Chen et al. show that XLF functions to limit fork reversal during DNA replication. H2AX prevents MRE11-dependent replication stress in XLF-deficient cells, suggesting that H2AX prevents the resection of regressed arms at reversed forks.
Abstract
XRCC4-like factor (XLF) is a non-homologous end joining (NHEJ) DNA double strand break repair protein. However, XLF deficiency leads to phenotypes in mice and humans that are not necessarily consistent with an isolated defect in NHEJ. Here we show that XLF functions during DNA replication. XLF undergoes cell division cycle 7–dependent phosphorylation; associates with the replication factor C complex, a critical component of the replisome; and is found at replication forks. XLF deficiency leads to defects in replication fork progression and an increase in fork reversal. The additional loss of H2AX, which protects DNA ends from resection, leads to a requirement for ATR to prevent an MRE11-dependent loss of newly synthesized DNA and activation of DNA damage response. Moreover, H2ax−/−:Xlf−/− cells exhibit a marked dependence on the ATR kinase for survival. We propose that XLF and H2AX function in series to prevent replication stress induced by the MRE11-dependent resection of regressed arms at reversed replication forks.
Introduction
Faithful replication of the genome in dividing cells relies on a network of sophisticated DNA replication mechanisms that are orchestrated in a temporally controlled manner (Masai et al., 2010; Fragkos et al., 2015). Defects in these events can hinder replication fork progression, leading to replication fork stalling and replication stress (Zeman and Cimprich, 2014; Berti and Vindigni, 2016). Replication fork stalling can lead to activation of the ataxia-telangiectasia and Rad3-related (ATR) kinase, a critical regulator of replication stress responses and the S-phase checkpoint (Zou and Elledge, 2003; Cimprich and Cortez, 2008; Nam and Cortez, 2011; Saldivar et al., 2017). Stalled replication forks can undergo fork reversal, where newly synthesized (nascent) DNA strands dissociate from template strands and anneal to each other, forming a regressed arm (Sogo et al., 2002; Quinet et al., 2017b). Several proteins are known to promote fork reversal, including RAD51, SMARCAL1, HLTF, and ZRANB3 (Zellweger et al., 2015; Kolinjivadi et al., 2017; Quinet et al., 2017b; Taglialatela et al., 2017; Vujanovic et al., 2017). ATR phosphorylates SMARCAL1, providing a link between ATR activation and fork reversal (Couch et al., 2013). While fork reversal may be a mechanism for limiting replication stress, failure to restart stalled replication forks can result in replication fork collapse, activation of a DNA damage response (DDR), and cell death (Ciccia and Elledge, 2010; Neelsen and Lopes, 2015; Quinet et al., 2017b).
Non-homologous end joining (NHEJ) is a major pathway of DNA double strand break (DSB) repair that directly joins broken DNA ends (Chang et al., 2017). The XRCC4-like factor (XLF) protein functions in NHEJ-mediated DNA DSB repair by forming a filament with XRCC4 that aligns and stabilizes broken DNA ends so they can be joined (Ahnesorg et al., 2006; Buck et al., 2006; Andres et al., 2007; Zha et al., 2007; Li et al., 2008; Hammel et al., 2011; Ropars et al., 2011; Fattah et al., 2014; Brouwer et al., 2016). Deficiency of XLF in humans leads to severe combined immunodeficiency consistent with a defect in lymphocyte antigen receptor gene assembly by V(D)J recombination, a reaction that requires the generation of DNA DSBs by the RAG endonuclease and their repair by NHEJ (Fugmann et al., 2000; Ahnesorg et al., 2006; Buck et al., 2006; Helmink and Sleckman, 2012). However, XLF-deficient murine lymphoid cells do not exhibit overt defects in RAG DSB repair, raising the possibility that XLF has additional functions that could contribute to the phenotype of XLF deficiency (Li et al., 2008). In this regard, cells derived from XLF-deficient patients have been reported to have increased sensitivity to replicative stress (Schwartz et al., 2009).
The histone variant H2AX is phosphorylated (forming γH2AX) by the DDR kinases ATM, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and ATR in chromatin flanking damaged DNA (Rogakou et al., 1998, 1999; Ward and Chen, 2001; Savic et al., 2009; Blackford and Jackson, 2017). γH2AX functions to retain DDR factors at DNA damage sites to repair damaged DNA and amplify DDR signaling (Celeste et al., 2002, 2003; Savic et al., 2009). γH2AX also protects broken DNA ends from nucleolytic resection mediated by CtIP, and presumably MRE11, in G1-phase cells (Helmink et al., 2011). γH2AX colocalizes with proliferating cell nuclear antigen (PCNA) foci and has been implicated in the responses to replication stress (Ward and Chen, 2001; Sirbu et al., 2011; Schmid et al., 2018). Indeed, H2AX-deficient cells exhibit increased sensitivity to the DNA replication inhibitor aphidicolin, especially when ATR is inhibited (Chanoux et al., 2009).
Like XLF, H2AX is not required for NHEJ during RAG DSB repair in murine lymphoid cells; however, a combined deficiency of XLF and H2AX leads to a severe block in RAG DSB repair in murine lymphoid cells, demonstrating that both of these proteins have activities in NHEJ during V(D)J recombination in these cells (Zha et al., 2011). Xlf−/− and H2ax−/− mice are both viable, but H2ax−/−:Xlf−/− mice exhibit embryonic lethality at a developmental stage much earlier than mice deficient in the core NHEJ factors, DNA ligase IV or XRCC4, suggesting that XLF and H2AX function in fundamental cellular processes other than NHEJ (Barnes et al., 1998; Frank et al., 1998; Gao et al., 1998; Zha et al., 2011). Here we demonstrate that XLF-deficient cells have DNA replication defects that lead to an increase in replication fork reversal. Moreover, loss of H2AX in XLF-deficient cells leads to a dependence on ATR to prevent a potent MRE11-dependent DDR that would otherwise lead to cell death. We propose that during DNA replication, XLF and H2AX function in series to limit the formation of reversed replication forks (XLF) and to protect regressed arms of reversed forks from being resected by MRE11 (H2AX) and activating a DDR that can lead to cell death. These important functions are independent of the activities of XLF and H2AX in NHEJ.
Results and discussion
Association of XLF with DNA replication machinery
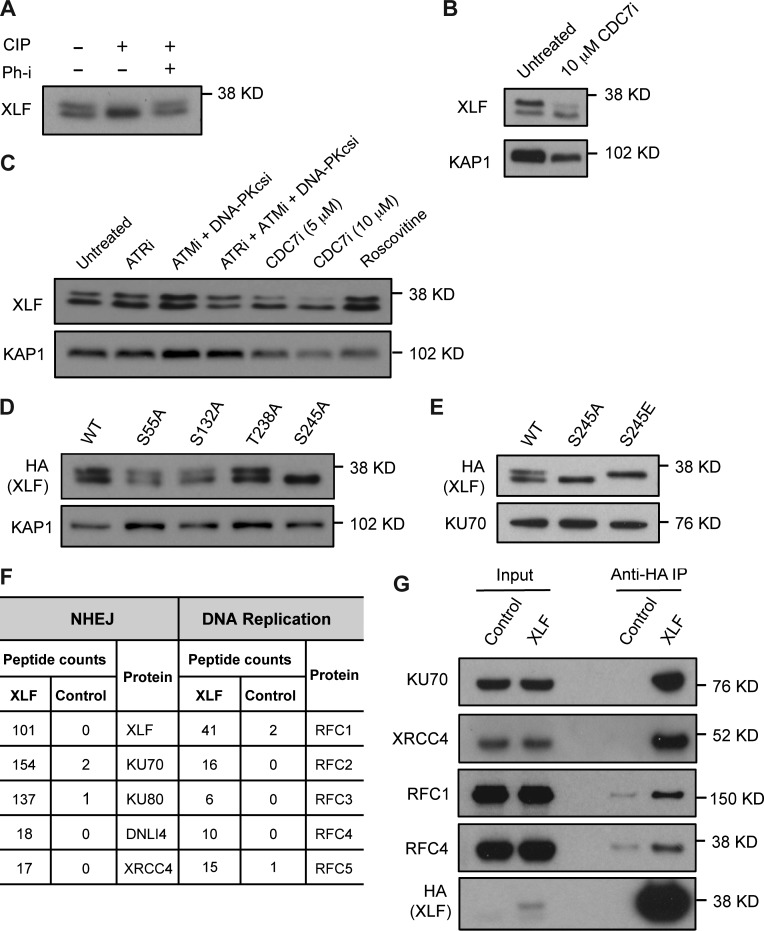
XLF from mouse embryonic fibroblasts (MEFs) or mouse Abelson virus–transformed preB cells (hereafter referred to as abl preB cells) migrates as a doublet slightly below 38 kD (Fig. 1, A–E). Phosphatase treatment leads to loss of the slower migrating form, and coincubation with a phosphatase inhibitor restores this form (Fig. 1 A). Neither inhibition of the ATR, ATM, and DNA-PKcs DDR kinases nor mutation of murine XLF SQ/TQ motifs (S55, S132, and T238) to AQ leads to loss of XLF phosphorylation (Fig. 1, C and D). However, mutation of serine 245 to alanine leads to a complete loss of XLF phosphorylation, and mutation of this serine to aspartic acid causes a complete shift to the slower migrating form (Fig. 1, D and E). The CDK inhibitor roscovitine had no effect on XLF phosphorylation (Fig. 1 C). However, an inhibitor of the cell division cycle 7 (CDC7) kinase, PHA-767491 (CDC7i), which is required to promote the initiation of DNA synthesis (Fragkos et al., 2015), leads to reduced XLF phosphorylation (Fig. 1, B and C).
Figure 1.
CDC7-dependent XLF phosphorylation and association with replication proteins. (A) Anti-XLF Western blot analysis of FLAG-HA-XLF immunoprecipitated with anti-HA antibody from MEFs untreated or treated with calf intestine phosphatase (CIP) or with phosphatase inhibitor (Ph-i). (B and C) Whole cell lysates from WT abl preB cells untreated or treated with CDC7 inhibitor (CDC7i) for 24 h (B) or 8 h (C), ATR inhibitor (ATRi), ATM inhibitor (ATMi), DNA-PKcs inhibitor (DNA-PKcsi), or roscovitine for 8 h were analyzed by Western blotting using the anti-XLF antibody. (D and E) FLAG-HA–tagged WT or mutant XLF expressed in Xlf−/− MEFs and analyzed Western blotting of whole cell lysates using anti-HA antibody. (F) Summary of mass spectrometry analysis of XLF interacting proteins isolated by immunoprecipitation of FLAG-HA-XLF from abl preB cell lysate. (G) FLAG-HA-XLF was immunoprecipitated (IP) from Xlf−/− MEF cells using anti-HA and associated proteins identified by Western blotting with the indicated antibodies. KAP1 or KU70 levels were assayed as protein loading controls. All Western blots shown here and in the rest of the figures were repeated in two independently generated cell lines, and representative images are shown.
We performed mass spectrometry of proteins that coimmunoprecipitate with a FLAG-HA–tagged XLF fusion protein expressed in Xlf−/− abl preB cells (Fig. 1 F and Table S1). As previously reported, these analyses revealed that XLF associates with the NHEJ factors Ku70, Ku80, DNA ligase IV, and XRCC4 (Fig. 1, F and G; Ahnesorg et al., 2006; Yano et al., 2008). Additionally, we found that XLF also associates with all five components of the replication factor C complex (RFC1–5), which loads PCNA at replication forks (Fig. 1 F; Shiomi and Nishitani, 2017). Immunoprecipitation of XLF from MEFs confirmed that XLF associates with RFC1 and RFC4 (Fig. 1 G). Thus, XLF is phosphorylated by CDC7 and associates with RFC, suggesting that XLF may function during DNA replication.
XLF associates with DNA replication forks
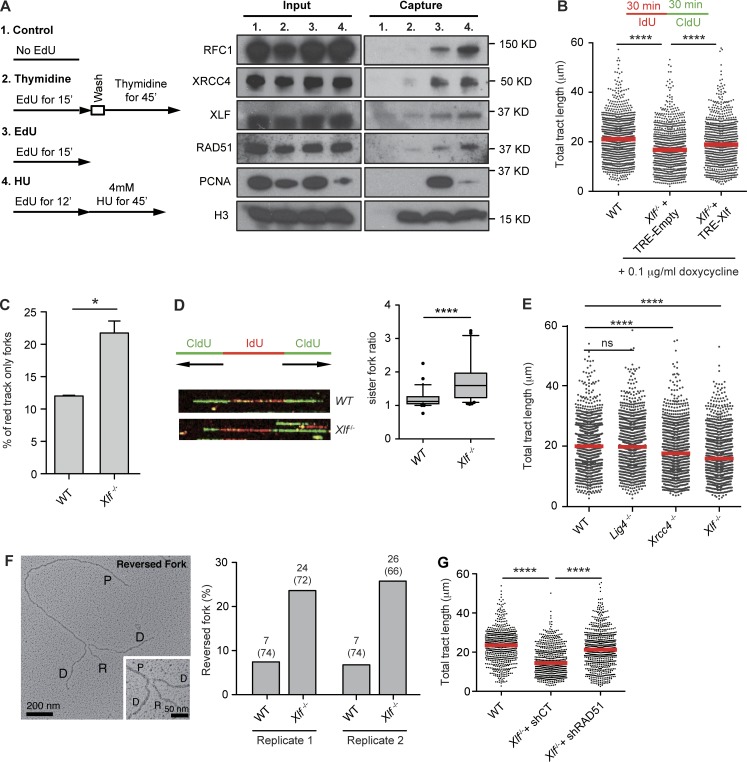
We determined whether XLF associates with active replication forks in HEK293T cells by accelerated native iPOND (aniPOND; Fig. 2 A; Sirbu et al., 2011, 2012; Leung et al., 2013; Wiest and Tomkinson, 2017). Similar to PCNA, RFC1, and RAD51, which associate with replication forks, XLF coprecipitates with nascent DNA labeled with the thymidine analogue EdU (Fig. 2 A; Sirbu et al., 2011). Moreover, XLF association decreases after thymidine chase, demonstrating that XLF specifically associates with active replication forks (Fig. 2 A). XRCC4 also associates with DNA at active replication forks (Fig. 2 A). Treatment of HEK293T cells with hydroxyurea (HU), which causes replication fork stalling, leads to decreased association of PCNA and increased association of RAD51 at replication forks (Fig. 2 A; Ragland et al., 2013; Dungrawala et al., 2015; Zellweger et al., 2015). HU treatment leads to a mild increase in XLF association with nascent DNA (Fig. 2 A). We conclude that XLF associates with replication forks.
Figure 2.
XLF regulates replication fork dynamics. (A) 293T cells not labeled (1), labeled with EdU followed by thymidine chase (2), labeled with EdU (3), or labeled with Edu followed by HU treatment (4) were subject to aniPOND, and the isolated proteins were analyzed by Western blotting. Shown is solubilized chromatin (Input) and proteins eluted from streptavidin beads (Capture). (B) WT and Xlf−/− MEFs transduced with a control lentivirus (TRE-Empty) or lentivirus with tetracycline-inducible XLF (TRE-Xlf) were consecutively labeled with IdU and CldU for 30 min each. Size distribution of total tract length (IdU+CldU) scored from bicolor DNA fibers is shown with red lines representing median. At least 450 tracts were scored for each dataset from three independent experiments (Mann–Whitney U; ****, P < 0.0001). (C) Frequency of IdU (red) only tracts in DNA fiber assay described in B (unpaired t test; *, P < 0.05). (D) Representative image of a symmetric (WT) and an asymmetric (Xlf−/−) fork, and quantification of the ratio of the lengths of sister forks. The top and bottom bars on the whiskers of the box-and-whisker plots represent the 90th and 10th percentiles. At least 40 bidirectional forks were analyzed in three independent experiments. (E) Size distribution of total tract length from indicated MEFs as described in B. At least 450 tracts were scored for each dataset from three independent experiments (Mann–Whitney U; ****, P < 0.0001). (F) Left: Electron micrograph of a representative reversed fork on enriched genomic DNA from Xlf−/− cells. Inset: Magnified four-way junction at the reversed replication fork. The Daughter (D) and Parental (P) strands are indicated as is the Reversed arm (R). The frequency of fork reversal in WT and Xlf−/− MEFs is shown for two independent experiments. The percentage of reversed forks and total number of replication intermediates analyzed are indicated in parentheses. (G) Size distribution of tract lengths of bicolor DNA fibers from Xlf−/− cells expressing control (shCT) and RAD51 (shRAD51) shRNAs labeled as described in B. At least 450 tracts were scored for each dataset from two independent experiments (Mann–Whitney U; ns, non-significant; ****, P < 0.0001).
XLF deficiency impairs replication fork dynamics
We analyzed DNA fibers in WT and Xlf−/− MEFs that were consecutively incubated with 5-iodo-2′-deoxyuridine (IdU; red) and 5-chloro-2′-deoxyuridine (CldU; green) to label newly synthesized DNA tracts (Fig. 2 B). As compared with WT MEFs, the lengths of the bicolor DNA tracts were significantly reduced in Xlf−/− MEFs, and this reduction was corrected by the ectopic expression of XLF (Fig. 2 B). There was a higher frequency of DNA tracts labeled only with IdU (red) in Xlf−/− MEFs as compared with WT MEFs (Fig. 2 C). The lengths of bidirectional newly synthesized DNA tracts emerging from single replication origins (two CldU tracts extending from a single IdU tract) were measured (Fig. 2 D). Under conditions of unperturbed DNA synthesis, the lengths of these newly synthesized DNA tracts should be approximately equal. Indeed, in WT MEFs, the ratio of the lengths of the bidirectional newly synthesized DNA tracts are close to one (1.2; Fig. 2 D). In contrast, this value is significantly greater (1.8) in Xlf−/− MEFs, indicating that loss of XLF leads to increased replication fork asymmetry (Fig. 2 D). Xrcc4−/− MEFs exhibit defects in DNA replication that are similar to those observed in Xlf−/− MEFs (Fig. 2 E). Analyses of DNA ligase IV–deficient (Lig4−/−) MEFs, which, like Xrcc4−/− MEFs, have a block in NHEJ, revealed no significant differences in newly synthesized DNA tracts as compared with WT MEFs (Fig. 2 E). Together, these data are indicative of a DNA replication defect due to diminished replication fork progression, replication fork stalling, or nucleolytic resection of newly synthesized DNA in XRCC4- and XLF-deficient MEFs. Moreover, that DNA fiber defects were not observed in Lig4−/− MEFs suggests that the replication defects in Xlf−/− and Xrcc4−/− MEFs are not due to the function of XLF and XRCC4 in promoting NHEJ.
Replication fork stalling can lead to fork regression, which is best visualized by EM (Sogo et al., 2002; Vindigni and Lopes, 2017). We visualized the fine architecture of the replication intermediates using a combination of in vivo psoralen cross-linking and EM (Fig. 2 F and Fig. S1; Neelsen et al., 2014). Treating WT MEFs with HU leads to a significant increase in the frequency of reversed forks (Fig. S1). Analysis of untreated Xlf−/− MEFs reveals an increase in the frequency of reversed forks of similar magnitude to that observed in WT MEFs treated with HU (Fig. 2 F and Fig. S1). Lig4−/− MEFs do not exhibit an increase in replication fork reversal, demonstrating that the reversed forks in Xlf−/− MEFs are not due to a defect in NHEJ per se (Fig. S1). Knockdown of RAD51, which is required for fork reversal, leads to partial recovery of newly synthesized DNA tracts in Xlf−/− MEFs (Fig. 2 G; Zellweger et al., 2015). Thus, XLF deficiency leads to increased fork reversal during DNA replication.
ATR inhibition leads to increased DDR in XLF-deficient cells
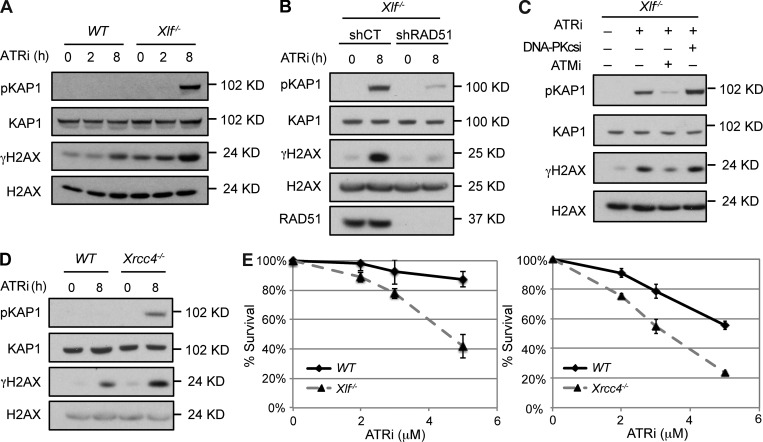
XLF-deficient MEFs appear to divide normally, suggesting that any replication defects are resolvable, likely through the activation of ATR. Indeed, as compared with WT MEFs, treatment of Xlf−/− MEFs with the ATR kinase inhibitor VE-821 leads to DDR activation, as indicated by increased γH2AX and phospho-KAP1 (pKAP1; Fig. 3 A and Fig. S2 A). This increased DDR is reduced when Rad51 is knocked down, consistent with the notion that it is due to replication fork reversal (Fig. 3 B). Treatment with the ATM kinase inhibitor KU55933, but not the DNA-PKcs kinase inhibitor NU7441, leads to abrogation of the DDR (Fig. 3 C). MEFs deficient in XRCC4 also exhibit an enhanced DDR in response to ATR inhibition (Fig. 3 D and Fig. S2 B). Moreover, both Xlf−/− and Xrcc4−/− MEFs exhibit diminished survival in response to ATR inhibition as compared with WT MEFs (Fig. 3 E and Fig. S2 C). Thus, XLF- and XRCC4-deficient MEFs have DNA replication defects that must be resolved by ATR to prevent the activation of an ATM-dependent DDR.
Figure 3.
ATR inhibition leads to DDR in Xlf−/− cells. (A) Cell lysates from Xlf−/− MEFs treated with ATRi for the indicated times were analyzed by Western blotting using the indicated antibodies. (B) Cell lysates from Xlf−/− MEFs transduced with Control (shCT) or RAD51 (shRAD51) shRNAs and treated with ATRi for 8 h were analyzed by Western blotting using the indicated antibodies. (C) Cell lysates from Xlf−/− MEFs untreated or pretreated with ATMi or DNA-PKcsi for 30 min before treatment with ATRi for 8 h were analyzed by Western blotting using the indicated antibodies. (D) Western blot analysis of whole cell lysates from WT and Xrcc4−/− MEFs treated with ATRi for indicated times using the indicated antibodies. (E) Cell viability of WT, Xlf−/−, or Xrcc4−/− MEFs treated with the ATRi at indicated concentrations for 4 d. Error bars indicating SD of three technical repeats from a representative experiment from analyses of two independent cell lines of each genotype analyzed in two experiments, each in triplicate.
H2AX deficiency amplifies the replication defect in Xlf−/− MEFs
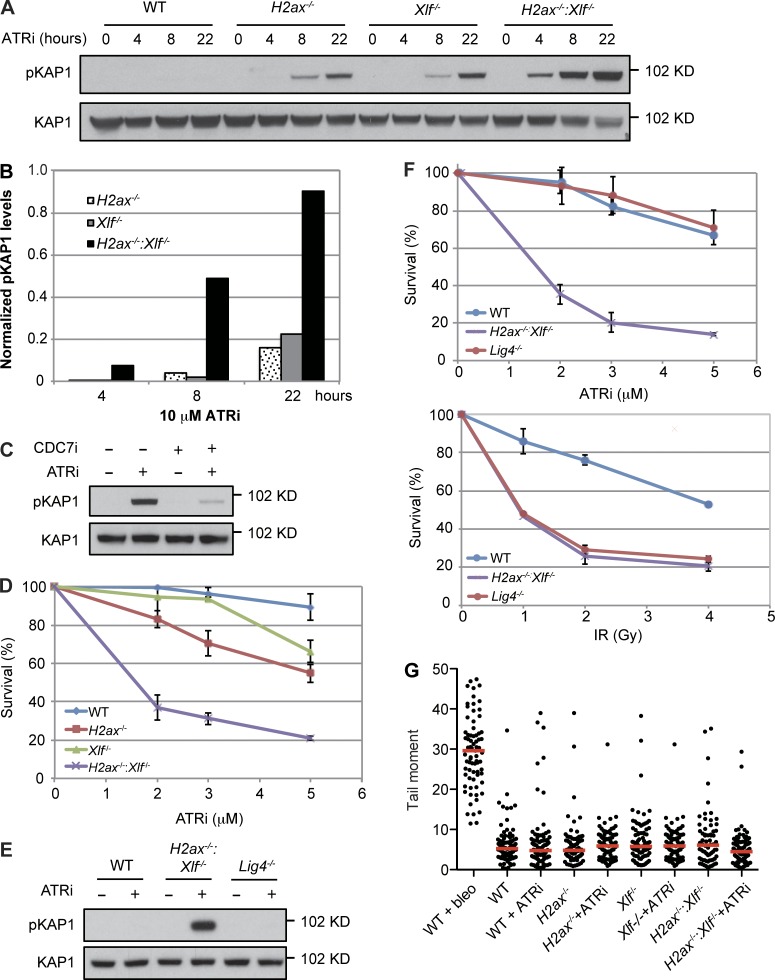
ATR inhibition of either H2ax−/− or Xlf−/− MEFs leads to activation of the DDR as indicated by pKAP1 (Fig. 4, A and B; and Fig. S2, D and E). However, ATR inhibition of H2ax−/−:Xlf−/− MEFs leads to a synergistic increase in pKAP1, suggesting that the combined loss of H2AX and XLF leads to significant defects in DNA replication that require ATR for resolution (Fig. 4, A and B; and Fig. S2, D and E). Indeed, inhibition of DNA replication initiation by CDC7 limits the DDR in H2ax−/−:Xlf−/− MEFs treated with the ATR inhibitor (Fig. 4 C). Moreover, H2ax−/−:Xlf−/− MEFs exhibit a significant loss of viability upon ATR inhibition as compared with WT, H2ax−/−, or Xlf−/− MEFs (Fig. 4 D and Fig. S2 F). This increased sensitivity and DDR to ATR inhibition exhibited by H2ax−/−:Xlf−/− MEFs is not due to XLF and H2AX function in NHEJ, as Lig4−/− MEFs do not exhibit increased pKAP1 upon ATR inhibition (Fig. 4 E). Moreover, while Lig4−/− and H2ax−/−:Xlf−/− MEFs both exhibit increased sensitivity to ionizing radiatio indicative of NHEJ defects, only H2ax−/−:Xlf−/− MEFs exhibit sensitivity to ATR inhibition (Fig. 4 F). Finally, a neutral comet assay of H2ax−/−:Xlf−/− MEFs treated with the ATR inhibitor did not reveal a detectable increase in unrepaired two-ended DSBs (Fig. 4 G). Together, these data demonstrate that XLF and H2AX have NHEJ-independent functions during DNA replication, preventing the formation of lesions that, if not resolved by ATR, lead to a robust DDR and cell death.
Figure 4.
H2AX deficiency enhances replication defects in Xlf−/− MEFs. (A) Whole cell lysates from MEFs of the indicated genotypes untreated or treated with ATRi for the indicated time were analyzed by Western blotting for pKAP1. (B) Quantification of pKAP1 levels normalized to KAP1 shown in A. (C) Western blot analysis of pKAP1 in H2ax−/−:Xlf−/− MEFs untreated or treated with ATRi, CDC7 inhibitor (CDC7i), or both. (D and F) MEFs of the indicated genotype were treated with ATRi and viability assayed as described in Fig. 3 E. IR, ionizing radiation. Error bars indicate SD from three replicates. (E) Western blot analysis of pKAP1 in whole cell lysates from MEFs of the indicated genotype untreated or treated with ATRi for 8 h. (G) Tail moments in a neutral comet assay of MEFs of the indicated genotype untreated or treated with ATRi for 8 h. Red lines indicate median. WT MEFs treated with 20 µg/ml of bleocin (bleo) for 2 h is shown as a positive control.
Replication defects in H2ax−/−:Xlf−/− MEFs
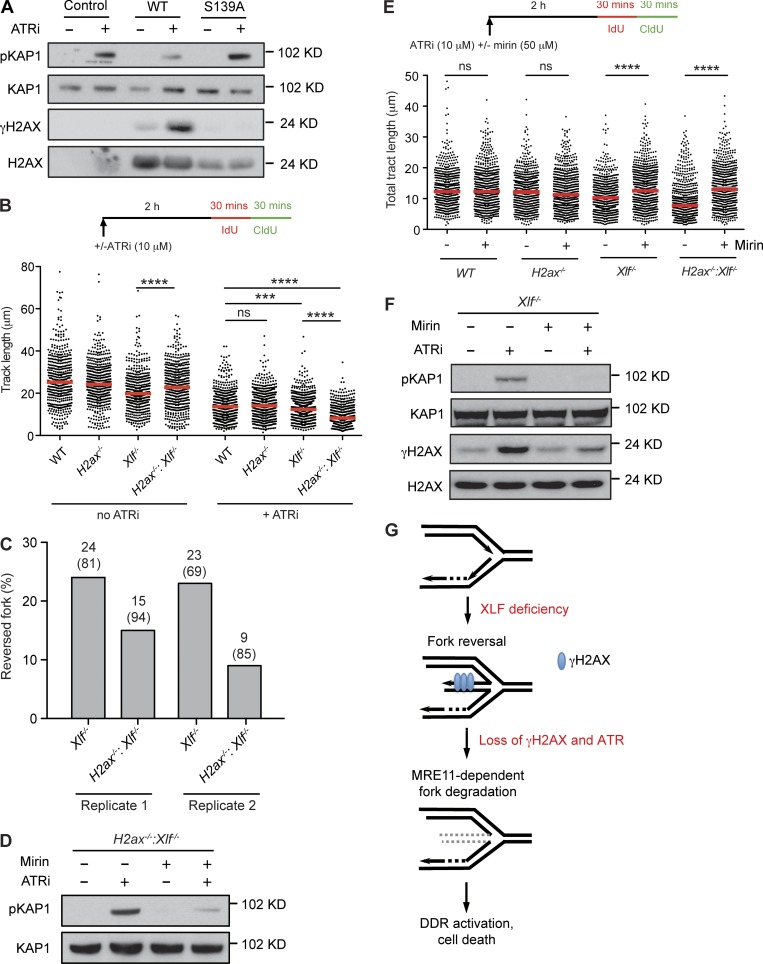
The regressed arm of a reversed fork generates a one-ended DNA DSB that could activate a DDR. In G1-phase cells, γH2AX protects DNA ends from resection; therefore, it is possible that γH2AX protects regressed arms from being resected, which would activate a DDR and prevent fork restart. In agreement, complementation of H2ax−/−:Xlf−/− MEFs with WT H2AX, but not a serine 139 to alanine mutant of H2AX (H2AXS139A), which cannot form γH2AX, leads to reduced pKAP1 formation in response to ATR inhibition (Fig. 5 A). To determine whether γH2AX may function by protecting the regressed arm from resection, we initially examined newly synthesized DNA tracts in MEFs treated with the ATR inhibitor (Fig. 5 B). There was no significant difference in these tracts when comparing WT and H2ax−/− MEFs treated with the ATR inhibitor (Fig. 5 B). In contrast, after ATR inhibition, Xlf−/− MEFs exhibited shorter tract lengths, and H2ax−/−:Xlf−/− MEFs markedly shorter tract lengths, when compared with WT MEFs (Fig. 5 B). Moreover, after ATR inhibition, H2ax−/−:Xlf−/− MEFs had significantly fewer reversed forks as compared with Xlf−/− MEFs by EM analysis (Fig. 5 C). Together, these findings are consistent with the notion that in the absence of H2AX, regressed arms of reversed replication forks may be resected. Notably, H2ax−/−:Xlf−/− MEFs have slightly longer newly synthesized DNA tract lengths than Xlf−/− MEFs, suggesting that H2AX could have additional roles in promoting replication fork reversal under unperturbed conditions (Fig. 5 B).
Figure 5.
MRE11-mediated replication defects in H2ax−/−:Xlf−/− MEFs. (A) Cell lysates from H2ax−/−:Xlf−/− MEFs expressing WT or S139A mutant H2AX, treated with ATRi for 8 h, were analyzed by Western blotting using the indicated antibodies. (B) DNA tract lengths in ATRi treated or untreated MEFs of the indicated genotype were assayed as described in Fig. 2 B. At least 450 tracts were scored for each dataset from three independent experiments (Mann–Whitney U; ***, P < 0.001; ****, P < 0.0001). (C) Frequency of reversed forks in Xlf−/− and H2ax−/−:Xlf−/− MEFs treated with ATRi for 3 h before EM analysis as described in Fig. 2 F. (D) pKAP1 analysis in whole cell lysates from H2ax−/−:Xlf−/− MEFs treated with ATRi, mirin, or both were analyzed by Western blotting. (E) DNA tract lengths in ATRi treated MEFs of the indicated genotype in the presence or absence of mirin were assayed as described in Fig. 2 B. At least 450 tracts were scored for each dataset from three independent experiments (Mann–Whitney U; ns, non-significant; ****, P < 0.0001). (F) Whole cell lysates from Xlf−/− MEFs treated with ATRi, mirin, or both were analyzed by Western blotting for the indicated proteins. (G) A model for replication defects caused by H2AX and XLF deficiency as described in the text.
DNA end resection is initiated by MRE11 and CtIP (Lengsfeld et al., 2007; Sartori et al., 2007). MRE11 mediates the degradation of reversed replication forks in BRCA1- and BRCA2-deficient cells (Schlacher et al., 2011; Ray Chaudhuri et al., 2016; Kolinjivadi et al., 2017; Lemaçon et al., 2017; Mijic et al., 2017; Taglialatela et al., 2017). If γH2AX protects regressed arms from resection, then the replication defects observed in H2ax−/−:Xlf−/− MEFs should be dependent on MRE11 nuclease activity. Indeed, inhibition of MRE11 nuclease activity with mirin leads to a reduction in the DDR in H2ax−/−:Xlf−/− MEFs treated with the ATR inhibitor (Fig. 5 D and Fig. S2 G). Moreover, DNA tracts were significantly longer in ATR inhibitor–treated H2ax−/−:Xlf−/− MEFs that were also treated with mirin (Fig. 5 E). Mirin treatment of ATR-treated Xlf−/− MEFs led to an increase in DNA tract length, suggesting that even in the presence of H2AX, regressed arms may undergo some resection (Fig. 5 E). Indeed, pKAP1 and γH2AX levels in Xlf−/− MEFs in response to ATR inhibition are also abrogated by mirin treatment (Fig. 5 F and Fig. S2 H). In agreement, another study found that the DNA tract length shortening in human cells expressing H2AX S139A also depends on Mre11 activity (Schmid et al., 2018). Notably, deficiency of 53BP1, which is recruited to DNA DSBs by γH2AX and antagonizes DNA end resection in mammalian cells, or deficiency in the yeast 53BP1 counterpart, Rad9, also leads to degradation of DNA at stalled replication forks (Bunting et al., 2010; Her et al., 2018; Schmid et al., 2018; Villa et al., 2018). We conclude that the defects in newly synthesized DNA tracts, and the resulting DDR, observed in H2ax−/−:Xlf−/− MEFs are dependent on MRE11 nuclease activity.
We have shown that XLF functions during DNA replication in a manner that is independent of NHEJ. Moreover, loss of XRCC4 leads to DNA replication defects that are similar to those observed in Xlf−/− cells, suggesting that XLF-XRCC4 filament formation, which functions during NHEJ, may also function during DNA replication to prevent or resolve reversed DNA replication forks. H2AX deficiency synergistically exacerbates the replication defects and DDR activation in Xlf−/− MEFs, suggesting that the defects imposed by the loss of these proteins are mechanistically in series. Moreover, this exacerbation of replication defects in H2ax−/−:Xlf−/− MEFs depends on MRE11 activity. We propose that XLF functions during DNA replication to prevent the accumulation of reversed replication forks and that when a reversed fork is generated γH2AX functions to prevent MRE11-dependent resection of the regressed arm and DDR activation (Fig. 5 G). Thus, the lymphopenia observed in XLF-deficient humans and embryonic lethality observed in H2ax−/−:Xlf−/− mice may be due, in part, to the requirement for XLF and H2AX during DNA replication. Although this seems at odds with the growth of H2ax−/−:Xlf−/− and Xlf−/− MEFs in cell culture, this growth is more dependent on ATR activity than WT MEFs. Moreover, different cell types could have differential requirements for pathways that promote efficient DNA replication. Indeed, embryonic stem cells exhibit decreased replication fork speed, increased fork reversal, and a higher level of associated DDR activation (Ahuja et al., 2016).
Materials and methods
Cells culture and chemicals
Abl preB cells were generated as described previously (Bredemeyer et al., 2006). MEFs were generated from embryonic day 13.5 embryos of mice that are Xlf+/+ or Xlf−/− carrying conditional H2ax alleles (H2axcon/con) flanked by loxP sites. Multiple H2axcon/con:Xlf+/+ and H2axcon/con:Xlf−/− primary MEFs were generated from two mice. Primary MEFs were propagated for two passages before being immortalized with SV40 large T antigen. Cre recombinase was transiently introduced to these cells to generate H2ax−/-:Xlf+/+ or H2ax−/−:Xlf−/− cells. MEFs used in this work were derived from embryos from each mouse: the first set (from mouse 1) includes H2axcon/con:Xlf+/+ (WT, AB-2), H2ax−/− (AB-4), H2axcon/con:Xlf−/− (Xlf−/−, AB-2), and H2ax−/−:Xlf−/− (AB-8); the second set (from mouse 2) includes H2axcon/con:Xlf+/+ (WT, 4A-8), H2ax−/− (4A-9), H2axcon/con:Xlf−/− (Xlf−/−, 2A-8), and H2ax−/−:Xlf−/− (2A-1). Ectopic expression of XLF in Xlf−/− MEFs was achieved by transducing cells with lentivirus expressing XLF inducibly under a tetracycline-regulated element (TRE) promoter and treating cells with 0.1 µg/ml of doxycycline for 7 d. All cells were cultured in DMEM supplemented with 10% fetal bovine serum and 0.4% β-mercaptoethanol. ATR inhibitor VE-821(10 µM; S8007), ATM inhibitor KU55933 (15 µM; S1092), DNA-PKcs inhibitor NU7441 (10 µM; S2638), CDC7 inhibitor PHA-767491 (5 µM or 10 µM; S2742), and CDK inhibitor roscovitine (10 µM; S1153) were purchased from Selleckchem. MRE11 inhibitor mirin was purchased from Sigma-Aldrich (50 µM; M9948).
Immunoprecipitation and Western blotting
For nuclear extract immunoprecipitation, cells were lysed in cytoplasmic extraction buffer (10 mM Tris-HCl, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, and 0.05% Triton X-100) supplemented with a protease and phosphatase inhibitor cocktail on ice for 15 min. After centrifugation at 3,000 rpm, 4°C for 5 min, the pellet was washed with cytoplasmic extraction buffer and incubated in 0.5× pellet volume of nuclease buffer (20 mM Tris-HCl, pH 7.4, 1.5 mM MgCl2, and 25% vol/vol glycerol) with 5 U/µl benzonase (E1014; Sigma-Aldrich) on ice for 1 h to digest genomic DNA. 1× pellet volume of nuclear extraction buffer (20 mM Tris-HCl, pH 7.4, 500 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 25% vol/vol glycerol) was then added, and the pellet was ground using a dounce tissue grinder (D8938; Sigma-Aldrich) before incubation with gentle rotation at 4°C for 1 h. The lysate was clarified by centrifugation at 15,000 rpm, 4°C for 30 min, and the supernatant was dialyzed in BC100 buffer (20 mM Tris-HCl, pH 7.4, 100 mM KCl, 0.2 mM EDTA, and 20% vol/vol glycerol) at 4°C overnight using the Pur-A-Lyzer Maxi 6000 Kit (PURX60015; Sigma-Aldrich). The dialyzed supernatant was clarified again by centrifugation at 15,000 rpm, 4°C for 30 min, and incubated with EZView Red HA affinity gel beads (E6779; Sigma-Aldrich) on a rotator at 4°C for 4 h. The beads were washed four times with TAP buffer (50 mM Tris-HCl, pH 7.4, 100 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 10% vol/vol glycerol, and 0.1% vol/vol Triton X-100). Protein complexes were eluted by boiling in LDS sample buffer (NP0007; Thermo Fisher Scientific) and resolved in 4–12% NuPAGE Bis-Tris gels for Western blotting. For detecting phosphorylated XLF, protein was resolved in 10% Tris-Glycine gels for Western blotting analysis. Anti-XLF, anti-PCNA, and anti-pKAP1 (S824) antibodies (all produced in rabbits) were obtained from Bethyl Laboratories (A7300-730A, A300-276A, and A300-767A, respectively). Anti-KAP1 and anti-RFC4 antibodies were obtained from Genetex (GTX10484, rabbit, and GTX104052, rabbit, respectively). Anti-KU70 (D10A7) antibody was obtained from Cell Signaling Technologies (4588S, rabbit). Anti-phospho-histone H2AX (S139), γH2AX, clone JBW301, anti-H2AX, and anti-RAD51 antibodies were obtained from EMD Millipore (05–636, mouse; 07–627, rabbit; and 07–1782, rabbit, respectively). Anti-histone H3 and anti-RFC1 antibodies were obtained from Abcam (ab1791, rabbit, and ab193559, rabbit, respectively). Anti-XRCC4 (C-20) was obtained from Santa Cruz Biotechnology (sc-8285, goat).
Mass spectrometry
Immunoprecipitation for mass spectrometry was described previously (Ciccia et al., 2009). Briefly, lentiviral construct TRE-FLAG-HA-XLF was used to transduce Xlf−/− abl-preB cells. 150 million transduced cells were treated with 3 µM imatinib and 0.2 µg/ml doxycycline for 48 h before harvesting for protein lysates in low-salt buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 1% NP-40) supplemented with protease inhibitor (P8340; Sigma-Aldrich) and phosphatase inhibitor cocktails 2 and 3 (P0044 and P5726; Sigma-Aldrich). Cell lysis was performed on a rotator at 4°C for 30 min and then centrifuged at 14,000 rpm for 20 min. Cell pellets were resuspended in high-salt buffer (50 mM Tris, pH 7.5, 500 mM NaCl, and 1% NP-40) and incubated on a rotator at 4°C for 1 h, followed by centrifugation at 14,000 rpm for 20 min. The high- and low-salt extracts were mixed together, and the salt concentration was adjusted to 150 mM NaCl. The final cell lysate was precleared with protein A/G agarose (sc-2003; Santa Cruz Biotechnology), and precleared lysate was immunoprecipitated with anti-HA agarose (sc-7392 AC; Santa Cruz Biotechnology) overnight at 4°C. Precipitated protein complex was washed five times in low-salt buffer and eluted with HA peptide (500 µg/ml; I2149; Sigma-Aldrich). Eluted proteins were TCA-precipitated and analyzed by tandem mass spectrometry at the Taplin Mass Spectrometry Facility at Harvard Medical School.
AniPOND analysis
AniPOND analysis was performed as previously described with some modifications (Leung et al., 2013; Wiest and Tomkinson, 2017). Cells were pulse-labeled with 10 µM EdU in DMEM for 15 min, washed with PBS, and then incubated in medium with 10 µM thymidine for 1 h or with 4 mM HU for 2 h. After labeling, cells were immediately lysed and harvested with nuclei extraction buffer (20 mM Hepes, pH 7.2, 40 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 0.5% NP-40). Nuclei pellets were washed with 1× PBS, resuspended in click reaction mix (in order of addition: 25 mM biotin picolyl azide [Click Chemistry Tools], 10 mM (+)-sodium l-ascorbate, and 2 mM CuSO4) and rotated at 4°C for 1 h. Samples were then washed with 1× PBS. The resulting pellets were resuspended in 500 µl Buffer B1 (25 mM NaCl, 2 mM EDTA, 50 mM Tris-HCl, pH 8.0, 1% NP-40, and protease inhibitors), rotated for 30 min at 4°C, and spun down at maximum speed for 10 min at 4°C. The above procedure was repeated once more, and the resulting nuclei were resuspended in 500 µl Buffer B1. Samples were then sonicated using a Model 50 Sonic Dismembrator (Thermo Fisher Scientific) 12 × 10 s at 20 amplitude to solubilize DNA-bound proteins. Samples were spun down at maximum speed for 10 min, and the supernatant was collected. 500 µl Buffer B2 (150 mM NaCl, 2 mM EDTA, 50 mM Tris-HCl, pH 8.0, 0.5% NP-40, and protease inhibitors) was added to the supernatant to bring the total sample size to ∼1 ml. Samples were rotated overnight (16–20 h) with streptavidin beads (Thermo Fisher Scientific). A chromatin input sample was collected immediately before streptavidin capture. Beads were washed extensively with Buffer B2, and captured proteins were eluted by boiling in Laemmli buffer.
EM of replication intermediates
EM analysis of replication intermediates was performed as previously described (Neelsen et al., 2014). Briefly, 5–10×106 asynchronously growing MEFs were collected, and genomic DNA was cross-linked by three rounds of incubation in 10 µg/ml 4,5′,8-trimethylpsoralen (T6137; Sigma-Aldrich) and 3 min of irradiation with 366 nm UV light on a precooled metal block. Cells were lysed, and genomic DNA was isolated from the nuclei by proteinase K digestion and phenol-chloroform extraction. DNA was purified by isopropanol precipitation, digested with PvuII High Fidelity for 3–5 h at 37°C, and replication intermediates were enriched on a benzoylated naphthoylated DEAE–cellulose (B6385; Sigma-Aldrich) column. EM samples were prepared by spreading the DNA on carbon-coated grids in the presence of benzyl dimethylalkylammonium chloride and visualized by platinum rotary shadowing. Images were acquired on a transmission electron microscope (JEOL 1200 EX) with side-mounted camera (AMTXR41 supported by AMT software v601) and analyzed with ImageJ (National Institutes of Health).
DNA fiber assay
The DNA fiber assay was performed as described previously (Quinet et al., 2017a). Briefly, MEFs were labeled with two thymidine analogues: 20 µM IdU (I7125; Sigma-Aldrich) followed by 200 µM CldU (C6891; Sigma-Aldrich) for the indicated times. Where indicated, ATR inhibitor (10 µM) and mirin (50 µM) were added 2 h before labeling and remained in culture during the course of the experiments. Labeled cells were resuspended in 1× PBS at 106 cells/ml. 2 µl of cell suspension was mixed with 6 µl of lysis buffer (200 mM Tris–HCl, pH 7.5, 50 mM EDTA, and 0.5% SDS) on a glass slide. After 5 min, the slides were tilted at a 15–45° angle, and the resulting DNA spreads were air-dried, fixed in 3:1 methanol/acetic acid for 5 min, and stored at 4°C. The DNA fibers were denatured with 2.5 M HCl for 1 h, washed with 1× PBS, and blocked with 5% BSA in 1× PBS for 1 h. DNA immunostaining was performed with rat anti-BrdU antibody (1:100; Ab6326; Abcam) for CldU and mouse anti-BrdU antibody (1:20; 347580; Becton Dickson) for IdU in a humid chamber at room temperature for 1 h. The following secondary antibodies were used: anti-rat Alexa Fluor 488 (1:100; A21470; Molecular Probes) and anti-mouse Alexa Fluor 546 (1:100; A21123; Molecular Probes) at room temperature for 1 h. The slides were air-dried and mounted with Prolong Gold Anti-Fade reagent (P36930; Invitrogen). Images were acquired with LAS AF software using a TCS SP5 confocal microscope (Leica). The DNA tract lengths were measured using ImageJ. Only bicolor fibers were scored, and the total length (IdU + CldU) per fiber is presented in micrometers. Statistical analysis (Mann-Whitney U) was performed using GraphPad Prism Software.
Cell survival assay
PrestoBlue Cell Viability Reagent (A13261; Thermo Fisher Scientific) was used to estimate the fraction of viable cells present in 24-well plates after treating 3,000 cells/well with ATR inhibitor VE-821 for 4 d. The drug-containing media were then replaced with 0.5 ml of 1× PrestoBlue in growth media followed by incubation for 3 h at 37°C. The absorbance was recorded at 560 nm (experimental wavelength) and 570 nm (reference wavelength) using a Multiskan Ascent plate reader (Thermo Fisher Scientific).
Online supplemental material
All the peptides identified in our mass spectrometry analysis of XLF interaction proteins are available in Table S1. Additional analyses of reversed replication forks by EM and images are shown in Fig. S1. Independent biological replicates of Western blots are shown in Fig. S2.
Supplementary Material
Acknowledgments
We thank Dr. John Petrini for insightful comments. We are grateful to Dr. Frederick Alt (Harvard Medical School, Boston, MA) for H2axcon/con:Xlf+/− mice. We thank the Research Microscopy Facility of Saint Louis University, St. Louis, MO, for technical support and Mr. Christopher Haddock for technical assistance.
A. Vindigni is supported by National Institutes of Health grant R01GM108648 and Department of Defense Award BC151728. B.P. Sleckman is supported by National Institutes of Health grants AI047829 and AI074953.
The authors declare no competing financial interests.
Author contributions: A. Quinet and S. Thangavel designed and conducted the DNA fiber experiments. J. Jackson conducted EM analysis. M. Berti and A.K. Byrum performed iPOND and aniPOND analysis. B-R. Chen conducted all experiments except those above. A.L. Bredemeyer and I. Hindi helped carry out experiments. B-R. Chen, A. Quinet, A.L. Bredemeyer, N. Mosammaparast, J.K. Tyler, A. Vindigni, and B.P. Sleckman designed experiments and interpreted the results. B-R. Chen, A. Vindigni, and B.P. Sleckman wrote the manuscript.
References
- Ahnesorg P., Smith P., and Jackson S.P.. 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 124:301–313. 10.1016/j.cell.2005.12.031 [DOI] [PubMed] [Google Scholar]
- Ahuja A.K., Jodkowska K., Teloni F., Bizard A.H., Zellweger R., Herrador R., Ortega S., Hickson I.D., Altmeyer M., Mendez J., and Lopes M.. 2016. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 7:10660 10.1038/ncomms10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres S.N., Modesti M., Tsai C.J., Chu G., and Junop M.S.. 2007. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell. 28:1093–1101. 10.1016/j.molcel.2007.10.024 [DOI] [PubMed] [Google Scholar]
- Barnes D.E., Stamp G., Rosewell I., Denzel A., and Lindahl T.. 1998. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8:1395–1398. 10.1016/S0960-9822(98)00021-9 [DOI] [PubMed] [Google Scholar]
- Berti M., and Vindigni A.. 2016. Replication stress: getting back on track. Nat. Struct. Mol. Biol. 23:103–109. 10.1038/nsmb.3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford A.N., and Jackson S.P.. 2017. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell. 66:801–817. 10.1016/j.molcel.2017.05.015 [DOI] [PubMed] [Google Scholar]
- Bredemeyer A.L., Sharma G.G., Huang C.Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., and Sleckman B.P.. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 442:466–470. 10.1038/nature04866 [DOI] [PubMed] [Google Scholar]
- Brouwer I., Sitters G., Candelli A., Heerema S.J., Heller I., de Melo A.J., Zhang H., Normanno D., Modesti M., Peterman E.J., et al. 2016. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature. 535:566–569. 10.1038/nature18643 [DOI] [PubMed] [Google Scholar]
- Buck D., Malivert L., de Chasseval R., Barraud A., Fondanèche M.C., Sanal O., Plebani A., Stéphan J.L., Hufnagel M., le Deist F., et al. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 124:287–299. 10.1016/j.cell.2005.12.030 [DOI] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254. 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M.J., et al. 2002. Genomic instability in mice lacking histone H2AX. Science. 296:922–927. 10.1126/science.1069398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A., Fernandez-Capetillo O., Kruhlak M.J., Pilch D.R., Staudt D.W., Lee A., Bonner R.F., Bonner W.M., and Nussenzweig A.. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679. 10.1038/ncb1004 [DOI] [PubMed] [Google Scholar]
- Chang H.H.Y., Pannunzio N.R., Adachi N., and Lieber M.R.. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18:495–506. 10.1038/nrm.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanoux R.A., Yin B., Urtishak K.A., Asare A., Bassing C.H., and Brown E.J.. 2009. ATR and H2AX cooperate in maintaining genome stability under replication stress. J. Biol. Chem. 284:5994–6003. 10.1074/jbc.M806739200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., and Elledge S.J.. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell. 40:179–204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A., Bredemeyer A.L., Sowa M.E., Terret M.E., Jallepalli P.V., Harper J.W., and Elledge S.J.. 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 23:2415–2425. 10.1101/gad.1832309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K.A., and Cortez D.. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616–627. 10.1038/nrm2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch F.B., Bansbach C.E., Driscoll R., Luzwick J.W., Glick G.G., Bétous R., Carroll C.M., Jung S.Y., Qin J., Cimprich K.A., and Cortez D.. 2013. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27:1610–1623. 10.1101/gad.214080.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H., Rose K.L., Bhat K.P., Mohni K.N., Glick G.G., Couch F.B., and Cortez D.. 2015. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol. Cell. 59:998–1010. 10.1016/j.molcel.2015.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattah F.J., Kweon J., Wang Y., Lee E.H., Kan Y., Lichter N., Weisensel N., and Hendrickson E.A.. 2014. A role for XLF in DNA repair and recombination in human somatic cells. DNA Repair (Amst.). 15:39–53. 10.1016/j.dnarep.2013.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkos M., Ganier O., Coulombe P., and Méchali M.. 2015. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 16:360–374. 10.1038/nrm4002 [DOI] [PubMed] [Google Scholar]
- Frank K.M., Sekiguchi J.M., Seidl K.J., Swat W., Rathbun G.A., Cheng H.L., Davidson L., Kangaloo L., and Alt F.W.. 1998. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 396:173–177. 10.1038/24172 [DOI] [PubMed] [Google Scholar]
- Fugmann S.D., Lee A.I., Shockett P.E., Villey I.J., and Schatz D.G.. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495–527. 10.1146/annurev.immunol.18.1.495 [DOI] [PubMed] [Google Scholar]
- Gao Y., Sun Y., Frank K.M., Dikkes P., Fujiwara Y., Seidl K.J., Sekiguchi J.M., Rathbun G.A., Swat W., Wang J., et al. 1998. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 95:891–902. 10.1016/S0092-8674(00)81714-6 [DOI] [PubMed] [Google Scholar]
- Hammel M., Rey M., Yu Y., Mani R.S., Classen S., Liu M., Pique M.E., Fang S., Mahaney B.L., Weinfeld M., et al. 2011. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J. Biol. Chem. 286:32638–32650. 10.1074/jbc.M111.272641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink B.A., and Sleckman B.P.. 2012. The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol. 30:175–202. 10.1146/annurev-immunol-030409-101320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink B.A., Tubbs A.T., Dorsett Y., Bednarski J.J., Walker L.M., Feng Z., Sharma G.G., McKinnon P.J., Zhang J., Bassing C.H., and Sleckman B.P.. 2011. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 469:245–249. 10.1038/nature09585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her J., Ray C., Altshuler J., Zheng H., and Bunting S.F.. 2018. 53BP1 Mediates ATR-Chk1 Signaling and Protects Replication Forks under Conditions of Replication Stress. Mol. Cell. Biol. 38:e00472-17 10.1128/MCB.00472-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinjivadi A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Techer H., Baldi G., Shen R., Ciccia A., Pellegrini L., et al. 2017. Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments. Mol. Cell. 67:867–881.e7. 10.1016/j.molcel.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaçon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., and Vindigni A.. 2017. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8:860 10.1038/s41467-017-01180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld B.M., Rattray A.J., Bhaskara V., Ghirlando R., and Paull T.T.. 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell. 28:638–651. 10.1016/j.molcel.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K.H., Abou El Hassan M., and Bremner R.. 2013. A rapid and efficient method to purify proteins at replication forks under native conditions. Biotechniques. 55:204–206. 10.2144/000114089 [DOI] [PubMed] [Google Scholar]
- Li G., Alt F.W., Cheng H.L., Brush J.W., Goff P.H., Murphy M.M., Franco S., Zhang Y., and Zha S.. 2008. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol. Cell. 31:631–640. 10.1016/j.molcel.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., and Oda M.. 2010. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79:89–130. 10.1146/annurev.biochem.052308.103205 [DOI] [PubMed] [Google Scholar]
- Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P., and Lopes M.. 2017. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 8:859 10.1038/s41467-017-01164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam E.A., and Cortez D.. 2011. ATR signalling: more than meeting at the fork. Biochem. J. 436:527–536. 10.1042/BJ20102162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen K.J., and Lopes M.. 2015. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 16:207–220. 10.1038/nrm3935 [DOI] [PubMed] [Google Scholar]
- Neelsen K.J., Chaudhuri A.R., Follonier C., Herrador R., and Lopes M.. 2014. Visualization and interpretation of eukaryotic DNA replication intermediates in vivo by electron microscopy. Methods Mol. Biol. 1094:177–208. 10.1007/978-1-62703-706-8_15 [DOI] [PubMed] [Google Scholar]
- Quinet A., Carvajal-Maldonado D., Lemacon D., and Vindigni A.. 2017a DNA Fiber Analysis: Mind the Gap! Methods Enzymol. 591:55–82. 10.1016/bs.mie.2017.03.019 [DOI] [PubMed] [Google Scholar]
- Quinet A., Lemaçon D., and Vindigni A.. 2017b Replication Fork Reversal: Players and Guardians. Mol. Cell. 68:830–833. 10.1016/j.molcel.2017.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland R.L., Patel S., Rivard R.S., Smith K., Peters A.A., Bielinsky A.K., and Brown E.J.. 2013. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 27:2259–2273. 10.1101/gad.223180.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J., et al. 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 535:382–387. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., and Bonner W.M.. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868. 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Boon C., Redon C., and Bonner W.M.. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916. 10.1083/jcb.146.5.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars V., Drevet P., Legrand P., Baconnais S., Amram J., Faure G., Márquez J.A., Piétrement O., Guerois R., Callebaut I., et al. 2011. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc. Natl. Acad. Sci. USA. 108:12663–12668. 10.1073/pnas.1100758108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar J.C., Cortez D., and Cimprich K.A.. 2017. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18:622–636. 10.1038/nrm.2017.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., and Jackson S.P.. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514. 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic V., Yin B., Maas N.L., Bredemeyer A.L., Carpenter A.C., Helmink B.A., Yang-Iott K.S., Sleckman B.P., and Bassing C.H.. 2009. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol. Cell. 34:298–310. 10.1016/j.molcel.2009.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Christ N., Siaud N., Egashira A., Wu H., and Jasin M.. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 145:529–542. 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J.A., Berti M., Walser F., Raso M.C., Schmid F., Krietsch J., Stoy H., Zwicky K., Ursich S., Freire R., et al. 2018. Histone Ubiquitination by the DNA Damage Response Is Required for Efficient DNA Replication in Unperturbed S Phase. Mol. Cell. 71:897–910.e8. 10.1016/j.molcel.2018.07.011 [DOI] [PubMed] [Google Scholar]
- Schwartz M., Oren Y.S., Bester A.C., Rahat A., Sfez R., Yitzchaik S., de Villartay J.P., and Kerem B.. 2009. Impaired replication stress response in cells from immunodeficiency patients carrying Cernunnos/XLF mutations. PLoS One. 4:e4516 10.1371/journal.pone.0004516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi Y., and Nishitani H.. 2017. Control of Genome Integrity by RFC Complexes; Conductors of PCNA Loading onto and Unloading from Chromatin during DNA Replication. Genes (Basel). 8:E52 10.3390/genes8020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu B.M., Couch F.B., Feigerle J.T., Bhaskara S., Hiebert S.W., and Cortez D.. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25:1320–1327. 10.1101/gad.2053211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu B.M., Couch F.B., and Cortez D.. 2012. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 7:594–605. 10.1038/nprot.2012.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., and Foiani M.. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 297:599–602. 10.1126/science.1074023 [DOI] [PubMed] [Google Scholar]
- Taglialatela A., Alvarez S., Leuzzi G., Sannino V., Ranjha L., Huang J.W., Madubata C., Anand R., Levy B., Rabadan R., et al. 2017. Restoration of Replication Fork Stability in BRCA1- and BRCA2-Deficient Cells by Inactivation of SNF2-Family Fork Remodelers. Mol. Cell. 68:414–430.e8. 10.1016/j.molcel.2017.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa M., Bonetti D., Carraro M., and Longhese M.P.. 2018. Rad9/53BP1 protects stalled replication forks from degradation in Mec1/ATR-defective cells. EMBO Rep. 19:351–367. 10.15252/embr.201744910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindigni A., and Lopes M.. 2017. Combining electron microscopy with single molecule DNA fiber approaches to study DNA replication dynamics. Biophys. Chem. 225:3–9. 10.1016/j.bpc.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujanovic M., Krietsch J., Raso M.C., Terraneo N., Zellweger R., Schmid J.A., Taglialatela A., Huang J.W., Holland C.L., Zwicky K., et al. 2017. Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity. Mol. Cell. 67:882–890.e5. 10.1016/j.molcel.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., and Chen J.. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759–47762. 10.1074/jbc.C100569200 [DOI] [PubMed] [Google Scholar]
- Wiest N.E., and Tomkinson A.E.. 2017. Optimization of Native and Formaldehyde iPOND Techniques for Use in Suspension Cells. Methods Enzymol. 591:1–32. 10.1016/bs.mie.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Yano K., Morotomi-Yano K., Wang S.Y., Uematsu N., Lee K.J., Asaithamby A., Weterings E., and Chen D.J.. 2008. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 9:91–96. 10.1038/sj.embor.7401137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., and Lopes M.. 2015. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208:563–579. 10.1083/jcb.201406099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman M.K., and Cimprich K.A.. 2014. Causes and consequences of replication stress. Nat. Cell Biol. 16:2–9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S., Alt F.W., Cheng H.L., Brush J.W., and Li G.. 2007. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc. Natl. Acad. Sci. USA. 104:4518–4523. 10.1073/pnas.0611734104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S., Guo C., Boboila C., Oksenych V., Cheng H.L., Zhang Y., Wesemann D.R., Yuen G., Patel H., Goff P.H., et al. 2011. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 469:250–254. 10.1038/nature09604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., and Elledge S.J.. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300:1542–1548. 10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.