Abstract
Wingless-related MMTV integration site 1 (WNT1)/β-catenin signaling plays a crucial role in the generation of mesodiencephalic dopaminergic (mdDA) neurons, including the substantia nigra pars compacta (SNc) subpopulation that preferentially degenerates in Parkinson's disease (PD). However, the precise functions of WNT1/β-catenin signaling in this context remain unknown. Stem cell-based regenerative (transplantation) therapies for PD have not been implemented widely in the clinical context, among other reasons because of the heterogeneity and incomplete differentiation of the transplanted cells. This might result in tumor formation and poor integration of the transplanted cells into the dopaminergic circuitry of the brain. Dickkopf 3 (DKK3) is a secreted glycoprotein implicated in the modulation of WNT/β-catenin signaling. Using mutant mice, primary ventral midbrain cells, and pluripotent stem cells, we show that DKK3 is necessary and sufficient for the correct differentiation of a rostrolateral mdDA neuron subset. Dkk3 transcription in the murine ventral midbrain coincides with the onset of mdDA neurogenesis and is required for the activation and/or maintenance of LMX1A (LIM homeobox transcription factor 1α) and PITX3 (paired-like homeodomain transcription factor 3) expression in the corresponding mdDA precursor subset, without affecting the proliferation or specification of their progenitors. Notably, the treatment of differentiating pluripotent stem cells with recombinant DKK3 and WNT1 proteins also increases the proportion of mdDA neurons with molecular SNc DA cell characteristics in these cultures. The specific effects of DKK3 on the differentiation of rostrolateral mdDA neurons in the murine ventral midbrain, together with its known prosurvival and anti-tumorigenic properties, make it a good candidate for the improvement of regenerative and neuroprotective strategies in the treatment of PD.
SIGNIFICANCE STATEMENT We show here that Dickkopf 3 (DKK3), a secreted modulator of WNT (Wingless-related MMTV integration site)/β-catenin signaling, is both necessary and sufficient for the proper differentiation and survival of a rostrolateral (parabrachial pigmented nucleus and dorsomedial substantia nigra pars compacta) mesodiencephalic dopaminergic neuron subset, using Dkk3 mutant mice and murine primary ventral midbrain and pluripotent stem cells. The progressive loss of these dopamine-producing mesodiencephalic neurons is a hallmark of human Parkinson's disease, which can up to now not be halted by clinical treatments of this disease. Thus, the soluble DKK3 protein might be a promising new agent for the improvement of current protocols for the directed differentiation of pluripotent and multipotent stem cells into mesodiencephalic dopaminergic neurons and for the promotion of their survival in situ.
Keywords: differentiation, DKK3, mouse, stem cell, substantia nigra dopamine neuron, WNT1
Introduction
Dopamine (DA)-synthesizing neurons located in the mammalian ventral midbrain (VM) play crucial roles in the control and modulation of motor, motivational/rewarding, and cognitive behaviors (Schultz, 2007). They comprise two major subpopulations: (1) a rostrolateral bilateral cluster, the substantia nigra pars compacta (SNc); and (2) a caudomedial cluster, the ventral tegmental area (VTA) (Björklund and Dunnett, 2007). The SNc DA neurons project predominantly to the dorsolateral striatum, in which they control the execution of voluntary movements (Björklund and Dunnett, 2007; Schultz, 2007). The preferential degeneration of these neurons is a hallmark of Parkinson's disease (PD; Dauer and Przedborski, 2003). The need of better therapies for this disease has fueled basic research to understand the precise molecular mechanisms underlying the generation and survival of SNc DA neurons (Arenas, 2010).
During mouse development, the progenitors of these “mesodiencephalic” dopaminergic (mdDA) neurons arise from the ventral midline, the floor plate (FP), of the midbrain and caudal diencephalon under the influence of the Sonic hedgehog (SHH), Wingless-related MMTV integration site 1 (WNT1), and fibroblast growth factor 8 (FGF8) signaling pathways (Ono et al., 2007; Bonilla et al., 2008; Joksimovic et al., 2009; Blaess et al., 2011; for review, see Hegarty et al., 2013). SHH is necessary for the establishment of the mesencephalic FP and basal plate (BP) progenitor domains between embryonic day 8.5 (E8.5) and E9.5 (Blaess et al., 2006; Perez-Balaguer et al., 2009), but its expression has to be repressed by WNT/β-catenin signaling to convey mdDA neurogenic potential to the midbrain FP cells at approximately E11.5 (Joksimovic et al., 2009). The in vivo generation of mdDA neurons depends crucially on WNT1 and its downstream (β-catenin-mediated) signaling pathway (Prakash et al., 2006; Tang et al., 2009), although increasing evidence indicates that WNT1/β-catenin signaling must be tightly regulated to ensure proper mdDA neuron differentiation (Joksimovic et al., 2009; Tang et al., 2010; Andersson et al., 2013). The secreted Dickkopf (DKK1–DKK4) glycoproteins are one class of WNT/β-catenin signaling modulators (Niehrs, 2006). DKK1/2/4 antagonize WNT/β-catenin signaling by binding to low-density lipoprotein receptor-related protein (LRP) and Kremen coreceptors and inducing their internalization, thereby inhibiting the formation of an active WNT/Frizzled receptor (FZD)/LRP coreceptor complex on the cell surface (Niehrs, 2006). The function of the more distantly related DKK3 remains unresolved (Niehrs, 2006; Veeck and Dahl, 2012): DKK3 does not bind to LRP6 and can act as both a repressor and activator of WNT/β-catenin signaling (Nakamura et al., 2007; Nakamura and Hackam, 2010; Das et al., 2013; Xiang et al., 2013). The importance of the WNT1/β-catenin signaling pathway in this context has also been recognized in recent years for the derivation of mdDA neurons from in vitro cultured murine and human pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced PSCs (iPSCs). Exposure of differentiating ESCs and iPSCs to WNT1 or a glycogen synthase kinase 3 β (GSK3b) inhibitor is now incorporated routinely in established mdDA differentiation protocols for human PSCs, which can be used for PD modeling, drug screening, and cell-replacement therapies (Yu et al., 2013; for review, see Li et al., 2013; Tabar and Studer, 2014).
We show here that Dkk3 is necessary for the correct differentiation of an mdDA precursor subset into rostrolateral (dorsomedial SNc and dorsolateral VTA) mdDA neurons in the mouse embryo. DKK3 appears to activate and/or maintain the expression of LIM homeobox transcription factor 1α (LMX1A) and paired-like homeodomain transcription factor 3 (PITX3) in these cells, two homeodomain (HD) transcription factors (TFs) required for the proper generation and survival of mdDA and in particular SNc DA neurons (for review, see Smidt and Burbach, 2007; Hegarty et al., 2013). We also show that the treatment of differentiating murine PSCs with DKK3 and WNT1 proteins promotes the generation of mdDA neurons with molecular SNc DA cell characteristics in vitro, suggesting that DKK3 might be an additional candidate for the improvement of cell differentiation strategies in the treatment of PD.
Materials and Methods
Mice.
Generation and genotyping of Dkk3−/− mice was described previously (del Barco-Barrantes et al., 2006). Generation of Pitx3GFP/+ mice was described by Zhao et al. (2004); these mice were PCR genotyped using the green fluorescent protein (GFP) and Pitx3 primer pairs listed in Table 1. Dkk3+/+, Dkk3+/−, and Dkk3−/− mice were kept as brother–sister matings on a mixed (129X1/SvJ × C57BL/6) genetic background. Pitx3GFP/+ embryos were derived from Pitx3GFP/GFP (on a mixed 129P2/OlaHsd × C57BL/6 background) × C57BL/6 intercrosses. CD-1 and C57BL/6 mice were provided by the Transgenic Unit, Helmholtz Center Munich. Pregnant dams were killed by CO2 asphyxiation or cervical dislocation. Collection of embryonic stages was done from timed-pregnant females; noon of the day of vaginal plug detection was designated as E0.5. Detection of the Y chromosome-specific Sry gene by genomic PCR (Table 1) was used for embryonic sex determination. Dkk3 mutant embryos of either sex were compared with their wild-type (WT; Dkk3+/+ and Dkk3+/−) littermates, and at least three embryos were analyzed for each probe, genotype, and stage, if not indicated otherwise. This study was performed in strict accordance with the recommendations in the European Union Directive 2010/63/EU and the Guide for the Care and Use of Laboratory Animals of the Federal Republic of Germany (German Animal Protection Law). The protocol was approved by the Institutional Animal Care and Use Committee (Committee for Animal Experiments and Laboratory Animal Facility) of the Helmholtz Center Munich. All efforts were made to minimize suffering.
Table 1.
Primer and PCR conditions used for genotyping, semi-qPCR, and qPCR
| Gene (application) | Primer (5′ → 3′) | Product length (bp) | Tm (°C) | Cycles |
|---|---|---|---|---|
| GFP (genotyping) | Forward, CCATGGTGAGCAAGGGCGA | 800 | 60 | 35 |
| Reverse, CTCGAGCTTGTACAGCTCGTCCATG | ||||
| Pitx3 (genotyping) | Forward, GGAGTTTGGGCTGCTTGGTG | 512 | 60 | 35 |
| Reverse, CCACACCGCGATCTCTTCG | ||||
| Sry (genotyping) | Forward, GAGAGCATGGAGGGCCAT | 282 | 62 | 30 |
| Reverse, CCACTCCTCTGTGACACT | ||||
| Lmx1a (semi-qPCR) | Forward, ACCAACAGAACACCCAGAGG | 285 | 62 | * |
| Reverse, CTTCTGAGGTTGCCAGGAAG | ||||
| Lmx1b (qPCR) | Forward, CACTTGGGCTGTTTCTGCTG | 296 | 60 | * |
| Reverse, TTGAAAGCTCTTCGCTGCTG | ||||
| Gapdh (semi-qPCR, qPCR) | Forward, ACCACAGTCCATGCCATCAC | 452 | 59 | 30* |
| Reverse, TCCACCACCCTGTTGCTGTA |
*As indicated in Materials and Methods for semi-qPCR and qPCR.
5-Ethynyl-2′-deoxyuridine treatments.
Embryos were dissected 1 h after intraperitoneal injection of 10 μg of 5-ethynyl-2′-deoxyuridine (EdU; Life Technologies) per gram body weight into pregnant dams on E11.5. Treated embryos were processed for EdU immunohistochemistry (IHC) as described previously (Castelo-Branco et al., 2010), and incorporated EdU was detected using the Click-iT EdU Alexa Fluor 488 imaging kit (Life Technologies) according to the instructions of the manufacturer.
Radioactive in situ hybridization.
Serial paraffin sections (8 μm) were hybridized with radioactive ([α-35S]UTP) riboprobes as described previously (Brodski et al., 2003). Riboprobes used were Dkk1, Dkk2, and Dkk3 (Monaghan et al., 1999), tyrosine hydroxylase (Th) and Pitx3 (Brodski et al., 2003), Wnt1 and Lmx1b (Stuebner et al., 2010), Fgf8, Engrailed 1 (En1), Shh, Neurog1 (Ngn1), and Neurog2 (Ngn2) (Castelo-Branco et al., 2010). Images were taken with an Axioplan2 microscope or StemiSV6 stereomicroscope using bright-field and dark-field optics, AxioCam MRc camera and Axiovision 4.6 software (Zeiss), and processed with Adobe Photoshop CS3 software (Adobe Systems). Dark-field images were pseudocolored in some cases as indicated in the figure legends.
Immunohistochemistry/immunocytochemistry.
IHC on 8 μm paraffin sections and immunocytochemistry (ICC) on cultured cells were performed as reported previously (Puelles et al., 2004; Peng et al., 2011). Monoclonal antibodies used were mouse anti-TH (1:600; Millipore MAB318), anti-EN1 (1:100; Developmental Studies Hybridoma Bank 4G11), and rat anti-dopamine transporter (DAT, also known as SLC6A3; 1:500; Millipore MAB369). Polyclonal antisera used were rabbit anti-TH (1:150; Millipore AB152; 1:1000; Pel_Freeze P40101), anti-PITX3 (1:300; Life Technologies 38-2850), anti-LMX1A (1:1000; Millipore ab10533), anti-phosphorylated Histone H3 (pH3) (1:500; Millipore 06-570), anti-cleaved (activated) Caspase 3 (cCASP3; 1:100; Cell Signaling Technology 9664), anti-nuclear receptor subfamily 4, group A, member 2 (NURR1, also known as NR4A2; 1:2000; Santa Cruz Biotechnology sc-990), anti-calbindin 1 [CALB1; 1:400 (embryos) and 1:5000 (cell cultures); Swant CB-38a], anti-potassium voltage-gated channel, ShaI-related family, member 3 (KCND3, also known as Kv4.3; 1:2000; Alomone Labs APC-017), and anti-potassium inwardly rectifying channel, subfamily J, member 6 (KCNJ6, also known as GIRK2; 1:2000; Alomone Labs APC-006). The guinea pig anti-LMX1B antibody (1:20,000) was a gift from Prof. C. Birchmeier-Kohler and Dr. T. Müller (Max Delbrück Center, Berlin, Germany). Secondary antibodies were either fluorescently labeled (Alexa Fluor 488/594; 1:500; Life Technologies) or coupled to biotin/streptavidin–horseradish peroxidase or alkaline phosphatase (AP; Jackson ImmunoResearch) and detected using the VECTASTAIN ABC or VECTOR Red AP Substrate kits (Vector Laboratories). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Fluorescent images were taken with an Axiovert 200M microscope and AxioCam HRc camera (Zeiss), a Keyence BZ-9000 microscope, or an Olympus IX81 confocal laser scanning microscope and processed with Adobe Photoshop CS3 or CS5 software. For the simultaneous detection of TH protein and Dkk3 mRNA on embryonic tissue sections, the Dkk3 in situ hybridization (ISH) was preceded by a TH IHC as described above, leaving out all extensive ethanol washes.
Primary VM cultures and DKK3/EdU treatments.
Primary VM cultures were prepared from E11.5 Dkk3+/+ (WT) and Dkk3−/− mouse embryos as described by Pruszak et al. (2009) with minor modifications. Briefly, VM tissues were trypsinized for 5 min at 37°C, and 1.5–2 × 105 cells per well were plated in a 24-well plate on poly-d-lysine (Sigma)-coated coverslips in DMEM/F-12 medium supplemented with 10% fetal calf serum (FCS), glutamine, and penicillin/streptomycin (Life Technologies). At 24 h after plating, cells were incubated in expansion medium for 1 d and then switched to differentiation medium (Pruszak et al., 2009) for the remaining culture time. Concomitant with the change to differentiation medium after 2 d in vitro (2 DIV), primary VM cells were treated with 100 ng/well recombinant human DKK3 protein [R&D Systems; 100 ng/μl DKK3 in 0.1% bovine serum albumin (BSA; Sigma)] or 1 μg/well BSA (control; 1 μg/μl) for the indicated time. DKK3 or BSA was added with each medium change every fourth day. In some cases, cells were incubated with 10 μm EdU for 1 h before fixation. Cells were harvested for ICC or RNA isolation after 4 d (6 DIV) or 7 d (9 DIV) of treatments.
ESC/iPSC culture and differentiation into mdDA neurons.
Generation, characterization, and culture of Pitx3GFP/+ iPSCtfs (line FM77A3) were described previously (Pertek et al., 2014). Pitx3GFP/+ iPSCtfs were cultured on mitomycin-C treated mouse embryonic fibroblasts (MEFs) in ESC medium before initiating their differentiation into mdDA neurons using a monolayer differentiation protocol that was adapted from published procedures for mouse and human PSCs (Chambers et al., 2009; Jaeger et al., 2011; Kriks et al., 2011). On day 0 of the differentiation procedure, cells were trypsinized, and 1.4 × 104 cells/cm2 were transferred to tissue culture dishes in serum replacement medium (SRM; DMEM/F-12 GlutaMAX, 10% KnockOut Serum Replacement, and 0.1 mm β-mercaptoethanol; Life Technologies) containing 10 μm Rho-dependent protein kinase inhibitor Y-27632 (4-[(1R)-1-aminoethyl]-N-pyridin-4-yl-cyclohexane-1-carboxamide) (Calbiochem). After incubation for 50 min at 37°C, the cell supernatant was plated at a density of 1.4 × 104 cells/cm2 on poly-dl-ornithine-coated (0.1 mg/ml in 300 mm borate buffer, pH 8.3; Sigma) and laminin-coated (125 μg/ml in PBS; Roche) coverslips in SRM containing 10 μm transforming growth factor β (TGFβ) type I receptor [activin receptor-like kinase (ALK) 4/5/7] inhibitor SB431542 [4-(4-(benzo[d][1,3]dioxol-5-yl)-5-(pyridin-2-yl)-1H-imidazol-2-yl)benzamide] (SB; in DMSO; Tocris Bioscience) and 100 nm bone morphogenetic protein type I receptor (ALK1/2/3/6) inhibitor LDN193189 [4-(6-(4-(piperazin-1-yl)phenyl)pyrazolo[1,5-a]pyrimidin-3-yl)quinoline] (LDN; in DMSO; Axon Medchem) to suppress alternative mesodermal, endodermal, and epidermal cell fates. On day 1, the medium was changed to 50% SRM/50% N2 medium [DMEM/F-12 GlutaMAX, 1× N2 supplement (Life Technologies) and 200 μm ascorbic acid (Sigma)] containing 10 μm SB, 100 nm LDN, and 1 μm mitogen-activated protein kinase kinase inhibitor PD0325901 [(R)-N-(2,3-dihydroxypropoxy)-3,4-difluoro-2-((2-fluoro-4-iodophenyl)amino)benzamide] (in DMSO; Stemgent) to promote the VM neural fate. On day 3, differentiation of the Pitx3GFP/+ iPSCtfs into mdDA neurons was initiated by treating the cells for 4 d with N2 medium containing 200 μm ascorbic acid, 10 μm SB, 100 nm LDN, and 100 ng/ml recombinant mouse FGF8b (R&D Systems) either alone or together with 3 μm GSK3b inhibitor CHIR99021 [6-((2-((4-(2,4-dichlorophenyl)-5-(5-methyl-1H-imidazol-2-yl)-2 pyrimidinyl)amino)ethyl)amino)-3-pyridinecarbonitrile] (CHIR99; in DMSO; Stemgent), 100 ng/ml recombinant human DKK3 (R&D Systems), and/or 100 ng/ml recombinant human WNT1 (Peprotech), as indicated. On day 5, the medium was changed to N2B27 medium (50% N2, 50% 1× B27 supplement in Neurobasal medium, 2 mm l-glutamine, and 200 μm ascorbic acid; Life Technologies) containing 100 nm LDN and 100 ng/ml FGF8b either alone or together with 3 μm CHIR99, 100 ng/ml DKK3, and/or 100 ng/ml WNT1, as indicated. On day 7, the medium was replaced with B27 medium containing 200 μm ascorbic acid, 100 nm LDN and 3 μm CHIR99, 100 ng/ml DKK3, and/or 100 ng/ml WNT1, as indicated. The LDN inhibitor was omitted from day 9 onward. Omission of the WNT/β-catenin signaling factors (CHIR99, DKK3, and/or WNT1) was used as a control. Addition of 20 ng/ml recombinant human brain-derived neurotrophic factor (BDNF; R&D Systems) to B27 medium containing 200 μm ascorbic acid from day 11 promoted the maturation and survival of the generated Pitx3GFP/+ mdDA neurons, which were analyzed routinely on day 21.
Unbiased stereology and cell countings.
For IHC, TH+ and PITX3+ single-labeled cells in WT and Dkk3−/− brains were evaluated on every fourth (E12.5), sixth (E14.5), or eighth (E18.5 and adult) serial coronal or horizontal midbrain section by the optical fractionator method using Stereo Investigator 5.05.4 software (MBF Bioscience). Single- and double-labeled cells were counted on every fourth (TH+/PITX3+, TH+/LMX1A+, LMX1A+/EdU+, or pH3+ cells) or eighth (TH+/cCASP3+ or TH+/CALB1+ cells) coronal midbrain section from E10.5, E11.5, E12.5, E14.5, or E18.5 WT and Dkk3−/− embryos using Stereo Investigator 5.05.4 software. For ICC on primary VM cells, TH+, PITX3+, EdU+, pH3+, and cCASP3+ single- or double-labeled cells were counted in 10 random fields per coverslip. For ICC on differentiating PSCs, TH+/PITX3+ double-labeled cell clusters and TH+, PITX3+, and NR4A2+ single- and double-labeled cells within double-positive cell clusters were counted on one coverslip and normalized to cluster area using Stereo Investigator 5.05.4 software. The proportion of TH+/PITX3+, TH+/NR4A2+, TH+/CALB1+, and TH+/KCND3+ double-labeled cells among the total number of TH+ cells was determined for at least three clusters per coverslip. TH+/KCND3+ double-labeled cells were counted on confocal images using Olympus Fluoview version 2.0b software. The numbers (n) of independent experiments evaluated in each case are indicated in the figure legends.
Semiquantitative and quantitative RT-PCR.
Total RNA was isolated from primary VM cells using TRIzol Reagent (Life Technologies) or RNeasy Mini kit (Qiagen) including a DNase I treatment. One microgram of total RNA was reverse transcribed using the iScript cDNA Synthesis kit [Bio-Rad; for semiquantitative (semi-q) PCR] or QuantiTect Reverse Transcription kit [Qiagen; for quantitative (q) PCR] according to the instructions of the manufacturer. One microliter (semi-qPCR) or 1 μl of a 1:3 diluted (qPCR) cDNA were amplified by PCR using the intron-spanning primers and conditions listed in Table 1. For semi-qPCR, 5 μl samples were taken from the PCR reactions after 22, 25, 28, 31, and 34 cycles and separated on a 2% agarose gel. Densitometric analysis of the gel pictures was done using NIH ImageJ software. The qPCR (20 μl reactions in a 384-well plate) was performed using SYBR green PCR Master Mix and the 7900HT Fast Real-Time PCR System (Applied Biosystems) according to the instructions of the manufacturer. For semi-qPCR and qPCR, signals from specific Lmx1a and Lmx1b PCR products were normalized against glyceraldehyde-3-phosphate dehydrogenase (Gapdh), and relative values were calculated by setting the normalized value of controls as 1. For qPCR, the threshold cycle (Ct) value was recorded for Lmx1b and Gapdh, and the relative Ct value was calculated for each reaction. All assays included negative controls, and samples were from three independent experiments.
Fluorescence-activated cell sorting and global transcriptome analysis.
Single-cell suspensions from microdissected VM tissues of E13.5 and E14.5 Pitx3GFP/+ embryos (one biological sample each) were prepared as described previously (Zhao et al., 2004). Differentiated Pitx3GFP/+ iPSCtfs (days 19–23 of the monolayer protocol; five biological replicates/independent experiments) were trypsinized, filtered through a 70 μm cell strainer, pelleted, and resuspended in ice-cold PBS. Fluorescence-activated cell sorting (FACS) of GFP+ cells was performed with a FACSAria III (BD Biosciences) device essentially as described previously (Fischer et al., 2011; Pertek et al., 2014). Sorted cells were collected directly in RLT buffer, and total RNA was purified with the RNeasy Micro kit (Qiagen), including a DNase I treatment and following the instructions of the manufacturer. The quality of the purified total RNA was assessed with the RNA 6000 Pico kit and 2100 Bioanalyzer (Agilent). Total RNA (∼1 ng) was amplified using the Ovation PicoSL WTA System V2 in combination with the Encore Biotin Module (Nugen). Amplified cDNAs were hybridized on Affymetrix Mouse Gene 1.0 ST arrays containing ∼28,000 probe sets. Staining and scanning was done according to the Affymetrix expression protocol including minor modifications as suggested in the Encore Biotin protocol. Expression console (version 1.2; Affymetrix) was used for quality control and to obtain annotated normalized robust multi-array average gene-level data (standard settings including median polish and sketch-quantile normalization). Statistical analyses were performed with R software (http://www.R-project.org) implemented in CARMAweb (Rainer et al., 2006). Gene-wise testing for differential expression was done using the limma paired t test and Benjamini–Hochberg multiple testing correction (false discovery rate <10%). The set of 460 probe sets was defined by p < 0.05 (p value of the limma t test), ratios >1.2-fold up or down, and an average expression level >50. Heat maps were generated with CARMAweb and cluster dendrograms with the R script hclust. Gene Ontology (GO) term enrichment analyses (p < 0.01) were done with GePS software (Genomatix). Array data have been submitted to GEO (accession number GSE58813).
Statistical analyses.
All values are mean ± SEM unless indicated otherwise. Cell numbers were averaged for each genotype, stage, sex, region, and/or treatment and subjected to tests for the estimation of statistical significance relative to the control. For comparison of two groups, the Welsh t test for unequal variances was applied. The effect of experimental factors with more than two levels and the effect of two or more experimental factors were analyzed with one-way, two-way, or three-way ANOVA, depending on the number of factors. In the case of two or more factors, the analysis started with an ANOVA with the highest possible interaction effect and excluded stepwise nonsignificant interactions to identify the simplest model. For factors involved in a significant interaction effect, subgroups were analyzed separately. Post hoc t tests were performed with p value adjustment according to Holm (1979). For cell countings with several sections per embryo and for semi-qPCR and qPCR, ANOVA for repeated measurements was applied. For all calculations, the R software with the nlme package was used.
Results
Dkk3 expression in the murine VM coincides with the initiation of mdDA neuron differentiation and persists in adulthood
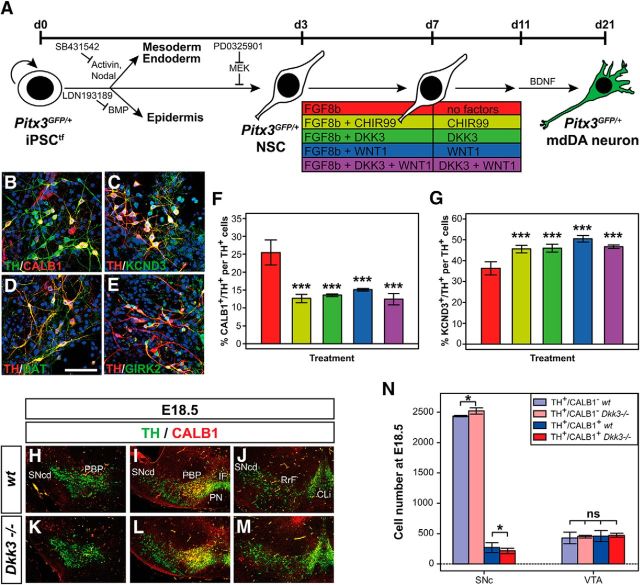
To establish a potential role of DKK3 in murine mdDA neuron development, we first determined the spatiotemporal pattern of Dkk3 expression in the developing mouse VM in relation to two other Dkk family members, Dkk1 and Dkk2, and two markers for differentiating mdDA neurons, the mdDA-specific HD TF Pitx3 and Th, the rate-limiting enzyme for DA biosynthesis (Smidt and Burbach, 2007). The strong expression of Dkk1 and Dkk2 in the midbrain FP at E10.5 (Fig. 1A–C) declined at E11.5 (Monaghan et al., 1999; Ribeiro et al., 2011) and became undetectable at E12.5 (Fig. 1E–G). In contrast, a weak Dkk3 signal was first detected at E11.5 in the medial FP of the rostral midbrain (Fig. 1D,D′,H). This Dkk3+ domain extended into the lateral FP and the BP in the caudal midbrain (Fig. 1H′) and overlapped with the Th+ domain along the anterior–posterior axis of the VM (Fig. 1I–I‴). At E12.5, Dkk3 expression in the VM was confined to the midbrain FP, in which it was transcribed in the Pitx3− ventricular and subventricular zone (VZ/SVZ) and in a mantle zone (MZ) domain overlapping with Pitx3+ mdDA precursors and neurons (Fig. 1J). At E14.5, Dkk3 was expressed in a very narrow FP domain from which Pitx3+ mdDA precursors/neurons emerged (Fig. 1L). A weak expression of Dkk3 became evident within a lateral Th+ domain at E15.5 (Fig. 1K–K‴). At E18.5 and in the adult mouse brain, Dkk3 transcription was detected in a rostrolateral Th+ and Pitx3+ mdDA domain comprising the parabrachial pigmented nucleus (PBP), part of the medial and dorsal SNc, and the paranigral nucleus, but the majority of the Dkk3+ cells were located outside of this mdDA domain (Fig. 1M–R‴). Of note, higher-magnification views revealed that some TH+ and Pitx3+ cells coexpressed Dkk3 in these regions (Fig. 1N–Q, inset in N″). We concluded that Dkk3 was expressed initially (at E11.5–E15.5) in mdDA progenitors and precursors located in the VZ/SVZ and MZ of the midbrain FP and later (after E15.5 into adulthood) in an mdDA neuronal subset comprising mostly the PBP, part of the dorsomedial SNc, and the paranigral nucleus.
Figure 1.
Dkk3 expression in the murine VM initiates with mdDA neuron differentiation and persists into adulthood. A–R‴, Representative close-up views of the VM (A–H′, I′–J, K′–L, M′–Q, R′–R‴) and brain overviews (I, K, M, R) on cresyl violet-stained coronal (A–H′, J–Q; dorsal top) and sagittal (I–I‴, R–R‴; anterior left) sections (coronal and sagittal level indicated by broken lines in I and K, respectively) from WT embryos at E10.5 (A–D′), E11.5 (H–I‴), E12.5 (E–G, J), E14.5 (L), E15.5 (K–K‴), E18.5 (M–Q) and adult brains (R–R‴), hybridized with riboprobes for Dkk1 (B, F), Dkk2 (C, G), Dkk3 (D, D′, H, H′, I′, J, K′, L, M′, N, O, P, Q, R′), Th (I″, K″, M″, R″), and Pitx3 (J, L, P, Q), or immunostained for TH (N′, O′). D and H are dark-field views of A and E, and I″, K″, M″, and R″ are higher-magnification dark-field views of the boxed areas in I, K, M, and R, respectively. D, D′, H, H′, and N–Q are different rostrocaudal levels of the VM. I‴, J, K‴, L, M‴, P, Q, and R‴ are pseudocolored overlays of consecutive sections hybridized with Dkk3 (red) and Th (green in I‴, K‴, M‴, R‴) or Pitx3 (green in J, L, P, Q); overlapping expression domains appear in yellow. N″ and O″ are superimposed dark-field images after Dkk3 ISH (green pseudocolor in N, O) and the corresponding bright-field views after TH IHC (VECTOR Red staining in N′, O′). Broken white lines in B–D′, F–H′, J, L outline the neuroepithelium and in J delimit the FP and the VZ/SVZ from the MZ. Broken black lines in E delimit the FP and BP. Red arrows in K′ point to the weak Dkk3+ domain in the E15.5 rostrolateral VM, and white arrows in N″ point to TH+ cells that coexpress Dkk3. Inset in N″ is a higher magnification of the framed cell. Cb, Cerebellum; CLi, caudal linear nucleus; Cx, cortex; Di, diencephalon; DR, dorsal raphe nucleus; IF, interfascicular nucleus; Mes, mesencephalon; Met, metencephalon; Ob, olfactory bulb; PN, paranigral nucleus; RLi, rostral linear nucleus; RrF, retrorubral field; SNcd, Substantia nigra pars compacta dorsalis; T, tectum; Tg, tegmentum; Tel, telencephalon; III, third ventricle.
Defective differentiation of rostrolateral mdDA neurons in Dkk3−/− mice
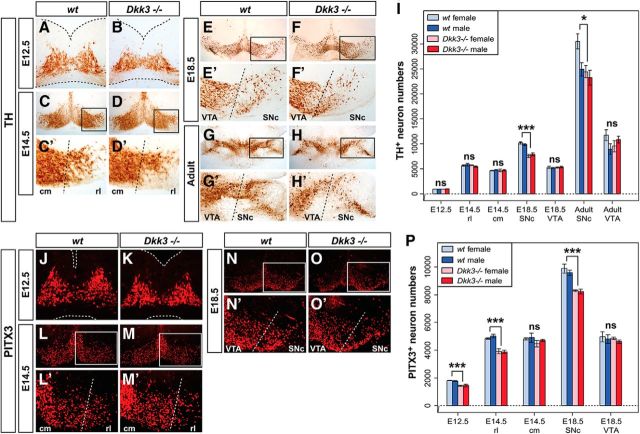
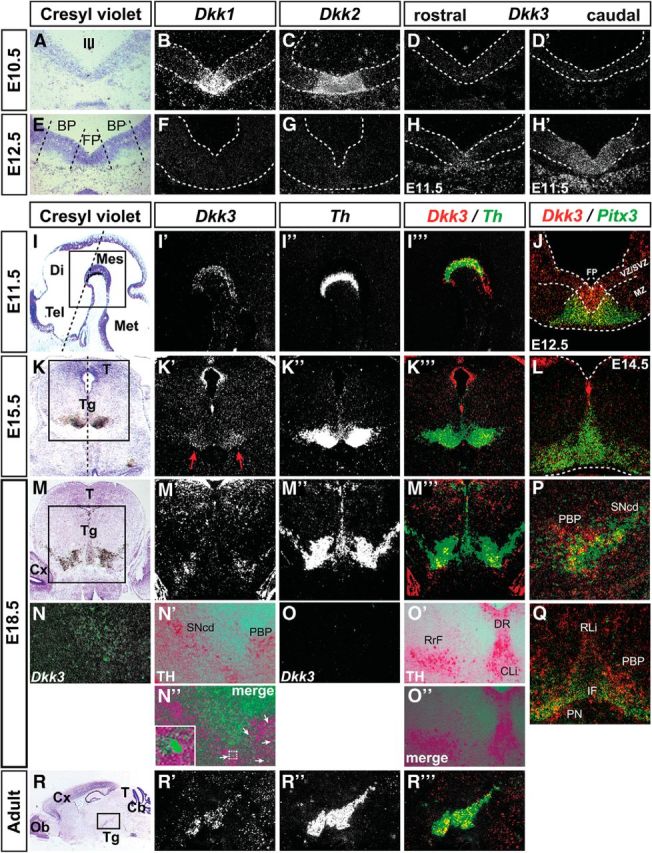
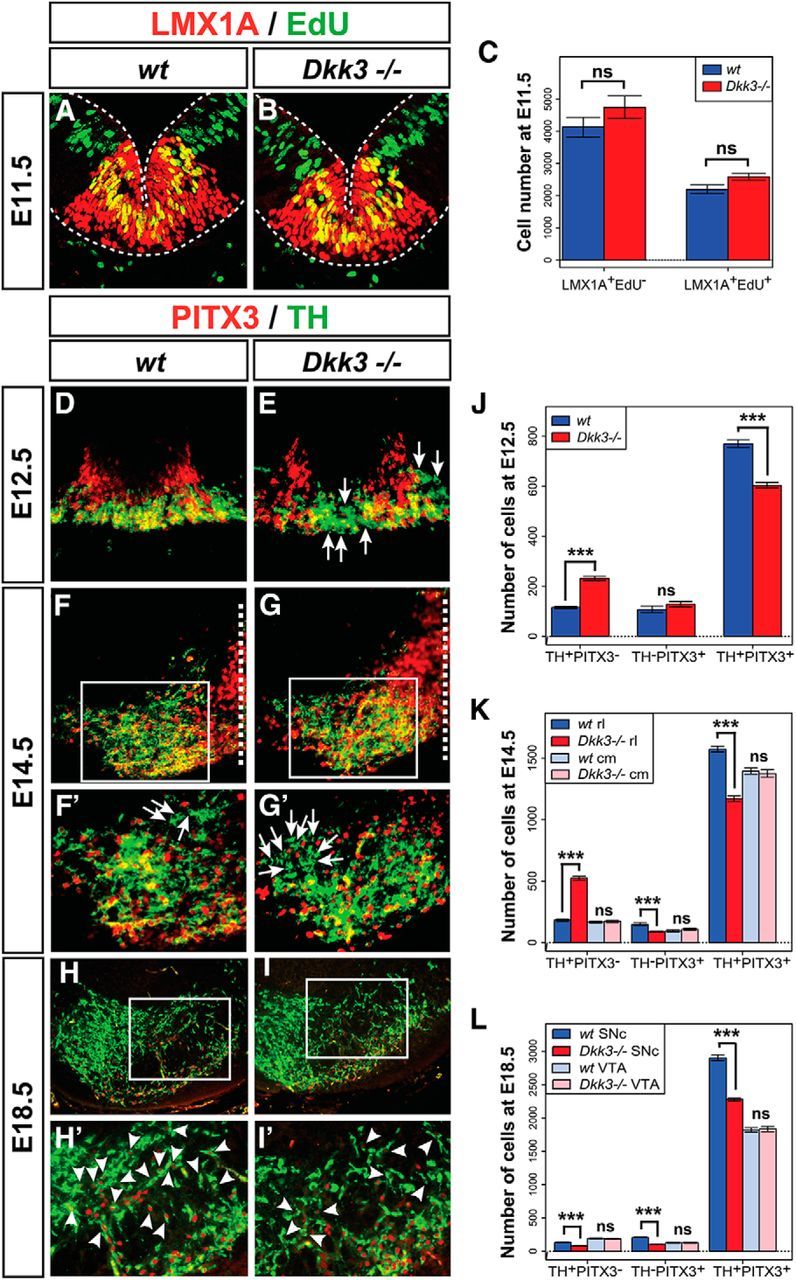
The peculiar Dkk3 expression pattern in the murine VM suggested a potential function of DKK3 in mdDA neuron development and/or survival in vivo. The transcription of genes required for proper patterning of the midbrain/hindbrain region and neurogenesis in this region, such as Dkk1/2, Fgf8, En1, Shh, Wnt1, Ngn1, Ngn2, and Lmx1b (Wurst and Bally-Cuif, 2001; Hegarty et al., 2013), was unaffected in the Dkk3−/− embryos (data not shown), and we thus examined whether the generation of mdDA neurons was altered in the absence of Dkk3. Because Dkk3−/− mice exhibit a sex-related behavioral phenotype (del Barco-Barrantes et al., 2006), we also determined the sex of the corresponding embryos. The total number of TH+ neurons in the Dkk3−/− VM was comparable with the WT VM at E12.5 and E14.5, but a significant and sex-independent reduction was detected in the rostrolateral (PBP and dorsomedial SNc) domain of the Dkk3−/− embryos at E18.5 (20–25% less TH+ neurons) and in adult Dkk3−/− mice (7–20% less TH+ neurons) (Fig. 2A–I). Notably, the total number of PITX3+ cells was already reduced significantly and sex independently by 18–21% in the Dkk3−/− embryos at E12.5 and by 14–23% in the rostrolateral (PBP and dorsomedial SNc) region of the mutant VM at E14.5 and E18.5 (Fig. 2J–P). In contrast to the rostrolateral (PBP and dorsomedial SNc) domain, the caudomedial (VTA) region was not affected in the Dkk3−/− embryos and mice (Fig. 2A–P). To assess this aberrant differentiation and loss of an mdDA neuron subset in more detail, we first determined the numbers of proliferating LMX1A+/EdU+ mdDA progenitors, postmitotic LMX1A+/EdU− mdDA precursors, and mitotic pH3+ VM neural progenitors in the Dkk3−/− VM. Because these numbers were not altered in the mutant VM (Fig. 3A–C and data not shown), we next determined the numbers of TH or PITX3 single- and double-positive cells at different developmental stages in the Dkk3−/− and WT embryos. TH+/PITX3− single-positive cells were increased significantly by ∼2-fold (at E12.5) to 2.8-fold (at E14.5) at the expense of TH+/PITX3+ double-positive cells, which were decreased by 22% (at E12.5) to 26% (at E14.5) in the mutant VM (Fig. 3D–G′,J,K). Although the numbers of TH−/PITX3+ single-positive cells were not altered at E12.5, they were significantly decreased by 39% in the rostrolateral region of the Dkk3−/− VM at E14.5 (Fig. 3D–G′,J,K). These results might account for the initial observation of decreased PITX3+ but normal TH+ cell numbers in the mutant VM at E12.5 and E14.5 (Fig. 2). However, the total number of TH+ cells was reduced in the mutant rostrolateral (PBP and dorsomedial SNc) domain at E18.5 (Fig. 2) and consistent with this finding, TH+/PITX3−, TH−/PITX3+, and TH+/PITX3+ single- and double-positive cells were decreased by 38, 50, and 21%, respectively, in the rostrolateral (PBP and dorsomedial SNc) mdDA domain of the E18.5 Dkk3−/− embryos (Fig. 3H–I′, L). The lack of these cells was most notable in the mutant PBP and dorsal tier of the SNc at this stage (Fig. 3H–I′). Because the loss of an mdDA neuron subset in the Dkk3−/− mice might also be attributable to a reduced survival of the corresponding cells, we determined the numbers of TH+/cCASP3+ apoptotic mdDA neurons and TH−/cCASP3+ VM cells in the WT and Dkk3 mutant VM at E18.5. These numbers were not altered in the Dkk3−/− VM (data not shown). Thus, our data indicated that a TH+/PITX3− mdDA precursor subpopulation failed to differentiate correctly into TH+/PITX3+ mdDA neurons in the absence of Dkk3, resulting in an initial accumulation of TH+/PITX3− single-positive cells at the expense of TH+/PITX3+ double-positive neurons in the mutant VM and the later loss of these TH+ rostrolateral (PBP and dorsomedial SNc) mdDA neurons. However, the proliferation and specification of the corresponding mdDA progenitors/precursors were not affected by the lack of Dkk3 in the murine VM.
Figure 2.
Midgestational PITX3+ cell loss and late gestational TH+ cell reduction in the Dkk3−/− midbrain. (A–H′, J–O′) Representative close-up views of the VM on coronal (A–F′, J–O′; dorsal top) and horizontal (G–H′; rostral top) sections from WT (A, C, E, G, J, L, N) and Dkk3−/− (B, D, F, H, K, M, O) embryos at E12.5 (A, B, J, K), E14.5 (C–D′, L–M′) and E18.5 (E–F′, N–O′) or adult brains (G–H′), immunostained for TH (A–H′) or PITX3 (J–O′). C′, D′, E′, F′, G′, H′, L′, M′, N′, and O′ are higher magnifications of the boxed areas in C, D, E, F, G, H, L, M, N, and O, respectively. Broken black or white lines in A, B and J, K outline the neuroepithelium and in C′, D′, E′, F′, G′, H′, L′, M′, N′, and O′ delimit the rostrolateral (rl) or SNc domain from the caudomedial (cm) or VTA domain, as indicated. I, P, Unbiased stereological quantification of TH+ (I) or PITX3+ (P) cells in female or male WT and Dkk3−/− embryos and adult brains. *p < 0.05; ***p < 0.001; ns, not significant in the Welsh t test for unequal variances.
Figure 3.
Dkk3 is required for the correct differentiation of rostrolateral mdDA neurons in vivo. A, B, D–I, Representative confocal close-up views of the VM on coronal sections (dorsal top) from WT (A, D, F, H) and Dkk3−/− (B, E, G, I) embryos at E11.5 (A, B), E12.5 (D, E), E14.5 (F, G), and E18.5 (H, I), immunostained for LMX1A (red) and EdU (green) (A, B), or PITX3 (red) and TH (green) (D–I). Broken white lines in A and B outline the neuroepithelium and in F and G indicate the midline. F′, G′, H′, and I′ are higher magnifications of the boxed areas in F, G, H, and I, respectively. White arrows in E, F′, and G′ point to TH+/PITX3− single-positive cells; white arrowheads in H′ and I′ point to TH+/PITX3+ double-positive cells. C, Quantification of LMX1A+/EdU− and LMX1A+/EdU+ cells in the WT and Dkk3−/− VM at E11.5 (n = 3 embryos/genotype). J–L, Quantification of TH+/PITX3−, TH−/PITX3+, and TH+/PITX3+ cells in WT and Dkk3−/− embryos at E12.5 (J), E14.5 (K), and E18.5 (L) (n ≥ 6 embryos/genotype/stage). ***p < 0.001; ns, not significant in the paired t test. cm, Caudomedial mdDA domain; rl, rostrolateral mdDA domain.
DKK3 promotes the differentiation and survival of an mdDA neuron subset in vitro
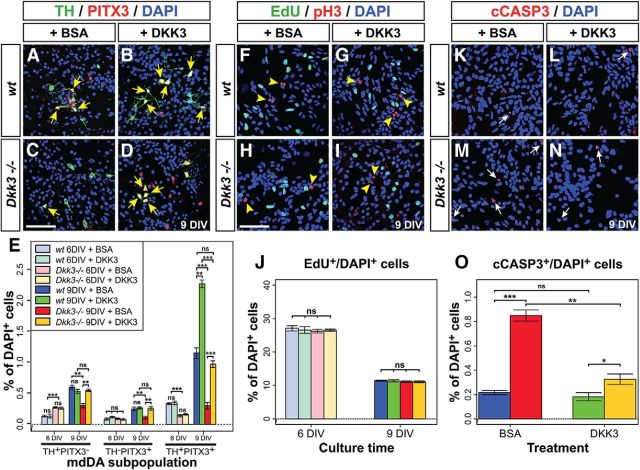
Our previous results indicated that the differentiation of an mdDA neuronal subset was compromised in the absence of Dkk3. Therefore, we investigated whether the exogenous application of DKK3 protein might rescue this phenotype in primary VM cells isolated from E11.5 Dkk3−/− embryos. After 4 d (i.e., 6 DIV; see Materials and Methods) of BSA (vehicle control) or DKK3 treatment, a significant and approximately twofold increase of TH+/PITX3− single-positive cells and an ∼56% decrease of TH+/PITX3+ double-positive cells were detected in the Dkk3−/− compared with the WT primary VM cultures (Fig. 4E), thus reflecting similar changes in the mutant VM at E12.5 (Fig. 3J). However, seven days of DKK3 treatment (i.e., after 9 DIV) led to a significant and almost twofold increase (relative to the BSA-treated cells) of TH+/PITX3+ double-positive cells in the WT cultures, whereas TH+/PITX3− or TH−/PITX3+ single-positive cell numbers were not altered in these cultures (Fig. 4A,B,E). The DKK3 treatment also rescued the numbers of TH/PITX3 single- and double-positive cells in the Dkk3−/− cultures to approximately the same numbers as in control (BSA)-treated WT cultures after 9 DIV (Fig. 4A,C–E). At this time point (9 DIV), BSA treatment of the Dkk3−/− cultures resulted in a significant reduction of TH/PITX3 single-positive (by 50–56%) and double-positive (by 74%) cells (Fig. 4A,C,E), thus reflecting similar changes in the mutant VM at E18.5 (Fig. 3L). These results indicated that exogenous DKK3 protein promoted the differentiation of an mdDA precursor subset into TH+/PITX3+ mdDA neurons under normal (WT) conditions and rescued the loss of TH/PITX3 single- and double-positive cells in the Dkk3−/− mutant cultures.
Figure 4.
DKK3 promotes the differentiation and survival of an mdDA neuron subset in vitro. A–D, F–I, K–N, Representative confocal overviews of primary VM cells isolated from E11.5 WT (A, B, F, G, K, L) and Dkk3−/− (C, D, H, I, M, N) embryos and treated for 7 d (9 DIV) with BSA (control; A, C, F, H, K, M) or DKK3 (B, D, G, I, L, N) protein. Cells were immunostained for TH (green) and PITX3 (red) (A–D), EdU (green) and pH3 (red) (F–I), or cCASP3 (red) (K–N) and counterstained with DAPI (blue). Yellow arrows in A–D point to TH+/PITX3+ mdDA neurons, yellow arrowheads in F–I point to pH3+ mitotic cells, and white arrows in K–N point to cCASP3+ apoptotic cells. E, J, O, Quantification of TH+/PITX3−, TH−/PITX3+, and TH+/PITX3+ (E), EdU+ (J), and cCASP3+ (O) cells per DAPI+ cells after 6 and 9 DIV (E, J) or 9 DIV (O) in BSA- or DKK3-treated WT and Dkk3−/− primary VM cultures (n ≥ 3 experiments/genotype/condition). *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant in the t test. Color coding of bars shown in J and O as in the legend for E. Scale bars: C, H, 50 μm.
The treatment of WT and Dkk3−/− primary VM cultures with DKK3 protein did not change the numbers of proliferating EdU+ (S-phase) and pH3+ (M-phase) cells after 4 d (6 DIV) or 7 d (9 DIV; Fig. 4F–J), indicating that DKK3 did not alter the proliferation of VM progenitors in vitro. However, the significant approximately fourfold increase of apoptotic (cCASP3+) cells in the BSA-treated Dkk3−/− cultures after 9 DIV was reduced by DKK3 treatment of these cultures to approximately one-third (∼38%) (Fig. 4K–O). Nevertheless, the numbers of apoptotic cells still remained significantly increased (∼1.8-fold) in the mutant cultures relative to the DKK3-treated WT cultures (Fig. 4K–O). This suggested that exogenous DKK3 protein might promote to a certain extent the survival of Dkk3-deficient VM cells in vitro.
DKK3 regulates the expression of LMX1A in mdDA precursors and neurons in vivo and in vitro
Our data so far indicated that DKK3 was required for the sustained expression of PITX3 in a developing mdDA neuronal subset. Dkk3 transcription initiates in the mouse VM at approximately the same time (E11.5) when Pitx3 starts to be expressed in postmitotic mdDA precursors and neurons (Fig. 1; Smidt and Burbach (2007)), and the two expression domains partly overlap in the developing mouse VM at this early developmental stage (Fig. 1J). However, previous data suggested that Pitx3 is an indirect target gene of WNT1/β-catenin signaling in the embryonic mouse VM, which is bound and activated by the direct WNT1/β-catenin target and HD TF LMX1A (Chung et al., 2009). The numbers of TH−/LMX1A+ single- and TH+/LMX1A+ double-positive cells were indeed decreased significantly by 40 and 12.5%, respectively, in the rostrolateral but not caudomedial region of the Dkk3−/− VM at E14.5 (Fig. 5A–C). In contrast, the numbers of TH+/LMX1A− single-positive cells were increased by ∼2.6-fold in the rostrolateral mdDA domain of the mutants at this stage (Fig. 5A–C). These changes corresponded well with a similar decrease of TH−/PITX3+ (by 39%) and increase of TH+/PITX3− (by ∼2.8-fold) single-positive cells in the mutant VM at this stage (Fig. 3K). Conversely, DKK3 treatment of primary VM cells for 7 d (9 DIV) increased the transcription of Lmx1a by ∼1.5-fold in the WT cultures and rescued the decreased expression of Lmx1a (∼33% reduction) in control (BSA)-treated Dkk3−/− cultures to levels similar to those of their BSA-treated WT counterparts (Fig. 5D). LMX1B is another LMX1 TF that also binds to the Pitx3 promoter and compensates partially for the loss of Lmx1a (Chung et al., 2009; Deng et al., 2011; Yan et al., 2011). In contrast to PITX3 and LMX1A, we did not detect any obvious changes of LMX1B expression in the Dkk3−/− VM at E12.5, except for an apparently slight reduction of TH+/LMX1B+ double-positive cells at this stage (Fig. 5E–J). In line with this finding, the treatment of primary VM cells with DKK3 protein for 7 d (9 DIV) did not affect the endogenous expression of Lmx1b in the mutant and WT cells (Fig. 5K). The selective failure of an mdDA precursor subset to differentiate into TH-, LMX1A-, and PITX3-expressing rostrolateral (PBP and dorsomedial SNc) mdDA neurons, and the later loss of these neurons, was therefore most likely attributable to the failure to sequentially activate and/or maintain the transcription of Lmx1a and Pitx3 in these cells in the absence of Dkk3.
Figure 5.
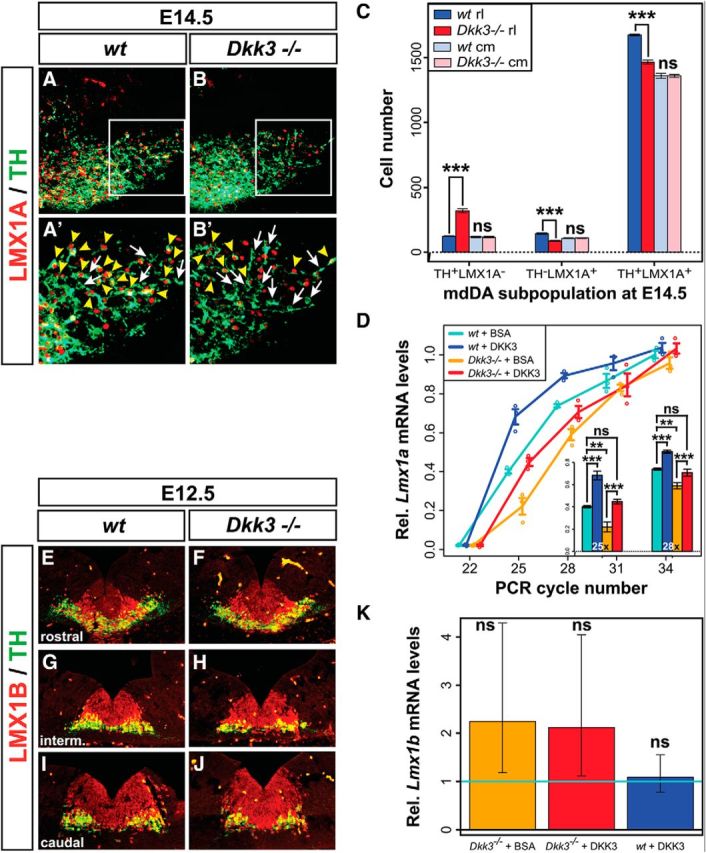
DKK3 regulates the expression of LMX1A in mdDA precursors and neurons in vivo and in vitro. A, B, Representative close-up views of the VM on coronal sections (dorsal top) from WT (A) and Dkk3−/− (B) embryos at E14.5, immunostained for LMX1A (red) and TH (green). A′ and B′ are higher magnifications of the boxed areas in A and B, respectively. White arrows in A′ and B′ point to TH+/LMX1A− single-positive cells; yellow arrowheads in A′ and B′ point to TH+/LMX1A+ double-positive cells. C, Quantification of TH+/LMX1A−, TH−/LMX1A+, and TH+/LMX1A+ cells in WT and Dkk3−/− embryos at E14.5 (n = 4 embryos/genotype). D, Relative Lmx1a mRNA levels (semi-qPCR) in Dkk3+/+ (WT) and Dkk3−/− primary VM cells treated for 7 d (9 DIV) with BSA (+BSA) or DKK3 (+DKK3) protein (n = 3 experiments/genotype/treatment). E–J, Representative close-up views of the VM on coronal sections (dorsal top) from WT (E, G, I) and Dkk3−/− (F, H, J) embryos at E12.5, immunostained for LMX1B (red) and TH (green). Sections are from anterior (E, F), intermediate (G, H), and posterior (I, J) levels of the VM. K, Relative Lmx1b mRNA levels (qPCR) in WT and Dkk3−/− primary VM cells treated for 7 d (9 DIV) with BSA or DKK3 protein [fold-change relative to the untreated WT control (wt + BSA) set as 1, light blue line; n = 3 experiments/genotype/treatment]. Error bars in K represent 95% confidence intervals; **p < 0.01; ***p < 0.001; ns, not significant in the unpaired t test (genotypes)/paired t test (treatments). cm, Caudomedial mdDA domain; rl, rostrolateral mdDA domain.
Combined DKK3/WNT1 treatment generates the highest proportion of TH+/PITX3+ mdDA neurons from differentiating murine PSCs
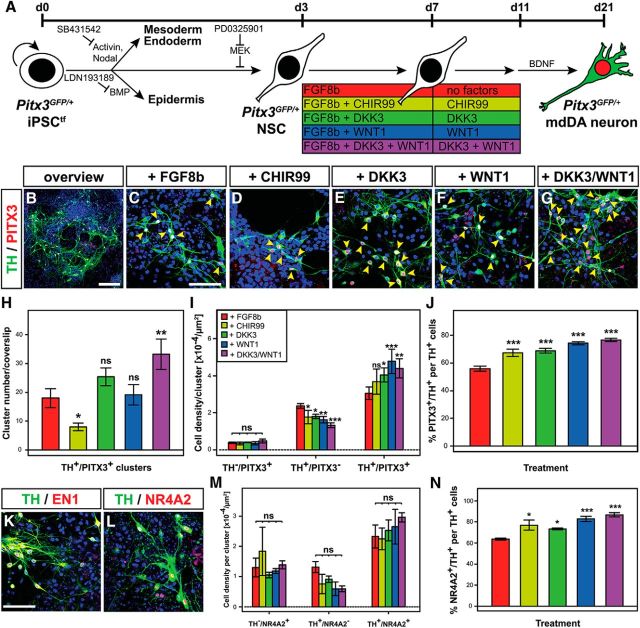
The previous results prompted us to investigate whether the treatment of PSCs (mouse ESCs and iPSCs) with DKK3 protein might enhance their differentiation into TH+ and PITX3+ mdDA neurons. We tested the function of DKK3 in the absence and presence of WNT1, because the WNT1 ligand might be necessary for the action of DKK3 in these cultures and/or because both soluble factors might act synergistically during PSC differentiation. The high variability of differentiation outcomes using the published embryoid body-based “five-stage” protocol (Lee et al., 2000) for the differentiation of mouse ESCs into mdDA neurons (data not shown) motivated us to develop an adherent monolayer differentiation protocol that was adapted from established procedures for the neural and mdDA differentiation of mouse and human PSCs (Chambers et al., 2009; Jaeger et al., 2011; Kriks et al., 2011). For this purpose, we also generated a transgene-free (tf) and “mdDA-reporter” iPSC line (Pitx3GFP/+ iPSCtfs) from Pitx3GFP/+ MEFs (Pertek et al., 2014). The treatment of differentiating Pitx3GFP/+ iPSCtfs with recombinant DKK3 and WNT1 proteins consistently led to the highest number of TH+/PITX3+ double-positive cell clusters compared with all other conditions and promoted the generation of TH+/PITX3+ double-positive cells at the expense of TH+/PITX3− single-positive cells in these clusters, reaching up to ∼77% TH+/PITX3+ double-positive cells among the generated TH+ neurons (Fig. 6A–J). This effect was reproduced independently with another murine PSC line [NesE–eGFP mouse ESCs (Friling et al., 2009); data not shown] and corresponded to similar changes observed after DKK3 treatment of primary VM cells (Fig. 4). Remarkably, the treatment of differentiating Pitx3GFP/+ iPSCtfs with the potent GSK3b inhibitor and WNT/β-catenin signaling activator CHIR99 (Kriks et al., 2011) reduced significantly the numbers of TH+/PITX3+ double-positive cell clusters and had no significant effect on double-positive cell densities in these clusters (Fig. 6D,H,I). The TH+ mdDA neurons generated after combined DKK3 + WNT1 treatment of differentiating Pitx3GFP/+ iPSCtfs also expressed other markers typical for these cells, such as the HD TF EN1 and NURR1 (Fig. 6K,L). The densities of TH+/NR4A2+ double-positive cells showed a similar tendency to be increased at the expense of TH+/NR4A2− single-positive cells after combined DKK3 + WNT1 treatment of differentiating Pitx3GFP/+ iPSCtfs, and the proportion of TH+/NR4A2+ double-positive cells among all TH+ neurons was also significantly increased to ∼87% under these conditions (Fig. 6M,N). Because the combined DKK3 + WNT1 treatment generated the highest proportions of PITX3+/TH+ and NR4A2+/TH+ double-positive cells compared with all other conditions tested (Fig. 6J,N), we concluded that DKK3 together with WNT1 indeed promoted the differentiation of murine PSCs into TH+, PITX3+, and NR4A2+ mdDA neurons.
Figure 6.
Combined DKK3/WNT1 treatment generates the highest proportion of TH+/PITX3+ and TH+/NR4A2+ mdDA neurons from differentiating murine iPSCs. A, Scheme of the monolayer protocol for the differentiation of Pitx3GFP/+ iPSCtfs into mdDA neurons. B–G, Representative confocal overview (B) and close-up views (C–G) of TH+ (green) and PITX3+ (red) cells (yellow arrowheads) after treatment of differentiating Pitx3GFP/+ iPSCtfs with FGF8b alone (C) or together with CHIR99 (D), DKK3 (E), WNT1 (F), or DKK3 + WNT1 (B, G). H, I, Quantification of TH+/PITX3+ double-positive cell clusters per coverslip (H) and TH/PITX3 single- and double-positive cell densities within these clusters (I) after treatment of differentiating Pitx3GFP/+ iPSCtfs with different factor combinations (n = 5 experiments/condition). J, Quantification of the proportion of PITX3+/TH+ double-positive cells among all TH+ cells in these cultures (n = 3 experiments/condition). K, L, Representative confocal close-up views of TH+ (green)/EN1+ (red) (K) and TH+ (green)/NR4A2+ (red) (L) cells after treatment of differentiating Pitx3GFP/+ iPSCtfs with DKK3 + WNT1. M, N, Quantification of TH/NR4A2 single- and double-positive cell densities within clusters (M) and the proportion of NR4A2+/TH+ double-positive cells among all TH+ cells (N) after treatment of differentiating Pitx3GFP/+ iPSCtfs with different factor combinations (n = 3 experiments/condition). Color coding of bars in H–J, M, and N as indicated in the scheme in A; statistical testing for significance in H–J, M, and N was always done in relation to the untreated (only FGF8b-treated) cells. *p < 0.05; **p < 0.005; ***p < 0.001; ns, not significant in one-way (H, J, N) or two-way (I, M) ANOVA relative to the FGF8b-treated cells. Scale bars: B, 100 μm; C, K, 50 μm.
Global gene expression changes in DKK3/WNT1-treated versus untreated iPSC-derived Pitx3GFP/+ mdDA neurons
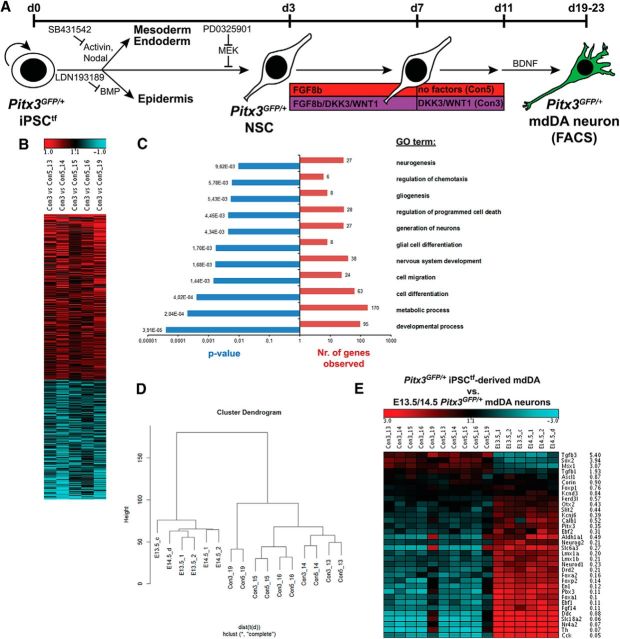
To assess the global effects of the combined DKK3 + WNT1 treatment on the transcriptome of the iPSC-derived Pitx3GFP/+ mdDA neurons in relation to the untreated (only FGF8b-treated) and the endogenous (in vivo differentiated) E13.5/E14.5 Pitx3GFP/+ mdDA neurons, we performed a microarray-based gene expression profiling of these cells. For this purpose, we took advantage of their endogenous expression of enhanced GFP (eGFP) under the control of the Pitx3 promoter and feasibility for direct FACS (Zhao et al., 2004; Pertek et al., 2014 and data not shown; Fig. 7A). Heat map comparison of five independent differentiation experiments showed that the expression of 460 probe sets corresponding to 422 distinct genes was consistently (p < 0.05) upregulated or downregulated in the DKK3 + WNT1-treated compared with the untreated iPSC-derived Pitx3GFP/+ mdDA neurons (Fig. 7B). GO term enrichment analyses of these differentially regulated genes revealed the overrepresentation of GO terms such as developmental/metabolic processes, cell differentiation, and cell migration (Fig. 7C). However, hierarchical clustering analysis showed that the primary Pitx3GFP/+ mdDA neurons isolated from the E13.5 and E14.5 VM formed a distinct group from the iPSC-derived Pitx3GFP/+ mdDA neurons (Fig. 6D). A closer inspection of selected mdDA progenitor- and neuron-specific genes in these groups revealed that some genes expressed in PSCs and implicated in neural induction and/or mdDA neurogenesis [such as Tgfb3, Sox2, and Tgfb1, (Oshimori and Fuchs, 2012; Sarkar and Hochedlinger, 2013; Hegarty et al., 2014)] were upregulated, whereas the majority of genes transcribed in maturing mdDA neurons [including Th, Nr4a2, En1, Drd2, Lmx1a/b, Slc6a3, Pitx3, Calb1, and Kcnj6, (Smidt and Burbach, 2007; Hegarty et al., 2013)] were expressed at lower levels in the iPSC-derived compared with the primary Pitx3GFP/+ mdDA neurons (Fig. 7E). Notably, some midbrain FP and mdDA progenitor marker genes [such as Corin, Ascl1, and Foxp1 (Kele et al., 2006; Ono et al., 2007; Konstantoulas et al., 2010)] were expressed at similar levels in primary and iPSC-derived mdDA neurons (Fig. 7E). There was little or no variation in the transcript levels of these genes between the DKK3 + WNT1-treated and untreated iPSC-derived Pitx3GFP/+ mdDA neurons relative to their in vivo counterparts (Fig. 7E). Moreover, the global transcriptome profiling of the DKK3 + WNT1-treated versus untreated iPSC-derived Pitx3GFP/+ mdDA neurons did not reveal any significant (p < 0.05) changes between both treatments in the expression levels of selected genes associated with WNT/β-catenin signaling and mdDA neuron development in vivo, except for Sox17, Sox10, Wnt4, Fzd2, and Sfrp4, which were significantly downregulated in the DKK3 + WNT1-treated cells (data not shown). These data showed that the DKK3 + WNT1 treatment had clear effects on the global gene expression profile of the in vitro-differentiated (iPSC-derived) mdDA neurons, but PSC-specific and especially mdDA-specific transcript levels in these cells appeared to be influenced by their derivation from reprogrammed MEFs (Roessler et al., 2014).
Figure 7.
Global gene expression changes in DKK3/WNT1-treated versus untreated iPSC-derived Pitx3GFP/+ mdDA neurons. A, Scheme of the monolayer protocol for the differentiation of Pitx3GFP/+ iPSCtfs with the conditions analyzed by microarray of FACS-sorted eGFP-expressing Pitx3GFP/+ mdDA neurons (5 independent differentiation experiments: 13, 14, 15, 16, and 19). B, C, Heat map (B) and GO term analysis (C) of differentially regulated genes in the DKK3 + WNT1-treated iPSC-derived Pitx3GFP/+ mdDA neurons (Con3) relative to the untreated cells (Con5) (red, upregulated; blue, downregulated genes). D, Hierarchical clustering analysis of the global gene expression profiles of E13.5/E14.5 primary and DKK3 + WNT1-treated (Con3) or untreated (Con5) iPSC-derived Pitx3GFP/+ mdDA neurons. Clustering was done with the R script hclust on probe sets with an average expression value >50 in at least one of the four groups. The ordinate in the hierarchical cluster dendrogram indicates the distance of samples. E, Expression levels of selected mdDA progenitor- or mdDA neuron-specific genes in DKK3 + WNT1-treated (Con3) and untreated (Con5) iPSC-derived Pitx3GFP/+ mdDA neurons, as well as their E13.5/E14.5 in vivo counterparts. Average ratios shown are Con3 (DKK3 + WNT1-treated) versus E13.5 Pitx3GFP/+ mdDA neurons.
WNT/β-catenin factor treatment promotes a rostrolateral (SNc) DA and suppresses a caudomedial (VTA) DA neuronal fate in differentiating PSCs in vitro
The selective requirement of DKK3 for the correct differentiation of a rostrolateral (PBP and dorsomedial SNc) mdDA neuron subset in vivo suggested that WNT/β-catenin factor treatment of differentiating PSCs might also promote the generation of this particular mdDA neuron subtype in vitro. Because of the lack of a specific and restricted marker for this mdDA neuronal subtype, we used CALB1 (also known as CalbindinD28K), a calcium-binding protein confined mostly to PITX3− VTA and dorsomedial SNc DA neurons (Liang et al., 1996; Fu et al., 2012; Luk et al., 2013), and KCND3 (also known as Kv4.3), an A-type potassium channel subunit expressed in mature SNc DA neurons (Serôdio and Rudy, 1998; Liss et al., 2001) to discriminate at least between these two neuronal subsets among the in vitro-differentiated (iPSC-derived) mdDA neurons. The treatment of differentiating Pitx3GFP/+ iPSCtfs with CHIR99, DKK3, and/or WNT1 led to a significant decrease (by ∼51%) of the proportion of TH+ neurons coexpressing CALB1 (Fig. 8A,B,F) and a significant increase (by ∼39%) of the proportion of TH+ neurons coexpressing KCND3 (Fig. 8A,C,G). Although the majority of the TH+ iPSC-derived Pitx3GFP/+ mdDA neurons also expressed the DAT (also known as SLC6A3; Fig. 8D), only a subset of these cells coexpressed the G-protein-activated inwardly rectifying potassium channel GIRK2 (also known as KCNJ6; Fig. 8E), which is detected in SNc and VTA DA neurons (Fu et al., 2012; Reyes et al., 2012). Altogether, our data indicated that WNT/β-catenin factor treatment of differentiating mouse PSCs indeed promoted, at least at the molecular level, a PITX3+/CALB1−/KCND3+/DAT+ identity (dorsomedial SNc and dorsolateral VTA) and suppressed a PITX3−/CALB1+/KCND3−/DAT− identity [central and ventromedial VTA (Di Salvio et al., 2010a,b)] of the generated mdDA neurons.
Figure 8.
WNT/β-catenin factor treatment promotes a rostrolateral (SNc) and suppresses a caudomedial (VTA) DA neuronal fate in differentiating PSCs in vitro. A, Scheme of the monolayer protocol for the differentiation of Pitx3GFP/+ iPSCtfs into mdDA neurons. B–E, Representative confocal close-up views of TH+ (green)/CALB1+ (red) (B), TH+ (red)/KCND3+ (green) (C), TH+ (red)/DAT+ (green) (D), and TH+ (red)/GIRK2+ (green) (E) cells after treatment of differentiating Pitx3GFP/+ iPSCtfs with DKK3 + WNT1. F, G, Quantification of the proportions of CALB1+/TH+ (F) or KCND3+/TH+ (G) double-positive cells among all TH+ cells after treatment of differentiating Pitx3GFP/+ iPSCtfs with factor combinations as color coded in the scheme in A (n = 3 experiments/condition). Statistical testing for significance in F and G was done in relation to the untreated (only FGF8b-treated) cells. H–M, Representative close-up views of the VM on coronal sections (dorsal top) from WT (H–J) and Dkk3−/− (K–M) embryos at E18.5, immunostained for TH (green) and CALB1 (red). Sections were taken from rostral (H, K), intermediate (I, L), and caudal (J, M) levels of the VM. N, Quantification of the amount of TH+/CALB1− single-positive and TH+/CALB1+ double-positive cells in the SNc and VTA of E18.5 WT or Dkk3−/− embryos (n = 3 embryos/genotype). *p < 0.05 by the t test; ***p < 0.001 by one-way ANOVA relative to the FGF8b-treated cells; ns, not significant in the t test. CLi, Caudal linear nucleus; IF, interfascicular nucleus; PN, paranigral nucleus; RrF, retrorubral field; SNcd, Substantia nigra pars compacta dorsalis. Scale bar in D, 50 μm.
The previous results prompted us to determine the amount of mdDA neurons expressing TH alone or together with CALB1 in the E18.5 Dkk3−/− brain. The numbers of TH+/CALB1− single-positive cells were increased significantly, whereas TH+/CALB1+ double-positive cells were decreased significantly, in the rostrolateral (SNc) but not caudomedial (VTA) domain of the mutant VM at E18.5 (Fig. 8H–N). In fact, the proportion of CALB1+/TH+ double-positive cells among all TH+ neurons was reduced by ∼21% in the Dkk3−/− SNc and remained unaffected in the Dkk3−/− VTA (data not shown), thus corresponding to a similar reduction (−21%) of PITX3+/TH+ double-positive mdDA neurons in the rostrolateral VM (PBP and dorsomedial SNc) of the mutant embryos at this stage (Fig. 3L). This finding indicated that the lack of Dkk3 in the murine VM resulted in the general failure of an mdDA precursor subset to differentiate correctly into TH+/PITX3+/LMX1A+/CALB1+ rostrolateral [dorsolateral VTA (PBP) and dorsomedial SNc] mdDA neurons and in the later loss of these neurons.
Discussion
We show that the secreted glycoprotein DKK3 was necessary and sufficient for the correct differentiation of an mdDA precursor subset into TH-, LMX1A-, and PITX3-expressing rostrolateral (PBP and dorsomedial SNc) mdDA neurons and their subsequent maintenance in the murine VM in vivo and primary VM cell cultures in vitro. We also show that the combined treatment of differentiating murine PSCs with DKK3 and WNT1 promoted their differentiation into mdDA neurons with molecular SNc DA neuron characteristics, indicating that DKK3 is an important factor for the in vivo and in vitro generation of these neurons.
The onset of Dkk3 transcription in the E11.5 mouse VM coincided with the extinction of Dkk1 and Dkk2 expression in this region of the brain (Monaghan et al., 1999; Ribeiro et al., 2011). DKK1 and DKK2 are known inhibitors of WNT/β-catenin signaling (Niehrs, 2006), whereas DKK3 appears to modulate both positively and negatively this signaling pathway (Nakamura et al., 2007; Nakamura and Hackam, 2010; Das et al., 2013; Xiang et al., 2013). The sharp transition from Dkk1/2 to Dkk3 expression in the murine VM might thus mark the transition from an inactive to active WNT/β-catenin signal transduction in neural progenitors and precursors located within the midbrain FP (Joksimovic et al., 2009). The onset of Dkk3 expression in the midbrain FP also coincided with the peak of mdDA neurogenesis between E10 and E11 in the mouse (Bayer et al., 1995; Bye et al., 2012) and suggested a requirement of DKK3 for the generation of these neurons. Despite its expression throughout the midbrain FP, Dkk3 was required only for the correct differentiation of a rostrolateral (PBP and dorsomedial SNc) mdDA neuronal subpopulation. Therefore, the responsiveness to DKK3 appears to be somehow restricted to the corresponding mdDA progenitor/precursor subset. The early loss of Lmx1b and concomitantly of Wnt1 expression in the mesencephalic lateral FP selectively affects mdDA neurogenesis from this lateral mdDA progenitor domain (Deng et al., 2011), suggesting that the restricted availability of the WNT1 ligand and of some of its downstream signaling effectors within this domain might be one limiting factor.
DKK3 was required specifically for the acquisition of PITX3 expression by a TH+ mdDA precursor subset during early mdDA neurogenesis (at E12.5; Fig. 9B) and for the later maintenance of PITX3, LMX1A, and TH expression in a rostrolateral (PBP and dorsomedial SNc) mdDA neuronal subset (at E14.5–E18.5; Fig. 9C,D). This effect was also recapitulated in WT and Dkk3−/− primary VM cells treated with DKK3 or BSA proteins in vitro. Although the overexpression of WNT1 in differentiating mouse ESCs induces the expression of Pitx3, this HD TF gene does not appear to be a direct target of WNT/β-catenin signaling (Chung et al., 2009). WNT1 overexpression strongly induces the expression of Lmx1a, and the LMX1A HD TF, in turn, appears to activate the transcription of Pitx3 in mouse ESCs (Chung et al., 2009). The Dkk3−/− embryos exhibited a similar defect in the acquisition of LMX1A expression by a TH+ mdDA precursor subset as detected for PITX3, and DKK3 treatment of primary VM cells derived from WT or Dkk3−/− embryos significantly increased or rescued, respectively, the transcription of Lmx1a in these cells. Therefore, the defective differentiation of an mdDA precursor subset in the Dkk3−/− embryos was very likely to be caused by their inability to induce and/or maintain the transcription of Lmx1a and consequently of Pitx3. Notably, the expression of LMX1B was not altered in the Dkk3−/− embryos or DKK3-treated VM cells, thus confirming previous evidence that Lmx1b is not a direct target of WNT1/β-catenin signaling (Chung et al., 2009; Joksimovic et al., 2009), and LMX1B alone is not sufficient for proper mdDA neuron differentiation (Yan et al., 2011).
Figure 9.
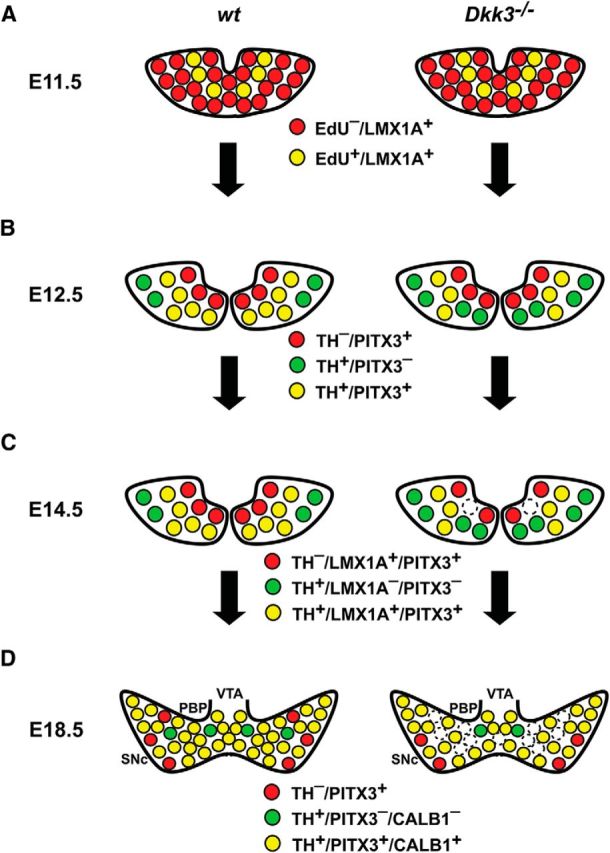
Selective differentiation defects and loss of a rostrolateral mdDA neuronal subset in Dkk3−/− mice. A–D, Schematic coronal views of the rostrolateral mdDA domain in the VM of E11.5 (A), E12.5 (B), E14.5 (C), and E18.5 (D) WT and Dkk3−/− embryos, depicting the phenotypic defects of the mutant embryos. A, Initially, at E11.5, the development of proliferating (EdU+) mdDA progenitors and postmitotic (EdU−) precursors expressing LMX1A (LMX1A+) proceeded normally in the Dkk3−/− embryos. B, At E12.5, the first defects became apparent in the mutant embryos: the numbers of TH+/PITX3− single-positive (green) cells were increased at the expense of TH+/PITX3+ double-positive (yellow) cells in the Dkk3−/− VM, whereas TH−/PITX3+ single-positive (red) cells remained unaffected at this stage. C, At E14.5, the numbers of TH+/LMX1A− or PITX3− single-positive (green) cells were still increased at the expense of TH+/LMX1A+ or PITX3+ double-positive (yellow) cells in the Dkk3−/− VM, but TH−/LMX1A+ or PITX3+ single-positive (red) cells were also decreased at this stage in the mutants. D, Shortly before birth, at E18.5, TH−/PITX3+ (red), TH+/PITX3−, or CALB1− (green), and TH+/PITX3+ or CALB1+ (yellow) single- and double-positive cells were reduced by at least one-fifth (∼20%) in the rostrolateral mdDA domain that comprises mostly the PBP and dorsomedial SNc. In vitro application of DKK3 protein rescued this selective differentiation defect in differentiating primary cells from the Dkk3−/− VM and promoted the differentiation of PSCs into TH+/PITX3+ double-positive mdDA neurons with molecular SNc DA characteristics (CALB1−/KCND3+/DAT+). Note that the coexpression of PITX3 and LMX1A or CALB1 in the same cell, as suggested by the scheme, has not been demonstrated in this work.
The lack of Dkk3 in the murine VM or the treatment of primary VM cells with DKK3 protein did not affect the proliferation of LMX1A+ mdDA and other VM neural progenitors or the initial generation of postmitotic LMX1A+ mdDA precursors (at E11.5; Fig. 9A), indicating that DKK3 had an exclusive prodifferentiation function in developing mdDA precursors. Although the apoptotic death of TH+ mdDA neurons and other VM cells did not appear to be altered in the absence of Dkk3, the treatment with DKK3 protein promoted the survival of primary VM cells in vitro. The loss of TH+ rostrolateral (PBP and dorsomedial SNc) mdDA neurons in the late gestational Dkk3−/− embryos (at E18.5; Fig. 9D) might be an indirect consequence of the lack of PITX3 expression in these cells, because PITX3 is required for the transcription of Bdnf and the retinoic acid-synthesizing and DA metabolizing enzyme Aldh1a1 (aldehyde dehydrogenase family 1, subfamily A1) in SNc DA neurons and thus for their neurotrophic support and neuroprotection (Jacobs et al., 2007; Peng et al., 2011). Alternatively or in addition, DKK3 might act in an autocrine/paracrine manner as an anti-apoptotic and/or prosurvival factor in the mouse VM. There is ample evidence for such a cell type- and context-dependent anti-apoptotic/prosurvival function of DKK3 (Nakamura et al., 2007; Veeck and Dahl, 2012), which might be mediated by a distinct molecular pathway, such as TGFβ/SMAD signaling known to be required for mdDA neuron survival and to interact with DKK3 (Pinho and Niehrs, 2007; Hegarty et al., 2014).
Analogous to the primary VM cells, the treatment of differentiating mouse PSCs with DKK3 and WNT1 proteins promoted the acquisition of the TH+/PITX3+/NR4A2+ mdDA phenotype by these cells. Remarkably, the treatment with WNT/β-catenin signaling factors generally decreased the proportion of TH+ cells coexpressing the predominant VTA DA neuron marker CALB1 (Di Salvio et al., 2010a; Fu et al., 2012) and increased the proportion of TH+ cells coexpressing KCND3, a molecular marker of SNc DA neurons (Serôdio and Rudy, 1998; Liss et al., 2001). Therefore, our data support the idea that WNT/β-catenin signaling is particularly important, at least in the mouse, for the generation of a rostrolateral (PBP and dorsomedial SNc) mdDA neuronal subpopulation in vivo and in vitro.
The treatment of differentiating Pitx3GFP/+ iPSCtfs with DKK3 and WNT1 significantly upregulated or downregulated a number of genes implicated in developmental processes, such as cell differentiation and migration. Despite significant changes in the numbers of cells expressing the corresponding marker protein between the DKK3/WNT1-treated and untreated cultures, the transcript levels of mature mdDA neuronal markers generally (independent of treatment) appeared to be lower, whereas the transcript levels of PSC markers were increased, in the iPSC-derived relative to the primary (E13.5/E14.5) Pitx3GFP/+ mdDA neurons. This suggested either that the iPSC-derived mdDA neurons were still in a relatively immature state after 19–23 DIV compared with their in vivo counterparts or that several of the mdDA-specific genes were epigenetically silenced in these iPSC-derived neurons, as shown recently by Roessler et al. (2014) using an experimental approach very similar to ours. Furthermore, genes associated with the WNT/β-catenin signaling pathway (either as components or direct/indirect targets of this pathway) were not regulated significantly between the DKK3/WNT1-treated and untreated Pitx3GFP/+ mdDA neurons except for five downregulated genes, including Wnt4, Sox10, and Sox17. Interestingly, Wnt4 is a WNT/β-catenin target gene that is repressed by the action of DKK3 (Das et al., 2013), whereas Sox10 and Sox17 are two genes implicated in oligodendrocyte differentiation and SOX17 is a direct antagonist of WNT/β-catenin signaling (Stolt et al., 2002; Sohn et al., 2006; Chew et al., 2011). These findings suggest that DKK3 might be required for the attenuation of active WNT/β-catenin signaling in developing mdDA precursors to facilitate their differentiation into rostrolateral (PBP and dorsomedial SNc) mdDA neurons and to suppress alternative (e.g., oligodendroglial) cell fates. In fact, the treatment of differentiating murine PSCs with the strong activator of WNT/β-catenin signaling CHIR99 rather decreased than increased the amount of TH+/PITX3+ mdDA neurons generated in these cultures, although we used the same CHIR99 concentration that was reported previously to be much more potent than recombinant WNT1 protein for the induction of mdDA neurons from human PSCs (Kriks et al., 2011). Because the constitutive activation of WNT/β-catenin signaling in mouse VM neural progenitors and precursors inhibits their differentiation into mature mdDA neurons in vivo (Joksimovic et al., 2009; Tang et al., 2010), our data also suggested that a difference between mice and humans might be the strength of the WNT/β-catenin signal required for proper mdDA neuron generation. However, the molecular details of this signal transduction pathway and its potential species-specific differences remain to be elucidated.
Altogether, our results showed that DKK3 enhanced specifically the differentiation of mdDA precursors in vivo and PSCs or primary VM cells in vitro into rostrolateral (PBP and dorsomedial SNc) mdDA neurons, without affecting the proliferation and specification of their progenitors. Additionally, DKK3 promoted the survival of primary VM cells in vitro. These particular properties of DKK3, together with its known anti-tumorigenic effects (Veeck and Dahl, 2012), make it another prime candidate for the directed differentiation of PSCs into mdDA (SNc DA) neurons, which can be used in stem cell-based regenerative therapies and disease models of PD.
Footnotes
This work was supported by Italian Association for Cancer Research Grant IG2013 (A.S.), the German Research Foundation within the framework of the Munich Cluster for Systems Neurology (EXC 1010 SyNergy), the Bavarian State Ministry for Education and Culture, Science, and Art within the Bavarian Research Network “Human Induced Pluripotent Stem Cells,” and the Systems Biology of Stem Cells and Reprogramming Project, which received funding from the European Union Seventh Framework Programme for Research, Technological Development, and Demonstration under Grant Agreement FP7-HEALTH-F4-2010-242129 (W.W.). We are particularly grateful to Prof. C. Birchmeier-Kohler and Dr. T. Müller for kindly providing the LMX1B antibody and to Prof. M. Li for providing the Pitx3GFP/+ mice. We thank Dr. M. Endele for help with the FACS of VM cells and A. Bettenbrock, A. Folchert, and S. Badeke for excellent technical assistance. The monoclonal En1 antibody was obtained through the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa (Iowa City, IA).
The authors declare no competing financial interests.
References
- Andersson ER, Saltó C, Villaescusa JC, Cajanek L, Yang S, Bryjova L, Nagy II, Vainio SJ, Ramirez C, Bryja V, Arenas E. Wnt5a cooperates with canonical Wnts to generate midbrain dopaminergic neurons in vivo and in stem cells. Proc Natl Acad Sci U S A. 2013;110:E602–E610. doi: 10.1073/pnas.1208524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas E. Towards stem cell replacement therapies for Parkinson's disease. Biochem Biophys Res Commun. 2010;396:152–156. doi: 10.1016/j.bbrc.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Wills KV, Triarhou LC, Ghetti B. Time of neuron origin and gradients of neurogenesis in midbrain dopaminergic neurons in the mouse. Exp Brain Res. 1995;105:191–199. doi: 10.1007/BF00240955. [DOI] [PubMed] [Google Scholar]
- Björklund A, Dunnett SB. Dopamine neuron systems in the brain: an update. Trends Neurosci. 2007;30:194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Blaess S, Bodea GO, Kabanova A, Chanet S, Mugniery E, Derouiche A, Stephen D, Joyner AL. Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev. 2011;6:29. doi: 10.1186/1749-8104-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla S, Hall AC, Pinto L, Attardo A, Götz M, Huttner WB, Arenas E. Identification of midbrain floor plate radial glia-like cells as dopaminergic progenitors. Glia. 2008;56:809–820. doi: 10.1002/glia.20654. [DOI] [PubMed] [Google Scholar]
- Brodski C, Weisenhorn DM, Signore M, Sillaber I, Oesterheld M, Broccoli V, Acampora D, Simeone A, Wurst W. Location and size of dopaminergic and serotonergic cell populations are controlled by the position of the midbrain–hindbrain organizer. J Neurosci. 2003;23:4199–4207. doi: 10.1523/JNEUROSCI.23-10-04199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bye CR, Thompson LH, Parish CL. Birth dating of midbrain dopamine neurons identifies A9 enriched tissue for transplantation into parkinsonian mice. Exp Neurol. 2012;236:58–68. doi: 10.1016/j.expneurol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco G, Andersson ER, Minina E, Sousa KM, Ribeiro D, Kokubu C, Imai K, Prakash N, Wurst W, Arenas E. Delayed dopaminergic neuron differentiation in Lrp6 mutant mice. Dev Dyn. 2010;239:211–221. doi: 10.1002/dvdy.22094. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew LJ, Shen W, Ming X, Senatorov VV, Jr, Chen HL, Cheng Y, Hong E, Knoblach S, Gallo V. SRY-box containing gene 17 regulates the Wnt/beta-catenin signaling pathway in oligodendrocyte progenitor cells. J Neurosci. 2011;31:13921–13935. doi: 10.1523/JNEUROSCI.3343-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Leung A, Han BS, Chang MY, Moon JI, Kim CH, Hong S, Pruszak J, Isacson O, Kim KS. Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway. Cell Stem Cell. 2009;5:646–658. doi: 10.1016/j.stem.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das DS, Wadhwa N, Kunj N, Sarda K, Pradhan BS, Majumdar SS. Dickkopf homolog 3 (DKK3) plays a crucial role upstream of WNT/beta-CATENIN signaling for Sertoli cell mediated regulation of spermatogenesis. PLoS One. 2013;8:e63603. doi: 10.1371/journal.pone.0063603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. Parkinson's disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/S0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- del Barco-Barrantes I, Montero-Pedrazuela A, Guadaño-Ferraz A, Obregon MJ, Martinez de Mena R, Gailus-Durner V, Fuchs H, Franz TJ, Kalaydjiev S, Klempt M, Hölter S, Rathkolb B, Reinhard C, Morreale de Escobar G, Bernal J, Busch DH, Wurst W, Wolf E, Schulz H, Shtrom S, et al. Generation and characterization of dickkopf3 mutant mice. Mol Cell Biol. 2006;26:2317–2326. doi: 10.1128/MCB.26.6.2317-2326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Andersson E, Hedlund E, Alekseenko Z, Coppola E, Panman L, Millonig JH, Brunet JF, Ericson J, Perlmann T. Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in ventral midbrain development. Development. 2011;138:3399–3408. doi: 10.1242/dev.065482. [DOI] [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Omodei D, Acampora D, Simeone A. Otx2 expression is restricted to dopaminergic neurons of the ventral tegmental area in the adult brain. Int J Dev Biol. 2010a;54:939–945. doi: 10.1387/ijdb.092974ms. [DOI] [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Acampora D, Prosperi R, Omodei D, Prakash N, Wurst W, Simeone A. Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP. Nat Neurosci. 2010b;13:1481–1488. doi: 10.1038/nn.2661. [DOI] [PubMed] [Google Scholar]
- Fischer J, Beckervordersandforth R, Tripathi P, Steiner-Mezzadri A, Ninkovic J, Götz M. Prospective isolation of adult neural stem cells from the mouse subependymal zone. Nat Protoc. 2011;6:1981–1989. doi: 10.1038/nprot.2011.412. [DOI] [PubMed] [Google Scholar]
- Friling S, Andersson E, Thompson LH, Jönsson ME, Hebsgaard JB, Nanou E, Alekseenko Z, Marklund U, Kjellander S, Volakakis N, Hovatta O, El Manira A, Björklund A, Perlmann T, Ericson J. Efficient production of mesencephalic dopamine neurons by Lmx1a expression in embryonic stem cells. Proc Natl Acad Sci U S A. 2009;106:7613–7618. doi: 10.1073/pnas.0902396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yuan Y, Halliday G, Rusznák Z, Watson C, Paxinos G. A cytoarchitectonic and chemoarchitectonic analysis of the dopamine cell groups in the substantia nigra, ventral tegmental area, and retrorubral field in the mouse. Brain Struct Funct. 2012;217:591–612. doi: 10.1007/s00429-011-0349-2. [DOI] [PubMed] [Google Scholar]
- Hegarty SV, Sullivan AM, O'Keeffe GW. Midbrain dopaminergic neurons: a review of the molecular circuitry that regulates their development. Dev Biol. 2013;379:123–138. doi: 10.1016/j.ydbio.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Hegarty SV, Sullivan AM, O'Keeffe GW. Roles for the TGFbeta superfamily in the development and survival of midbrain dopaminergic neurons. Mol Neurobiol. 2014;50:559–573. doi: 10.1007/s12035-014-8639-3. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Statistics. 1979;6:65–70. [Google Scholar]
- Jacobs FM, Smits SM, Noorlander CW, von Oerthel L, van der Linden AJ, Burbach JP, Smidt MP. Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development. 2007;134:2673–2684. doi: 10.1242/dev.02865. [DOI] [PubMed] [Google Scholar]
- Jaeger I, Arber C, Risner-Janiczek JR, Kuechler J, Pritzsche D, Chen IC, Naveenan T, Ungless MA, Li M. Temporally controlled modulation of FGF/ERK signaling directs midbrain dopaminergic neural progenitor fate in mouse and human pluripotent stem cells. Development. 2011;138:4363–4374. doi: 10.1242/dev.066746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joksimovic M, Yun BA, Kittappa R, Anderegg AM, Chang WW, Taketo MM, McKay RD, Awatramani RB. Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat Neurosci. 2009;12:125–131. doi: 10.1038/nn.2243. [DOI] [PubMed] [Google Scholar]
- Kele J, Simplicio N, Ferri AL, Mira H, Guillemot F, Arenas E, Ang SL. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development. 2006;133:495–505. doi: 10.1242/dev.02223. [DOI] [PubMed] [Google Scholar]
- Konstantoulas CJ, Parmar M, Li M. FoxP1 promotes midbrain identity in embryonic stem cell-derived dopamine neurons by regulating Pitx3. J Neurochem. 2010;113:836–847. doi: 10.1111/j.1471-4159.2010.06650.x. [DOI] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, Yang L, Beal MF, Surmeier DJ, Kordower JH, Tabar V, Studer L. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- Liang CL, Sinton CM, German DC. Midbrain dopaminergic neurons in the mouse: co-localization with Calbindin-D28K and calretinin. Neuroscience. 1996;75:523–533. doi: 10.1016/0306-4522(96)00228-X. [DOI] [PubMed] [Google Scholar]
- Li W, Li K, Wei W, Ding S. Chemical approaches to stem cell biology and therapeutics. Cell Stem Cell. 2013;13:270–283. doi: 10.1016/j.stem.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liss B, Franz O, Sewing S, Bruns R, Neuhoff H, Roeper J. Tuning pacemaker frequency of individual dopaminergic neurons by Kv4.3L and KChip3.1 transcription. EMBO J. 2001;20:5715–5724. doi: 10.1093/emboj/20.20.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Rymar VV, van den Munckhof P, Nicolau S, Steriade C, Bifsha P, Drouin J, Sadikot AF. The transcription factor Pitx3 is expressed selectively in midbrain dopaminergic neurons susceptible to neurodegenerative stress. J Neurochem. 2013;125:932–943. doi: 10.1111/jnc.12160. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Kioschis P, Wu W, Zuniga A, Bock D, Poustka A, Delius H, Niehrs C. Dickkopf genes are co-ordinately expressed in mesodermal lineages. Mech Dev. 1999;87:45–56. doi: 10.1016/S0925-4773(99)00138-0. [DOI] [PubMed] [Google Scholar]
- Nakamura RE, Hackam AS. Analysis of Dickkopf3 interactions with Wnt signaling receptors. Growth Factors. 2010;28:232–242. doi: 10.3109/08977191003738832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura RE, Hunter DD, Yi H, Brunken WJ, Hackam AS. Identification of two novel activities of the Wnt signaling regulator Dickkopf 3 and characterization of its expression in the mouse retina. BMC Cell Biol. 2007;8:52. doi: 10.1186/1471-2121-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- Ono Y, Nakatani T, Sakamoto Y, Mizuhara E, Minaki Y, Kumai M, Hamaguchi A, Nishimura M, Inoue Y, Hayashi H, Takahashi J, Imai T. Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E. The harmonies played by TGF-beta in stem cell biology. Cell Stem Cell. 2012;11:751–764. doi: 10.1016/j.stem.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Aron L, Klein R, Li M, Wurst W, Prakash N, Le W. Pitx3 is a critical mediator of GDNF-induced BDNF expression in nigrostriatal dopaminergic neurons. J Neurosci. 2011;31:12802–12815. doi: 10.1523/JNEUROSCI.0898-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Balaguer A, Puelles E, Wurst W, Martinez S. Shh dependent and independent maintenance of basal midbrain. Mech Dev. 2009;126:301–313. doi: 10.1016/j.mod.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Pertek A, Meier F, Irmler M, Beckers J, Skylaki S, Endele M, Wurst W, Prakash N, Kühn R. Simple derivation of transgene-free iPS cells by a dual recombinase approach. Mol Biotechnol. 2014;56:697–713. doi: 10.1007/s12033-014-9748-y. [DOI] [PubMed] [Google Scholar]
- Pinho S, Niehrs C. Dkk3 is required for TGF-beta signaling during Xenopus mesoderm induction. Differentiation. 2007;75:957–967. doi: 10.1111/j.1432-0436.2007.00185.x. [DOI] [PubMed] [Google Scholar]
- Prakash N, Brodski C, Naserke T, Puelles E, Gogoi R, Hall A, Panhuysen M, Echevarria D, Sussel L, Weisenhorn DM, Martinez S, Arenas E, Simeone A, Wurst W. A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development. 2006;133:89–98. doi: 10.1242/dev.02181. [DOI] [PubMed] [Google Scholar]
- Pruszak J, Just L, Isacson O, Nikkhah G. Isolation and culture of ventral mesencephalic precursor cells and dopaminergic neurons from rodent brains. Curr Protoc Stem Cell Biol Chapter. 2009;2 doi: 10.1002/9780470151808.sc02d05s11. Unit 2D.5. [DOI] [PubMed] [Google Scholar]
- Puelles E, Annino A, Tuorto F, Usiello A, Acampora D, Czerny T, Brodski C, Ang SL, Wurst W, Simeone A. Otx2 regulates the extent, identity and fate of neuronal progenitor domains in the ventral midbrain. Development. 2004;131:2037–2048. doi: 10.1242/dev.01107. [DOI] [PubMed] [Google Scholar]
- Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes S, Fu Y, Double K, Thompson L, Kirik D, Paxinos G, Halliday GM. GIRK2 expression in dopamine neurons of the substantia nigra and ventral tegmental area. J Comp Neurol. 2012;520:2591–2607. doi: 10.1002/cne.23051. [DOI] [PubMed] [Google Scholar]
- Ribeiro D, Ellwanger K, Glagow D, Theofilopoulos S, Corsini NS, Martin-Villalba A, Niehrs C, Arenas E. Dkk1 regulates ventral midbrain dopaminergic differentiation and morphogenesis. PLoS One. 2011;6:e15786. doi: 10.1371/journal.pone.0015786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler R, Smallwood SA, Veenvliet JV, Pechlivanoglou P, Peng SP, Chakrabarty K, Groot-Koerkamp MJ, Pasterkamp RJ, Wesseling E, Kelsey G, Boddeke E, Smidt MP, Copray S. Detailed analysis of the genetic and epigenetic signatures of iPSC-derived mesodiencephalic dopaminergic neurons. Stem Cell Rep. 2014;2:520–533. doi: 10.1016/j.stemcr.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W. Multiple dopamine functions at different time courses. Annu Rev Neurosci. 2007;30:259–288. doi: 10.1146/annurev.neuro.28.061604.135722. [DOI] [PubMed] [Google Scholar]
- Serôdio P, Rudy B. Differential expression of Kv4 K+ channel subunits mediating subthreshold transient K+ (A-type) currents in rat brain. J Neurophysiol. 1998;79:1081–1091. doi: 10.1152/jn.1998.79.2.1081. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- Sohn J, Natale J, Chew LJ, Belachew S, Cheng Y, Aguirre A, Lytle J, Nait-Oumesmar B, Kerninon C, Kanai-Azuma M, Kanai Y, Gallo V. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci. 2006;26:9722–9735. doi: 10.1523/JNEUROSCI.1716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuebner S, Faus-Kessler T, Fischer T, Wurst W, Prakash N. Fzd3 and Fzd6 deficiency results in a severe midbrain morphogenesis defect. Dev Dyn. 2010;239:246–260. doi: 10.1002/dvdy.22127. [DOI] [PubMed] [Google Scholar]
- Tabar V, Studer L. Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nat Rev Genet. 2014;15:82–92. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Miyamoto Y, Huang EJ. Multiple roles of beta-catenin in controlling the neurogenic niche for midbrain dopamine neurons. Development. 2009;136:2027–2038. doi: 10.1242/dev.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Villaescusa JC, Luo SX, Guitarte C, Lei S, Miyamoto Y, Taketo MM, Arenas E, Huang EJ. Interactions of Wnt/beta-catenin signaling and sonic hedgehog regulate the neurogenesis of ventral midbrain dopamine neurons. J Neurosci. 2010;30:9280–9291. doi: 10.1523/JNEUROSCI.0860-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeck J, Dahl E. Targeting the Wnt pathway in cancer: the emerging role of Dickkopf-3. Biochim Biophys Acta. 2012;1825:18–28. doi: 10.1016/j.bbcan.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Wurst W, Bally-Cuif L. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci. 2001;2:99–108. doi: 10.1038/35053516. [DOI] [PubMed] [Google Scholar]
- Xiang T, Li L, Yin X, Zhong L, Peng W, Qiu Z, Ren G, Tao Q. Epigenetic silencing of the WNT antagonist Dickkopf 3 disrupts normal Wnt/beta-catenin signalling and apoptosis regulation in breast cancer cells. J Cell Mol Med. 2013;17:1236–1246. doi: 10.1111/jcmm.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CH, Levesque M, Claxton S, Johnson RL, Ang SL. Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J Neurosci. 2011;31:12413–12425. doi: 10.1523/JNEUROSCI.1077-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DX, Marchetto MC, Gage FH. Therapeutic translation of iPSCs for treating neurological disease. Cell Stem Cell. 2013;12:678–688. doi: 10.1016/j.stem.2013.05.018. [DOI] [PubMed] [Google Scholar]
- Zhao S, Maxwell S, Jimenez-Beristain A, Vives J, Kuehner E, Zhao J, O'Brien C, de Felipe C, Semina E, Li M. Generation of embryonic stem cells and transgenic mice expressing green fluorescence protein in midbrain dopaminergic neurons. Eur J Neurosci. 2004;19:1133–1140. doi: 10.1111/j.1460-9568.2004.03206.x. [DOI] [PubMed] [Google Scholar]