Abstract
Epithelial-mesenchymal transition (EMT) is widely-considered to be a modulating factor of anoikis and cancer metastasis. We found that, in MDA-MB-231 cells, TP53I11 (tumor protein P53 inducible protein 11) suppressed EMT and migration in vitro, and inhibited metastasis in vivo. Our findings showed that hypoxic treatment upregulated the expression of HIF1α, but reduced TP53I11 protein levels and TP53I11 overexpression reduced HIF1α expression under normal culture and hypoxicconditions, and in xenografts of MDA-MB-231 cells. Considering HIF1α is a master regulator of the hypoxic response and that hypoxia is a crucial trigger of cancer metastasis, our study suggests that TP53I11 may suppress EMT and metastasis by reducing HIF1α protein levels in breast cancer cells.
Keywords: EMT, HIF1α, Hypoxia, Metastasis, TP53I11
INTRODUCTION
Metastasis, an important characteristic of malignant tumors, is one of the most life-threatening pathological events (1), and is the end-product of a multi-step cell-biological process (2). Epithelial-mesenchymal transition (EMT), which has been well-characterized in embryonic development, is widely-considered to be a modulating factor for anoikis and cancer metastasis (3–7). Many studies have suggested that the loss of epithelial features and the acquisition of mesenchymal properties are the major characteristics necessary for the progression of breast cancer (8).
Hypoxia is a hallmark of solid tumors, and can activate hypoxia inducible factors (HIFs). As a group of DNA binding proteins, HIFs participate in the regulation of angiogenesis, glycolysis, metabolic adaptation, erythropoiesis, cell survival, EMT, and metastasis (9, 10). HIF1α was reported to induce EMT and metastasis by regulating EMT regulators, including TWIST, vimentin, Slug, and E-cadherin (11).
TP53I11 (tumor protein P53-inducible protein 11) was first reported as one of the early transcriptional targets of TP53 (12). So far, the studies on TP53I11 were mostly focused on the positive effect of TP53I11 on apoptosis as a tumor suppressor in cancer cells (13, 14). However, the underlying mechanisms of TP53I11 are still unclear. We found that TP53I11 suppressed EMT and the metastasis of breast cancer cells. Hypoxic stress decreased endogenous TP53I11 protein levels in MDA-MB-231 cells. The overexpression of TP53I11 suppressed HIF1α protein levels in cells under both normal and hypoxic culture conditions. The results suggest that TP53I11 may affect EMT and metastasis through regulation of HIF1α.
RESULTS
TP53I11 inhibits mesenchymal traits
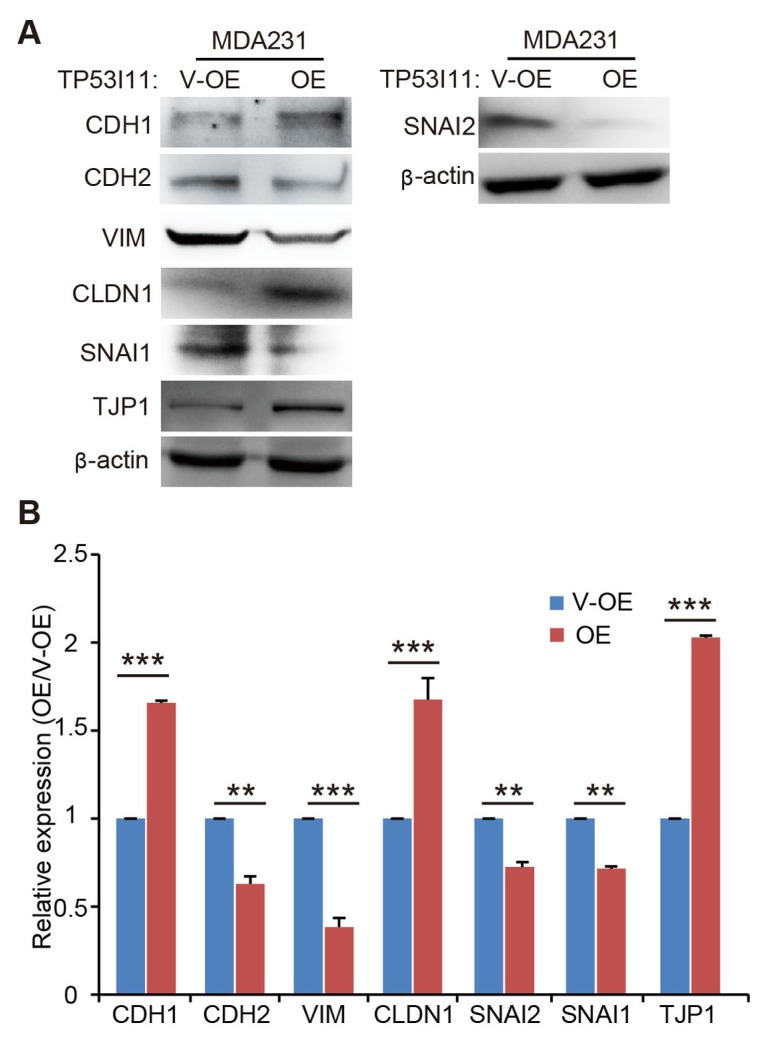
MCF10A is an immortalized human non-tumorigenic mammary epithelial cell line and is widely-used for in vitro studies of normal breast cell function and transformation. MDA-MB-231 is a highly-metastatic breast cancer cell line with typical mesenchymal traits. Our previous study showed that TP53I11 promoted anoikis in MCF10A and MDA-MB-231 cells (15). Anoikis is regarded as a hallmark of oncogenic EMT. To determine if TP53I11 was associated with EMT, we constructed a TP53I11-overexpressing breast cancer MDA-MB-231 cell line (M231-P11-OE) and its vector control cells (M231-V-OE) (Fig. S1A). The effect of TP53I11 on the expression of EMT markers was examined using MDA-MB-231 cells (Fig. 1A and B). We found that TP53I11 overexpression reduced the expression of CDH2 (N-cadherin), VIM (vimentin), SNAI2 (Slug), and SNAI1 (Snail), and enhanced the expression of CDH1 (E-cadherin), CLDN1 (claudin-1), and TJP1 (zonaoccludens 1, ZO-1) (Fig. 1A and B). These results showed the negative effect of TP53I11 on mesenchymal transition.
Fig. 1.
TP53I11 reduces the protein levels of mesenchymal markers in MDA-MB-231. (A) Immunoblotting assay of EMT markers in MDA-MB-231 derived cells. (B) Quantification of the relative protein expression (normalized to β-actin) in immunoblots. V-OE, vector control for overexpression; OE, overexpression; MDA231, MDA-MB-231 cell line. **P < 0.01, ***P < 0.005. P values were determined using a two-tailed student’s t-test. Error bars represent the S.D.s from the average of at least three biological replicates.
TP53I11 inhibits cell migration
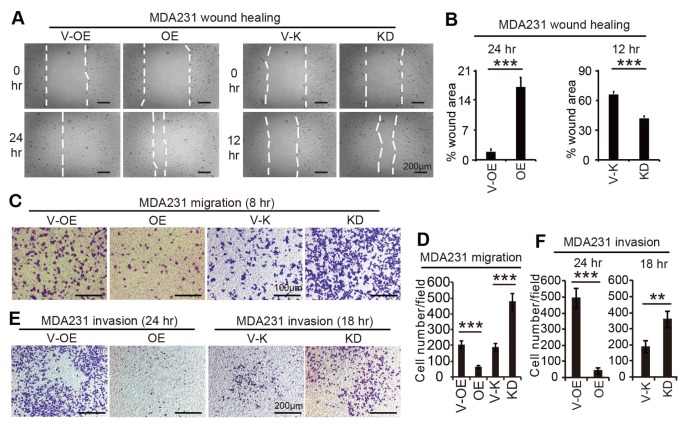
Considering mesenchymal transition is required for cell migration, we therefore investigated if TP53I11 also affected on the migration and invasion of MDA-MB-231 cells (Fig. 2). The results showed that overexpression of TP53I11 suppressed, and loss of TP53I11 promoted, wound closure, cell migration, and invasion in MDA-MB-231 cells (Fig. 2).
Fig. 2.
TP53I11 suppresses migration and invasion in MDA-MB-231. Representative images of wound healing assays (A), transwell migration assays (C), and invasion assays (E) of MDAMB-231 derived cells. The images were captured at the indicated times of cell culture. The percentages of wound areas were calculated and shown in (B). The number of migrated cells and invaded cells in each visual field were counted and shown in (D) and (F), respectively. V-OE, vector control for overexpression; OE, overexpression; V-K, vector control for knockdown or knockout; KD, knockdown; **P < 0.01, ***P < 0.005. P values were determined using a two-tailed student’s t-test. Error bars represent the S.D.s from the average value of at least three biological replicates.
TP53I11 inhibits tumor metastasis in vivo
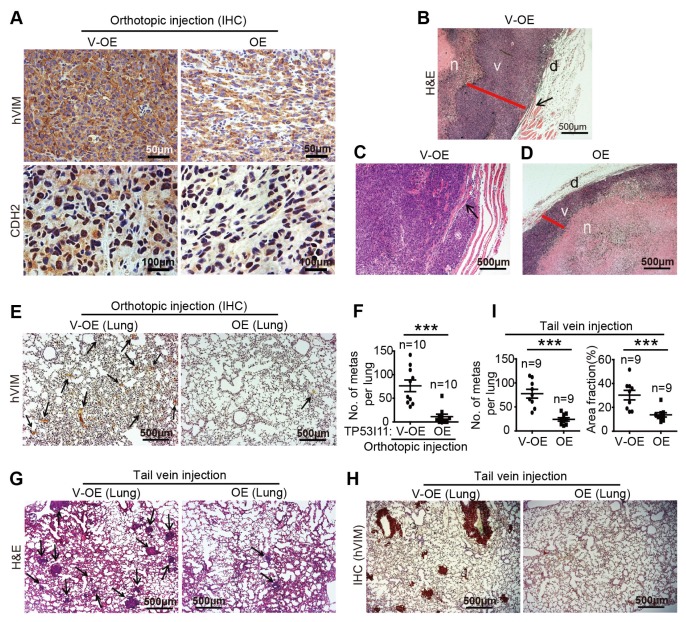
We transplanted M231-V-OE and M231-P11-OE cells into BALB/c female nude mice by orthotopic injection into the mammary fat pads or by tail-vein injection. According to the immunohistochemical (IHC) staining results of tumor sections in the orthotopic injections, we found that TP53I11 overexpression reduced the expression of mesenchymal markers hVIM and CDH2 (Fig. 3A). Three distinct layers in primary tumors were detected by hematoxylin and eosin (H&E) staining. The necrotic tumor center (n) was surrounded by a healthy, viable rim (v), and the outermost layer was dermis (Fig. 3B and D). Compared with the control M231-V-OE tumors, the M231-P11-OE tumors generally demonstrated a smaller viable tumor rim (Fig. 3B and D). We found that TP53I11 overexpression substantially suppressed local invasion (Fig. 3B and C) and significantly reduced the metastatic burden in mice lungs (Fig. 3E and F). The colonization of breast cancer cells into the lungs was also measured by H&E and IHC for human VIM in mice with tail-vein injections (Fig. 3G and H). We found that TP53I11 overexpression significantly reduced the number and the size of tumor colonies in the lungs (Fig. 3I). These in vivo results confirmed our in vitro findings that TP53I11 suppressed EMT, cell migration, and invasion.
Fig. 3.
TP53I11 overexpression reduces MDA-MB-231 cells metastasis in vivo. Orthotopic injection: 231-P11-OE (OE) and vector control 231-V-OE (V-OE) cells were injected into the mammary fat pads of immunodeficient mice. Mice were euthanized 48 days later. (A) The tumors were sectioned and stained for hVIM and CDH2, and with hematoxylin and eosin (H&E). The necrotic center “n” is surrounded by a section of viable tumor rim “v” with an outer dermal layer “d” (B-D). TP53I11 overexpression reduced tumor invasion into nearby tissues. Black arrows point to the invasion locations. Red lines show the thickness of the viable tumor rim. Scale bar = 500 μm. (E) IHC staining for hVIM in lung tissue sections and (F) hVIM-positive nodules in the lungs of mice with orthotopic injections. Tail vein injection: 231-P11-OE (OE) and vector control 231-V-OE (V-OE) cells were injected into immunodeficient mice through the tail veins and the mice were sacrificed 35 days later. (G) H&E and (H) IHC (hVIM) staining of lung tissue sections. (I) The number and size of metastatic nodules. V-OE, vector control for overexpression; OE, overexpression. ***P < 0.005. P values were determined using a two-tailed student’s t-test. Results are expressed as mean ± SD from the indicated number of mice.
Hypoxia treatment decreases TP53I11 protein levels and TP53I11 reduces HIF1α protein levels
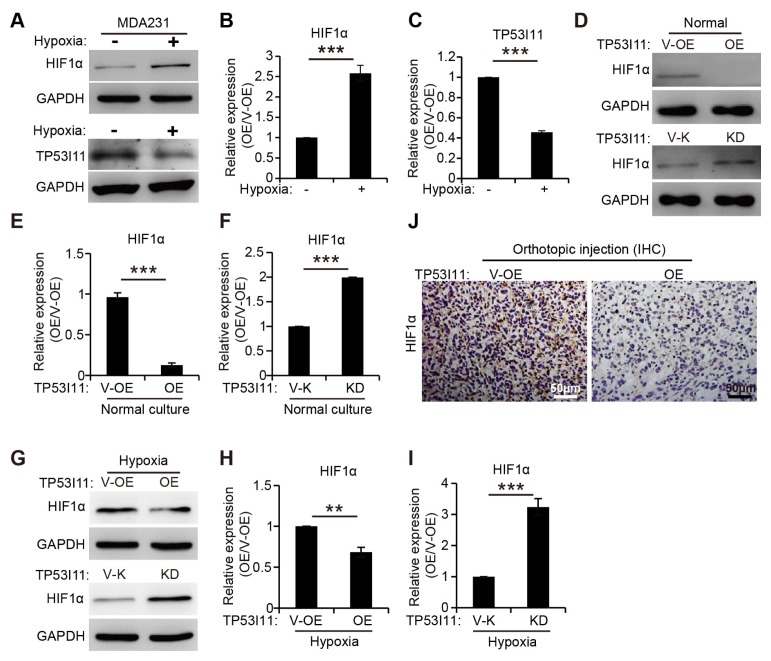
In our study, under hypoxic conditions, HIF1α expression was upregulated, as previously reported (Fig. 4A and B) (16), but, the expression level of TP53I11 in MDA-MB-231 cells was markedly decreased (Fig. 4A and C). A few studies have reported that TP53I11 functioned as a tumor suppressor in some cancer cells (13, 14), and HIF1α acted a crucial role in EMT and metastasis (11, 17). Here, we found that overexpression of TP53I11 decreased, and loss of TP53I11 increased, HIF1α expression under normal culture and hypoxic conditions (Fig. 4D–I). IHC staining of tumor sections showed that overexpression of TP53I11 reduced the HIF1α expression in MDA-MB-231 xenografts (Fig. 4J). The results in Fig. 4 indicate that TP53I11 may suppress the EMT and metastasis by inhibiting HIF1α expression in MDA-MB-231 cells.
Fig. 4.
TP53I11 reduces the HIF1α protein level. (A) Immunoblotting assays showing decreased protein levels of TP53I11 and increased HIF1α after hypoxic treatment. (B and C) Quantification of the relative protein expression (normalized to GAPDH) in immunoblots. The HIF1α protein levels were also examined in MDA-MB-231-derived cells under normal culture (D), hypoxic treatment (G) and in vivo (J). (E, F, H, and I) Quantification of the relative protein expression (normalized to GAPDH) in immunoblots. V-OE, vector control for overexpression; OE, overexpression; V-K, vector control forknockdown or knockout; KD, knockdown. **P < 0.01, ***P < 0.005. P values were determined using a two-tailed student’s t-test. Error bars represent the S.D.s from the average of at least three biological replicates.
DISCUSSION
Our previous research demonstrated that TP53I11 promoted anoikis in MCF10A and MDA-MB-231 cells (15). The fact that anoikis has been widely-considered to be a hallmark of oncogenic EMT motivated us to investigate if, and how TP53I11 affected EMT and cell migration in MDA-MB-231 cells. In this study, we reported that TP53I11 suppressed EMT and cell migration in MDA-MB-231 cells, and inhibited metastasis to mice lungs in vivo. This work supports other findings in which TP53I11 was regarded to be a tumor suppressor gene (13, 18).
As a key mediator of cellular adaptation to hypoxia, HIF1α enhances cell EMT and tumor metastasis (17, 19). Under hypoxic conditions, HIF1α was stabilized and translocated into the nucleus, which resulted in formation of an active HIF1α transcription factor (20). TP53I11 has been reported to be a target gene of Mir-210 (21), and Mir-210 was directly regulated by HIF1α (22). This suggests that HIF1α may regulate TP53I11 expression by regulation of Mir-210. We found that the expression level of HIF1α was upregulated, but TP53I11 expression was reduced by hypoxic treatment. Moreover, HIF1α expression was upregulated by the loss of TP53I11 and downregulated by the overexpression of TP53I11. We also found that TP53I11 exerted no effect on HIF1α transcription (data are not shown). These findings suggest that the TP53I11 may have regulated HIF1α protein accumulation in MDA-MB-231 cells, which then affected EMT and cancer metastasis. Of course, whether HIF1α regulates TP53I11 expression by regulation of Mir-210, and how TP53I11 affects HIF1α accumulation needs more research in the future. Overall, for the first time, we reported the effect of TP53I11 on EMT and breast cancer metastasis and provided new insight into the mechanism of TP53I11 as a tumor suppressor.
MATERIALS AND METHODS
Cell culture
MDA-MB-231 (ATCC, Manassas, VA, USA) cells were grown in complete Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin under humidified conditions in 95% air and 5% CO2 at 37°C (3). For hypoxic treatment, MDAMB-231 were cultured for 12 hours in 1% oxygen, 5% carbon dioxide, and 94% nitrogenin a hypoxia chamber as previously described (15).
Plasmids construction, viral production, and transduction
The human TP53I11 cDNA was cloned from MDA-MB-231 cells by RT-PCR with primers (Table S1) and inserted into a pQCXIH retrovector (Clontech Laboratories, Mountain View, CA). TP53I11 and a control vector retrovirus were produced and used to infect cells for 48 hr. After infecting, the cells were selected with 100 μg/ml hygromycin B. The shRNA for TP53I11 knockdown (Table S1) was cloned into a pLKO.1 vector. Cells were infected with TP53I11-shRNA lentivirus for 48 hr and selected with 1 μg/ml puromycin. Overexpression or knockdown of genes was evaluated by qRT-PCR using the primers shown in Table S1 and by immunoblotting.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The qRT-PCR was performed according to our previous study (3). The qRT-PCR primers for TP53I11 (F: 5’-GGCTCAGGG TCTGGCAGTT-3’; R: 5’-CCATCAAAGACCGCATCATAGA-3’) and GAPDH (F: 5’-ACCCACTCCTCCACCTTTGA-3’; R: 5’-TGTTGCTGTAGCCAAATTCGTT-3’) were designed using Primer Express software (version 2.0, Applied Biosystems).
Immunoblotting
Cell lysates were prepared in lysis buffer (25 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Nonidet P-40, 0.1% SDS, and 1% sodium deoxycholate) supplemented with 1 × protease cocktail. The lysates were cleared by centrifugation at 21,000 × g for 15 minutes at 4°C, separated on SDS-PAGE gels, and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% non-fat milk for 1 hr and incubated with primary antibodies overnight at 4°C, followed by incubation with the corresponding HRP-conjugated secondary antibody. Finally, the membranes were visualized by chemiluminescence. The band intensities were analyzed with Image J software and normalized to β-actin or GAPDH. Antibodies against the following proteins were purchased from Cell Signaling Technology (Beverly, MA, USA): GAPDH, β-actin,N-cadherin (CDH2), E-cadherin (CDH1), vimentin (VIM), Slug (SNAI2), Snail (SNAI1), claudin-1 (CLDN1), ZO-1 (TJP1), and TCF8/ZEB1 (ZEB1).The antibodies against N-cadherin (CDH2) and vimentin (VIM) used for IHC were purchased from ImmunoWay (Plano, TX, USA). The goat anti-rabbit or anti-mouse HRP-conjugated secondary antibodies were also purchased from Cell Signaling Technology (Beverly, MA, USA). The primary antibody against TP53I11 was generated by GL Biochem Ltd (Shanghai, China). Briefly, three mice were immunized three times with the YDAVFDGAQVTSKTP polypeptide. Serum was obtained from blood samples and the specificity of the antibody against TP53I11 was confirmed by immunoblotting using a FLAG antibody against TP53I11-FLAG fusion protein.
Wound healing, Transwell migration, and Matrigel invasion assays
For wound healing, cells were seeded into 6-well plates at a density of 2 × 105 cells per well and cultured at 37°C in growth medium. Monolayers with 90% confluency were starved overnight and a single scratch wound was created using a 200 μl micropipette tip. Wounds were visualized and images were captured by phase-contrast microscopy (Nikon Eclipse TS100) at the indicated hours post-wounding. Areas of the wounds were measured by ImageJ software and normalized to the initial wound area. The migration and invasion assays were performed according to our previous study (3). For the migration assay, MDA-MB-231-derived cells were starved overnight, then plated in the top upper transwell chambers (8 μm pore size; Corning) and cultured in growth medium. Growth medium (500 μl) was added to each bottom lower chamber as an attractant for migration. At the indicated times, cells on the basal side of the membrane were fixed with 100% methanol and stained with 0.1% crystal violet solution. For the invasion assay, each upper chamber was first coated with 50 μl basement membrane growth factor-reduced Matrigel, then seeded with cells and incubated for the indicated hours. The number of cells on the basolateral microporous membrane was determined as described above for the transwell migration assay.
Metastasis assays
The metastasis assays were conducted with our previous methods (3). For orthotopic inoculation, cells (6 × 106 in 60 μl) were injected orthotopically into the mammary fat pad of 6-week-old female nude mice. All animals were sacrificed 48 days after the initial implantation. Primary tumors and lung tissues were collected and fixed in 4% paraformaldehyde for further analyses. For tail inoculation, 1 × 106 viable cells were injected into the lateral tail vein. All animals were sacrificed after 35 days of inoculation. Lung tissues were collected and fixed in 4% paraformaldehyde for further analyses. Paraformaldehyde-fixed tumors and lungs were paraffin-embedded and sectioned (5 μm). Hematoxylin and eosin (H&E) staining and immunohistochemical (IHC) assays using antibodies, such as anti-N-cadherin (CDH2) and anti-human vimentin (hVIM) were performed. The stained sections were observed and the images were acquired using a microscope (ZEISS AXIO microscope, Axio Scope A1). The number of metastatic nodules in each lung was calculated. ImageJ Processing and Analysis software was used to quantify areas of abnormal tissue, which were defined as tissue that appeared as areas of solid mass compared to normal sparsely-looking tissue. The percentage of abnormal lung tissue was calculated using the following formula: ((area of abnormal tissue / total tissue area) × 100). Three sections were used for each animal.
Statistical analysis
All data are expressed as mean ± the standard error of the mean from at least three independent experiments. The mean and S.D. were calculated using Microsoft Excel or GraphPad Prism 5 software. Statistical significance was determined by one-way ANOVA or unpaired two-tailed student’s t-test and P < 0.05 was considered statistically significant.
SUPPLEMENTARY INFORMATION
ACKNOWLEDGEMENTS
We thank Jianwu Dai, Qiangbin Wang, Zhijun Zhang, Renjun Pei, and Guosheng Cheng for insightful comments on the study, Xiang He for her excellent technical assistance. Nano-Bio-Chem Centre in Suzhou Institute of Nano-Tech and Nano-Bionics (SINANO) is acknowledged for professional assistance with cell imaging and FACS assays. This work was supported by the National Natural Science Foundation of China (Grant No. 31471307); and the Ministry of Science and Technology (MOST) (Grant No. 2017YFA0104301 and 2014CB965003). G. Suo was also supported by the Hundred Talent Program of the Chinese Academy of Sciences.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 2.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Y, Xu Z, Quan D, et al. Nuclear respiratory factor 1 promotes spheroid survival and mesenchymal transition in mammary epithelial cells. Oncogene. 2018;37:6152–6165. doi: 10.1038/s41388-018-0349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Au SH, Storey BD, Moore JC, et al. Clusters of circulating tumor cells traverse capillary-sized vessels. Proc Natl Acad Sci U S A. 2016;113:4947–4952. doi: 10.1073/pnas.1524448113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimshaw MJ, Cooper L, Papazisis K, et al. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10:R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kokkinos MI, Wafai R, Meng KW, et al. Vimentin and Epithelial-Mesenchymal Transition in Human Breast Cancer – Observations in vitro and in vivo. Cells Tissues Organs. 2007;185:191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 9.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon Y, Park B, Park H. Hypoxic repression of CYP7A1 through a HIF-1alpha- and SHP-independent mechanism. BMB Rep. 2016;49:173–178. doi: 10.5483/BMBRep.2016.49.3.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh Y, Byeon H, Kim W, et al. 458 Hypoxia-inducible Factor 1a (HIF-1a) Induces Epithelialmesenchymal Transition (EMT) in Thyroid Cancer Cell Lines. Eur J Cancer. 2012;48:142–142. doi: 10.1016/S0959-8049(12)72256-6. [DOI] [Google Scholar]
- 12.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran C, Rodriguez S, Ramachandran R, et al. Expression profiles of apoptotic genes induced by curcumin in human breast cancer and mammary epithelial cell lines. Anticancer Res. 2005;25:3293–3302. [PubMed] [Google Scholar]
- 14.Wu Y, Liu XM, Wang XJ, et al. PIG11 is involved in hepatocellular carcinogenesis and its over-expression promotes Hepg2 cell apoptosis. Pathol Oncol Res. 2009;15:411–416. doi: 10.1007/s12253-008-9138-5. [DOI] [PubMed] [Google Scholar]
- 15.Xiao T, Xu Z, Zhou Y, et al. Loss of TP53I11 Enhances the Extracellular Matrix-independent Survival by Promoting Activation of AMPK. IUBMB Life. 2019;71:183–191. doi: 10.1002/iub.1949. [DOI] [PubMed] [Google Scholar]
- 16.Ghoshal P, Teng Y, Lesoon LA, Cowell JK. HIF1A induces expression of the WASF3 metastasis-associated gene under hypoxic conditions. Int J Cancer. 2012;131:E905–915. doi: 10.1002/ijc.27631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Naggar AM, Veinotte CJ, Cheng H, et al. Translational Activation of HIF1alpha by YB-1 Promotes Sarcoma Metastasis. Cancer Cell. 2015;27:682–697. doi: 10.1016/j.ccell.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Liang XQ, Cao EH, Zhang Y, Qin JF. P53-induced gene 11 (PIG11) involved in arsenic trioxide-induced apoptosis in human gastric cancer MGC-803 cells. Oncol Rep. 2003;10:1265–1269. [PubMed] [Google Scholar]
- 19.Higgins DF, Kimura K, Bernhardt WM, et al. Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J Clin Invest. 2007;117:3810–3820. doi: 10.1172/JCI30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 21.Noman MZ, Buart S, Romero P, et al. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res. 2012;72:4629–4641. doi: 10.1158/0008-5472.CAN-12-1383. [DOI] [PubMed] [Google Scholar]
- 22.Huang X, Ding L, Bennewith KL, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.