Abstract
Radial glial cells are stem cell-like populations of glial nature that supply neurons either directly or indirectly via basal progenitors that give rise to neurons. Here we show that signal transducer and activator of transcription-3 (STAT3) signaling, a cytokine signaling mediated by Janus tyrosine kinase (Jak), is active during neurogenesis in radial glia (RG) but not in basal progenitors. Enhanced STAT3 signaling in cortical progenitors caused more RG to persist rather than become neurons. Targeted deletion or RNAi-mediated knockdown of Stat3 resulted in fewer radial glial cells and more basal progenitors and led to premature neurogenesis. The neuronal populations affected in Stat3 mutant mice were the late-born neurons that constitute the upper cortical layers rather than early-born neurons, thus supporting the view that the role of STAT3 at mid-neurogenesis is layer specific. Analysis of dividing RG revealed that STAT3 selectively increased the proportion of dividing RG, whereas downregulation of STAT3 reduced the proportion. Consistent with this, STAT3 activity in dividing RG was associated frequently with vertical cleavage. Pair-cell analysis showed that elevated STAT3 activity correlated with symmetric division of RG, producing more RG, whereas elimination of STAT3 generated more neurogenic cells. Together, our results suggest that STAT3 maintains the stemness of RG and inhibits their transition to basal progenitors at mid-neurogenesis, so probably preserving a pool of RG for later neurogenesis or gliogenesis.
Keywords: neurogenesis, radial glia, STAT3
Introduction
Radial glial cells are widespread and unique precursor cells in the developing central nervous system that were once thought to merely support migrating newborn neurons as structural scaffolds. Initially, a thin layer of neuroepithelial cells comprising the ventricular zone (VZ) undergoes symmetric divisions to expand a pool of neural stem cells. At the onset of neurogenesis, at approximately embryonic day 9 (E9) and E10 in mice, the transition from neuroepithelium to RG occurs, revealed by the appearance of distinctively long radial fibers that span the entire width of the cortex (Götz and Huttner, 2005). RG mostly undergo asymmetric divisions to self-renew and generate intermediate progenitor cells (IPCs), transient amplifying cells that give rise to neurons. IPCs in the VZ are also called short neural precursors, distinct from the IPCs in the subventricular zone (SVZ; Gal et al., 2006). Both neuroepithelium and RG sequentially supply a diverse repertoire of neurons and glia that constitute the entire brain. Early progenitors tend to produce lower-layer neurons, whereas late progenitors generate upper-layer neurons, creating a cortical lamination in an inside-out manner. At the end of neurogenesis, RG cease to generate neurons and transform into astrocytes or provide glial progenitors (Götz and Huttner, 2005; Kriegstein and Alvarez-Buylla, 2009). Thus, neuroepithelium and RG contribute complementary components of cortical development: the initial expansion of the neuroepithelium dictates overall brain size, whereas the later propagation of RG establishes the balance between layer-specific neurons and glia (Götz and Huttner, 2005; Kriegstein and Alvarez-Buylla, 2009; Pinto et al., 2009).
We hypothesized that STAT3, a cytokine signaling that is important in gliogenesis in perinatal brains, played a key role in early progenitors (Levy and Darnell, 2002). STAT3 and leukemia inhibitory factor (LIF), a cytokine that activates STAT3 and maintains neural stem cells, are present during the neurogenic periods of brain development, especially in the VZ in which progenitors reside (Kamakura et al., 2004; He et al., 2005; Yoshimatsu et al., 2006). Also, LIF is used widely to propagate embryonic stem cells and neural stem cells that maintain their pluripotency (Shimazaki et al., 2001; Pitman et al., 2004). In agreement with this, a reduction of dividing progenitors was reported in mice that lacked the cytokine receptors that initiate the Jak-STAT3 pathway (Shimazaki et al., 2001; Hatta et al., 2002; Gregg and Weiss, 2005; Bauer and Patterson, 2006; Gallagher et al., 2013).
We here describe a novel role of STAT3 as a major determinant of the generation and maintenance of radial glial cells. We show that STAT3 is expressed in RG at the onset of initial neurogenesis and enriched in dividing RG destined to become upper-layer neurons but not in neurogenic basal progenitors. Enhanced STAT3 activity combined with exposure to LIF resulted in the persistence of more RG, whereas absence of STAT3 resulted in a reduction of RG and premature neurogenesis, mainly producing upper-layer neurons. In agreement with this, we found that STAT3 promoted the symmetric division of RG so increasing the overall proliferating population. Together, our findings show that STAT3 is essential for assigning and maintaining RG identity, which is critical for maintaining a balance between neurogenesis and gliogenesis.
Materials and Methods
Mouse lines.
The production of Stat3 floxed mice was described previously (Takeda et al., 1997). Stat3fl/fl;nestin::Cre double-mutant mice were obtained by crossing a male Stat3fl/+;nestin::Cre mouse and a female Stat3fl/fl mouse. The day of vaginal plug formation was designated E0.5. Littermates of Stat3fl/+or Stat3fl/fl genotypes and no Cre were used as controls, unless indicated otherwise. Animals were housed in specific pathogen-free barrier facilities and used in accordance with protocols approved by the Animal Care and Ethics Committees of the Gwangju Institute of Science and Technology.
Immunoblotting.
For Western blot assays, mouse embryonic brains were harvested and samples were processed for immunoblotting as described previously (Kang and Song, 2010). Antibodies used were as follows: rabbit anti-Stat3 (New England Biolabs), rabbit anti-Stat1 (Santa Cruz Biotechnology), mouse anti-GFAP (Sigma), rabbit anti-Sox2 (Millipore), and mouse anti-β-tubulin (Sigma). Secondary goat anti-mouse or anti-rabbit IgG–HRP antibodies (Santa Cruz Biotechnology) were used. Pierce ECL Western Blotting Substrate (Pierce) was used for detection.
Immunohistochemistry and in situ hybridization.
Embryos were fixed in 4% paraformaldehyde (PFA) for immunohistochemistry. Transverse sections of 12 μm thickness or primary cells grown on glass coverslips were incubated with primary antibodies. The following antibodies were used: rabbit, mouse, or chick anti-GFP (Invitrogen, Abcam), mouse anti-RC2 (Developmental Studies Hybridoma Bank), mouse anti-vimentin (Developmental Studies Hybridoma Bank), mouse anti-Nestin (BD Pharmingen), rabbit anti-Tuj1 (Covance), rabbit anti-doublecortin (DCX; Cell Signaling Technology), rabbit anti-Pax6 (Covance), mouse and rabbit anti-Sox2 (Abcam, Millipore), rabbit anti-Cux1 (Santa Cruz Biotechnology), rabbit anti-Ngn2 (Dr. M. Greenberg, Harvard Medical School, Boston, MA), rabbit anti-Tbr2 (Abcam), rabbit anti-Tbr1 (Abcam), rat anti-Ctip2 (Abcam), and mouse anti-γ-tubulin (Sigma). Fluorophore-conjugated species-specific secondary antibodies were used as recommended (The Jackson Laboratory and Invitrogen). To detect STAT3, we used anti-Stat3 antibody (catalog #4904; Cell Signaling Technology), which detects both phosphorylated and nonphosphorylated STAT3, along with autoclaved antigen retrieval (121°C in 0.01 m tri-sodium citrate buffer, pH 6.0) and a TSA kit (Invitrogen), as described previously (Kang et al., 2013). For in situ hybridization, transverse sections were hybridized with digoxigenin-labeled probes specific for Stat3 that were amplified from mouse embryonic cDNA using an Advantage cDNA PCR kit (Clontech).
DNA constructs and in utero electroporation.
Timed pregnant mice were ethically anesthetized with isoflurane combined with oxygen/nitric oxide. DNAs were injected into the lateral ventricles of embryos and electroporated using a squared wave electroporator (BTX; for E13.5 embryos, 5 pulses, 30 V, 50 ms, 950 ms intervals; for E11.5 embryos, 3 pulses, 30 V, 50 ms, 950 ms intervals). To construct short-hairpin RNA (shRNA)-expressing vectors, oligonucleotides targeting the coding sequence and their complementary sequences were inserted into pCAG mir-30 plasmid. The targeting sequences for Stat3 were as follows: Stat3 shRNA 551 (5′-CATGCAGGATCTGAATGGAAAC-3′) and Stat3 shRNA 551 scrambled (5′-GAACCTGAGATATGCGACAAGT-3′). To generate a STAT3-responsive SBS8–H2BdGFP reporter, eight repeats of the STAT3 binding element of the GFAP promoter (TTCCGAGAA, −1518 to −1510 for mouse) were subcloned into pCS2–miniCMV–H2BdGFP, containing the minimal CMV promoter and destabilized nuclear GFP (dGFP) with nuclear localization signal H2B (Takizawa et al., 2001). CMV–H2BdGFP was made by fusing the full-length CMV promoter and dGFP. The Stat3 CA (contains A662C, N664 C mutations) plasmid was generated by site-directed mutagenesis using primer pairs reported in previous studies (Bromberg et al., 1999; Hong and Song, 2014).
P19 cell cultures.
P19 cells were grown with 1 μm retinoic acid (Sigma) to induce embryonic bodies, and ciliary neurotrophic factor (CNTF; 50 ng/ml), a ligand that activates the JAK–STAT signaling pathway, was applied for an additional 3 d. Cells were dissociated with 0.25% trypsin-EDTA (Invitrogen) and replated. Each set of cells was harvested after 2 or 4 days in vitro (DIV), immunostained for brain lipid-binding protein (BLBP), and expressed in RG during development using rabbit anti-BLBP (Abcam) antibody.
Neurosphere cultures.
Cortical cells dissected from E13.5 mouse embryos were dissociated into single-cell suspensions. Primary cells were plated at low density in DMEM/F-12 (Sigma) supplemented with FGF-2 (20 ng/ml; R&D Systems), EGF (20 ng/ml; R&D Systems), and B27 (Invitrogen). CNTF (50 ng/ml; R&D Systems) or LIF (50 ng/ml; Millipore) was added at 3 d after from the start of incubation. For differentiation, neurospheres were dissociated at 7 DIV and plated on poly-d-lysine-coated coverslips in DMEM/F-12 supplemented with 2 ng/ml FGF-2, 2 ng/ml EGF, 2% heat-inactivated horse serum, and B27.
Ex vivo slice culture.
Cortices were isolated from E13.5 mouse embryos. Brains were injected with CAG-based plasmid DNA using microcapillary pipettes with aspirator tube assemblies (Sigma). The electrodes were placed next to the brains and electroporated ex utero using a squared wave electroporator (BTX; 5 pulses, 30 V, 50 ms, 950 ms interval). Electroporated cortices were embedded in 3% agarose gel and sectioned coronally at 300 μm thickness using a vibratome (Ted Pella). Brain slices were placed onto membrane inserts (Corning) and incubated with slice culture medium (basal medium eagle, complete HBSS, 20 mm d-glucose, 1 mm l-glutamine, and 1% penicillin/streptomycin). After 2 d, brain slices were fixed with 4% PFA and underwent immunohistochemistry procedures. To immunostain the slices with anti-DCX and Tuj1 antibodies, fixed slices were cryoprotected with 20% sucrose in PBS and embedded in OCT compound (CellPath Labs). Transverse sections of 16 μm thickness of cortical slices underwent immunohistochemistry procedures (Zhao et al., 2008). Cells were imaged with an FV1000 confocal microscope to test colocalization.
BrdU and ethynyl deoxyuridine incorporation assay.
For cell-cycle analysis, mice were electroporated at E13.5 or E11.5, and ethynyl deoxyuridine (EdU; 50 mg/kg; Life Technologies) was injected intraperitoneally at E15.5 or E13.5, 48 h after electroporation. Embryos were harvested either 2 h or 1 d thereafter, as indicated. Brain sections were stained with a Click-iT EdU Imaging kit (Invitrogen). To obtain labeling indices, we counted the number of EdU+GFP+/GFP+ cells in areas of 100 × 100 μm2 on photographic images. To measure cell-cycle exit and reentry, in utero electroporation was performed in pregnant mice carrying E13.5 embryos. After 2 d, BrdU (50 mg/kg; Sigma) and EdU (50 mg/kg) were administered sequentially intraperitoneally 10 h apart. Embryos were harvested 2 h after EdU injection (Cubelos et al., 2008). BrdU was detected using mouse anti-BrdU (BD Biosciences, Sigma) antibody using an antigen-retrieval method (0.5N HCl, protease K treatment).
Retroviral infection and pair-cell analysis.
For retroviral production, the pBMN–GFP retroviral expression vector, group-specific antigen (GAG), and the G-protein of vesicular stomatitis virus plasmid were transfected into retrovirus-packaging Phoenix ecotrophic cells (American Type Culture Collection). For pair-cell analysis in vivo, we electroporated a minimal amount of DNA (10 ng/μl) at E13.5 and harvested brains at E15.5. We chose regions of the VZ in which labeled cells were at least 50 μm apart from each other. Serial sections of brains were analyzed by z-stack confocal microscopy to locate two-cell clones whose cell bodies contacted each other and were located at least 20 μm above the ventricle within the VZ.
Cleavage plane angle analysis.
For cleavage plane analysis, E13.5 mouse embryos were electroporated with CMV–H2BdGFP or SBS8–H2BdGFP. After 24 h, embryo cortices were prepared and processed for immunostaining. To label mitotic radial glial cells, we used anti-phospho-vimentin (MBL International), γ-tubulin (Sigma), and phospho-histone H3 (PH3; Millipore), a specific marker for condensed chromatin during mitosis and meiosis. Because of the limited choice of antibodies, we used the same channels to detect phospho-vimentin and γ-tubulin, which occupy distinct locations (Postiglione et al., 2011; Xie et al., 2013). For quantification, we considered H2BdGFP, phospho-vimentin, and PH3 triple-positive cells to be dividing RG. The angle of the cleavage plane between the ventricular surface and equatorial plate of dividing radial glial cells in anaphase or telophase was measured with NIH ImageJ software.
Time-lapse image acquisition.
Brain slices were prepared from CMV–H2BdGFP or SBS8–H2BdGFP-electroporated ICR mice. For time-lapse imaging, acute brain slices (300 μm thick) were placed on glass-bottomed 35 mm plastic dishes (SPL Life Science) in 5% horse serum MEM (Invitrogen) at 37°C. The slices were viewed through the 20× objective lens (UPLSAPO, 0.75 numerical aperture) of an Olympus FV1000 inverted confocal microscope. Images were collected in 10 z-steps (∼10 μm intervals) every 15 min. Divisions of RG were observed by monitoring H2BdGFP-labeled nuclei. z-series images were reconstructed with the Olympus FLUOVIEW imaging software.
Quantification analysis.
Cells were counted on z-series of cortical slice images with a 20× objective on a 2× digital zoom using an FV1000 confocal microscope and FLUOVIEW imaging software (Olympus). For each experiment, at least three brains were quantified from each group, and three to four images were collected from each brain. Quantification of cell numbers were performed as follows: for Pax6, Cux1, Ctip2, and Tbr1, the number of cells within the columnar regions of 100 μm width was quantified; for Tbr2 and Ngn2, the one within 2500 μm2 area of the VZ and SVZ were counted. The border between VZ and SVZ was determined by Pax6 expression. The number of phospho-vimentin+Sox2+ outer RG (oRG) cells was counted in the entire one hemisphere because of their limited number. For Cux1/Ctip2/Tbr2 immunostaining images, two adjacent sections were immunostained with anti-Cux1 and Ctip2 antibodies and with anti-Tbr1 antibody, respectively, because of the limited choice of primary antibodies. The images of the adjacent sections were overlaid manually. To exclude RG in the VZ, only Cux1+ neurons located above the Ctip2+ layer were counted (Franco et al., 2012). To quantify RG in organotypic culture, we only counted cells, within 200 × 200 μm2 areas, whose cell bodies were in the VZ and that had GFP+Nestin+ processes in the cortical plate (CP). Typically, three squares were counted in each slice. For SBS8–dGFP labeling, STAT3 overexpression from E13.5 to E17.5 for the EdU and BrdU labeling experiment, cells in a 100 × 100 μm2 area from the ventricular surface or from the pial surface (localization of GFP or STAT3-overexpressed cells in the CP) were quantified.
Results
STAT3 is selectively expressed in RG in the neocortex
To see whether STAT3 signaling was active during neurogenesis, we examined STAT3 expression by Western blotting (Fig. 1A). STAT3 was detected in E10.5 and E12.5 mouse brain lysates when neurogenesis was ongoing. It continued to be present in the brains of E14.5, E16.5, and E18.5 embryos but at a lower level (Fig. 1A). This sequence coincided with the expression of the progenitor markers nestin and Sox2. GFAP, a marker for mature astrocytes directly induced by STAT3, only began to appear at E18.5 when gliogenesis began (Bonni et al., 1997; Barnabé-Heider et al., 2005). Thus, the role of STAT3 may not be limited to glial differentiation; it may also be involved in aspects of neurogenesis. In the E10.5 mouse cortex when most cells are progenitors, STAT3 immunoreactivity was found in the neuroepithelium labeled by nestin (Fig. 1B,C). At E14.5, Stat3 mRNA and protein were restricted to the VZ in which there are radial glial cells labeled by Sox2 and Pax6, especially on the apical side of the VZ (Fig. 1D–G). The basal progenitors (or IPCs) labeled by Tbr2 were found mostly on the basal side of the VZ and SVZ (Fig. 1H). In higher-magnification images, STAT3 expression overlapped with that of nestin and was prominent from the VZ to the intermediate zone (IZ) but was low in the processes close to the pial surface (Fig. 1I,J) (Yoshimatsu et al., 2006). Thus, STAT3 appears to be located in both the nucleus and cytoplasm of RG, suggesting the possibility of a dynamic translocation between nucleus and cytoplasm.
Figure 1.
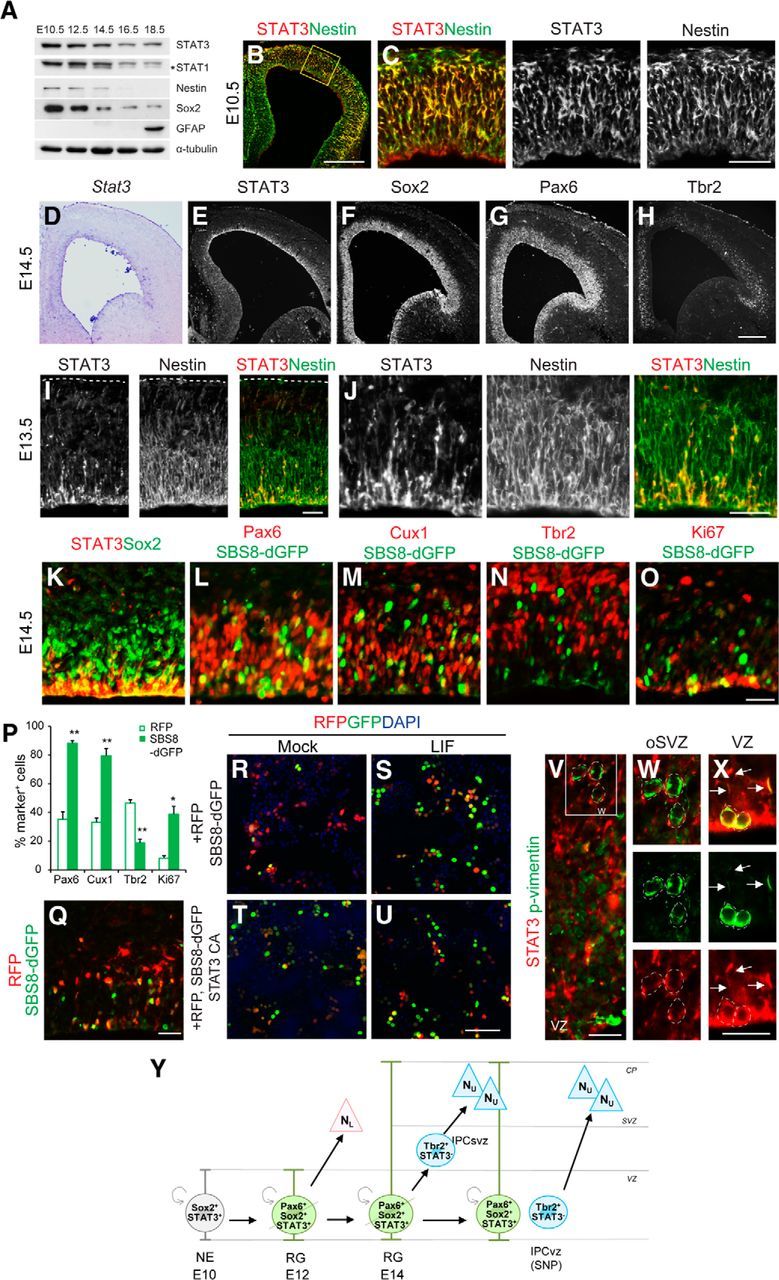
STAT3 expression in mouse embryonic brains. A, Expression of STAT3, STAT1, nestin, Sox2, GFAP, and α-tubulin in embryonic mouse brains assessed by Western blotting. The asterisk marks a splice variant of STAT1. B, C, STAT3 is expressed in the VZ of E10.5 brains marked by Nestin. D–H, At E14.5, Stat3 transcripts and protein are located in the VZ in which Sox2 and Pax6 are expressed. The IPC marker Tbr2 is more broadly expressed. I, STAT3 expression (red) partially overlaps with that of nestin (green) in E13.5 brains. J, Higher-magnification views of I. K–O, STAT3 protein or STAT3 reporter expression is merged with Sox2, Pax6, Cux1, Tbr2, or Ki67. P, Percentages of STAT3 reporter-labeled cells that express the indicated markers (n = 2–4 embryos, n = 3–5 sections per group). Q, Differential expression of CAG–RFP and STAT3 reporter in the coelectroporated cortex. R–U, The SBS8–H2BdGFP reporter and CAG–RFP were transfected into HEK293T cells. The cells were incubated with LIF for 1 d before harvest. The SBS8–H2BdGFP reporter was activated in response to LIF or when STAT3 CA was cotransfected. V–X, oRG cells and phospho-vimentin + cells (green) in the oSVZ do not express STAT3 (red; W). Mitotic RG labeled by phospho-vimentin on the apical side of the VZ express STAT3 in their nuclei and radial processes (X, arrows). Y, A schematic diagram summarizing the expression of STAT3 and other markers among progenitors. For simplicity, only asymmetrically dividing radial glial cells are shown. NE, Neuroepithelium; SNP, short neural precursor; NL, lower-layer neurons; NU, upper-layer neurons. Error bars represent SEM. *p < 0.05, **p < 0.01, unpaired Student's t test. Scale bars: B, 200 μm; C, and (in H) D–H, Q, 50 μm; I, J, 10 μm; (in O) K–O, V and (in X) W–X, 20 μm; (in U) R–U, 100 μm.
To define the cell types containing STAT3 in more details, we examined the overlaps between progenitor markers and STAT3, and a STAT3 reporter introduced by in utero electroporation. In the E14.5 cortex, only ∼30% of Sox2+ progenitors expressed STAT3, consistent with the heterogeneous expression of STAT3 in the VZ (Fig. 1K). We next constructed the SBS8–dGFP, STAT3 reporter containing eight consecutive copies of the STAT binding site fused to dGFP, which is believed to reflect STAT activity in vivo (Takizawa et al., 2001). The selective responsiveness of the STAT3 reporter was confirmed in HEK293T cells, in which GFP expression was induced in response to LIF or when the STAT3 active form was cotransfected (Fig. 1R–U). When a ubiquitous CAG promoter-based RFP and the SBS8–GFP reporter were coelectroporated, most RFP+GFP− cells migrated toward the CP in which newborn neurons reside, whereas cells that expressed both RFP and GFP remained in the VZ (Fig. 1Q). This suggests that STAT3-active cells are more likely to be progenitors. In the VZ, 35% (18 of 52 cells) and 47% (24 of 52 cells) of RFP+ cells expressed both Pax6 and Tbr2, whereas STAT3 reporter-expressing cells mainly expressed Pax6 (88% of GFP cells, 50 of 57 cells) rather than Tbr2 (19% of GFP cells, 6 of 30 cells; Fig. 1L,N,P). More GFP+ cells (80%, 37 of 46 cells) expressed Cux1, which labels a subset of RG for upper-layer neurons, than did CAG-RFP-expressing cells (33%, 17 of 52 cells; Franco et al., 2012; Fig. 1M,P). Thirty-nine percent (13 of 34 cells) of STAT3 reporter-labeled cells expressed Ki67 (a G1- to S-phase marker) compared with 8% (6 of 74 cells) of the CAG–RFP controls, respectively. This indicates that STAT3 is relatively enriched in dividing cells (Fig. 1O,P,Y). Stat3 protein was not present in oRG cells, the basal progenitors located in the outer SVZ (oSVZ) marked by phospho-vimentin (Fig. 1V–X). These findings indicate that STAT3 is present and active in RG rather than in basal progenitors (Fig. 1Y).
LIF–STAT3 signaling induces RG identity
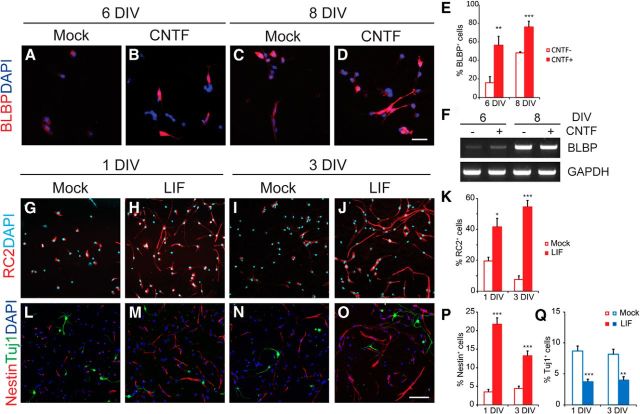
To investigate the potential role of STAT3 in RG, we tested the STAT3 responsiveness of P19 cells, which readily expressed the radial glial marker BLBP in the presence of STAT3 ligands in vitro (Fig. 2A–F). Dissociated neurospheres derived from E13.5 mouse cortical cells also contained an increased number of cells expressing RC2, nestin, and Sox2 protein but a reduced number of Tuj1-labeled neurons in the presence of LIF at 1 and 3 DIV (Fig. 2G–P,Q). This led us to reason that STAT3 activity may induce the RG population. We electroporated GFP or the Stat3 plasmid into E13.5 brains ex vivo and incubated the brain slices in organotypic culture, which permitted better visualization of the morphology of RG and allowed us to examine the effects of ligands added to the culture medium (Fig. 3A–I). At 2 DIV, most GFP-expressing cells were found in the VZ, whereas others were in the IZ and CP (Fig. 3A). At high magnification, the cells in the IZ and CP resembled typical migrating neurons with leading processes (Fig. 3B). They expressed the neuronal marker DCX but not nestin, indicating that they were migrating neurons (Fig. 3C). When LIF was added, most of the GFP+ cell bodies were seen within the VZ, but their processes spanned the entire width of the cortical wall and expressed nestin, a hallmark of RG (Fig. 3D,E). When STAT3 was introduced by electroporation, nestin+ radial processes appeared even without LIF treatment (Fig. 3F,G), and their presence was further enhanced by LIF (Fig. 3H,I). Thus, compared with GFP-electroporated cells in the absence of LIF (5%, 46 of 944), more cells (29%, 216 of 776) became RG in the presence of LIF and in the STAT3-electroporated groups (Fig. 3J).
Figure 2.
Enhanced STAT3 activity promotes generation of RG-like cells in in vitro culture. A–D, P19 cells were grown as embryonic bodies with retinoic acid and CNTF. Cells were replated and harvested at 2 and 4 DIV. Cells were labeled with BLBP (red) and DAPI (blue). E, Quantification of BLBP-expressing cells (n = 3 cultures, n = 4–5 coverslips for each condition). F, Measurement of BLBP transcript in cultures at 6 and 8 DIV by RT-PCR. G–J, L–O, Immunolabeling for RC2, nestin, Sox2, and Tuj1 in dissociated neurospheres at 1 or 3 DIV. K, P, Q, Quantification of cells expressing RC2 (K), nestin (P), and Tuj1 (Q) among the DAPI-stained cells (n = 3–4 cultures, n > 6 coverslips per condition). Only cells with processes longer than 50 μm were counted to ensure their identity. Error bars represent SEM. *p < 0.05, **p < 0.01, ***p < 0.001, unpaired Student's t test. Scale bars: (in D) A–D, 20 μm; (in O) G–J, L–O, 50 μm.
Figure 3.
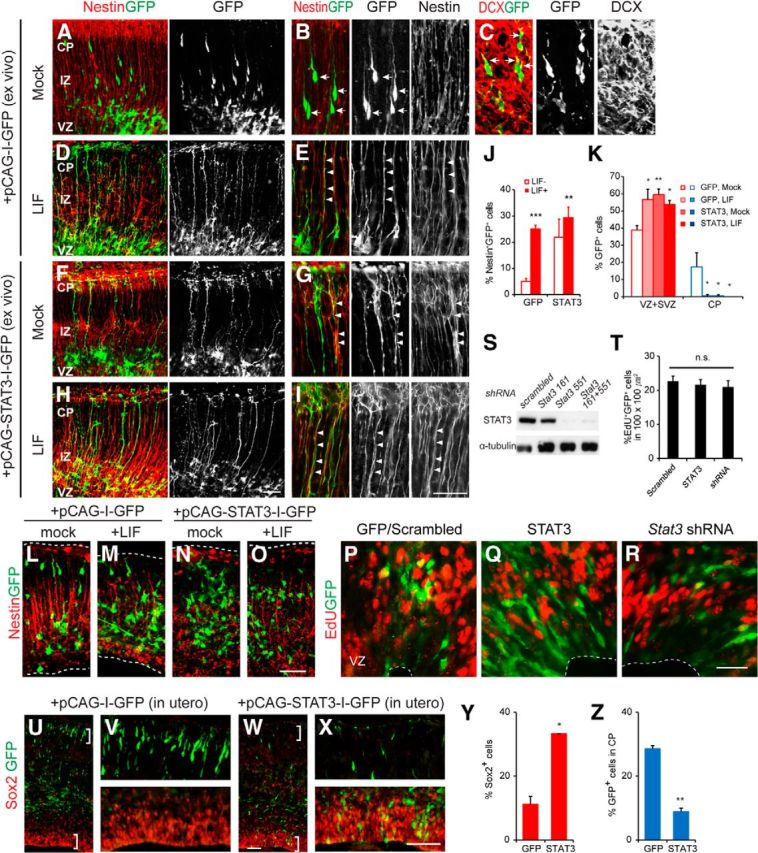
RG is induced by enhanced LIF–STAT3 signaling. A–I, Brain slices from E13.5 brains were harvested at 2 DIV and stained for nestin or DCX (red) and GFP (green) as indicated. The cortical layers were identified by DAPI counterstaining. In the GFP-electroporated group without LIF, GFP+ cells in the CP express the neuronal marker DCX (arrows, C) but lack nestin expression (arrows, B). Nestin+ radial processes appeared when LIF is present or Stat3 is introduced by electroporation (arrowheads, E, G, I). J, K, Quantification of nestin+GFP+ cells and distribution of cell soma (n = 3 embryos, n = 4–6 slices per group). L–O, Slices of E12.5 brains from mice were electroporated with GFP or Stat3 in vitro. No nestin expression was induced by LIF. P–T, In utero electroporation of gfp, Stat3, or Stat3 shRNA into E11.5 brains, injected EdU at E13.5 before harvest. Validation of Stat3 shRNA in HEK293T cells transfected with STAT3 by Western blotting (S). No obvious change in percentage EdU+GFP+ cells (T; n = 3–4 embryos, n = 6–7 sections per group). U–X, In utero electroporation of gfp or Stat3 into E13.5 brains and analysis at E17.5. Y, Z, Quantification of Sox2+GFP+ cells and GFP+ cells in the CP (n = 3 embryos, n = 3 sections per group). Error bars represent SEM. *p < 0.05, **p < 0.01, ***p < 0.001, unpaired Student's t test. Scale bars: (in H) A, D, F, H and (in O) L–O, 50 μm; (in I) B, C, E, G, I and (in R) P–R, 20 μm; (in W) U, W, and (in X) V, X, 200 μm.
Next, we compared the distribution of GFP-labeled cell bodies within the cortex in each condition. In the case of dividing progenitors, the cell bodies remain in the VZ; in the case of migrating newborn postmitotic neurons, the cell bodies should be found in the IZ/CP. In the GFP-electroporated slices with no LIF treatment, 39% of the GFP-labeled cells (213 of 556 cells) were located in the VZ/SVZ, whereas 17% reached the CP (96 of 556 cells; Fig. 3K). When LIF was added, more GFP-labeled cells (57%, 202 of 342 cells) remained in the VZ/SVZ, and fewer were found in the IZ/CP (0.3%, 1 of 342 cells). When STAT3 was introduced, more cells were found in the VZ regardless of the presence of LIF (59% in the VZ/SVZ, 226 of 379 cells without LIF; 54% in the VZ/SVZ, 215 of 409 cells with LIF). Thus, when STAT3 is active, more cells stay in the VZ. Interestingly, we noted no obvious production of RG in response to LIF in cultures of earlier brains (Fig. 3L–O). We also introduced STAT3 and Stat3 shRNA into E11.5 cortices using in utero electroporation and injected EdU before harvest at E13.5. However, STAT3 and Stat3 shRNA did not alter the proportion of dividing cells, indicating that STAT3 activity is not critical for cell division in the early progenitors (Fig. 3P–T). Thus, STAT3 mainly acts on radial glial cells at mid-neurogenesis.
To define the identity of the STAT3-electroporated cells in vivo, we electroporated GFP or Stat3 at E13.5 in utero and examined the brains at E17.5. In the control, more GFP-labeled cells were found in the CP than in the VZ and probably became newborn neurons (Fig. 3U,V,Y,Z). When Stat3 was introduced, more cells remained in the VZ and expressed Sox2, and fewer cells were found in the CP (Fig. 3W–Z). Thus, STAT3 maintains the identity of RG.
The apparent increase of RG in response to STAT3 could be attributable to the production of more RG or more rapid production or prolonged maintenance of the same number of RG. We first monitored the time course of radial fiber formation in organotypic slice culture (Fig. 4A–P). In the absence of LIF, most GFP+ RG were found at 16–20 h but had disappeared by 26 h. When LIF was applied, the maximum number of RG was already present at 16 h and persisted until 26 h (Fig. 4Q). Thus, STAT3 activity allows RG to persist for longer.
Figure 4.
Temporal responsiveness of STAT3 and its ligands in in vitro organotypic brain slices. A–P, Time course of RG formation. Brain slices from E13.5 brains were electroporated with GFP plasmid and harvested at the indicated times. Nestin+ or Vimentin+ radial processes were marked by arrows. At 26 h, radial processes had disappeared (bracket, C, G) in the mock-treated group, whereas they persisted in the LIF-treated group (bracket, K, L). E–H, M–P, Higher-magnification images of A–D, I–L from the pial surface. Arrows indicate Nestin+GFP+ radial processes. Q, The percentage of RG labeled by GFP and nestin among GFP-labeled cells at each time point indicated (n = 3–5 embryos, n = 4–7 slices per condition). Error bars represent SEM. **p < 0.01, unpaired Student's t test. Scale bars: (in L) A–D, I–L, 50 μm; (in O) E–G, M–O and (in P) H, P, 20 μm.
STAT3 is required for maintaining radial glial cells
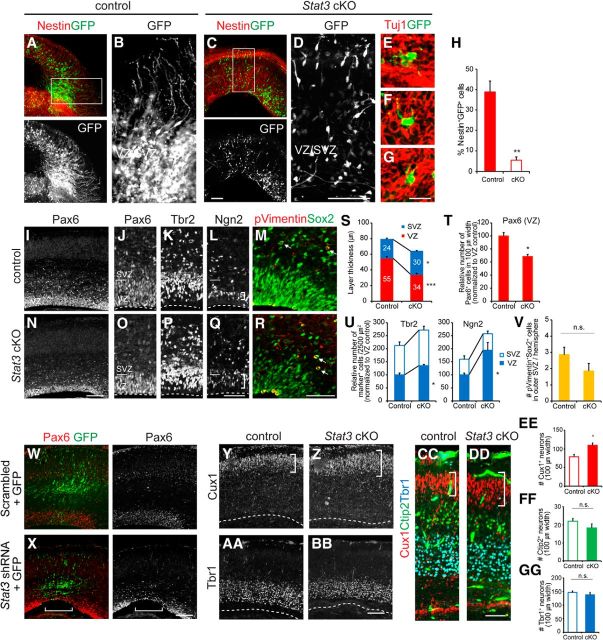
To see whether STAT3 is required for RG formation, we eliminated or reduced STAT3 using mouse genetics or knockdown approaches. To avoid the embryonic lethality of Stat3 null mice, we used Stat3 conditional mutant mice crossed with Nestin–Cre mice; this selectively removes Stat3 within neural progenitors (Takeda et al., 1997). To selectively trace radial glial cells in these animals, we electroporated GFP plasmids into E13.5 Stat3 conditional knock-out (cKO) mice and incubated brain slices in the presence of CNTF. The GFP-labeled cells in mice heterozygous for the Stat3 flox allele displayed the characteristics of RG, with long RG processes and cell bodies in the VZ (Fig. 5A,B). However, the GFP+ cells in slices from the Stat3 cKO mice developed significantly fewer RG, even in the presence of CNTF (Fig. 5C,D,H). No significant apoptotic cell death was observed compared with the control (data not shown). The somata of these cells were mostly located in the IZ and expressed an early neuronal marker, Tuj1, indicating that they are immature neurons (Fig. 5E–G).
Figure 5.
Depletion of RG and precocious neurogenesis in the absence of STAT3. A–G, Organotypic brain slice cultures from E13.5 Stat3 cKO mice or littermate controls and immunostaining of nestin or Tuj1 (red) and GFP (green) as indicated. The brains were electroporated with GFP, and the slices were grown for 2 DIV in the presence of CNTF. H, The proportion of Nestin+GFP+ cells among GFP-labeled cells (n = 3–5 embryos, n > 4 slices for each group). I–R, Immunostaining for Pax6, Ngn2, Tbr2, phospho-vimentin, and Sox2 in E16.5 Stat3 cKO mice or littermate controls. Thickness of Ngn2-expressing layer was greater in Stat3 cKO (brackets, L, Q). Phospho-vimentin+Sox2+ oRG cells were located (arrows, M, R). S, Thickness of the VZ and SVZ determined by Pax6 and Tbr2 expression (n = 3–5 embryos, n > 7 sections for each group). T, Quantification of Pax6 expression in 100-μm-wide segments of the VZ or SVZ (n = 3–5 embryos, n = 7 sections per group). U, Quantification of Ngn2 and Tbr2 expression in 2500 μm2 of the VZ or SVZ (n = 3–5 embryos, n > 4 sections per group). V, Quantification of phospho-vimentin+Sox2+ oRG cells (n = 4–5 embryos, n = 7 sections per group). W, X, Immunostaining for Pax6 and GFP of E17.5 brains electroporated with either scrambled or Stat3 shRNA at E13.5. Note that Pax6 has been depleted in the electroporated area (brackets, X) but not in the adjacent non-electroporated area. Y–DD, Expression of Cux1, Ctip2, and Tbr1 in E18.5 Stat3 mutants and controls. Separate images from adjacent sections were merged for easier comparison (CC, DD). Cux1-expressing layer was expanded in Stat3 cKO (brackets, Y, Z, CC, DD). White dotted lines are the ventricular surface. EE–GG, The numbers of Cux1+, Ctip2+, and Tbr1+ cells were measured in 100-μm-wide segments. Cux1+ cells above the Ctip2+ layer were counted (n = 3–5 embryos, n = 5–10 sections per group). Error bars represent SEM. *p < 0.05, **p < 0.01, ***p < 0.001, unpaired Student's t test. Scale bars: (in C) A, C, (in D) B, D, and (in BB) Y–BB, 100 μm; (in G) E–G, 20 μm; (in N) I, N and (in X) W, X, 400 μm; (in Q) O–Q, (in R) M, R, and (in DD) CC, DD, 50 μm.
To further define the nature of Stat3-null cells, we examined the distribution of progenitors in the Stat3 cKO mice (Fig. 5I–R). In the E16.5 Stat3 cKO cortex, the thickness of the VZ was reduced, whereas the thickness of the SVZ was increased (Fig. 5I,J,N,O,S). The number of Pax6+ cells per unit width of VZ was reduced accordingly to 70% of the control in the absence of STAT3 (95 cells in the control, 65 cells in the Stat3 cKO; Fig. 5T). The number of Ngn2+ or Tbr2+ progenitors per unit area destined to become neurons was higher in the VZ of the Stat3 cKO mice than in that of the controls (Fig. 5K,L,P,Q,U). The number of oRG cells marked by phospho-vimentin and Sox2 was unchanged in the Stat3 cKO (2.86 ± 0.46 in control; 1.86 ± 0. 46 in Stat3 cKO; Fig. 5M,R,V). A similar reduction of Pax6+ cells was also found in brains when Stat3 shRNA was introduced by electroporation (Fig. 5W,X). These results imply that, when STAT3 activity is low, progenitors fail to become RG and instead become neurogenic IPCs.
Next we tested whether the additional neurons produced in the absence of STAT3 constitute specific layers of the CP. We examined laminar formation in E18.5 Stat3 cKO brains, which were distinguished by expression of Cux1 (layers II–IV), Ctip2 (layer V), or Tbr1 (layers II–VI; Hevner et al., 2001; Zimmer et al., 2004; Arlotta et al., 2005). Lower-layer neurons are born early in neurogenesis (E10–E12), and upper-layer neurons are born in mid-neurogenesis (E13–E17; Takahashi et al., 1999; Hevner et al., 2001; Nieto et al., 2004). The number of Cux1+ neurons in the upper layer of the brain increased, whereas those in the Ctip2+ or Tbr1+ lower layers were relatively unchanged (Fig. 5Y–GG). We conclude that, in the absence of STAT3, more upper-layer neurons are produced, whereas the number of neurons in other layers is not affected.
STAT3 promotes division of RG
To investigate the cell division of radial glial cells in relation to STAT3, we introduced Stat3 or Stat3 shRNA by electroporation at E13.5 and analyzed the cells at E15.5 when the electroporated DNA was fully expressed. Scrambled shRNA coelectroporated with GFP (referred to as GFP/scrambled) was used as a control. We injected EdU to label dividing cells at E15.5 and harvested embryos 2 h later. The EdU labeling index (percentage EdU+GFP+/GFP+ cells) was ∼20% in the GFP/scrambled-electroporated cells, and this increased to 31% (5 of 16 cells) when Stat3 was overexpressed and decreased to 13% (3 of 19 cells) in Stat3 knockdown conditions (Fig. 6A–G).
Figure 6.
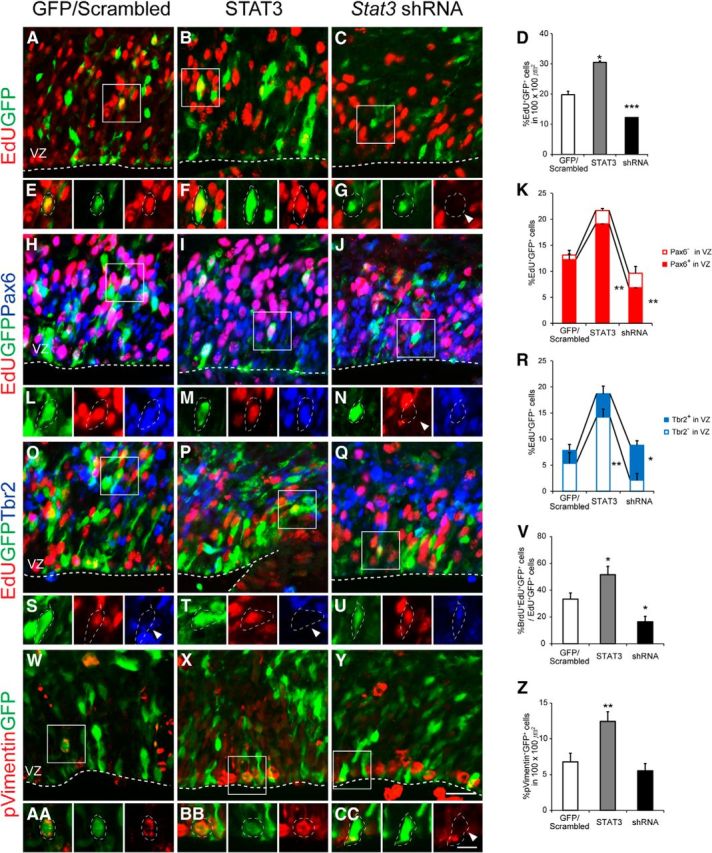
Proliferation of RG controlled by STAT3. A–C, H–J, O–Q, W–Y, Immunostaining for GFP, EdU, Pax6, Tbr2 or GFP, phospho-vimentin in E15.5 brains electroporated with GFP/scrambled, STAT3, or Stat3 shRNA at E13.5. EdU was administered 2 h before harvest. Images were taken in the VZ. E–G, L–N, S–U, AA–CC, High-magnification images of squares in A–C, H–J, O–Q, W–Y. D, K, R, Percentages of EdU+GFP+ cells that express Pax6 or Tbr2 (n = 3–5 embryos, n > 5 sections for each group). V, Percentages of BrdU+EdU+GFP+ cells among EdU+GFP+ cells (n = 3–5 embryos, n > 5 sections for each group). Z, Percentages of phospho-vimentin+GFP+ cells among GFP+ cells (n = 4–7 embryos, n > 7 sections for each group). Arrowheads are examples of GFP-labeled cells lacking expression of EdU (G, N), Tbr2 (S, T), and phospho-vimentin (CC). *p < 0.05, **p < 0.01, ***p < 0.001, unpaired Student's t test. Scale bars: (in Y) A–C, H–J, O–Q, W–Y, 20 μm; (in CC) E–G, L–N, S–U, AA–CC, 10 μm.
To test whether altered STAT3 activity selectively affects particular cell types, we examined the expression of Pax6 and Tbr2 among the EdU+GFP+ dividing cells (Fig. 6H–U). Compared with the GFP/scrambled-electroporated group, Pax6-expressing cells in the VZ increased when STAT3 was overexpressed (Fig. 6K). The number of cells in the VZ that did not express Tbr2 and that could be Pax6+ was elevated (Fig. 6R). Thus, when STAT3 is overexpressed, the proportion of dividing RG rises. Next, we analyzed the proportion of dividing cells in Stat3 knockdown conditions. Compared with the GFP/scrambled-electroporated group, Pax6-expressing cells in the VZ were reduced and Tbr2+ cells increased. Measurement of cell-cycle reentry by BrdU/EdU double labeling at 10 h intervals showed that the proportion of cells that reentered the cell cycle was greater (1.5-fold) in the STAT3-overexpressed group than in the control (52 vs 33%) and reduced (0.5-fold) in the Stat3 knockdown condition (16%; Fig. 6V). Furthermore, the proportion of phospho-vimentin-expressing RG, which are mitotically active, increased when STAT3 was overexpressed (Fig. 6W–CC). Together, our analysis suggests that STAT3 augments the number of dividing RG at the expense of IPC. Conversely, when STAT3 activity is low, dividing RG decrease and dividing IPCs increase, probably because of a transition from RG to IPCs.
STAT3 drives symmetric divisions of RG
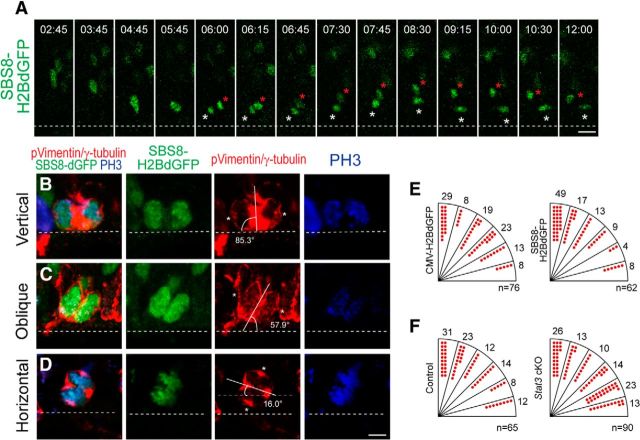
One way that STAT3 might increase progenitors is by promoting symmetric cell division of RG. Because the fate of daughter cells is correlated with the orientation of the cleavage plane, we measured this parameter in dividing RG (Chenn and McConnell, 1995). Time-lapse imaging of living slices showed that SBS8-H2BdGFP-labeled radial glial cells often underwent vertical division, and the characteristic interkinetic nuclear movement could be seen thereafter in their progeny (Fig. 7A). We divided the angles of the cleavage planes into vertical (60°-90°), oblique (30°-60°), and horizontal (0°-30°; Fig. 7B–D); if STAT3-active RG were more prone to divide symmetrically, the proportion of vertical divisions would be increased in cells electroporated with the STAT3 reporter SBS8–H2BdGFP. We electroporated CMV–H2BdGFP and the SBS8–H2BdGFP STAT3 reporter in utero at E13.5 and examined GFP expression in dividing RG in fixed brains labeled with phospho-vimentin, γ-tubulin, and PH3 at E14.5 (Bultje et al., 2009; Fig. 7E). Vertical division was more frequent in the SBS8–H2BdGFP-expressing cells (66%) than in the CMV–H2BdGFP-expressing cells (37%; Fig. 7E). Conversely, instances of oblique and horizontal divisions decreased in the SBS8–H2BdGFP-positive cells (22% oblique, 12% horizontal) compared with the control group (42% oblique, 21% horizontal). These results suggest that STAT3-active RG tend to divide vertically and are thus likely to undergo symmetric divisions and give rise to progenitors. To test whether STAT3 determines the orientation of the cleavage plane, we electroporated CMV–H2BdGFP into Stat3 cKO cortices. Compared with the control embryos (20% horizontal, 54% vertical), more horizontal (37%) and lesser vertical (39%) divisions were found in Stat3 cKO embryos (Fig. 7F). Together, these observations suggest that STAT3 activity is associated with vertically dividing radial glial cells.
Figure 7.
STAT3-expressing radial glial cells undergo vertical division. A, Time-lapse stills of vertically dividing single mitotic RG labeled with SBS8–H2BdGFP in E14.5 mouse cortical slice. SBS8–H2BdGFP was electroporated into the brains of E13.5 embryos, and the acute brain slices were made after 24 h. Asterisks indicate two daughter cells after cell division. Dashed line marks the ventricular surface. B–D, Examples of mitotic RG in fixed mouse brain sections labeled with SBS8–H2BdGFP (green), phospho-vimentin (pVimentin, red), γ-tubulin (red), and PH3 (blue). Each mitotic radial glial cell showed vertical (B), oblique (C), and horizontal (D) cleavage plane orientations at the ventricular surface. The ventricular surface is marked by the dotted line, and γ-tubulin-labeled centrosomes are marked with asterisks. E, F, Quantification of cleavage plane orientation in CMV–H2BdGFP and SBS8–H2BdGFP expression conditions (E) and CMV–H2BdGFP expression in the Stat3 cKO condition (F) (E, n = 76 cells, 5 embryos for CMV–H2BdGFP and n = 62 cells, 8 embryos for SBS8–H2BdGFP; F, n = 65 cells, 4 embryos for control and n = 90 cells, 5 embryos for Stat3 cKO). Scale bars: A, (in D) B–D, 10 μm.
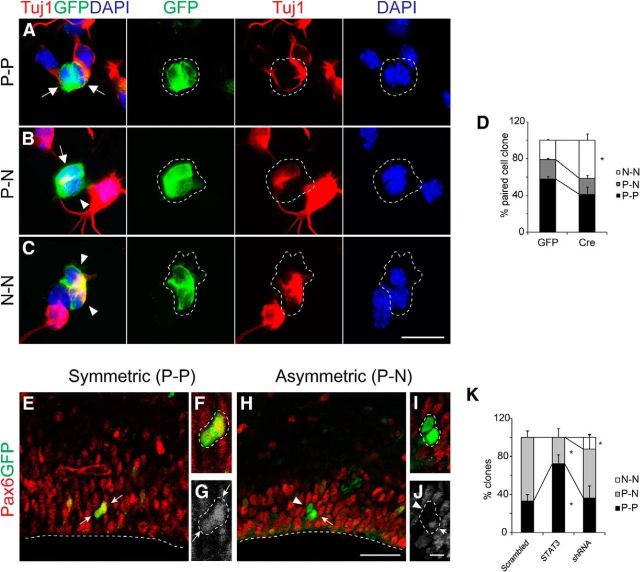
Next we traced the fate of daughter cells when STAT3 activity was altered, using markers for progenitors and neurons. We first conducted pair-cell analysis in vitro. Cre–GFP or GFP retrovirus was injected into the ventricles of E12.5 Stat3fl/fl brains, and the brains were harvested at E13.5 for low-density culture. There are three possible modes of cell division: (1) progenitor–progenitor (P–P); (2) progenitor–neuron (P–N); and (3) neuron–neuron (N–N; Fig. 8A–C). The early neuronal marker Tuj1 was used to determine the identity of daughter cells at the first cell division. The proportion of P–P clones was reduced and that of N–N clones increased after Cre–GFP injection (Fig. 8D).
Figure 8.
Requirement for STAT3 for symmetric division of RG. A–C, Examples of two-cell clones (P–P, P–N, and N–N) seen in the paired-cell analysis using expression of GFP (green) and Tuj1 (red). GFP and Cre retrovirus was injected into E12.5 brains in utero. The brains were harvested at E13.5, cultured for 24 h to allow one cell division, and immunostained before clonal analysis (arrows, Tuj1− progenitors; arrowheads, Tuj1+ neurons). Broken lines indicate the appearance of each clone. D, Clonal analysis of STAT3 knockdown and control groups. E, H, Representative examples of two-cell clones in E15.5 brains labeled with GFP (green) and Pax6 (red). F, G, I, J, Higher-magnification images of radial clone in E and H. Arrowheads, Pax6-neurons; arrows, Pax6+progenitors. K, The proportion of each clone in GFP, STAT3-expressing and Stat3 knockdown conditions (n = 29 clones, 8 embryos for scrambled; n = 20 clones, 6 embryos for STAT3; n = 26 clones, 8 embryos for Stat3 shRNA). Error bars represent SEM. *p < 0.05, unpaired Student's t test. Scale bars: (in C) A–C, 20 μm; (in H) E, H, 30 μm; (in J) F, G, I, J, 10 μm.
To test whether STAT3 also affected the mode of cell division of RG in vivo, we electroporated a minimal amount of GFP or Stat3 DNA into E13.5 mouse cortices in utero and harvested brains at E15.5. The average distance between GFP-labeled cells was ∼50 μm, large enough to identify two-cell clones of RG (data not shown; Bultje et al., 2009). Individual clones whose cell bodies contacted each other and lay in the VZ were confirmed by z-stack confocal microscopy. Because the fates of daughter cells indicated by marker expression become obvious several hours after mitosis, we only examined Pax6 asymmetry in clones located 20 μm above the ventricle (Ochiai et al., 2009). The mode of cell division was determined by Pax6 expression in these cells (Fig. 8E–J). When scrambled shRNA was electroporated, 35% of the clones underwent symmetric (Pax6+–Pax6+) divisions (8 of 23 clones), and this increased to 70% when STAT3 was overexpressed (14 of 20 clones; Fig. 8K). Conversely, downregulation of STAT3 by shRNA resulted in the appearance of N–N doublets (Pax6−–Pax6−) not seen in the scrambled or STAT3-overexpressing groups (Fig. 8K). Together, our observations suggest that STAT3 activity is associated with symmetric P–P division and indicate that STAT3-expressing RG tend to undergo vertical cleavage and symmetric divisions.
Discussion
Over the past several decades, the role of STAT3 in glial differentiation has been studied extensively, but our understanding of its role in the production of cortical progenitors remains limited. Here we show that STAT3 is expressed in Pax6+ RG rather than Tbr2+ basal progenitors and promotes the symmetric division of RG to maintain their pool size.
Role of STAT3 in RG generating upper-layer neurons
Our detailed spatiotemporal assessment of STAT3 expression and activity suggests that STAT3 influences a specific lineage of RG. Only 30% of Sox2+ radial glial cells expressed STAT3, and STAT3 was relatively enriched in dividing RG. The RG with elevated STAT3 activity divided symmetrically, producing two progenitors. Conversely, those with low STAT3 activity failed to maintain their identity and became prematurely differentiated upper-layer neurons. Interestingly, only RG at mid-neurogenesis responded to STAT3 by persisting as RG. The progenitors of lower-layer (deep) neurons did not respond to STAT3 by forming RG, and production of deep-layer neurons was normal in Stat3 mutant mice. IPCs or other recently identified basal progenitors, such as oRG cells in the oSVZ, did not express STAT3, suggesting that STAT3 does not directly control these populations (Shitamukai et al., 2011; Wang et al., 2011). Thus, STAT3 is likely to act selectively on the actively dividing RG that produce upper-layer neurons.
There is a great similarity between the roles of STAT3 and Cux factors in cortical development. STAT3 appears to be enriched in Cux1-expressing RG (62% of STAT3-active cells; Franco et al., 2012). Expression of STAT3 is more transient or restricted because it is enriched on the apical side, in contrast to Cux1 expression that is sparsely but evenly distributed over the VZ. Second, both have been implicated in cell-cycle progression by repressing cyclin-dependent kinase inhibitors (Coqueret et al., 1998; Fukada et al., 1998). For instance, Cux2 promotes cell-cycle progression and mediates cell-cycle exit, leading to neuronal differentiation in the spinal cord (Iulianella et al., 2008). Third, both STAT3 and Cux1/2 are involved in the formation of upper-layer neurons because the absence of either gene leads to an expansion of Tbr2+ IPCs and upper-layer neurons without affecting the birth of lower-layer neurons (Cubelos et al., 2008). Last, symmetric divisions that generate more RG are prominent among Cux2+ RG (Franco et al., 2012) or when STAT3 activity was increased in this study. Together, these findings suggest that STAT3 has a role in maintaining RG subsets.
Symmetric division of RG induced by STAT3
At the beginning of neurogenesis, symmetric divisions prevail to expand progenitors, but soon asymmetric divisions arise that generate neurons and at the same time preserve progenitors. Time-lapse imaging has suggested that both asymmetric and symmetric divisions occur in RG with an increasing tendency to divide asymmetrically as neurogenesis proceeds (Noctor et al., 2001, 2004). In this study, our pair-cell analysis of RG both in vivo and in vitro suggested that STAT3 promotes symmetric division so that more radial glial cells persist instead of becoming neurons. Consistent with this, our time-lapse imaging analysis demonstrated that vertical divisions are more frequent in STAT3 active RG, whereas horizontal divisions are more prevalent when STAT3 is eliminated. This is similar to what occurs with Cux factors or Pax6, which drive symmetric divisions (Asami et al., 2011; Franco et al., 2012). Although it remains to be determined whether these factors interact to maintain the heterogeneity of RG subsets or diversify their multipotency, we propose that STAT3 is one definite molecular determinant that drives radial glial fate.
Recently, it has been suggested that several structural factors, such as inheritance of the apical membrane or spindle orientation, are correlated with the mode of cell division (Zhong and Chia, 2008). In this study, we found a correlation between STAT3 activity and the orientation of the cleavage plane, with greater STAT3 activity in vertically dividing cells. Furthermore, STAT3 activity was highest but heterogeneous at the most apical side within the VZ in which RG undergo mitosis. Although the ultimate fates of daughter cells that have different STAT3 activities need to be determined, the dynamic distribution of STAT3 is reminiscent of oscillating notch activity and the asymmetric distribution of numb protein, a component of the Notch pathway (Zhong et al., 1996; Shimojo et al., 2008). Interestingly, the oscillatory behavior of gene expression in neural progenitors was also found in several key bHLH transcription factors known to drive neurogenesis, and STAT3 was also reported to oscillate in certain conditions in vitro (Yoshiura et al., 2007; Imayoshi et al., 2013). Although we did not detect any obvious oscillations because of the technical limitations of our system, it is likely that STAT3 coordinates with other oscillating factors to dictate the ultimate cell fates of RG.
Footnotes
Support for M.-R.S. and this research was provided by National Research Foundation (NRF) Grant NRF-2013R1A1A2058548, Cell Dynamics Research Center/NRF Grant 2007-0056157, and the BioImaging Research Center at the Gwangju Institute of Science and Technology (GIST). We are grateful to Dr. Sohyun Ahn for valuable comments and members of the Song laboratory for helpful discussion and encouragement. We also thank the BioImaging Research Center and Systems Biology Research Center at GIST for use of the confocal imaging facility.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Asami M, Pilz GA, Ninkovic J, Godinho L, Schroeder T, Huttner WB, Götz M. The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. Development. 2011;138:5067–5078. doi: 10.1242/dev.074591. [DOI] [PubMed] [Google Scholar]
- Barnabé-Heider F, Wasylnka JA, Fernandes KJ, Porsche C, Sendtner M, Kaplan DR, Miller FD. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron. 2005;48:253–265. doi: 10.1016/j.neuron.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Bauer S, Patterson PH. Leukemia inhibitory factor promotes neural stem cell self-renewal in the adult brain. J Neurosci. 2006;26:12089–12099. doi: 10.1523/JNEUROSCI.3047-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notchl immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Coqueret O, Bérubé G, Nepveu A. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 1998;17:4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Sebastián-Serrano A, Kim S, Moreno-Ortiz C, Redondo JM, Walsh CA, Nieto M. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cereb Cortex. 2008;18:1758–1770. doi: 10.1093/cercor/bhm199. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, Müller U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada T, Ohtani T, Yoshida Y, Shirogane T, Nishida K, Nakajima K, Hibi M, Hirano T. STAT3 orchestrates contradictory signals in cytokine-induced G1 to S cell-cycle transition. EMBO J. 1998;17:6670–6677. doi: 10.1093/emboj/17.22.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher D, Norman AA, Woodard CL, Yang G, Gauthier-Fisher A, Fujitani M, Vessey JP, Cancino GI, Sachewsky N, Woltjen K, Fatt MP, Morshead CM, Kaplan DR, Miller FD. Transient maternal IL-6 mediates long-lasting changes in neural stem cell pools by deregulating an endogenous self-renewal pathway. Cell Stem cell. 2013;13:564–576. doi: 10.1016/j.stem.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Gregg C, Weiss S. CNTF/LIF/gp130 receptor complex signaling maintains a VZ precursor differentiation gradient in the developing ventral forebrain. Development. 2005;132:565–578. doi: 10.1242/dev.01592. [DOI] [PubMed] [Google Scholar]
- Hatta T, Moriyama K, Nakashima K, Taga T, Otani H. The Role of gp130 in cerebral cortical development: in vivo functional analysis in a mouse exo utero system. J Neurosci. 2002;22:5516–5524. doi: 10.1523/JNEUROSCI.22-13-05516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Ge W, Martinowich K, Becker-Catania S, Coskun V, Zhu W, Wu H, Castro D, Guillemot F, Fan G, de Vellis J, Sun YE. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat Neurosci. 2005;8:616–625. doi: 10.1038/nn1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/S0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Hong S, Song MR. STAT3 but not STAT1 is required for astrocyte differentiation. PLoS One. 2014;9:e86851. doi: 10.1371/journal.pone.0086851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, Fujiwara T, Ishidate F, Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- Iulianella A, Sharma M, Durnin M, Vanden Heuvel GB, Trainor PA. Cux2 (Cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development. 2008;135:729–741. doi: 10.1242/dev.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. 2004;6:547–554. doi: 10.1038/ncb1138. [DOI] [PubMed] [Google Scholar]
- Kang K, Song MR. Diverse FGF receptor signaling controls astrocyte specification and proliferation. Biochemical and biophysical research communications. 2010;395:324–329. doi: 10.1016/j.bbrc.2010.03.174. [DOI] [PubMed] [Google Scholar]
- Kang K, Lee D, Hong S, Park SG, Song MR. The E3 ligase Mind bomb-1 (mib1) modulates Delta-Notch signaling to control neurogenesis and gliogenesis in the developing spinal cord. J Biol Chem. 2013;288:2580–2592. doi: 10.1074/jbc.M112.398263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Ochiai W, Nakatani S, Takahara T, Kainuma M, Masaoka M, Minobe S, Namihira M, Nakashima K, Sakakibara A, Ogawa M, Miyata T. Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Mol Cell Neurosci. 2009;40:225–233. doi: 10.1016/j.mcn.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Pinto L, Drechsel D, Schmid MT, Ninkovic J, Irmler M, Brill MS, Restani L, Gianfranceschi L, Cerri C, Weber SN, Tarabykin V, Baer K, Guillemot F, Beckers J, Zecevic N, Dehay C, Caleo M, Schorle H, Götz M. AP2gamma regulates basal progenitor fate in a region- and layer-specific manner in the developing cortex. Nat Neurosci. 2009;12:1229–1237. doi: 10.1038/nn.2399. [DOI] [PubMed] [Google Scholar]
- Pitman M, Emery B, Binder M, Wang S, Butzkueven H, Kilpatrick TJ. LIF receptor signaling modulates neural stem cell renewal. Mol Cell Neurosci. 2004;27:255–266. doi: 10.1016/j.mcn.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Postiglione MP, Jüschke C, Xie Y, Haas GA, Charalambous C, Knoblich JA. Mouse inscuteable induces apical-basal spindle orientation to facilitate intermediate progenitor generation in the developing neocortex. Neuron. 2011;72:269–284. doi: 10.1016/j.neuron.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki T, Shingo T, Weiss S. The ciliary neurotrophic factor/leukemia inhibitory factor/gp130 receptor complex operates in the maintenance of mammalian forebrain neural stem cells. J Neurosci. 2001;21:7642–7653. doi: 10.1523/JNEUROSCI.21-19-07642.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Goto T, Miyama S, Nowakowski RS, Caviness VS., Jr Sequence of neuron origin and neocortical laminar fate: relation to cell cycle of origin in the developing murine cerebral wall. J Neurosci. 1999;19:10357–10371. doi: 10.1523/JNEUROSCI.19-23-10357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–758. doi: 10.1016/S1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Jüschke C, Esk C, Hirotsune S, Knoblich JA. The phosphatase PP4c controls spindle orientation to maintain proliferative symmetric divisions in the developing neocortex. Neuron. 2013;79:254–265. doi: 10.1016/j.neuron.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu T, Kawaguchi D, Oishi K, Takeda K, Akira S, Masuyama N, Gotoh Y. Non-cell-autonomous action of STAT3 in maintenance of neural precursor cells in the mouse neocortex. Development. 2006;133:2553–2563. doi: 10.1242/dev.02419. [DOI] [PubMed] [Google Scholar]
- Yoshiura S, Ohtsuka T, Takenaka Y, Nagahara H, Yoshikawa K, Kageyama R. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc Natl Acad Sci U S A. 2007;104:11292–11297. doi: 10.1073/pnas.0701837104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Heng JI-T, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18:4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/S0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]