Abstract
Background
Paracoccidioidomycosis (PCM) is a systemic mycosis commonly found in Latin America that is caused by distinct species of Paracoccidioides genus: Paracoccidioides brasiliensis complex (S1, PS2, PS3 and PS4) and Paracoccidioides lutzii. Its pathobiology has been recently explored by different approaches to clarify the mechanisms of host-pathogen interactions underpinning PCM. The diversity of clinical forms of this disease has been attributed to both host- and fungus-related factors.
Methodology/Principal findings
For better understanding of the molecular underpinnings of host-fungus interactions, we evaluated in vivo virulence of nine Paracoccidioides brasiliensis complex isolates and correlated it to protein expression profiles obtained by two-dimensional gel electrophoresis. Based on the recovery of viable fungi from mouse organs, the isolates were classified as those having low, moderate, or high virulence. Highly virulent isolates overexpressed proteins related to adhesion process and stress response, probably indicating important roles of those fungal proteins in regulating the colonization capacity, survival, and ability to escape host immune system reaction. Moreover, highly virulent isolates exhibited enhanced expression of glycolytic pathway enzymes concomitantly with repressed expression of succinyl-CoA ligase beta chain, a protein related to the tricarboxylic acid cycle.
Conclusions/Significance
Our findings may point to the mechanisms used by highly virulent P. brasiliensis isolates to withstand host immune reactions and to adapt to transient iron availability as strategies to survive and overcome stress conditions inside the host.
Introduction
Paracoccidioidomycosis (PCM) is the most frequent endemic systemic mycosis in Latin America with high incidence in Brazil, Argentina, Colombia, and Venezuela [1, 2]. It is caused by the thermally dimorphic species of the genus Paracoccidioides. Until recently, Paracoccidioides was considered a monotypic taxon typified by Paracoccidioides brasiliensis [3]. However, the introduction of molecular phylogenetics shed light on the taxonomy of Paracoccidioides, leading to the description of new cryptic entities. To date, four phylogenetic species are recognized inside the P. brasiliensis complex: S1, PS2, PS3 and PS4 [4, 5]. P. brasiliensis sensu stricto (s. str.), formerly known as S1, is the most widely distributed agent of PCM, occurring in Brazil, Argentina, Paraguay, Uruguay, Peru and Venezuela [6]. Paracoccidioides americana (formerly known as PS2) occurs in Venezuela and Brazil, in sympatry with P. brasiliensis s. str. [6]. Paracoccidioides restrepiensis (PS3) and P. venezuelensis (PS4) are geographically restricted to Colombia and Venezuela, respectively [6]. Finally, P. lutzii, an ancient divergent of the P. brasiliensis complex occurs in Brazil with its epicenter in the Central-West region [7, 8]. A recent speciation event is assumed for species embedded in the P. brasiliensis complex (especially PS3 and PS4), whereas it seems that P. brasiliensis sensu lato (s.l.) and P. lutzii are reproductively isolated in nature [6].
The ecological niche or exact habitat of these species remains poorly understood [9]. Paracoccidioides brasiliensis s. str. and P. americana has been described in armadillos, but not P. lutzii. The nine-banded armadillo (Dasypus novemcinctus) is a natural reservoir of the fungus and animal infection due to Paracoccidioides spp. has been observed repeatedly in several endemic areas of Brazil and Colombia [10]. Also, culture-independent surveys based on DNA detection techniques revealed that P. brasiliensis complex and P. lutzii are present in the soil [11]. It is accepted that conidia present in nature are inhaled by patients, transformed into budding yeast cells in the lungs, and then, these cells spread to different organs [12]. PCM may manifest itself in a variety of clinical forms, ranging from a benign and localized condition to a more severe and disseminated disease, depending on the extent of the depression of cellular immunity [12–14].
Paracoccidioides spp. are able to cause disease symptoms in the murine model. However, different isolates are not homogeneous in their virulence characteristics, a fact that could explain different clinical forms of PCM. The virulence profile of Paracoccidioides spp. isolates has been shown to depend upon mycological properties [15], isolate origins [16, 17], species identity [18], genetic patterns [19–21], adhesion process [22–24], activation of immune response [25, 26], culture conditions [27–29], antigenic characteristics [17], and protein levels [17, 30].
Cell-mediated immunity is the predominant host defense mechanism against fungal infections [31]. The role of antibodies in protective immunity during fungal infections can be achieved through many mechanisms, such as neutralization of fungal PAMPS (Pathogen Associated Molecular Patterns), opsonization of fungi and facilitation of their phagocytosis by recognition by Fc receptors present in phagocytes, activation of the classic pathway of the complement system, antibody dependent cell cytotoxicity and direct inhibition of fungal growth.
Though, many breakthrough studies have dissected the role of antibodies in antifungal immunity, as reviewed by Casadevall & Pirofski [32], it is not well characterized how the specific antibodies can mediate this protection [33]. In paracoccidioidomycosis, a robust set of evidences points towards the conclusion that cell-mediated immunity is the main host defense during P. brasiliensis infection, as stated as early as in 1988 by Castaneda and colleagues [34].
Although the role of specific antibodies as effector molecules of the adaptive immune response is classically known and accepted, their role in providing protective immunity in fungal infections, restricting fungal burden and enhancing their clearance is not a consensus because antibodies are frequently associated with severe PCM. Therefore, it is in dispute whether specific anti-P. brasiliensis contribute to susceptibility or merely constitute a marker of infection severity or even are protective.
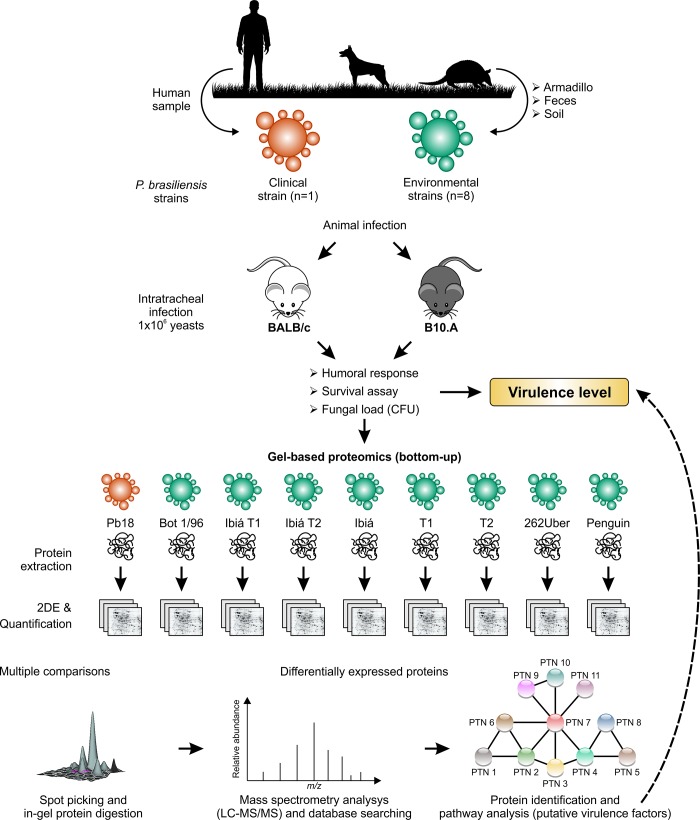
For host invasion and colonization, Paracoccidioides spp. must be able to survive in hostile environments, which requires the presence of various regulatory mechanisms and expression of different virulence factors. These mechanisms allow the fungus to grow at body temperature, adhere to host cells, evade host defenses, and spread to other organs and tissues [23]. The identification of key virulence factors required for disease progression is critical for understanding the biology of Paracoccidioides spp. infection. Thus, in the present study, we determined protein levels of P. brasiliensis complex isolates and correlated them with their virulence characteristics in a murine model. In particular, we evaluated eight P. brasiliensis complex strains obtained from different environmental and animal sources and compare them to the highly virulent Pb18. The isolates were grouped according to virulence levels, and protein profiles were determined by a two-dimensional (2D) proteomic approach in an attempt to identify possible virulence factors that could correlate with the extent of immunological disturbances observed in the experimental murine PCM model (Fig 1).
Fig 1. Schematic depicting a two-dimensional (2D) proteomic approach used in this study in an attempt to identify possible virulence factors of Paracoccidioides spp.
Isolates were selected according to their source (clinical and environmental) and submitted to virulence assays in BALB/c and B10.A mice. Afterwards, the yeasts were recovered, proteins were extracted and then resolved by 2D gel electrophoresis. Proteins were deemed to have differential abundance levels if their spot volumes were changed at least twofold compared to the normalized spot volume, and based on statistical significance. Proteins with differential abundance levels were identified by mass spectrometry analysis. Finally, based on virulence and proteomic assays we classified isolates according to their virulence level.
Materials and methods
Ethical approval
The study was performed in strict accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the Institutional Ethics in Research Committee of the Federal University of São Paulo (protocol number 0692/05).
Fungal isolates
Table 1 shows the origin and location of P. brasiliensis complex isolates used in this study. These isolates were characterized down to species level by PCR-RFLP of the alpha-tubulin gene as described earlier [35]. Yeast-form samples from all isolates were cultivated in semi-solid Fava Netto's medium [36] at 36°C. The fungi were used at the 7th day in culture, which corresponds to the exponential phase of growth [37]. Attenuation of virulence may occur more rapidly in some Paracoccidioides strains than others when subjected to successive in vitro subculturing [27, 28] and therefore impact multiple comparisons as proposed here. To prevent any bias among Paracoccidioides spp. isolates at the start of in vitro culturing, all isolates were passed through BALB/c and then re-isolated prior to challenge experiments.
Table 1. Origin and location of P. brasiliensis complex isolates used in this study.
| Isolate | EPM code1 | Genetic group/ Phylogenetic species2 | Origin | Location | References |
|---|---|---|---|---|---|
| Pinguim | EPM 34 | PS3 (P. restrepiensis) | Animal (Penguin feces) | Uruguay | [38] |
| Ibiá T1 | EPM 101 | S1 (P. brasiliensis s. str.) | Animal (Armadillo) | Ibiá (Minas Gerais), Brazil | [39] |
| Ibiá T2 | EPM 102 | S1 (P. brasiliensis s. str.) | Animal (Armadillo) | Ibiá (Minas Gerais), Brazil | [39] |
| Bot 1/96 | EPM 11 | S1 (P. brasiliensis s. str.) | Animal (Armadillo) | Botucatu (São Paulo), Brazil | [40] |
| T1 | EPM 53 | S1 (P. brasiliensis s. str.) | Environment (Soil) | Miranda (Paracotos), Venezuela | [41] |
| T2 | EPM 54 | PS3 (P. restrepiensis) | Environment (Soil) | Miranda (Paracotos), Venezuela | [41] |
| 262Uber | EPM 28 | S1 (P. brasiliensis s. str.) | Dog food contaminated with soil | Uberlândia (Minas Gerais), Brazil | [42] |
| Ibiá | EPM 30 | PS3 (P. restrepiensis) | Environment (Soil) | Ibiá (Minas Gerais), Brazil | [43] |
| Pb18 | EPM 16 | S1 (P. brasiliensis s. str.) | Human | São Paulo, Brazil | [44] |
1Paulista School of Medicine (EPM), Federal University of São Paulo
2Molecular characterization based on TUB1-RFLP.
Mice and infection
B10.A and BALB/c isogenic male mice (8 to 12 week-old) were provided by the animal facility of the Federal University of São Paulo, Brazil. Animals were divided into 10 groups of 5 mice each (one group for each Paracoccidioides isolate and one negative control group). Animals were housed in temperature-controlled rooms at 23–25°C, five per cage, in standard boxes with ad libitum access to food and water. Mice were infected intratracheally (i.t.) with 106 P. brasiliensis yeast cells/animal. The yeast cells were washed three times in phosphate-buffered saline (PBS), and fungal suspensions were used at a concentration of 1 × 106 cells per 50 μL, adjusted after counting with a hemocytometer. The viability of fungal cells was evaluated using the vital dye Trypan blue as previously described [45] and was always higher than 95%. The control group received 50 μl of PBS only.
Fungal loads
The severity of the infection was determined by the Colony Forming Unit (CFU) assay using a total of 90 mice. Thirty days after i.t. administration of fungal cells, the animals (n = 5) were euthanized by CO2 anesthesia and organs such as the lungs, liver, and spleen were macerated, seeded on Petri dishes containing Brain Heart Infusion agar and incubated at 36°C. The colonies were counted on the 10th day of plating, when the number of colonies was no longer increasing [46].
Purification of gp43 antigen
Mycelial form samples of P. brasiliensis isolate B-339 (ATCC 32069; PS3) were cultivated in solid Sabouraud dextrose agar (Difco Laboratories, Detroit, MI, USA) at room temperature. The fungus was converted to the yeast form on modified Sabouraud dextrose agar (Sab-T-A) containing 0.01% thiamine and 0.14% asparagine (Difco Laboratories, Detroit, MI, USA) at 35°C. Exoantigen was produced according to the method of de Camargo et al. [47, 48], and gp43 antigen was purified from that exoantigen [49]. Concentrations of purified protein were determined by the Bradford method [50]. Protein fractions were submitted to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) [51] and silver stained [52] to confirm the purification. Gp43 antigen was stored at -20°C until use.
Antibody detection
B10.A and BALB/c mice were infected as described above. Serum samples were obtained by bleeding the tail vein at the 30th day post-infection and stored at -20°C until use. For the indirect enzyme-linked immunosorbent assay (ELISA), polystyrene 96-well microplates (Costar, Corning Inc., Cambridge, MA, USA) were coated with purified gp43 (250 ng/well) diluted in 0.1 M carbonate-bicarbonate buffer, pH 9.6 and incubated at 37°C for 2 h and overnight at 4°C. The plates were washed three times with a 0.05% solution of Tween-20 in PBS (PBS-T), and free sites were blocked with 5% skim milk in PBS-T (200 μL/well) at 37°C for 2 h. After three washes, 100 μL of mouse serum samples diluted at 1:50 in PBS-T (containing 0.25% gelatin—PBS-T-G) were added to the wells in duplicates. The plates were incubated at 37°C for 1 h and washed again. Then, 100 μL of peroxidase conjugated anti-mouse IgG (y-chain specific; 1:1000 dilution in PBS-T-G; Sigma Chemical Co., St Louis, MO, USA) was added to each well. The plates were incubated at 37°C for 1 h, washed, and then, 100 μL of the substrate solution (5 mg of O-phenylenediamine in 10 mL of 0.1 M citrate phosphate buffer, pH 4.5, plus 10 μL of 30% H2O2) was added to the wells. After color development, the reaction was stopped by the addition of 50 μL of 4 N H2SO4. The optical density (OD) values were measured at 492 nm using an ELISA microplate reader (Sunrise absorbance reader, Tecan, Mannedorf, Switzerland).
Protein extraction
P. brasiliensis yeast cells were grown for 7 days at 36°C in triplicate on Fava-Netto’s medium, and protein extract was obtained as previously described by Rodrigues et al. [53]. Briefly, yeast cells were washed in PBS, centrifuged (5,000×g, 5 min, 4°C), frozen in liquid nitrogen, and disrupted by mechanical maceration. Then, 2 mL of buffer extraction medium (20 mM Tris-HCl, pH 8.8, 2 mM CaCl2) containing a cocktail of protease and nuclease inhibitors (1:100; GE-healthcare, Uppsala, Sweden) was added. The extract was vortexed, centrifuged (8,000×g, 15 min), and the supernatant was kept at −80°C until use. Protein concentrations were determined by the Bradford method [50].
Two-dimensional gel electrophoresis
Before two-dimensional gel electrophoresis (2-DE), proteins were concentrated using a 2-D Clean-up Kit (GE Healthcare, Uppsala, Sweden). Next, 300 μg of total protein was diluted in 250 μL of rehydration solution (7 M urea, thiourea 2 M, 2% CHAPS, DeStreak 1,2%, 1% vol/vol isoeletric focusing buffer pH 3–10, applied into 13-cm immobilized pH gradient (IPG 3–10) (GE Healthcare, Uppsala, Sweden), and rehydrated at 20°C for 12 h using an Ettan IPGphor III system (GE Healthcare, Uppsala, Sweden). The rehydrated strips were focused at 20°C as follows: 1 h at 500 V, 1 h at 1000 V, 4 h at 8000 V, 6 h at 8000 V, and 12 h at 1000 V. Focused IPG strips were sequentially incubated for 2×20 min in two equilibration buffer solutions (6 M Urea, 50 mM Tris HCl, pH 6.8, 30% glycerol, 2% SDS), containing 10 mg/mL dithiothreitol and 25 mg/mL iodoacetamide, respectively. The second dimensional separation was performed on 10% polyacrylamide gels (45 mA per gel, 10°C) using a Hoefer SE 600 system (GE Healthcare, Uppsala, Sweden). The gels were stained with Coomassie Brilliant Blue G-250 [54]. The experiments were carried out in triplicate. 2-DE gels were scanned on an Image Scanner III (GE Healthcare, Uppsala, Sweden) and analyzed using Image Master 2D platinum 7.0 software (GE Healthcare, Uppsala, Sweden).
2-DE gels image and data analysis
The images of the 2-DE gels were captured by ImageScanner at 300 dots/inch, and spots were quantitatively analyzed using Image Master 2D platinum 7.0 software. After automated matching, manual matching was carried out to correct the mismatched or unmatched spots by adding, splitting and removing spots. To compare spots across gels in each Paracoccidioides spp., a match set was obtained with images from all gels, and only well-resolved spots in all three biological replicates were considered reproducible. For the matched protein spots in each 2-DE gel, their volumes were normalized to the total spot volume using the software ImageMaster 2D Platinum 7.0, in order to eliminate the possible variations due to staining. The normalized volume of each protein spot was used as its expression abundance. All values are presented as mean ± S.E.M (standard error of the mean). Proteins were deemed to have differential abundance levels if their spot volumes were changed at least twofold (>2.0-folds) compared to the normalized spot volume, and based on analysis of variance (ANOVA, P < 0.05), implemented in the ImageMaster 2D Platinum 7.0.
Protein digestion and peptide extraction
Digestion was performed according to Pitarch et al. [55]. Briefly, the spots were excised from 2-DE gels, destained twice with 50% acetonitrile (ACN) in 25 mM NH4HCO3, and vacuum-dried. The proteins were then reduced with 10 mM dithioerythritol in 25 mM NH4HCO3 for 30 min at 56°C and alkylated with 55 mM iodoacetamide in 25 mM NH4HCO3 for 20 min in the dark. Afterwards, gel pieces were washed with 25 mM NH4HCO3 and ACN, and dried under vacuum. All gel pieces were incubated with 12.5 ng/mL sequencing grade trypsin (Promega, Madison, WI, USA) in 25 mM NH4HCO3 overnight at 37°C. Peptides were then extracted from the gel pieces with 50% ACN, 1% trifluoroacetic acid solution in 25 mM NH4HCO3, and finally with 100% ACN. The combined extracts were dried in a SpeedVac concentrator (Thermo Fisher Scientific, Waltham, MA, USA). Samples were then subjected to mass spectrometry analysis.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
A 4.5-μL aliquot of digested proteins was injected into a C18 1.7 μm BEH 130 (100 μm × 100 mm) RP-ULC analytic column (nanoAcquity UPLC, Waters Corporation, Manchester, UK) coupled with nano-electrospray tandem mass spectrometry system on a Q-Tof Ultima API mass spectrometer (MicroMass/Waters, Corporation, Manchester, UK) at a flow rate of 600 nL/min. A trapping Symmetry C18 column (180 μm × 20 mm) was used for sample desalting at a flow rate of 20 μL/min for 1 min. The gradient was 0–50% ACN (acetonitrile) in 0.1% formic acid over 45 min. The instrument was operated in the MS positive mode, data continuum acquisition from m/z 100 to 2,000 Da at a scan rate of 1 s and inter-scan delay of 0.1 s.
Database search
Database searches for the identification of peptides from LC MS-MS experiments were done with Mascot Distiller v.2.3.2.0, 2009 (Matrix Science, Boston, MA) using carbamidomethyl-cys as fixed modification (monoisotopic mass 57.02015 Da), oxidation (HW) and oxidation (M) as variable modification (monoisotopic mass 15.0215 15.9949), and 0.1 Da MS and MS/MS fragment tolerances. After the analysis, the data from each spot were exported in a text file format. Sequence database search was carried out with MASCOT search engine (Matrix Science Ltd., London, UK). The results were compared to known sequences from Paracoccidioides database (http://www.broadinstitute.org/) [56, 57]. The default significance threshold was P < 0.05.
Statistical analysis
CFU assay and ELISA results were assessed statistically by using the two-way ANOVA followed by pairwise comparisons by the post hoc Tukey’s test. Differences were considered statistically significant if corresponding P-values were below 0.05.
Results
Experimental paracoccidioidomycosis
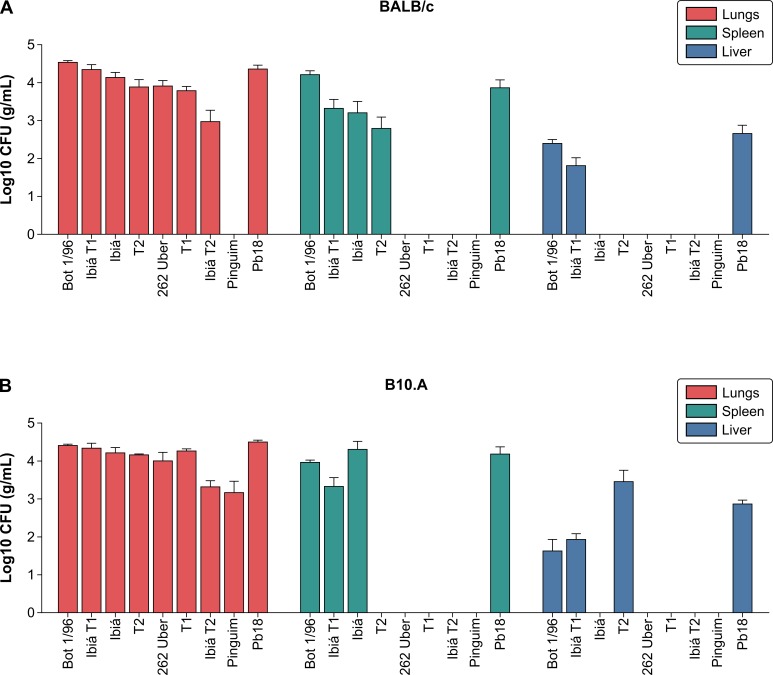
The fungal tissue burden in the liver, spleen, and lungs for the BALB/c (Fig 2A) and B10.A mice (Fig 2B) inoculated with one of the nine P. brasiliensis complex isolates studied is shown in Fig 2. All isolates except Pinguim isolate were able to colonize the lungs, the target organ of inoculation in both mouse lines. Bot 1/96 and Ibiá T1 isolates that colonized mouse spleen and liver caused disseminated disease and, therefore, were considered highly virulent and comparable to control Pb18 P. brasiliensis strain. Ibiá and T2 isolates colonized only the spleen besides the lungs and induced a less severe form of the disease. These isolates were therefore classified as moderately virulent. The remaining isolates (262Uber, T1, and Ibiá T2) caused limited disease manifestations and affected only the lungs, being unable to disseminate to other organs. These isolates were considered to possess low virulence. It was not possible to recover the fungi from any examined organs of BALB/c mice infected with Pinguim isolate. At the same time, viable fungi could be recovered from the lungs of infected B10.A mice (Fig 2B). Similar patterns of fungal dissemination were observed in both mouse strains, except for those of T2 and Pinguim isolates. T2 isolate disseminated to the spleen in BALB/c mice and to the liver in B10.A mice. During the thirty days post inoculation with P. brasiliensis complex isolates, no deaths were observed in the two mouse strains.
Fig 2. Fungal loads in different mouse tissues.
Colony-forming units (CFUs) were obtained from samples of the spleen, liver, and lungs of BALB/c (A) and B10.A (B) mice infected intratracheally with 1×106 P. brasiliensis yeast cells. Mice were euthanized at 30 days after inoculation. Data are representative of two independent experiments and values are expressed in mean ± SD. Horizontal bars indicate statistical significance of differences between numbers of CFUs obtained from isolate samples and those of virulent control Pb18: *P < 0.05, Tukey’s test.
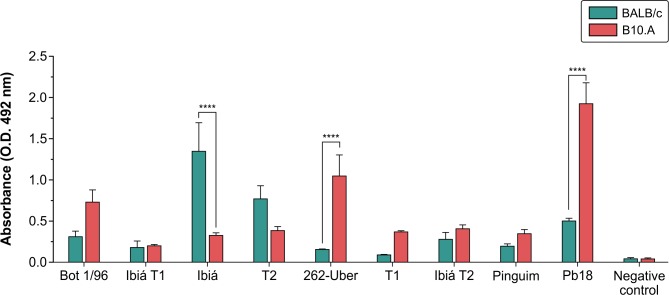
We then evaluated the humoral immune response of BALB/c and B10.A mice infected with Paracoccidioides isolates as described above (Fig 2). Fig 3 shows that all P. brasiliensis complex isolates induced the production of anti-gp43 antibodies. However, the isolates elicited distinct serological responses during infection. We observed that BALB/c mice infected with moderately virulent Ibiá and T2 isolates had higher antibody titers than highly virulent control Pb18 P. brasiliensis strain. Statistical analysis revealed that there was no correlation between the degree of P. brasiliensis complex isolate virulence (Fig 2) and antibody production in mice. The data on the virulence and dissemination of P. brasiliensis complex isolates are summarized in Table 2.
Fig 3. Anti-gp43 antibody detection.
The levels of anti-gp43 antibody were determined in sera from B10.A and BALB/c mice by ELISA. Groups of five mice each were inoculated intratracheally with 106 P. brasiliensis yeast cells. Serum samples were obtained by tail vein bleeding at the 30th day post-infection. Serum samples from non-infected mice were used as negative control. Data are representative of two independent experiments and values are expressed in mean ± SD. Statistical significance of differences is indicated as follows: ****P <0.0001, Tukey’s test.
Table 2. Virulence characteristics of P. brasiliensis complex isolates based on murine model of infection.
| P. brasiliensis isolate | Dissemination in BALB/c mice | Dissemination in B10.A mice |
Mortality | Virulence level |
|---|---|---|---|---|
| Bot 1/96 | Spleen and liver | Spleen and liver | No | High |
| Ibiá T1 | Spleen and liver | Spleen and liver | No | High |
| Ibiá | Spleen | Spleen | No | Medium |
| T2 | Spleen | Liver | No | Medium |
| 262 Uber | None | None | No | Low |
| T1 | None | None | No | Low |
| Ibiá T2 | None | None | No | Low |
| Pinguim | None | None | No | Low |
| Pb18 | Spleen and liver | Spleen and liver | No | High |
Comparative proteomic analysis of P. brasiliensis complex isolates
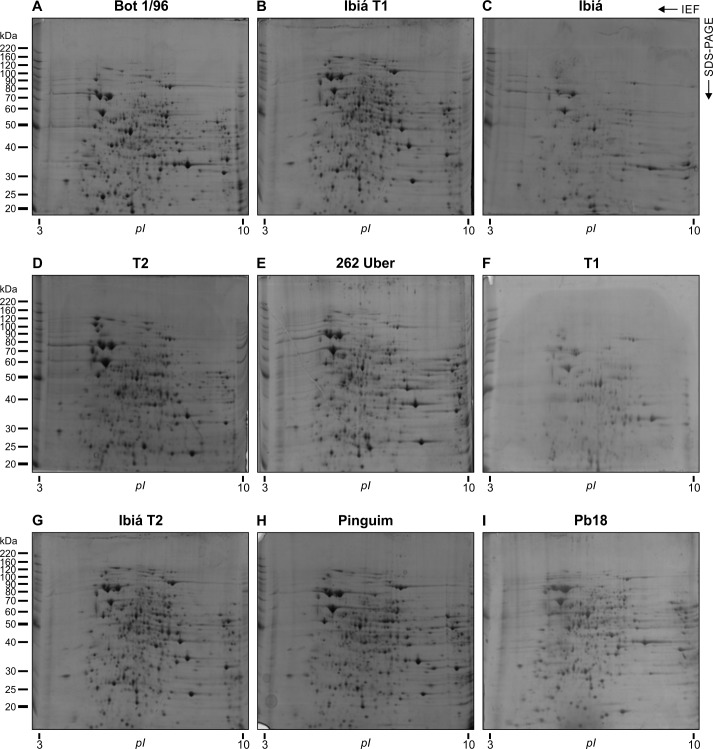
Comparative analysis of proteins up- or down-regulated in P. brasiliensis complex isolates with highly, moderate, and low virulence was performed in order to determine the proteins whose levels correlates with the extent of virulence. For this purpose, protein extracts of the nine isolates (Table 1) were fractionated by 2D electrophoresis in triplicate (pH 3–10). Representative images of proteome maps from P. brasiliensis complex isolates are shown in Fig 4.
Fig 4. Proteomic maps of P. brasiliensis complex isolates.
Cell extracts (originated from three biological replicates) were subjected to 2D electrophoresis on 13-cm immobilized pH gradient strips in the range of pI values from 3 to 10 (GE Healthcare, USA), and the proteins were developed by Coomassie blue staining. A representative 2D gel is shown for each Paracoccidioides isolate; a: Bot 1/96, b: Ibiá T1, c: Ibiá, d: T2, e: 262 Uber, f: T1, g: Ibiá T2, h: Pinguim, i: Pb18. Molecular masses of standard proteins are given on the left side of the gel (BenchMark Protein Ladder, Invitrogen). Further information about protein levels using 1D gel can be found in the S1 Fig.
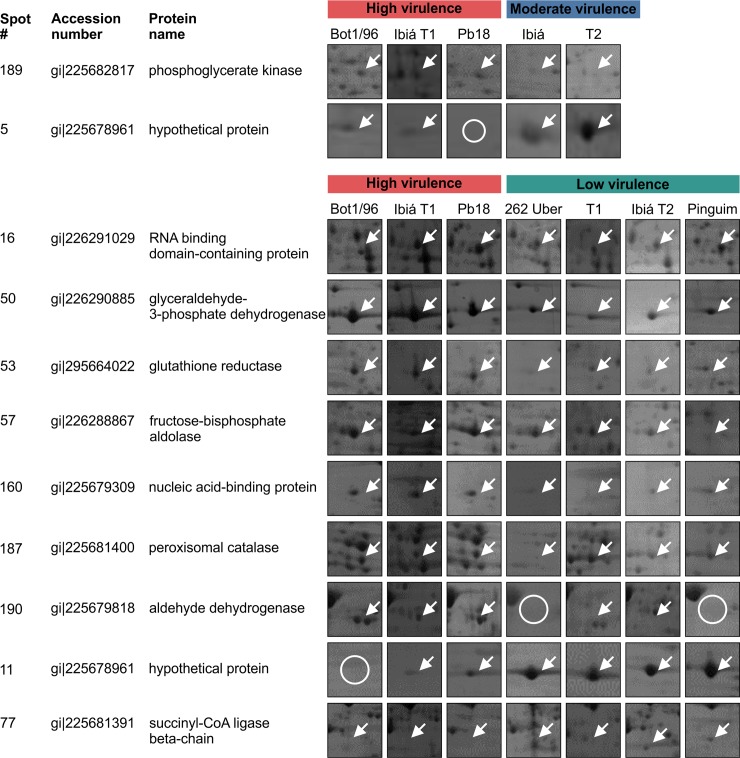
Gel image analyses were conducted using protein profiles of highly virulent isolates (Bot 1/96, Ibiá, and Pb18) as references that were compared to profiles of isolates with moderate (Ibiá and T2) and low (262-Uber, T1, Ibiá T2, and Pinguim) virulence. Qualitative analyses were performed considering the presence or absence of particular protein spots, depending on biological properties, i.e., by noting whether the corresponding isolate had high, moderate, or low virulence. Quantitative comparisons were carried out by focusing on spots that exhibited two-fold variations in intensity. By comparing highly and moderately virulent isolates, we revealed 25 spots corresponding to the proteins that were strongly expressed by highly virulent Bot 1/96, Ibiá, and Pb18 isolates, as well as five protein spots that were denser in moderately virulent Ibiá and T2 isolates (S1 Table). The comparative analysis of highly virulent Bot 1/96, Ibiá, and Pb18 isolates with 262 Uber, T1, and Ibiá T2 strains with low virulence showed that 29 proteins were up-regulated in the former isolates and five proteins had higher density in isolates of low virulence (S2 Table). Protein spots up-regulated in both biological conditions were analyzed by mass spectrometry (Table 3). The identified proteins were located in the proteomic maps of P. brasiliensis complex isolates (Table 1) and are shown in Fig 5.
Table 3. Identification of protein spots from 2D electrophoresis gels from P. brasiliensis complex isolates by LC-MS/MS.
| Experimental condition | Isolate virulence | Spot number1 | Accession number2 | Protein name |
Experimental MM3 (kDa)/pI4 |
Theoretical MM3 (kDa)/pI4 | % seq. cov.5 | Protein score |
|---|---|---|---|---|---|---|---|---|
| Highly versus moderately virulent isolates | High | 189 | gi|225682817 | phosphoglycerate kinase | 43.37/5.24 | 45.13/6.28 | 10% | 153 |
| Moderate | 5 | gi|225678961 | predicted protein | 23.37/8.21 | 24.61/8.49 | 17% | 203 | |
| Highly versus slightly virulent isolates | High | 16 | gi|226291029 | RNA binding domain-containing protein | 25.5/5.95 | 30.53/9.20 | 24% | 395 |
| 50 | gi|226290885 | glyceraldehyde-3-phosphate dehydrogenase | 33.71/8.07 | 36.57/7.12 | 49% | 687 | ||
| 53 | gi|295664022 | glutathione reductase | 35.09/4.98 | 51.95/6.74 | 10% | 302 | ||
| 57 | gi|226288867 | fructose-bisphosphate aldolase | 37.5/6.90 | 39.74/6.28 | 26% | 371 | ||
| 160 | gi|225679309 | nucleic acid-binding protein | 43.27/4.49 | 10.88/5.77 | 2% | 44 | ||
| 187 | gi|225681400 | peroxisomal catalase | 54.70/7.30 | 57.65/6.42 | 4% | 102 | ||
| 190 | gi|225679818 | aldehyde dehydrogenase | 55.00/5.37 | 54.24/5.30 | 2% | 38 | ||
| Low | 11 | gi|225678961 | predicted protein | 24.00/8.20 | 24.61/8.49 | 12% | 146 | |
| 77 | gi|225681391 | succinyl-CoA ligase beta-chain | 46.25/5.11 | 48.56/5.61 | 15% | 310 |
Fig 5. Identification of proteins by LC-MS/MS.
Proteins corresponding to the spots of interest in 2D electrophoresis gels were identified by LC-MS/MS. An arrow indicates the presence of the protein, whereas an open circle indicates the absence of the protein. A: Proteins with differential abundance levels between highly and moderately virulent isolates. B: Proteins with differential abundance levels between highly and slightly virulent isolates. The data presented represent the results of three biological replicates.
Discussion
In this study, we used a proteomic approach to identify proteins differentially expressed by P. brasiliensis complex isolates that exhibited different levels of virulence. Our main objective was to correlate biological roles of these proteins to respective virulence level. A total of eight up-regulated proteins were successfully identified by LC-MS/MS in highly virulent isolates. One up-regulated protein was identified in a moderately virulent isolate, and two—in isolates of low virulence. Unfortunately, other proteins with differential abundance levels could not be identified, probably because their amounts were too low to produce a good spectrum, or because the confidence levels of the database search were insufficient to yield unambiguous results.
According to CFU assays, the isolates were classified as those having high (Bot 1/96, Ibiá T1 and Pb 18), moderate (Ibiá and T2), or low (262Uber, T1, Ibiá T2, and Pinguim) virulence. A very similar pattern of fungal dissemination in BALB/c and B10.A mice was observed for the majority of isolates except for that of Pinguim isolate, which was unable to colonize any organs of BALB/c mice, but could be recovered from the lungs of B10.A mice. In relevance to this result, it has been shown previously that B10.A mice were highly susceptible, whereas BALB/C mice were moderately susceptible to P. brasiliensis Pb18 infection by intraperitoneal route [58]. In addition, the isolate Pinguim, which was previously considered a new Paracoccidioides species, P. antarcticus [MB#492297], was isolated from Pygoscelis adeliae faeces (a penguin found along the entire Antarctic coast and at some of its nearby islands), is capable of producing experimental orchids in guinea pigs [59]. In vitro cultivation at 36°C reveals marked morphological variation in the yeast phase [59], but antigenic preparations displayed total immunological identity with classical P. brasiliensis strains, mainly regarding the specific gp43 component [60], as detected here using ELISA. From a phylogenetically point of view, Pinguim is classified as P. brasiliensis S1, a paraphyletic group, and shares genetic similarity with other atypical isolates (e.g. JT-2, 1430) [61]. The diversity of morphology, genetic and proteomic characteristics may support the variety of responses during interaction with different hosts as observed here for the BALB/C and B10.A mice.
In our experiments, we evaluated both the ability of P. brasiliensis complex isolates to induce immune response in mice and possible correlation between the level of isolate virulence and antibody titer. We observed that all isolates, including Pinguim isolate, were able to induce the production of anti-gp43 antibodies in both BALB/c and B10.A strains of infected mice, indicating that the infection occurs even if the corresponding isolate cannot be readily recovered from mouse organs.
Furthermore, the statistical analysis revealed that there was no correlation between isolate virulence and induced antibody response. Our results showed that although the isolates had varying immunogenicity levels, these differences were not sufficient to explain either the severity of infection or tissue tropism. In a fact, our results are in disagreement with several influential publications on human PCM, which posited that severe forms of the disease are associated with the highest antibody titers [12, 62]. Discrepant results regarding the relationship between antibody response and severity of clinical forms of PCM have been also documented by other researchers [63–65]. Singer-Vermes et al. [66] examined six P. brasiliensis s.l. isolates obtained from samples of patients that presented with distinct and well-defined clinical forms of PCM and compared their virulence, tissue tropism, and humoral immune response in susceptible B10.A mice. They found that in general, pathogenicity and immunogenicity parameters in humans and in susceptible mice were not analogous. The authors argued that in human body, unlike in experimental laboratory conditions, PCM has a very slow evolution. In addition, factors related to the host, such as genetic pattern, sex, age, nutritional and immunological status may be of greater relevance to the evolution and outcome of PCM.
The reports showing that humoral immune responses play an important role in conferring protection against PCM are still scarce. In experimental PCM, B lymphocytes-knockout mice were shown to be more susceptible to P. brasiliensis infection than their wild-type controls, presenting higher mortality rate and numbers of viable P. brasiliensis in the lungs. The granulomas of the knock-out mice were larger than those of the control mice, and as the size and organization of P. brasiliensis granulomas reflect the control or not of the infection [67], the results point towards a protective effect of B lymphocytes. The absence of B cells leads to increased levels of IL-10, confirming experimental data that links this observation with more severe disease. Therefore, this data suggests that in experimental PCM B lymphocytes are paramount to effectively control both P. brasiliensis growth and the organization of the granulomatous lesions [68].
A report by Montagnoli and colleagues revealed that antibodies have a critical role in the generation of memory antifungal immunity [69]. Also working with mice deficient in B lymphocytes, Montagnoli et al. showed that although passive administration of antibodies increased the fungal clearance, the innate and Th1-mediated resistance to the primary and secondary infections were both heightened in mu MT mice with candidiasis and aspergillosis. However, although capable of efficiently restricting the fungal growth, mu MT mice did not survive the re-infection with Candida albicans, and this was concurrent with the failure to generate IL-10-producing dendritic cells and regulatory CD4(+)CD25(+) T cells. Antifungal opsonizing antibodies restored IL-10 production by dendritic cells from mu MT mice, a finding suggesting that the availability of opsonizing antibodies may condition the nature of the dendritic cell interaction with fungi, possibly impacting on the development of long-lasting antifungal immunity [69].
IgG2a and IgG2b monoclonal antibodies against the major diagnostic antigen of Paracoccidioides brasiliensis, gp43 were shown to reduce fungal burden and was associated with the enhanced phagocytosis of P. brasiliensis by macrophages leading to increased nitric oxide production. The monoclonal antibody against the major diagnostic antigen of P. brasiliensis mediates immune protection in infected BALB/c mice challenged intratracheally with the fungus [70].
Although humoral immunity might not have a major role in conferring protection against fungal infections in human, passive administration of specific protective antibodies proved to be beneficial in drug resistance cases, to reduce the dosage and associated toxic symptoms of antifungal drugs.
It was experimentally demonstrated that antibodies produced against gp70, a circulating antigen detected during PCM, prevented the establishment of the disease in mice [71]. Also, the adaptive transference of WT immune or non-immune serum to B-lymphocyte knock-out mice is associated with better clinical features, including diminished infiltration of inflammatory cells and formation of organized granuloma. The authors conclude that B cells are effectively involved in the control of P. brasiliensis growth and participate in the organization of the granulomatous lesion observed in the lungs from Pb18-infected mice [68].
Our proteomic analysis indicates that highly virulent isolates probably expressed a higher amount of phosphoglycerate kinase than isolates of low virulence. Furthermore, highly virulent isolates had higher levels of RNA binding domain-containing protein (RBP), glyceraldehyde-3-phosphate dehydrogenase, glutathione reductase, fructose-bisphosphate aldolase, nucleic acid-binding protein, peroxisomal catalase, and aldehyde dehydrogenase (ALDH) than isolates of low virulence. Judging from the proteins above, there is a connection between pathogenicity, metabolism, and redox homeostasis. Energy metabolism is largely from glycolysis, a metabolic pathway that is fundamental to the assimilation of carbon for either respiration or fermentation, and therefore is critical for the growth of Paracoccidioides and other fungal pathogens [72, 73]. Our results support the up-regulation of genes involved in gluconeogenesis in highly virulent Paracoccidioides, as the protein levels related to carbohydrate metabolism increase. Interestingly, glucose-6-phosphate dehydrogenase, the first enzyme of the pentose phosphate cycle, has the interesting property of reducing NADP+ to NADPH(H)+ and thus is the key enzyme that provides the reducing power of the cell [74]. It has been demonstrated that HeLa cells expressing high levels of glucose-6-phosphate dehydrogenase display an increased level of reduced glutathione and show oxidoresistance [74–76]. In addition, aldehyde dehydrogenase may consolidate intracellular redox homeostasis in Paracoccidioides by detoxifying stress-generated aldehydes, an important feature to survive within the human host [73].
It has been shown that proteins may have multiple independent functions. This phenomenon occurs in both eukaryotes and prokaryotes, including the representatives of Paracoccidioides genus [77]. In our experiments, we found three glycolytic enzymes whose expression varied with Paracoccidioides virulence: phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, and fructose-bisphosphate aldolase. Despite their role in carbohydrate metabolism, it has been demonstrated that the relevance of these proteins for PCM pathogenesis stems from their role in adhesion [78, 79], oxidative stress [73], synthesis of extracellular vesicles and cell wall [80, 81], and as immunogenicity [82, 83]. Those reports corroborate our findings, because the above mentioned enzymes were overexpressed in isolates that caused severe and disseminated disease in mice. Thus, the enzymes up-regulated in highly virulent isolates were probably important for shaping virulent phenotype of these strains.
During the infection with P. brasiliensis, macrophages and neutrophils constitute one of the primary defense mechanisms. These cells generate reactive oxygen and reactive nitrogen species that can damage amino acids, lipids, DNA, and ultimately lead to cell death [84]. To overcome this defense system, intracellular pathogens must have adaptive mechanisms to survive in this hostile environment. In our experiments, we revealed two molecules with protective antioxidant activity, glutathione reductase and peroxisomal catalase, which were up-regulated in highly virulent isolates. Glutathione reductase has been implicated in the virulence of Cryptococcus neoformans [85] and Candida albicans [86]. In Paracoccidioides spp., the importance of glutathione reductase during infection has not been studied in detail. However, this protein has been shown to be up-regulated in mycelial secretome [87] and expressed with differential abundance levels in Paracoccidioides species [87, 88]. Peroxisomal catalase has been identified as a typical monofunctional enzyme highly expressed at the yeast phase [82, 89] and up-regulated when the fungus is phagocytosed by macrophages [90, 91]. The exposure of Paracoccidioides yeast cells to hydrogen peroxide induced overexpression of peroxisomal catalase [73, 89, 92]. In our study, up-regulation of these proteins in highly virulent isolates may be related to the mechanism by which P. brasiliensis evades immune system as a strategy for its survival within infected host cells and dissemination to other organs. In contrast, lower expression of these enzymes in isolates with low virulence seems to correspond to their limited ability to cause fungal infection at the inoculation site, the lungs.
Representatives of the ALDH protein superfamily are expressed by species of all three taxonomic domains and are involved in a variety of biological processes, including metabolism of toxic aldehydes and maintenance of the cellular homeostasis [93]. It has been shown that exposure of organisms to stress conditions leads to the increase in ALDH expression [94]. In Paracoccidioides genus, expression ALDH has not been extensively studied. However, de Arruda Grossklaus et al. [73] demonstrated that yeast cells treated with hydrogen peroxide had higher expression of ALDH than non-treated cells. Recently, Chaves et al. [79] demonstrated that ALDH binds to plasminogen, a fact that indicates a potential role of ALDH in the pathogen-host interaction. In our study, the up-regulation of ALDH in highly virulent isolates could be an indication of the involvement of this enzyme in the protection of the fungus from general stress generated by host defense mechanisms.
In the present study, we also detected an increased expression of the RBP and nucleic acid binding protein in highly virulent isolates. RBPs that regulate gene expression at all levels, are numerous and widely distributed in nature. They are key modulators of gene expression and are involved in cell differentiation, cellular response to environmental changes, and cell death [95]. The relationship between RBPs and post-transcriptional regulation/ stress response was observed in several organisms, such as viruses [96], bacteria [97], protozoa [95], plants [98], and mammals [99]. RBPs have been implicated in the regulation of stress response in pathogenic fungi such as Aspergillus fumigatus [100], Cryptococcus neoformans [101], and C. albicans [102–104]. The mechanism by which RBP contributes to the virulence of Paracoccidioides is unclear. However, Parente et al. [105] showed that Paracoccidioides yeast cells exposed to nitrosative stress overexpressed the posttranscriptional regulator mRNA binding protein. Our present study suggests that RBP is important to Paracoccidioides resistance to host defense system, because its presence appears to enhance Paracoccidioides virulence in vivo. Moreover, during the infection, Paracoccidioides species become exposed to a very hostile environment, and the adaptation processes require global reorganization of gene expression, a fact that could explain RBP overexpression in highly virulent isolates.
Iron is essential for supporting infectiveness of many microorganisms, including P. brasiliensis [106], due to its role in electron transfer and acid-base reactions, and because it acts as cofactor in a variety of biological processes [107]. For these reasons, the host needs to maintain a balance in iron bioavailability to ensure sufficient levels for own cellular metabolism and, at the same time, to limit the availability of iron to pathogens, as a defense measure [108]. In this regard, Paracoccidioides spp. developed a mechanism to obtain iron from high-affinity iron-binding proteins, such as hemoglobin [109]. In addition to its iron uptake system, P. brasiliensis is able to alter its metabolism according to iron availability, e.g., by inducing the expression of glycolytic pathway proteins at high iron concentrations or by repressing the expression of tricarboxylic acid cycle (TCA) proteins under conditions of limited iron availability, as TCA reactions are mediated by enzymes containing Fe/S [110]. In our experiments, we found that succinyl-CoA ligase beta chain, a protein related to TCA, was down-regulated in highly virulent isolates whereas phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, and fructose-bisphosphate aldolase were overexpressed by highly virulent isolates. These findings may indicate a mechanism used by virulent P. brasiliensis complex isolates to adapt to transient iron availability as a strategy to survive and overcome stress conditions inside the host. Interestingly, glyceraldehyde-3-phosphate dehydrogenase and fructose-bisphosphate aldolase, which were overexpressed by highly virulent isolates in our experiments, were also found in Paracoccidioides extracellular vesicles preparations of Pb18 isolate [111]. Such elegant vesicular transport may deliver substances across Paracoccidioides cell wall, possibly modulating the host’s immune response and supporting the high virulence phenotype observed in our murine models.
Another potential virulence factor detected in our analysis was a hypothetical protein (accession number: gi|225678961) with a theoretical molecular mass of 24.61 kDa and pI of 8.49 that was overexpressed in isolates of moderate and low virulence when compared to its level in highly virulent isolates. In fact, Desjardins et al. [57] reported that nearly 60% of Paracoccidioides genes were annotated as those encoding hypothetical proteins with unknown cellular functions, but for which no evidence of in vivo expression exists [112]. To the best of our knowledge, this is the first report describing the role of such a protein in Paracoccidioides-induced mouse infection. Thus, further studies will be needed to characterize this protein and understand its role in PCM infection.
In summary, the data suggest that highly virulent P. brasiliensis complex isolates that caused disseminated disease in a murine model of PCM expressed high levels of common proteins, such as phosphoglycerate kinase, RNA binding protein, glyceraldehyde-3-phosphate dehydrogenase, glutathione reductase, fructose-bisphosphate aldolase, nucleic acid-binding protein, peroxisomal catalase, and aldehyde dehydrogenase. These proteins were probably critical to the ability of P. brasiliensis to colonize the host, to survive in its hostile environment, and to escape host immune system, because they appear to be more abundant in highly virulent isolates than in isolates with low virulence. Although our data are not sufficiently complete to create an integral model of P. brasiliensis pathogenicity, however, they provide important clues towards understanding how fungi adapt to host immune response. In light of our present observations, these proteins levels with differential abundance levels need to be validated using more isolates from human cases of PCM as well as reverse genetic function analysis [113]. The roles of these proteins in different clinical forms of PCM should be studied in order to determine either their potential to be used as biomarkers or targets of therapeutic treatment.
Supporting information
Quantitative analysis of proteins with differential expression in highly virulent isolates (Bot 1/96, Ibiá, Pb18) and moderately virulent isolates (Ibiá and T2) is shown. Positive and negative values indicate higher or lower expression of the protein, respectively. Positive (+) and negative (−) symbols mean the presence or absence of the spot, respectively. Proteins were deemed to have differential abundance levels if their spot volumes were changed at least twofold compared to the normalized spot volume.
(PDF)
Quantitative analysis of proteins with differential expression in highly virulent isolates (Bot 1/96, Ibiá, Pb18) and isolates of low virulence (262Uber, T1 and Ibiá T2). Positive and negative values indicate higher or lower expression of the protein, respectively. Positive (+) and negative (−) symbols mean the presence or absence of the spot, respectively. Proteins were deemed to have differential abundance levels if their spot volumes were changed at least twofold compared to the normalized spot volume.
(PDF)
(A) Paracoccidioides brasiliensis yeast cells were grown for 7 days at 36°C in triplicate on Fava-Netto’s medium, and protein extract was obtained as previously described [1]. Protein concentrations were determined by the Bradford method [2] and 5 μg of each extract were subjected to 1D SDS-PAGE (10%). The molecular masses (in kDa) of standard proteins are given to the left of the gel (BenchMarkTM Protein Ladder, Invitrogen). From left to right: Bot 1/96, Ibiá T1, Ibiá, T2, 262 Uber, T1, Pinguim, Ibiá T2 and Pb18. (B) The completed electrophoresis gel was imaged on an Image Scanner III (GE Healthcare, Uppsala, Sweden) and the comparison was carried out by densitometry measurements of scanned image (8-bit image) along the ROI (arrow) using a 256 grey level scale to determine the average gray value using Adobe Photoshop CC. This region was chosen because it did not present proteins with different abundance levels in the comparative analyzes (2D-GE). (C) Lowest and highest gray values were used to set the ratio between each of the extreme values and the ROI revealing minimum variation across the different samples.
(PDF)
Acknowledgments
We thank Brazilian Synchrotron Light Laboratory (LNLS) for the use of Mass Spectrometry Laboratory under proposal MAS-11262 and Dr. Ricardo Bertolla for his assistance with ImageMaster 7.0 software.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported, in part, by grants from São Paulo Research Foundation (FAPESP), the National Council for Scientific and Technological Development (CNPq) and Coordination for the Improvement of Higher Education Personnel (CAPES). ZPdC acknowledges the financial support of FAPESP (2009/54024-2) and CNPq (429594/2018-6). AMR acknowledges the financial support of FAPESP (2017/27265-5) and CAPES (88887.177846/2018-00). GFF is a fellow and acknowledges the financial support of CNPq (150605/2015-3). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bocca AL, Amaral AC, Teixeira MM, Sato PK, Shikanai-Yasuda MA, Soares Felipe MS. Paracoccidioidomycosis: eco-epidemiology, taxonomy and clinical and therapeutic issues. Future Microbiology. 2013;8(9):1177–91. 10.2217/fmb.13.68 . [DOI] [PubMed] [Google Scholar]
- 2.Muñoz JF, Farrer RA, Desjardins CA, Gallo JE, Sykes S, Sakthikumar S, et al. Genome diversity, recombination, and virulence across the major lineages of Paracoccidioides. mSphere. 2016;1(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutz A. Uma micose pseudococídica localizada na boca e observada no Brasil: contribuição ao conhecimento das hifoblastomicoses americanas. Brasil Med. 1908;22:121–4. [Google Scholar]
- 4.Teixeira MM, Theodoro RC, Nino-Vega G, Bagagli E, Felipe MSS. Paracoccidioides species complex: Ecology, phylogeny, sexual reproduction, and virulence. PLoS Pathog. 2014;10(10):e1004397 10.1371/journal.ppat.1004397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turissini DA, Gomez OM, Teixeira MM, McEwen JG, Matute DR. Species boundaries in the human pathogen Paracoccidioides. Fungal Genetics and Biology. 2017;106(Supplement C):9–25. 10.1016/j.fgb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theodoro RC, Teixeira MdM, Felipe MSS, Paduan KdS, Ribolla PM, San-Blas G, et al. Genus Paracoccidioides: Species recognition and biogeographic aspects. PLoS ONE. 2012;7(5):e37694 10.1371/journal.pone.0037694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn RC, Rodrigues AM, Fontes CJ, Nery AF, Tadano T, de Padua Queiroz Junior L, et al. Fatal Fungemia due to Paracoccidioides lutzii. Am J Trop Med Hyg. 2014;91(2):394–8. 10.4269/ajtmh.13-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gegembauer G, Araujo LM, Pereira EF, Rodrigues AM, Paniago AM, Hahn RC, et al. Serology of paracoccidioidomycosis due to Paracoccidioides lutzii. PLoS Negl Trop Dis. 2014;8(7):e2986 10.1371/journal.pntd.0002986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arantes TD, Theodoro RC, Da Graça Macoris SA, Bagagli E. Detection of Paracoccidioides spp. in environmental aerosol samples. Med Mycol. 2013;51(1):83–92. 10.3109/13693786.2012.698444 . [DOI] [PubMed] [Google Scholar]
- 10.Hrycyk MF, Garcia Garces H, Bosco SdMG, de Oliveira SL, Marques SA, Bagagli E. Ecology of Paracoccidioides brasiliensis, P. lutzii and related species: infection in armadillos, soil occurrence and mycological aspects. Med Mycol. 2018:myx142-myx. 10.1093/mmy/myx142 [DOI] [PubMed] [Google Scholar]
- 11.Arantes TD, Theodoro RC, Teixeira MdM, Bosco SdMG, Bagagli E. Environmental mapping of Paracoccidioides spp. in Brazil reveals new clues into genetic diversity, biogeography and wild host association. PLoS Negl Trop Dis. 2016;10(4):e0004606 10.1371/journal.pntd.0004606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shikanai-Yasuda MA, Mendes RP, Colombo AL, Queiroz-Telles F, Kono ASG, Paniago AMM, et al. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Rev Soc Bras Med Trop. 2017;50(5):715–40. 10.1590/0037-8682-0230-2017 . [DOI] [PubMed] [Google Scholar]
- 13.Fortes MR, Miot HA, Kurokawa CS, Marques ME, Marques SA. Immunology of paracoccidioidomycosis. An Bras Dermatol. 2011;86(3):516–24. 10.1590/s0365-05962011000300014 . [DOI] [PubMed] [Google Scholar]
- 14.Hahn RC, Rodrigues AM, Della Terra PP, Nery AF, Hoffmann-Santos HD, Góis HM, et al. Clinical and epidemiological features of paracoccidioidomycosis due to Paracoccidioides lutzii. PLoS Negl Trop Dis. 2019; 13(6): e0007437 10.1371/journal.pntd.0007437 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macoris SA, Sugizaki MF, Peracoli MT, Bosco SM, Hebeler-Barbosa F, Simoes LB, et al. Virulence attenuation and phenotypic variation of Paracoccidioides brasiliensis isolates obtained from armadillos and patients. Mem Inst Oswaldo Cruz. 2006;101(3):331–4. 10.1590/s0074-02762006000300019 . [DOI] [PubMed] [Google Scholar]
- 16.Nishikaku AS, Peracoli MT, Bagagli E, Sugizaki MF, Sartori A. Experimental infections with Paracoccidioides brasiliensis obtained from armadillos: Comparison to clinical isolates. The Brazilian Journal of Infectious Diseases. 2008;12(1):57–62. . [DOI] [PubMed] [Google Scholar]
- 17.Costa PF, Fernandes GF, dos Santos PO, Amaral CC, Camargo ZP. Characteristics of environmental Paracoccidioides brasiliensis isolates. Mycopathologia. 2010;169(1):37–46. 10.1007/s11046-009-9228-2 . [DOI] [PubMed] [Google Scholar]
- 18.Siqueira IM, Fraga CL, Amaral AC, Souza AC, Jeronimo MS, Correa JR, et al. Distinct patterns of yeast cell morphology and host responses induced by representative strains of Paracoccidioides brasiliensis (Pb18) and Paracoccidioides lutzii (Pb01). Med Mycol. 2016;54(2):177–88. 10.1093/mmy/myv072 . [DOI] [PubMed] [Google Scholar]
- 19.Kurokawa CS, Lopes CR, Sugizaki MF, Kuramae EE, Franco MF, Peracoli MT. Virulence profile of ten Paracoccidioides brasiliensis isolates: association with morphologic and genetic patterns. Rev Inst Med Trop S Paulo. 2005;47(5):257–62. 10.1590/s0036-46652005000500004 . [DOI] [PubMed] [Google Scholar]
- 20.Borba Cde M, Correia J, Vinhas E, Martins A, Alves BC, Unkles S, et al. Genetic characterization of morphologically variant strains of Paracoccidioides brasiliensis. Mem Inst Oswaldo Cruz. 2008;103(3):306–9. 10.1590/s0074-02762008005000013 . [DOI] [PubMed] [Google Scholar]
- 21.Tamayo D, Munoz JF, Lopez A, Uran M, Herrera J, Borges CL, et al. Identification and analysis of the role of superoxide dismutases isoforms in the pathogenesis of Paracoccidioides spp. PLoS Negl Trop Dis. 2016;10(3):e0004481 10.1371/journal.pntd.0004481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendes-Giannini MJ, Andreotti PF, Vincenzi LR, da Silva JL, Lenzi HL, Benard G, et al. Binding of extracellular matrix proteins to Paracoccidioides brasiliensis. Microbes Infect. 2006;8(6):1550–9. 10.1016/j.micinf.2006.01.012 . [DOI] [PubMed] [Google Scholar]
- 23.de Oliveira HC, Assato PA, Marcos CM, Scorzoni L, de Paula ESAC, Da Silva Jde F, et al. Paracoccidioides-host Interaction: An overview on recent advances in the paracoccidioidomycosis. Front Microbiol. 2015;6:1319 10.3389/fmicb.2015.01319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andre DC, Lopes JD, Franco MF, Vaz CA, Calich VL. Binding of laminin to Paracoccidioides brasiliensis induces a less severe pulmonary paracoccidioidomycosis caused by virulent and low-virulence isolates. Microbes Infect. 2004;6(6):549–58. 10.1016/j.micinf.2004.02.010 . [DOI] [PubMed] [Google Scholar]
- 25.Toledo RG, Da Silva WD, Calich VL, Kipnis TL. Mannose-binding lectin complement pathway plays a key role in complement activation by Paracoccidioides brasiliensis. Molecular Immunology. 2010;48(1–3):26–36. 10.1016/j.molimm.2010.09.015 . [DOI] [PubMed] [Google Scholar]
- 26.Spencer LM, Mateu G, Magaldi S, Garcia F, Mata-Essayag S. Humoral response of paracoccidioidomycosis sera in hamsters with different Venezuelan isolates. Revista de Biología Tropical. 2009;57(3):505–13. . [DOI] [PubMed] [Google Scholar]
- 27.Brummer E, Restrepo A, Hanson LH, Stevens DA. Virulence of Paracoccidiodes brasiliensis: the influence of in vitro passage and storage. Mycopathologia. 1990;109(1):13–7. . [DOI] [PubMed] [Google Scholar]
- 28.Castilho DG, Chaves AF, Xander P, Zelanis A, Kitano ES, Serrano SM, et al. Exploring potential virulence regulators in Paracoccidioides brasiliensis isolates of varying virulence through quantitative proteomics. Journal of Proteome Research. 2014;13(10):4259–71. 10.1021/pr5002274 . [DOI] [PubMed] [Google Scholar]
- 29.Tashima AK, Castilho DG, Chaves AF, Xander P, Zelanis A, Batista WL. Data in support of quantitative proteomics to identify potential virulence regulators in Paracoccidioides brasiliensis isolates. Data Brief. 2015;5:155–60. 10.1016/j.dib.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallejo MC, Nakayasu ES, Longo LV, Ganiko L, Lopes FG, Matsuo AL, et al. Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS One. 2012;7(6):e39463 10.1371/journal.pone.0039463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romani L. Immunity to Candida albicans: Th1, Th2 cells and beyond. Curr Opin Microbiol. 1999;2(4):363–7. 10.1016/S1369-5274(99)80064-2 . [DOI] [PubMed] [Google Scholar]
- 32.Casadevall A, Pirofski LA. Immunoglobulins in defense, pathogenesis, and therapy of fungal diseases. Cell Host & Microbe. 2012;11(5):447–56. 10.1016/j.chom.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casadevall A. Antibody immunity and invasive fungal infections. Infect Immun. 1995;63(11):4211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castaneda E, Brummer E, Pappagianis D, Stevens DA. Impairment of cellular but not humoral immune responses in chronic pulmonary and disseminated paracoccidioidomycosis in mice. Infect Immun. 1988;56(7):1771–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberto TN, Rodrigues AM, Hahn RC, de Camargo ZP. Identifying Paracoccidioides phylogenetic species by PCR-RFLP of the alpha-tubulin gene. Med Mycol. 2016;54(3):240–7. 10.1093/mmy/myv083 . [DOI] [PubMed] [Google Scholar]
- 36.Fava-Netto C. Contribuição para o estudo imunológico da blastomicose de Lutz (blastomicose sul-americana). Revista do Instituto Adolfo Lutz. 1961;21(1–2):99–194. [Google Scholar]
- 37.San-Blas F, Cova LJ. Growth curves of the yeast-like form of Paracoccidioides brasiliensis. Sabouraudia. 1975;13 Pt 1:22–9. . [PubMed] [Google Scholar]
- 38.Gezuele E, editor Aislamiento de Paracoccidioides sp. de heces de pinguino de la Antartida. Proceedings IV International Symposium on Paracoccidioidomycosis; 1989; Caracas, Venezuela.
- 39.Silva-Vergara ML, Martinez R, Camargo ZP, Malta MH, Maffei CM, Chadu JB. Isolation of Paracoccidioides brasiliensis from armadillos (Dasypus novemcinctus) in an area where the fungus was recently isolated from soil. Med Mycol. 2000;38(3):193–9. 10.1080/mmy.38.3.193.199 . [DOI] [PubMed] [Google Scholar]
- 40.Hebeler-Barbosa F, Montenegro MR, Bagagli E. Virulence profiles of ten Paracoccidioides brasiliensis isolates obtained from armadillos (Dasypus novemcinctus). Med Mycol. 2003;41(2):89–96. 10.1080/mmy.41.2.89.96 . [DOI] [PubMed] [Google Scholar]
- 41.De Albornoz MB. Isolation of Paracoccidioides brasiliensis from rural soil in Venezuela. Sabouraudia. 1971;9(3):248–53. . [PubMed] [Google Scholar]
- 42.Ferreira MS, Freitas LH, Lacaz Cda S, del Negro GM, de Melo NT, Garcia NM, et al. Isolation and characterization of a Paracoccidioides brasiliensis strain from a dogfood probably contaminated with soil in Uberlandia, Brazil. Med Mycol. 1990;28(3):253–6. . [DOI] [PubMed] [Google Scholar]
- 43.Silva-Vergara ML, Martinez R, Chadu A, Madeira M, Freitas-Silva G, Leite Maffei CM. Isolation of a Paracoccidioides brasiliensis strain from the soil of a coffee plantation in Ibia, State of Minas Gerais, Brazil. Med Mycol. 1998;36(1):37–42. . [PubMed] [Google Scholar]
- 44.Teixeira HC, Calich VL, Singer-Vermes LM, D'Imperio-Lima MR, Russo M. Experimental paracoccidioidomycosis: early immunosuppression occurs in susceptible mice after infection with pathogenic fungi. Braz J Med Biol Res. 1987;20(5):587–9. . [PubMed] [Google Scholar]
- 45.Cano LE, Singer-Vermes LM, Vaz CA, Russo M, Calich VL. Pulmonary paracoccidioidomycosis in resistant and susceptible mice: relationship among progression of infection, bronchoalveolar cell activation, cellular immune response, and specific isotype patterns. Infect Immun. 1995;63(5):1777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribeiro LR, Loures FV, de Araujo EF, Feriotti C, Costa TA, Serezani CH, et al. Lipoxin inhibits fungal uptake by macrophages and reduces the severity of acute pulmonary infection caused by Paracoccidioides brasiliensis. Mediators of Inflammation. 2015;2015:852574 10.1155/2015/852574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Camargo ZP, Unterkircher C, Campoy SP, Travassos LR. Production of Paracoccidioides brasiliensis exoantigens for immunodiffusion tests. J Clin Microbiol. 1988;26(10):2147–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Camargo ZP, Berzaghi R, Amaral CC, Silva SHM. Simplified method for producing Paracoccidioides brasiliensis exoantigens for use in immunodiffusion tests. Med Mycol. 2003;41(6):539–42. 10.1080/13693780310001615358 [DOI] [PubMed] [Google Scholar]
- 49.Puccia R, Schenkman S, Gorin PA, Travassos LR. Exocellular components of Paracoccidioides brasiliensis: identification of a specific antigen. Infect Immun. 1986;53(1):199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–54. 10.1016/0003-2697(76)90527-3 . [DOI] [PubMed] [Google Scholar]
- 51.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–5. 10.1038/227680a0 . [DOI] [PubMed] [Google Scholar]
- 52.Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8(2):93–9. 10.1002/elps.1150080203 [DOI] [Google Scholar]
- 53.Rodrigues AM, Kubitschek-Barreira PH, Fernandes GF, de Almeida SR, Lopes-Bezerra LM, de Camargo ZP. Immunoproteomic analysis reveals a convergent humoral response signature in the Sporothrix schenckii complex. Journal of Proteomics. 2015;115:8–22. 10.1016/j.jprot.2014.11.013 . [DOI] [PubMed] [Google Scholar]
- 54.Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, et al. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25(9):1327–33. 10.1002/elps.200305844 [DOI] [PubMed] [Google Scholar]
- 55.Pitarch A, Sanchez M, Nombela C, Gil C. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol Cell Proteomics. 2002;1(12):967–82. 10.1074/mcp.m200062-mcp200 . [DOI] [PubMed] [Google Scholar]
- 56.Munoz JF, Gallo JE, Misas E, Priest M, Imamovic A, Young S, et al. Genome update of the dimorphic human pathogenic fungi causing paracoccidioidomycosis. PLoS Negl Trop Dis. 2014;8(12):e3348 10.1371/journal.pntd.0003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Desjardins CA, Champion MD, Holder JW, Muszewska A, Goldberg J, Bailao AM, et al. Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis. PLoS Genet. 2011;7(10):e1002345 10.1371/journal.pgen.1002345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Calich VL, Singer-Vermes LM, Siqueira AM, Burger E. Susceptibility and resistance of inbred mice to Paracoccidioides brasiliensis. Br J Exp Pathol. 1985;66(5):585–94. [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia NM, Del Negro GMB, Heins-Vaccari EM, Melo NTd, Assis CMd, Lacaz CdS. [Paracoccidioides brasiliensis, a new strain isolated from a fecal matter of a penguin (Pygoscelis adeliae)]. Rev Inst Med Trop S Paulo. 1993;35:227–35. 10.1590/s0036-46651993000300003 [DOI] [PubMed] [Google Scholar]
- 60.Taborda CP, Camargo ZP. Antigenic relationship between Paracoccidioides brasiliensis isolated from faeces of a penguin and a human isolate of P. brasiliensis. Med Mycol. 1993;31(5):347–52. 10.1080/02681219380000441 [DOI] [Google Scholar]
- 61.Hahn RC, Macedo AM, Santos NL, Resende JC, Hamdan JS. Characterization of Paracoccidioides brasiliensis atypical isolates by random amplified polymorphic DNA analysis. Revista Iberoamericana de Micologia. 2002;19(1):49–51. . [PubMed] [Google Scholar]
- 62.Camargo ZP. Serology of paracoccidioidomycosis. Mycopathologia. 2008;165(4–5):289–302. 10.1007/s11046-007-9060-5 [DOI] [PubMed] [Google Scholar]
- 63.Barbosa S, Takeda A, Chacha J, Cuce L, Fava Netto C. Anticorpos especificos das classes IgG, IgM E IgA para Paracoccidioide brasiliensis dosados atraves da reacao de imunofluorescencia indireta no soro de pacientes e sua correlacao com o tempo de evolucao e forma clinica da doenca. Rev Inst Adolfo Lutz. 1981;41(2):121–6. [Google Scholar]
- 64.Biagioni L, Souza MJ, Chamma LG, Mendes RP, Marques SA, Mota NG, et al. Serology of paracoccidioidomycosis. II. Correlation between class-specific antibodies and clinical forms of the disease. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1984;78(5):617–21. 10.1016/0035-9203(84)90220-7 . [DOI] [PubMed] [Google Scholar]
- 65.Bueno JP, Mendes-Giannini MJ, Del Negro GM, Assis CM, Takiguti CK, Shikanai-Yasuda MA. IgG, IgM and IgA antibody response for the diagnosis and follow-up of paracoccidioidomycosis: comparison of counterimmunoelectrophoresis and complement fixation. Med Mycol. 1997;35(3):213–7. . [DOI] [PubMed] [Google Scholar]
- 66.Singer-Vermes LM, Burger E, Calich VL, Modesto-Xavier LH, Sakamoto TN, Sugizaki MF, et al. Pathogenicity and immunogenicity of Paracoccidioides brasiliensis isolates in the human disease and in an experimental murine model. Clinical and Experimental Immunology. 1994;97(1):113–9. 10.1111/j.1365-2249.1994.tb06588.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xidieh CF, Lenzi HL, Calich VL, Burger E. Influence of the genetic background on the pattern of lesions developed by resistant and susceptible mice infected with Paracoccidioides brasiliensis. Medical Microbiology and Immunology. 1999;188(1):41–9. . [DOI] [PubMed] [Google Scholar]
- 68.Tristao FS, Panagio LA, Rocha FA, Cavassani KA, Moreira AP, Rossi MA, et al. B cell-deficient mice display enhanced susceptibility to Paracoccidioides brasiliensis Infection. Mycopathologia. 2013;176(1–2):1–10. 10.1007/s11046-013-9671-y . [DOI] [PubMed] [Google Scholar]
- 69.Montagnoli C, Bozza S, Bacci A, Gaziano R, Mosci P, Morschhauser J, et al. A role for antibodies in the generation of memory antifungal immunity. Eur J Immunol. 2003;33(5):1193–204. 10.1002/eji.200323790 . [DOI] [PubMed] [Google Scholar]
- 70.Buissa-Filho R, Puccia R, Marques AF, Pinto FA, Muñoz JE, Nosanchuk JD, et al. The monoclonal antibody against the major diagnostic antigen of Paracoccidioides brasiliensis mediates immune protection in infected BALB/c mice challenged intratracheally with the fungus. Infection and Immunity. 2008;76(7):3321–8. 10.1128/IAI.00349-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Mattos Grosso D, de Almeida SR, Mariano M, Lopes JD. Characterization of gp70 and anti-gp70 monoclonal antibodies in Paracoccidioides brasiliensis pathogenesis. Infect Immun. 2003;71(11):6534 10.1128/IAI.71.11.6534-6542.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Askew C, Sellam A, Epp E, Hogues H, Mullick A, Nantel A, et al. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 2009;5(10):e1000612 10.1371/journal.ppat.1000612 PubMed Central PMCID: PMC2749448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Arruda Grossklaus D, Bailao AM, Vieira Rezende TC, Borges CL, de Oliveira MA, Parente JA, et al. Response to oxidative stress in Paracoccidioides yeast cells as determined by proteomic analysis. Microbes Infect. 2013;15(5):347–64. 10.1016/j.micinf.2012.12.002 . [DOI] [PubMed] [Google Scholar]
- 74.Arrigo AP, Virot S, Chaufour S, Firdaus W, Kretz-Remy C, Diaz-Latoud C. Hsp27 consolidates intracellular redox homeostasis by upholding glutathione in its reduced form and by decreasing iron intracellular levels. Antioxidants & Redox Signaling. 2005;7(3–4):414–22. 10.1089/ars.2005.7.414 . [DOI] [PubMed] [Google Scholar]
- 75.Preville X, Salvemini F, Giraud S, Chaufour S, Paul C, Stepien G, et al. Mammalian small stress proteins protect against oxidative stress through their ability to increase glucose-6-phosphate dehydrogenase activity and by maintaining optimal cellular detoxifying machinery. Exp Cell Res. 1999;247(1):61–78. 10.1006/excr.1998.4347 . [DOI] [PubMed] [Google Scholar]
- 76.Salvemini F, Franze A, Iervolino A, Filosa S, Salzano S, Ursini MV. Enhanced glutathione levels and oxidoresistance mediated by increased glucose-6-phosphate dehydrogenase expression. J Biol Chem. 1999;274(5):2750–7. 10.1074/jbc.274.5.2750 . [DOI] [PubMed] [Google Scholar]
- 77.Marcos CM, de Oliveira HC, da Silva Jde F, Assato PA, Fusco-Almeida AM, Mendes-Giannini MJ. The multifaceted roles of metabolic enzymes in the Paracoccidioides species complex. Front Microbiol. 2014;5:719 10.3389/fmicb.2014.00719 ; PubMed Central PMCID: PMC4271699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barbosa MS, Bao SN, Andreotti PF, de Faria FP, Felipe MS, dos Santos Feitosa L, et al. Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect Immun. 2006;74(1):382–9. 10.1128/IAI.74.1.382-389.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chaves EG, Weber SS, Bao SN, Pereira LA, Bailao AM, Borges CL, et al. Analysis of Paracoccidioides secreted proteins reveals fructose 1,6-bisphosphate aldolase as a plasminogen-binding protein. BMC Microbiology. 2015;15:53 10.1186/s12866-015-0393-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Longo LVG, da Cunha JPC, Sobreira TJP, Puccia R. Proteome of cell wall-extracts from pathogenic Paracoccidioides brasiliensis: Comparison among morphological phases, isolates, and reported fungal extracellular vesicle proteins. EuPA Open Proteomics. 2014;3:216–28. 10.1016/j.euprot.2014.03.003. [DOI] [Google Scholar]
- 81.Nimrichter L, de Souza MM, Del Poeta M, Nosanchuk JD, Joffe L, Tavares Pde M, et al. Extracellular vesicle-associated transitory cell wall components and their impact on the interaction of fungi with host cells. Front Microbiol. 2016;7:1034 10.3389/fmicb.2016.01034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.da Fonseca CA, Jesuino RSA, Felipe MSS, Cunha DA, Brito WA, Soares CMA. Two-dimensional electrophoresis and characterization of antigens from Paracoccidioides brasiliensis. Microbes Infect. 2001;3(7):535–42. 10.1016/S1286-4579(01)01409-5. [DOI] [PubMed] [Google Scholar]
- 83.da Silva Santos R, Martelli de Paula N, Santiago Barbosa M, de Almeida Soares CM. Caracterização imunológica da proteína recombinante gliceraldeido-3-fosfato desidrogenase do patógeno humano Paracoccidioides brasiliensis. SaBios-Revista de Saúde e Biologia. 2012;7(2). [Google Scholar]
- 84.Missall TA, Lodge JK, McEwen JE. Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Eukaryot Cell. 2004;3(4):835–46. 10.1128/EC.3.4.835-846.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Missall TA, Pusateri ME, Donlin MJ, Chambers KT, Corbett JA, Lodge JK. Posttranslational, translational, and transcriptional responses to nitric oxide stress in Cryptococcus neoformans: Implications for virulence. Eukaryot Cell. 2006;5(3):518–29. 10.1128/EC.5.3.518-529.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tillmann AT, Strijbis K, Cameron G, Radmaneshfar E, Thiel M, Munro CA, et al. Contribution of Fdh3 and Glr1 to glutathione redox state, stress adaptation and virulence in Candida albicans. PLoS One. 2015;10(6):e0126940 10.1371/journal.pone.0126940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weber SS, Parente AF, Borges CL, Parente JA, Bailao AM, de Almeida Soares CM. Analysis of the secretomes of Paracoccidioides mycelia and yeast cells. PLoS One. 2012;7(12):e52470 10.1371/journal.pone.0052470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pigosso LL, Parente AF, Coelho AS, Silva LP, Borges CL, Bailao AM, et al. Comparative proteomics in the genus Paracoccidioides. Fungal Genet Biol. 2013;60:87–100. 10.1016/j.fgb.2013.07.008 . [DOI] [PubMed] [Google Scholar]
- 89.Moreira SF, Bailao AM, Barbosa MS, Jesuino RS, Felipe MS, Pereira M, et al. Monofunctional catalase P of Paracoccidioides brasiliensis: identification, characterization, molecular cloning and expression analysis. Yeast (Chichester, England). 2004;21(2):173–82. 10.1002/yea.1077 . [DOI] [PubMed] [Google Scholar]
- 90.Chagas RF, Bailao AM, Pereira M, Winters MS, Smullian AG, Deepe GS Jr, et al. The catalases of Paracoccidioides brasiliensis are differentially regulated: protein activity and transcript analysis. Fungal Genet Biol. 2008;45(11):1470–8. 10.1016/j.fgb.2008.08.007 . [DOI] [PubMed] [Google Scholar]
- 91.Parente-Rocha JA, Parente AF, Baeza LC, Bonfim SM, Hernandez O, McEwen JG, et al. Macrophage interaction with Paracoccidioides brasiliensis yeast cells modulates fungal metabolism and generates a response to oxidative stress. PLoS One. 2015;10(9):e0137619 10.1371/journal.pone.0137619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dantas AS, Andrade RV, de Carvalho MJ, Felipe MS, Campos EG. Oxidative stress response in Paracoccidioides brasiliensis: assessing catalase and cytochrome c peroxidase. Mycological Research. 2008;112(Pt 6):747–56. 10.1016/j.mycres.2007.11.018 . [DOI] [PubMed] [Google Scholar]
- 93.Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, et al. Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Human genomics. 2011;5(4):283–303. 10.1186/1479-7364-5-4-283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, et al. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free radical biology & medicine. 2013;56:89–101. 10.1016/j.freeradbiomed.2012.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alves LR, Goldenberg S. RNA-binding proteins related to stress response and differentiation in protozoa. World Journal of Biological Chemistry. 2016;7(1):78–87. 10.4331/wjbc.v7.i1.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Z, Nagy PD. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011;8(2):305–15. 10.4161/rna.8.2.15391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Van Assche E, Van Puyvelde S, Vanderleyden J, Steenackers HP. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front Microbiol. 2015;6:141 10.3389/fmicb.2015.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee K, Kang H. Emerging roles of RNA-Binding proteins in plant growth, development, and stress responses. Molecules and Cells. 2016;39(3):179–85. 10.14348/molcells.2016.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Harvey R, Dezi V, Pizzinga M, Willis AE. Post-transcriptional control of gene expression following stress: the role of RNA-binding proteins. Biochem Soc Trans. 2017;45(4):1007–14. 10.1042/BST20160364 PMC5655797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Krishnan K, Ren Z, Losada L, Nierman WC, Lu LJ, Askew DS. Polysome profiling reveals broad translatome remodeling during endoplasmic reticulum (ER) stress in the pathogenic fungus Aspergillus fumigatus. BMC Genomics. 2014;15:159 10.1186/1471-2164-15-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Glazier VE, Panepinto JC. The ER stress response and host temperature adaptation in the human fungal pathogen Cryptococcus neoformans. Virulence. 2014;5(2):351–6. 10.4161/viru.27187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ariyachet C, Solis NV, Liu Y, Prasadarao NV, Filler SG, McBride AE. SR-like RNA-binding protein Slr1 affects Candida albicans filamentation and virulence. Infect Immun. 2013;81(4):1267–76. 10.1128/IAI.00864-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dagley MJ, Gentle IE, Beilharz TH, Pettolino FA, Djordjevic JT, Lo TL, et al. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol Microbiol. 2011;79(4):968–89. 10.1111/j.1365-2958.2010.07503.x . [DOI] [PubMed] [Google Scholar]
- 104.Gank KD, Yeaman MR, Kojima S, Yount NY, Park H, Edwards JE Jr., et al. SSD1 is integral to host defense peptide resistance in Candida albicans. Eukaryot Cell. 2008;7(8):1318–27. 10.1128/EC.00402-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parente AF, Naves PE, Pigosso LL, Casaletti L, McEwen JG, Parente-Rocha JA, et al. The response of Paracoccidioides spp. to nitrosative stress. Microbes Infect. 2015;17(8):575–85. 10.1016/j.micinf.2015.03.012 . [DOI] [PubMed] [Google Scholar]
- 106.Dias-Melicio LA, Moreira AP, Calvi SA, Soares AM. Chloroquine inhibits Paracoccidioides brasiliensis survival within human monocytes by limiting the availability of intracellular iron. Microbiology and Immunology. 2006;50(4):307–14. . [DOI] [PubMed] [Google Scholar]
- 107.Schrettl M, Haas H. Iron homeostasis—Achilles’heel of Aspergillus fumigatus?. Curr Opin Microbiol. 2011;14(4):400–5. 10.1016/j.mib.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host & Microbe. 2013;13(5):509–19. 10.1016/j.chom.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bailao EF, Parente JA, Pigosso LL, de Castro KP, Fonseca FL, Silva-Bailao MG, et al. Hemoglobin uptake by Paracoccidioides spp. is receptor-mediated. PLoS Negl Trop Dis. 2014;8(5):e2856 10.1371/journal.pntd.0002856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Parente AF, Bailao AM, Borges CL, Parente JA, Magalhaes AD, Ricart CA, et al. Proteomic analysis reveals that iron availability alters the metabolic status of the pathogenic fungus Paracoccidioides brasiliensis. PLoS One. 2011;6(7):e22810 10.1371/journal.pone.0022810 ; PubMed Central PMCID: PMC3145762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vallejo MC, Nakayasu ES, Matsuo AL, Sobreira TJ, Longo LV, Ganiko L, et al. Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: comparative analysis with other pathogenic fungi. Journal of Proteome Research. 2012;11(3):1676–85. 10.1021/pr200872s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Silva PF, Novaes E, Pereira M, Soares CM, Borges CL, Salem-Izacc SM. In silico characterization of hypothetical proteins from Paracoccidioides lutzii. Genetics And Molecular Research: GMR. 2015;14(4):17416–25. 10.4238/2015.December.21.11 . [DOI] [PubMed] [Google Scholar]
- 113.Fernandes FF, Oliveira AF, Landgraf TN, Cunha C, Carvalho A, Vendruscolo PE, et al. Impact of paracoccin gene silencing on Paracoccidioides brasiliensis virulence. mBio. 2017;8(4). 10.1128/mBio.00537-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative analysis of proteins with differential expression in highly virulent isolates (Bot 1/96, Ibiá, Pb18) and moderately virulent isolates (Ibiá and T2) is shown. Positive and negative values indicate higher or lower expression of the protein, respectively. Positive (+) and negative (−) symbols mean the presence or absence of the spot, respectively. Proteins were deemed to have differential abundance levels if their spot volumes were changed at least twofold compared to the normalized spot volume.
(PDF)
Quantitative analysis of proteins with differential expression in highly virulent isolates (Bot 1/96, Ibiá, Pb18) and isolates of low virulence (262Uber, T1 and Ibiá T2). Positive and negative values indicate higher or lower expression of the protein, respectively. Positive (+) and negative (−) symbols mean the presence or absence of the spot, respectively. Proteins were deemed to have differential abundance levels if their spot volumes were changed at least twofold compared to the normalized spot volume.
(PDF)
(A) Paracoccidioides brasiliensis yeast cells were grown for 7 days at 36°C in triplicate on Fava-Netto’s medium, and protein extract was obtained as previously described [1]. Protein concentrations were determined by the Bradford method [2] and 5 μg of each extract were subjected to 1D SDS-PAGE (10%). The molecular masses (in kDa) of standard proteins are given to the left of the gel (BenchMarkTM Protein Ladder, Invitrogen). From left to right: Bot 1/96, Ibiá T1, Ibiá, T2, 262 Uber, T1, Pinguim, Ibiá T2 and Pb18. (B) The completed electrophoresis gel was imaged on an Image Scanner III (GE Healthcare, Uppsala, Sweden) and the comparison was carried out by densitometry measurements of scanned image (8-bit image) along the ROI (arrow) using a 256 grey level scale to determine the average gray value using Adobe Photoshop CC. This region was chosen because it did not present proteins with different abundance levels in the comparative analyzes (2D-GE). (C) Lowest and highest gray values were used to set the ratio between each of the extreme values and the ROI revealing minimum variation across the different samples.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.