An antibiotic cocktail leads to reductions in brain amyloidosis, altered morphology, and gene expression in microglia of male but not female transgenic mice that exhibit Aβ deposits. The microbiome plays a causal role in modulating Aβ burden and microglia phenotypes.
Abstract
We demonstrated that an antibiotic cocktail (ABX)-perturbed gut microbiome is associated with reduced amyloid-β (Aβ) plaque pathology and astrogliosis in the male amyloid precursor protein (APP)SWE/presenilin 1 (PS1)ΔE9 transgenic model of Aβ amyloidosis. We now show that in an independent, aggressive APPSWE/PS1L166P (APPPS1-21) mouse model of Aβ amyloidosis, an ABX-perturbed gut microbiome is associated with a reduction in Aβ pathology and alterations in microglial morphology, thus establishing the generality of the phenomenon. Most importantly, these latter alterations occur only in brains of male mice, not in the brains of female mice. Furthermore, ABX treatment lead to alterations in levels of selected microglial expressed transcripts indicative of the “M0” homeostatic state in male but not in female mice. Finally, we found that transplants of fecal microbiota from age-matched APPPS1-21 male mice into ABX-treated APPPS1-21 male restores the gut microbiome and partially restores Aβ pathology and microglial morphology, thus demonstrating a causal role of the microbiome in the modulation of Aβ amyloidosis and microglial physiology in mouse models of Aβ amyloidosis.
Introduction
Alzheimer’s disease (AD), the most common age-associated chronic progressive neurodegenerative disorder, is pathologically characterized by the extracellular deposition of amyloid-β (Aβ) peptides in senile plaques and the intracellular accumulation of hyperphosphorylated tau protein in neurofibrillary tangles. In addition, positron emission tomography studies showed that activated microglia and neuroinflammation are highly correlated with cognitive decline in AD patients (Cagnin et al., 2001; Hamelin et al., 2016). Furthermore, genome-wide association studies have identified numerous polymorphisms within genes critical for innate immune responses (i.e., CD33 and TREM-2) that confer an elevated risk of late-onset AD (Griciuc et al., 2013; Guerreiro et al., 2013; Naj et al., 2014; Efthymiou and Goate, 2017). It is now well established that neuroinflammation, instead of being simply a bystander that is activated by central stimuli (such as senile plaques and neurofibrillary tangles; Heneka et al., 2015) and peripheral stimuli (such as endotoxins; Wendeln et al., 2018), plays an important role in AD pathogenesis.
Recent evidence suggests that intestinal microbiota could play a major role in peripheral as well as central immune activation and inflammation in various neurological conditions including autism spectrum disorders, multiple sclerosis, Parkinson’s disease, and AD (Cryan and O’Mahony, 2011; Sherwin et al., 2018). Specifically, patients with cognitive impairment and brain amyloidosis exhibit altered gut microbiota, which in turn was significantly associated with pro-inflammatory cytokine gene profiles in the blood of these subjects (Cattaneo et al., 2017). In this regard, other studies have reported decreased Firmicutes, increased Bacteroidetes, and decreased Bifidobacterium in AD subjects (Vogt et al., 2017; Zhuang et al., 2018). Besides these human studies, microbiome changes have also been reported in transgenic (Tg) mouse models of Aβ amyloidogenesis (Brandscheid et al., 2017; Shen et al., 2017; Bäuerl et al., 2018). More recently, Harach et al. (2017) demonstrated that germ-free (GF) APPPS1-21 Tg mice that express familial AD–linked APPSWE and PS1L166P transgenes driven by the neuron-specific Thy1 promoter (Radde et al., 2006) exhibit reduced Aβ deposition in the brain compared with their specific pathogen-free counterparts (Harach et al., 2017). Importantly, we have documented that in the APPSWE/PS1ΔE9 mouse model of Aβ amyloidosis, postnatal and long-term antibiotic cocktail (ABX) administration in male mice leads to reduced Aβ deposition and associated gliosis (Minter et al., 2016, 2017) and that long-term ABX treatment leads to reductions in Aβ deposition selectively in males but not in females (Minter et al., 2016). While these studies collectively establish a strong link between the gut microbiome and AD pathogenesis, the mechanism(s) by which microbial alterations impact on pathogenesis remain silent.
Several investigations of the microbiome-gut-brain axis have shown that the microbiome plays a critical role in the development of brain circuitry, neurophysiology, behavior, and most importantly, the physiology of brain microglial cells (Diaz Heijtz et al., 2011; Braniste et al., 2014; Sharon et al., 2016). Microglia, also known as “the resident macrophages of the brain,” constitute the first line of defense against injury and infection. Microglia also contribute to brain development throughout embryonic and adult life by regulating aspects of synaptic transmission, synapse formation and pruning, cell death, cell survival, and embryonic brain wiring (Schafer and Stevens, 2015; Hong et al., 2016; Ransohoff and El Khoury, 2016; Reemst et al., 2016; Tay et al., 2017). The gut microbiome is an important environmental cue for microglial function in immune and neuronal responses throughout the host’s life span. In this regard, microglia respond to environmental challenges in a sex- and time-dependent manner beginning at prenatal stages (Thion et al., 2018), and significant differences in the transcriptomes of microglia obtained from adult male versus female mice have been documented (Villa et al., 2018). Furthermore, microglial maturation and function are impaired both in the absence of the microbiome in GF or ABX-treated mice (Erny et al., 2015; Matcovitch-Natan et al., 2016). For example, microglial cells in GF and ABX-treated mice display significantly altered inflammatory gene expression profiles that influence their surveillance function that is accompanied by altered cellular morphologies (Erny et al., 2015).
To these aforementioned aspects of microbiome-mediated effects on microglial homeostasis, clinical and preclinical studies have revealed that sex has a major impact on a wide range of autoimmune disorders (i.e., females are 2–10 times more susceptible than males; Zandman-Goddard et al., 2007; Fish, 2008). In this regard, while sex-specific differences in Aβ burden and tauopathy are not apparent in the brains of aged individuals and AD patients, sex differences in the clinical phenotype and progression of AD have been well documented (Scheinin et al., 2014; Gottesman et al., 2016; Johnson et al., 2016; Jack et al., 2017; Vemuri et al., 2017; Cavedo et al., 2018).
While compelling, our published studies on the role of the gut microbiome on Aβ amyloidosis in male mice (Minter et al., 2016, 2017) was limited to assessments in a single strain of mice, namely, the amyloid precursor protein (APP)SWE/presenilin 1 (PS1)ΔE9 mouse model (Jankowsky et al., 2002) in which expression of cDNA encoding familial AD–linked human APPSWE and PS1ΔE9 variants are transcriptionally driven by the ubiquitously expressed mouse prion protein promoter. In view of potential caveats associated with transgene insertion and mis- and/or overexpression of the prion protein promoter-driven transgenes, we examined the impact of long-term ABX treatment in male and female APPPS1-21 mice that exhibit Aβ amyloidosis by the age of 6–7 wk (Radde et al., 2006). Confirming our earlier findings, we now report that long-term ABX treatment resulted in reductions of Aβ deposition and microglial morphological alterations in male but not female APPPS1-21 mice. Transcriptomic profiling revealed that males exhibited profound changes in microglial gene expression compared with ABX-treated female mice. Finally, transplants of fecal microbiota from age-matched APPPS1-21 male mice into ABX-treated male mice partially restored Aβ amyloidosis and microglial phenotypes that are observed in vehicle-treated APPPS1-21 mice. Collectively, our studies reveal that in two independent Tg mouse models of Aβ amyloidosis ABX-mediated microbiome perturbations influence brain Aβ amyloidosis and microglial homeostasis in a sex-specific manner.
Results
Thy1 promoter-driven APPSWE/PS1L166P transgenes are selectively expressed in the brain
To assess the tissue-level expression of human APPswe in the Tg mouse line, we performed Western blot analysis of detergent-soluble extracts using the human APP-specific 6E10 antibody. These studies revealed that in 4- to 5-mo-old APPPS1-21 mice, human APPswe is expressed selectively in the brain (Fig. S1, lanes 3 and 4), while in 8-mo-old APPSWE/PS1ΔE9 Tg mice, APPswe is expressed both in the brain and in a variety of sampled peripheral tissues (Fig. S1), as previously described (Jankowsky et al., 2002). Importantly, Aβ peptides are only detectable in extracts prepared from the brains of either mouse line (Fig. S1, lanes 1–4).
Postnatal antibiotic treatment results in similar microbiome changes in male and female APPPS1-21 mice at postnatal day (P) 22
The fecal microbiota at P22 was evaluated by using Illumina MiSeq amplicon sequencing. The β diversity differences between bacterial communities found in the vehicle- and ABX-treated groups were calculated with the weighted UniFrac metric and visualized by principal coordinate analysis (PCoA). Microbial β diversity was significantly different between vehicle- and ABX-treated groups and showed clear clustering by using PCoA (Fig. S2 A). There was no significant difference in β diversity between male and female samples, but there was significantly reduced richness and evenness in ABX-treated males and ABX-treated females compared with their vehicle-treated counterparts (Fig. S2, B and C).
Analysis of composition of microbiomes (ANCOM) analyses of 16S ribosomal RNA (rRNA) sequencing data at the DADA2 inferred exact sequence variant (ESV) level (taxonomy assigned via GreenGenes) and clustered species levels were performed to determine the taxa that described the most variance between groups. Vehicle-treated male and female mice showed no differences at P22. Likewise, ABX-treated mice did not differ significantly in ESVs or species by sex. However, as expected, ABX treatment showed significant changes in microbial composition in both sexes (Table S1) compared with their respective vehicle controls.
Similarly, as anticipated, predicted metagenome analysis through Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) showed significant differences in many pathways between ABX-treated male and female mice compared with their vehicle-treated counterparts (Table 1), and there were no significant differences between male and female mice irrespective of the vehicle or ABX treatments. Taken together, these data indicate that the microbiome changes in both male and female mice after P14–P21 ABX gavage are highly comparable.
Table 1. List of significantly different Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using PICRUSt analysis in fecal samples.
| Pathways | Abundance mean | FDR-P | |
|---|---|---|---|
| 22-d-old mice (n = 10/group) | |||
| M_Vehicle | M_ABX | ||
| M_Vehicle and M_ABX | |||
| Bacterial toxins | 13,235.5 | 6,249.4 | 0.000866 |
| Lipopolysaccharide biosynthesis | 49,150.4 | 13,221.2 | 0.000866 |
| Oxidative phosphorylation | 150,609.4 | 55,329.3 | 0.000809 |
| Protein digestion and absorption | 4,943 | 677.2 | 0.000866 |
| F_Vehicle | F_ABX | ||
| F_Vehicle and F_ABX | |||
| Bacterial invasion of epithelial cells | 1.7 | 280.4 | 0.016666 |
| Bacterial secretion system | 76,012.7 | 2,6207.1 | 0.000365 |
| Bacterial toxins | 14,961.8 | 6,209.4 | 0.000365 |
| Lipopolysaccharide biosynthesis | 58,487.2 | 8,127.1 | 0.000365 |
| Lipopolysaccharide biosynthesis proteins | 73,421.5 | 13,334.1 | 0.000365 |
| Primary immunodeficiency | 6,215.7 | 3813 | 0.001623 |
| Protein digestion and absorption | 4,994.6 | 354.6 | 0.000599 |
| Protein folding and associated processing | 80,495.2 | 2,8776.3 | 0.000365 |
| 7-wk-old mice (n = 10/group) | |||
| M_Vehicle | M_ABX | ||
| M_Vehicle vs. M_ABX | |||
| Bacterial chemotaxis | 46,114.3 | 7,472.1 | 0.001933 |
| Bacterial secretion system | 102,269.1 | 59,364.3 | 0.006067 |
| Bacterial toxins | 21,094 | 13,546.8 | 0.009024 |
| Primary immunodeficiency | 8,583 | 4,712.8 | 0.002232 |
| Protein folding and associated processing | 115,193 | 64,080.4 | 0.006067 |
| Secretion system | 168,262.9 | 84,057.7 | 0.006067 |
| Ubiquitin system | 509.6 | 1,545.5 | 0.005581 |
| F_Vehicle | F_ABX | ||
| F_Vehicle vs. F_ABX | |||
| Bacterial chemotaxis | 31,193 | 144,38.1 | 0.023303 |
| Ubiquitin system | 585.7 | 2,040.6 | 0.002061 |
| M_ABX | F_ABX | ||
| M_ABX vs. F_ABX | |||
| Bacterial chemotaxis | 7,472.1 | 14,438.1 | 0.007826 |
| Bacterial secretion system | 59,364.3 | 106,765.7 | 0.006385 |
| Bacterial toxins | 13,546.8 | 20,489.3 | 0.012471 |
| Lipopolysaccharide biosynthesis | 50,195.1 | 82,159 | 0.007149 |
| Lipopolysaccharide biosynthesis proteins | 65,304.7 | 103,956.6 | 0.007149 |
| Primary immunodeficiency | 4,712.8 | 6,936.5 | 0.006385 |
| Protein folding and associated processing | 64,080.4 | 110,534.4 | 0.006385 |
| Secretion system | 84,057.7 | 156,112.4 | 0.006385 |
Significantly different KEGG pathways were analyzed by using PICRUSt analysis at both P22 (22-d-old) and 7 wk of age. The false discovery rate P (FDR-P) < 0.05 was selected for significance. Fecal bacterial relevant KEGG pathways are reported to understand the predicted functional pathways. F_ABX, ABX-treated female mice; F_Vehicle, vehicle-treated female mice; M_ABX, ABX-treated male mice; M_Vehicle, vehicle-treated male mice. n = 10/group.
Long-term ABX treatment results in sex-specific microbiome changes in APPPS1-21 mice at the time of cull
It is well documented that ABX treatment results in cecum enlargement (Savage and Dubos, 1968; Minter et al., 2016). To confirm this finding in our mice, we showed that at the time of cull, ABX-treated male and female APPPS1-21 mice exhibited larger ceca compared with the vehicle-treated controls (Fig. S2 D). Cecal weight analyses showed significantly larger ceca in ABX-treated male compared with vehicle-treated male mice, in ABX-treated female compared with vehicle-treated female mice, and ABX-treated male compared with ABX-treated female mice (Fig. S2 E).
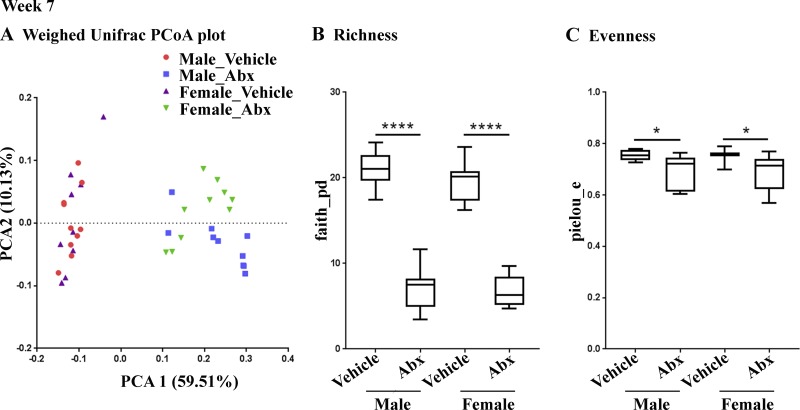
Amplicon sequencing was performed to analyze the fecal microbiota in mice at necropsy. As before, microbial β diversity at 7 wk was significantly different between ABX-treated mice and their vehicle-treated counterparts (Fig. 1 A), but no significant differences were observed for male versus female groups. We observed significantly reduced diversity in ABX-treated male and ABX-treated female mice compared with their vehicle-treated counterparts (Fig. 1, B and C).
Figure 1.
ABX treatment results in microbiome changes in male and female APPPS1-21 mice (n = 10/group). (A) The PCoA plot at week 7 generated by using weighted versions of the UniFrac distance metric. The two components explained 59.51% of the variance. (B and C) At week 7 also, both α diversity values (species richness and evenness) were significantly lower in the ABX treatment group compared with the control (Faith’s phylogenetic diversity [B]: two-way ANOVA, F[1,35] = 367.6, P < 0.0001; Pielou’s evenness [C]: two-way ANOVA, F[1,35] = 14.73, P = 0.0005). Sex or an interaction between sex and ABX treatment showed no significant difference in either richness or evenness (P > 0.05). Post hoc comparisons showed reduced diversity in ABX-treated male (Faith: P < 0.0001, Pielou: P = 0.049) and ABX-treated female (Faith: P < 0.0001, Pielou: P = 0.0462) mice compared with their vehicle-treated counterparts. Data are mean ± SEM. *, P < 0.05; ****, P < 0.0001. n = 10/group. PCA1, principal coordinate 1; PCA2, principal coordinate 2.
ANCOM comparison of ESVs and clustered species at 7 wk again showed large numbers of taxa whose proportions were significantly different between vehicle- and ABX-treated groups in both males and females (Table 2) with some overlapping features. Among ABX-treated mice, an Akkermansia muciniphila ESV and an Allobaculum spp. cluster were significantly enriched in females compared with males. In vehicle-treated mice, a Mollicutes RF39 ESV was significantly enriched in males.
Table 2. ANCOM significant differences in fecal microbiota at week 7 (FDR-P < 0.05).
| Higher in F_Vehicle | Lower in F_Vehicle | |
|---|---|---|
| M_Vehicle vs. F_Vehicle | ||
| ESV | NS | p_Tenericutes; c_Mollicutes; o_RF39; f_; g_; s_ |
| L7 clustered taxa | NS | NS |
| Higher in F_ABX | Lower in F_ABX | |
| M_ABX vs. F_ABX | ||
| ESV | p_Verrucomicrobia;c_Verrucomicrobiae;f_Verrucomicrobiaceae;g_Akkermansia;s_muciniphila | NS |
| L7 clustered taxa | p_Firmicutes;c_Erysipelotrichi;f_Erysipelotrichaceae;g_Allobaculum;s_ | NS |
| Higher in M_ABX | Lower in M_ABX | |
| M_Vehicle vs. M_ABX | ||
| ESV | p_Bacteroidetes; c_Bacteroidia; f_Bacteroidaceae; g_Bacteroides; s_ovatus | p_Bacteroidetes; c_Bacteroidia; f_S24-7; g_; s_ (×8) |
| p_Firmicutes; c_Erysipelotrichi; f_Erysipelotrichaceae; g_; s_ | p_Proteobacteria; c_Epsilonproteobacteria; f_Helicobacteraceae; g_Helicobacter; s_ | |
| p_Actinobacteria; c_Actinobacteria; f_Bifidobacteriaceae; g_Bifidobacterium | ||
| p_Bacteroidetes; c_Bacteroidia; f_Porphyromonadaceae; g_Parabacteroides; s_ | ||
| p_Firmicutes; c_Bacilli; f_Lactobacillaceae; g_Lactobacillus; s_ | ||
| L7 clustered taxa | p_Bacteroidetes; c_Bacteroidia; f_Bacteroidaceae; g_Bacteroides; s_ovatus | p_Actinobacteria;c_Actinobacteria;f_Bifidobacteriaceae;g_Bifidobacterium;_ |
| p_Bacteroidetes;c_Bacteroidia;f_Porphyromonadaceae;g_Parabacteroides;s_distasonis | p_Firmicutes;c_Bacilli;f_Lactobacillaceae;g_Lactobacillus;s_ | |
| p_Firmicutes;c_Erysipelotrichi;f_Erysipelotrichaceae;_;_ | p_Firmicutes;c_Clostridia;o_Clostridiales;_;_;_ | |
| p_Firmicutes;c_Erysipelotrichi;f_Erysipelotrichaceae;g_;s_ | p_Firmicutes;c_Clostridia;o_Clostridiales;f_;g_;s_ | |
| p_Firmicutes;c_Clostridia;f_Ruminococcaceae;g_Ruminococcus;s_ | ||
| p_Firmicutes;c_Erysipelotrichi;f_Erysipelotrichaceae;g_Allobaculum;s_ | ||
| p_Proteobacteria;c_Deltaproteobacteria;f_Desulfovibrionaceae;g_Desulfovibrio;s_C21_c20 | ||
| p_Proteobacteria;c_Epsilonproteobacteria;f_Helicobacteraceae;g_Helicobacter;s_ | ||
| Higher in M_ABX | Lower in M_ABX | |
| F_Vehicle vs. F_ABX | ||
| ESV | p_Bacteroidetes; c_Bacteroidia; f_Bacteroidaceae; g_Bacteroides; s_uniformis | p_Bacteroidetes; c_Bacteroidia; f_S24-7; g_; s_ (×11) |
| p_Verrucomicrobia; c_Verrucomicrobiae; f_Verrucomicrobiaceae; g_Akkermansia; s_muciniphila | p_Actinobacteria; c_Actinobacteria; f_Bifidobacteriaceae; g_Bifidobacterium | |
| p_Bacteroidetes; c_Bacteroidia; f_S24-7; g_; s_ | p_Bacteroidetes; c_Bacteroidia; f_Porphyromonadaceae; g_Parabacteroides; s_ | |
| p_Proteobacteria; c_Betaproteobacteria; f_Alcaligenaceae; g_Sutterella; s_ | p_Firmicutes; c_Bacilli; f_Lactobacillaceae; g_Lactobacillus; s_ | |
| p_Firmicutes; c_Erysipelotrichi; f_Erysipelotrichaceae | ||
| L7 clustered taxa | p_Bacteroidetes;c_Bacteroidia;f_Bacteroidaceae;g_Bacteroides;s_uniformis | p_Actinobacteria;c_Actinobacteria;f_Bifidobacteriaceae;g_Bifidobacterium;_ |
| p_Bacteroidetes;c_Bacteroidia;f_Porphyromonadaceae;g_Parabacteroides;s_distasonis | p_Firmicutes;c_Clostridia;o_Clostridiales;_;_;_ | |
| p_Firmicutes;c_Erysipelotrichi;f_Erysipelotrichaceae;_;_ | p_Firmicutes;c_Clostridia;f_Ruminococcaceae;g_Ruminococcus;s_ | |
| p_Verrucomicrobia;c_Verrucomicrobiae;f_Verrucomicrobiaceae;g_Akkermansia;s_muciniphila | p_Proteobacteria;c_Epsilonproteobacteria;f_Helicobacteraceae;g_Helicobacter;s_ |
To determine the taxa that described the most variance between groups, ANCOM analyses of 16S rRNA sequencing data at the DADA2-inferred ESV level (taxonomy assigned via Greengenes) and clustered species levels were performed between vehicle- or ABX-treated groups in a sex-dependent manner. F_ABX, ABX-treated female mice; F_Vehicle, vehicle-treated female mice; M_ABX, ABX-treated male mice; M_Vehicle, vehicle-treated male mice. n = 10/group.
As expected, PICRUSt showed significant differences in many pathways between ABX-treated male and female mice compared with their vehicle-treated controls and between ABX-treated male and ABX-treated female mice but not between vehicle-treated male and vehicle-treated female mice (Table 1). Pro-inflammatory pathways, such as bacterial secretion system, bacterial toxins, lipopolysaccharide biosynthesis, and lipopolysaccharide synthesis protein, were significantly enriched in ABX-treated female compared with ABX-treated male mice. In this regard, higher levels of microbial endotoxins could affect the peripheral and central inflammatory processes as has been shown recently (Wendeln et al., 2018). Collectively, these data indicate that long-term ABX treatment results in distinct microbiome changes in both male and female APPPS1-21 mice at 7 wk of age and that ABX-treated female mice exhibit an enrichment of pro-inflammatory metabolic pathways compared with ABX-treated male mice.
Long-term ABX treatment results in sex-specific alterations of circulating inflammatory mediators in APPPS1-21 mice
It is well established that the commensal bacteria can modulate the peripheral innate immune system (Lathrop et al., 2011) and impact cytokine production (Fung et al., 2016). We quantified the levels of 62 circulating cytokines and chemokines in the plasma samples of the vehicle- and ABX-treated mice using commercially available cytokine array membrane kits. Numerous cytokines were altered with ABX-treatment, and this pattern was different in ABX-treated male versus female mice (Fig. S3, A and B). ABX-treated male mice exhibited elevated levels of anti-inflammatory/neuroprotective factors, including insulin-like growth factor binding protein 3, Interleukin 6 (IL-6), and IL-10, and decreased levels of pro-inflammatory cytokines/chemokines, including Eotaxin1, IL-1B, IL-2, IL-3, IL-17A, and RANTES (Regulated upon Activation, Normal T cell Expressed, and Secreted; also known as Chemokine [C-C motif] ligand 5 [CCL5]), relative to vehicle-treated controls. In contrast, ABX-treated female mice showed up-regulation of pro-inflammatory cytokines/chemokines, including IL-1B, IL-5, IL-9, and IL-17A. Importantly, ABX-treated female mice exhibited elevated levels of pro-inflammatory cytokines/chemokines, such as CCL11 (Eotaxin1; 7.11-fold), IL-1B (6.92-fold), IL-2 (6.35-fold), IL-3 (7.01-fold), IL-5 (7.32-fold), IL-9 (8.51-fold), and IL-17A (7.44-fold), relative to ABX-treated male mice. Moreover, ABX-treated female mice also exhibited decreased levels of IL-12 (11.16-fold), chemokine (C-X-C motif) ligand 1 (CXCL1; also known as KC; 13.02-fold), and P-selectin (11.94-fold) cytokines/chemokines compared with ABX-treated male mice. These findings suggest that the long-term ABX-altered microbiome has distinct effects on the circulating chemokines and cytokines in a sex-specific manner with female mice exhibiting higher levels of pro-inflammatory mediators.
Long-term ABX treatment results in reduced Aβ deposition only in male APPPS1-21 mice
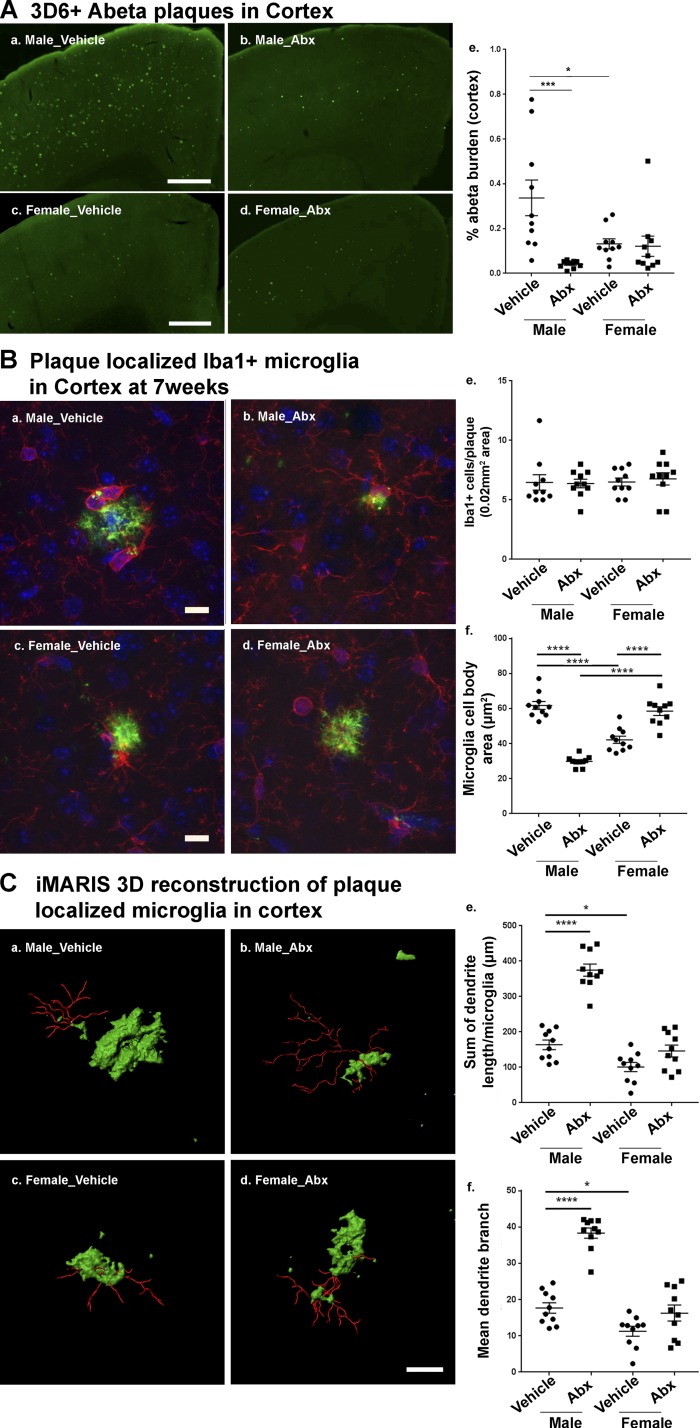
To investigate the effect of long-term ABX treatment on Aβ amyloidosis in APPPS1-21 mice, we sacrificed animals at 7 wk of age. Using Aβ-specific monoclonal antibody 3D6, we confirmed that Aβ deposition is prominent in the cortex, with negligible levels in the hippocampus (Radde et al., 2006; Fig. 2 A). ABX-treated male APPPS1-21 animals exhibited significantly reduced cortical Aβ burden compared with vehicle-treated male mice. Notably, vehicle-treated female mice showed significantly lower Aβ burden compared with vehicle-treated male mice, but ABX-treated female mice did not demonstrate a reduction in Aβ burden compared with vehicle-treated female mice (Fig. 2 A). We also observed trends of lower amyloid plaque size in ABX-treated male compared with vehicle-treated male mice (Fig. S4). Taken together, these data indicate that ABX-induced perturbations in the microbiome are associated with significantly lower amyloidosis only in male APPPS1-21 mice.
Figure 2.
Reductions in Aβ pathology and altered plaque-localized microglial phenotypes are only observed in ABX-treated male APPPS1-21 mice. (A) Representative images of Aβ plaque burden in the cortex of vehicle (a and c) and ABX-treated (b and d) in 7-wk-old APPPS1-21 mice using anti-Aβ monoclonal antibody, 3D6. (a and b) Male vehicle vs. ABX, respectively; (c and d) female vehicle vs. ABX, respectively. (e) Quantification of plaque burden in vehicle- and ABX-treated APPPS1-21 mice using threshold-limited particle analysis of 3D6+-positive staining from six sections per case and expressed relative to the total cortical area of each slice. Aβ burden analysis revealed a significant effect of ABX (two-way ANOVA: F[1, 36] = 10.58, P = 0.0025) and an interaction (F[1, 36] = 9.145, P = 0.0046) while sex showed no significant effect (F[1, 36] = 1.76, P = 1929). (B) Representative images of microglial cells using Iba1 antibody. No differences in total Iba1-positive cells were observed in vehicle versus ABX treatment irrespective of sex (male: a, b, and e; female: c, d, and e; two-way ANOVA—ABX treatment: F[1, 36] = 0.036, P = 0.85; or sex: F[1,36] = 0.19, P = 0.668; or an interaction between ABX treatment and sex: F[1,36] = 0.131, P = 0.718). In contrast, the area of Iba1-positive cell bodies was significantly reduced in ABX treated male mice (b and f) but not in females (d and f; two-way ANOVA—ABX treatment: F[1, 36] = 14.77, P = 0.0005; sex: F[1, 36] = 4.97, P = 0.0321; and an interaction: F[1, 36] = 142.2, P < 0.0001). Control male mice showed larger cell bodies compared with control female mice (P < 0.0001). ABX treatment resulted in significantly smaller cell bodies in males (P < 0.0001) and larger cell bodies in females (P < 0.0001) compared with their respective controls. (C) Imaris 3D reconstruction showed significantly different microglia morphology characteristics in males (a and b) compared with females (c and d) after ABX treatment (microglial branch length—two-way ANOVA: F[1, 36] = 70.77, P < 0.0001; microglial branch end points: F[1, 36] = 61.24, P < 0.0001), with sex (microglial branch length: F[1, 36] = 91.42, P < 0.0001; microglial branch end points: F[1, 36] = 75.38, P < 0.0001), and an interaction (microglial branch length: F[1, 36] = 29.63, P < 0.0001; microglial branch end points: F[1, 36] = 22.73, P < 0.0001). Vehicle-treated male mice showed significantly longer branch lengths (a; quantified in e, P = 0.029) and number of branch points (a; quantified in f, P = 0.04) compared with vehicle-treated females (c). ABX treatment resulted in significantly longer branch lengths (b; quantified in e, P < 0.0001) and increased numbers of branch points (b; quantified in f, P < 0.0001) in male mice. Scale bars in A (a–d), B (a–d), and C (a–d) represent 500 µm, 10 µm, and 10 µm, respectively. Data are mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; n = 10/group.
The morphology of plaque-localized microglia show sex-specific characteristics in APPPS1-21 mice at the age of 7 wk
In view of earlier studies demonstrating microbiome-dependent influences on microglial gene expression and morphological phenotypes (Erny et al., 2015), we sought to evaluate the morphological characteristics of microglia in the brains of vehicle- or ABX-treated APPPS1-21 mice. We evaluated plaque-localized microglia (0.02 mm2 area containing a 3D6+ Aβ plaque) in the cerebral cortex using immunofluorescence staining with antibodies raised to the microglial antigen, ionized calcium binding adaptor molecule 1 (Iba1). High-magnification images containing Iba1+ microglia around the 3D6+ Aβ plaque structures were collected and analyzed by using ImageJ software. Microglial cell body area and the total number of microglia were quantified (Fig. 2 B). The total number of plaque-localized microglia showed no significant differences with ABX-treatment or sex (Fig. 2 B). In contrast, we observed a significantly reduced area of microglial cell bodies in ABX-treated male mice compared with vehicle-treated male mice. Notably, vehicle-treated female mice showed reduced microglial cell body area compared with vehicle-treated male mice. More importantly, ABX-treated female mice showed significantly larger cell body area compared with vehicle-treated female mice (Fig. 2 B).
To assess the morphology of plaque-localized microglia, we employed an Imaris software platform to generate 3D reconstructions (Fig. 2 C). The total dendrite branch length and total number of branch points of plaque-localized microglia were evaluated in vehicle- or ABX-treated mice. ABX treatment resulted in significantly longer dendritic branch lengths in male but not female mice compared with their vehicle-treated counterparts. Notably, vehicle-treated female mice showed shorter dendritic branch lengths compared with vehicle-treated male mice (Fig. 2 C, e). In addition, vehicle-treated female mice showed significantly lower dendritic branch points compared with vehicle-treated male mice. However, ABX treatment resulted in a higher number of dendritic branch points in male but not in female mice compared with their control counterparts (Fig. 2 C, f).
Collectively, these data indicate that in male mice an ABX-perturbated microbiome is associated with lower numbers of phagocytic, plaque-localized microglia, as defined by smaller cell bodies, longer dendritic branch lengths, and increased dendritic branches. On the other hand, female mice exhibited a phagocytic microglial phenotype as defined by larger cell bodies, shorter branch lengths, and fewer branch points that are independent of vehicle or ABX treatment.
ABX treatment results in sex-specific gene expression profiles in the cerebral cortex of APPPS1-21 mice at the age of 7 wk
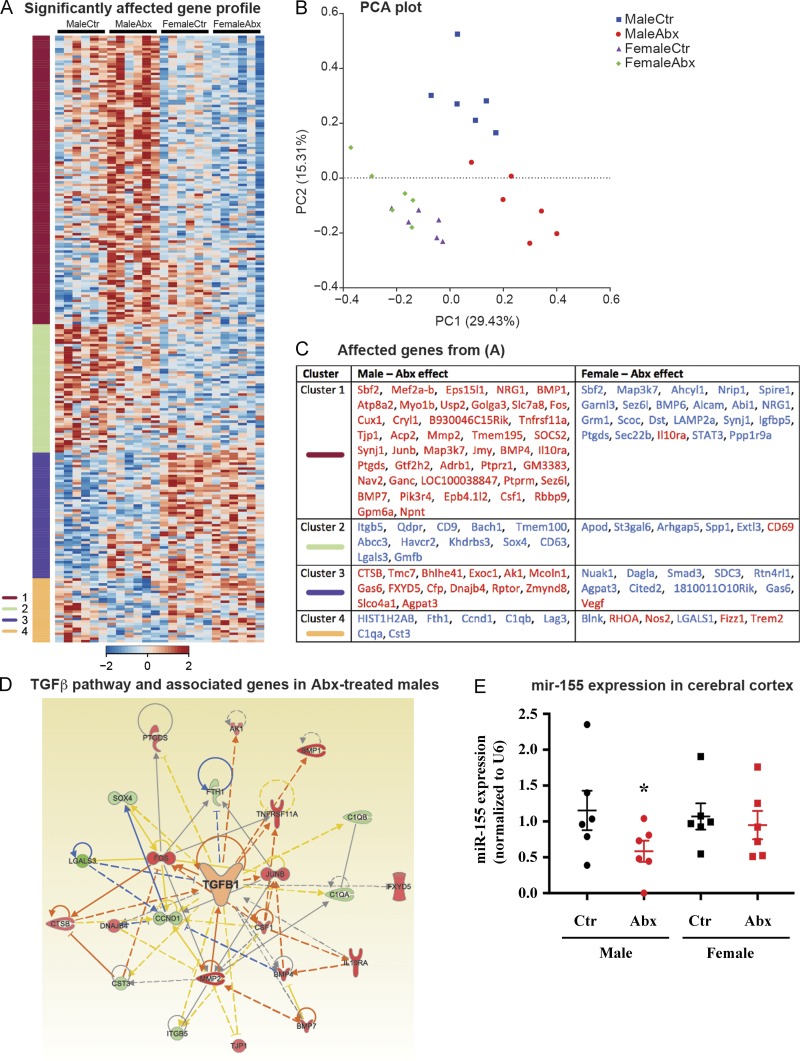
In addition to microglia morphology (Fig. 2, B and C), we also investigated transcriptome profiles that might reflect relevant functional outcomes. We purified total cortical RNA and then hybridized these preparations to a NanoString MG550 chip that harbors targets of microglia-specific transcripts and a panel of additional inflammation and phagocytosis-related genes (Krasemann et al., 2017). Of 482 targets, 160 showed a significant sex effect, while 57 showed a significant treatment effect, and 108 showed significant sex and treatment interactions. The comparison of the gene patterns via K-means clustering of significantly affected genes revealed four gene clusters (Fig. 3 A), and a principal component analysis plot of these genes revealed distinct sex-specific differences in ABX- versus vehicle-treated APPPS1-21 mice (Fig. 3 B). Here, we observed marked differences in gene expression between control and ABX-treated male mice, while the female mice did not show major changes between groups.
Figure 3.
ABX treatment restores TGFβ signaling and the microglial homeostatic signature in the cerebral cortex of APPPS1-21 male mice. (A) K-means clustering of 246 significantly affected genes in RNA samples extracted from cortex tissue lysates from vehicle and Abx-treated APPPS1-21 male and female mice by using NanoString analysis. Vertical lanes are biological replicates per treatment and sex. (B) A principal component analysis (PCA) plot based on significantly affected genes between treatment and sex groups. (C) Student’s t tests within each sex and between ABX-treated and -untreated mice were conducted on the genes within each cluster from A to reveal significantly affected genes based on ABX treatment (P < 0.05). Genes in red are up-regulated, and genes in blue are down-regulated. (D) Ingenuity pathway analysis shows up-regulation of the TGFβ pathway–related molecules in tissue from ABX-treated male mice. (E) Quantitative PCR analysis of ABX-treated male and female mice revealed a significant decrease (P < 0.05) of miR-155 expression in ABX-treated male but not female mice. Data are mean ± SEM. *, P < 0.05. n = 6/group. Ctr, control.
We extracted the genes from each cluster (Fig. 3 A) in order to identify the effects of ABX treatment within sexes (Fig. 3 C). Microglia switch from an "M0 (homeostatic molecular and functional signature)" phenotype (Butovsky et al., 2014) to a microglia neurodegenerative phenotype (MGnD) during the course of aging and disease (Krasemann et al., 2017). We observed that ABX-treated male mice exhibited increase in the homeostatic microglial transcriptomic signature in clusters 1 and 3, including Mef2a, Junb, Bhlhe41, Fos, and Tnfrsf11a, and decreased expression of MGnD genes, including Lgals3, C1qa, C1qb, CD63, and Lag3 in clusters 2 and 4. Interestingly, ABX treatment had opposite effects in male and female mice on eight genes consisting of Sbf2, NRG1, Synj1, Map3k7, Ptgds, Sez6l, Gas6, and Agpat3.
Ingenuity pathway analysis revealed that the TGFβ pathway was a major upstream regulator predicted to be increased in ABX-treated males (Fig. 3 D). Up-regulated genes in the pathway included Il10ra, Fos, Csf1, Bmp4, and Bmp7 while the suppressed genes included Lgals3 and Ccnd1. Quantitative PCR analysis of miR-155, a microRNA that is induced in MGnD microglia, showed a decrease in expression in male mice treated with ABX compared with vehicle, while the female mice did not show a significant difference between groups (Fig. 3 E). Taken together, these results suggest that treatment of male APPPS1-21 mice with ABX restores microglial homeostasis, likely via induction of TGFβ signaling and several key transcriptional regulators of M0 microglia including Mef2a, Fos, and other homeostatic and anti-inflammatory modulators such as Il10ra and Gas6.
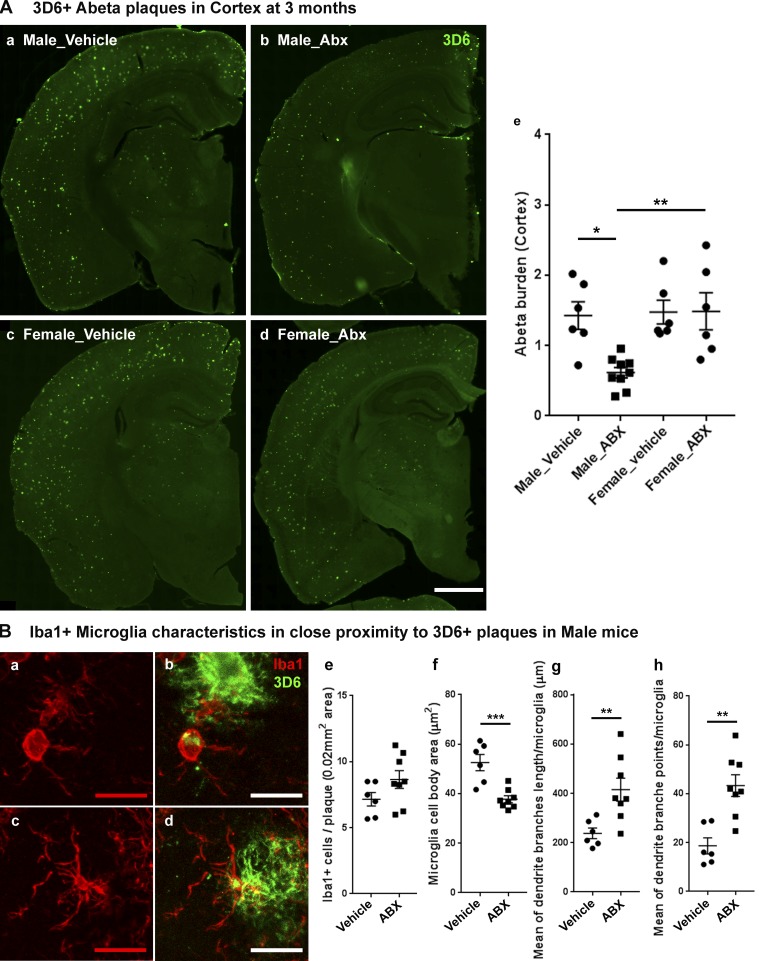
Longer-term ABX treatment results in reduced Aβ deposition only in male APPPS1-21 mice
In studies presented in Fig. 4 A, we interpreted these results to suggest that Aβ burden was selectively decreased in male mice compared with female mice by ABX. The caveat of this interpretation is that the Aβ burden in female mice treated with vehicle was far too low to make an assessment of ABX-mediated changes in Aβ burden. To circumvent this potential caveat, we evaluated the impact of ABX on male and female APPPS1-21 mice at 3 mo of age. As expected, Aβ burden was elevated in vehicle-treated, 3-mo-old male and female mice (Fig. 2 A and Fig. 4 A), and more importantly, cortical Aβ burden was comparable between male and female mice. In 3-mo-old male APPPS1-21 mice, ABX lead to a significant lowering of amyloid burden compared with vehicle-treated male mice, but this was not the case in female mice (Fig. 4 A).
Figure 4.
Reduced Aβ pathology and altered microglial phenotypes in cerebral cortex of ABX-treated male APPPS1-21 mice at 3 mo of age. (A) Representative images of Aβ plaque burden in the vehicle (a and c) and ABX (b and d) treated male (a and b) and female (c and d) mice using 3D6 antibody (e). Aβ burden was significantly lower in ABX-treated male mice (n = 8) compared with vehicle-treated male mice (P = 0.0116) while female mice showed no significant differences in Aβ burden (P > 0.05) with ABX treatment (two-way ANOVA—ABX treatment: F[1, 23] = 5.419, P = 0.0291; sex: F[1, 23] = 7.173, P = 0.0134, and an interaction: F[1, 23] = 5.707, P = 0.0255). (B) Representative images (a–d) of Iba1-positive microglial cells in close proximity to 3D6+ immunostained Aβ plaque (<0.07-mm radial distance from plaque) in 3-mo-old male mice. (e) Cell count analysis showed no significant differences between vehicle- or ABX-treated male mice; (unpaired Student’s t test: t[12] = 1.661, P = 0.1225). ABX treatment (c and d) resulted in smaller cell bodies compared with vehicle-treated (a and b) male mice (f; unpaired Student’s t test: t[12] = 4.573, P = 0.0006). Imaris 3D reconstruction (g and h) showed significantly different microglial characteristics between vehicle- and ABX-treated male APPPS1 mice. Branch length/microglia (g) showed significantly higher branch length in ABX-treated mice (unpaired Student’s t test: t[12] = 3.096, P < 0.0093). Moreover, ABX-treated mice also showed significantly higher numbers of branch points per plaque localized microglia (h; unpaired Student’s t test: t[12] = 4.178, P < 0.0013). Scale bars in A (a, b, c, and d) and B (a, b, c, and d) represent 1,000 µm and 10 µm, respectively. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Male_vehicle: n = 6. Male_ABX: n = 8. Female_vehicle: n = 6. Female_ABX: n = 6.
We extended these latter analyses to characterize the morphological phenotypes of plaque-localized microglia in male mice at 3 mo of age. While the total number of plaque-localized microglia showed no significant differences with ABX treatment (Fig. 4 B), the area of microglial cell bodies in ABX-treated male mice was significantly reduced compared with vehicle-treated male mice. Furthermore, plaque-localized microglial morphology showed significantly longer dendritic branch lengths and increased numbers of dendritic branch points in ABX-treated male mice compared with vehicle-treated male mice (Fig. 4 B), similar to the observations in 7-wk-old animals (Fig. 4 C). Collectively, ABX leads to reductions in Aβ burden in male but not in female mice.
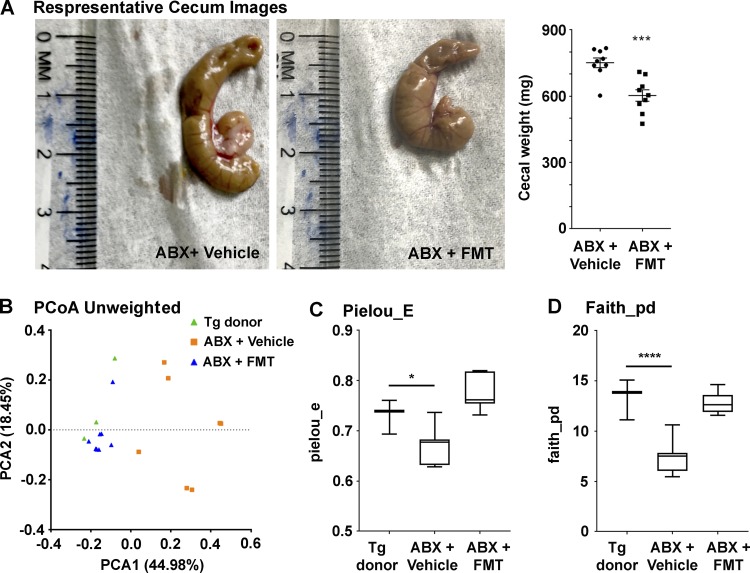
Fecal microbiota transplantation (FMT) from APPPS1-21 donor into ABX-treated male mice reduces cecal enlargement and restores fecal microbiome profiles
To confirm the causal role of the microbiome in Aβ amyloidosis and gliosis, we performed FMT from age-matched APPPS1-21 male mice into our ABX-treated male mice every day from P25 till the time of cull (7 wk of age).
As above, cecal-weight analysis confirmed the effectiveness of FMT in ABX-treated male mice (Fig. 5 A); compared with ABX+vehicle–treated mice, ABX+FMT–treated mice showed significantly lower cecum weight, thus suggesting recolonization of the host (Fig. 5 A).
Figure 5.
FMT reverts ABX-associated changes in the gut microbiome (n = 9/group). (A) Cecum weights from vehicle- or FMT-treated ABX-treated male mice. FMT introduced into ABX-treated APPPS1-21 male mice lead to reduced cecum weight (unpaired Student’s t test: t[16] = 4.372, P = 0.0005). (B) PCoA plot generated by using unweighted versions of the UniFrac distance metric. The two components explained 44.98% of the variance. FMT-treated ABX-treated male mice showed no clear separation compared with the donor fecal microbiome while vehicle-treated ABX-treated male mice showed a clear separation compared with the other two groups. The α diversity was measured by using Pielou’s evenness (Pielou_E; C) and Faith's phylogenetic diversity (Faith_pd; D) indices. FMT treatment restored the microbiome diversity as measured by both Faith’s phylogenetic diversity (one-way ANOVA: F[2,16] = 32.91, P < 0.0001) and Pielou’s evenness (one-way ANOVA: F[2,16] = 18.7, P < 0.0001). Data are mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. n = 9/group. PCA1, principal coordinate 1; PCA2, principal coordinate 2.
We then profiled the microbiome at 7 wk to confirm the effectiveness of the FMT protocol. The β diversity using unweighted UniFrac was significantly different between Tg-donor and ABX+Vehicle–treated mice and showed a clear separation on a PCoA visualization. FMT resulted in a clear separation between ABX+FMT–treated mice compared with ABX+Vehicle–treated mice while Tg-donor and FMT-gavaged ABX-treated mice were clustered very closely, thus suggesting that FMT-treatment in ABX-treated mice showed similar microbiome profiles as the Tg-donor mice (Fig. 5 B). Unweighted β diversity was significantly different although with a low effect size, while weighted was not significant, indicating that lower diversity taxa had a larger effect on differentiating the microbial profiles. α diversity indices showed significantly lower richness and evenness in ABX+Vehicle–treated mice compared with the Tg-donor while ABX+FMT mice showed no significant differences compared with the Tg-donor (Fig. 5, C and D).
ANCOM comparison at the ESV and species levels showed significant differences between both Tg-donor and ABX+FMT–treated groups and between ABX+FMT–treated and ABX+Vehicle–treated groups but with far more differences observed in the latter pairing, as expected. Tg-donor groups only differed from ABX+FMT–treated mice in two Bacteroidales S24-7 family member ESVs and a Bacteroides species. At the ESV level, the ABX+FMT–treated group compared with ABX+Vehicle–treated group had a significantly higher abundance of several members of the Bacteroidales order, including Bacteroides, Prevotella, and S24-7, while ABX+Vehicle–treated mice showed a higher abundance of a single S24-7 ESV. At the L7 level, a number of varying taxa were found, but only those higher in abundance in the ABX+FMT–treated group were significant (Table 3). Taken together, ABX+Vehicle treatment resulted in reduced diversity and altered microbiome, whereas the FMT protocol in ABX-treated mice resulted in the restoration of diversity and a microbiome profile more similar to the transgenic donor.
Table 3. ANCOM significant differences in fecal microbiota at week 7 (FDR P < 0.05).
| Higher in ABX+FMT | Lower in ABX+FMT | |
|---|---|---|
| Tg-donor vs. ABX+FMT | ||
| ESV | p_Bacteroidetes; c_Bacteroidia; f_S24-7; g_ (×2) | |
| L7 clustered taxa | p_Bacteroidetes;c_Bacteroidia;f_Bacteroidaceae;g_Bacteroides;s_ | |
| ABX+Vehicle vs. ABX+FMT | ||
| ESV | p_Bacteroidetes; c_Bacteroidia; f_S24-7; g_ (×2) | p_Bacteroidetes; c_Bacteroidia; f_S24-7; g_; s_ |
| p_Bacteroidetes; c_Bacteroidia; f_Prevotellaceae; g_Prevotella; s_ | ||
| p_Bacteroidetes; c_Bacteroidia; f_Bacteroidaceae; g_Bacteroides | ||
| p_Bacteroidetes; c_Bacteroidia; f_Bacteroidaceae; g_Bacteroides; s_ | ||
| L7 clustered taxa | p_Bacteroidetes;c_Bacteroidia;f_Bacteroidaceae;g_Bacteroides;s_ | |
| p_Bacteroidetes;c_Bacteroidia;f_Bacteroidaceae;g_Bacteroides;s_g_Odoribacter;s_ | ||
| p_Bacteroidetes;c_Bacteroidia;f_[Paraprevotellaceae];g_Paraprevotella;s_ | ||
| p_Deferribacteres;c_Deferribacteres;f_Deferribacteraceae;g_Mucispirillum;s_schaedleri | ||
| p_Firmicutes;c_Clostridia;f_Ruminococcaceae;g_Ruminococcus;s_ | ||
| p_Proteobacteria;c_Alphaproteobacteria;o_RF32;f_;g_;s_ | ||
| p_Proteobacteria;c_Deltaproteobacteria;f_Desulfovibrionaceae;g_;s_ |
To determine the taxa that described the most variance between groups, ANCOM analyses of 16S rRNA sequencing data at the DADA2-inferred ESV level (taxonomy assigned via Greengenes) and clustered species levels were performed between vehicle-treated or FMT-treated ABX-treated male mice at the age of 7 wk. ABX+FMT, FMT (from Tg-donor)-gavaged ABX-treated male mice; ABX+Vehicle, vehicle-gavaged ABX-treated male mice.
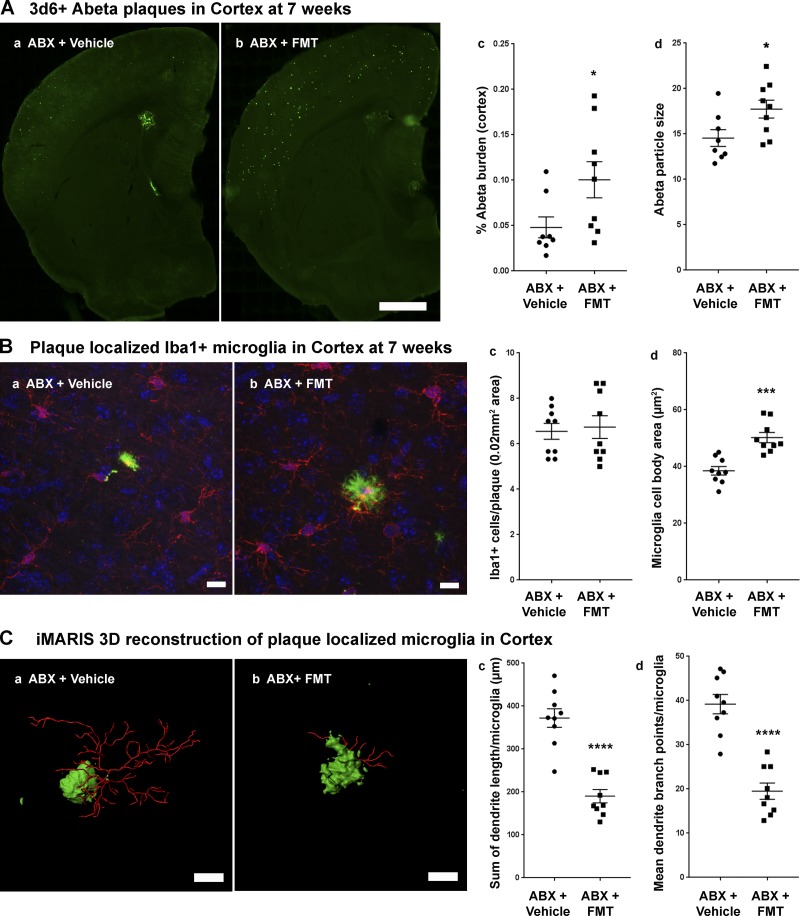
FMT from APPPS1-21 Tg-donor partially restores Aβ plaque burden and microglial morphologies in ABX-treated male mice at the age of 7 wk
Having demonstrated that the microbiota in 7-wk-old mice subject to the ABX+FMT protocol is similar to the Tg-donor, we then asked whether this would translate to a restoration in plaque burden and microglial phenotypes. The ABX+FMT–treated group showed significantly higher Aβ burden and larger plaque size in the cortex compared with the ABX+Vehicle–treated group (Fig. 6 A). Moreover, The ABX+FMT–treated group showed significantly larger microglial cell body area compared with ABX+Vehicle–treated group (Fig. 6 B), but no changes in total microglia numbers were observed (Fig. 6 B). Finally, microglia showed significant shortening of dendritic branch lengths and reductions of dendritic branch points in the ABX+FMT–treated group compared with the ABX+Vehicle–treated group (Fig. 6 C). These data suggest that FMT from Tg-donor into ABX-treated male mice partially restored Aβ amyloidosis and microglia morphology.
Figure 6.
FMT from APPPS1-21 male mice to ABX-treated APPPS1-21 male mice resulted in restoration of Aβ pathology and microglial phenotypes. (A) Representative images of Aβ plaque burden in the cortex of vehicle- (a) and FMT-treated ABX-treated mice (b) using 3D6 antibody. (c) Quantification of plaque burden in vehicle- and FMT-treated ABX-treated male mice. Aβ burden was significantly higher in ABX+FMT–treated mice compared with ABX+vehicle–treated mice (two-tailed Student’s t test: t[15] = 2.209, P = 0.0431). (d) Aβ particle size was also significantly higher in ABX+FMT–treated mice compared with ABX+vehicle–treated mice (two-tailed Student’s t test: t[15] = 2.355, P = 0.0326). (B) Representative images of Iba1-positive microglial cells (a and b). (c) Cell count analysis showed no significant differences between ABX+vehicle–treated mice (a) versus ABX+FMT–treated mice (b; unpaired Student’s t test: t[16] = 0.3026, P = 0.7661). However, FMT resulted in larger cell bodies compared with vehicle-treated ABX-treated male mice (d; unpaired Student’s t test: t[16] = 4.968, P = 0.0001). (C) Imaris 3D reconstruction showed significantly different microglial characteristics between vehicle-treated (a) and FMT-treated (b) ABX-treated male mice. Branch length/microglia (c) showed significantly lower branch length in ABX+FMT–treated mice (unpaired Student’s t test: t[16] = 6.823, P < 0.0001). Moreover, ABX+FMT–treated mice showed significantly lower numbers of branch points per plaque-localized microglia (d; unpaired Student’s t test: t[16] = 6.876, P < 0.0001). Scale bars in A (a and b), B (a and b), and C (a and b) represent 1,000 µm, 10 µm, and 10 µm, respectively. Data are mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. n = 8–9/group.
Discussion
Dysregulation in the microbiome-gut-brain axis is now emerging for a variety of neurological diseases, including Parkinson’s disease and AD and developmental and psychiatric conditions. We have explored the impact of the microbiome in Tg mouse models of Aβ amyloidosis. Extending our earlier findings using the APPSWE/PS1ΔE9 mouse model showing a significant impact of ABX-mediated alterations of the microbiome on Aβ pathology and microglial phenotypes (Minter et al., 2016, 2017), we have examined these parameters in another well-established APPPS1-21 mouse model and now offer several important insights. First, we demonstrated that long-term ABX treatment of male APPPS1-21 mice results in distinct alterations in peripheral cytokines/chemokines, changes in the composition of the fecal microbiome, and a significant reduction in Aβ deposits. In contrast, ABX-treated female mice exhibited a greater abundance of bacteria and predicted metabolic pathways that are considered pro-inflammatory, higher levels of plasma pro-inflammatory cytokines/chemokines, and no apparent effects on the levels of Aβ deposits compared with vehicle-treated animals. Second, we show that ABX-treated male mice exhibited alterations in microglial morphologies similar to those observed in GF wild-type mice (Erny et al., 2015) and transcriptional signatures that are consistent with activation of the TGFβ signaling pathway that is a hallmark of M0 (homeostatic) microglia (Butovsky et al., 2014). In sharp contrast, the microglia in ABX-treated female mice exhibited retracted processes and larger cell bodies that closely mimicked that observed in MGnD microglia, findings supported by transcriptional readouts. Finally, transplants of feces from age-matched APPPS1-21 male donor mice into long-term ABX-treated male mice resulted in the restoration of microbiota profiles, elevated MGnD-type microglia, and partial restoration in Aβ deposition, thus confirming a causal relationship between the gut microbiome and AD-associated inflammation and Aβ pathology in this model.
While it has been reported that the microbiome can modulate disease phenotypes in a sex-specific manner (Markle et al., 2013; Yurkovetskiy et al., 2013; Sampson et al., 2016), our current studies are the first to report sex-specific alterations in AD-related phenotypes by the gut microbiome using a mouse model of Aβ amyloidosis. Our results are consistent with published studies that included GF and ABX-treated wild-type mice, wherein the microbiome exerts sex-specific effects on microglial maturation leading to altered immune responses to environmental stimuli such as the bacterial endotoxin (Erny et al., 2015; Hanamsagar et al., 2017; Thion et al., 2018). Taken together with our current findings, it is highly probable that the microbiome can have a significant impact on the maturation and function of brain immune cells and in a sex-specific manner.
The mechanism(s) underlying “clearance” of Aβ aggregates/oligomers/plaques are largely silent. In this regard, our microglial transcriptome studies indicate that microglia in vehicle-treated mice exhibit an MGnD signature and that ABX-treated male mice exhibit an M0 homeostatic signature, leading to the suggestion that ABX alters the cellular response of microglia to Aβ plaques (or oligomeric Aβ species). Alternatively, and a model that we favor, is that at the time of culling, microglia exhibit an M0 signature because these cells have been clearing plaques/oligomers throughout the lifetime of the animal and there is very little left to be cleared. Future studies using real-time, simultaneous two-photon confocal imaging of GFP-labeled microglia and plaque intercalating dyes during the mouse lifetime will be essential to resolving this important lacuna in our understanding of the role and mechanism(s) by which microglia in ABX-treated animals mediate clearance of Aβ in the mouse models.
Peripheral inflammation plays a major role in central inflammation (Perry et al., 2007; Wendeln et al., 2018). We observed relatively lower expression of pro-inflammatory cytokines/chemokines, including cluster of differentiation 30 (CD30), CD40, CCL11 (Eotaxin1), IL-1B, IL-2, IL-3, IL-17A, LIX (CXC5), RANTES (CCL5), and SCF, and higher anti-inflammatories, such as IL10 in ABX-treated male mice. In contrast, ABX-treated female mice showed relatively higher levels of pro-inflammatory cytokines (CCL11, IL-1B, IL-2, IL-3, IL-5, IL-9, IL-17, LIX, and SCF) compared with ABX-treated male mice. In this regard, microbiome-derived metabolites can influence peripheral cytokine production (Wikoff et al., 2009). Microbiota can also influence the relative populations and function of subsets of immune cells (Ivanov et al., 2009; Dorrestein et al., 2014) modulating mucosal immune responses (Mazmanian et al., 2005; Kamada et al., 2013) and systemic immunity (Belkaid and Naik, 2013). It is possible that altered peripheral inflammation due to microbiome perturbations in our model could impact brain inflammation that ultimately affects microglia. Future investigations of the effect of plasma samples on microglial Aβ phagocytosis from our mice will allow us to address the role of peripheral inflammation.
Our microbiome data reveal no significant differences comparing sexes in vehicle and ABX treatment at P22, but the microbiome profiles at the time of culling (7 wk of age) represented a significant effect of sex in ABX-treated mice. Importantly, we observed ABX-treated female mice showed a greater abundance of Allobaculum (s_unknown) and Akkermansia (s_muciniphila), as well as higher levels of peripheral pro-inflammatory cytokines and phagocytic microglia and no changes in Aβ amyloidosis. Certain Allobaculum and Akkermansia species are known mucin degraders, a greater proportion of which could lead to a decrease the mucin-layer thickness, which may not only result in greater inflammatory activation but also could increase the likelihood of translocation of bacteria or bacterial metabolites from the gut to circulation. We are not able to prove this with the current data, but it remains a compelling hypothesis to test. Increased microbial or metabolite translocation can increase endotoxin levels in the systemic circulation, affecting peripheral and central inflammation as reported (de Punder and Pruimboom, 2015). Akkermansia has previously been associated with various neurodegenerative disorders (Keshavarzian et al., 2015; Jangi et al., 2016; Berer et al., 2017; Engen et al., 2017). Similarly, Allobaculum has been negatively correlated with anti-inflammatory genes Forkhead box protein P3 (FOXP3), IL-10, and intestinal barrier proteins ZO-1 and occludin (Lee et al., 2015). In the FMT study, a large number of organisms of order Bacteroidales were found to be higher in ABX+FMT–treated mice, possibly driving a restoration in pathology. Significant changes in Akkermansia and Allobaculum were not observed via ANCOM in this cohort. Other taxa that were increased in ABX+FMT–treated mice also may have contributed to pathology; in particular Mucispirillum schaedleri, a possible pathobiont (Nguyen et al., 2015), was in a higher abundance in the FMT treatment group. It thus remains unclear if changes in diversity, changes in overall bacterial communities, or changes in mentioned individual genera result in observed sex-specific immune and pathological outcomes in our model. Future transplantation studies involving full FMT or selected bacteria in GF mice will help resolve these issues.
Finally, we acknowledge several limitations of our studies. First, we used long-term ABX treatment to evaluate microbiome perturbations and effects on amyloidosis and microglial activity and our FMT protocol to prove causation was performed simultaneously with diluted ABX. It is plausible that diluted ABX in drinking water could have an impact on FMT efficiency that affects our final outcomes of amyloidosis and microglia morphology measures. Future work using established short-term (P14–P21) ABX gavage will allow us to address this concern in our model. Second, our NanoString studies were aimed at investigating microglial inflammatory gene profiles, but because the RNA was prepared from the entire cerebral cortex our data could certainly be influenced by the expression of the identified genes in other cell types represented in the cortex. Future transcriptomic studies of purified microglia and other cortical cell types in our experimental paradigm will be essential to clarify the underlying cellular and physiological basis underlying the observations reported herein.
In conclusion, our findings are fully consistent with the conclusion that ABX-mediated perturbations of the microbiome have selective, sex-specific influences on brain Aβ amyloidosis and microglial homeostasis.
Materials and methods
Animal housing and handling
APPPS1-21 mice were maintained on a C57BL6Cj background and obtained from M. Jucker (University of Tubingen, Tubingen, Germany). Mice were housed in sterile micro-isolator cage and fed ad libitum on standard chow. All animal experimental procedures were performed in accordance with the approved Animal Care and Use Protocols by the Institutional Animal Care and Use Committee at the University of Chicago.
ABX treatment
Pups receiving the ABX were gastric gavaged (200 µl ABX by using animal feeding needles; catalog number 7901; Cadence) with established ABX (Stefka et al., 2014; 4 mg/ml kanamycin, Sigma-Aldrich K4000-5g; 0.35 mg/ml gentamicin, Sigma-Aldrich G1914-250mg; 8,500 U/ml colistin, Sigma-Aldrich C4461-1g; 2.15 mg/ml metronidazole, Sigma-Aldrich M1547-25g; 0.45 mg/ml vancomycin, Sigma-Aldrich V2002-1g) in autoclaved water from P14 to P21 followed by an ad libitum access to freshly prepared 1:50 diluted ABX water until the time of sacrifice. During 7 d of postnatal ABX gavage, all mice were transferred to a new sterile cage after each gavage to avoid microbial contamination from accumulated fecal pellets in the cages. Parents from the same cage as pups receiving ABX treatment were euthanized after weaning the pups and were not used for any breeding or future experiments. Water with ABX was changed every 5–6 d.
Fecal pellet collection
To evaluate the microbiome profile after high-dose ABX treatment (P14–P21), fresh fecal pellet samples from each pup were collected in sterile 1.5-ml centrifuge tubes at P22 and stored in −80°C. Fresh fecal pellets were also collected at the time of sacrifice and immediately frozen and stored at −80°C until the time of bacterial DNA extraction.
Necropsy and tissue harvesting
The necropsy was performed according to procedures approved by Animal Care and Use Protocols. Briefly, a mixture of ketamine and xylazine was introduced i.p., and after confirmation of deep anesthesia, the heart was accessed through abdominal surgery, and blood was collected from the right ventricle by using a 25-gauge needle and stored in buffered sodium citrate blood collection tubes (catalog number 366393; BD Vacutainer) on ice. After the blood collection, the descending aorta was clamped, and mice were perfused by using cold saline (pH 7.4) for 3 min. Brains were then excised and dissected into two hemispheres (one hemisphere was postfixed with 4% paraformaldehyde, and the other was frozen for RNA extraction). Cecum was collected and weighed after careful removal from the small and large intestine. After the necropsy, plasma was separated from blood cells by centrifugation at 2,000 rpm for 10 min at 4°C by using a Beckman Coulter centrifuge and stored at −80 for the cytokine measurements.
Microbiome analysis
The V4 region of the bacterial 16S rRNA gene was amplified by using the Earth Microbiome Project primer set (515f-806r). Each 25-ml PCR reaction contained 12.5 ml of AccuStart II PCR ToughMix (Quantabio), 1 µl of 5 µM forward primer, 1 µl of 5 µM reverse primer, 9.5 µl of water, and 1 µl of DNA extraction. The PCR program was 94°C for 3 min to denature the DNA, with 35 cycles at 94°C for 45 s, 50°C for 60 s, 72°C for 90 s, and then a final 72°C step. Amplification was quantified by using PicoGreen (Invitrogen), and each sample was pooled at 70 ng per sample. Pools were cleaned by using Agencourt AMPure XP beads (Beckman Coulter), and the clean pools were combined. This final pool was quantified and sent to the Environmental Sample Preparation and Sequencing Facility at Argonne National Laboratory for sequencing on an Illumina Miseq by using v3 chemistry following the Earth Microbiome Project protocol.
Raw 16S reads were demultiplexed, chimera and phiX filtered through the QIIME2 pipeline, and denoised with DADA2 (Callahan et al., 2016). The resulting representative sequences were aligned and masked with MAFFT (Katoh et al., 2002). A phylogenetic tree was constructed via FastTree 2 (Price et al., 2010). Samples were subsequently clustered and taxonomically classified into operational taxonomic units (OTUs) with QIIME2 by using the Greengenes 99% database (DeSantis et al., 2006). Diversity metrics were calculated after rarefying reads at a depth of 10,000 sequences. β diversity was calculated as unweighted and weighted UniFrac distances. Faith’s phylogenetic diversity and Pielou’s evenness were used to measure α diversity. ESVs are reported with Greengenes-assigned taxonomy and species level clustered to OTUs by the QIIME2 Bayesian classifier. Differences in relative abundances were calculated via ANCOM (Mandal et al., 2015) and considered significant at the P < 0.05 level. Correlations of metadata with OTU abundances were calculated as Spearman rank correlations with raw P values determined via bootstrapping (1,000 permutations). All reported P values are false discovery rate–corrected for multiple comparisons.
Cytokine antibody array
At the time of necropsy, blood samples were collected from the right ventricles of anesthetized mice. Plasma was separated by using 10-min centrifugation at 2,000 rpm at 4°C. Plasma (50 µl) from 10 mice in each treatment group was pooled into one sample and diluted 1:1 (vol/vol) in blocking buffer per the manufacturer’s suggestion. Diluted plasma was then probed for the cytokine profile by using the Mouse Cytokine Antibody Array 3.1 Kit according to the manufacturer’s protocol (RayBiotech). Briefly, membranes were blocked with a blocking buffer, and then 1 ml of diluted serum sample per group was added to each membrane and incubated at 4°C overnight. The next day, membranes were washed and incubated in biotinylated antibody cocktail at room temperature for 1.5 h. The membranes were then incubated with HRP-streptavidin concentrate at room temperature for 2 h, and cytokine presence was detected by chemiluminescence. Films of array dots were scanned and converted to TIFF format to import into ImageJ software. Densitometry was performed for each circle by using ImageJ software. The background was subtracted by using blank optical density, and data were normalized against controls among membranes. These data were then transformed into Log2 to calculate Log2fold change values. Heatmaps were generated using R software for Log2fold change data.
Immunocytochemistry
To perform immunofluorescence staining, a full series was selected for Aβ (3D6, 1:10,000) staining, and a half series was selected for microglia (Iba1, 1:500) marker. Briefly, cryoprotectant-stored brain tissue sections (40-µm thick) were washed with dilution media for 60 min (10 min/wash, six washes). After blocking with serum solution for 1 h, tissue sections were incubated in primary antibodies for overnight in a 4°C refrigerator. The next day, the sections were brought to room temperature. Tissue sections were washed with dilution media for 60 min (10 min/wash, six washes) and incubated in secondary fluorescence antibodies (DAM488 and DARab594; Invitrogen; 1:500) for 1 h at room temperature in the dark. Sections were then washed with dilution media again and incubated in DAPI (Invitrogen, 1:10,000) for counterstaining. After three washes (10 min each) of Tris-buffered saline, the tissue samples were then mounted on glass slides and coverslipped by using Fluoromount aqueous mounting medium (catalog number F4680, Sigma-Aldrich). 3D6+ Aβ plaque images were captured by using Pannoramic SCAN BF plus FL Optimum slide scanner (PerkinElmer/3DHistech). Microglia images were captured by using a laser confocal microscope (SP6, Leica) under 40 × 1.5 (for cell counts and diameter measures) or 60 × 2 (for 3D reconstruction) magnification.
Aβ load (3D6 counting) analysis
Six sections of cortical cerebrum at an equidistance of 240 µm were selected for 3D6+ Aβ plaque analysis. Total plaque number and Aβ area fraction were calculated by using the Fiji ImageJ software (ImageJ 1.51n, National Institutes of Health). In particular, images were normalized, and an automatic thresholding based on the entropy of the histogram (“MaxEntropy”) was used to identify the plaques. Pictures were converted into 8-bit format, followed by binary conversion and the “fill holes” and “watershed” algorithm were applied. Finally, plaque number, plaque area, and region of interest were calculated by using the “analyze particles” plugin of ImageJ. The Aβ area fraction (Aβ burden) was determined by dividing the total plaque area by the area of the cerebral cortex (region of interest).
Microglia morphology characterization
Cell counting and cell body area measurements were collected by using National Institutes of Health ImageJ software. In brief, z-stack images of individual plaques from 40-µm-thick sections were obtained under 40 × 1.5 magnification (oil immersion lens) with 0.35-µm-step increments in the z plane. A total of four to five immunofluorescence images per case containing 3D stacks of 3D6 (Aβ plaque), Iba1 (microglia), and DAPI (nucleus) were collected and imported into ImageJ. After individual channels were separated, the Iba1 staining containing z-stack was processed with maximum-intensity projections. These maximum-intensity projection images were used to measure the number of microglia cells and cell body area. The total area for the region of interest and number of microglia cells around the plaque were quantified manually in each section. Iba1+ individual cell body or cell overlaps were confirmed with DAPI containing the z-stack. The average number of microglia cells/plaque per 0.02-µm2 area was collected for each case. By using freehand selection, each microglial cell body was outlined to measure the cell body area. The average cell body area was collected for each case.
Three-dimensional reconstruction by using Imaris (Bitplane) was performed to measure microglia cell branch lengths and the number of branch point parameters as published previously (Minter et al., 2017). Sections co-labeled with 3D6 (Aβ plaque), Iba1 (microglia), and DAPI (nucleus) were prepared as above. The z-stack images of individual plaques from 40-µm-thick sections were obtained under 60× magnification with 0.35-µm-step increments in the z plane. These z-stack images were then analyzed by using Imaris software. By using the filament tool, all microglial dendrite-like processes were mapped and recorded. The surface tool was used to establish the realm of the Aβ plaque. Quantification of average dendrite length and the total number of dendrite branch points was collected by using an in-built analysis program. A total of four Aβ plaque-containing microenvironments were analyzed per mouse.
FMT
Control age-matched APPPS1-21 male donor mice were used to collect the fresh fecal pellets. Donor mice (n = 4) were housed under specific pathogen-free conditions in a separate cage from experimental groups with ad libitum food and water access. Fresh fecal pellets from individual donor mice were collected and immediately mixed in sterile water (5 mg fecal content dissolved in 1 ml sterile water). The fecal slurry supernatant was collected after allowing the suspension to settle down by using gravity for 5 min. Each postnatally treated high-dose ABX-recipient mouse in the ABX+FMT–treated group received 200 µl of freshly prepared fecal slurry from the age-matched donor mouse by gastric gavage daily starting at on P25 until the time of sacrifice simultaneously with low-dose ABX in the water (original experimental design). The ABX+Vehicle–treated group received 200 µl of sterile water in a similar fashion to nullify the everyday gavage effect.
RNA isolation and NanoString analysis
Total RNA was isolated from the cortex of 7-wk-old mice by using TRIzol reagent (Ambion) followed by RNeasy Micro clean-up (QIAGEN) procedure according to the manufacturer’s protocol. All RNAs were analyzed on an Agilent Bioanalyzer for quality assessment. 100 ng of total RNA was used in NanoString analysis by using MG550 Chip, which contains 400 unique and enriched microglial genes and 150 inflammation- and phagocytosis-related genes (Butovsky et al., 2014). NanoString data were analyzed by using nSolver software. RNA counts for target genes were normalized to the mean of housekeeping genes (Cltc, Gapdh, Gusb, Hprt, Pgk1, and Tubb5) to account for differences in the RNA content. The value of the highest negative control on the chip was used to set the cutoff, and the fold changes were calculated by comparing the ABX-treated group to the respective vehicle-treated group.
Statistical analysis
Statistical analysis was performed by using GraphPad Prism software. Multiple-group comparisons were performed by using two-way ANOVA, and group differences were evaluated by using the post hoc Bonferroni multiple-comparison test. For three-group comparisons (Tg_donor versus ABX+Vehicle versus ABX+FMT), one-way ANOVA was performed followed by Bonferroni multiple-comparison post hoc analysis. Two-group comparisons (ABX+Vehicle and ABX+FMT) were performed by using the two-tailed unpaired Student’s t test. Significance was accepted at values of P < 0.05. NanoString data (RNA counts) were normalized by the geometric mean of six housekeeping genes: Cltc, Gapdh, Gusb, Hprt, Pgk1, and Tubb5. A cutoff was introduced at the value of the highest negative control present on the chip. A two-way ANOVA was used to identify significantly affected genes (P < 0.05) among four treatment/sex groups and Student’s t tests were used to identify ABX treatment effects. Heatmaps and clustering were generated in R by using heatmap.2 from the gplots package and the prcomp from the stats package. For clustering, z-scores were calculated by using the mean expression of biological replicates per treatment group and sex and then subsequently clustered by using K-means. A scree plot was used to assess the number of clusters.
Online supplemental material
Fig. S1 presents Western blot results by using human APP)-specific 6E10 antibody to detect steady-state levels of full-length human APP (APP-FL), β-carboxyl-terminal fragment (β-CTF), and Aβ peptides comparing both APPPS1-21 and APPSWE/PS1ΔE9 mice. Fig. S2 presents microbial diversity data at P22 and cecal weight analysis at 7-wk of age. Fig. S3 presents immunoblots and heatmap analysis of fold-changes in levels of plasma cytokines/chemokines from vehicle- or ABX-treated mice in a sex-dependent manner. Fig. S4 shows Aβ plaque size analysis in the cerebral cortex of 7-wk-old APPPS1-21 mice. Table S1 presents ANCOM significant differences in fecal microbiota at P22.
Supplementary Material
Acknowledgments
We thank Ms. Shirley Bond and Dr. Christine Labno for the image processing at the University of Chicago Integrated Light Microscopic Core Facility.
This study was supported by the Cure Alzheimer’s Fund (to S.S. Sisodia and to O. Butovsky), Open Philanthropy Project and Good Ventures Foundation (to S.S. Sisodia), the Edward H. Levi Fund (to S.S. Sodia), National Institutes of Health National Institute of Neurological Disorders and Stroke (1R01NS088137, R21NS104609, and R21NS101673 to O. Butovsky) and National Institute on Aging (R01AG051812 and R01AG054672 to O. Butovsky), National Multiple Sclerosis Society (to O. Butovsky), and Amyotrophic Lateral Sclerosis Association (to O. Butovsky).
The authors declare no competing financial interests.
Author contributions: H.B. Dodiya conceived and performed ABX experiments and FMT experimental studies, microbiome data interpretation, cytokine assessments, immunocytochemistry experiments, confocal microscope and Imaris evaluation, final figure preparation, and manuscript preparation. T. Kuntz assessed the gut microbiota profile and performed bioinformatics analysis. S.M. Shaik, C. Baufeld, and J. Leibowitz performed NanoString experiments and data analysis, interpretation, and preparation of a figure. X. Zhang performed Western blots of tissue samples. N. Gottel supervised and processed microbial DNA extraction and the 16S sequencing process. X. Zhang maintained and genotyped APPPS1-21 mice. O. Butovsky reviewed and critiqued the manuscript. J.A. Gilbert reviewed and critiqued the manuscript. S.S. Sisodia developed the microbiota-related hypothesis, co-designed the experimental study, and reviewed and critiqued the manuscript.
References
- Bäuerl C., Collado M.C., Diaz Cuevas A., Viña J., and Pérez Martínez G.. 2018. Shifts in gut microbiota composition in an APP/PSS1 transgenic mouse model of Alzheimer’s disease during lifespan. Lett. Appl. Microbiol. 66:464–471. 10.1111/lam.12882 [DOI] [PubMed] [Google Scholar]
- Belkaid Y., and Naik S.. 2013. Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 14:646–653. 10.1038/ni.2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer K., Gerdes L.A., Cekanaviciute E., Jia X., Xiao L., Xia Z., Liu C., Klotz L., Stauffer U., Baranzini S.E., et al. . 2017. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. USA. 114:10719–10724. 10.1073/pnas.1711233114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandscheid C., Schuck F., Reinhardt S., Schäfer K.H., Pietrzik C.U., Grimm M., Hartmann T., Schwiertz A., and Endres K.. 2017. Altered Gut Microbiome Composition and Tryptic Activity of the 5xFAD Alzheimer’s Mouse Model. J. Alzheimers Dis. 56:775–788. 10.3233/JAD-160926 [DOI] [PubMed] [Google Scholar]
- Braniste V., Al-Asmakh M., Kowal C., Anuar F., Abbaspour A., Tóth M., Korecka A., Bakocevic N., Ng L.G., Kundu P., et al. . 2014. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 6:263ra158 10.1126/scitranslmed.3009759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E., et al. . 2014. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17:131–143. 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnin A., Brooks D.J., Kennedy A.M., Gunn R.N., Myers R., Turkheimer F.E., Jones T., and Banati R.B.. 2001. In-vivo measurement of activated microglia in dementia. Lancet. 358:461–467. 10.1016/S0140-6736(01)05625-2 [DOI] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., and Holmes S.P.. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13:581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo A., Cattane N., Galluzzi S., Provasi S., Lopizzo N., Festari C., Ferrari C., Guerra U.P., Paghera B., Muscio C., et al. INDIA-FBP Group . 2017. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging. 49:60–68. 10.1016/j.neurobiolaging.2016.08.019 [DOI] [PubMed] [Google Scholar]
- Cavedo E., Chiesa P.A., Houot M., Ferretti M.T., Grothe M.J., Teipel S.J., Lista S., Habert M.O., Potier M.C., Dubois B., et al. Alzheimer Precision Medicine Initiative (APMI) . 2018. Sex differences in functional and molecular neuroimaging biomarkers of Alzheimer’s disease in cognitively normal older adults with subjective memory complaints. Alzheimers Dement. 14:1204–1215. 10.1016/j.jalz.2018.05.014 [DOI] [PubMed] [Google Scholar]
- Cryan J.F., and O’Mahony S.M.. 2011. The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterol. Motil. 23:187–192. 10.1111/j.1365-2982.2010.01664.x [DOI] [PubMed] [Google Scholar]
- de Punder K., and Pruimboom L.. 2015. Stress induces endotoxemia and low-grade inflammation by increasing barrier permeability. Front. Immunol. 6:223 10.3389/fimmu.2015.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., and Andersen G.L.. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R., Wang S., Anuar F., Qian Y., Björkholm B., Samuelsson A., Hibberd M.L., Forssberg H., and Pettersson S.. 2011. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA. 108:3047–3052. 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrestein P.C., Mazmanian S.K., and Knight R.. 2014. Finding the missing links among metabolites, microbes, and the host. Immunity. 40:824–832. 10.1016/j.immuni.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiou A.G., and Goate A.M.. 2017. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener. 12:43 10.1186/s13024-017-0184-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen P.A., Dodiya H.B., Naqib A., Forsyth C.B., Green S.J., Voigt R.M., Kordower J.H., Mutlu E.A., Shannon K.M., and Keshavarzian A.. 2017. The Potential Role of Gut-Derived Inflammation in Multiple System Atrophy. J. Parkinsons Dis. 7:331–346. 10.3233/JPD-160991 [DOI] [PubMed] [Google Scholar]
- Erny D., Hrabě de Angelis A.L., Jaitin D., Wieghofer P., Staszewski O., David E., Keren-Shaul H., Mahlakoiv T., Jakobshagen K., Buch T., et al. . 2015. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18:965–977. 10.1038/nn.4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish E.N. 2008. The X-files in immunity: sex-based differences predispose immune responses. Nat. Rev. Immunol. 8:737–744. 10.1038/nri2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.C., Bessman N.J., Hepworth M.R., Kumar N., Shibata N., Kobuley D., Wang K., Ziegler C.G.K., Goc J., Shima T., et al. . 2016. Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism. Immunity. 44:634–646. 10.1016/j.immuni.2016.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman R.F., Schneider A.L., Zhou Y., Chen X., Green E., Gupta N., Knopman D.S., Mintz A., Rahmim A., Sharrett A.R., et al. . 2016. The ARIC-PET amyloid imaging study: Brain amyloid differences by age, race, sex, and APOE. Neurology. 87:473–480. 10.1212/WNL.0000000000002914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A., Serrano-Pozo A., Parrado A.R., Lesinski A.N., Asselin C.N., Mullin K., Hooli B., Choi S.H., Hyman B.T., and Tanzi R.E.. 2013. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 78:631–643. 10.1016/j.neuron.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S., Younkin S., et al. Alzheimer Genetic Analysis Group . 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368:117–127. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin L., Lagarde J., Dorothée G., Leroy C., Labit M., Comley R.A., de Souza L.C., Corne H., Dauphinot L., Bertoux M., et al. Clinical IMABio3 team . 2016. Early and protective microglial activation in Alzheimer’s disease: a prospective study using 18F-DPA-714 PET imaging. Brain. 139:1252–1264. 10.1093/brain/aww017 [DOI] [PubMed] [Google Scholar]
- Hanamsagar R., Alter M.D., Block C.S., Sullivan H., Bolton J.L., and Bilbo S.D.. 2017. Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia. 65:1504–1520. 10.1002/glia.23176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harach T., Marungruang N., Duthilleul N., Cheatham V., Mc Coy K.D., Frisoni G., Neher J.J., Fåk F., Jucker M., Lasser T., and Bolmont T.. 2017. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci. Rep. 7:41802 10.1038/srep41802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka M.T., Carson M.J., El Khoury J., Landreth G.E., Brosseron F., Feinstein D.L., Jacobs A.H., Wyss-Coray T., Vitorica J., Ransohoff R.M., et al. . 2015. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14:388–405. 10.1016/S1474-4422(15)70016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Dissing-Olesen L., and Stevens B.. 2016. New insights on the role of microglia in synaptic pruning in health and disease. Curr. Opin. Neurobiol. 36:128–134. 10.1016/j.conb.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. . 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R. Jr., Wiste H.J., Weigand S.D., Therneau T.M., Knopman D.S., Lowe V., Vemuri P., Mielke M.M., Roberts R.O., Machulda M.M., et al. . 2017. Age-specific and sex-specific prevalence of cerebral β-amyloidosis, tauopathy, and neurodegeneration in cognitively unimpaired individuals aged 50-95 years: a cross-sectional study. Lancet Neurol. 16:435–444. 10.1016/S1474-4422(17)30077-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi S., Gandhi R., Cox L.M., Li N., von Glehn F., Yan R., Patel B., Mazzola M.A., Liu S., Glanz B.L., et al. . 2016. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7:12015 10.1038/ncomms12015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky J.L., Savonenko A., Schilling G., Wang J., Xu G., and Borchelt D.R.. 2002. Transgenic mouse models of neurodegenerative disease: opportunities for therapeutic development. Curr. Neurol. Neurosci. Rep. 2:457–464. 10.1007/s11910-002-0073-7 [DOI] [PubMed] [Google Scholar]
- Johnson K.A., Schultz A., Betensky R.A., Becker J.A., Sepulcre J., Rentz D., Mormino E., Chhatwal J., Amariglio R., Papp K., et al. . 2016. Tau positron emission tomographic imaging in aging and early Alzheimer disease. Ann. Neurol. 79:110–119. 10.1002/ana.24546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N., Seo S.U., Chen G.Y., and Núñez G.. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13:321–335. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., and Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzian A., Green S.J., Engen P.A., Voigt R.M., Naqib A., Forsyth C.B., Mutlu E., and Shannon K.M.. 2015. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 30:1351–1360. 10.1002/mds.26307 [DOI] [PubMed] [Google Scholar]
- Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. . 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 47:566–581. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop S.K., Bloom S.M., Rao S.M., Nutsch K., Lio C.W., Santacruz N., Peterson D.A., Stappenbeck T.S., and Hsieh C.S.. 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature. 478:250–254. 10.1038/nature10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Han H.W., and Yim S.Y.. 2015. Beneficial effects of soy milk and fiber on high cholesterol diet-induced alteration of gut microbiota and inflammatory gene expression in rats. Food Funct. 6:492–500. 10.1039/C4FO00731J [DOI] [PubMed] [Google Scholar]
- Mandal S., Van Treuren W., White R.A., Eggesbø M., Knight R., and Peddada S.D.. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle J.G., Frank D.N., Mortin-Toth S., Robertson C.E., Feazel L.M., Rolle-Kampczyk U., von Bergen M., McCoy K.D., Macpherson A.J., and Danska J.S.. 2013. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 339:1084–1088. 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- Matcovitch-Natan O., Winter D.R., Giladi A., Vargas Aguilar S., Spinrad A., Sarrazin S., Ben-Yehuda H., David E., Zelada González F., Perrin P., et al. . 2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 353:aad8670 10.1126/science.aad8670 [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Liu C.H., Tzianabos A.O., and Kasper D.L.. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 122:107–118. 10.1016/j.cell.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Minter M.R., Zhang C., Leone V., Ringus D.L., Zhang X., Oyler-Castrillo P., Musch M.W., Liao F., Ward J.F., Holtzman D.M., et al. . 2016. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci. Rep. 6:30028 10.1038/srep30028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter M.R., Hinterleitner R., Meisel M., Zhang C., Leone V., Zhang X., Oyler-Castrillo P., Zhang X., Musch M.W., Shen X., et al. . 2017. Antibiotic-induced perturbations in microbial diversity during post-natal development alters amyloid pathology in an aged APPSWE/PS1ΔE9 murine model of Alzheimer’s disease. Sci. Rep. 7:10411 10.1038/s41598-017-11047-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj A.C., Jun G., Reitz C., Kunkle B.W., Perry W., Park Y.S., Beecham G.W., Rajbhandary R.A., Hamilton-Nelson K.L., Wang L.S., et al. . 2014. Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol. 71:1394–1404. 10.1001/jamaneurol.2014.1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.L., Vieira-Silva S., Liston A., and Raes J.. 2015. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 8:1–16. 10.1242/dmm.017400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry V.H., Cunningham C., and Holmes C.. 2007. Systemic infections and inflammation affect chronic neurodegeneration. Nat. Rev. Immunol. 7:161–167. 10.1038/nri2015 [DOI] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., and Arkin A.P.. 2010. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 5:e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radde R., Bolmont T., Kaeser S.A., Coomaraswamy J., Lindau D., Stoltze L., Calhoun M.E., Jäggi F., Wolburg H., Gengler S., et al. . 2006. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 7:940–946. 10.1038/sj.embor.7400784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff R.M., and El Khoury J.. 2016. Microglia in Health and Disease. Cold Spring Harb. Perspect. Biol. 8:a020560 10.1101/cshperspect.a020560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reemst K., Noctor S.C., Lucassen P.J., and Hol E.M.. 2016. The Indispensable Roles of Microglia and Astrocytes during Brain Development. Front. Hum. Neurosci. 10:566 10.3389/fnhum.2016.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson T.R., Debelius J.W., Thron T., Janssen S., Shastri G.G., Ilhan Z.E., Challis C., Schretter C.E., Rocha S., Gradinaru V., et al. . 2016. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell. 167:1469–1480. 10.1016/j.cell.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D.C., and Dubos R.. 1968. Alterations in the mouse cecum and its flora produced by antibacterial drugs. J. Exp. Med. 128:97–110. 10.1084/jem.128.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.P., and Stevens B.. 2015. Microglia Function in Central Nervous System Development and Plasticity. Cold Spring Harb. Perspect. Biol. 7:a020545 10.1101/cshperspect.a020545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinin N.M., Wikman K., Jula A., Perola M., Vahlberg T., Rokka J., Någren K., Viitanen M., and Rinne J.O.. 2014. Cortical 11C-PIB uptake is associated with age, APOE genotype, and gender in “healthy aging”. J. Alzheimers Dis. 41:193–202. 10.3233/JAD-132783 [DOI] [PubMed] [Google Scholar]
- Sharon G., Sampson T.R., Geschwind D.H., and Mazmanian S.K.. 2016. The Central Nervous System and the Gut Microbiome. Cell. 167:915–932. 10.1016/j.cell.2016.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Liu L., and Ji H.F.. 2017. Alzheimer’s Disease Histological and Behavioral Manifestations in Transgenic Mice Correlate with Specific Gut Microbiome State. J. Alzheimers Dis. 56:385–390. 10.3233/JAD-160884 [DOI] [PubMed] [Google Scholar]
- Sherwin E., Dinan T.G., and Cryan J.F.. 2018. Recent developments in understanding the role of the gut microbiota in brain health and disease. Ann. N. Y. Acad. Sci. 1420:5–25. 10.1111/nyas.13416 [DOI] [PubMed] [Google Scholar]
- Stefka A.T., Feehley T., Tripathi P., Qiu J., McCoy K., Mazmanian S.K., Tjota M.Y., Seo G.Y., Cao S., Theriault B.R., et al. . 2014. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA. 111:13145–13150. 10.1073/pnas.1412008111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay T.L., Savage J.C., Hui C.W., Bisht K., and Tremblay M.E.. 2017. Microglia across the lifespan: from origin to function in brain development, plasticity and cognition. J. Physiol. 595:1929–1945. 10.1113/JP272134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thion M.S., Low D., Silvin A., Chen J., Grisel P., Schulte-Schrepping J., Blecher R., Ulas T., Squarzoni P., Hoeffel G., et al. . 2018. Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell. 172:500–516. 10.1016/j.cell.2017.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P., Knopman D.S., Lesnick T.G., Przybelski S.A., Mielke M.M., Graff-Radford J., Murray M.E., Roberts R.O., Vassilaki M., Lowe V.J., et al. . 2017. Evaluation of Amyloid Protective Factors and Alzheimer Disease Neurodegeneration Protective Factors in Elderly Individuals. JAMA Neurol. 74:718–726. 10.1001/jamaneurol.2017.0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A., Gelosa P., Castiglioni L., Cimino M., Rizzi N., Pepe G., Lolli F., Marcello E., Sironi L., Vegeto E., and Maggi A.. 2018. Sex-Specific Features of Microglia from Adult Mice. Cell Reports. 23:3501–3511. 10.1016/j.celrep.2018.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N.M., Kerby R.L., Dill-McFarland K.A., Harding S.J., Merluzzi A.P., Johnson S.C., Carlsson C.M., Asthana S., Zetterberg H., Blennow K., et al. . 2017. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 7:13537 10.1038/s41598-017-13601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendeln A.C., Degenhardt K., Kaurani L., Gertig M., Ulas T., Jain G., Wagner J., Häsler L.M., Wild K., Skodras A., et al. . 2018. Innate immune memory in the brain shapes neurological disease hallmarks. Nature. 556:332–338. 10.1038/s41586-018-0023-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., and Siuzdak G.. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA. 106:3698–3703. 10.1073/pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Burrows M., Khan A.A., Graham L., Volchkov P., Becker L., Antonopoulos D., Umesaki Y., and Chervonsky A.V.. 2013. Gender bias in autoimmunity is influenced by microbiota. Immunity. 39:400–412. 10.1016/j.immuni.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandman-Goddard G., Peeva E., and Shoenfeld Y.. 2007. Gender and autoimmunity. Autoimmun. Rev. 6:366–372. 10.1016/j.autrev.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Zhuang Z.Q., Shen L.L., Li W.W., Fu X., Zeng F., Gui L., Lü Y., Cai M., Zhu C., Tan Y.L., et al. . 2018. Gut Microbiota is Altered in Patients with Alzheimer’s Disease. J. Alzheimers Dis. 63:1337–1346. 10.3233/JAD-180176 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.