Abstract
The aims of the present study were to screen differentially expressed genes (DEGs) in breast cancer (BC) and investigate NDC80 kinetochore complex component (NUF2) as a prognostic marker of BC in detail. A total of four BC microarray datasets, downloaded from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases, were used to screen DEGs. A total of 190 DEGs with the same expression trends were identified in the 4 data-sets, including 65 upregulated and 125 downregulated DEGs. Functional and pathway enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery. The upregulated DEGs were enriched for 10 Gene Ontology (GO) terms and 7 pathways, and the downregulated DEGs were enriched for 10 GO terms and 10 pathways. A protein-protein interaction network containing 149 nodes and 930 edges was constructed using the Search Tool for the Retrieval of Interacting Genes, and 2 functional modules were identified using the MCODE plugin of Cytoscape. Based on an in-depth analysis of module 1 and literature mining, NUF2 was selected for further research. Oncomine database analysis and reverse transcription-quantitative PCR showed that NUF2 is significantly upregulated in BC tissues. In analyses of correlations between NUF2 and clinical pathological characteristics, NUF2 was significantly associated with the malignant features of BC. Using 5 additional datasets from GEO, it was demonstrated that NUF2 has a significant prognostic role in both ER-positive and ER-negative BC. A Gene Set Enrichment Analysis indicated that NUF2 may regulate breast carcinogenesis and progression via cell cycle-related pathways. The results of the present study demonstrated that NUF2 is overexpressed in BC and is significantly associated with its multiple pathological features and prognosis.
Keywords: bioinformatics, breast cancer, differentially expressed gene, NUF2, prognosis
Introduction
Breast cancer (BC) is the most common female malignancy and the second leading cause of mortality in women worldwide (1). According to the World Health Organization in 2012, one-third of Asian women develop BC (2,3). Currently, BC treatment includes partial excision with or without radiotherapy and systemic therapies such as endocrine therapy, chemotherapy, molecular targeted therapy, and a combination of them (4). Although advanced therapeutic techniques based on surgery have considerably improved the survival of patients with BC and the five-year survival has increased from 75% in 1976 to 91% in 2017, high rates of metastasis and recurrence remain (1,5,6). Recently, molecular targeted therapy has been shown to play an important role in individualized treatment of BC. For instance, a monoclonal antibody against HER2, trastuzumab, has been demonstrated to improve survival of patients with BC; however, the prognosis remains poor (7,8). Therefore, to improve BC prognosis, effective therapeutic targets and prognostic biomarkers are needed.
NDC80 kinetochore complex component (NUF2), also known as CDCA1, is a centromere-related protein (9). It regulates the binding of centromeres to spindle microtubules, participates in cell cycle regulation and has important roles in cell proliferation and apoptosis (10). NUF2 is overexpressed in a number of cancers, including lung cancer, cholangiocarcinoma, renal cell carcinoma and bladder cancer (11). Although the expression and prognostic significance of NUF2 in BC have been suggested (12,13), its precise role and underlying molecular mechanisms of action remain to be investigated.
In the present study, 4 mRNA microarray datasets were analyzed from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases to identify differentially expressed genes (DEGs) between BC tissues and normal breast tissues. The bioinformatics analysis and literature mining suggested that NUF2 is a key gene in the progression of BC. The expression of NUF2 in BC samples and its correlation with clinical pathological characteristics were then analyzed. In addition, the prognostic value of NUF2 was analyzed using individual and pooled methods. In a gene set enrichment analysis (GSEA), it was demonstrated that NUF2 might be involved in cell-cycle related pathways. The results of the present study suggest that NUF2 is a prognostic indicator of BC.
Materials and methods
Microarray data
GSE42568 (14), GSE45827 (15), GSE65194 (16) and TCGA BC microarray datasets, downloaded from GEO (17) and TCGA (18), were used to screen DEGs in BC. The TCGA dataset was used for analyses of clinical pathological characteristics associated with NUF2 in patients with BC. The following 5 additional BC microarray datasets were selected for prognostic analyses: GSE1456 (19), GSE22220 (20), NKI (21), GSE4299 (22) and GSE20685 (23). To normalize mRNA levels, patients for each dataset were reclassified into four subsets (X1, X2, X3 and X4) based on the quartile for expression values. The datasets were then reclassified into a new dataset for a pooled analysis.
DEG identification
BC-related microarray data downloaded from the GEO and TCGA databases were processed using R software (version 3.4.3; https://cran.r-project.org/). DEGs between BC tissues and normal breast tissues were identified using the limma package in R. Fold-change (FC) values were calculated and the DEGs were further selected based on the following cutoff criteria: P<0.01 and log |FC|>2. Overlapping DEGs among the four datasets were identified using Funrich (version 3.1.3; http://www.funrich.org).
Functional and pathway enrichment analyses of DEGs
Gene Ontology (GO) is used to identify enriched functions of genes in three independent categories: Biological process (BP), molecular function (MF) and cellular component (CC) (24). Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to identify relevant pathways for the genes (25). GO BP and KEGG signaling pathway analyses of the DEGs were performed using the Database for Annotation Visualization and Integrated Discovery (DAVID) online tool (https://david.ncifcrf.gov/) (26) with P<0.05 as the threshold for significance.
Protein-protein interaction (PPI) network analysis
The Search Tool for the Retrieval of Interacting Genes (STRING; https://string-db.org/) was used to develop a PPI network. Using the STRING database, DEGs with a combined score ≥0.4 were chosen to construct the network, which was visualized using Cytoscape (version 3.6.1) (27). Molecular Complex Detection (MCODE), a plugin for Cytoscape, was used to construct functional modules in the PPI network.
Gene set enrichment analysis (GSEA)
A GSEA was conducted based on protocols obtained from the website (http://software.broadinstitute.org/gsea/index.jsp) and a previous study (28). GSEA (version 3.0) was run for the KEGG gene sets (c2.cp.kegg.v.6.0.symbols.gmt). The number of permutations was set to 1,000 and the phenotype labels were NUF2-high and NUF2-low. FDR <0.25 and NOM P<0.05 indicated statistical significance.
Oncomine analysis
Oncomine (https://www.oncomine.org/) is an online cancer microarray database, aiming to facilitate the discovery of novel biomarkers from genome-wide expression analyses. In the present study, the mRNA expression differences of NUF2 between BC and normal breast tissues were explored using the Oncomine database.
Patients and samples
BC and matched adjacent tissues were collected from the Pathology Department of Shaoxing People's Hospital (Shaoxing, China). Samples were obtained from 42 patients at initial diagnosis and were immediately frozen in liquid nitrogen. The present study was authorized by the Hospital Ethics Committee and informed consent was obtained from all patients.
Reverse transcription (RT)-quantitative (q)PCR
Total RNA was isolated from the BC and matched adjacent tissues using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The Nanodrop 2000 (Thermo Fisher Scientific, Inc.) was used to detect the purity and concentration of the total RNA. According to the manufacturer's protocol, RT-qPCR was performed using the LightCycler® 480 PCR apparatus (Roche Diagnostics, Basel, Switzerland) and the One Step SYBR® PrimeScript™ RT-PCR kit II (Takara Bio, Inc., Otsu, Japan). Amplification was performed under the following conditions: 42°C for 5 min, 95°C for 10 sec; 40 cycles of 95°C for 5 sec and 60°C for 20 sec; and 65°C for 15 sec. The primers used were as follows: NUF2 forward primer 5′-TACCATTCAGCAATTTAGTTACT-3′ and reverse primer 5′-TAGAATATCAGCAGTCTCAAAG-3′; and β-actin forward primer 5′-CATGTACGTTGCTATCCAGGC-3′ and reverse primer 5′-CTCCTTAATGTCACGCACGAT-3′. The relative levels of NUF2 expression were evaluated by the 2−ΔΔCq (29) method using β-actin as the control.
Statistical analyses
All statistical analyses were performed using SPSS 20.0 (IBM Corps., Armonk, NY, USA). An independent t-test was used for analyzing the continuous data. The χ2 test and χ2 test with continuity correction were performed to analyze the association of NUF2 with clinical pathological characteristics. Bonferroni's post hoc test was used to analyze the clinical pathological characteristics between more than 2 groups. Survival curves were generated by the Kaplan-Meier method and significance was determined using the log-rank test. Bonferroni's post hoc test was used for pairwise comparisons. Multivariable survival analysis was performed using the Cox proportional hazards regression model and significance was determined using the likelihood ratio test. P<0.05 was considered to indicate statistically significant differences, while for Bonferroni's test, P<0.05/N was considered to indicate statistically significant differences, where N=the number of pairwise comparisons.
Results
Identification of DEGs in BC
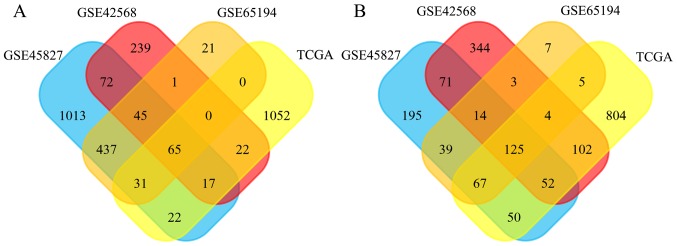
DEGs between the BC and normal breast tissues were screened using the GEO and TCGA databases. As shown in Fig. 1A, 1,702, 461, 600 and 337 DEGs were upregulated in the GSE45827, GSE42568, GSE65194, and TCGA datasets, and 613, 715, 264, and 872 DEGs were downregulated, respectively (Fig. 1B). In total, 190 DEGs exhibited the same expression trends in all datasets, including 65 upregulated and 125 downregulated genes.
Figure 1.
Identification of DEGs in breast cancer microarray datasets. In total, (A) 65 DEGs were upregulated and (B) 125 DEGs were downregulated in the intersection of the GSE45827, GSE42568, GSE65194, and TCGA datasets. DEGs, differentially expressed genes; TCGA, the Cancer Genome Atlas.
Functional and pathway enrichment for the DEGs
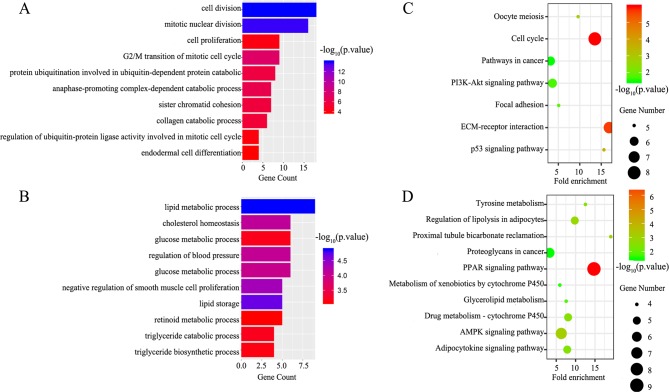
GO BP and KEGG signaling pathway analyses of the DEGs were performed using DAVID. The upregulated DEGs were mainly enriched for the BP terms cell division, mitotic nuclear division and G2/M transition of mitotic cell cycle (Fig. 2A), while downregulated DEGs were significantly associated with lipid metabolic process, cholesterol homeostasis, and glucose metabolic process (Fig. 2B). Additionally, seven KEGG pathways were identified for the upregulated genes, including the p53 signaling pathway, cell cycle and extracellular matrix (ECM)-receptor interaction (Fig. 2C). The peroxisome proliferator-activated receptor (PPAR) signaling pathway, AMP-activated protein kinase (AMPK) signaling pathway and proximal tubule bicarbonate reclamation were associated with the downregulated DEGs (Fig. 2D). The detailed results are presented in Table I.
Figure 2.
GO BP analysis and KEGG signaling pathway analysis of DEGs in breast cancer. GO BP analysis of (A) upregulated and (B) downregulated DEGs. KEGG signaling pathway enrichment analysis of (C) upregulated and (D) downregulated DEGs. DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto encyclopedia of genes and genomes; ECM, extracellular matrix.
Table I.
Significantly enriched GO biological process terms and KEGG pathways.
| A, Upregulated
| |||
|---|---|---|---|
| Terms | Description | Number of genes | P-value |
| GO Terms | |||
| GO:0051301 | Cell division | 18 | 1.02×10−14 |
| GO:0007067 | Mitotic nuclear division | 16 | 1.90×10−14 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 9 | 4.82×10−08 |
| GO:0031145 | Anaphase-promoting complex-dependent catabolic process | 7 | 5.40×10−07 |
| GO:0042787 | Protein ubiquitination involved in Ubiquitin-dependent protein catabolic process | 8 | 1.83×10−06 |
| GO:0007062 | Sister chromatid cohesion | 7 | 2.58×10−06 |
| GO:0030574 | Collagen catabolic process | 6 | 4.40×10−06 |
| GO:0008283 | Cell proliferation | 9 | 7.20×10−05 |
| GO:0051439 | Regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 4 | 8.86×10−05 |
| GO:0035987 | Endodermal cell differentiation | 4 | 1.45×10−04 |
| KEGG pathways | |||
| hsa04110 | Cell cycle | 8 | 1.17×10−06 |
| hsa04512 | ECM-receptor interaction | 7 | 2.33×10−06 |
| hsa04115 | p53 signaling pathway | 5 | 0.000237 |
| hsa04114 | Oocyte meiosis | 5 | 0.001499 |
| hsa04510 | Focal adhesion | 5 | 0.014347 |
| hsa04151 | PI3K-Akt signaling pathway | 6 | 0.019962 |
| hsa05200 | Pathways in cancer | 6 | 0.032832 |
|
B, Downregulated | |||
| Terms | Description | Number of genes | P-value |
|
GO Terms | |||
| GO:0006629 | Lipid metabolic process | 9 | 1.32×10−05 |
| GO:0019915 | Lipid storage | 5 | 2.06×10−05 |
| GO:0048662 | Negative regulation of smooth muscle cell proliferation | 5 | 4.49×10−05 |
| GO:0042632 | Cholesterol homeostasis | 6 | 7.95×10−05 |
| GO:0008217 | Regulation of blood pressure | 6 | 8.56×10−05 |
| GO:0006006 | Glucose metabolic process | 6 | 9.90×10−05 |
| GO:0019433 | Triglyceride catabolic process | 4 | 6.61×10−04 |
| GO:0042593 | Glucose homeostasis | 6 | 6.74×10−04 |
| GO:0019432 | Triglyceride biosynthetic process | 4 | 7.44×10−04 |
| GO:0001523 | Retinoid metabolic process | 5 | 8.33×10−04 |
| KEGG pathways | |||
| hsa03320 | PPAR signaling pathway | 9 | 1.14×10−07 |
| hsa04152 | AMPK signaling pathway | 7 | 7.34×10−04 |
| hsa04964 | Proximal tubule bicarbonate reclamation | 4 | 0.001072 |
| hsa04923 | Regulation of lipolysis in adipocytes | 5 | 0.001523 |
| hsa00982 | Drug metabolism-cytochrome P450 | 5 | 0.003116 |
| hsa04920 | Adipocytokine signaling pathway | 5 | 0.003462 |
| hsa00350 | Tyrosine metabolism | 4 | 0.00367 |
| hsa00561 | Glycerolipid metabolism | 4 | 0.014956 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 4 | 0.028412 |
| hsa05205 | Proteoglycans in cancer | 6 | 0.033006 |
KEGG, Kyoto Encyclopedia of genes and genomes; GO, Gene Ontology.
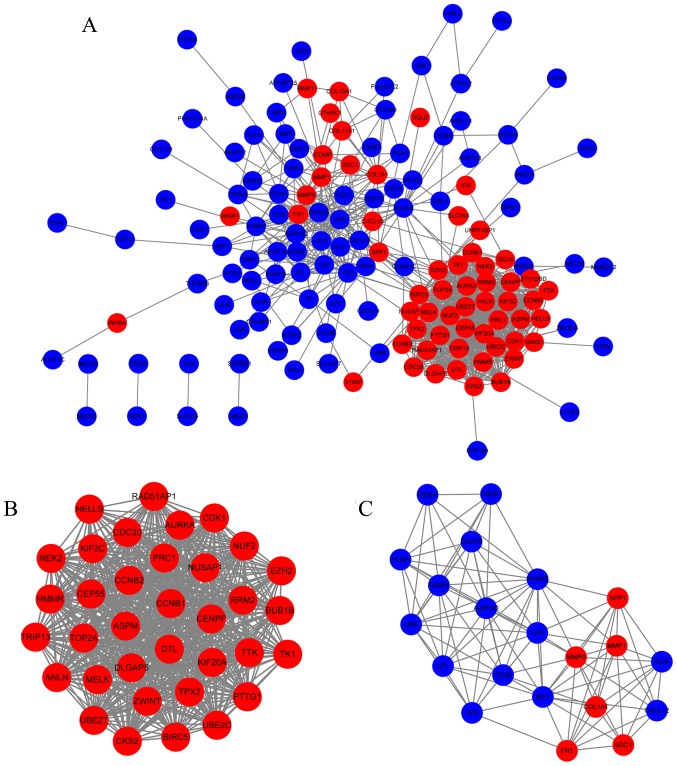
PPI network analysis and the selection of NUF2
Protein interactions often play important roles in cancer progression. A PPI network analysis was performed using the STRING database and Cytoscape. The PPI network was constructed using 149 DEGs (57 upregulated and 92 downregulated DEGs) with combined scores ≥0.4, and contained 149 nodes and 930 edges (Fig. 3A). A total of two functional modules were identified using the MCODE plugin. Module 1 consisted of 35 nodes and 573 edges including NUF2, TOP2A, ASPM, and CCNB1 (Fig. 3B). Module 2 included 21 nodes and 104 edges including COL1A1, MMP1, MMP9, and LPL (Fig. 3C). Based on the degree of importance, module 1 was chosen for further analysis.
Figure 3.
PPI network analysis of DEGs. (A) PPI network containing 149 nodes and 930 edges. (B) Module 1 consisted of 35 nodes and 573 edges, as identified by the MCODE plugin in Cytoscape. (C) Module 2 consisted of 21 nodes and 104 edges, as identified by the MCODE plugin in Cytoscape. Blue nodes represent downregulated genes in BC tissues; red nodes represent upregulated genes in BC tissues. PPI, protein-protein interaction; DEG, differentially expressed genes; BC, breast cancer; MCODE, Molecular Complex Detection.
The 35 genes in module 1 were ranked based on log |FC| values in the TCGA database and selected the top 10 hub genes for further analysis. The expression levels of the 10 hub genes in the BC tissues were >10-fold (log |FC|≥3.42) increased compared with those in the normal breast tissues. Through literature mining, it was identified that UBE2C, ASPM, BIRC5, TOP2A, KIF20A, CEP55, TPX2, NEK2 and ANLN, but not NUF2, have been reported extensively in BC-related studies. Therefore, NUF2 was selected as the focus of subsequent analyses.
NUF2 expression in BC
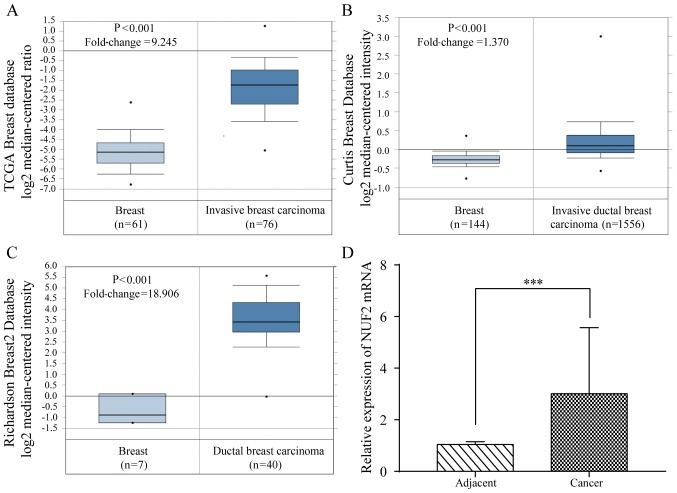
The expression of NUF2 mRNA in the BC tissues was evaluated using Oncomine (https://www.oncomine.org/) (30). The results indicated that the NUF2 expression level is significantly increased in the BC tissues compared with in the normal breast tissues (P<0.01; Fig. 4A-C). To further verify these results, 42 pairs of BC tissues and adjacent tissues were analyzed by RT-qPCR. Consistent with the results of the database analysis, the expression of NUF2 mRNA in the BC tissues was significantly increased (P<0.001) compared with in the adjacent tissues (Fig. 4D).
Figure 4.
Expression of NUF2 in BC. (A) NUF2 mRNA was overexpressed in human BC tissues compared with normal breast tissues in the TCGA, (B) Curtis and (C) Richardson microarray databases, as evaluated by Oncomine analysis. (D) NUF2 mRNA expression was increased in most BC lesions compared with para-carcinoma tissues, as determined by reverse transcription-quantitative PCR. ***P<0.001. BC, breast cancer; NUF2, NDC80 kinetochore complex component.
Association of NUF2 with clinical pathological characteristics and survival of patients with BC
To further validate the clinical value of NUF2, the association between its expression and the clinical pathological characteristics of the 42 patients with BC recruited from Shaoxing People's Hospital were assessed. The expression of NUF2 was only significantly associated with age (P<0.05). Using the data in the TCGA database (contains data on 1,090 patients with BC), NUF2 expression was found to be significantly associated with age (P<0.001), estrogen receptor (ER) status (P<0.001), progesterone receptor (PR) status (P<0.001), histological type (P<0.001), TNM stage (P<0.05), and molecular subtype (P<0.001). The results are shown in Tables II and III. Furthermore, the clinical pathological characteristics that have multiple groups (>2) need a post hoc test to determine exactly what groups exhibit a difference. Therefore, Bonferroni's post hoc test was used for pairwise comparison. The results showed that NUF2 expression is statistically different between TNM stage 1 and 2 (P<0.01), and tumor stage T1 and T2 (P<0.001). In terms of molecular subtype, NUF2 expression was also significantly different in all pairwise comparisons (P<0.001) except between luminal A and normal-like, and luminal B and basal-like. The detailed results are shown in Tables IV and V.
Table II.
Association of NUF2 with clinical pathological characteristics of breast cancer patients from Shaoxing People's Hospital.
| Pathological characteristics | Number of patients (%) |
NUF2 (%)
|
P-valuea | |
|---|---|---|---|---|
| Low | High | |||
| Age | 0.031 | |||
| <60 | 21 (50.0) | 14 (66.7) | 7 (33.3) | |
| ≥60 | 21 (50.0) | 7 (33.3) | 14 (66.7) | |
| HER2 status | 0.05 | |||
| Positive | 14 (33.3) | 10 (71.4) | 4 (29.6) | |
| Negative | 28 (66.7) | 11 (39.3) | 17 (60.7) | |
| ER status | 0.679b | |||
| Positive | 35 (83.3) | 17 (48.6) | 18 (51.4) | |
| Negative | 7 (16.7) | 4 (57.1) | 3 (42.9) | |
| PR status | 0.107 | |||
| Positive | 27 (64.3) | 11 (40.7) | 16 (59.3) | |
| Negative | 15 (35.7) | 10 (66.7) | 5 (33.3) | |
| TNM stage | 0.751 | |||
| 1 | 12 (28.6) | 5 (41.7) | 7 (58.3) | |
| 2 | 25 (59.5) | 13 (52.0) | 12 (48.0) | |
| 3 | 5 (11.9) | 3 (60.0) | 2 (40.0) | |
| Tumor stage | 0.333 | |||
| T1 | 19 (45.2) | 8 (42.1) | 11 (57.9) | |
| T2 | 22 (52.4) | 13 (59.1) | 9 (40.9) | |
| T3 | 1 (2.4) | 0 (0.0) | 1 (100.0) | |
| Lymph node stage | 0.946 | |||
| N0 | 27 (64.3) | 13 (48.1) | 14 (51.9) | |
| N1 | 10 (23.8) | 5 (50.0) | 5 (50.0) | |
| N2 | 3 (7.1) | 2 (66.7) | 1 (33.3) | |
| N3 | 2 (4.3) | 1 (50.0) | 1 (50.0) | |
| Node metastasis | 0.533 | |||
| Yes | 18 (42.9) | 10 (55.6) | 8 (44.4) | |
| No | 24 (57.1) | 11 (45.8) | 13 (54.2) | |
Unless otherwise noted, χ2 tests were used for comparisons between groups.
χ2 test with continuity correction was used. ER, estrogen receptor; PR, progesterone receptor; TNM, tumor node metastasis; NUF2, NDC80 kinetochore complex component.
Table III.
Association of NUF2 with clinical pathological characteristics of breast cancer patients derived from TCGA database.
| Pathological characteristics | Number of patients (%) |
NUF2 (%)
|
P-valuea | |
|---|---|---|---|---|
| Low | High | |||
| Age | <0.001 | |||
| <60 | 579 (53.2) | 255 (44.0) | 324 (66.0) | |
| ≥60 | 510 (46.8) | 289 (56.7) | 221 (43.3) | |
| HER2 status | 0.296 | |||
| Positive | 90 (21.4) | 39 (43.3) | 51 (56.7) | |
| Negative | 331 (78.6) | 164 (49.5) | 167 (50.5) | |
| ER status | <0.001 | |||
| Positive | 803 (77.2) | 467 (58.2) | 336 (41.8) | |
| Negative | 237 (22.8) | 60 (25.3) | 177 (74.7) | |
| PR status | <0.001 | |||
| Positive | 694 (66.9) | 415 (60.0) | 279 (40.0) | |
| Negative | 343 (33.1) | 110 (32.1) | 233 (67.9) | |
| Histology type | <0.001 | |||
| IDC | 779 (79.3) | 326 (41.8) | 453 (58.2) | |
| ILC | 203 (20.7) | 156 (76.8) | 47 (23.2) | |
| TNM stage | 0.032 | |||
| 1 | 181 (17.0) | 108 (59.7) | 73 (40.3) | |
| 2 | 619 (58.0) | 293 (47.3) | 326 (52.7) | |
| 3 | 247 (23.1) | 119 (48.2) | 128 (51.8) | |
| 4 | 20 (1.9) | 10 (50.0) | 10 (50.0) | |
| Tumor stage | <0.001 | |||
| T1 | 279 (25.7) | 171 (61.3) | 108 (38.7) | |
| T2 | 631 (58.0) | 283 (44.8) | 348 (55.2) | |
| T3 | 137 (12.6) | 71 (51.8) | 66 (48.2) | |
| T4 | 40 (3.7) | 19 (47.5) | 21 (52.5) | |
| Lymph node stage | 0.095 | |||
| N0 | 514 (48.0) | 255 (49.6) | 259 (50.4) | |
| N1 | 360 (33.6) | 185 (51.4) | 175 (48.6) | |
| N2 | 120 (11.2) | 48 (40.0) | 72 (60.0) | |
| N3 | 76 (7.2) | 43 (56.6) | 33 (43.4) | |
| Metastasis stage | 0.82 | |||
| M0 | 906 (97.6) | 434 (47.9) | 472 (52.1) | |
| M1 | 22 (2.4) | 10 (45.5) | 12 (54.5) | |
| Molecular subtype | <0.001 | |||
| Luminal A | 419 (0.5) | 305 (72.8) | 114 (27.2) | |
| Luminal B | 190 (0.23) | 26 (13.7) | 164 (86.3) | |
| HER2+ | 67 (0.08) | 26 (38.8) | 41 (61.2) | |
| Basal-like | 139 (0.16) | 14 (10.1) | 125 (89.9) | |
| Normal-like | 23 (0.03) | 21 (91.3) | 2 (8.7) | |
χ2 tests were used for comparisons between groups. TNM, tumor node metastasis; ER, estrogen receptor; PR, progesterone receptor; IDC, infiltrating ductal carcinoma; ILC, infiltrating lobular carcinoma; NUF2, NDC80 kinetochore complex component.
Table IV.
Comparison of clinical pathological characteristics of breast cancer patients from Shaoxing People's Hospital among multiple groups.
| Pathological characteristics | Pairwise comparisons (P-values) | |||
|---|---|---|---|---|
| TNM stage | 1 | 2 | 3 | |
| 1 | N/A | 0.556 | 0.620 | |
| 2 | 0.556 | N/A | 1.000 | |
| 3 | 0.620 | 1.000 | N/A | |
| Tumor stage | T1 | T2 | T3 | |
| T1 | N/A | 0.278 | 1.000 | |
| T2 | 0.278 | N/A | 0.435 | |
| T3 | 1.000 | 0.435 | N/A | |
| Lymph node stage | N0 | N1 | N2 | N3 |
| N0 | N/A | 1.000 | 1.000 | 1.000 |
| N1 | 1.000 | N/A | 1.000 | 1.000 |
| N2 | 1.000 | 1.000 | N/A | 1.000 |
| N3 | 1.000 | 1.000 | 1.000 | N/A |
P<0.05/N was considered statistically significant, where N was the number of pairwise comparisons. TNM, tumor node metastasis.
Table V.
Comparison of clinical pathological characteristics of breast cancer patients in The Cancer Genome Atlas database among multiple groups.
| Pathological characteristics | Pairwise comparisons (P-values) | ||||
|---|---|---|---|---|---|
| TNM stage | 1 | 2 | 3 | 4 | |
| 1 | N/A | 0.004a | 0.019 | 0.405 | |
| 2 | 0.004a | N/A | 0.822 | 0.814 | |
| 3 | 0.019 | 0.822 | N/A | 0.875 | |
| 4 | 0.405 | 0.814 | 0.875 | N/A | |
| Tumor stage | T1 | T2 | T3 | T4 | |
| T1 | N/A | 0.000a | 0.066 | 0.097 | |
| T2 | 0.000a | N/A | 0.138 | 0.744 | |
| T3 | 0.066 | 0.138 | N/A | 0.630 | |
| T4 | 0.097 | 0.744 | 0.630 | N/A | |
| Lymph node stage | N0 | N1 | N2 | N3 | |
| N0 | N/A | 0.605 | 0.058 | 0.257 | |
| N1 | 0.605 | N/A | 0.031 | 0.410 | |
| N2 | 0.058 | 0.031 | N/A | 0.023 | |
| N3 | 0.257 | 0.410 | 0.023 | N/A | |
| Molecular subtype | Luminal A | Luminal B | HER2+ | Basal-like | Normal-like |
| Luminal A | N/A | 0.000a | 0.000a | 0.000a | 0.049 |
| Luminal B | 0.000a | N/A | 0.000a | 0.322 | 0.000a |
| HER2+ | 0.000a | 0.000a | N/A | 0.000a | 0.000a |
| Basal-like | 0.000a | 0.322 | 0.000a | N/A | 0.000a |
| Normal-like | 0.049 | 0.000a | 0.000a | 0.000a | N/A |
P<0.05/N was considered statistically significant, where N was the number of pairwise comparisons.
P<0.05/N. TNM, tumor, node and metastasis.
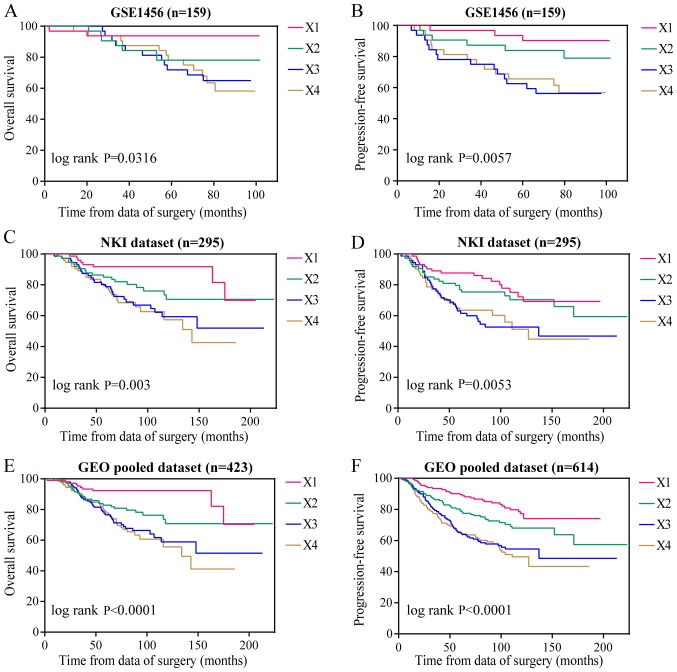
Furthermore, to elucidate the correlation between the expression of NUF2 and patient survival, 5 GEO datasets were used. Samples from each dataset were reclassified into four subsets (X1, X2, X3 and X4) according to the quartile of NUF2 expression. The X1 subset was set with the lowest expression as the reference to calculate the hazard ratio (HR). Each dataset was analyzed by Kaplan-Meier analysis and Cox proportional hazard analysis. High NUF2 expression was associated with shorter overall survival (OS) and progression-free survival (PFS) compared with low NUF2 expression in the GSE1456 dataset (Fig. 5A and B) and NKI dataset (Fig. 5C and D). Similar results were obtained in the GEO pooled analysis, as shown in Fig. 5E and F. Based on a further GEO pooled analysis, it was demonstrated that NUF2 expression levels are significantly associated with poor OS and PFS in both ER-positive (P<0.01; Fig. 6A and B) and ER-negative (P<0.01; Fig. 6C and D) BC, and the association is more obvious in ER-positive BC. The results of Cox proportional hazards analysis are shown in Table VII. In addition, the results of the Bonferroni's post hoc tests used to compare the Kaplan-Meier survival curves corresponding to >2 groups are shown in Table VI. These results indicated that NUF2 might be a prognostic factor for BC.
Figure 5.
Role of NUF2 in the prognosis of BC patients. (A) NUF2 expression was significantly associated with OS of BC patients in the GSE1456 dataset, (C) NKI dataset, and (E) GEO pooled dataset. NUF2 expression was significantly associated with PFS of BC patients in the (B) GSE1456 dataset, (D) NKI dataset and (F) GEO pooled dataset. GEO, gene expression omnibus; BC, breast cancer; OS, overall survival; PFS, progression free survival; ER, estrogen receptor; PR, progesterone receptor; NUF2, NDC80 kinetochore complex component.
Figure 6.
Prognostic value of NUF2 in ER-positive and ER-negative BC. Association of NUF2 expression with (A) OS and (B) PFS in ER-positive BC in the GEO pooled dataset. Association of NUF2 expression with (C) OS and (D) PFS in ER-negative BC in the GEO pooled dataset. GEO, gene expression omnibus; BC, breast cancer; OS, overall survival; PFS, progression free survival; ER, estrogen receptor; PR, progesterone receptor; NUF2, NDC80 kinetochore complex component.
Table VII.
Univariate and multivariate analyses of NUF2 and survival in GEO datasets.
| Dataset | Overall survival
|
Progression-free survival
|
||
|---|---|---|---|---|
| HR (95% CI) | Adjusted HR (95% CI) | HR (95% CI) | Adjusted HR (95% CI) | |
| GSE1456 | ||||
| X1 | Reference | Reference | Reference | Reference |
| X2 | 3.94 (0.82-18.95) | 3.66 (0.75-17.75) | 2.19 (0.55-8.74) | 1.99 (0.49-8.05) |
| X3 | 6.18 (1.37-27.90)a | 4.6 (0.91-23.28) | 5.7 (1.64-19.83)b | 3.94 (1.00-15.45)a |
| X4 | 6.69 (1.50-29.91)a | 5.11 (1.03-25.24)a | 5.21 (1.48-18.29)b | 3.76 (0.97-14.57) |
| GSE22220 | ||||
| X1 | N/A | N/A | Reference | Reference |
| X2 | N/A | N/A | 2.05 (0.95-4.43) | 1.69 (0.77-3.69) |
| X3 | N/A | N/A | 2.31 (1.07-4.96)a | 1.79 (0.82-3.92) |
| X4 | N/A | N/A | 3.34 (1.60-6.95)b | 2.30 (1.05-5.02)a |
| NKI dataset | ||||
| X1 | Reference | Reference | Reference | Reference |
| X2 | 2.39 (1.04-5.51)a | 2.01 (0.87-4.68) | 1.35 (0.72-2.54) | 1.29 (0.68-2.46) |
| X3 | 3.97 (1.80-8.77)b | 2.88 (1.28-6.48)a | 2.39 (1.33-4.30)b | 1.97 (1.07-3.61)a |
| X4 | 4.48 (2.03-9.89)c | 2.63 (1.13-6.12)a | 2.31 (1.27-4.21)b | 1.84 (0.96-3.52) |
| GSE4299 | ||||
| X1 | N/A | N/A | Reference | Reference |
| X2 | N/A | N/A | 1.95 (0.99-3.85) | 1.93 (0.97-3.81) |
| X3 | N/A | N/A | 2.16 (1.09-4.27)a | 2.13 (1.07-4.24)a |
| X4 | N/A | N/A | 2.11 (1.07-4.17)a | 2.09 (1.04-4.19)a |
| GSE20685 | ||||
| X1 | Reference | Reference | Reference | Reference |
| X2 | 1.11 (0.57-2.14) | 1.1 (0.57-2.14) | 1.17 (0.67-2.05) | 1.16 (0.66-2.03) |
| X3 | 1.58 (0.85-2.95) | 1.58 (0.85-2.95) | 1.12 (0.64-1.98) | 1.13 (0.64-2.00) |
| X4 | 1.59 (0.85-2.96) | 1.58 (0.84-2.97) | 1.46 (0.84-2.52) | 1.42 (0.82-2.48) |
| GEO pooled | ||||
| X1 | Reference | Reference | Reference | Reference |
| X2 | 2.69 (1.29-5.60)b | 2.18 (1.04-4.57)a | 1.67 (1.05-2.65)a | 1.5 (0.95-2.39) |
| X3 | 4.43 (2.20-8.91)c | 3 (1.46-6.15)b | 2.72 (1.77-4.19)c | 2.13 (1.36-3.32)b |
| X4 | 4.94 (2.46-9.92)c | 2.87 (1.38-5.94)b | 2.98 (1.94-4.59)c | 2.1 (1.33-3.32)b |
| GEO pooled ER(+) | ||||
| X1 | Reference | Reference | Reference | Reference |
| X2 | 3.84 (1.41-10.48)b | 3.23 (1.18-8.88)a | 1.57 (0.79-3.13) | 1.39 (0.70-2.79) |
| X3 | 5.48 (2.08-14.49)b | 3.93 (1.46-10.61)b | 3.03 (1.62-5.69)b | 2.32 (1.22-4.44)a |
| X4 | 5.60 (2.06-15.23)b | 3.74 (1.34-10.46)a | 2.83 (1.45-5.52)b | 2.09 (1.04-4.16)a |
| GEO pooled ER(−) | ||||
| X1 | Reference | Reference | Reference | Reference |
| X2 | 1.40 (0.46-4.20) | 1.19 (0.39-3.61) | 1.28 (0.46-3.62) | 1.09 (0.38-3.11) |
| X3 | 2.96 (1.07-8.20)a | 1.88 (0.65-5.45) | 2.60 (1.01-6.72)a | 1.58 (0.58-4.34) |
| X4 | 3.23 (1.22-8.54)a | 1.96 (0.70-5.52) | 2.44 (0.97-6.10) | 1.46 (0.54-3.91) |
For multivariate analysis, HR was adjusted by ER status and Elston grade in GSE1456. In GSE22220, HR was adjusted by age and Elston grade. In NKI and GSE4299, HR was adjusted by age, Elston grade, and ER status. For GSE20685, HR was adjusted by age. HR was adjusted by ER status and Elston grade in the pooled analysis. For GEO pooled ER(+) and GEO pooled ER(−), HR was adjusted by Elston grade.
P<0.05,
P<0.01 and
P<0.001 vs. the X1 group. HR, hazard ratio; ER, estrogen receptor; GEO, gene expression omnibus; CI, confidence interval.
Table VI.
Comparison of Kaplan-Meier curves among multiple groups.
| Datasets | Pairwise comparisons (P-values) | ||||
|---|---|---|---|---|---|
| GSE1456 OS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.088 | 0.008a | 0.003a | |
| X2 | 0.088 | N/A | 0.356 | 0.253 | |
| X3 | 0.008a | 0.356 | N/A | 0.851 | |
| X4 | 0.003a | 0.253 | 0.851 | N/A | |
| GSE1456 PFS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.259 | 0.002a | 0.004a | |
| X2 | 0.259 | N/A | 0.044 | 0.068 | |
| X3 | 0.002a | 0.044 | N/A | 0.800 | |
| X4 | 0.004a | 0.068 | 0.800 | N/A | |
| NKI OS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.036 | 0.000a | 0.000a | |
| X2 | 0.036 | N/A | 0.113 | 0.041 | |
| X3 | 0.000a | 0.113 | N/A | 0.663 | |
| X4 | 0.000a | 0.041 | 0.663 | N/A | |
| NKI PFS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.361 | 0.002a | 0.003a | |
| X2 | 0.361 | N/A | 0.034 | 0.057 | |
| X3 | 0.002a | 0.034 | N/A | 0.840 | |
| X4 | 0.003a | 0.057 | 0.840 | N/A | |
| GEO pooled OS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.007a | 0.000a | 0.000a | |
| X2 | 0.007a | N/A | 0.063 | 0.017 | |
| X3 | 0.000a | 0.063 | N/A | 0.630 | |
| X4 | 0.000a | 0.017 | 0.630 | N/A | |
| GEO pooled PFS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.027 | 0.000a | 0.000a | |
| X2 | 0.027 | N/A | 0.010 | 0.002a | |
| X3 | 0.000a | 0.010 | N/A | 0.639 | |
| X4 | 0.000a | 0.002a | 0.639 | N/A | |
| GEO pooled ER(+) OS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.006a | 0.000a | 0.000a | |
| X2 | 0.006a | N/A | 0.307 | 0.314 | |
| X3 | 0.000a | 0.307 | N/A | 0.925 | |
| X4 | 0.000a | 0.314 | 0.925 | N/A | |
| GEO pooled ER(+) PFS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.197 | 0.000a | 0.001a | |
| X2 | 0.197 | N/A | 0.022 | 0.065 | |
| X3 | 0.000a | 0.022 | N/A | 0.730 | |
| X4 | 0.001a | 0.065 | 0.730 | N/A | |
| GEO pooled ER(−) OS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.495 | 0.031 | 0.016 | |
| X2 | 0.495 | N/A | 0.079 | 0.028 | |
| X3 | 0.031 | 0.079 | N/A | 0.762 | |
| X4 | 0.016 | 0.028 | 0.762 | N/A | |
| GEO pooled ER(−) PFS | Subsets | X1 | X2 | X3 | X4 |
| X1 | N/A | 0.679 | 0.029 | 0.055 | |
| X2 | 0.679 | N/A | 0.095 | 0.106 | |
| X3 | 0.029 | 0.095 | N/A | 0.834 | |
| X4 | 0.055 | 0.106 | 0.834 | N/A | |
P<0.05/N was considered statistically significant, where N was the number of pairwise comparisons.
P<0.05/N. OS, overall survival; PFS, progression-free survival; ER, estrogen receptor.
Signaling pathways associated with NUF2
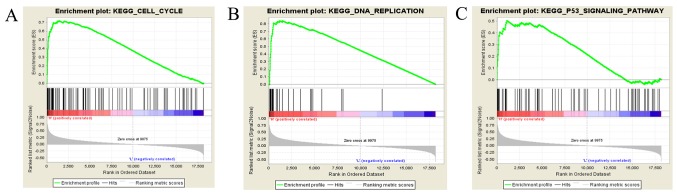
Single-gene differential expression analyses in the study of biological processes are limited (28). To effectively reveal the biological significance of microarray datasets, a GSEA was performed to predict gene sets and signaling pathways associated with NUF2 using the data obtained from the TCGA database. As shown in Fig. 7A-C, NUF2 may function in cell cycle-related pathways, including the cell cycle, DNA replication, and p53 signaling pathway.
Figure 7.
Signaling pathways associated with NDC80 kinetochore complex component predicted by gene set enrichment analysis. (A) Cell cycle (P=0.000; FDR=0.000; ES=0.72). (B) DNA replication (P<0.001; FDR<0.001; ES=0.84). (C) p53 signaling pathway (P=0.000; FDR=0.004; ES=0.51). FDR, false discovery rate; KEGG, Kyoto Encyclopedia of genes and genomes; ES, enrichment score.
Discussion
BC is the most common malignant tumor in women, accounting for 25% of female tumors (3). Despite advanced treatment techniques, BC remains the leading cause of death among women (31). Therefore, there is a pressing need for more effective molecular biomarkers to prevent, diagnose, and treat BC. With the development of microarray technology, hundreds of molecules have been found to be abnormally expressed during breast carcinogenesis and progression. The TCGA and GEO databases provide a large number of publicly available microarray datasets for biomarker identification.
In this study, 190 DEGs with the same expression trends were identified in four datasets, and a GO BP analysis showed that the upregulated DEGs were mainly enriched in the biological processes of cell division, mitotic nuclear division, and G2/M transition of mitotic cell cycle, while the downregulated DEGs were related to lipid metabolic process, cholesterol homeostasis, and glucose metabolic process. Cell division and cell cycle are the basic processes in cell proliferation, and their abnormalities contribute to carcinogenesis and tumor progression (32). Furthermore, the activation of key regulators of lipid and cholesterol metabolism drives the estrogen-independent growth of invasive lobular breast carcinoma cells (33). A KEGG signaling pathway analysis of the DEGs in this study revealed the importance of the cell cycle, ECM-receptor interaction, PPAR signaling pathway, and AMPK signaling pathway in BC. Previous studies have reported that ECM could regulate tissue homeostasis, and its dysregulation could promote tumor progression by affecting adhesion, migration, differentiation, proliferation, and apoptosis of tumor cells (34,35). In addition, an increase in the rigidity of the ECM due to the local accumulation of crosslinked collagen matrix is associated with cancer progression (36). Yao et al (37) found that the PPAR signaling pathway is involved in breast carcinogenesis. Song et al (38) suggested that activation of the AMPK signaling pathway may be beneficial for the promotion of tumor necrosis factor-related apoptosis-inducing ligand-mediated anti-BC treatment. a PPI network was constructed in this study and two functional modules were identified. According to the MCODE scores, which represent importance, module 1 was found to play a major role in the PPI network. By combining the log |FC| values of the DEGs in the TCGA database and literature mining, NUF2 was selected for further research as a key gene in BC.
Although Xiang et al (12) found that NUF2 is upregulated in BC, using cDNA microarray data of BC patients, further experiments have not been performed to verify this finding. In this study, by Oncomine analysis and RT-qPCR assay, it was verified that NUF2 is overexpressed in BC tissues, further confirming the results obtained by data mining. Shiraishi et al (39) found that NUF2 expression is significantly associated with prostate cancer recurrence, and patients with high NUF2 expression have significantly shorter survival times without biochemical recurrence. Hu et al (10) showed that the overexpression of NUF2 could be related to poor prognosis in pancreatic cancer. Zhang et al (13) found that NUF2 expression has prognostic values for BC patients, by simply using the BC-GenExMiner online analysis tool. However, further analysis has not been conducted. To this end, the present study analyzed the precise roles and underlying molecular mechanisms of NUF2 in BC. By stratified analysis and pooled analysis of five GEO datasets, it was found that patients with BC and high NUF2 expression had worse prognosis than patients with low NUF2 expression in both ER-positive and ER-negative BC. Using clinical data for 42 patients, it was demonstrated that NUF2 expression was only associated with age. Small sample size, erroneous tissue sampling, RNA degradation, and fluctuating efficiency of RT-qPCR may affect the results of the analysis. This hypothesis can be tested through the following methods: Increasing sample size, determining the type of tissue with pathological examination, detecting RNA degradation by RNA electrophoresis, and verifying amplification efficiency of RT-qPCR by the standard curve method. The lack of additional experiments to test these possibilities is a limitation to the present study. Therefore, the relationship was further analyzed using clinical data for patients with BC in the TCGA database. The expression of NUF2 was positively correlated with tumor stage and negatively correlated with ER status, consistent with the results from a number of studies showing that advanced tumors and ER-negative tumors are probably related to poor survival (40-42), suggesting that NUF2 plays an important role in tumor progression and prognosis. To elucidate the molecular mechanisms by which NUF2 mediates breast carcinogenesis and progression, GSEA was performed in this study. The results revealed that NUF2 is involved in cell cycle-related pathways.
In conclusion, the present data analysis and RT-qPCR validation indicated that NUF2 is highly expressed in BC and is associated with its multiple pathological features and prognosis. NUF2 is therefore a potential therapeutic target and prognostic indicator of BC. However, this study had several limitations. First, only mRNA data were obtained from the databases and RT-qPCR, and this single-gene-level analysis cannot fully capture the expression of NUF2 in BC. Second, experimental validation of the results was not performed. Therefore, further research is required to determine the roles of NUF2 in BC.
NUF2 is overexpressed in BC and is associated with its multiple pathological features. More importantly, NUF2 may play an important role in predicting the clinical outcomes of different BC subgroups.
Acknowledgments
Not applicable.
Funding
This study was supported by the Public Technology Research Project of Zhejiang Province (grant no. LGF18H200006) and the Medicines Health Technology Plan Project of Zhejiang Province (grant no. 2018PY073).
Availability of data and materials
The datasets analyzed during the current study are available in the TCGA (https://cancergenome.nih.gov/) and GEO (https://www.ncbi.nlm.nih.gov/geo/) databases.
Authors' contributions
WJX, YNW, and XJD participated in the study design. WJX, YNW, and XPX contributed to data collection and analysis. WJX, YZW, and SML were involved in the collection of samples and RT-qPCR. All authors were involved in the writing of the article. XJD critically reviewed the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved and supervised by the Ethics Committee of Shaoxing People's Hospital (Shaoxing, China). Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Donepudi MS, Kondapalli K, Amos SJ, Venkanteshan P. Breast cancer statistics and markers. J Cancer Res Ther. 2014;10:506–511. doi: 10.4103/0973-1482.137927. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Tan W, Yang M, Yang H, Zhou F, Shen W. Predicting the response to neoadjuvant therapy for early-stage breast cancer: Tumor-, blood-, and imaging-related biomarkers. Cancer Manag Res. 2018;10:4333–4347. doi: 10.2147/CMAR.S174435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salem Y, Yacov N, Propheta-Meiran O, Breitbart E, Mendel I. Newly characterized motile sperm domain-containing protein 2 promotes human breast cancer metastasis. Int J Cancer. 2019;144:125–135. doi: 10.1002/ijc.31665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinsella MD, Nassar A, Siddiqui MT, Cohen C. Estrogen receptor (ER), progesterone receptor (PR), and HER2 expression pre- and post-neoadjuvant chemotherapy in primary breast carcinoma: A single institutional experience. Int J Clin Exp Pathol. 2012;5:530–536. [PMC free article] [PubMed] [Google Scholar]
- 7.Piao J, Chen L, Jin T, Xu M, Quan C, Lin Z. Paip1 affects breast cancer cell growth and represents a novel prognostic biomarker. Hum Pathol. 2018;73:33–40. doi: 10.1016/j.humpath.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 8.Sandoo A, Kitas GD, Carmichael AR. Breast cancer therapy and cardiovascular risk: Focus on trastuzumab. Vasc Health Risk Manag. 2015;11:223–228. doi: 10.2147/VHRM.S69641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: A link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- 10.Hu P, Chen X, Sun J, Bie P, Zhang LD. siRNA-mediated knockdown against NUF2 suppresses pancreatic cancer proliferation in vitro and in vivo. Biosci Rep. 2015;35: pii:e00170. doi: 10.1042/BSR20140124. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Fu HL, Shao L. Silencing of NUF2 inhibits proliferation of human osteosarcoma Saos-2 cells. Eur Rev Med Pharmacol Sci. 2016;20:1071–1079. [PubMed] [Google Scholar]
- 12.Xiang YJ, Fu Q, Ma ZB, Gao DZ, Zhang Q, Li YY, Li L, Liu L, Ye CM, Yu ZG, Guo MM. Screening for candidate genes related to breast cancer with cDNA microarray analysis. Chronic Dis Transl Med. 2015;1:65–72. doi: 10.1016/j.cdtm.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Mao JH, Zhu W, Jain AK, Liu K, Brown JB, Karpen GH. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun. 2016;7:12619. doi: 10.1038/ncomms12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke C, Madden SF, Doolan P, Aherne ST, Joyce H, O'Driscoll L, Gallagher WM, Hennessy BT, Moriarty M, Crown J, et al. Correlating transcriptional networks to breast cancer survival: A large-scale coexpression analysis. Carcinogenesis. 2013;34:2300–2308. doi: 10.1093/carcin/bgt208. [DOI] [PubMed] [Google Scholar]
- 15.Gruosso T, Mieulet V, Cardon M, Bourachot B, Kieffer Y, Devun F, Dubois T, Dutreix M, Vincent-Salomon A, Miller KM, Mechta-Grigoriou F. Chronic oxidative stress promotes H2AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO Mol Med. 2016;8:527–549. doi: 10.15252/emmm.201505891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maubant S, Tesson B, Maire V, Ye M, Rigaill G, Gentien D, Cruzalegui F, Tucker GC, Roman-Roman S, Dubois T. Transcriptome analysis of Wnt3a-treated triple-negative breast cancer cells. PLoS One. 2015;10:e0122333. doi: 10.1371/journal.pone.0122333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee GY, Haverty PM, Li L, Kljavin NM, Bourgon R, Lee J, Stern H, Modrusan Z, Seshagiri S, Zhang Z, et al. Comparative oncogenomics identifies PSMB4 and SHMT2 as potential cancer driver genes. Cancer Res. 2014;74:3114–3126. doi: 10.1158/0008-5472.CAN-13-2683. [DOI] [PubMed] [Google Scholar]
- 18.Tomczak K, Czerwińska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, Ragoussis J. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 21.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 22.Jones SB, DePrimo SE, Whitfield ML, Brooks JD. Resveratrol-induced gene expression profiles in human prostate cancer cells. Cancer Epidemiol Biomarkers Prev. 2005;14:596–604. doi: 10.1158/1055-9965.EPI-04-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 27.Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue D, Cheng P, Han M, Liu X, Xue L, Ye C, Wang K, Huang J. An integrated bioinformatical analysis to evaluate the role of KIF4A as a prognostic biomarker for breast cancer. OncoTargets Ther. 2018;11:4755–4768. doi: 10.2147/OTT.S164730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Yuan L, Qian K, Qian G, Zhu Y, Wu CL, Dan HC, Xiao Y, Wang X. Identification of biomarkers associated with pathological stage and prognosis of clear cell renal cell carcinoma by co-expression network analysis. Front Physiol. 2018;9:399. doi: 10.3389/fphys.2018.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du T, Sikora MJ, Levine KM, Tasdemir N, Riggins RB, Wendell SG, Van Houten B, Oesterreich S. Key regulators of lipid metabolism drive endocrine resistance in invasive lobular breast cancer. Breast Cancer Res. 2018;20:106. doi: 10.1186/s13058-018-1041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaushik N, Kim S, Suh Y, Lee SJ. Proinvasive extracellular matrix remodeling for tumor progression. Arch Pharm Res. 2019;42:40–47. doi: 10.1007/s12272-018-1097-0. [DOI] [PubMed] [Google Scholar]
- 35.Walker C, Mojares E, Del Rio Hernández A. Role of extracellular matrix in development and cancer progression. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19103028. pii: E3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tilghman RW, Cowan CR, Mih JD, Koryakina Y, Gioeli D, Slack-Davis JK, Blackman BR, Tschumperlin DJ, Parsons JT. Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS One. 2010;5:e12905. doi: 10.1371/journal.pone.0012905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao PL, Morales JL, Zhu B, Kang BH, Gonzalez FJ, Peters JM. Activation of peroxisome proliferator-activated receptor-β/δ (PPAR-β/δ) inhibits human breast cancer cell line tumorigenicity. Mol Cancer Ther. 2014;13:1008–1017. doi: 10.1158/1535-7163.MCT-13-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song W, Yan CY, Zhou QQ, Zhen LL. Galangin potentiates human breast cancer to apoptosis induced by TRAIL through activating AMPK. Biomed Pharmacother. 2017;89:845–856. doi: 10.1016/j.biopha.2017.01.062. [DOI] [PubMed] [Google Scholar]
- 39.Shiraishi T, Terada N, Zeng Y, Suyama T, Luo J, Trock B, Kulkarni P, Getzenberg RH. Cancer/testis antigens as potential predictors of biochemical recurrence of prostate cancer following radical prostatectomy. J Transl Med. 2011;9:153. doi: 10.1186/1479-5876-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bagaria SP, Ray PS, Sim MS, Ye X, Shamonki JM, Cui X, Giuliano AE. Personalizing breast cancer staging by the inclusion of ER, PR, and HER2. JAMA Surg. 2014;149:125–129. doi: 10.1001/jamasurg.2013.3181. [DOI] [PubMed] [Google Scholar]
- 41.Ryu JM, Choi HJ, Kim I, Lee SK, Yu J, Kim JE, Kang BI, Lee JE, Nam SJ, Kim SW. Only estrogen receptor 'positive' is not enough to predict the prognosis of breast cancer. Breast Cancer Res Treat. 2018;172:627–636. doi: 10.1007/s10549-018-4948-y. [DOI] [PubMed] [Google Scholar]
- 42.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: A prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9:R6. doi: 10.1186/bcr1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available in the TCGA (https://cancergenome.nih.gov/) and GEO (https://www.ncbi.nlm.nih.gov/geo/) databases.