observed, and images were captured using a fluorescent microscope (Leica Microsystems GmbH). The images were analyzed using Image-Pro Plus version 6.0 software (Media Cybernetics, Inc.).
Bioinformatic analysis
The online software TargetScan (version 7.1; www.targetscan.org/vert_71/) was utilized to predict the miR-1277-5p binding sites on MMP-13 and ADAMTS5 3′-untranslated regions (3′UTRs). In addition, another online software LncBase (version 2; carolina.imis. athena-innovation.gr) was applied to predict the miR-1277-5p binding sites on XIST.
Dual luciferase reporter assay
XIST, MMP-13 and ADAMTS5 reporter plasmids wt-XIST, mut-XIST, wt-MMP-13, mut-MMP-13, wt-ADAMTS5 and mut-ADAMTS5 were purchased from Shanghai GenePharma Co., Ltd. When the cells reached 60-80% confluence, the aforementioned reporter plasmids as well as the miR-1277-5p mimics and mimic control were co-transfected into the cells using Lipofectamine 2000™. The luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega Corporation) 48 h after co-transfection, according to the manufacturer's protocol.
RNA immunoprecipitation (RIP) assay
An RIP assay was conducted as previously described, using the Magna RNA-binding protein immunoprecipitation kit (Merck KGaA) (27). Briefly, whole-cell lysate was incubated with RIP buffer containing magnetic beads conjugated with human anti-argonaute-2 antibody (cat. no. ab156870; dilution 1:200; Abcam), or normal mouse immunoglobulin G (cat. no. ab188776; dilution 1:200; Abcam) as a negative control. The samples were incubated with proteinase K and the immunoprecipitated RNA was then isolated. The RNA concentration was measured by spectrophotometry (wavelength, A260 nm) and the RNA quality was assessed using a BioAnalyzer (Agilent Technologies, Inc.). Furthermore, the purified RNA was analyzed by qPCR to confirm the presence of the binding targets.
Establishment of an OA rat model
Lentivirus-mediated XIST silencing plasmids (lenti-siXIST) and the corresponding negative control plasmids (lenti-vector) were chemically synthesized by Hanbio (Hanbio Biotechnology Co., Ltd.). A total of 30 male rats (~8 weeks old) with bodyweight of 300-325 g were purchased from Shanghai SLAC Laboratory Animal Co., Ltd and housed at 24°C with 40-50% humidity, a 12-h light/dark cycle and free access to food and water. The Institute Research Medical Ethics Committee of the Central Hospital Affiliated to Shenyang Medical College approved the animal experiments. A destabilization of the medial meniscus (DMM) method was used to simulate OA in rats, as previously reported (28). Briefly, 30 rats were randomly divided into 3 groups: Sham surgery group (n=10, non-DMM and intra-articular injection of sterile saline), lenti-vector group (n=10, DMM and intra-articular injection of lenti-vector) and lenti-siXIST group (n=10, DMM and intra-articular injection of lenti-siXIST). A 3-mm longitudinal incision was made over the distal patella to the proximal tibial plateau on the right knee of each rat, the fat pad over the inter-condylar area was bluntly resected, and the medial meniscus was transected using a #15 surgical blade. The medial retinaculum was repaired, and the capsule and skin were layer-sutured. The rats were injected with 1×108 plaque-forming units of lenti-vector, lenti-siXIST or the equivalent volume of sterile saline into the operated knee joint cavities fortnightly. All the rats were housed in groups of five mice per cage and fed with free water and food in room temperature normal atmosphere. After 8 weeks, the rats were sacrificed for further experimentation.
Sample collection and immunohistochemistry (IHC)
Samples were collected from the rats and IHC was performed as reported previously (24). Briefly, cartilage samples of the tibial plateau containing 2-cm thick subchondral bone were collected and sliced following 4% paraformaldehyde fixation for 4 h at room temperature, 10% EDTA (pH 7.3) decalcification, dehydration and paraffin embedding. For Safranin-O staining, the slices (5-µm thickness) were stained with 1% Safranin-O for 1 min at room temperature using a Safranin-O staining kit (Nanjing KeyGen Biotech Co., Ltd.) according to the manufacturer's protocol. The expression of MMP-13/ADAMTS5 proteins were detected by IHC staining as previously described (29). Briefly, slices were incubated with primary antibodies anti-MMP-13 (cat. no. ab39012; dilution 1:200), anti-ADAMTS5 (cat. no. ab231595; dilution 1:200; both Abcam) at 4°C overnight, and secondary antibodies at 37°C for 30 min consecutively. The samples were then incubated with streptavidin horseradish peroxidase, stained with 3, 3-diaminobenzidine for 1 min at room temperature, counterstained with hematoxylin for 30 sec at room temperature, dehydrated in a graded ethanol series (absolute ethyl alcohol, 95% ethanol and 85% ethanol, for 3 min each), and finally mounted.
Statistical analysis
All experiments were repeated in triplicate and all data from three independent experiments are expressed as the mean ± standard deviation. GraphPad Prism version 5.0 (GraphPad Software, Inc.) software and SPSS version 19.0 (IBM Corp.) were used for the statistical analysis. The association between miR-1277-5p and XIST was assessed using a Pearson's correlation analysis. Means of different groups were compared using a one-way analysis of variance test and a followed Bonferroni's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of XIST in OA tissues, AC/OA cells and IL-1β-treated ACs
The expression of XIST in the 40 clinical OA and 20 non-OA cartilage specimens was detected by RT-qPCR. The levels of this lncRNA were significantly elevated in the OA samples compared with those measured in the non-OA samples (P<0.001; Fig. 1A). Furthermore, the expression of XIST was determined in the cultured AC/OA, AC/IL-1β-1.0, AC/IL-1β-5.0 and normal ACs. As observed in Fig. 1B, compared with the normal ACs, the XIST expression was significantly increased in AC/OA, AC/IL-1β-1.0 and AC/IL-1β-5.0 cells (P<0.001).
Figure 1.
Expression of XIST in OA tissues, OA chondrocytes and IL-1β-treated ACs. (A) Expression of XIST in the collected non-OA (n=20) and OA (n=40) tissue specimens was determined by RT-qPCR using GAPDH as an internal control. (B) Expression of XIST in ACs, AC/OA, and ACs treated with 1.0 and 5.0 ng/ml IL-1β (AC/IL-1β-1.0 and AC/IL-1β-5.0) was also detected by RT-qPCR. All data are normalized to the AC group. The error bars represent standard deviation, (n=3). ***P<0.001. XIST, X-inactive-specific transcript; OA, osteoarthritis; IL-1β, interleukin-1β; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; AC, articular chondrocytes.
Downregulation of XIST inhibits ECM degradation in AC/OA and AC/IL-1β-5.0 cells
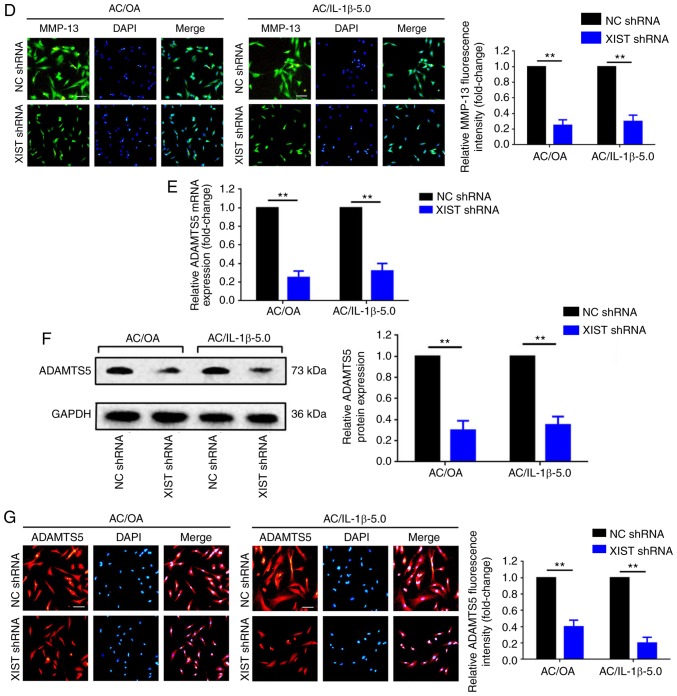
The findings that XIST was overex-pressed in OA tissues and in OA chondrocytes (AC/OA and AC/IL-1β-5.0) indicated that XIST may contribute to the pathological mechanism of OA. Therefore, the function of XIST in AC/OA cells was explored. First, the expression of XIST was significantly downregulated using XIST shRNA in AC/OA and AC/IL-1β-5.0 cells, which was verified by RT-qPCR (P<0.01; Fig. 2A). Secondly, the expression of MMP-13, a marker of ECM degradation, was separately measured by RT-qPCR, western blot analysis and immunofluorescence staining. The downregulation of XIST led to the inhibition of MMP-13 expression in the AC/OA and in AC/IL-1β-5.0 cells (Fig. 2B-D). Thirdly, another marker of ECM degradation, ADAMTS5, was also investigated. Similar to the results of MMP-13, the knockdown of XIST resulted in a decrease in the expression of ADAMTS5 (Fig. 2E-G). Overall, these findings imply that the downregulation of XIST decreases ECM degradation in AC/OA and AC/IL-1β-5.0 cells.
Figure 2.
Downregulation of XIST decreases extracellular matrix degradation in AC/OA and AC/IL-1β-5.0 cells. (A) Downregulation of XIST by an RNA interference assay was confirmed by RT-qPCR. MMP-13 expression decreased following knockdown of XIST, as determined by (B) RT-qPCR, (C) western blot analysis (lysates were analyzed by immunoblotting with MMP-13 and GAPDH antibodies; the left panel presents data from 3 independent experiments) and (D) immunofluorescence analysis. The knockdown of XIST also downregulated the expression of ADAMTS5 as determined by (E) RT-qPCR, (F) western blot analysis (lysates were analyzed by immunoblotting with ADAMTS5 and GAPDH antibodies; the left panel presents data from 3 independent experiments) and (G) immunofluorescence analysis. All data were normalized to the negative control shRNA group. Magnification, ×100; scale bar, 100 µm. The error bars represent standard deviation (n=3). **P<0.01. XIST, X-inactive-specific transcript; AC, articular chondrocyte; OA, osteoarthritis; IL-1β, interleukin-1β; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; MMP-13, matrix metalloproteinase 13; ADAMTS5, ADAM metallopeptidase with thrombospondin type 1 motif 5; NC, negative control; sh, short hairpin.
miR-1277-5p attenuates ECM degradation by directly targeting MMP-13 and ADAMTS5 in AC/OA and AC/IL-1β-5.0 cells
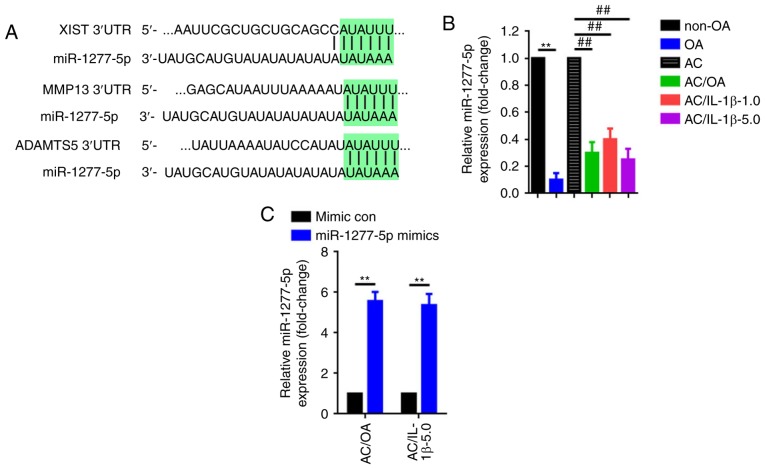
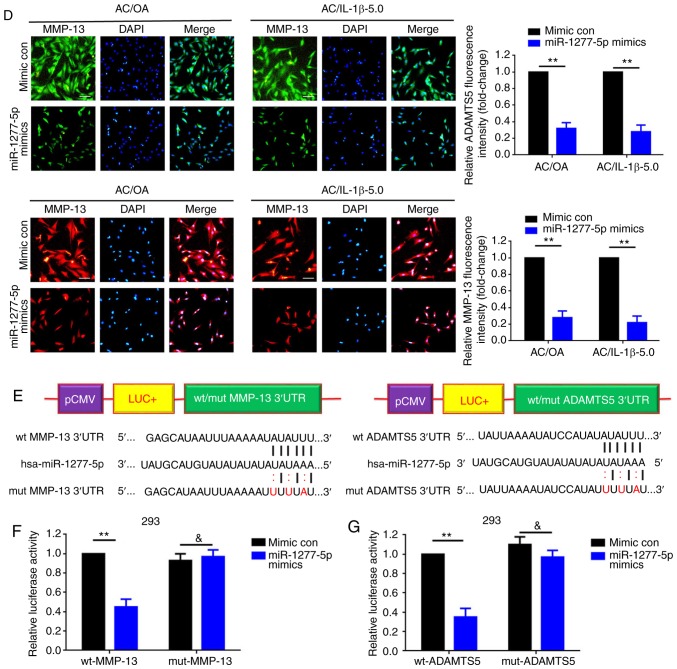
The predominant theory of how lncRNAs work is that they function as ceRNAs. The present study questioned whether XIST exerts its function through certain miRNAs. Through an online bioinformatic prediction via TargetScan and LncBase, miR-1277-5p was selected due to its potential binding for MMP-13, ADAMTS and XIST (Fig. 3A). The expression levels of miR-1277-5p were confirmed to be significantly decreased in OA tissues (P<0.01) and in AC/OA, AC/IL-1β-1.0 and AC/IL-1β-5.0 cells compared with non-OA tissue samples and ACs, respectively (Fig. 3B). Furthermore, the upregulation of miR-1277-5p led to the attenuation of the expression of MMP-13 and ADAMTS5, two markers of ECM degradation, in AC/OA and AC/IL-1β-5.0 cells (Fig. 3C and D). In addition, wild-type and mutant luciferase plasmids of MMP-13 (wt-MMP-13 and mut-MMP-13) and ADAMTS5 (wt-ADAMTS5 and mut-ADAMTS5) were constructed, containing wild-type and mutant miR-1277-5p binding sites, respectively (Fig. 3E). Finally, the luciferase reporter assay confirmed that miR-1277-5p directly targets MMP-13 and ADAMTS5 via the same binding site (Fig. 3F and G).
Figure 3.
miR-1277-5p attenuates extracellular matrix degradation by directly targeting MMP-13 and ADAMTS5 in AC/OA and AC/IL-1β-5.0 cells. (A) XIST, MMP-13 and ADAMTS5 share the same miR-1277-5p binding site within their 3′UTRs. (B) Downregulation of miR-1277-5p in OA tissue specimens and in AC/OA, AC/IL-1β-1.0 and AC/IL-1β-5.0 cells, as confirmed by reverse transcription-quantitative polymerase chain reaction. **P<0.01 vs. non-OA group; ##P<0.01 vs. AC group. (C) miR-1277-5p was upregulated by transfection of miR-1277-5p mimics as determined by reverse transcription-quantitative polymerase chain reaction. **P<0.01 vs. mimic con group. (D) Upregulation of miR-1277-5p (transfection of miR-1277-5p mimics) led to a notable decrease in MMP-13 and ADAMTS5 expression, as measured by immunofluorescence analysis. **P<0.01 vs. mimic control group; magnification, ×100; scale bar, 100 µm. (E) Diagram of the luciferase reporter plasmids with the wt- or mut-MMP-13 and -ADAMTS5 3′UTRs. (F) Compared with the effect of the mimic con, co-transfection of miR-1277-5p mimic and wt-MMP-13 led to a distinct decrease in fluorescence, but the suppressive effect was rescued by a mutation in the putative miR-1277-5p binding sites in the MMP-13 3′UTR (co-transfection of mut-MMP-13 and miR-1277-5p mimic), as verified by dual luciferase assays. **P<0.01 and &P>0.05 vs. mimic con group. (G) The same effects were observed during the verification of the interaction between miR-1277-5p and ADAMTS5 3′UTR. **P<0.01 and &P>0.05 vs. mimic con group. All error bars represented standard deviation (n=3). miR, microRNA; MMP-13, matrix metallopro-teinase 13; ADAMTS5, ADAM metallopeptidase with thrombospondin type 1 motif 5; AC, articular chondrocyte; OA, osteoarthritis; IL-1β, interleukin-1β; UTR, untranslated region; wt, wild type; mut, mutant; con, control; LUC+, luciferase.
XIST and miR-1277-5p exhibit reciprocal repression and XIST is a target of miR-1277-5p
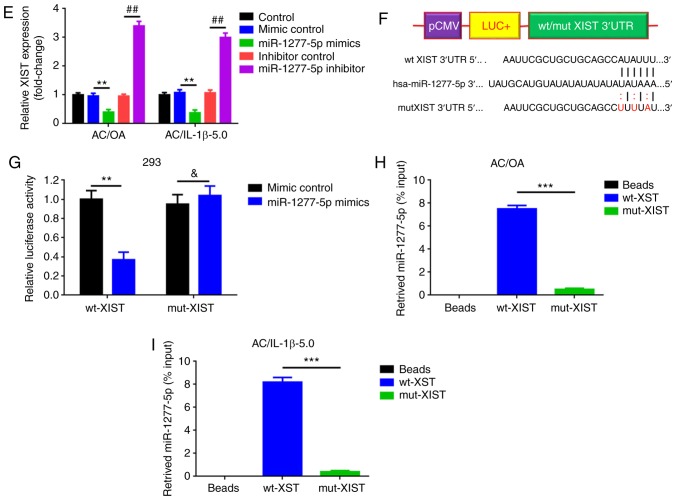
The downregulation of XIST and the upregulation of miR-1277-5p were revealed to present a similar protective effect on ECM degradation. This finding highlighted the question of the nature of the association between the two RNAs. Initially, a negative correlation was demonstrated (Fig. 4A). Subsequently, the upregulation and downregulation of XIST were revealed to significantly inversely regulate miR-1277-5p expression (P<0.01; Fig. 4B and C). An increase or a decrease of miR-1277-5p also affected XIST expression in the opposite manner (Fig. 4D and E). These results indicate that XIST and miR-1277-5p repress each other in a reciprocal manner. As predicted, a binding site was confirmed between XIST 3′-UTR and miR-1277-5p. Furthermore, a luciferase reporter assay was used to investigate whether miR-1277-5p targets XIST. The luciferase reporting plasmids containing a wild-type and mutant miR-1277-5p binding sites (wt-XIST and mut-XIST; Fig. 4F) were constructed as follows: Wt-XIST or mut-XIST and miR-1277-5p mimics were co-transfected into 293 cells while the mimic control was used as a negative control, and luciferase activity changes were measured. Co-transfection of wt-XIST and miR-1277-5p mimics led to a significant decrease in luminescence, compared with that detected in the mimic control (P<0.01; Fig. 4G). However, the luminescence signal was significantly recovered in the co-transfection of mut-XIST and miR-1277-5p mimics (P<0.05). Ultimately, an RNA pull-down assay was conducted to confirm the interaction between XIST and miR-1277-5p. As demonstrated in Fig. 4H and I, wt-XIST, but not mut-XIST, could pull down miR-1277-5p. Together, these findings reveal that miR-1277-5p directly targets XIST.
Figure 4.
XIST and miR-1277-5p repress each other and XIST is a target of miR-1277-5p. (A) The expression of XIST was inversely correlated with that of miR-1277-5p. The overexpression and downregulation of (B) XIST negatively regulated (C) miR-1277-5p expression, as determined by RT-qPCR. **P<0.01 and ##P<0.01 vs. the corresponding control group (n=3). Increasing and decreasing (D) miR-1277-5p levels negatively affected (E) XIST expression, as indicated by RT-qPCR. **P<0.01 and ##P<0.01 vs. control group (n=3). (F) Diagram of the luciferase reporter plasmids with the wt- or mut-XIST 3′UTR. (G) The targeted binding between XIST 3′UTR and miR-1277-5p was verified by a dual luciferase assay. **P<0.01 and &P>0.05 vs. mimic control group. (H) wt-XIST, but not mut-XIST, could pull down miR-1277-5p, as determined by an RNA pull-down assay in AC/OA and (I) AC/IL-1b-5.0. ***P<0.001 vs. the wt-XIST group (normalized to the beads group). The error bars represent standard deviation. (n=3). XIST, X-inactive-specific transcript; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; wt, wild type; mut, mutant; UTR, untranslated region.
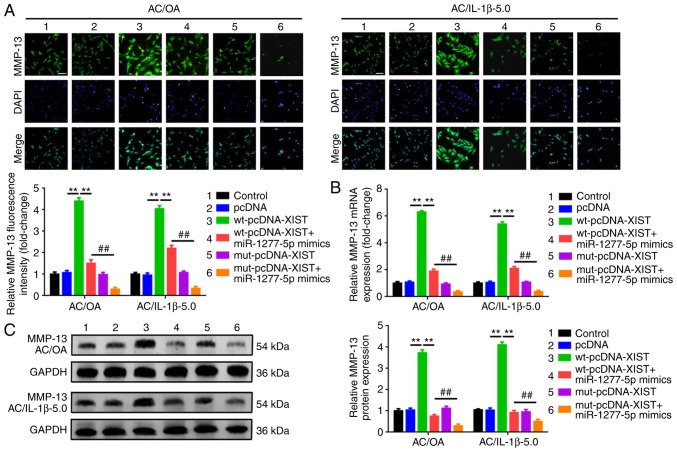
XIST promotes ECM degradation by acting as a ceRNA of miR-1277-5p in AC/OA and AC/IL-1β-5.0 cells
To confirm whether the effect of XIST on ECM degradation is achieved through the miR-1277-5p pathway with a ceRNA mechanism, the expression of MMP-13 and ADAMTS5 was determined following a dual intervention with XIST and miR-1277-5p. Wild-type and mutant XIST overexpression plasmids wt-pcDNA-XIST and mut-pcDNA-XIST were co-transfected with an miR-1277-5p mimic into AC/OA and AC/IL-1β-5.0 cells. The expression of MMP-13 was determined by immunofluorescence staining, RT-qPCR and western blotting. As observed in Fig. 5A-C, the transfection of wt-pcDNA-XIST led to a notable elevation of MMP-13 mRNA and protein expression. On the other hand, when the putative miR-1277-5p binding site in XIST was mutated, the facilitative effect was not present. Even more convincingly, when the miR-1277-5p mimic was co-transfected, the facilitative effect of wt-pcDNA-XIST on the MMP-13 expression was significantly weakened (P<0.01). The expression of ADAMTS5 presented a similar tendency to MMP-13 (Fig. 5D-F). Combined with the reciprocally repressing association between XIST and miR-1277-5p, XIST was confirmed to promote ECM degradation by acting as a ceRNA of miR-1277-5p in AC/OA and AC/IL-1β-5.0 cells.
Figure 5.
XIST promotes extracellular matrix degradation by acting as a competing endogenous RNA of miR-1277-5p in AC/OA and AC/IL-1β-5.0 cells. Normal XIST overexpression plasmids (wt-pcDNA-XIST) led to a notable upregulation of MMP-13, but the effect was attenuated by the overexpression of miR-1277-5p (co-transfection of wt-pcDNA-XIST and miR-1277-5p mimics). When the putative miR-1277-5p binding site in XIST was mutated (transfection of mut-pcDNA-XIST), the facilitative effect was eliminated. The phenomenon was detected by (A) immunofluorescence analysis, (B) RT-qPCR and (C) western blot analysis (lysates were analyzed by immunoblotting with MMP-13 and GAPDH antibodies; the left panel presents data from 3 independent experiments). **P<0.01 vs. wt-pcDNA-XIST; ##P<0.01 vs. wt-pcDNA-XIST + miR-1277-5p mimic group. wt-pcDNA-XIST, but not mut-pcDNA-XIST, promoted ADAMTS5 expression, and the effect was repressed when the putative miR-1277-5p binding site in XIST was mutated (transfection of mut-pcDNA-XIST). As with MMP-13, elevation of miR-1277-5p also reversed the facilitative effect of wt-pcDNA-XIST on ADAMTS5 expression, as determined by (D) immunofluorescence analysis, (E) RT-qPCR and (F) western blot analysis (lysates were analyzed by immunoblotting with ADAMTS5 and GAPDH antibodies; the left panel presents data from 5 independent experiments). **P<0.01 vs. wt-pcDNA-XIST; ##P<0.01 vs. wt-pcDNA-XIST + miR-1277-5p mimic group. All data were normalized to the control group, and all error bars represent standard deviation (n=3). XIST, X-inactive-specific transcript; miR, microRNA; AC, articular chondrocyte; OA, osteoarthritis; IL-1β, interleukin-1β; wt, wild type; mut, mutant; MMP-13, matrix metalloproteinase 13; ADAMTS5, ADAM metallopeptidase with thrombospondin type 1 motif 5; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
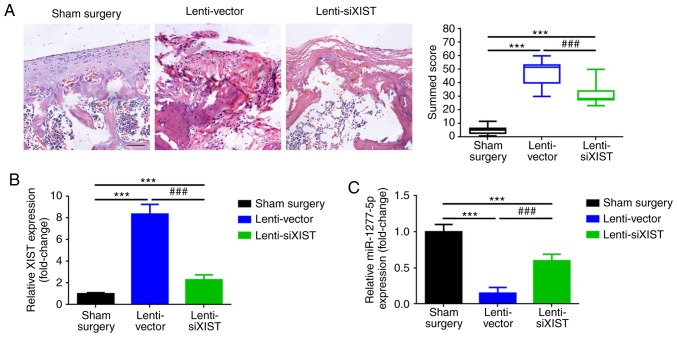
Depression of XIST inhibits ECM degradation in vivo
A DMM mouse model was used to further investigate the function of XIST in ECM degradation in vivo, according to a previous study (27). Lenti-vector and lenti-siXIST were injected into the joint cavities of the animals. The results of Safranin-O staining revealed that, compared with the results of the sham surgery group, the resection of the medial meniscus led to an elevated Safranin-O staining, a rougher articular surface and a significantly increased summed score (P<0.001; Fig. 6A). On the other hand, compared with the lenti-vector group, the group with silenced XIST expression presented with decreased Safranin-O staining, smoother articular surface and a significantly decreased summed score (P<0.001), indicating that the knockdown of XIST led to a protective effect on the development of OA. In each animal group, the expression levels of XIST and miR-1277-5p were measured by RT-qPCR (Fig. 6B-C), and the level of ECM degradation (expression of MMP-13 and ADAMTS5) was determined by IHC, RT-qPCR and western blot analysis. DMM resulted in a significant elevation of MMP-13 and ADAMTS5, compared with that in the sham surgery group (P<0.001; Fig. 6D and E), whereas the knockdown of XIST led to a decrease in the expression of MMP-13 and ADAMTS5 (ECM degradation), compared with that observed in the lenti-vector group. In conclusion, as presented in the schematic of the mechanism in Fig. 7, the present results suggest that the downregulation of XIST inhibits ECM degradation during the formation of OA.
Figure 6.
Depression of XIST inhibited extracellular matrix degradation in vivo. (A) DMM (lenti-vector and lenti-siXIST group) resulted in increased Safranin-O staining and an elevated summed score compared with the sham surgery group, whereas a certain cartilage-protective effect was presented in the lenti-siXIST group compared with the lenti-vector group. (B) Elevated XIST and (C) depressed miR-1277-5p levels were attributed to DMM, and the injection of lenti-siXIST led to the inhibition of XIST and the promotion of miR-1277-5p compared with the lenti-vector group, as determined by RT-qPCR. (D) In contrast with the sham surgery group, increased MMP-13 expression was detected in the DMM groups (lenti-vector and lenti-siXIST group). In addition, downregulation of XIST (injection of lenti-siXIST) inhibited MMP-13 expression, as detected by IHC, RT-qPCR and western blot analysis (lysates were analyzed by immunoblotting with MMP-13 and GAPDH antibodies; the left panel presents data from 5 independent experiments). (E) Upregulated ADAMTS5 expression was also observed in the DMM groups (lenti-vector and lenti-siXIST group) compared with the surgery group, whereas, compared with the lenti-vector group, the injection of lenti-siXIST led to a decrease in ADAMTS5 expression, as confirmed by IHC, RT-qPCR and western blot analysis (lysates were analyzed by immunoblotting with ADAMTS5 and GAPDH antibodies; the left panel presents data from 5 independent experiments). ***P<0.001 vs. the sham surgery group; ###P<0.001 vs. the lenti-vector group. All data were normalized to the control group, and all error bars represent standard deviation (n=3). XIST, X-inactive-specific transcript; DMM, destabilization of the medial meniscus; siXIST, XIST silencing; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; ADAMTS5, ADAM metallopeptidase with thrombospondin type 1 motif 5; MMP-13, matrix metalloproteinase 13; IHC, immu-nohistochemistry.
Figure 7.
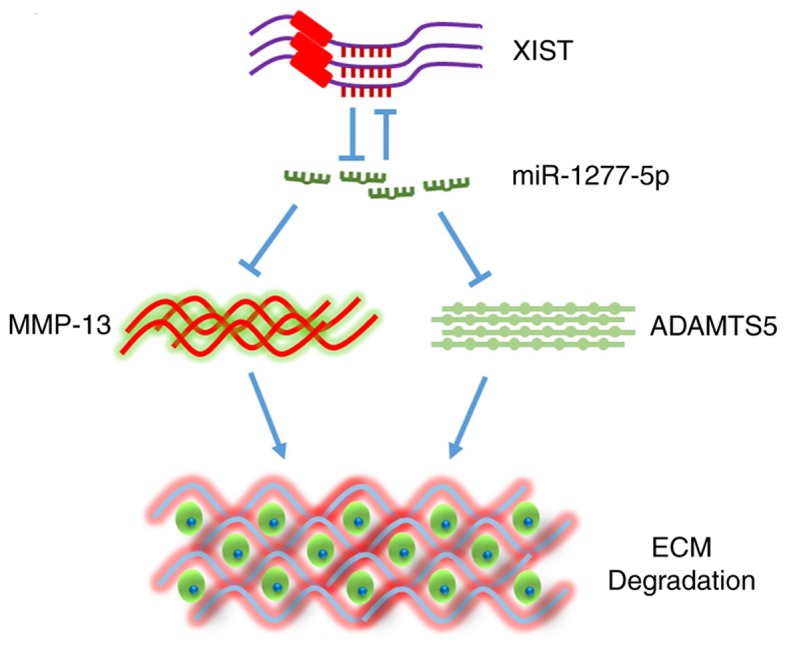
Schematic diagram of the mechanism concluded from the present study. XIST, X-inactive-specific transcript; ADAMTS5, ADAM metallopeptidase with thrombospondin type 1 motif 5; MMP-13, matrix metalloproteinase 13; ECM< extracellular matrix; miR, microRNA.
Discussion
OA is a common degenerative disease of the joints, characterized by cartilage loss, subchondral bone remodeling and osteophyte development. A number of factors contribute to the composition and structural changes of the ECM during the development and progression of OA. As the only cell type in cartilage, chondrocytes participate in the catabolic activities that ultimately lead to the degradation of cartilaginous ECM (30). During ECM degradation, catabolic factors including MMP-13 and ADAMTS5 mediating the loss of type II collagen/aggrecan, contribute significantly to the process (4). As an ECM-degrading enzyme, MMP-13 is structurally characterized among the zinc-dependent endopeptidases that degrade various components of the ECM (31). Numerous studies have demonstrated that MMP-13 is a key regulator in the pathology of OA (32-34). ADAMTS5, a member of the ADAMTS family, contains a domain arrangement that consists of a pro-domain, a catalytic metalloproteinase domain, a disintegrin-like domain, a cysteine-rich domain and a spacer domain (5). ADAMTS5, which can degrade the cartilage proteoglycan aggrecan, is widely reported as the most efficient aggrecanase and has been generally considered to be the most likely candidate for its role in the pathological mechanisms of OA (35-38). In the present study, MMP-13 and ADAMTS5 were used as markers of ECM degradation, as previously reported.
miR-1277-5p is a miRNA encoded by a site located on chromosome Xq24. The function of miR-1277-5p is presently not well understood. Budak et al (39) reported that miR-1277-5p was downregulated in chronic brucellosis, but its function in this infectious disease remains unknown. In a study of colorectal cancer, Motieghader et al (40) revealed that miR-1277-5p was downregulated in adenocarcinoma compared with high-grade intraepithelial neoplasia. Through deep sequencing and northern blot analyses, Crowe et al (41) demonstrated that miR-1277-5p was a novel miRNA which was downregulated in OA. The results of the present study revealed that miR-1277-5p expression was downregulated in OA specimens and in AC/OA, AC/IL-1β-1.0 and AC/IL-1β-5.0 cells. Furthermore, the detection of markers MMP-13 and ADAMTS5 demonstrated that the upregulation of miR-1277-5p could reverse ECM degradation. It is well known that miRNAs exert their functions by directly or indirectly regulating their targets (42-44). In the present study, the results of the constructed luciferase assay verified that MMP-13 and ADAMTS5 are targets of miR-1277-5p by sharing the same binding sites within their 3′UTRs. These outcomes indicate that miR-1277-5p may function as a protective factor in OA development.
It is widely accepted that non-coding RNAs lncRNAs, circular RNAs and miRNAs are involved in numerous diseases, including OA (45-48). The lncRNA XIST has been extensively studied in a number of types of cancer, including colorectal cancer, pancreatic cancer, osteosarcoma, non-small cell lung cancer and bladder cancer (9,49-52). To date, studies on XIST and OA are rare. In the present study, XIST was revealed to be upregulated in OA in clinical tissue samples as well as cell lines. Subsequently, a loss-of-function experiment demonstrated that the knockdown of XIST suppressed ECM degradation in AC/OA and AC/IL-1β-5.0 cells, as determined by measuring the levels of markers MMP-13 and ADAMTS5, indicating that XIST promotes OA development. Now a prevalent theory in the field of non-coding RNAs, the mechanism of ceRNAs between lncRNAs and miRNAs was first proposed by Salmena et al (20) in 2011. Li et al (25) reported that lncRNA CIR promoted chondrocyte ECM degradation in OA by acting as a ceRNA of miR-27b. Li et al (53) revealed that lncRNA plasmacytoma variant translocation 1 regulated the apoptosis of chondrocytes by acting as a sponge for miR-488-3p in OA. In the present study, an online bioinformatic analysis predicted that XIST contains a similar binding site for miR-1277-5p as MMP-13 and ADAMTS5, implying that XIST may be a ceRNA of miR-1277-5p. The ensuing overexpression and downregulation experiments revealed a reciprocal suppression effect between XIST and miR-1277-5p. The subsequent co-localization and luciferase assays verified that XIST is a direct target of miR-1277-5p. Furthermore, through a series of antisense experiments, XIST was confirmed to exert its effect on ECM degradation (increased expression of MMP-13 and ADAMTS5) by acting as a ceRNA of miR-1277-5p. Finally, the constructed OA mouse model confirmed the facilitative effect of XIST on ECM degradation in vivo. Based on the present outcomes, it was concluded that XIST promotes ECM degradation by functioning as a ceRNA of miR-1277-5p in OA.
The development of OA is a complicated process covering broad mechanisms and a number of molecules. The results of the present study suggest that XIST promotes ECM degradation by working as a ceRNA of miR-1277-5p. These findings may provide a novel molecular target in treating OA.
Acknowledgments
Not applicable.
Abbreviations
- OA
osteoarthritis
- ECM
extracellular matrix
- lncRNA
long non-coding RNA
- XIST
X-inactive-specific transcript
- miR/miRNA
microRNA
- AC
articular chondrocyte
- MMP-13
matrix metalloproteinase 13
- ADAMTS5
ADAM metallopeptidase with thrombospondin type 1 motif 5
- ceRNA
competing endogenous RNA
- IL-1β
interleukin-1β
- RIP
RNA immunoprecipitation
- DMM
destabilization of the medial meniscus
Funding
The present study was supported by the Foundation for Innovative Talent in Higher Education of Liaoning (grant no. LR2017056) and the China Postdoctoral Science Foundation (grant no. 2016M591437).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YW and ZZ conceived the experiments; TW, YL and XH performed the experiments; YW and WZ analyzed the data; YW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study and the associated experimental protocols (both human and animal experiments) were performed in compliance with ethical guidelines and approved by the Institute Research Medical Ethics Committee of the Central Hospital Affiliated to Shenyang Medical College, Shenyang, China (approval no. 2016JULY15-7). All OA tissues and non-OA tissues were also used in accordance with the Helsinki declaration. Written informed consent was signed by each patient before the study. All patients agreed that the data from their samples could be used for experimental studies and paper presentations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.van der Kraan PM. Osteoarthritis year 2012 in review: Biology. Osteoarthritis Cartilage. 2012;20:1447–1450. doi: 10.1016/j.joca.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Mobasheri A. Osteoarthritis year 2012 in review: Biomarkers. Osteoarthritis Cartilage. 2012;20:1451–1464. doi: 10.1016/j.joca.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Lee AS, Ellman MB, Yan D, Kroin JS, Cole BJ, van Wijnen AJ, Im HJ. A current review of molecular mechanisms regarding osteoarthritis and pain. Gene. 2013;527:440–447. doi: 10.1016/j.gene.2013.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sofat N. Analysing the role of endogenous matrix molecules in the development of osteoarthritis. Int J Exp Pathol. 2009;90:463–479. doi: 10.1111/j.1365-2613.2009.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verma P, Dalal K. ADAMTS-4 and ADAMTS-5: Key enzymes in osteoarthritis. J Cell Biochem. 2011;112:3507–3514. doi: 10.1002/jcb.23298. [DOI] [PubMed] [Google Scholar]
- 6.Chen WX, Shan FJ, Jin HT, Wang PE, Xiao LW, Tong PJ. Research on application of determination of MMP-13 in osteoarthritis. Zhongguo Gu Shang. 2014;27:617–620. In Chinese. [PubMed] [Google Scholar]
- 7.Huynh NP, Anderson BA, Guilak F, McAlinden A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect Tissue Res. 2017;58:116–141. doi: 10.1080/03008207.2016.1194406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang SD, Lu J, Deng ZH, Li YS, Lei GH. Long noncoding RNAs in osteoarthritis. Joint Bone Spine. 2017;84:553–556. doi: 10.1016/j.jbspin.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Deng C, Zhang H, Zhang J, Peng B, Hu C. Long non-coding RNA XIST promotes cell growth and metastasis through regulating miR-139-5p mediated Wnt/β-catenin signaling pathway in bladder cancer. Oncotarget. 2017;8:94554–94568. doi: 10.18632/oncotarget.21791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Sun N, Zhang G, Liu Y. Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/β-catenin signaling pathway. Gene. 2018;665:141–148. doi: 10.1016/j.gene.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Zheng R, Lin S, Guan L, Yuan H, Liu K, Liu C, Ye W, Liao Y, Jia J, Zhang R. Long non-coding RNA XIST inhibited breast cancer cell growth, migration, and invasion via miR-155/CDX1 axis. Biochem Biophys Res Commun. 2018;498:1002–1008. doi: 10.1016/j.bbrc.2018.03.104. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Q, Hu W, Zhu W, Zhang F, Lin-Lin L, Liu C, Songyang YY, Sun CC, Li D. Long non coding RNA XIST as a prognostic cancer marker - A meta-analysis. Clin Chim Acta. 2018;482:1–7. doi: 10.1016/j.cca.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Fu M, Huang G, Zhang Z, Liu J, Zhang Z, Huang Z, Yu B, Meng F. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis Cartilage. 2015;23:423–432. doi: 10.1016/j.joca.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanduri C. Long noncoding RNAs: Lessons from genomic imprinting. Biochim Biophys Acta. 2016;1859:102–111. doi: 10.1016/j.bbagrm.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Wang L, Ding Y, Lu X, Zhang G, Yang J, Zheng H, Wang H, Jiang Y, Xu L. LncRNA structural characteristics in epigenetic regulation. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan SX, Zhang J, Xu QG, Yang Y, Zhou WP. Long noncoding RNA, the methylation of genomic elements and their emerging crosstalk in hepatocellular carcinoma. Cancer Lett. 2016;379:239–244. doi: 10.1016/j.canlet.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Zampetaki A, Mayr M. Long noncoding RNAs and angiogenesis: Regulatory information for chromatin remodeling. Circulation. 2017;136:80–82. doi: 10.1161/CIRCULATIONAHA.117.028398. [DOI] [PubMed] [Google Scholar]
- 20.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu S, Xie R, Liu X, Shou J, Gu W, Che X. Long coding RNA XIST contributes to neuronal apoptosis through the downregulation of AKT phosphorylation and is negatively regulated by miR-494 in rat spinal cord injury. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18040732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang R, Xia T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol. 2017;51:1460–1470. doi: 10.3892/ijo.2017.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat Protoc. 2008;3:1253–1260. doi: 10.1038/nprot.2008.95. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Yang T, Liu Y, Zhao W, Zhang Z, Lu M, Zhang W. Decrease of miR-195 promotes chondrocytes proliferation and maintenance of chondrogenic phenotype via targeting FGF-18 pathway. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18050975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YF, Li SH, Liu Y, Luo YT. Long noncoding RNA CIR promotes chondrocyte extracellular matrix degradation in osteoarthritis by acting as a sponge for Mir-27b. Cell Physiol Biochem. 2017;43:602–610. doi: 10.1159/000480532. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Cai H, Liu X, Zheng J, Xue Y, Ma J, Li Z, Xi Z, Li Z, Bao M, Liu Y. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36:318–331. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 28.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15:1061–1069. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Zhang Y, Yang T, Zhao W, Wang N, Li P, Zeng X, Zhang W. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8:59417–59434. doi: 10.18632/oncotarget.19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoff P, Buttgereit F, Burmester GR, Jakstadt M, Gaber T, Andreas K, Matziolis G, Perka C, Röhner E. Osteoarthritis synovial fluid activates pro-inflammatory cytokines in primary human chondrocytes. Int Orthop. 2013;37:145–151. doi: 10.1007/s00264-012-1724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chowdhury TT, Schulz RM, Rai SS, Thuemmler CB, Wuestneck N, Bader A, Homandberg GA. Biomechanical modulation of collagen fragment-induced anabolic and catabolic activities in chondrocyte/agarose constructs. Arthritis Res Ther. 2010;12:R82. doi: 10.1186/ar3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Pizio A, Agamennone M, Tortorella P. Non-Zinc-binding inhibitors of MMP-13: GRID-based approaches to rationalize the binding process. Curr Top Med Chem. 2016;16:449–459. doi: 10.2174/1568026615666150813150631. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Wang D, Yuan Y, Min J. New insights on the MMP-13 regulatory network in the pathogenesis of early osteoarthritis. Arthritis Res Ther. 2017;19:248. doi: 10.1186/s13075-017-1454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie XW, Wan RZ, Liu ZP. Recent research advances in selective matrix metalloproteinase-13 inhibitors as anti-osteoarthritis agents. ChemMedChem. 2017;12:1157–1168. doi: 10.1002/cmdc.201700349. [DOI] [PubMed] [Google Scholar]
- 35.Apte SS. Anti-ADAMTS5 monoclonal antibodies: Implications for aggrecanase inhibition in osteoarthritis. Biochem J. 2016;473:e1–e4. doi: 10.1042/BJ20151072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bondeson J, Wainwright S, Hughes C, Caterson B. The regulation of the ADAMTS4 and ADAMTS5 aggrecanases in osteoarthritis: A review. Clin Exp Rheumatol. 2008;26:139–145. [PubMed] [Google Scholar]
- 37.Tortorella MD, Burn TC, Pratta MA, Abbaszade I, Hollis JM, Liu R, Rosenfeld SA, Copeland RA, Decicco CP, Wynn R, et al. Purification and cloning of aggrecanase-1: A member of the ADAMTS family of proteins. Science. 1999;284:1664–1666. doi: 10.1126/science.284.5420.1664. [DOI] [PubMed] [Google Scholar]
- 38.Zeng W, Corcoran C, Collins-Racie LA, Lavallie ER, Morris EA, Flannery CR. Glycosaminoglycan-binding properties and aggrecanase activities of truncated ADAMTSs: Comparative analyses with ADAMTS-5, -9, -16 and -18. Biochim Biophys Acta. 2006;1760:517–524. doi: 10.1016/j.bbagen.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Budak F, Bal SH, Tezcan G, Akalın H, Goral G, Oral HB. Altered expressions of miR-1238-3p miR-494, miR-6069, and miR-139-3p in the formation of chronic brucellosis. J Immunol Res. 2016;2016:4591468. doi: 10.1155/2016/4591468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Motieghader H, Kouhsar M, Najafi A, Sadeghi B, Masoudi-Nejad A. mRNA-miRNA bipartite network reconstruction to predict prognostic module biomarkers in colorectal cancer stage differentiation. Mol Biosyst. 2017;13:2168–2180. doi: 10.1039/C7MB00400A. [DOI] [PubMed] [Google Scholar]
- 41.Crowe N, Swingler TE, Le LT, Barter MJ, Wheeler G, Pais H, Donell ST, Young DA, Dalmay T, Clark IM. Detecting new microRNAs in human osteoarthritic chondrocytes identifies miR-3085 as a human, chondrocyte-selective, microRNA. Osteoarthritis Cartilage. 2016;24:534–543. doi: 10.1016/j.joca.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nugent M. MicroRNAs: Exploring new horizons in osteoarthritis. Osteoarthritis Cartilage. 2016;24:573–580. doi: 10.1016/j.joca.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 43.Sondag GR, Haqqi TM. The role of MicroRNAs and their targets in osteoarthritis. Curr Rheumatol Rep. 2016;18:56. doi: 10.1007/s11926-016-0604-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang M, Lygrisse K, Wang J. Role of MicroRNA in osteoarthritis. J Arthritis. 2017;6 doi: 10.4172/2167-7921.1000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kopańska M, Szala D, Czech J, Gabło N, Gargasz K, Trzeciak M, Zawlik I, Snela S. MiRNA expression in the cartilage of patients with osteoarthritis. J Orthop Surg Res. 2017;12:51. doi: 10.1186/s13018-017-0542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peffers MJ, Balaskas P, Smagul A. Osteoarthritis year in review 2017: Genetics and epigenetics. Osteoarthritis Cartilage. 2018;26:304–311. doi: 10.1016/j.joca.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: A novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 48.Zhao C, Wang Y, Jin H, Yu T. Knockdown of microRNA-203 alleviates LPS-induced injury by targeting MCL-1 in C28/I2 chondrocytes. Exp Cell Res. 2017;359:171–178. doi: 10.1016/j.yexcr.2017.07.034. [DOI] [PubMed] [Google Scholar]
- 49.Li C, Wan L, Liu Z, Xu G, Wang S, Su Z, Zhang Y, Zhang C, Liu X, Lei Z, Zhang HT. Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018;418:185–195. doi: 10.1016/j.canlet.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Cui L, Hua D. Long non-coding RNA XIST regulates miR-137-EZH2 axis to promote tumor metastasis in colorectal cancer. Oncol Res. 2018;27:99–106. doi: 10.3727/096504018X15195193936573. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Sun Z, Zhang B, Cui T. Long non-coding RNA XIST exerts oncogenic functions in pancreatic cancer via miR-34a-5p. Oncol Rep. 2018;39:1591–1600. doi: 10.3892/or.2018.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang C, Wu K, Wang S, Wei G. Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J Cell Biochem. 2018;119:5646–5656. doi: 10.1002/jcb.26743. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Li S, Luo Y, Liu Y, Yu N. LncRNA PVT1 regulates chondrocyte apoptosis in osteoarthritis by acting as a sponge for miR-488-3p. DNA Cell Biol. 2017;36:571–580. doi: 10.1089/dna.2017.3678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.