Which antigen-presenting cells are involved in Tfh responses in humans remains unclear. Durand et al. show that human tonsil cDC2 and CD14+ macrophages provide Tfh polarizing signals to CD4+ T cells in distinct locations within tonsil, playing sequential roles in Tfh induction.
Abstract
CD4+ T follicular helper (Tfh) cells are essential for inducing efficient humoral responses. T helper polarization is classically orientated by dendritic cells (DCs), which are composed of several subpopulations with distinct functions. Whether human DC subsets display functional specialization for Tfh polarization remains unclear. Here we find that tonsil cDC2 and CD14+ macrophages are the best inducers of Tfh polarization. This ability is intrinsic to the cDC2 lineage but tissue dependent for macrophages. We further show that human Tfh cells comprise two effector states producing either IL-21 or CXCL13. Distinct mechanisms drive the production of Tfh effector molecules, involving IL-12p70 for IL-21 and activin A and TGFβ for CXCL13. Finally, using imaging mass cytometry, we find that tonsil CD14+ macrophages localize in situ in the B cell follicles, where they can interact with Tfh cells. Our results indicate that human lymphoid organ cDC2 and macrophages play complementary roles in the induction of Tfh responses.
Graphical Abstract
Introduction
CD4+ T follicular helper (Tfh) cells are essential for inducing germinal center (GC) and plasma cell formation, and for supporting efficient humoral responses (Vinuesa et al., 2016). Tfh cells represent promising therapeutic targets for improving the efficacy of vaccines or for down-modulating production of auto-antibodies in autoimmune diseases, but this approach has been hindered by a limited understanding of the Tfh differentiation process in humans.
Tfh cells are classically characterized by their phenotype (CXCR5+PD-1+ICOS+), the expression of transcription factors Bcl6 and Ascl2, and their ability to provide help to B cells via the secretion of IL-21 (Chtanova et al., 2004; Johnston et al., 2009; Liu et al., 2014). In addition, human Tfh cells secrete CXCL13, the ligand for CXCR5 (Kim et al., 2004). Mouse Tfh cell differentiation is a multi-step process (Crotty, 2014). Tfh differentiation is primed by dendritic cells (DCs) in the T cell zone (Goenka et al., 2011), before migration of activated “preTfh” cells toward the border of the B and T cell zones (B-T border), where they interact with antigen presenting B cells and receive additional signals for Tfh polarization. Tfh cells then enter the GC, where they support B cell selection and proliferation. In addition, it has been proposed that interactions between GC Tfh and B cells maintain the Tfh cell phenotype and production of effector molecules (Qi, 2016). Whether similar stages exist in human Tfh cell differentiation and which APCs are involved have remained unclear.
DCs are composed of distinct subsets that can be distinguished by their ontogeny: plasmacytoid DC (pDC), Batf3-dependent “classical” DC 1 (cDC1), and Batf3-independent cDC2 (Guilliams et al., 2014). While a functional specialization of human DC subsets has been reported for inducing T helper (Th) 2 and Th17 cell differentiation (Schlitzer et al., 2013; Yu et al., 2014), whether such specialization exists for Tfh cells remains unknown. Mouse skin-derived cDC1s, but not cDC2s, induce Tfh cells in skin-draining lymph nodes (Yao et al., 2015). cDC1s are also efficient inducers of antibody responses when targeted for antigen delivery via Clec9a (Caminschi et al., 2008; Kato et al., 2015) or XCR1 (Gudjonsson et al., 2017), but not via CD205 (Shin et al., 2015). By contrast, cDC2s induce robust Tfh responses when targeted with anti-DCIR2 (Shin et al., 2015). In addition, in a model of alloimmunization, depletion of cDC2, but not of cDC1, abrogated humoral responses (Calabro et al., 2016). A preferential role for cDC2 has been demonstrated in favoring Tfh maturation in the outer T cell zone where only cDC2s are positioned (Li et al., 2016). Among spleen cDC2s, only the Notch2-dependent subset is required for Tfh responses (Briseño et al., 2018). Finally, upon intranasal immunization, migratory cDC2s, but not cDC1s, are responsible for Tfh priming (Krishnaswamy et al., 2017). In humans, some studies have concluded that skin CD14+ CD206+ DCs are the most efficient skin-derived DC subset for Tfh polarization (Klechevsky et al., 2008; Segura et al., 2012), while another reports that Langerhans cells and dermal CD1a+ cDC2s are the best at inducing IL-21 production by CD4+ T cells (Penel-Sotirakis et al., 2012). The ability of other human DC subsets to induce Tfh cells is not known.
To determine which human APCs are involved in Tfh polarization, we analyzed DCs and macrophages directly purified from human tissues. We found that tonsil cDC2s and macrophages play complementary roles in Tfh induction, with cDC2s being the best inducers of Tfh polarization among DC subsets, while macrophages are uniquely positioned in the B cell follicles, where they can interact with Tfh cells to stimulate the secretion of effector molecules. This function is tissue specific, as macrophages from peripheral tissues were not able to polarize Tfh cells. Our results enable a better understanding of the tissue specialization of human macrophages and of the mechanisms of Tfh differentiation in humans.
Results
Tonsil cDC2s and macrophages are the most efficient for inducing Tfh polarization
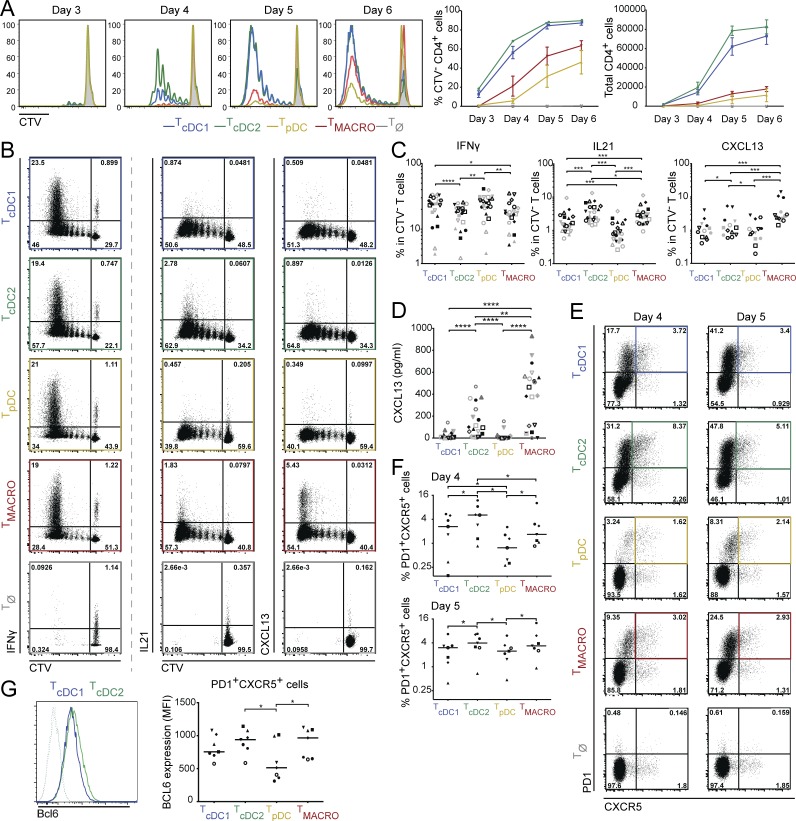
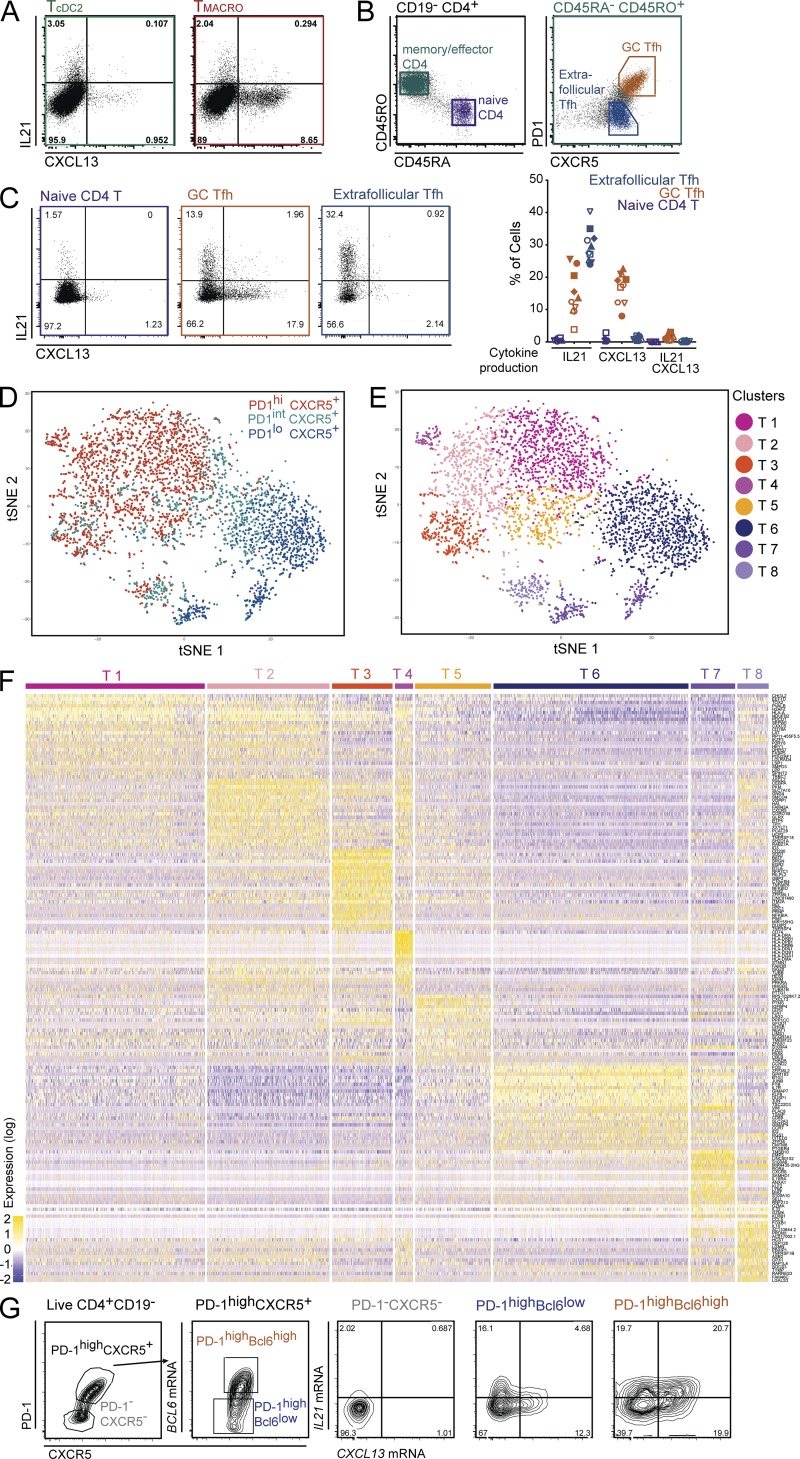
To address which human DC subsets can induce Tfh polarization, we analyzed the ability of DCs, and macrophages for comparison, from human nonpathological tonsils to polarize allogenic naive CD4+ T cells. We isolated pDCs, CD141/BDCA3+ DCs (cDC1s), CD1c/BDCA1+ DCs (cDC2s), and CD14+ macrophages (Fig. S1 A). cDC1 and cDC2 were the most efficient for stimulating naive CD4+ T cell proliferation (Fig. 1 A). To determine Th cell profiles induced by the different APCs, we analyzed cytokine production in CD4+ T cells by intracellular staining after PMA and ionomycin stimulation (Fig. 1, B and C; and Fig. S1, B and C). We found that all APC subsets could induce IFN-γ–producing cells, with cDC1 and pDC inducing the highest proportion among CD4+ T cells that had proliferated (Fig. 1 C). Regarding Tfh induction, cDC2 and macrophages induced the highest proportion of IL-21–producing CD4+ T cells, and macrophages were the most efficient inducers of CXCL13-producing CD4+ T cells in this assay (Fig. 1, B and C). It has been reported that detection of intracellular CXCL13 after PMA and ionomycin stimulation is suboptimal (Rao et al., 2017). Therefore, to complement this analysis, we analyzed cytokine secretion in the supernatant after anti-CD3/CD28 stimulation (Fig. 1 D and Fig. S1 D). In these conditions, both cDC2s and macrophages induced the secretion of CXCL13. Because APCs are not viable in the culture at this time point (Fig. S1 E), we excluded that CXCL13 was secreted by DCs or macrophages.
Figure 1.
Tonsil cDC2 and macrophages are the best inducers of Tfh polarization. Purified human tonsil DC subsets and macrophages were co-cultured with allogeneic naive CD4+ T cells. T cells polarized with cDC1s, cDC2s, pDCs, or CD14+ macrophages are termed TcDC1, TcDC2, TpDC, and TMACRO, respectively. (A) T cell proliferation was assessed by dilution of a proliferation dye (CTV). Histogram representative of three independent experiments. Graphs show mean ± SEM (n = 3). (B and C) Cytokine production was analyzed at day 5 (cDC1s and cDC2s) or day 6 (pDCs and macrophages), by intracellular staining after restimulation with PMA and ionomycin in presence of brefeldin A. (B) Representative staining, gated on live CD4+ T cells. TØ corresponds to T cells cultured without APCs. (C) Percentage of divided cells (CTV−) producing IFN-γ (n = 22), IL-21 (n = 22), and CXCL13 (n = 13). Each symbol represents an individual donor. (D) Cytokine secretion was analyzed by ELISA after 6 d of culture and restimulation with anti-CD3/CD28 beads. Each symbol represents an individual donor (n = 19). (E and F) Expression of PD-1 and CXCR5 was analyzed by flow cytometry at day 4 or day 5. Each symbol represents an individual donor (n = 7). (E) Representative staining, gated on total live CD4+ T cells. (F) Percentage of PD-1+CXCR5+ cells. (G) Mean fluorescence intensity (MFI) of Bcl6 staining in PD-1+CXCR5+ cells at day 4 (n = 7). Representative staining is shown for T cells cultured with cDC1 or cDC2. Dashed line represents isotype control staining. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; Wilcoxon test.
To confirm the induction of Tfh cells in the culture, we analyzed the expression of classical Tfh markers CXCR5, PD-1, and Bcl6. Because of the difference in T cell proliferation kinetics between APC subsets (Fig. 1 A), we measured CXCR5 and PD-1 expression on CD4+ T cells at day 4 and day 5 of the culture (Fig. 1, E and F). At day 4, cDC2 induced the highest proportion of CXCR5+PD-1+ T cells, while at day 5, cDC2s and macrophages induced similar proportions of CXCR5+PD-1+ T cells. In addition, CXCR5+PD-1+ T cells induced by cDC2s and macrophages had the highest expression of Bcl6, as evaluated by intracellular staining (Fig. 1 G and Fig. S1 F).
Collectively, these results indicate that cDC2s and macrophages are the most efficient for inducing Tfh polarization in CD4+ T cells.
The ability to polarize Tfh cells is intrinsic to the cDC2 lineage, but is tissue dependent for macrophages
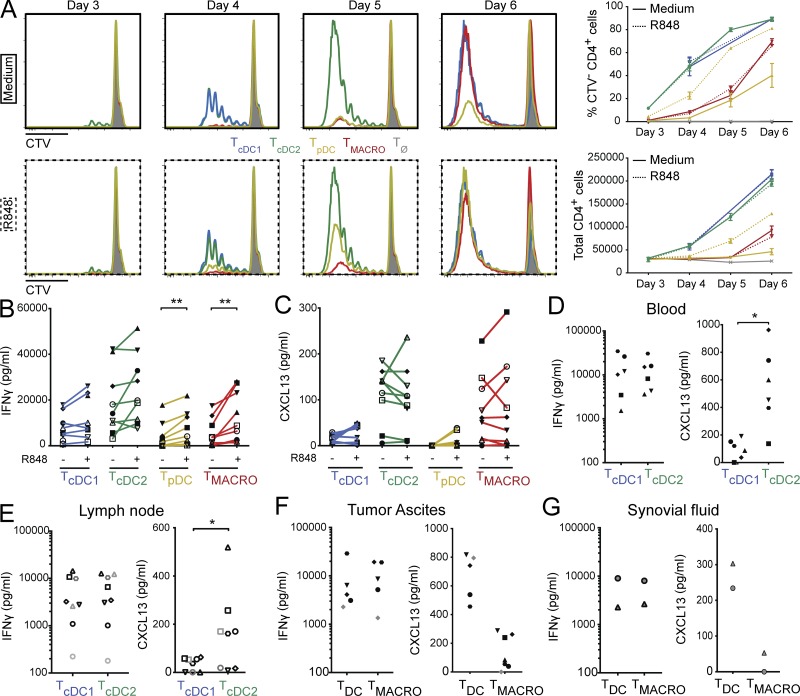
APC properties can be modified upon exposure to pathogen-derived products. In particular, TLR8 activation of human APC has been shown to favor Tfh cell differentiation (Ugolini et al., 2018). To address whether differential capacity for Tfh induction was conserved after activation, we performed the same co-culture experiment with tonsil DCs and macrophages that had been preactivated during 3 h with R848, a ligand for TLR7 and TLR8. pDCs express TLR7, while cDC1s, cDC2s, and macrophages express TLR8 (Fig. S1 G) and secrete cytokines in response to R848 exposure (Fig. S1 H). Stimulation with R848 did not modify the ability of cDC1 and cDC2 to induce naive CD4+ T cell proliferation, while activated pDCs became more efficient (Fig. 2 A). R848-exposed macrophages remained poor stimulators of naive CD4+ T cell proliferation compared with DC. When analyzing cytokine secretion by CD4+ T cells, we found that APC activation by R848 increased IFN-γ secretion by CD4+ T cells cultured with pDCs or macrophages (Fig. 2 B). However, there was no significant impact on CXCL13 secretion (Fig. 2 C). We conclude that cDC2s and macrophages are the most efficient inducers of CXCL13 secretion, irrespective of their activation status.
Figure 2.
The ability to polarize Tfh cells is intrinsic for cDC2 but is tissue dependent for macrophages. (A–C) Purified human tonsil DC subsets and macrophages were cultured for 3 h with or without R848. After washing, preactivated DCs or macrophages were co-cultured with allogeneic naive CD4+ T cells for 6 d. T cells polarized with cDC1s, cDC2s, pDCs, or CD14+ macrophages are termed TcDC1, TcDC2, TpDC, and TMACRO, respectively. TØ corresponds to T cells cultured without APCs. (A) T cell proliferation was assessed by dilution of a proliferation dye (CTV). Histogram representative of three independent experiments. Graphs show mean ± SEM (n = 3). (B and C) Cytokine secretion was analyzed by cytometric bead array (CBA) (B) or ELISA (C) after restimulation with anti-CD3/CD28 beads. Each symbol represents an individual donor (n = 9). (D–G) Purified cDC1 and cDC2 from blood (D) or skin-draining lymph node (E), or DCs and macrophages from peritoneal tumor ascites (F) or synovial fluid of rheumatoid arthritis patients (G) were co-cultured with allogeneic naive CD4+ T cells for 6 d. IFN-γ and CXCL13 secretion was measured in the supernatant after restimulation with anti-CD3/CD28 beads. Each symbol represents an individual donor (n = 6 for blood, 8 for lymph node, 5–7 for ascites, and 2 for synovial fluid). *, P < 0.05; **, P < 0.01; Wilcoxon test.
To address whether the higher ability of cDC2 for Tfh induction was specific to tonsil DC, we compared cDC1 and cDC2 isolated from the blood of healthy donors and from uninvaded lymph nodes from breast cancer patients for their ability to induce CXCL13 secretion (Fig. 2, D and E). We found that cDC2s were again the most efficient for inducing CXCL13-producing CD4+ T cells.
We also examined whether macrophages from other tissues could efficiently induce Tfh polarization. We first compared macrophages and in vivo–generated monocyte-derived DC (mo-DC) purified from peritoneal tumor ascites (Tang-Huau et al., 2018; Fig. 2 F). Ascites macrophages stimulated IFN-γ secretion to a similar level as ascites mo-DC, but were poor inducers of CXCL13 secretion compared with mo-DC. Similarly, macrophages isolated from the synovial fluid of rheumatoid arthritis patients were inefficient for inducing CXCL13 secretion compared with DC from the same samples (Fig. 2 G). Finally, we observed that lung macrophages were very poor inducers of naive CD4+ T cell proliferation (Fig. S1 I). We conclude that only macrophages from tonsils, but not from peritoneal ascites, synovial fluid, or lung, can induce Tfh polarization.
Collectively, these results show that the ability to polarize Tfh cells is an intrinsic property of the human cDC2 lineage. By contrast, this feature is tissue dependent in macrophages.
Tonsil CD14+ cells do not contain a population of DC
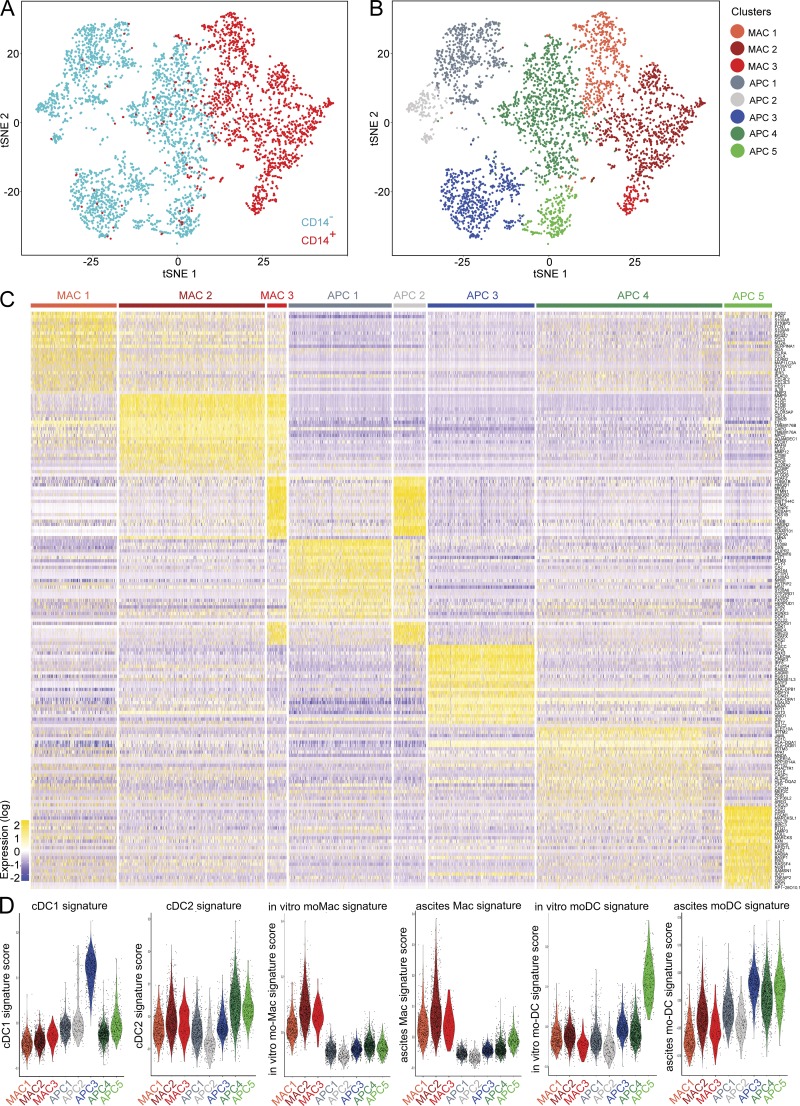
In vivo–generated mo-DCs express CD14 (Segura et al., 2013b) and efficiently induce CXCL13 secretion (Fig. 2, F and G). To exclude the possibility that our tonsil macrophage preparations contain a population of CD14+ DC, we addressed the heterogeneity of tonsil CD14+ cells using single-cell RNA sequencing (RNA-seq) analysis. We purified tonsil macrophages (gated as HLA-DR+CD11c+CD14+ cells) and tonsil HLA-DR+CD11c+CD14− cells (containing cDC populations) from the same sample (Fig. S2 A), and analyzed single-cell transcriptomes using a droplet-based method enabling 3′ mRNA counting (Zheng et al., 2017). To evaluate the heterogeneity of these cells, we merged the two datasets and performed unsupervised clustering based on a graph-based approach using the Seurat package (Satija et al., 2015). Unsupervised clustering identified eight main clusters (Fig. 3, A and B), three corresponding to cells from the “CD14+ macrophage” sample (annotated MAC) and five corresponding to cells from the CD14− sample (to avoid confusion, these clusters are annotated APC). We first performed hierarchical ordering of the meta-cells corresponding to these clusters, where gene expression is averaged across all cells from each cluster to construct an “average” cell (Fig. S2 B). This analysis identified three pairs of clusters that were more closely related to each other than to other clusters: MAC2 and MAC3, APC1 and APC2, and APC3 and APC4. We then analyzed differential gene expression between clusters (Fig. 3 C; note that clusters are ordered based on the hierarchical ordering tree). Cluster MAC1 expressed high levels of monocyte- and macrophage-related genes such as S100A8, FCN1, S100A9, and CTSS. Clusters MAC2 and MAC3 expressed high levels of macrophage genes such as C1QA, CTSD, CD14, MAFB, and LMNA. Clusters APC1 and APC2 shared expression of genes expressed on progenitors or related to cDC development such as LTB, PRDM16, LST1, RUNX3, and CD164. In addition, clusters MAC3 and APC2 expressed cell cycle genes including STMN1, MKI67, TOP2A, and CDK1. Cluster APC3 expressed cDC1 genes such as CLEC9A, IRF8, C1orf54, RAB32, and CADM1. Cluster APC4 expressed cDC2 genes including CLEC10A, CD1C, FCER1A, and CD1E. Finally, cluster APC5 expressed DC activation genes such as CCR7, LAMP3, CCL19, MARCKSL1, and CD83. The complete list of top differentially expressed genes for this dataset is available in Table S1.
Figure 3.
Tonsil CD14+ cells do not contain a population of DC. Purified tonsil HLA-DR+CD11c+CD14− cells and HLA-DR+CD11c+CD14+ cells were analyzed by single-cell RNA-seq using a Drop-seq approach. Combined single-cell transcriptomes were analyzed. (A and B) t-SNE representation of cell clusters identified using unsupervised clustering. Each dot represents an individual cell. (A) Colors represent sample origin. (B) Colors represent identified clusters. Clusters are manually ordered. (C) Heatmap of scaled expression (log values of UMI) for the top 25 differentially expressed genes of each cluster (based on log fold change). (D) Signature scores (arbitrary units) in individual cells for indicated gene signatures.
We then analyzed putative conserved genes between clusters, which may not be visible in the differential gene expression analysis. We have previously evidenced a convergent transcriptional program between human cDC2 and in vivo–generated mo-DC (Tang-Huau et al., 2018). We interrogated the dataset to identify genes whose expression would be conserved between clusters MAC1 (potential monocyte-related population) and APC4 (putative cDC2 cluster) compared with all other clusters. The algorithm failed to identify genes that would be common to these two clusters without being also expressed by cluster MAC2 (Fig. S2 C). By contrast, the same analysis for conserved genes between MAC1 and MAC2 identified genes highly expressed by all clusters from the “macrophage” sample (Fig. S2 D), such as FTL, PSAP, FCER1G, SERPINA1, and CTSS. The complete gene lists for these heatmaps is available in Table S1.
Finally, to confirm the identity of these clusters, we analyzed signature scores in individual cells using transcriptomic gene signatures (Fig. 3 D). For each cell, we calculated the average expression of each signature, subtracted by the aggregated expression of control gene sets (Tirosh et al., 2016). We used published signatures for blood cDC1 and cDC2 (Carpentier et al., 2016), and signatures we had previously generated for in vitro–generated monocyte-derived macrophages (mo-Macs) and mo-DCs, and peritoneal ascites macrophages and mo-DCs (Tang-Huau et al., 2018). Complete lists of genes for each signature are shown in Table S2. Cluster APC3 had the highest score for the cDC1 signature. Clusters APC4 and APC5 had the highest score for the cDC2 signature. Clusters MAC1, MAC2, and MAC3 had high scores for the in vitro mo-Mac and the ascites macrophage signatures, suggesting that these three clusters are bona fide macrophages. Finally, in vitro–derived and ascites mo-DC signatures were enriched in clusters APC5, and APC3, APC4, and APC5, respectively, suggesting shared transcriptomic programs between DC populations. Importantly, MAC1, MAC2, or MAC3 clusters had a low score for these signatures, confirming they do not correspond to mo-DC.
Based on these results, we identified two populations of macrophages within tonsil CD14+ cells, with one population comprising actively cycling cells, corresponding to clusters MAC1 and MAC2+MAC3, respectively. Within HLA-DR+CD11c+CD14− cells, we identified cDC precursors, with a proportion of them actively cycling (clusters APC1+APC2), cDC1 (cluster APC3), cDC2 (cluster APC4), and activated cDC2 (cluster APC5). We conclude that tonsil CD14+ cells are macrophages, and do not contain a population of DC, neither cDC nor mo-DC.
Tfh cells can be separated based on production of IL-21 versus CXCL13, corresponding to functional states rather than distinct Tfh cell subsets
Our results show that tonsil cDC2s and macrophages can induce similar proportions of IL-21 production by activated CD4+ T cells, but macrophages are more efficient for inducing CXCL13 production (Fig. 1, C and D). While both molecules are considered hallmarks of human Tfh cells, whether they are produced by the same cells remains unclear. To address this, we analyzed coexpression of these molecules in our in vitro assay by intracellular staining after PMA and ionomycin stimulation (Fig. 4 A). IL-21 and CXCL13 were expressed by distinct populations. To address whether this observation holds true in vivo, we measured cytokine production in CD4+ T cells directly purified from tonsils using the same procedure (Fig. 4, B and C). We analyzed extra-follicular Tfh cells (CXCR5+PD-1low) and GC Tfh cells (CXCR5+PD-1high; Fig. 4 B) and also included naive CD4+ T cells for comparison. As expected, very few naive CD4+ T cells produced either cytokine. GC Tfh cells produced either CXCL13 or IL-21, while a large proportion of extra-follicular Tfh cells produced only IL-21, with hardly any cell producing CXCL13 (Fig. 4 C). In both cases, we did not detect a population coexpressing IL-21 and CXCL13. These results suggest that tonsil CD4+CXCR5+ T cells contain distinct populations producing either IL-21 or CXCL13.
Figure 4.
Human Tfh cells comprise two distinct effectors states characterized by IL-21 or CXCL13 production. (A) Purified tonsil DC subsets and macrophages were co-cultured with allogeneic naive CD4+ T cells. IL-21 and CXCL13 production was analyzed by intracellular staining after restimulation with PMA and ionomycin in presence of brefeldin A. Representative results, gated on live CD4+ CTV− cells (n = 13). T cells polarized with cDC2 or CD14+ macrophages are termed TcDC2 and TMACRO, respectively. (B–G) CD4+ T cells were extracted from tonsils. (B) Gating strategy. Naive CD4+ T cells were CD4+CD45RA+CD45RO−, GC Tfh cells CD4+CD45RO+PD-1highCXCR5+, and extra-follicular Tfh cells CD4+CD45RO+PD-1lowCXCR5+. (C) IL-21 and CXCL13 production was analyzed as in A. Percentage of cells expressing IL-21, CXCL13, or double positive. Each symbol represents an individual donor. Cells were analyzed either directly after enrichment (filled symbols, n = 5) or after cell sorting (open symbols, n = 5). (D–F) Tonsil CD4+CD45RA−CD45RO+ cells were purified as CXCR5+PD-1low, CXCR5+PD-1int, and CXCR5+PD-1high cells and were analyzed by single-cell RNA-seq using a Drop-seq approach. Combined single-cell transcriptomes were analyzed. (D and E) t-SNE representation of cell clusters identified using unsupervised clustering. Each dot represents an individual cell. (D) Colors represent sample origin. (E) Colors represent identified clusters. Clusters are manually ordered. (F) Heatmap of scaled expression (log values of UMI) for the top 25 differentially expressed genes of each cluster (based on log fold change). (G) IL21 and CXCL13 mRNA expression was analyzed in tonsil CD4+CD45RA−CD45RO+ cells by in situ hybridization coupled to flow cytometry. Cells were gated based on Bcl6 and CXCR5 expression. Representative results (n = 4).
To address the potential existence of distinct Tfh subsets, we examined the heterogeneity of tonsil CD4+CXCR5+ T cells in an unsupervised way using single-cell RNA-seq analysis. We generated single-cell transcriptomes of CD4+CD45RA−CD45RO+ T cells by purifying CXCR5+PD-1low, CXCR5+PD-1int, and CXCR5+PD-1high cells (Fig. S3 A), and combining the sequencing data for downstream analysis. Unsupervised analysis grouped cells into eight main clusters, five comprising mostly cells from the CXCR5+PD-1high sample with some cells from the CXCR5+PD-1int sample, one cluster with a mix of cells from the CXCR5+PD-1int and CXCR5+PD-1high samples, and two with mostly cells from the CXCR5+PD-1low sample (Fig. 4, D and E). Hierarchical ordering of the meta-cells corresponding to these clusters identified three groups of clusters that were more related to each other than to other clusters: T1, T2, T3, and T4 (with T1 and T2 being even more closely related), T5 and T6, and T7 and T8 (Fig. S3 B). Differential gene expression analysis showed conserved gene expression patterns between several clusters (Fig. 4 F). In particular for Tfh genes, PDCD1 (encoding PD-1) was highly expressed in clusters T1, T2, T3, T4, T5, and T8, ASCL2 in clusters T1, T2, T3, T4, and T8, CXCR5 in clusters T1, T2, T4, T5, and T8, and CXCL13 in clusters T2, T3, T4, and T5. By contrast, GC Tfh markers BATF and BTLA were most highly expressed in cluster T3. The complete list of top differentially expressed genes for each cluster is available in Table S3.
Unsupervised analysis of conserved genes between clusters T1, T2, T3, T4, and T5 compared with other clusters confirmed the existence of transcriptional patterns common to clusters T1, T2, T3, T4, T5, and T8, including known Tfh cell genes such as PDCD1, POU2AF1 (encoding the transcription factor BOB1), ASCL2, CXCL13, CXCR5, SH2D1A (encoding SAP), and CD200 (Fig. S3 C). The complete list of genes for this heatmap is available in Table S3. Of note, IL21 was detected only at very low levels (Fig. S3 D). These results suggest the existence of several states of Tfh cells, possibly corresponding to maturation stages, rather than transcriptionally distinct subsets.
Cluster T6 expressed effector or memory T cell genes such as IL7R, CCR7, and ID2. Clusters T5 and T6 shared the expression of some genes including IL7R and GPR183 (encoding EBI2; Fig. S3 E), which is involved in the positioning of mouse memory Tfh cells at the B-T border (Suan et al., 2015). Cluster T7 expressed regulatory T cell (T reg) genes such as LAG3, RORA, IL10RA, and PRDM1 (encoding BLIMP1). Finally, cluster T8 expressed both Tfh cell genes and regulatory genes such as LAG3, IL10, and PRDM1.
Based on these results, we identified clusters T1, T2, T3, T4, and T5 as states of Tfh cells, with T5 being memory Tfh cells, cluster T6 as effector/memory T cells of other lineages, cluster T7 as T reg and cluster T8 as T follicular regulatory cells.
In addition, our analysis suggests that IL21 and CXCL13 expression is not specific to a particular Tfh state. To confirm this, we directly assessed the mRNA expression of IL21 and CXCL13 in tonsil CXCR5+PD-1+ cells at the single-cell level using in situ hybridization coupled to flow cytometry (Fig. 4 G). Cells were separated into three gates based on Bcl6 expression. In contrast to protein expression (Fig. 4 C), IL21 and CXCL13 mRNA was coexpressed by a proportion of GC Tfh cells (Bcl6highCXCR5+).
We conclude that Tfh cells comprise different effector states that cannot be distinguished by their transcriptional profile, and that are characterized by the production of IL-21 or CXCL13.
cDC2 and macrophages are the most efficient producers of Tfh-polarizing signals
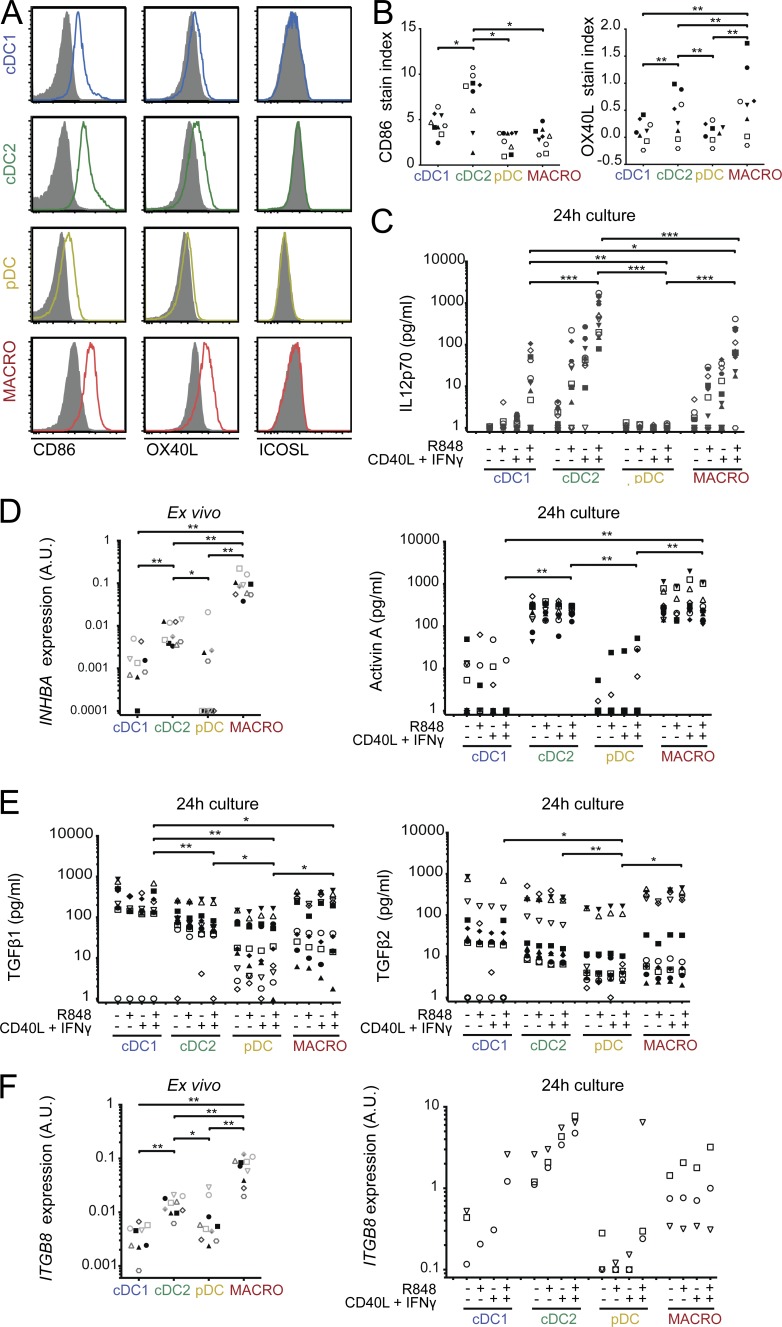
Several molecules have been identified as regulators of human Tfh polarization, including IL-12p70, activin A, and TGFβ (Schmitt et al., 2009, 2014; Locci et al., 2016) and costimulatory molecule OX40L (Jacquemin et al., 2015; Pattarini et al., 2017). Consistent with published reports, we found that IL-12p70, but not activin A or TGFβ, could induce IL-21 production in naive CD4+ T cells (Fig. S4 A), while activin A and TGFβ, but not IL-12p70, induced CXCL13 secretion (Fig. S4 B). To address the potential role of these molecules in the induction of effector cytokines by tonsil cDC2s and macrophages, we first analyzed differential expression of costimulatory molecules CD86, OX40L, and ICOSL (Fig. 5, A and B). cDC2s had the highest expression of CD86, and cDC2s and macrophages had the highest expression of OX40L. We could not detect ICOSL expression in this assay. At the mRNA level, pDCs had the highest expression of ICOSL (Fig. S4 C). To assess the ability of DCs and macrophages to produce soluble mediators, we stimulated them ex vivo with R848, or a combination of CD40L and IFN-γ to mimic T cell encounter, or all three stimuli at the same time. As a control, all APC subsets could secrete CXCL9 and CXCL10 in these conditions (Fig. S4 D). cDC2s were the best producers of IL-12p70 upon stimulation, with macrophages being also more efficient IL-12p70 producers than cDC1s, both at the protein and mRNA levels (Fig. 5 C and Fig. S4 E). While both cDC2s and macrophages secreted activin A and expressed INHBA (encoding for activin A) upon culture (Fig. 5 D and Fig. S4 F), macrophages expressed ex vivo the highest levels of INHBA (Fig. 5 D), suggesting that macrophages are the best producers of activin A in situ. Finally, to analyze the production of TGFβ, we first measured the expression of total TGFβ (both latent and active forms) in the supernatant. We found that all populations could produce TGFβ1 and TGFβ2 (Fig. 5 E), while TGFβ3 was undetectable (Fig. S4 G). We then analyzed the expression of integrin β8, which cleaves TGFβ precursor to release the active form (Travis et al., 2007; Fenton et al., 2017; Kelly et al., 2018). In freshly purified cells, macrophages expressed the highest levels of ITGB8, while both cDC2s and macrophages expressed ITGB8 upon culture (Fig. 5 F). This suggests that macrophages, and to a lesser extent cDC2s, are the most efficient for producing bioavailable TGFβ. Collectively, these results indicate that cDC2s and macrophages are the most efficient producers of Tfh-polarizing signals.
Figure 5.
cDC2 and macrophages are the best producers of Tfh cell–polarizing signals. (A and B) Tonsil DCs and macrophages were stained ex vivo for CD86, OX40L, and ICOSL. (A) Representative results. Filled gray histograms represent isotype control staining. (B) Stain index quantification for CD86 and OX40L. Each symbol represents an individual donor (n = 9 for CD86, n = 8 for OX40L). (C–F) Purified DC and macrophages were analyzed ex vivo or after 24 h culture with or without R848, CD40L, and IFN-γ. Cytokine secretion was analyzed by CBA (IL12p70) or ELISA (activin A and TGFβ), and mRNA expression by RT-qPCR. (C) n = 12 for IL12p70. (D) n = 10 for INHBA expression and for activin A. (E) n = 10 for TGFβ. (F) n = 10 for ITGB8 expression ex vivo, n = 3 after 24 h culture except for cDC1 (n = 1–3). Each symbol represents an individual donor. A.U., arbitrary units. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Wilcoxon test.
Production of IL-21 or CXCL13 is induced by distinct mechanisms
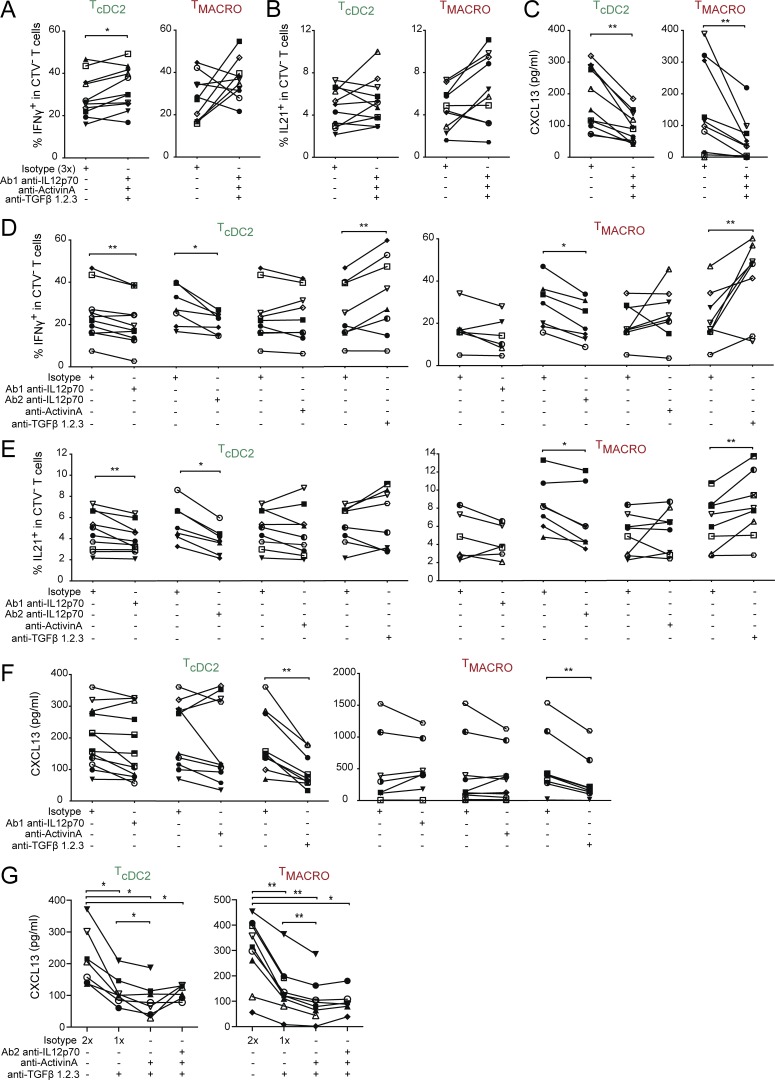
To test whether IL-12, activin A, or TGFβ was involved in the induction of effector molecules by tonsil cDC2s and macrophages, we analyzed Th cell polarization in the presence of blocking antibodies for IL-12, activin A, and TGFβ. We confirmed the efficacy of blocking antibodies (Fig. S4, H and I) and the absence of effect on T cell numbers in the co-culture (Fig. S4 J). A cocktail of the three blocking antibodies had a minimal effect on the production of IFN-γ (Fig. S4 K and Fig. 6 A) and increased the secretion of Th2 cytokines (Fig. S4 L). We did not observe any significant impact of these blocking antibodies on the production of IL-21 (Fig. 6 B), while the cocktail of blocking antibodies significantly decreased the production of CXCL13 by CD4+ T cells activated by cDC2 or macrophages (Fig. 6 C). We then blocked cytokines individually. The production of IFN-γ and IL-21 was decreased by both anti–IL-12 clones, remained unaffected by blocking activin A, and was increased when blocking TGFβ (Fig. 6, D and E). By contrast, CXCL13 secretion was significantly decreased by blocking TGFβ but not IL-12p70 or activin A (Fig. 6 F). Finally, to address whether activin A and TGFβ could act in a redundant or synergistic way, we blocked both activin A and TGFβ and observed an enhanced inhibition of CXCL13 secretion compared with TGFβ blockade alone (Fig. 6 G). Addition of anti–IL-12 did not further inhibit CXCL13 secretion, confirming it had a negligible role.
Figure 6.
IL-12 or activin A and TGFβ are involved in inducing IL-21 or CXCL13 production, respectively. Purified cDC2s and macrophages were co-cultured with naive CD4+ T cells in the presence of blocking antibodies, in combination or individually, against IL-12 (two different clones termed Ab1 and Ab2), activin A, and TGFβ1.2.3, or isotype control at the same total concentration. T cells polarized with cDC2 or CD14+ macrophages are termed TcDC2 and TMACRO, respectively. (A, B, D, and E) IFN-γ and IL-21 production was analyzed by intracellular staining after restimulation with PMA and ionomycin in presence of brefeldin A. (C, F, and G) CXCL13 secretion was analyzed by ELISA after restimulation with anti-CD3/CD28 beads. Each symbol represents an individual donor. (A–C) n = 11 for cDC2s and n = 10 for macrophages. (D and E) For cDC2: n = 10 for anti–IL-12 Ab1, n = 7 for anti–IL-12 Ab2, n = 9 for anti–activin A, and n = 8 for anti-TGFβ. For macrophages: n = 6 for anti–IL-12 Ab1, n = 7 for anti–IL-12 Ab2, n = 8 for anti–activin A, and n = 8 for anti-TGFβ. (F) For cDC2: n = 12 for anti–IL-12, n = 10 for anti–activin A, and n = 9 for anti-TGFβ. For macrophages: n = 7 for anti–IL-12, n = 10 for anti–activin A, and n = 8 for anti-TGFβ. (G) n = 7 for cDC2, n = 8 for macrophages. *, P < 0.05; **, P < 0.01; Wilcoxon test.
These results indicate the existence of two different mechanisms for the production of Tfh effector molecules, activin A and TGFβ being involved in CXCL13 production, while IL-12 is involved in IL-21 production.
Tonsil macrophages are localized in close proximity to Tfh cells in situ
To understand the respective roles of cDC2s and macrophages in Tfh cell induction in vivo, we analyzed the in situ localization of APC in tonsils, relative to the T cell zone, the B cell follicles, and Tfh cells. The chemoattractant receptor EBI2/GPR183 controls activated CD4+ T cells and cDC2 positioning at the B-T zone border in mouse lymphoid organs (Gatto et al., 2013; Yi and Cyster, 2013; Li et al., 2016). To determine whether tonsil DC subsets and macrophages displayed differential expression of EBI2/GPR183, we analyzed mRNA expression (Fig. S5 A). pDCs had the highest expression of GPR183, while cDC2s and macrophages had comparable expression and cDC1s the lowest expression level. Results from single-cell RNA-seq also indicated that macrophages and cDC2s had higher expression than cDC1s.
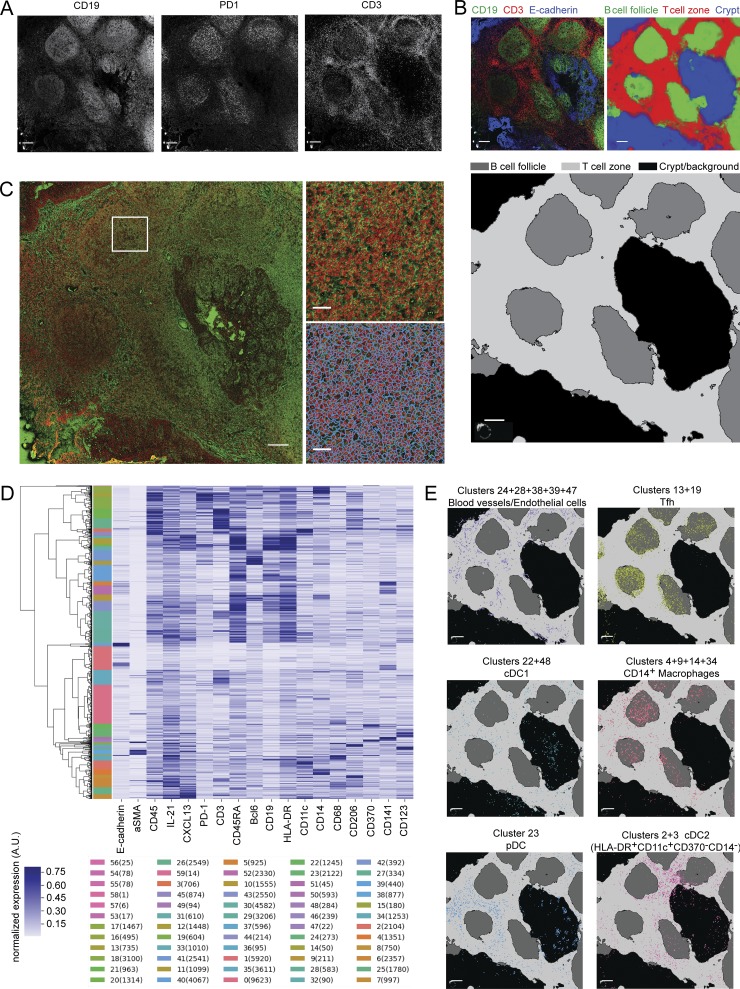
We and others have shown that human lymphoid organ cDCs are localized in the T cell zone (Segura et al., 2013a; Granot et al., 2017). However, the localization of tonsil macrophages is unclear. To address this question, we performed imaging on tonsil sections using a technique enabling simultaneous detection of multiple antibodies: imaging mass cytometry (Giesen et al., 2014; Chang et al., 2017). To assess in situ positioning within the tonsil in an unbiased way, we performed automated image analysis. We first normalized signal for each marker (Fig. 7 A). We performed image segmentation in order to define B cell follicles (CD19+), T cell zone (CD3+), and crypt (E-cadherin+) in the image (Fig. 7 B). We then segmented individual cells using DNA staining signal as well as merged signals for membrane molecules (Fig. 7 C). We then performed unsupervised hierarchical clustering to identify groups of cells with similar staining patterns (Fig. 7 D). We annotated these clusters for cell identity and projected the position of individual cells on the segmented map for visualization (Fig. 7 E). Finally, we performed distance analysis relative to the B cell follicles, the T cell zone, and the crypt/background region.
Figure 7.
Analysis pipeline for imaging mass cytometry. Tonsil sections were analyzed by imaging mass cytometry. (A) Signal for each marker is normalized. Example images are shown. Bars, 200 μm. (B) The image is segmented into B cell follicles, T cell zone, and crypt/background region. Input image for pixel classification is shown: CD3 in red, CD19 in green, and E-cadherin in blue (artificial colors). Posterior probability of the Random Forest trained by the Ilastik Software is shown. Segmentation result: dark gray is B cell follicles, light gray is T cell zone, and black is crypt and background. Bars, 200 µm. (C) Individual cells are detected using DNA intercalator staining and merged signals from membrane markers. Zoom-in of DNA staining in red and membrane markers in green (top) and result of the Watershed algorithm (bottom). Bars, 200 μm (left); 40 μm (right). (D) Individual cells displaying similar staining patterns are detected using unsupervised hierarchical clustering. 60 clusters are extracted in order to obtain overclustering. Individual clusters for one dataset with number of cells in each cluster are shown. (E) After cell identity assignment and merging of clusters with the same identity, individual cells are projected on the segmentation map. Example images are shown. Bars, 200 μm.
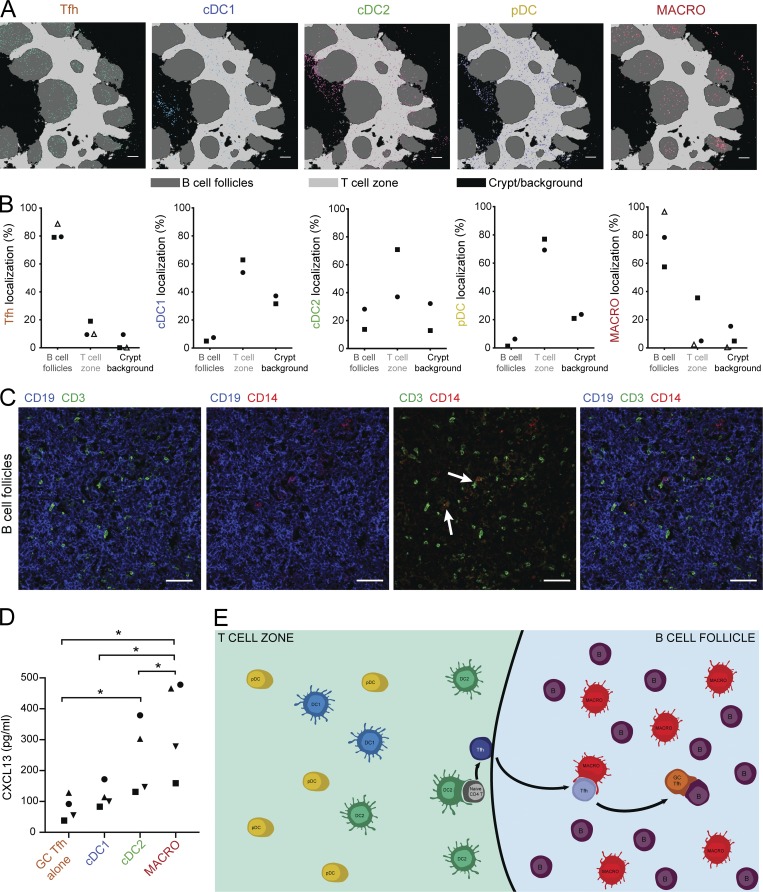
In a first imaging experiment, we found that the majority of macrophages, identified as CD11c+CD14+ cells (Fig. S5 B), were localized in B cell follicles where Tfh cells are also detected (Fig. S5 C). However, some of the clusters with high CD14+ staining also displayed high CD19+ staining (Fig. S5 B), suggesting neighboring effects for membrane markers, i.e., distinct cells localized next to each other for which membrane staining from one cell was assigned to both cells. Follicular DCs (FDCs), a type of stromal cells of the B cell follicle, have been reported to express CD14 (Liu et al., 1997). To rule out the detection of these cells and to confirm macrophage localization, we designed an improved panel including additional macrophage markers as well as DC subset markers. In the second imaging series, Tfh cells were found in B cell follicles as expected (Fig. 8 A), while pDC and cDC1 were positioned in the T cell zone as previously reported (Fig. 8, A and B). Because of low signal for CD1c, we localized cDC2 by analyzing clusters composed of HLA-DR+CD11c+CD14−Clec9A− cells. cDC2s were localized mostly in the T cell zones and at the border of the T cell zone and B cell follicles (Fig. 8 A), consistent with published work (Granot et al., 2017). Cells from macrophage clusters (identified as CD45+CD11c+CD14+CD68+) were localized mainly in the B cell follicles. Of note, we also observed in the T cell zone a population of CD11c+CD14−CD206+ cells (Fig. S5, D and E), which may represent perivascular macrophages and which would have been absent from the CD14+ macrophage population we analyzed. Other cell populations were found as expected in the crypt region for crypt epithelial cells, B cell follicle for B cells, and T cell zone for blood vessels (Fig. S5, D and E). Distance analysis of the three different tonsil donors confirmed the preferential positioning of CD14+ macrophages in the B cell follicles (Fig. 8 B).
Figure 8.
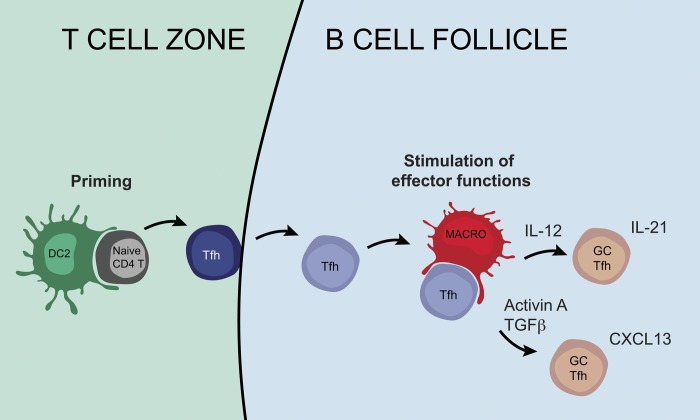
Tonsil CD14+ macrophages colocalize with Tfh cells in the B cell follicles. (A and B) Tonsil sections were analyzed by imaging mass cytometry. (A) Topographic representation of cells identified as Tfh cells, cDC1s, cDC2s, pDCs, and CD14+ macrophages. Representative of two different donors stained with the same antibody panel. Bars, 200 µm. (B) Proportion of each population in the different topographic regions. Each symbol represents an individual donor. Open symbols are from a first experiment, and black symbols are from a second series using additional markers. (C) Immunostaining of tonsil sections by confocal microscopy. Representative of three different donors. Arrows indicate cellular contacts between CD3+ and CD14+ cells. Bars, 50 µm. (D) Purified GC Tfh cells were co-cultured with autologous purified cDC1s, cDC2s, or macrophages. CXCL13 secretion in the supernatant was measured by ELISA. Each symbol represents an individual donor (n = 4). *, P < 0.05; paired t test. (E) Working model: cDC2s interact with naive CD4+ T cells at the border of the T cell zone and B cell follicle to initiate Tfh cell differentiation, while macrophages interact with maturing Tfh cells in the B cell follicle to further instruct their polarization and the production of effector molecules.
Consistent with their localization in the B cell follicles and their involvement in Tfh responses and B cell help, we found that tonsil macrophages were the best producers ex vivo of B-cell activating factor (encoded by TNFSF13B; Fig. S5 F), a survival and proliferation factor for B cells.
To confirm the in situ positioning of tonsil CD14+ macrophages, we performed immunostaining and confocal microscopy on tonsil sections. We costained for CD19, CD3, and CD14. CD14+ cells were mainly localized in the B cell follicles (marked by CD19 staining), and we observed instances of cell–cell contact between CD14+ and CD3+ cells in the B cell follicles (Fig. 8 C), suggesting that macrophages and Tfh cells can interact in vivo. Finally, to directly address whether macrophages can stimulate GC Tfh cells, we co-cultured purified cDCs and macrophages with autologous GC Tfh cells (Fig. 8 D). Macrophages were the most efficient for inducing CXCL13 secretion by GC Tfh cells.
Collectively, these results indicate a differential localization of cDC and CD14+ macrophages within tonsils, and suggest that, in a physiological setting, cDC2 would interact with naive CD4+ T cells at the border of the T cell zone and B cell follicles, while CD14+ macrophages would preferentially interact with maturing Tfh cells in the B cell follicles (Fig. 8 E).
Discussion
In this work, we have analyzed the ability of human lymphoid organ-resident DCs and macrophages to polarize Tfh cells. We demonstrated that cDC2s and CD14+ macrophages are the most potent for inducing Tfh polarization. We evidenced that human Tfh cells comprise two different effector states with similar transcriptional profiles and producing either IL-21 or CXCL13. Our results showed that activin A and TGFβ were involved in the induction of CXCL13 production, while IL-12 was involved in the induction of IL-21 production. Further, we found that tonsil CD14+ macrophages are located close to Tfh cells in situ, supporting the physiological relevance of our findings. We therefore propose a model in which cDC2s and CD14+ macrophages have complementary and sequential roles in the induction of Tfh responses, with cDC2s priming “preTfh” in the T cell zone and macrophages interacting with maturing Tfh cells in the B cell follicles to further instruct their polarization and the production of effector molecules.
A wealth of studies have shown that DC subsets possess differential functional abilities, in particular for antigen presentation and T cell polarization. We showed that among human resident DC subsets, cDC2s are the most efficient for secreting IL-12 and activin A and for inducing Tfh polarization. Regardless of their tissue localization, DCs from a given lineage share a core transcriptional program that is likely key to their functional abilities (Miller et al., 2012; Heidkamp et al., 2016). We would therefore predict that the functional specialization of tonsil cDC2 for Tfh induction will be a feature shared with all human cDC2. This idea is supported by our finding that blood and lymph node cDC2s are more efficient at inducing CXCL13 production than cDC1s, and by reports of cDC2s being the best producers of IL-12 among blood and spleen DC subsets (Mittag et al., 2011; Nizzoli et al., 2013). However, previous work has shown that the skin equivalent of cDC2s, dermal CD1a+CD1c+ DCs, are poor inducers of CXCL13 secretion compared with skin CD14+ DCs (Klechevsky et al., 2008; Segura et al., 2012). By contrast, human skin cDC2s efficiently induce IL-21–producing CD4+ T cells (Penel-Sotirakis et al., 2012) and are able to produce IL-12 in situ (Yawalkar et al., 2009). Therefore, we propose that human cDC2s are specialized for inducing Tfh cells, with a superior ability for stimulating IL-21 production. In vivo targeted delivery to DC of vaccine antigens or modulating agents has become the subject of intense investigation. Our results indicate that cDC2 should be preferentially targeted for vaccine delivery when aiming for efficient Tfh responses.
In contrast to DC, mouse macrophage functions are strongly imprinted by their tissue of residence (Lavin et al., 2015; Varol et al., 2015). Our results extend this concept to human macrophages. While tonsil macrophages are efficient inducers of Tfh polarization and CXCL13 secretion, we show that macrophages from peritoneal tumor ascites, synovial fluid of rheumatoid arthritis patients, or lung are not endowed with the same capacity. Consistent with this, we have previously shown that peritoneal ascites macrophages do not secrete IL-12p70 (Tang-Huau et al., 2018), in contrast to tonsil CD14+ macrophages. Tonsils contain a population of tingible body macrophages, which phagocytose apoptotic B cells in the GC. We did not determine whether Tfh-inducing macrophages are tingible body macrophages. What signals derived from the tonsil microenvironment drive these functional features also remains to be investigated.
Here we also evidence that human in vivo–differentiated mo-DCs, isolated from tumor ascites or synovial fluid of rheumatoid arthritis patients, are potent inducers of CXCL13 secretion, suggesting they efficiently induce Tfh polarization. Similar ability has been evidenced for human skin CD14+ DCs (Klechevsky et al., 2008; Segura et al., 2012), which are thought to be monocyte derived (McGovern et al., 2014). Consistent with this, mouse mo-DCs enhance Tfh cell differentiation during adjuvant-induced inflammation (Chakarov and Fazilleau, 2014). Tfh cells are deleterious in chronic inflammatory and autoimmune diseases (Ueno, 2016). These findings suggest that mo-DCs could contribute to the physiopathology by inducing Tfh cells or stimulating peripheral CXCL13-producing Th cells (Rao et al., 2017), in addition to previously described mechanisms (Tang-Huau and Segura, 2019).
The existence of functionally different Tfh cell populations is consistent with previous work describing two pools of spatially segregated Tfh cells in human tonsils (Bentebibel et al., 2011). Extra-follicular Tfh cells (CXCR5low) produce high levels of IL-21 and low levels of CXCL13 compared with GC CXCR5high Tfh cells (Bentebibel et al., 2011). These IL-21–producing Tfh cells were proposed to be specialized for helping naive and memory B cells, while GC Tfh cells would be involved in helping GC B cells. An alternative hypothesis would be that IL-21– and CXCL13-producing human Tfh cells represent two distinct stages of differentiation or two different effector states. Given that we failed to detect, in our single-cell RNA-seq analysis, the specific expression of IL21 versus CXCL13 in distinct Tfh cell populations, we favor the idea that effector Tfh cells are instructed by signals from their microenvironment to secrete either IL-21 to directly help B cells, or CXCL13 to promote cell migration toward the GC. We propose that macrophages are preferentially involved in the latter, due to their localization in the B cell follicles and the production of high levels of activin A and bioactive TGFβ.
The cytokine requirements for human Tfh induction have been mostly studied by in vitro stimulation of CD4+ T cells with purified cytokines or using in vitro–derived model DCs. In this setting, IL-12p70, activin A, and TGFβ were identified as regulators of human Tfh differentiation (Schmitt et al., 2009, 2014; Locci et al., 2016). We show that these findings also apply to Tfh polarization by tonsil APCs. IL-12p70 is involved in the induction of IL-21 production, while activin A and TGFβ are involved in the induction of CXCL13. This is consistent with previous reports using purified individual cytokines and showing the induction of CXCL13 production by activin A or TGFβ but not by IL-12p70 (Jacquemin et al., 2015; Kobayashi et al., 2016; Locci et al., 2016), and of IL-21 by IL-12p70 but not activin A (Locci et al., 2016). Blocking IL-12p70 in our assay had only a partial effect on IL-21 production, which could indicate that other molecules may be involved. In particular, the role of costimulatory molecules, such as OX40L (Jacquemin et al., 2015; Pattarini et al., 2017), in the induction of IL-21 by human APC will need further investigation. Whether activin A and TGFβ are redundant and which cytokine is the most important in vivo also remain to be determined.
Accumulating evidence indicates that disregulated Tfh responses are involved in the physiopathology of autoimmune diseases such as lupus, type 1 diabetes, or rheumatoid arthritis, and molecules inhibiting Tfh function or differentiation are tested in clinical trials (Ueno et al., 2015; Ueno, 2016). By contrast, robust Tfh responses are essential for the development of protective antibodies in response to vaccines (Crotty, 2014). Tfh cells therefore represent a promising target for modulating antibody responses for the treatment of autoimmune disorders or the enhancement of vaccine efficacy. By enabling a better understanding of human Tfh differentiation, our results should provide new opportunities for the therapeutic manipulation of Tfh cells.
Materials and methods
Human samples
Tonsils from healthy patients undergoing tonsillectomy were obtained from Hôpital Necker (Paris, France). Uninvaded lymph nodes from breast cancer patients were obtained from Hôpital de l’Institut Curie (Paris, France) in accordance with hospital ethical guidelines. Samples of ovarian or breast tumor ascites from untreated patients were obtained from Hôpital de l’Institut Curie. Samples of synovial fluid were obtained from Hôpital Cochin (Paris, France) in accordance with hospital ethical guidelines. Buffy coats from healthy donors were obtained from Etablissement Français du Sang (Paris, France) in accordance with Institut National de la Santé et de la Recherche Médicale ethical guidelines. According to French Public Health Law (art L 1121–1-1, art L 1121–1-2), written consent and Institutional Review Board approval are not required for human noninterventional studies.
Lung resection tissue was obtained from Sahlgrenska University Hospital (Gothenburg, Sweden). Samples were obtained in accordance with hospital and AstraZeneca ethical guidelines and with written consent.
Tonsil and lymph node APCs isolation
Tonsil or lymph node samples were digested as described previously (Durand and Segura, 2016). In brief, DC subsets and macrophages were selected by density gradient, enriched by negative selection, and isolated by cell sorting on a FACS Aria instrument (BD Biosciences). Antibodies used were anti-CD11c (clone Bu15; BioLegend), HLA-DR (clone LN3; eBioscience), CD14 (clone 61D3; eBioscience), CD1c (clone L161; BioLegend), CD141 (clone AD5-14H12; Miltenyi Biotec), and CD123 (clone AC145; Miltenyi Biotec).
Ascites and synovial fluid APCs isolation
Cells were isolated after centrifugation on a Ficoll gradient (Lymphoprep; Greiner Bio-One) followed by cell sorting on a FACS Aria instrument (BD Biosciences) as described previously (Segura et al., 2013b). In brief, DCs were gated as HLA-DR+CD11c+CD16−CD1c+ and macrophages as HLA-DR+CD11c+CD16+CD1c−. Anti-CD16 antibody was clone 3G8 (BD Biosciences).
Lung macrophage isolation
Macroscopically healthy lung resection tissue was obtained from four patients undergoing lobectomy or pneumonectomy due to lung cancer (three males and one female; two ex-smokers and two nonsmokers; average age, 72 ± 8 yr). Lung tissue was extensively flushed with PBS to remove excessive blood contamination and alveolar cells. The flushed tissue was subsequently cut into small pieces (0.5 cm × 0.5 cm) and incubated in a digestion buffer, containing 1 mg/ml Collagenase D (Sigma-Aldrich) in Roswell Park Memorial Institute medium (Invitrogen), for 30 min at 37°C. Afterward, the lung tissue pieces were minced over a 100-µm cell strainer (Miltenyi Biotec) to obtain a single cell suspension. Subsequently, the HLA-DR+ cell fraction was prepurified by HLA-DR microbeads according to manufacturer’s protocol (Miltenyi Biotec). HLA-DR+ cells were then incubated with Aqua Live Dead (Thermo Fisher Scientific); Fc Block; antibodies against CD45 (clone HI30), CD16 (clone 3G8), and CD11c (clone B-ly6; all from BD Biosciences); CD14 (clone 63D3); and CD1c (clone L161; both from BioLegend) in PBS for 30 min at 4°C. Macrophages were sorted via FACS Aria III (BD Biosciences) to a purity of >95%.
Blood naive CD4 T cell isolation
Peripheral blood mononuclear cells were prepared by blood centrifugation on a Ficoll gradient (Lymphoprep). Naive CD4+ T cells were isolated by negative selection using the Human Naive CD4+ T Cell Isolation Kit according to the manufacturer’s instructions (Stem Cell). Cell purity (defined as CD4+ CD45 RA+ CD45 RO−) was evaluated after each naive CD4 T cell isolation by flow cytometry on a Macsquant analyzer (Miltenyi Biotec), and only cells with a purity >95% were used.
Tonsil Tfh cell isolation
Tonsils were mechanically disrupted by first cutting the tissue samples with a scalpel into small pieces, and then dissociating into single cells using gentleMACS technology (C tube and gentleMACS; Miltenyi Biotec). Alternatively, cells were dissociated by forcing through a 40-µm cell strainer. Single-cell suspension was then enriched for light density cells by a Ficoll gradient centrifugation (Lymphoprep). Tonsil CD4+ T cells were enriched using CD4+ T cell isolation kit (Miltenyi Biotec) and used for intracellular flow cytometry or further purified by cell sorting. Naive CD4+ T cells, GC Tfh cells, and extra-follicular Tfh cells were sorted using antibodies against CD4 (clone RPA-T4; BioLegend), CD19 (clone HIB19; eBioscience), CD45RO (clone UCHL1; BD Biosciences), CXCR5 (clone RF8B2 from BD Biosciences or clone J252D4 from BioLegend), and PD-1 (clone EH12.1 or clone EH12.2H7 from BD Biosciences) on a FACS Aria instrument (BD Biosciences).
Th cell polarization by APCs
APCs (20,000 cells) from one donor were cultured with blood naive CD4+ T cells (50,000 cells) from another donor, for 5 or 6 d in Yssel medium supplemented with 10% FCS. Each allogeneic culture was performed with different pairs of donors. To analyze T cell polarization, total cells from each culture well were washed and incubated with anti-CD3/CD28 beads (Thermo Fisher Scientific) for 24 h in X-VIVO 15 serum free medium (Lonza). Supernatants were collected, and cytokine secretion was assessed by Cytometric Bead Array for IFN-γ, IL-4, IL-5, IL-13, and IL-17A (BD Biosciences) or by ELISA for CXCL13 (R&D Bio-techne). Note that cell numbers were not normalized before restimultion with anti-CD3/CD28 beads, and that ELISA for CXCL13 has a detection limit of 30 pg/ml. To analyze T cell proliferation, naive CD4+ T cells were stained with Cell Trace Violet (CTV; Thermo Fisher Scientific) before culture. For proliferation kinetics, the cells were harvested at days 3, 4, 5, and 6, stained for CD4 and viability (Live/dead eFluor 780; Thermo Fisher Scientific) and analyzed on a FACS Verse instrument (BD Biosciences). In some experiments, DCs and macrophages were preactivated with R848 for 3 h in Yssel medium with 1 µg/ml of R848 (InvivoGen) at 37°C, then washed prior to the co-culture with naive T cells. For blocking experiments with antibodies against IL-12p70 (clone 24910; R&D Bio-Techne), IL-12p35 (clone B-T21; eBioscience), activin A (clone 69403; R&D Bio-techne), and TGFβ1,2,3 (clone 1D11; R&D Bio-techne), two doses of 20 µg/ml of each antibody or isotype control (mouse IgG1, clone 11711; R&D Bio-techne) were added to the co-culture, one at the beginning of the culture and the second 24 h later.
For lung macrophages, mixed leukocyte reactions were set up with 20,000 macrophages and 50,000 allogeneic naive blood CD4+ T cells, stained with 0.25 µM CFSE (eBioscience). Expression of intracellular cytokines was assessed after 7 d of co-culture.
Autologous co-culture
For autologous culture, three fourths of tonsils were processed for isolation of APCs as described in Tonsil and lymph node APCs isolation. One fourth of tonsils was mechanically disrupted by first cutting the tissue samples with a scalpel into small pieces, and cells were dissociated by forcing through a 40-µm cell strainer. GC Tfh cells were then isolated by cell sorting as described in Tonsil Tfh cell isolation (gated as CD45RO+PD-1+CXCR5high). Purified GC Tfh cells (50,000 cells) from one donor were cultured for 24 h alone or with purified APCs (20,000 cells) from the same donor in Yssel medium supplemented with 5% FCS. CXCL13 secretion was assessed in the culture supernatant by ELISA.
Th cell polarization with recombinant cytokines
Blood naive CD4+ T cells (75,000 cells) were cultured with anti-CD3/CD28 beads (2 µl/well) for 5 d in X-VIVO 15 serum-free medium, in the presence or absence of recombinant human IL-12p70 (2 ng/ml; R&D Bio-techne), activin A (100 ng/ml; R&D Bio-techne), and/or TGFβ1 (2 ng/ml; Peprotech). For blocking experiments with antibodies against IL-12p70 (clone 24910 from R&D Bio-techne or clone B-T21 from eBioscience), activin A (clone 69403; R&D Bio-techne), and TGFβ1,2,3 (clone 1D11; R&D Bio-techne), one dose of 20 µg/ml of each antibody or isotype control (mouse IgG1, clone 11711; R&D Bio-techne) was added at the start of the culture. For analysis of CXCL13 secretion, cells were washed and restimulated with anti-CD3/CD28 beads for 18 h in X-VIVO 15 serum-free medium.
Intracellular staining for flow cytometry
To assess the expression of intracellular cytokines, except for lung macrophage co-cultures, T cells were stimulated with PMA (50 ng/ml), ionomycin (1 µg/ml), and BFA (4 µg/ml; all from Sigma-Aldrich) for 4 h (for tonsil T cells) or 5.5–6 h (for co-cultures). After stimulation, cells were stained for surface PD-1 and CD45RO (for tonsil T cells) or CD4 (for co-cultures) for 30 min at 4°C, washed, and stained with Live/dead eFluor780 (Thermo Fisher Scientific) for 10 min at 4°C. Then the cells were fixed and permeabilized (Intracellular Fixation & Permeabilization Buffer Set; eBioscience) and stained for intracellular cytokines at room temperature for 60 min in a buffer containing 2% of normal mouse serum and 10 µg/ml human Fc block (BD Biosciences). Antibodies were anti–IFN-γ (clone 4S.B3; eBioscience), CXCL13 (clone 53610; R&D Bio-techne), IL-21 (clone 3A3-N2.1; BD Biosciences), IL-17A (clone BL168; BioLegend), or IL-4 (clone 8D4-8; eBioscience). For tonsil T cells, cells were also stained for CXCR5 by intracellular staining. The samples were acquired on a FACS Verse instrument (BD Biosciences).
For lung macrophage co-cultures, T cells were stimulated with PMA (30 ng/ml) and ionomycin (1 µg/ml) for 6 h, in the presence of GolgiPug and GolgiStop (both from BD Biosciences) for 4 h. After stimulation, cells were stained with Aqua Live Dead and anti-CD3 (clone UCHT1; BD Biosciences) for 30 min at 4°C. Then cells were fixed and permeabilized (Fixation/Permeabilization Buffer Set; BD Biosciences) and stained against IL-21 (clone 3A3-N2; eBioscience) and CXCL13 (clone 53610; R&D Systems). The samples were acquired on a FACS Fortessa instrument (BD Biosciences).
To assess Bcl6 expression, cells were stained for surface CD4, CXCR5, and PD-1 for 30 min at 37°C, washed, and stained with Live/dead eFluor780 for 20 min at 4°C. Then the cells were fixed and permeabilized (Intracellular Fixation & Permeabilization Buffer Set; eBioscience) and stained for Bcl6 (clone K112-91; BD Biosciences) at room temperature for 60 min in a buffer containing 2% of normal mouse serum. The samples were acquired on a FACS Verse instrument (BD Biosciences).
Fluorescent in situ hybridization coupled to flow cytometry
Cells were analyzed using a PrimeFlow RNA Assay (eBioscience) as previously described (Coillard and Segura, 2018), using human Bcl6 type 1 probe, human IL-21 type 4 probe, and human CXCL13 type 6 probe (Thermo Fisher Scientific). Cells were also stained for extracellular CD19, CXCR5, and PD-1. Dead cells were identified using fixable viability dye eFluor 506 (eBioscience). Cells were analyzed on a FACS Verse instrument (BD Biosciences).
Cytokine secretion by APCs
Sorted APCs were cultured for 24 h in Yssel medium in the presence or absence of R848 (1 µg/ml; InvivoGen), dimerized CD40-ligand (2.5 µg/ml; Enzo Life Science) and recombinant human IFN-γ1b (0.25 µg/ml; Miltenyi Biotec). Supernatants were collected, and cytokine secretion was assessed by Cytometric Bead Array for IFN-α, IL-1β, IL-6, IL-12p70, CXCL9, and CXCL10 (BD Biosciences) or ELISA for activin A (R&D Bio-techne) and TGFβ1, TGFβ2, and TGFβ3 (MSD).
Costimulatory molecules expression
Enriched APCs were stained with antibodies recognizing OX40L (clone ANC10G1; Ancell), ICOSL (clone 136726; R&D Bio-techne), or CD86 (clone FUN-1; R&D Bio-techne), or isotype control, and HLA-DR (clone LN3; eBiosciences), CD14 (clone 61D3; eBiosciences), CD11c (clone Bu15; BioLegend), CD1c (clone L161; BioLegend), CD304 (clone 12C2; BioLegend), and CD141 (clone AD5-14H12; Miltenyi Biotec). Samples were acquired on a FACS Verse instrument (BD Biosciences). Stain index was calculated to quantify the level of expression as follows: stain index = (MFIsample − MFIisotype)/(2 × SDisotype), where MFI indicates mean fluorescence intensity, and SD indicates standard deviation.
Reverse transcription quantitative PCR (RT-qPCR)
Cells were lysed in RLT buffer (QIAGEN) after sorting (ex vivo) and/or after 24 h of in vitro activation. The RNA extraction was performed with the RNeasy micro kit (QIAGEN) according to the manufacturer’s instructions. Reverse transcription was performed on total RNA using superscript II polymerase (Invitrogen) with random hexamers and oligo deoxythymine and deoxynucleotide triphosphates (Promega). Transcript quantification was done by real-time PCR on a 480 LightCycler instrument (Roche) using a master mix (Eurogentec), and the following TaqMan Assays (Life Technologies): TLR7 (Hs00152971_m1), TLR8 (Hs00152972_m1), ITGB8 (Hs00174456_m1), INHBA (Hs01081598_m1), IL12A (Hs01073447_m1), IL12B (Hs01011518_m1), TGFB1 (Hs00998133_m1), and TNFSF13B (Hs00198106_m1). The second derivative maximum method was used to get the Crossing points from each analyte. The relative expression of transcripts was quantified by comparison to the mean of the two housekeeping genes: HPRT1 (Hs02800695_m1) and GAPDH (Hs99999905_m1).
Immunofluorescence analysis
Tonsils were fixed in 4% paraformaldehyde, washed with PBS, incubated overnight in 30% sucrose at 4°C, embedded in Tissue-Tek O.C.T. (Sakura), and stored at −80°C. Sections 5 µm in thickness were cut with a Leica CM 1850 UV cryomicrotome (Leica Microsystems). For staining, sections were incubated 30 min in Tris-NaCl blocking (TNB) buffer (0.1 M Tris-HCl, 0.15 M NaCl, and 0.5% Blocking Reagent; Perkin Elmer), then incubated overnight with primary antibodies in TNB at 4°C. After five washes in PBS, sections were incubated 1 h with fluorochrome-conjugated secondary antibodies or fluorochrome-conjugated streptavidin and DAPI (Roche) in TNB. Samples were mounted on coverslips with fluorescent mounting medium (Dako) and visualized using a Zeiss LSM710 confocal microscope using a 25×/0.8 numerical aperture objective. Images were analyzed with ImageJ. Primary antibodies used were biotinylated or purified anti-human CD19 (clone HIB19; eBioscience), purified anti-human CD3 (clone UCHT1; Invitrogen), biotinylated anti-human BDCA1 (clone AD5-8E7; Miltenyi Biotec), biotinylated anti-human CD14 (clone HCD14; BioLegend), or purified anti-human Clec9A (clone 8F9; Miltenyi Biotec). The corresponding streptavidin conjugate and secondary antibodies (all from Molecular Probes) were Alexa Fluor 488 anti-rabbit, Alexa Fluor 568 or Alexa Fluor 647 anti-mouse, and Alexa Fluor 568 or Alexa Fluor 647 streptavidin.
Imaging mass cytometry
Human tonsils were fixed in 4% paraformaldehyde, washed with PBS, incubated overnight in 30% sucrose at 4°C, embedded in Tissue-Tek O.C.T. (Sakura), and stored at −80°C. Sections 5 µm in thickness were cut with a Leica CM 1850 UV cryomicrotome. Sections were stored at −80°C. Before use, slides were warmed up to −20°C during 1 h and equilibrated to room temperature for 10 min. Slides were rinsed three times in PBS thoroughly to remove the tissue-freezing matrix. Tissue sections were blocked with 3% BSA (Rockland) in PBS for 45 min at room temperature. After washing, the slides were incubated with metal-conjugated antibodies in PBS 0.5% BSA. Tissue sections were incubated with the antibody cocktail overnight at 4°C in a humidified chamber, then washed several times in 0.1% Triton X-100 (Thermo Fisher Scientific) in PBS. Slides were rinsed in PBS and stained with DNA intercalator-Ir (1:2,000 dilution; Fluidigm) for 30 min at room temperature. Slides were washed in distilled deionized water and air-dried for ∼30 min. Slides were inserted into the Hyperion Imaging System (Fluidigm) for data acquisition (Chang et al., 2017).
We used two different metal-conjugated antibody cocktails (all antibodies were from Fluidigm, unless otherwise indicated, in which case they were conjugated in-house). The first experiment was performed with Rutenium Red (R&D Systems), Avanti-Lipid (R&D Systems), anti-alphaSMA-141Pr (clone IA4), CD8a-146Nd (clone RPA-T8), CD123-148Nd (clone 45016), CD1c/BDCA1-152Sm (clone AD5-8E7; Miltenyi Biotec), CD44-153Eu (clone 691534), CD45-154Sm (clone MEM-28), CD45RA-155Gd (clone HI100), CD14-156Gd (clone HCD14), CXCL13-157Gd (polyclonal; R&D Biotechne), CD324/E-cadherin-158Gd (clone 24 E10), CD11c-159Tb (clone Bu15), CTLA4-161Dy (clone OTI1G10; Origene), Foxp3-162Dy (clone 259/C7), Bcl6-163Dy (clone K112-91), IL-21-164Dy (polycolonal; Novus Bio), Bcl2-166Er (clone 124; NeoBioTech), Ki67-168Er (clone B56), CD19-169Tm (clone HIB19), CD3-170Er (clone UCHT1), CD185/CXCR5-171Yb (clone 51505), collagen14-173Yb (polyclonal; Thermo Fisher Scientific), CD279/PD-1-175Lu (clone EH12.2H7), histoneH3-176Yb (clone D1H2), and DNA-191/193Ir. We excluded from analysis several markers showing low staining: CD123, CD1c, CD44, CTLA4, CXCR5, and collagen14. The second series was performed with anti–alphaSMA-141Pr (clone IA4), CD303/BDCA2-147Sm (clone 201A), CD123-143Nd (clone 6H6), ICOS-148Nd (clone D1K2T), CD11b-149Sm (clone EPR1344), CD1c/BDCA1-biotin (clone AD5-8E7; Miltenyi Biotec), CD45-152Sm (clone CD45-2B11), CD45RA-155Gd (clone HI100), CD14-156Gd (clone HCD14), CXCL13-157Gd (polyclonal; R&D Biotechne), CD324/E-cadherin-158Gd (clone 24 E10), CD11c-159Tb (clone Bu15), CD370/Clec9A-161Dy (clone 8F9), Bcl6-163Dy (clone K112-91), IL-21-164Dy (polycolonal; Novus Bio), CD206-168Er (clone 15–2), CD19-169Tm (clone HIB19), CD68-171Yb (clone Y1/82A), CD141/BDCA3-165Ho (clone AD5-14H12; Miltenyi Biotec), CD3-170Er (clone UCHT1), CD185/CXCR5-151Eu (clone MM0225), HLA-DR-174Yb (clone L243), CD279/PD-1-175Lu (clone EH12.2H7), CD56-176Yb (clone HCD56), and DNA-191/193Ir. The second series was subsequently incubated with Neutravidin-173Yb. We excluded from analysis several markers showing low staining: CD303, ICOS, CD11b, CD1c, CXCR5, and CD56.
Analysis of imaging mass cytometry data
First, images were segmented by pixel classification into three regions: B cell follicles, T cell zone, and background/crypt. For this, we used the open-source software Ilastik (Sommer, C., C. Strähle, U. Köthe, F. A. Hamprecht. 2011. Eighth IEEE International Symposium on Biomedical Imaging). We manually annotated pixels inside each of the three regions, and the software then learned to perform segmentation in the entire image. Second, we detected individual nuclei from the DNA staining with in-house–developed software. After removal of salt-and-pepper noise and normalization of the image, we applied a LoG filter (sigma = 0.004), thereby enhancing spot-like features, which appear as local minima. We then applied the h-minima transform (Soille, 2003) to detect local minima with a reasonable local contrast (h = 0.001), thereby avoiding oversegmentation into nuclei fragments. The cellular region is calculated by applying the watershed transformation to a linear combination of the distance map of the segmentation background and the average image of all membrane-bound marker proteins, and by further restricting the cellular regions by a circle of a defined radius (r = 8 pixels) around each selected local minimum in order to avoid oversized cells in regions of low density. From this, we could now measure the intensity in each channel, and thus a proxy of the expression level of the protein in each individual cell. The dynamic ranges of the different channels vary considerably between each other. As there is no prior information on the overall abundance of proteins, we normalized the individual channels, such that the first percentile was assigned to 0 and the 99th percentile to 1. We then performed clustering of single-cell expression profiles and extracted 60 clusters. The number of clusters was chosen (arbitrarily) in order to obtain an oversegmentation of cell types. As the expression also varied spatially, we then manually checked different clusters and the corresponding cell localizations; the cell types correspond to fusions of clusters. We identified clusters based on phenotypic markers: epithelial cells (E-cadherin+), blood vessels and endothelial cells (α-SMA+), B cells (CD19+CD45RA+HLA-DR+), B cells and FDC (CD19+CD45RA+HLA-DR+CD14+CD11c−CD68−), B cells and Tfh cells (CD19+CD45RA+HLA-DR+PD1+Bcl6+), CD14−CD206+ macrophages (CD14−CD206+CD11c+), Tfh cells (CD3+PD1+Bcl6+), cDC1 (CD370+CD141+HLA-DR+), cDC2 (HLA-DR+CD11c+CD14−CD370−), pDCs (CD123+HLA-DR+), and macrophages (CD11c+HLA-DR+CD14+CD68+). This provided us with coordinates for each population studied and their distance distribution to the border of the corresponding regions. All scripts were written in Python, and made use of the open-source library scikit-image (van der Walt et al., 2014).
Normalized images for each staining were deposited in BioStudies (https://www.ebi.ac.uk/biostudies/studies/S-BSST221).
The software is publicly available at https://thomaswalter.github.io/ImageMassCytometry/.
Single cell RNA-seq library preparation
Cellular suspension (3,500 cells) was loaded on a 10× Chromium instrument (10× Genomics) according to the manufacturer’s protocol based on the 10× GEMCode proprietary technology. For the APC analysis, tonsil HLA-DR+CD11c+CD14− and tonsil HLA-DR+CD11c+CD14+ cells were sorted and loaded separately. For the CD4+ T cell analysis, CXCR5+PD-1low T cells, CXCR5+PD-1int T cells, and CXCR5+PD-1high T cells were sorted and loaded separately. Single-cell RNA-seq libraries were prepared using the Chromium Single Cell 3′ v2 Reagent Kit (10× Genomics) according to the manufacturer’s protocol. Briefly, the initial step consisted in performing an emulsion where individual cells were isolated into droplets together with gel beads coated with unique primers bearing 10× cell barcodes, unique molecular identifiers (UMIs), and poly(dT) sequences. Reverse transcription reactions were engaged to generate barcoded full-length cDNA followed by the disruption of emulsions using the recovery agent and cDNA cleanup with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific). Bulk cDNA was amplified using a GeneAmp PCR System 9700 with 96-Well Gold Sample Block Module (Applied Biosystems; 98°C for 3 min; cycled 14 times: 98°C for 15 s, 67°C for 20 s, and 72°C for 1 min; 72°C for 1 min; held at 4°C). Amplified cDNA product was cleaned up with the SPRIselect Reagent Kit (Beckman Coulter). Indexed sequencing libraries were constructed using the reagents from the Chromium Single Cell 3′ v2 Reagent Kit, following these steps: (1) fragmentation, end repair, and A-tailing; (2) size selection with SPRIselect; (3) adaptor ligation; (4) postligation cleanup with SPRIselect; and (5) sample index PCR and cleanup with SPRIselect beads. Library quantification and quality assessment were performed using a Qubit fluorometric assay (Invitrogen) with double stranded DNA HS (High Sensitivity) Assay Kit and Bioanalyzer Agilent 2100 using a High Sensitivity DNA chip (Agilent Genomics). Indexed libraries were equimolarly pooled and sequenced on an Illumina HiSeq2500 using paired-end 26 × 98 bp as the sequencing mode.
Single-cell RNA-seq data analysis
Single-cell expression was analyzed using the Cell Ranger Single Cell Software Suite (v2.0.1) to perform quality control, sample de-multiplexing, barcode processing, and single-cell 3′ gene counting (Zheng et al., 2017). Sequencing reads were aligned to the UCSC hg38 transcriptome using the Cell Ranger suite with default parameters. Samples for APC or CD4+ T cells were merged using the Cellranger aggregate function with default parameters. A total of 4,402 single cells were analyzed for the APC dataset, and 4,342 single cells were analyzed for the CD4+ T cell dataset. Mean raw reads per cell were 77,372 for the tonsil HLA-DR+CD11c+CD14− sample, 119,822 for the tonsil HLA-DR+CD11c+CD14+ sample, 162,772 for the CXCR5+PD-1low T cell sample, 110,381 for the CXCR5+PD-1int T cell sample, and 64,291 for the CXCR5+PD-1high Tfh cell sample. Further analysis was performed in R (v3.5) using the Seurat package (v2.3.3; Satija et al., 2015). The gene-cell-barcode matrix of the samples was log-transformed and filtered based on the number of genes detected per cell (any cell with <400 genes or >5,000 genes per cell was filtered out). Regression in gene expression was performed based on the number of UMIs and the percentage of mitochondrial genes. Only genes detected in ≥3 cells were included. Cells were then scaled to a total of 1e4 molecules. Any cell with >6% of mitochondrial UMI counts and >50% of ribosomal UMI was filtered out. Altogether, 4,330 cells for the APC dataset and 4,090 cells for the CD4+ T cell dataset were kept for statistical analysis. To reduce data dimensionality, 17,581 variable genes for the APC dataset and 15,535 variable genes for the CD4+ T cell dataset were selected based on their expression and dispersion (expression cutoff = 0, and dispersion cutoff = 0.5). Principal component analysis was run on the normalized gene-barcode matrix. Barnes-hut approximation to t-distributed stochastic neighbor embedded (t-SNE; van der Maaten, L. 2013. Proceedings of the International Conference on Learning Representations) was then performed on the first 15 principal components to visualize cells in a two-dimensional space. The first 15 principal components were used for the t-SNE projection and clustering analysis using the Elbow Plot approach. Clusters were identified using the “FindClusters” function in Seurat with a resolution parameter of 0.6 for the APC dataset and 0.8 for the CD4+ T cell dataset. This graph-based clustering method relies on a clustering algorithm based on shared nearest neighbor modularity optimization. Unique cluster-specific genes were identified by running the Seurat “FindAllMarkers” function, and putative conserved markers running the “FindMarkers” function, both using the Model-based Analysis of Single-cell Transcriptomics (MAST) framework (Finak et al., 2015). Hierarchical clustering between meta-cells was performed running the Seurat “BuildClusterTree” function using default parameters. Clusters containing contaminating cells were removed from the analysis: one cluster of 72 cells from the APC dataset corresponding to natural killer T cells (top genes: KLRB1, CTSW, CD7, TRDC, XCL2, XCL1, AC092580.4, TNFRSF18, TRBC1, CD247, KLRC1, and GNLY), and two clusters from the CD4+ T cell dataset of 262 cells corresponding to natural killer cells (top genes: MALAT1, N4BP2L2, PTPRC, POLR2J3, MBNL1, PLCG2, ATM, MT-ND4L, HSPA1B, NKTR, PCSK7, and ANKRD36C) and of 77 cells corresponding to B cells (top genes: ISG15, MX1, IFI6, LY6E, IFI44L, OAS1, IFIT1, MX2, EPSTI1, IFI16, IFI44, and EIF2AK2). Heatmaps and violin plots were plotted using Seurat. Data are available at GEO (accession no. GSE115007 for tonsil HLA-DR+CD11c+CD14− cells, GSE119506 for tonsil HLA-DR+CD11c+CD14− cells, and GSE119507 for tonsil CD4+ T cells).
Analysis of gene signatures at the single-cell level
Signature scores were computed using the Seurat function “AddModuleScore” using the gene signature of interest. This function calculates for each individual cell the average expression of each gene signature, subtracted by the aggregated expression of control gene sets (Tirosh et al., 2016). All analyzed genes are binned into 25 bins based on averaged expression, and for each gene of the gene signature, 100 control genes are randomly selected from the same bin as the gene. We used published gene signatures for blood cDC1s, blood cDC2s (Carpentier et al., 2016), ascites macrophages, and in vitro–generated mo-Macs (Tang-Huau et al., 2018). Complete gene signatures are included in Table S2.
Software and statistical analysis
Flow cytometry data were analyzed using FlowJo software (Tree Star).
Statistical analyses were performed using the Prism software v7 (GraphPad). Paired Wilcoxon tests were applied in all analysis to compare two groups, except for Fig. 8 D, where a paired t test was applied. Significance was retained for P < 0.05.
The software designed for analysis of imaging mass cytometry data is publicly available at https://github.com/ThomasWalter/ImageMassCytometry.
Online supplemental material
Fig. S1 shows Th cell polarization by DC subsets and macrophages. Fig. S2 shows that tonsil CD14+ cells do not contain a population of DCs. Fig. S3 shows that Tfh cells comprise two distinct effector states. Fig. S4 shows an analysis of IL-12, activin A, and TGFβ in Th cell polarization. Fig. S5 shows an analysis of in situ localization in tonsils. Table S1 shows top genes per cluster, related to Fig. 3. Table S2 shows gene signatures used in Fig. 3. Table S3 shows top genes per cluster, related to Fig. 4.
Supplementary Material
Acknowledgments
The authors wish to thank the Flow Cytometry Platform of Institut Curie for cell sorting, L. Pattarini and C. Trichot for helpful discussions, and L. Ardouin-Bataille and A. Coillard for critical reading of the manuscript.
This work was supported by the Institut National de la Santé et de la Recherche Médicale, Agence Nationale de la Recherche (ANR-10-LABX-0043, ANR-CHIN-0002, ANR-10-IDEX-0001-02 PSL, and HI-FISH ANR-14-CE10-0018-01), Institut Curie (CIC IGR-Curie 1428), the European Research Council (2013-AdG N° 340046 DCBIOX to S. Amigorena), La Ligue Nationale contre le Cancer (EL2014.LNCC/SA to S. Amigorena), and the Fondation ARC pour la recherche contre le cancer (to E. Segura). M. Durand was supported by a PhD scholarship from Université Paris Descartes. P. Gueguen was supported by a PhD scholarship from Ligue contre le Cancer. High-throughput sequencing performed by the Institut Curie Génomique d'Excellence Next Generation Sequencing platform of the Institut Curie was supported by grants from Equipex (ANR-10-EQPX-03), the France Génomique Consortium (ANR-10-INBS-09-08), Cancéropôle Ile-de-France, the SESAME program from Région Ile-de-France, and the Site de Recherche Intégrée sur le Cancer–Curie program (Site de Recherche Intégrée sur le Cancer grant INCa-DGOS-4654).
The authors declare no competing financial interests.
Author contributions: M. Durand, T. Naessens, C. Goudot, S. Amigorena, and E. Segura designed experiments. M. Durand, T. Pirnay, T. Naessens, S. Lameiras, Q. Chang, N. Talaei, O. Ornatsky, and E. Segura performed experiments. T. Walter designed the pipeline and software for analysis of imaging mass cytometry data. M. Durand, T. Walter, T. Naessens, P. Gueguen, C. Goudot, and E. Segura analyzed the data. M. Durand, T. Walter, and E. Segura prepared the figures. S. Baulande provided expertise on sequencing. Q. Chang, N. Talaei, T. Vassilevskaia, and O. Ornatsky provided expertise on imaging mass cytometry. M. Durand and E. Segura wrote the manuscript, with input from all authors. E. Segura supervised the project.
References
- Bentebibel S.E., Schmitt N., Banchereau J., and Ueno H.. 2011. Human tonsil B-cell lymphoma 6 (BCL6)-expressing CD4+ T-cell subset specialized for B-cell help outside germinal centers. Proc. Natl. Acad. Sci. USA. 108:E488–E497. 10.1073/pnas.1100898108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseño C.G., Satpathy A.T., Davidson J.T. IV, Ferris S.T., Durai V., Bagadia P., O’Connor K.W., Theisen D.J., Murphy T.L., and Murphy K.M.. 2018. Notch2-dependent DC2s mediate splenic germinal center responses. Proc. Natl. Acad. Sci. USA. 115:10726–10731. 10.1073/pnas.1809925115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabro S., Gallman A., Gowthaman U., Liu D., Chen P., Liu J., Krishnaswamy J.K., Nascimento M.S., Xu L., Patel S.R., et al. . 2016. Bridging channel dendritic cells induce immunity to transfused red blood cells. J. Exp. Med. 213:887–896. 10.1084/jem.20151720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminschi I., Proietto A.I., Ahmet F., Kitsoulis S., Shin Teh J., Lo J.C., Rizzitelli A., Wu L., Vremec D., van Dommelen S.L., et al. . 2008. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood. 112:3264–3273. 10.1182/blood-2008-05-155176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier S., Vu Manh T.P., Chelbi R., Henri S., Malissen B., Haniffa M., Ginhoux F., and Dalod M.. 2016. Comparative genomics analysis of mononuclear phagocyte subsets confirms homology between lymphoid tissue-resident and dermal XCR1(+) DCs in mouse and human and distinguishes them from Langerhans cells. J. Immunol. Methods. 432:35–49. 10.1016/j.jim.2016.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarov S., and Fazilleau N.. 2014. Monocyte-derived dendritic cells promote T follicular helper cell differentiation. EMBO Mol. Med. 6:590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q., Ornatsky O.I., Siddiqui I., Loboda A., Baranov V.I., and Hedley D.W.. 2017. Imaging Mass Cytometry. Cytometry A. 91:160–169. 10.1002/cyto.a.23053 [DOI] [PubMed] [Google Scholar]
- Chtanova T., Tangye S.G., Newton R., Frank N., Hodge M.R., Rolph M.S., and Mackay C.R.. 2004. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173:68–78. 10.4049/jimmunol.173.1.68 [DOI] [PubMed] [Google Scholar]
- Coillard A., and Segura E.. 2018. Visualization of RNA at the Single Cell Level by Fluorescent in situ Hybridization Coupled to Flow Cytometry. Bio Protoc. 8:e2892 10.21769/BioProtoc.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity. 41:529–542. 10.1016/j.immuni.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand M., and Segura E.. 2016. Dendritic cell subsets purification from human tonsils and lymph nodes. Methods Mol Biol. 1423:89–99. 10.1007/978-1-4939-3606-9_6 [DOI] [PubMed] [Google Scholar]
- Fenton T.M., Kelly A., Shuttleworth E.E., Smedley C., Atakilit A., Powrie F., Campbell S., Nishimura S.L., Sheppard D., Levison S., et al. . 2017. Inflammatory cues enhance TGFβ activation by distinct subsets of human intestinal dendritic cells via integrin αvβ8. Mucosal Immunol. 10:624–634. 10.1038/mi.2016.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. . 2015. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16:278 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto D., Wood K., Caminschi I., Murphy-Durland D., Schofield P., Christ D., Karupiah G., and Brink R.. 2013. The chemotactic receptor EBI2 regulates the homeostasis, localization and immunological function of splenic dendritic cells. Nat. Immunol. 14:446–453. 10.1038/ni.2555 [DOI] [PubMed] [Google Scholar]
- Giesen C., Wang H.A., Schapiro D., Zivanovic N., Jacobs A., Hattendorf B., Schüffler P.J., Grolimund D., Buhmann J.M., Brandt S., et al. . 2014. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods. 11:417–422. 10.1038/nmeth.2869 [DOI] [PubMed] [Google Scholar]
- Goenka R., Barnett L.G., Silver J.S., O’Neill P.J., Hunter C.A., Cancro M.P., and Laufer T.M.. 2011. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J. Immunol. 187:1091–1095. 10.4049/jimmunol.1100853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot T., Senda T., Carpenter D.J., Matsuoka N., Weiner J., Gordon C.L., Miron M., Kumar B.V., Griesemer A., Ho S.H., et al. . 2017. Dendritic Cells Display Subset and Tissue-Specific Maturation Dynamics over Human Life. Immunity. 46:504–515. 10.1016/j.immuni.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson A., Lysén A., Balan S., Sundvold-Gjerstad V., Arnold-Schrauf C., Richter L., Bækkevold E.S., Dalod M., Bogen B., and Fossum E.. 2017. Targeting Influenza Virus Hemagglutinin to Xcr1+ Dendritic Cells in the Absence of Receptor-Mediated Endocytosis Enhances Protective Antibody Responses. J. Immunol. 198:2785–2795. 10.4049/jimmunol.1601881 [DOI] [PubMed] [Google Scholar]
- Guilliams M., Ginhoux F., Jakubzick C., Naik S.H., Onai N., Schraml B.U., Segura E., Tussiwand R., and Yona S.. 2014. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14:571–578. 10.1038/nri3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidkamp G.F., Sander J., Lehmann C.H.K., Heger L., Eissing N., Baranska A., Lühr J.J., Hoffmann A., Reimer K.C., Lux A., et al. . 2016. Human lymphoid organ dendritic cell identity is predominantly dictated by ontogeny, not tissue microenvironment. Sci. Immunol. 1:eaai7677 10.1126/sciimmunol.aai7677 [DOI] [PubMed] [Google Scholar]
- Jacquemin C., Schmitt N., Contin-Bordes C., Liu Y., Narayanan P., Seneschal J., Maurouard T., Dougall D., Davizon E.S., Dumortier H., et al. . 2015. OX40 Ligand Contributes to Human Lupus Pathogenesis by Promoting T Follicular Helper Response. Immunity. 42:1159–1170. 10.1016/j.immuni.2015.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Zaid A., Davey G.M., Mueller S.N., Nutt S.L., Zotos D., Tarlinton D.M., Shortman K., Lahoud M.H., Heath W.R., and Caminschi I.. 2015. Targeting Antigen to Clec9A Primes Follicular Th Cell Memory Responses Capable of Robust Recall. J. Immunol. 195:1006–1014. 10.4049/jimmunol.1500767 [DOI] [PubMed] [Google Scholar]
- Kelly A., Gunaltay S., McEntee C.P., Shuttleworth E.E., Smedley C., Houston S.A., Fenton T.M., Levison S., Mann E.R., and Travis M.A.. 2018. Human monocytes and macrophages regulate immune tolerance via integrin αvβ8-mediated TGFβ activation. J. Exp. Med. 215:2725–2736. 10.1084/jem.20171491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.H., Lim H.W., Kim J.R., Rott L., Hillsamer P., and Butcher E.C.. 2004. Unique gene expression program of human germinal center T helper cells. Blood. 104:1952–1960. 10.1182/blood-2004-03-1206 [DOI] [PubMed] [Google Scholar]
- Klechevsky E., Morita R., Liu M., Cao Y., Coquery S., Thompson-Snipes L., Briere F., Chaussabel D., Zurawski G., Palucka A.K., et al. . 2008. Functional specializations of human epidermal Langerhans cells and CD14+ dermal dendritic cells. Immunity. 29:497–510. 10.1016/j.immuni.2008.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Watanabe T., Suzuki R., Furu M., Ito H., Ito J., Matsuda S., and Yoshitomi H.. 2016. TGF-β induces the differentiation of human CXCL13-producing CD4(+) T cells. Eur. J. Immunol. 46:360–371. 10.1002/eji.201546043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy J.K., Gowthaman U., Zhang B., Mattsson J., Szeponik L., Liu D., Wu R., White T., Calabro S., Xu L., et al. . 2017. Migratory CD11b+ conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Sci. Immunol. 2:eaam9169 10.1126/sciimmunol.aam9169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Mortha A., Rahman A., and Merad M.. 2015. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 15:731–744. 10.1038/nri3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lu E., Yi T., and Cyster J.G.. 2016. EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature. 533:110–114. 10.1038/nature17947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen X., Zhong B., Wang A., Wang X., Chu F., Nurieva R.I., Yan X., Chen P., van der Flier L.G., et al. . 2014. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 507:513–518. 10.1038/nature12910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J., Xu J., de Bouteiller O., Parham C.L., Grouard G., Djossou O., de Saint-Vis B., Lebecque S., Banchereau J., and Moore K.W.. 1997. Follicular dendritic cells specifically express the long CR2/CD21 isoform. J. Exp. Med. 185:165–170. 10.1084/jem.185.1.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M., Wu J.E., Arumemi F., Mikulski Z., Dahlberg C., Miller A.T., and Crotty S.. 2016. Activin A programs the differentiation of human TFH cells. Nat. Immunol. 17:976–984. 10.1038/ni.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern N., Schlitzer A., Gunawan M., Jardine L., Shin A., Poyner E., Green K., Dickinson R., Wang X.N., Low D., et al. . 2014. Human dermal CD14+ cells are a transient population of monocyte-derived macrophages [erratum in Immunity. 2015. 42:391]. Immunity. 41:465–477. 10.1016/j.immuni.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., et al. Immunological Genome Consortium . 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13:888–899. 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag D., Proietto A.I., Loudovaris T., Mannering S.I., Vremec D., Shortman K., Wu L., and Harrison L.C.. 2011. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J. Immunol. 186:6207–6217. 10.4049/jimmunol.1002632 [DOI] [PubMed] [Google Scholar]
- Nizzoli G., Krietsch J., Weick A., Steinfelder S., Facciotti F., Gruarin P., Bianco A., Steckel B., Moro M., Crosti M., et al. . 2013. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood. 122:932–942. 10.1182/blood-2013-04-495424 [DOI] [PubMed] [Google Scholar]
- Pattarini L., Trichot C., Bogiatzi S., Grandclaudon M., Meller S., Keuylian Z., Durand M., Volpe E., Madonna S., Cavani A., et al. . 2017. TSLP-activated dendritic cells induce human T follicular helper cell differentiation through OX40-ligand. J. Exp. Med. 214:1529–1546. 10.1084/jem.20150402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penel-Sotirakis K., Simonazzi E., Péguet-Navarro J., and Rozières A.. 2012. Differential capacity of human skin dendritic cells to polarize CD4+ T cells into IL-17, IL-21 and IL-22 producing cells. PLoS One. 7:e45680 10.1371/journal.pone.0045680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H. 2016. T follicular helper cells in space-time. Nat. Rev. Immunol. 16:612–625. 10.1038/nri.2016.94 [DOI] [PubMed] [Google Scholar]
- Rao D.A., Gurish M.F., Marshall J.L., Slowikowski K., Fonseka C.Y., Liu Y., Donlin L.T., Henderson L.A., Wei K., Mizoguchi F., et al. . 2017. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 542:110–114. 10.1038/nature20810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., and Regev A.. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33:495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W., See P., Shin A., Wasan P.S., et al. . 2013. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 38:970–983. 10.1016/j.immuni.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N., Morita R., Bourdery L., Bentebibel S.E., Zurawski S.M., Banchereau J., and Ueno H.. 2009. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 31:158–169. 10.1016/j.immuni.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N., Liu Y., Bentebibel S.-E., Munagala I., Bourdery L., Venuprasad K., Banchereau J., and Ueno H.. 2014. The cytokine TGF-β co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat. Immunol. 15:856–865. 10.1038/ni.2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Valladeau-Guilemond J., Donnadieu M.H., Sastre-Garau X., Soumelis V., and Amigorena S.. 2012. Characterization of resident and migratory dendritic cells in human lymph nodes. J. Exp. Med. 209:653–660. 10.1084/jem.20111457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Durand M., and Amigorena S.. 2013a Similar antigen cross-presentation capacity and phagocytic functions in all freshly isolated human lymphoid organ-resident dendritic cells. J. Exp. Med. 210:1035–1047. 10.1084/jem.20121103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Touzot M., Bohineust A., Cappuccio A., Chiocchia G., Hosmalin A., Dalod M., Soumelis V., and Amigorena S.. 2013b Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. 38:336–348. 10.1016/j.immuni.2012.10.018 [DOI] [PubMed] [Google Scholar]
- Shin C., Han J.A., Koh H., Choi B., Cho Y., Jeong H., Ra J.S., Sung P.S., Shin E.C., Ryu S., and Do Y.. 2015. CD8α(-) Dendritic Cells Induce Antigen-Specific T Follicular Helper Cells Generating Efficient Humoral Immune Responses. Cell Reports. 11:1929–1940. 10.1016/j.celrep.2015.05.042 [DOI] [PubMed] [Google Scholar]
- Soille P. 2003. Morphological Image Analysis: Principles and Applications. Second edition. Springer-Verlag, Berlin. [Google Scholar]
- Suan D., Nguyen A., Moran I., Bourne K., Hermes J.R., Arshi M., Hampton H.R., Tomura M., Miwa Y., Kelleher A.D., et al. . 2015. T follicular helper cells have distinct modes of migration and molecular signatures in naive and memory immune responses. Immunity. 42:704–718. 10.1016/j.immuni.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Tang-Huau T.L., and Segura E.. 2019. Human in vivo-differentiated monocyte-derived dendritic cells. Semin. Cell Dev. Biol. 86:44–49. 10.1016/j.semcdb.2018.02.018 [DOI] [PubMed] [Google Scholar]
- Tang-Huau T.L., Gueguen P., Goudot C., Durand M., Bohec M., Baulande S., Pasquier B., Amigorena S., and Segura E.. 2018. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat. Commun. 9:2570 10.1038/s41467-018-04985-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H. II, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. . 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352:189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis M.A., Reizis B., Melton A.C., Masteller E., Tang Q., Proctor J.M., Wang Y., Bernstein X., Huang X., Reichardt L.F., et al. . 2007. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 449:361–365. 10.1038/nature06110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H. 2016. T follicular helper cells in human autoimmunity. Curr. Opin. Immunol. 43:24–31. 10.1016/j.coi.2016.08.003 [DOI] [PubMed] [Google Scholar]
- Ueno H., Banchereau J., and Vinuesa C.G.. 2015. Pathophysiology of T follicular helper cells in humans and mice. Nat. Immunol. 16:142–152. 10.1038/ni.3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini M., Gerhard J., Burkert S., Jensen K.J., Georg P., Ebner F., Volkers S.M., Thada S., Dietert K., Bauer L., et al. . 2018. Recognition of microbial viability via TLR8 drives TFH cell differentiation and vaccine responses. Nat. Immunol. 19:386–396. 10.1038/s41590-018-0068-4 [DOI] [PubMed] [Google Scholar]
- van der Walt S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T., and scikit-image contributors . 2014. scikit-image: image processing in Python. PeerJ. 2:e453 10.7717/peerj.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C., Mildner A., and Jung S.. 2015. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 33:643–675. 10.1146/annurev-immunol-032414-112220 [DOI] [PubMed] [Google Scholar]
- Vinuesa C.G., Linterman M.A., Yu D., and MacLennan I.C.M.. 2016. Follicular Helper T Cells. Annu. Rev. Immunol. 34:335–368. 10.1146/annurev-immunol-041015-055605 [DOI] [PubMed] [Google Scholar]
- Yao C., Zurawski S.M., Jarrett E.S., Chicoine B., Crabtree J., Peterson E.J., Zurawski G., Kaplan D.H., and Igyártó B.Z.. 2015. Skin dendritic cells induce follicular helper T cells and protective humoral immune responses. J. Allergy Clin. Immunol. 136:1387–1397.e7. 10.1016/j.jaci.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawalkar N., Tscharner G.G., Hunger R.E., and Hassan A.S.. 2009. Increased expression of IL-12p70 and IL-23 by multiple dendritic cell and macrophage subsets in plaque psoriasis. J. Dermatol. Sci. 54:99–105. 10.1016/j.jdermsci.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Yi T., and Cyster J.G.. 2013. EBI2-mediated bridging channel positioning supports splenic dendritic cell homeostasis and particulate antigen capture. eLife. 2:e00757 10.7554/eLife.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.I., Becker C., Metang P., Marches F., Wang Y., Toshiyuki H., Banchereau J., Merad M., and Palucka A.K.. 2014. Human CD141+ dendritic cells induce CD4+ T cells to produce type 2 cytokines. J. Immunol. 193:4335–4343. 10.4049/jimmunol.1401159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. . 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8:14049 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.