Singh et al. show that MYC affects mRNA translation efficiency and start-site selection. Notable examples include the translation of mRNAs encoding most proteins of the electron transport chain and aberrant translation initiation site usage in the CD19 receptor that allows escape from CD19-directed CAR-T therapy.
Abstract
The oncogenic c-MYC (MYC) transcription factor has broad effects on gene expression and cell behavior. We show that MYC alters the efficiency and quality of mRNA translation into functional proteins. Specifically, MYC drives the translation of most protein components of the electron transport chain in lymphoma cells, and many of these effects are independent from proliferation. Specific interactions of MYC-sensitive RNA-binding proteins (e.g., SRSF1/RBM42) with 5′UTR sequence motifs mediate many of these changes. Moreover, we observe a striking shift in translation initiation site usage. For example, in low-MYC conditions, lymphoma cells initiate translation of the CD19 mRNA from a site in exon 5. This results in the truncation of all extracellular CD19 domains and facilitates escape from CD19-directed CAR-T cell therapy. Together, our findings reveal MYC effects on the translation of key metabolic enzymes and immune receptors in lymphoma cells.
Graphical Abstract
Introduction
The oncogenic c-MYC (MYC) transcription factor has broad effects on normal and malignant cell physiology. MYC’s immediate transcriptional activity has been the main focus, and initial studies showed selective effects on consensus E-boxes as the immediate targets of MYC action (Land et al., 1983; Blackwood and Eisenman, 1991). More recently, MYC has been described as a “global amplifier,” and augmentation of the expression of most active genes has been reported (Lin et al., 2012; Nie et al., 2012). Refinements of this concept take into account MYC interactions with coactivators and inhibitors that contribute to specific transcriptional effects (Ouyang et al., 2009; Walz et al., 2014; Kress et al., 2015). However, MYC has also been implicated in the control of mRNA translation. These effects are secondary and include changes in the expression of ribosomal proteins, key translation factors such as eIF4E and eIF4A, or cotranscriptional changes in mRNA capping (Schlosser et al., 2003; Arabi et al., 2005; Grandori et al., 2005; Cole and Cowling, 2009; van Riggelen et al., 2010).
Cancer-relevant effects on specific mRNAs involved in metabolism, migration, and metastasis have also been reported (Topisirovic and Sonenberg, 2011; Hsieh et al., 2012; Pourdehnad et al., 2013; Elkon et al., 2015; Truitt et al., 2015; Lindqvist et al., 2018). These have been largely attributed to activation of the mTOR/4EBP1/eIF4E signaling axis that acts as a driver of an anabolic and growth-promoting translation program that includes the translation of a subset of mRNA encoding mitochondrial proteins in an eIF4E-dependent manner and that is opposed by the catabolic effects of adenosine monophosphate–activated protein kinase (Lin et al., 2008; Hardie et al., 2012; Bhat et al., 2015; Morita et al., 2017; Saxton and Sabatini, 2017). In the present study, we report how MYC affects global mRNA translation efficiency (TE), and, using harringtonine to arrest the initiating ribosomes, we can precisely map changes in translation start-site usage that result in abnormal proteins.
Results
MYC has global and specific effects on mRNA translation in lymphoma cells
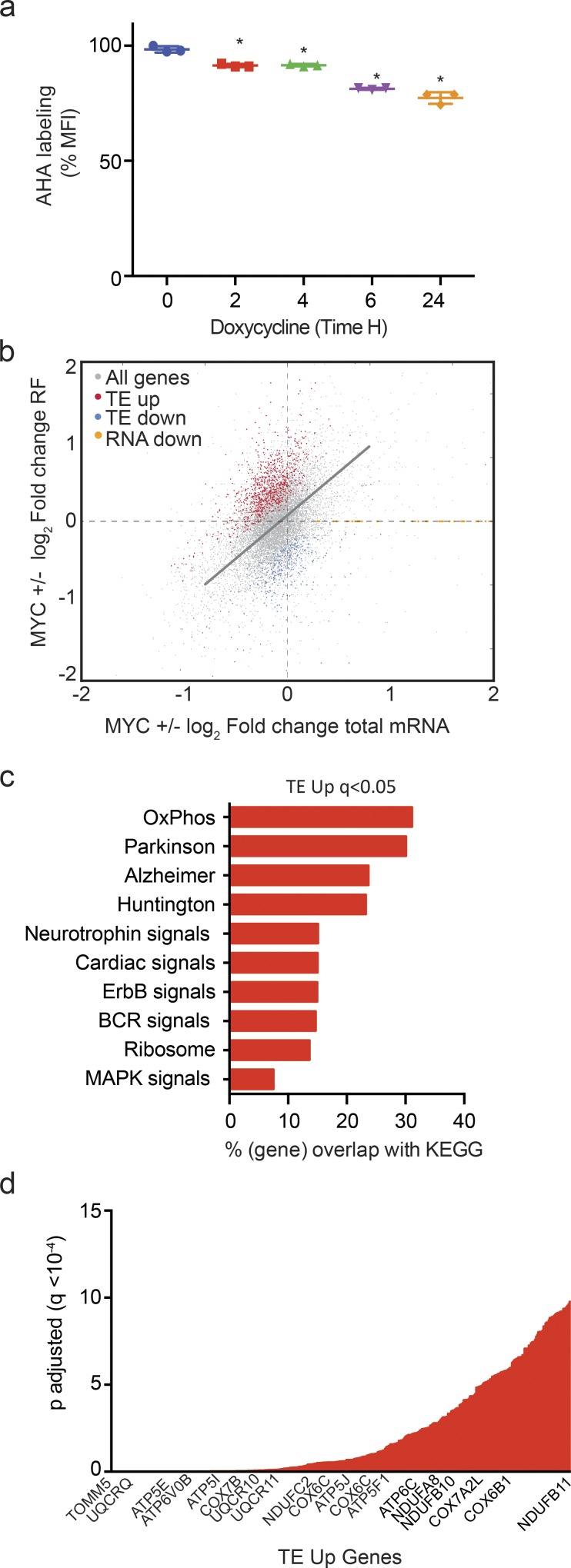
MYC inactivation (24 h) causes an overall 20% decrease in mRNA translation by metabolic labeling with l-azidohomoalanine (AHA) in P493-6 lymphoma cells that have an inducible MYC gene (P < 0.0001; n = 3; Fig. 1 a). We performed transcriptome-scale ribosome profiling to identify precisely which mRNAs are affected translationally by MYC. This method isolates changes in translation from changes in transcription by relating ribosome-protected fragment (RF) reads to total mRNA levels (Pajic et al., 2000; Ingolia et al., 2009; Wolfe et al., 2014). We performed the study in triplicates on P493-6 human B lymphoma cells that express MYC in a doxycycline-sensitive manner and compared high- and low-MYC states at a 24-h time point (Fig. S1, a and b). Important quality-control data are shown in Fig. S1, c–k, and described in the figure legend. For most transcripts, the change in translation as indicated by ribosome coverage (RF reads) was proportional to the change in mRNA abundance (r = 0.41; Fig. 1 b, indicated by the gray diagonal line). However, using a strict statistical cutoff at q < 0.01, we identified mRNAs whose translation was disproportionally affected by MYC. Specifically, the TE was up-regulated in the high-MYC state for 882 mRNAs (TE up) and decreased (TE down) for 315 mRNAs (Fig. 1 b and Fig. S1 l, marked in red and blue, respectively). MYC affected the total cellular mass (Schuhmacher and Eick, 2013); however, at the 24-h time point, we did not observe changes in total protein, RNA content, or cell size in P493 cells (Fig. S1, m–o).
Figure 1.
MYC has specific effects on mRNA translation. (a) Metabolic labeling of newly synthesized proteins with AHA in P493-6 cells treated with doxycycline (0.1 µg/ml) for the indicated time period measured by FACS (n = 3 biological replicates, mean ± SD from three replicates, data representative of three independent experiments). P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05. (b) Change in total mRNA levels versus change in RF reads in the presence or absence of MYC in P493-6 cells. The linear function indicates proportional changes in both: genes with a significantly (q < 0.01) disproportional increase in TE (TE up, red) or decrease (TE down, blue). Ribosome footprinting was performed in three biological replicates for each group. (c) GO analysis of pathways enriched in TE-up genes (q < 0.05); n = 3 biological replicates in each group; q values were calculated using the FDR method of Benjamini and Hochberg. (d) mRNAs whose translation was stimulated by MYC (TE up) ranked by significance (q < 0.05); n = 3 biological replicates in each group.
MYC induces the translation of mitochondrial respiration genes
Gene ontology (GO) analysis of genes whose translation is stimulated in high-MYC states (TE up) indicated oxidative phosphorylation as the most significantly affected category (P = 4.88 × 10−44). Others were the related, mitochondrial gene sets implicated in Parkinson’s, Alzheimer’s, and Huntington’s (Fig. 1 c). Ranking of transcripts by significance readily identified many components of the mitochondrial respiratory complex among the genes whose translation is stimulated by MYC (Fig. 1 d). Notably, most of these genes showed no significant change in transcript levels, indicating their expression at this early (24-h) time point was primarily responsive to changes in their translation.
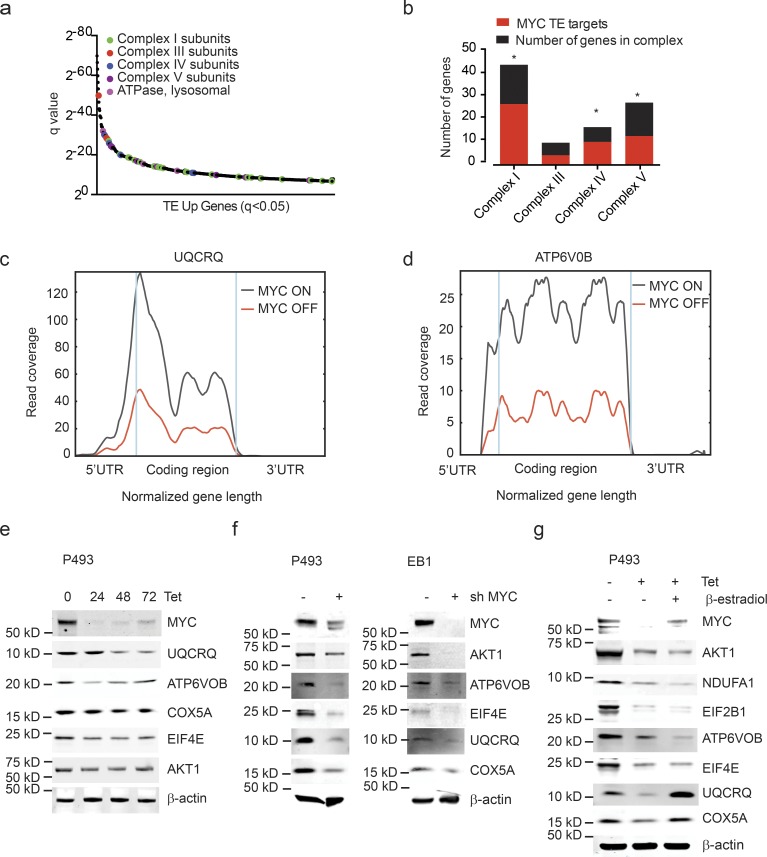
Specifically, MYC augmented the translation of the majority of proteins of the electron transport complexes I, III, IV, and V (enrichment statistics for complex I: P = 0.000001; complex IV: P = 0.004; and complex V: P = 0.01; Fig. 2 a). Complex I was rate limiting for cellular respiration. MYC increased the translation mRNAs encoding 27 of the 45 protein subunits of complex I (Fig. 2 b). Conversely, MYC inactivation caused a striking loss of ribosome coverage across the 5′ untranslated regions (UTRs) and coding sequence (CDS) of UQCRQ and ATP6V0B (Fig. 2, c and d). Immunoblot and quantitative real-time PCR (qRT-PCR) were consistent with increased translation in the absence of increased mRNA for UQCRQ, ATP6VOB, COX5A, EIF4E, AKT1, and many other targets (Fig. 2 e and Fig. S2 a).
Figure 2.
MYC stimulates the translation of ETC proteins. (a) MYC-dependent mRNAs (TE up; ranked by significance) reveal a preponderance of ETC components: complex I genes (green), complex III genes (red), complex IV genes (blue), complex V genes (purple), and lysosomal ATPase (pink); n = 3 biological replicates in each group. (b) Proportion of MYC-dependent mRNAs for each ETC complex. n = 3 biological replicates in each group. P values were calculated using a hypergeometric test; *, P < 0.05. (c and d) RF tracks exemplify MYC-dependent translation of UQCRQ (c) and ATP6V0B (d) transcripts in presence/absence of MYC. n = 3 biological replicates in each group. (e–g) Confirmation by immunoblot for indicated protein components of the ETC in lysates of P493-6 cells treated with doxycycline (0.1 μg/ml; e), P493-6 and EB1 cells treated with shMYC (f), and P493-6 cells treated with doxycycline (0.1 μg/ml) and β-estradiol (1 μM) for 72 h (g). The immunoblotting experiment was performed more than three times as biological replicates. A representative experiment is shown.
We confirmed the MYC effect on selected proteins in P493 and a second lymphoma line (EB1) using shRNA knockdown of MYC. We readily detected down-regulation of the MYC mRNA and protein in both P493 and EB1 cells (Fig. 2 f and Fig. S2 b). Comparison of immunoblot and qRT-PCR confirmed a translational effect on key proteins (Fig. 2 f and Fig. S2, c and d). As expected, MYC knockdown also caused a decrease in proliferation in both lines (Fig. S2, e and f). Changes in cell growth affect mRNA translation (Cornelis et al., 2000; Stumpf et al., 2013; Marques-Ramos et al., 2017), and in order to separate MYC effects on translation form its effects on cell proliferation, we treated P493 cells with β-estradiol, which restores proliferation in that cell line in the absence of MYC (Kempkes et al., 1995; Fig. S2, g and h). Estradiol treatment was unable to restore the translation of key translational targets (AKT, NDUFA1, EIF2B1, ATP6VOB); it did, however, restore expression of the UQCRQ and COX5 proteins in the absence of mRNA changes, suggesting that effects on some proteins reflected cell-cycle changes. However, we also noticed that estradiol treatment increased MYC expression (Fig. 2 g and Fig. S2 i). A related experiment using FBS to block growth in the MYC ON state showed a decrease in endogenous MYC levels (not shown). Together, these data illustrate the experimental difficulties of separating cell-cycle effects of MYC from changes in translation.
5′UTR motifs mark MYC sensitive mRNAs
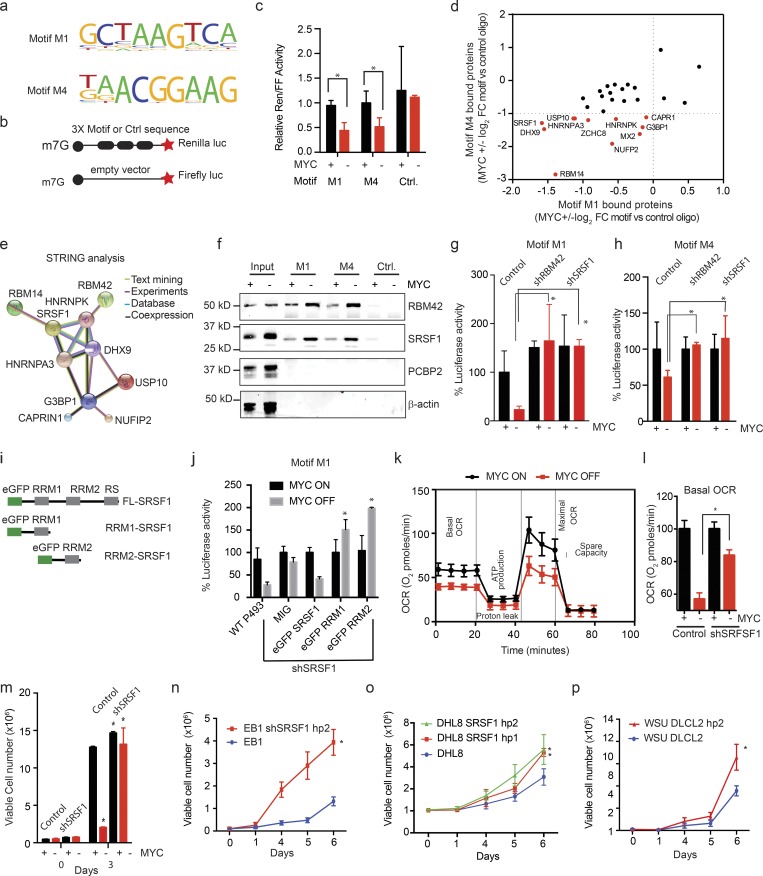
To understand their translational control, we searched for mRNA sequence motifs that were over- or underrepresented among MYC-dependent transcripts. We compared the TE-up group (882 mRNAs; 865 with annotated 5′UTRs) and the TE-down group (315 mRNAs; 308 with annotated 5′UTRs) to each other and to a background list of 1,537 mRNAs with annotated 5′UTRs that were equally expressed and that showed no significant change in their TE (Table S1). Our unbiased search using the DREME (Discriminative Regular Expression Motif Elicitation) algorithm (Grant et al., 2011) identified four 5′UTR sequence elements that were significantly enriched in the TE-up group (M1: P = 0.001; M2: P = 0.00004; M3: P = 0.0002; M4: P = 0.008; Fig. S3 a and Table S2). Motifs M1 and M4 showed corresponding and significant depletion in the TE-down group (M1: P = 0.005; M4: P = 0.01; Fig. 3 a and Table S2). We did not observe any differentially represented motifs in the coding sequences or 3′UTRs and saw no significant differences in 5′UTR length, guanidine/cytidine (GC) content, or known translation regulatory elements (Pelletier and Sonenberg, 1988; Meyuhas, 2000; Thoreen et al., 2012; Wolfe et al., 2014; Fig. S3, b–d). We tested how MYC affected these 5′UTR sequences in a dual luciferase translation reporter assay using a construct with three repeats of each motif compared with a scrambled control sequence and a cap-firefly luciferase reporter to normalize transcriptional effects. Briefly, translation from motifs M1 and M4 was sensitive to MYC, whereas scrambled sequences or motifs M2 and M3 were not affected by MYC (P < 0.05; n = 3; Fig. 3, b and c; and Fig. S3 e). Comparison with a reported list of cell cycle (G1)–regulated mRNAs (Stumpf et al., 2013) showed little overlap, and motifs M1 and M4 were present in only a small fraction (17 of 362 and 8 of 362, respectively; Table S2 e). Hence, MYC expression stimulates the translation of mRNAs with specific 5′UTR sequence elements.
Figure 3.
MYC-dependent mRNAs have 5′UTR SRF1/RBM42 binding elements. (a) Motifs M1 and M4 are significantly enriched in MYC-dependent mRNAs and confer MYC sensitivity in reporter assays; motifs M2 and M3 are enriched and did not perform in the reporter assay (sequence in Table S2); n > 3 replicates. (b) Schematic of the translation reporter assay to test MYC responsiveness of 5′UTR motifs relative to a capped firefly luciferase reporter construct of equal length and GC content. (c) Translation reporter comparing the ratio of Renilla luciferase expressed under control of a 5′UTR with three repeats of the indicated motif and a capped firefly luciferase reporter construct of equal length and GC content in P493-6 cells under MYC ON and OFF (mean ± SD; n = 3 biological replicates, performed as five independent experiments). P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05. (d) Mass-spectrometric identification of proteins with MYC-dependent binding to motifs M1 and M4 compared with random RNA sequence (fold change [FC] of proteins showing >5 peptides detected at an FDR <0.1% are included); n = 1 biological sample in each group. FDR values were calculated using the original Benjamini and Hochberg method. (e) STRING analysis of interactions between proteins identified in mass spectrometry analysis shown in panel d. (f) Biotin pull down and immunoblot show MYC-dependent binding of RBM42 and SRSF1 to RNA oligomers encoding motif M1/M4 versus the control sequence in P493-6 cells; PCBP2 and β-actin are controls. n = 3 biological replicates. (g and h) Effect of knockdown of SRSF1 or RBM42 versus control shRNA on MYC-dependent luciferase translation controlled by motif M1 (g) or motif M4 (h); assay as shown in panel b. Values are mean ± SD. n > 3 replicates, representative data are shown from three independent experiments. (i) Schematic of wild-type and RRM-deleted versions of SRSF1 expression constructs fused with enhanced GFP (eGFP). (j) Ectopic expression of wild-type SRSF1 could rescue the luciferase activity mediated by motif M1 in SRSF1 knockdown P493 cells while the RRM1- and RRM2-deleted SRSF1 protein could not mediate the translation repression. n = 3 replicates, mean ± SD from three replicates, representative data are shown from five independent experiments. P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05. (k) Mitochondrial stress assay to measure oxygen consumption rate (OCR) in MYC ON and OFF P493 cells. n = 4 replicates, mean ± SD from four replicates. Representative data are shown from three independent experiments. (l) Basal OCR in P493 control and shSRSF1-stable cells treated with tetracycline (0.1 μg/ml; 24 h). n = 4 replicates, mean ± SD from four replicates. Representative data are shown from three independent experiments. P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05. (m–p) Cell proliferation assays in indicated lymphoma cell lines, MYC ON and OFF in control and shSRSF1 P493-6 cells (m), control and shSRSF1 EB1 (n), DHL8 (o), and WSU-DLCL2 cells (p). n = 3 replicates in each group, mean ± SD from three replicates. Representative data are shown from three independent experiments. P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05. Ctrl., control.
An MYC-sensitive translation repressor complex affects lymphoma cell behavior
To identify specific and MYC-dependent proteins bound to sequences M1 and M4, we performed RNA immunoprecipitation followed by mass spectrometry. We identified 55 and 51 proteins that show MYC-sensitive binding to M1 and M4, respectively. Notably, 29 of these proteins bound both sequences and did not interact with a control oligomer (Fig. 3 d and Fig. S3, f–h). A STRING analysis (Functional Protein Association Network; Szklarczyk et al., 2017) predicted that 10 of the 29 proteins are components of an SRSF1/RBM42 protein complex (Fig. 3 e). Consistently, the TOMTOM motif comparison tool available at MEME Suite predicted binding sequences for SRSF1 and RBM42 that include motifs M1 and M4 (Gu et al., 2009; Byron et al., 2012; Table S3). We experimentally tested these by RNA immunoprecipitation using biotinylated RNA oligomers encoding M1 and M4 followed by immunoblot and confirmed MYC-sensitive binding of SRSF1 and RBM42 to M1 and M4, while an unrelated RNA-binding protein (PCBP2) showed no binding (Fig. 3 f). SRSF1 has been shown to cooperate with MYC and mTOR to induce tumorigenesis by positively regulating the translation; however, it has been suggested in many reports that under stress conditions SRSF1 can associate with mRNP complex and act as translation repressor (Sun, 1987; Karni et al., 2007; Delestienne et al., 2010; Anczuków et al., 2012; Das and Krainer, 2014). We did not observe any changes in total SRSF1 protein following MYC inactivation in P493 cells (Fig. S3 i). Moreover, knockdown of either SRSF1 or RBM42 with two independent shRNAs significantly reduced MYC-dependent luciferase reporter translation from M1 (P < 0.05; n = 3 MYC OFF control vs. shRNA) and M4 sequence elements (P < 0.05; n = 3; MYC OFF control vs. shRNA) without affecting a scrambled 5′UTR reporter (Fig. 3, g and h; and Fig. S3, j–o).
To explore the role of different domains within SRFS1, we generated truncation mutants lacking the RRM1 and RRM2 domains. We expressed either wild-type or the mutant e-GFP-SRSF1 constructs in P493 cells transduced with an shRNA specific for the endogenous for SRSF1 and measured the translation mediated by motif M1 and M4 (Fig. 3 i). While ectopic expression of wild-type e-GFP-SRSF1 rescued translation inhibition, neither RRM deletion was able to restore translation repression, indicating that RRM domains are required for translation repression (Fig. 3 j and Fig. S3 p).
We next explored the metabolic implications of MYC-dependent translation. As expected, MYC inactivation readily resulted in a loss of mitochondrial respiration as measured by the oxygen consumption rate in MYC ON and OFF states in P493 cells (Fig. 3 k). This effect depended on SRSF1 and RBM42 such that shRNA-mediated knockdown of either factor largely restored cellular respiration and cell growth (Fig. 3, l and m; and Fig. S3 q). We confirmed these effects in additional lymphoma cell lines (EB1, DHL8, WSU, and DLCL2) and consistently observed an increase in cell numbers upon SRSF1 knockdown (Fig. 3, n–p; and Fig. S3 r). Hence, MYC-sensitive 5′UTR binding of a translation repressor complex including SRSF1 and RBM42 affects cell metabolism and proliferation in lymphoma cells.
MYC changes translation initiation sites (TISs) and open reading frames (ORFs)
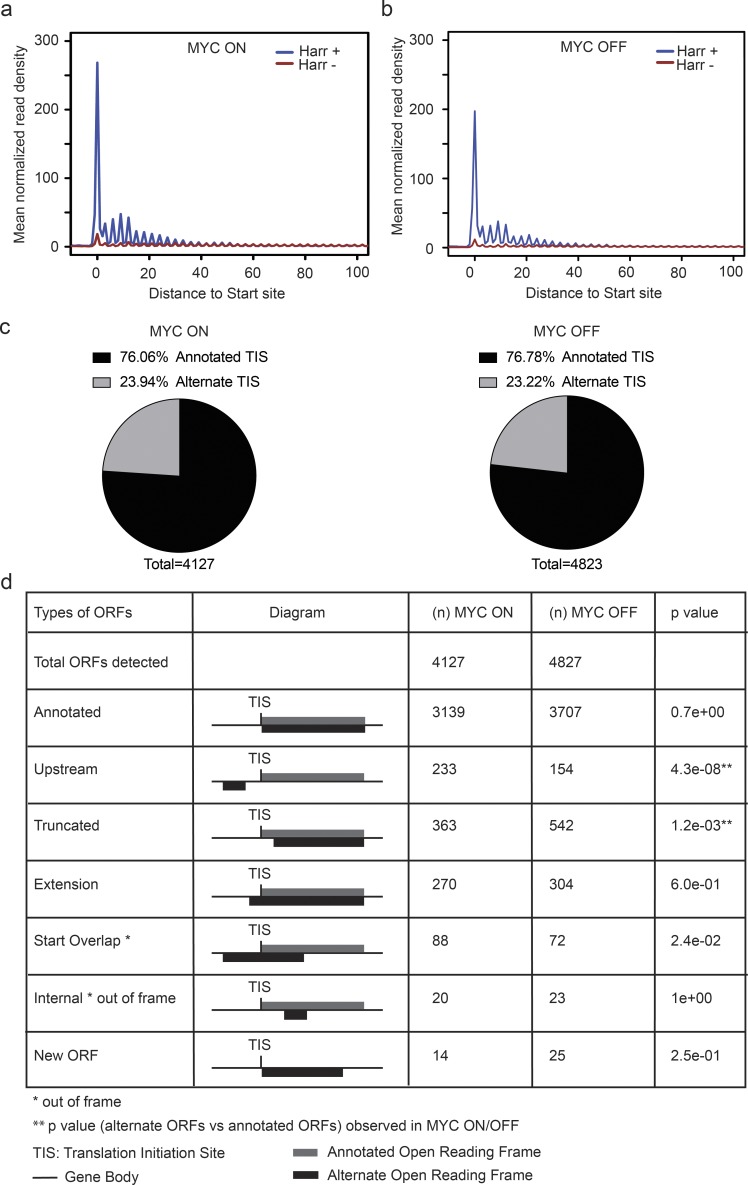
Next, we wanted to explore to what extent MYC affects translation start sites and potentially the integrity of ORFs. We experimentally mapped TISs in the presence and absence of MYC by performing ribosome profiling in the presence of harringtonine. Briefly, harringtonine arrests the initiating ribosomes, and this is readily detectable as an RNase I–protected sequence peak overlaying the actual start site (Fresno et al., 1977; Robert et al., 2009; Fig. S1 a). We performed the experiment in triplicates, removed one outlier sample from further analyses (Fig. S4 a), and discarded irrelevant reads; the final read counts are summarized in Table S4. A metagene analysis (for positions −2 to +90) confirmed a robust harringtonine-induced arrest (Fig. 4, a and b). Briefly, we determined the peptidyl (P-site) offset for different read lengths by aligning the ribosome-protected reads to the annotated AUG start codons (Fig. S4 b). On average the P-site offset was 12 nucleotides, and we used this number to identify alternate TISs (ATISs; Fig. S4 c). In both conditions, most transcripts initiated from a single TIS (Fig. S4, d and e). We used the ORF-RATER algorithm to identify all consensus and variant TIS in each condition; the program identifies TISs based on a ribosome-protected RNA sequence peak and the presence of a potential start site NUG, where N represent A/T/G/C. Briefly, we grouped annotated RNA isoforms that share a genomic position on the same strand into “transcript families.” We used an ORF-RATER score > 0.8 as a significant cutoff and used only these ORFs for further analyses (Fields et al., 2015; Table S5).
Figure 4.
MYC affects TIS choice. (a and b) Metagene analysis of TIS detection in the presence and absence of MYC under harringtonine (Harr)-induced translation arrest (2 μg/ml; 2 min). Ribosome densities were averaged after aligning the gene density profile at the TIS to obtain the mean normalized read density. n = 3 biological replicates in each group. (c) Annotated and ATIS in all ORFs detected in MYC ON and OFF samples. n = 3 biological replicates in each group. (d) Annotated and alternate ORFs detected in the presence/absence of MYC; significance by Fisher’s exact test. n = 3 biological replicates in each group.
We noticed a surprising variation in actual versus predicted TISs. The predicted TIS was the first consensus AUG start codon and gave rise to a functional ORF and protein; the ATIS reflects actual ribosome accumulation upon initiation arrest with harringtonine. Overall, we detected ∼23% of ATISs in both MYC conditions (Fig. 4 c). Generally, in the presence of MYC, we detected a significant (P = 4.3 × 10−08) increase in the usage of 5′ upstream ATISs that corresponded to upstream ORFs (uORFs) and new ORFs that overlapped with the annotated start sites (Fig. 4 d, Fig. S4 f, and Table S5). This change was also detected by increased 80S ribosome coverage across 5′ upstream mRNA sequences (Fig. S4 g). Conversely, as a general rule with some exceptions, we saw a significant (P = 1.2 × 10−03) shift to an ATIS downstream (3′) from the annotated site in the absence of MYC; the latter is expected to give rise to N-terminal truncations (Fig. 4 d, Fig. S4 f, and Table S5). This change was also reflected in the start codon choice, and alternate ORFs typically initiated from near cognate CUG, GUG, or UUG codons instead of the annotated AUG codon (P < 0.05; Fig. S4, h and i). Hence, high- and low-MYC conditions lead to surprising usage of up- and downstream ATIS, respectively.
Variant ORFs result in abnormal proteins
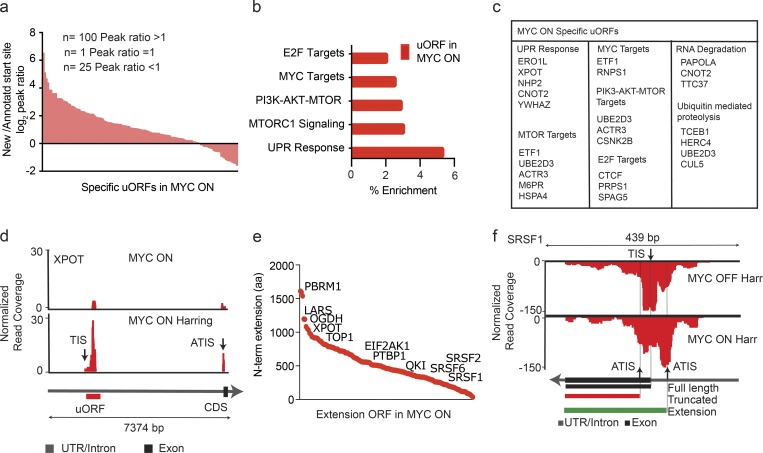
First, we examined ATIS usage in high-MYC conditions. Read count ratios from the annotated TIS and ATIS indicated that, when both were present in a transcript, the ATISs were preferred (80%) over the annotated TIS (20%; Fig. 5 a). We do not know the biological relevance of these extended variant ORFs (ntotal = 233, nspecific = 157; Fig. 4 d, Fig. S4 f, and Table S5). However, GO analysis indicated a significant enrichment for genes that were also MYC and E2F transcriptional targets (Fig. 5, b and c). Recent studies indicate that under stress conditions uORFs enhance the translation of the downstream ORF (Vattem and Wek, 2004; Sendoel et al., 2017). Consistently, we observed that the presence of a uORF is linked to increased or unchanged, but never to reduced, TE (Fig. S4 j). One example is the XPOT gene encoding exportin-T, a nuclear exporter of aminoacylated transfer RNAs, which gains a prominent uORF in high-MYC states (Arts et al., 1998; Fig. 5 d). In other instances, MYC activation leads to 5′ extended ORFs that encode N-terminally extended proteins. This group includes many RNA-binding proteins including SRSF family members (Fig. 5 e). The SRSF1 transcript showed loss of initiation from the annotated TIS and relative increases of usage of both 5′ and 3′ ATIS (Fig. 5 f). The variable use of ATISs likely affected production of the functional protein.
Figure 5.
High-MYC conditions favor upstream translation initiation. (a) The peak height ratio of RF reads across the annotated TIS versus the uORF TIS indicates preferential uORF initiation for most uORF-containing genes in the MYC ON condition. n = 3 biological replicates in each group. (b) GO identifies categories of genes with MYC-activated uORFs. n = 3 biological replicates in each group. (c) List of genes harboring uORFs in the MYC ON state by KEGG category. n = 3 biological replicates in each group. (d) RF distribution with and without harringtonine for XPOT indicates uORF usage in the MYC ON state; black and red arrows indicate the predicted (TIS) and ATIS, respectively. n = 3 biological replicates in each group. (e) MYC-induced 5′ extended ORFs ranked by the number of additional N-terminal amino acids. n = 3 biological replicates in each group. (f) RF read distribution across the SRSF1 transcript in high and low MYC indicates variable ATIS usage. Harr indicates harringtonine arrest. Black arrows indicate predicted TIS and ATIS, respectively. Exons shown as black squares. n = 3 biological replicates in each group.
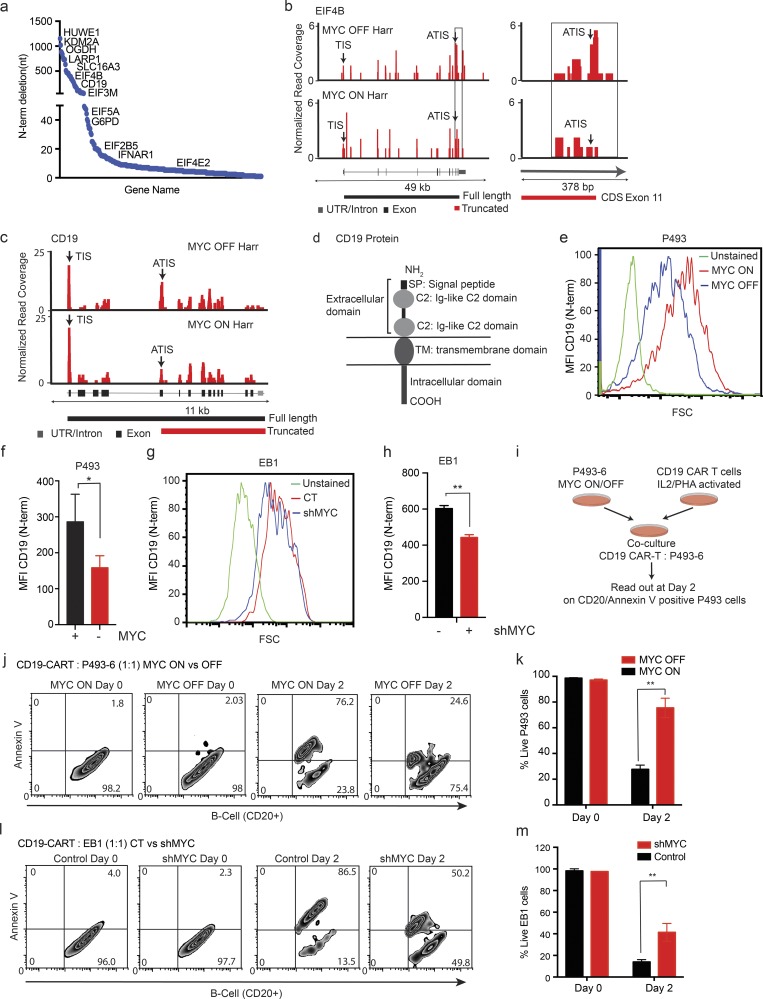
The absence of MYC favored initiation from an ATIS downstream (3′), and this would cause N-terminal truncations (Fig. 4 d, Fig. S4 f, and Table S5). Generally, the ATIS in low-MYC conditions were typically located between 3 and 5,038 nucleotides downstream from the annotated AUG, they were typically CUG or GUG codons, and in low-MYC states, the ATIS was preferred (70%) over the annotated TIS (30%; Fig. S5, a–c). Some examples of N-terminally truncating start sites include mTOR and translation regulators such as LARP1, eIF4B, eIF3M, eIF2B5, eIF4E2, and eIF4B (Fig. 6 a and Table S5). The ORF-RATER identified the new ATIS based on an RF peak, and the presence of a potential start site; for example, eIF4B acquired a new ATIS in exon 11 resulting in a truncated protein that loses all relevant RNA-binding domains (Shahbazian et al., 2010; Fig. 6 b).
Figure 6.
Low-MYC expression favors downstream start sites that lead to functional N-terminal truncations. (a) Genes ranked by the distance of ATIS from TIS resulting in truncated ORFs. n = 3 biological replicates in each group. (b) RF distribution across the eIF4B transcript under harringtonine (Harr) treatment in MYC ON and OFF states. Arrows indicate the predicted TIS and the ATIS, the zoomed-in region on the right illustrates differential usage of the ATIS in MYC ON and OFF states. n = 3 biological replicates in each group. (c) RF distribution across the CD19 transcript in MYC-high and -low states shows prevalent use of the ATIS in exon 5 in the MYC OFF state. Representative samples are shown from three biological replicates analyzed. (d) Schematic of the CD19 receptor protein; exons 1–5 encode the ectodomains of CD19. (e–h) FACS analysis of surface CD19 using an N-terminal–specific antibody in the presence/absence of MYC in P493-6 and EB1 cells. MFI represents CD19 expression across the cell population in P493-6 (f) and EB1 (h). n = 3 biological replicates in each group, mean ± SD from three replicates. Representative data are shown from three independent experiments. P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05; **, P ≤ 0.001. (i) Schematic of the co-culture experiment of P493-6 cells (CD20 positive) and CAR-T cells (CD3 positive). (j) Example of FACS analysis of CD20-PE/Cy5, Annexin V (Alexa Fluor 647), and CD3-FITC staining to measure P493-6 cell death induced by CD19-directed CAR-T cells in the co-culture experiment. n = 3 biological replicates in each group. Representative data are shown from five independent experiments. (k) Quantification of CD20/Annexin V–positive P493-6 cells on day 0 and day 2 of the co-culture experiment showing eradication of MYC high P493-6 cells and significant protection of MYC low P493-6 lymphoma cells. n = 3 biological replicates in each group, mean ± SD from three replicates. Representative data are shown from five independent experiments. P values were calculated using an unpaired Student’s t test: **, P ≤ 0.001. (l) Example of FACS analysis of CD20-PE/Cy5, Annexin V (Alexa Fluor 647), and CD3-FITC staining to measure cell death induced in control and shMYC EB1 cells by CD19-directed CAR-T cells in the co-culture experiment. n = 3 biological replicates in each group. Representative data are shown from five independent experiments. (m) Quantification of CD20/Annexin V–positive EB1 cells on day 0 and day 2 of the co-culture experiment showing eradication of control EB1 cells with significant protection of shMYC EB1 lymphoma cells. n = 3 biological replicates in each group, mean ± SD from three replicates. Representative data are shown from five independent experiments. P values were calculated using an unpaired Student’s t test: **, P ≤ 0.001.
The B cell surface receptor CD19 illustrates the biological and clinical relevance of an alternate MYC-dependent start-site choice in lymphoma cells. Briefly, in the low-MYC state, the ORF-RATER detected a candidate new ATIS in exon 5 that showed an RF peak and a potential start codon and that would lead to loss of all extracellular domains (Fig. 6, c and d). We confirmed the prediction by FACS using an antibody that recognizes the N-terminal CD19 ectodomain, and that revealed a loss in mean fluorescence intensity (MFI) by 50%; moreover, 20% of MYC low P493-6 and EB1 cells were completely FACS negative for the CD19 ectodomain (P < 0.05, n = 3, MYC ON vs. OFF, control vs. shRNA-mediated knockdown of MYC (shMYC); Fig. 6, e–h; and Fig. S5, d and e). This change is potentially important because immunotherapies for lymphoma target the CD19 receptor that is expressed on malignant and mature B cells but absent on precursor cells, thus enabling regeneration of the lineage (Kochenderfer et al., 2015). Loss of CD19 expression on lymphoma cells has been implicated in resistance to CD19 chimeric antigen receptor (CAR)–T cells (Otero et al., 2003; Sotillo et al., 2015; Perna and Sadelain, 2016). We tested the effect of high- and low-MYC (with or without doxycycline) expression on lymphoma cell killing by CAR-T cells in a co-culture experiment (Fig. 6 i). CD19-directed CAR-T cells (CD3 positive) and P493 lymphoma cells (CD20 positive) were readily detected and separated by FACS, and neither showed significant cell death in the initial co-culture by Annexin V stain (Fig. S5, f and g). After 2 d of co-culture, >75% of MYC low P493-6 cells were still viable, whereas in the high-MYC condition, only ∼25% remained viable (shown as CD20+/Annexin V+ population gated on CD20+ population; Fig. 6, j and k; and Fig. S5 h). We observed the same difference in EB1 cells with an shRNA against MYC that impaired CD19–CAR-T cell–induced cell death (Fig. 6, l and m; and Fig. S5, i and j). Together, these results indicate that reduced MYC levels lead to translation of a truncated CD19 receptor protein that enables escape from CD19-directed immunotherapy for lymphoma.
Discussion
Our findings provide new insight into the biological activity of MYC and its effect on mRNA translation. We mapped the global and gene-selective effects of MYC activation on mRNA translation. Global changes largely parallel transcript abundance and may contribute to the proliferation-related effects and the dauer-like state of biosynthetic dormancy observed upon MYC inactivation in murine stem cells (Scognamiglio et al., 2016). Intriguingly, MYC also has selective effects on the translation of specific RNAs that exceed the increase in their transcription. MYC is known to induce expression of key translation factors such as eIF4E (Lin et al., 2008), and we expected to see this reflected as an “MYC-eIF4E translational regulon”; however, the mRNAs that respond to MYC and the predominant sequence motif indicate different or additional mechanisms. MYC also has broad effects on cell behavior, growth, and proliferation, and while it is difficult to completely separate these effects experimentally, we find that MYC directly affects populations of mRNAs that are overlapping but different from cell cycle–responsive mRNAs, and this is reflected in their enrichment for different sequence motifs (Polymenis and Schmidt, 1997; Thoreen et al., 2012; Stumpf et al., 2013; Truitt et al., 2015; Marques-Ramos et al., 2017).
MYC-dependent mRNAs are marked by specific sequence elements that bind an SRSF1/RBM42-containing complex in an MYC-dependent manner. The role of SRSF1 in translation is complex, and prior work suggests that it acts as a translation in an enhancer when mTOR is active (Sanford et al., 2005; Karni et al., 2008; Anczuków et al., 2012; Das and Krainer, 2014); by contrast, when mTOR is inactive or under stress, SRSF1 is thought to repress translation in a 3′UTR-dependent manner (Delestienne et al., 2010; Sun et al., 2010; Twyffels et al., 2011). Our data indicate that in low-MYC conditions, a complex containing SRSF1, RBM42, and others binds to specific 5′UTR sequences and represses the translation of those mRNAs. We do not yet know exactly how MYC regulates the SRSF1 protein complex, although we find MYC affects the SRSF1 TIS, and this may affect SRSF1 activity.
MYC’s effect on translation impacts cell metabolism and proliferation. Specifically, MYC affects the translation of the majority of nuclear-encoded protein components of the mitochondrial electron transport chain (ETC), indicating an important role in metabolic adaptation. This may reflect coordinated regulation of proteins within these complexes. Prior work has focused on transcriptional control of mitochondrial genes in response to MYC (Li et al., 2005; Dang et al., 2006; Das et al., 2012); however, at an early (24-h) time point, we did not observe significant changes in transcript levels. Instead, we detected increased translation of existing mRNAs corresponding to increased and SRSF1/RBM42-sensitive cellular respiration and lymphoma cell proliferation. Together, in lymphoma cells, a regulatory model emerges where MYC controls the translation of key metabolic genes through a repressive SRSF1/RBM42 protein complex that binds specific 5′UTR sequences.
MYC has surprising and profound effects on the choice of translation start sites in mammalian cells. We found that TIS choice affects the integrity of ORFs and, therefore, the production of functional proteins. In general, we observed that MYC activation shifts TIS upstream from the annotated site, whereas low-MYC states correspond to the opposite effect. The 5′ shift activates uORFs and produces overlapping and 5′ extended ORFs, whose biological activities are not yet known. The 5′ truncation seen in low-MYC conditions deletes important functional domains and may also affect protein stability. For example, the eIF4B translation initiation factor loses all of its RNA-binding domains and is unlikely to retain activity. Immediately relevant to lymphoma therapy is the effect on the CD19 cell surface receptor. Under low-MYC conditions, we observed predominant usage of a downstream TIS that results in loss of all receptor ectodomains. This change impairs detection by antibodies against the CD19 N-terminus, and it also protects lymphoma cells from attack by CD19-directed CAR-T cells. Clinically, loss of surface CD19 has been linked to resistance to CAR-T cell therapy for lymphoma although the molecular mechanism has not been defined (Sotillo et al., 2015; Perna and Sadelain, 2016). Our results indicate that alternate translation start-site choice can lead to the expression of abnormal cell surface receptors. Altogether, we found physiologically relevant effects of MYC levels on the efficiency of mRNA translation and the integrity of ORFs and proteins.
Materials and methods
Human cell lines and plasmids
The lymphoma cell lines P493, EB1, DHL8, and WSU-DLCL2 were maintained in Roswell Park Memorial Institute 1640 supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin-streptomycin. When indicated, respective cell lines were transduced with lentiviruses expressing empty vector (pLKO.1) or shRNA against MYC (pLKO.1; Sigma-Aldrich, shMYC: TRCN0000174055), SRSF1 (shSRSF1 hp1: TRCN0000001095; and shSRSF1 hp2: TRCN0000001096), and RBM42 (shRBM42 hp1: TRCN0000148512; and shRBM42 hp2: TRCN0000180790). cDNA constructs for SRSF1 were previously described (Paz et al., 2015). Full-length and mutant cDNA for SRSF1 was expressed by transient transfection using FuGENE reagent. Cell lines were authenticated by short tandem repeat DNA profiling by Bio-Synthesis (https://www.biosyn.com/cell-line-authentication.aspx). Mycoplasma contamination is routinely tested with the Universal Mycoplasma Detection Kit (American Type Culture Collection).
Ribosome footprinting
Human B cell lymphoma P493-6 cells were treated with doxycycline (0.1 μg/ml) for 24 h and/or harringtonine (1 μg/ml) for 2 min followed by cycloheximide treatment for 10 min. Total RNA and RFs were isolated following published protocol (Ingolia et al., 2009). Deep sequencing libraries were generated from these fragments and sequenced on the HiSeq 2000 platform. Genome annotation was from the Ensembl database release 75.
Metabolic labeling of nascent protein (non-radioactive)
P493-6 cells were labeled for nascent protein synthesis using Click-iT AHA metabolic labeling reagent (C10102; Invitrogen) per the manufacturer’s instructions. Briefly, following doxycycline treatment (0.1 μg/ml), cells were incubated in methionine-free medium for 30 min before AHA labeling for 1 h. Cells were fixed with 4% paraformaldehyde in PBS for 15 min, permeabilized with 0.25% Triton X-100 in PBS for 15 min followed by one wash with 3% BSA. Cells were then stained using Alexa Fluor 488 Alkyne (A10267; Invitrogen) with Click-iT Cell Reaction Buffer Kit (C10269; Invitrogen). Changes in MFI as a measure of newly synthesized protein were detected by Flow cytometric analysis.
Sequence alignment
The human genome sequence GRCh37 was downloaded from the Ensembl public database. RF reads were aligned to reference genome GRCh37 using STAR (Grant et al., 2011). STAR clips the linker sequence (5′-CTGTAGGCACCATCAAT-3′), which is technically introduced during RF library construction, and trims the remaining sequence from the 3′ end while aligning the reads to reference sequence. Briefly, we set the parameters for PALMapper as follows: maximum number of mismatches: 2; minimum aligning length: 15; maximum intron length (splice alignment): 10,000. We used only the uniquely aligned reads for further analysis. To remove ribosomal RNA, the footprint reads were also aligned to a ribosome sequence database using STAR with the same parameters except not allowing splice alignment. We retrieved the human ribosome sequences from SILVA (Quast et al., 2013) databases. The FASTA file was used as a reference sequence to align against. The rRNA-aligned reads were filtered out from GRCh37-aligned reads. Briefly, we removed reads mapping to ribosomal and noncoding RNAs, library linker oligomers, and sequences that aligned incompletely to the reference genome (Wolfe et al., 2014; Zhong et al., 2017).
After removing the ribosomal RNA, we still observed a portion of reads that was dominated by the linker sequence and Illumina P7 adapter. These reads can also be trimmed during mapping and cause false alignment. Therefore, we searched a string of 1∼8 nt from linker sequence around the trimming site (±2 bp) allowing 1 nt mismatch. We removed the read if there was no such linker sequence. Finally, we filtered out reads ≤24 bp and ≥36 bp, and the remaining reads with an aligned length from 25 to 35 bp were used to analyze the translational effects of MYC. The total number of RF reads mapped to exons was 11–12 million in control (MYC ON) and 13–15 million in doxycycline-treated (MYC OFF) samples, corresponding to 20,356 proteins coding genes.
Total mRNA sequencing reads were aligned to the GRCh37 reference using STAR (Dobin et al., 2013). We performed the splice alignment and used only the uniquely aligned reads with a maximum of three mismatches. rRNA-contaminating reads were also filtered out using the same strategy described before.
ORF analysis using harringtonine data
We predicted the annotated and alternative ORFs for MYC ON and OFF samples using ORF-RATER (Fields et al., 2015). Both untreated and harringtonine-treated samples were used to perform the prediction analysis, in which harringtonine ON samples contributed to the prediction of the TIS, and harringtonine OFF samples contributed to the prediction of both the TIS and translation termination sites. We extracted the reliable uORFs, truncated ORFs, extension ORFs, start-overlap ORFs, internal ORFs, new ORFs, and annotated ORFs using 0.8 as the score cutoff suggested by Fields et al. (2015). MYC ON– and MYC OFF–specific ORFs are those ORFs that were detected only in MYC ON or MYC OFF samples, respectively.
Alternative and annotated initiation site peak ratio analysis
For genes with ATISs, we compared the relative translation level of the alternative ORF and its corresponding annotated ORF using the peak ratio of the translation start site. We extracted the sum of the footprint read counts in the region of −30 nt to +30 nt relative to the ATIS and annotated TIS in the harringtonine-treated samples. We calculated the peak ratio of an alternative ORF as the reads ratio of the ATIS to the annotated TIS.
Metagene analysis
For each gene, we calculated the mean ribosome footprint density across the positions on the longest transcript of a gene with ≥64 footprint read counts. We normalized the positional footprint density of each gene by the average footprint density. Then we scaled both the 5′UTRs and CDSs of genes to an equal number of windows and calculated the averaged signals across all genes as the metagene profile. We plotted the final metagene profile by averaging the metagene profiles across replicates.
CDS ribosome pause site analysis
Similarly, we used the normalized footprint density of the longest transcript of each gene that has ≥64 ribosome footprint read counts for the CDS ribosome pause-site analysis. We defined the normalized codon density as the sum of the normalized footprint density of the nucleotides at positions −1, 0, and +1 relative to the first nucleotide of that codon. We considered a codon as a ribosome pause if the codon density was ≥150. This cutoff was decided by the 0.1% quantile of the normalized codon density distribution along the CDSs. CDS ribosome pause sites were excluded if they are within 5 codons to the TIS or the translation stop sites. We aligned the metagene profile of the flanking region around the ribosome pause sites and plotted the averaged signals.
CDS ribosome pause-site motif analysis
We extracted the peptide sequences of the flanking region around the pause sites as the positive sequence set for the motif analysis. We choose the 5-peptide upstream and downstream flanking region to the pause site, which is 11 peptides in total including the pause-site peptide. We generated a set of random regions with the same size outside the flanking regions of the pause sites and extracted the peptide sequences of these random regions as the negative sequence set. Then, we did the motif analysis based on the positive and negative sequence sets and plotted the motif probability logos using kpLogo (Wu and Bartel, 2017).
Footprint profile analysis using Ribodiff
For each gene, we only counted the number of aligned reads that were mapped within exonic regions. The genome annotation was downloaded from the Ensembl database. We used our recently developed tool Ribodiff to analyze the ribosome footprinting data. Ribodiff uses a negative binomial-based generalized linear model to detect the significant difference of footprint read counts in two conditions, taking the RNA sequencing transcriptional measurements as the confounding factor. Therefore, it is capable of identifying the genes having translational control that is independent of the transcriptional regulation. The statistical test is a χ2 test on the deviances of the H0 and H1 model fitting. The Benjamini–Hochberg procedure was used to obtain the false discovery rate (FDR). The ribosomal distribution curves for a single gene were plotted in a similar way but without normalizing the read coverage, and the coverage was smoothed using the “moving average” smoothing algorithm.
UTR motif analysis
The transcripts of each gene were quantified based on the total mRNA sequencing data using MISO (Katz et al., 2010; Grant et al., 2011). The 5′UTR of the most abundant transcript was collected for predicting motifs. Both the significant genes with increased or decreased TE and altered ribosomal distribution and the corresponding background gene sets were predicted by DREME (Grant et al., 2011). We considered the predicted consensus sequences with P < 1 × 10−4 as significant motifs. Motif occurrences were called using FIMO (Grant et al., 2011) with default parameters for strand-specific prediction in human and other species using gene orthologues. 5′UTR sequences for the respective group of targets were subjected to motif prediction using the online available program RegRNA (a regulatory RNA motifs and elements finder; http://regrna.mbc.nctu.edu.tw/html/prediction.html) and looked specifically for motifs that occur in 5′UTR. Statistical significance for the results obtained was calculated using Fisher’s exact test for count data.
Luciferase assays
Three tandem repeats of the identified motifs or random sequence were cloned into the 5′UTR of Renilla luciferase plasmid pGL4.73. Empty firefly luciferase plasmid pGL4.13 or empty firefly plasmid was used as an internal control. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (E1960; Promega) following the manufacturer’s instructions.
Motif sequences were as follows: 3X TE DOWN M1: 5′-AGCTTTCCCAGGCACGCTAAGTCATCCAGGACTATCCCAGGCACGCTAAGTCATCCAGGACTATCCCAGGCACGCTAAGTCATCCAGGACTAC-3′; 3X TE DOWN M2: 5′-AGCTTTCCCAGGCACGACCAGATTCCAGGACTATCCCAGGCACGACCAGATTCCAGGACTATCCCAGGCACGACCAGATTCCAGGACTAC-3′; 3X TE DOWN M3: 5′-AGCTTTCCCAGGCACTAGGAACTCCAGGACTATCCCAGGCACTAGGAACTCCAGGACTATCCCAGGCACTAGGAACTCCAGGACTAC-3′; 3X TE DOWN M4: 5′-AGCTTTCCCAGGCACTAACGGAAGTCCAGGACTATCCCAGGCACTAACGGAAGTCCAGGACTATCCCAGGCACTAACGGAAGTCCAGGACTAC-3′; 3X NC: 5′-AGCTTTCCCAGGCACACGGCGGTCCAGGACTATCCCAGGCACACGGCGGTCCAGGACTATCCCAGGCACACGGCGGTCCAGGACTAC-3′. Underlined sequences show the motif M1, M4, and NC sequences.
Biotin pull down and mass spectrometry analysis
Biotinylated RNA oligos containing three tandem repeats of the identified motifs or random sequence were used for pull down. Briefly, 5 μg of denatured RNA biotinylated oligos were incubated with 500 μg total protein lysates obtained from P493-6 cells that were the control or treated with doxycycline (0.1 μg/ml; 24 h) in RNA immunoprecipitation buffer (150 mM KCl, 2.5 mM Tris [pH 7.5], 0.5 mM dithiothreitol, 0.5% NP-40, protease inhibitor cocktail [Roche], and SUPERase In [Invitrogen]) and streptavidin magnetic beads for 1 h at room temperature. RNA-bound proteins were eluted in 2× Lamelli buffer following three washes with RNA immunoprecipitation buffer (150 mM KCl, 25 mM Tris [pH 7.4], 0.5 mM dithiothreitol, 0.5% NP-40, supplemented with protease inhibitor cocktail; Roche). RNA-bound proteins were quantitatively detected with mass spectrometry analysis.
RNA oligo sequences were as follows: 3X TE DOWN M1 5′-biotin 3′biotin: 5′-TCCCAGGCACGCTAAGTCATCCAGGACTATCCCAGGCACGCTAAGTCATCCAGGACTATCCCAGGCACGCTAAGTCATCCAGGACTA-3′; 3X TE DOWN M6 5′-biotin 3′biotin: 5′-TCCCAGGCACTAACGGAAGTCCAGGACTATCCCAGGCACTAACGGAAGTCCAGGACTATCCCAGGCACTAACGGAAGTCCAGGACTA-3′; 3X NC 5′-biotin 3′biotin: 5′-TCCCAGGCACACGGCGGTCCAGGACTATCCCAGGCACACGGCGGTCCAGGACTATCCCAGGCACACGGCGGTCCAGGACTA-3′. Underlined sequences show the motif M1, M4, and NC sequences.
Mitochondrial stress analysis
The oxygen consumption rate was measured using Seahorse XF Cell Mito stress test kit (103015–100; Seahorse Bioscience) and XF-24 extracellular flux analyzer (Seahorse Bioscience) following the manufacturer’s protocol. Briefly, P493-6 cells untreated or treated with doxycycline (0.1 μg/ml; 24 h) were seeded in a Seahorse XF96 microplate coated with Cell-Tak (354240; Corning) in the XF assay medium. Half a million cells were incubated in a 37°C incubator without CO2 for 1 h to allow them to pre-equilibrate with the assay medium. The final concentrations of 1.0 µM oligomycin was used to inhibit mitochondrial respiration. A final concentration of 0.25 and 0.125 µM FCCP (trifluoromethoxy carbonylcyanide phenylhydrazone) was used to deplete mitochondrial membrane potential and measure maximum mitochondrial respiration capacity for control and doxycycline-treated samples, respectively.
qRT-PCR
Total RNA was extracted using the miRNeasy Mini Kit (217004; Qiagen). cDNA was made using SuperScript III First-Strand (18080–400; Invitrogen). Analysis was performed by ΔΔCt: Applied Biosystems Taqman GeneExpression assays: human SRSF1 (Hs00199471_m1), RBM42 (Hs00225667_m1), PCBP2 (Hs01590472_mH), ATP6VOB (Hs01072387_g1), UQCRQ (Hs00429571_g1), COX5A (Hs00362067_m1), NDUFA1 (Hs00244980_m1), AKT1 (Hs00178289_m1), EIF2B (hs00426752_m1), EIF4E (Hs04978771_m1), MYC (Hs00153408_m1), and actin (4352667).
Immunoblots
Lysates were made using TNN lysis buffer (50 mM Tris-Cl, 250 mM NaCl, 5 mM EDTA, and 0.5% NP-40 supplemented with protease inhibitor). 60 μg of protein was loaded onto SDS-PAGE gels and then transferred onto Immobilon-FL transfer membranes (IPFL00010; Millipore). The antibodies used were MYC, SRSF1, RBM42, PCBP2, UQCRQ, ATP6V0B, COX5A, EIF4E, AKT1, and NDUFA1 (Cell Signaling Technology) and β-actin (A5316; Sigma-Aldrich).
Cell proliferation assay
Human lymphoma cell lines P493-6, EB1, DHL8, and WSU-DLCL2 were used for the cell proliferation assay. Viable cells were quantified using Via-Count reagent (4000–0040; Millipore) detected by FACS analysis.
Cell cycle analysis
The human lymphoma cell line P493-6 was used for cell-cycle analysis following treatment with doxycycline (0.1 μg/ml) and β-estradiol (1 μM) for 72 h. Cell cycle reagent from Millipore (4700–0160) was used, and stained cells were acquired and analyzed using the Guava easyCyte instrument (EMD Millipore).
Flow cytometry analysis
We performed surface staining to detect CD19 expression using an N-terminal–specific antibody in P493-6 cells treated with doxycycline (0.1 μg/ml) for 24 h. Briefly, one million P493-6 cells per replicate were harvested, washed twice with 1× PBS, and stained with rabbit anti–human CD19 polyclonal antibody (MBS9204578; MyBioSource, LLC; 1:500 dilutions) for 10 min at room temperature followed by goat anti–rabbit Alexa Flour 594 secondary antibody (A11012; Life Technologies; 1:1,000 dilutions) incubation for 10 min at room temperature. Staining was done in 10% FBS in 1× PBS. For CD3 staining we used mouse anti–CD3-FITC antibody (555339; BD Pharmingen), and for CD20 staining we used mouse anti–CD20-PE/Cy5 antibody (302308; BioLegend). To detect apoptotic cell death, we used Annexin V Alexa Fluor 647 conjugate (A23204; Invitrogen). Live cells were stained with CD3-FITC antibody, CD20-PE/Cy5 antibody, and Annexin V Alexa Fluor 647 conjugate in Annexin binding buffer (79998; BioLegend) for 20 min at room temperature. Flow cytometry detection of CD19 staining was performed using the Guava easyCyte instrument. Data were analyzed using FlowJo software. We used two experimental replicates and performed experiments in three biological replicates. Statistical significance was calculated using a two-tailed Student’s t test.
CD19 CAR-T cell generation and co-culture experiment
Human blood samples were obtained from healthy donors after written informed consent and approved by the Human Biospecimen Utilization Committee (HBUC number HBS2009024 IRB; waiver number WA0210-09) and Institutional Review Board at Memorial Sloan Kettering Cancer Center on July 22, 2009. Human T cells were isolated from human peripheral blood mononuclear cells by density centrifugation and activated and expanded by culturing with CD3/CD28 Dynabeads (11131D; Gibco) in the presence of human recombinant IL-2 (PHC0023; Life Technologies) and phytohemagglutinin (11249738001; Sigma-Aldrich). The CD19-directed CAR construct was used to transduce T cells (Brentjens et al., 2013; Boice et al., 2016). Transduction was performed on RectroCectin (T201; Takara)-coated plates following the manufacturer’s instructions. Following T cell transduction, activated T cells were further used for the co-culture experiment. The cytolytic capacity of transduced T cells was determined by co-culturing them with target P493-6 or EB1 cells at particular cell ratios (Pegram et al., 2012). After 48 h of co-culture, cells were harvested and stained for CD3-FITC, CD20-PE/Cy5 antibody, and Annexin V Alexa Fluor 647 and assayed by flow cytometry to detect changes in CD20/Annexin V–positive P493-6 and EB1 viable cells.
Statistical analysis
RT-PCRs were analyzed with two-tailed Student’s t tests. The significance of motif enrichments was determined using a one-tailed binomial test with correction for differences in 5′UTR lengths. The P value for this test is defined as
where TPos respective to TNeg is the number of genes in the positive respective to negative (background) set, LPos respective to LNeg is the average length of the 5′UTR of the positive respective to negative set of genes, HPos respective to HNeg is the number of genes with the motif under consideration, and B (neg,pos)(i) is the probability for observing i events drawn from the distribution B (neg,pos). A hypergeometric test was performed to test for the significance in the enrichment of the gene overlap in the KEGG pathway. The total number of genes taken in the hypergeometric test was 8,000.
Accession number
The ribosome footprinting and total mRNA sequencing raw and processed data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession no. GSE79864.
Online supplemental material
Fig. S1 shows the experimental design, the cell line model, and the quality-control analysis of ribosome footprinting sequencing data. Fig. S2 shows the effect of MYC on the mRNA expression of mitochondrial respiration genes, cell proliferation, and cell cycle. Fig. S3 shows the sequence analysis of 5′UTR to identify MYC-regulated motifs, proteins binding to these motifs, and reporter assays to evaluate the role of SRSF1 and RBM42 on MYC-dependent translation. Fig. S4 shows the quality-control data, P-site analysis, types of ORFs detected, and distribution of start-site usage in the harringtonine-treated samples. Fig. S5 shows the truncated ORFs specific to the MYC OFF condition. It also shows the staining controls and the gating strategy for the CD20/CD3 costaining FACS experiment to determine CAR-T cell–induced cell death in P493-6 and EB1 lymphoma cells shown in Fig. 6. Table S1 contains lists of translational targets of MYC in MYC ON versus MYC OFF P493 cells and their 5′UTR sequences. Table S2 lists motifs identified in 5′UTR of mRNA transcripts that are translationally affected by MYC. Table S3 lists the prediction for RNA-binding proteins for motif M1 and M4 identified in 5′UTR of mRNA transcripts that are translationally affected by MYC. Table S4 lists ribosome footprinting read counts in MYC OFF and MYC ON P493 cells treated with or without harringtonine. Table S5 lists translational start sites detected in MYC OFF and MYC ON P493 cells treated with or without harringtonine.
Supplementary Material
Acknowledgments
We thank Anne Quy Hoa Le Thi (Johns Hopkins University School of Medicine, Baltimore, MD) for providing the P493-6 cell line. We thank Renier J. Brentjens (Memorial Sloan Kettering Cancer Center, New York, NY) for providing the CD19 CAR construct. We thank Massimo Caputi (Charles E. Schmidt College of Medicine, Florida Atlantic University, Boca Raton, FL) for providing the c-DNA constructs for SRSF1. We also thank Liz Chang for expert technical assistance with LC-MS experiments.
H.-G. Wendel is supported by the National Institutes of Health grants RO1CA183876-03, R01CA207217-01, R01CA190384, and 5R01 1P50CA192937-01; the Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center (GC229409); the Lymphoma Research Foundation (GC227729); the Starr Cancer Consortium grant I10-0064; the Geoffrey Beene Cancer Research Center; Leukemia and Lymphoma Society Specialized Center of Research grant; New York State Stem Cell Science II IRP grant C028131; National Institutes of Health SPORE in Soft Tissue Sarcoma grant P50 CA217694; and the Memorial Sloan Kettering Cancer Center core grant (P30 CA008748). H.-G. Wendel is a scholar of the Leukemia and Lymphoma Society. G. Rätsch and Y. Zhong were supported by core funding of the Sloan Kettering Institute. G. Rätsch is supported by core funding of the Swiss Federal Institute of Technology in Zürich. Z. Ouyang is partially supported by National Institutes of Health/National Institute of General Medical Sciences grant R35 GM124998. We acknowledge the use of the Integrated Genomics Operation Core, funded by the National Cancer Institute Cancer Center Support Grant (P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
The authors declare no competing financial interests.
Author contributions: K. Singh designed, performed, and analyzed experiments and co-wrote the manuscript. J. Lin, Y. Zhong, and A. Burčul performed the computational analysis. Z. Ouyang and G. Rätsch supervised the computational analysis. P. Mohan and M. Jiang provided technical assistance. A. Viale and L. Sun performed the HiSeq-2000. J.R. Cross and V. Yong-Gonzalez performed metabolic studies. R.C. Hendrickson performed mass spectrometry studies. H.-G. Wendel designed the study and co-wrote the paper.
References
- Anczuków O., Rosenberg A.Z., Akerman M., Das S., Zhan L., Karni R., Muthuswamy S.K., and Krainer A.R.. 2012. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19:220–228. 10.1038/nsmb.2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi A., Wu S., Ridderstråle K., Bierhoff H., Shiue C., Fatyol K., Fahlén S., Hydbring P., Söderberg O., Grummt I., et al. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7:303–310. 10.1038/ncb1225 [DOI] [PubMed] [Google Scholar]
- Arts G.J., Fornerod M., and Mattaj I.W.. 1998. Identification of a nuclear export receptor for tRNA. Curr. Biol. 8:305–314. 10.1016/S0960-9822(98)70130-7 [DOI] [PubMed] [Google Scholar]
- Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., and Topisirovic I.. 2015. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 14:261–278. 10.1038/nrd4505 [DOI] [PubMed] [Google Scholar]
- Blackwood E.M., and Eisenman R.N.. 1991. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 251:1211–1217. 10.1126/science.2006410 [DOI] [PubMed] [Google Scholar]
- Boice M., Salloum D., Mourcin F., Sanghvi V., Amin R., Oricchio E., Jiang M., Mottok A., Denis-Lagache N., Ciriello G., et al. 2016. Loss of the HVEM Tumor Suppressor in Lymphoma and Restoration by Modified CAR-T Cells. Cell. 167:405–418.e13. 10.1016/j.cell.2016.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens R.J., Davila M.L., Riviere I., Park J., Wang X., Cowell L.G., Bartido S., Stefanski J., Taylor C., Olszewska M., et al. 2013. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med. 5:177ra38 10.1126/scitranslmed.3005930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byron A., Humphries J.D., Craig S.E., Knight D., and Humphries M.J.. 2012. Proteomic analysis of α4β1 integrin adhesion complexes reveals α-subunit-dependent protein recruitment. Proteomics. 12:2107–2114. 10.1002/pmic.201100487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M.D., and Cowling V.H.. 2009. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene. 28:1169–1175. 10.1038/onc.2008.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S., Bruynooghe Y., Denecker G., Van Huffel S., Tinton S., and Beyaert R.. 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell. 5:597–605. 10.1016/S1097-2765(00)80239-7 [DOI] [PubMed] [Google Scholar]
- Dang C.V., O’Donnell K.A., Zeller K.I., Nguyen T., Osthus R.C., and Li F.. 2006. The c-Myc target gene network. Semin. Cancer Biol. 16:253–264. 10.1016/j.semcancer.2006.07.014 [DOI] [PubMed] [Google Scholar]
- Das S., and Krainer A.R.. 2014. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 12:1195–1204. 10.1158/1541-7786.MCR-14-0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Anczuków O., Akerman M., and Krainer A.R.. 2012. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Reports. 1:110–117. 10.1016/j.celrep.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delestienne N., Wauquier C., Soin R., Dierick J.F., Gueydan C., and Kruys V.. 2010. The splicing factor ASF/SF2 is associated with TIA-1-related/TIA-1-containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression. FEBS J. 277:2496–2514. 10.1111/j.1742-4658.2010.07664.x [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R., Loayza-Puch F., Korkmaz G., Lopes R., van Breugel P.C., Bleijerveld O.B., Altelaar A.F., Wolf E., Lorenzin F., Eilers M., and Agami R.. 2015. Myc coordinates transcription and translation to enhance transformation and suppress invasiveness. EMBO Rep. 16:1723–1736. 10.15252/embr.201540717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields A.P., Rodriguez E.H., Jovanovic M., Stern-Ginossar N., Haas B.J., Mertins P., Raychowdhury R., Hacohen N., Carr S.A., Ingolia N.T., et al. 2015. A Regression-Based Analysis of Ribosome-Profiling Data Reveals a Conserved Complexity to Mammalian Translation. Mol. Cell. 60:816–827. 10.1016/j.molcel.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno M., Jiménez A., and Vázquez D.. 1977. Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem. 72:323–330. 10.1111/j.1432-1033.1977.tb11256.x [DOI] [PubMed] [Google Scholar]
- Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., and White R.J.. 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7:311–318. 10.1038/ncb1224 [DOI] [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., and Noble W.S.. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics. 27:1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Sato Y., Kariya Y., Isaji T., Taniguchi N., and Fukuda T.. 2009. A mutual regulation between cell-cell adhesion and N-glycosylation: implication of the bisecting GlcNAc for biological functions. J. Proteome Res. 8:431–435. 10.1021/pr800674g [DOI] [PubMed] [Google Scholar]
- Hardie D.G., Ross F.A., and Hawley S.A.. 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13:251–262. 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh A.C., Liu Y., Edlind M.P., Ingolia N.T., Janes M.R., Sher A., Shi E.Y., Stumpf C.R., Christensen C., Bonham M.J., et al. 2012. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 485:55–61. 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Ghaemmaghami S., Newman J.R., and Weissman J.S.. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324:218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., and Krainer A.R.. 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14:185–193. 10.1038/nsmb1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R., Hippo Y., Lowe S.W., and Krainer A.R.. 2008. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proc. Natl. Acad. Sci. USA. 105:15323–15327. 10.1073/pnas.0801376105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y., Wang E.T., Airoldi E.M., and Burge C.B.. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 7:1009–1015. 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempkes B., Spitkovsky D., Jansen-Dürr P., Ellwart J.W., Kremmer E., Delecluse H.J., Rottenberger C., Bornkamm G.W., and Hammerschmidt W.. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88–96. 10.1002/j.1460-2075.1995.tb06978.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenderfer J.N., Dudley M.E., Kassim S.H., Somerville R.P., Carpenter R.O., Stetler-Stevenson M., Yang J.C., Phan G.Q., Hughes M.S., Sherry R.M., et al. 2015. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J. Clin. Oncol. 33:540–549. 10.1200/JCO.2014.56.2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress T.R., Sabò A., and Amati B.. 2015. MYC: connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer. 15:593–607. 10.1038/nrc3984 [DOI] [PubMed] [Google Scholar]
- Land H., Parada L.F., and Weinberg R.A.. 1983. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 304:596–602. 10.1038/304596a0 [DOI] [PubMed] [Google Scholar]
- Li F., Wang Y., Zeller K.I., Potter J.J., Wonsey D.R., O’Donnell K.A., Kim J.W., Yustein J.T., Lee L.A., and Dang C.V.. 2005. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 25:6225–6234. 10.1128/MCB.25.14.6225-6234.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.J., Cencic R., Mills J.R., Robert F., and Pelletier J.. 2008. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 68:5326–5334. 10.1158/0008-5472.CAN-07-5876 [DOI] [PubMed] [Google Scholar]
- Lin C.Y., Lovén J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., and Young R.A.. 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 151:56–67. 10.1016/j.cell.2012.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist L.M., Tandoc K., Topisirovic I., and Furic L.. 2018. Cross-talk between protein synthesis, energy metabolism and autophagy in cancer. Curr. Opin. Genet. Dev. 48:104–111. 10.1016/j.gde.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Ramos A., Candeias M.M., Menezes J., Lacerda R., Willcocks M., Teixeira A., Locker N., and Romão L.. 2017. Cap-independent translation ensures mTOR expression and function upon protein synthesis inhibition. RNA. 23:1712–1728. 10.1261/rna.063040.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O. 2000. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 267:6321–6330. 10.1046/j.1432-1327.2000.01719.x [DOI] [PubMed] [Google Scholar]
- Morita M., Prudent J., Basu K., Goyon V., Katsumura S., Hulea L., Pearl D., Siddiqui N., Strack S., McGuirk S., et al. 2017. mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol. Cell. 67:922–935.e5. 10.1016/j.molcel.2017.08.013 [DOI] [PubMed] [Google Scholar]
- Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D.R., Tessarollo L., Casellas R., et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 151:68–79. 10.1016/j.cell.2012.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero D.C., Anzelon A.N., and Rickert R.C.. 2003. CD19 function in early and late B cell development: I. Maintenance of follicular and marginal zone B cells requires CD19-dependent survival signals. J. Immunol. 170:73–83. 10.4049/jimmunol.170.1.73 [DOI] [PubMed] [Google Scholar]
- Ouyang Z., Zhou Q., and Wong W.H.. 2009. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 106:21521–21526. 10.1073/pnas.0904863106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic A., Spitkovsky D., Christoph B., Kempkes B., Schuhmacher M., Staege M.S., Brielmeier M., Ellwart J., Kohlhuber F., Bornkamm G.W., et al. 2000. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int. J. Cancer. 87:787–793. [DOI] [PubMed] [Google Scholar]
- Paz S., Lu M.L., Takata H., Trautmann L., and Caputi M.. 2015. SRSF1 RNA Recognition Motifs Are Strong Inhibitors of HIV-1 Replication. J. Virol. 89:6275–6286. 10.1128/JVI.00693-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegram H.J., Lee J.C., Hayman E.G., Imperato G.H., Tedder T.F., Sadelain M., and Brentjens R.J.. 2012. Tumor-targeted T cells modified to secrete IL-12 eradicate systemic tumors without need for prior conditioning. Blood. 119:4133–4141. 10.1182/blood-2011-12-400044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., and Sonenberg N.. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 334:320–325. 10.1038/334320a0 [DOI] [PubMed] [Google Scholar]
- Perna F., and Sadelain M.. 2016. Myeloid leukemia switch as immune escape from CD19 chimeric antigen receptor (CAR) therapy. Transl. Cancer Res. 5(S2, Suppl 2):S221–S225. 10.21037/tcr.2016.08.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenis M., and Schmidt E.V.. 1997. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 11:2522–2531. 10.1101/gad.11.19.2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourdehnad M., Truitt M.L., Siddiqi I.N., Ducker G.S., Shokat K.M., and Ruggero D.. 2013. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc. Natl. Acad. Sci. USA. 110:11988–11993. 10.1073/pnas.1310230110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., and Glöckner F.O.. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(Database issue, D1):D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F., Carrier M., Rawe S., Chen S., Lowe S., and Pelletier J.. 2009. Altering chemosensitivity by modulating translation elongation. PLoS One. 4:e5428 10.1371/journal.pone.0005428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Ellis J.D., Cazalla D., and Cáceres J.F.. 2005. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl. Acad. Sci. USA. 102:15042–15047. 10.1073/pnas.0507827102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., and Sabatini D.M.. 2017. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 168:960–976. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser I., Hölzel M., Mürnseer M., Burtscher H., Weidle U.H., and Eick D.. 2003. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 31:6148–6156. 10.1093/nar/gkg794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M., and Eick D.. 2013. Dose-dependent regulation of target gene expression and cell proliferation by c-Myc levels. Transcription. 4:192–197. 10.4161/trns.25907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scognamiglio R., Cabezas-Wallscheid N., Thier M.C., Altamura S., Reyes A., Prendergast A.M., Baumgärtner D., Carnevalli L.S., Atzberger A., Haas S., et al. 2016. Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell. 164:668–680. 10.1016/j.cell.2015.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A., Dunn J.G., Rodriguez E.H., Naik S., Gomez N.C., Hurwitz B., Levorse J., Dill B.D., Schramek D., Molina H., et al. 2017. Translation from unconventional 5′ start sites drives tumour initiation. Nature. 541:494–499. 10.1038/nature21036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian D., Parsyan A., Petroulakis E., Hershey J., and Sonenberg N.. 2010. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle. 9:4106–4109. 10.4161/cc.9.20.13630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo E., Barrett D.M., Black K.L., Bagashev A., Oldridge D., Wu G., Sussman R., Lanauze C., Ruella M., Gazzara M.R., et al. 2015. Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer Discov. 5:1282–1295. 10.1158/2159-8290.CD-15-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf C.R., Moreno M.V., Olshen A.B., Taylor B.S., and Ruggero D.. 2013. The translational landscape of the mammalian cell cycle. Mol. Cell. 52:574–582. 10.1016/j.molcel.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.C. 1987. Allergic contact dermatitis of the face from contact with nickel and ammoniated mercury in spectacle frames and skin-lightening creams. Contact Dermat. 17:306–309. 10.1111/j.1600-0536.1987.tb01483.x [DOI] [PubMed] [Google Scholar]
- Sun S., Zhang Z., Sinha R., Karni R., and Krainer A.R.. 2010. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat. Struct. Mol. Biol. 17:306–312. 10.1038/nsmb.1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. 2017. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45(D1):D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., and Sabatini D.M.. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 485:109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I., and Sonenberg N.. 2011. mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 pathways. Cold Spring Harb. Symp. Quant. Biol. 76:355–367. 10.1101/sqb.2011.76.010785 [DOI] [PubMed] [Google Scholar]
- Truitt M.L., Conn C.S., Shi Z., Pang X., Tokuyasu T., Coady A.M., Seo Y., Barna M., and Ruggero D.. 2015. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell. 162:59–71. 10.1016/j.cell.2015.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyffels L., Gueydan C., and Kruys V.. 2011. Shuttling SR proteins: more than splicing factors. FEBS J. 278:3246–3255. 10.1111/j.1742-4658.2011.08274.x [DOI] [PubMed] [Google Scholar]
- van Riggelen J., Yetil A., and Felsher D.W.. 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 10:301–309. 10.1038/nrc2819 [DOI] [PubMed] [Google Scholar]
- Vattem K.M., and Wek R.C.. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA. 101:11269–11274. 10.1073/pnas.0400541101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz S., Lorenzin F., Morton J., Wiese K.E., von Eyss B., Herold S., Rycak L., Dumay-Odelot H., Karim S., Bartkuhn M., et al. 2014. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 511:483–487. 10.1038/nature13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A.L., Singh K., Zhong Y., Drewe P., Rajasekhar V.K., Sanghvi V.R., Mavrakis K.J., Jiang M., Roderick J.E., Van der Meulen J., et al. 2014. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 513:65–70. 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., and Bartel D.P.. 2017. kpLogo: positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res. 45(W1):W534–W538. 10.1093/nar/gkx323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Karaletsos T., Drewe P., Sreedharan V.T., Kuo D., Singh K., Wendel H.G., and Rätsch G.. 2017. RiboDiff: detecting changes of mRNA translation efficiency from ribosome footprints. Bioinformatics. 33:139–141. 10.1093/bioinformatics/btw585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.