Matson et al. find that human cells re-entering the cell cycle from quiescence have an impaired p53-dependent DNA replication origin licensing checkpoint and slow origin licensing. This combination makes every first S phase underlicensed and hypersensitive to replication stress.
Abstract
To maintain tissue homeostasis, cells transition between cell cycle quiescence and proliferation. An essential G1 process is minichromosome maintenance complex (MCM) loading at DNA replication origins to prepare for S phase, known as origin licensing. A p53-dependent origin licensing checkpoint normally ensures sufficient MCM loading before S phase entry. We used quantitative flow cytometry and live cell imaging to compare MCM loading during the long first G1 upon cell cycle entry and the shorter G1 phases in the second and subsequent cycles. We discovered that despite the longer G1 phase, the first G1 after cell cycle re-entry is significantly underlicensed. Consequently, the first S phase cells are hypersensitive to replication stress. This underlicensing results from a combination of slow MCM loading with a severely compromised origin licensing checkpoint. The hypersensitivity to replication stress increases over repeated rounds of quiescence. Thus, underlicensing after cell cycle re-entry from quiescence distinguishes a higher-risk first cell cycle that likely promotes genome instability.
Introduction
Proliferating mammalian cells initiate DNA replication at thousands of DNA replication origins every cell cycle. Replication origins are chromosomal loci where DNA synthesis initiates in S phase. The minichromosome maintenance complex (MCM) is an essential component of the helicase that unwinds DNA to initiate replication (Bell and Labib, 2016). Cells prepare for DNA replication in S phase by loading MCMs at replication origins in the preceding G1 phase, a process called “origin licensing.” The amount of DNA-loaded MCM increases as cells progress through G1 until reaching a maximum at the G1/S transition (Remus and Diffley, 2009; Siddiqui et al., 2013). Once cells enter S phase, multiple mechanisms block any new MCM loading to restrict origin licensing activity to G1 phase (Arias and Walter, 2007; Truong and Wu, 2011). Cells block MCM loading outside of G1 phase to prevent genome instability caused by rereplication (Arias and Walter, 2007; Truong and Wu, 2011). MCMs unwind DNA in S phase and travel with replication forks, and MCMs are unloaded throughout S phase as replication forks terminate (Maric et al., 2014; Moreno et al., 2014).
Replication forks can stall or slow during S phase from a variety of endogenous and exogenous sources. A stalled replication fork can be rescued if MCM at a nearby licensed origin initiates a new fork to replicate the intervening DNA (Yekezare et al., 2013; Alver et al., 2014). Since MCM loading is restricted to G1 phase, but the location of stalled forks in S is unpredictable, cells license many more origins than they would require to complete S phase if there were no replication stress. These excess licensed origins function as “dormant origins” and are activated where needed (Woodward et al., 2006; Ge et al., 2007; Ibarra et al., 2008). Cells with considerably less loaded MCM can still complete a normal S phase under ideal growth conditions (Ge et al., 2007). Nonetheless, if cells enter S phase “underlicensed” with fewer dormant origins, they are hypersensitive to replication stress. In addition, animal models illustrate the long-term consequences of underlicensing. Mice heterozygous for MCM null alleles or homozygous for hypomorphic MCM alleles have less MCM loading, increased replication stress, and defects in highly proliferative tissues (Pruitt et al., 2007; Alvarez et al., 2015). In addition, these mice are prone to genomic instability, premature aging, and cancer (Pruitt et al., 2007; Shima et al., 2007; Kunnev et al., 2010).
Since dormant origins are critical to protect cells during replication stress, a control mechanism ensures sufficient origin licensing. An origin licensing cell cycle checkpoint in untransformed mammalian cells ensures abundant licensing in G1 phase before S phase entry (Shreeram et al., 2002; Liu et al., 2009; Nevis et al., 2009). The checkpoint was revealed by artificially reducing MCM loading, which delayed the late G1 activation of cyclin E/CDK2 (Nevis et al., 2009). Delayed cyclin E/CDK2 activation delays the phosphorylation of substrates that drive S phase entry (Giacinti and Giordano, 2006). Delaying CDK2 activity lengthens G1 phase and ensures that cells do not enter S phase underlicensed. Moreover, this checkpoint is p53 dependent (Nevis et al., 2009), meaning that a common genetic perturbation in transformed cancer cells compromises the normal coordination of origin licensing and S phase onset.
Given the importance of coordinating G1 length with the progress of origin licensing for robust S phase completion, we considered natural circumstances where G1 length changes. We previously found that stem cells with short G1 phases load MCM faster than differentiated cells with longer G1 phases to achieve the same amount of loaded MCM at S phase entry (Matson et al., 2017). An alternative example is the long G1 after cell cycle re-entry from quiescence (Coller, 2007). Cell cycle quiescence, or “G0,” is a reversible cell cycle exit to a nondividing state. G0 is distinct from a G1 arrest; it is an active state requiring up-regulation of anti-apoptotic, anti-senescent, and anti-differentiation genes as well as repression of cell cycle genes (Coller et al., 2006; Litovchick et al., 2007). The longer G1 phase during re-entry likely reflects the need to reactivate and express genes repressed in G0 and other fundamental differences in G1 regulation.
The unique features of G0 and the first G1 phase suggest that origin licensing may be distinctly regulated during cell cycle re-entry. We used single cell flow cytometry to measure the amount of loaded MCM at S phase entry and live cell imaging to measure cell cycle timing to determine if the amount of loaded MCM differs between cell cycle re-entry and active proliferation. We discovered that cells re-entering the cell cycle from G0 are routinely and significantly underlicensed, rendering them hypersensitive to replication stress. This finding is consistent with a recent report that the first S phase experiences higher spontaneous replication stress, though the source of that stress was not identified (Daigh et al., 2018). We report that MCM loading is slow in the first G1 phase and furthermore that the first cell cycle has a severely compromised origin licensing checkpoint relative to the robust checkpoint in actively proliferating cells. To our knowledge, these results demonstrate the first naturally underlicensed cell cycle and suggest that repeated cell cycle re-entry from G0 is particularly hazardous for long-term genome stability.
Results
G0 cells re-entering the cell cycle are underlicensed compared with actively proliferating cells
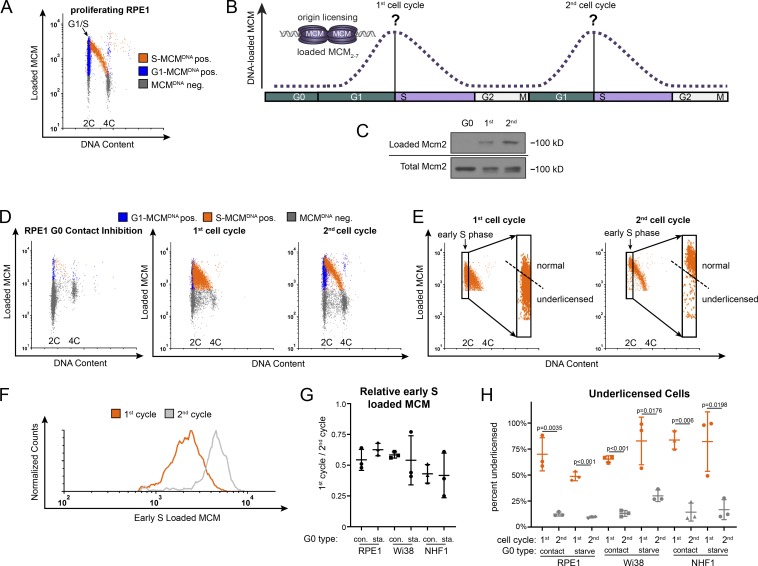
Cells re-entering the cell cycle from G0 have a long G1 before S phase entry relative to actively proliferating cells (Coller, 2007). We hypothesized that the difference in G1 length and unique aspects of the G0 to G1 transition could alter the timing or amount of loaded MCM at S phase entry. We therefore focused on quantifying how much MCM is loaded at the onset of S phase (G1/S transition). Standard synchronization and immunoblotting is an inadequate approach, however, because G0 cells re-enter G1 phase semi-synchronously. Even in the first cycle after re-entry, there is no time point near the G1/S transition that is not a mix of different phases (e.g., G1 and S phase cells), and the following second cell cycle is nearly asynchronous (Kwon et al., 2017; Wang et al., 2017). We therefore used a previously published and validated single cell flow cytometry assay to measure DNA-loaded MCM at the G1/S transition (Håland et al., 2015; Moreno et al., 2016; Matson et al., 2017). We examined proliferating retinal pigmented epithelial cells (RPE) immortalized with telomerase (RPE1-hTert, also referred to hereafter as RPE1 for simplicity). We extracted soluble proteins with nonionic detergent so that DNA-bound proteins were retained, then fixed and stained with anti-Mcm2 antibody as a marker for the whole MCM2-7 complex, with DAPI to measure DNA content, and with the nucleotide analogue 5-ethynyl-2´-deoxyuridine (EdU) to measure DNA synthesis (Fig. 1 A). We defined MCM-positive cells based on an antibody-negative control; gray cells in the Fig. 1 plots are below this background for detecting DNA-loaded MCM (MCMDNA negative). Fig. S1 A shows the flow cytometry gating scheme. We marked cells blue for 2C DNA content (G1) that are negative for DNA synthesis but positive for DNA-loaded MCM (G1-MCMDNA positive). MCM loading is presumably unidirectional in G1, and loaded MCM complexes are very stable and are only unloaded in S phase as replication terminates (Kuipers et al., 2011; Maric et al., 2014; Moreno et al., 2014). We marked cells orange for S phase based on EdU incorporation plus DNA-loaded MCM (S-MCMDNA positive). In actively proliferating populations, we consistently observe that the majority of cells reach a similar maximum loaded MCM per cell in G1 (blue) before becoming EdU positive (orange) in very early S phase. In this way, we accurately measured loaded MCM in single cells at very early S phase even in fully asynchronous populations (Fig. 1 A, G1/S marked by the arrow).
Figure 1.
The first S phase after cell cycle re-entry from quiescence (G0) is underlicensed. (A) Flow cytometry of proliferating RPE1-hTert cells extracted with nonionic detergent to measure DNA-bound protein. Cells were labeled with 10 µM EdU for 30 min before harvesting then stained with MCM2 antibody to measure DNA loaded MCM, DAPI to measure DNA content, and EdU to measure DNA synthesis. Gates to define colors are provided in Fig. S1 A. Orange cells are S phase: EdU-positive + DNA-loaded MCM-positive; blue cells are G1 phase: EdU-negative + DNA-loaded MCM-positive; gray cells are all MCM-negative cells. Arrow indicates the G1/S transition. (B) MCM loading in the first and second cell cycles. At the start of this study, relative MCM loading in the long G1 of cell cycle re-entry relative to MCM loading in second and subsequent cell cycles was unknown. (C) RPE1 cells were synchronized in G0 by contact inhibition in serum and released from G0 into the cell cycle. Cells were analyzed in G0, 24 h after release (first cell cycle), and 48 h after release (second cell cycle). Samples were fractionated into DNA-loaded chromatin fraction and total lysate for immunoblot. (D) Cells treated as in C were analyzed by chromatin-bound flow cytometry as in A. (E) Gating of early S cells from D, showing only the S phase, MCM DNA-positive cells. Rectangles define early S phase cells as MCM DNA-positive with 2C DNA content. Inset: Only early S phase cells. The dashed line defines normally licensed versus underlicensed cells. (F) Histogram measuring DNA-loaded MCM per cell in early S phase cells from E. The orange line is early S phase loaded MCM from the first cell cycle, and the gray line is early S phase loaded MCM from the second cell cycle. (G) Relative mean MCM loaded in early S phase in the first cycle divided by the second cycle. RPE1, Wi38, and NHF1 were synchronized by contact inhibition or mitogen starvation and release; see also E; Fig. S1, F–I; and Fig. S2, A–F. n = 3, mean with SD. (H) Percentage of underlicensed cells from the first and second cell cycles in G. n = 3, mean with SD.
We then asked how much MCM had been loaded by the time of the G1/S transition in the first cell cycle compared with the amount loaded at G1/S in the second cell cycle (Fig. 1 B). We arrested RPE cells in G0 by contact inhibition (in the presence of growth serum) for 48 h. Contact-inhibited G0 cells showed a robust cell cycle exit to G0 with very little loaded MCM; 94% of cells were G0/G1, but only 1% were still in S phase (Fig. S1 B). G0 cells also expressed the expected high levels of p27 and low cyclin D1 compared with cells in the first cycle (Fig. S1 C). We then released cells to re-enter the cell cycle by plating at sub-confluent cell density. For comparison to the flow cytometry analysis, we also used biochemical chromatin fractionation and immunoblotting to observe MCM loading after cell cycle re-entry (Fig. 1 C). We pulse-labeled cells with EdU and harvested some cells in G0 or at 24 h when they were a mix of first G1 and ∼50–70% first S phase cells or at 48 h when they had proceeded to the second cell cycle (Fig. 1, C and D). In a separate continuous EdU labeling experiment in which EdU was added at the time of release, we determined that nearly 100% of cells had entered S phase by 28 h (Fig. S1 D), and almost no cells remained in G0/G1. Thus, we concluded that additional cells we harvested 48 h after release had completed their first S phase and were into their second cell cycle.
Strikingly, the amount of loaded MCM at the G1/S transition was markedly different between the first and second cell cycles after G0. Cells in the first cell cycle progressed into S phase (Fig. 1 D, orange cells) with substantially less MCM loaded than cells in the second cell cycle. By the second cell cycle, G1 cells progressed into S phase as a tight group with relatively high amounts of loaded MCM (transition from blue to orange). In contrast, many cells in the first cell cycle entered S phase with low amounts of loaded MCM. This behavior creates the filled orange triangle on flow cytometry plots of the first S phase instead of the normally clear region under a high arc characteristic of the second S phase.
To quantify the amount of loaded MCM at the G1/S transition, we analyzed the S phase cells from Fig. 1 D and defined very early S phase cells as EdU positive with ∼2C DNA content (Fig. 1 E, black rectangles; and see Fig. S1 E for early S phase on EdU plots). We focused on the early S population because they report the amount of MCM that had been loaded by the G1/S transition without confounding contributions from replication termination. Early S phase cells in the first cycle had a broad range of loaded MCM levels that included many cells with low MCM, whereas early S phase cells in the second cycle primarily had high loaded MCM levels. The second cell cycle was nearly indistinguishable from asynchronously proliferating cells. We thus used the second cell cycle to define the normal licensing levels and classified cells with less MCM loaded in early S as underlicensed (Fig. 1 E, dashed line). To visualize and directly compare the full MCM loading distribution in different populations, we generated histograms of loaded MCM levels in early S phase cells (Fig. 1 F). We noted on the one-dimensional histograms that the maximum MCM loading in the first cycle rarely reached the same values as in the second cycle (x axis positions). We also quantified the differences as the ratio of mean MCM loaded in the first versus second early S phases for multiple biological replicates. Cells in the first cycle after re-entry from G0 loaded only half as much MCM as cells in the second cell cycle (Fig. 1 G). We then used this same comparison to test if this underlicensed first cell cycle is common among different untransformed cell lines or methods of quiescence induction. We observed similar underlicensing after quiescence induction in RPE by serum starvation and restimulation and in two fibroblast cell lines arrested by contact inhibition or serum starvation (Fig. 1 G is the ratio of the means within each experiment, and representative replicates are shown in Fig. S1 [F–I] and Fig. S2 [A–F]). We noted some variation in the shapes of the one-dimensional histograms among individual replicates, but the relative means of these populations were consistently lower in the first cycle. We observed not only that the mean MCM loaded by the first S phase was half that of subsequent S phases but also that the majority of first S phase cells were underlicensed (Fig. 1 H). Additionally, the earliest cells re-entering S phase before 24 h after G0 release were similarly underlicensed (Fig. S2, G–K). Thus, re-entry from G0 is characterized by substantial underlicensing in the first cell cycle.
Cells re-entering S phase from G0 are hypersensitive to replication stress
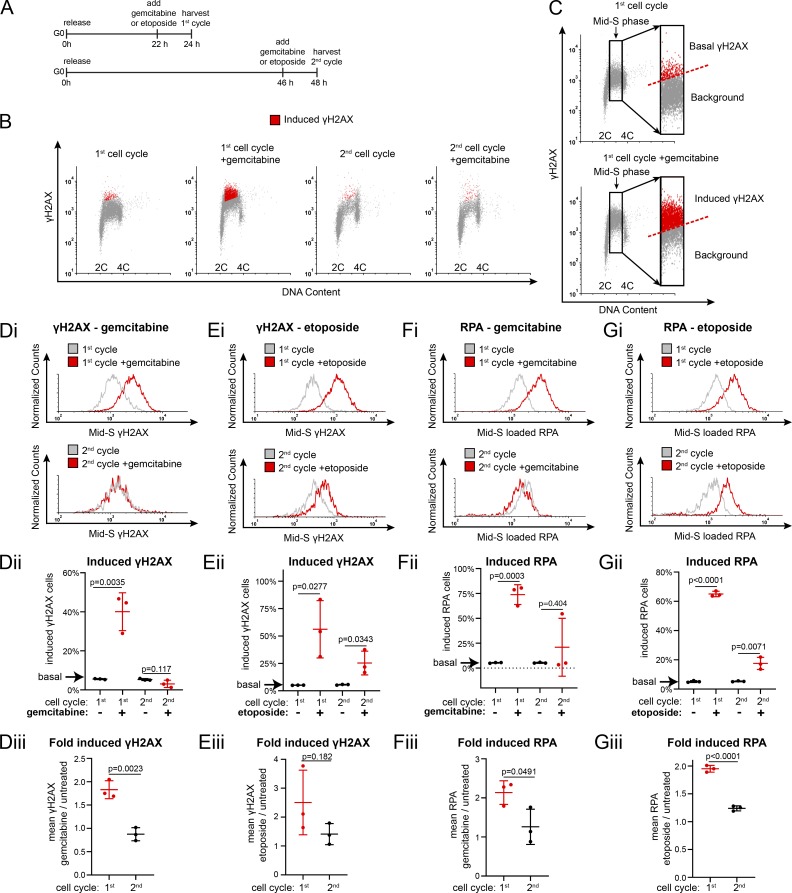
Cells typically load extra MCM in G1 to license dormant origins so they can tolerate replication stress in S phase (Woodward et al., 2006). Given that the first S phase is underlicensed, we hypothesized that cells re-entering the cell cycle from G0 would be hypersensitive to replication stress in the first S phase. To test that idea, we treated cells in the first or second cell cycle after G0 with gemcitabine, a chemotherapeutic drug that perturbs nucleotide pools to cause replication stress (Zhang et al., 2016; Fig. 2 A). We used flow cytometry to measure the expression of γH2AX, a common replication stress marker (Ewald et al., 2007; Fig. 2 B and Fig. S3 A). We specifically analyzed mid-S phase cells because the outcome of underlicensing on replication forks is expected when many forks are active and to account for differences in cell cycle distribution. We scored the number of γH2AX-positive S phase cells with expression equal to or greater than the top 5% of basal γH2AX levels in untreated cells (Fig. 2 C, dashed line). By this measurement, cells in the first S phase after G0 are significantly more sensitive to replication stress than cells in the second S phase (Fig. 2, B and Di). Moreover, gemcitabine-treated first S phase cells expressed double the amount of γH2AX per cell than cells in the second S phase, suggesting that not only were more total cells exhibiting a replication stress response but also that there was more replication stress per cell (Fig. 2, Dii and Diii). Cells in the first S phase were similarly hypersensitive to the topoisomerase inhibitor etoposide (Fig. 2, Ei–Eiii). We also measured the ssDNA binding protein replication protein A (RPA) as an alternative replication stress marker and found that both gemcitabine and etoposide increased DNA-loaded RPA in the first cell cycle (Fig. 2, Fi–Giii; and Fig. S3 B). This hypersensitivity to replication stress in the underlicensed first cell cycle suggests that cell cycle re-entry is an inherently higher-risk cycle with respect to genome stability compared with subsequent cell cycles.
Figure 2.
The first S phase after G0 is hypersensitive to replication stress. (A) Diagram of experimental workflow. RPE1 cells were synchronized in G0 by contact inhibition and released into the first cell cycle (24 h) or second cell cycle (48 h). Cells were treated with gemcitabine, etoposide, or vehicle for the 2 h before harvesting. (B) Flow cytometry of chromatin-bound proteins in cells treated as in A with 50 nM gemcitabine and stained for DNA and γH2AX. Red cells are replication stress–induced γH2AX-positive, as defined in C. (C) Gating of cells from B to define the threshold of γH2AX signal induced in S phase by gemcitabine or etoposide. Inset: Replication stress–induced γH2AX-positive cells (red) are defined as those equal to or above the top 5% of the corresponding untreated cells and marked red in B. (Di) Histograms of mid-S phase γH2AX intensity per cell from B. (Dii) Percentage of replication stress–induced γH2AX from B. n = 3, mean with SD. (Diii) Relative mean γH2AX intensity per cell plotted as gemcitabine-treated divided by untreated cells from B. n = 3, mean with SD. (Ei) Histograms of mid-S phase γH2AX intensity per cell treated with 0.5 μM etoposide as in A. (Eii) Percentage of replication stress–induced γH2AX from cells in Ei. n = 3, mean with SD. (Eiii) Comparison of mean γH2AX intensity, etoposide-treated divided by untreated cells from cells in Ei. n = 3, mean with SD. (Fi) Histograms of mid-S phase RPA intensity per cell from cells treated with 100 nM gemcitabine as in A. (Fii) Percentage of replication stress–induced RPA from Fi. n = 3, mean with SD. (Fiii): Comparison of mean RPA intensity per cell, gemcitabine-treated divided by untreated cells from cells in Fi. n = 3, mean with SD. (Gi) Histograms of mid-S phase RPA intensity per cell from cells treated with 5 μM etoposide as in A. (Gii) Percentage of replication stress–induced RPA from Gi. n = 3, mean with SD. (Giii) Comparison of mean RPA intensity per cell, mean gemcitabine-treated divided by untreated cells from cells in Gi. n = 3, mean with SD.
Many cells in vivo switch between periods of active proliferation and periods of G0, repeatedly re-entering the cell cycle to proceed through this presumably underlicensed first cell cycle (van Velthoven and Rando, 2019). We considered the possibility that repeated cell cycle exit and re-entry would lead to additional replication stress sensitivity. To measure accumulation of replication stress during repeated cell cycle re-entry, we synchronized RPE cells in G0 by contact inhibition for 48 h and released them into the cell cycle for ∼48 h (∼2–3 cycles) until they became contact inhibited again. We reiterated this procedure for a total of three rounds of G0 arrest and re-entry. In the third release, we treated with gemcitabine and measured the induction of γH2AX (Fig. S3 C). Cells re-entering S phase from three repeated arrests in G0 (3xG0) were more likely to induce γH2AX than cells that been arrested only once (1xG0), and these cells were also more intensely γH2AX positive per cell (Fig. S3, D and E). The difference was not due to, however, increased passage number or a change in MCM loading after repeated rounds of G0 (Fig. S3, F–K). The increased replication stress sensitivity after three rounds of G0 varied somewhat due to inherent variations in the G0 synchronization over multiple rounds (Kwon et al., 2017; Wang et al., 2017). This variability was reflected by unpaired t test P values of approximately P = 0.15 for both the number of γH2AX-positive cells and the γH2AX intensity (Fig. S3, D and E). Nevertheless, we observed a clear trend that repeated cell cycle re-entry from G0 increased sensitivity to replication stress.
Proliferating epithelial cells have a robust p53-dependent origin licensing checkpoint
Our findings raised a larger question about the relationship between origin licensing and S phase entry. The consistently high amount of loaded MCM we observed before G1/S in the second and subsequent proliferating cell cycles is consistent with an origin licensing cell cycle checkpoint (e.g., Fig. 1 A, arrow). Checkpoints enforce the order of events in the cell cycle, delaying progression through a phase until important events are completed (Hartwell and Weinert, 1989). The origin licensing checkpoint extends G1 and delays S phase entry when the amount of loaded MCM is low by delaying the activation of G1 CDKs until MCM loading is completed (McIntosh and Blow, 2012). Evidence for this checkpoint is the response to reducing MCM loading, which causes lower CDK activity and lengthening of G1 in proliferating cells (Shreeram et al., 2002; Liu et al., 2009; Nevis et al., 2009). Because we observed underlicensed cells in the first S phase after G0 and higher licensing in the second cell cycle, we hypothesized that the origin licensing checkpoint is primarily active in only the second and subsequent cell cycles.
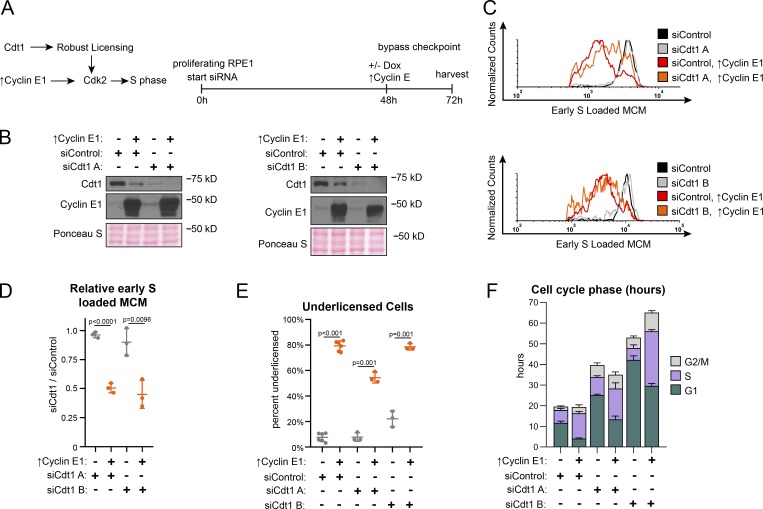
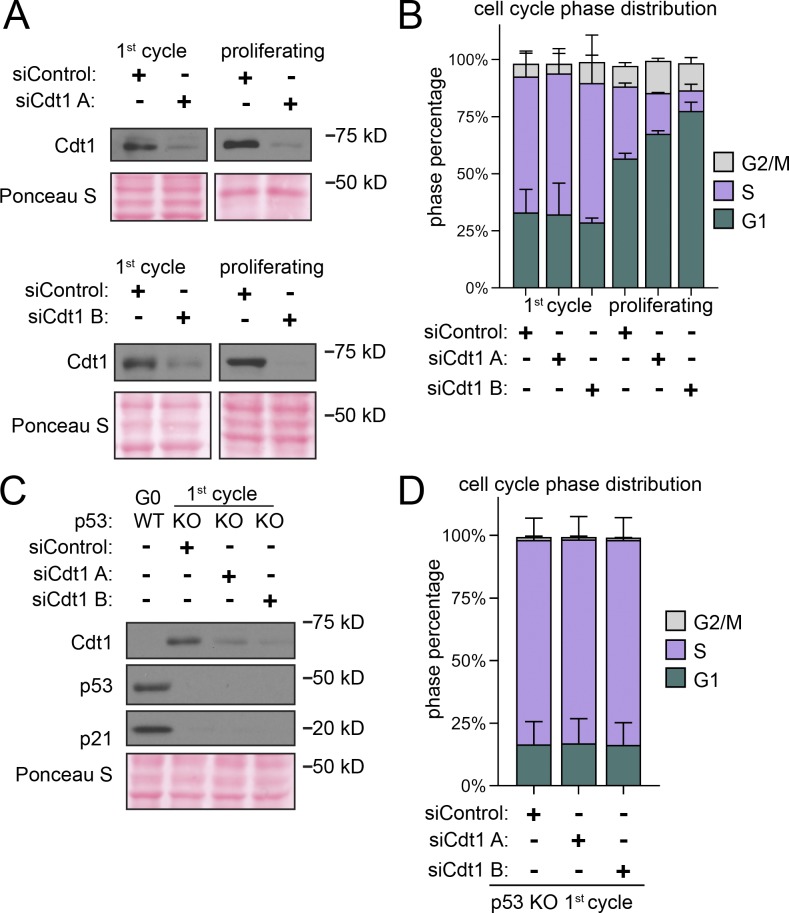
Origin licensing checkpoint activity is cell type dependent, however (Shreeram et al., 2002; Nevis et al., 2009). Therefore, we assessed checkpoint status in proliferating untransformed RPE1 (Fig. 3 A). We decreased origin licensing with siRNA targeting Cdc10-dependent transcript 1 (Cdt1), an essential MCM loading protein (Pozo and Cook, 2016). We used either a pool of four siRNAs (siCdt1 A) or a single independent siCdt1 (siCdt1 B) and analyzed Cdt1 protein by immunoblotting (Fig. 3 B). We then tested both MCM loading and the length of G1 phase. Cdt1 depletion induced both a reduction in the rate of MCM loading during G1 phase (Fig. S4, A and B; note the left-shifted histograms for siCdt1 in Fig. S4 B) and also a striking G1 lengthening. G1 length dramatically increased by several-fold, and the more profound Cdt1 depletion by siCdt1 B caused a greater G1 delay (Fig. 3 F, green bars). Importantly, Cdt1 depletion alone changed neither the amount of early S loaded MCM (compare black and gray lines in Fig. 3 C, and relative loading in Fig. 3 D) nor the percentage of underlicensed early S phase cells (Fig. 3 E). By the time cells finally entered S phase, they had achieved normal amounts of loaded MCM. These observations indicate that actively proliferating RPE1 cells wait for the normal amount of loaded MCM in G1 phase before entering S phase.
Figure 3.
Cyclin E1 overproduction bypasses the licensing checkpoint in proliferating cells. (A) Left: Cdt1 promotes robust licensing, cyclin E/CDK2 activation, and S phase entry. Right: Plan: Proliferating RPE1 cells containing integrated doxycycline inducible cyclin E1 were treated with siControl, or independent siRNAs targeting Cdt1 (siCdt1 A or siCdt1 B) for 72 h. 100 ng/ml doxycycline to overproduce cyclin E1 and directly activate CDK2 was added at 48 h. (B) Immunoblots of total protein lysates of cells treated according to A. ↑Cyclin E1 indicates addition of doxycycline to overproduce cyclin E1. (C) Loaded MCM in early S phase of cells treated according to A determined by flow cytometric analysis as in Fig. 1 E. See Fig. S3 A for complete flow cytometry plots. (D) Relative early S phase DNA-loaded MCM per cell from C. Values are the ratio of mean loaded MCM in Cdt1-depleted versus control cells. Left to right: 1: SiCdt1 A alone/siControl alone. 2: SiCdt1 A, ↑Cyclin E1/siControl alone. 3: SiCdt1 B alone/siControl alone. 4: SiCdt1 B, ↑cyclin E1/siControl alone. n = 3, mean with SD. (E) Percentage of underlicensed cells in C. n = 3, mean with SD. (F) Mean cell cycle phase length in hours in C. n = 3, mean with SD.
A cell cycle checkpoint that delays progression to the next phase is distinct from a simple incapacity to proceed to the next phase because it is possible for genetic alterations to bypass a checkpoint and induce premature cell cycle progression (Hartwell and Weinert, 1989). The origin licensing checkpoint delays the activation of cyclin E/CDK2 (Nevis et al., 2009). To test if the G1 phase delay can be bypassed in Cdt1-depleted RPE1 cells, we overproduced cyclin E1 in a polyclonal line to prematurely activate CDK2. We used flow cytometry to measure early S phase loaded MCM as before (Fig. 3 C and Fig. S3 A). As expected from prior reports, cyclin E1 overproduction shortened G1 phase (Fig. 3 F, green bars, siControl versus ↑cyclin E1; Resnitzky et al., 1994; Matson et al., 2017). In addition, cyclin E1 overproduction alone induced underlicensing as measured by the amount of early S loaded MCM (compare black and red lines, Fig. 3 C, and the percentage of underlicensed cells, Fig. 3 E; Ekholm-Reed et al., 2004; Matson et al., 2017). Strikingly, when we overproduced cyclin E1 after Cdt1 depletion (Fig. 3 A), early S phase cells became severely underlicensed, quantified by both MCM loaded per cell (compare gray and orange lines, Fig. 3 C and relative loading in Fig. 3 D), and the percentage of underlicensed cells (Fig. 3 E). Relative to the single manipulations, many Cdt1-depleted cyclin E–overproducing cells even entered S phase with MCM levels below the detection limit of the antibody (Fig. S4 A, gray MCMDNA-negative cells in S between 2C and 4C DNA). Moreover, G1 length decreased even in the Cdt1-depleted cells (Fig. 3 F). We previously determined that the apparent decease in Cdt1 protein upon cyclin E1 overproduction is an indirect effect of cell cycle phase distribution (Matson et al., 2017). Thus, cyclin E1 overproduction bypassed both the normal relationship between MCM loading and G1 length and the strong G1 delay in Cdt1-depleted cells to cause underlicensed S phases.
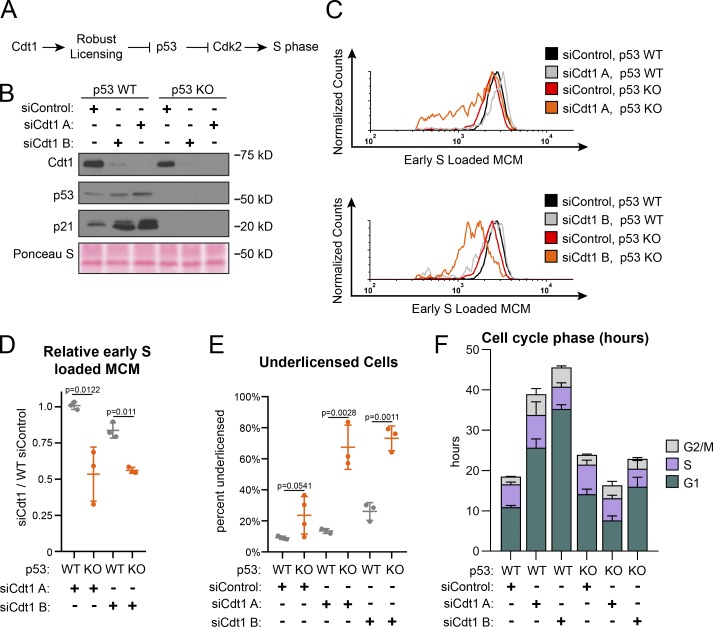
Full origin licensing checkpoint activity in fibroblasts requires p53 (Fig. 4 A; Nevis et al., 2009). To test if p53-deficient epithelial cells are also more likely to enter S phase underlicensed, we compared isogenic WT and p53 homozygous null RPE1 (Rodriguez-Rodriguez et al., 2018). We first noted by immunoblotting that Cdt1 depletion induced accumulation of both p53 and the CDK2 inhibitor p21, the product of a p53-inducible gene, whereas p53 null cells lacked both p53 and detectable levels of p21 (Fig. 4 B). The absence of p53 had little effect on G1 phase length, MCM loading during G1 (Fig. 4 F and Fig. S4, C and D, respectively), or the amount of MCM loaded by early S phase in otherwise unperturbed cells (compare black and gray lines in Fig. 4 C, relative loading in Fig. 4 D, and percentage of underlicensed cells in Fig. 4 E). As before, Cdt1 depletion slowed MCM loading in WT cells and increased G1 length by several-fold (Fig. 4 F and Fig. S3 D). In contrast, Cdt1-depleted cells lacking p53 entered S phase with significantly less MCM loaded (compare red and orange lines in Fig. 4 C, relative loading in Fig. 4 D, and percentage of underlicensed cells in Fig. 4 E). The p21 increase in Fig. 4 B is likely attributable to the increased G1 and G2 phase percentages at the expense of S phase, and p21 is actively degraded in S phase (Abbas and Dutta, 2011). A secondary DNA damage response could also have been triggered. Moreover, unlike p53 WT cells, Cdt1 depletion in p53 null cells did not cause G1 lengthening (Fig. 4 F). Thus, loss of p53 cripples the origin licensing checkpoint in proliferating cells, allowing premature S phase entry of underlicensed cells. We note that even in control cells that had not been depleted of Cdt1, the p53 null cells entered S phase at a slightly lower amount of loaded MCM on average compared with p53 WT cells (compare black and red lines in Fig. 4 C). We also detected a modest increase in the percentage of underlicensed p53 null siControl cells (Fig. 4 E). Overall we conclude that a p53-dependent checkpoint couples S phase entry to the status of origin licensing.
Figure 4.
p53 loss cripples the licensing checkpoint and promotes underlicensing. (A) Model. Cdt1 promotes robust origin licensing, blocking p53-dependent inhibition of CDK2 which delays S phase entry. (B) Immunoblot of total protein lysates of RPE1 p53 WT or p53 null, KO cells treated with siControl, siCdt1 A, or siCdt1 B for 72 h. (C) Loaded MCM in early S phase determined by flow cytometric analysis as in Fig. 1 E of cells treated as in B. The black siControl + p53 WT and red siControl + p53 KO are the same data on both histograms. (D) Relative early S phase DNA-loaded MCM per cell from C. Values are the ratio of mean loaded MCM per cell in Cdt1-depleted versus control cells. Left to right: 1: WT siCdt1 A/WT siControl. 2: KO siCdt1 A/WT siControl. 3: WT siCdt1 B/WT siControl. 4: KO siCdt1 B/WT siControl. n = 3 or 4, mean with SD. (E) Percentage of underlicensed cells in C. n = 3 or 4, mean with SD. (F) Mean cell cycle phase length in hours in C. n = 3 or 4, mean with SD.
The first G1 phase after G0 has an impaired origin licensing checkpoint
We established that cells re-entering the first cell cycle after G0 are underlicensed relative to subsequent cycles. We hypothesized that the first cell cycle has an impaired origin licensing checkpoint that poorly couples the length of G1 phase to the status of MCM loading. To test that hypothesis, we compared actively proliferating cells treated with siCdt1 to G0 cells re-entering the first cycle treated with siCdt1 (Fig. 5 A). We measured cell cycle phase distribution by DNA content (DAPI) and DNA synthesis (EdU; Fig. 5 B). The actively proliferating cells treated with siCdt1 increased the percentage of G1 cells as expected. However, cells re-entering the first cell cycle from G0 while treated with siCdt1 did not extend G1 and entered S phase to the same degree as siControl-treated cells 24 h after G0 release (Fig. 5 B). This difference is consistent with an impaired licensing checkpoint in the first cell cycle. We also tested if the loss of p53 in the first cell cycle enhanced this apparent checkpoint deficiency (Fig. 5 C). Unlike the proliferating cells in Fig. 4 F, first cell cycle p53 null cells were no worse than first cell cycle WT cells (Fig. 5 D). We note that siControl p53 null cells re-entering the first cycle also started S phase sooner than their corresponding p53 WT cells based on cell cycle distributions at the same time after G0 release (Fig. 5, B and D). The faster S phase entry by the p53 null cells could be due to both the impaired licensing checkpoint and the general loss of basal p21 protein, among other possible p53-dependent effects on G1/S progression (Overton et al., 2014; Fig. 5 C). The ability of actively proliferating cells to rely on the licensing checkpoint to extend G1 combined with the observation that cells re-entering the first cycle do not extend G1 but instead enter S phase underlicensed strongly suggest that cells in the first cell cycle after G0 have an impaired origin licensing checkpoint.
Figure 5.
Cells re-entering the first G1 after G0 lack a licensing checkpoint-induced G1 delay. (A) Immunoblot of total protein lysates of cells released from G0 into the first cell cycle with siControl or siCdt1 for 24 h and of proliferating cells treated with siControl or siCdt1 for 72 h. (B) Mean cell cycle phase distribution of cells treated as A defined by flow cytometry (DAPI and EdU). n = 3, mean with SD. (C) Immunoblot of total protein lysate of RPE1 WT and p53 KO synchronized in G0 or released into the first cell cycle with siControl or siCdt1 for 24 h as indicated. (D) Cell cycle phase distribution of cells treated as in C defined by flow cytometry. n = 3, mean with SD.
G0 cells re-entering the first cell cycle load MCM to license origins slowly
We considered two explanations for underlicensing after a long G1 in the first S phase: Cells may start loading MCM later in the first G1 and have less time to load than cells in the second cycle. Alternatively, cells in the first cycle may load MCM more slowly than cells in the second cycle. In both cases, the compromised licensing checkpoint is unable to extend G1, resulting in an underlicensed first S phase. To distinguish between those explanations, we measured the nuclear accumulation of the cell division cycle 6 (Cdc6) protein, an essential MCM loading protein. Cdc6 is degraded by the anaphase-promoting complex-Cdh1 (APCCdh1) in G1 phase, both in cells re-entering the cell cycle and in proliferating cells (Petersen et al., 2000; Mailand and Diffley, 2005). Cyclin E/CDK2 phosphorylates Cdc6 in late G1 to protect it from APCCdh1, allowing Cdc6 protein to accumulate (Mailand and Diffley, 2005). Because Cdc6 is essential for MCM loading, Cdc6 accumulation is one of the limiting steps for MCM loading in G1. The time for MCM loading ends when S phase starts (Petersen et al., 1999; Truong and Wu, 2011). Therefore, the time Cdc6 is detectable in nuclei is one proxy for the length of available MCM loading time in G1 phase.
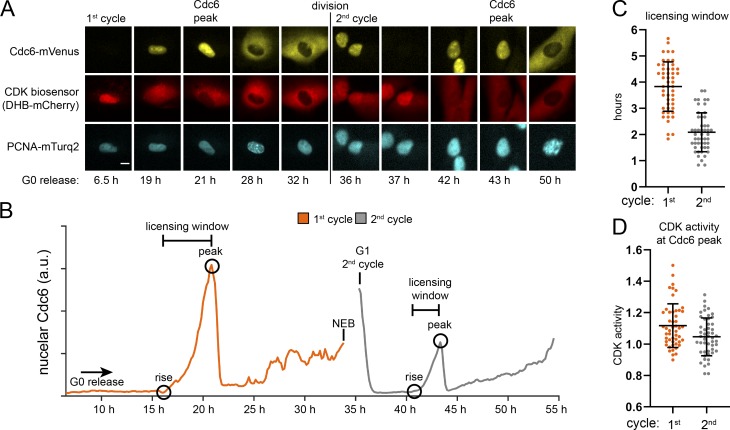
We used live cell imaging of fluorescently tagged proteins to compare the maximum length of available MCM loading time between the first and second cell cycles after G0. We imaged RPE1s stably expressing three fluorescent fusion proteins: (1) full-length Cdc6 fused to mVenus (Segev et al., 2016), (2) proliferating cell nuclear antigen (PCNA) fused to mTurq2 to track cell nuclei and the borders of S phase (Burgess et al., 2012; Grant et al., 2018), and (3) a CDK kinase activity sensor fused to mCherry (Hahn et al., 2009; Spencer et al., 2013; Schwarz et al., 2018). CDK2 phosphorylates the reporter beginning in late G1 phase to induce export from the nucleus to the cytoplasm. The higher the ratio of cytoplasmic/nuclear signal, the higher the kinase activity. We imaged these RPE1 cells synchronized in G0 and released into the cell cycle for 72 h, capturing an image every 10 min, starting 6.5 h after release. PCNA-mTurq2 was present throughout the cell cycle and become punctate during S phase (Fig. 6 A). Nuclear Cdc6 first appeared in G1 and increased until S phase, when it was lost from the nuclei and accumulated in the cytoplasm. After mitosis, Cdc6 was degraded in early G1 phase; then nuclear Cdc6 increased again later in the second G1 (Fig. 6 A). The CDK reporter (DHB-mCherry) was nuclear in G0 (low CDK activity) and gradually became cytoplasmic beginning in late G1 (high CDK activity; Fig. 6 A). We tracked 50 cells through the first and second cell cycles after G0 release. Fig. 6 B shows an individual cell trace for nuclear Cdc6 in which the first full cell cycle took nearly 35 h, whereas the second cycle took only 20 h, mostly from the difference in G1 length. The nuclear Cdc6 trace for the first cycle ends at nuclear envelope breakdown (NEB), and the second cycle trace begins in G1 (Fig. 6 B). The time Cdc6 first appeared in G1 is marked “rise,” and the time of maximum nuclear Cdc6 in G1 is marked “peak” (Fig. 6 B). Cells began S phase as measured by the first appearance of PCNA foci ∼20–30 min after this peak (data not shown). The length of time Cdc6 was present in G1 is the time between nuclear Cdc6 rise and peak, and represents the maximum “licensing window” for MCM loading in G1 (Fig. 6 B). Cells re-entering the first cell cycle after G0 had about double the licensing window of cells in the second cell cycle (Fig. 6 C). Cells in the first G1 have more potential time to load MCM and yet enter S phase with less loaded MCM than cells in the second cycle. Taken together, these data suggest that cells in the first G1 after G0 load MCM slowly.
Figure 6.
Cdc6 dynamics in the first and second cycles after G0. (A) RPE1 cells expressing Cdc6-mVenus, PCNA mTurq2, and a CDK activity sensor (DHB-mCherry) were synchronized in G0 by contact inhibition and release. Images were captured every 10 min for two cell cycles, and selected frames from one of 50 cells are shown. The scale bar is 10 µm and applies to all images. Images brightness/contrast adjusted. (B) An individual cell trace of mean nuclear Cdc6 intensity from A. Hours indicate hours from G0 release beginning at 6.5 h after release. “Rise” is the first appearance of nuclear Cdc6; “peak” is maximum nuclear Cdc6 before S phase and cytoplasmic translocation. The “licensing window” is the difference between peak and rise. NEB indicates NEB in mitosis, no nuclear Cdc6 quantification. (C) Quantification of licensing window times (Cdc6 peak time minus Cdc6 rise time) for cells imaged in A. n = 50, mean with SD. (D) CDK activity. Ratio of mean cytoplasmic DHB-mCherry divided by mean nuclear DHB-mCherry at the time of Cdc6 peak for cells imaged in A. n = 50, mean with SD.
Cells in both the first and second cell cycles also reached a similar CDK activity (cytoplasmic/nuclear ratio >1) at the time of peak nuclear Cdc6 and S phase entry (Fig. 6 D) close to previously reported values of 0.84–1.0 in other untransformed cell lines (Spencer et al., 2013; Schwarz et al., 2018). In other words, the underlicensed first G1 cells achieved the same level of CDK activity in late G1 as normally licensed second G1 cells. These equivalent CDK activity indicators suggest that the licensing checkpoint does not delay CDK2 activation in the first G1.
Extending the first G1 after G0 substitutes for the impaired licensing checkpoint
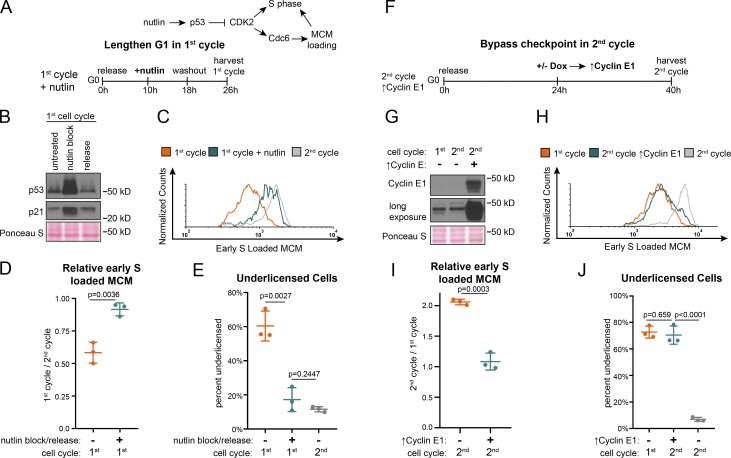
Cells re-entering the cell cycle from G0 license origins slowly and begin S phase underlicensed. We hypothesized that artificially extending the first G1 phase could rescue the underlicensing by extending time for MCM loading. If successful, cells would enter S phase with the high amount of loaded MCM typical of proliferating cells. We chose to extend G1 phase using nutlin-3a, a p53-stabilizing drug previously shown to lengthen G1 (Tovar et al., 2006). We treated cells re-entering the first G1 phase with nutlin-3a beginning in mid-G1 for 8 h, then washed off the drug to permit passage into S phase and measured the amount of loaded MCM at S phase entry (Fig. 7, A and B). Nutlin-3a stabilized p53, causing p21 protein accumulation. This effect is known to inhibit CDK2 kinase activity and delay S phase entry (Giono and Manfredi, 2007). A secondary effect of low CDK2 activity is failure to protect Cdc6 in late G1 phase, which would prevent MCM loading. To allow MCM loading during the nutlin-induced G1 arrest, we constitutively expressed a mutant form of Cdc6 that does not require CDK2 activity for stability (Matson et al., 2017). We then compared licensing and cell cycle progression in these treated cells to untreated cells from the first and second cell cycles (Fig. 7 C and Fig. S5 A). Strikingly, transiently extending the first G1 phase by several hours almost fully rescued licensing in the first cell cycle to the same high level as in the second cell cycle (compare green and gray lines, Fig. 7 C, and fold change, Fig. 7 D) and decreased the percentage of underlicensed cells to the same low level as the second cell cycle (Fig. 7 E). Unlike the effects of stable Cdc6 on the rate of MCM loading in proliferating cells, neither stable Cdc6 alone nor in combination with Cdt1 overproduction increased the rate of MCM loading or changed underlicensing in the first G1 phase (Fig. S5, B–F; Matson et al., 2017). Therefore, the improved licensing by early S phase from nutlin-induced G1 extension is due to the increased time for MCM loading, rather than improving MCM loading itself.
Figure 7.
G1 lengths of the first and second cell cycles impacts licensing at S phase entry. (A) Plan. RPE1 constitutively producing 5myc-Cdc6-mut (resistant to APCCDH1) was synchronized in G0 by contact inhibition, then released into the first cell cycle. Cells were treated with 10 µM nutlin-3a 10 h after release, followed by nutlin washout at 18 h, and then harvesting of cells at 26 h. Untreated cells were harvested at 24 h (first cell cycle) and 48 h (second cell cycle) after release from G0. (B) Immunoblot of total protein lysate from cells treated according to A with the exception that untreated is 18 h after G0 without nutlin-3a to examine cells in G1 (Fig. S2 G). (C) Loaded MCM in early S phase determined by flow cytometric analysis as in Fig. 1 E from cells treated as in A. See also Fig. S5 A. (D) Relative mean MCM loaded in early S phase in the first cycle divided by the second cycle for cells in C as indicated. n = 3, mean with SD. (E) Percentage of underlicensed cells in C. n = 3, mean with SD. (F) Plan. RPE1 cells with inducible cyclin E1 (as in Fig. 3) were synchronized in G0 by contact inhibition, then released adding 100 ng/ml of doxycycline at 24 h to overproduce cyclin E1 and shorten G1 of the second cell cycle; cells were harvested at 40 h. Untreated cells were harvested at 24 h (first cell cycle) and 40 h (second cell cycle) after release from G0. (G) Immunoblot of total protein lysate from cells treated according to F. (H) Loaded MCM in early S phase determined by flow cytometry as in Fig. 1 E of cells treated according to F. See also Fig. S5 G. (I) Relative mean MCM loaded in early S phase in the first cycle divided by the second cycle for cells in F as indicated. n = 3, mean with SD. (J) Percentage of underlicensed cells in F. n = 3, mean with SD.
Finally, we predicted that since the second cell cycle has a robust origin licensing checkpoint, bypassing the checkpoint and artificially shortening the second G1 would induce a phenotype similar to the underlicensed first cell cycle. To test this prediction, we overproduced cyclin E1 as cells approached the second cell cycle to shorten the second G1 phase and bypass the checkpoint. We then compared licensing in the second S phase in these cyclin E1–overproducing cells to control second S phase and first S phase cells (Fig. 7, F and G; and Fig. S5 G). The second cell cycle with a bypassed licensing checkpoint strongly resembled the first cell cycle (compare green and orange lines, Fig. 7 H, fold change in Fig. 7 I, and percentage of underlicensed cells in Fig. 7 J). Taken together, we conclude that cells re-entering the cell cycle from G0 are underlicensed, and the second and subsequent cell cycles are fully licensed because of a robust p53-dependent checkpoint that ultimately controls the timing of cyclin E/CDK2-mediated S phase entry.
Discussion
The origin licensing checkpoint protects cells from premature S phase entry that could lead to genome instability (Fig. 8). This checkpoint couples the activity of CDK to MCM loading such that the G1 CDKs are not activated while the amount of loaded MCM is low. Cyclin E1 overproduction can bypass the checkpoint-induced delay and induce an underlicensed premature S phase entry (Fig. 3; Ekholm-Reed et al., 2004; Matson et al., 2017). Moreover, p53 loss clearly impairs the normal coupling of MCM loading and S phase entry, whereas p53 activation lengthens G1 to give enough time to complete licensing (Figs. 4, 5, and 7). These observations fit the classic definition by Hartwell and Weinert (1989) for a cell cycle checkpoint. A checkpoint enforces the dependence of one event on a previous event, but the dependency can be bypassed by mutation to occur prematurely. The ability of mutations to induce inappropriate cell cycle progression indicates that in WT cells progression could have occurred but was instead restrained by the checkpoint. This relationship is in contrast to a simple inability to progress. Importantly, we induced S phase entry when the amount of loaded MCM was low but not completely absent. If we could have entirely prevented MCM loading, there would have been no DNA synthesis even in p53 null or cyclin E–overproducing cells because MCM is an essential component of the replicative helicase (Lemmens et al., 2018).
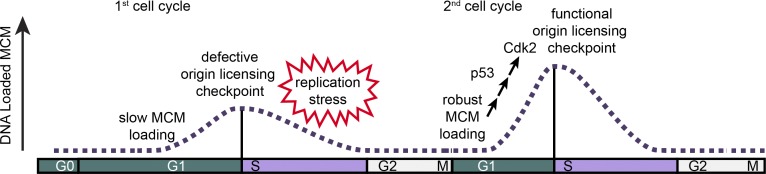
Figure 8.
Model of MCM loading in first and second cycles after G0. Cells in the first cell cycle have slow MCM loading and an impaired origin licensing checkpoint. These defects cause underlicensing and make the first S phase hypersensitive to replication stress. Cells in the second cycle have a functional origin licensing checkpoint and load MCM normally. The checkpoint is p53 dependent, and robust MCM loading promotes CDK2 activation and normal S phase entry.
We note that licensing checkpoint activity is only readily detected in cells with slow MCM loading. In previous studies, MCM loading was artificially slowed to test the checkpoint (McIntosh and Blow, 2012). Normally however, both checkpoint-proficient and -impaired proliferating cells enter S phase with high levels of MCM loading. We were surprised to find that the first cell cycle after re-entry from G0 is naturally underlicensed, due to a combination of slow MCM loading and an impaired licensing checkpoint. It is unclear why cells re-entering G0 load MCM slowly, since overexpressing MCM loading proteins Cdt1 and Cdc6 together did not increase MCM loading rate (Fig. S5, B–F). It is striking, however, that both the checkpoint defect and slow MCM loading are restored just one cell cycle later. This behavior suggests that MCM loading is usually fast enough to achieve high levels of loaded MCM in advance of the other rate-limiting events necessary to trigger CDK2 activation and S phase entry. It is only slow-loading cells that are affected by the checkpoint. In that sense, the origin licensing checkpoint resembles other cell cycle checkpoints that operate in all cell cycles in that “elimination of the checkpoint may have catastrophic or subtle consequences depending on the prevailing conditions” (Hartwell and Weinert, 1989). For example, the mitotic spindle assembly checkpoint is most obvious when attachments are severely compromised (Lara-Gonzalez et al., 2012). The effect of the spindle assembly checkpoint on the rate of unperturbed mitotic progression is relatively minor except in individual cells that fail to form kinetochore–microtubule attachments on time (Meraldi et al., 2004). Similarly, we detected a modest increase in the number of underlicensed S phase cells in otherwise unperturbed proliferating p53 null cells compared with p53 WT cells (Fig. 4). We infer that these individual cells loaded MCM slowly, but then could not delay S phase entry.
Previous analysis by Daigh et al. (2018) indicated that cells released from G0 had increased endogenous replication stress in the first S phase compared with proliferating cells. We propose that the underlicensed S phase after the first G1 has higher endogenous replication stress because fewer dormant origins are available. While we did not detect frank stress markers in the absence of exogenous replication stress, we presume that underlicensing generated endogenous stress because these cells were particularly sensitive to both gemcitabine and etoposide. This hypersensitivity is a hallmark of cells that enter S phase while underlicensed (Woodward et al., 2006).
How frequent is this naturally underlicensed cell cycle in vivo? Cell cycle re-entry from G0 is common in tissues that naturally turn over cell populations (Wells et al., 2013; Sosa et al., 2014). Additionally, recent cell culture studies have detected subpopulations of proliferating cells that appear to spontaneously and transiently exit to a G0-like state (Spencer et al., 2013; Overton et al., 2014). Such transient cell cycle exit may be even more common in tissues than in cell culture that has been optimized for maximal growth. Whether or not that transient G0 also causes underlicensing is unknown.
Quiescent hematopoietic stem cells re-enter the cell cycle in response to stresses such as viral infection and then return to quiescence (Cheung and Rando, 2013). Interestingly, hematopoietic stem cells acquire DNA damage in the first cycle after G0, accumulate DNA damage, and become depleted as they age from repeated cell cycle re-entry (Beerman et al., 2014; Walter et al., 2015). Additionally, quiescent human T cells re-enter the first S phase even when treated with siRNA to reduce the amount of loaded MCM to ∼5–10% of their normal loaded amount (Orr et al., 2010). Taken together, these data suggest that at least some quiescent cells in vivo lack a licensing checkpoint in the first cycle, although it is unknown if the first cell cycle is naturally underlicensed compared with the second cycle in vivo. Old hematopoietic cells experience more replication stress in the first S phase after G0 compared with young cells. In that regard, a defective licensing checkpoint would be especially toxic to old quiescent cells re-entering G1 (Flach et al., 2014). These in vivo studies are consistent with our observation that repeated rounds of quiescence and cell cycle re-entry enhanced replication stress sensitivity (Fig. S3). We suggest that over many rounds of quiescence and re-entry, incremental DNA damage accrues from unresolved replication stress or incomplete replication that may pass through mitosis into the next cell generation. Unresolved replication stress can carry forward to future cell cycles, and previous work shows that reducing loaded MCM by 50% (similar to the first cycle from G0) increases the amount of unresolved damage carrying over to following cell cycles (Ahuja et al., 2016; Moreno et al., 2016; Arora et al., 2017; Yang et al., 2017). Other effects intrinsic to G0 could also accumulate over multiple G0s, including attenuated expression of DNA repair genes, error-prone repair, and reactive oxygen species damage (Mohrin et al., 2010; Beerman et al., 2014). Repeated rounds of underlicensed cell cycle re-entry could contribute to the genome damage that drives both aging and oncogenesis.
Materials and methods
Cell culture and synchronization
RPE1-hTERT (i.e., RPE1; CRL-4000; ATCC), 293T (CRL3216; ATCC), NHF1-hTERT (Boyer et al., 1991; Heffernan et al., 2002), and RPE1-hTERT p53 CRISPR null (gift from P. Jallepalli, Memorial Sloan Kettering Cancer Center, New York, NY; gRNA target: GGCAGCTACGGTTTCCGTC) cells were grown in DMEM (Sigma-Aldrich) with 10% FBS (Seradigm) and 2 mM L-glutamine (Gibco) at 37°C with 5% CO2. Wi38 cells (CCL-75; ATCC) were grown in MEM with 10% FBS (Seradigm), 2 mM L-glutamine (Gibco), and MEM nonessential amino acids (NEAA; Gibco) at 37°C with 5% CO2. Cells were passaged every 3 d with trypsin (Sigma-Aldrich) and not allowed to reach confluency except where intentionally arrested.
To synchronize RPE1 cells in G0 by contact inhibition, cells were grown to 100% confluency, washed with PBS, and incubated in DMEM with 10% FBS and 2 mM L-glutamine for 48 h. To release cells into the first or second cell cycles, G0 cells were restimulated by passaging 1:10 for the first cycle, harvesting 24 h later, or 1:20 for the second cycle, harvesting 48 h later, in DMEM with 10% FBS and 2 mM L-glutamine. To synchronize RPE1 cells in G0 by serum starvation, cells were plated to be sub-confluent, washed three times with PBS, and starved in DMEM with 0% FBS and 2 mM L-glutamine for 72 h. To release cells into the first or second cell cycles, cells were washed with PBS and restimulated by washing once with PBS and adding DMEM with 10% FBS and 2 mM L-glutamine for 24 h (first cycle) or 48 h (second cycle). To synchronize NHF1-hTERT cells in G0 by contact inhibition, cells were grown to 100% confluency, washed with PBS, and incubated in DMEM with 0.1% FBS and 2 mM L-glutamine for 72 h. To release cells into the first or second cell cycles, G0 cells were restimulated by passaging 1:4 for the first cycle, harvesting 24 h later, or 1:8 for the second cycle, harvesting 48 h later, in DMEM with 10% FBS and 2 mM L-glutamine. To synchronize NHF1 cells in G0 by serum starvation, cells were plated to be subconfluent, washed three times with PBS, and starved in DMEM with 0.1% FBS and 2 mM L-glutamine for 72 h. To release cells into the first or second cell cycles, cells were washed with PBS and restimulated by washing once with PBS and adding DMEM with 10% FBS and 2 mM L-glutamine for 24 h (first cycle) or 48 h (second cycle). To synchronize Wi38 cells in G0, cells were grown to 100% confluency, washed with PBS, and incubated in MEM with 0.1% FBS, 2 mM L-glutamine, and NEAA for 72 h. To release cells into the first or second cell cycles, G0 cells were restimulated by passaging 1:4 for the first cycle, harvesting 24 h later, or 1:8 for the second cycle, harvesting 48 h later, in MEM with 10% FBS, 2 mM L-glutamine, and NEAA. To synchronize Wi38 cells in G0 by serum starvation, cells were plated to be sub-confluent, washed three times with PBS, and starved in MEM with 0.1% FBS, NEAA, and 2 mM L-glutamine for 72 h. To release cells into the first or second cell cycles, cells were washed with PBS and restimulated by washing once with PBS and adding MEM with 10% FBS, NEAA, and 2 mM L-glutamine for 24 h (first cycle) or 48 h (second cycle).
To synchronize RPE1 in three repeated G0s, cells were synchronized in G0 by contact inhibition (above), restimulated into the cell cycle by passaging 1:6 in DMEM with 10% FBS and 2 mM L-glutamine, and grown to 100% confluency within 48–72 h to start the second G0 by contact inhibition for 48 h. Cells were restimulated as before and grown to 100% confluency to start the third G0 for 48 h. To release cells for the experiment, G0 cells were restimulated by passaging 1:10 in DMEM with 10% FBS and 2 mM L-glutamine, harvesting the first cell cycle at 24 h after restimulation.
For the passage number experiment in Fig. S3, cells were passaged every 3 d. Passage 9 cells (counting from thawing the cells from frozen stocks) were grown for 24 d more than passage 1 cells.
DNA cloning and cell lines
The RPE1 CRISPR p53 knockout (KO) was a gift of P. Jallepalli and described in the section Cell culture and synchronization (Rodriguez-Rodriguez et al., 2018). The polyclonal RPE1 cells with doxycycline-inducible cyclin E1 and the RPE1 cells with doxycycline-inducible Cdt1 and stable Cdc6 WT or Cdc6-mut were described previously. The plasmids are available on Addgene (109332, 109333, 109335, and 109348; Matson et al., 2017). The pLenti-PGK hygro PCNA-mTurq2 was described previously and is available on Addgene (118617; Grant et al., 2018). The CSII-EF zeo DHB-mCherry CDK activity reporter was a gift from S. Spencer (University of Colorado-Boulder, Boulder, CO) and encodes retrovirus packaging sequences to package a DNA helicase B fragment fused to mCherry for expression under the EF1 promoter. The CSII-EF zeo Cdc6-mVenus was a gift from M. Brandeis (The Hebrew University of Jerusalem, Safra Campus, Jerusalem, Israel) and encodes retrovirus packaging sequences to package Cdc6 WT fused to mVenus for expression under the EF1 promoter. Each reporter is under control of a constitutive heterologous promoter.
To make the RPE1 line containing PCNA-mTurq2, Cdc6-mVenus, and DHB-mCherry, the plasmids were transfected into 293T with pCI-GPZ or ΔNRF and Vesicular stomatitis virus G packaging plasmids with 50 μg/ml polyethylenimine (Aldrich Chemistry). Viral supernatants were transduced onto RPE1 cells in the presence of 8 µg/ml polybrene (Millipore). A clonal cell line was picked based on fluorescence of all three biosensors.
siRNA transfections and drug treatment
siRNA concentration and sequences were as follows:
siControl (siLuciferase), 100 nM (synthesized by Life Technologies), 5′-CUUACGCUGAGUACUUCGA-3′; siCdt1 A, mixture of four sequences, 25 nM each (siGENOME CDT1 siRNA; Dharmacon); 5′-CCAAGGAGGCACAGAAGCA-3′; 5′-GCUUCAACGUGGAUGAAGU-3′; 5′-UCUCCGGGCCAGAAGAUAA-3′; 5′-GGACAUGAUGCGUAGGCGU-3′; siCdt1 B, 50 nM (synthesized by Life Technologies); 5′-CCUACGUCAAGCUGGACAATT-3′.
For siRNA transfections, siRNA was spotted into plates with DharmaFECT 4 transfection reagent (Dharmacon) and Opti-MEM (Gibco), incubating for 20 min before adding cells in DMEM with 10% FBS and 2 mM L-glutamine (final concentrations), harvesting cells 72 h later for proliferating experiments, and 24 h later for G0 release, as described above. To overproduce cyclin E1 or Cdt1-HA, 100 ng/ml of doxycycline (CalBiochem) was added at times indicated in the legends of Figs. 3, 7, S4, and S5. To block and release cells with nutlin-3a (Sigma-Aldrich), RPE1 was synchronized in G0 by contact inhibition and released into the first cell cycle, adding 10 µM nutlin-3a 10 h after release, washing off nutlin-3a 18 h after release by washing three times with PBS and adding back DMEM with 10% FBS and 2 mM L-glutamine. To treat cells with gemcitabine (Sigma-Aldrich) or etoposide (Cayman Chemical) in Fig. 2, cells were synchronized in G0 by contact inhibition, released into the first cycle, and treated with 50 or 100 nM gemcitabine from 22 to 24 h or released into the second cycle and treated with 50 or 100 nM gemcitabine from 44 to 46 h. See legends of Fig. 2 and Fig. S3 for concentrations. Etoposide treatment was the same as gemcitabine, with 0.5 µM or 5 µM etoposide. See legends of Fig. 2 and Fig. S3 for concentrations. To treat cells with gemcitabine in Fig. S3, cells were synchronized in one or three G0 (above), adding 5 nM gemcitabine at the time of restimulation from 0 to 24 h.
Total protein lysate and chromatin fractionation
To prepare total protein lysate for immunoblot, cells were harvested with trypsin and frozen in dry ice, and then lysed in cold cytoskeletal buffer (CSK; 10 mM Pipes, pH 7.0, 300 mM sucrose, 100 mM NaCl, and 3 mM MgCl2 hexahydrate) with 0.5% Triton X-100 (Sigma-Aldrich) and protease and phosphatase inhibitors (0.1 mM Pefabloc, 1 µg/ml pepstatin A, 1 µg/ml leupeptin, 1 µg/ml aprotinin, 10 µg/ml phosvitin, 1 mM β-glycerol phosphate, and 1 mM sodium orthovanadate) on ice for 15 min. The lysate was centrifuged at 13,000 g at 4°C for 10 min, and a Bradford assay (Biorad) was done on the supernatant to load equal amounts of protein per sample.
To prepare chromatin fractions for immunoblot, cells were harvested with trypsin and frozen in dry ice, then lysed in cold CSK with 0.5% Triton X-100, 1 mM ATP, 5 mM CaCl2, and protease and phosphatase inhibitors (complete CSK) on ice for 20 min. Then a Bradford assay for equal loading was done on the lysate, a small aliquot was removed from each sample as total lysate, and complete CSK was added to each sample, mixed, and centrifuged for 5 min, 1,000 g. The supernatant was removed, and the pellet was washed again with complete CSK, incubated for 10 min on ice, and then centrifuged for 5 min, 1,000 g. The supernatant was removed, and DNA loaded proteins were released by incubation with S7 nuclease (Sigma-Aldrich) in complete CSK at RT for 10 min. Samples were centrifuged again, keeping the supernatant as the chromatin fraction.
Immunoblotting
Samples were diluted with loading buffer to final concentrations: 1% SDS, 2.5% 2-mercaptoethanol, 0.1% bromophenol blue, 50 mM Tris, pH 6.8, and 10% glycerol; and then boiled. Samples were separated on SDS-PAGE gels and transferred to nitrocellulose (GE Healthcare) or polyvinylidene difluoride membranes (Thermo Fisher Scientific). After transferring, samples were blocked in 5% milk in TBS with 0.1% Tween 20 (TBST) and incubated overnight at 4°C in primary antibody with 2.5% milk in TBST. Then membranes were washed with TBST, incubated in HRP-conjugated secondary antibody for 1 h at RT, washed with TBST, and imaged with ECL Prime (Amersham) on autoradiography film (Denville). Antibodies used were Mcm2 (1:10,000; mouse; 610700; BD Biosciences), cyclin E1 (1:2,000; mouse; 4129S; Cell Signaling Technology), p53 (1:2,000; mouse; sc-126; Santa Cruz Biotechnology), p21 (1:6,000; rabbit; 2947S; Cell Signaling Technology) Cdc6 (1:2,000; mouse; sc-9964; Santa Cruz Biotechnology), Cdt1 (1:3,000; mouse; sc-365305; Santa Cruz Biotechnology), donkey-anti-mouse-HRP (1:10,000; 715-035-150; Jackson ImmunoResearch), and donkey-anti-rabbit-HRP (1:10,000; 711–035-152; Jackson ImmunoResearch). Membranes were stained with Ponceau S (Sigma-Aldrich) to visualize protein loading.
Flow cytometry
For DNA synthesis detected, cells were incubated with 10 µM EdU for 30 min before harvesting, except in Fig. S1 D, when cells were incubated with 1 µM EdU from restimulation to harvesting. Cells were harvested with trypsin to measure DNA loaded proteins by flow cytometry. Cells were washed once with PBS and centrifuged at 2,000 g for 3 min. Then the supernatant was aspirated and pellets lysed on ice in cold CSK with 0.5% Triton X-100 and protease and phosphatase inhibitors for 5 min. After incubation, 1% BSA (Thermo Fisher Scientific) in PBS (B-PBS) was added to each sample, mixed, and centrifuged at 2,000 g for 3 min. The supernatant was aspirated, and pellets were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS for 15 min at RT. B-PBS was then added and mixed, and samples were centrifuged (2,000 g for 7 min, the same centrifuge conditions were used for all subsequent flow cytometry steps). The supernatant was aspirated, B-PBS was added, and samples were stored at 4°C before labeling.
Processing for EdU detection was performed before antibody staining. Cells were centrifuged, the supernatant was aspirated, and cells were incubated in PBS with 1 mM CuSO4, 100 mM ascorbic acid (fresh), and 1 µM Alexa Fluor 647–azide (Life Technologies) for 30 min at RT in the dark. Then B-PBS with 0.5% NP-40 (United States Biochemical) was added, mixed, and centrifuged. For antibody staining, the supernatant was aspirated and cells were incubated in primary antibodies anti-Mcm2 (1:200; mouse; 610700; BD Biosciences), anti-γH2AX phospho-S139 (1:200; rabbit; 9718S; Cell Signaling Technologies), and anti-RPA2 (1:200; mouse; ab2175; Abcam) in B-PBS with 0.5% NP-40 for 1 h at 37°C in the dark. Next, B-PBS with 0.5% NP-40 was added and mixed and samples were centrifuged. The supernatant was aspirated and cells were incubated in secondary antibodies donkey anti-mouse–Alexa Fluor 488 (1:1,000; for Mcm2 or RPA2; 715–545-150; Jackson ImmunoResearch) and donkey anti-rabbit–Alexa Fluor 647 (1:1,000; for γH2AX; 711–605-152; Jackson ImmunoResearch) in B-PBS with 0.5% NP-40 for 1 h at 37°C in the dark. Then B-PBS with 0.5% NP-40 was added and mixed and samples were centrifuged. Finally, the supernatant was aspirated and cells were incubated in 1 µg/ml DAPI (Sigma-Aldrich) and 100 ng/ml RNase A (Sigma-Aldrich) in B-PBS with 0.5% NP-40 overnight at 4°C in the dark. For negative control samples used to draw positive/negative gates, cells were not incubated with EdU but were labeled with Alexa Fluor 647–azide and were not incubated with primary antibody but were labeled with secondary antibody and DAPI.
Data were acquired primarily on an Attune NxT flow cytometer (Thermo Fisher Scientific), with some data acquired on a CyAn ADP flow cytometer (Beckman Coulter). Data were analyzed using FCS Express 6 (De Novo Software). Gates are shown in Fig. S1. The gate to isolate cells from debris was from forward scatter area versus side scatter area. The gate to isolate singlets from doublets was from DAPI area versus DAPI height (parent gate: cells). The gate to isolate cell cycle phases was from DAPI area (DNA content) versus 647 area (DNA synthesis, EdU; parent gate: singlets). Color gates to isolate S-MCMDNA–positive, G1-MCMDNA–positive, and MCMDNA-negative were on 647 area (DNA synthesis, EdU) versus 488 area (loaded MCM), using a negative control sample to mark positive cells (parent gate: singlets). The early S phase gate was on DAPI area (DNA content) versus 488 area (loaded MCM), gating cells with 2C DNA content, S-MCMDNA–positive in early S phase (parent gate: S-MCMDNA–positive). The mid-S phase gate was on DAPI area (DNA content) versus 647 area (γH2AX), cells between 2C and 4C DNA content (parent gate: singlets). Replication stress–induced γH2AX was gated as cells equal to or greater than the top 5–6% of γH2AX signal from untreated cells (parent gate: mid-S). Each flow cytometry plot typically has 9,000–11,000 total single cells. Histogram counts were normalized to the peak value of the second cell cycle or siControl. The normalization allows visual comparison of cell distributions between populations with different numbers of cells due to changes in synchrony in the second cell cycle. The quantification of relative loaded MCM or underlicensed cells was not normalized.
Doubling time and cell cycle phases
Cells were plated with siRNA and counted 48 h or 72 h later after dissociating with trypsin using a Luna II automated cell counter (Logos Biosystems). Each treatment was done with three biological replicates, and each dish in the replicate was counted twice as technical replicates. Doubling time was calculated using Prism 8 (GraphPad) regression analysis: exponential growth equation. Doubling times from the three biological replicates were averaged, and then the average doubling time was multiplied by the cell cycle phase percentages determined by DNA content and DNA synthesis (flow cytometry) to obtain cell cycle phase hours.
Live cell imaging
Cells were plated for live cell imaging from G0 by contact inhibition and restimulation in Flourobrite DMEM (Gibco) with 10% FBS and 2 mM L-glutamine in #1.5 glass bottom plates (Cellvis) in a humidified enclosure at 37°C with 5% CO2. Image collection started 6.5 h after plating, and cells were imaged for 72 h with images collected every 10 min. Cells were imaged on a Nikon Ti Eclipse inverted microscope with a 20× (NA 0.75) Apochromat dry objective lens with the Nikon Perfect Focus System. The camera was an Ando Zyla 4.2 sCMOS detector with 12-bit resolution, and filters were obtained from Chroma (excitation; beam splitter; emission filter): CFP, 436/20 nm, 455 nm, and 480/40 nm; YFP, 500/20 nm, 515 nm, and 535/30 nm; and mCherry, 560/40 nm, 585 nm, and 630/75 nm. Images were collected with Nikon NIS-Elements AR software.
Images were analyzed with Fiji version 1.51n (ImageJ). Briefly, images were background subtracted and tracked with custom ImageJ scripts based on the PCNA signal, and the nuclear signals quantified in a region of interest as described previously (Grant et al., 2018). Images were brightness/contrast adjusted. Cdc6 traces of mean nuclear intensity were scored as follows: Cdc6 peak time, frame with highest nuclear Cdc6 intensity before the sharp drop in intensity indicating export to the cytoplasm; Cdc6 rise time, frame with Cdc6 nuclear intensity two SDs greater than the lowest Cdc6 nuclear intensity before Cdc6 peak time; licensing window time, Cdc6 peak time minus Cdc6 rise time; and relative licensing window time, licensing window time divided by Cdc6 peak time.
CDK biosensor (DHB-mCherry) cytoplasmic measurement was described previously (Chao et al., 2019). Fluorescence signal of the cytoplasm was approximated by measuring signal within a ring-shaped region (5 pixels wide) around the nucleus. The ratio between the cytoplasm and the nucleus was calculated using mean signals of the ring-shaped cytoplasmic region and the full nuclear region.
Statistical analysis
All summary data are plotted as means with error bars showing SDs, with a minimum of three biological replicates for all experiments. All P values were calculated with an unpaired, two-tailed t test.
Online supplemental material
Figs. S1 and S2 define flow cytometry gating and representative raw data for additional cell lines summarized in Fig. 1, G and H. Fig. S3 demonstrates replication stress sensitivity over repeated rounds of G0 and cell cycle re-entry. Fig. S4 is representative complete flow cytometry color dot plots and histograms of G1 MCM loading with siCdt1 treatment. Fig. S5 shows representative flow cytometry color dot plots for the nutlin-3a block and release as well as cyclin E1 overproduction in the second cell cycle. It also includes the minimal effects of Cdc6 and Cdt1 overproduction in the first cell cycle on underlicensing.
Supplementary Material
Acknowledgments
We thank Jeffrey Jones for managerial assistance and the Cook laboratory for helpful discussion. We thank Dr. Sam Wolff, Seraphina Wong, Aimee Littlejohn, and Walli Driggers for support. We thank Dr. Kasia Kedziora for help with imaging analysis. The DHB-mCherry plasmid was a gift from Dr. Sabrina Spencer. The Cdc6-mVenus plasmid was a gift from Dr. Michael Brandeis. The RPE1_hTert p53 CRIPSR null line was a gift from Dr. Prasad Jallepalli.
This work was supported by a fellowship from the National Science Foundation (DGE-1144081) to J.P. Matson, a University of North Carolina Dissertation Completion Fellowship to J.P. Matson, and grants from the National Institutes of Health/National Institute of General Medical Sciences to J.G. Cook (GM083024, GM102413, and R25GM089569; R25GM089569 also supported J. Perez). The University of North Carolina Flow Cytometry Core Facility is supported in part by P30 CA016086.
The authors declare no competing financial interests.
Author contributions: J.P. Matson designed and performed the experiments and analyzed the results. A.M. House tracked cells from live cell imaging and processed data. G.D. Grant tracked cells and ran the microscope. H. Wu performed some immunoblots. J. Perez created the RPE1 Cdc6-mVenus, PCNA-mTurq2, DHB-mCherry cell line. J.G. Cook designed experiments and supervised the project. J.P. Matson and J.G. Cook wrote the manuscript with input from the other authors.
References
- Abbas T., and Dutta A.. 2011. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle. 10:241–249. 10.4161/cc.10.2.14530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja A.K., Jodkowska K., Teloni F., Bizard A.H., Zellweger R., Herrador R., Ortega S., Hickson I.D., Altmeyer M., Mendez J., and Lopes M.. 2016. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 7:10660 10.1038/ncomms10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez S., Díaz M., Flach J., Rodriguez-Acebes S., López-Contreras A.J., Martínez D., Cañamero M., Fernández-Capetillo O., Isern J., Passegué E., and Méndez J.. 2015. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nat. Commun. 6:8548 10.1038/ncomms9548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alver R.C., Chadha G.S., and Blow J.J.. 2014. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA Repair (Amst.). 19:182–189. 10.1016/j.dnarep.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E.E., and Walter J.C.. 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21:497–518. 10.1101/gad.1508907 [DOI] [PubMed] [Google Scholar]
- Arora M., Moser J., Phadke H., Basha A.A., and Spencer S.L.. 2017. Endogenous Replication Stress in Mother Cells Leads to Quiescence of Daughter Cells. Cell Reports. 19:1351–1364. 10.1016/j.celrep.2017.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I., Seita J., Inlay M.A., Weissman I.L., and Rossi D.J.. 2014. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 15:37–50. 10.1016/j.stem.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P., and Labib K.. 2016. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 203:1027–1067. 10.1534/genetics.115.186452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J.C., Kaufmann W.K., and Cordeiro-Stone M.. 1991. Role of postreplication repair in transformation of human fibroblasts to anchorage independence. Cancer Res. 51:2960–2964. [PubMed] [Google Scholar]
- Burgess A., Lorca T., and Castro A.. 2012. Quantitative live imaging of endogenous DNA replication in mammalian cells. PLoS One. 7:e45726 10.1371/journal.pone.0045726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao H.X., Fakhreddin R.I., Shimerov H.K., Kedziora K.M., Kumar R.J., Perez J., Limas J.C., Grant G.D., Cook J.G., Gupta G.P., and Purvis J.E.. 2019. Evidence that the human cell cycle is a series of uncoupled, memoryless phases. Mol. Syst. Biol. 15:e8604 10.15252/msb.20188604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung T.H., and Rando T.A.. 2013. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 14:329–340. 10.1038/nrm3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller H.A. 2007. What’s taking so long? S-phase entry from quiescence versus proliferation. Nat. Rev. Mol. Cell Biol. 8:667–670. 10.1038/nrm2223 [DOI] [PubMed] [Google Scholar]
- Coller H.A., Sang L., and Roberts J.M.. 2006. A new description of cellular quiescence. PLoS Biol. 4:e83 10.1371/journal.pbio.0040083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigh L.H., Liu C., Chung M., Cimprich K.A., and Meyer T.. 2018. Stochastic Endogenous Replication Stress Causes ATR-Triggered Fluctuations in CDK2 Activity that Dynamically Adjust Global DNA Synthesis Rates. Cell Syst. 7:17–27.e3. 10.1016/j.cels.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekholm-Reed S., Méndez J., Tedesco D., Zetterberg A., Stillman B., and Reed S.I.. 2004. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell Biol. 165:789–800. 10.1083/jcb.200404092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald B., Sampath D., and Plunkett W.. 2007. H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol. Cancer Ther. 6:1239–1248. 10.1158/1535-7163.MCT-06-0633 [DOI] [PubMed] [Google Scholar]
- Flach J., Bakker S.T., Mohrin M., Conroy P.C., Pietras E.M., Reynaud D., Alvarez S., Diolaiti M.E., Ugarte F., Forsberg E.C., et al. . 2014. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 512:198–202. 10.1038/nature13619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Q., Jackson D.A., and Blow J.J.. 2007. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 21:3331–3341. 10.1101/gad.457807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C., and Giordano A.. 2006. RB and cell cycle progression. Oncogene. 25:5220–5227. 10.1038/sj.onc.1209615 [DOI] [PubMed] [Google Scholar]
- Giono L.E., and Manfredi J.J.. 2007. Mdm2 is required for inhibition of Cdk2 activity by p21, thereby contributing to p53-dependent cell cycle arrest. Mol. Cell. Biol. 27:4166–4178. 10.1128/MCB.01967-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant G.D., Kedziora K.M., Limas J.C., Cook J.G., and Purvis J.E.. 2018. Accurate delineation of cell cycle phase transitions in living cells with PIP-FUCCI. Cell Cycle. 17:2496–2516. 10.1080/15384101.2018.1547001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A.T., Jones J.T., and Meyer T.. 2009. Quantitative analysis of cell cycle phase durations and PC12 differentiation using fluorescent biosensors. Cell Cycle. 8:1044–1052. 10.4161/cc.8.7.8042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håland T.W., Boye E., Stokke T., Grallert B., and Syljuåsen R.G.. 2015. Simultaneous measurement of passage through the restriction point and MCM loading in single cells. Nucleic Acids Res. 43:e150 10.1093/nar/gkv744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., and Weinert T.A.. 1989. Checkpoints : Controls that Ensure the Order of Cell Cycle Events. Science. 246:629–634. [DOI] [PubMed] [Google Scholar]
- Heffernan T.P., Simpson D.A., Frank A.R., Heinloth A.N., Paules R.S., Cordeiro-Stone M., and Kaufmann W.K.. 2002. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol. Cell. Biol. 22:8552–8561. 10.1128/MCB.22.24.8552-8561.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A., Schwob E., and Méndez J.. 2008. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. USA. 105:8956–8961. 10.1073/pnas.0803978105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers M.A., Stasevich T.J., Sasaki T., Wilson K.A., Hazelwood K.L., McNally J.G., Davidson M.W., and Gilbert D.M.. 2011. Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J. Cell Biol. 192:29–41. 10.1083/jcb.201007111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnev D., Rusiniak M.E., Kudla A., Freeland A., Cady G.K., and Pruitt S.C.. 2010. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene. 29:3630–3638. 10.1038/onc.2010.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J.S., Everetts N.J., Wang X., Wang W., Della Croce K., Xing J., and Yao G.. 2017. Controlling Depth of Cellular Quiescence by an Rb-E2F Network Switch. Cell Reports. 20:3223–3235. 10.1016/j.celrep.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P., Westhorpe F.G., and Taylor S.S.. 2012. The spindle assembly checkpoint. Curr. Biol. 22:R966–R980. 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Lemmens B., Hegarat N., Akopyan K., Sala-Gaston J., Bartek J., Hochegger H., and Lindqvist A.. 2018. DNA Replication Determines Timing of Mitosis by Restricting CDK1 and PLK1 Activation. Mol. Cell. 71:117–128.e3. 10.1016/j.molcel.2018.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S.K., Velmurugan S., Chen R., Washburn M.P., Liu X.S., and DeCaprio J.A.. 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 26:539–551. 10.1016/j.molcel.2007.04.015 [DOI] [PubMed] [Google Scholar]
- Liu P., Slater D.M., Lenburg M., Nevis K., Cook J.G., and Vaziri C.. 2009. Replication licensing promotes cyclin D1 expression and G1 progression in untransformed human cells. Cell Cycle. 8:125–136. 10.4161/cc.8.1.7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N., and Diffley J.F.X.. 2005. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 122:915–926. 10.1016/j.cell.2005.08.013 [DOI] [PubMed] [Google Scholar]
- Maric M., Maculins T., De Piccoli G., and Labib K.. 2014. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 346:1253596 10.1126/science.1253596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson J.P., Dumitru R., Coryell P., Baxley R.M., Chen W., Twaroski K., Webber B.R., Tolar J., Bielinsky A.-K., Purvis J.E., and Cook J.G.. 2017. Rapid DNA replication origin licensing protects stem cell pluripotency. eLife. 6:e30473 10.7554/eLife.30473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh D., and Blow J.J.. 2012. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb. Perspect. Biol. 4:a012955 10.1101/cshperspect.a012955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., and Sorger P.K.. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60. 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Mohrin M., Bourke E., Alexander D., Warr M.R., Barry-Holson K., Le Beau M.M., Morrison C.G., and Passegué E.. 2010. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 7:174–185. 10.1016/j.stem.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A., Carrington J.T., Albergante L., Al Mamun M., Haagensen E.J., Komseli E.-S., Gorgoulis V.G., Newman T.J., and Blow J.J.. 2016. Unreplicated DNA remaining from unperturbed S phases passes through mitosis for resolution in daughter cells. Proc. Natl. Acad. Sci. USA. 113:E5757–E5764. 10.1073/pnas.1603252113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S.P., Bailey R., Campion N., Herron S., and Gambus A.. 2014. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science. 346:477–481. 10.1126/science.1253585 [DOI] [PubMed] [Google Scholar]
- Nevis K.R., Cordeiro-Stone M., and Cook J.G.. 2009. Origin licensing and p53 status regulate Cdk2 activity during G(1). Cell Cycle. 8:1952–1963. 10.4161/cc.8.12.8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr S.J., Gaymes T., Ladon D., Chronis C., Czepulkowski B., Wang R., Mufti G.J., Marcotte E.M., and Thomas N.S.B.. 2010. Reducing MCM levels in human primary T cells during the G(0)-->G(1) transition causes genomic instability during the first cell cycle. Oncogene. 29:3803–3814. 10.1038/onc.2010.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton K.W., Spencer S.L., Noderer W.L., Meyer T., and Wang C.L.. 2014. Basal p21 controls population heterogeneity in cycling and quiescent cell cycle states. Proc. Natl. Acad. Sci. USA. 111:E4386–E4393. 10.1073/pnas.1409797111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B.O., Lukas J., Sørensen C.S., Bartek J., and Helin K.. 1999. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18:396–410. 10.1093/emboj/18.2.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B.O., Wagener C., Marinoni F., Kramer E.R., Melixetian M., Lazzerini Denchi E., Gieffers C., Matteucci C., Peters J.M., and Helin K.. 2000. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14:2330–2343. 10.1101/gad.832500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo P.N., and Cook J.G.. 2016. Regulation and Function of Cdt1; A Key Factor in Cell Proliferation and Genome Stability. Genes (Basel). 8:2 10.3390/genes8010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt S.C., Bailey K.J., and Freeland A.. 2007. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 25:3121–3132. 10.1634/stemcells.2007-0483 [DOI] [PubMed] [Google Scholar]
- Remus D., and Diffley J.F.X.. 2009. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21:771–777. 10.1016/j.ceb.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Resnitzky D., Gossen M., Bujard H., and Reed S.I.. 1994. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14:1669–1679. 10.1128/MCB.14.3.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez J.A., Lewis C., McKinley K.L., Sikirzhytski V., Corona J., Maciejowski J., Khodjakov A., Cheeseman I.M., and Jallepalli P.V.. 2018. Distinct Roles of RZZ and Bub1-KNL1 in Mitotic Checkpoint Signaling and Kinetochore Expansion. Curr. Biol. 28:3422–3429.e5. 10.1016/j.cub.2018.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz C., Johnson A., Kõivomägi M., Zatulovskiy E., Kravitz C.J., Doncic A., and Skotheim J.M.. 2018. A Precise Cdk Activity Threshold Determines Passage through the Restriction Point. Mol. Cell. 69:253–264.e5. 10.1016/j.molcel.2017.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev H., Zenvirth D., Simpson-Lavy K.J., Melamed-Book N., and Brandeis M.. 2016. Imaging Cell Cycle Phases and Transitions of Living Cells from Yeast to Woman. In Cell Cycle Oscillators: Methods and Protocols. Coutts A.S., and Weston L., editors. Humana Press, New York, NY: 321–336. [DOI] [PubMed] [Google Scholar]
- Shima N., Alcaraz A., Liachko I., Buske T.R., Andrews C.A., Munroe R.J., Hartford S.A., Tye B.K., and Schimenti J.C.. 2007. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 39:93–98. 10.1038/ng1936 [DOI] [PubMed] [Google Scholar]
- Shreeram S., Sparks A., Lane D.P., and Blow J.J.. 2002. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 21:6624–6632. 10.1038/sj.onc.1205910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui K., On K.F., and Diffley J.F.X.. 2013. Regulating DNA replication in eukarya. Cold Spring Harb. Perspect. Biol. 5:a012930 10.1101/cshperspect.a012930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa M.S., Bragado P., and Aguirre-Ghiso J.A.. 2014. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer. 14:611–622. 10.1038/nrc3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S.L., Cappell S.D., Tsai F.-C., Overton K.W., Wang C.L., and Meyer T.. 2013. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 155:369–383. 10.1016/j.cell.2013.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C., Rosinski J., Filipovic Z., Higgins B., Kolinsky K., Hilton H., Zhao X., Vu B.T., Qing W., Packman K., et al. . 2006. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl. Acad. Sci. USA. 103:1888–1893. 10.1073/pnas.0507493103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong L.N., and Wu X.. 2011. Prevention of DNA re-replication in eukaryotic cells. J. Mol. Cell Biol. 3:13–22. 10.1093/jmcb/mjq052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velthoven C.T.J., and Rando T.A.. 2019. Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling. Cell Stem Cell. 24:213–225. 10.1016/j.stem.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P., et al. . 2015. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 520:549–552. 10.1038/nature14131 [DOI] [PubMed] [Google Scholar]
- Wang X., Fujimaki K., Mitchell G.C., Kwon J.S., Della Croce K., Langsdorf C., Zhang H.H., and Yao G.. 2017. Exit from quiescence displays a memory of cell growth and division. Nat. Commun. 8:321 10.1038/s41467-017-00367-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells A., Griffith L., Wells J.Z., and Taylor D.P.. 2013. The dormancy dilemma: quiescence versus balanced proliferation. Cancer Res. 73:3811–3816. 10.1158/0008-5472.CAN-13-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.M., Göhler T., Luciani M.G., Oehlmann M., Ge X., Gartner A., Jackson D.A., and Blow J.J.. 2006. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 173:673–683. 10.1083/jcb.200602108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.W., Chung M., Kudo T., and Meyer T.. 2017. Competing memories of mitogen and p53 signalling control cell-cycle entry. Nature. 549:404–408. 10.1038/nature23880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekezare M., Gómez-González B., and Diffley J.F.X.. 2013. Controlling DNA replication origins in response to DNA damage - inhibit globally, activate locally. J. Cell Sci. 126:1297–1306. 10.1242/jcs.096701 [DOI] [PubMed] [Google Scholar]
- Zhang J., Dai Q., Park D., and Deng X.. 2016. Targeting DNA replication stress for cancer therapy. Genes (Basel). 7:E51 10.3390/genes7080051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.